SUMMARY
Pluripotent stem cells (PSCs) offer unprecedented opportunities for disease modeling and personalized medicine. However, PSC-derived cells exhibit fetal-like characteristics and remain immature in a dish. This has emerged as a major obstacle for their application for late-onset diseases. We previously showed that there is a neonatal arrest of long-term cultured PSC-derived cardiomyocytes (PSC-CMs). Here, we demonstrate that PSC-CMs mature into adult CMs when transplanted into neonatal hearts. PSC-CMs became similar to adult CMs in morphology, structure, and function within a month of the transplantation into rats. The similarity was further supported by single-cell RNA-sequencing analysis. Moreover, this in vivo maturation allowed patient-derived PSC-CMs to reveal the disease phenotype of arrhythmogenic right ventricular cardiomyopathy, which predominantly manifests in adults. This study lays a foundation for understanding human CM maturation and pathogenesis and can be instrumental in PSC-based modeling of adult heart diseases.
INTRODUCTION
It has been a decade since Yamanaka and colleagues found a way to induce formation of pluripotent stem cells (iPSCs) from adult cells (Takahashi and Yamanaka, 2006). PSCs are capable of becoming any cell type in principle, so there is tremendous enthusiasm for their use in disease modeling, drug discovery, and regenerative medicine as well as understanding human development. Consequently, numerous iPSCs have been generated from patients harboring various mutations or diseases (Fox et al., 2014; Tabar and Studer, 2014). While they have the potential to model and treat a broad spectrum of human diseases, PSC-derived cells are morphologically and functionally similar to fetal cells. This has become a major and common impediment for their application to model and treat late-onset disorders (Cho et al., 2014; Svendsen, 2013; Tabar and Studer, 2014).
The PSC field is intensely focused on heart disease because of its worldwide prevalence causing high morbidity and mortality. In particular, with methodological advances in differentiating PSCs into cardiomyocytes (CMs), current cardiac PSC research is centering on modeling cardiomyopathy (Kamdar et al., 2015; Lalit et al., 2014), a leading cause of heart failure. However, cardiomyopathy occurs predominantly in adult stages, making it difficult to recapitulate the true disease phenotype and to validate the efficacy of drugs discovered using PSC-derived CMs (PSC-CMs). For this reason, extensive tissue engineering efforts are underway with the goal of mature PSC-CMs in vitro. Studies demonstrated that electrical and mechanical stimulation promote the structural and functional maturation of PSC-CMs (Nunes et al., 2013; Ruan et al., 2015). Substrate properties were also shown to play an important role in their maturation (Feaster et al., 2015; Ribeiro et al., 2015). For instance, myofibril alignment and contractility were significantly enhanced in PSC-CMs grown in micropatterned polyacrylamide (Ribeiro et al., 2015). These studies evince the critical role of microenvironment in PSC-CM maturation.
Developmentally, the maturation of CMs begins at an early embryonic stage and continues throughout postnatal stages. PSC-CMs resemble early embryonic CMs in structure, function, and gene expression (Robertson et al., 2013). Transcriptional analysis revealed that PSC-CMs undergo maturation in culture, but are arrested at late embryonic/neonatal stages (Uosaki et al., 2015). In the present study, we leveraged the potential of the neonatal heart environment to demonstrate that neonatal hearts are capable of maturing PSC-CMs to adult CMs.
RESULTS
In Vivo-Matured PSC-CMs Are Morphologically and Structurally Indistinguishable from Adult CMs
To examine the morphology of PSC-CMs in long-term culture, we differentiated mouse embryonic stem cells (mESCs) into CMs by sequential differentiation of ESCs into mesoderm, cardiac progenitor cells (CPCs), and CMs (Cheng et al., 2013; Uosaki et al., 2012). The resulting CMs were cultured in conditions shown to enhance CM maturation (Lundy et al., 2013). The mESC-derived CMs (mESC-CMs) increased in size over time, but remained mononucleated and irregular in shape with cytoskeletal disarray, which is similar to neonatal CMs, but distinct from adult CMs that are cylindrical with a well-organized cytoskeleton (Figure 1A). This is consistent with our previous finding that in vitro-matured PSC-CMs are arrested at a neonatal stage at the molecular level (Uosaki et al., 2015).
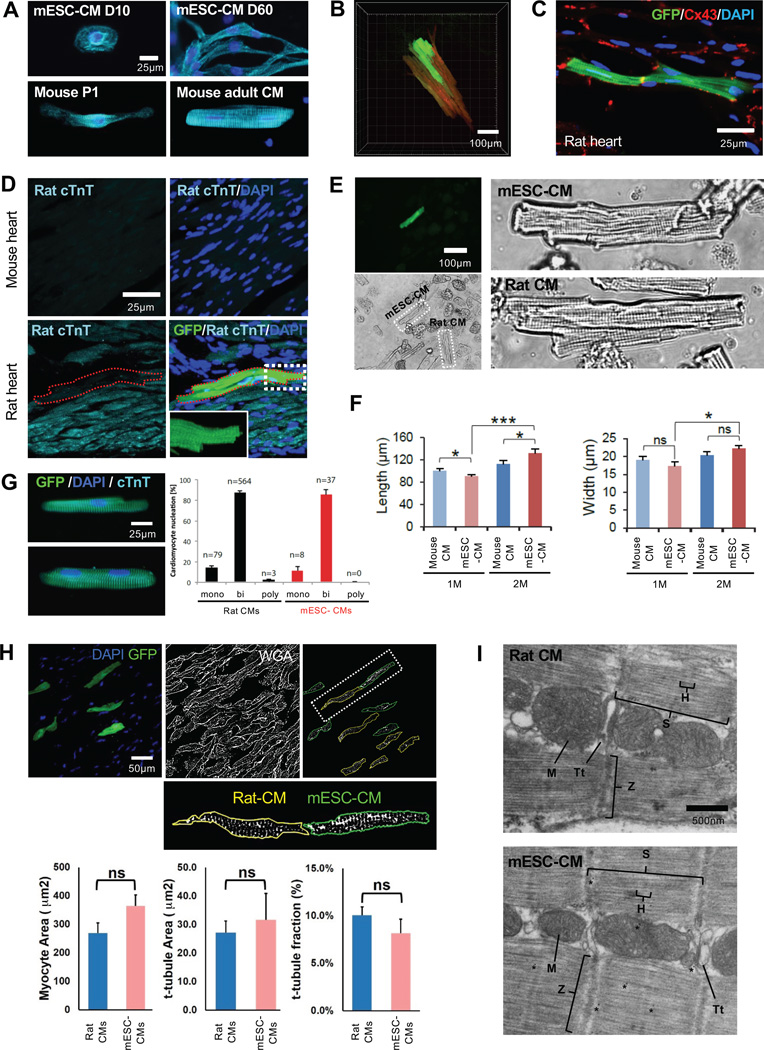
Figure 1. In Vivo-Matured PSC-CMs Show Adult CM Morphology.
(A) α-Actinin (cyan) staining of mESC-CMs matured in vitro for 10 or 60 days (top) and endogenous mouse CMs at postnatal day 1 and 2 months (adult) (bottom). DAPI (blue) was used to counterstain nuclei. (B) 3D image of mESC-CMs matured in the rat heart for 2 months (Movie 1). (C) CX43 staining (red) of mESC-CMs matured in the rat heart. (D) Adult mouse heart section stained with Rat cTnT (top) and mESC-CMs (GFP+) matured in the rat heart (bottom). The red dotted line indicates mESC-CMs. Inset (bottom right) shows a magnified image of the white box. (E) In vivo-matured mESC-CM (GFP+) isolated from the rat heart (top) and adult rat CMs (bottom). (F) Average cell length and width of mouse CMs and in vivo-matured mESC-CMs at indicated stages. Data are mean ± SD; n=7 per group; *p<0.05; ***p<0.001; ns, not significant (p>0.05). p values were determined using the paired Student t test. (G) Binucleation % of adult rat CMs (n=3 hearts) and in vivo-matured mESC-CMs (n=3 hearts). (H) In vivo-matured mESC-CM (GFP+) in rat heart (top, left). WGA binary image and selected t-tubule network excluding surface membrane of rat-CM (yellow line) and mESC-CMs (green line) (top, middle and right). Segmentation and particle analysis of rat CMs and in vivo-matured mESC-CMs (bottom). (I) Transmission electron micrographs of adult rat CM and in vivo-matured mESC-CM. D, day; M, month; p, postnatal day; .H, H band; M, mitochondria; S, sarcomere; Tt, t-tubule; Z, Z-line. Student’s t test and one way-ANOVA were used for statistical analyses.
Based on the neonatal arrest, we reasoned that a neonatal environment in situ might possess the capability of maturing engrafted PSC-CMs. Since neonatal hearts contain Isl1+ CPCs that were shown to become adult CMs without cell fusion (Laugwitz et al., 2005; Zaruba et al., 2010), we hypothesized that PSC-derived Isl1+ CPCs (PSC-CPCs) could mature along with their host CPCs/CMs when inserted into neonatal myocardium. To test this, we generated a mESCIsl1-Cre; Rosa-RFP; aMHC-GFP line that expresses red fluorescent protein (RFP) constitutively in Isl1+ CPCs and green fluorescent protein (GFP) in CMs, which allows tracing mESC-CPCs and monitoring their differentiation into CMs (Shenje et al., 2014). We purified RFP+ CPCs at day 6 by fluorescence-activated cell sorting (FACS) and monitored their development following their intraventricular delivery (~2×105 cells/injection) at postnatal days (P) 1–3 (Figure S1). To avoid immune rejection, NIH nude rats (Liang et al., 1997) were used. On average, we obtained ~2,000 RFP+ cells per injection site, which was sufficient for subsequent in vivo and in vitro analyses. The CPCs expressed GFP at 1 week post-injection, suggesting their differentiation into CMs, but they were spherical with a single nucleus (Figure S2A). However, they became similar to adult CMs in morphology after 1 month of incubation (Figure S2A). A 3D reconstruction of the heart, generated with a tissue-clearing method (Susaki et al., 2014), revealed that the incubated cells form adult CM-like patches (Figure 1B and Movie S1). Connexin43, a gap junction protein, was expressed in the mESC-CMs, indicating coupling with neighboring CMs (Figure 1C). The mESC-CMs were not detected by rat cardiac troponin T (cTnT) antibody that does not cross-react with mouse CMs (Figures 1D, S2C), excluding the possibility of cell fusion. These data suggest that early postnatal hearts are capable of maturing mESC-CMs. However, the capability appears to be lost when mESC-CPCs are transplanted at P14 (Figure S2B). This may imply the presence of a window similar to that observed for neonatal heart regeneration following tissue excision (Porrello et al., 2011).
To analyze the morphology of in vivo-incubated PSC-CMs in detail, they were isolated via enzymatic digestion and compared with endogenous CMs. The mESC-CMs, identified by GFP expression, had asarcomere structure that was as well-organized as control adult CMs (Figure 1E). The length and width of 1-month-incubated mESC-CMs were similar to those of 1-month-old mouse CMs. 2-month-old mESC-CMs were slightly bigger than 1-month-old ones, but similar to 2-month-old mouse CMs in size (Figure 1F). Similar to adult CMs, the mESC-CMs exhibited a high level of binucleation, an indicator of CM maturation (Figures 1G, S2D).
Formation of transverse (t)-tubules, invaginations of the plasma membrane essential for excitation-contraction coupling in adult CMs, is considered a structural hallmark of CM maturation (Yang et al., 2014). In rats, t-tubules appear sparsely around 2 weeks after birth and fully develop by the first month (Ziman et al., 2010). To determine if the in vivo incubation is accompanied by t-tubule formation, we stained myocardial sections with Alexa Fluor® 568-conjugated wheat germ agglutinin (WGA), which binds to sialic acid and N-acetylglucosaminyl residues on the surface membranes of the myocytes. WGA images were thresholded to create binary images of the t-tubule network after excluding the surface membrane from the analysis and mESC-CMs, identified by the GFP marker, were compared with control rat heart myocytes from the same image field to determine total cell and t-tubule areas and fraction of the cell occupied by t-tubules (Figure 1H). Contrary to mESC-CMs matured in vitro (Figure S2E), in vivo-matured mESC-CMs displayed a t-tubule density that was not significantly different from native control myocytes (Figure 1H). The presence of t-tubules was further confirmed by transmission electron micrographs, which revealed additional adult CM ultrastructures such as well-developed mitochondria and sarcomeres (Figure 1I). The mature CMs were also generated by transplanting immature mESC-CMs isolated at day 8 of ESC differentiation.
In Vivo-Matured PSC-CMs Exhibit Calcium Transients and Contractility of Adult CMs
To determine if the morphological maturation of mESC-CMs is accompanied by functional maturation, we measured Ca2+ transients of mESC-CMs matured in the heart versus in culture, using the ratiometric dye Fura-2 AM. The peak Ca2+ transient amplitude of in vivo-matured mESC-CMs was similar to that of cultured mESC-CMs. However, the time to peak amplitude and rate of return to baseline was significantly shorter in in vivo-matured versus cultured mESC-CMs (Figure 2A, 2B and Table S1). The faster kinetics supports greater maturation of the calcium cycling apparatus required for Ca2+ release and re-sequestration compared to that obtained in mESC-CMs cultured in vitro for 10 or 30 days. We further compared Ca2+ transients of 1–2 month matured mESC-CMs to freshly isolated adult myocytes from similar aged mice. The peak amplitude and kinetics of Ca2+ rise and decay did not significantly differ between these cells (Figure2C and 2D and Table S1).
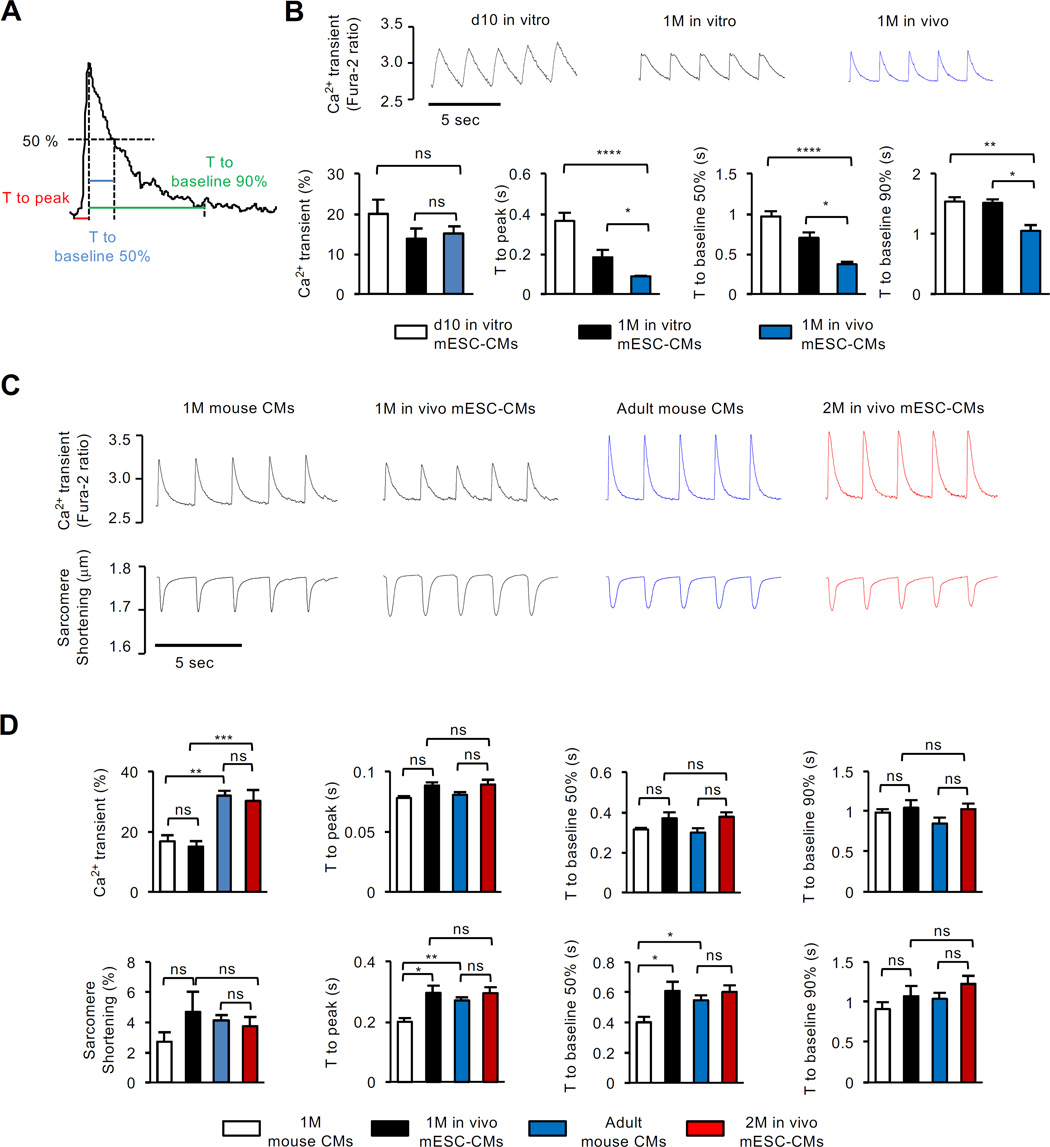
Figure 2. In Vivo-Matured mESC-CMs Show Adult CM Function.
(A) Definitions for Ca2+ transient analysis. (B) Representative trace and quantification of Ca2+ transients, time to peak and baseline 50% and 90% for in vitro-matured mESC-CMs at day 10 (n=13) and 1 month (n=10) and in vivo-matured mESC-CMs at 1 month (n=14). (C) Representative Ca2+ transients and sarcomere shortening of endogenous mouse CMs and in vivo-matured mESC-CMs at indicated stages, stimulated at 0.5 Hz with pulse. (D) Quantifications of peak amplitude of Ca2+ transients and sarcomere shortening, time to peak, and time to baseline 50% and 90% measured with endogenous mouse CMs at 1 month (n=10) or adult stage (n=7) and with in vivo-matured mPSC-CMs at 1 month (n=8–14) or 2 months (n=13). Data are mean ± SEM; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; ns, not significant (p>0.05). p values were determined using the One-way ANOVA (B) or Two-way ANOVA (D) with non-parametric multiple comparison.
Contractile properties of isolated vivo-matured mESC-CMs were assessed by video microscopy and compared to those of freshly isolated adult cells. This assay cannot be performed in cultured PSC-CMs due to their disorganized sarcomere structure. By contrast, the mESC-CMs and freshly isolated mouse CMs had similar well-defined sarcomere shortening behavior. The one-month old cells of either type showed somewhat faster kinetics in mESC-CMs, but this disappeared by 2 months of maturation (Figure 2C, 2D and Table S1).
In Vivo-Matured PSC-CMs Show Similar Gene Expression to Adult CMs
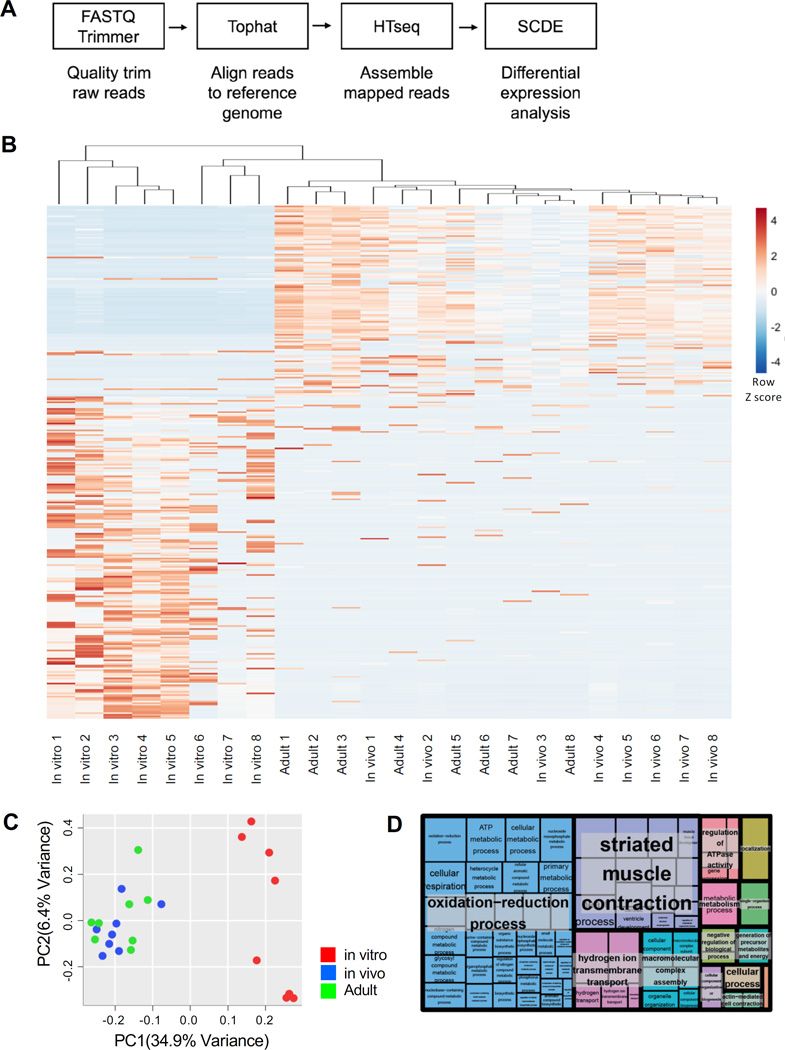
We next sought to characterize the transcriptomes of mESC-CMs matured in vivo and in vitro. To do this, the CMs were isolated from hearts or culture after 1 month of incubation and subjected to single-cell RNA-sequencing analysis (Figure 3A). Mouse adult CMs were used as a control group. We identified 312 differentially expressed genes (> twofold change) based on a p-value less than 0.01. Hierarchical clustering analysis revealed that in vivo-matured CMs clustered closer to adult CMs than in vitro-matured CMs (Figure 3B). Consistently, adult and in vivo-matured CMs were grouped closer than to in vitro-matured CMs in principal component analysis (Figure 3C). These suggest similarity in gene expression between adult and in vivo-matured CMs. Notably, in vitro-matured CMs showed a more dispersed pattern (Figure 3C). This may be attributed to variability in their differentiation and maturation in vitro. Gene ontology (GO) analysis indicated that groups of the differentially expressed genes were related to mitochondrial function and muscle contraction (Figure 3D, Table S2). This may reflect the increase in mitochondrial biogenesis and sarcomeric organization during maturation.
Figure 3. Single-cell RNA-Seq Analysis.
(A) Outline of RNA-Seq pipeline for data analysis. (B) Heatmap visualization of hierarchically clustered samples showing high (red) and low (blue) expression of 8 in vitro-matured mESC-CMs, 8 in vivo-matured mESC-CMs and 8 adult mouse CMs. (C) PCA of gene expression of in vitro (red), in vivo (blue), and adult (green) CMs. (D) Treemap plot of gene ontology (GO) analysis of differentially expressed genes showing superclusters of related terms.
Postnatal Extracellular Factors Can Promote PSC-CM Maturation In Vitro
The fact that PSC-CMs do not mature beyond neonatal stages in culture (Uosaki et al., 2015) but can be further matured by incubating within neonatal myocardium implies environmental differences between postnatal and prenatal hearts. To gain insights into the environmental potential of postnatal hearts, we analyzed genes expressed differentially during CM maturation and identified 89 genes using our Affymetrix® array datasets (Uosaki et al., 2015). Of those, 25 were highly downregulated and 16 were highly upregulated in the postnatal heart. We focused on genes encoding secreted or membrane-bound proteins and found that 6 of those [CXCL14 (C-X-C Motif Chemokine Ligand 14), IL-15 (Interleukin 15), CCL6 (C-C Chemokine ligand 6), Adipoq (Adiponectin), Grm1 (Glutamate Metabotropic Receptor 1), Nampt (Nicotinamide phosphoribosyltransferase)] were increased in postnatal hearts, determined by qPCR (Figure S3A). We subsequently treated mESC-CMs with activators (P7C3- activator of Nampt and DHPG -Glutamate Receptor agonist) and recombinant proteins (CXCL14, IL-15, CCL16 and ADIPOQ) of those factors for different time intervals and assessed their maturation status (Figure S3C). To generate a genetic readout for the assessment, we utilized Ingenuity pathway analysis and identified a group of nuclear receptors (NRs) strongly associated with the postnatal heart maturation (Figure S3B). Upon activating the factors, the majority of NRs were significantly upregulated after 7 days of culture (Figure S3D, E). More NRs were increased over time and after 14 days of culture (Figure S3F). These suggest that the extracellular proteins may be an environmental component of postnatal hearts utilized for PSC-CM maturation.
Human PSC-CMs Are Matured to Adult CMs in Rodent Neonatal Hearts
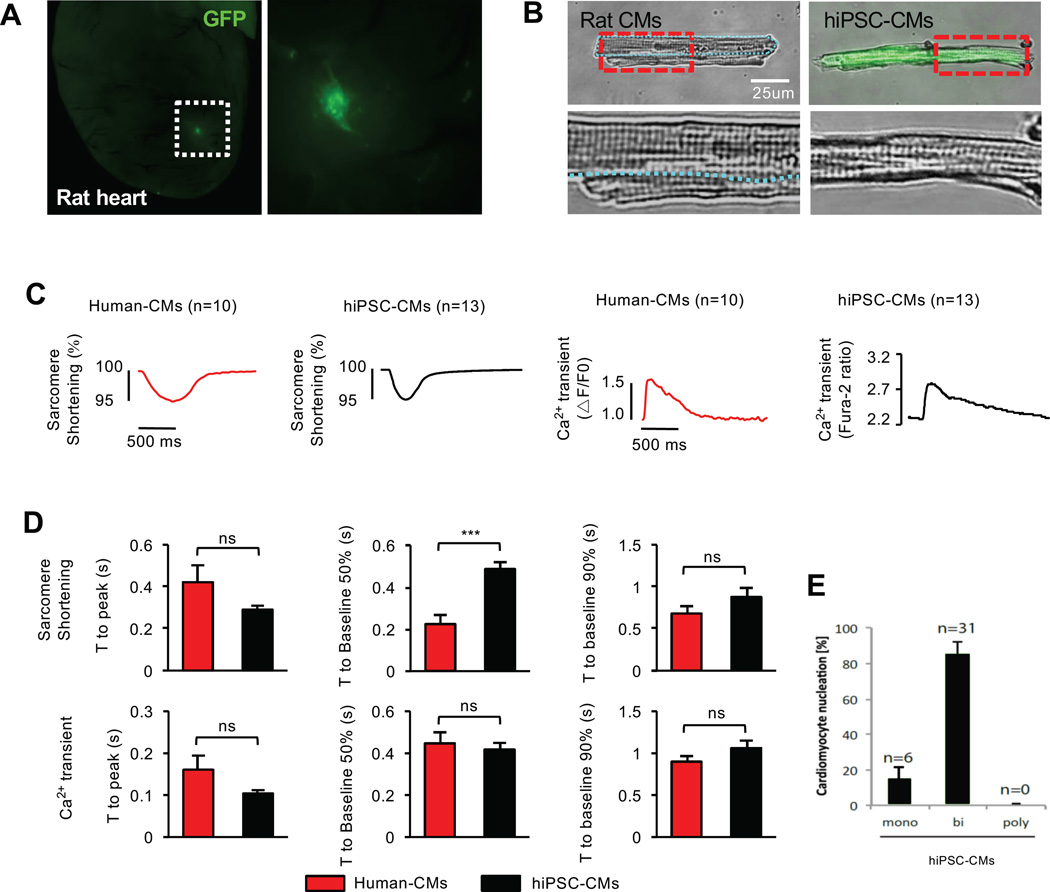
Human CMs are analogous to rodent CMs in size and structure, but their maturation transpires over a decade. Since CM development is a conserved process in mammals, we hypothesized that human iPSC-CMs (hiPSC-CMs) can be matured in rat postnatal hearts. To test this, hiPSCs (Takahashi et al., 2007) were labeled with GFP, differentiated into CMs, and incubated in neonatal rat hearts as described earlier (Figure 4A). Similar to mESC-CMs, hiPSC-CMs exhibited adult CM-like features after 1 month of incubation. They were rod-shaped with highly organized sarcomeres (Figure 4B) and functionally similar to human adult CMs with the exception of a faster time to 50% cell relengthening in the hiPSC-CMs (Figure 4C, D and Table S3). These data demonstrate that rodent neonates are capable of generating adult CMs from human PSCs. The hiPSC-CMs showed ~80% binucleation (Figure 4E), which is higher than that of human CMs (25–57%) reported (Olivetti et al., 1996; Schmid and Pfitzer, 1985).
Figure 4. In Vivo-Matured hiPSC-CMs Become Adult-like CMs.
(A) hiPSC-CMs (GFP) engrafted in the rat heart for 1 month. (B) High resolution images of adult rat CMs (two CMs) and in vivo-matured hiPSC-CM showing well-organized sarcomeric structure. Boxed regions are enlarged in the bottom. (C) Representative sarcomere shortening and Ca2+ transients for in vivo-matured hiPSC-CMs compared to adult human CMs. (D) Quantifications of time to peak, time to baseline 50% and 90% of sarcomere shortening and Ca2+ transients of adult human CMs (n=10, red) and in vivo-matured hiPSC-CMs (n=12, black). (E) Binucleation % of in vivo-matured hiPSC-CMs. Data are mean ± SEM; ns, not significant (p>0.05). p values were determined using the non-parametric Mann-Whitney test.
In Vivo-Maturation System Allows Modeling Human Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC)
ARVC is an inherited form of cardiomyopathy that manifests in adolescence/adulthood in humans, and is characterized by fibro-fatty replacement, apoptosis and intercalated disc abnormalities (Basso et al., 2006; Calkins and Marcus, 2008). This adult-phenotype is recapitulated in our mouse ARVC model (Chelko et al., 2016) (Figure 5A, B). We next examined ARVC hiPSCs generated from patients with the syndrome (Kim et al., 2013a). The resulting ARVC hiPSC-CMs remained fetal-like in both morphology and function, and required a lipogenic medium to partially mimic ARVC properties in vitro (Kim et al., 2013a). Consistently, we found that ARVC hiPSC-CMs matured in vitro did not replicate the adult disease phenotype (Figure 5C). To test if the neonatal maturation system can be used to model human ARVC, we transplanted GFP-labeled ARVC hiPSC-CMs into neonatal rat hearts and analyzed them after 1 month of incubation. The incubation resulted in the accumulation of lipids/adipocytes, accompanied by markedly increased apoptosis (Figure 5D–F), recapitulating the disease phenotype. Moreover, electron microscopy showed abnormal intercalated discs (intercellular gap widening) observed in ARVC human patients (Figure 5G).
Figure 5. in Vivo-Matured ARVC-hiPSC-CMs Exhibit Human ARVC Disease Phenotype.
(A) Adult WT/ARVC mouse heart sections stained with antibodies against Perilipin (yellow), cTnT (green) and DAPI. Perilipin is a lipid droplet-associated protein. (B) Adult WT/ARVC mouse heart sections stained with TUNEL (red), showing apoptotic cells. (C) In vitro-matured GFP-labeled ARVC hiPSC-CMs stained with Perilipin (red), GFP (green), and α-Actinin (cyan) antibody. (D) In vivo-matured GFP labeled control hiPSC-CMs (left) and ARVC hiPSC-CMs (right) stained with Perilipin (red) and human-specific mitochondria (cyan) antibodies. DAPI (blue) was used to counterstain nuclei. (E) TUNEL staining of control hiPSC-CMs and ARVC hiPSC-CMs matured in vivo. (F) Quantification of TUNEL positive CMs. Data are mean ± SD; section number=3; hiPSC CMs (n=391), ARVC hiPSC-CMs (n= 430); *p<0.05; p values were determined using the paired Student t test. (G) Transmission electron micrographs of human control, ARVC patient CMs, and in vivo-matured ARVC hiPSC-CMs. n=10 Rats. Blue arrows indicate intercalated disc abnormalities (widening of intercellular space).
DISCUSSION
Generation of mature adult CMs from hPSCs has remained intractable. This suggests substantial complexity to the signaling and stimuli needed for CM maturation that normally takes place over a decade in humans. The current study demonstrates that hPSC-CMs can mature to adult CMs in a month when incubated in rat neonatal myocardium, the latter providing a bio-incubator for CM maturation. The resulting CMs can be studied at the single cell level both in vivo and in vitro, allowing the study of mature human CMs to address the pathogenesis of adult-onset CM diseases. This system may also be used for in vivo drug testing.
A number of studies have demonstrated that PSC-CMs develop more adult-like phenotype over time in vitro (Robertson et al., 2013; Yang et al., 2014). While the precise status of their maturation remains to be determined, a multi-stage, genome-wide analysis indicated that early or late PSC-CMs in culture resemble early embryonic or late embryonic/neonatal CMs, respectively (Uosaki et al., 2015). The absence of the extensive t-tubule network also supports their immaturity (Kane et al., 2015; Knollmann, 2013). The in vivo-incubated PSC-CMs, however, displayed features very similar to adult CMs in morphology, function, and gene expression. For example, T-tubules were formed with a regular pattern and invagination as adult CMs, and this is consistent with their adult calcium transients and sarcomere shortening. Moreover, in vivo-incubated ARVC hiPSC-CMs showed disease phenotypes that appear in adults. This suggests that the in vivo-matured PSCs are highly analogous to adult CMs and that this system can be used to study and model human CM development and late-onset CM-autonomous diseases.
Early postnatal hearts contain endogenous CPCs and immature CMs, and we found that exogenously introduced PSC-CPCs/CMs can give rise to mature CMs in the hearts. This finding suggests that young hearts provide the environmental cues necessary to guide PSC-CPCs/CMs to become adult CMs. The cues might come from extracellular factors enriched in postnatal hearts, as they were able to promote the molecular maturation of PSC-CMs in vitro. Further analysis will be needed to identify the source of the factors and to determine their effects on morphological and functional maturation. Curiously, PSC-CMs remain immature when transplanted into adult hearts (Shiba et al., 2012), suggesting the presence of a critical time window required for PSC-CM maturation. It will be of great importance to investigate temporal factors present in early postnatal hearts that mediate the process.
It is worth noting that hPSC-CMs mature into adult-like CMs after a month of incubation in rodent hearts. This suggests that the machinery needed for CM maturation might be conserved in rodents and humans. In fact, although rodents have a shorter lifespan, comparative transcriptional analyses revealed that genes involved in mouse CM maturation are similarly regulated during human heart maturation (Uosaki and Taguchi, 2016). It will be interesting to test if hPSC-CMs can also be matured in larger animal models. This approach may be extended for generating other types of adult cells prone to disease, such as skeletal muscle cells, pancreatic cells, renal cells etc., from hiPSCs, which would allow us to study and model adult-onset human diseases.
EXPERIMENTAL PROCEDURES
Animals
All animals were housed at the Johns Hopkins Medical Institutions. All protocols involving animals followed US National Institutes of Health guidelines and were approved by the animal and care use committee of the Johns Hopkins Medical Institutions. The animals were randomly allocated to experimental groups and both male and female pups were used for cell delivery. No inclusion or exclusion parameters were used for animal experiments. We were not blinded to the group allocation during the experiment.
Cell Culture, Differentiation, and Delivery
mESCs and hESC/iPSCs (Table S4) were obtained, maintained, and differentiated as described (Cheng et al., 2013; Kim et al., 2013a; Uosaki et al., 2012; Uosaki et al., 2011). For CPC purification, cells were dissociated at day 7 and resuspended in PBS containing 0.1% FBS, 20 mM Hepes and 1 mM EDTA. RFP+ CPCs were isolated with SH800 sorter (Sony Biotechnology, Japan). For cell delivery, RNU Rats (Charles River Laboratories) were used as host animals. Postnatal rats were anesthetized by cooling on an ice bed for 5min, and a hole was made between 4th and 5th rib. Before injection, cells were mixed with IMDM and matrigel at 60:1 ratio (Laflamme et al., 2007) and injected into ventricle wall with Eppendorf FemtoJet® Microinjector (10µl/injection). To close hole, we used tissue glue (3M Vetbond tissue adhesive). After injection, pups were recovered on heating pad for 10min and returned to mother. To test mycoplasma contamination, we used MycoProbe® Mycoplasma Detection Kit (R&D systems, Catalog# CULoo1B). To assess the effect of secreted/membrane-bound factors, mESC-CMs were either incubated with the six factors: CXCL14 (50ng/ml, Abcam), IL-15 (10ng/ml, Novus), CCL6 (100ng/ml, Novus) and Adipoq (250ng/ml, Sigma), DHPG (50nM, Abcam), P7C3 (10nM, Abcam) or DMSO (control) for 4, 7 and 14 days. The medium was changed every 2–3 days. Four independent experiments were performed.
Immunohistochemistry and t-tubule analysis
For immunohistochemistry, cultured cells and dissected hearts were fixed in 4% paraformaldehyde, blocked for 1 hour with 1% BSA, and incubated overnight with the following primary antibodies: α-Actinin (Sigma A7732), RFP (Clontech Laboratories 5f8), GFP (Life Technologies A11122, A10262), Perilipin (Cell Signaling 9349), cTnT (Thermo Scientific MS 295-P1), Rat and human specific cTnT (Abcam ab45932), Connexin 43 (Sigma, C6219) and human mitochondria (Abcam ab92824). Alexa fluor secondary antibodies (488, 564, 648, Life Technologies) were used for secondary detection. For fluorescent t-tubule staining, nuclei (DAPI), GFP, and t-tubules (Alexa Fluor® 568-conjugated wheat germ agglutinin WGA) were imaged using a confocal microscope (Leica DM2500) with a 40× lens (1.15 NA) oil immersion lens. WGA images were analyzed using ImageJ (Version 1.50e; Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/, 1997–2016.). A median filter (pixel radius 1) was applied and the plugin Auto_Local_Threshold (Niblack method) was used to create a binary mask of the WGA signal and the mask eroded one time. A region of interest was created for each cell that excluded the boundary membrane to isolate the t-tubule network for segmentation analysis. The Analyze_Particles plugin was then used to measure t-tubule area (µm2), which was normalized to the total cell area to calculate the fractional area (expressed as % total). A two-tailed t-test was then performed to compare the two groups. Control and mESC-derived myocytes from the same optical field were used for statistical comparisons to eliminate errors that could arise due to variable staining efficiency or imaging conditions.
Whole-Organ Optical Clearing and Imaging
To visualize the extent of CM differentiation and incorporation, the heart of CPC-injected mice was perfused with ice-cold saline followed by 4% PFA in PBS and post-fixed overnight. The heart was subsequently subjected to Scale CUBIC-1 tissue clearing solution (Susaki et al., 2014) at 37°C for 7 days with mild shaking. The solution was exchanged with fresh reagents twice. The optically transparent heart was then mounted in the same solution and imaged using a Zeiss LSM 510 or 710 laser scanning confocal microscope with a 10× 0.3 NA or 20× 0.5 NA objective. 3D renderings of the Z-stack images were made using Imaris (Bitplane).
Measurement of Calcium Transients and Sarcomere Shortening
CMs were freshly isolated from mouse or rat hearts. Hearts were quickly excised under anesthesia, and the aorta retroperfused with an enzymatic perfusion solution containing collagenase, as described in (Bassani and Bers, 1994; Lee et al., 2010). The isolated CMs were incubated in 1 mM of the ratiometric Ca2+ indicator dye Fura-2AM (Invitrogen, Molecular Probes) containing 1 mM Ca2+ 1× Tyrode solution, and then cells were placed in a perfusion chamber and stimulated at 0.5 Hz with pulses. Sarcomere length and whole cell Ca2+ transients were recorded using an inverted fluorescence microscope (Nikon, TE2000) with IonOptix (Myocam®) software. To measure Ca2+ transients with embryonic and postnatal CMs, hearts were minced and enzymatically dissociated with collagenase and trypsin. Cells were then seeded on Gelatin or Laminin coated cover glasses in 10% serum supplemented SFD medium and analyzed the following day. Differentiated PSC-CMs were replated at day 9 for 10-day studies, or day 20 for 30-day studies. Whole cell Ca2+ transients were measured as described above. Human left ventricular myocytes were isolated from donor hearts that were not suitable for transplantation as described previously (Chen et al., 2002) and isolated myocytes loaded with Fluo-3 AM for measurements. Myocytes were placed in a heated chamber on the stage of an inverted microscope (Nikon Diaphot), superfused with Tyrode’s solution, and shortening detected by video-edge detection and intracellular Fluo-3 fluorescence was recorded with Clampex (Molecular Devices) as described (Piacentino et al., 2003). Data were analyzed offline with pClampfit.
Transmission Electron Microscopy
Transmission electron microscopy (TEM) was performed with 1-month-old in vivo-matured mESC-CMs and ARVC iPSC-CMs. Sources of human control and ARVC heart sections were described (Basso et al., 2006). Heart tissue was fixed with freshly made EM grade 1% glutaraldehyde (Pella), 80 mM phosphate buffer (Sorenson’s) and 3 mM magnesium chloride pH 7.2 at 4C for 1 hour. To ensure complete fixation, tissues were then microwaved in a Pelco 3400 laboratory grade microwave oven. Tissues were placed in 4 ml uncapped glass vials containing 2 mls of fixative. The vials were then placed in a shallow ice bucket with the top of the fixative level equal to the top ice level. Two 600 ml beakers containing room temperature D-H2O were positioned on either side of the ice bucket to serve as heat traps. Samples were microwaved pulsed for 10 sec, paused for 20 sec, then pulsed again for 10 sec. Tissues were allowed to sit in fixative for 5 min., then microwaved again in the same manner. Fixative temperatures never exceeded 27 C. Samples were rinsed in buffer containing 3% sucrose (3 × 15 min.), then microwaved twice again as before in secondary fixative. This osmication was performed in 1.5% potassium ferrocyanide reduced 1% osmium tetroxide in 100 mM phosphate buffer, containing 3 mM magnesium chloride. All subsequent steps were performed at 4C. Tissues were then rinsed in 100 mM maleate buffer (3 × 5 min) containing 3% sucrose, then en-bloc stained with 1% filtered uranyl acetate in the same buffer for 1 hr. Samples were dehydrated at 4C up to 70% ethanol when they were brought to room temp and further dehydrated to 100% ethanol. Samples were embedded with Eponate 12 after a brief acetone transition, and finally cured in a 60C oven for two days. 80 nm ultra-thin sections were picked up on formvar coated 200 mesh nickel grids. Sections were floated on all subsequent steps. All solutions were filtered except for antibodies, which were centrifuged at 13K for 5 min. Grids were placed on 3% sodium metaperiodate (aq) for 20 min. After a 15 min D-H2O rinse, grids were placed on 50 mM NH4Cl in TBS for 10 min, followed by 20 min triple serum block (3% NGS, 3% BSA 1% fish gelatin) in TBST (blocking solution). GFP antibody (mouse) incubation was done at 1:200 with no primary antibody as negative controls. Incubations were carried out at 4C overnight. After 1 hr to equilibrate to room temperature, grids were placed on blocking solution for 10 min, followed by a 1 min rinse in TBS. 12nm GAM (Gold conjugated secondary antibody, Jackson Immunoresearch) were diluted 1:40 in TBS and grids were incubated for 2 hrs at room temperature in a humidity chamber. After a 10 min TBS incubation followed by a quick D-H20 rinse, grids were hard fixed in 1% glutaraldehyde in 100 mM sodium cacodylate buffer for 5 min. After a brief D-H2O rinse, grids were stained with 2% uranyl acetate (aq.) for 20 min, rinsed again with D-H20; blot dried an allowed to sit in grid boxes overnight before viewing.
Quantitative PCR
mESC-CMs were isolated using Trypsin and placed in Trizol (Life Technologies). RNA isolation was performed following the manufacturer’s instructions and cDNA was generated using the High-Capacity cDNA reverse transcription kit (Applied Biosystems). All quantitative PCR reactions were performed using the Sybr Select qPCR mix (Thermo Fisher) with indicated primers (Table S5). Gene expression levels were normalized to GAPDH.
Library Preparation and Sequencing
Single CMs (αMHC-GFP) were either FACS sorted (SH800, Sony Technologies) or manually picked under the microscope into 96-plates containing water (2.4 µL) with RNase-free DNase I (0.2 µL; NEB) and RNase inhibitor (0.25 µL; NEB). DNase I was inactivated by increasing the temperature (72°C for 3 min), and samples were then stored on ice. Custom designed 2A oligo 1 µL primer (12 µM, Integrated DNA Technologies (Shin et al., 2015) was added and annealed to the polyadenylated RNA by undergoing a temperature increase (72°C for 2 min) and being quenched on ice. A mixture of 1 µL SMARTscribe reverse transcriptase (Clontech Laboratories, Inc), 1 µL custom designed TS oligo (12 µM, Integrated DNA Technologies (Shin et al., 2015)), 0.3 µL MgCl2 (200 mM, Sigma), 0.5 µL RNase inhibitor (Neb), 1 µL dNTP (10 mM each, Thermo), 0.25 µL DTT (100 mM, Invitrogen) were incubated at 42°C for 90 min, which was followed by enzyme inactivation at 70°C for 10 min. A mixture of 29 µL water, 5 µL Advantage2 taq polymerase buffer, 2 µL dNTP (10 mM each, Thermo), 2 µL custom-designed PCR primer (12 µM, Integrated DNA Technologies (Shin et al., 2015)), and 2 µL Advantage2 taq polymerase was directly added to the reverse transcription product and the amplification was performed for 19 cycles. The amplification product was purified using Ampure XP beads (Beckman-Coulter). Libraries and transposome assembly where made using a previously published protocol (Picelli et al., 2014). Briefly, 100 pg of total cDNA was added to a 2× tagment DNA Buffer (TD) (2xTAPS buffer: 20 mM TAPS-NaOH, 10 mM MgCl2 (pH 8.5) at 25°C, 16% w/v PEG 8000) then spiked with 0.5 µL of 1:64 diluted Tn5 (Epicenter) and incubated for 8 min at 55°C. Tn5 was stripped off from the tagmented DNA, by adding 0.2% SDS for a final concentration of 0.05%. Libraries were enriched used KAPAHiFi which included 5× Kappa Fidelity Buffer, 10mM dNTPs, HIFI Polymerase and 1ul of index primers was used directly in the enrichment PCR amplification of libraries for the Illumina sequencers for a 50µL reaction. The PCR program was as follows: 5 min at 72°C, 1min at 95°C, then 16 cycles at 30 sec at 95°C, 30 sec at 55°C, 30 sec at 72° and 5 min at 72°. Successful libraries were multiplexed and sequenced using NextSeq 500. For analysis, trimmed reads (Figure 3A) were mapped to the mouse reference genome (GRCm38/mm10) using Tophat (2.1.0) (Kim et al., 2013b). Cells with >100,000 aligned reads were assembled in python package HTseq (Anders et al., 2015) then analyzed for differential expression in the Single Cell Differential Expression (SCDE) package (Kharchenko et al., 2014) and DESeq2 package (Love et al., 2014) in R. Gene ontology analysis results were visualized using Revigo (Supek et al., 2011).
Binucleation Analysis
CMs were isolated from rat hearts using the Langendorff technique. Cardiomyocytes in single cell dispersion were stained using antibodies against RFP (ChromoTeck), GFP and cTnT (Thermo Fisher) and DAPI. Cells were analyzed by FACS (SH800, Sony Biotechnologies) and by microscopy (EVOSfl, AMG) to determine nuclear content percentage and nucleation status.
TUNEL Staining
In Situ Cell Death Detection Kit, TMR red (Roche Applied Science, cat# 12156792910) was used for TUNEL staining.
Statistical Analyses
For all analyses, sample size is described in the Figure legends. Most of in vitro studies were done with two to three sets of independent experiments. Two-group analysis used either Student’s t-test or non-parametric Mann-Whitney test. Comparisons of multiple groups were performed using either One-way or Two-way ANOVA (if appropriate). If normality or equal variance tests failed, then a Kruskal Wallis test was used. Post-hoc multiple comparisons testing used either a Turkey’s or Dunn’s test. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
The authors thank Kwon and Kass laboratory members for critical reading and discussions. We also thank Dr. Soroosh Solhjoo for t-tubule analysis, Dr. Shinya Yamanaka for providing hiPSCs, and Drs. Mark Anderson and Marc Halushka for helpful suggestions. E.T. was supported by the Johns Hopkins School of Medicine Clinician Scientist Award. C.B. was supported by TRANSAC Strategic Research Grant CPDA133979/13, University of Padua, Italy. D.A.K. was supported by National Health Service – NHLBI grants HL-119012, HL-107153, and Fondation Leducq. This work was supported by the Magic that Matters Fund and grants from NHLBI/NIH (R01HL111198), NICHD/NIH (R01HD086026) and MSCRF (2015-MSCRFI-1622) to C.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, G.S.C. and C.K.; Methodology, G.S.C., D.I.L., D.A.K., C.K.; Software, S.M. and H.U.; Investigation, G.S.C., D.I.L., E.T., P.A., S.C., K.C., I.H., K.S., X.C., C.B., and B.O.; Writing – Original Draft, G.S.C., D.I.L.; Resources, H.V.C., S.R.H., G.F.T., and D.P.J.; Writing – Review & Editing, B.O., D.A.K, and C.K.; Supervision, D.A.K and C.K.; Funding Acquisition, D.A.K and C.K.
ACCESSION NUMBERS
The RNA-seq data have been deposited to GEO (Accession number: GSE92247)
COMPETING FINANCIAL INTERESTS
Authors declare no competing financial interests.
REFERENCE
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassani RA, Bers DM. Na-Ca exchange is required for rest-decay but not for rest-potentiation of twitches in rabbit and rat ventricular myocytes. J Mol Cell Cardiol. 1994;26:1335–1347. doi: 10.1006/jmcc.1994.1152. [DOI] [PubMed] [Google Scholar]
- Basso C, Czarnowska E, Della Barbera M, Bauce B, Beffagna G, Wlodarska EK, Pilichou K, Ramondo A, Lorenzon A, Wozniek O, et al. Ultrastructural evidence of intercalated disc remodelling in arrhythmogenic right ventricular cardiomyopathy: an electron microscopy investigation on endomyocardial biopsies. Eur Heart J. 2006;27:1847–1854. doi: 10.1093/eurheartj/ehl095. [DOI] [PubMed] [Google Scholar]
- Calkins H, Marcus F. Arrhythmogenic right ventricular cardiomyopathy/dysplasia: an update. Curr Cardiol Rep. 2008;10:367–375. doi: 10.1007/s11886-008-0059-4. [DOI] [PubMed] [Google Scholar]
- Chelko SP, Asimaki A, Andersen P, Bedja D, Amat-Alarcon N, DeMazumder D, Jasti R, MacRae CA, Leber R, Kleber AG, et al. Central role for GSK3beta in the pathogenesis of arrhythmogenic cardiomyopathy. JCI insight. 2016;1 doi: 10.1172/jci.insight.85923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Piacentino V, 3rd, Furukawa S, Goldman B, Margulies KB, Houser SR. L-type Ca2+ channel density and regulation are altered in failing human ventricular myocytes and recover after support with mechanical assist devices. Circ Res. 2002;91:517–524. doi: 10.1161/01.res.0000033988.13062.7c. [DOI] [PubMed] [Google Scholar]
- Cheng P, Andersen P, Hassel D, Kaynak BL, Limphong P, Juergensen L, Kwon C, Srivastava D. Fibronectin mediates mesendodermal cell fate decisions. Development. 2013;140:2587–2596. doi: 10.1242/dev.089052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho GS, Fernandez L, Kwon C. Regenerative medicine for the heart: perspectives on stem-cell therapy. Antioxid Redox Signal. 2014;21:2018–2031. doi: 10.1089/ars.2014.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaster TK, Cadar AG, Wang L, Williams CH, Chun YW, Hempel JE, Bloodworth N, Merryman WD, Lim CC, Wu JC, et al. Matrigel Mattress: A Method for the Generation of Single Contracting Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circ Res. 2015;117:995–1000. doi: 10.1161/CIRCRESAHA.115.307580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox IJ, Daley GQ, Goldman SA, Huard J, Kamp TJ, Trucco M. Stem cell therapy. Use of differentiated pluripotent stem cells as replacement therapy for treating disease. Science. 2014;345:1247391. doi: 10.1126/science.1247391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdar F, Klaassen Kamdar A, Koyano-Nakagawa N, Garry MG, Garry DJ. Cardiomyopathy in a Dish: Using Human Inducible Pluripotent Stem Cells to Model Inherited Cardiomyopathies. J Card Fail. 2015 doi: 10.1016/j.cardfail.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane C, Couch L, Terracciano CM. Excitation-contraction coupling of human induced pluripotent stem cell-derived cardiomyocytes. Front Cell Dev Biol. 2015;3:59. doi: 10.3389/fcell.2015.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, Kan NG, Forcales S, Puri PL, Leone TC, et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013a;494:105–110. doi: 10.1038/nature11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013b;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knollmann BC. Induced pluripotent stem cell-derived cardiomyocytes: boutique science or valuable arrhythmia model? Circ Res. 2013;112:969–976. doi: 10.1161/CIRCRESAHA.112.300567. discussion 976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- Lalit PA, Hei DJ, Raval AN, Kamp TJ. Induced pluripotent stem cells for post-myocardial infarction repair: remarkable opportunities and challenges. Circ Res. 2014;114:1328–1345. doi: 10.1161/CIRCRESAHA.114.300556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DI, Vahebi S, Tocchetti CG, Barouch LA, Solaro RJ, Takimoto E, Kass DA. PDE5A suppression of acute beta-adrenergic activation requires modulation of myocyte beta-3 signaling coupled to PKG-mediated troponin I phosphorylation. Basic Res Cardiol. 2010;105:337–347. doi: 10.1007/s00395-010-0084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang SC, Lin SZ, Yu JF, Wu SF, Wang SD, Liu JC. F344-rnu/rnu athymic rats: breeding performance and acceptance of subcutaneous and intracranial xenografts at different ages. Lab Anim Sci. 1997;47:549–553. [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundy SD, Zhu WZ, Regnier M, Laflamme MA. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev. 2013;22:1991–2002. doi: 10.1089/scd.2012.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, Jiang J, Masse S, Gagliardi M, Hsieh A, et al. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat Methods. 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivetti G, Cigola E, Maestri R, Corradi D, Lagrasta C, Gambert SR, Anversa P. Aging, cardiac hypertrophy and ischemic cardiomyopathy do not affect the proportion of mononucleated and multinucleated myocytes in the human heart. J Mol Cell Cardiol. 1996;28:1463–1477. doi: 10.1006/jmcc.1996.0137. [DOI] [PubMed] [Google Scholar]
- Piacentino V, 3rd, Weber CR, Chen X, Weisser-Thomas J, Margulies KB, Bers DM, Houser SR. Cellular basis of abnormal calcium transients of failing human ventricular myocytes. Circ Res. 2003;92:651–658. doi: 10.1161/01.RES.0000062469.83985.9B. [DOI] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro MC, Tertoolen LG, Guadix JA, Bellin M, Kosmidis G, D'Aniello C, Monshouwer-Kloots J, Goumans MJ, Wang YL, Feinberg AW, et al. Functional maturation of human pluripotent stem cell derived cardiomyocytes in vitro--correlation between contraction force and electrophysiology. Biomaterials. 2015;51:138–150. doi: 10.1016/j.biomaterials.2015.01.067. [DOI] [PubMed] [Google Scholar]
- Robertson C, Tran DD, George SC. Concise review: maturation phases of human pluripotent stem cell-derived cardiomyocytes. Stem Cells. 2013;31:829–837. doi: 10.1002/stem.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan JL, Tulloch NL, Saiget M, Paige SL, Razumova MV, Regnier M, Tung KC, Keller G, Pabon L, Reinecke H, et al. Mechanical Stress Promotes Maturation of Human Myocardium From Pluripotent Stem Cell-Derived Progenitors. Stem Cells. 2015;33:2148–2157. doi: 10.1002/stem.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid G, Pfitzer P. Mitoses and binucleated cells in perinatal human hearts. Virchows Archiv B, Cell pathology including molecular pathology. 1985;48:59–67. doi: 10.1007/BF02890115. [DOI] [PubMed] [Google Scholar]
- Shenje LT, Andersen P, Uosaki H, Fernandez L, Rainer PP, Cho GS, Lee DI, Zhong W, Harvey RP, Kass DA, et al. Precardiac deletion of Numb and Numblike reveals renewal of cardiac progenitors. Elife. 2014;3:e02164. doi: 10.7554/eLife.02164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y, Fernandes S, Zhu WZ, Filice D, Muskheli V, Kim J, Palpant NJ, Gantz J, Moyes KW, Reinecke H, et al. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature. 2012;489:322–325. doi: 10.1038/nature11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, et al. Whole-Brain Imaging with Single-Cell Resolution Using Chemical Cocktails and Computational Analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- Svendsen CN. Back to the future: how human induced pluripotent stem cells will transform regenerative medicine. Hum Mol Genet. 2013;22:R32–R38. doi: 10.1093/hmg/ddt379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabar V, Studer L. Pluripotent stem cells in regenerative medicine: challenges and recent progress. Nat Rev Genet. 2014;15:82–92. doi: 10.1038/nrg3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Uosaki H, Andersen P, Shenje LT, Fernandez L, Christiansen SL, Kwon C. Direct Contact with Endoderm-like Cells Efficiently Induces Cardiac Progenitors from Mouse and Human Pluripotent Stem Cells. PLoS One. 2012 doi: 10.1371/journal.pone.0046413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uosaki H, Cahan P, Lee DI, Wang S, Miyamoto M, Fernandez L, Kass DA, Kwon C. Transcriptional Landscape of Cardiomyocyte Maturation. Cell Rep. 2015;13:1705–1716. doi: 10.1016/j.celrep.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uosaki H, Fukushima H, Takeuchi A, Matsuoka S, Nakatsuji N, Yamanaka S, Yamashita JK. Efficient and scalable purification of cardiomyocytes from human embryonic and induced pluripotent stem cells by VCAM1 surface expression. PLoS One. 2011;6:e23657. doi: 10.1371/journal.pone.0023657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uosaki H, Taguchi YH. Comparative Gene Expression Analysis of Mouse and Human Cardiac Maturation. Genomics Proteomics Bioinformatics. 2016;14:207–215. doi: 10.1016/j.gpb.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Pabon L, Murry CE. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res. 2014;114:511–523. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaruba MM, Soonpaa M, Reuter S, Field LJ. Cardiomyogenic potential of C-kit(+)-expressing cells derived from neonatal and adult mouse hearts. Circulation. 2010;121:1992–2000. doi: 10.1161/CIRCULATIONAHA.109.909093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman AP, Gomez-Viquez NL, Bloch RJ, Lederer WJ. Excitation-contraction coupling changes during postnatal cardiac development. J Mol Cell Cardiol. 2010;48:379–386. doi: 10.1016/j.yjmcc.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.