Significance
Noncoding RNAs are an underexplored reservoir of regulatory molecules in eukaryotes. We analyzed the environmental response of roots to phosphorus (Pi) nutrition to understand how a change in availability of an essential element is managed. Pi availability influenced translational regulation mediated by small upstream ORFs on protein-coding mRNAs. Discovery, classification, and evaluation of long noncoding RNAs (lncRNAs) associated with translating ribosomes uncovered diverse new examples of translational regulation. These included Pi-regulated small peptide synthesis, ribosome-coupled phased small interfering RNA production, and the translational regulation of natural antisense RNAs and other regulatory RNAs. This study demonstrates that translational control contributes to the stability and activity of regulatory RNAs, providing an avenue for manipulation of traits.
Keywords: long noncoding RNA, ribosome footprint profiling, small peptides, phosphate deficiency, Arabidopsis thaliana
Abstract
Eukaryotic transcriptomes contain a major non–protein-coding component that includes precursors of small RNAs as well as long noncoding RNA (lncRNAs). Here, we utilized the mapping of ribosome footprints on RNAs to explore translational regulation of coding and noncoding RNAs in roots of Arabidopsis thaliana shifted from replete to deficient phosphorous (Pi) nutrition. Homodirectional changes in steady-state mRNA abundance and translation were observed for all but 265 annotated protein-coding genes. Of the translationally regulated mRNAs, 30% had one or more upstream ORF (uORF) that influenced the number of ribosomes on the principal protein-coding region. Nearly one-half of the 2,382 lncRNAs detected had ribosome footprints, including 56 with significantly altered translation under Pi-limited nutrition. The prediction of translated small ORFs (sORFs) by quantitation of translation termination and peptidic analysis identified lncRNAs that produce peptides, including several deeply evolutionarily conserved and significantly Pi-regulated lncRNAs. Furthermore, we discovered that natural antisense transcripts (NATs) frequently have actively translated sORFs, including five with low-Pi up-regulation that correlated with enhanced translation of the sense protein-coding mRNA. The data also confirmed translation of miRNA target mimics and lncRNAs that produce trans-acting or phased small-interfering RNA (tasiRNA/phasiRNAs). Mutational analyses of the positionally conserved sORF of TAS3a linked its translation with tasiRNA biogenesis. Altogether, this systematic analysis of ribosome-associated mRNAs and lncRNAs demonstrates that nutrient availability and translational regulation controls protein and small peptide-encoding mRNAs as well as a diverse cadre of regulatory RNAs.
A large fraction of the transcripts produced from the nuclear genomes of eukaryotes do not code for proteins (1). In the case of the model flowering plant Arabidopsis thaliana, an estimated 80–90% of the genome is transcribed at some point during development (2) with an estimated 38% encoding proteins of ≥100 aa. The other transcripts encode non–protein-coding housekeeping RNAs (tRNAs, ribosomal RNAs, small nuclear and nucleolar RNAs), regulatory RNAs involved in gene silencing [microRNAs (miRNAs), small interfering RNAs (siRNAs) (3)], and the diverse cohort called long noncoding RNAs (lncRNAs). lncRNAs are subclassified according to their site of origin, orientation relative to neighboring genes, cellular compartmentation, and function (reviewed for plants by ref. 4). These include lncRNAs transcribed from intergenic or intronic regions (lincRNAs), natural antisense transcripts (NATs) complementary to protein-coding transcripts, and precursors of small RNAs (sRNAs) including trans-acting phased small interfering RNAs (tasiRNA/phasiRNA) and microRNAs (miRNA). NATs originate from the reverse strand of sense (protein) coding regions (cis-NATs) or from distinct genomic loci (trans-NATs) and are often associated with regulation of development (5). There is a profusion of NATs of cognate protein-coding genes in eukaryotes [Saccharomyces cerevisiae (27%), Drosophila melanogaster (17%), mice (72%), humans (61–72%)] (6). A survey of A. thaliana transcriptomes from multiple tissues and growth conditions identified 37,238 sense–antisense transcript pairs, corresponding to 70% of annotated mRNAs (7). Many NATs are detectable only in mutants defective in mRNA degradation (8), indicating that their abundance is tightly regulated.
lncRNAs can function in the nucleus or cytoplasm. Nuclei of Arabidopsis seedlings accumulate over 200 lncRNAs, including over 30 that are protein-bound and evolutionarily conserved (9). One of these is AUXIN REGULATED PROMOTER LOOP RNA (APOLO), which controls a chromatin loop and DNA methylation at the neighboring PINOID locus, thereby modulating its transcription (10). The cis-NATs collectively named COOLAIR are generated from the opposite strand of the FLOWERING LOCUS C (FLC) of A. thaliana and its relative A. alpina (8). COOLAIR RNAs mediate deposition of the repressive chromatin mark trimethylated histone H3 Lys27 at FLC to control the process of vernalization (2). Thus, nuclear lncRNAs can determine epigenetic or transcriptional regulation in plants.
Cytoplasmic lncRNAs serve diverse roles in the regulation of mRNA stability and translation. For example, a mammalian mRNA with an ALU motif in its 3′-untranslated region (UTR) is recognized by a lncRNA that facilitates association of an RNA binding protein that activates mRNA decay when the transcript is ribosome-associated (11). Some plant cytoplasmic lncRNAs participate in molecular mimicry that controls the activity of miRNAs (12, 13). For example, Arabidopsis INDUCED BY PHOSPHATE STARVATION 1 and 2 (IPS1, IPS2/AT4) and their orthologs in other species are strongly induced in roots deficient in inorganic phosphate (H2PO4−; Pi) (4, 12). IPS1/2 bind miR399 but are a poor substrate for miRNA-mediated cleavage. It is the elevation of these endogenous miRNA target mimics (eTMs) under Pi deficiency that fine-tunes regulation of the true miR399 target PHOSPHATE 2 (PHO2). This gene encodes a ubiquitin-conjugating enzyme E2 that promotes turnover of Pi transporters. IPS1/2 up-regulation ultimately reduces PHO1 (14) and related transporters to maintain correct Pi homeostasis (15). Beyond these, the regulation and function of the vast majority of cytoplasmic lncRNAs in plants remain uncharacterized (4, 7, 16, 17).
The development of high-throughput ribosome profiling (ribo-seq) methods that map the position of individual 80S ribosome footprints (RFs) on gene transcripts has greatly enhanced the resolution of translation dynamics in eukaryotes (18), including plants (19–21). Several studies have noted that small upstream ORFs (uORFs) can dampen translation of the main protein-coding ORF (mORF) and may modulate the stability of mRNAs (22, 23). Ribo-seq studies in plants (19, 24) and other eukaryotes (mice, humans, zebrafish, yeast) (25) have noted lncRNAs with ribosomes situated on small ORFs (sORFs). Scrutiny of ribosome occupancy on sORFs has aided identification of putative small peptides (sPEPs) (15, 26), including micropeptides involved in zebrafish embryo development (27) and mammalian heart muscle contraction (28, 29). Studying Arabidopsis, we performed ribo-seq analyses on immunopurified ribosomes and identified over 200 putatively translated lncRNAs (19), whereas others used trinucleotide codon periodicity of RFs to identify 15 conserved and translated sORFs (<100 aa), of which 3 were supported by detection of epitope-tagged sPEPs (24). To date, proteomic datasets have not been used to validate lncRNA translation in plants.
The presence of RFs on lncRNAs may be fortuitous, reflecting a default level of preinitiation complex scanning on 5′-7mG-capped and 3′-polyadenylated RNAs (26). A possibility that is not mutually exclusive with this proposal is that translation of a lncRNA may facilitate its trans-regulation of an mRNA. Indeed, ribosome association of mammalian lincRNA-p21 selectively represses the translation of TRANSCRIPTION FACTOR JUN-B and CATENIN BETA 1 mRNAs (30). An antithetical scenario exists for mammalian UBIQUITIN CARBOXY TERMINAL HYDROLASE L1 (UCHL1) mRNA, which is regulated by a cis-NAT that is shuttled to the cytoplasm in response to inhibition of the target of rapamycin (TOR) kinase pathway. There, direct base-pairing between the cis-NAT and the sense transcript enhances UCHL1 transcript translation (31). In rice, a cis-NAT up-regulated under Pi deficiency trans-regulates PHOSPHATE 1;2 (PHO1;2) mRNA translation through a direct cis-NAT–mRNA interaction (32). Given these examples, we reasoned that the coupled analysis of RFs and double-stranded RNA (dsRNA) formation could enable identification of cis-NATs or other lncRNAs involved in trans-regulation of mRNAs.
Here, we combine genome-wide technologies to explore the importance of translational regulation and lncRNA function in controlling the plasticity of seedling roots to Pi availability in A. thaliana. We found that translational regulation impacted a subset of protein-coding mRNAs under low Pi. More remarkable was the extensive and dynamic association of one-half of the detected lncRNAs with ribosomes, including Pi-regulated lncRNAs. A systematic analysis identified (i) lncRNAs that produce detectable peptides, demonstrating they are a reservoir of conserved and differentially regulated small peptide-coding genes; (ii) cis-NATs whose ribosome association is linked to stability and translation of the sense protein-coding mRNA, and (iii) a plethora of well-known lncRNAs such as endogenous miRNA target mimics and tasiRNAs/phasiRNAs. We also demonstrate that translation of the TAS3a sORF enhances tasiRNA production.
Results and Discussion
Phosphate Deficiency Impacts the Translational Regulation of a Subset of mRNAs.
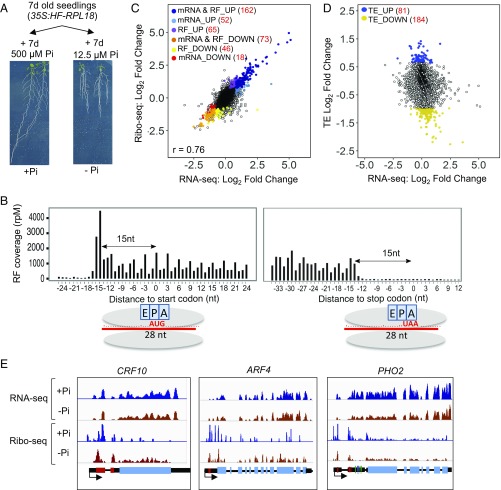
A comparative transcriptome and ribosome-footprinting study was performed on the complete root system of seedlings initiated on replete Pi (500 µM Pi, +P) medium for 7 d and then transferred to replete or deficient (12.5 µM Pi, −P) medium for an additional 7 d. The long-term low-Pi treatment triggered the typical adaptative response of the Arabidopsis root system with attenuated growth of the primary root and stimulation of the emergence and elongation of lateral roots (Fig. 1A), a plasticity in development that enhances capture of mineral reserves in the rhizosphere near the soil surface (33). Root tissue was used to isolate total cellular poly(A)+ mRNA as well as to determine the position of individual cytoplasmic 80S ribosomes on transcripts (Fig. S1A). For the latter, ribosome–mRNA complexes were captured by translating ribosome affinity purification (TRAP), which takes advantage of a 60S subunit ribosomal protein L18 that is FLAG-epitope tagged and expressed under the control of the near-constitutive CaMV 35S promoter (34). The polyribosome complexes that were purified were treated with RNase I to generate 80S ribosomes, which were processed to obtain 26- to 32-nt RFs for synthesis of a strand-specific ribo-seq library. These were sequenced to a depth of 54–81 million reads. After filtering out fragments of housekeeping RNAs [rRNA, tRNA, small nucleolar RNA (snoRNA)], 81–87% of the RFs mapped uniquely to protein-coding sequences (CDSs), ≤2.7% to 5′-UTRs, and ≤1% to 3′-UTRs. Strand-specific mRNA-seq libraries were prepared with chemically fragmented poly(A)+ mRNA and sequenced to a depth of 28–43 million reads. There was a high correlation (r ≥ 0.98) between biological replicates of both library types (Dataset S1a), confirming reproducible affinity purification and processing of mRNA–ribosome complexes.
Fig. 1.
Transcriptome mRNA-seq and ribo-seq expose selective translational regulation in response to phosphate starvation. (A) Pi starvation promotes lateral root development. (B) Coverage values [reads per million reads (rpM)] of the first nucleotide of 28-nt ribosome footprints (RFs) in the start and stop codon regions of expressed protein-coding genes at the same scale. Inferred ribosome position relative to the acyl (A), peptidyl (P), and exit (E) sites of the ribosome with the AUG in the P and stop codon in the A site. (C) Comparison of RNA-seq [total poly(A)+ mRNA] and ribo-seq (RFs) log2 FC in response to Pi deficiency (+Pi roots relative to −Pi roots). (D) Translational efficiency (TE) relative to RNA-seq log2 fold change in response to Pi deficiency. Based on mRNAs with ≥5 RF reads per kilobase million reads in all replicates. Genes regulated at RNA and/or RF and TE levels (FDR < 0.01) are indicated with colored dots. Coefficient of determination is indicated in C. (E) Gene view of coverage of mRNA and RF reads on selected genes: CYTOKININ RESPONSE FACTOR 10 (CRF10) (AT1G68550), AUXIN RESPONSE FACTOR 4 (ARF4) (AT5G60450), and PHOSPHATE 2 (PHO2) (AT2G33770). Scales are identical for each data type per gene. Gene structure is diagrammed at the Bottom: red boxes mark upstream ORFs in the 5′ leader of protein-coding genes; light blue boxes represent the main ORF; and dark blue and green lines in the PHO2 5′-UTR represent noncleavable and cleavable miR399 binding sites, respectively. Black arrows indicate direction of transcription.
To evaluate the efficiency and precision of the RNase I digestion, a metagene analysis was performed to map the position of the 5′-end of 28-nt RFs and their number on annotated protein-coding regions (Fig. 1B). The distance between the ribosome peptidyl site and the 5′-end of RFs was 14–15 nt, a signature that commenced at the AUG codon, with only a fraction of RFs mapping to 5′-UTRs (Fig. 1B). The increase in RF coverage at the AUG and decline at the stop codon is consistent with the rate-limiting nature of initiation and termination of translation (35). Finally, over one-half (56%) of the RF 5′-termini mapped to the first nucleotide of individual codons (Fig. S1B), a signature of the trinucleotide periodicity of ribosome decoding.
We first used our data to compare dynamics in transcript abundance and translation in response to Pi starvation. The response to 7-d Pi starvation by 15,297 protein-coding mRNAs was highly correlated between mRNA-seq and ribo-seq datasets (Fig. 1C; r = 0.76; Dataset S1b), and the differentially expressed genes defined by both datasets had similar Gene Ontology (GO) term enrichment (Dataset S1, c and d). This was not unanticipated because sucrose density gradient fractionation of polysomes indicated no difference in global levels of protein synthesis (Fig. S1B). There was an overlap of 84 up-regulated and 163 down-regulated genes between analyses [Fig. S1D and Dataset S1b; |log2 fold change (FC)| ≥1; false-discovery rate (FDR) ≤ 0.05]. Of the mRNAs previously reported to be differentially regulated in roots by short-term (3–12 h), medium-term (24–48 h), and long-term (12-d) Pi depletion (5 µM Pi) (36), 86 and 84 of these were differentially expressed in our mRNA-seq and ribo-seq data, respectively, including 70 recognized in both studies (Dataset S1b).
Our data enabled the evaluation of differences in translational efficiency (TE) for individual mRNAs, defined as the proportion of ribo-seq to mRNA-seq reads on each protein-coding ORF. In contrast to the strong concordance between changes in mRNA-seq and ribo-seq transcript abundance in response to Pi deficiency, 81 and 184 mRNAs displayed a significant increase or decrease in TE, respectively (FDR < 0.01; Fig. 1D and Fig. S1D and Dataset S1b). These translationally regulated mRNAs were present but not highly responsive to hormonal, nutrient, or abiotic stresses based on public transcriptome datasets (Dataset S1i), indicating that they are not generally regulated at the level of transcript abundance (Fig. S1E). Our data confirm independent modulation of mRNA accumulation and ribosome association by Pi availability, as seen under other environmental responses [photomorphogenesis (20), hypoxia (19), water deficit (21), and heat stress (37)].
mRNAs with a significantly higher TE under Pi depletion were overrepresented for “transporter activity” (P < 7.88E-04) and “kinase activity” (P < 4.19E-05) (Dataset S1e), whereas those with reduced TE were enriched for “transcription” (P < 8.54E-05), with a bias for cellular component “ribosome” (P < 5.34E-06). The reduced translation of ribosomal protein mRNAs under abiotic stress is well established (reviewed by refs. 23 and 38). The decline in TE was more limited under low Pi than under hypoxia (Fig. S1F). Collectively, these data demonstrate that Pi homeostasis selectively influences translation of a subset of cellular mRNAs.
Phosphate Availability Selectively Alters Ribosome Occupancy on Upstream ORF-Containing mRNAs.
The translation of individual mRNAs is often mediated by the presence of uORFs that may be sensitive to cellular metabolites or other factors (23, 38). To discern whether Pi-mediated changes in TE involved the presence and translation of uORFs, we identified mRNAs with nonoverlapping uORFs (>60 nt) located 5′ to the main protein-coding ORF (mORF) (Dataset S1f). Sixty-three of the 4,505 uORF-possessing mRNAs displayed Pi-modulated TE (Fig. S2A), of which 16 had significantly more RFs on the uORF relative to the mORF under Pi-replete conditions (Dataset S1g; repressive uORFs; Fig. S2B). Most of these displayed limited change in mRNA accumulation in response to Pi availability. Of the uORFs identified, 51 were evolutionarily conserved peptide (CP) uORFs (23, 38) (Fig. S2C). Over one-half of the CPuORFs had fewer ribosomes on the downstream mORF under Pi-replete or -deficient conditions, consistent with the propensity of translated uORFs to limit ribosome (re)initiation at a downstream start codon. However, the TE of eight CPuORF mRNAs increased significantly under Pi deficiency, concomitant with a decrease in uORF-to-mORF ratio of RFs and without a change in mRNA abundance (Fig. S2D and Dataset S1h). Two of these encode putative transcriptional regulators, CYTOKININ RESPONSIVE FACTOR 10 (CRF10) (39, 40) and AUXIN RESPONSE FACTOR 4 (ARF4) (41) (Fig. 1E). In both cases, the increase in mORF translation exceeded log2 1.3-fold under Pi deficiency. Although CRF10 function is unknown, other CRFs promote lateral root growth (39, 40). The regulation and function of ARF4 is also less known. The uORFs of ARF3, 4, and 5 limit translation in protoplasts (42), with the control of ARF3 and ARF5 mRNAs translation requiring TOR kinase and a specific subunit of the translation initiation factor eIF3 (43). These data provide strong evidence that Pi homeostasis influences uORF-mediated regulation of genes involved in root system architecture.
Our data also revealed that Pi homeostasis influenced the translation of the uORF-containing PHO2 mRNA. mORF-to-uORF ratios for PHO2 suggest similar and significant uORF-mediated translational repression under both conditions (log2 2.35 [+Pi], log2 2.43 [−Pi]; FDR < 7.0E-9) (Dataset S1b). Consistently, RFs were abundant in the PHO2 5′ leader before the mORF, particularly in the region of three small nonoverlapping uORFs of 15, 9, and 13 codons (Fig. 1E). These precede a cluster of five miR399 binding sites including two sites that are uncleavable (44). Most plant miRNAs guide transcript cleavage, although some mediate bona fide translational repression involving localization to the endoplasmic reticulum (45–47). Based on data from in vitro translation systems and uncapped RNA-sequencing analysis, RNA-induced silencing complex (RISC) bound to cleavable miRNA binding sites does not impair the elongation phase of translation (45–47).
We reasoned that Pi-regulated changes in RISC bound to the cleavable or uncleavable miR399 sites could limit PHO2 mRNA translation. To address this hypothesis, we compared the number of RFs on the full PHO2 5′-leader relative to the mORF. This ratio was greater under Pi-replete conditions (log2 2.19 ± 0.26 [+Pi], log2 1.56 ± 0.31 [−Pi]). Taking both the decrease in RF coverage on the full PHO2 5′ leader and the twofold decline in PHO2 mRNA under Pi deficiency (Dataset S1b) into consideration, repression of mORF translation may be less under low-Pi conditions and this could be due to elevated miR399-mediated cleavage (48). The increase in the eTMs IPS1/IPS2 (log2 5–10; FDR < 3.88E-5; Dataset S1b) and miR399 under Pi deficiency (48) likely modulates the balance between translational inhibition and cleavage to fine-tune PHO2 synthesis according to availability and use of Pi. These results suggest a connection between miRNA cleavage and translational initiation in the control of a key regulator of Pi transporter abundance.
A Subset of Root lncRNAs Are Associated with Ribosomes.
Next, we combined our mRNA-seq and ribo-seq data with sRNA-seq and double-stranded RNA (dsRNA)-seq data obtained with the same root samples to classify lncRNAs expressed under the two conditions. Annotated and de novo predicted lncRNAs were combined as input into a classification pipeline (Fig. 2A). After removal of lncRNAs with insufficient mRNA-seq and ribo-seq reads, 2,382 poly(A)+ lncRNAs were identified. Remarkably, over one-half (1,234) were candidate ribosome-associated lncRNAs (ribo-lncRNAs). The dsRNA-sequencing (dsRNA-seq) data were used to filter out highly structured RNAs with regions resistant to RNase I, a step we find beneficial to exclude snoRNAs and other structured RNAs that contaminate RF libraries (19). In this manner, 1,140 ribo-lncRNAs were identified, corresponding to ∼48% of all detected in seedling roots. These were further subclassified as lincRNAs (intergenic), cis-NATs (>50-nt overlap with a protein-coding gene exon), and phasiRNA/siRNA precursors (sRNA clusters enriched for a specific read length and/or showing a phased distribution) (Fig. 2B and Dataset S2a).
Fig. 2.
Nearly one-half of all detected lncRNAs are associated with ribosomes. (A) Discovery pipeline for ribosome-associated lncRNAs (ribo-lncRNAs) and their classification. lncRNAs were predicted de novo from compiled mRNA-seq data (Pi-replete and -deficient roots) (Cufflinks) and merged with annotated lncRNAs (Araport11). Classification as a ribo-lncRNA required ribosome footprint (RF) read coverage above background (HTSFilter). Small RNA (sRNA)-seq data from Pi-replete roots were used to identify sRNA and phasiRNA precursors (88). sORF translation was recognized using two analytics: ribosome release score (RRS) (15) and RiboTaper (50), and supported by evolutionary conservation of sORFs and proteomic mass spectra data of encoded small peptides. (B) Summary of classified lncRNAs. Canonical lncRNAs are all those not classified as a ribo-lncRNA. Numbers in red indicate the number of differentially expressed lncRNAs (FDR < 0.01) in each subclass. (C) Normalized read coverage of mRNA-seq, ribo-seq, and 21- to 22-nt-long sRNA-seq reads on representatives of subclasses. Nuclear lncRNA ASCO (AT1G67105); miRNA target mimic INDUCED BY PHOSPHATE STARVATION 1 (IPS1) (AT3G09922); putative small peptide-coding gene AT1G20724; ribo-cis-NAT generating 21- to 22-nt siRNAs [SUPPRESSOR OF PHYB 3 (SOB3) cis-NAT AT1G76500]; precursor of 21-nt phased sRNA (phasiRNA) AT1G62860; and TRANS-ACTING SIRNA 3a (TAS3a) (AT3G17185). Scales are identical for each data type per gene. Gene structure is diagrammed at the Bottom: light blue boxes represent coding regions including ORFs with ribosome footprints; red box on IPS1 is a miR399 binding site. Black arrows indicate direction of transcription of sense (protein-coding) ORFs and NATs.
The majority of lincRNAs (572 of 710; ∼80%) were associated with RFs. As an example, RFs mapped toward the 5′-terminus and largely overlapped with the first sORF on the Pi-regulated eTMs IPS1 and IPS2 (Fig. 2C and Fig. S3A). Notably, 12 of 15 predicted eTMs were ribo-lncRNAs (Dataset S2a). By contrast, the nuclear lncRNAs ASCO (Fig. 2C) and APOLO (Fig. S3B) lacked RFs. NATs comprised the largest class of lncRNAs (1,676), including 568 with a significant number of RFs. Similar processing of three published ribo-seq datasets from seedlings (20, 24, 49) detected 567 ribo-lncRNAs overall, including 114 ribo-lncRNAs detected in all analyses (81 lincRNAs, 33 NATs), including 8 out of 11 detectable phasiRNA precursors (Fig. S3C and Dataset S2a). Pi-regulated accumulation was evident for 67 lncRNAs (|log2 FC| ≥ 1, FDR ≤ 0.05), including 56 ribo-lncRNAs (44 lincRNAs, 12 NATs) (Dataset S2a). However, we found no evidence of ribosome-associated miRNA precursors. Taken together, many lincRNAs and NATs may be translated in plants.
Translated sORF Are Hidden in Plant lncRNAs.
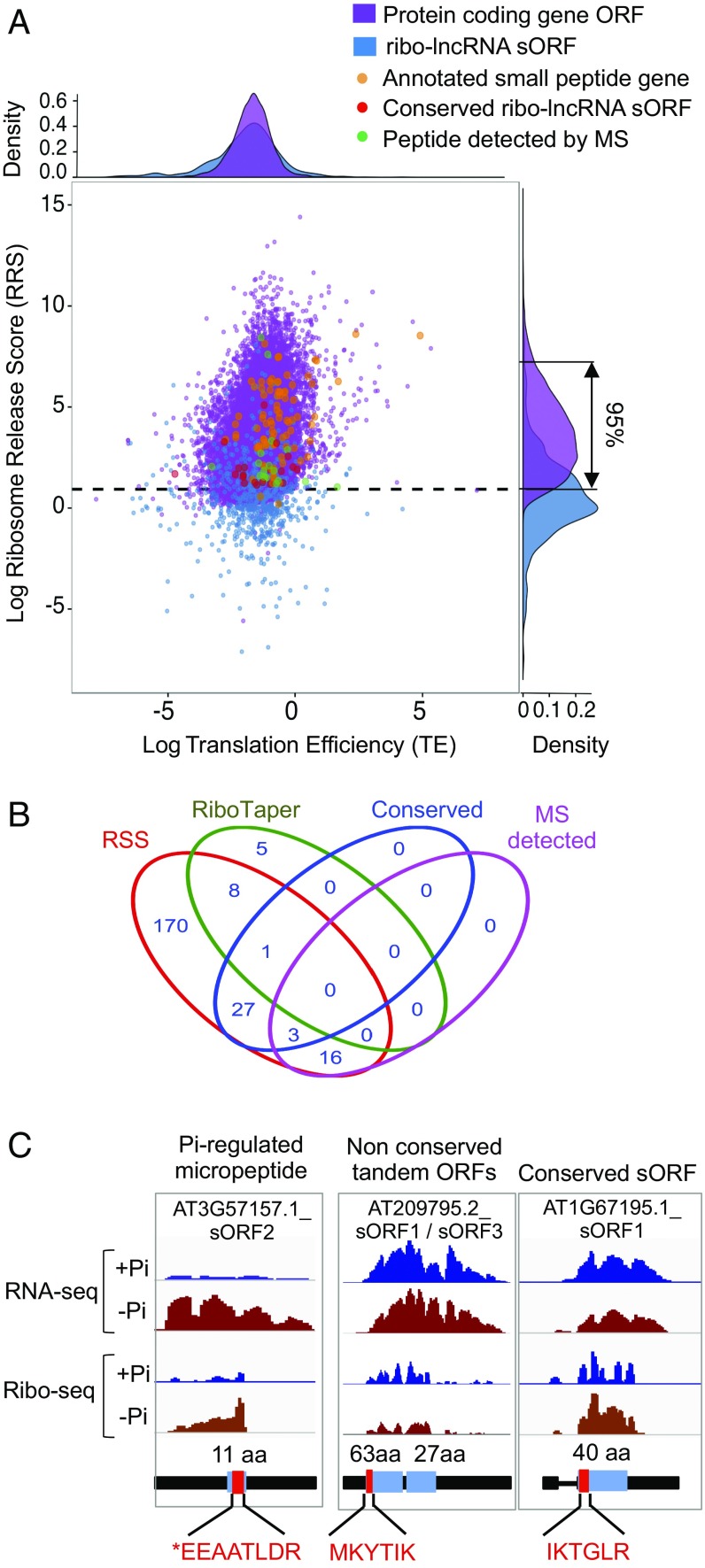
All 1,140 ribo-lncRNAs contained at least one sORF encoding ≥10 aa, but only 397 met the threshold of ≥1 rpkM for RFs on at least one sORF of ≥30 nt (Dataset S2b). To seek evidence of sORF translation, we determined the ribosome release score (RRS) (15) (Fig. 2A), which quantifies the decline in RF number (drop-off) seen after the termination codon (Fig. 1C). Comparison of TE and RRS values of protein-coding and ribo-lncRNA ORFs identified 225 putative lncRNA_sORFs based on a depletion of RFs in the 3′-UTR, as seen for annotated protein-coding ORFs (Fig. 3A). Moreover, RRS values of characterized sPEP mRNAs almost completely overlapped with those encoding proteins of >100 aa (Fig. 3A).
Fig. 3.
Translated sORF are hidden in lncRNAs. (A) Translation efficiency (TE) and ribosome release score (RSS) for protein-coding genes and ribosome-associated lncRNAs with small ORFs (ribo-lncRNA_sORFs). Annotated small peptide-coding genes, conserved ribo-lncRNA sORFs and mass spectra validated sORFs are indicated as orange, red, and green dots, respectively. The vertical arrow on the RSS density plot (Right) marks the region between the upper and lower 5% quantiles of RRS distribution of protein-coding genes. Ribo-lncRNA sORFs with an RSS within this range are the putative translated lncRNA_sORFs. (B) Venn diagram of ribo-lncRNA sORFs predicted using RRS or RiboTaper, having significant conservation across plants and evidence of peptide production by mass spectrometry. (C) Example sORFs with peptidic support. Normalized transcript coverage plots of mRNA-seq and ribo-seq reads from Pi replete and deficient roots. An 11-aa micropeptide (AT3G57157). Tandem sORFs of 63 and 27 aa (AT2G20724). A translated conserved sORF (AT1G67195). Scales are identical for each data type per gene. Gene structure is diagramed as in Fig. 1, with amino acid length of predicted ORF. Red box indicates peptide region detected in proteomic spectra; asterisk, amino acid acetylation.
As a second metric, we monitored the trinucleotide periodicity of RF 5′-termini on individual transcripts as seen at the global level (Fig. 1B). This reflects the precision in codon decoding and is evident in ribo-seq datasets if RNase I digestion is taken to completion. Recognition of translated ORFs based on codon trinucleotide periodicity quantified using RiboTaper (50) recognized 14 lncRNA_sORFs (P < 0.05; Dataset S2b), including 9 with a significant RSS score (Fig. 3B). This indicates that RiboTaper was more restrictive in sORF discovery for our dataset. Previous RiboTaper analysis of root RFs identified 23 sORFs (24), including 21 that we detected here using RRS or RiboTaper.
Exploration of the evolutionary conservation of the putative lncRNA_sORFs identified 31 present in Brassicaceae outside of the Arabidopsis genus. Of these, 9 were broadly conserved in angiosperms, 13 were recognizable in some eudicots and monocots, and 9 were limited to Brassicaceae (Fig. S4 and Dataset S2b). Three putative lncRNA_sORFs were unannotated members of conserved sPEP families [C-terminally encoded (CEP), CLAVATA-like (CLE), PAMP-induced secreted peptide-like (PIPL)] and one was a new member of an uncharacterized family of sPEPs with a conserved 13-aa C-terminal domain (Fig. S5).
To establish whether lncRNA_sORF-encoded peptides are synthesized and accumulate in planta, we searched 1,653 mass spectrometry files from five previously established proteomic datasets (51–55) from diverse Arabidopsis tissues against a TAIR10 protein database extended with predicted sPEP sequences. Bona fide translation of lncRNA_sORFs was supported by mass spectra of 19 predicted sPEPs supported by RSS (Fig. 3C and Fig. S6 and Dataset S2b). These included sPEP fragments of three conserved sORFs supported by RSS but not RiboTaper. The sPEPs can produce limited tryptic peptides, and therefore it is unsurprising that data mining yielded N-terminal spectra, including three with the initial Met removed and an acetylated penultimate residue (Fig. 3C). Altogether, the validated lncRNA_sORFs ranged from an 11-residue low-Pi–induced micropeptide to a 100-residue polypeptide. An sPEP closely related to CLE26 was detected in all five independent datasets with a total of 32 spectra, providing strong support of its synthesis. Another peptide was from the first of two sORFs encoded by ribo-lncRNA AT2G09795.2 (Fig. 3C and Dataset S2b). The RF coverage and RSS score of the second sORF of this transcript support its translation, indicating this may be a polycistronic mRNA. The proteomic spectra also confirmed synthesis of 14 sPEPs encoded by uORFs (Dataset S2c), including a CPuORF of SUPPRESSOR OF ACAULIS 51 (SAC51) that has uORF-regulated translation mediated by availability of thermospermine (56).
Further support of the biological relevance of ribo-lncRNA_sORFs was obtained with surveys of evolutionary conservation. First, using the PhastCons score derived from whole-genome sequence alignments from 20 angiosperms, a method amenable to evaluation of the conservation of plants coding and noncoding sequences at the nucleotide level (57), we confirmed high conservation near the start codon of predicted ribo-lncRNA_sORFs with high but not low RSS values (Fig. S7A). Second, we monitored the sequence variation in ribo-lncRNA_sORFs and uORFs in 1,135 resequenced Arabidopsis accessions (58). As many of these ORFs had no missense mutations across the accessions, we calculated the proportion of ORFs lacking high-impact nucleotide variants for each class (Fig. S7B) and then determined the ratio of synonymous to missense mutations in the genes with high-impact variants (Fig. S7C). The occurrence of missense mutations was low in the conserved sORFs as well as in sORFs with detected sPEPs. As anticipated, there were few missense mutations in CPuORFs but also in the uORFs that repressed mORF translation under Pi-replete conditions, confirming positive selection on conserved sORFs and uORFs regulating translation.
Our data expand the sORFs repertoire predicted by ribo-seq (24) by nearly 10-fold and confirm the synthesis and detection of 19 sPEPs. Of particular interest was that translation was significantly up-regulated for eight and down-regulated for four lncRNA_sORFs by Pi deficiency based on RSS and RF values (Dataset S2b). One of the up-regulated lncRNA_sORFs encodes an 11-aa sPEP supported by six mass spectra (Fig. 3C). These results indicate lncRNA_sORFs can encode conserved peptides of biological relevance (59) and raise the possibility that sPEP signaling is modulated by Pi homeostasis.
Ribo-cis-NAT Regulation Is Associated with Translational Enhancement.
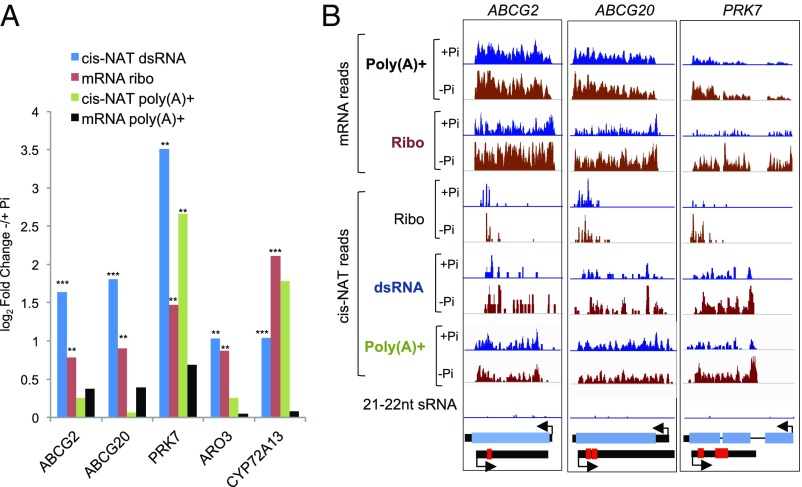
Many cis-NATs are differentially regulated by environmental conditions in plants, but very few have been functionally characterized (reviewed by refs. 60 and 61). Our lncRNA classification pipeline identified 568 ribo-cis-NATs that are associated with dsRNA but not siRNA production, including 12 with significantly different accumulation under the two Pi conditions (Fig. 2B and Dataset S2 a and d). Among ribo-cis-NATs, 167 had ribosome-bound ORFs including 49 with an RRS score supportive of active translation. To explore whether these cis-NATs might regulate their sense mRNAs under low Pi, we quantified dsRNA-seq reads in overlapping cis-NAT and sense mRNA regions under the two conditions. This identified 143 cis-NATs regulated by Pi availability (dsRNA-seq; log2FC > 1; FDR < 0.05; Dataset S2d), including 41 ribo-cis-NATs. The corresponding sense mRNAs of five of these displayed significantly increased TE (log2FC TE > 0.71; FDR ≤ 0.01; Dataset S1a) concomitant with elevation of their cis-NAT under Pi deficiency (Fig. 4A). Three of the five cis-NATs had an sORF with a high RRS score, and all five had RFs that mapped toward their 5′ termini (Fig. 4B), a characteristic subsequently recognized across all ribo-cis-NATs (Fig. S8A). The mRNAs displaying ribo-cis-NAT regulation included two ATP-BINDING CASSETTE SUBFAMILY G transporters (ABCG2, ABCG20), required for endodermal suberization that influences nutrient uptake and lateral root formation (62, 63), and a POLLEN-SPECIFIC RECEPTOR-LIKE KINASE 7 (PRK7) family member associated with root cell elongation (64). PRK7 mRNA is enriched in root-hair cells (65), which elongate under low Pi to increase the surface area for nutrient uptake. Increased translation under Pi deficiency of all five mRNAs was concomitant with a significant elevation in dsRNA production (Fig. 4B and Fig. S8B), indicating sense–antisense interaction. With the exception of the PRK7 cis-NAT, levels of poly(A)+ cis-NAT RNA was not influenced by Pi deficiency, indicating regulation occurs at multiple levels in addition to ribosome association.
Fig. 4.
Regulation of a subset of ribo-cis-NATs correlates with translation of their cognate sense mRNAs. (A) Log2 FC of ribo-cis-NAT (cisNAT dsRNA-seq), mRNA RF (mRNA Ribo), poly(A)+ cis-NAT, and poly(A)+ mRNA values for the five translationally regulated NAT/sense mRNA pairs (**FDR < 0.05; ***FDR < 0.01). (B) Normalized read coverage of RNA-seq, ribo-seq, dsRNA-seq, and 21- to 22-nt sRNA-seq reads of representative sense–antisense pairs displaying low Pi regulation. ATP-BINDING CASSETTE SUB-FAMILY G transporter (ABCG2) (AT2G37360), ABCG20 (AT3G53510), and POLLEN-SPECIFIC RECEPTOR-LIKE KINASE 7 (PRK7) (AT4G31250). Blue and red boxes, coding regions based on annotation (protein-coding gene) or RFs (cis-NAT), respectively; thin black lines, introns; thick black boxes, noncoding regions. Arrows mark direction of transcription.
We propose that translation may stabilize or appropriately target cis-NATs and other ribo-lncRNAs within cells for interaction with their sense target. The five ribo-cis-NATs described above had limited nucleotide variation within their ORF with the highest RSS score across sequenced Arabidopsis accessions (Fig. S7 D and E). Nucleotide variants of significance were overall higher in NAT sORFs than in ORFs of sense transcripts. Nonetheless, point mutations in ribo-cis-NAT 5′-sORFs could impact their abundance or interaction with sense mRNAs, explaining phenotypic variability between Arabidopsis ecotypes (8) and contributing to the diversity of developmental responses to Pi starvation among ecotypes and species (66).
sORF Translation Enhances TAS3a Stability.
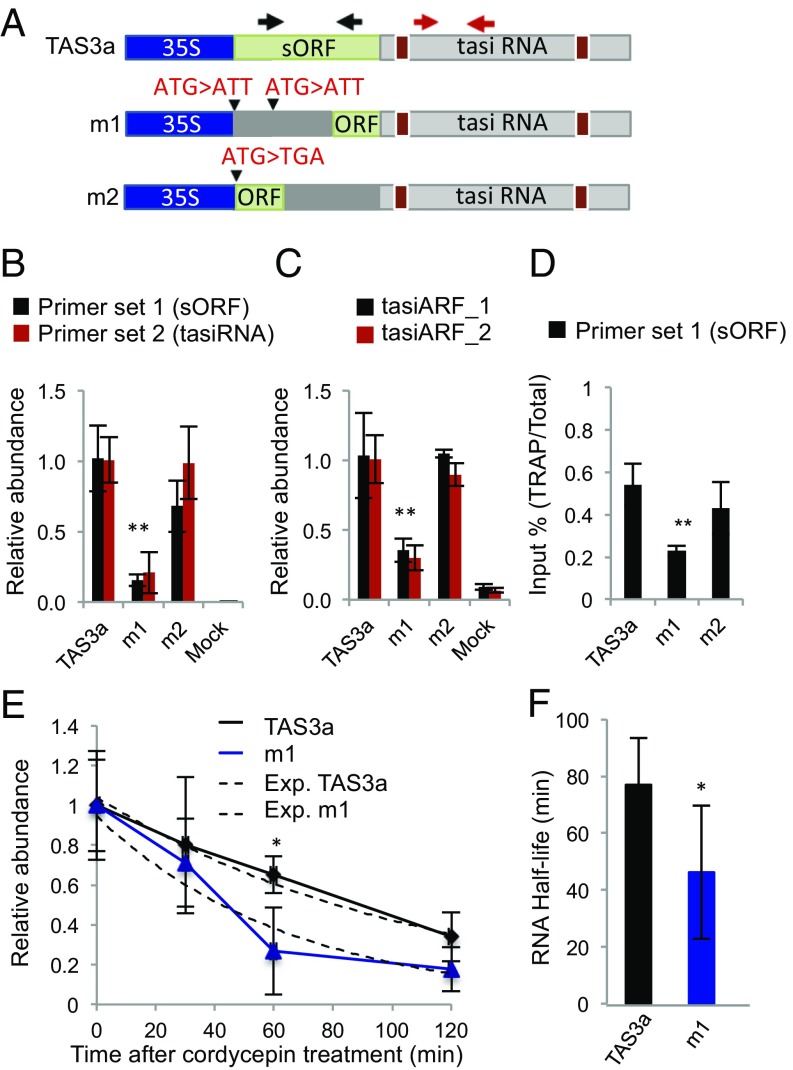
The ribo-lncRNAs discovered included known precursors of tasiRNAs (67–69). As an example, RFs mapped just 5′ of the tasiRNA-generating regions of TAS1b, TAS2a, TAS3a, and TAS4 (Fig. 2C and Fig. S9). sRNA-seq data were used to explore the relationship between ribosome occupancy and siRNA biogenesis (Fig. 2A). This yielded 54 and 179 ribo-lncRNAs that coincided with 21- to 22- and 24-nt sRNA clusters, respectively (Dataset S2a). Notably, the RFs on 13 of 14 ribo-lncRNAs that produced 21- to 22-nt phasiRNAs (phase score > 20) were positioned on sORFs located just 5′ of the region that generated the sRNAs (Fig. 2 B and C; AT1G62860, TAS3a; Fig. S9; other tasiRNA loci). Of the 13, 4 had an RRS score highly supportive of translation in seedling roots (Dataset S2c). TAS3a was particularly notable. First, the termination codon of its 52-aa sORF is positioned just 5′ of the noncleavable miR390/ARGONAUTE 7 (AGO7) binding site (67) (Fig. 5A and Fig. S10A). Second, RFs map precisely to the TAS3a sORF in all ribo-seq datasets we generated or reevaluated (19, 20, 24, 49) (Fig. S10B). Third, TAS3s from diverse plants have a similar codon length (50–58 aa) but poorly conserved sORF positioned 8–20 nt 5′ of the miR390 binding site (Fig. S10 C and D). Based on this, we hypothesized that TAS3a sORF translation may promote the production of its phasiRNAs (tasiARF1/2).
Fig. 5.
Translation of the TAS3a sORF promotes tasiRNA production. (A) Constructs used to evaluate the coupling of TAS3a translation and tasiRNA production. Green boxes show sORF region, and black triangles show targeted mutations. Brown boxes show miR390 binding sites. Black arrows represent the position of primers used for quantification of transcripts with the sORF region (black arrows, primer set 1) and tasiRNA-generating region (red arrows, primer set 2). TAS3a precursor transcript detection by RT-qPCR (B) and tasiARF_1 and tasiARF_2 by RT-Stem Loop-qPCR (C), 48 h after transfection of N. benthamiana leaves with Agrobacterium-containing TAS3a constructs. Relative abundance (mean ± SD) with Ct values normalized against the BAR gene expressed from the same T-DNA. (D) Quantification of ribosome association of TAS3a, TAS3a-m1, and TAS3a-m2 transcripts in Nicotiana benthamiana (35S:FLAG-AtRPL18) leaves 48 h after Agrobacterium infiltration using TRAP. Results are shown as percentage of input (total RNA) and represent the mean ± SD of three independent replicates. Student’s t test, *P < 0.05; **P < 0.01. (E) RNA decay time course of TAS3a precursor (primer set 1) abundance in N. benthamiana leaves infiltrated with constructs and treated with cordycepin 24 h after transfection. Relative abundance values were calculated as for C. Solid lines, data points; dashed line, exponential regression (Exp.) used to calculate RNA half-life. (F) RNA half-life derived from exponential regression (mean ± SD). Student’s t test, *P < 0.05.
To test this, a series of mutations in the TAS3a sORF were made, and levels of TAS3a and tasiARF1/ARF2 were monitored in Nicotiana benthamiana (Fig. 5 A–C). The removal of the first and second in-frame AUG (TAS3a-m1), so that the sORF was reduced to 22 codons commencing just upstream of the miR390 binding site, dramatically reduced TAS3a abundance and tasiARF1/2 production. By contrast, a nonsense mutation at the second in-frame AUG (Met29 → STOP) that reduced the sORF to 28 codons (TAS3a-m2) had no effect. To evaluate the impact of these mutations on TAS3a association with ribosomes, the constructs were tested in N. benthamiana that produces FLAG-tagged AtRPL18B. TRAP was used to isolate ribosomes to avoid contamination with other ribonucleoprotein complexes. The proportion of TAS3a RNA associated with ribosomes ranged from 55% for the nonmutated sORF to just over 20% for TAS3a-m1 (Fig. 5D). This suggests that altering the start site but not necessarily the length of the TAS3a sORF significantly decreased the proportion of RNA that copurified with ribosomes.
These results indicate that translation from the first in-frame AUG coordinately elevates TAS3a abundance, and subsequently phasiRNA accumulation. Monitoring the rate of decline in TAS3a and TAS3a-m1 abundance following inhibition of transcription with the nonfunctional adenosine analog cordycepin revealed that TAS3a-m1 RNA was significantly less stable than TAS3a (Fig. 5 E and F). These data support the conclusion that evolutionarily conserved translation of a nonconserved sORF enhances TAS3 RNA stability and consequently promotes phasiRNA accumulation.
Other studies point to TAS RNA translation as a critical factor in its production of phasiRNAs. First, the mapping of 5′ ends of truncated RNAs identified a signature of ribosome association of TAS3 sORF which was lost in the absence of endomembrane-localized AGO7 (70, 71). Second, TAS RNAs required localization in a subset of polysomes anchored to the rough endoplasmic reticulum to properly trigger phasiRNAs biogenesis (47). Our observation that manipulation of TAS3a sORF position can decrease TAS3a RNA stability indicates that translating ribosomes limit its degradation. The 5′- to 3′-EXORIBONUCLEASE 4 (XRN4) is involved in degradation of numerous mRNAs and competes with RNA-DEPENDENT RNA POLYMERASE 2 and 6 to process miRNA targets after mRNA cleavage (72). However, TAS transcript abundance is not affected in xrn4 mutants, and therefore how they escape XRN4-mediated degradation is unclear (73). As miRNA-mediated cleavage of TAS2 was recently reported to occur in association with ribosomes and to require a translatable sORF (74), it will be of interest to see whether cytosolic lncRNA stability and function is generally linked to the position and translation of an sORF.
Conclusions
Here, comparative transcriptome and ribosome footprint profiling with root tissue of Arabidopsis seedlings grown under Pi-replete and -deficient conditions has unveiled that many lncRNAs, nearly all of which are uncharacterized, undergo translation. These include small-peptide encoding mRNAs, eTMs that regulate miRNA activity, cis-NATs, and precursors of tasiRNAs. Translated lncRNAs that displayed dynamics in abundance and translation in response to altered Pi homeostasis include uncharacterized small peptide-coding genes as well as cis-NATs that interact with and enhance translation of their sense transcript. The translation of regulatory lncRNAs may foster their function, as demonstrated for TAS3a. The TAS3a sORF position relative to the noncleavable miR390 binding and its translation appeared to augment transcript stability and tasiRNA biogenesis. These results indicate that translation of a lncRNA should be considered even when its major function is linked to the RNA molecule itself, either through complementarity with its sense mRNA or via production of siRNAs. The propensity for ribosome association of regulatory RNAs illustrates that translational control extends to regulatory RNAs. Further exploration of the evolutionary conservation, accumulation, and biological activity of the cache of ribo-lncRNAs is likely to identify targets for genetic manipulation of phenotypes.
Materials and Methods
Detailed procedures are given in SI Materials and Methods.
Plant Growth and Manipulation.
Arabidopsis thaliana (Col-0) expressing 35S:His6FLAG-RPL18B (34), grown on Murashige and Skoog (MS/10) medium (Pi-replete, 500 µM), were transferred to Pi-replete (500 µM) or Pi-deficient (12.5 µM) MS/10 for 7 d. Growth was at 23 °C, 16-h light (80 μE⋅m−2⋅s−1)/8-h dark cycle. For transient assays, leaves of 2- to 3-wk-old Nicotiana benthamiana expressing 35S:His6FLAG-AtRPL18B (75) were infiltrated with Agrobacterium tumefaciens (AGL-0) (76) carrying T-DNA plasmid pB7GW2D (77) with TAS3a constructs. To assay mRNA decay, one fully expanded leaf per plant per construct and per bioreplicate was Agro infiltrated; 48 h later, leaf discs were prepared, vacuum infiltrated with 1 mM cordycepin, and sampled after 0, 30, 60, and 120 min.
RNA Analyses.
Total RNA was isolated using TRIzol (Invitrogen), treated with DNase I and reverse transcribed. Real-time (RT)-PCR was used for mRNA quantitation and stem-loop RT-qPCR (78) using two technical replicates each with five leaf discs was used for tasiRNA quantification. Relative differences were calculated by the ΔΔCt method (79) and RNA half-life was determined from the exponential regression of decay (80). To assay mRNAs associated with ribosomes, TRAP was performed (81) using magnetic anti-FLAG–coupled protein G Dynabeads. Results were analyzed using a percent-of-input method and Student’s t tests using three bioreplicates.
Ribo-Seq, mRNA-Seq, dsRNA-Seq, and sRNA-Seq Library Synthesis and Sequencing.
Ribo-seq libraries were prepared with ribosomes obtained by TRAP using root tissue (81) and EZview Red anti-FLAG M2 magnetic beads. RFs were generated by RNase I, purified, size selected, depleted of contaminating rRNA, and processed into libraries (82). This included poly(A)+ mRNA-seq libraries that were prepared from DNase I-treated RNA extracted from the clarified supernatant (S-18 fraction) of the TRAP protocol (81) as described (83). sRNA of 18–26 nt from total RNA was prepared for sequencing using the NEXTflex Small RNA Sequencing Kit (BIOO Scientific). dsRNA-seq libraries were prepared as described (84).
Bioinformatic Analyses.
Short read sequencing data were analyzed using a combination of Unix software and R packages from Bioconductor. Read count data (rpkM) for protein-coding gene features were generated as described (19, 85). Statistical identification of differentially expressed genes and feature types was by the generalized linear model, applying FC and FDRs. Enrichment analysis of GO terms was performed with the Classification SuperViewer tool (bar.utoronto.ca). Translational efficiency of protein-coding genes and other ribo-seq statistics were obtained using systemPipeR (85). uORFs were predicted from 5′-UTR sequences from Araport 11 (86) with the predORF function using ATG as start codon and ≥60 nt. RFs on uORFs and mORFs were determined as in ref. 19. For gene feature analysis, we computed coverage from the first nucleotide of each 28-nt RF (87). Data were visualized using normalized and merged datasets. Detailed procedures of the lncRNA pipeline, prediction of sORFs translation with RRS (15) and RiboTaper (50), and analysis of published proteomic data are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Thomas Girke, Mauricio Reynoso, Reed Sorenson, and Maureen Hummel for many helpful discussions. This research was supported by Marie Curie European Economic Community Fellowship PIOF-GA-2012-327954 (to J.B.), National Science Foundation (NSF) Grant MCB-1021969 (to J.B.-S.), and the Eidgenössische Technische Hochschule, Zurich (K.B.). This research used the Genomics Core instrumentation and the Bioinformatics Core computing facilities of the Integrative Institute of Genome Biology at the University of California, Riverside, supported by NSF Grant DBI-1429826 and National Institutes of Health Grant S10-OD016290-01A1.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE98610).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708433114/-/DCSupplemental.
References
- 1.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ariel F, Romero-Barrios N, Jégu T, Benhamed M, Crespi M. Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci. 2015;20:362–371. doi: 10.1016/j.tplants.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25:2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazin J, Bailey-Serres J. Emerging roles of long non-coding RNA in root developmental plasticity and regulation of phosphate homeostasis. Front Plant Sci. 2015;6:400. doi: 10.3389/fpls.2015.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, et al. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 2014;24:444–453. doi: 10.1101/gr.165555.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castaings L, et al. Evolutionary conservation of cold-induced antisense RNAs of FLOWERING LOCUS C in Arabidopsis thaliana perennial relatives. Nat Commun. 2014;5:4457. doi: 10.1038/ncomms5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gosai SJ, et al. Global analysis of the RNA-protein interaction and RNA secondary structure landscapes of the Arabidopsis nucleus. Mol Cell. 2015;57:376–388. doi: 10.1016/j.molcel.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ariel F, et al. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol Cell. 2014;55:383–396. doi: 10.1016/j.molcel.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 11.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franco-Zorrilla JM, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 13.Karakülah G, Yücebilgili Kurtoğlu K, Unver T. PeTMbase: A database of plant endogenous target mimics (eTMs) PLoS One. 2016;11:e0167698. doi: 10.1371/journal.pone.0167698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu T-Y, et al. PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell. 2012;24:2168–2183. doi: 10.1105/tpc.112.096636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, et al. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012;24:4333–4345. doi: 10.1105/tpc.112.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Wang H, Chua N-H. Long noncoding RNA transcriptome of plants. Plant Biotechnol J. 2015;13:319–328. doi: 10.1111/pbi.12336. [DOI] [PubMed] [Google Scholar]
- 18.Ingolia NT. Ribosome profiling: New views of translation, from single codons to genome scale. Nat Rev Genet. 2014;15:205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- 19.Juntawong P, Girke T, Bazin J, Bailey-Serres J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:E203–E212. doi: 10.1073/pnas.1317811111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu M-J, et al. Translational landscape of photomorphogenic Arabidopsis. Plant Cell. 2013;25:3699–3710. doi: 10.1105/tpc.113.114769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei L, et al. Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J. 2015;84:1206–1218. doi: 10.1111/tpj.13073. [DOI] [PubMed] [Google Scholar]
- 22.Wethmar K. The regulatory potential of upstream open reading frames in eukaryotic gene expression. Wiley Interdiscip Rev RNA. 2014;5:765–778. doi: 10.1002/wrna.1245. [DOI] [PubMed] [Google Scholar]
- 23.Roy B, von Arnim AG. Translational regulation of cytoplasmic mRNAs. Arabidopsis Book. 2013;11:e0165. doi: 10.1199/tab.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu PY, et al. Super-resolution ribosome profiling reveals novel translation events in Arabidopsis. Proc Natl Acad Sci USA. 2016;113:E7126–E7135. doi: 10.1073/pnas.1614788113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu Q, Ma J, Saghatelian A. Identification and characterization of sORF-encoded polypeptides. Crit Rev Biochem Mol Biol. 2015;50:134–141. doi: 10.3109/10409238.2015.1016215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulitsky I, Bartel DP. lincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pauli A, et al. Toddler: An embryonic signal that promotes cell movement via Apelin receptors. Science. 2014;343:1248636. doi: 10.1126/science.1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson BR, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson DM, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon J-H, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrieri C, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 32.Jabnoune M, et al. A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell. 2013;25:4166–4182. doi: 10.1105/tpc.113.116251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Péret B, Clément M, Nussaume L, Desnos T. Root developmental adaptation to phosphate starvation: Better safe than sorry. Trends Plant Sci. 2011;16:442–450. doi: 10.1016/j.tplants.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 34.Zanetti ME, Chang I-F, Gong F, Galbraith DW, Bailey-Serres J. Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 2005;138:624–635. doi: 10.1104/pp.105.059477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Browning KS, Bailey-Serres J. Mechanism of cytoplasmic mRNA translation. Arabidopsis Book. 2015;13:e0176. doi: 10.1199/tab.0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Misson J, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukoszek R, Feist P, Ignatova Z. Insights into the adaptive response of Arabidopsis thaliana to prolonged thermal stress by ribosomal profiling and RNA-seq. BMC Plant Biol. 2016;16:221. doi: 10.1186/s12870-016-0915-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jorgensen RA, Dorantes-Acosta AE. Conserved peptide upstream open reading frames are associated with regulatory genes in angiosperms. Front Plant Sci. 2012;3:191. doi: 10.3389/fpls.2012.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J. CYTOKININ RESPONSE FACTORs gating environmental signals and hormones. Trends Plant Sci. 2016;21:993–996. doi: 10.1016/j.tplants.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Jeon J, Cho C, Lee MR, Van Binh N, Kim J. CYTOKININ RESPONSE FACTOR2 (CRF2) and CRF3 regulate lateral root development in response to cold stress in Arabidopsis. Plant Cell. 2016;28:1828–1843. doi: 10.1105/tpc.15.00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marin E, et al. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell. 2010;22:1104–1117. doi: 10.1105/tpc.109.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou F, Roy B, von Arnim AG. Translation reinitiation and development are compromised in similar ways by mutations in translation initiation factor eIF3h and the ribosomal protein RPL24. BMC Plant Biol. 2010;10:193. doi: 10.1186/1471-2229-10-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schepetilnikov M, et al. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 2013;32:1087–1102. doi: 10.1038/emboj.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Iwakawa H-O, Tomari Y. Molecular insights into microRNA-mediated translational repression in plants. Mol Cell. 2013;52:591–601. doi: 10.1016/j.molcel.2013.10.033. [DOI] [PubMed] [Google Scholar]
- 46.Li S, et al. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell. 2013;153:562–574. doi: 10.1016/j.cell.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S, et al. Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. Elife. 2016;5:1–24. doi: 10.7554/eLife.22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol. 2005;15:2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 49.Merchante C, et al. Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell. 2015;163:684–697. doi: 10.1016/j.cell.2015.09.036. [DOI] [PubMed] [Google Scholar]
- 50.Calviello L, et al. Detecting actively translated open reading frames in ribosome profiling data. Nat Methods. 2016;13:165–170. doi: 10.1038/nmeth.3688. [DOI] [PubMed] [Google Scholar]
- 51.Bischof S, et al. Plastid proteome assembly without Toc159: Photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins. Plant Cell. 2011;23:3911–3928. doi: 10.1105/tpc.111.092882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reiland S, et al. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 2009;150:889–903. doi: 10.1104/pp.109.138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Svozil J, Hirsch-Hoffmann M, Dudler R, Gruissem W, Baerenfaller K. Protein abundance changes and ubiquitylation targets identified after inhibition of the proteasome with syringolin A. Mol Cell Proteomics. 2014;13:1523–1536. doi: 10.1074/mcp.M113.036269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reiland S, et al. Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF) Proc Natl Acad Sci USA. 2011;108:12955–12960. doi: 10.1073/pnas.1104734108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Svozil J, Gruissem W, Baerenfaller K. Proteasome targeting of proteins in Arabidopsis leaf mesophyll, epidermal and vascular tissues. Front Plant Sci. 2015;6:376. doi: 10.3389/fpls.2015.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cai Q, et al. The SAC51 family plays a central role in thermospermine responses in Arabidopsis. Plant Cell Physiol. 2016;57:1583–1592. doi: 10.1093/pcp/pcw113. [DOI] [PubMed] [Google Scholar]
- 57.Hupalo D, Kern AD. Conservation and functional element discovery in 20 angiosperm plant genomes. Mol Biol Evol. 2013;30:1729–1744. doi: 10.1093/molbev/mst082. [DOI] [PubMed] [Google Scholar]
- 58.1001 Genomes Consortium 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell. 2016;166:481–491. doi: 10.1016/j.cell.2016.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tavormina P, De Coninck B, Nikonorova N, De Smet I, Cammue BPA. The plant peptidome: An expanding repertoire of structural features and biological functions. Plant Cell. 2015;27:2095–2118. doi: 10.1105/tpc.15.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsui A, Nguyen AH, Nakaminami K, Seki M. Arabidopsis non-coding RNA regulation in abiotic stress responses. Int J Mol Sci. 2013;14:22642–22654. doi: 10.3390/ijms141122642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Charon C, Moreno AB, Bardou F, Crespi M. Non-protein-coding RNAs and their interacting RNA-binding proteins in the plant cell nucleus. Mol Plant. 2010;3:729–739. doi: 10.1093/mp/ssq037. [DOI] [PubMed] [Google Scholar]
- 62.Yadav V, et al. ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell. 2014;26:3569–3588. doi: 10.1105/tpc.114.129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barberon M, et al. Adaptation of root function by nutrient-induced plasticity of endodermal differentiation. Cell. 2016;164:447–459. doi: 10.1016/j.cell.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 64.Takeuchi H, Higashiyama T. Tip-localized receptors control pollen tube growth and LURE sensing in Arabidopsis. Nature. 2016;531:245–248. doi: 10.1038/nature17413. [DOI] [PubMed] [Google Scholar]
- 65.Bruex A, et al. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 2012;8:e1002446. doi: 10.1371/journal.pgen.1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T. Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ. 2006;29:115–125. doi: 10.1111/j.1365-3040.2005.01405.x. [DOI] [PubMed] [Google Scholar]
- 67.Montgomery TA, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 68.Felippes FF, Weigel D. Triggering the formation of tasiRNAs in Arabidopsis thaliana: The role of microRNA miR173. EMBO Rep. 2009;10:264–270. doi: 10.1038/embor.2008.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Borges F, Martienssen RA. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol. 2015;16:727–741. doi: 10.1038/nrm4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hou C-Y, et al. Global analysis of truncated RNA ends reveals new insights into ribosome stalling in plants. Plant Cell. 2016;28:2398–2416. doi: 10.1105/tpc.16.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jouannet V, et al. Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J. 2012;31:1704–1713. doi: 10.1038/emboj.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gregory BD, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 73.Vazquez F, Hohn T. Biogenesis and biological activity of secondary siRNAs in plants. Scientifica (Cairo) 2013;2013:783253. doi: 10.1155/2013/783253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoshikawa M, et al. A short open reading frame encompassing the microRNA173 target site plays a role in trans-acting small interfering RNA biogenesis. Plant Physiol. 2016;171:359–368. doi: 10.1104/pp.16.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meteignier L-V, et al. NB-LRR signaling induces translational repression of viral transcripts and the formation of RNA processing bodies through mechanisms differing from those activated by UV stress and RNAi. J Exp Bot. 2016;67:2353–2366. doi: 10.1093/jxb/erw042. [DOI] [PubMed] [Google Scholar]
- 76.Li X. Infiltration of Nicotiana benthamiana protocol for transient expression via Agrobacterium. Bio-Protoc. 2011;101:e95. [Google Scholar]
- 77.Karimi M, Depicker A, Hilson P. Recombinational cloning with plant gateway vectors. Plant Physiol. 2007;145:1144–1154. doi: 10.1104/pp.107.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Varkonyi-Gasic E, Hellens RP. Quantitative stem-loop RT-PCR for detection of microRNAs. Methods Mol Biol. 2011;744:145–157. doi: 10.1007/978-1-61779-123-9_10. [DOI] [PubMed] [Google Scholar]
- 79.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen H, Shiroguchi K, Ge H, Xie XS. Genome-wide study of mRNA degradation and transcript elongation in Escherichia coli. Mol Syst Biol. 2015;11:808. doi: 10.15252/msb.20159000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mustroph A, Juntawong P, Bailey-Serres J. Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol Biol. 2009;553:109–126. doi: 10.1007/978-1-60327-563-7_6. [DOI] [PubMed] [Google Scholar]
- 82.Juntawong P, Hummel M, Bazin J, Bailey-Serres J. Ribosome profiling: A tool for quantitative evaluation of dynamics in mRNA translation. Methods Mol Biol. 2015;1284:139–173. doi: 10.1007/978-1-4939-2444-8_7. [DOI] [PubMed] [Google Scholar]
- 83.Wang L, et al. A low-cost library construction protocol and data analysis pipeline for Illumina-based strand-specific multiplex RNA-seq. PLoS One. 2011;6:e26426. doi: 10.1371/journal.pone.0026426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zheng Q, et al. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 2010;6:e1001141. doi: 10.1371/journal.pgen.1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Juntawong P, Bazin J, Hummel M, Bailey-Serres J, Girke T. 2015 systemPipeR workflow for Ribo-seq and polyRibo-seq experiments. Available at www.bioconductor.org/. Accessed October 1, 2017.
- 86.Cheng C-Y, et al. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017;89:789–804. doi: 10.1111/tpj.13415. [DOI] [PubMed] [Google Scholar]
- 87.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Axtell MJ. ShortStack: Comprehensive annotation and quantification of small RNA genes. RNA. 2013;19:740–751. doi: 10.1261/rna.035279.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mustroph A, et al. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:18843–18848. doi: 10.1073/pnas.0906131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morgan M, et al. ShortRead: A bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25:2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.H Backman TW, Girke T. systemPipeR: NGS workflow and report generation environment. BMC Bioinformatics. 2016;17:388. doi: 10.1186/s12859-016-1241-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Quinlan AR, Hall IM. The BEDTools manual. Genome. 2010;16:1–77. [Google Scholar]
- 94.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ben Amor B, et al. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009;19:57–69. doi: 10.1101/gr.080275.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rau A, Gallopin M, Celeux G, Jaffrézic F. Data-based filtering for replicated high-throughput transcriptome sequencing experiments. Bioinformatics. 2013;29:2146–2152. doi: 10.1093/bioinformatics/btt350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hirsch-Hoffmann M, Gruissem W, Baerenfaller K. pep2pro: The high-throughput proteomics data processing, analysis, and visualization tool. Front Plant Sci. 2012;3:123. doi: 10.3389/fpls.2012.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baerenfaller K, et al. pep2pro: A new tool for comprehensive proteome data analysis to reveal information about organ-specific proteomes in Arabidopsis thaliana. Integr Biol. 2011;3:225–237. doi: 10.1039/c0ib00078g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.