SUMMARY
While the intrinsic antiviral cell defenses of many kingdoms utilize pathogen-specific small RNAs, the antiviral response of chordates is primarily protein-based and not uniquely tailored to the incoming microbe. In an effort to explain this evolutionary bifurcation, we determined whether antiviral RNA interference (RNAi) was sufficient to replace the protein-based type I interferon (IFN-I) system of mammals. To this end, we recreated an RNAi-like response in mammals and determined its effectiveness to combat influenza A virus in vivo in the presence and absence of the canonical IFN-I system. Mammalian antiviral RNAi, elicited by either host- or virus-derived small RNAs, effectively attenuated virus and prevented disease independently of the innate immune response. These data find that chordates could have utilized RNAi as their primary antiviral cell defense and suggest that the IFN-I system emerged as a result of natural selection imposed by ancient pathogens.
INTRODUCTION
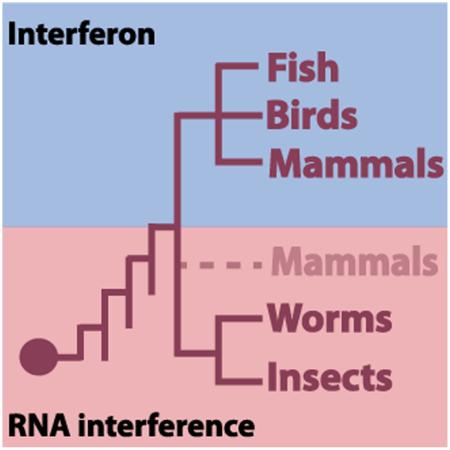
The cellular response to virus infection is of fundamental importance for survival and can differ dramatically within the tree of life. Prokaryotes defend themselves against virus infection through the utilization of clustered regularly interspaced short palindromic repeats (CRISPR), which provides a small pathogen-specific RNA template to guide a Cas nuclease towards the incoming virus (Barrangou et al., 2007; Wiedenheft et al., 2012). Similarly, many eukaryotes use small RNAs to target virus (Ding and Voinnet, 2007; Hutvagner and Zamore, 2002). Like CRISPR, the antiviral RNA interference (RNAi) system relies on the generation of pathogen-derived small RNAs to provide specificity to a nuclease, in this case, a member of the Argonaute (Ago) family (Ding and Voinnet, 2007; Hutvagner and Zamore, 2002). In this antiviral defense mechanism, an RNAseIII nuclease called Dicer is responsible for processing viral RNA into short 21-24 nucleotide fragments called short interfering RNAs (siRNAs) which are subsequently loaded into an Ago-containing, RNA induced silencing complex (RISC) (Ding and Voinnet, 2007). Curiously, while chordates have retained much of the small RNA machinery to enable an antiviral RNAi response, this activity is seemingly limited to plants, arthropods, and nematodes (Cullen et al., 2013). In place of RNAi, chordates utilize a small RNA-independent, protein-based defense called the Type I interferon (IFN-I) system as the major antiviral cellular defense (Platanias, 2005). In this system, cellular recognition of viral RNA culminates in the transcriptional activation of a family of IFN-I genes, cytokines that induce the subsequent upregulation of hundreds of IFN-I stimulated genes (ISGs) which work together to inhibit the cellular processes required by the virus to replicate and spread (Platanias, 2005).
Interestingly, chordates do utilize a form of RNAi to target transposable elements through the generation of a class of short RNAs known as PIWI-interacting short RNAs (piRNAs) but this activity is limited to vertebrate germ cells (Aravin et al., 2007). While some experimental results support the notion that pluripotent cells also elicit a small RNA-mediated antiviral response, evidence for such activity is lacking from differentiated cells (Cullen et al., 2013; Li et al., 2013; Maillard et al., 2013). In fact, ablation of Dicer expression from mammalian fibroblasts has been investigated and found to have no impact on virus replication levels with the exception of those viruses that produce their own miRNAs (Bogerd et al., 2014a). Moreover, evidence is mounting that the IFN and RNAi responses may be incompatible with each other. Stem cells have been shown to process double stranded RNA (dsRNA) and not generate IFN-I in contrast to differentiated cells that do not generate siRNAs but instead produce high levels of IFN-I (Wang et al., 2014). The idea that these two systems are mutually exclusive with each other is also supported by the fact that the IFN-I–mediated antiviral response shuts down the RNA induced silencing complex whereas expression of antiviral Dicer induces the IFN-I response (Girardi et al., 2015; Seo et al., 2013). While it remains controversial as to whether stem cells can employ a piRNA-independent antiviral RNAi defense, it is clear that the dominant intrinsic response to virus infection in mammals is IFN-I-based (Backes et al., 2014).
Collectively, these data suggest that IFN-I may have replaced a small RNA-mediated antiviral defense at some point in evolution. While our understanding of the long arms race between chordate hosts and their ever-present pathogenic neighbors remains far from complete, data from chickens suggests that the IFN system arose before the divergence of mammals and birds, ~350 million years ago (Hedges et al., 1996). This framework is further supported by fish, which also generate IFN-I following virus infection (Langevin et al., 2013). Moreover, as the biology of DNA recombination to diversify immune receptors arose in ancestors of jawed vertebrates, the utilization of IFN-I in fish allows us to infer that this defense system appeared prior to the evolution of the more sophisticated innate and adaptive immune responses observed in mammals (Zhang and Gui, 2004). The basis for why chordates seemingly abandoned RNAi in place of IFN-I remains unknown.
In an effort to test whether RNAi could function as the mammalian antiviral defense system, we set out to reconstitute this environment by exploiting the presence of endogenous microRNAs (miRNAs) and repurpose them to be virus-specific siRNAs (tenOever, 2013). To this end, we incorporated species-specific miRNA targets into a non-coding region of influenza A virus (IAV) and characterized the virus in the presence and absence of an IFN-I response. These data found that small RNA-mediated silencing elicited a potent inhibition of virus infection that resulted in strains that could be attenuated by more than five logs. Moreover, we could induce small RNA-mediated virus attenuation by generating strains that produced virus-derived siRNAs. In all, we demonstrate that RNAi is an effective antiviral strategy in mammals and suggest an evolutionary event may have caused the transition from RNAi to IFN-I as either system could have functioned in chordates.
RESULTS
Defining the durability of miRNA-mediated virus targeting
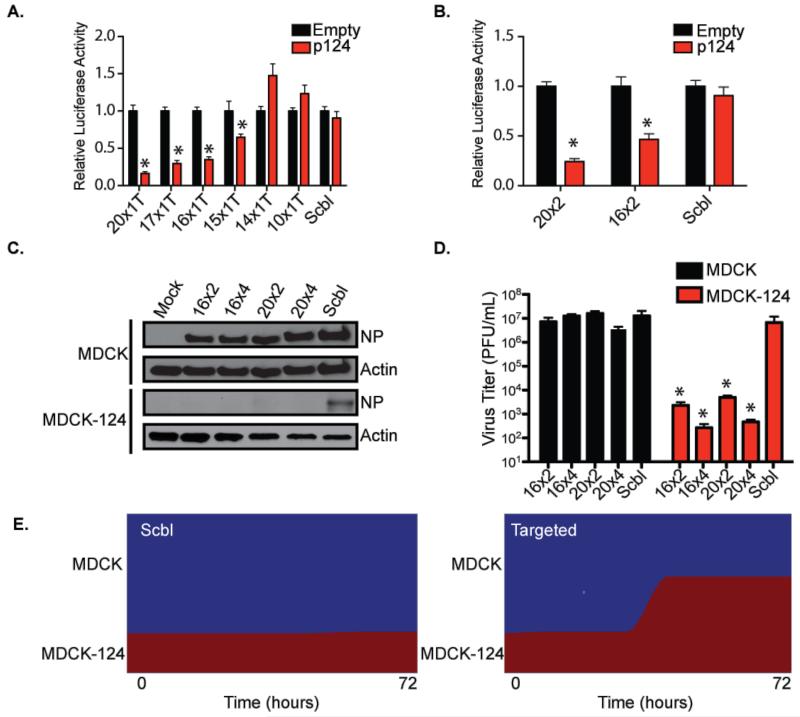
We, and others, have demonstrated that mammalian viruses can be effectively silenced by introducing complementary targets of host miRNAs into the viral genome (Barnes et al., 2008; Kelly et al., 2008; Langlois et al., 2012; Perez et al., 2009; Pham et al., 2012). In an effort to address the durability of this surrogate RNAi response, we first set out to define the minimal complementarity required to induce silencing. To this end, we generated luciferase constructs containing a miR-124 target site in the 3’ untranslated region (UTR) of the mRNA. miR-124 is a neural-specific miRNA of 20 nucleotides (nts) that has been implicated in influencing host splicing (Makeyev et al., 2007). Incorporation of one miR-124 target site, composed of 20nts of perfect complementarity, into the 3’UTR of Gaussia luciferase can successfully silence greater than 80% of its activity following exogenous expression of this neuronal miRNA (Figure 1A). In contrast, when the target was composed of 14nts or less of complementarity, repression was completely lost suggesting the potency of silencing was directly proportional to the strength at which the cognate miRNA could engage its target. In agreement with this, target sites of 17nt, 16nt, and 15nt (measured from the 5’ end of miR-124) demonstrated a clear relationship that correlated binding strength to silencing efficiency (Figure 1A). Interestingly, comparing one vs. two target sites did not impact overall silencing potential in the context of this biological readout (Figure 1B).
Figure 1. Determining miRNA-mediated silencing potential.
(A) Gaussia Luciferase activity from 293T cells co-transfected with a vector expressing miR124 (p124) and a luciferase reporter containing a single miR-124 site with complementarity ranging from 10nts (10×1T) to 20nts (20×1T). Renilla luciferase was used to control for transfection efficiency. Error bars, SD, *p<0.05. (B) As described in (A), where the Gaussia Luciferase construct had two sites of either 16 or 20nts of complementarity. (C) Western blot of protein derived from MDCK or MDCK-124 cells infected with IAV (MOI=0.01) containing two or four target sites described in (A). Top two panels denote NP and actin from MDCK cells, bottom two panels depict the same from MCDK-124 cells. (D) Virus titers derived from multi-cycle growth curves in MDCK or MDCK-124 cells treated with the viruses described in (C). Error bars, SD, *p<0.05. (E) Flow cytometry-based determination of MDCK and MDCK-124 levels following mock infection or treatment with the scrambled (Scbl) or miRNA-targeted (Targeted) virus at 72 hpi.
Drawing upon these results, we generated IAV strains containing miR-124 target sites with 20 or 16nt of complementarity in the 3’ UTR of the nucleoprotein (NP) segment; these target lengths represent perfect binding sites that would be generated in the context of a bona fide RNAi response or the minimal site length required to maintain silencing, respectively (Figure 1A). We reasoned that including these two distinct small RNA targeting site lengths would allow us to evaluate the emergence of escape mutants. Furthermore, despite only needing a single target site to achieve potent silencing of an artificial luciferase reporter assay, we incorporated either two or four sites to ascertain whether the abundance of targets was significant in the context of virus infection. These viruses are referred to as 16×2, 16×4, 20×2, and 20×4 to denote the extent of complementarity and number of target sites, respectively (Figure 1C). Furthermore we generated a control virus (Scbl), which contained a scrambled UTR of identical length and A/U composition to the miR-124 target sites. To determine the impact of lengthening the 3’ UTR of NP, we compared wild type virus (A/Puerto Rico/8/34 or PR8) to our Scbl virus at the level of replication and protein production. These data demonstrated that IAV and IAV-Scbl showed no discernable difference that could be associated with the modified segment as both replicated to levels between 107-108 plaque forming units (pfu)/ml and generated robust levels of NP (Figure S1A and S1B). Next, we chose to compare our Scbl virus to 16×2, 16×4, 20×2, and the 20×4 engineered strains in the presence and absence of miR-124. To this end, we developed a Madin-Darby Canine Kidney (MDCK) cell population expressing miR-124 (MDCK-124) to allow us to compare replication in a model system that differed only in the expression of a single miRNA. Northern blot of MDCK and MDCK-124 cells demonstrated robust production of the desired miRNA (Figure S1C). Consequently, while virus replication of the Scbl, 16×2, 16×4, 20×2, and 20×4 strains demonstrated robust NP expression and titers of ~107pfu/ml in MDCK cells, virus administration of MDCK-124 cells produced no detectable NP and a precipitous drop in titers ranging from 102-104pfu/ml in all but the control untargeted strain (Figure 1C and 1D). Interestingly, in the context of virus infection, we note that attenuation increased with the number of target sites but was not significantly different between a target of complete complementarity vs. one of only 16 contiguous bases.
In an effort to determine whether virus infection could rapidly escape targeting and thus explain the lack of mammalian antiviral RNAi, we passaged the aforementioned viruses in MDCK-124 cells. However, after ~10 passages, we were unable to isolate virus suggesting a lack of escape variants (data not shown). Therefore, we next infected co-cultures of MDCK and MDCK-124 cells at an approximate ratio of 3:1, respectively, to determine if the capacity to replicate in the absence of small RNA-mediated selection would result in the emergence of escape variants. Interestingly, this experimental methodology also failed to generate escape mutants but instead resulted in the selective death of MDCK cells and the generation of an MDCK-124 population majority in just 72hrs (Figure 1E). Taken together, these results illustrate the potency of RNAi and suggest that small RNA-targeting can be achieved with as little as 16nt of complementarity.
Self-targeting demonstrates lack of NS1 antagonism and RNAi potency
While the inability to generate an escape mutant may illustrate the potency of the RNAi system, this phenotype could also be explained by the reported antagonistic RNAi activity of NS1 (Hutvagner and Zamore, 2002; Li et al., 2004). Alternatively, as exploiting host miRNAs provides the cell with the “antiviral siRNAs” prior to infection, the kinetics of this model system, coupled to the incorporation of multiple target sites, may impose a greater selective pressure as would be observed during a bona fide RNAi response where the siRNAs are generated following pathogen recognition.
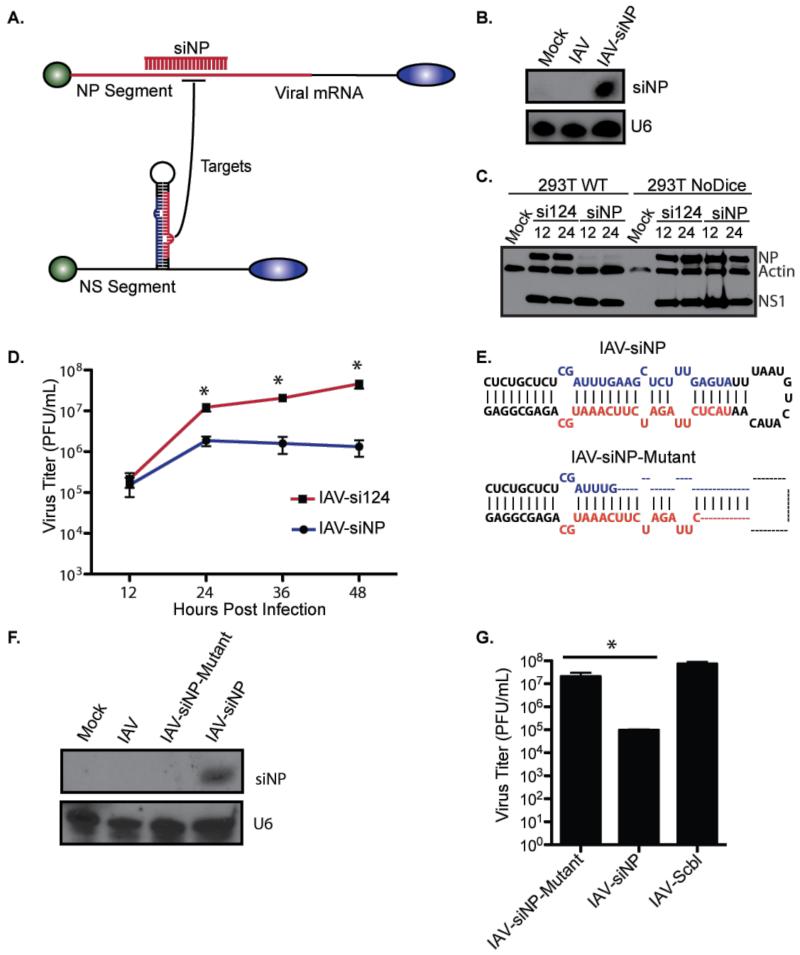
To better discern between these distinct possibilities, we generated a self-targeted IAV strain that expressed an siRNA from the NS segment (encoding wild type NS1) and targeting the ORF of the NP segment at just a single site (Figure 2A). As prior studies had already established that miRNA insertion did not inherently impact IAV biology (Benitez et al., 2015a; Varble et al., 2010), we reasoned this virus design would behave in one of two ways. Either the engineered virus would fail to self-target given the expression of wild type NS1 or the kinetics of siRNA synthesis, or we would force the emergence of an escape mutant through either mutation of the hairpin or the target site. To this end, we infected A549 cells and performed small-RNA northern blotting. The results showed that the virus effectively generated the artificial miRNA against NP (herein referred to as siNP, Figure 2B). Infection of wild-type or Dicer-deficient fibroblasts (termed NoDice cells, (Bogerd et al., 2014b)), with a control virus expressing miR-124 (IAV-si124) or the self-targeted virus (IAV-siNP) showed that the self-targeted virus was dramatically attenuated in wild-type cells but not the absence of Dicer as evidenced by NP protein levels (Figure 2C). Furthermore, IAV-siNP demonstrated more than a ten-fold attenuation within 48hrs of infection compared to IAV124 both in vitro and in vivo at which time escape mutants emerged (Figure 2D, 2E and S2A). Sequencing of plaque-purified variants from self-targeted virus growth curves identified a mutant containing a large deletion in the artificial miRNA responsible for the NP siRNA, herein referred to as IAV-siNP-Mutant (Figure 2E). Northern blot of IAV-siNP-Mutant corroborated that this deletion terminated the production of siNP and restored levels of WT replication (Figure 2F and 2G). Interestingly, we were unable to identify any viruses that mutated the NP target in the ORF, despite the presence of only a single site.
Figure 2. Self-targeting IAV demonstrates the selective pressure imposed by miRNA-mediated silencing.
(A) Schematic depicting a self-targeted virus producing a siRNA against the NP segment (siNP). (B) Northern blot of RNA derived from MDCK cells infected (MOI=1) with IAV-siNP or IAV-si124. Top panel depicts siNP, and lower panel shows U6 as a loading control. (C) Western blot of extracts derived from 293T or NoDice cells infected (MOI=0.1) with IAV-siNP or IAV-si124 for 12 and 24 hpi. Panel depicts NP, NS1, and actin as a loading control. (D) Multi-cycle growth curve in MDCK cells infected with the viruses described in (B). Supernatants were collected at 12, 24, 36, and 48 hpi and plaqued on MDCK cells. Error bars, SD, *p<0.05. (E) Schematic depicting the hairpin in the parental virus (top) and the mutant virus (bottom). (F) Northern blot in MDCK cells infected (MOI=1) with WT IAV and the viruses described in (D). Top panel depicts siNP and lower panel shows U6 as a loading control. (G) Multi-cycle growth in MDCK cells with the viruses in (F). Supernatants were collected and plaqued on MDCK cells. Error bars, SD, *p<0.05.
In an effort to determine how much time the host cell would have to generate a successful RNAi response, we next transfected cells with plasmids expressing either miR-124 or the siNP at 3, 6, 12, or 24 hours prior to infection to determine the degree of attenuation as it relates to the availability of the virus-specific siRNA. As transfection time does not equate to the immediate production of miRNAs, we independently analyzed uninfected samples by northern blot to estimate the levels of miR-124 or siNP at the time of inoculation. These data found that miR-124 and siNP were clearly visible by small RNA northern blot by 12 hrs post transfection (Figure S2B). Comparing miR-124 and siNP signal intensity to the ubiquitous miR-93 loading control, we estimate that plasmid transfection induces approximately 100-1000 copies/cell within 12 hours based on the average quantification of other constitutively expressed miRNAs (Bissels et al., 2009). Cells infected under these same conditions found only samples transfected 12 or 24 hrs prior to virus treatment successfully induced attenuation (Figure S2C). As transfection 6hrs prior to infection failed to target virus, we estimate that the cell must accumulate 100-1000 siRNAs/cell within the initial six hours of infection to successfully result in attenuation. These data are also consistent with the successful self-targeting of the IAV-siNP strain as this virus design generates 100-1000 copies/cell within the first three hours of infection (Varble et al., 2010).
To further corroborate the results for the NP self-targeted virus strain, we generated alternate versions of this virus design where targeting was miR-124-based. To this end, we again used a segment eight that expressed miR-124 and coupled this to the various segment five constructs described in Figure 1C. As anticipated, all but the virus containing a scrambled (Scbl) NP 3’ UTR demonstrated a self-targeting phenotype resulting in a 2-3 log attenuation (Figure S3A-B and Figure 3A). After passaging and sequencing these viruses, we again isolated escape mutants and, as previously observed, found them to be limited to miRNA hairpin mutants that lost the capacity to produce the siRNA. The self-targeting virus with four siRNA binding sites composed of 16nts of complementary (16×4) generated a segment whereby the miR-124 hairpin contained six nt deletions dispersed throughout the hairpin (Figure 3B). This mutated strain (herein referred to as 16×4-Mutant) lost the capacity to generate miR-124 and demonstrated a replication capacity that matched wild type virus (Figure 3C-D). Comparably, passaging a self-targeting virus with four siRNA binding sites composed of 20nts of complementarity also generated an escape mutant. This mutant (20×4-Mutant) excised the complete hairpin and also demonstrated the loss of miR-124 and self-induced viral attenuation (Figure 3E-F).
Figure 3. Determining the role of NS1 during self-targeting.
(A) Multi-cycle growth in MDCK cells infected (MOI=0.01) with the corresponding viruses. Supernatants were collected 24 hpi and plaqued on MDCK cells. Error bars, SD, *p<0.05. (B) Schematic depicting the hairpin in the parental virus (top) and the mutant virus (bottom). The siRNA against NP (siNP) is depicted in red. (C) Northern blot of MDCK cells infected with IAV, 16×4-mutant, or 16×4-control viruses. Top panel depicts miR-124 and lower shows U6 as loading control. (D) Virus titers from MDCK cells infected with the viruses in (C). (E) Northern blot of MDCK cells infected with IAV, 20×4-mutant, or 20×4-control viruses. Top panel depicts miR-124 and lower shows U6 as loading control. (F) Virus titers from MDCK cells infected with the viruses in (E). Error bars, SD, *p<0.05. (G) Virus titers from Wt or Irf3−/−/Irf7−/− MEF infected (MOI=1) with IAV, IAV-124, mIAV-124, IAV-124t/wtNS-124, and IAV-124t/mNS-124. Supernatants were collected 24 hpi and plaqued on MDCK cells. Error bars, SD, *p<0.05. (H) Fold titer of the viruses used in (G). Error bars, SD, *p<0.05.
Next, to further address the role of the intrinsic antiviral response and NS1 antagonistic function during self-targeting, we repeated similar studies in wild type murine embryonic fibroblasts (MEFs) and MEFs lacking Interferon Regulatory Factors 3 and 7 (Irf3−/−/Irf7−/−) which are essential for the establishment of the antiviral response (Daffis et al., 2009). To this end, we infected wt and double knock out cells with either self-targeting or control viruses encoding either wild type NS1 or an RNA binding mutant of NS1 (herein referred to as mNS1, (Donelan et al., 2003)) and ascertained silencing potential (Figure 3G-H). Infection of MEFs with either IAV or IAV-124 yielded comparable titers of 105 pfu/ml and 106 pfu/ml in wild type and Irf3−/−/Irf7−/− cells, respectively, supporting established findings that NS1 effectively antagonizes IRF signaling and that miRNA inclusion does not impact IAV replication in vitro (Garcia-Sastre et al., 1998; Varble et al., 2010). This latter concept could be further corroborated in vivo, as titers of IAV and IAV-124 were indistinguishable in mice at two days post infection (Figure S3C).
In agreement with past studies (Donelan et al., 2003), we also found that loss of NS1 function significantly impaired virus replication in wild type cells but replicated to comparable titers as wild type virus in the absence of an IRF3/7-dependent antiviral response (Figure 3G). Next, to address a report that has suggested NS1 antagonizes small RNA silencing in mammals (Li et al., 2004), we compared self-targeting in these two in vitro systems with viruses encoding either wild type or the RNA binding mutant of NS1. In agreement with the results obtained from our siNP- and 124-based self-targeting viruses, we found that NS1 functionality had minimal impact on the extent of attenuation when the IRF-mediated defenses of the cell are eliminated. Comparing genotype matched viruses to their self-targeting counterparts, encoding either wild type or mutant NS1, demonstrated a ~20-50 fold loss of virus titers during a single cycle of replication in the absence of the intrinsic antiviral cell response (Figure 3H). Interestingly, in an environment when antiviral RNAi was enabled in parallel with a functional IFN-I system, virus titers demonstrated a ~50-100 fold loss suggesting the two pathways can complement each other. Taken together, these results illustrate the effectiveness of small RNA silencing as an antiviral mechanism and suggest that neither NS1 nor the IFN-I response dramatically disrupt the processing, loading, or silencing potential of small RNAs.
Species-specific miRNA-mediated attenuation of influenza A virus
In an effort to generate an attenuated virus that was unchanged at the protein level but could be inhibited independently of the intrinsic defenses of mammalian cells, we incorporated five unique miRNA targets sites into the NP transcript encoded from segment five. In an effort to maintain the capacity to grow the virus, we chose to exploit mammalian-specific miRNAs to permit uninhibited replication in eggs. To this end, we incorporated miR-93 and -192 targets as previously described (Langlois et al., 2013; Perez et al., 2009) in addition to individual targets for miR-21, -31, -and -29b which small RNA deep sequencing identified as being low in embryonated eggs but high in human and mouse lung (Perez et al., 2009).
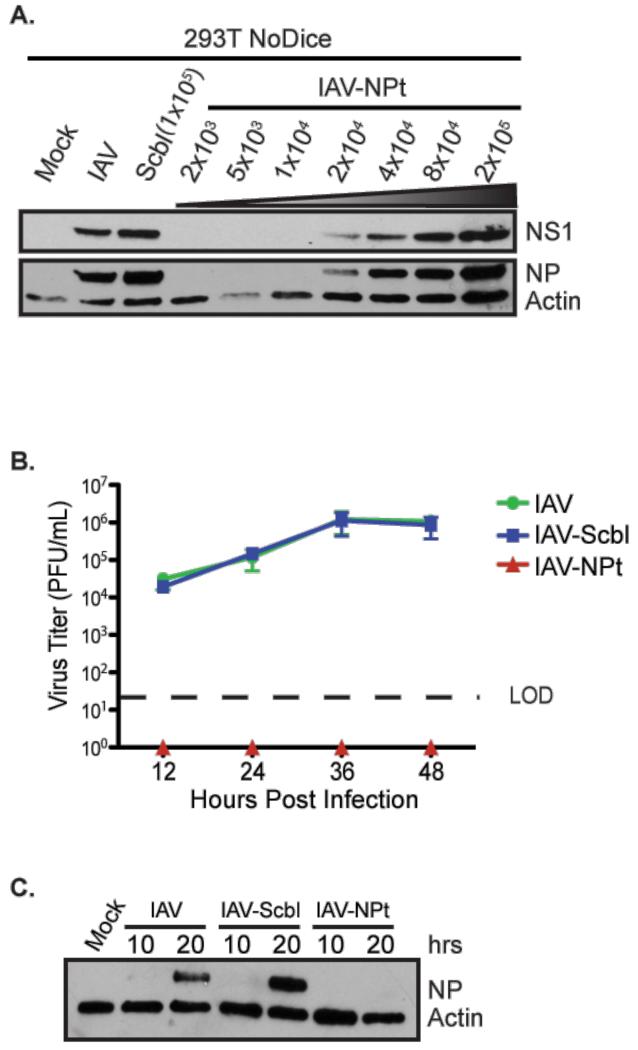
Upon confirming differential expression of miR-21, -31, and -29b (Figure S4A), we inserted individual target sites as an artificial 3’ UTR of NP as previously described (Langlois et al., 2012). As a control, we also generated a virus containing five tandem targets (corresponding to a position in the ORF of enhanced green fluorescent protein (EGFP) that was previously targeted by an artificial miRNA (Varble et al., 2013). In agreement with past studies, the egg-derived stocks of virus demonstrated indistinguishable hemaggluttination (HA) activity, presumably owing to the absence of the aforementioned miRNAs (Figure S4B). In contrast, attempts to quantify virus by standard plaque assay in MDCK cells revealed that the miRNA-targeted strain failed to produce plaques, even when NP was probed with an antibody in place of standard crystal violet staining (Data not shown). As a result, we determined the egg infectious dose (EID) to infect ~50% of the eggs (EID50) for both the control and targeted strains and used this to estimate plaque forming units (pfu) based on the control virus standards. These data suggested the miRNA-targeted virus grew comparably in eggs (2×105 EID50/mL as compared to 6 × 105 EID50/mL for the control strain), corresponding to approximately 1×105 pful/ml and 3×105 pful/ml, respectively). These titers could also be corroborated by semi-quantitative western blot in cells lacking Dicer (Figure 4A).
Figure 4. Engineered antiviral RNAi can potently inhibit virus replication in mammals.
(A) Western blot of extracts derived from NoDice cells infected (MOI=0.1) with IAV, IAV-Scbl, or with increasing amounts of the NP-targeted viruses (IAV-NPt) for 24hrs. Top panel depicts NS1, and the lower panel shows NP and actin as a loading control. Bracketed values represent egg infectious dose 50 (EID50) units for Scbl and IAV-NPt. (B) Multi-cycle growth curve in A549s cells infected (MOI=0.01) with the viruses used in (A). Supernatants were collected 12, 24, 36, and 48 hpi and plaqued on MDCK cells. LOD denotes the level of detection. (C) Western blot of extracts derived from A549 cells infected (MOI=0.1) with the viruses in (A). Lysates were collected 10, and 20 hpi and blotted for NP and actin.
In an effort to better characterize the replication properties of the miRNA-targeted strain, we performed multi-cycle growth curves in A549 cells and measured titers at 12, 24, 36, and 48hrs post infection (Figure 4B). Interestingly, while the control and parental viruses replicated to the same titers with comparable kinetics, the NP-targeted virus failed to produce a single plaque or demonstrate HA activity (Figure 4B and S4C). These data could be further corroborated by western blot demonstrating a complete lack of NP expression (Figure 3C). Taken together, these results illustrate the silencing potential of miRNA-mediated targeting to attenuate virus infection in vitro in a host-specific manner.
Assessing miRNA-mediated virus attenuation in vivo
In an effort to ascertain whether miRNA-mediated virus attenuation maintained its in vitro potency in vivo, we administered 6-week old C57BL/6 mice intranasally with 250 pfu of the control and targeted viruses and monitored morbidity (Figure 5A). Given that the 50% lethal dose (LD50) of the wild type virus is ~50pfu, inoculation of ~5LD50 of the control virus resulted in dramatic weight loss ultimately leading to 100% mortality (Figure 5A). In contrast, the miRNA-targeted strain demonstrated no signs of morbidity or mortality at this inoculating dose or at doses of 2500 and 25000 pfu (Figure 5A). Moreover, hematoxylin and eosin staining of lungs derived from mice infected with 250 pfu of the miRNA-targeted strain demonstrated no signs of inflammation or damage at either two or nine days post infection (Figure 5B). In contrast, the same treatment with untargeted IAV found severe lesions in the bronchiolar epithelium consistent with highly virulent viruses with additional evidence for focal perivascular and alveolar edema at both time points monitored (Figure 5B).
Figure 5. in vivo demonstration of small RNA-mediated attenuation of virus in both wild type and IFN-defect animals.
(A) Graph depicting change in body mass of C57BL mice following intranasal inoculation with PBS, IAV, or IAV-NPt. Error bars, SD, *p<0.05. (B) Histology of lungs from mice infected with IAV, or IAV-NPt. Lungs were harvested 2 and 9 dpi, sectioned and slides were stained with H&E. (C) Graph depicting change in body mass of Ifnar1−/− mice following intranasal inoculation with PBS, IAV, or IAV-NPt. Error bars, SD, *p<0.05.
Finally, in an effort to determine whether mammals could have maintained the evolutionary usage of antiviral small RNAs in place of our IFN-I system, we characterized our virus strains in the context of Ifnar1−/− mice. To this end, we treated knockout mice in a manner identical to that performed in wild type animals, administrating 250, 2500, or 25000 pfu of the miRNA-targeted strain intranasally (Figure 5C). Remarkably, these data found that even at a dose of 25000 pfu (a dose more than 2500 times greater than the LD50 (Koerner et al., 2007) there were no signs of morbidity or mortality despite the absence of IFN-I signaling. Consistent with these findings, virus transcripts on day two showed no detectable levels of virus mRNA (Figure S5A). Attenuation in both WT and Ifnar1−/− mice could be further confirmed at both day two and nine post infection by NP-based plaque assays (Figure S5B and S5C).
miRNA-mediated attenuation can elicit antiviral protection that negates the requirement for IFN-I
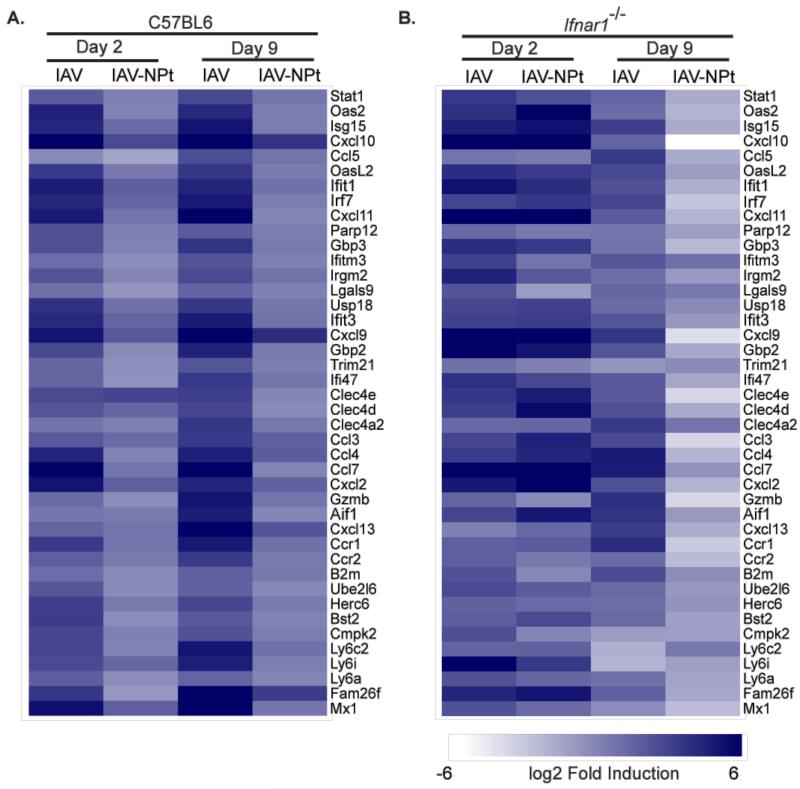
To better ascertain how the intrinsic and adaptive antiviral responses are coordinated under conditions where small RNA silencing has replaced the intrinsic antiviral defenses of the cell, we next used RNA-Seq to compare and contrast the host transcriptome to infection in either the presence or absence of the IFN-I response (Figure 6A and 6B). Strikingly, comparing mock treated mice to either the scrambled control or the miRNA-targeted strain illustrated the potency of small RNA-mediated attenuation. In response to the untargeted strain, the host showed a robust induction of canonical IFN-I stimulated genes (Figure 6A). For example, levels of myxovirus resistance 1 (Mx1) were induced more than 40 fold in response to the control virus but only two fold in response to the miRNA-targeted strain (Table S1). Additional canonical ISGs, such as Ifit1, Oas1b, Oas2, and Isg15 were induced ~10 fold in response to the control infection but remained at baseline levels in response to the miRNA-targeted virus. These trends were also evident at day 9 and included chemokines such as: Ccl12, Cxcl10, Cxcl9, Cscl11, Cxcl13, Ccr5, Ccr1, and Ccl4 (Table S1).
Figure 6. Attenuation of miRNA-mediated viruses does not require the intrinsic antiviral response.
(A and B) Heat maps depicting the transcriptomes of WT or Ifnar1−/− mice infected for 2 and 9 days with IAV or the IAV-NPt viruses based on biological replicates of RNA-Seq data.
The antiviral transcriptional response observed in C57BL6 mice was significantly muted when the same experiment was performed in Ifnar1−/− mice (Figure 6B). Profiling of mRNA in these mice demonstrated the diminished antiviral response that occurs in the absence of any feedback loop elicited by IFN-I signaling. For example, Mx1 levels demonstrated only a three-fold induction in response to wild type virus infection 48hrs post infection (Table S2). Comparable to the transcriptional profiling in wild type mice, the response in Ifnar1−/− also demonstrated a muted response following administration of the miRNA-targeted strain. However, in this analysis, differential expression of ISGs between the two experimental cohorts was less pronounced with genes such as Ifit1. This result is not surprising given that many “ISG”s can also be induced directly in response to the activation of IRF3 and/or IRF7 (Schmid et al., 2010). The modest induction of known IRF-mediated antiviral genes on day two suggests that the intrinsic cellular response to either wild type or miRNA-targeted viruses was comparable – showing an ~5fold induction of antiviral transcription factors (i.e. Stat1), antiviral effectors (i.e. Oas and Isg15), and proinflammatory cytokines (i.e. Cxcl9, Ccl4, and Ccl7). In contrast, on day nine, the elevated levels of these antiviral transcripts remain high in response to untargeted virus yet are completely absent following infection with the miRNA-targeted strain. While this trend is evident for the complete antiviral transcriptome, it is most striking with regards to the proinflammatory cytokines suggesting miRNA-targeted strains did not induce sustained inflammation. Furthermore, qPCR analysis of IAV matrix demonstrated that wild type viruses had not been completely cleared by day nine in either wild type or Ifnar1−/− mice in contrast to the virus targeted via RNAi which yielded only background levels under all conditions tested (Figure S6A and S6B). In all, these data suggest that a small RNA-mediated antiviral defense strategy can effectively replace the intrinsic protein-based response normally utilized by mammals.
DISCUSSION
The capacity to inhibit virus infection using pathogen-derived small RNAs has proven an effective strategy in both prokaryotes and eukaryotes (tenOever, 2013). While it remains unknown whether we replaced an ancient RNAi-based antiviral response with something that was more suited to our evolutionary needs, it is clear that mammals predominantly utilize a cytokine-based system that relies on paracrine signaling and the upregulation of hundreds of antiviral effector proteins (Schoggins et al., 2011). Here we attempt to determine if there is something unique to mammals that precludes our use of a small RNA antiviral defense system by recreating a comparable environment in mice. Interestingly, by exploiting the presence of host or virus-derived siRNAs, we can demonstrate potent attenuation that imposes a strong selective pressure and fails to generate virus escape mutants, even in the complete absence of an IFN-I response.
Given the evolutionary success of the RNAi-mediated antiviral platform, the question remains as to why mammals do not employ a small RNA-based strategy to inhibit virus infection. There is strong circumstantial evidence that self-replicating RNAs were necessary for the emergence of life and that these virus-like elements would have therefore evolved with the earliest common ancestors before branching off into prokaryote- and eukaryote-specific lineages (Prangishvili et al., 2006; Rice et al., 2004). It therefore seems a fair assumption that all life required a common means of responding to these pathogenic RNA elements. As our origins stem from prokaryotes, this would further imply that our defenses at one time involved coupling a nuclease to a specific pathogen-derived sequence, general themes observed in both the CRISPR and RNAi systems. Interestingly, while germline cells have a CRISPR-like activity in the form of piRNAs to combat transposable elements, the general response to virus infection in all other cells is quite distinct (Loo and Gale, 2011).
Explanations for why mammals do not utilize RNAi are numerous. Arguably the most logical explanation would be that strong selective pressure imposed by the capacity to inhibit RNAi may have accelerated the emergence of a new antiviral strategy. Interestingly, mammalian viruses have been suggested to encode suppressors of RNAi (Guo and Steitz, 2014). Amongst RNA viruses, Flock House virus, Nodavirus, Ebola virus, and Influenza A virus have all been suggested to inhibit small RNA-mediated silencing (Fabozzi et al., 2011; Li et al., 2004; Li et al., 2013; Maillard et al., 2013). While this work cannot speak to the claims of the Nodaviridae or Ebola virus antagonistic activity, we do not find that the NS1 protein of IAV is an effective inhibitor of small RNA-mediated silencing. While we do note that attenuation of a self-targeting virus was marginally elevated when comparing wild type and mutant NS1 genotypes, the small difference in targeting efficiency is unlikely to reflect direct antagonism of this pathway but rather is more likely due to an unrelated mechanism such as increased defective interfering particles that form in the absence of a functional NS1. This is supported by independent studies that found NS1 overexpression had no impact on RNAi activity in mammalian cells in the presence or absence of virus infection (Perez et al., 2009). It should also be noted that Ebola virus infections have been successfully treated, even three days post infection, with exogenous delivery of siRNAs (Thi et al., 2015; Thi et al., 2014). Taken together, these results suggest that should these viruses have some capacity to block the RNAi response, viruses such as these would be unlikely to exert the type of evolution pressure needed to demand the emergence of the IFN-I system. In contrast, many RNA viruses readily escape RNAi targeting and thus do not require a mechanism to inhibit such a response. Experiments applying exogenous siRNAs as a means to treat virus infection, while generally effective, often result in the emergence of escape mutants (Barnes et al., 2008; Boden et al., 2003; Das et al., 2004; Gitlin et al., 2002; Heiss et al., 2011; Heiss et al., 2012; Teterina et al., 2014; Wilson and Richardson, 2005). While virus escape was sometimes the result of an excision event (Pham et al., 2012), in general, single mutations in a given small RNA target were sufficient to relieve silencing (van Rij and Andino, 2006). For viruses such as poliovirus, which encodes an error-prone RNA-dependent RNA polymerase (RdRp), a spectrum of viruses encoding numerous single mutations is generated at every round of infection, inevitably allowing for escape of RISC-mediated silencing (Gitlin et al., 2005). Given this, an error-prone RNA virus might provide the evolutionary pressure needed to select for the emergence of the IFN-I system.
In contrast to RNA viruses, there is strong evidence that DNA viruses encode antagonists to the small RNA silencing pathways of mammals. DNA viruses that disrupt aspects of miRNA biology include: poxviruses (Backes et al., 2012), adenoviruses (Lu and Cullen, 2004), and herpesviruses (Cazalla et al., 2010). Given these dynamics, one could envision that perhaps a DNA pathogen in our evolutionary past rendered our RNAi response ineffective and thus would have demanded an evolutionary need for the emergence of IFN-I.
Lastly, it is possible that the intrinsic response to virus infection in chordates never included small RNAs. This concept is supported by the lack of evolutionary duplication with regards to components of the small RNA machinery. In contrast to mammals, plants show evidence of many Dicer-like proteins, each one with the unique capacity to recognize specific virus genomes (Xie et al., 2004) much like the expansion of IRFs and/or IFN alpha loci in many chordate species (Langevin et al., 2013; Manry et al., 2011). Furthermore, given the necessity to generate high levels of siRNAs in a very short time period, it would seem that chordates would also require an RdRp to amplify virus-derived small RNAs and an RNA transporter to provide systemic protection. Interestingly, while chordates do express a homolog of SID-1 (systemic RNAi defective 1), a gene product responsible for small RNA movement between cells, we do not express an RdRp (Jose and Hunter, 2007). While the function of SID-1 in mammals remains unknown, expression of an exogenous RdRp has been associated with the induction of IFN-I suggesting these two systems may be inherently incompatible (Yu et al., 2012).
Regardless of the RNAi history in chordate biology, it is clear that the small RNA machinery of mammalian cells has the capacity to potently target RNA viruses when exploited. While this may be universally true for all viruses, one possible explanation for the emergence of the more general IFN-I response could have derived from pathogens whose biology is not amenable to small RNA targeting. For example, present day pathogens such as malaria and cholera have been implicated in imposing strong selective pressure in humans in a very short evolutionary time span (Siddle and Quintana-Murci, 2014), so perhaps it was an ancient protozoan or bacterium of early chordates that rendered the specificity of the RNAi response useless and demanded the emergence of the more general IFN-I response. While finding answers to such evolutionary questions are near impossible to address, what is clear based on this study is that mammals could have utilized RNAi in place of IFN-I and that the existing machinery can be exploited for therapeutic benefit.
EXPERIMENTAL PROCEDURES
Virus Engineering and Generation
HEK293T cells were transfected with plasmids expressing bidirectional vRNA and mRNA as previously described (Hoffmann et al., 2000). All of the constructs encode for components of A/Puerto Rico/8/34 (H1N1). To generate the self-targeted viruses, a modified NS segment was used which has been described elsewhere (Varble et al., 2010). Viruses encoding self-targeting siRNAs were rescued in NoDice cells (Bogerd et al., 2014b). Cells were collected 48 hours post transfection and injected into the allantoic fluid of 8-day old embryonated eggs. Allantoic fluid was extracted 48 hours post injection and viruses were quantified by hemagglutinin assay using chicken red blood cells (Lampire Biological Pharmaceuticals) in Alsevers and tittered by plaque assay on MDCK cells. Influenza plaque assays were performed with media containing EMEM (Lonza), L-glutamine (Corning), NaHCO3, BSA, and 2% agar (Oxoid).
Small-RNA Northern Blot
Total RNA was harvested at the indicated time points using the Trizol method and Northern blotting was performed as previously described (Pall and Hamilton, 2008). The following probes were used: anti-miR-21: TCAACATCAGTCTGATAAGCTA, anti-miR-31: AGCTATGCCAGCATCTTGCCT, miR-29b: AACACTGATTTCAAATGGTGCTA, anti-miR-192: GGCTGTCAATTCATAGGTCAG, anti-miR-93: CTACCTGCACGAACAGCACTTTG, anti-miR-124: TGGCATTCACCGCGTGCCTTAA, anti-siNP: GCATTTGAAGATCTAAGAGTA, and anti-U6: GCCATGCTAATCTTCTCTGTATC.
Deep Sequencing
mRNA-Seq libraries were prepared using 1ug total RNA. The TruSeq RNA Library Preparation Kit v2 was used according to the manufacture’s instructions. Briefly, mRNA was purified via oligo-dT beads, fragmented, and reverse transcribed with SuperScript II (Invitrogen), followed by second strand synthesis, end repair, A-tailing, and adapter ligation. Quantification of barcoded samples and pooled libraries were assessed using the Universal complete KAPA Library Quantification Kit (KAPA Biosystems). Pooled libraries were run on either an Illumina MiSeq platform using the MiSeq Reagent Kit v3 (Illuminia) or an Illumina HiSeq. Read mapping, statistical analysis, and dot plot generation was preformed using BaseSpace RNA Express application (Illumina). The publicly available platform www.interferome.org was used to select the interferon-stimulated genes (ISGs) displayed in the heat maps. RNA-Seq results were visualized using the Microarray Software Suite TM4 (http://www.tm4.org/mev.html). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE73698.
Supplementary Material
Acknowledgements
We wish to thank Dr. B. Cullen (Duke University) and Michael Gale (Washington University), for NoDice and Irf3−/−Irf7−/− knockout cells, respectively. We would also like to acknowledge the technical contribution of the flow cytometry Core at the Icahn School of Medicine at Mount Sinai. This material is based upon work supported in part by the Burroughs Wellcome Fund. BRT is also supported by the National Institute of Allergy and Infectious Diseases grants R01AI110575 and 5R01AI093571. AAB is supported by the National Institute of Health grant 1F31AI110130-01A1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contribution
AAB and LAS designed and conducted all virus-based experiments and wrote the manuscript. MB designed and performed miR-124-mediated silencing of luciferase. DS analyzed all RNA-Seq data. BRT designed experiments and wrote the manuscript.
REFERENCES
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Backes S, Langlois RA, Schmid S, Varble A, Shim JV, Sachs D, tenOever BR. The Mammalian response to virus infection is independent of small RNA silencing. Cell Rep. 2014;8:114–125. doi: 10.1016/j.celrep.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes S, Shapiro JS, Sabin LR, Pham AM, Reyes I, Moss B, Cherry S, tenOever BR. Degradation of host microRNAs by poxvirus poly(A) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell Host Microbe. 2012;12:200–210. doi: 10.1016/j.chom.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes D, Kunitomi M, Vignuzzi M, Saksela K, Andino R. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe. 2008;4:239–248. doi: 10.1016/j.chom.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Benitez AA, Panis M, Xue J, Varble A, Shim JV, Frick AL, Lopez CB, Sachs D, tenOever BR. In Vivo RNAi Screening Identifies MDA5 as a Significant Contributor to the Cellular Defense against Influenza A Virus. Cell Rep. 2015a doi: 10.1016/j.celrep.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez AA, Panis M, Xue J, Varble A, Shim JV, Frick AL, Lopez CB, Sachs D, tenOever BR. In Vivo RNAi Screening Identifies MDA5 as a Significant Contributor to the Cellular Defense against Influenza A Virus. Cell Rep. 2015b;11:1714–1726. doi: 10.1016/j.celrep.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissels U, Wild S, Tomiuk S, Holste A, Hafner M, Tuschl T, Bosio A. Absolute quantification of microRNAs by using a universal reference. RNA. 2009;15:2375–2384. doi: 10.1261/rna.1754109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden D, Pusch O, Lee F, Tucker L, Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Skalsky RL, Kennedy EM, Furuse Y, Whisnant AW, Flores O, Schultz KL, Putnam N, Barrows NJ, Sherry B, et al. Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J Virol. 2014a;88:8065–8076. doi: 10.1128/JVI.00985-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Whisnant AW, Kennedy EM, Flores O, Cullen BR. Derivation and characterization of Dicer- and microRNA-deficient human cells. RNA. 2014b;20:923–937. doi: 10.1261/rna.044545.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR, Cherry S, tenOever BR. Is RNA interference a physiologically relevant innate antiviral immune response in mammals? Cell Host Microbe. 2013;14:374–378. doi: 10.1016/j.chom.2013.09.011. [DOI] [PubMed] [Google Scholar]
- Daffis S, Suthar MS, Szretter KJ, Gale M, Jr., Diamond MS. Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent signal that does not require IRF-3 and IRF-7. PLoS Pathog. 2009;5:e1000607. doi: 10.1371/journal.ppat.1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, Berkhout B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelan NR, Basler CF, Garcia-Sastre A. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol. 2003;77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabozzi G, Nabel CS, Dolan MA, Sullivan NJ. Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J Virol. 2011;85:2512–2523. doi: 10.1128/JVI.01160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Girardi E, Lefevre M, Chane-Woon-Ming B, Paro S, Claydon B, Imler JL, Meignin C, Pfeffer S. Cross-species comparative analysis of Dicer proteins during Sindbis virus infection. Sci Rep. 2015;5:10693. doi: 10.1038/srep10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L, Karelsky S, Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- Gitlin L, Stone JK, Andino R. Poliovirus escape from RNA interference: short interfering RNA-target recognition and implications for therapeutic approaches. J Virol. 2005;79:1027–1035. doi: 10.1128/JVI.79.2.1027-1035.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YE, Steitz JA. Virus meets host microRNA: the destroyer, the booster, the hijacker. Mol Cell Biol. 2014;34:3780–3787. doi: 10.1128/MCB.00871-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Parker PH, Sibley CG, Kumar S. Continental breakup and the ordinal diversification of birds and mammals. Nature. 1996;381:226–229. doi: 10.1038/381226a0. [DOI] [PubMed] [Google Scholar]
- Heiss BL, Maximova OA, Pletnev AG. Insertion of microRNA targets into the flavivirus genome alters its highly neurovirulent phenotype. J Virol. 2011;85:1464–1472. doi: 10.1128/JVI.02091-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss BL, Maximova OA, Thach DC, Speicher JM, Pletnev AG. MicroRNA targeting of neurotropic flavivirus: effective control of virus escape and reversion to neurovirulent phenotype. J Virol. 2012;86:5647–5659. doi: 10.1128/JVI.07125-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. RNAi: nature abhors a double-strand. Curr Opin Genet Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- Jose AM, Hunter CP. Transport of sequence-specific RNA interference information between cells. Annu Rev Genet. 2007;41:305–330. doi: 10.1146/annurev.genet.41.110306.130216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly EJ, Hadac EM, Greiner S, Russell SJ. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat Med. 2008;14:1278–1283. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]
- Koerner I, Kochs G, Kalinke U, Weiss S, Staeheli P. Protective role of beta interferon in host defense against influenza A virus. J Virol. 2007;81:2025–2030. doi: 10.1128/JVI.01718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin C, Aleksejeva E, Passoni G, Palha N, Levraud JP, Boudinot P. The antiviral innate immune response in fish: evolution and conservation of the IFN system. J Mol Biol. 2013;425:4904–4920. doi: 10.1016/j.jmb.2013.09.033. [DOI] [PubMed] [Google Scholar]
- Langlois RA, Albrecht RA, Kimble B, Sutton T, Shapiro JS, Finch C, Angel M, Chua MA, Gonzalez-Reiche AS, Xu K, et al. MicroRNA-based strategy to mitigate the risk of gain-of-function influenza studies. Nat Biotechnol. 2013;31:844–847. doi: 10.1038/nbt.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois RA, Varble A, Chua MA, Garcia-Sastre A, tenOever BR. Hematopoietic-specific targeting of influenza A virus reveals replication requirements for induction of antiviral immune responses. Proc Natl Acad Sci U S A. 2012;109:12117–12122. doi: 10.1073/pnas.1206039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia-Sastre A, Ball LA, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci U S A. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. Antiviral RNA interference in mammalian cells. Science. 2013;342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manry J, Laval G, Patin E, Fornarino S, Itan Y, Fumagalli M, Sironi M, Tichit M, Bouchier C, Casanova JL, et al. Evolutionary genetic dissection of human interferons. J Exp Med. 2011;208:2747–2759. doi: 10.1084/jem.20111680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- Perez JT, Pham AM, Lorini MH, Chua MA, Steel J, tenOever BR. MicroRNA-mediated species-specific attenuation of influenza A virus. Nat Biotechnol. 2009;27:572–576. doi: 10.1038/nbt.1542. [DOI] [PubMed] [Google Scholar]
- Pham AM, Langlois RA, TenOever BR. Replication in cells of hematopoietic origin is necessary for Dengue virus dissemination. PLoS Pathog. 2012;8:e1002465. doi: 10.1371/journal.ppat.1002465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: a unifying view. Nat Rev Microbiol. 2006;4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- Rice G, Tang L, Stedman K, Roberto F, Spuhler J, Gillitzer E, Johnson JE, Douglas T, Young M. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc Natl Acad Sci U S A. 2004;101:7716–7720. doi: 10.1073/pnas.0401773101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid S, Mordstein M, Kochs G, Garcia-Sastre A, Tenoever BR. Transcription factor redundancy ensures induction of the antiviral state. J Biol Chem. 2010;285:42013–42022. doi: 10.1074/jbc.M110.165936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Kincaid RP, Phanaksri T, Burke JM, Pare JM, Cox JE, Hsiang TY, Krug RM, Sullivan CS. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe. 2013;14:435–445. doi: 10.1016/j.chom.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddle KJ, Quintana-Murci L. The Red Queen’s long race: human adaptation to pathogen pressure. Curr Opin Genet Dev. 2014;29:31–38. doi: 10.1016/j.gde.2014.07.004. [DOI] [PubMed] [Google Scholar]
- tenOever BR. RNA viruses and the host microRNA machinery. Nat Rev Microbiol. 2013;11:169–180. doi: 10.1038/nrmicro2971. [DOI] [PubMed] [Google Scholar]
- Teterina NL, Liu G, Maximova OA, Pletnev AG. Silencing of neurotropic flavivirus replication in the central nervous system by combining multiple microRNA target insertions in two distinct viral genome regions. Virology. 2014;456-457:247–258. doi: 10.1016/j.virol.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi EP, Mire CE, Lee AC, Geisbert JB, Zhou JZ, Agans KN, Snead NM, Deer DJ, Barnard TR, Fenton KA, et al. Lipid nanoparticle siRNA treatment of Ebola-virus-Makona-infected nonhuman primates. Nature. 2015;521:362–365. doi: 10.1038/nature14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi EP, Mire CE, Ursic-Bedoya R, Geisbert JB, Lee AC, Agans KN, Robbins M, Deer DJ, Fenton KA, MacLachlan I, et al. Marburg virus infection in nonhuman primates: Therapeutic treatment by lipid-encapsulated siRNA. Sci Transl Med. 2014;6:250ra116. doi: 10.1126/scitranslmed.3009706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij RP, Andino R. The silent treatment: RNAi as a defense against virus infection in mammals. Trends Biotechnol. 2006;24:186–193. doi: 10.1016/j.tibtech.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Varble A, Benitez AA, Schmid S, Sachs D, Shim JV, Rodriguez-Barrueco R, Panis M, Crumiller M, Silva JM, Sachidanandam R, et al. An in vivo RNAi screening approach to identify host determinants of virus replication. Cell Host Microbe. 2013;14:346–356. doi: 10.1016/j.chom.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Varble A, Chua MA, Perez JT, Manicassamy B, Garcia-Sastre A, tenOever BR. Engineered RNA viral synthesis of microRNAs. Proc Natl Acad Sci U S A. 2010;107:11519–11524. doi: 10.1073/pnas.1003115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Wang J, Acharya D, Paul AM, Bai F, Huang F, Guo YL. Antiviral responses in mouse embryonic stem cells: differential development of cellular mechanisms in type I interferon production and response. J Biol Chem. 2014;289:25186–25198. doi: 10.1074/jbc.M113.537746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- Wilson JA, Richardson CD. Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J Virol. 2005;79:7050–7058. doi: 10.1128/JVI.79.11.7050-7058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu GY, He G, Li CY, Tang M, Grivennikov S, Tsai WT, Wu MS, Hsu CW, Tsai Y, Wang LH, et al. Hepatic expression of HCV RNA-dependent RNA polymerase triggers innate immune signaling and cytokine production. Mol Cell. 2012;48:313–321. doi: 10.1016/j.molcel.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gui J. Molecular characterization and IFN signal pathway analysis of Carassius auratus CaSTAT1 identified from the cultured cells in response to virus infection. Dev Comp Immunol. 2004;28:211–227. doi: 10.1016/s0145-305x(03)00138-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.