Significance
Reservoirs of HIV-infected cells persist during antiretroviral therapy, and understanding persistence is essential to develop HIV curative strategies. During replication, HIV integrates into the host genome; most proviruses are not infectious, but some with replication-competent HIV persist. Cells with integrated HIV can proliferate, potentially expanding the reservoir, but whether cells with replication-competent HIV actually undergo expansion is unknown. HIV reactivation is often lethal to infected cells, and others have reported finding no replication-competent HIV in expanded populations. We describe a highly expanded clone containing infectious HIV that was the source of viremia for years in a patient. Clonally expanded populations can represent a long-lived reservoir of HIV. Curative strategies will require targeting this persistence mechanism.
Keywords: HIV persistence, clonal expansion of infected cells, replication-competent HIV
Abstract
Reservoirs of infectious HIV-1 persist despite years of combination antiretroviral therapy and make curing HIV-1 infections a major challenge. Most of the proviral DNA resides in CD4+T cells. Some of these CD4+T cells are clonally expanded; most of the proviruses are defective. It is not known if any of the clonally expanded cells carry replication-competent proviruses. We report that a highly expanded CD4+ T-cell clone contains an intact provirus. The highly expanded clone produced infectious virus that was detected as persistent plasma viremia during cART in an HIV-1–infected patient who had squamous cell cancer. Cells containing the intact provirus were widely distributed and significantly enriched in cancer metastases. These results show that clonally expanded CD4+T cells can be a reservoir of infectious HIV-1.
HIV-1 infection can be controlled, but not cured, by combination antiretroviral therapy (1); cells infected with replication-competent HIV-1 persist despite many years of combination antiretroviral therapy (cART). Recently, we and others (2, 3) showed that there is frequent clonal expansion of HIV-1–infected cells in patients and that these clones can persist for more than 10 y. Most of the proviruses that persist in patients on cART are defective (4), and it is not known whether clonally expanded cells can harbor a replication-competent provirus. It has been suggested that CD4+ T-cell clones are not likely to contain replication-competent HIV-1 because expression of viral proteins is cytotoxic, and the host’s immune system would preferentially eliminate infected cells that express viral proteins (4). Only a small proportion of integrated proviruses are intact (4), and it has been proposed that clonally expanded CD4+ T cells contain only defective proviruses and that intact proviruses that make up the reservoir are found only in unexpanded cells (5). Previous studies have reported that populations of virus with identical sequences emerge in the plasma after several years of suppressive cART (6, 7), but it is not known whether the identical viruses found in the blood of patients are infectious. We report here the identification of a highly expanded clone of HIV-infected CD4+ T cells that produced infectious virus at a level that caused persistent plasma viremia. This result shows that cells containing replication-competent HIV-1 proviruses can clonally expand and persist in vivo, presenting a challenge for achieving a cure of HIV-1 infection.
Results
Clinical Summary.
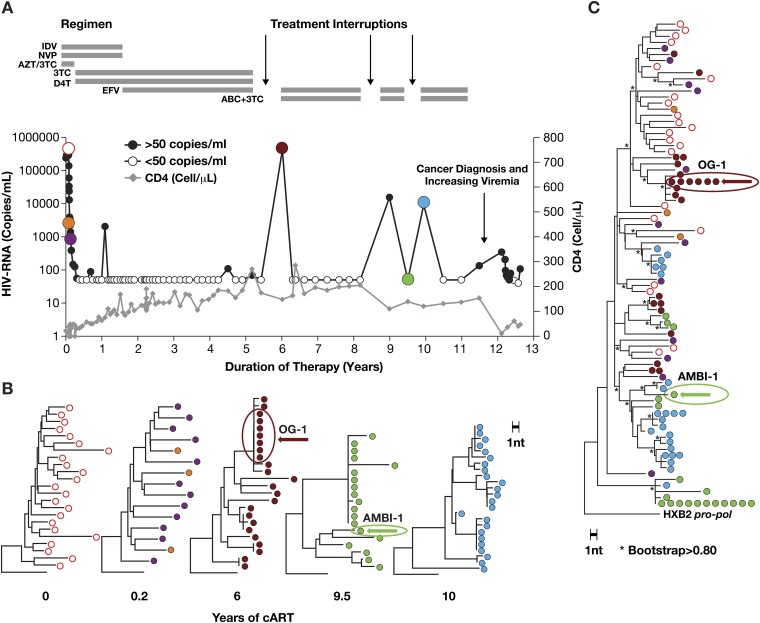
The patient was a 58-y-old African-American man diagnosed with an HIV-1 infection in May 2000. We characterized the virus present in samples obtained from this patient over 13 y, from the time cART was initiated until his death from metastatic squamous cell carcinoma. At the time of his initial HIV-1 diagnosis, his CD4+ T-cell count was 16 cells/µL, and plasma HIV-1 RNA was 5.4 log10 copies/mL. Analysis of HIV genetic variation using single genome sequencing (SGS) of the HIV pro–pol region (7) revealed that the virus present at the time of diagnosis was genetically diverse (nucleotide average pairwise distance of 1.4%; Fig. S1). cART was started, and plasma HIV-1 RNA decreased to <50 copies/mL within 4 mo, with typical decay kinetics (8), and remained <50 copies/mL for 5 y, with transient viremic periods due to nonadherence to medication. Despite a good virological response to cART, immune recovery was incomplete (1), and the CD4+ T-cell count never exceeded 350 cells/µL (Fig. S1).
Fig. S1.
Clonally expanded cells responsible for low-level viremia emerged from a diverse population of HIV-1–infected cells. (A) Time course of HIV-1 viremia (circles) and CD4+ T-cell numbers (gray diamonds) before and following the initiation of combination antiretroviral therapy. (B) Plasma samples obtained at the time points indicated by colored circles were subjected to SGS (p6-RT) as described, and neighbor joining phylogenetic analysis was performed. (C) Composite neighbor joining tree of all sequences in Fig. S2B. *Bootstrap support ≥80% (16).
About 12 y after cART was initiated, plasma HIV-1 RNA increased to 100–200 copies/mL (Fig. 1). SGS of the viral RNA in plasma revealed two distinct populations: a genetically diverse group of drug-resistant viruses and a second population of identical viruses that lacked drug resistance mutations (Fig. 1 and Fig. S1). At the time of the increase in viremia, the patient was diagnosed with human papilloma virus (HPV)-negative squamous cell carcinoma of the tongue. The cART regimen was changed to optimize efficacy against the drug-resistant variants, and viremia decreased to 50–100 copies/mL but did not become undetectable in commercial assays (Fig. 1 and Fig. S1). The reduction in viremia was associated with a decline of the drug-resistant viral variants to below the limit of detection, but the population of identical viruses that did not carry resistance mutations persisted for more than 8 mo at a level similar to that seen before the change in cART (Fig. 1B). During the subsequent period when the patient was treated with radiation and chemotherapy, the size of the primary tumor decreased, the level of plasma HIV-1 RNA declined gradually to less than 40 copies/mL, and the proportion of virus without resistance mutations increased to >90% (Fig. 1 A and C).
Fig. 1.
Persistent low-level viremia during cART is due, in part, to a clonal population of HIV-1 that has no drug resistance mutations. (A) Profile of plasma HIV-1 RNA levels starting 9.5 y after initiation of therapy, with annotations for clinical events (arrows), and sampling for SGS of plasma viral RNA (∼1,100 nt p6-RT) indicated by colored circles (14) and triangles for cell-associated HIV-1 DNA. The cART regimen is indicated by the horizontal bars (see Fig. S1 for treatment details). (B) Phylogenetic analysis of plasma viral RNA and cell DNA. (C) Abundance of the AMBI-1 variant in plasma. The percentage of proviruses that are AMBI-1 at the times indicated is shown below the graph.
Clonal Expansion of the Cell Lineage Responsible for Plasma Viremia.
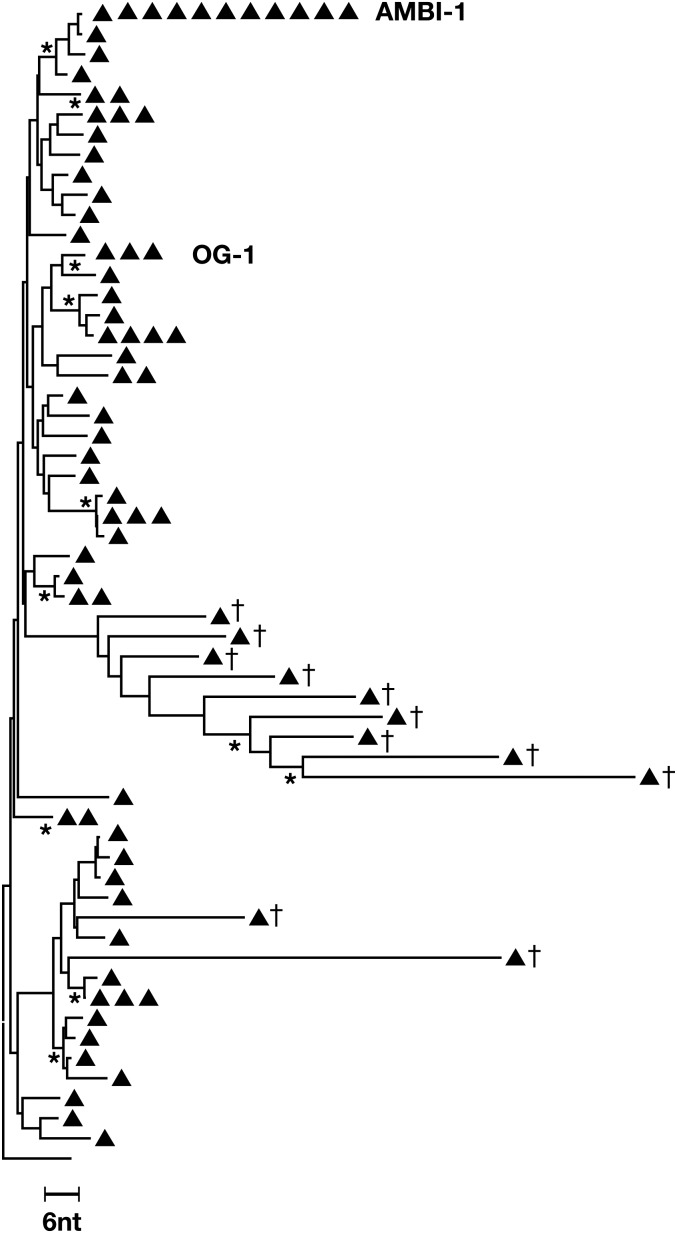
We previously showed that these identical viral sequences were produced by a clonally expanded cell containing a provirus, which was integrated into a region of the human genome that cannot be mapped to a unique location; thus, the location is ambiguous (3) and the provirus was designated AMBI-1. AMBI-1 viral RNA was first detected (as 1 of 19 sequences obtained) in a plasma sample taken when plasma HIV-1 RNA was <50 copies/mL at year 9.5 of cART, 2 y before viremia increased to a level that was detectable by routine clinical HIV-1 RNA assay. AMBI-1 viral RNA was not detected in several samples taken earlier, between 0.2 and 7.8 y after the initiation of cART. At the time of the increase in viremia, after 12 y of cART, the cells that contained the AMBI-1 provirus comprised one of the most highly expanded HIV-infected clones detected in this patient; analysis of peripheral blood mononuclear cells (PBMCs) taken after 12.1 y of cART (about 11% of the PBMCs were CD4+ T cells) revealed that 13% of the pro–pol sequences were AMBI-1 (Fig. 1C). We had previously estimated, based on integration site analysis, that AMBI-1 constituted a smaller fraction of the proviral DNA (at least 3.2%) (3). These data imply that a substantial fraction of the proviruses detected in the integration site analysis were defective and may have had large internal deletions, or hypermutations, which would have prevented their detection by PCR for pro–pol DNA. Quantitative analysis of PBMCs taken after 12.1 y of cART revealed 209 HIV-1 DNA copies/million PBMC; thus, we estimate that there were ∼9 million cells containing the AMBI-1 provirus in the patient at the time of the viral rebound at year 12 (Fig. S2). AMBI-1 proviruses were not detected in PBMC obtained after 3.6 or 7.8 y on cART (Fig. 1C and Dataset S1), suggesting that extensive expansion of this clone occurred after 7.8 y on therapy.
Fig. S2.
Cells from the AMBI-1 clone represent a significant fraction of the infected peripheral lymphocytes. PBMCs from the 9 December 2011 (12.1 y after therapy) time point were subjected to SGS (p6-RT) and neighbor-joining phylogenetic analyses were performed. AMBI-1 represented ∼13% of the total p6-RT sequences recovered from PBMCs at this time point (there were 83 HIV-1 sequences of which 11 were AMBI-1). Real-time PCR amplification for total HIV-1 DNA (17) revealed that there were 209 HIV-1 DNA copies per 106 PBMCs, of which ∼13% were AMBI-1, which would correspond to 27 × 106 PBMCs. The peripheral T-cell count was 1,279 cells/µL, and the total number of PBMCs in this patient was estimated to be 3.3 × 1011, based on total blood volume (Nadler formula) = 5.12 L, assuming that 2% of the total T cells are in the blood. From these estimates, the total number of expanded cells containing AMBI-1 proviruses is calculated to be ∼9 × 106. DNA sequences that correspond to a second clonal virus (OG-1) detected in the ex vivo infectious virus recovery assay were also present. †Hypermutants (5).
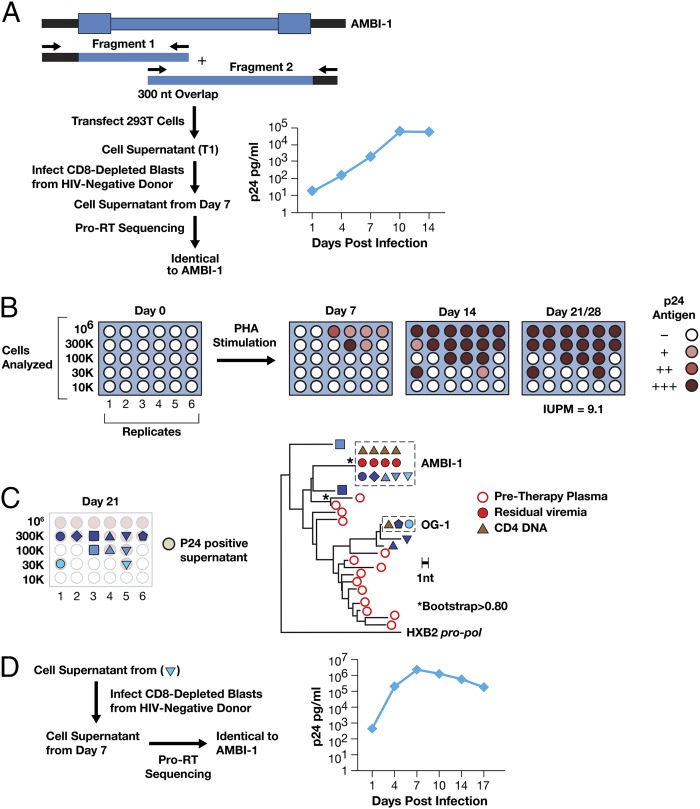
To obtain the full-length sequence of the AMBI-1 integrated provirus, we selectively PCR-amplified two overlapping DNA fragments from CD8-depleted CD4+ T cells (12.1 y on cART), using primers that matched the flanking host and internal HIV-1 sequences (Fig. 2A and SI Materials and Methods). Sequence analysis showed that the reading frames for all viral genes were open and that there were no obvious disabling mutations; env sequence analyses predicted that AMBI-1 was CCR5-tropic (15% false-positive rate by Geno2Pheno; GENAFOR).
Fig. 2.
Recovery of infectious HIV-1 from a provirus present in a clonally expanded CD4+ T cells. (A) The AMBI-1 provirus was amplified from CD8-depleted CD4+ T cells in two fragments that overlapped in the IN-vif region, using primers in the flanking host sequence and in HIV (primers named with HXB2 coordinates are listed in Table S3). Sequence analysis revealed ORFs for all HIV-1 genes with no obvious debilitating mutations. Amplified fragments were mixed 1:1 and used to transfect 293T cells with lipofectamine 2000, and the supernatant was used to infect CD8-depleted blasts from a healthy, HIV-negative donor; p24 was measured in culture supernatants by ELISA (Alliance HIV-1 p24 ELISA Kit; Perkin-Elmer). Viral sequences from the culture supernatants were identical to AMBI-1. (B) CD4+ T cells were purified from patient 1 PBMCs, serially diluted, PHA treated, and added to cocultures with irradiated feeder cells and CD8-depleted allogeneic blasts from healthy HIV-negative donors, which were maintained for 28 d as described (SI Materials and Methods); p24 antigen in the culture supernatants was determined weekly. p24-positive wells are shown. p24-negative wells were also negative by an assay for HIV-1 RNA (Roche Taqman v2.0). Infectious units per million (IUPM) were calculated using maximum likelihood estimate (5, 9). (C) HIV-1 sequences obtained from the p24-positive wells on day 21, as indicated by the symbols, were obtained by bulk sequencing of the p6-RT region and subjected to neighbor-joining phylogenetic analysis. Virus recovered from the supernatant (inverted triangle) also underwent full-length sequencing and was identical to AMBI-1. (D) HIV-1 recovered by in vitro cultivation is infectious in subsequent infections. Cell supernatant from one of the p24-containing wells with AMBI-1() was used to infect fresh CD8-depleted blasts from a healthy, HIV-negative donor. p24 levels were measured in culture supernatants at days 1, 7, 10, and 14. Viral RNA sequence (p6-RT) obtained from culture supernatants obtained at day 7 was identical to AMBI-1.
Infectivity of AMBI-1 Provirus.
To determine whether the AMBI-1 provirus was infectious, we cotransfected the two overlapping amplified fragments into 293T cells using lipofectamine 2000 (Fig. 2B). Supernatants from the transfection were used to infect CD8-depleted lymphoblasts from a healthy HIV-negative donor. As shown in Fig. 2A, HIV-1 p24 antigen levels increased exponentially, and sequencing of the corresponding viral RNA showed that it was identical to AMBI-1. The same transfection supernatants were serially diluted and used to infect TZM-bl indicator cells, in the presence and absence of 300 nm efavirenz, a nonnucleoside RT inhibitor. Luciferase activity above background was detected in cultures in the absence of efavirenz but not in its presence, again demonstrating the presence of replication-competent HIV-1 (Table S1).
Table S1.
Infection of TZM-bl cells with viral supernatants
| 2-Fold diluations | ||||||||||||
 | ||||||||||||
| Template | Virus dilution in plate | |||||||||||
| A | ||||||||||||
| No drug | B | 1:2 | 1:4 | 1:8 | 1:16 | 1:32 | 1:64 | 1:128 | 1:256 | 1:512 | Cell control | |
| C | ||||||||||||
| D | ||||||||||||
| 300 nM EFV | E | 1:2 | 1:4 | 1:8 | 1:16 | 1:32 | 1:64 | 1:128 | 1:256 | 1:512 | Cell control | |
| F | ||||||||||||
| G | ||||||||||||
| H | ||||||||||||
| pNL4-3 Plasmid T1 RLU Minus Cell Control | ||||||||||||
| Average | 377.62 | 572.45 | 692.80 | 736.23 | 772.31 | 566.81 | 368.27 | 216.19 | 108.36 | |||
| A | ||||||||||||
| No drug | B | 375.52 | 530.12 | 649.92 | 708.90 | 721.49 | 531.50 | 371.36 | 201.55 | 105.19 | ||
| C | 363.75 | 571.90 | 696.14 | 747.73 | 794.75 | 585.42 | 389.93 | 222.46 | 113.21 | |||
| D | 393.58 | 615.35 | 732.36 | 752.07 | 800.70 | 583.52 | 343.51 | 224.55 | 106.68 | |||
| 300 nM EFV | E | −1.09 | 0.74 | 0.95 | 0.83 | 0.74 | 0.86 | 0.66 | 0.43 | 0.25 | ||
| F | −1.09 | 0.04 | 0.40 | 0.21 | 0.02 | 0.14 | 0.05 | 0.04 | 0.06 | |||
| G | −1.09 | −0.16 | 0.60 | 0.01 | 0.01 | 0.13 | 0.02 | −0.02 | 0.02 | |||
| Ambiclone T1 RLU Minus Cell Control | ||||||||||||
| Average: | 36.38 | 30.34 | 14.53 | 6.78 | 2.78 | 1.60 | 0.75 | 0.23 | 0.06 | |||
| A | ||||||||||||
| No drug | B | 29.97 | 29.22 | 15.57 | 7.29 | 2.29 | 1.45 | 0.57 | 0.20 | 0.02 | ||
| C | 41.45 | 31.68 | 14.52 | 7.16 | 3.28 | 1.33 | 0.86 | 0.32 | −0.02 | |||
| D | 37.71 | 30.12 | 13.49 | 5.89 | 2.78 | 2.03 | 0.83 | 0.17 | 0.19 | |||
| 300 nM EFV | E | −0.30 | 0.05 | −0.05 | 0.05 | −0.01 | −0.08 | −0.03 | −0.08 | −0.11 | ||
| F | −0.03 | 0.20 | 0.26 | 0.14 | −0.02 | −0.04 | −0.03 | 0.06 | 0.09 | |||
| G | −0.26 | −0.01 | 0.05 | −0.02 | −0.17 | −0.16 | −0.14 | −0.03 | −0.10 | |||
| H | ||||||||||||
Infection of TZM-bl cells with serially diluted HEK293T AMBI-1 T1 supernatants was performed as described in SI Materials and Methods. Triplicate infections were done with each of the supernatant dilutions in the presence and absence of 300 nm of efavirenz. The data from each infection are shown as relative light units (RLUs) minus the uninfected cell control (sham infection). Virus dilution are color coded: 300 nM Efavirenz (light orange); no drug control (grey). Virus production (average RLU): light green.
Recovery of Replication-Competent HIV from Clonally Expanded Cells.
To determine whether AMBI-1 could be directly recovered from PBMCs, we performed an infectious virus recovery assay on cells obtained after 12.2 y of cART (SI Materials and Methods). Serial dilutions of purified total CD4+ T cells (CD4+ cells were selected without regard to their activation state) were phytohemagglutinin (PHA) treated, cocultured, in replicates, with γ-irradiated feeder cells and CD8-depleted allogeneic blasts from healthy HIV-negative donors for 28 d, and the culture supernatants were assayed weekly for virus production by measuring p24 antigen. As shown in Fig. 2C, infectious virus was recovered from multiple coculture replicates; none of the p24-negative wells contained HIV-1 RNA. The titer of infectious HIV-1 in the starting sample, calculated by a maximum likelihood estimate (9), was 9.1 infectious units per million CD4+ T cells. Thus, positive cocultures containing 3 × 104 or 1 × 105 donor CD4+ T cells were likely to have arisen from a single patient-derived cell that produced infectious virus. SGS analysis of p6-RT sequences obtained from virus-positive wells that had been seeded with 3 × 105, 1 × 105, and 3 × 104 CD4+ T cells showed that there was a single viral variant in each well. As shown in Fig. 2C, 5 of the 11 positive coculture wells contained sequences identical to AMBI-1, including 2 wells with 100,000 cells and a well with 30,000. Full-length sequencing of one of the isolates (derived from the well containing 30,000 cells) showed that the virus was identical to AMBI-1 and not a mixture or a recombinant. Culture supernatant from this well was used to infect CD8-depleted lymphoblasts from a healthy HIV-negative donor, resulting in rapid increases in HIV-1 p24 antigen (Fig. 2D). Sequence analysis of the HIV-1 RNA from this viral passage was identical to AMBI-1. Analysis of the HIV-1 sequences in the supernatants from the other positive wells revealed that a second variant, referred to as outgrowth 1 (OG-1), was also present in multiple wells (Fig. 2C), suggesting that there is a second clonally expanded cell in this patient that can produce infectious virus, although integration site analysis will be needed to confirm that OG-1 was present in a clonally expanded cell. OG-1 was transiently detected in rebound viremia during a treatment interruption (Fig. S1B) ∼6 y before the sample used for the in vitro virus recovery assay. Approximately half of the proviruses that were induced produced AMBI-1. We estimated that there were ∼250 AMBI-1 proviruses per million CD4+ cells (discussed above) and a total of ∼9 million AMBI-1–infected cells in the infected individual (Fig. S2). Thus, only a small fraction (<5%) of the cells that carry the AMBI-1 provirus produced virus in this assay. It is possible that this calculation may underestimate the abundance of AMBI-1 virus-producing cells present in the infected individual. p24 accumulation in AMBI-1 containing wells was slower than in some of the other wells (Fig. 2 B and C), and it is possible that some of the AMBI-1–positive wells might not have been detected after 28 d in culture in our assay.
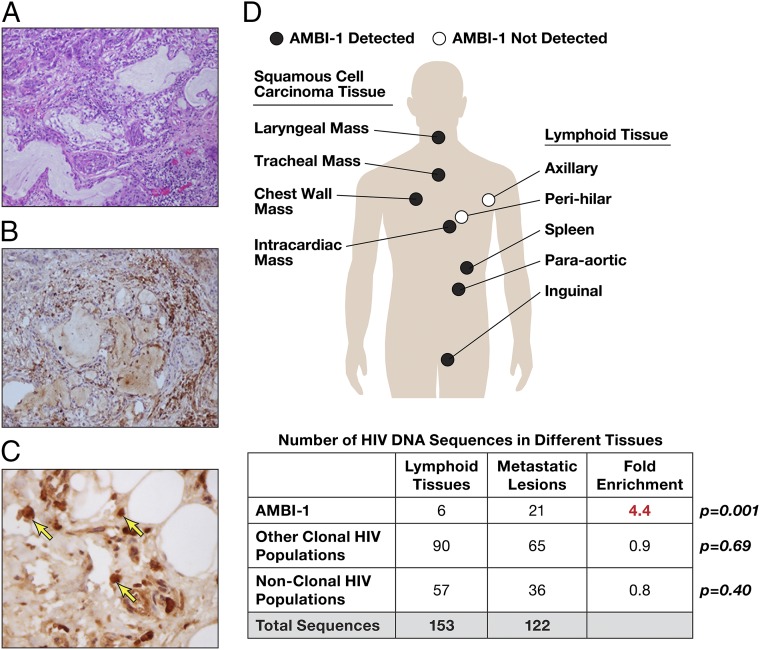
Wide Anatomic Distribution of Clonally Expanded Cells and Enrichment in Tumors.
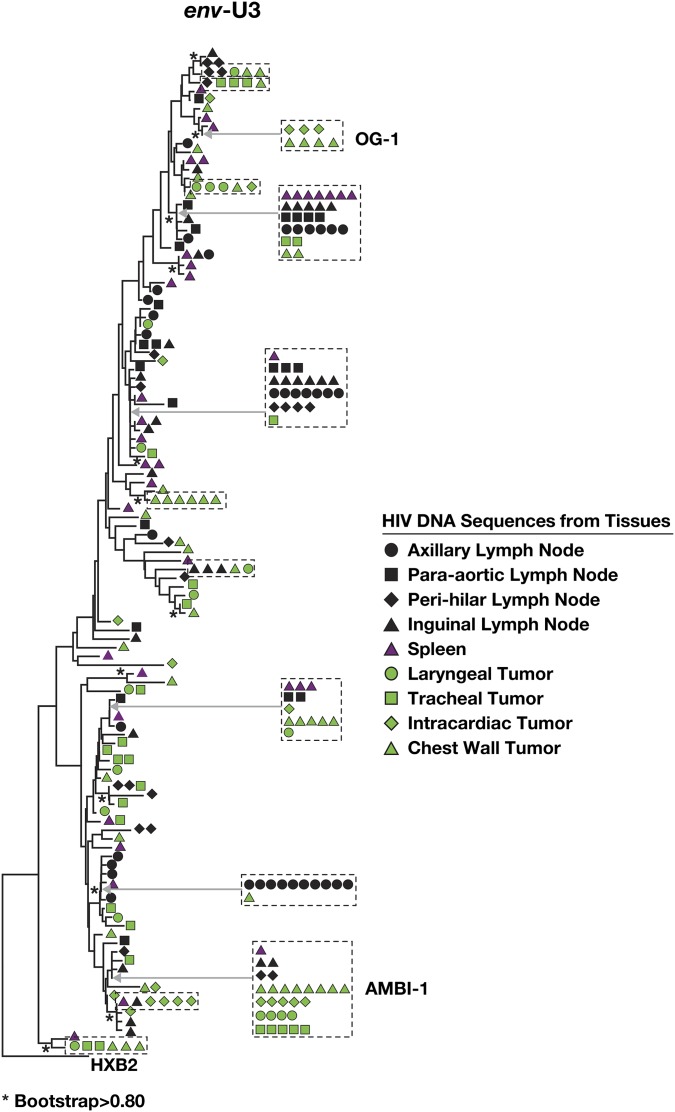
The level of AMBI-1 RNA in plasma declined after treatment of the patient’s cancer and then increased with its recurrence (Fig. 1), suggesting a connection between the extent of the tumor and the abundance of the clone carrying the AMBI-1 provirus. We examined samples obtained at autopsy from the region of the primary squamous cell carcinoma and some of the metastatic lesions (Fig. 3A). These samples showed infiltration by CD4+ lymphoid cells (Fig. 3 B and C). HIV-1–infected cells were detected in all of the lymphoid tissues and metastatic lesions tested (summarized in Fig. 3D, Table S2, and Dataset S1); the levels of HIV-1 DNA present in cancer tissue (∼12–20 copies per million cells) were, in general, about 10-fold lower than that detected in lymphoid tissue (Table S2). However, the AMBI-1 provirus and the integration site were detected in DNA derived from both tumor and lymphoid tissues (Table S2 and Dataset S1) and were present as a significantly larger fraction of all of the HIV-1 DNA sequences in the cancer metastases than in lymphoid tissue (Fig. 3D; P = 0.001). Other clonal populations of infected cells, as well as proviruses encoding the replication competent variant OG-1, were detected in both tumor and lymphoid tissues (Fig. S3).
Fig. 3.
Cells carrying the AMBI-1 proviruses are widely distributed anatomically and enriched in tumor metastases. (A) H&E-stained section of a metastasis of the invasive squamous cell carcinoma shows lymphocytic infiltration. (Magnification, 200×.) (B) Sections were stained for CD4 expression to demonstrate that the infiltrate includes CD4+ lymphocytes. CD8 staining was negative. (Magnification, 200×.) (C) High magnification (12) of CD4 expressing lymphocytes (arrows) (D) Samples from autopsy specimens were subjected to env-U3 SGS. HIV-1 sequences were amplified from individual metastatic lesions and lymphoid tissues and used for phylogenetic analyses (Fig. S3). The table shows the number of AMBI-1 and other HIV-1 DNA sequences in metastatic lesions and lymphoid tissues; P values were derived from the Fisher exact test.
Table S2.
Summary of biopsy and autopsy tissues
| Procedure | Date | Tissue | Type | Anatomic location | Histology | Squamous cell carcinoma markers | Cell equivalents | HIV copies per million cell equivalents | Total SGS | Identical sequences | AMBI-1 |
| Biopsy | 7/23/12 | Tongue | Squamous cell carcinoma invading tongue | ND | ND | ND | ND | ND | ND | ||
| Autopsy | 7/2/13 | Lymphoid | Spleen | Spleen | p63– | 4.4 × 108 | 37.8 | 37 | 19 | 1 | |
| Lymph node | Peri-hilar | Sinus histiocytosis | p63– | 2.4 × 108 | 161.20 | 24 | 12 | 0 | |||
| Axillary | Sinus histiocytosis | p63– | 2 × 108 | 154.00 | 36 | 24 | 0 | ||||
| Para-aortic | Sinus histiocytosis | p63– | 2.5 × 107 | 21.50 | 25 | 20 | 3 | ||||
| Inguinal | Sinus histiocytosis | p63– | 1.7 × 108 | 264.30 | 31 | 21 | 2 | ||||
| Neoplastic | Residual local disease | Laryngeal | Invasive squamous cell carcinoma | p63+ | 3.3 × 108 | 18.60 | 18 | 12 | 4 | ||
| Tracheal | Invasive squamous cell carcinoma | p63+ | 9.3 × 106 | 12.60 | 27 | 20 | 5 | ||||
| Metastasic lesion | Skin: chest wall | Invasive squamous cell carcinoma | p63+ | 2.0 × 107 | 15.80 | 55 | 38 | 7 | |||
| Intracardiac | Invasive squamous cell carcinoma | p63+ | 2.0 × 10 | 19.50 | 22 | 16 | 5 |
Samples were obtained from a biopsy done at the time the tumor was diagnosed, or at autopsy. ND, not done.
Fig. S3.
Phylogenetic analysis of HIV from autopsy tissues. Frozen samples obtained from the indicated autopsy material were subjected to SGS (env-U3) and neighbor-joining phylogenetic analysis. For ease of identification, large groups of expanded clones are grouped (boxes), and their position in the tree is noted by arrows.
SI Materials and Methods
The patient was a 58-y-old African-American man with an MSM risk factor and a 30-y history of smoking and alcohol use who was in good health until April 2000, when he developed thrush and was diagnosed with HIV-1 infection. Duration of the HIV-1 infection is uncertain; a prior HIV-1 ELISA performed in 1985 was negative. At diagnosis, he had 16 CD4+ T cells/µL and viral RNA of 283,186 copies/mL plasma. He enrolled in an NIH study of antiretroviral therapy and viral decay kinetics (protocol 97-I-0027). On enrollment in an NIH natural history protocol of HIV infection in June of 2000, CD4+ T-cell count was 22 cells/µL, his viral RNA level was 5.4 log10 copies/mL plasma, and therapy with azidothymidine, lamivudine, indinavir, and nevirapine was initiated.
The patient tolerated antiretroviral therapy, and viral RNA in the blood underwent typical initial first- and second-phase decay kinetics to <50 copies HIV RNA/mL within 4 mo; his CD4+ T-cell count increased but never exceeded 350 cells/µL (Fig. S1). His course was marked by an episode of Kaposi sarcoma that developed within 6 mo of the initiation of cART. After 5 y of continuous therapy, he underwent several interruptions of cART due to insurance lapses. During these interruptions, he experienced rebound viremia, but there was prompt suppression of viremia when the therapy was restarted (Fig. S1A).
The patient remained well until December 2011, when he developed persistent low-level viremia (134 copies/mL) measured by bDNA, Versant 3.0. His CD4+ T-cell count was stable at 153 cells/µL (12%). and he reported >95% adherence to cART. Commercial drug resistance testing (TRUGENE) revealed the presence of the following mutations in RT: L74V, M184V, K103N, and V108I; these mutations are known to confer some degree of resistance to all of the antiretroviral drugs in the regimen he was taking at that time (abacavir, lamivudine, and efavirenz).
At the time the persistent low-level viremia emerged, a physical examination identified several macerated papules on the dorsum of his tongue that were asymptomatic; shortly thereafter, the patient reported the presence of small mass on the dorsum of his tongue, which was subsequently diagnosed as stage III, T3N0 moderately differentiated squamous cell carcinoma, which was human papilloma virus DNA negative (Quest Diagnostics), EBV negative, and p61 positive. CD4+ and CD8+ cells were present in the tumor-containing regions of the biopsy.
In preparation for oncology treatment, he was admitted and underwent a change in cART to a regimen that included tenofovir, emtricitabine, raltegravir, and etravirine, which he received as an inpatient by directly observed therapy. Viral RNA levels promptly declined but did not fall below the limit of detection of 50 copies/mL (Fig. 1). At that time, cisplatin chemotherapy was initiated, and tenofovir was switched to abacavir to avoid potential renal toxicity. He underwent radiation therapy with 70 Gy total delivered to the primary tumor and a comprehensive neck field encompassing levels IA, IB, II, II, and IV. Radiaton therapy was given during an 8-wk course starting 6 September 2012. He received cisplatin 100 mg/m2 every 3 wk for total of three doses over 6 wk. Repeated measurements of his HIV-1 RNA levels revealed persistently detectable viremia, with slow viral decay, declining to <50 copies/mL with a half-life of ∼6 mo. CD4+ cell numbers declined in response to radiation therapy, as previously reported (21, 22).
The patient did well and had remission in the size of the local lesion. Approximately 6 wk after his last cisplatin therapy, he had recurrence of discomfort in his tongue. In January 2013, he was diagnosed with an overt recurrence of the cancer; CT scans and MRI showed no evidence of local metastatic disease. He underwent radical subtotal glossectomy with reconstructive surgery in March 2013. He tolerated the surgery well and did not have further chemotherapy. He did well, did not require percutaneous endoscopic gastrostomy for feeding, and maintained some speech, which improved after speech therapy. In May–June 2013, there was a local recurrence of the cancer and the development of skin lesions, which gradually increased in size. He initiated do-not-resuscitate status. There were additional symptoms that included difficulty breathing, and he died due to respiratory insufficiency in the emergency room while undergoing evaluation. A limited autopsy showed that he had residual disease at the site of surgery, as well as multiple discrete metastases (skin, larynx, right atrium). Microscopic examination of material from the spleen and several lymph nodes (laryngeal, para-aortic, inguinal, axillary nodes) revealed no histologic or immunohistochemical evidence of squamous cell carcinoma. Local tumor and metastatic lesions showed metastatic invasive squamous cell carcinoma, and all lesions contained lymphoid cells infiltrating the periphery of the tumor. The local tumor contained both CD4+ and CD8+ cells, and metastatic lesions obtained at autopsy contained predominantly CD4+ cells (CD8+ cells were not detected by immunohistochemistry).
The initial description of this patient has been reported (ref. 3, patient 1).
Discussion
It is widely held that resting memory T cells carrying infectious proviruses constitute the major reservoir of infectious HIV-1 (10, 11). We show here that clonally expanded CD4+ T cells can harbor an infectious provirus and produce virus at levels that are detectable in blood over a period of several years. The AMBI-1 clone was able to persist and expand despite the potential cytopathic effects of the viral proteins or immune clearance of virus-producing cells. The fact that cells carrying infectious proviruses can expand by cell division complicates the problem of eliminating all of the HIV-1 proviruses that make up the HIV-1 reservoir. The patient we describe had advanced HIV infection at presentation and, although clinically stable for a number of years without opportunistic infections, he never achieved robust CD4 recovery while undergoing cART; it is possible that overall, immune surveillance was weak in this patient because of the poor recovery of his T cells during cART (12, 13), which may have impaired tumor surveillance. It is likely that only a fraction of the cells in the clone were producing virus at any given time, which could help the clone escape from HIV-1–specific immune surveillance. We recovered the AMBI-1 virus in vitro using total PBMCs, and the activation state of the cells before in vitro stimulation is not known. Quiescent cells are known to be an important reservoir of HIV persistence. Whether AMBI-1 virus was produced from quiescent or partially activated CD4+ T cells is not known. We have estimated that the proportion of AMBI-1–positive cells producing HIV was relatively small (∼5%). AMBI-1 emerged slowly in vitro relative to other viruses recovered (Fig. 2C), and it is possible that we may have underestimated the frequency of AMBI-1 proviruses producing HIV, because some AMBI-1 viruses could have been outcompeted by more rapidly growing viruses in vitro or because we did not monitor cultures long enough to detect AMBI-1 from slowly reactivating cells present in some of the wells.
Because virus is rapidly cleared from the blood (8), the persistence of detectable levels of AMBI-1 virus in the blood for more than 3.1 y indicates that at least a fraction of the cells in the clone were producing virus during this period. The overall level of plasma virus derived from the clone was relatively constant for more than 8 mo before cancer treatment, and the level was not affected by a change in cART. Thus, the stimulus that caused the cells in the AMBI-1 clone to divide (perhaps encounter with an antigen or homeostatic proliferation) was sufficient to drive expansion to ∼9 million cells (Fig. S2).
These data indicate that a clonally expanded population is capable of producing infectious HIV. Because HIV infection is often cytopathic and because HIV proteins, such as vpr, can block cell division, most of the infected cells that expand are unlikely to produce virus. We estimated that only a fraction of AMBI clones were producing HIV at a given time. There are a number of mechanisms that could explain this result. These mechanisms include the AMBI-1 provirus is transcriptionally silent in most cells, but is expressed after cells have divided or AMBI-1 may be produced only at low levels that are insufficient to induce cytopathicity or trigger killing by cells of the host’s immune system. AMBI-1 cells are likely to represent a long-lived cell CD4+ T-cell type, and some specific T-cell subtypes may favor HIV production. Further research will be essential in determining the mechanisms of HIV production during persistence and clonal expansion, the CD4 subsets responsible for expansions, and whether these expansions are the product of homeostatic proliferation of HIV infected cells, and/or response to antigen.
We found strong evidence that proviral integration into the proto-oncogenes BACH-2 and MKL-2 resulted in clonal expansion of CD4+ T cells in this patient, probably because the integrated proviruses affected the expression of these genes (3). We have no evidence that the site of AMBI-1 provirus integration contributed to the clonal expansion of the cells that carry AMBI-1. The AMBI-1 integration junction sequence [chrUn_gl000220; University of California Santa Cruz (UCSC) genome hg19] does not have an unequivocal assignment in the human genome but matches several highly related sequences present at distinct chromosomal locations. However, the AMBI-1 clone did expand in parallel with the growth of the squamous cell cancer, decreased as the tumor underwent regression after radiation and chemotherapy, recurred with disease relapse (Fig.1), and was also enriched in tumor tissue at autopsy (Fig. 3D), suggesting that expansion of AMBI-1–infected cells may have been in response to a tumor antigen.
The data presented here demonstrate that clonally expanded cells containing an intact, infectious provirus can persist and produce virus for many years in a patient on cART. Studies of additional patients will be necessary to estimate the frequency of clonally expanded populations with replication competent HIV. Given that clonal expansion of HIV-infected cells is common, our results suggest an important mechanism for persistence of HIV reservoirs. To develop successful curative strategies, a better understanding is needed of the fraction of the replication-competent reservoir that is composed of clonally expanded cells and how to specifically target and eliminate them.
Materials and Methods
Experimental Design.
We conducted a virologic, immunologic, and anatomic analysis of samples from a single HIV-infected patient from the time of initiating antiretroviral therapy to death 13 y later. The patient was enrolled in research studies of HIV replication (97-I-0082 and 08-I-0221) approved by the intramural National Institute of Allergy and Infectious Diseases (NIAID) Institutional Review Board, FWA00005897, at the National Institutes of Health (NIH) Clinical Center (Bethesda). The study participant underwent an informed consent process and provided written consent for research studies, including genetic testing.
Single Genome Sequencing.
Viral RNA was extracted from plasma and was used to synthesize cDNA with a primer in pol (HXB2 position 3500). Genomic DNA (gDNA) was extracted directly from PBMCs. The cDNA or gDNA was end-point diluted and PCR amplified, and single-genome sequences were obtained from an 1,100-bp region that encodes a segment from p6 through the first 660 nt of RT as previously described (7). Frozen pieces of tissue from autopsy-derived samples were disrupted with disposable pestles and lysed using proteinase K, and gDNA was extracted with DNeasy Blood & Tissue Kit (Qiagen). Isolated gDNA was then end-point diluted and subjected to single genome sequencing applying the same cycling conditions previously described (7) but with primers amplifying the env-U3 region (HXB2 positions from 8694 to 9486). Primer sequences are given in Table S3. PCR conditions were as follows: 94 °C for 2 min, 94 °C for 30 s, 50 °C for 30 s, 72 °C for 1 min 30 s (44 cycles), 72 °C for 3 min, 4 °C hold (14); 94 °C for 2 min, 94 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min (15), and 72 °C for 3 min, 4 °C hold (second step).
Table S3.
List of primers used to amplify HIV
| Primer set | Primer position | Primer name | Sequence 5'→3' |
| ENVU3 SGS | Outer | NEF8513FO | GTGCCTCTTCAGCTACCACC |
| U39496RO | CTTATATGCAGCTTCTGAGGGC | ||
| Nested | NEF8694FN | GAGGGGACAGATAGGGTTATAGAAG | |
| U39486RN | TCTGAGGGCTCGCCACTCC | ||
| AMBI-1 Provirus | 5′ End | UN5140034FO | CATTAGGGCGTGAGCTTGTCTTC |
| 5264RO | CCTGTATGCAGACCCCAATATG | ||
| UN5140034FN | CTATCAGGGTCAATCCTAGACATTGTG | ||
| 5070RN | CACAATCATCACCTGCCATC | ||
| 3′ End | 4813FO | GTGCAGGGGAAAGAATAGTAGAC | |
| UN3140034RO | CGTGACAATAAAGAGCTATCTAACCAGAA | ||
| 4930FN | TTTGGAAAGGACCAGCAAAGCT | ||
| UN3140034RN | GCACCATCCATAGGGCTTTGTCTTTAG | ||
| AMBI-1 RNA | U5-RT | HIVGENF1 | CCTTGAGTGCTTCAAGTAGTGTGTGCCCGTCTGT |
| 3500– | CTATTAAGTATTTTGATGGGTCATAA | ||
| HIVGENF2 | AGTAGTGTGTGCCCGTCTGTTGTGTGACTC | ||
| 3410– | CAGTTAGTGGTATTACTTCTGTTAGTGCTT | ||
| RT-ENV | 3090+ | TATCAATACATGGATGATTTGTATGTAG | |
| 6950- | ATGTGTACATTGTACTGTGCTGACAT | ||
| 3150+ | CTGACTTAGAAATAGGGCAGCATAG | ||
| 6850– | GGCACAATAATGTATGGGAATTGG | ||
| ENV-U3 | E20F | CAGTTAGTGGTATTACTTCTGTTAGTGCTT | |
| U39496RO | CTTATATGCAGCTTCTGAGGGC | ||
| E30F | GTGTACCCACAGACCCCAGCCCACAAG | ||
| U39486RN | TCTGAGGGCTCGCCACTCC |
Primers used to amplify HIV from DNA and RNA, the env-U3 region, and to amplify the full genome of AMBI-1.
Sequences were aligned using Clustal W, G > A hypermutants were identified with Hypermut 2.0 (16), and neighbor-joining trees were constructed in MEGA 6.0 (14) with HXB2 HIV-1 as the outgroup. Phylogenetic structure was tested by bootstrap analysis (1,000 replicates); branches with bootstrap support ≥80% are indicated in figures with an asterisk (16).
Specific Provirus Sequencing.
To amplify and sequence specific proviruses from CD8-depleted CD4+ T cells, ad hoc primer sets were designed based on sequences in the human genome near the integration site. Complementary primers spanning different regions of the HIV-1 genome were used. A nested PCR was performed using Phusion hot start flex polymerase (NEB), following the manufacturer’s instructions; annealing temperatures were calculated with the NEB Tm calculator available online (tmcalculator.neb.com). The full-length AMBI-1 genome was isolated using two amplicons that overlapped in the IN/vif region. PCR products were isolated by gel electrophoresis and directly sequenced. Primer sequences are listed in Table S3. The PCR products were used to transfect HEK293T cells to generate infectious virus (see Transfection Analysis).
Quantitation of HIV DNA and RNA.
HIV-1 DNA copy numbers were determined, in DNA extracted from PBMCs, using HIV-1 integrase as the target for PCR amplification, as previously reported (17). To determine HIV-1 DNA levels in lymphoid and tumor tissue, samples were collected in 2 mL SPEX SamplePrep (Metuchen) tubes with 1.4-mm acid-washed zirconium beads. With the samples on dry ice, 1.0 mL TriReagent (Molecular Research Center) was added, the tubes were loaded into a Precellys 24 (Bertin Technologies) tissue homogenizer, homogenized twice at 6,000 rpm for 30 s, frozen for 3 min on dry ice, and then homogenized twice more at 6,000 rpm for 30 s. RNA was isolated by adding 0.1 mL 1-bromo-3-chloropropane (Molecular Research Center), followed by vortexing or homogenization for 15 s, and then centrifugation at 14,000 rpm for 15 min at 4 °C. The aqueous phase, which contained the RNA, was collected and transferred to a new tube containing 240 ng glycogen (Roche Life Sciences). The RNA was precipitated with 0.5 mL 2-propanol, and the pellet was washed with 70% ethanol. DNA was extracted from the remaining organic phase by adding 0.5 mL 4 M guanidine thiocyanate, 1 M Tris base, and 50 mM sodium citrate and repeating the phase separation steps described above. The aqueous phase with the DNA was transferred to a new tube containing 240 ng glycogen and precipitated with 0.4 mL 2-propanol, the DNA was collected, and the pellet was washed with 70% (vol/vol) ethanol. HIV gag DNA copy numbers were determined using a multiplexed qPCR assay, as described previously (18). Samples were tested undiluted and diluted 1:10 with spiked-in RCAS (replication competent avian sarcoma virus) controls incorporated in the multiplexed assay to monitor any inhibition of the assay. Results were normalized to diploid genome cell equivalents of DNA, using quantitative PCR for the CCR5 gene as previously described (19). Results were reported as cell associated HIV-1 copies per 106 cell equivalents (Table S2). No HIV RNA was detected in any of the autopsy tissue samples; however, this probably reflects a prolonged delay in obtaining the autopsy.
Isolation of Integration Sites.
Integration site analysis was performed as previously described (3). Briefly, DNA was isolated from Ficoll-purified PBMCs (5–10 × 106), negatively selected CD4+ T cells, or autopsy samples. gDNA was fragmented by random shearing into 300- to 500-bp fragments. Linker-mediated nested PCR was performed to amplify the human genomic regions and the linked viral sequences from both the 5′ and 3′ long terminal repeats (LTRs). Paired end-sequencing was carried out using the MiSEq. 2 × 150-bp paired end kit (Illumina). The sequences of the host-viral junctions and the host DNA breakpoints were determined. The host DNA sequences were then mapped to human genome (hg19) with BLAT. A stringent filter was used to check quality of recovered integration sites. Sequences that have exactly the same integration site but different host DNA breakpoints came from different cells; this test identifies proviruses in clonally amplified cells (3).
Transfection of Amplified Viral DNA.
HEK293T cells were cotransfected (using Lipofectamine 2000) with two overlapping AMBI-1 DNA fragments generated by PCR (10 µg of each). Forty-eight hours later, supernatants from transfected HEK293T cells (referred to as T1 supernatants) were harvested. T1 supernatants were used to infect CD8-depleted lymphoblasts obtained from a healthy HIV-negative donor. The cells were isolated using Ficoll-hypaque density-gradient centrifugation. Cells were incubated in 60 mL T-cell growth medium [phenol red-free RPMI 1640, 10% (vol/vol) heat-inactivated FBS, 60 U/mL penicillin, 60 µg/mL streptomycin, 100 U/mL recombinant human-IL-2, and T-cell conditioned medium] + 0.5 µg/mL PHA for 4 d. On day 4, blasts were CD8 depleted using BD IMag CD8 depletion beads, and two million CD8-depleted blasts were resuspended in medium with 400 µL T1 supernatant and cultured for 2 h. Cells were washed in RPMI and cultured in new medium. Supernatants were sampled at 0, 4, 7, 10, and 14 d after infection and assayed for p24 antigen following the manufacturer’s instruction (Alliance HIV-1 p24 ELISA Kit; Perkin-Elmer). HIV-1 RNA was extracted from the day 7 supernatant, and the pro-RT region was amplified by bulk one-step RT-PCR and bidirectionally sequenced for comparison with the sequence of AMBI-1. To exclude DNA carryover from transfection, the remaining RNA was DNase-treated, reextracted, and subjected to cDNA synthesis, and the pro-RT region was amplified by bulk PCR with or without RT, using primers described for SGS in Table S3.
Infectivity in TZM-bl Cells.
AMBI-1 infectivity was determined using the TZM-bl reporter cell line. TZM-bl cells were infected with serial dilutions of T1 supernatants in the presence or absence of 300 nM efavirenz and were assayed, after 48 h, for luciferase activity. Studies were done in triplicate, and results are shown in Table S1. The luciferase activity of uninfected cells (sham infection) was measured, and this background activity was subtracted from the data.
Ex Vivo Recovery of Infectious Virus.
Virus culture assays were performed as previously described (9, 20) with the modifications described below. Cryopreserved PBMCs were enriched for CD4+ T cells by negative selection, serially diluted from 1,000,000 to 10,000 cells per well, and seeded in individual wells in sets of 6. Cells were stimulated with PHA in the presence of a 10-fold excess of γ-irradiated allogeneic PBMCs from healthy HIV-negative donors. Replication-competent viruses were amplified by adding CD8-depleted lymphoblasts from healthy HIV-negative donors. Virus cultures were maintained for 4 wk; supernatants were collected weekly and used to measure HIV-1 p24 antigen by ELISA. The p24-negative wells were tested for the presence of HIV-1 RNA using Roche Taqman v2.0. The frequency of HIV-1–infected cells was determined by a maximum likelihood method as previously described (9). HIV-1 RNA was extracted from the supernatants and subjected to bulk RT-PCR or to p6-RT SGS (as described above), yielding at least 20 sequences per supernatant. Sequences were then aligned with MEGA v6.0 (14) and subjected to phylogenetic analysis. The complete AMBI-1 genome was recovered from the supernatant and the sequence compared with the sequence of the AMBI-1 provirus. RT-PCR was performed with SuperScriptIII and OligoDT (Life Technologies), and cDNA was then amplified as described above to produce three overlapping fragments spanning U5-RT, RT-env, and env-U3 (Table S3). PCR products were subjected to direct sequencing.
Passage of Infectious Virus.
Supernatant from a culture well (replicate 5 of 30,000 patient cells per well) from AMBI-1 was recovered as described above and was used to infect CD8-depleted lymphoblasts from a healthy HIV-negative donor. HIV-1 p24 antigen was measured in supernatants collected on days 1, 4, 7, 10, 14, and 17. HIV-1 RNA in the supernatant from days 7, 10, and 14 was extracted, and the pro-RT region was amplified by bulk RT-PCR and sequenced for comparison with AMBI-1 sequence.
Statistical Analysis.
Differences in numbers of copies of HIV proviruses in lymphoid and tumor tissues were analyzed using Fisher exact test statistics.
Supplementary Material
Acknowledgments
We thank the study participant, his family, and the clinical staff of the National Institute of Allergy and Infectious Diseases/Critical Care Medicine Department clinic who cared for him. We thank C. Lane, H. Masur, and H. Imamichi, for insightful discussions and A. Kane for help with the figures. This work was supported in part with federal funds from National Cancer Institute, NIH, Contract HHSN261200800001E (to R.G., B.F.K., and M.P.), the NIH Bench to Bedside Program (J.W.M. and F.M.), and Contracts 12XS547 and 13XS110 through Leidos Biomedical Research (to J.W.M. and J.M.C., respectively). J.M.C. was a Research Professor of the American Cancer Society, with support from the F. M. Kirby Foundation.
Footnotes
Conflict of interest statement: J.W.M. is a consultant for Gilead Sciences and owns shares in Co-Crystal, Inc.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KU644730–KU645194 and KU641402).
See Commentary on page 1692.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522675113/-/DCSupplemental.
References
- 1.Baker JV, et al. Terry Beirn Community Programs for Clinical Research on AIDS (CPCRA) CD4+ count and risk of non-AIDS diseases following initial treatment for HIV infection. AIDS. 2008;22(7):841–848. doi: 10.1097/QAD.0b013e3282f7cb76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner TA, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345(6196):570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maldarelli F, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345(6193):179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho YC, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohn LB, et al. HIV-1 integration landscape during latent and active infection. Cell. 2015;160(3):420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey JR, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80(13):6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kearney MF, et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog. 2014;10(3):e1004010. doi: 10.1371/journal.ppat.1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perelson AS, et al. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387(6629):188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 9.Cillo AR, et al. Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc Natl Acad Sci USA. 2014;111(19):7078–7083. doi: 10.1073/pnas.1402873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun TW. Tracking replication-competent HIV reservoirs in infected individuals. Curr Opin HIV AIDS. 2013;8(2):111–116. doi: 10.1097/COH.0b013e32835d6e1c. [DOI] [PubMed] [Google Scholar]
- 11.Siliciano JD, Siliciano RF. Recent developments in the search for a cure for HIV-1 infection: Targeting the latent reservoir for HIV-1. J Allergy Clin Immunol. 2014;134(1):12–19. doi: 10.1016/j.jaci.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 12.Baker JV, et al. Terry Beirn Community Programs for Clinical Research on AIDS (CPCRA) Poor initial CD4+ recovery with antiretroviral therapy prolongs immune depletion and increases risk for AIDS and non-AIDS diseases. J Acquir Immune Defic Syndr. 2008;48(5):541–546. doi: 10.1097/QAI.0b013e31817bebb3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson EM, Sereti I. Immune restoration after antiretroviral therapy: The pitfalls of hasty or incomplete repairs. Immunol Rev. 2013;254(1):343–354. doi: 10.1111/imr.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stecher G, et al. MEGA-MD: Molecular evolutionary genetics analysis software with mutational diagnosis of amino acid variation. Bioinformatics. 2014;30(9):1305–1307. doi: 10.1093/bioinformatics/btu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joos B, et al. Swiss HIV Cohort Study HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci USA. 2008;105(43):16725–16730. doi: 10.1073/pnas.0804192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rose PP, Korber BT. 2015 HYPERMUT analysis and detection of APOBEC-induced hypermutation. Available at www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html. Accessed July 15, 2015.
- 17.Besson GJ, et al. HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin Infect Dis. 2014;59(9):1312–1321. doi: 10.1093/cid/ciu585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Somsouk M, et al. The immunologic effects of mesalamine in treated HIV-infected individuals with incomplete CD4+ T cell recovery: A randomized crossover trial. PLoS One. 2014;9(12):e116306. doi: 10.1371/journal.pone.0116306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas JA, et al. Human immunodeficiency virus type 1 nucleocapsid zinc-finger mutations cause defects in reverse transcription and integration. Virology. 2006;353(1):41–51. doi: 10.1016/j.virol.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 20.Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 21.Alfa-Wali M, et al. Chemoradiotherapy for anal cancer in HIV patients causes prolonged CD4 cell count suppression. Ann Oncol. 2012;23(1):141–147. doi: 10.1093/annonc/mdr050. [DOI] [PubMed] [Google Scholar]
- 22.Harshany ML, Polhemus ME. Effects of adjuvant radiotherapy for testicular cancer on CD4+ cell count in HIV-positive patients: A case report. AIDS Read. 2004;14(4):189–190, 192–193. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.