Significance
Histone H2B monoubiquitination is important for gene expression. It is mediated by the yeast E3 ligase Bre1 (human RNF20/40) and the E2 conjugating enzyme Rad6. These enzymes associate with RNA polymerase II and their activity is counteracted by the H2B deubiquitinase of the Spt–Ada–Gcn5 acetyltransferase (SAGA) complex. In contrast to many other E3 ligases, the Rad6–Bre1 machinery exclusively modifies a single lysine on the nucleosome. Given the number of other nearby lysines, it was unknown how such specificity could be achieved. Our study shows how Rad6–Bre1 recognition of the nucleosome is mechanistically coupled to ubiquitin transfer, how a single target lysine is chosen, and how Rad6–Bre1 compete with SAGA for regulatory access to a specific region on the nucleosome.
Keywords: nucleosome, ubiquitin, Bre1–Rad6, cross-linking mass spectrometry, RING E3 ligase
Abstract
Cotranscriptional ubiquitination of histone H2B is key to gene regulation. The yeast E3 ubiquitin ligase Bre1 (human RNF20/40) pairs with the E2 ubiquitin conjugating enzyme Rad6 to monoubiquitinate H2B at Lys123. How this single lysine residue on the nucleosome core particle (NCP) is targeted by the Rad6–Bre1 machinery is unknown. Using chemical cross-linking and mass spectrometry, we identified the functional interfaces of Rad6, Bre1, and NCPs in a defined in vitro system. The Bre1 RING domain cross-links exclusively with distinct regions of histone H2B and H2A, indicating a spatial alignment of Bre1 with the NCP acidic patch. By docking onto the NCP surface in this distinct orientation, Bre1 positions the Rad6 active site directly over H2B Lys123. The Spt–Ada–Gcn5 acetyltransferase (SAGA) H2B deubiquitinase module competes with Bre1 for binding to the NCP acidic patch, indicating regulatory control. Our study reveals a mechanism that ensures site-specific NCP ubiquitination and fine-tuning of opposing enzymatic activities.
Eukaryotes organize their DNA into chromatin, which packages DNA while allowing coordinated gene expression. The basic unit of the chromatin polymer is the nucleosome core particle (NCP), formed by an octameric complex of core histones (two copies each of H2A, H2B, H3, and H4) that is encircled by 145–147 bp of DNA (1). A large set of chromatin factors “write,” “read,” or “erase” histone posttranslational modifications and thereby alter the transcriptional properties of chromatin (2). The ∼200 kDa NCP provides a varied interaction surface for chromatin factors through the flexible histone N- and C-terminal tails, the rigid disk faces of the histone octamer, and the nucleosomal DNA (3). The recognition of unstructured histone tails by numerous protein domains is well studied, but far less is known about how chromatin factors recognize the disk face of the NCP. Monoubiquitination of histone H2B occurs at lysine 123 (H2B Lys123∼Ub) in yeast (equivalent to mammalian Lys120). The enzymatic reaction is carried out by the E3 ligase Bre1 (human RNF20/40) together with the E2 enzyme Rad6 (4–7). This highly site-specific ubiquitination of H2B controls various aspects of gene expression, which include transcription initiation and elongation (8), DNA replication (9) and repair (10), and kinetochore function (11). H2B Lys123∼Ub is thought to exert its effects through altering chromatin compaction and by promoting specific histone H3 methylations (12, 13). H2B ubiquitination is reversed by a heterotetrameric deubiquitinase module, which is part of the Spt–Ada–Gcn5 acetyltransferase (SAGA) complex (14, 15). Aberrant H2B Lys123∼Ub levels are observed in various disease states (16), suggesting that the fine-tuning of opposing ubiquitin ligase and deubiquitinase activities is critical for normal cell function.
Ubiquitination involves a three-step enzymatic reaction requiring ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin-ligating (E3) enzymes (17). Most E3 ligases use a RING domain to activate an E2∼ubiquitin thioester and mediate ubiquitin transfer to a substrate (“∼” denotes a covalent and “–” denotes a noncovalent interaction). The E3 RING domain, the E2, and ubiquitin interact with each other to stabilize a conformation that primes the thioester bond for nucleophilic attack by the substrate lysine residue (18). The Rad6–Bre1 machinery is unusual insofar as Rad6 interacts canonically with the Bre1 RING and through its “backside” with a separate non-RING domain of Bre1, which can potentiate ubiquitin transfer to the substrate (19).
After decades of research on H2B Lys123∼Ub (20), it is still unclear how the Rad6–Bre1 machinery exclusively targets H2B Lys123 but no other histone lysine residues on the NCP (21). It was previously suggested that acidic histone residues are required to maintain normal cellular H2B∼Ub levels in Saccharomyces cerevisiae (22, 23). However, it has remained unclear whether those residues mediate a direct physical interaction with Rad6–Bre1, or whether their mutation indirectly perturbs chromatin architecture or affects other transcription components. Now, by using cross-linking and mass spectrometry (XL-MS) we have succeeded in biochemically capturing the transient Bre1–NCP interaction and elucidating how this dictates Rad6 lysine specificity. We further establish how Bre1 directly recognizes the NCP disk surface in opposition to the SAGA H2B deubiquitinase module. Our study is fundamental for understanding the molecular basis of H2B ubiquitination in yeast and metazoa.
Results
Bre1–NCP Interaction Topology Determined by XL-MS.
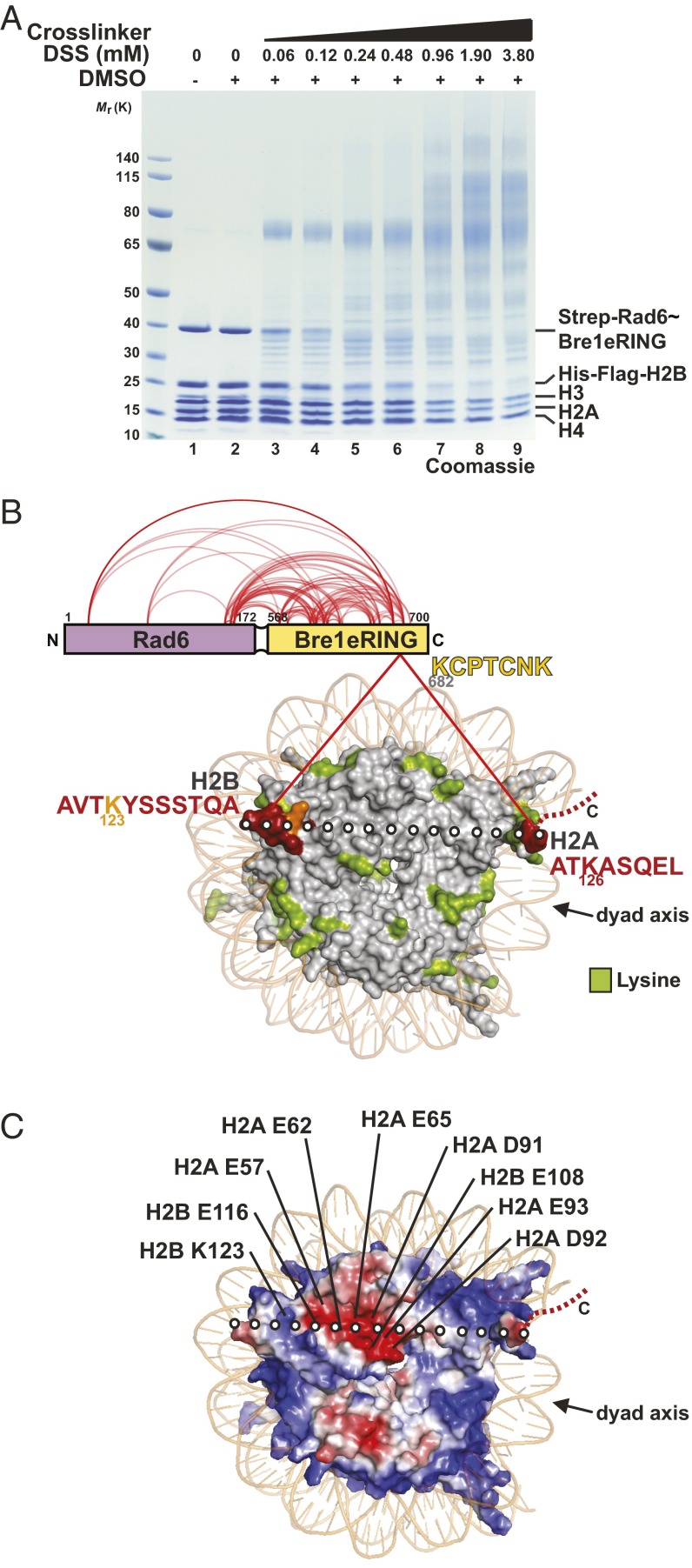
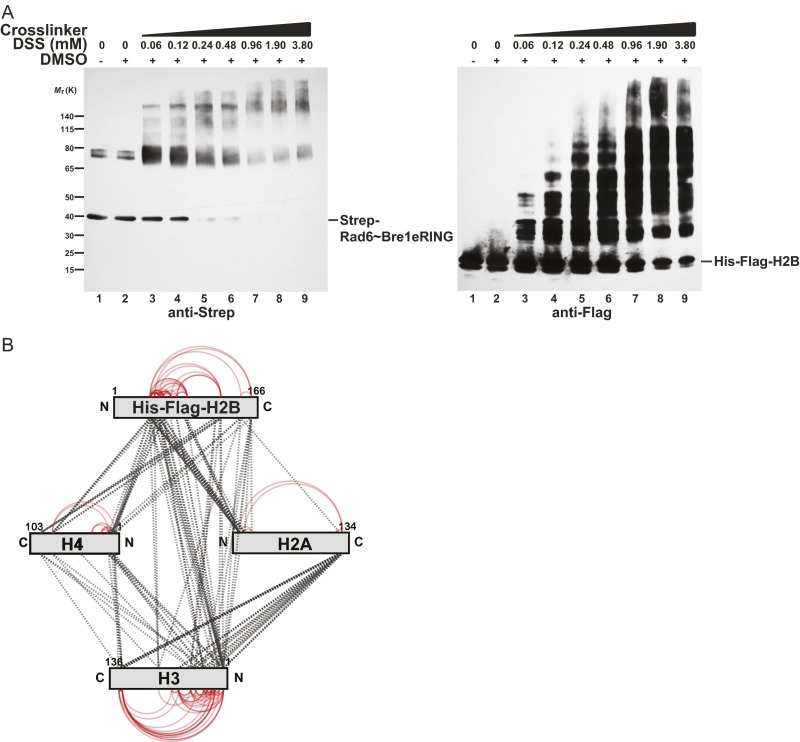
To map the NCP interaction sites of Rad6 and Bre1, we used a recombinant Rad6∼Bre1 fusion, in which the C terminus of Rad6 was connected to the N terminus of the Bre1 eRING domain (extended RING; amino acids 568–700) via a flexible linker (Fig. 1A, lane1). This minimal construct specifically modifies H2B Lys123 and is biochemically stable (19). The fusion protein is more active than full-length Bre1 and Rad6 alone in an in vitro NCP ubiquitination assay, likely because the transient RING–E2 interaction is stabilized. We used NCPs reconstituted from polycistronically expressed yeast histones in Escherichia coli and the Widom 601 DNA sequence as a substrate. Titrating increasing amounts of the homobifunctional cross-linker DSS (disuccinimidyl suberate) to a 1:1 molar mixture of Rad6∼Bre1 and NCP lead to the formation of high molecular weight complexes (Fig. 1A and Fig. S1A). DSS induces nucleophilic attacks on primary amines and thereby couples lysine residues. Notably, of all histone proteins, histone H2B was preferentially cross-linked with increasing amounts of DSS (compare histone stoichiometry, e.g., lane 2 vs. 5 in Fig. 1A), which parallels the reduction and upward shift of Rad6∼Bre1. This finding indicates a preferential cross-linking and hence spatial proximity between Rad6∼Bre1 and the target protein. Subsequent MS analysis revealed 94 high-confidence interlinks and 100 intralinks among histones (Fig. S1B and Dataset S1). Ninety-eight intralinks were identified within Rad6∼Bre1 (Fig. 1B and Dataset S1), consistent with the previously reported direct interaction between Rad6 and the Bre1 eRING domain (19) as well as Bre1 homodimerization (24). Of significance, only two unique interlinks between Rad6∼Bre1 and the NCP were identified (Fig. 1B). These high-confidence linkages correspond to a single peptide within the Bre1 RING domain that cross-links exclusively to two different peptides within histone H2B and H2A. No cross-links between Rad6 and histones were found. The H2B peptide maps to the C-terminal α-helix of the protein, which harbors the Lys123 ubiquitination site, and is located surface-exposed at the DNA edge opposite of the NCP dyad. The cross-linked H2A peptide maps to a C-terminal extension at the opposite DNA edge next to the NCP dyad. Notably, despite the large number of available lysine residues in the NCP (114 Lys in total, including 54 Lys in the tails), only two lysines were specifically cross-linked to Bre1. This finding suggests that the E3 binds in a defined orientation on the disk face of the NCP. The cross-linking of one Bre1 peptide to two different histone peptides is consistent with the symmetric arrangement and the overall size of the dimeric RING domain (24), as one RING monomer could contact H2B and the other H2A. A line drawn between the identified H2A and H2B peptides (Fig. 1B) runs directly over a prominent feature of the NCP surface, the acidic patch, a groove formed by conserved aspartate and glutamate residues from H2A and H2B (Fig. 1C). Remarkably, the Bre1 RING domain features four highly conserved basic residues, which are surface-exposed (24) and could readily direct Bre1 to the NCP acidic patch. Taken together, the XL-MS–derived spatial restraints suggested a specific interaction topology that is encoded in the electrostatic complementarity of basic RING and acidic NCP residues, a hypothesis that we tested further, as described below.
Fig. 1.
Bre1–NCP interaction topology determined by XL-MS. (A) Titration of DSS to recombinant Rad6∼Bre1 and NCP (S. cerevisiae proteins mixed at a 1:1 molar ratio). Samples were separated by SDS/PAGE (4–12%, MOPS buffer) and stained by Coomassie. DSS at 0.24 mM was chosen for further XL-MS analysis. (B) XL-MS analysis revealed Rad6∼Bre1 intralinks and interlinks with NCP (red lines). Yeast NCP (PDB ID code 1ID3) is depicted in surface representation when viewed down the DNA superhelical axis. Sequences of cross-linked Bre1 and histone peptides with amino acid position of Lys residues are indicated. Peptides (red) were mapped onto the NCP surface. H2B Lys123 is labeled in orange; all other NCP Lys residues visible in the structure are in green. The C terminus of H2A is only partially resolved (dashed line). Dotted white line represents the axis between cross-linked H2B and H2A peptides. NCP dyad axis is indicated as a reference. (C) NCP electrostatic surface potential (negative in red, positive in blue) highlights the NCP acidic patch formed by H2A/H2B residues. Axis of cross-linked peptides as in B. H2B Lys123 is also indicated.
Fig. S1.
(A) Titration of DSS to recombinant Rad6∼Bre1 and NCP (Saccharomyces cerevisiae proteins mixed at a 1:1 M ratio). Samples were separated by SDS/PAGE (4–12%, MOPS buffer), analyzed by immunoblotting against Strep-Rad6∼Bre1 and His-Flag–H2B and show the presence of cross-linked high molecular weight complexes. DSS at 0.24 mM was chosen for further XL-MS analysis. (B) XL-MS analysis of intralinks (red) and interlinks (dotted black lines) among yeast histones.
The NCP Acidic Patch Directly Recruits Bre1 to Monoubiquitinate H2B.
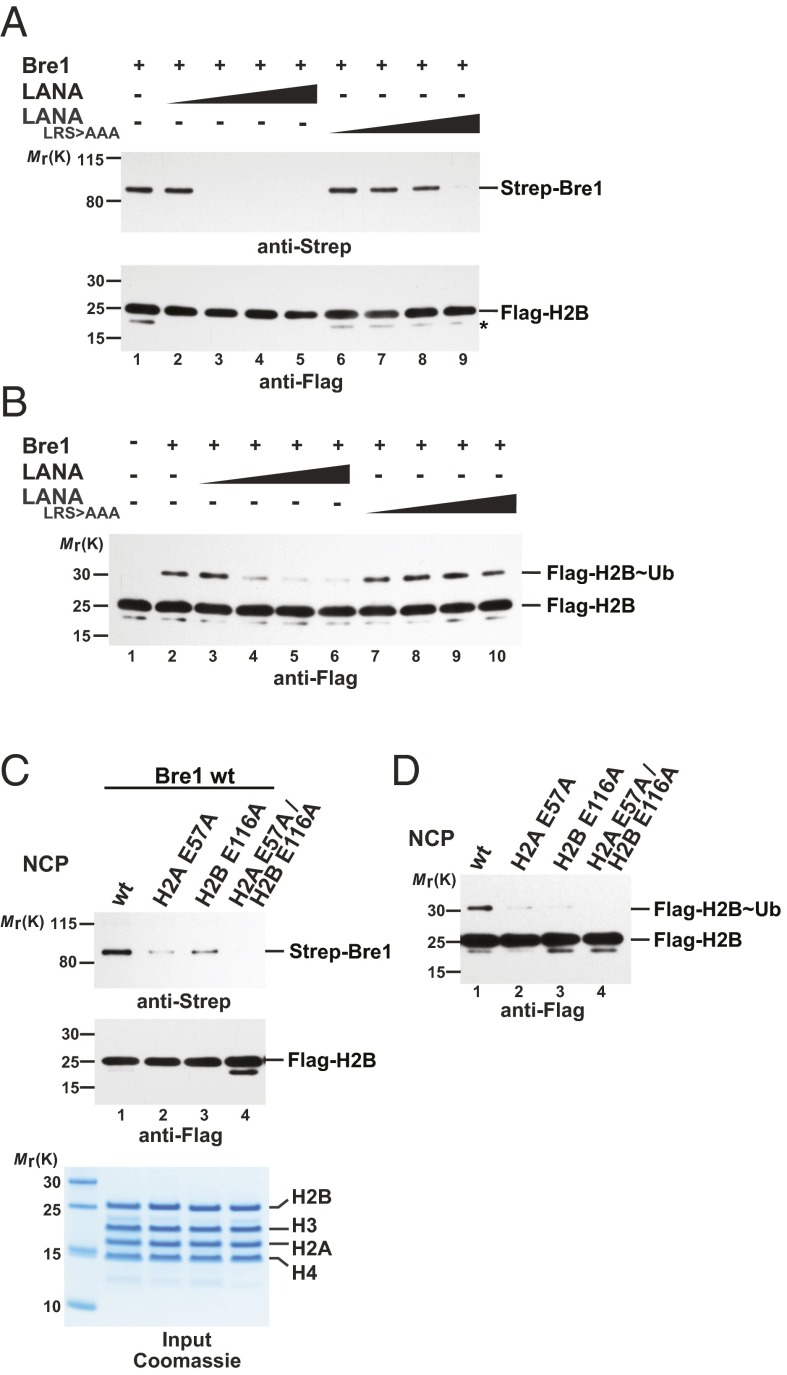
To determine whether Bre1 directly binds the NCP acidic patch, we first attempted to inhibit the interaction with a viral peptide that targets the same region. The N terminus of the herpes virus latency-associated nuclear antigen (LANA) forms a hairpin that inserts into the H2A/H2B acidic patch as seen in the LANA–NCP cocrystal (25). Of note, the overall structure of the NCP is not perturbed by LANA binding. Increasing amounts of a 23-residue LANA peptide blocked binding of full-length Bre1 to the NCP in pull-down assays, consistent with LANA obstructing Bre1 access to the acidic patch. In contrast, a mutant LANA peptide (L8R9S10 > AAA) with a reduced acidic patch affinity (25) was less efficient in competing with Bre1 for NCP binding (Fig. 2A). As a result of the impaired NCP interaction, Bre1 exhibited a reduced H2B monoubiquitination activity in an in vitro assay, whereas the mutant LANA peptide had little inhibitory effect (Fig. 2B). These results demonstrate that an accessible acidic patch is required for efficient NCP recognition and ubiquitination of H2B Lys123. To corroborate these findings, we reconstituted NCPs carrying point mutations in key acidic residues. Notably, alanine substitutions of H2A Glu57 and H2B Glu116, which lie close to the target Lys123 (Fig. 1C), impaired Bre1 binding to the mutant NCP and this effect was enhanced when both mutations were combined (Fig. 2C). Consequently, all these acidic patch mutations inhibited H2B monoubiquitination in vitro (Fig. 2D). This finding demonstrates that an accessible and intact acidic patch is required for Bre1 recruitment to the NCP in agreement with the interaction topology obtained by XL-MS.
Fig. 2.
The NCP acidic patch directly recruits Bre1. (A) The LANA peptide competes with Bre1 for interaction with the NCP acidic patch. In vitro binding assays with recombinant NCPs immobilized on anti-Flag beads and recombinant full-length Strep-tagged Bre1 incubated in a 1:3 molar ratio (NCP:Bre1). LANA (lanes 2–5) or a LANA_LRS > AAA peptide (lanes 6–9), mutated in critical NCP-interacting residues (25), were added with increasing concentrations (20, 40, 60, 80 µM). Following elution, anti-Strep immunoblotting was used to detect NCP-bound Strep–Bre1. Anti-Flag detection confirmed equal input of NCPs. The asterisk indicates an H2B degradation product. (B) H2B Lys123∼Ub by Bre1 requires an accessible NCP acidic patch. In vitro NCP ubiquitination assay was performed in the presence of LANA (lanes 3–6) or a LANA_LRS > AAA peptide (lanes 7–10) (20, 40, 60, 80 µM peptide). Anti-Flag immunoblotting detects both H2B and H2B Lys123∼Ub. (C) NCP acidic patch mutations impair Bre1 recognition. In vitro binding assay with NCP wild-type or different NCP mutants carried out as in A. Input of mutant NCPs shows their intact stoichiometry. (D) NCP acidic patch mutations affect H2B Lys123∼Ub. In vitro NCP ubiquitination assay with wild-type or mutant NCPs. Reactions were incubated for 20 min and analyzed by SDS/PAGE and immunoblotting.
XL-MS Captures the Transient Bre1 RING–Rad6 Interaction.
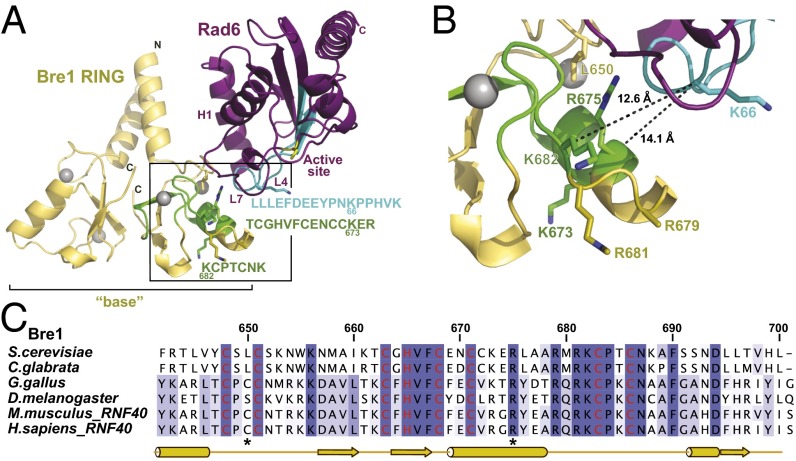
A key question that emerges is how electrostatic interactions between Bre1 and the NCP, spatially distinct from the site of catalysis, would position Rad6 such that it targets Lys123, yet no other histone Lys residue. The Bre1 RING domain directly interacts with Rad6 and is sufficient to monoubiquitinate H2B Lys123 in vitro (19). We used the known crystal structures of the Bre1 RING domain and Rad6 to generate a homology model of the E2–E3 complex (24, 26). Superposition of Bre1 RING and Rad6 onto the cCbl–UbcH7 crystal structure (27), followed by superposition onto RNF4–UbcH5∼Ub (18) generated a dimeric Bre1 RING–Rad6∼Ub model with no severe steric collisions (Fig. 3A and Fig. S2A). Importantly, the interface between Rad6 and the Bre1 RING in our model is in agreement with XL-MS–derived contact points. Notably, a single Rad6 peptide that maps to loop 4 (L4), cross-linked to two peptides located on the opposite Bre1 RING surface. The measured distance between cross-linked lysine residues in the Bre1–Rad6 model satisfies the maximal distance restraints imposed by the DSS cross-linker (<26 Å between Cα atoms) (28) (Fig. 3B) and supports the validity of the model. Other cross-links between Rad6 and Bre1 include peptides that are not present in the partial Bre1 structure (e.g., Bre1 residues 568–623) (24). Taking these data together, we find that XL-MS enabled us to monitor the highly dynamic E3–E2 interaction, providing concise spatial restraints for the Rad6–Bre1 interface.
Fig. 3.
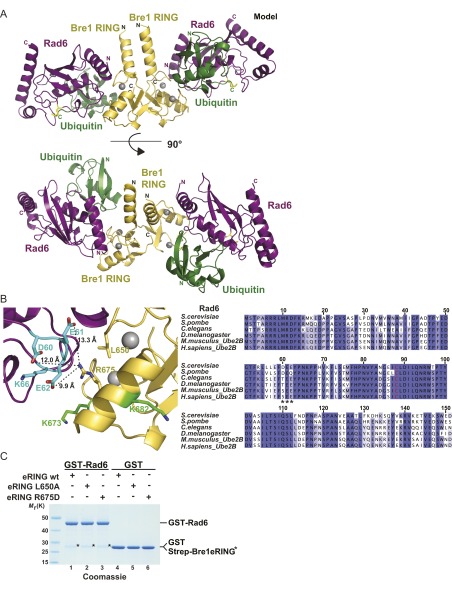
Bre1 RING-Rad6 complex based on XL-MS and homology modeling. (A) Ribbon representation of the modeled S. cerevisiae Bre1 RING domain (yellow) interaction with Rad6 (purple). See Fig. S2A for further details on modeling templates and a version that includes ubiquitin. Zinc ions are shown as gray spheres. The Rad6 active site is marked as a yellow stick. The Rad6 N-terminal helix 1 (H1) and loops L4 and L7 comprise the canonical E3-binding site. Cross-linked peptides (Bre1 in green; Rad6 in cyan) were mapped onto the structure and their sequences are indicated with the same color code, including amino acid position of cross-linked Lys residues. Boxed region is magnified in B. (B) Close-up of the predicted Bre1 RING–Rad6 interface highlighting cross-linked peptides in Bre1 (green) and Rad6 (cyan). Side-chains of relevant residues are shown in stick representation and measured distances between cross-linked lysines are indicated. (C) Sequence alignment of Bre1 RING domain orthologs from S. cerevisiae, Candida glabrata, Gallus gallus, Drosophila melanogaster, Mus musculus, and Homo sapiens. Zinc-coordinating residues are labeled in red. Asterisks indicate the position of residues involved in Bre1 RING-Rad6 interaction. The alignment was generated with ClustalW and colored in Jalview by identity. Numbers refer to S. cerevisiae Bre1 residues.
Fig. S2.
(A) Homology model of the dimeric S. cerevisiae Bre1 RING domain (yellow) (PDB ID code 4R7E) bound to S. cerevisiae Rad6 (purple) (PDB ID code 1AYZ) charged with ubiquitin (green) is shown in ribbon representation. Gray spheres are zinc ions. The Rad6 active site Cys88 is marked as a yellow stick. A BLAST search with the S. cerevisiae Bre1 RING sequence against sequences of known RING structures deposited in PDB identified the Cbl RING domain from the Cbl–UbcH7 complex (PDB ID code 1FBV) as the highest-scoring homolog. Superposition of the Cbl–RING with Bre1–RING resulted in an excellent spatial alignment (0.7 Å rmsd). This alignment brought UbcH7 to the Bre1–RING dimer. Superposition of S. cerevisiae Rad6 to UbcH7 resulted in a reasonable spatial alignment (1.5 Å rmsd). These steps were repeated for the second copy of the Bre1RING in the homodimer. The RNF4–UbcH5∼Ub (PDB ID code 4AP4) was superimposed onto Rad6, bringing ubiquitin to Rad6. This was repeated for the second copy of Rad6 in the Bre1 dimer. The end result is a dimeric form of a Bre1 RING–Rad6∼Ub complex. (B) Close-up of the Bre1 RING–Rad6 model highlighting an oppositely charged interface. Three Rad6 acidic residues located on a loop are shown in stick representation (cyan) and measured distances to Bre1 Arg675 are indicated. Cross-linked Lys residues on Bre1 (Lys673, Lys682 in green) and Rad6 (Lys66 in cyan) are also indicated as a reference. (Right) Sequence alignment of Rad6 orthologs in the fungi/metazoa group. The Rad6 catalytic Cys88 is labeled in red. Asterisks mark the position of Rad6 acidic residues potentially interacting with the Bre1 RING domain. The alignment was generated with ClustalW and colored in Jalview by identity. Numbers refer to S. cerevisiae Rad6 residues. (C) In vitro binding assay with GST–Rad6 immobilized on GSH beads and incubated with Bre1 eRING constructs in 1:5-M ratio. Recombinant GST was used as negative control. After elution, proteins were analyzed by SDS/PAGE (4–12%, MOPS buffer) and Coomassie staining. Same eluates were immunoblotted with anti-Strep antibody (see Fig. 4C).
Specific Bre1 Residues Required for Rad6 Activation and NCP Recognition.
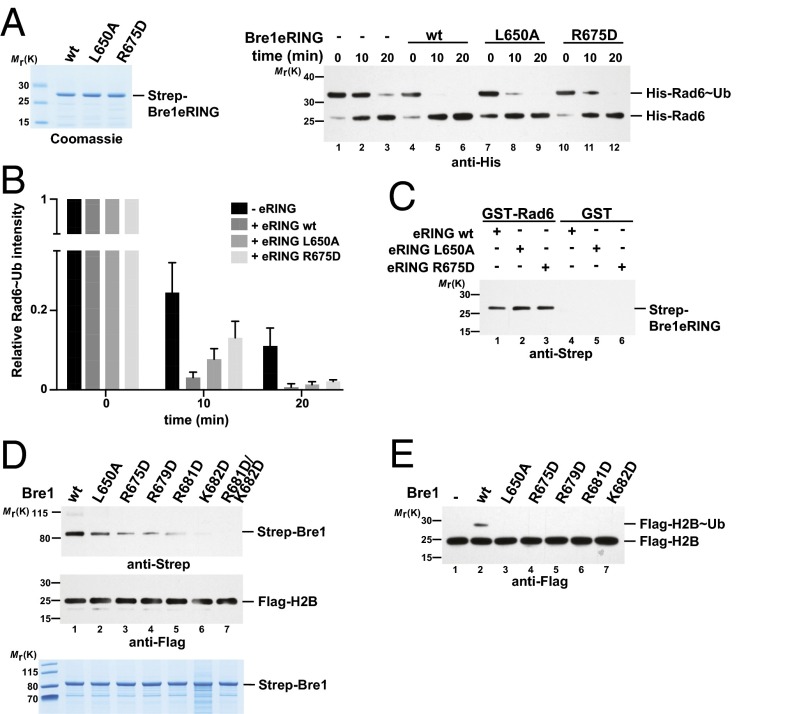
We next sought to validate the E2–E3 interface by mutating conserved Bre1 amino acids while leaving the structurally relevant zinc-coordinating residues intact. After screening an extensive number of recombinant constructs, we finally succeeded in generating stable Leu650 > Ala and Arg675 > Asp mutants of Bre1, both of which map to the predicted Rad6 interface (Figs. 3B and 4A). To directly measure their effect on Rad6 activation, we analyzed the discharge of the ubiquitin thioester (∼Ub) from the active site of Rad6 in single turnover assays (Fig. 4A, Right, and Fig. 4B). To this end, Rad6 was charged with ubiquitin by the E1 enzyme and ATP and then treated with EDTA to inhibit recharging. Rad6 is known to undergo spontaneous Ub discharge independently of Bre1 over time (Fig. 4A, Right, compare lane 1 and 3; and Fig. 4B) (19, 29). However, adding a wild-type Bre1 eRING (amino acids 494–700) significantly accelerated the reaction. In contrast, Ub discharge was impaired in the Bre1 Arg675 > Asp and to a lesser extent in the Leu650 > Ala mutant. The involvement of Arg675 suggests a complementary charge on the Rad6 side. Indeed, three acidic Rad6 residues (Asp60, Glu61, and Glu62) are located within the cross-linked peptide on loop L4, ∼10–13 Å away from Arg675 in our model (Fig. S2B). Reduced Ub discharge from Rad6 could arise from a decreased affinity of Rad6 toward the Bre1 RING mutants. Alternatively, the mutant RING domains may specifically affect Rad6 activation. To distinguish between these possibilities, we performed in vitro binding assays. Notably, Rad6 binding to the Bre1 RING mutants was not perturbed (Fig. 4C and Fig. S2C). Thus, we conclude that these Bre1 RING residues are required for stimulating Rad6 catalytic activity, possibly through an allosteric effect. Whereas Bre1 Arg675 is located on a short α-helix with its side chain pointing laterally toward Rad6, three other conserved basic residues (Arg679, Arg681, Lys682) are part of a surface-exposed loop with their side chains oriented toward the “base” of the Bre1 RING homodimer (24) (Fig. 3 B and C). To pinpoint the function of single residues to Rad6 activation and NCP recognition, we analyzed them individually by inverting their charge. Notably, mutation of the basic Bre1 RING residues affected NCP recognition and the combined Arg681/Lys682 > Asp mutant exhibited the strongest loss of affinity (Fig. 4D), likely caused by an electrostatic repulsion with the NCP acidic patch. Interestingly, the Leu650 > Ala and Arg675 > Asp mutants were defective in both Rad6 activation and NCP recognition. This result could reflect an allosteric coupling of substrate recognition with ubiquitin discharge or other structural rearrangements. As a consequence of perturbing the NCP–RING interface, H2B ubiquitination was impaired in an in vitro assay (Fig. 4E). In sum, we could identify specific Bre1 residues required for Rad6 activation and NCP recognition, which are located at the protein interfaces suggested by XL-MS.
Fig. 4.
Bre1 residues required for Rad6 activation and NCP recognition. (A) Representative immunoblot of a single turnover ubiquitin discharge experiment, in which precharged Rad6∼Ub was incubated without Bre1 or with 100 µM of wild-type or mutant Bre1 eRING. Reactions were stopped after 0, 10, and 20 min by denaturation. Samples were separated by nonreducing SDS/PAGE (12% gel, MES buffer) followed by anti-His immunoblotting. Bre1 proteins are shown (Left). (B) Quantification of single turnover experiments as shown in A. Intensity of the Rad6∼Ub and Rad6 bands were quantified with the ImageLab 1.5.2 software. The ratio between Rad6∼Ub and Rad6 was calculated for every condition and time 0 was used for normalization. Four independent experiments were performed. Mean and SD are indicated. (C) Bre1 RING mutants are not impaired in Rad6 binding. GST–Rad6 was immobilized on GSH beads and incubated with Bre1 eRING constructs in a 1:5 molar ratio. Recombinant GST was used as negative control. After elution, bound proteins were detected by anti-Strep immunoblotting. See Fig. S2C for protein input. (D) Specific RING domain mutations impair NCP recognition. In vitro binding assay with NCPs immobilized on anti-FLAG beads and Bre1 constructs added in 1:3 molar ratio (NCP:Bre1). After elution, immunoblotting was used to detect NCP-bound Bre1 and equal NCP loading. Input for recombinant Bre1 proteins is shown (Lower). (E) In vitro NCP ubiquitination assay performed with the indicated Bre1 mutants.
A Model for the E3–E2∼Ub–NCP Complex.
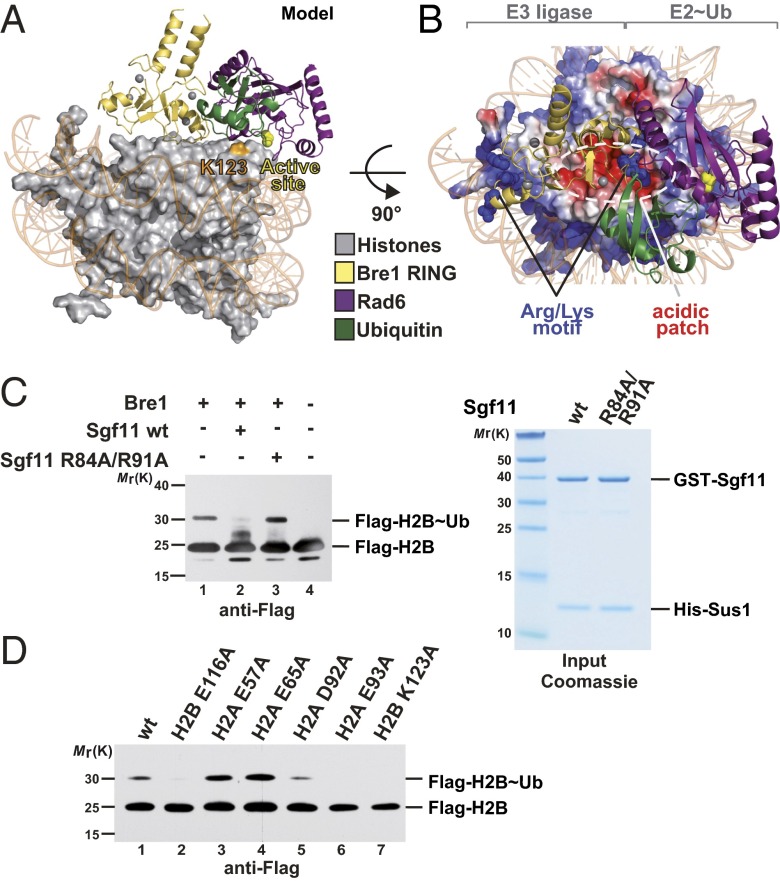
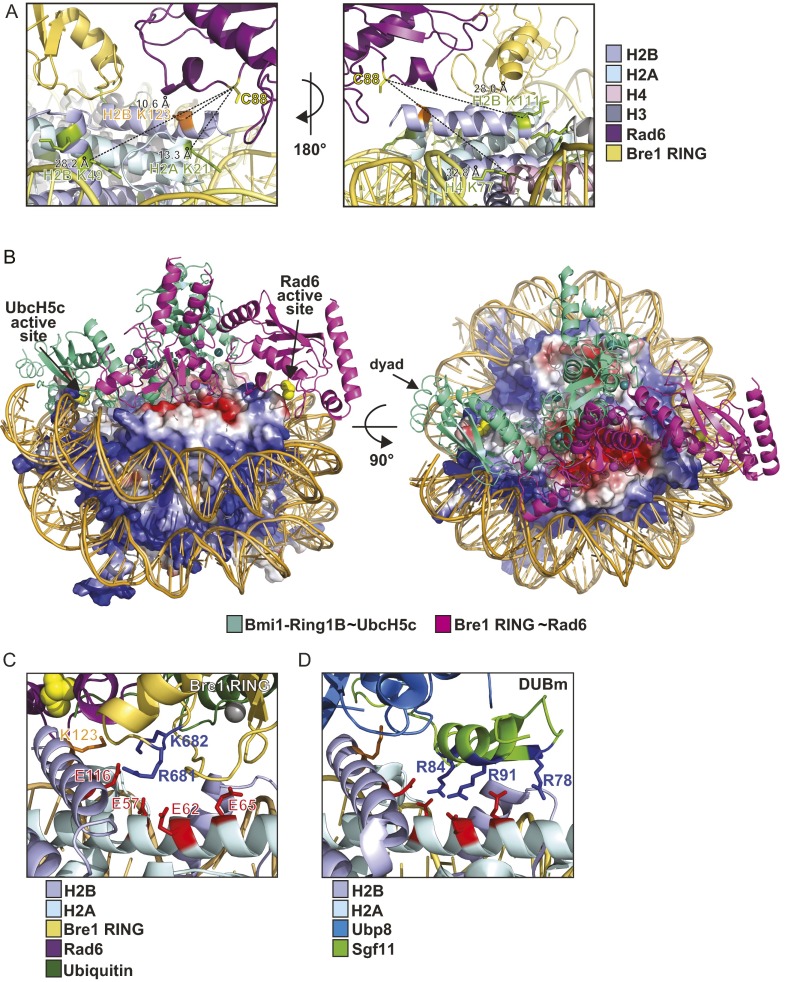
To integrate our experimental data, we sought to develop a structural model of the enzyme-substrate complex that fulfills the constraints derived from XL-MS, mutational analyses, and the general E2 catalytic mechanism. We used the Bre1 RING homodimer modeled in contact with one Rad6∼Ub under the assumption that only a single Rad6 will target H2B Lys123 (18). The complex was manually docked onto the NCP disk surface by apposing the critical basic Bre1 Arg679 and Arg681 residues to H2B Glu116, an acidic patch residue required for the direct interaction with Bre1 (Fig. 2C). Next, the Bre1 RING–Rad6∼Ub was rotated around the DNA superhelical axis of the NCP to approach the catalytic center of Rad6 (Cys88) toward the H2B Lys123 target residue (Fig. 5 A and B). This model was refined using all-atom energy minimization and subsequent molecular dynamics (MD) equilibration in explicit solvent. Notably, the resulting structural model brings the Rad6 catalytic Cys88 within a 10.6-Å distance to the H2B Lys123 (Fig. S3A). This is the closest distance of Cys88 to any histone Lys residue in its vicinity. Taking the intrinsic flexibility of the complex into account, this distance is reasonably close to a 3–3.5 Å upper limit for a ubiquitin transfer reaction to happen. Importantly, our model is in excellent agreement with all XL-MS distance restraints and lends strong support to the suggested mechanism for H2B Lys123 ubiquitination and recognition of the acidic patch.
Fig. 5.
Model of Bre1 RING–Rad6∼Ub interaction with the NCP. (A) Orthogonal view of the E3–E2∼Ub complex that was manually docked onto the NCP acidic patch and subjected to energy minimization and MD equilibration. H2B Lys123 is labeled in orange. The Rad6 active site (Cys88) is depicted as a yellow sphere. (B) View of the complex looking down on the DNA superhelical axis. The electrostatic surface potential of the histone octamer (negative in red, positive in blue) is indicated. Basic residues at the “base” of the RING domain (Arg679, Arg681, and Lys682) are labeled as blue spheres. The estimated Cα cross-linking distances between Bre1 Lys682 and the H2A peptide are 22.6 Å (Bre1 Lys682–H2A Thr125; note that cross-linked H2A Lys126 is not resolved in the NCP structure) and 15.1 Å for Bre1 Lys682 and H2B Lys123. (C) Bre1 competes with the Sgf11 ZnF for recognition of the NCP acidic patch. In vitro NCP ubiquitination assay was performed in the presence of wild-type or mutant GST–Sgf11–Sus1 (60 µM). Reactions were incubated for 60 min and analyzed by SDS/PAGE and immunoblotting. Input for Sgf11–Sus1 constructs is shown (Right). (D) Global H2B Lys123∼Ub levels depend on an intact NCP acidic patch. Cell lysates from S. cerevisiae strains with Flag-tagged histone H2B were subjected to anti-Flag immunoprecipitation. Recovered proteins were detected by immunoblotting.
Fig. S3.
(A) Distance measurements between the Rad6 active site and histone Lys residues on the NCP surface. Structure is based on manual-docking/MD simulations. The active site Cys88 of Rad6 is highlighted as a yellow stick, Ubiquitin was omitted for clarity. All nucleosomal Lys residues are shown in green stick mode, only H2B Lys123 is marked in orange. Distances between the Cα atom of Rad6 Cys88 and adjacent histone Lys Cα atoms were measured in PyMol. H2B Lys123 lies closest to the Rad6 active site. (B) Superposition analysis of the Bmi1–Ring1B∼UbcH5c–NCP complex (PDB ID code 4R8P) with the Bre1–Rad6∼Ub–NCP model using the NCP for alignment. Both E3 RING domains insert their Arg anchors into the NCP acidic patch. However, the resulting E3–E2 geometry is entirely different as the E2 enzymes are rotated by ∼180° when viewed down the DNA superhelical axis. The electrostatic surface potential of the yeast histone octamer (negative in red, positive in blue) is indicated. The active sites of the E2 enzymes (Cys88 for Rad6; Cys85 for UbcH5) are labeled as yellow spheres. (C) Close-up view of the S. cerevisiae Bre1 RING domain in contact with the S. cerevisiae NCP (XL-MS/homology model). Critical basic residues on the Bre1 RING (blue) and acidic NCP residues (red) are shown in stick mode. The catalytic center of Rad6 appears as a yellow sphere and the target Lys123 is highlighted as an orange stick. (D) Close-up view of the S. cerevisiae DUB module Sgf11 ZnF in complex with the Xenopus laevis NCP (PDB ID code 4ZUX). Criticial basic and acidic residues as well as the target lysine are indicated in stick mode (color code as in C). The highlighted X. laevis acidic patch residues correspond to the yeast acidic NCP residues in a sequence alignment.
The NCP Acidic Patch Recruits Opposing Enzymatic Activities.
Cellular H2B Lys123∼Ub levels likely reflect the balance of ubiquitin ligase and deubiquitinase activities. This finding raises the question whether the NCP acidic patch can regulate recruitment of competing enzymes. We previously implicated solvent-exposed basic residues on the SAGA H2B DUB module (Ubp8–Sgf11–Sgf73–Sus1) in NCP recognition (14). These basic residues map to a small ZnF domain of the DUB subunit Sgf11 and were recently confirmed to target the NCP surface, specifically the NCP acidic patch (30). In light of our findings on Rad6–Bre1, we sought to test whether basic residues on Bre1 and Sgf11 indeed compete for binding to the NCP acidic patch in solution. Thus, we carried out NCP ubiquitination reactions in the presence of Sgf11, which forms a stable heterodimer with Sus1 (14) (Fig. 5C). Notably, this DUB subcomplex could effectively inhibit H2B ubiquitination by Bre1–Rad6. In contrast, Sgf11 failed to inhibit the reaction when Arg84 and Arg91 were mutated, consistent with their requirement for NCP acidic patch recognition (Fig. 5C). These observations led us to test whether the NCP acidic patch mediates the functional opposition between Rad6–Bre1 and the SAGA DUB module in vivo. To this end, we assessed global histone H2B∼Ub levels in selected acidic patch mutants in S. cerevisiae. The H2B Glu116 > Ala mutant recapitulates the ubiquitination defect seen in vitro. This result is also observed upon mutating H2A Glu93, a residue located toward the center of the NCP disk (Figs. 1C and 5D, lanes 2 and 6), and is consistent with a previous report (23). In marked contrast, H2A Glu57 and Glu65 mutations showed increased H2B Lys123∼Ub levels. This result is different from our in vitro finding in which the H2A Glu57 mutant exhibited reduced Bre1 binding and H2B ubiquitination (compare Figs. 2D, lane 2, and 5D, lane 3). This interesting observation can be explained by an interference between H2B DUB and E2/E3 activities, which are only uncovered in an in vitro system that lacks DUB activity. Remarkably, both enzymes target the NCP acidic patch, but some residues, like H2A Glu57 and Glu65, appear to be more important for DUB than E2/E3 recruitment (Fig. S3 C and D). Of note, our in vivo result on H2A Glu65 is consistent with a key structural role of this residue in contacting the basic surface of the Sgf11 ZnF, which is also important for H2B deubiquitination in vitro (30) (Fig. S3D). In sum, we establish a molecular mechanism for Rad6–Bre1 recruitment to the NCP acidic patch that occurs in competition with specific elements of the SAGA deubiquitinase.
Discussion
Despite the importance of cotranscriptional H2B ubiquitination for gene expression, the most basic aspects of the reaction, including substrate recognition and site-specific ubiquitin transfer, were poorly understood. Here, we used XL-MS to capture topologically complex relationships between the NCP and the Rad6–Bre1 ubiquitination machinery in their native state, which allowed us to identify the E3, E2, and NCP surfaces critical for specificity and activity. By integrating different experimental constraints, we derive a precise molecular mechanism in which the Bre1 RING homodimer uses an arginine motif to interact with the NCP acidic patch. This positions Rad6 directly above the ubiquitination site and enables exclusive modification of H2B Lys123, despite the large number of lysine residues on the NCP disk surface and flexible histone tails.
A comparison of Bre1–Rad6 with the H2A Lys119 ubiquitinating module of the polycomb repressive complex 1 (PRC1) highlights interesting similarities and differences (31). This study reported the crystal structure of the human Ring1B–Bmi1 E3 ligase fused to the E2 enzyme UbcH5c in complex with the Xenopus laevis NCP. Remarkably, the E3 also employs an arginine motif to interact with the NCP acidic patch and this orients the E2’s active site toward the target lysine. Although the principle of NCP recognition looks similar for Bre1, the overall geometry of the E3–E2–NCP complex is entirely different. H2B Lys123 (Lys120 in humans) is located on the opposite DNA edge of the NCP disk, compared with the Lys residue targeted by PRC1. To reach Lys123, Bre1–Rad6 is rotated by about 180° against the PRC1 E3–E2 complex, when viewed down the DNA superhelical axis (Fig. S3B), which is remarkable given that both E3–E2 complexes use the same acidic groove as an attachment site. The large-scale reorientation of Rad6–Bre1 versus PRC1 clearly shows that acidic patch interactions per se do not dictate the selection of an NCP ubiquitination site. In fact, all NCP–protein complexes that have been structurally determined so far use positively charged residues on multiple types of secondary structures as molecular anchors to bind to the NCP acidic patch (3).
All currently available cocrystal structures depicting proteins bound to the NCP acidic patch are derived from functionally unrelated factors from divergent species (3). A key insight of our study is that interference can exist in the same pathway and species. We find that the yeast H2B ubiquitination machinery uses an overlapping NCP binding surface with the yeast SAGA H2B deubiquitinating module (Fig. S3 C and D). Intriguingly, the DUB also contacts the NCP acidic patch via an arginine cluster on the Sgf11 zinc finger (30) and we show that Rad6–Bre1 effectively compete with the Sgf11 zinc finger for binding to the NCP acidic patch. This finding raises important questions: Why do both enzymes use the same binding surface? And how is the interaction of opposing enzymatic activities with the NCP regulated? An overlapping recognition surface may serve to avoid unproductive interference of enzymatic activities and also to enable the fine-tuning of cellular H2B∼Ub levels, which is important for gene expression. Conceivably, the genome wide-occupancy of Bre1 on chromatin is determined by the kinetics of RNA polymerase (Pol) II elongation. The prevailing model is that Rad6 and Bre1 are physically tethered to the transcribing Pol II through the conserved Paf1 adaptor complex (32). SAGA is primarily considered as a promoter-bound coactivator, but a fraction may also associate with elongating RNA Pol II (33). In vivo measurements of eukaryotic transcription elongation have estimated a Pol II speed of up to 5,000 bases per minute (34). This finding implies that Pol II elongation rate could be a key rate-limiting step of H2B ubiquitination as each encounter between Pol II and an NCP opens only a short window of opportunity for substrate recognition and ubiquitin transfer (35). The highly specific substrate-targeting mechanism discovered in this study should be important to ensure efficient H2B∼Ub in the context of a moving polymerase and in competition with SAGA DUB activity. Moreover, a SAGA-occupied and previously deubiquitinated nucleosome (e.g., at the promoter) may only be available for reubiquitination upon dissociation of SAGA.
In this study, we have identified the basic mechanism of NCP recognition and modification by Bre1–Rad6. Given the conservation of critical residues, the mechanism is likely conserved between eukaryotes. The future challenge is to understand how Bre1 and Rad6 execute a constrained enzymatic reaction while “riding” on the Pol II machine and to elucidate how opposing enzymatic activities are regulated to achieve precise spatiotemporal control of gene expression.
Materials and Methods
Constructs of Rad6, Bre1, Sgf11, ubiquitin, and histones were cloned, expressed, and purified as described previously (14, 19). NCPs were reconstituted from histone octamers and the 167-bp 601 WIDOM positioning sequence. Point mutations were generated by PCR-based methods and confirmed by sequencing. Cross-linking analysis of Rad6∼Bre1 RING:NPC was performed by mixing purified components in a 1:1 molar ratio with an equimolar mixture of light and heavy-labeled (deuterated) disuccinimidyl suberate DSS-H12/D12 (Creative Molecules). A final concentration of 0.24 mM DSS was used for cross-linking at 30 °C for 30 min. The reaction was quenched by the addition of final 100 mM ammonium bicarbonate and samples were processed by MS. In vitro ubiquitination assays, binding assays and E2 discharging assays were performed essentially as described previously (19). Immunoprecipitation of FLAG-tagged histone H2B was performed as described previously (14). A manually assembled configuration of the Bre1–Rad6/nucleosome core particle complex was refined using all-atom energy minimization and subsequent MD equilibration. Full experimental details are available in SI Materials and Methods.
SI Materials and Methods
Protein Expression and Purification.
His6–tobacco etch virus (TEV)–Rad6, Strep–Bre1 constructs, GST–TEV–Sgf11 constructs, ubiquitin, and histones were cloned, expressed, and purified as described previously (14, 19). Mutations and deletions were generated by PCR-based methods and confirmed by sequencing. Strep–Rad6∼Bre1 RING fusion protein was affinity purified on a 5-mL Strep–Tactin Superflow column (GE Healthcare) in buffer containing 50 mM Tris, pH 8.0, 100 mM NaCl, 5 mM MgCl2, and eluted by 2.5 mM desthiobiotin. Protein was further purified by size-exclusion chromatography on a Superdex 200 HiLoad 16/60 column (GE Healthcare) equilibrated with 50 mM Hepes, pH 8.0, 100 mM NaCl, 5 mM MgCl2.
Histone Octamer Purification and NCP Reconstitution.
A single-step purification strategy for the histone octamer was used as described previously (19). Mutations on histones were generated by PCR-based methods and confirmed by sequencing. NCPs were reconstituted from histone octamers and the 167-bp 601 WIDOM positioning sequence by salt dialysis and further purified by size-exclusion chromatography on a Superdex 200 HiLoad 16/60 column (GE Healthcare) equilibrated in 20 mM Tris⋅HCl, pH 7.5. NCPs used for cross-linking experiments were purified in 20 mM Hepes, pH 8.0.
Chemical XL-MS.
Cross-linking analysis of Rad6∼Bre1 RING:NPC was performed by mixing purified components in 1:1 molar ratio with an equimolar mixture of light and heavy-labeled (deuterated) disuccinimidyl suberate DSS-H12/D12 (Creative Molecules). A final concentration of 0.24 mM DSS was used for the cross-linking reaction at 30 °C for 30 min. The reaction was quenched by adding final 100 mM of ammonium bicarbonate incubating for 10 min at 30 °C. Digestion with lysyl endopeptidase (Wako) was performed at 35 °C, 6 M Urea for 2 h (at enzyme-substrate ratio of 1:50 wt/wt) and was followed by a second digestion with trypsin (Promega) at 35 °C overnight (at 1:50 ratio wt/wt). Digestion was stopped by the addition of 1% (vol/vol) trifluoroacetic acid (TFA). Acidified peptides were purified using C18 columns (Sep-Pak, Waters). The eluate was dried by vacuum centrifugation and reconstituted in water/acetonitrile/TFA, 75:25:0.1. Cross-linked peptides were enriched on a Superdex Peptide PC 3.2/30 column (300 × 3.2 mm) at a flow rate of 50 μL/min and water/acetonitrile/TFA, 75:25:0.1 as a mobile phase. Fractions of 100 μL were collected, dried, and reconstituted in 2% acetonitrile and 0.2% FA, and further analyzed by liquid chromatography coupled to tandem MS using an LTQ Orbitrap Elite instrument (Thermo Scientific). Cross-linked peptides were identified using xQuest (36). False-discovery rates (FDRs) were estimated by using xProphet (37) and results were filtered according to the following parameters: FDR < 0.05, minimum δ score = 0.85, MS1 tolerance window of –4 –4 ppm, ld-score > 22. The cross-links were visualized using the webserver xVis (38).
NCP Ubiquitination Assay.
For NCP ubiquitination reactions, 1.2 µM NCP, 100 nM E1, 3 µM Rad6, 36 µM ubiquitin, 18 µM Bre1, 3 mM ATP, and 0.1 mM DTT were mixed in a reaction volume of 20 µL (50 mM Tris⋅HCl, pH 8.0, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2) and incubated for 90 min at 30 °C (unless otherwise indicated in figure legends). Reactions were stopped by addition of 4× SDS loading buffer and analyzed by SDS/PAGE (NuPAGE 4–12% gels; Invitrogen, MOPS buffer) followed by immunoblotting with anti-FLAG M2 antibody (Sigma, F1804).
In Vitro Binding Assays.
NCP–Bre1.
Reconstituted NCPs were mixed with Bre1 constructs at a 1:3 molar ratio in binding buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 1.5 mM MgCl2) and incubated with 30 µL of anti-FLAG M2 beads (Sigma). After washing, proteins were eluted by incubation with FLAG peptide (0.1 mg/mL in binding buffer) for 45 min at 4 °C, TCA-precipitated, and analyzed by SDS/PAGE (NuPAGE 4–12% gel, MOPS buffer) and immunoblotting.
Rad6-Bre1eRING.
Binding assays were performed as previously described (19) by mixing eRING constructs in a 1:5 molar ratio with GST–Rad6 or GST as negative control.
E2 Discharging Assay.
Rad6 precharging was performed as described previously (19). Then, 100 µM of Bre1 eRING constructs were added to the charged E2 in buffer containing 50 mM CHES, pH 8.0, 100 mM NaCl. Samples were incubated at 30 °C for 0, 10, and 20 min. Reactions were terminated by adding stop buffer [60 mM Tris⋅HCl, pH 6.8, 2% (wt/vol) SDS, 10% (vol/vol) glycerol, 0.005% Bromphenol blue] and kept on ice until analysis by nonreducing SDS/PAGE (12% NuPAGE gels, MES buffer) and by immunoblotting with anti-His IgG2a antibody (GE Healthcare, 27471001). Blots were visualized with a ChemiDoc Touch Imaging System (Bio-Rad), and quantification of band intensities was performed with ImageLab 5.2.1 software (Bio-Rad).
Measurement of Global H2B∼Ubiquitin Levels.
FLAG-H2B yeast strains were from a mutant collection (22). Immunoprecipitation of FLAG-tagged histone H2B was performed as described previously (14).
Energy Minimization and MD Equilibration.
Two manually assembled configurations of the Bre1–Rad6/NCP complex were refined using all-atom energy minimization and subsequent MD equilibration. The yeast NCP structure was taken from the Protein Databank (PDB ID code 1ID3) together with the yeast Bre1–Rad6 homology model (based on PDB ID codes 4R7E and 1AYZ) and were protonated and repaired for the missing atoms using PDB2PQR utility (39). All further calculations were performed using GROMACS 4.5.1 package (40) and Amber99SB-ILDN force-field parameters (41). The system was placed into a 13 × 13 × 13-nm3 rectangular box, initially energy minimized and solvated with TIP3P water molecules (42). Additionally, Na+ and Cl− ions were added to achieve electroneutrality and at the final salt concentration of 80 mM. After this step, the complete system was energy-minimized using position restraints placed on protein Ca-atoms, Zn2+ ions in the Bre1 cysteine pockets and DNA P-atoms, and subjected to 30,000 steps of MD with a 0.5-fs time step and subsequent 250,000 steps of MD with a 1-fs time step. A twin-range (10/14 Å) spherical cut-off function was used to truncate the van der Waals interactions. Electrostatic interactions were treated using the particle-mesh Ewald summation (real space cut-off 10 and 1.2-Å grid with fourth-order spline interpolation). MD simulations were carried out using 3D periodic boundary conditions, in the isothermal−isobaric (NPT) ensemble with an isotropic pressure of 1.013 bar and a constant temperature of 310 K. The pressure and temperature were controlled using a Nose-Hoover thermostat (43) and a Parrinello–Rahman barostat (44) with 0.5- and 10-ps relaxation parameters, respectively, and a compressibility of 4.5 × 10−5 bar−1 for the barostat. Bond lengths were constrained using LINCS (45).
Supplementary Material
Acknowledgments
We thank D. Finley and G. Warren for comments on the manuscript, and E. Turco and A. Kahraman for help in the early stages of the project. L.D.G. is recipient of a DOC Fellowship of the Austrian Academy of Sciences; B.Z. is funded by European Research Council (ERC) Grant 279408, PROTINT; N.Z. is funded by the Howard Hughes Medical Institute and National Institutes of Health Grant R01-CA107134; Institute of Molecular Pathology is funded by Boehringer Ingelheim; F.H. is supported by ERC Grant MolStruKT StG 638218 and by the German Research Foundation (GRK 1721); and A.K. is funded by ERC Grant 281354, NPC GENEXPRESS, START Grant FWF, Y557-B11 from the Austrian Science Fund, and Doctoral Program RNA Biology W1207-B09.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606863113/-/DCSupplemental.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14(11):1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGinty RK, Tan S. Nucleosome structure and function. Chem Rev. 2015;115(6):2255–2273. doi: 10.1021/cr500373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood A, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11(1):267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 5.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287(5452):501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 6.Kim J, et al. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137(3):459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J, Roeder RG. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J Biol Chem. 2009;284(31):20582–20592. doi: 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weake VM, Workman JL. Histone ubiquitination: Triggering gene activity. Mol Cell. 2008;29(6):653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 9.Trujillo KM, Osley MA. A role for H2B ubiquitylation in DNA replication. Mol Cell. 2012;48(5):734–746. doi: 10.1016/j.molcel.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moyal L, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell. 2011;41(5):529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Latham JA, Chosed RJ, Wang S, Dent SY. Chromatin signaling to kinetochores: Transregulation of Dam1 methylation by histone H2B ubiquitination. Cell. 2011;146(5):709–719. doi: 10.1016/j.cell.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vitaliano-Prunier A, et al. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol. 2008;10(11):1365–1371. doi: 10.1038/ncb1796. [DOI] [PubMed] [Google Scholar]
- 13.Lee JS, et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131(6):1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 14.Köhler A, Zimmerman E, Schneider M, Hurt E, Zheng N. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell. 2010;141(4):606–617. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samara NL, et al. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science. 2010;328(5981):1025–1029. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole AJ, Clifton-Bligh R, Marsh DJ. Histone H2B monoubiquitination: Roles to play in human malignancy. Endocr Relat Cancer. 2015;22(1):T19–T33. doi: 10.1530/ERC-14-0185. [DOI] [PubMed] [Google Scholar]
- 17.Deshaies RJ, Joazeiro CAP. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 18.Plechanovová A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489(7414):115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turco E, Gallego LD, Schneider M, Köhler A. Monoubiquitination of histone H2B is intrinsic to the Bre1 RING domain-Rad6 interaction and augmented by a second Rad6-binding site on Bre1. J Biol Chem. 2015;290(9):5298–5310. doi: 10.1074/jbc.M114.626788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.West MHP, Bonner WM. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 1980;8(20):4671–4680. doi: 10.1093/nar/8.20.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattiroli F, Sixma TK. Lysine-targeting specificity in ubiquitin and ubiquitin-like modification pathways. Nat Struct Mol Biol. 2014;21(4):308–316. doi: 10.1038/nsmb.2792. [DOI] [PubMed] [Google Scholar]
- 22.Nakanishi S, et al. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol. 2008;15(8):881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cucinotta CE, Young AN, Klucevsek KM, Arndt KM. The nucleosome acidic patch regulates the H2B K123 monoubiquitylation cascade and transcription elongation in Saccharomyces cerevisiae. PLoS Genet. 2015;11(8):e1005420. doi: 10.1371/journal.pgen.1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar P, Wolberger C. Structure of the yeast Bre1 RING domain. Proteins. 2015;83(6):1185–1190. doi: 10.1002/prot.24812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbera AJ, et al. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science. 2006;311(5762):856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 26.Worthylake DK, Prakash S, Prakash L, Hill CP. Crystal structure of the Saccharomyces cerevisiae ubiquitin-conjugating enzyme Rad6 at 2.6 A resolution. J Biol Chem. 1998;273(11):6271–6276. doi: 10.1074/jbc.273.11.6271. [DOI] [PubMed] [Google Scholar]
- 27.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102(4):533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 28.Merkley ED, et al. Distance restraints from crosslinking mass spectrometry: Mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 2014;23(6):747–759. doi: 10.1002/pro.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruneda JN, Stoll KE, Bolton LJ, Brzovic PS, Klevit RE. Ubiquitin in motion: Structural studies of the ubiquitin-conjugating enzyme∼ubiquitin conjugate. Biochemistry. 2011;50(10):1624–1633. doi: 10.1021/bi101913m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan MT, et al. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science. 2016;351(6274):725–728. doi: 10.1126/science.aac5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514(7524):591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piro AS, Mayekar MK, Warner MH, Davis CP, Arndt KM. Small region of Rtf1 protein can substitute for complete Paf1 complex in facilitating global histone H2B ubiquitylation in yeast. Proc Natl Acad Sci USA. 2012;109(27):10837–10842. doi: 10.1073/pnas.1116994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyce A, et al. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell. 2007;27(2):275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 34.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16(3):167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuchs G, Hollander D, Voichek Y, Ast G, Oren M. Cotranscriptional histone H2B monoubiquitylation is tightly coupled with RNA polymerase II elongation rate. Genome Res. 2014;24(10):1572–1583. doi: 10.1101/gr.176487.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herzog F, et al. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337(6100):1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 37.Walzthoeni T, et al. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat Methods. 2012;9(9):901–903. doi: 10.1038/nmeth.2103. [DOI] [PubMed] [Google Scholar]
- 38.Grimm M, Zimniak T, Kahraman A, Herzog F. xVis: A web server for the schematic visualization and interpretation of crosslink-derived spatial restraints. Nucleic Acids Res. 2015;43(W1):W362–W369. doi: 10.1093/nar/gkv463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dolinsky TJ, et al. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35(Web Server):W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pronk S, et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jorgensen WL, Madura JD. Solvation and conformation of methanol in water. Am Chem Soc. 1983;105(6):1407–1413. [Google Scholar]
- 43.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys Rev A Gen Phys. 1985;31(3):1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 44.Parrinello M. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys. 1981;52(12):7182–7190. [Google Scholar]
- 45.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18(12):1463–1472. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.