Abstract
Prostate-associated gene 4 (PAGE4) is a remarkably prostate-specific Cancer/Testis Antigen that is highly upregulated in the human fetal prostate and its diseased states but not in the adult normal gland. PAGE4 is an intrinsically disordered protein (IDP) that functions as a stress-response protein to suppress reactive oxygen species as well as prevent DNA damage. In addition, PAGE4 is also a transcriptional regulator that potentiates transactivation by the oncogene c-Jun. c-Jun forms the AP-1 complex by heterodimerizing with members of the Fos family and plays an important role in the development and pathology of the prostate gland, underscoring the importance of the PAGE4/c-Jun interaction. HIPK1, also a component of the stress-response pathway, phosphorylates PAGE4 at T51 which is critical for its transcriptional activity. Phosphorylation induces conformational and dynamic switching in the PAGE4 ensemble leading to a new cellular function. Finally, bioinformatics evidence suggests that the PAGE4 mRNA could be alternatively spliced resulting in four potential isoforms of the polypeptide alluding to the possibility of a range of conformational ensembles with latent functions. Considered together, the data suggest that PAGE4 may represent the first molecular link between stress and prostate cancer (PCa). Thus, pharmacologically targeting PAGE4 may be a novel opportunity for treating and managing patients with PCa, especially patients with low-risk disease.
Keywords: prostate-associated Gene 4, cancer/testis antigen, intrinsically disordered protein, prostate cancer, c-Jun, AP-1, protein interaction networks, homeodomain-interacting protein 1
INTRODUCTION
In this article, in the Special Issue on Intrinsically Disordered Proteins and Prostate Cancer, we review the cumulative progress in research on Prostate-associated Gene 4 (PAGE4), a remarkably prostate-specific tumor antigen that is upregulated in prostate cancer (PCa). We begin with the biology of PAGE4 and review data regarding its expression at the mRNA as well as protein level that allude to its potentially pleotropic roles. These data indicate the following main points. First, while PAGE4 is upregulated in the developing prostate1,2 but is undetectable in the normal adult gland,1,3 it is aberrantly expressed in the diseased gland.1,3,4,5 Second, PAGE4 is upregulated in prostatic lesions that are infiltrated with inflammatory cells1,5 and are believed to represent precursors of frank PCa.6 Third, although mostly cytoplasmic,1,5,7,8 the protein is also detected in the nucleus5,8 and mitochondrial fraction.1 Finally, PAGE4 is a stress-response protein that is upregulated in response to a variety of stress factors including inflammatory stress.1
We then discuss the biochemistry of PAGE4. Biochemical data9 demonstrate that PAGE4 is a highly intrinsically disordered protein (IDP) that lacks a rigid 3D structure and exists as a conformational ensemble instead. In light of the salient features of IDPs and the important roles they play in various cellular processes such as signaling and transcriptional regulation by occupying hub positions in the cell's protein interaction network (PIN) (see article by Landau et al. in the Special Issue),10,11,12 we highlight the role of PAGE4 as a potentiator of the oncogene c-Jun reinforcing its nuclear localization.2 Next, we discuss how this activity is regulated by site-specific phosphorylation by Homeodomain-Interacting Protein Kinase 1 (HIPK1), also a component of the cell's stress-response pathway.8
Finally, we discuss the biophysics of PAGE4 summarizing data that were obtained employing single molecule Fluorescence Resonance Energy Transfer (smFRET) microscopy and multidimensional Nuclear Magnetic Resonance (NMR) spectroscopy. The smFRET data revealed that nonphosphorylated PAGE4 interacts with c-Jun2 but phosphorylation attenuates this interaction.8 NMR experiments helped elucidate, at the single-residue level, how site-specific phosphorylation of PAGE4 leads to shifts in the conformational ensemble with large functional consequences.13 Finally, we point to alternative splicing events that can result in four different isoforms of the PAGE4 protein with potentially latent functions. We conclude by emphasizing that this endeavor serves as a paradigm for discerning the role of IDPs in cancer given that ~80% of the proteins dysregulated in the disease are IDPs.14
THE BIOLOGY OF PAGE4
PAGE4 is a cancer/testis antigen (CTA)
The CTAs are a heterogeneous group of tumor antigens that are typically restricted to the testicular germ cells, remain silent in most somatic tissues, but are aberrantly expressed in several types of cancer.15,16 To date, more than 250 CTAs have been reported in the CT Database17,18 that is broadly divided into two groups, namely, the CT-X antigens located on the X chromosome and the non-X CT antigens located on the autosomes. While the non-X CT antigens are well conserved evolutionarily, the CT-X antigens, with the possible exception of the MAGE family, appear to lack orthologs in lower vertebrates beyond primates,18,19 suggesting that these proteins have evolved relatively recently. The CT-X antigens not only constitute ~50% of all CTAs but also are significantly more testis-restricted than the non-X CT antigens.20 However, because of the lack of phylogenetic conservation, the functions of most, if not all CT-X antigens, both in gametogenesis and tumorigenesis remain poorly understood.16,21,22 Furthermore, no activating mutations have been observed for the CTAs in any cancer thus far. Nonetheless, an overwhelming majority of the CTAs in general, and the CT-X antigens in particular (including PAGE4), are predicted to be IDPs23 alluding to their potential roles in signaling and transcriptional regulation (see article by Landau et al. 2016 in this Issue).
PAGE4 appears to be a proto-oncogene
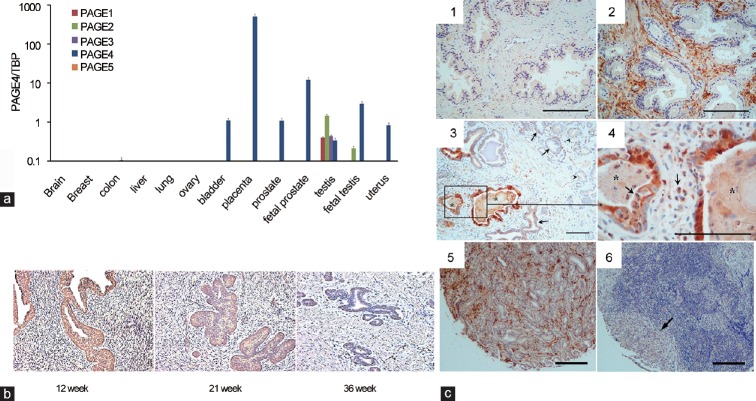
The PAGE family is a subgroup of the CT-X antigens and consists of five members (PAGE1-5) that share significant sequence homology. Although all PAGE genes are expressed in the testes of the adult human, PAGE4 is the only member of this family that is expressed in the prostate1 (Figure 1a). Furthermore, as shown in Figure 1a, PAGE4 expression is remarkably prostate-specific in human male yet highly dynamic.1,3 Immunohistochemistry data using a PAGE4-specific antibody generated against a synthetic peptide corresponding to amino acids 44–60 revealed that PAGE4 is highly upregulated within the human prostate gland. Upregulation occurs both spatially and temporally in the prostatic epithelium and stromal cells of the fetal prostate up to 21 weeks but is undetectable by term (36 weeks) (Figure 1b). In contrast, PAGE4 is undetectable in the normal adult prostate at the level of sensitivity afforded by this specific antibody1,3 (Figure 1c1). However, in the malignant prostate, the PAGE4 protein is highly expressed in the epithelial cells in Proliferative Inflammatory Atrophy (PIA) lesions1 (Figure 1c3 and 1c4) and in high-grade Prostatic Intraepithelial Neoplasia (PIN) lesions,5 both of which are thought to be precursors of frank PCa.6 Furthermore, while PAGE4 protein is upregulated in frank PCa (Figure 1c5), metastatic PCa specimens showed no reaction to the PAGE4 antibody indicating the lack of PAGE4 expression in advanced disease1 (Figure 1c6). However, Sampson et al. reported weak expression of PAGE4 protein in metastatic PCa, using an in-house PAGE4-specific antibody to stain a PCa progression Tissue Microarray.5 Regardless, these data suggest that PAGE4 may actually function as a proto-oncogene that is important in early development but is aberrantly expressed in the diseased prostate. Consistent with this hypothesis, (1) PAGE4 mRNA is significantly upregulated when somatic cells are reprogrammed to form induced pluripotent stem (iPS) cells but is undetectable in the parent somatic cells used in reprogramming (Figure 2a) and (2) while knocking down PAGE4 expression results in cell death in vitro (Figure 2b), its overexpression results in a growth advantage of PCa cells (Figure 2c).9
Figure 1.
PAGE expression at the mRNA and protein levels. (a) Expression of mRNAs encoding PAGE1-5 in different normal human tissue samples obtained from healthy donors. (b) Immunohistochemistry analysis of PAGE4 in the human fetal prostate. Samples were stained for PAGE4 at gestational weeks 12, 21 and 36. (c) Immunohistochemistry analysis of PAGE4 in prostate cancer. (1) Negative staining in the normal prostate. (2) Intense staining shown in the stromal tissue in BPH. (3) Intense staining shown in cancer adjacent “normal” glands (asterisk) associated with inflammation but only moderate staining in the cancer cells (arrowhead). (4) High-power view of boxed area in (3). (5) PAGE4 protein expression in organ-confined prostate cancer. (6) Loss of PAGE4 protein expression in metastatic prostate cancer. Asterisk: PIA lesions; arrows indicate inflammatory cells. Scale bars in all panels: 100 μm. Data presented are reproduced with permission from original publications by the authors.
Figure 2.
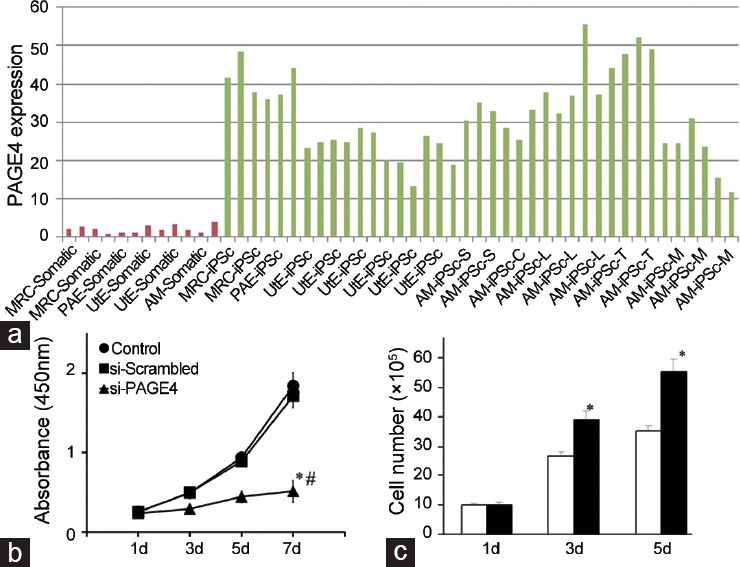
PAGE4 expression stem cells and the effect of silencing and overexpression. (a) PAGE4 mRNA is upregulated in transcription factor-induced pluripotent stem cells (iPS cells) compared to the parental somatic cells. The various somatic cells used are indicated in red bars and the resulting iPS cells in green bars. Data were obtained from GEO (http://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID = GDS3842:15390). (b) Silencing PAGE4 expression inhibits cell survival and enhances chemocytotoxicity in prostate cancer cells. Prostate cancer LNCaP cells were transfected with 50 nmol l−1 PAGE4 SMARTpool siRNA. Cell viability was evaluated by WST-1 assay 3–7 days after transfection. (c) Overexpression of PAGE4 protected cells from stress-induced death. Prostate cancer CWR22rv1 cells were seeded in 6-well plates and transfected with 4 µg of pCMV6-PAGE4-GFP vector (black bar) or empty vector pCMV6-GFP (white bar). The number of living cells was counted using a hemocytometer after excluding dead cells by trypan blue staining 1–5 days after transfection. Data presented in b and c are reproduced with permission from original publications by the authors.
PAGE4 is a stress-response protein
In addition to PCa, the PAGE4 protein is also found to be expressed in benign prostatic hyperplasia (BPH).5 However, while the protein is not expressed in asymptomatic BPH (also referred to as histologic or incidental BPH), it is highly overexpressed in the symptomatic disease (Figure 1c2) which shares similarities with PCa at the molecular level.3 In addition, unlike in the case of PCa, in BPH, PAGE4 protein is predominantly expressed in the stromal and smooth muscle cells.3,5 Considered together, the extensive spatiotemporal and cell-type specific expression data in the fetal and diseased gland suggest that PAGE4 appears to play important roles in both benign symptomatic BPH and malignant disease of the prostate perhaps by modulating the effects of inflammatory stress. The fact that PAGE4 is upregulated in PCa precursor lesions affected by inflammatory stress lends further credence to this argument. Indeed, inflammation appears to play an important role in both BPH3,24,25,26 and PCa.27,28 Current epidemiological data indicate that over 25% of all cancers are related to chronic infections and other types of unresolved inflammation27 and chronic inflammation is now regarded as an “enabling characteristic” of all human cancers including PCa.28
Consistent with its role as a stress-response protein, PAGE4 is upregulated in response to a variety of stress inducers including inflammatory stress. Thus, exposing PCa cells to environmental (drug treatment) or nutrient stress (glucose deprivation), or treating them with the proinflammatory cytokine tumor necrosis factor alpha (TNF-α), results in upregulation of PAGE4 both at the mRNA and protein level.1 Perhaps the most tantalizing evidence supporting PAGE4 as a molecular link between stress and PCa onset comes from the observation that PAGE4 is upregulated in normal prostate epithelial cells (PrEC) when exposed to the same stress inducers.1
PAGE4 suppresses ROS production and protects DNA from damage
Additional evidence supporting its stress-response function stems from the fact that in cells overexpressing PAGE4, levels of the CDK inhibitor p21 that serves as a checkpoint protein involved in DNA damage response were increased compared to control cells.1 The increase was pronounced when cells were subjected to glucose deprivation or Adriamycin treatment although it was not dependent on p53 activation. Furthermore, the DNA damage response marker γ-H2A. X was less activated by the stress stimulants in those cells overexpressing PAGE4, underscoring the protective function of PAGE4. On the other hand, the activation of the cell survival-related signaling molecule phospho-Akt (pAkt) was higher in the PAGE4 overexpressing cells than nonexpressing control cells, indicating that PAGE4 overexpression attenuated the stress-induced damage and enhanced cell survival. However, when these cells were treated with the highly reactive oxygen species (ROS), hydrogen peroxide, the protective effects of PAGE4 overexpression were diminished. This suggests that PAGE4 overexpression is insufficient to inhibit ROS-induced cellular stress once ROS is generated. Finally, cells overexpressing PAGE4 showed an inverse correlation between PAGE4 expression and ROS levels when cultured in medium without glucose supplement. Treating cells with Adriamycin readily induced ROS while this process was inhibited by PAGE4 overexpression.1 All together, these results strongly suggest that one of the mechanisms by which PAGE4 protects stress-induced cellular damage is by inhibiting ROS generation.
PAGE4 translocates to mitochondria upon stress stimulation
PAGE4 has been shown to be a predominantly cytoplasmic protein in prostatic tissue samples as well as in the cell lines that overexpress PAGE4.1,5,7,8 In light of the importance of mitochondria in ROS production and the cellular response to stress, it is imperative to discern whether PAGE4 expression in the cytoplasm is related to mitochondrial function. Thus, Zeng et al. interrogated mitochondrial fractions isolated from CWR22rv1 PCa cells grown with or without glucose supplement for the presence of PAGE4 by immunoblotting. Interestingly, the authors found that ectopically expressed PAGE4 was detected in the mitochondrial fraction although its expression level remained high in the cytosol fraction. More importantly, however, when the cells were cultured in medium without glucose supplement, increased PAGE4 protein was detectable in the mitochondrial fraction with a concomitant decrease in the cytosolic fraction. Consistent with subcellular fractionation results, confocal microscopy revealed that the PAGE4 protein accumulated in the mitochondria when cells were subjected to glucose deprivation, indicating that PAGE4 translocates to the mitochondria in response to stress.1
When considered together, these observations on the biology of PAGE4 indicate that it may represent the first molecular link between stress, especially inflammatory stress, and PCa. The results also suggest that attenuating the stress-response pathway by targeting PAGE4 may be a rational approach to treat PCa. However, additional research will be needed to warrant this strategy. For example, direct evidence linking PAGE4 phosphorylation by HIPK1 in response to stress is lacking. Further, how phosphorylation of PAGE4 affects its intracellular trafficking and subcellular localization is unclear. While bioinformatics predicts a nuclear localization signal supporting its transcriptional regulatory role,9 how PAGE4 associates with the mitochondrion in response to stress and whether it interacts with the membrane by being translocated into the matrix or residing in the intermembrane space is not known. The availability of information regarding the expression of PAGE4 in various prostate cell lines9 should greatly aid in designing experiments to elucidate further the biology of PAGE4.
THE BIOCHEMISTRY OF PAGE4
PAGE4 is a highly intrinsically disordered protein
Despite the data implicating PAGE4 and the dynamic expression patterns that parallel the proliferative growth of the prostate gland during development and disease, the function(s) of PAGE4 remained poorly understood until recently. The first clue regarding its potential function came from examining the amino acid sequence of the PAGE4 polypeptide. Intriguingly, while the sequence is enriched in polar and charged amino acids (~53%), there are few bulky hydrophobic residues such as L, M, F, W, and Y (<10%), suggesting that PAGE4 may be an IDP. Indeed, bioinformatics algorithms predicted strongly that PAGE4 is highly intrinsically disordered.9 In agreement with these predictions, biochemical experiments such as denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analytical size exclusion chromatography revealed that PAGE4 polypeptide behaved anomalously, which is typical of IDPs. In SDS-PAGE, their mobility appears to be retarded because of abnormally low SDS binding to hydrophilic residues. In size exclusion columns, IDPs elute at retention times corresponding to higher molecular weight globular proteins due to their larger Stokes radius. Circular dichroism (CD) and one-dimensional 1H Nuclear Magnetic Resonance (NMR) spectroscopy over a range of temperatures (5–25°C) further confirmed that PAGE4 is an IDP.9 The CD spectra showed that the PAGE4 polypeptide chain contains no significant α-helical or β-strand secondary structural elements over this temperature range as evidenced by low ellipticity values in the 215–230 nm region. NMR spectra displayed a narrow chemical shift dispersion of amide proton signals, with most resonances in the 7.0–8.5 ppm range. Narrow dispersion of peaks was also present in the aliphatic methyl region of the spectrum. Resonances in the amide and methyl regions of folded proteins are more spread out due to ordered packing and hydrogen bonding effects. Thus, the lack of signal dispersion was strongly indicative of a polypeptide chain with little or no defined tertiary structure.9
IDPs and PIN rewiring
Comprehensive analyses of protein interaction networks (PINs) from yeast to humans29 revealed that proteins that occupy hub positions (nodes with multiple interactions) in a PIN are significantly more disordered compared to proteins that constitute edges, highlighting the role of IDPs in signaling.30 Consistent with the preference for IDPs to occupy hub positions, many IDPs rapidly undergo disorder-to-order transitions upon binding to their biological target (coupled folding and binding) to perform their function.31 Thus, conformational dynamics is believed to represent a major functional advantage for the IDPs, enabling them to interact with a broad range of biological targets under normal physiological conditions where their expression is tightly regulated.32,33
Intrinsic disorder also appears to be an important determinant of dosage-sensitive effects. Thus, IDPs are prone to initiate promiscuous interactions when overexpressed, suggesting that this is the likely cause of the resulting toxicity/pathology. Indeed, studies in model organisms provide compelling evidence supporting this causality.34 Interestingly, the same properties are strongly associated with dosage-sensitive oncogenes as well as several other cancer-associated genes,14,23 suggesting that mass action driven molecular interactions may be a frequent cause of the observed pathology.35 In fact, numerous IDPs are also associated with several other human diseases,36,37 highlighting the link between intrinsic protein disorder, promiscuity, and dosage sensitivity.
The structural flexibility of IDPs could also contribute significantly to “noise” in the PINs especially when IDP expression is dysregulated. Indeed, recent evidence indicates that the information transduced in cellular signaling pathways is significantly affected by noise.38 In fact, it has been proposed that noise in these pathways is generated by the interconnected and promiscuous nature of the PINs and that this source of noise significantly influences the way that signals are transmitted. Therefore, noise may be considered as an integral part of the correct transmission of signals and signaling cascades. Mahmoudabadi et al. recently hypothesized that noise due to IDP conformational dynamics can rewire PINs to cause phenotypic switching such as the transformation of a normal cell to a cancer cell. Because noise affects central regulatory switches in cell functions, it is plausible that alterations in noise level could rewire PIN by switching interacting partners by IDPs to induce pathological states such as cancer.39 Although this theoretical framework helps envision the role of IDPs in general, experimental evidence discerning noise caused by the conformational dynamics of PAGE4 and actually identifying partners that interact with it could provide much needed insight on the role of this IDP in PCa.
PAGE4 potentiates c-Jun transcriptional activity
Toward this end, Rajagopalan et al. identified the proto-oncogene c-Jun as a PAGE4 interacting partner using a yeast two-hybrid system.2 Employing a cell-based reporter system, the authors then went on to demonstrate that PAGE4 dramatically potentiates c-Jun transactivation (Figure 3a). While these observations implied a direct interaction between the two proteins, evidence that indicating PAGE4 physically interacts with c-Jun was lacking. Thus, there remained the possibility that PAGE4 interacts indirectly with c-Jun/AP-1 complex by binding to yet another factor in the transcription factor machinery assembled on the promoter. To discriminate between the two possibilities, Rajagopalan et al. employed single-molecule Förster resonance energy transfer (smFRET) microscopy and demonstrated that PAGE4 binds to c-Jun and changes conformations upon binding (see section on the Biophysics of PAGE4 below for details). Taken together, these results suggested that conformational dynamics may underlie the observed pleiotropic functions of PAGE4 during prostatic development and disease.
Figure 3.
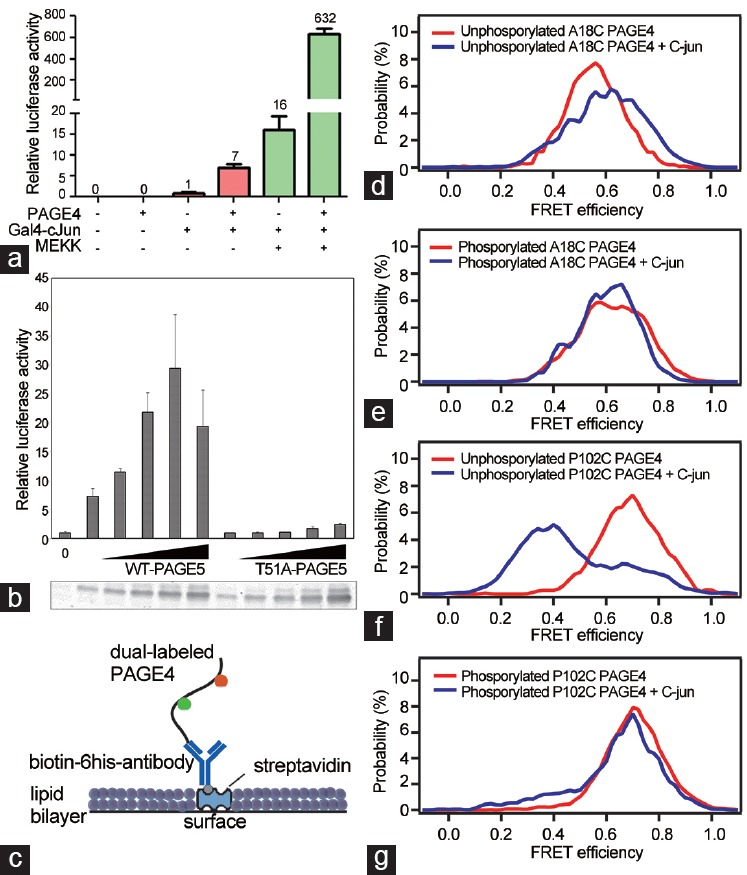
PAGE4 is a powerful potentiator of c-Jun and phosphorylation of PAGE4 is critical for its activity. (a) PAGE4 potentiates c-Jun transcriptional activity in a cell-based assay. Luciferase is used as the reporter. MEKK represents MAP kinase. (b) PAGE4/c-Jun transactivation requires T51. Top panel, luciferase assay of nV5-PAGE4 WT/T51A with GAL4-c-Jun1−231 in PC3 cells. Lower panel shows a representative V5-PAGE4 western blot of the respective wells in the luciferase assay. FRET from phosphorylated PAGE4 is minimally altered by exposure to c-Jun. (c) Cartoon showing how single PAGE4 molecules were tethered directly to a surface bound antibody to the 6His tag. Gaussian fits of the FRET histograms show c-Jun causes larger FRET changes in nonphosphorylated PAGE4 than kinase-treated PAGE4. Details of the fits are as follows. For the A18C nonphosphorylated (d) without c-Jun, FRET = 0.56 (width 0.15), and with c-Jun, 65% molecules are FRET = 0.56 (width 0.15) while 35% are FRET = 0.72 (width 0.12). For A18C kinase-treated PAGE4 (e), without c-Jun, 58% have FRET = 0.56 (width 0.15), while 42% have FRET = 0.72 (width 0.13), whereas with c-Jun, fits used only single peak of FRET = 0.61 (width 0.16). FRET was 0.69 (width 0.16) for P102C PAGE4 nonphosphorylated (f) without c-Jun but dramatically changed to a double Gaussian with 70% having FRET 0.37 (width 0.16) and 30% having FRET 0.69 (width 0.16) when c-Jun was added. Kinase-treated P102C PAGE4 (g) samples were dominated by a FRET population >80% at FRET 0.71 (width 0.15) both with and without c-Jun. Data presented are reproduced with permission from original publications by the authors.
PAGE4 is phosphorylated at T51 in vivo and this is critical for its transcriptional activity
In general, across both plant and animal kingdoms, IDPs appear to be more prone to alternative splicing and posttranslational modifications (particularly phosphorylation) than are ordered proteins.32,40,41,42 Therefore, Mooney et al. examined the possibility that PAGE4 is also subjected to posttranslational modification by phosphorylation.8 Utilizing mass spectrometry, the authors discovered that PAGE4 isolated from PC3 cells is phosphorylated predominantly at T51. To discern the functional significance of this covalent modification, the authors mutated T51 to an alanine residue (T51A). They observed that the mutant version was not phosphorylated and failed to potentiate c-Jun transactivation in a cell-based reporter assay, indicating that phosphorylation at T51 is critical for this process8 (Figure 3b). To rule out the possibility that the mutant protein is not expressed or is unstable, the cell lysates are probed with the PAGE4 antibody in an immunoblotting experiment. These experiments confirmed that the T51A mutant is indeed expressed and to the same extent as the “wild-type” (WT) (nonphosphorylated) PAGE4 but that the mutant protein is not phosphorylated by the kinase that robustly phosphorylates the WT PAGE4 polypeptide (Figure 3b).8
HIPK1 phosphorylates PAGE4 in vitro primarily at T51
To identify the kinase that phosphorylates PAGE4 in PCa cells, a panel of 190 serine and threonine (S/T) kinases was surveyed by Mooney et al.8 Homeodomain-interacting Protein Kinase 1 (HIPK1), a component of the cell's stress-response pathway, was identified as a kinase that phosphorylates PAGE4 in vitro. Phosphorylation of bacterially produced recombinant PAGE4 was achieved by incubation with commercially available HIPK1 in the presence of 32P-γATP. While the WT recombinant PAGE4 was robustly phosphorylated, the T51A mutant of PAGE4 had only ~10% of the phosphorylation level by comparison, indicating that HIPK1 phosphorylates PAGE4 primarily at T51.
Together, these and other results1 implicated PAGE4 in the HIPK1-JNK1-c-Jun activated stress-response pathway. The amplifying effect on c-Jun transactivation suggested that PAGE4 may modulate the activity of AP-1, a family of early response transcription factors with important roles in the control of cell growth, apoptosis, and stress response.43 Notably, c-Jun is also upregulated in the developing44 and diseased prostate45,46,47,48 while the AP-1 complex is upregulated in symptomatic but not in asymptomatic BPH49 as well as in prostate cancer.50 These patterns are coincident with PAGE4 expression3 and underscore the importance of the PAGE4/c-Jun interactions in prostatic development and disease.
While these data afford the first clue to the function of PAGE4 at least in the prostate, more work is needed to fully comprehend its function(s). For example, PAGE4 may be subjected to additional posttranslational modifications that could potentially remodel the conformational ensemble and thus, uncover a latent function(s). Indeed, preliminary data indicate that PAGE4 is covalently modified by O-GlcNAc-transferase (OGT) that adds O-linked β-N-acetylglucosamine (O-GlcNAc) on to S/T residues (Dr. Steven Mooney, personal communication). O-GlcNAc modification is encountered with nucleo-cytoplasmic proteins involved in nutrient sensing51 and often competes for the same S/T residues recognized by kinases.52 In fact, alternative O-GlcNAcylation/O-Phosphorylation at the same residue can induce different conformations in the same polypeptide53 underscoring the critical role of posttranslational modifications in protein conformational dynamics. In light of the role of PAGE4 in stress-response including nutrient stress, it may be important to discern the effect of this posttranslational modification on its function. In addition, c-Jun (as well as its heterodimeric partners from the Fos family of transcription factors in the AP-1 complex) is intrinsically disordered and subjected to phosphorylation by JNK1 and other kinases.54,55 Therefore, how these modifications affect its interaction with nonphosphorylated PAGE4 and how phosphorylated PAGE4 interacts with the AP-1 complex remain poorly understood.
THE BIOPHYSICS OF PAGE4
Single-molecule fluorescence resonance energy transfer (smFRET) microscopy
Rajagopalan et al. employed smFRET microscopy, a powerful technique used to measure distances at the 1–10 nm scale in single molecules, to demonstrate a direct interaction between nonphosphorylated PAGE4 and c-Jun.2 A unique advantage of smFRET is that it can capture information normally lost through ensemble averaging of heterogeneous and dynamic samples. Furthermore, immobilization of single molecules under conditions that retain their biological activity allows for extended observation of the same molecule for tens of seconds, facilitating the capture of slow conformational transitions or protein binding and unbinding cycles. Finally, the technique permits direct observation of the response to changing solution conditions or adding ligands.56
WT PAGE4 interacts with c-Jun
To discern whether or not PAGE4 physically interacts with c-Jun, Rajagopalan et al.2 generated two different cysteine (Cys) mutants of PAGE4 that were labeled with donor and acceptor fluorophores. Cysteine residues were introduced in the N- or C-terminus of the 102 residue PAGE4 molecule at positions 18 or 102 by replacing an alanine (A18C construct) or proline (P102C construct) residue by site-directed mutagenesis, respectively. These mutants alternately combined with the single native Cys residue at position 63 in the PAGE4 polypeptide to generate two double mutant constructs that could be used to simultaneously label with fluorescent dye resulting in random attachment of donor and acceptor on the cysteines. The constructs were used to express recombinant proteins in E. coli, which were purified and labeled with Alexa Fluor 555 (FRET donor) and Alexa Fluor 647 (FRET acceptor). The labeled proteins were separated from excess unincorporated dye by size exclusion chromatography, and then liposome encapsulated together with unlabeled recombinant c-Jun (residues 1–241). Using this technique, the authors showed that a) nonphosphorylated WT PAGE4 binds to c-Jun and b) upon binding to c-Jun, PAGE4 changes conformation.
Phosphorylation of PAGE4 changes the interaction with c-Jun
To examine the effect of phosphorylation on the conformational dynamics of PAGE4, recombinant PAGE4 coexpressed with HIPK1 was purified and used in smFRET experiments.8 FRET efficiency measured from purified PAGE4 A18C encapsulated inside liposomes that were surface immobilized was increased, which suggested that phosphorylation causes compaction of the N-terminal region of PAGE4. In contrast, essentially no change in FRET was observed in the C-terminal pair of labels in PAGE4 P102C due to phosphorylation.
Direct immobilization of 6-His tagged PAGE4 on a surface coated with 6-His antibodies allowed c-Jun to be added to the solution around the protein and possible conformational changes to be observed (Figure 3c). Without kinase exposure, FRET from PAGE4 A18C increased from 0.56 to 0.72 upon exposure to full-length c-Jun while FRET from PAGE4 P102C decreased from 0.69 to 0.37 in the same conditions. The A18C PAGE4 protein was not well suited for testing the interaction of c-Jun with kinase exposed PAGE4 because individually, both A18C and P102C mutant proteins caused similar shifts in FRET compared to the wild-type protein. Therefore, the authors used P102C-PAGE4 protein to determine the interaction between c-Jun and PAGE4. When exposed to c-Jun, only 17% of kinase-coexpressed P102C PAGE4 had low FRET efficiency similar in value to that of c-Jun interacting with nonphosphorylated PAGE4. In comparison, 70% of the population of nonphosphorylated P102C PAGE4 shifted to low FRET upon exposure to c-Jun (Figure 3d–3g). Thus, the absence of the large change in FRET for the C-terminal PAGE4 fluorophore pair suggested either that phosphorylation weakens the ability of PAGE4 to interact with c-Jun or that if it binds c-Jun, the induced conformational changes were substantially different from those in the case of nonphosphorylated PAGE4.8 Regardless, the smFRET experiments pointed to a conformational switching mechanism upon phosphorylation of PAGE4.
NMR spectroscopy
The structurally and dynamically heterogeneous nature of IDPs makes NMR spectroscopy an ideal tool for their characterization at high resolution. Therefore, to elucidate how phosphorylation perturbs the structural features of PAGE4 and to provide a possible explanation for the attenuation in binding to c-Jun at the single-residue level, He et al.13 employed multidimensional NMR spectroscopy.
PAGE4 becomes more compact upon phosphorylation at T51
The authors first carried out the backbone and side chain assignments for PAGE4. Although PAGE4 is an IDP, NMR analyses using chemical shifts, Nuclear Overhauser Enhancement (NOE), coupling constants, paramagnetic relaxation enhancement (PRE), and dynamics (heteronuclear NOE, R1, R2, R1ρ) data revealed that regions of the polypeptide chain have distinct local- and long-range conformational preferences. These preferences were perturbed by phosphorylation at T51, which increases the population of transient turn-like structures in the central acidic region. The central region thus becomes more compact and more negatively charged upon phosphorylation, resulting in increased long-range contacts to basic sequence motifs near the N- and C-termini of the PAGE4 polypeptide chain as determined by PRE measurements. Dynamics data indicated that chain flexibility is decreased in T51 phosphorylated PAGE4 relative to WT-PAGE4, particularly in the central and N-terminal regions.
Based on these results, He et al.13 used a set of conservative PRE and NOE restraints as inputs for calculating ensemble conformations for both WT-PAGE4 and phosphorylated PAGE4. The resulting models provide a useful framework for visualizing preferred states of the highly flexible ensemble. The WT-PAGE4 on average populate conformations where the highly basic N-terminal motif (RVRSRSRGR) is within 20 Å of the central acidic region (E44, E47, E49, E55, E56, E60, and D62) neighboring C63. Phosphorylation at T51 increases the negative charge in this central acidic region and induces turn-like structures that provide a more compact transient interaction with the N-terminal motif (Figure 4a–4d). Secondary phosphorylation at S9 lowers the net positive charge of the N-terminal basic motif, and this leads to a weakening of the PRE effect. The interaction between these two regions therefore appears to be driven mostly by favorable electrostatic effects. In addition, other interactions between C63 and a C-terminal motif centered on residue N88 contribute to the overall conformational ensemble. These long-range contacts may also be at least partly due to electrostatic interactions of several basic residues in this region (K82, K84, K90, K93, and K95) with the central acidic amino acids. However, phosphorylation at T51 does not appear to affect this interaction as much as the N-terminal motif as judged both by changes in PRE and dynamics data, consistent with the smFRET results by Mooney et al.8
Figure 4.
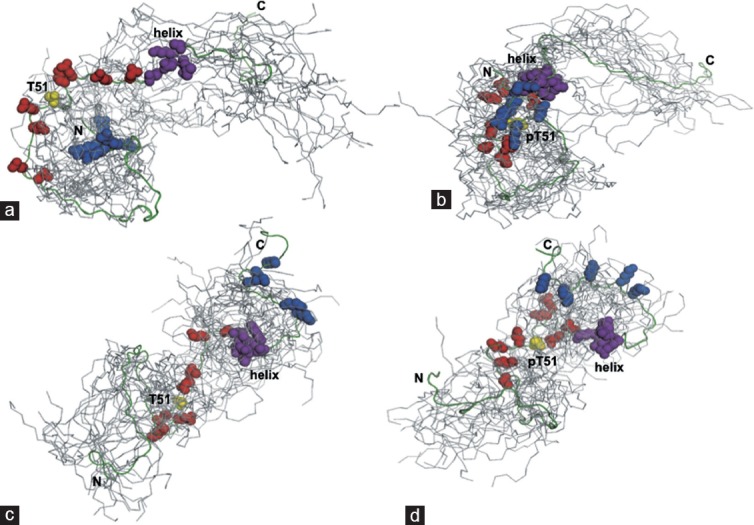
A model for phosphorylation-induced conformational ensemble switching seen in PAGE4. (a) The nonphosphorylated PAGE4 adopts preferred transient structures such as the one highlighted from an ensemble of the 20 lowest energy conformers, where, on average, the N-terminal basic motif (blue spheres: Arg-4, Arg-6, Arg-8, Arg-10, and Arg-12) interacts weakly with the central acidic region (red spheres: Glu-43, Glu-47, Glu-49, Glu-55, Glu-56, Glu-60, and Asp-62) neighboring Thr-51 (yellow). (b) Upon phosphorylation at Thr-51, the central region becomes more compact and more negatively charged, decreasing the average distance between Thr(P)-51, the basic motif, and the transient helix (magenta). (c and d) Models of the transient interaction between the central acidic region and the C-terminal basic motif (blue spheres; Lys-82, Lys-84, Lys-90, Lys-93, and Lys-95) in nonphosphorylated PAGE4 (c) and Thr(P)-51 PAGE4 (d). The total number of distance restraints used was as follows: (a) 51; (b) 55; (c) 53; (d) 61. Data presented are reproduced with permission from original publications by the authors.
Functional studies
NMR-based ligand binding experiments were used to define the PAGE4 binding interface with human c-Jun.13 A series of unlabeled c-Jun fragments, 1–61, 1–90, 1–150, 1–223, and full-length c-Jun, were tested individually with 15N-labeled WT-PAGE4 and phospho-PAGE4. Chemical shift perturbation experiments with the shorter c-Jun fragments, 1–61, 1–90, and 1–150, showed no change in 2D 15N HSQC spectra of WT-PAGE4 with up to 6 equivalents of c-Jun fragment added. In contrast, addition of either unlabeled full-length c-Jun or c-Jun (1–223) perturbed a specific set of WT-PAGE4 residues in a similar way albeit weakly. The largest chemical shift changes occurred in the transient helix region of PAGE4 from residues 69 to 73 (Figure 5a–5c). The binding interaction between WT-PAGE4 and c-Jun was estimated at a KD of >50 μmol l−1 from the NMR titration data, with precipitation of c-Jun at higher concentrations preventing a more accurate determination. Phosphorylation of PAGE4 on T51 significantly attenuates binding at the helical interface with c-Jun, consistent with lower resolution studies using smFRET (18).8
Figure 5.
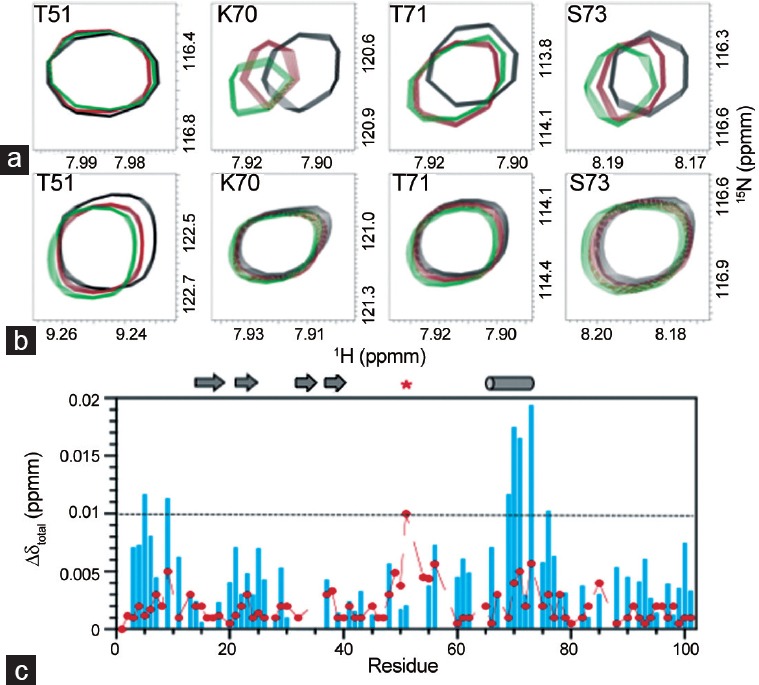
Summary of PAGE4 functional studies. (a) Regions from two-dimensional 1H-15N HSQC spectra in the titration of 15N-labeled WT PAGE4 with unlabeled c-Jun showing superimposed control (black), 1:1.5 PAGE4/c-Jun (red), and 1:3 PAGE4/c-Jun (green) spectra. (b) Similar regions using 15N-labeled Thr(P)-51 PAGE4 with unlabeled c-Jun at the same molar ratios as for a. (c) Chemical shift perturbation plot for WTPAGE4 (blue) and Thr(P)-51 PAGE4 (red) as a function of residue number. Values of δΔtotal below the dashed line are estimated to be within the experimental error for measuring chemical shift changes. Arrows and cylinders represent regions with transient β-strand and α-helical preferences, respectively. The red star indicates the position of phosphorylation. Data presented are reproduced with permission from original publications by the authors.
Since the c-Jun (1–223) fragment, but not shorter constructs, gave similar NMR results to full-length c-Jun, residues, He et al. surmised that 150–223 must be important for the interaction with PAGE4. This region is adjacent to the basic DNA-binding domain of c-Jun, which extends from residues 257 to 276. Consistent with the NMR binding data, results from a luciferase assay in the PC3 prostate cancer cell line with truncated c-Jun fragments demonstrated that PAGE4-mediated amplification of c-Jun transactivation was negligible and required inclusion of the region from 150 to 223. An additional mutant of PAGE4, lacking most of the N-terminal basic motif from residues 5 to 11, had a small (~3-fold) but significant increase in c-Jun transactivation. The N-terminal motif therefore appears to have a partial auto-inhibitory effect on the cellular function of PAGE4, possibly through its long-range interactions with the central region.13
ALTERNATIVE SPLICING
According to AceView,57 the PAGE4 gene contains 4 exons, and RNAseq experiments have identified PAGE4 transcripts that could code for PAGE4 isoforms having 102 amino acids, which corresponds to the full-length protein, as well as additional mRNAs that likely arose from alternative splicing (AS) encoding isoforms with 93, 44, and 43 amino acids (for PAGE4's AceView entry, see www.ncbi.nlm.nih.gov/gene/9506). However, so far, to the best of our knowledge, there is no experimental evidence demonstrating the existence of PAGE4 isoforms resulting from AS. However, as discussed below, there are several reasons for making efforts to determine whether the indicated AS isoforms or other PAGE4 AS isoforms do indeed exist, and if so, to determine the functions of these AS isoforms and whether they contribute to the development of PCa.
In general, increasing amounts of AS isoforms are associated with increasing organism complexity, suggesting that AS isoforms likely play important roles in the development of complexity among the multicellular eukaryotes.58 Also, using deep sequencing to estimate AS complexity suggests that approximately 95% of human multiexon genes undergo alternative splicing,59 but it is unclear what fraction of these AS events actually leads to different protein isoforms, so this point needs to be tested experimentally for each different and potentially important RNA transcript indicated to exist by RNAseq or EST experiments.
At the protein translation level, pre-mRNA segments that are observed to undergo AS are found to code for IDP regions much more often than for structured regions and these AS-determined IDP regions often contain sites for binding to nucleic acids or to other proteins, so these AS events often lead to the “rewiring” of the networks involving genes or protein-protein interactions.60,61 Interestingly, studies of tissue-specific AS indicate that this rewiring or remodeling of protein-protein interaction networks is likely an important factor leading to different functional pathways in different types of cells,62,63,64 which in turn is important for the different biochemistry that occurs in the different types of cells in multicellular organisms.61
Although this manuscript deals with PAGE4 and the functional alterations that arise from phosphorylation, which is a particularly important type of posttranslational modification (PTM), we would like to point out that for other important proteins, such as p53,65 Bcl-2,66 and many others,61 their biological impact arises from functional IDP regions that are modulated in their activities by both AS and PTMs. Furthermore, bioinformatics studies have suggested that proteins involved with cellular differentiation are especially enriched in IDP regions.67 Combining these observations has led to the proposal that synergy among IDP, PTM, and AS was crucial for both the evolution of multicellular organisms61 and thus also for the gene regulatory networks that underlie the development of complex multicellular organisms.68 For these reasons, we believe that it would be worthwhile to investigate whether PAGE4 contains isoforms that arise from AS, and if so, whether the modulation of PAGE4 function by PTMs (phosphorylation or other modifications) and also by AS combine to contribute to the development of the cellular alterations that lead to cancer.
CONCLUSIONS
PAGE4 is one of the few PCa-related IDPs, and the only CTA overexpressed in PCa, that has been studied in considerable detail. Progress in research over the last decade has uncovered this IDP as an important player in prostatic development and disease. The cumulative data suggest that PAGE4 may represent the first molecular link between stress and PCa. In addition, experimental evidence supports the idea that the pleotropic functions of PAGE4 result from its interactions with different partners. Such interactions appear to be facilitated by the conformational plasticity of the PAGE4 protein(s), which may also uncover novel functionalities latent in the PAGE4 ensemble expanding its functional utility through new binding interactions. For example, phosphorylation of PAGE4 leads to conformational shifts in the dynamic ensemble with large functional consequences. Such changes in the structural ensemble are similar conceptually to the conformational switching events seen in some marginally stable (“metamorphic”) folded proteins in response to mutational or environmental triggers.69
In summary, PAGE4 is a prototypical IDP involved in PCa that exploits its conformational flexibility for functional gain. While questions remain, much progress has been made in understanding PAGE4 structure and molecular function. As such, PAGE4 should be given further consideration as a novel therapeutic target for treating and managing PCa.
COMPETING FINANCIAL INTERESTS
The authors declared that they had no competing financial interests.
ACKNOWLEDGMENTS
Research in the PK and JO laboratories was supported by the National Institutes of Health Grant CA181730. PK Would like to thank Dr. Steve Mooney for sharing unpublished information on the posttranslation modifications of PAGE4 and Dr. Mooney and Ms. Krithika Rajagopalan for helpful discussions.
REFERENCES
- 1.Zeng Y, Gao D, Kim JJ, Shiraishi T, Terada N, et al. Prostate-associated gene 4 (PAGE4) protects cells against stress by elevating p21 and suppressing reactive oxygen species production. Am J Clin Exp Urol. 2013;1:39–52. [PMC free article] [PubMed] [Google Scholar]
- 2.Rajagopalan K, Qiu R, Mooney SM, Rao S, Shiraishi T, et al. The stress-response protein prostate-associated gene 4, interacts with c-Jun and potentiates its transactivation. Biochim Bophys Acta. 2014;1842:154–63. doi: 10.1016/j.bbadis.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prakash K, Pirozzi G, Elashoff M, Munger W, Waga I, et al. Symptomatic and asymptomatic benign prostatic hyperplasia: molecular differentiation by using microarrays. Proc Natl Acad Sci U S A. 2002;99:7598–603. doi: 10.1073/pnas.112191399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinkmann U, Vasmatzis G, Lee B, Yerushalmi N, Essand M, et al. PAGE-1, an X chromosome-linked GAGE-like gene that is expressed in normal and neoplastic prostate, testis, and uterus. Proc Natl Acad Sci U S A. 1998;95:10757–62. doi: 10.1073/pnas.95.18.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sampson N, Ruiz C, Zenzmaier C, Bubendorf L, Berger P. PAGE4 positivity is associated with attenuated AR signaling and predicts patient survival in hormone-naive prostate cancer. Am J Pathol. 2012;181:1443–54. doi: 10.1016/j.ajpath.2012.06.040. [DOI] [PubMed] [Google Scholar]
- 6.Nelson WG, De Marzo AM, Isaacs WB. Prostate cancer. N Engl J Med. 2003;349:366–81. doi: 10.1056/NEJMra021562. [DOI] [PubMed] [Google Scholar]
- 7.Iavarone C, Wolfgang C, Kumar V, Duray P, Willingham M, et al. PAGE4 is a cytoplasmic protein that is expressed in normal prostate and in prostate cancers. Mol Cancer Ther. 2002;1:329–35. [PubMed] [Google Scholar]
- 8.Mooney SM, Qiu R, Kim JJ, Sacho EJ, Rajagopalan K, et al. Cancer/testis antigen PAGE4, a regulator of c-Jun transactivation, is phosphorylated by homeodomain-interacting protein kinase 1, a component of the stress-response pathway. Biochemistry. 2014;53:1670–9. doi: 10.1021/bi500013w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng Y, He Y, Yang F, Mooney SM, Getzenberg RH, et al. The cancer/testis antigen prostate-associated gene 4 (PAGE4) is a highly intrinsically disordered protein. J Biol Chem. 2011;286:13985–94. doi: 10.1074/jbc.M110.210765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uversky VN. Unusual biophysics of intrinsically disordered proteins. Biochim Biophys Acta. 2013;1834:932–51. doi: 10.1016/j.bbapap.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–84. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 12.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y, Chen Y, Mooney SM, Rajagopalan K, Bhargava A, et al. Phosphorylation-induced conformational ensemble switching in an intrinsically disordered cancer/testis antigen. J Biol Chem. 2015;290:25090–102. doi: 10.1074/jbc.M115.658583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–84. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 15.Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 16.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–25. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 17.Almeida LG, Sakabe NJ, deOliveira AR, Silva MC, Mundstein AS, et al. CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 2009;37:D816–9. doi: 10.1093/nar/gkn673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobrynin P, Matyunina E, Malov SV, Kozlov AP. The novelty of human cancer/testis antigen encoding genes in evolution. Int J Genomics 2013. 2013:105108. doi: 10.1155/2013/105108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevenson BJ, Iseli C, Panji S, Zahn-Zabal M, Hide W, et al. Rapid evolution of cancer/testis genes on the X chromosome. BMC Genomics. 2007;8:129–39. doi: 10.1186/1471-2164-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann O, Caballero OL, Stevenson BJ, Chen YT, Cohen T, et al. Genome-wide analysis of cancer/testis gene expression. Proc Natl Acad Sci U S A. 2008;105:20422–7. doi: 10.1073/pnas.0810777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalejs M, Erenpreisa J. Cancer/testis antigens and gametogenesis: a review and “brain-storming” session. Cancer Cell Int. 2005;5:4–14. doi: 10.1186/1475-2867-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitehurst AW. Cause and consequence of cancer/testis antigen activation in cancer. Annu Rev Pharmacol Toxicol. 2014;54:251–72. doi: 10.1146/annurev-pharmtox-011112-140326. [DOI] [PubMed] [Google Scholar]
- 23.Rajagopalan K, Mooney SM, Parekh N, Getzenberg RH, Kulkarni P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J Cell Biochem. 2011;112:3256–67. doi: 10.1002/jcb.23252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anim JT, Udo C, John B. Characterisation of inflammatory cells in benign prostatic hyperplasia. Acta Histochem. 1998;100:439–49. doi: 10.1016/S0065-1281(98)80040-8. [DOI] [PubMed] [Google Scholar]
- 25.Elsasser-Beile U, Przytulski B, Gierschner D, Grussenmeyer T, Katzenwadel A, et al. Comparison of the activation status of tumor infiltrating and peripheral lymphocytes of patients with adenocarcinomas and benign hyperplasia of the prostate. Prostate. 2000;45:1–7. doi: 10.1002/1097-0045(20000915)45:1<1::aid-pros1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 26.Robert G, Salagierski M, Schalken JA, de La Taille A. Inflammation and benign prostatic hyperplasia: cause or consequence? Prog Urol. 2010;20:402–7. doi: 10.1016/j.purol.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Vendramini-Costa DB, Carvalho JE. Molecular link mechanisms between inflammation and cancer. Curr Pharm Des. 2012;18:3831–52. doi: 10.2174/138161212802083707. [DOI] [PubMed] [Google Scholar]
- 28.Sfanos KS, De Marzo AM. Prostate cancer and inflammation: the evidence. Histopathology. 2012;60:199–215. doi: 10.1111/j.1365-2559.2011.04033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patil A, Kinoshita K, Nakamura H. Hub promiscuity in protein-protein interaction networks. Int J Mol Sci. 2010;11:1930–43. doi: 10.3390/ijms11041930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gsponer J, Babu MM. The rules of disorder or why disorder rules. Prog Biophys Mol Biol. 2009;99:94–103. doi: 10.1016/j.pbiomolbio.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–75. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 32.Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science. 2008;322:1365–8. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards YJ, Lobley AE, Pentony MM, Jones DT. Insights into the regulation of intrinsically disordered proteins in the human proteome by analyzing sequence and gene expression data. Genome Biol. 2009;10:R50. doi: 10.1186/gb-2009-10-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 35.Marcotte EM, Tsechansky M. Disorder, promiscuity, and toxic partnerships. Cell. 2009;138:16–8. doi: 10.1016/j.cell.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–46. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 37.Uversky VN. Wrecked regulation of intrinsically disordered proteins in diseases: pathogenicity of deregulated regulators. Front Mol Biosci. 2014;1:6–29. doi: 10.3389/fmolb.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ladbury JE, Arold ST. Noise in cellular signaling pathways: causes and effects. Trends Biochem Sci. 2012;37:173–8. doi: 10.1016/j.tibs.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahmoudabadi G, Rajagopalan K, Getzenberg RH, Hannenhalli S, Rangarajan G, et al. Intrinsically disordered proteins and conformational noise: implications in cancer. Cell Cycle. 2013;12:26–31. doi: 10.4161/cc.23178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–49. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collins MO, Yu L, Campuzano I, Grant SG, Choudhary JS. Phosphoproteomic analysis of the mouse brain cytosol reveals a predominance of protein phosphorylation in regions of intrinsic sequence disorder. Mol Cell Proteomics. 2008;7:1331–48. doi: 10.1074/mcp.M700564-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114:6589–631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leppa S, Bohmann D. Diverse functions of JNK signaling and c-Jun in stress response and apoptosis. Oncogene. 1999;18:6158–62. doi: 10.1038/sj.onc.1203173. [DOI] [PubMed] [Google Scholar]
- 44.Li W, Wu CL, Febbo PG, Olumi AF. Stromally expressed c-Jun regulates proliferation of prostate epithelial cells. Am J Pathol. 2007;171:1189–98. doi: 10.2353/ajpath.2007.070285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato N, Sadar MD, Bruchovsky N, Saatcioglu F, Rennie PS, et al. Androgenic induction of prostate-specific antigen gene is repressed by protein-protein interaction between the androgen receptor and AP-1/c-Jun in the human prostate cancer cell line LNCaP. J Biol Chem. 1997;272:17485–94. doi: 10.1074/jbc.272.28.17485. [DOI] [PubMed] [Google Scholar]
- 46.Edwards J, Krishna NS, Mukherjee R, Bartlett JM. The role of c-Jun and c-Fos expression in androgen-independent prostate cancer. J Pathol. 2004;204:153–8. doi: 10.1002/path.1605. [DOI] [PubMed] [Google Scholar]
- 47.Cai C, Hsieh CL, Shemshedini L. c-Jun has multiple enhancing activities in the novel cross talk between the androgen receptor and Ets variant gene 1 in prostate cancer. Mol Cancer Res. 2007;5:725–35. doi: 10.1158/1541-7786.MCR-06-0430. [DOI] [PubMed] [Google Scholar]
- 48.Kajanne R, Miettinen P, Tenhunen M, Leppa S. Transcription factor AP-1 promotes growth and radioresistance in prostate cancer cells. Int J Oncol. 2009;35:1175–82. doi: 10.3892/ijo_00000434. [DOI] [PubMed] [Google Scholar]
- 49.Lin-Tsai O, Clark PE, Miller NL, Fowke JH, Hameed O, et al. Surgical intervention for symptomatic benign prostatic hyperplasia is correlated with expression of the AP-1 transcription factor network. Prostate. 2014;74:669–79. doi: 10.1002/pros.22785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ouyang X, Jessen WJ, Al-Ahmadie H, Serio AM, Lin Y, et al. Activator protein-1 transcription factors are associated with progression and recurrence of prostate cancer. Cancer Res. 2008;68:2132–44. doi: 10.1158/0008-5472.CAN-07-6055. [DOI] [PubMed] [Google Scholar]
- 51.Zachara NE, Hart GW. O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim Biophys Acta. 2004;1673:13–28. doi: 10.1016/j.bbagen.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 52.Golks A, Guerini D. The O-linked N-acetylglucosamine modification in cellular signalling and the immune system. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:748–53. doi: 10.1038/embor.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen YX, Du JT, Zhou LX, Liu XH, Zhao YF, et al. Alternative O-GlcNAcylation/O-phosphorylation of Ser16 induce different conformational disturbances to the N terminus of murine estrogen receptor beta. Chem Biol. 2006;13:937–44. doi: 10.1016/j.chembiol.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 54.Minden A, Lin A, Smeal T, Derijard B, Cobb M, et al. c-Jun N-terminal phosphorylation correlates with activation of the JNK subgroup but not the ERK subgroup of mitogen-activated protein kinases. Mol Cell Biol. 1994;14:6683–8. doi: 10.1128/mcb.14.10.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whitmarsh AJ, Davis RJ. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J Mol Med. 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- 56.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–16. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl 1):S121, 14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen L, Bush SJ, Tovar-Corona JM, Castillo-Morales A, Urrutia AO. Correcting for differential transcript coverage reveals a strong relationship between alternative splicing and organism complexity. Mol Biol Evol. 2014;31:1402–13. doi: 10.1093/molbev/msu083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 60.Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci U S A. 2006;103:8390–5. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dunker AK, Bondos SE, Huang F, Oldfield CJ. Intrinsically disordered proteins and multicellular organisms. Semin Cell Dev Biol. 2015;37:44–55. doi: 10.1016/j.semcdb.2014.09.025. [DOI] [PubMed] [Google Scholar]
- 62.Buljan M, Chalancon G, Eustermann S, Wagner GP, Fuxreiter M, et al. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol Cell. 2012;46:871–83. doi: 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buljan M, Chalancon G, Dunker AK, Bateman A, Balaji S, et al. Alternative splicing of intrinsically disordered regions and rewiring of protein interactions. Curr Opin Struct Biol. 2013;23:443–50. doi: 10.1016/j.sbi.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 64.Ellis JD, Barrios-Rodiles M, Colak R, Irimia M, Kim T, et al. Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol Cell. 2012;46:884–92. doi: 10.1016/j.molcel.2012.05.037. [DOI] [PubMed] [Google Scholar]
- 65.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–64. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 66.Rautureau GJ, Day CL, Hinds MG. Intrinsically disordered proteins in bcl-2 regulated apoptosis. Int J Mol Sci. 2010;11:1808–24. doi: 10.3390/ijms11041808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, et al. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res. 2007;6:1882–98. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niklas KJ, Bondos SE, Dunker AK, Newman SA. Rethinking gene regulatory networks in light of alternative splicing, intrinsically disordered protein domains, and post-translational modifications. Front Cell Dev Biol. 2015;3:1–13. doi: 10.3389/fcell.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bryan PN, Orban J. Proteins that switch folds. Curr Opin Struct Biol. 2010;20:482–8. doi: 10.1016/j.sbi.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]