ABSTRACT
Previous studies identified the nuclear domain 10 (ND10) components promyelocytic leukemia protein (PML), hDaxx, and Sp100 as factors of an intrinsic immune response against human cytomegalovirus (HCMV). This antiviral function of ND10, however, is antagonized by viral effector proteins like IE1p72, which induces dispersal of ND10. Furthermore, we have shown that both major immediate early proteins of HCMV, IE1p72 and IE2p86, transiently colocalize with ND10 subnuclear structures and undergo modification by the covalent attachment of SUMO. Since recent reports indicate that PML acts as a SUMO E3 ligase, we asked whether the SUMOylation of IE1p72 and IE2p86 is regulated by PML. To address this, PML-depleted fibroblasts, as well as cells overexpressing individual PML isoforms, were infected with HCMV. Western blot experiments revealed a clear correlation between the degree of IE1p72 SUMO conjugation and the abundance of PML. On the other hand, the SUMOylation of IE2p86 was not affected by PML. By performing in vitro SUMOylation assays, we were able to provide direct evidence that IE1p72 is a substrate for PML-mediated SUMOylation. Interestingly, disruption of the RING finger domain of PML, which is proposed to confer SUMO E3 ligase activity, abolished PML-induced SUMOylation of IE1p72. In contrast, IE1p72 was still efficiently SUMO modified by a SUMOylation-defective PML mutant, indicating that intact ND10 bodies are not necessary for this effect. Thus, this is the first report that the E3 ligase PML is capable of stimulating the SUMOylation of a viral protein which is supposed to serve as a cellular mechanism to compromise specific functions of IE1p72.
IMPORTANCE The major immediate early proteins of human cytomegalovirus, termed IE1p72 and IE2p86, have previously been shown to undergo posttranslational modification by covalent coupling to SUMO moieties at specific lysine residues. However, the enzymatic activities that are responsible for this modification have not been identified. Here, we demonstrate that the PML protein, which mediates an intrinsic immune response against HCMV, specifically serves as an E3 ligase for SUMO modification of IE1p72. Since SUMO modification of IE1p72 has previously been shown to interfere with STAT factor binding, thus compromising the interferon-antagonistic function of this viral effector protein, our finding highlights an additional mechanism through which PML is able to restrict viral infections.
KEYWORDS: human cytomegalovirus, immediate early protein IE1, nuclear domain 10, promyelocytic leukemia protein PML, sumoylation
INTRODUCTION
Covalent conjugation to small ubiquitin-like modifier (SUMO) proteins is a posttranslational modification (PTM) that is emerging as a critical regulatory event of host-virus interactions (1–3). SUMO exists in five paralogs termed SUMO-1 to -4 and the recently detected SUMO-5 (4, 5). While SUMO-2 and SUMO-3 are nearly identical in their sequences and functions, they share only approximately 50% sequence identity with SUMO-1. Analogous to the ubiquitin pathway, the 11-kDa SUMO moiety is covalently attached to the ε-amino group of the lysine residue of its target protein through a four-step enzymatic cascade (6). First, immature SUMO is processed by SUMO-specific proteases (SENPs) to expose two C-terminal glycine residues required for conjugation. After processing, mature SUMO is activated in an ATP-dependent manner by the E1-activating enzyme, which is a heterodimer consisting of Aos1/SAE1 and Uba2/SAE2. Next, SUMO is transferred to the E2-conjugating enzyme UBC9 and finally conjugated to its target protein by SUMO E3 ligases. The ligases play an important role by conferring specificity and efficiency on the SUMOylation reaction.
While over 600 ubiquitin E3 ligases are currently known, only a limited number of SUMO E3 ligases have been identified (7). Interestingly, a recent report showed that several members of the tripartite motif (TRIM) family of proteins, including the promyelocytic leukemia protein (PML) (alternatively termed TRIM19), act as SUMO E3 ligases (8). These proteins are defined by the TRIM/RBCC motif, which consists of a RING finger domain, one or two B boxes (cysteine/histidine-rich motifs), and a predicted coiled-coil region. As for certain TRIM proteins that have been demonstrated to exhibit ubiquitin E3 activity, the SUMO E3 activity of the identified TRIM proteins could also be attributed to the RING domain (8–10).
PML, which has previously been proposed to function as a SUMO E3 ligase due to its capacity to interact with Ubc9 and SUMO, as well as to stimulate SUMO conjugation in Saccharomyces cerevisiae, represents the main structural determinant of the subnuclear structure nuclear domain 10 (ND10) (alternatively termed PML nuclear bodies [PML-NBs] or PML oncogenic domains [PODs]) (11–13). ND10, which are defined by the presence of the three major ND10 factors PML, hDaxx, and Sp100, are implicated in the coordinated regulation of diverse cellular processes, like DNA damage repair, apoptosis, and senescence (reviewed in reference 14). Interestingly, recent studies support the hypothesis that ND10 may also provide a nuclear platform for posttranslational modifications, like phosphorylation, acetylation, and SUMOylation (15–17). The last has been suggested by Van Damme and colleagues, who set up a manually curated network of the PML-NB interactome. This bioinformatics approach showed an association of all essential components of the SUMO conjugation pathway with ND10, along with enrichment of SUMOylated proteins at these sites (17).
In addition to its function as a nuclear hot spot for posttranslational modifications, ND10 is known to play an important role in the intrinsic defense against viral infections (18, 19). In the case of the betaherpesvirus human cytomegalovirus (HCMV), we and others have shown that the ND10-instituted antiviral state has to be antagonized by the virus in order to efficiently start the viral gene expression program for lytic replication (reviewed in references 20 and 21). In this regard, the HCMV immediate early protein IE1p72 (IE1) plays an important role, as it is able to induce the destruction of this subnuclear structure by abrogating the SUMOylation of PML and Sp100 and it was recently shown that this is due to a blockage of de novo SUMOylation (22–25). Since covalent as well as noncovalent SUMO interactions constitute the basis for ND10 formation, this ultimately leads to a loss of PML-NB integrity (26–29).
Besides IE1, the immediate early protein IE2p86 (IE2) of HCMV, which is an important activator of viral early and late gene expression, is also known to transiently localize to ND10 during the initial stage of infection (30, 31). Intriguingly, both HCMV regulatory proteins are among the most prominent examples of viral proteins that undergo posttranslational modification by the covalent attachment of SUMO (IE1, Lys450; IE2, Lys175 or Lys180) (32–34). In light of the fact that ND10 are regarded as sites for nuclear SUMOylation events, we wondered whether the SUMO modification of IE1 or IE2 might be regulated by PML and/or ND10.
Here, we report that IE1, but not IE2, represents a substrate for the SUMO E3 ligase PML, as shown by in vivo, as well as in vitro, experiments, thus illustrating the first viral target of PML. The activity of PML requires an intact RING finger domain but is not dependent on the structural integrity of ND10. Our results imply that PML affects the regulatory function of IE1 during infection by modulating its posttranslational SUMO modification status.
RESULTS
PML positively affects the SUMOylation of IE1, but not of IE2.
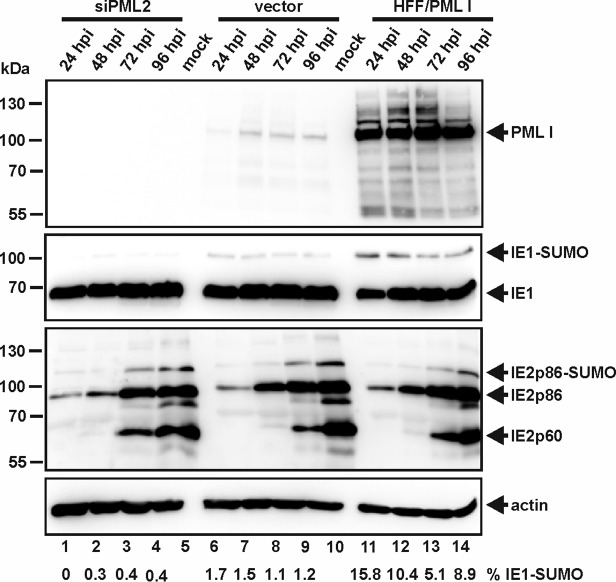
ND10 bodies are hypothesized to function as SUMOylation hot spots within the nucleus, with PML itself representing one of the various SUMO E3 ligases that can be found in these subnuclear structures (8, 17). Given the fact that the two important viral regulatory proteins IE1p72 (IE1) and IE2p86 (IE2), which are both known to be posttranslationally modified by SUMO, transiently localize at ND10 during the initial stage of infection, we questioned whether their SUMO modification might be affected by PML. To address this question, we infected PML knockdown (kd) human foreskin fibroblasts (HFFs) with a small interfering RNA-mediated knockdown of PML (siPML2 cells) in which PML expression was highly reduced, along with control cells (vector) expressing endogenous levels of PML, as well as HFFs overexpressing PML isoform I (HFF/PMLI), with HCMV at a multiplicity of infection (MOI) of 0.1 (Fig. 1). In accordance with previous data, we found that the extent of IE1 SUMOylation is directly correlated with the abundance of PML (Fig. 1, second gel from top) (34). The IE2 SUMOylation status, on the other hand, remained unaffected irrespective of whether PML was present or absent (Fig. 1, third gel from top). Thus, PML specifically enhances SUMOylation of IE1. This correlates with the observation of a direct protein interaction between PML and IE1 (23).
FIG 1.
Direct correlation between the extent of IE1 SUMOylation and PML abundance. PML-kd cells (siPML2) and control HFFs (vector), along with HFFs overexpressing PML isoform I (HFF/PMLI), were either mock infected or infected with HCMV AD169 at an MOI of 0.1. At the indicated times postinfection (24 to 96 h postinfection [hpi]), the cell lysates were harvested and analyzed by Western blotting for PML, IE1, and IE2 protein levels. Beta-actin was included as an internal loading control. The ratio of IE1 SUMOylation was quantitated and is shown below the blot (% IE1-SUMO).
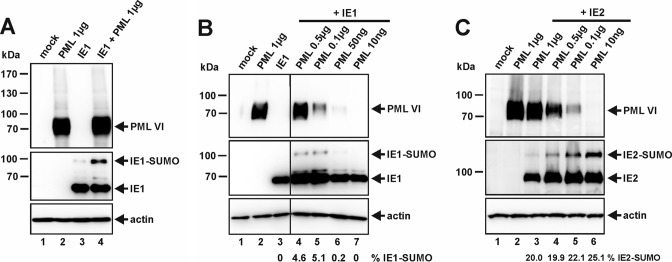
Similar results were obtained in transient-transfection experiments with HEK293T cells, where IE1 was expressed alone or in combination with PML isoform VI (Fig. 2). As shown in Fig. 2A, coexpression of PML significantly augmented the steady-state level of SUMOylated IE1. In addition, we observed that the enhanced conjugation of SUMO to IE1 exhibited PML dose dependency; however, at high concentrations of PML, SUMOylation of IE1 could not be further increased (Fig. 2B, lanes 4 to 7). Since E3 ligases are thought to serve as adaptor proteins, limiting amounts of additional components of the SUMO conjugation machinery may explain this effect. In contrast, quantification revealed that SUMO conjugation of IE2 did not increase but was slightly diminished after coexpression of large amounts of PML (Fig. 2C). Hence, PML expression, intriguingly, has contrary effects on the SUMOylation status of IE1 and IE2.
FIG 2.
PML stimulates SUMO conjugation of IE1 in vivo. (A to C) Western blot analyses of cell extracts of HEK293T cells transfected with expression constructs for PML VI in combination with IE1 (A and B) or IE2 (C). For all Western blot experiments, lysates from nontransfected HEK293T cells (mock) served as a negative control, and detection of beta-actin was included as an internal loading control. (B and C) Coexpression of IE1, together with decreasing amounts of PML. The quantity of transfected PML-encoding DNA ranged from 1 μg to 10 ng, as indicated. The ratio of protein SUMOylation was quantitated and is shown below panels B and C (% IE1-SUMO and % IE2-SUMO, respectively).
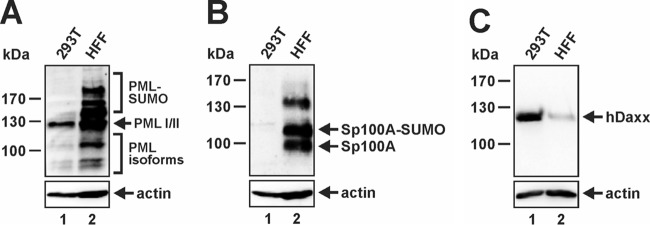
In this context, it is notable that we regard HEK293T cells as an advantageous cell line to study the effects of exogenously introduced PML, since in these cells very little endogenous PML is present compared to primary HFFs (Fig. 3A). In line with this finding, knockdown of PML in HEK293T cells did not alter the outcome of any of our transfection experiments (see Fig. 9C). In addition, HEK293T cells are nearly devoid of the major ND10 factor Sp100 (Fig. 3B), while hDaxx protein levels, on the other hand, are clearly elevated in comparison to HFFs (Fig. 3C). Thus, the transformed phenotype of HEK293T cells goes along with dramatic changes in ND10 protein abundance and composition, and as a consequence, no genuine ND10 structures are detectable by immunofluorescence staining in these cells (data not shown).
FIG 3.
HEK293T cells exhibit altered ND10 protein abundance in comparison to HFFs. (A to C) Equal amounts of protein lysates (prepared from 600.000 cells) of HEK293T cells and HFFs were compared by immunoblotting for PML (A), Sp100 (B), and hDaxx (C) protein expression levels. Beta-actin served as an internal loading control.
FIG 9.
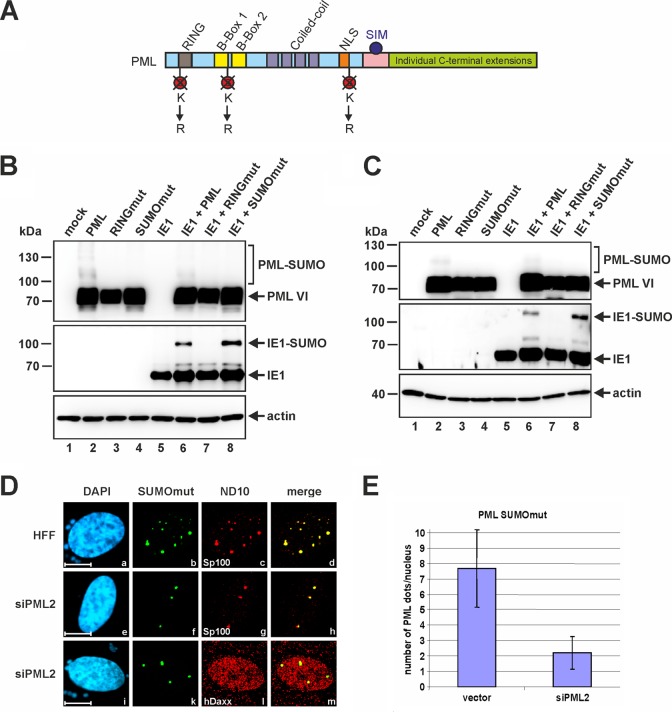
PML-promoted SUMOylation of IE1 in the absence of intact ND10 bodies. (A) Schematic representation of the SUMOylation-deficient PML mutant (SUMOmut) based on PML isoform VI, where the lysine residues of the covalent SUMO attachment sites at positions 65, 160, and 490 were exchanged for arginines. (B and C) Cotransfection of normal HEK293T cells (B) or siPML2-transduced HEK293T cells (C) with expression plasmids coding for IE1 in combination with either wt PML, RINGmut, or SUMOmut. Lysates of transfected and nontransfected cells (mock) were analyzed by Western blotting for PML, IE1, and beta-actin protein levels. (D) Immunostaining of normal HFFs or PML-kd HFFs (siPML2) transfected with a Myc-tagged version of PML SUMOmut. Two days posttransfection, the cells were fixed for detection of the ND10 factors Sp100 and hDaxx. PML SUMOmut was identified by its Myc tag moiety. Cell nuclei were visualized by DAPI staining (scale bars = 10 μm). (E) Direct quantification of the PML dots formed by PML SUMOmut upon transfection of vector- or siPML2-transduced HFFs with a SUMOmut expression construct. The graph shows mean values for 50 analyzed cells; standard deviations are indicated.
PML is capable of stimulating IE1 SUMO modification in vitro.
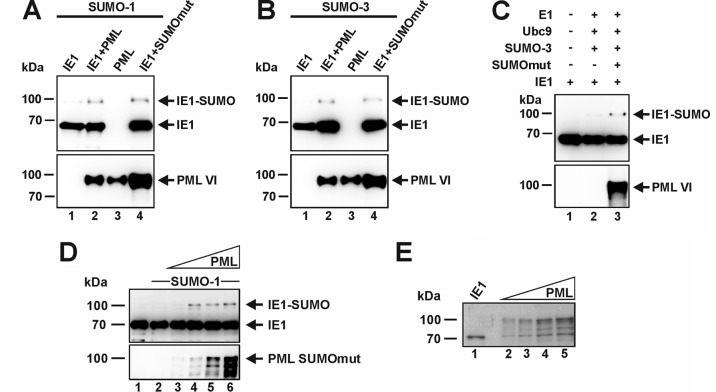
To determine whether PML has a direct effect on the SUMO modification of IE1, an in vitro SUMOylation assay was performed (Fig. 4). To this end, glutathione S-transferase (GST)-tagged variants of wild-type (wt) PML or a SUMOylation-negative PML mutant (both PML isoform VI), as well as of IE1, were expressed in Escherichia coli, which lacks a SUMOylation system. Thereafter, prokaryotically expressed PML and IE1 were affinity purified, and in the case of IE1, the GST tag was removed by enzymatic cleavage followed by chromatographic purification. When incubated in a SUMOylation reaction mixture, the addition of either wt PML (PML) or the SUMO-deficient PML mutant (SUMOmut) upregulated IE1 modification by both SUMO-1 and SUMO-3 (Fig. 4A and B). In contrast, after incubation of IE1 alone without any components of the SUMO conjugation pathway, no higher-molecular-mass band of IE1 was detectable (Fig. 4C, lane 1). The effect of PML on IE1 SUMOylation in these in vitro reactions was comparable to the previously reported E3 ligase function of PML on p53 (8). In a reaction with increasing amounts of PML, the enhanced SUMOylation of IE1 was confirmed; however, no linear correlation of the intensity of SUMOylation signals with the amount of PML was detected (Fig. 4D, lanes 3 to 6, and E). This is consistent with our observations from cotransfection analyses (Fig. 2B). Taken together, these data indicate that PML has the ability to stimulate the SUMOylation of IE1, not only in vivo, but also in vitro. This further strengthens the assumption that IE1 is a direct target protein for the SUMO E3 ligase activity of PML.
FIG 4.
PML promotes SUMOylation of IE1 in vitro. (A to D) Bacterially purified IE1 was incubated with E1 (SAE1/SAE2), Ubc9, ATP, and either SUMO-1 (A and D) or SUMO-3 (B and C) in the presence or absence of GST-PML or GST-PML-SUMOmut (both isoform VI), as described in Materials and Methods. In panel D, increasing concentrations of PML-SUMOmut were used. The reaction mixtures were analyzed by Western blotting using anti-IE1 and anti-PML antibodies. (E) Coomassie blue-stained gel showing the purified proteins (15-fold amounts) that were used for the blots in panel D. Lanes: 1, purified IE1; 2 to 5, increasing amounts of PML-SUMOmut as used in lanes 3 to 6 of panel D.
PML isoforms differ in their abilities to SUMOylate IE1.
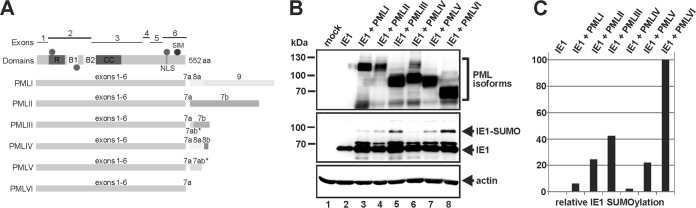
The PML gene consists of nine exons, and by alternative splicing, six nuclear PML isoforms (PML I to VI) are expressed (35). All these PML variants contain a common N terminus harboring the TRIM motif, but they vary in their C termini due to individual C-terminal extensions (Fig. 5A). Consequently, the question arose as to whether the observed stimulation of IE1 SUMOylation by PML isoform VI is a shared feature of all nuclear PML species. To assess the roles of the individual PML isoforms for IE1 SUMOylation, we expressed IE1 alone or in combination with PML I to VI in vivo (Fig. 5B). As shown in Fig. 5B, irrespective of which PML isoform was expressed (Fig. 5B, top), in each case an IE1-SUMO band was detectable (Fig. 5B, middle, lanes 3 to 8), which was absent in the PML-negative sample (Fig. 5B, middle, lane 2). Interestingly, however, although all PML variants showed rather comparable expression levels (Fig. 5B, top), the degree of IE1 SUMOylation varied significantly depending on the PML isoform expressed (Fig. 5B, middle; quantified in panel C). While PML III and VI induced large amounts of SUMO-modified IE1 (Fig. 5B, lanes 5 and 8), the presence of PML IV resulted in only very weak IE1-SUMO signals (Fig. 5B, lane 6), whereas expression of PML I, II, or V gave rise to an intermediate IE1 SUMOylation phenotype (Fig. 5B, lanes 3, 4, and 7).
FIG 5.
PML isoform-specific SUMOylation of IE1. (A) Schematic representation of exon organization and structural domains of the nuclear-localized PML isoforms I to VI, which all share the N-terminal region containing the RBCC motif (exons 1 to 6). The different C termini are generated by alternative use of 3′ exons. R, RING domain; B1 and B2, zinc binding boxes; CC, coiled-coil domain; S, SUMO; SIM, SUMO interaction motif; NLS, nuclear localization signal. (B) Cotransfection of HEK293T cells with plasmids encoding IE1 and the various nuclear-localized PML isoforms (FLAG-PML I to VI), followed by Western blotting detection of IE1, beta-actin, and PML I to VI. The last were detected by their FLAG moieties. (C) Relative IE1 SUMOylation from panel B (normalized for the expression level of the respective PML isoform). IE1 SUMOylation by PML isoform VI was set to 100%.
Direct protein interaction is required for PML-mediated SUMOylation of IE1.
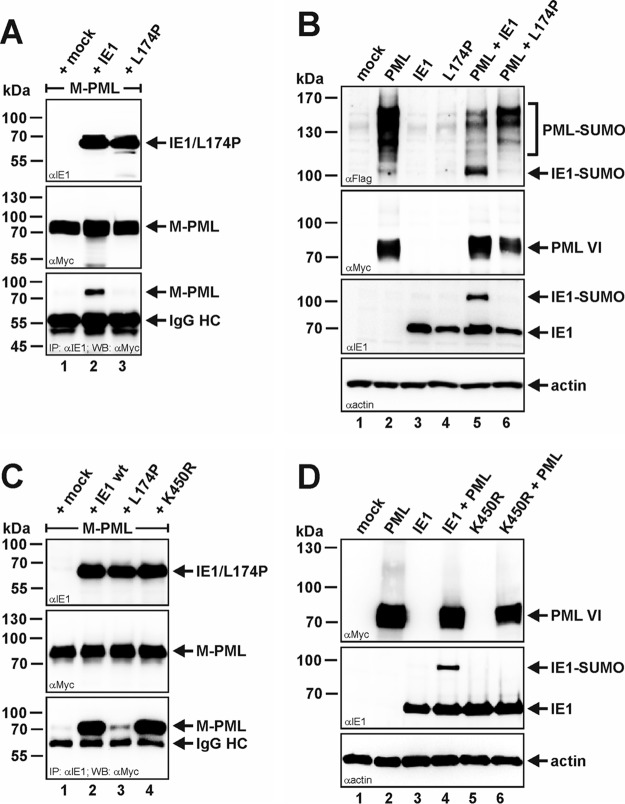
To investigate whether the physical interaction between PML and IE1 is a prerequisite for PML-induced posttranslational SUMO modification of IE1, we took advantage of an IE1 point mutant (L174P) that has been reported to fail to associate with PML (36, 37). By performing coimmunoprecipitation experiments, we could confirm a loss of interaction between PML and the IE1 mutant L174P, since, in contrast to wt IE1, the mutant was no longer able to coprecipitate PML (isoform VI) (Fig. 6A, bottom, compare lanes 2 and 3). Furthermore, the mutant was deficient in the deSUMOylation of PML (Fig. 6B, top, compare lanes 2, 5, and 6). As can be seen in Fig. 6B, the IE1 mutant L174P could also no longer be SUMOylated by PML compared to wt IE1 (Fig. 6B, third panel from top, compare lanes 5 and 6). This indicates that direct interaction is essential for PML-induced SUMOylation of IE1. In addition, by mutating the lysine residue at position 450 of IE1 to arginine (K450R), which has been mapped as the SUMO acceptor site of IE1, we could likewise abolish the appearance of the high-molecular-weight form of IE1 upon coexpression of PML (Fig. 6D, middle, lane 6), although this K450R mutant interacted as efficiently with PML as wt IE1 (Fig. 6C, bottom, compare lanes 2 and 4). Thus, this unambiguously identifies the slower-migrating IE1 band that runs at around 100 kDa as the SUMO-modified version of IE1.
FIG 6.
Disturbance of the PML-IE1 interaction interface abrogates IE1 SUMOylation. (A and C) Coimmunoprecipitation analysis of PML-kd/293T cells cotransfected with plasmids encoding Myc-tagged PML isoform VI (M-PML), together with either wt IE1, the PML interaction-defective IE1 mutant L174P, or the SUMOylation-deficient IE1 mutant K450R, as indicated. Two days posttransfection, the cells were lysed, and immunoprecipitation was performed using an anti-IE1 antibody. After electrophoresis, coprecipitated PML (M-PML) was visualized using an anti-Myc antibody (bottom). Immunoglobulin heavy chain (IgG HC) served as an internal control for the presence of the precipitating antibody. (B and D) Western blot analysis of extracts from HEK293T cells transfected with PML isoform VI alone (myc tagged) or in combination with either wt IE1 or the IE1 mutants L174 and K450R. Lysates from mock-transfected HEK293T cells served as specificity controls and detection of beta-actin expression as an internal loading control. (B) A FLAG-SUMO-1 expression construct was included in the cotransfection reaction, and SUMOylated PML was detected using an anti-FLAG antibody (top).
PML SUMOylates IE1 in a RING finger-dependent manner.
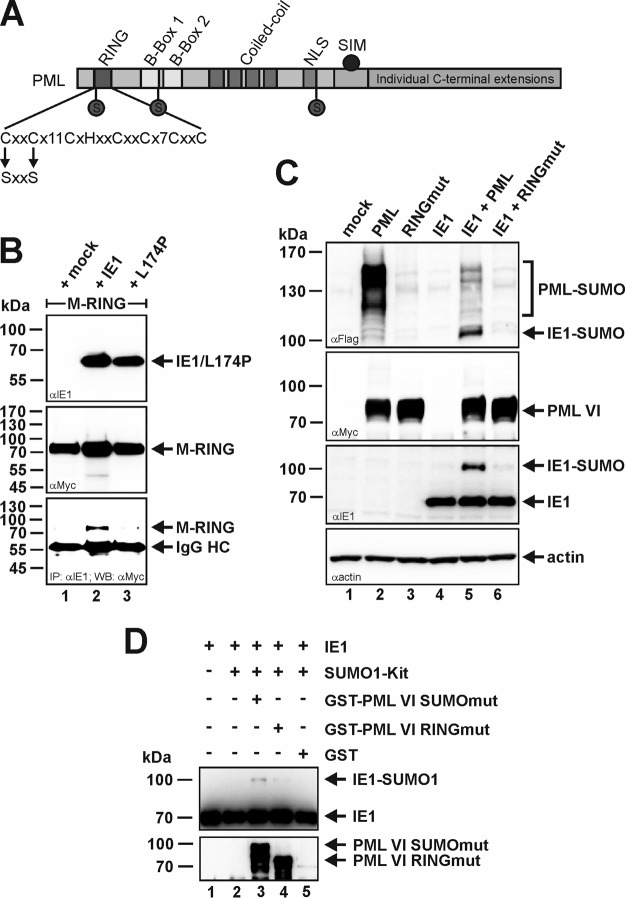
Next, we set out to elucidate the structural determinant for PML-mediated SUMOylation of IE1 (Fig. 7). Since the RING finger domain of PML is proposed to confer the E3 ligase activity, we disrupted the RING structure by mutating two key zinc-chelating cysteine residues at positions 57 and 60 to serines (Fig. 7A). As shown by Shen and coworkers, such a PML mutant is still able to interact with wt PML (27). This led the authors to conclude that introduction of the respective mutations does not disturb the overall structure of the protein. In addition, we could demonstrate that wt IE1, but not the L174P mutant, was still capable of interacting with the PML RING finger mutant (PML RINGmut) (Fig. 7B). This further strengthens the assumption of correct folding of the PML RING finger mutant. Inactivation of the SUMO E3 ligase activity of PML abrogated PML-induced SUMOylation of IE1 in vivo (Fig. 7C): comparably to the situation where IE1 was expressed alone (Fig. 7C, lane 4), no IE1-SUMO variant could be detected upon coexpression of PML RINGmut (Fig. 7C, lane 6). Furthermore, the experiment revealed that SUMOylation of the RING mutant PML protein was drastically reduced compared to wt PML (Fig. 7C, top, compare lanes 2 and 3 or 5 and 6). This is in agreement with previous findings by Shen and coworkers, which fostered the idea that PML acts as a SUMO E3 ligase that is also responsible for catalyzing its auto-SUMOylation (27). By performing an in vitro SUMOylation assay, we additionally confirmed the importance of PML's RING domain for IE1 SUMOylation (Fig. 7D). In contrast to the positive-control PML SUMOmut (Fig. 7D, lane 3), a GST-tagged PML mutant (isoform VI) with a deletion of the RING domain failed to stimulate IE1 SUMO conjugation above levels seen without PML or in the presence of GST alone (Fig. 7D). Notably, deletion of the PML RING domain does not affect the interaction between PML and IE1, as previously demonstrated by coimmunoprecipitation analyses (38). Taken together, these data demonstrate that an intact RING finger domain is not only necessary for PML self-SUMOylation, but also absolutely essential for PML-based SUMO conjugation of IE1.
FIG 7.
An intact RING finger domain of PML is required to stimulate IE1 SUMO conjugation. (A) Schematic representation of RINGmut based on PML isoform VI, where the key cysteine amino acids at positions 57 and 60 were mutagenized to similar nonpolar serine residues. (B) Coimmunoprecipitation analysis of PML-kd/293T cells cotransfected with plasmids encoding Myc-tagged PML RINGmut VI (M-RING) and wt IE1 or the PML interaction-negative mutant L174P. Two days posttransfection, the cells were lysed and immunoprecipitation was performed using an anti-IE1 antibody. After electrophoresis, coprecipitated PML RINGmut (M-RING) was visualized using an anti-Myc antibody (bottom). Immunoglobulin heavy chain (IgG HC) served as an internal control for the presence of the precipitating antibody. (C) Extracts from HEK293T cells transfected with IE1 and FLAG–SUMO-1 in combination with either wt PML or RINGmut (both isoform VI; myc tagged) were analyzed by Western blotting. Lysates from nontransfected HEK293T cells (mock) served as a specificity control. (D) Bacterially purified IE1 was incubated together with an in vitro SUMOylation kit containing E1 (SAE1/SAE2), Ubc9, ATP, and SUMO-1 in the presence or absence of PML GST-SUMOmut, GST-RINGmut (both isoform VI), or GST alone, as described in Materials and Methods. The reaction mixtures were analyzed by Western blotting using anti-IE1 and anti-PML antibodies.
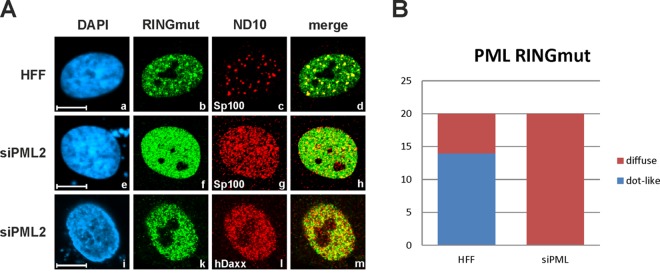
A PML RING finger mutant fails to orchestrate ND10 formation.
Since SUMOylation of PML is a prerequisite for the nucleation of ND10 structures, we next tested the capacity of the SUMOylation-defective RING domain mutant (PML RINGmut) to assemble ND10 bodies. To this end, the subnuclear localization pattern of PML RINGmut was analyzed in normal HFFs, as well as PML-kd cells (siPML2), by confocal immunofluorescence microscopy (Fig. 8). Consistent with the results of Western blot experiments demonstrating a lack of PML auto-SUMOylation, introduction of PML RINGmut into siPML2 cells lacking endogenous PML did not result in a re-formation of ND10 structures (Fig. 8A, e to m, and B). Instead, PML RINGmut and other ND10-related factors, like Sp100 or hDaxx, displayed a microdispersed distribution throughout the nucleus (Fig. 8A, f, g, k, and l). In contrast, expression of PML RINGmut in normal HFFs resulted in a partial recruitment of the mutant into the preexisting ND10 bodies, presumably through interaction with endogenous PML via the coiled-coil domain (Fig. 8A, a to d, and B). Collectively, our data demonstrate that the PML RING domain is critical for PML auto-SUMOylation and ND10 formation.
FIG 8.
The PML RING finger mutant fails to orchestrate ND10. (A) Indirect immunofluorescence analysis of siPML2-transduced HFFs (siPML2), as well as nontransduced control HFFs (HFF), transfected with plasmid DNA coding for Myc-tagged PML RINGmut (isoform VI). Two days posttransfection, the cells were fixed and stained for the ND10 marker Sp100 or hDaxx, along with PML RINGmut. The last was visualized by applying an anti-Myc antibody. Cell nuclei were identified by detection of DAPI signals (scale bars = 10 μm). (B) Numbers of cells with either a diffuse or a dot-like subnuclear distribution of PML RINGmut. Twenty cells were evaluated for either transfected control HFFs or transfected siPML2 cells.
Intact ND10 structures are not necessary for PML-based SUMOylation of IE1.
In light of the fact that the PML RING finger mutant lacks the ability to assemble ND10 bodies, the question arose as to whether intact ND10 structures are required for PML-mediated SUMOylation of IE1. This is important, since other components of the SUMO conjugation machinery that are involved in the modification of IE1, like the E1-activating enzyme, the E2-conjugating enzyme, or SUMO itself, accumulate at ND10. To investigate this, we took advantage of a SUMOylation-negative mutant of PML (SUMOmut) with lysine-to-arginine substitutions at residues 65, 160, and 490 (Fig. 9A) that has been reported to be defective in ND10 formation (28). In the next step, we coexpressed IE1, together with either wt PML, PML RINGmut, or PML SUMOmut in HEK293T cells (Fig. 9B). Interestingly, although the auto-SUMOylation of both PML mutants was highly compromised (Fig. 9B, top), only the RING finger mutant failed to stimulate IE1 SUMO conjugation (Fig. 9B, middle, lane 7). In contrast, IE1 was as efficiently SUMOylated by SUMOmut as by wt PML (Fig. 9B, middle, compare lanes 6 and 8), which is in accordance with the findings obtained by in vitro SUMOylation reactions (Fig. 4). In this context, it is noteworthy, that we ruled out the possibility that the small amount of endogenous PML present in HEK293T cells (Fig. 3) has any impact on the outcome of this experiment. Identical experiments were performed in both PML-kd HEK293T (PML-kd/293T) and genuine HEK293T cells, yielding identical results (compare Fig. 9B and C). Therefore, we conclude that SUMOylation of PML and the accompanying capacity to form genuine ND10 structures are not required for SUMOylation of IE1, while an intact RING finger domain of PML is indispensable for the activity. This assumption is consistent with the observation that IE1 SUMOylation is detectable throughout the HCMV replication cycle, even after ND10 bodies are disrupted (Fig. 1, second panel from top).
Finally, we investigated the subnuclear localization pattern of PML SUMOmut in normal HFFs and PML-kd cells (siPML2) (Fig. 9D). In cells expressing endogenous PML (HFFs), the SUMO-deficient mutant (SUMOmut) localized exclusively to ND10 (Fig. 9D, top row). Hence, the targeting of PML SUMOmut to ND10 bodies occurs much more efficiently than that of PML RINGmut (Fig. 8). Intriguingly, even in a PML knockdown background (siPML2), PML SUMOmut was capable of aggregating in distinct foci (Fig. 9D, middle and bottom rows), which is in clear contrast to the microdispersed distribution pattern of PML RINGmut in siPML2 cells (Fig. 8A, e to m). However, the number of PML SUMOmut foci formed in PML-depleted cells was clearly reduced compared to normal HFFs (Fig. 9E), indicating that these SUMOmut aggregations do not represent genuine ND10. The latter notion is further supported by our observation that these nuclear foci are completely devoid of hDaxx while Sp100 still efficiently localized to these SUMOmut-formed structures (Fig. 9D, e to m). Consequently, PML RINGmut and SUMOmut seem to differ in their capabilities to nucleate punctate PML-based complexes and to associate with other ND10-related factors.
DISCUSSION
PML and PML-NBs have attracted the attention of many researchers over the years due to their involvement in numerous aspects of cellular processes, including antiviral defense (19–21). Evidence is growing that ND10 can be regarded as subnuclear sites that provide a platform for the regulation of multiple PTM by hosting components of various PTM pathways (15–17). The idea of ND10 as a nuclear hot spot for PTM might explain how PML and PML-NBs are able to participate in such a plethora of diverse cellular processes. In line with this hypothesis, Cuchet-Lourenco and colleagues, for instance, demonstrated that the ND10-instituted antiviral response against HSV-1 depends on and is regulated by the SUMO modification pathway (39). Besides the E1-activating enzyme, the E2-conjugating enzyme UBC9, and SUMO itself, a number of SUMO E3 ligases, like PIAS1, RANBP2, or Topors, can be found in association with ND10 (17, 40). Recently, PML itself was confirmed to function as a SUMO E3 ligase, as it possesses the ability to promote the conjugation of SUMO to many of its cellular interaction partners, like p53, Mdm2, or c-Jun (8).
In the present study, we identified the effector protein IE1 of HCMV as the first viral target of PML's SUMO E3 ligase activity. In vitro experiments showed that PML is capable of stimulating IE1 SUMOylation by both SUMO-1 and -3. In addition, PML also enhanced IE1 SUMO conjugation in mammalian cells upon cotransfection, as well as during HCMV infection, which further underlines the in vivo relevance of this finding. Although IE1 SUMOylation was clearly stimulated under a variety of conditions by the presence of PML, no clear dose response of IE1 SUMOylation was detected in these experiments. This is consistent with the function of PML as an E3 adaptor protein, which depend in their activity on the presence of additional components of the conjugation machinery that may be rate limiting for the reaction.
The IE1 protein is known to play an important role during HCMV replication due to its ability to disrupt PML-NBs, thereby interfering with the intrinsic antiviral response of the host. Through a direct physical interaction with PML, IE1 initially localizes to ND10 in order to induce a loss of the SUMOylated forms of PML and Sp100 (22, 24). By destroying the PML-IE1 binding interface, we abolished PML-mediated SUMO modification of IE1. Furthermore, we showed that IE1 SUMOylation occurs in a PML isoform-dependent manner. This observation suggests that the individual PML variants may differ in their abilities to associate with IE1 due to their distinct C-terminal extensions, which are known to be responsible for PML isoform-specific interactions (41). Finally, as for other PML substrates, we prevented PML-based SUMOylation of IE1 by mutating the RING finger domain of PML, which constitutes an important structural determinant of PML's SUMO ligase activity (8). Since PML is thought to possess auto-SUMOylation activity, such a mutant exhibited a severe SUMOylation defect (27). Consequently, PML RINGmut failed to form ND10 structures when introduced into PML-kd cells. However, intact ND10 bodies are not a prerequisite for PML-induced SUMO conjugation of IE1, as a SUMOylation-deficient mutant of PML promoted IE1 SUMOylation as efficiently as wt PML. In addition, we showed that IE1 SUMOylation is sustained throughout the HCMV replication cycle, even after ND10 domains are dispersed by IE1, which further strengthens the finding that ND10 integrity is not required for PML-mediated attachment of SUMO to IE1.
Intriguingly, although for both PML mutants, RINGmut and SUMOmut, the posttranslational modification by SUMO is highly compromised, we found that they differ in their abilities to form PML-based aggregates and to associate with other ND10 factors. In the absence of endogenous PML, the RING finger mutant was evenly distributed throughout the nucleus and showed no further association with Sp100 or hDaxx. In contrast, and consistent with previous work by Cuchet-Lourenco and colleagues (39), PML SUMOmut was still capable of forming intense nuclear aggregates in PML-depleted cells. However, the number of SUMOmut-formed foci was drastically reduced compared to normal ND10 bodies in control cells. In addition, these PML SUMOmut foci contained only Sp100, but no hDaxx, thus representing no genuine ND10 structures. On the contrary, in cells expressing endogenous PML, the RING mutant exhibited partial relocalization and the SUMOylation-negative mutant exhibited complete relocalization into the preexisting ND10 structures, which is probably mediated through interactions of the mutants with endogenous PML via their coiled-coil domains.
Therefore, it was of major importance to exclude the possibility that endogenously present PML influences the experimental outcome when we evaluated the effect of exogenously introduced PML mutants on IE1 SUMOylation. To circumvent this problem, we performed the transfection experiments in HEK293T cells, which express clearly reduced levels of PML compared to HFFs and obviously no Sp100. Hence, it is not surprising, that HEK293T cells do not show genuine ND10 accumulations when analyzed by immunofluorescence staining (data not shown). Our finding that the PML RING finger mutant failed to stimulate IE1 SUMOylation compared to wt PML or PML SUMOmut indicates that the trace amount of wt PML that is still present in HEK293T cells indeed has no impact on the outcome of the experiment. This assumption is further strengthened by the finding that we obtained identical results when transfection experiments were performed in PML knockdown HEK293T cells, where the residual amount of endogenous PML was depleted by expression of the respective short hairpin RNA (shRNA) (22).
Besides IE1, HCMV encodes another prominent IE gene product, termed IE2, which plays a key role in initiating the lytic replication cycle of the virus and is well known to undergo posttranslational modification by SUMO (32, 42, 43). However, we found that SUMOylation of IE2, which likewise temporarily resides at ND10, was not influenced by PML during infection, indicating that IE1 represents a specific target of PML (31). This correlates with the finding that only IE1 has the capacity to directly interact with PML (23).
In this context, it is noteworthy that modification of IE1 and IE2 by SUMO has opposing functional consequences for the two viral regulatory proteins. In the case of IE2, this posttranslational modification is necessary for efficient IE2 function. Previous reports showed that the transactivating capacity of IE2 is dependent on SUMO interactions, and viruses expressing SUMOylation-negative IE2 reveal strongly impaired replication efficacy due to reduced viral DNA and protein accumulation, as well as diminished initiation of immediate early gene expression (32, 44). On the other hand, SUMOylation of IE1, which is elevated by PML, seems to negatively affect IE1's function to modulate STAT signaling. Besides annihilating the repressive effect of ND10, the IE1 protein is also known to be responsible for inhibiting the activation of interferon (IFN)-stimulated genes (ISGs) by, for instance, binding to STAT2 (45, 46). Interestingly, this IE1-STAT2 interaction and the concomitant inhibition of IFN-mediated gene expression, which is required for efficient viral growth, are negatively regulated by SUMO (46). A previous study demonstrated that SUMOylated IE1 no longer binds to STAT2. This was shown using an IE1-SUMO fusion protein, thus ensuring that all the IE1 was attached to SUMO (46). During infection, however, only a small fraction of IE1 undergoes SUMO modification, and our own efforts to modulate the IFN-antagonizing function of IE1 by overexpression of PML were not successful due to the fact that less than 20% of the IE1 was modified by SUMO under these conditions (data not shown). Nevertheless, one might speculate that PML has the potential to negatively influence the activity of viral regulatory proteins by modulating their posttranslational modification status. This, however, may be abrogated by the propensity of IE1 to block PML E3 ligase functions (25).
Taken together, our experiments establish that PML acts as a SUMO E3 ligase for IE1, and we suggest that SUMOylation of IE1 by PML constitutes a cellular attempt to compromise specific functions of this viral effector protein. Interestingly, since PML-based SUMOylation of IE1 is detectable throughout HCMV replication, this antiviral property of PML does not seem to be restricted to the initial stage of infection with intact ND10 but is still active even after ND10 are disrupted by IE1. Thus, it is tempting to speculate that, comparably to Sp100, where we could demonstrate a dual antiviral function during the immediate early and late phases of the HCMV replicative cycle, PML likewise seems to be able to negatively interfere with HCMV infection at different steps of the viral life cycle (22).
MATERIALS AND METHODS
Oligonucleotides and plasmid constructs.
The oligonucleotide primers used for this study were purchased from Biomers GmbH (Ulm, Germany) and are listed in Table 1. The PML isoform VI plasmid was a generous gift from Gerd Maul (Philadelphia, PA, USA). Lentivirus vectors expressing FLAG-tagged versions of PML isoforms I to VI were kindly provided by Roger Everett (Glasgow, United Kingdom) (47). All cDNAs encoding the PML isoforms were made resistant to the anti-PML shRNA siPML2 (48) by introduction of five silent point mutations within the siPML2 target sequence (47). The altered sequence is 5′-AGATGCTGCAGTTAGCAAG-3′. Derivatives of Myc-PML isoform VI with point mutations in the RING domain (RINGmut; C57S and C60S) or of the SUMO modification sites (SUMOmut; K65R, K160R, and K490R) and IE1 mutants L174P (a PML interaction-negative mutant) and K450R (a SUMOylation-deficient mutant) were constructed by site-directed mutagenesis using a QuikChange site-directed mutagenesis kit, as instructed by the manufacturer (Stratagene, La Jolla, CA). For bacterial expression of GST-tagged variants of wt PML or PML SUMOmut, the respective coding sequences were cloned into the prokaryotic expression vector pGEX-6P-1 (GE Healthcare, Munich, Germany) via BamHI/EcoRI using primers 5′PML-1597 and 3′PML-1597/2691. GST-IE1 was obtained by cloning of a codon-optimized version of the IE1 cDNA sequence into pGEX-6P-1 (GE Healthcare, Munich, Germany) via BamHI/XhoI using the primer pair 5′IE1codonopt_BamH1 and 3′IE1codonopt_Xho1. For generation of GST-RINGmut containing a deletion of the RING domain (amino acids [aa] 1 to 102), a codon-optimized version of PML isoform VI was used for PCR amplification of aa 103 to 560 using primers 5′Bam_PMLco_aa103 and 3′Sal_PMLVIco, followed by cloning of the PCR product into pGEX-6P-1 via BamHI and SalI. The integrity of all the newly generated plasmids was confirmed by automated DNA sequence analysis.
TABLE 1.
Oligonucleotides used for plasmid construction
| Oligonucleotide | Sequence |
|---|---|
| 5′PML-RINGmut | CCAGTTTCTGCGCAGCCAGCAAAGCCAGGCGGAAGCC |
| 3′PML-RINGmut | GGCTTCCGCCTGGCTTTGCTGGCTGCGCAGAAACTGG |
| 5′PML-SUMO65-mut | CCAGGCGGAAGCCAGGTGCCCGAAGCTGC |
| 3′PML-SUMO65-mut | GCAGCTTCGGGCACCTGGCTTCCGCCTGG |
| 5′PML-SUMO160-mut | CCAGTGGTTCCTCAGGCACGAGGCCCGGC |
| 3′PML-SUMO160-mut | GCCGGGCCTCGTGCCTGAGGAACCACTGG |
| 5′PML-SUMO490-mut | CCAGGAAGGTCATCAGGATGGAGTCTGAGGAGG |
| 3′PML-SUMO490-mut | CCTCCTCAGACTCCATCCTGATGACCTTCCTGG |
| 5′IE1mut-L174P | GGCTTGTATTAAGGAGCCGCATGATGTGAGCAAGG |
| 3′IE1mut-L174P | CCTTGCTCACATCATGCGGCTCCTTAATACAAGCC |
| 5′IE1K450R | GACACTGTGTCTGTCCGGTCTGAGCCAGTGTCTG |
| 3′IE1K450R | CAGACACTGGCTCAGACCGGACAGACACAGTGTC |
| 5′PML-1597 | CATAGGATCCATGGAGCCTGCACCCGCCCG |
| 3′ PML-1597/2691 | CATAGAATTCTCACCACAACGCGTTCCTCT |
| 5′IE1codonopt_BamH1 | CATAGGATCCATGGAGAGTTCGGCAAAACGT |
| 3′IE1codonopt_Xho1 | CATACTCGAGTCACTGATCAGCTTTGGAGCG |
| 5′Bam_PMLco_aa103 | CATAGGATCCGCACTGGATAATGTTTTTTTTG |
| 3′Sal_PMLVIco | CATAGTCGACTCACCACAGTGCATTGCG |
Cells and viruses.
HEK293T cells were cultivated in Dulbecco's minimal essential medium (DMEM) containing 10% fetal calf serum. HEK293T cells with a small interfering RNA-mediated knockdown of PML (PML-kd/293T) were cultured in DMEM supplemented with 10% fetal calf serum and 5 μg/ml puromycin (22). Primary HFFs were prepared from human foreskin tissue as described previously and were maintained in Eagle's minimal essential medium (GIBCO/BRL, Eggenstein, Germany) supplemented with 5% fetal calf serum (32). siPML2 cells and the respective control cells (designated vector) were cultured in Dulbecco's minimal essential medium (GIBCO/BRL, Eggenstein, Germany) supplemented with 10% fetal calf serum and 5 μg/ml puromycin (48). Infection experiments were performed with the HCMV laboratory strain AD169. Viral stocks were titrated via IE1p72 fluorescence (49). For this purpose, HFFs were infected with various dilutions of virus stocks. After 24 h of incubation, the cells were fixed and stained with monoclonal antibody (MAb) p63-27, directed against IE1p72 (50). Subsequently, the number of IE1-positive cells was determined and used to calculate viral titers, expressed as IE protein-forming units (IEU).
Retrovirus transduction and selection of stably transduced cells.
HFFs stably expressing FLAG-PML isoform I were generated via retrovirus transduction using the ViraPower lentiviral system (Invitrogen, Karlsruhe, Germany) according to the manufacturer's protocol and maintained in Dulbecco's minimal essential medium supplemented with 10% fetal calf serum plus 50 μg/ml Geneticin.
Antibodies.
MAb p63-27, which recognizes IE1, has been described elsewhere (50). The polyclonal antiserum raised against exon 5 of IE2 (referred to as anti-pHM178) was generated by immunizing rabbits with the prokaryotically expressed protein. PML was detected by using either the rabbit polyclonal antibody H-238 (Santa Cruz Biotechnology, Santa Cruz, CA) or the mouse monoclonal antibody 5E10 (kindly provided by Roel van Driel, University of Amsterdam, Amsterdam, The Netherlands). For the detection of Myc- or FLAG-tagged versions of PML or SUMO-1, the monoclonal antibodies MAb-Myc (1-9E10.2; ATCC) and MAb-FLAG M2 (Sigma-Aldrich, Deisenhofen, Germany) were utilized. Sp100 was detected by applying either the rabbit polyclonal antiserum GH3 (a kind gift from Hans Will, Heinrich Pette Institute for Experimental Virology and Immunology, University of Hamburg, Hamburg, Germany) or a rabbit polyclonal antibody raised against aa 209 to 227 of Sp100 (kindly provided by Peter Hemmerich, Leibniz Institute for Age Research, Fritz Lipmann Institute, Jena, Germany). A rabbit monoclonal antibody from Epitomics (Burlingame, CA) or the mouse monoclonal antibody MCA2143 (Serotec, Duesseldorf, Germany) was used for the detection of hDaxx. Monoclonal antibody AC-15, which recognizes beta-actin, was purchased from Sigma-Aldrich (Deisenhofen, Germany). Horseradish peroxidase-conjugated anti-mouse and anti-rabbit secondary antibodies for Western blot analysis were obtained from Dianova (Hamburg, Germany), while Alexa Fluor 488- or Alexa Fluor 555-conjugated secondary antibodies for indirect immunofluorescence experiments were purchased from Molecular Probes (Karlsruhe, Germany).
Indirect immunofluorescence analysis.
For indirect immunofluorescence analysis, 3 × 105 HFFs were grown on coverslips for transfection using the transfection reagent FuGene HD (Promega, Mannheim, Germany) according to the manufacturer's instructions. At 48 h posttransfection, the cells were washed three times with phosphate-buffered saline (PBS), followed by fixation with 4% paraformaldehyde for 10 min at room temperature. Then, the cells were permeabilized with PBS–0.2% Triton X-100 on ice for 20 min, followed by incubation with the respective primary or secondary antibodies for 30 min at 37°C. Finally, the cells were mounted by using Vectashield mounting medium plus 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA). The samples were examined by using a Leica TCS SP5 confocal microscope with 488-nm and 543-nm laser lines, scanning each channel separately under image capture conditions that eliminated channel overlap. The images were exported as tagged-image file format (TIFF) files and were then processed using Photoshop.
Western blotting and co-IP assay.
For Western blot analysis, extracts from infected HFFs or transfected HEK293T cells were lysed, diluted in sodium dodecyl sulfate (SDS)-Laemmli buffer, and boiled at 95°C for 10 min (32). Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) on 8% to 12.5% polyacrylamide gels and transferred onto nitrocellulose membranes (GE Healthcare, Munich, Germany), followed by chemiluminescence detection according to the manufacturer's protocol (ECL Western blotting detection kit; Amersham Pharmacia Europe, Freiburg, Germany). The expression of SUMOylated and non-SUMOylated proteins was quantified using Image J and AIDA software, followed by the calculation of the ratio of protein SUMOylation relative to the overall protein expression. Coimmunoprecipitation (co-IP) analysis was performed as described previously (51). Briefly, HEK293T cells were transfected in 6-well plates (5.0 × 105 cells/well) via calcium phosphate coprecipitation. Two days posttransfection, the cells were lysed for 20 min at 4°C in 800 μl of co-IP buffer (50 mM Tris-HCl [pH 8.0], 150 to 300 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 μg/ml of aprotinin, 2 μg/ml of leupeptin, and 2 μg/ml of pepstatin). After centrifugation, the supernatant was incubated with the appropriate antibody (MAb p63-27) coupled to protein A Sepharose beads for 1.5 h at 4°C. The Sepharose beads were collected and washed six times in co-IP lysis buffer. Antigen-antibody complexes were recovered by boiling in SDS sample buffer and analyzed via Western blotting.
Protein generation and purification.
PML, SUMOmut, RINGmut, and IE1 were expressed in fusion with GST using E. coli strain BL21(DE3). The fusion proteins were purified from crude lysates by affinity chromatography using glutathione–Sepharose columns according to the specifications of the manufacturer (GE Healthcare, Bio-Sciences AB, Uppsala, Sweden). In the case of IE1, the GST tag was cleaved by addition of PreScission protease (GE Healthcare) at a ratio of 1:100 (wt/wt). Free GST and GST-tagged PreScission protease was removed by glutathione Sepharose affinity chromatography. Afterward, IE1 was purified by gel filtration using a HiLoad 16/60 Superdex 200 Prep Grade column (GE Healthcare, Bio-Sciences AB, Uppsala, Sweden) according to the manufacturer's protocol. Eluted proteins were analyzed by SDS-PAGE and Coomassie blue staining.
In vitro SUMOylation assays.
In vitro SUMOylation reactions were performed at 37°C for 1 h in 20-μl volumes containing prokaryotically expressed IE1 or GST-PML/GST-SUMOmut/GST-RINGmut (based on PML isoform VI), along with 50 nM the heterodimeric human E1-activating enzyme (SAE1/SAE2), 62.5 μM the E2-conjugating enzyme UbcH9, 1 mM Mg-ATP, 25 μM either SUMO-1 or SUMO-3, and reaction buffer (500 mM HEPES, pH 8, 1,000 mM NaCl, 10 mM dithiothreitol [DTT]; BostonBiochem, Cambridge, MA) according to the manufacturer's protocol. After termination of the reaction using SDS sample buffer containing beta-mercaptoethanol, the reaction products were fractionated by SDS-8% PAGE for immunoblot detection of IE1, PML, and GST.
ACKNOWLEDGMENTS
We thank Roger Everett (Glasgow, United Kingdom), Peter Hemmerich (Jena, Germany), and the late Gerd Maul (Philadelphia, PA, USA) for providing reagents for this study.
REFERENCES
- 1.Hannoun Z, Maarifi G, Chelbi-Alix MK. 2016. The implication of SUMO in intrinsic and innate immunity. Cytokine Growth Factor Rev 29:3–16. doi: 10.1016/j.cytogfr.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Gareau JR, Lima CD. 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilkinson KA, Henley JM. 2010. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J 428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flotho A, Melchior F. 2013. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 5.Liang YC, Lee CC, Yao YL, Lai CC, Schmitz ML, Yang WM. 2016. SUMO5, a novel poly-SUMO isoform, regulates PML nuclear bodies. Sci Rep 6:26509. doi: 10.1038/srep26509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hay RT. 2005. SUMO: a history of modification. Mol Cell 18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu Rev Biochem 78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 8.Chu Y, Yang X. 2011. SUMO E3 ligase activity of TRIM proteins. Oncogene 30:1108–1116. doi: 10.1038/onc.2010.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Napolitano LM, Jaffray EG, Hay RT, Meroni G. 2011. Functional interactions between ubiquitin E2 enzymes and TRIM proteins. Biochem J 434:309–319. doi: 10.1042/BJ20101487. [DOI] [PubMed] [Google Scholar]
- 10.Meroni G, Diez Roux G. 2005. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays 27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 11.Duprez E, Saurin AJ, Desterro JM, Lallemand-Breitenbach V, Howe K, Boddy MN, Solomon E, de Thé H, Hay RT, Freemont PS. 1999. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J Cell Sci 112:381–393. [DOI] [PubMed] [Google Scholar]
- 12.Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS. 1996. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13:971–982. [PubMed] [Google Scholar]
- 13.Quimby BB, Yong-Gonzalez V, Anan T, Strunnikov AV, Dasso M. 2006. The promyelocytic leukemia protein stimulates SUMO conjugation in yeast. Oncogene 25:2999–3005. doi: 10.1038/sj.onc.1209335. [DOI] [PubMed] [Google Scholar]
- 14.Borden KL, Culjkovic B. 2009. Perspectives in PML: a unifying framework for PML function. Front Biosci 14:497–509. [DOI] [PubMed] [Google Scholar]
- 15.Schmitz ML, Grishina I. 2012. Regulation of the tumor suppressor PML by sequential post-translational modifications. Front Oncol 2:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng X, Kao HY. 2012. Post-translational modifications of PML: consequences and implications. Front Oncol 2:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Damme E, Laukens K, Dang TH, Van Ostade X. 2010. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int J Biol Sci 6:51–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geoffroy MC, Chelbi-Alix MK. 2011. Role of promyelocytic leukemia protein in host antiviral defense. J Interferon Cytokine Res 31:145–158. doi: 10.1089/jir.2010.0111. [DOI] [PubMed] [Google Scholar]
- 19.Tavalai N, Stamminger T. 2008. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim Biophys Acta 1783:2207–2221. doi: 10.1016/j.bbamcr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Tavalai N, Stamminger T. 2011. Intrinsic cellular defense mechanisms targeting human cytomegalovirus. Virus Res 157:128–133. doi: 10.1016/j.virusres.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Tavalai N, Stamminger T. 2009. Interplay between herpesvirus infection and host defense by PML nuclear bodies. Viruses 1:1240–1264. doi: 10.3390/v1031240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tavalai N, Adler M, Scherer M, Riedl Y, Stamminger T. 2011. Evidence for a dual antiviral role of the major nuclear domain 10 component Sp100 during the immediate-early and late phases of the human cytomegalovirus replication cycle. J Virol 85:9447–9458. doi: 10.1128/JVI.00870-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahn JH, Brignole EJ III, Hayward GS. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol 18:4899–4913. doi: 10.1128/MCB.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HR, Kim DJ, Lee JM, Choi CY, Ahn BY, Hayward GS, Ahn JH. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J Virol 78:6527–6542. doi: 10.1128/JVI.78.12.6527-6542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schilling EM, Scherer M, Reuter N, Schweininger J, Muller YA, Stamminger T. 2017. The human cytomegalovirus IE1 protein antagonizes PML nuclear body-mediated intrinsic immunity via the inhibition of PML de novo SUMOylation. J Virol 91:e02049-16. doi: 10.1128/JVI.02049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, Ho CC, Chen YC, Lin TP, Fang HI, Hung CC, Suen CS, Hwang MJ, Chang KS, Maul GG, Shih HM. 2006. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell 24:341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 27.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. 2006. The mechanisms of PML-nuclear body formation. Mol Cell 24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong S, Muller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748–2753. [PubMed] [Google Scholar]
- 29.Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ETH, Strauss JF, Maul GG. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J Cell Biol 147:221–233. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchini A, Liu H, Zhu H. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J Virol 75:1870–1878. doi: 10.1128/JVI.75.4.1870-1878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sourvinos G, Tavalai N, Berndt A, Spandidos DA, Stamminger T. 2007. Recruitment of human cytomegalovirus immediate-early 2 protein onto parental viral genomes in association with ND10 in live-infected cells. J Virol 81:10123–10136. doi: 10.1128/JVI.01009-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hofmann H, Floss S, Stamminger T. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J Virol 74:2510–2524. doi: 10.1128/JVI.74.6.2510-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spengler ML, Kurapatwinski K, Black AR, Azizkhan-Clifford J. 2002. SUMO-1 modification of human cytomegalovirus IE1/IE72. J Virol 76:2990–2996. doi: 10.1128/JVI.76.6.2990-2996.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, Ahn JH, Cheng M, ap Rhys CM, Chiou CJ, Zong J, Matunis MJ, Hayward GS. 2001. Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression. J Virol 75:10683–10695. doi: 10.1128/JVI.75.22.10683-10695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nisole S, Maroui MA, Mascle XH, Aubry M, Chelbi-Alix MK. 2013. Differential roles of PML isoforms. Front Oncol 3:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller S, Dejean A. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol 73:5137–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scherer M, Otto V, Stump JD, Klingl S, Muller R, Reuter N, Muller YA, Sticht H, Stamminger T. 2015. Characterization of recombinant human cytomegaloviruses encoding IE1 mutants L174P and 1-382 reveals that viral targeting of PML bodies perturbs both intrinsic and innate immune responses. J Virol 90:1190–1205. doi: 10.1128/JVI.01973-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scherer M, Klingl S, Sevvana M, Otto V, Schilling EM, Stump JD, Muller R, Reuter N, Sticht H, Muller YA, Stamminger T. 2014. Crystal structure of cytomegalovirus IE1 protein reveals targeting of TRIM family member PML via coiled-coil interactions. PLoS Pathog 10:e1004512. doi: 10.1371/journal.ppat.1004512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuchet-Lourenco D, Boutell C, Lukashchuk V, Grant K, Sykes A, Murray J, Orr A, Everett RD. 2011. SUMO pathway dependent recruitment of cellular repressors to herpes simplex virus type 1 genomes. PLoS Pathog 7:e1002123. doi: 10.1371/journal.ppat.1002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown JR, Conn KL, Wasson P, Charman M, Tong L, Grant K, McFarlane S, Boutell C. 2016. SUMO ligase protein inhibitor of activated STAT1 (PIAS1) is a constituent promyelocytic leukemia nuclear body protein that contributes to the intrinsic antiviral immune response to herpes simplex virus 1. J Virol 90:5939–5952. doi: 10.1128/JVI.00426-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jensen K, Shiels C, Freemont PS. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223–7233. doi: 10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- 42.Ahn JH, Xu Y, Jang WJ, Matunis MJ, Hayward GS. 2001. Evaluation of interactions of human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9. J Virol 75:3859–3872. doi: 10.1128/JVI.75.8.3859-3872.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim ET, Kim YE, Kim YJ, Lee MK, Hayward GS, Ahn JH. 2014. Analysis of human cytomegalovirus-encoded SUMO targets and temporal regulation of SUMOylation of the immediate-early proteins IE1 and IE2 during infection. PLoS One 9:e103308. doi: 10.1371/journal.pone.0103308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berndt A, Hofmann-Winkler H, Tavalai N, Hahn G, Stamminger T. 2009. Importance of covalent and noncovalent SUMO interactions with the major human cytomegalovirus transactivator IE2p86 for viral infection. J Virol 83:12881–12894. doi: 10.1128/JVI.01525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paulus C, Krauss S, Nevels M. 2006. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc Natl Acad Sci U S A 103:3840–3845. doi: 10.1073/pnas.0600007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huh YH, Kim YE, Kim ET, Park JJ, Song MJ, Zhu H, Hayward GS, Ahn JH. 2008. Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J Virol 82:10444–10454. doi: 10.1128/JVI.00833-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cuchet D, Sykes A, Nicolas A, Orr A, Murray J, Sirma H, Heeren J, Bartelt A, Everett RD. 2011. PML isoforms I and II participate in PML-dependent restriction of HSV-1 replication. J Cell Sci 124:280–291. doi: 10.1242/jcs.075390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol 80:8006–8018. doi: 10.1128/JVI.00743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lorz K, Hofmann H, Berndt A, Tavalai N, Mueller R, Schlotzer-Schrehardt U, Stamminger T. 2006. Deletion of open reading frame UL26 from the human cytomegalovirus genome results in reduced viral growth, which involves impaired stability of viral particles. J Virol 80:5423–5434. doi: 10.1128/JVI.02585-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andreoni M, Faircloth M, Vugler L, Britt WJ. 1989. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J Virol Methods 23:157–167. doi: 10.1016/0166-0934(89)90129-8. [DOI] [PubMed] [Google Scholar]
- 51.Bannister AJ, Kouzarides T. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]