Three-dimensional visualization identifies structural remodeling in chloroplasts during barley stripe mosaic virus infection.
Abstract
Chloroplasts are multifunctional organelles whose morphology is affected by environmental stresses. Although the three-dimensional (3D) architecture of thylakoid membranes has been reported previously, a 3D visualization of chloroplast under stress has not been explored. In this work, we used a positive-strand RNA ((+)RNA) virus, barley stripe mosaic virus (BSMV) to observe chloroplast structural changes during infection by electron tomography. The analyses revealed remodeling of the chloroplast membranes, characterized by the clustering of outer membrane-invaginated spherules in inner membrane-derived packets. Diverse morphologies of cytoplasmic invaginations (CIs) were evident with spherules at the periphery and different sized openings connecting the CIs to the cytoplasm. Immunoelectron microscopy of these viral components verified that the aberrant membrane structures were sites for BSMV replication. The BSMV αa replication protein localized at the surface of the chloroplasts and played a prominent role in eliciting chloroplast membrane rearrangements. In sum, our results have revealed the 3D structure of the chloroplasts induced by BSMV infection. These findings contribute to our understanding of chloroplast morphological changes under stress conditions and during assembly of plant (+)RNA virus replication complexes.
Chloroplasts are responsible for the eukaryotic photosynthesis and carbon fixation, thus providing energy for much of life on the earth. Chloroplast biogenesis is a complex process and is highly integrated with cellular and plant development (Yang et al., 2010; Pogson et al., 2015). Although three-dimensional (3D) models of the thylakoid membrane architecture have been created using electron tomography of higher plant chloroplasts (Shimoni et al., 2005; Daum et al., 2010; Austin and Staehelin, 2011; Kowalewska et al., 2016) and Chlamydomonas reinhardtii chloroplasts (Engel et al., 2015), a 3D visualization of chloroplast changes in response to external stresses (e.g. viral attack) has not been investigated.
Positive-strand RNA viruses often manipulate host membrane systems to form microenvironments for replication and use diverse intracellular membranes to assemble viral replication complexes (VRCs; Salonen et al., 2005; Laliberté and Sanfaçon, 2010; Verchot, 2011; Romero-Brey and Bartenschlager, 2014). The VRCs, on one hand, are thought to shield viruses from host defense systems such as RNA silencing, and on the other hand, provide a microenvironment for enriching viral replication proteins, diverse host factors, and energy needed for efficient replication of progeny viral RNAs (Schwartz et al., 2004; Novoa et al., 2005; Miller and Krijnse-Locker, 2008; Verchot, 2011). VRCs have been detected in mitochondrial membranes (Kopek et al., 2007), endoplasmic reticulum membranes (Restrepo-Hartwig and Ahlquist, 1996), chloroplast membranes (Hatta et al., 1973), and peroxisomal membranes (Fernández de Castro et al., 2017). Intriguingly, some viruses, such as tomato bushy stunt virus, can utilize alternative organelles for replication (Jonczyk et al., 2007; Xu and Nagy, 2014). Despite the different locations of VRCs, the viral replication compartments share some common features such as invaginated vesicles/spherules or double membrane vesicle structures and a pore-like opening to the cytoplasm (den Boon et al., 2010; Paul and Bartenschlager, 2013).
Random sectioning used in conventional electron microscopy often misses crucial and subtle structural dimensions such as the sizes and shapes of vesicles (Baumeister, 2002). To more fully understand membrane structures within organelles and their functions, electron tomography based on transmission electron microscopy (TEM) or dual beam focused ion beam scanning electron microscopy (FIB-SEM) are increasingly used for 3D reconstructions of altered membranes during viral infection (Laliberté and Zheng, 2014; Risco et al., 2014; Harak and Lohmann, 2015; Fernández de Castro et al., 2017). Kopek et al. reported the first 3D architecture of the membrane-bound (+)RNA virus replication compartments in flock house virus-infected Drosophila cells (Kopek et al., 2007). A recent electron tomographic analysis of turnip mosaic virus (TuMV)-induced intracellular rearrangements revealed that the vesicle-like structures apparent in two-dimensional (2D) TEM images are, in fact, tubules (Wan et al., 2015). In 2015, a reconstruction analysis of ER-derived membranous spherules induced during beet black scorch virus infection provided the first 3D model of VRCs of a plant (+)RNA virus (Cao et al., 2015). Gómez-Aix et al. also determined the 3D architecture of remodeled mitochondria in melon necrotic spot virus-infected cells by focused ion beam-field emission scanning electron microscopy (Gómez-Aix et al., 2015). Collectively, electron tomography has become a powerful tool that can accurately define the overall architecture of intracellular membranes as well as changes in organelles during virus infection.
Viral proteins often play crucial roles in mediating the rearrangement of host endomembrane systems during virus infection (Paul and Bartenschlager, 2013). For example, hepatitis C virus NS5A and NS4B can induce the formation of double- and single-membrane vesicles, respectively (Romero-Brey et al., 2012). Brome mosaic virus (BMV) expression of different ratios of the 1a and 2a proteins induces distinct spherules with altered sizes (Schwartz et al., 2004). The closterovirus 1a and 1ab proteins also facilitate membrane remodeling and formation of multivesicular replication platforms (Gushchin et al., 2017). TuMV 6K2 is responsible for endomembrane alterations (Beauchemin et al., 2007). In addition, it has also become evident that remodeling of host endomembrane systems involves participation of viral RNAs and diverse host factors (Kallio et al., 2013; Wang, 2015; Kovalev et al., 2016).
Barley stripe mosaic virus (BSMV) is the type member of the genus Hordeivirus and infects numerous monocots and dicots (Jackson et al., 2009). BSMV has a tripartite positive-strand RNA genome encoding seven major proteins. The αa and γa proteins are encoded by the RNAα and RNAγ genomes, respectively, and are essential for BSMV RNA replication. RNAβ encodes the coat protein (CP) for viral RNA encapsidation, and subgenomic (sg) RNAs derived from RNAβ serve as mRNAs for the triple gene block proteins responsible for cell-to-cell movement. The multifunctional γb protein is a BSMV pathogenicity determinant that is translated from sgRNAγ (Jackson et al., 2009). During BSMV infection and VRC formation, chloroplast abnormalities are induced that include cytoplasmic invaginations (CIs) and peripheral vesicles in the chloroplasts (Carroll, 1970; McMullen et al., 1978; Lin and Langenberg, 1984, 1985; Torrance et al., 2006). Although previous studies indicated that chloroplasts are deformed in BSMV-infected plant cells, aberrant chloroplast structures arising during infection, particularly the 3D architecture and the molecular mechanisms underlying formation of the chloroplast-derived membranous structures, have not been investigated in detail. To clarify details of the chloroplast architecture in BSMV-infected Nicotiana benthamiana, we performed electron tomography to unravel the 3D structure of abnormal chloroplast-derived VRCs and CIs and carried out experiments to explore the functions of viral factors functioning in remodeling the chloroplasts.
RESULTS
Ultrastructural Analysis of BSMV-Infected Cells by Transmission Electron Microscopy
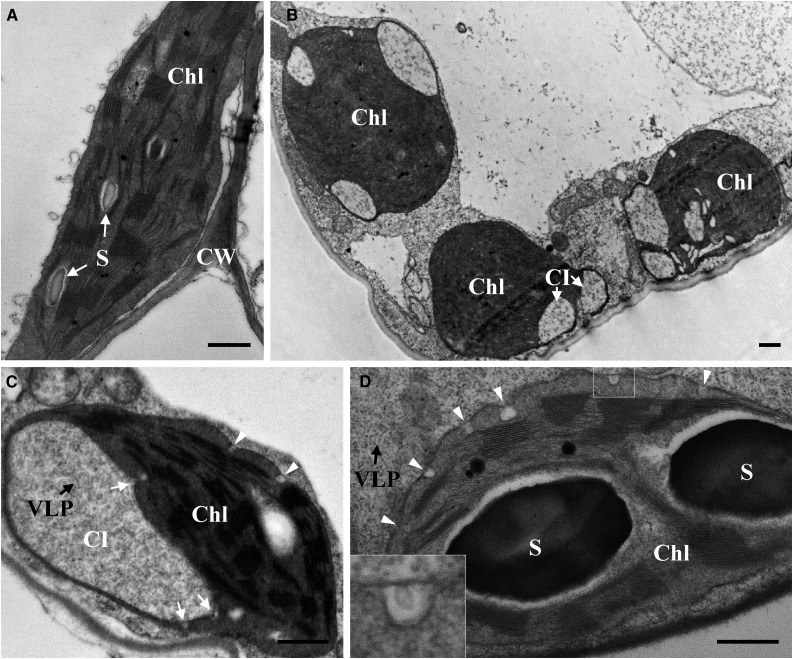
To investigate the ultrastructural changes in cells during BSMV infection, leaf tissues from healthy or BSMV-infected N. benthamiana leaves were chemically fixed and embedded in resin for TEM analysis. During BSMV infection, dramatic alterations of chloroplast structure occurred as CIs containing large amounts of virus-like particles (VLPs), and peripheral invaginations appeared (Fig. 1, B–D), whereas similar structures were absent in uninfected N. benthamiana chloroplasts (Fig. 1A). Interestingly, membrane invaginations connected to CIs were also observed at the periphery of the CIs (Fig. 1C, white arrows), and they were very similar to the other invaginations at the chloroplast periphery (Fig. 1, C and D, white arrowheads). In addition, spherules with an ∼50 nm diameter were observed occasionally within chloroplast envelope invaginations (Fig. 1D, boxed region). Moreover, other cellular changes, including membrane inclusions and elongated mitochondria, were observed occasionally (Supplemental Fig. S1, A and B). At the late stages of BSMV infection, cytoplasm containing large amounts of VLPs often protruded into the vacuole (Supplemental Fig. S1C). Taken together, these results reveal that numerous dramatic cytopathological changes are induced during BSMV infection, in particular chloroplast membrane remodeling.
Figure 1.
Ultrastructural changes in chloroplasts during BSMV infection of N. benthamiana. A, Healthy N. benthamiana leaves served as a control. B to D, Deformed chloroplasts in BSMV-infected N. benthamiana leaf tissues. B, CIs present in chloroplasts within a BSMV-infected cell. C, Virus-like particles (black arrows) were present within a CI, and membrane invaginations were observed at both the periphery of CI and the chloroplast envelope membranes (white arrows and arrowheads). D, Invaginations present around the chloroplast envelope (white arrowheads). Inset area shows magnification of the spherule that was occasionally observed inside the invagination. Chl, Chloroplast; S, starch granule; CI, cytoplasmic invagination; VLP, virus-like particle; CW, cell wall. Scale bars, 500 nm.
Localization of BSMV Double-Stranded RNAs (dsRNAs) and Proteins in BSMV-induced Chloroplast Membrane Structures
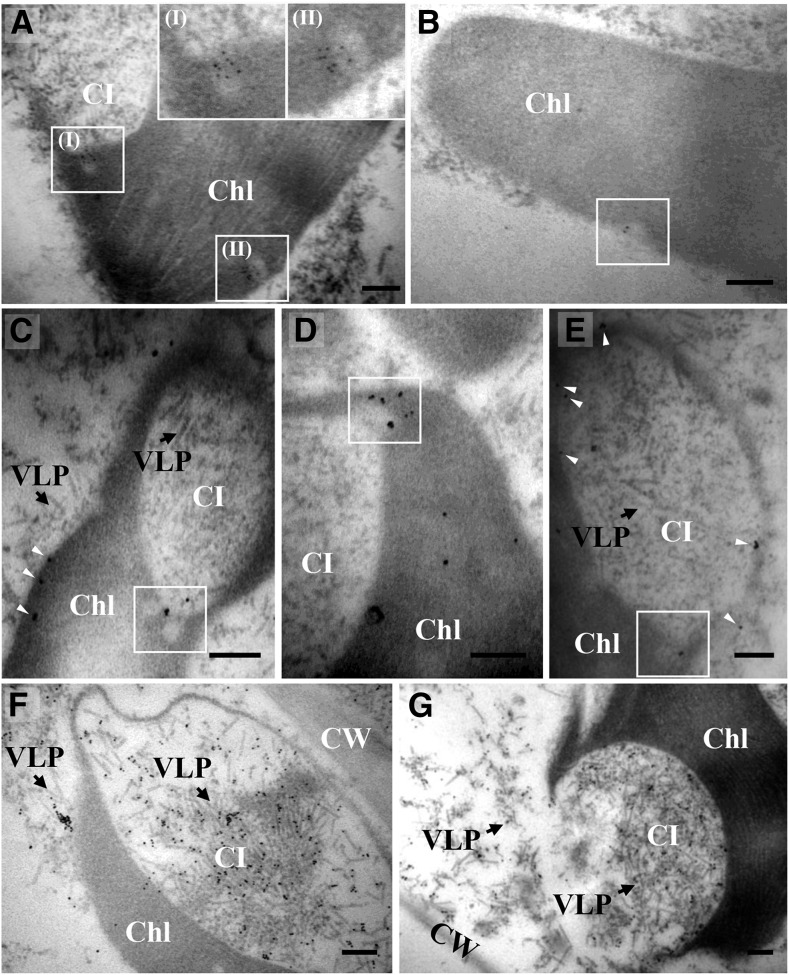
Our previous results revealed that chloroplasts are sites for replication of BSMV (Zhang et al., 2017). To further determine whether the BSMV replication protein αa and dsRNA replicative intermediates associate with chloroplast-derived membrane structures, immunoelectron microscopy was performed. Gold particles conjugated to dsRNA antibodies specifically labeled chloroplast invaginations of the chloroplast envelope or at the periphery of the CIs (Fig. 2, A and B, boxed regions). Meanwhile, antibodies against the αa protein showed specific labeling of the chloroplasts, which was characterized by the distribution of gold particles on the envelope membranes, invaginations, and the CI membranes (Fig. 2, C–E, boxed regions and arrowheads). In addition, labeling with a BSMV CP-specific antiserum revealed specific decoration of VLPs within the CIs, as well as the cytoplasm (Fig. 2, F and G), demonstrating that the abundant VLPs present within the CIs are indeed the BSMV virions. For control healthy leaf tissue, only a very low background labeling was detected in the chloroplasts or other cellular regions (Supplemental Fig. S2).
Figure 2.
Immunoelectron microscopy of viral dsRNA and proteins in BSMV-infected N. benthamiana cells. A and B, Immunogold labeling of dsRNA in BSMV-infected N. benthamiana cells. Insets I and II show magnifications of the corresponding boxed regions. C to E, αa localization at envelope membranes (white arrowheads), small invaginations (boxed regions), and CI periphery (white arrowheads) in BSMV-infected N. benthamiana cells. Black arrows point to virus-like particles. Boxed areas show the invaginations at the CI periphery. F and G, CP-specific antiserum showed specific labeling of virus-like particles (black arrows) in the cytoplasm and CIs. Note: OsO4 fixation of the leaf tissues was omitted to preserve antigenicity of target molecules. Hence, the fine structure of cellular membranes could not be unambiguously visualized. Chl, Chloroplast; CI, cytoplasmic invagination; VLP, virus-like particle; CW, cell wall. Scale bars, 200 nm.
Moreover, chloroplasts from mock- and BSMV-inoculated N. benthamiana leaves were purified and subjected to western blot and reverse transcription (RT)-PCR analyses (Supplemental Fig. S3). The results showed that the αa protein and the CP can be readily detected in chloroplasts purified from the BSMV-infected leaf tissues, but not from mock-inoculated plants (Supplemental Fig. S3A). As a control, potential contamination of the purified chloroplasts by cytoplasmic constituents was eliminated because the cytoplasmic-localized phosphoenolpyruvate carboxylase (PEPC) was only detected in total protein extracts from leaf tissues, but not from isolated chloroplasts (Supplemental Fig. S3A). RT-PCR analysis also showed that both plus- and minus-strand BSMV RNAs were present in the isolated chloroplasts. Controls also indicated that chloroplast PSII core protein C was restricted to the chloroplast fraction, whereas cytoplasmic 18S rRNA (18S rRNA) control was difficult to detect in the isolated chloroplasts (Supplemental Fig. S3B). Collectively, these data indicate that viral RNA synthesis occurs in the chloroplast membrane invaginations.
3D Reconstruction of BSMV-Induced Chloroplast Membrane Structures
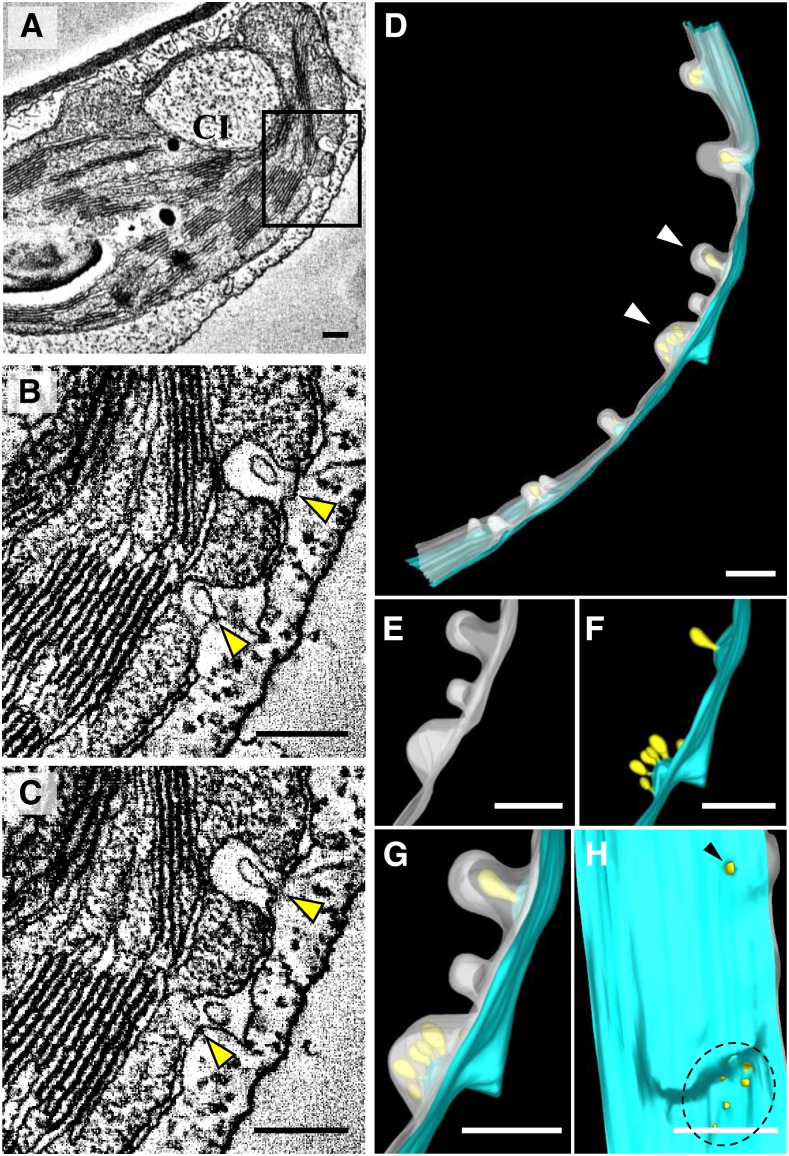
To characterize the 3D architecture of BSMV replication factories, and the CIs of chloroplasts induced by BSMV, we performed electron tomography analysis. The results showed that peripheral invaginations and spherules from the envelope membranes and large CIs are present in the remodeled chloroplasts (Fig. 3A), whereas such structural abnormalities in chloroplasts were absent in healthy N. benthamiana cells (Supplemental Movie 1). The peripheral invaginations were continuous with the inner chloroplast membrane, but not the outer membrane (Fig. 3, B and C, arrowheads), and spherules were frequently observed inside the inner membrane-invaginated packets (Fig. 3, B and C; Supplemental Movie 2). These spherules were connected to the outer membrane in some slices (Fig. 3, B and C), indicating that the smaller spherules are invaginations of the outer membrane. In addition, one or more spherules were surrounded by inner membrane-derived packets (Fig. 3, B and C; Supplemental Movie 2), and the CIs with large amounts of virus particles inside were double-membrane bounded (Fig. 3A; Supplemental Fig. S4, A and B). A 3D surface rendering of the chloroplast membranes clearly demonstrates that all of the spherules (Fig. 3D, light yellow) originate from the outer membranes (Fig. 3D, light blue) and have a tight neck-like opening to the cytoplasm (Fig. 3H, arrowhead and black dashed circle). The inner chloroplast membrane-derived invaginations (Fig. 3, D, E, and G, translucent white) form packets of varying sizes that enclose one or more outer membrane-derived spherules (Fig. 3, D–G; Supplemental Movies 3 and 4). The inner membrane invaginations had an average diameter of 112 ± 41 nm (n = 16). The average length and width of the spherules were 55 ± 8 nm and 42 ± 7 nm (n = 20), respectively, and their necks had an average diameter of 11 ± 4 nm (n = 20). Taken together, 3D electron tomography clearly revealed that the chloroplast invaginations are composed of inner chloroplast membrane-derived packets containing variable numbers of outer membrane-derived spherules. Each of the spherules has a narrow opening to the cytoplasm, supporting a role in BSMV replication.
Figure 3.
Three-dimensional visualization of remodeled chloroplast membranes in BSMV-infected cells by transmission electron tomography. A, Representative tomogram slice generated from a 250-nm-thick section of systemically infected N. benthamiana leaves. BSMV-induced peripheral invaginations and large CI can be observed in chloroplast. Boxed area shows the invaginations at the chloroplast periphery. B and C, Different slices of the boxed area shown in A. Yellow arrowheads show peripheral invaginations with internal spherules. D, 3D model of chloroplast membranes shown in A. Light blue, outer chloroplast membrane; translucent white, inner chloroplast membrane; light yellow, spherules derived from the outer membrane; white arrowheads, peripheral invaginations of the chloroplast. E, Close-up view of the area indicated by white arrowheads in D without showing the outer chloroplast membrane. F, Close-up view of the area indicated by white arrowheads in D without showing the inner chloroplast membrane. G, Close-up view of the area indicated by white arrowheads in D showing both the outer and inner chloroplast membranes. H, A 90° rotation of G highlighting spherule connections (arrowhead and black dashed circle) to the cytoplasm. CI, Cytoplasmic invagination. Scale bars, 200 nm. This tomogram is shown in Supplemental Movies 2 to 4.
FIB-SEM Tomography of the Chloroplasts Reveals Diverse Morphology of CIs During BSMV Infection
In addition to peripheral invaginations on the chloroplast surfaces (Fig. 3D), another remarkable feature of BSMV-induced chloroplast abnormalities is the formation of CIs (Figs. 1 and 3). Electron tomography was also performed to characterize the CIs. Transmission electron tomographic analyses revealed that CIs are bound by double membranes and that some invaginations are present at the periphery of the CIs (Supplemental Fig. S4, A and B). Furthermore, spherules were also observed in these invaginations and were connected to the CI (Supplemental Fig. S4B, arrowheads) and are similar to spherules within the chloroplast membrane packets as described in Figure 3B. A 3D surface rendering of the CI confirmed that these spherules are identical to the chloroplast outer membrane-invaginated spherules, as evidenced by their similar sizes and external connections (Supplemental Fig. S4, C–F; Figure 3D; Supplemental Movies 4 and 5). Due to the large sizes of the CIs, we could only obtain partial CI structures by transmission electron tomography (Supplemental Fig. S4, C and D; Supplemental Movie 5). Nevertheless, these preliminary data imply that the CIs resemble cavern-like structures generated by continuous invagination of the chloroplast envelope membranes.
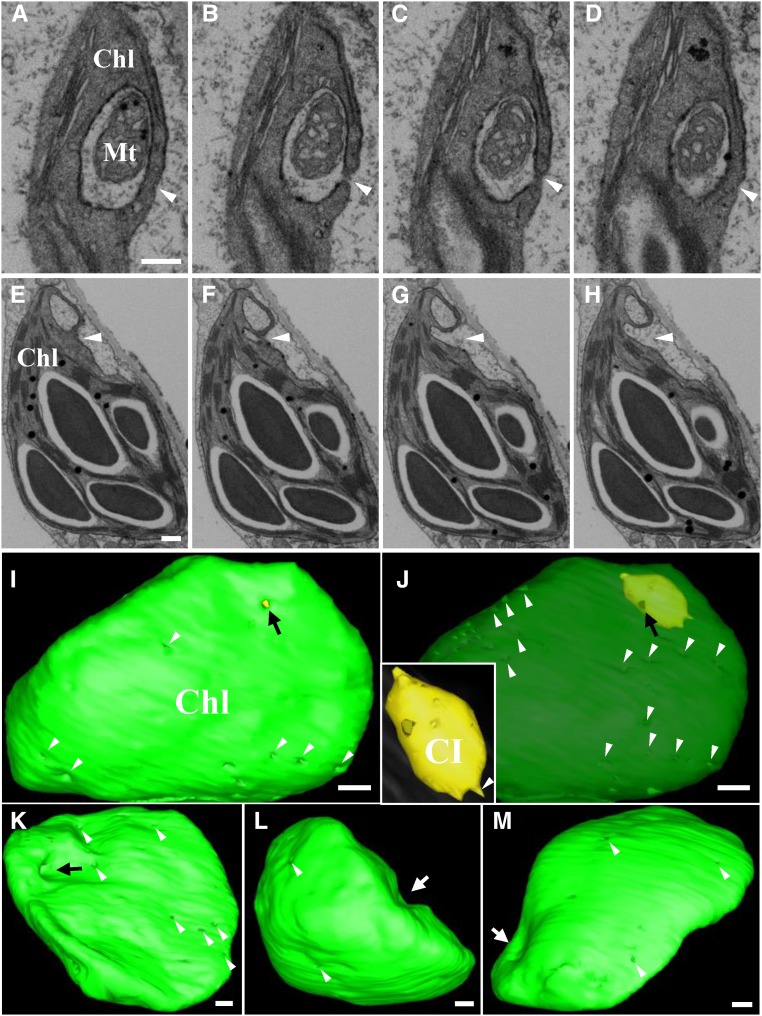
In order to reconstruct whole chloroplast CI structures, FIB-SEM, a powerful tool to determine the structures of large organelles, was performed to reconstruct the whole chloroplast with CIs. Figure 4, A to D, shows different slices from the same chloroplast. The images clearly show that a small CI opening was present in some slices (Fig. 4, B and C, arrowheads), but that the openings were not evident in contiguous slices (Fig. 4, A and D, arrowheads). These results indicate that the above CI has a very small opening because the distance between the two slices is only 50 nm. In addition, a mitochondrion was enclosed in the CI of this chloroplast (Fig. 4, A–D). However, in some chloroplasts, the opening of the CI is very large (Fig. 4, E–H, arrowheads). A 3D reconstruction of the chloroplast shown in Figure 4, A–D reveals a large number of small invaginations (Fig. 4, I and J, white arrowheads) at the surface of the chloroplast (Fig. 4, I and J, green envelope) that correspond to the membrane invaginations shown in Figure 3D. Meanwhile, small opening of the CI was present on the surface of the chloroplast (Fig. 4J, black arrow). In order to visualize the 3D architecture of the CI in more detail, the color of the chloroplast membrane was changed to translucent (Fig. 4J; Supplemental Fig. S5B). The results of this manipulation show that the CIs (yellow) have irregular shapes (Fig. 4J; Supplemental Fig. S5, A and B), with many protrusions present on the CI surface (Fig. 4J; Supplemental Fig. S5B; Supplemental Movies 6 and 7), that correspond to the small invaginations around the CIs. These data suggest that the double-membrane bound CIs with openings are also generated by the invagination of the chloroplast membranes. A 3D reconstruction of the chloroplast shown in Figure 4, E to H, was also performed. In addition to small invaginations on the surface of the chloroplast envelope membranes (Fig. 4, K–M, white arrowheads), the big opening shown in Figure 4, E to H (white arrowheads), was, in fact, a deep invagination of chloroplast envelope (Fig. 4K, black arrow; Supplemental Movie 8). Intriguingly, cavern-like invaginations were frequently observed on the surface of the chloroplast (Fig. 4, L and M, white arrows), indicating the irregular appearance of chloroplast membranes under viral attack (Supplemental Movie 8). The experiments described above provide a high-resolution model of chloroplast remodeling occurring during BSMV infection and suggest that the complex invaginations provide sites for virus replication.
Figure 4.
Three-dimensional visualization of BSMV-induced cytoplasmic invaginations by FIB-SEM. A to D, Four different slices from the same chloroplast selected to illustrate CI structures. Arrowheads indicate 3D variations in the CI opening. E to H, A series of slices from another chloroplast showing morphological changes of the CI. Arrowheads indicate 3D variations in the CI opening. I to J, A 3D surface rendering of the chloroplast tomogram in A to D. Green, chloroplast; Yellow, CI. To emphasize the CI (yellow) inside the chloroplast, the appearance of chloroplast shown in I was changed to translucent (J). White arrowheads indicate chloroplast envelope invaginations similar to those in Fig. 3D; black arrow point to the CI opening. The image inset in J is an enlarged view of the CI. Note: the 3D model in I to J represent partial 3D architecture of the chloroplast shown in A to D; see Supplemental Movies 6 and 7 for the complete 3D model. K to M, Different views of the 3D model corresponding to the chloroplast shown in E to H. White arrowheads indicate chloroplast envelope invaginations similar to those in Fig. 3D; black arrow points to the CI apertures; white arrows indicate cavern-like invaginations present on the surface of the chloroplasts. This tomogram is shown in Supplemental Movie 8. Chl, Chloroplast; CI, cytoplasmic invagination; Mt, mitochondria. Scale bars, 500 nm.
The αa Replication Protein Plays a Major Role in Initiating Chloroplast Remodeling during BSMV Infection
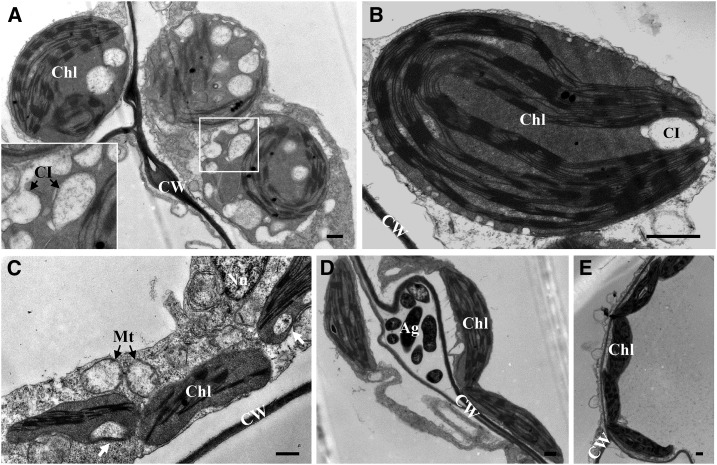
To identify BSMV components contributing to chloroplast remodeling, Agrobacterium tumefaciens derivatives harboring pCass4-Rz-BSMV (pCaBS)-α and pCaBS-γ, which are sufficient to support BSMV replication (Jackson et al., 2009), were coinfiltrated into N. benthamiana leaves. TEM analysis was performed at 3 d postinoculation (dpi). As shown in Figure 5, expression of RNAα and RNAγ is sufficient to induce peripheral invaginations and large CIs in chloroplasts (Fig. 5, A and B), and more than 70% of the chloroplasts observed in the infiltrated regions developed structural anomalies similar to those of wild-type BSMV-infected plants (Fig. 1). Some CIs had large openings to the cytosol (Fig. 5A), in agreement with the feature of the CIs as described in Figure 4. Next, each component encoded by RNAα and RNAγ was transiently expressed in N. benthamiana leaves. In order to achieve high expression levels and easy detection of protein expression, the αa and γa genes were fused to the 5′ terminus of the GFP gene and cloned under control of the CBF3 super promoter (Chinnusamy et al., 2003; Yang et al., 2010). The results revealed that αa-GFP can induce formation of CIs similar to those observed in BSMV-infected cells (Fig. 5C, white arrows), and the altered chloroplasts accounted for about 20% of the observed chloroplasts. In contrast, CIs were not observed in either γa-GFP or γb-infiltrated leaves (Fig. 5, D and E). Expression of these three proteins was confirmed by western blotting (data not shown). However, we should point out that despite the ability of αa-GFP to induce CI formation, spherules were difficult to observe in αa-GFP-expressing leaf tissues (Fig. 5C). This may be explained by a requirement for coexpression of the γa, γb, or perhaps BSMV RNA components for complete formation of membranous replication vesicles. Irrespectively, our results indicate that αa is a major factor mediating chloroplast membrane rearrangements during BSMV infection.
Figure 5.
Function of αa in chloroplast remodeling during BSMV infection. TEM of agroinfiltrated leaf tissues infected with BSMV. A and B, TEM of N. benthamiana leaf tissues coexpressing BSMV RNAα and RNAγ stands. Note the altered chloroplast membrane structures like cytoplasmic invaginations and peripheral invaginations. Inset: enlarged box in A shows cytoplasmic invaginations with openings facing the cytoplasm. C to E, TEM of N. benthamiana leaf tissues agroinfiltrated with bacteria harboring αa-GFP (C), γa (D), and γb (E) plasmids. Arrows indicate the CIs. Chl, Chloroplast; CI, cytoplasmic invagination; CW, cell wall; Mt, mitochondria; Nu, nucleus; Ag, Agrobacteria in intercellular spaces. Scale bars, 500 nm.
αa Localizes to the Surface of the Chloroplast
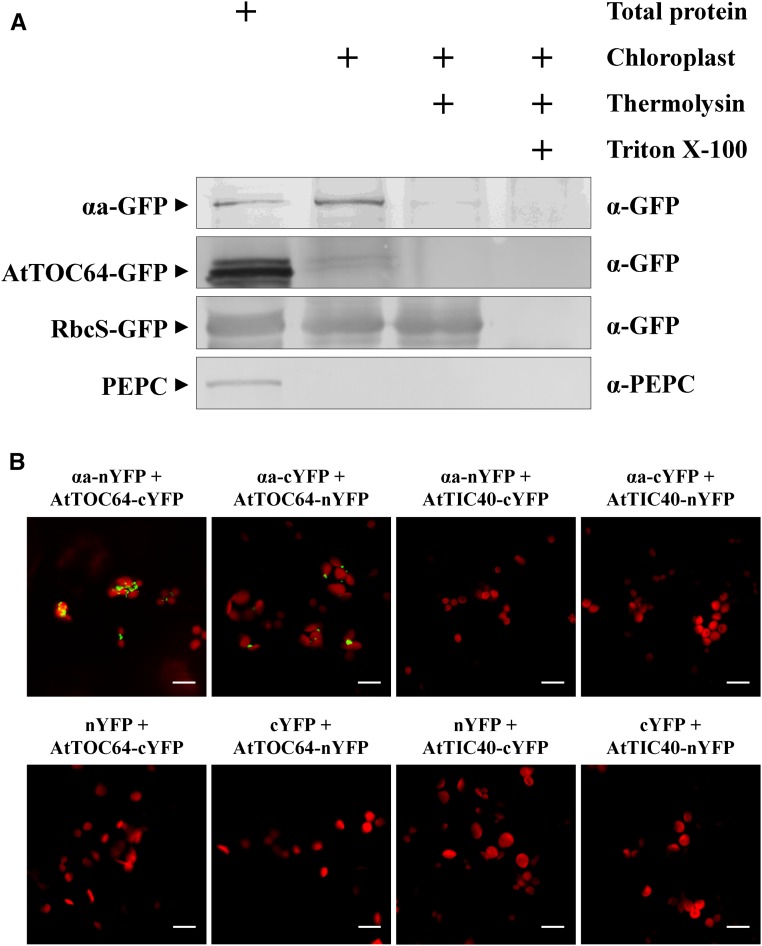
Our previous reports revealed that αa-GFP localizes to the chloroplasts (Zhang et al., 2017), and our current immuno-EM analyses demonstrate that αa also is targeted to the membranous invaginations (Fig. 2, C–E). To investigate whether αa localizes to the surface or the interior of the chloroplasts, protease protection assays were performed as previously described (Ham et al., 2006). Intact chloroplasts were isolated from N. benthamiana leaves infiltrated with Agrobacterium harboring αa-GFP, AtTOC64-GFP (Breuers et al., 2012), or chloroplast stroma marker ribulose-1,5-bisphosphate carboxylase small subunit GFP fusion (RbcS-GFP) plasmids (Lee et al., 2002), and the chloroplasts were treated with thermolysin in the presence or absence of Triton X-100. The results showed that GFP-fused αa (αa-GFP) was only detected in purified chloroplasts prior to thermolysin incubation (Fig. 6A). Control leaves expressing AtTOC64-GFP, a protein localized to the outer membrane of the chloroplast (Breuers et al., 2012), also had similar results. In contrast, the internal RbcS-GFP protein (Lee et al., 2002) was digested by thermolysin only after the chloroplast membrane was permeabilized by Triton X-100 (Fig. 6A).
Figure 6.
αa localizes to the surface of the chloroplasts. A, Protease protection assay to determine chloroplast localization of αa. Intact chloroplasts were isolated from N. benthamiana leaves expressing αa-GFP, AtTOC64-GFP, or RbcS-GFP and subjected to thermolysin digestion with or without Triton X-100 treatment. Protein samples were prepared from 5-μg chlorophyll equivalent of chloroplasts and subjected to western blot analyses using antibodies specified on the right of the panels. Phosphoenolpyruvate carboxylase (PEPC), a cytoplasmic enzyme, served as a control to assess cytosolic contaminations of isolated chloroplasts. Arrowheads indicate the specific band of the target protein. B, BiFC analysis of interactions between the BSMV αa protein and the chloroplast outer membrane protein AtTOC64 or the inner membrane protein AtTIC40. YFP signals are depicted as a false-green color, and chlorophyll autofluorescence is displayed in red. Scale bars, 10 μm.
In addition, a bimolecular fluorescence complementation (BiFC)-based method described previously was used to confirm the localization of αa to the cytosolic surface of the chloroplast (Zamyatnin et al., 2006; Chen et al., 2016). AtTOC64 is an outer-membrane-targeting protein whose C-terminal domain is exposed on the cytosolic surface (Qbadou et al., 2007), which provides putative binding to the αa protein. In contrast, contact between the αa and AtTIC40 proteins is unlikely in living cells, as AtTIC40 localizes to the chloroplast inner membranes (Chou et al., 2003). These protein sequences were engineered into BiFC vectors to provide C-terminal fusions with split YFP derivatives and transformed into A. tumefaciens. At 3 dpi, confocal laser-scanning microscopic analysis of the infiltrated leaf tissues revealed that YFP signals could be observed only for the combinations of AtTOC64 and αa, and the interaction between AtTOC64 and αa occurs on the chloroplast surface, in agreement with the subcellular localization of either AtTOC64 and αa (Fig. 6B). In contrast, coexpression of the αa-nYFP and AtTIC40-cYFP or the αa-cYFP and AtTIC40-nYFP combinations failed to result in fluorescence signals (Fig. 6B), and control leaves coexpressing split YFP-fused AtTOC64 or AtTIC40 with the corresponding empty BiFC vectors also did not elicit fluorescence signals (Fig. 6B, bottom). These results indicate that the C terminus of αa faces the cytoplasm, like AtTOC64, and are consistent with the results of the protease protection assay showing that αa-GFP is sensitive to thermolysin. Furthermore, total protein extracts from N. benthamiana leaves agroinfiltrated with the various BiFC constructs were subjected to western blot analyses with GFP antibodies to demonstrate that all of the split YFP-fused proteins are expressed in the infiltrated leaf tissues (Supplemental Fig. S6). Collectively, these data demonstrated that αa localizes at the surface of the chloroplasts.
DISCUSSION
Chloroplasts are one of the most important plant cell organelles because they are responsible for photosynthesis, synthesis of major phytohormones, participation in defense response, and are crucial for interorganelle communication. Chloroplast biogenesis is a complex process that is affected by environmental factors such as salt, high temperature, light intensity, and other stress factors (Ballantine and Forde, 1970; Gounaris et al., 1984; Salama et al., 1994). In addition, the subject of chloroplast-virus interplay has been recently attracting more and more attention (Zhao et al., 2016; Bhattacharyya and Chakraborty, 2017). In this study, electron tomography of the remodeled chloroplasts in BSMV-infected cells provided a model for the 3D architecture of the reorganized chloroplasts, and the viral proteins involved in the BSMV-induced chloroplast structural changes were also investigated.
BSMV-Induced Chloroplast Rearrangements Are Intimately Associated with Virus Replication
Lin and Langenberg (1985) first used root tips from BSMV-infected wheat cells to probe the location of dsRNA by immunoelectron microscopy, and revealed that peripheral vesicles in proplastids contain dsRNA (Lin and Langenberg, 1985). Our results consistently showed the intimate association of dsRNA with chloroplast peripheral invaginations in BSMV-infected N. benthamiana leaf tissues (Fig. 2, A and B). Our results, in conjunction with those of Lin and Langenberg (1985), suggest that the chloroplast changes induced by BSMV are evolutionarily conserved among monocot and dicot families. In addition, BSMV αa replication protein was also found in close proximity to membranous invaginations (Fig. 2, C–E). These data, together with the pore-like openings presented in the 3D architecture of spherules inside these invaginations (Fig. 3H), unequivocally verifying the chloroplast membrane-derived spherules are BSMV replication sites.
3D Reconstructions of the Remodeled Chloroplasts During BSMV Infection
Although diverse plant-virus interactions are able to cause chloroplast malformations (Zhao et al., 2016), only a limited amount of detailed information is available about structural alterations in chloroplasts during virus infection. Previous studies have shown that turnip yellow mosaic virus (TYMV) and wild cucumber mosaic virus (WCMV) infections lead to the ultrastructural changes in chloroplasts, including peripheral vesicles and cytoplasmic invaginations (Ushiyama and Matthews, 1970; Allen, 1972). However, 3D electron tomography revealed that deformation of chloroplasts induced by BSMV is distinct from that of TYMV or WCMV. The BSMV-induced invaginations of the chloroplast inner membrane are larger than those of TYMV and WCMV, and the enlarged space could frequently encompass more than one spherule (Fig. 3), rather than one in the cases of TYMV and WCMV (Ushiyama and Matthews, 1970; Allen, 1972).
The 3D architecture of BSMV replication compartments also reveals an intriguing parallel among membranous vesicles induced by distantly related viruses (Kopek et al., 2007; Cao et al., 2015), even though these membranes are derived from different organelles (Verchot, 2011; Romero-Brey and Bartenschlager, 2014). A representative example is the neck-like channel that connects the interior of the spherule to the cytosol. It is assumed that these pore-like openings permit entry of virus and cellular components required for replication, and exit of progeny RNAs to the cytoplasm. The structural similarity among membranous vesicles induced by different viruses supports the hypothesis that an evolutionary conserved mechanism underlies the assembly of virus replication factories.
Although the formation of CIs on the chloroplasts has been described in previous 2D EM analyses (McMullen et al., 1978; Lin and Langenberg, 1984, 1985; Torrance et al., 2006), the overall architecture of the CIs and detailed mechanisms whereby CIs are generated during virus infection are obscure. Therefore, electron tomography was performed to reconstruct the 3D structure of BSMV CIs. The results show that abundant virus particles are present in the CIs (Figs. 1 and 2) and that spherules are present at the periphery of the CIs (Supplemental Fig. S4), arguing that replication and packaging of BSMV replicon RNA are tightly coupled processes. Considering the fact that both the BSMV replication proteins αa and γa localize to the chloroplast, targeting of viral replication machinery to the chloroplast is initially mediated by the expression of αa and γa, followed by recruitment of the γb protein, BSMV RNAs, and diverse host factors to the chloroplast to help membranes to curve, stabilize, and activate the replication complexes for replication (Zhang et al., 2017). In addition, a replication-coupled packaging strategy widely utilized in other positive-strand RNA viruses is also applicable to BSMV, as suggested by virus particles present in the chloroplast CIs.
FIB-SEM analyses indicated that the BSMV-induced CIs are irregular cavern-like structures with an opening of varied sizes that have invaginated into the chloroplast interior (Fig. 4). Previous studies reported that potato mop-top virus (PMTV) also elicited formation of cytoplasmic invaginations into chloroplasts during infection (Cowan et al., 2012). Serial sections of the chloroplast in PMTV-infected cells revealed open flask-shaped invaginations (Cowan et al., 2012), similar to those generated by BSMV. However, the PMTV invaginations differ from the BSMV-induced CIs, in that few PMTV particles were observed in these CIs and that peripheral spherules were also absent (Cowan et al., 2012). In addition, in TYMV-infected cells, enlarged membrane spaces between two clumped chloroplasts with large numbers of virus particles inside were observed (Ushiyama and Matthews, 1970), which may have similar nature with BSMV-induced CIs.
In addition, our studies revealed that the shapes of the BSMV-induced CIs differ and that the sizes of the CIs’ openings varied greatly, from large to small openings (Fig. 4), and in some cases, it was difficult to discern openings of the CIs (Fig. 4; Supplemental Movie 6). CIs with varied sizes of openings present in the FIB-SEM analysis allowed us to devise a model whereby invagination of chloroplast membranes proceeds gradually during BSMV infection, leading to the appearance of CIs with diverse morphologies. This can be verified to some extent by the presence of mitochondrion within the CIs (Fig. 4, A–D).
Electron tomography analyses of BSMV-induced structural changes in the chloroplast indicated that spherules were often observed at the periphery of the CIs (Supplemental Fig. S4), suggesting a potential relationship during morphogenesis of these two structures. In addition, chloroplasts with CIs are more prevalent in the late stage of BSMV infection than in the early infected leaf tissues (data not shown), suggesting that the formation of CIs may be resulted from combined effects of BSMV replication and the physiological responses of the chloroplasts to virus attack. Also, abundant viral particles present inside the CIs might imply that involvement of BSMV virions contributes to some extent to the formation of CIs with diverse morphologies.
The αa Protein Is Essential But not Sufficient for the Formation of Chloroplast Membrane-Derived Vesicles
Virus-induced remodeling of host endomembranes is a complicated process that has been shown to involve participation of diverse viral and host factors. For example, formation of ER-derived spherules during BMV infection requires the viral 1a’s amphipathic α-helix, helix A, (Liu et al., 2009), and 1a-1a interactions (O’Reilly et al., 1997; Diaz et al., 2012). In addition, several host proteins, including membrane-shaping reticulon proteins (Diaz et al., 2010), endosomal sorting complex required for transport proteins (Diaz et al., 2015), and a lipid synthesis-related choline requiring2 protein (Zhang et al., 2016), are recruited by 1a to form spherules.
Our studies reveal that the αa replication protein has a significant role in rearranging the chloroplast membranes. This finding is in agreement with many previous reports showing that one or more virus-encoded proteins contribute predominately to the ultrastructural changes in cellular endomembrane systems (Schwartz et al., 2002; Liu et al., 2009; Welsch et al., 2009; Romero-Brey et al., 2012). However, this does not mean that the roles of other viral components in remodeling cellular membranes are negligible. In fact, we detected the BSMV αa replication protein and abundant BSMV virions in the CIs and chloroplast-derived spherules (Figs. 1 and 2). We also previously detected colocalization of the αa and γa proteins, as well as γb, in chloroplasts (Jackson et al., 2009; Zhang et al., 2017), implying that these viral components, together with the αa, may coordinately contribute to chloroplast remodeling and formation of malformed chloroplasts with diverse morphologies (Fig. 5). Hence, these findings provide the basis for further studies underway to investigate the detailed molecular mechanisms responsible for the chloroplast remodeling during BSMV infection.
In conclusion, our results enhance our understanding of chloroplast biogenesis under stress conditions and present the first 3D reconstruction of aberrant chloroplast structures induced in response to a (+)RNA virus infection and provide new insights into the assembly of (+)RNA virus replication complex.
MATERIALS AND METHODS
Plasmid Construction
All plasmids were constructed according to the standard molecular cloning methods (Sambrook and Russell, 2001). For transient expression assays, the γa and γb genes were amplified from pCaBS-γ (Yuan et al., 2011; Hu et al., 2015) and cloned into pSuper1300-GFP containing the CBF3 super promoter (Chinnusamy et al., 2003; Yang et al., 2010) and pGD as described previously (Zhang et al., 2017). For BiFC assays, AtTOC64 and AtTIC40 were cloned from the plasmids AtTOC64-GFP and AtTIC40-GFP (Breuers et al., 2012), respectively, and inserted into pSPYCE-35S or pSPYNE-35S (Walter et al., 2004). The plasmids pSPYCE-35S-NbrbcL and pSPYNE-35S-NbrbcL were described in a previous report (Cao et al., 2015). For protease protection assays, the NbRbcS gene was amplified from Nicotiana benthamiana cDNA and inserted into pSuper1300-GFP to generate pSuper1300-NbRbcS-GFP. Primers used for constructing these plasmids are listed in Supplemental Table S1, and sequence analyses were performed to confirm the accuracy of all plasmids.
TEM
TEM was performed as described previously (Cao et al., 2015). In brief, N. benthamiana leaves were cut into pieces and fixed overnight at 4°C in fixation buffer (2.5% glutaraldehyde, 0.05 M phosphate, pH 7.2). Samples were then washed three times with fixation buffer followed by postfixation in 2% osmium tetroxide at 4°C for 2 h. After dehydration in a graded ethanol series (50%, 70%, 80%, 90%, 95%, and 100%), the tissue samples were embedded in Spurr’s resin. Ultrathin sections (70–90 nm) were cut using a Leica EM UC7 ultramicrotome and sequentially stained with uranyl acetate for 20 min and Reynolds’ lead citrate for 5 min. The sections were then viewed with a Hiltachi H-7650 or a JEM-1230 transmission electron microscope operated at 80 kV.
Immunoelectron Microscopy
Immunogold labeling was performed according to previously described methods with minor modifications (Liu et al., 2015). Leaf tissues from healthy or BSMV-infected N. benthamiana were vacuum-infiltrated with a mixture of 3% formaldehyde, 0.1% glutaraldehyde, 4% Suc in 0.1 M phosphate buffer (pH 7.2) and fixed at 4°C for 2 to 3 h. After washing with phosphate buffer, the samples were dehydrated in increasing ethanol concentrations (30%, 50%, 70%, 80%, 95%, and 100%), infiltrated in increasing concentrations of Lowicryl K4M resin (50%, 75% in methanol, and pure resin), and polymerized under UV light (360 nm) for 72 h at −20°C, and then for 48 h at room temperature. After polymerization, ultrathin sections were cut from blocks and collected on Formvar-coated nickel grids. The grids were incubated in blocking solution (0.01 M phosphate-buffered saline [PBS], pH 7.2, with 1% bovine serum albumin, 0.05% Triton X-100, 0.05% Tween 20) for 5 min at room temperature to reduce nonspecific binding, and incubated with primary mouse monoclonal anti-dsRNA antibody (J2, English & Scientific Consulting Kft), rabbit polyclonal anti-CP or anti-αa antibodies, respectively. Polyclonal αa-specific antibodies were prepared from the sera of rabbits immunized with the purified αa protein helicase domain (Zhang et al., 2017). After washing six times with 0.01 M PBS buffer, the grids were incubated with goat anti-mouse or goat anti-rabbit secondary antibodies conjugated with 10 nm gold particles, followed by rinsing with 0.01 M PBS buffer. Finally, sections were stained with uranyl acetate and Reynolds’ lead citrate prior to viewing with a Hiltachi H-7650 or JEM-1230 transmission electron microscope.
Transmission Electron Tomography
All procedures were performed as described previously with minor modification (Kang et al., 2011; Toyooka and Kang, 2014). Serial sections (250 nm) were cut from blocks of BSMV-infected or control N. benthamiana leaf tissues, and collected on slot grids followed by staining with uranyl acetate and lead citrate as described above. Fiducial markers consisting of 15 nm gold particles were deposited on both sides of the sections to facilitate image tracing and alignment. For BSMV-infected and control samples, double-axis tilt series was collected with a cooled slow-scan charge-coupled-device camera (4K Eagle) of FEI TF20 (200 kV) at 1.5° tilt increments from −60° to 60° to obtain 81 images. After one double-axis tilt series finished, the grid was turned 90° and started the second double-axis tilt series. For each tilt series, two sections were collected and joined using the IMOD software package (Kremer et al., 1996), with 324 images were recorded for two sections in one sample in total. The magnification was 14,500×, corresponding to a pixel size of 1.50 nm. The 3D surface renderings of the tomograms were performed with the IMOD software package (http://bio3d.colorado.edu/imod/doc/3dmodguide.html)
FIB-SEM
Samples for FIB-SEM analysis were prepared as described for TEM with minor modifications. To enhance the contrast, samples were postfixed in a mixture of 2% osmium tetroxide and 1.5% potassium hexacyanoferrate at 4°C for 2 h, followed by block staining with 2% aqueous uranyl acetate overnight at 4°C. Dehydration and embedding were performed as described above.
For FIB-SEM observations, blocks were trimmed to obtain a smooth flat surface, interested areas were exposed, and the samples were glued on a stage and sputtered with gold (HITACHI, E1010) for 90 s, followed by placing in the FIB-SEM microscope (FEI Helios NanoLab G3 UC). A 500-nm-thick layer of platinum was deposited on the top surface of the block with the gas injector system. The focused gallium ion beam was used with the slice thickness of 50 nm, and an accelerating voltage of 30 kV (a current of 9.3 nA). After FIB milling, the freshly milled block face was tilted to 7° in order to be vertical to the electron beam. The accelerating voltage of electron beam was 2 kV and the current to 0.4 nA (10 μs pixel dwell time). Images were taken at a magnification of 11,477× with a resolution of 4096 × 3536 pixels, and the horizontal field of view was 24.1 μm corresponding to a pixel size of 5.88 nm. The SEM images were aligned, processed, and 3D surface-rendered using the IMOD software package (http://bio3d.colorado.edu/imod/doc/3dmodguide.html).
Agroinfiltration
Infiltration of A. tumefaciens into the N. benthamiana was described previously (Yang et al., 2000). To enhance protein expression, suspensions of Agrobacterium carrying plasmids were often coinfiltrated with Agrobacterium harboring the p19 expression cassette. At 3 dpi, the infiltrated leaves were viewed under confocal microscopy or used for transmission electron microscopic analysis.
Confocal Laser-Scanning Microscopy
Confocal analysis was performed as described previously (Cao et al., 2015). YFP or chlorophyll autofluorescence was observed using an Olympus FV1000 laser scanning confocal microscope with an excitation wavelength of 514 or 633 nm, respectively. Images were processed with Imaris 7.4.2 software (Bitplane).
Protease Protection Assays
Protease protection assays were performed according to previously described methods with minor modifications (Ham et al., 2006). In brief, intact chloroplasts were isolated from N. benthamiana leaves as described previously (Ling and Jarvis, 2015). Chloroplast yields were calculated using a unit chlorophyll basis. Ten microliters of chloroplast suspension was added to 1 mL of 80% acetone prior to centrifugation at 3,000g for 2 min. Absorbance of the supernatant was measured at 652 nm with a spectrophotometer (METASH) and inserted into the Sigma chloroplast isolation kit formula: mg chlorophyll/mL = A652 × 100/36. To obtain the same concentration of chloroplast suspension, chloroplast suspension was diluted to 1 mg/mL chlorophyll with the treating buffer containing 1 mM CaCl2, 300 mM sorbitol, 50 mM HEPES[4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid]-KOH (pH 8.0). Two hundred micrograms thermolysin was added for every 1 mg chlorophyll in treating buffer with or without the addition of 0.5% Triton X-100 followed by incubation on ice for 0.5 h. After thermolysin treatment, protease inhibitor cocktail (Sigma) and 5 mM ethylenediamine tetraacetic acid (EDTA) were added to terminate the reaction. For the treatment with Triton X-100, the suspension was directly subjected to western blot analysis. For the treatment without Triton X-100, intact chloroplasts were reisolated using 40%/80% Percoll density gradient centrifugation followed by western blot analyses.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Ultrastructural changes in BSMV-infected N. benthamiana cells.
Supplemental Figure S2. Immunoelectron microscopic analysis of healthy N. benthamiana leaf tissues.
Supplemental Figure S3. Analysis of viral components in agroinfiltrated N. benthamiana leaves and isolated chloroplasts.
Supplemental Figure S4. Transmission electron tomography showing three-dimensional visualization of the partial structure of BSMV-induced cytoplasmic invaginations.
Supplemental Figure S5. 3D model of the BSMV-induced cytoplasmic invaginations generated from FIB-SEM.
Supplemental Figure S6. Western blot analysis of the total protein samples from N. benthamiana leaves expressing split YFP-fused proteins.
Supplemental Table S1. List of primers used for plasmid constructions.
Supplemental Table S2. List of primers used for RT-PCR.
Supplemental Movie S1. Representative electron tomograms of chloroplast in a healthy N. benthamiana cell.
Supplemental Movie S2. Representative electron tomograms of chloroplast in a BSMV-infected N. benthamiana cell.
Supplemental Movie S3. 3D surface rendering of rearranged chloroplast envelope membranes based on the transmission electron tomograms.
Supplemental Movie S4. Observation of the 3D architecture of rearranged chloroplast envelope membranes at different angles.
Supplemental Movie S5. 3D surface rendering of BSMV-induced chloroplast CI based on the transmission electron tomograms.
Supplemental Movie S6. Merged images of BSMV-induced cytoplasmic invaginations from FIB-SEM analysis.
Supplemental Movie S7. 3D surface rendering of BSMV-induced cytoplasmic invaginations based on the merged FIB-SEM images.
Supplemental Movie S8. Observation of the 3D architecture of an aberrant chloroplast from a BSMV-infected N. benthamiana cell.
Acknowledgments
The authors would like to thank Drs. Jialin Yu, Chenggui Han, Xianbing Wang, and Ying Wang at China Agricultural University for valuable discussions during the course of this work. They also thank Dr. Andrew O. Jackson (University of California, Berkeley) for his great help in editing and proofreading of the manuscript and Dr. Jian Hong (Zhejiang University, China) for helpful suggestions on our project. We thank Drs. Ying Li and Xiaomin Li at Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for technical assistance of preparing the ultrathin sections and FIB-SEM analysis, and Drs. Wei Ding, Jianguo Zhang, and Lei Sun (HPC-Service Station in Center for Biological Imaging, Institute of Biophysics, Chinese Academy of Sciences) for their help in FIB-SEM analysis and data processing. We thank Dr. Andreas P.M. Weber (Heinrich Heine University Düsseldorf, Germany) for providing the AtTOC64-GFP and AtTIC40-GFP plasmids. The authors thank the editor and three anonymous reviewers for their constructive comments and suggestions on earlier drafts of this article.
Glossary
- VRC
viral replication complex
- TEM
transmission electron microscopy
- FIB-SEM
focused ion beam scanning electron microscopy
- CP
coat protein
- CI
cytoplasmic invagination
- BiFC
bimolecular fluorescence complementation
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31570143 and 31270184 to D.L.), the Project for Extramural Scientists of SKLAB (grant no. 2017SKLAB1-6 to D.L. and B.-H.K.), the Fundamental Research Funds for the Central Universities (grant no. 2017SY003 to Y.Z.), the Research Grants Council of Hong Kong (grant nos. GRF14126116, AoE/M-05/12, and C4011-14R to B.-H.K.), and the Cooperative Research Program for Agriculture Science and Technology Development of the Rural Development Administration, Republic of Korea (grant no. PJ010953092015 to B.-H.K.).
References
- Allen TC. (1972) Subcellular responses of mesophyll cells to Wild cucumber mosaic virus. Virology 47: 467–474 [DOI] [PubMed] [Google Scholar]
- Austin JR 2nd, Staehelin LA (2011) Three-dimensional architecture of grana and stroma thylakoids of higher plants as determined by electron tomography. Plant Physiol 155: 1601–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballantine JEM, Forde BJ (1970) The effect of light intensity and temperature on plant growth and chloroplast ultrastructure in soybean. Am J Bot 57: 1150–1159 [Google Scholar]
- Baumeister W. (2002) Electron tomography: Towards visualizing the molecular organization of the cytoplasm. Curr Opin Struct Biol 12: 679–684 [DOI] [PubMed] [Google Scholar]
- Beauchemin C, Boutet N, Laliberté J-F (2007) Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of Turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in planta. J Virol 81: 775–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya D, Chakraborty S (2017) Chloroplast: The Trojan horse in plant-virus interaction. Mol Plant Pathol [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon JA, Diaz A, Ahlquist P (2010) Cytoplasmic viral replication complexes. Cell Host Microbe 8: 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuers FKH, Bräutigam A, Geimer S, Welzel UY, Stefano G, Renna L, Brandizzi F, Weber APM (2012) Dynamic remodeling of the plastid envelope membranes—a tool for chloroplast envelope in vivo localizations. Front Plant Sci 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jin X, Zhang X, Li Y, Wang C, Wang X, Hong J, Wang X, Li D, Zhang Y (2015) Morphogenesis of endoplasmic reticulum membrane-invaginated vesicles during Beet black scorch virus infection: Role of auxiliary replication protein and new implications of three-dimensional architecture. J Virol 89: 6184–6195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll TW. (1970) Relation of Barley stripe mosaic virus to plastids. Virology 42: 1015–1022 [DOI] [PubMed] [Google Scholar]
- Chen YL, Chen LJ, Li HM (2016) Polypeptide transport-associated domains of the Toc75 channel protein are located in the intermembrane space of chloroplasts. Plant Physiol 172: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ML, Fitzpatrick LM, Tu SL, Budziszewski G, Potter-Lewis S, Akita M, Levin JZ, Keegstra K, Li HM (2003) Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J 22: 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan GH, Roberts AG, Chapman SN, Ziegler A, Savenkov EI, Torrance L (2012) The Potato mop-top virus TGB2 protein and viral RNA associate with chloroplasts and viral infection induces inclusions in the plastids. Front Plant Sci 3: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum B, Nicastro D, Austin J II, McIntosh JR, Kühlbrandt W (2010) Arrangement of photosystem II and ATP synthase in chloroplast membranes of spinach and pea. Plant Cell 22: 1299–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A, Gallei A, Ahlquist P (2012) Bromovirus RNA replication compartment formation requires concerted action of 1a’s self-interacting RNA capping and helicase domains. J Virol 86: 821–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A, Wang X, Ahlquist P (2010) Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. Proc Natl Acad Sci USA 107: 16291–16296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A, Zhang J, Ollwerther A, Wang X, Ahlquist P (2015) Host ESCRT proteins are required for bromovirus RNA replication compartment assembly and function. PLoS Pathog 11: e1004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Schaffer M, Kuhn Cuellar L, Villa E, Plitzko JM, Baumeister W (2015) Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4: e04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández de Castro I, Fernández JJ, Barajas D, Nagy PD, Risco C (2017) Three-dimensional imaging of the intracellular assembly of a functional viral RNA replicase complex. J Cell Sci 130: 260–268 [DOI] [PubMed] [Google Scholar]
- Gómez-Aix C, García-García M, Aranda MA, Sánchez-Pina MA (2015) Melon necrotic spot virus replication occurs in association with altered mitochondria. Mol Plant Microbe Interact 28: 387–397 [DOI] [PubMed] [Google Scholar]
- Gounaris K, Brain APR, Quinn PJ, Williams WP (1984) Structural reorganisation of chloroplast thylakoid membranes in response to heat-stress. Biochim Biophys Acta 766: 198–208 [Google Scholar]
- Gushchin VA, Karlin DG, Makhotenko AV, Khromov AV, Erokhina TN, Solovyev AG, Morozov SY, Agranovsky AA (2017) A conserved region in the Closterovirus 1a polyprotein drives extensive remodeling of endoplasmic reticulum membranes and induces motile globules in Nicotiana benthamiana cells. Virology 502: 106–113 [DOI] [PubMed] [Google Scholar]
- Ham BK, Park JM, Lee SB, Kim MJ, Lee IJ, Kim KJ, Kwon CS, Paek KH (2006) Tobacco Tsip1, a DnaJ-type Zn finger protein, is recruited to and potentiates Tsi1-mediated transcriptional activation. Plant Cell 18: 2005–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harak C, Lohmann V (2015) Ultrastructure of the replication sites of positive-strand RNA viruses. Virology 479-480: 418–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta T, Bullivant S, Matthews RE (1973) Fine structure of vesicles induced in chloroplasts of Chinese cabbage leaves by infection with Turnip yellow mosaic virus. J Gen Virol 20: 37–50 [DOI] [PubMed] [Google Scholar]
- Hu Y, Li Z, Yuan C, Jin X, Yan L, Zhao X, Zhang Y, Jackson AO, Wang X, Han C, et al. (2015) Phosphorylation of TGB1 by protein kinase CK2 promotes Barley stripe mosaic virus movement in monocots and dicots. J Exp Bot 66: 4733–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AO, Lim HS, Bragg J, Ganesan U, Lee MY (2009) Hordeivirus replication, movement, and pathogenesis. Annu Rev Phytopathol 47: 385–422 [DOI] [PubMed] [Google Scholar]
- Jonczyk M, Pathak KB, Sharma M, Nagy PD (2007) Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology 362: 320–330 [DOI] [PubMed] [Google Scholar]
- Kallio K, Hellström K, Balistreri G, Spuul P, Jokitalo E, Ahola T (2013) Template RNA length determines the size of replication complex spherules for Semliki Forest virus. J Virol 87: 9125–9134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Nielsen E, Preuss ML, Mastronarde D, Staehelin LA (2011) Electron tomography of RabA4b- and PI-4Kβ1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12: 313–329 [DOI] [PubMed] [Google Scholar]
- Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P (2007) Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol 5: e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalev N, de Castro Martín IF, Pogany J, Barajas D, Pathak K, Risco C, Nagy PD (2016) Role of viral RNA and co-opted cellular ESCRT-I and ESCRT-III factors in formation of tombusvirus spherules harboring the tombusvirus replicase. J Virol 90: 3611–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalewska Ł, Mazur R, Suski S, Garstka M, Mostowska A (2016) Three-dimensional visualization of the tubular-lamellar transformation of the internal plastid membrane network during runner bean chloroplast biogenesis. Plant Cell 28: 875–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Laliberté JF, Sanfaçon H (2010) Cellular remodeling during plant virus infection. Annu Rev Phytopathol 48: 69–91 [DOI] [PubMed] [Google Scholar]
- Laliberté JF, Zheng H (2014) Viral manipulation of plant host membranes. Annu Rev Virol 1: 237–259 [DOI] [PubMed] [Google Scholar]
- Lee KH, Kim DH, Lee SW, Kim ZH, Hwang I (2002) In vivo import experiments in protoplasts reveal the importance of the overall context but not specific amino acid residues of the transit peptide during import into chloroplasts. Mol Cells 14: 388–397 [PubMed] [Google Scholar]
- Lin N-S, Langenberg WG (1984) Chronology of appearance of Barley stripe mosaic virus protein in infected wheat cells. J Ultrastruct Res 89: 309–323 [DOI] [PubMed] [Google Scholar]
- Lin NS, Langenberg WG (1985) Peripheral vesicles in proplastids of Barley stripe mosaic virus-infected wheat cells contain double-stranded RNA. Virology 142: 291–298 [DOI] [PubMed] [Google Scholar]
- Ling Q, Jarvis P (2015) Regulation of chloroplast protein import by the ubiquitin E3 ligase SP1 is important for stress tolerance in plants. Curr Biol 25: 2527–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Westler WM, den Boon JA, Wang X, Diaz A, Steinberg HA, Ahlquist P (2009) An amphipathic alpha-helix controls multiple roles of Brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog 5: e1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X-Y, Yang J, Xie L, Li J, Song X-J, Chen J-P, Zhang H-M (2015) P5-2 of Rice black-streaked dwarf virus is a non-structural protein targeted to chloroplasts. Arch Virol 160: 1211–1217 [DOI] [PubMed] [Google Scholar]
- McMullen CR, Gardner WS, Myers GA (1978) Aberrant plastids in barley leaf tissue infected with Barley stripe mosaic virus. Phytopathology 68: 317–325 [Google Scholar]
- Miller S, Krijnse-Locker J (2008) Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6: 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C (2005) Virus factories: Associations of cell organelles for viral replication and morphogenesis. Biol Cell 97: 147–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly EK, Paul JD, Kao CC (1997) Analysis of the interaction of viral RNA replication proteins by using the yeast two-hybrid assay. J Virol 71: 7526–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D, Bartenschlager R (2013) Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol 2: 32–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Ganguly D, Albrecht-Borth V (2015) Insights into chloroplast biogenesis and development. Biochim Biophys Acta 1847: 1017–1024 [DOI] [PubMed] [Google Scholar]
- Qbadou S, Becker T, Bionda T, Reger K, Ruprecht M, Soll J, Schleiff E (2007) Toc64—a preprotein-receptor at the outer membrane with bipartide function. J Mol Biol 367: 1330–1346 [DOI] [PubMed] [Google Scholar]
- Restrepo-Hartwig MA, Ahlquist P (1996) Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J Virol 70: 8908–8916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco C, de Castro IF, Sanz-Sánchez L, Narayan K, Grandinetti G, Subramaniam S (2014) Three-dimensional imaging of viral infections. Annu Rev Virol 1: 453–473 [DOI] [PubMed] [Google Scholar]
- Romero-Brey I, Bartenschlager R (2014) Membranous replication factories induced by plus-strand RNA viruses. Viruses 6: 2826–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, et al. (2012) Three-dimensional architecture and biogenesis of membrane structures associated with Hepatitis C virus replication. PLoS Pathog 8: e1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama S, Trivedi S, Busheva M, Arafa A, Garab G, Erdei L (1994) Effects of NaCl salinity on growth, cation accumulation, chloroplast structure and function in wheat cultivars differing in salt tolerance. J Plant Physiol 144: 241–247 [Google Scholar]
- Salonen A, Ahola T, Kaariainen L (2005) Viral RNA replication in association with cellular membranes. In Marsh M, ed, Membrane Trafficking in Viral Replication, Vol 285 Springer, Berlin, Germany, pp 139–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell D, editors (2001). Molecular Cloning: A Laboratory Manual, Ed 3 Cold Spring Harbor Laboratory Press, Long Island, New York [Google Scholar]
- Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, Ahlquist P (2002) A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell 9: 505–514 [DOI] [PubMed] [Google Scholar]
- Schwartz M, Chen J, Lee WM, Janda M, Ahlquist P (2004) Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc Natl Acad Sci USA 101: 11263–11268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni E, Rav-Hon O, Ohad I, Brumfeld V, Reich Z (2005) Three-dimensional organization of higher-plant chloroplast thylakoid membranes revealed by electron tomography. Plant Cell 17: 2580–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrance L, Cowan GH, Gillespie T, Ziegler A, Lacomme C (2006) Barley stripe mosaic virus-encoded proteins triple-gene block 2 and γb localize to chloroplasts in virus-infected monocot and dicot plants, revealing hitherto-unknown roles in virus replication. J Gen Virol 87: 2403–2411 [DOI] [PubMed] [Google Scholar]
- Toyooka K, Kang B-H (2014) Reconstructing plant cells in 3D by serial section electron tomography. In Žárský V, Cvrčková F, eds, Plant Cell Morphogenesis: Methods and Protocols, Vol 1080 Humana Press, New York, pp 159-170 [DOI] [PubMed] [Google Scholar]
- Ushiyama R, Matthews REF (1970) The significance of chloroplast abnormalities associated with infection by Turnip yellow mosaic virus. Virology 42: 293–303 [DOI] [PubMed] [Google Scholar]
- Verchot J. (2011) Wrapping membranes around plant virus infection. Curr Opin Virol 1: 388–395 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wan J, Basu K, Mui J, Vali H, Zheng H, Laliberté J-F (2015) Ultrastructural characterization of Turnip mosaic virus-induced cellular rearrangements reveals membrane-bound viral particles accumulating in vacuoles. J Virol 89: 12441–12456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. (2015) Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu Rev Phytopathol 53: 45–66 [DOI] [PubMed] [Google Scholar]
- Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R (2009) Composition and three-dimensional architecture of the Dengue virus replication and assembly sites. Cell Host Microbe 5: 365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Nagy PD (2014) Expanding use of multi-origin subcellular membranes by positive-strand RNA viruses during replication. Curr Opin Virol 9: 119–126 [DOI] [PubMed] [Google Scholar]
- Yang Y, Li R, Qi M (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22: 543–551 [DOI] [PubMed] [Google Scholar]
- Yang H, Shi Y, Liu J, Guo L, Zhang X, Yang S (2010) A mutant CHS3 protein with TIR-NB-LRR-LIM domains modulates growth, cell death and freezing tolerance in a temperature-dependent manner in Arabidopsis. Plant J 63: 283–296 [DOI] [PubMed] [Google Scholar]
- Yuan C, Li C, Yan L, Jackson AO, Liu Z, Han C, Yu J, Li D (2011) A high throughput Barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS One 6: e26468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamyatnin AA Jr., Solovyev AG, Bozhkov PV, Valkonen JPT, Morozov SY, Savenkov EI (2006) Assessment of the integral membrane protein topology in living cells. Plant J 46: 145–154 [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang Z, Chukkapalli V, Nchoutmboube JA, Li J, Randall G, Belov GA, Wang X (2016) Positive-strand RNA viruses stimulate host phosphatidylcholine synthesis at viral replication sites. Proc Natl Acad Sci USA 113: E1064–E1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Zhang Y, Yang M, Liu S, Li Z, Wang X, Han C, Yu J, Li D (2017) The Barley stripe mosaic virus γb protein promotes chloroplast-targeted replication by enhancing unwinding of RNA duplexes. PLoS Pathog 13: e1006319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang X, Hong Y, Liu Y (2016) Chloroplast in plant-virus interaction. Front Microbiol 7: 1565. [DOI] [PMC free article] [PubMed] [Google Scholar]