Abstract
Detecting pathogenic DNA by intracellular receptors termed “sensors” is critical toward galvanizing host immune responses and eliminating microbial infections. Emerging evidence has challenged the dogma that sensing of viral DNA occurs exclusively in sub-cellular compartments normally devoid of cellular DNA. The interferon-inducible protein IFI16 was shown to bind nuclear viral DNA and initiate immune signaling, culminating in antiviral cytokine secretion. Here, we review the newly characterized nucleus-originating immune signaling pathways, their links to other crucial host defenses, and unique mechanisms by which viruses suppress their functions. We frame these findings in the context of human pathologies associated with nuclear replicating DNA viruses.
Keywords: cytokine induction, infectious disease, protein-DNA interaction, signaling, viral immunology, viral DNA sensing, IFI16, IFIX, cGAS, STING
Introduction
Existing under the constant risk of invasion by pathogenic microorganisms, mammalian cells employ an array of constitutively expressed, germ-line-encoded receptors to survey the intra- and extracellular milieu for pathogen- or damage-associated molecules. Binding of these cellular receptors to their specific ligands evokes intracellular immune signaling cascades that culminate in the robust expression and secretion of antiviral cytokines, such as type I interferons (IFNs). Upon their release from the cell, these induced signaling factors operate in both an autocrine and a paracrine manner, inciting nearby cells to assume antiviral transcriptional programs. Cytokines further stimulate mammalian innate and adaptive immune responses, including the recruitment of antigen-presenting or cytotoxic leukocytes and the production of microbe-specific antibodies. Altogether, these cytokine-coordinated events promote the elimination of the invading pathogen. Thus, the molecular mechanisms mediating the recognition of and response to microbial molecular signatures are critically important.
The DNA genomes of DNA viruses, a class of prevalent human pathogens, serve as one such immunogenic molecular signature. For preventing spurious auto-activation with cellular “self” DNA, the intracellular surveillance of viral DNA was thought to occur exclusively in cytosolic and endosomal compartments. However, this model fails to reconcile the fact that nearly all DNA viruses deposit and replicate their DNA genomes exclusively within host nuclei. As part of a shifting paradigm, recent studies have established the existence of cellular DNA sensors that detect viral DNA within the nucleus to trigger immune signaling. Here, we review the recent discoveries and ongoing challenges within the emerging field of nuclear DNA sensing.
The Impact of Nuclear Replicating DNA Viruses on Human Health
Nuclear replicating DNA viruses are at the root of many major human public health concerns worldwide, triggering a range of symptomatic diseases and irreversible disorders (Table 1). Many of these viruses manifest with high morbidity and mortality, particularly in the immunocompromised and young. Epstein-Barr virus (EBV),2 Kaposi sarcoma-associated herpesvirus (KSHV), human papillomavirus, hepatitis B virus, and Merkel cell polyomavirus are responsible for a staggering majority of virus-associated human cancers globally (1–3). Human papillomavirus is considered the most prevalent sexually transmitted pathogen in the United States (4). Hepatitis B virus has infected nearly 2 billion individuals globally, 350 million of whom are persistently infected, and thus, prone to developing hepatitis, hepatocellular carcinoma, and cirrhosis (4). As a family, adenoviruses account for ∼8% of symptomatic viral diseases worldwide and are common causes of febrile illness, acute respiratory diseases, and gastrointestinal diseases (5).
TABLE 1.
Human nuclear replicating DNA viruses, associated diseases, and implicated DNA sensors
Only nuclear replicating DNA viruses known to infect humans are listed. Viruses that infect other organisms, such as nuclear polyhedrosis virus and polyoma virus, are not listed. ND, not determined; DNA-PK, DNA-dependent protein kinase.
| Virus family and type | Associated human disease(s) | Implicated DNA sensor(s) |
|---|---|---|
| Adenoviridae | ||
| Adenovirus A (Types 12, 18, and 31) | Cryptic enteric infection | ND |
| Adenovirus B (Types 3, 7, 11, and 14) | Conjunctivitis, acute respiratory disease, hemorrhagic cystitis | ND |
| Adenovirus C (Types 1, 2, 5, and 6) | Endemic infection, respiratory symptoms | cGAS (87), TLR9 (88) |
| Adenovirus D (Types 8–10, 13, 15, 17, 19, 20, 22–30, 32, 33, 36–39, 42–49, 51, 53, and 54) | Keratoconjunctivitis | ND |
| Adenovirus E (Type 4) | Conjunctivitis, acute respiratory disease | ND |
| Adenovirus F (Types 40 and 41) | Infantile diarrhea | ND |
| Adenovirus G (Type 52) | Gastroenteritis | ND |
| Circoviridae | ||
| Transfusion-transmitted virus | Largely asymptomatic | ND |
| Hepadnaviridae | ||
| Hepatitis B virus | Hepatitis B | AIM2 (89), cGAS (90) |
| Herpesviridae | ||
| Cytomegalovirus | Mucoepidermoid carcinoma, retinitis, colitis, hepatitis, pneumonitis, myelitis, encephalitis in immunocompromised patients, mononucleosis-like syndrome in immunocompetent adults, congenital neurodevelopmental sequelae | AIM2, DAI/ZBP1, IFI16,a TLR7, TLR9 (91–93) |
| Epstein-Barr virus | Mononucleosis, cancer (Hodgkin lymphoma, Burkitt lymphoma, nasopharyngeal carcinoma) | RNA Pol III, IFI16a (43), TLR9 (94) |
| Herpes simplex virus type 1 | Orofacial disease (cold sores), keratitis, genital herpes | cGASa (58), DAI/ZBP1 (95), DDX41, DDX60, DHX9, DHX36, DNA-PK, RNA Pol III, IFI16,a TLR9 (96), IFIXa (29) |
| Herpes simplex virus type 2 | Orofacial disease (cold sores), keratitis, genital herpes | DNA-PK, TLR9 (96, 97) |
| Herpesvirus 6 variant A | Mononucleosis-like syndrome | ND |
| Herpesvirus 6 variant B | Infantile seizures, roseola infantum, exanthema subitum, respiratory disease, febrile seizures | ND |
| Herpesvirus 7 | Exanthema subitum | ND |
| Herpesvirus 8 (Kaposi sarcoma-associated virus) | Kaposi sarcoma, multicentric Castleman disease, primary effusion lymphoma, | IFI16a (39, 44) |
| Varicella zoster virus | Chicken pox, herpes zoster (shingles) | NLRP3 (98), TLR9 (99) |
| Papillomaviridae | ||
| Papillomavirus (>170 types) | Warts, epidermodysplasia verruciformis, focal epithelial hyperplasia and papillomas (oral), anal dysplasia, oropharyngeal and genital cancers, verrucous cyst | TLR9 (100) |
| Parvoviridae | ||
| Adeno-associated virus | Possible role in male sterility | ND |
| Parvovirus B19 | Erythema infectiosum (Fifth disease), arthropathy, hydrops fetalis, persistent anemia, transient aplastic crisis in patients with underlying hemolytic disorders | ND |
| Polyomaviridae | ||
| BK virus | Largely asymptomatic in healthy individuals, ureteric stenosis in renal transplant patients, hemorrhagic cystitis, nephropathy | ND |
| JC virus | Largely asymptomatic in healthy individuals, progressive multifocal leukoencephalopathy, ureteric stenosis in renal transplant patients | ND |
| Merkel cell polyomavirus | Merkel cell carcinoma | ND |
a DNA sensors with implicated functions in the nucleus.
Among DNA viruses, herpesviruses have evolved sophisticated strategies to establish lifelong latent infections in humans. Periodic virus reactivation within an individual allows for the continual spread of infectious particles to other susceptible hosts. For example, EBV persists in ∼90% of adults, irrespective of geographical location, and is linked to the development of B-cell lymphomas (6). Human cytomegalovirus (HCMV) can cause retinitis, myelitis, and pneumonitis upon reactivation from latency, but also incites serious health threats in pregnant women, neonates, and the immunocompromised. In the developed world, congenital CMV presents itself symptomatically in 22–38% of infants born with CMV (7, 8) and is the leading infectious cause of mental retardation and hearing loss (9).
Despite significant efforts, current therapeutic strategies for controlling nuclear replicating DNA viruses have limitations. Many current antiviral drugs either have teratogenic side effects or lead to the emergence of resistant viral strains. Nucleoside analogues that inhibit viral DNA polymerases are current standard therapies. Ganciclovir, for instance, is used for treating CMV-infected individuals, yet it induces leukocytopenia in immunosuppressed patients undergoing tissue transplantation. Acyclovir is another widely employed nucleoside analogue for managing herpes simplex virus (HSV)-associated pathologies, yet acquired viral drug resistance is observed in immunocompromised patients (10). Thus, in addition to targeting viral factors, rational drug design efforts have also focused on targeting cellular factors that influence viral replication. One promising strategy is to activate the cellular receptors mediating the immunological responses in infected cells. For antiviral therapeutics against DNA viruses, these targets include DNA sensors (11).
Cellular DNA-sensing Pathways
For over 50 years, it has been understood that mammalian cells can discriminate foreign DNA to elicit immunological responses. Early seminal work demonstrated that cellular uptake of exogenous DNA or ultraviolet-inactivated DNA viruses induced production of IFNs (12–15). Subsequently, when challenged with live viruses, these IFN-stimulated cells could drastically limit viral replication (16, 17). Only within the past 10 years, however, has there been significant progress in delineating the responsible cellular pathways. Screens of human and murine cDNA libraries led three independent research groups to simultaneously identify the endoplasmic reticulum adapter protein STING (also known as MITA, ERIS, and TMEM173) to spontaneously induce expression of IFNs and IFN-stimulated genes (18–20). Additional studies revealed that cells derived from Sting−/− mice or depleted of STING by RNA interference fail to produce IFN responses to various exogenous DNA substrates and DNA viruses. Furthermore, Sting−/− mice more readily succumb to HSV-1 infection and fail to generate appropriate adaptive immune responses to DNA vaccines (21). Importantly, STING is widely expressed across many tissues and cell types (18, 19) and phylogenetically conserved within vertebrates (22). Furthermore, the demonstration that its C terminus recruits the kinase TANK-binding kinase 1 (TBK-1) (23), in turn phosphorylating and activating the transcription factor IRF3 (24–27), mechanistically linked STING to established type I IFN signaling factors and solidified its central role as an immune signaling hub (27).
The characterization of the STING-TBK-1-IRF3 signaling axis has spawned significant efforts to identify upstream DNA sensors, which could engage foreign DNA and propagate immune signaling via STING. To date, a myriad of putative DNA sensors has been reported based on their ability to bind foreign or pathogenic DNA and contribute to cytokine induction (Table 1). Although the actions of these DNA sensors have been predominantly studied in the cytosol and endosomes, emerging evidence is shifting focus to a new sub-cellular compartment: the nucleus.
Emergence of Nuclear DNA Sensing
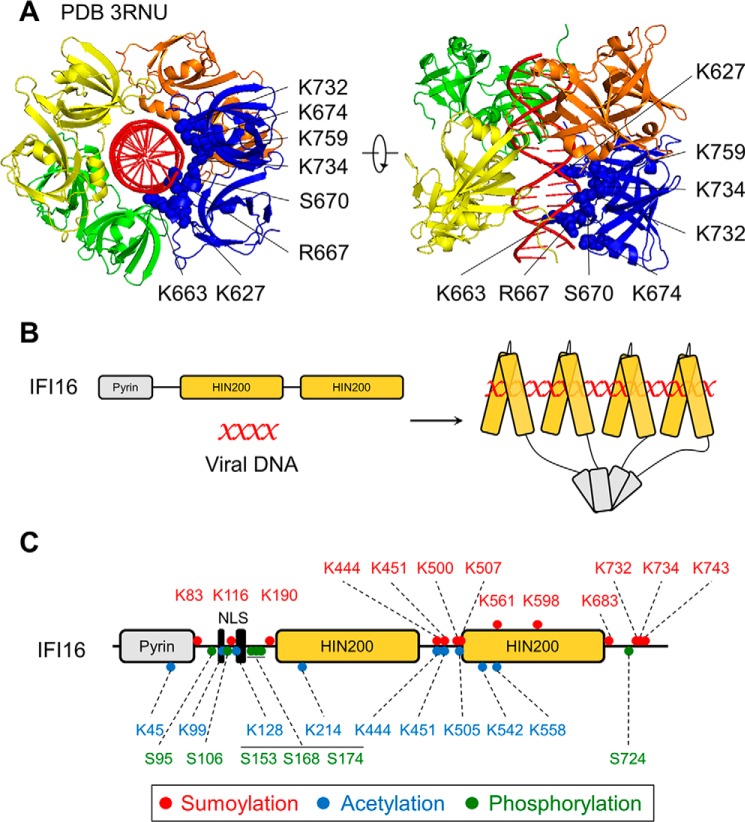
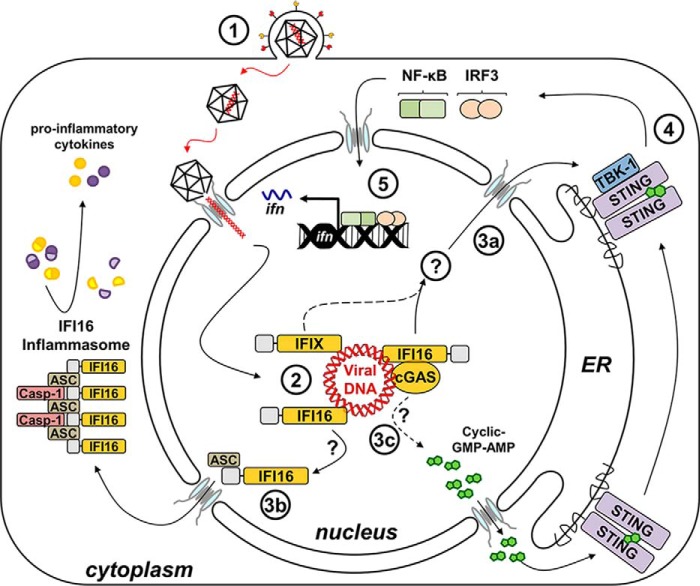
The human interferon-inducible protein IFI16 was recently characterized as the first viral DNA sensor to function within the nucleus. A member of the interferon-inducible human PYHIN protein family (28, 29), IFI16 possesses two C-terminal HIN200 domains that bind to DNA in a sequence-independent manner (30) and an N-terminal pyrin (PY) domain that mediates intramolecular homotypic interactions and cooperative assembly of IFI16 oligomers (31, 32) (Fig. 1, A and B). Originally, IFI16 was shown to bind to cytosolic viral DNA, physically engage STING, and induce IFN production in macrophage-like THP-1 cells (33–35). However, across non-immune cell types, IFI16 predominantly localizes to the nucleus (29, 31, 36, 37). We defined a multi-partite nuclear localization signal on IFI16 and observed that its deletion triggers the localization of IFI16 exclusively to the cytoplasm (36). This mis-localization compromises the ability of IFI16 to both bind to genomic HSV-1 DNA in the nucleus and promote an appropriate immune response to HSV-1 infection in epithelial cells (36). In a subsequent study, Orzalli et al. (37) demonstrated that in human primary fibroblasts, both STING and IFI16 are required for inducing expression of IFN and IFN-stimulated genes in response to the HSV-1 d109 strain, an HSV-1 mutant that deposits its DNA genome into the nucleus but lacks the ability to express viral gene products (38). Protease inhibitor treatment blocks the release of the genome from the d109 nucleocapsid and precludes IFI16- and STING-dependent immune signaling (37). Thus, both studies provide compelling evidence that the deposition of the HSV-1 DNA genome into the nuclear milieu and its subsequent detection by nuclear IFI16 initiate an immune signaling cascade derived from within the nucleus (Fig. 2). Corroborating this model, we have more recently demonstrated that infection with HCMV also induces the expression of host antiviral cytokines via an IFI16- and STING-dependent signaling pathway in human fibroblasts (31).
FIGURE 1.
Cooperative binding of IFI16 is mediated by HIN domains binding viral dsDNA and pyrin domains oligomerizing. A, crystal structure of HIN-b domains (yellow, orange, blue, and green) binding to viral dsDNA (red). Mutations at highlighted amino acids impaired DNA binding (adapted from Ref. 30). PDB 3RNU, Protein Data Bank ID 3RNU. B, model of IFI16 binding to viral DNA in a length-dependent manner (one IFI16 molecule per 15 bp of viral DNA) (adapted from Ref. 32). C, post-translational modifications identified on IFI16 (36, 85, 86).
FIGURE 2.
Nucleus-originating immune signaling is activated upon sensing of herpesviral dsDNA. Following viral entry into a host cell, the capsid extrudes viral dsDNA into the nucleus (1). Nuclear viral DNA is bound directly to DNA sensors IFI16 and IFIX (2). IFI16 signals to STING via a mechanism that has yet to be elucidated (3a). cGAS was also observed in the nucleus following HSV-1 infection and shown to stabilize IFI16. Upon activation and dimerization of STING, TBK-1 is phosphorylated (4), resulting in the phosphorylation of IRF3 and NF-κB, which translocate back into the nucleus to induce the expression of antiviral cytokines (5). Upon binding nuclear viral dsDNA, IFI16 also associates with ASC to form mature inflammasomes in the cytoplasm, processing pro-inflammatory cytokines (3b). It remains to be investigated whether cGAS may also sense viral DNA within the nucleus to produce cyclic GMP-AMP, which binds directly to STING (3c). Solid arrows depict experimentally demonstrated pathways, and dotted arrows illustrate other possible mechanisms. Casp-1, caspase-1; ER, endoplasmic reticulum.
The possibility that the integrity of the herpesvirus nucleocapsid is compromised in the cytosolic environment, leading to leakage of the encapsulated DNA, cannot be entirely excluded. Polyubiquitination and proteasomal degradation of the major HSV-1 capsid protein VP5 have been reported in primary macrophages (34). This mechanism explains the recruitment of IFI16 to HSV-1 and HCMV DNA foci within the cytosol of macrophages (33, 34). However, these processes do not appear to exist in non-immune cell types. In fact, microscopy and biochemical fractionation studies have demonstrated that IFI16 co-localizes with herpesviral DNA strictly within the nucleus (36, 39, 40) and maintains nuclear localization throughout infection (36, 37, 40, 41). These discrepancies in IFI16 DNA-sensing models are likely due to intrinsic cell type-dependent processes. As macrophages and dendritic cells act at the onset of innate and adaptive immune responses, DNA-sensing components may be localized to the cytoplasm to provide maximum sensitivity and response rapidity to foreign nucleic acids. In contrast, non-immune cells have divergent functions and may maintain IFI16 in the nucleus to focus its immunological activities on successful viral infection events or perform other housekeeping tasks. We found that acetylation of its nuclear localization signal (Fig. 1C) targets IFI16 to the cytosol, providing a mechanism by which cells fine-tune IFI16 sub-cellular distribution (36). Aside from its role in innate immune signaling, IFI16 has also been implicated in mounting inflammatory and apoptotic responses to foreign DNA via a multi-protein assembly known as the “inflammasome” (39, 42–45). Other pyrin domain-containing cellular receptors, including the related PYHIN protein, AIM2, and members of the NOD-like receptor (NLR) family, were previously shown to initiate inflammasome formation (as reviewed in Ref. 28). Although these receptors are activated by diverse pathogen- and damage-associated molecular signatures (microbial DNA in the case of AIM2), they ubiquitously oligomerize upon stimulation. The resulting aggregates recruit and activate the caspase-1 protease, which consequently drives the maturation of pro-inflammatory cytokines IL-1β and IL-18. Ultimately, these inflammatory responses culminate in a caspase-dependent form of programmed cell death, termed “pyroptosis” (reviewed in Refs. 46 and 47).
Until recently, inflammasome activity was thought to be restricted to the cytosol for surveying foreign lysosomal content or mitochondrial aberrations. However, Kerur and colleagues (39, 44) demonstrated that IFI16 co-localizes with replicating KSHV DNA in the nuclei of endothelial cells, consequently assembling an ASC- and caspase-1-containing inflammasome (Fig. 2). Interestingly, IFI16 association with ASC is detected during early infection within the nucleus; however, by later stages, mature inflammasome structures are thought to re-distribute to the cytosol (39). How these large multimeric structures translocate through the nuclear pore is unclear. However, de novo assembly in the cytoplasm remains a possibility. Furthermore, B cell lines harboring latent, episomal forms of either the KSHV or the EBV genomes exhibit constitutively high IFI16-dependent caspase-1 activity and expression of pro-inflammatory interleukins, relative to their uninfected parental cell lines (43, 44). As EBV and KSHV are tightly linked to the development of certain lymphomas, these processes could contribute to the inflammatory microenvironments found in tumors related to these pathologies. IFI16 inflammasome activity was also reported during HSV-1 infection in fibroblasts (42). Thus, these inflammatory responses may reflect generalized cellular reactions to herpesvirus infections. Most recently, IFI16 was shown to mediate CD4+ T cell death during HIV infection, implicating its inflammasome functions in the progression of AIDS (45, 48).
The role of IFI16 in eliciting both type I IFNs and inflammatory responses seems contradictory given the demonstrated anti-inflammatory properties of the former (49–51). Indeed, varied incongruences regarding IFI16 as an inflammasome initiator exist within the literature. Earlier seminal studies that characterized the AIM2 inflammasome did not observe maturation of caspase-1 and IL-1β following DNA stimulation or co-expression of IFI16 with critical inflammasome components (52, 53). In agreement, IFI16 was not observed to associate with ASC, a requirement for the recruitment and maturation of caspase-1 (31, 53). More recently, comprehensive proteomic analyses of IFI16 interaction networks did not detect interactions between endogenous IFI16 and ASC during HSV-1 infections in primary fibroblasts (29, 54). Several studies that probed IFI16 functions during early stages of herpesvirus infections have observed neither IFI16 inflammasome assembly nor caspase-1 maturation (31, 36, 37, 40, 41, 54). Finally, IFI16 was demonstrated to have a direct role in inhibiting the formation of both AIM2 and NOD-like receptor family inflammasomes (55). It remains possible that the role of IFI16 in DNA-dependent inflammasome responses is cell type-dependent and that other, yet uncharacterized factors are required. These disparate observations necessitate further study.
Until recently, AIM2 and IFI16 were the only members of the human PYHIN family with established pathogen DNA-sensing capabilities. The other PYHIN members, IFIX and MNDA, although also localized predominantly to the nucleus, had few characterized immunological functions. Our recent proteomic investigation of the PYHIN family protein interaction networks revealed that IFIX associates with antiviral factors and that its expression is inversely correlated with herpesvirus replicative capacity (29). IFIX was further shown to bind DNA substrates in a sequence-independent manner and to contribute to type I IFN production (29). Moreover, during HSV-1 infection, IFIX associates with viral genomic DNA and remains localized to the nucleus (29). Altogether, these data implicate IFIX as the second identified nuclear sensor of viral DNA to date (Fig. 2). Given that IFIX is differentially expressed across cell and tissue types, it is likely to have cell type-specific functions that remain to be determined (56, 57).
The discovery that viral DNA can be sensed within host nuclei has incited a re-evaluation of other previously characterized DNA sensors and the sub-cellular compartments in which they function. Upon binding to foreign DNA, the cyclic-GMP-AMP (cGAMP) synthase (cGAS) catalyzes the production of the cyclic dinucleotide (CDN) cGAMP from intracellular ATP and GTP, which, in turn, binds and activates STING to induce type I IFNs (58–63) (Fig. 2). As previous work had already determined that CDNs generated by intracellular bacterial pathogens directly activate STING (64–66), the discovery of cGAS demonstrated a cell-intrinsic mechanism exploiting bacterial CDN second messenger systems for mobilizing host defenses. Two critical observations underscore a central role for cGAS in DNA sensing. First, introduction of exogenous DNA paired with co-expression of cGAS and STING is sufficient to elicit robust interferon responses in HEK293T cells inherently devoid of these components (58, 60, 62, 63). Second, cGAS−/− mice and explanted cGAS−/− monocytes fail to mount proper IFN responses to DNA viruses, such as poxviruses and herpesviruses (61, 67). Interestingly, fibroblasts derived from cGAS−/− mice are attenuated in their production of IFN in response to both WT and d109 HSV-1 infection, relative to their cGAS+/+ counterparts (61). Even in human fibroblasts, depletion of either IFI16 or cGAS by RNA interference attenuates IFN induction in response to HSV-1 infection (68).
Although cGAS was shown to co-localize with foreign DNA in the cytosol of macrophages (58, 69), cGAS-dependent IFN responses to HSV-1 in non-immune cells implicate its function in nuclear sensing. In fact, in human fibroblasts, endogenous cGAS seems equally distributed between the cytosol and the nucleus and can co-localize with IFI16 and transfected plasmid DNA at foci in either sub-cellular compartment (68). Furthermore, we observed nuclear IFI16 to interact with cGAS in lysates from these cells (68). Therefore, IFI16 and cGAS may function synergistically to elicit immune signaling to nuclear foreign DNA. However, HSV-1 infection of human fibroblasts elicits little cGAMP production. Moreover, cGAS depletion triggers IFI16 de-stabilization via the proteasome (68). These results suggest that IFI16 functions as the dominant nuclear DNA sensor, whereas cGAS performs auxiliary functions, such as stabilization of IFI16 to enable or prolong signal potentiation. It will be interesting to test whether this is the case for other DNA viruses or whether these are directly sensed by nuclear pools of cGAS, which may stimulate STING at the endoplasmic reticulum via diffusion of cGAMP through the nuclear pores. In agreement, cGAMP derived from DNA-stimulated cells can diffuse through gap junctions to engage distal STING in unstimulated neighboring cells (70). Future studies are crucial for delineating cGAS nuclear functions and the intersection of IFI16- and cGAS-dependent signaling pathways.
Aside from regulating immune and inflammatory signaling pathways, cell-intrinsic nuclear factors have also been demonstrated to regulate chromatinization and transcription of viral DNA genomes. For example, IFI16 represses expression of HCMV DNA polymerase genes UL54 and UL44 by reducing the occupancy of the transcription factor Sp1 at promoter elements (71). Furthermore, IFI16 mediates nucleosomal loading and epigenetic addition of heterochromatin-associated markers to HSV-1 genomic DNA during infection (72, 73). These mechanisms are thought to limit recruitment of RNA Pol II and associated transcriptional activation complexes to restrict viral gene expression and genome replication. Emerging evidence suggests that these IFI16 functions are intertwined with those of sub-nuclear multi-protein complexes, known as nuclear domain 10 (ND10) bodies (also known as promyelocytic leukemia (PML) nuclear bodies). Like IFI16, ND10 body components are IFN-inducible and rapidly associate with deposited viral DNA genomes in the nucleus, acting as suppressors of viral gene expression (74–76). Stable knockdowns of ND10 body components, including PML, death domain-associated protein 6 (hDAXX), ATRX, or Sp100, increase titers of an HSV-1 strain that lacks the ubiquitin ligase activity ICP0, a viral protein that targets ND10 bodies for degradation (77, 78). In agreement, ATRX and hDaxx were shown to cooperate as part of a repressive chromatin-remodeling complex, inducing a transcriptionally inactive chromatin state around early-acting HCMV promoters with the aid of chromatin-modifying histone deacetylases (79, 80). Additionally, we have recently demonstrated a direct interaction between both IFI16 and IFIX with major ND10 body components and mediators of chromatin assembly and transcription (29, 54).
Broad Impact of DNA-sensing Mechanisms
Recent studies have indicated that, in addition to defense against DNA viruses, DNA-sensing mechanisms play critical roles following infection with RNA viruses. Although their genomes are composed of RNA, retroviruses, such as HIV, utilize DNA intermediates to replicate. Upon entry into the cell, the HIV RNA genome is reverse-transcribed to dsDNA in the cytosol, whereby it is imported into the nucleus and integrated into the host cell genome. Because of this unique replication strategy, “pro-viral” DNA is also vulnerable to cellular DNA-sensing mechanisms. Jakobsen et al. (35) tested an array of HIV-derived DNA substrates with varying secondary structures and found that DNA-dependent type I IFN responses require IFI16 in human monocytes. Moreover, cells with reduced expression of the 3′-5′ DNA exonuclease Trex enhanced these IFI16-dependent cytokine responses upon introduction of viral DNA. Finally, HIV infection in IFI16-depleted primary human macrophages exhibited higher levels of viral gene products relative to non-depleted cells. Together, these results suggest that IFI16 limits HIV replication through detection of HIV pro-viral DNA. As mentioned earlier, Monroe et al. (45) subsequently found that cell death induced in CD4+ T cells during HIV infection is specifically mediated by IFI16, rather than other characterized DNA sensors, and may account for the progression of acquired immunodeficiency in HIV-positive patients. Although the exact mechanisms by which IFI16 exerts its functions in this context remain undefined, it may be that sensing of the HIV provirus induces apoptotic cellular responses to limit viral propagation. Altogether, these studies have stirred interest in understanding the impact of IFI16 and other DNA sensors on retrovirus infections.
Viral Immune Evasion of DNA-sensing Pathways
Considering pathogen-host co-evolution and the prevalence of DNA viruses, the limited capacity of the host cell to detect and elicit immunity to viral DNA during infection implies the existence of viral immune evasion strategies. Although the concept of nuclear DNA sensing is still in its infancy, some mechanisms utilized by viruses to inhibit IFI16-dependent immune functions have been elucidated (Fig. 3). Recent studies from our laboratory identified a physical interaction between IFI16 and the major HCMV tegument protein pUL83 within HCMV-infected nuclei (31, 41). We identified distinct N- and C-terminal domains of pUL83 that directly bind the IFI16 PY domain and block its homo-oligomerization, respectively (31). Furthermore, infection with a pUL83-deficient HCMV strain induces both IFI16 multimerization and robust antiviral cytokine production, which is dependent on the presence IFI16, STING, and TBK-1 (31). Together, these results support a model in which IFI16 binding to nuclear viral DNA catalyzes the PY-mediated self-assembly of IFI16 aggregates that serve to initiate immune signaling through the STING axis (Fig. 3). In turn, these IFI16 assemblies are obscured by pUL83 during infection, attenuating immune responses (Fig. 3, right). We have also demonstrated that the pUL83-IFI16 complex can function to stimulate HCMV gene expression at low viral loads (41). In this way, HCMV both sequesters and repurposes IFI16 to serve as a viral gene transactivator.
FIGURE 3.

Herpesviruses have evolved distinct strategies for suppressing IFI16 functions during infection. The HSV-1 E3 ubiquitin ligase ICP0, directly or indirectly, catalyzes the proteasomal degradation of IFI16 (left). The HCMV major tegument protein pUL83 binds directly to the PY domain of IFI16, inhibiting IFI16 oligomerization and consequential immune signaling (right).
In contrast to HCMV, HSV-1 has evolved an alternative strategy for countering IFI16 antiviral activities. Several groups, including ours, have reported that IFI16 is rapidly degraded during early stages of HSV-1 infection and maintained at low levels onwards (37, 40, 42, 54). This degradation occurs exclusively at the protein level in a proteasome-dependent manner and requires transcription of viral gene products (54). Given these data and its previously characterized ability to target cellular proteins for proteasomal degradation (81, 82), the HSV-1 E3 ubiquitin ligase ICP0 was implicated. Indeed, HSV-1 mutants deficient for ICP0 (37, 42) or lacking ICP0 ubiquitin ligase activity (54) are unable to degrade IFI16 to the same extent as the wild-type virus. Furthermore, upon HSV-1 infection, IFI16 is recruited to nuclear ICP0-containing foci prior to its degradation (37, 40, 54) and physically associates with ICP0 (54). However, it has been argued that, because ICP0 is also the immediate-early transactivator of viral gene expression and its absence drastically slows the kinetics of viral productive infection, compromising ICP0 activity may mask the function of the true direct effector of IFI16 de-stabilization (40). Using the HCMV transactivating protein IE1 to complement an ICP0-deficient HSV-1 mutant, Cuchet-Lourenço et al. (40) demonstrated IFI16 degradation even in the absence of ICP0. However, these studies were performed in HepaRG cells, rather than primary fibroblasts used in similar studies. Considering that HSV-1-induced IFI16 degradation is not observed in all cell types (36, 40), the interpretation of these results is difficult. Nevertheless, ICP0 expression alone cannot induce loss of IFI16 (40, 54), and ubiquitylation of IFI16 upon HSV-1 infection remains to be reported. Despite these inconsistencies, there is substantial evidence that nuclear IFI16 is able to elicit robust immune signaling to HSV-1 in the absence of ICP0 (37, 54). Thus, ICP0, either directly or indirectly, functions to suppress this important signaling pathway (Fig. 3, left). Aside from these few studies, our knowledge of the strategies utilized by nuclear replicating DNA viruses to circumvent the aforementioned sensing pathways remains extremely limited.
Future Perspectives
Forthcoming challenges in DNA sensing rest upon elucidating several principal questions: What are the cellular mechanisms distinguishing viral DNA from cellular DNA within the nucleus? How are the nucleus-originating immune signaling pathways propagated through the endoplasmic reticulum STING axis? Specifically, identifying the molecular mechanisms governing nucleus-originating signal transmission to the cytosol, as well as back to the nucleus, remains an outstanding question. An uncertainty that arises from these gaps in fundamental knowledge is how DNA-sensing pathways differentiate between commensal microbiota and invading pathogens. Insight into these aspects of host-virus interactions will serve to catalyze the development of rational antiviral drug therapies.
In light of the immunological potential of DNA, its incorporation as an adjuvant in vaccines continues to be considered to promote antiviral cellular and humoral immune responses (11). Typically, a DNA vaccine consists of plasmid DNA encoding viral antigens. To date, despite repeated clinical trials in humans, the efficacy of such vaccines remains disappointingly poor, largely because of low inductions of immunogenicity, due in part to species-specific expression of TLR9 (83, 84). However, the growing success of DNA-based treatments encoding disease-specific antigens in veterinary medicine has added new incentive to revisit the use of DNA adjuvants in humans (83). Understanding how DNA activates sensing pathways is particularly paramount to developing novel prophylactic therapeutics against nuclear replicating DNA viruses.
This work was supported by National Institutes of Health Grants DP1DA026192, R01 GM114141, and R21AI102187 (to I. M. C.) and by an American Heart Association (AHA) predoctoral fellowship 14PRE18890044 (to B. A. D.). This is the third article in the Thematic Minireview series “Protein Interactions, Structures, and Networks.” The authors declare that they have no conflicts of interest with the contents of this article.
- EBV
- Epstein-Barr virus
- KSHV
- Kaposi sarcoma-associated herpesvirus
- HCMV
- human cytomegalovirus
- HSV
- herpes simplex virus
- Pol
- polymerase
- IFIX
- interferon inducible protein X
- MNDA
- myeloid cell nuclear differentiation antigen
- cGAMP
- cyclic GMP-AMP
- cGAS
- cGAMP synthase
- CDN
- cyclic dinucleotide
- ND10
- nuclear domain 10.
References
- 1.Arzumanyan A., Reis H. M., and Feitelson M. A. (2013) Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat. Rev. Cancer 13, 123–135 [DOI] [PubMed] [Google Scholar]
- 2.McFadden K., and Luftig M. A. (2013) Interplay between DNA tumor viruses and the host DNA damage response. Curr. Top. Microbiol. Immunol. 371, 229–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moens U., Rasheed K., Abdulsalam I., and Sveinbjørnsson B. (2015) The role of Merkel cell polyomavirus and other human polyomaviruses in emerging hallmarks of cancer. Viruses 7, 1871–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinstock H., Berman S., and Cates W. Jr. (2004) Sexually transmitted diseases among American youth: incidence and prevalence estimates, 2000. Perspect. Sex. Reprod. Health 36, 6–10 [DOI] [PubMed] [Google Scholar]
- 5.Kojaoghlanian T., Flomenberg P., and Horwitz M. S. (2003) The impact of adenovirus infection on the immunocompromised host. Rev. Med. Virol. 13, 155–171 [DOI] [PubMed] [Google Scholar]
- 6.Crawford D. H. (2001) Biology and disease associations of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vancíková Z., and Dvorák P. (2001) Cytomegalovirus infection in immunocompetent and immunocompromised individuals: a review. Curr. Drug Targets Immune Endocr. Metabol. Disord. 1, 179–187 [PubMed] [Google Scholar]
- 8.Cheeran M. C., Lokensgard J. R., and Schleiss M. R. (2009) Neuropathogenesis of congenital cytomegalovirus infection: disease mechanisms and prospects for intervention. Clin. Microbiol. Rev. 22, 99–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiaofei E., and Kowalik T. F. (2014) The DNA damage response induced by infection with human cytomegalovirus and other viruses. Viruses 6, 2155–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frobert E., Burrel S., Ducastelle-Lepretre S., Billaud G., Ader F., Casalegno J. S., Nave V., Boutolleau D., Michallet M., Lina B., and Morfin F. (2014) Resistance of herpes simplex viruses to acyclovir: an update from a ten-year survey in France. Antiviral Res. 111, 36–41 [DOI] [PubMed] [Google Scholar]
- 11.Coban C., Tozuka M., Jounai N., Kobiyama K., Takeshita F., Tang C. K., and Ishii K. J. (2014) Chapter 11, DNA vaccine: does it target the double stranded-DNA sensing pathway? in Biological DNA Sensor (Tang K. J. I. K., ed), pp. 257–270, Academic Press, Amsterdam [Google Scholar]
- 12.Waddell G. H. (1962) Factors Controlling Infection with Herpes Simplex Virus with Special Emphasis on Tissue Culture Systems. Ph. D. thesis, University of Miami [Google Scholar]
- 13.Glasgow L. A., and Habel K. (1962) The role of interferon in vaccinia virus infection of mouse embryo tissue culture. J. Exp. Med. 115, 503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isaacs A., Cox R. A., and Rotem Z. (1963) Foreign nucleic acids as the stimulus to make interferon. Lancet 2, 113–116 [DOI] [PubMed] [Google Scholar]
- 15.Rotem Z., Cox R. A., and Isaacs A. (1963) Inhibition of virus multiplication by foreign nucleic acid. Nature 197, 564–566 [DOI] [PubMed] [Google Scholar]
- 16.Isaacs A., Lindenmann J., and Valentine R. C. (1957) Virus interference. II. Some properties of interferon. Proc. R. Soc. Lond. B Biol. Sci. 147, 268–273 [PubMed] [Google Scholar]
- 17.Isaacs A., and Lindenmann J. (1957) Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147, 258–267 [PubMed] [Google Scholar]
- 18.Ishikawa H., and Barber G. N. (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong B., Yang Y., Li S., Wang Y. Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., and Shu H. B. (2008) The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 [DOI] [PubMed] [Google Scholar]
- 20.Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., and Jiang Z. (2009) ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U.S.A. 106, 8653–8658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa H., Ma Z., and Barber G. N. (2009) STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X., Wu F. H., Wang X., Wang L., Siedow J. N., Zhang W., and Pei Z. M. (2014) Molecular evolutionary and structural analysis of the cytosolic DNA sensor cGAS and STING. Nucleic Acids Res. 42, 8243–8257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pomerantz J. L., and Baltimore D. (1999) NF-κB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 18, 6694–6704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoneyama M., Suhara W., Fukuhara Y., Fukuda M., Nishida E., and Fujita T. (1998) Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17, 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato M., Tanaka N., Hata N., Oda E., and Taniguchi T. (1998) Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-β gene. FEBS Lett. 425, 112–116 [DOI] [PubMed] [Google Scholar]
- 26.Lin R., Heylbroeck C., Pitha P. M., and Hiscott J. (1998) Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18, 2986–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka Y., and Chen Z. J. (2012) STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5, ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schattgen S. A., and Fitzgerald K. A. (2011) The PYHIN protein family as mediators of host defenses. Immunol. Rev. 243, 109–118 [DOI] [PubMed] [Google Scholar]
- 29.Diner B. A., Li T., Greco T. M., Crow M. S., Fuesler J. A., Wang J., and Cristea I. M. (2015) The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA. Mol. Syst. Biol. 11, 787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin T., Perry A., Jiang J., Smith P., Curry J. A., Unterholzner L., Jiang Z., Horvath G., Rathinam V. A., Johnstone R. W., Hornung V., Latz E., Bowie A. G., Fitzgerald K. A., and Xiao T. S. (2012) Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36, 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li T., Chen J., and Cristea I. M. (2013) Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe 14, 591–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrone S. R., Wang T., Constantoulakis L. M., Hooy R. M., Delannoy M. J., and Sohn J. (2014) Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc. Natl. Acad. Sci. U.S.A. 111, E62–E71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unterholzner L., Keating S. E., Baran M., Horan K. A., Jensen S. B., Sharma S., Sirois C. M., Jin T., Latz E., Xiao T. S., Fitzgerald K. A., Paludan S. R., and Bowie A. G. (2010) IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11, 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horan K. A., Hansen K., Jakobsen M. R., Holm C. K., Søby S., Unterholzner L., Thompson M., West J. A., Iversen M. B., Rasmussen S. B., Ellermann-Eriksen S., Kurt-Jones E., Landolfo S., Damania B., Melchjorsen J., Bowie A. G., Fitzgerald K. A., and Paludan S. R. (2013) Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J. Immunol. 190, 2311–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakobsen M. R., Bak R. O., Andersen A., Berg R. K., Jensen S. B., Tengchuan J., Jin T., Laustsen A., Hansen K., Ostergaard L., Fitzgerald K. A., Xiao T. S., Mikkelsen J. G., Mogensen T. H., and Paludan S. R. (2013) IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc. Natl. Acad. Sci. U.S.A. 110, E4571–E4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li T., Diner B. A., Chen J., and Cristea I. M. (2012) Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc. Natl. Acad. Sci. U.S.A. 109, 10558–10563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orzalli M. H., DeLuca N. A., and Knipe D. M. (2012) Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl. Acad. Sci. U.S.A. 109, E3008–E3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samaniego L. A., Neiderhiser L., and DeLuca N. A. (1998) Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72, 3307–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerur N., Veettil M. V., Sharma-Walia N., Bottero V., Sadagopan S., Otageri P., and Chandran B. (2011) IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe 9, 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cuchet-Lourenço D., Anderson G., Sloan E., Orr A., and Everett R. D. (2013) The viral ubiquitin ligase ICP0 is neither sufficient nor necessary for degradation of the cellular DNA sensor IFI16 during herpes simplex virus 1 infection. J. Virol. 87, 13422–13432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cristea I. M., Moorman N. J., Terhune S. S., Cuevas C. D., O'Keefe E. S., Rout M. P., Chait B. T., and Shenk T. (2010) Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J. Virol. 84, 7803–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson K. E., Chikoti L., and Chandran B. (2013) Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J. Virol. 87, 5005–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ansari M. A., Singh V. V., Dutta S., Veettil M. V., Dutta D., Chikoti L., Lu J., Everly D., and Chandran B. (2013) Constitutive interferon-inducible protein 16-inflammasome activation during Epstein-Barr virus latency I, II, and III in B and epithelial cells. J. Virol. 87, 8606–8623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh V. V., Kerur N., Bottero V., Dutta S., Chakraborty S., Ansari M. A., Paudel N., Chikoti L., and Chandran B. (2013) Kaposi's sarcoma-associated herpesvirus latency in endothelial and B cells activates γ interferon-inducible protein 16-mediated inflammasomes. J. Virol. 87, 4417–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monroe K. M., Yang Z., Johnson J. R., Geng X., Doitsh G., Krogan N. J., and Greene W. C. (2014) IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science 343, 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergsbaken T., Fink S. L., and Cookson B. T. (2009) Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 7, 99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miao E. A., Rajan J. V., and Aderem A. (2011) Caspase-1-induced pyroptotic cell death. Immunol. Rev. 243, 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doitsh G., Galloway N. L., Geng X., Yang Z., Monroe K. M., Zepeda O., Hunt P. W., Hatano H., Sowinski S., Muñoz-Arias I., and Greene W. C. (2014) Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 505, 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Theofilopoulos A. N., Baccala R., Beutler B., and Kono D. H. (2005) Type I interferons (α/β) in immunity and autoimmunity. Annu. Rev. Immunol. 23, 307–336 [DOI] [PubMed] [Google Scholar]
- 50.Billiau A. (2006) Anti-inflammatory properties of Type I interferons. Antiviral Res. 71, 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guarda G., Braun M., Staehli F., Tardivel A., Mattmann C., Förster I., Farlik M., Decker T., Du Pasquier R. A., Romero P., and Tschopp J. (2011) Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity 34, 213–223 [DOI] [PubMed] [Google Scholar]
- 52.Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D. R., Latz E., and Fitzgerald K. A. (2009) AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandes-Alnemri T., Yu J. W., Datta P., Wu J., and Alnemri E. S. (2009) AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diner B. A., Lum K. K., Javitt A., and Cristea I. M. (2015) Interactions of the antiviral factor IFI16 mediate immune signaling and herpes simplex virus-1 immunosuppression. Mol. Cell. Proteomics 14, 2341–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Veeranki S., Duan X., Panchanathan R., Liu H., and Choubey D. (2011) IFI16 protein mediates the anti-inflammatory actions of the type-I interferons through suppression of activation of caspase-1 by inflammasomes. PLoS One 6, e27040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding Y., Wang L., Su L. K., Frey J. A., Shao R., Hunt K. K., and Yan D. H. (2004) Antitumor activity of IFIX, a novel interferon-inducible HIN-200 gene, in breast cancer. Oncogene 23, 4556–4566 [DOI] [PubMed] [Google Scholar]
- 57.Haque A., Koide N., Odkhuu E., Tsolmongyn B., Naiki Y., Komatsu T., Yoshida T., and Yokochi T. (2014) Mouse pyrin and HIN domain family member 1 (pyhin1) protein positively regulates LPS-induced IFN-β and NO production in macrophages. Innate Immun. 20, 40–48 [DOI] [PubMed] [Google Scholar]
- 58.Sun L., Wu J., Du F., Chen X., and Chen Z. J. (2013) Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu J., Sun L., Chen X., Du F., Shi H., Chen C., and Chen Z. J. (2013) Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diner E. J., Burdette D. L., Wilson S. C., Monroe K. M., Kellenberger C. A., Hyodo M., Hayakawa Y., Hammond M. C., and Vance R. E. (2013) The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 3, 1355–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li X. D., Wu J., Gao D., Wang H., Sun L., and Chen Z. J. (2013) Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341, 1390–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Röhl I., Hopfner K. P., Ludwig J., and Hornung V. (2013) cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X., Shi H., Wu J., Zhang X., Sun L., Chen C., and Chen Z. J. (2013) Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 51, 226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McWhirter S. M., Barbalat R., Monroe K. M., Fontana M. F., Hyodo M., Joncker N. T., Ishii K. J., Akira S., Colonna M., Chen Z. J., Fitzgerald K. A., Hayakawa Y., and Vance R. E. (2009) A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J. Exp. Med. 206, 1899–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woodward J. J., Iavarone A. T., and Portnoy D. A. (2010) c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burdette D. L., Monroe K. M., Sotelo-Troha K., Iwig J. S., Eckert B., Hyodo M., Hayakawa Y., and Vance R. E. (2011) STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schoggins J. W., MacDuff D. A., Imanaka N., Gainey M. D., Shrestha B., Eitson J. L., Mar K. B., Richardson R. B., Ratushny A. V., Litvak V., Dabelic R., Manicassamy B., Aitchison J. D., Aderem A., Elliott R. M., García-Sastre A., Racaniello V., Snijder E. J., Yokoyama W. M., Diamond M. S., Virgin H. W., and Rice C. M. (2014) Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Orzalli M. H., Broekema N.M., Diner B.A., Hancks D.C., Elde N. C., Cristea I. M., and Knipe D. M. (2015) cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc. Natl. Acad. Sci. U.S.A. 112, E1773–E1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hansen K., Prabakaran T., Laustsen A., Jørgensen S. E., Rahbæk S. H., Jensen S. B., Nielsen R., Leber J. H., Decker T., Horan K. A., Jakobsen M. R., and Paludan S. R. (2014) Listeria monocytogenes induces IFNβ expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 33, 1654–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ablasser A., Schmid-Burgk J. L., Hemmerling I., Horvath G. L., Schmidt T., Latz E., and Hornung V. (2013) Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503, 530–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gariano G. R., Dell'Oste V., Bronzini M., Gatti D., Luganini A., De Andrea M., Gribaudo G., Gariglio M., and Landolfo S. (2012) The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 8, e1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Orzalli M. H., Conwell S. E., Berrios C., DeCaprio J. A., and Knipe D. M. (2013) Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc. Natl. Acad. Sci. U.S.A. 110, E4492–E4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnson K. E., Bottero V., Flaherty S., Dutta S., Singh V. V., and Chandran B. (2014) IFI16 restricts HSV-1 replication by accumulating on the hsv-1 genome, repressing HSV-1 gene expression, and directly or indirectly modulating histone modifications. PLoS Pathog. 10, e1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Everett R. D., and Murray J. (2005) ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79, 5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Everett R. D., and Chelbi-Alix M. K. (2007) PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89, 819–830 [DOI] [PubMed] [Google Scholar]
- 76.Negorev D. G., Vladimirova O. V., and Maul G. G. (2009) Differential functions of interferon-upregulated Sp100 isoforms: herpes simplex virus type 1 promoter-based immediate-early gene suppression and PML protection from ICP0-mediated degradation. J. Virol. 83, 5168–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lukashchuk V., and Everett R. D. (2010) Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J. Virol. 84, 4026–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Glass M., and Everett R. D. (2013) Components of promyelocytic leukemia nuclear bodies (ND10) act cooperatively to repress herpesvirus infection. J. Virol. 87, 2174–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Woodhall D. L., Groves I. J., Reeves M. B., Wilkinson G., and Sinclair J. H. (2006) Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J. Biol. Chem. 281, 37652–37660 [DOI] [PubMed] [Google Scholar]
- 80.Newhart A., Rafalska-Metcalf I. U., Yang T., Negorev D. G., and Janicki S. M. (2012) Single-cell analysis of Daxx and ATRX-dependent transcriptional repression. J. Cell Sci. 125, 5489–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chelbi-Alix M. K., and de Thé H. (1999) Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18, 935–941 [DOI] [PubMed] [Google Scholar]
- 82.Parkinson J., Lees-Miller S. P., and Everett R. D. (1999) Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73, 650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ferraro B., Morrow M. P., Hutnick N. A., Shin T. H., Lucke C. E., and Weiner D. B. (2011) Clinical applications of DNA vaccines: current progress. Clin. Infect. Dis. 53, 296–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tudor D., Dubuquoy C., Gaboriau V., Lefèvre F., Charley B., and Riffault S. (2005) TLR9 pathway is involved in adjuvant effects of plasmid DNA-based vaccines. Vaccine 23, 1258–1264 [DOI] [PubMed] [Google Scholar]
- 85.Hendriks I. A., D'Souza R. C., Yang B., Verlaan-de Vries M., Mann M., and Vertegaal A. C. (2014) Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 21, 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Blomster H. A., Imanishi S. Y., Siimes J., Kastu J., Morrice N. A., Eriksson J. E., and Sistonen L. (2010) In vivo identification of sumoylation sites by a signature tag and cysteine-targeted affinity purification. J. Biol. Chem. 285, 19324–19329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lam E., Stein S., and Falck-Pedersen E. (2014) Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade. J. Virol. 88, 974–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhu J., Huang X., and Yang Y. (2007) Innate immune response to adenoviral vectors is mediated by both Toll-like receptor-dependent and -independent pathways. J. Virol. 81, 3170–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhen J., Zhang L., Pan J., Ma S., Yu X., Li X., Chen S., and Du W. (2014) AIM2 mediates inflammation-associated renal damage in hepatitis B virus-associated glomerulonephritis by regulating caspase-1, IL-1β, and IL-18. Mediators Inflamm. 2014, 190860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Verrier E. R., Gack M. U., and Baumert T. F. (2014) Cyclic guanosine monophosphate/adenosine monophosphate synthase (cGAS), innate immune responses, and viral hepatitis. Hepatology 60, 1098–1100 [DOI] [PubMed] [Google Scholar]
- 91.Tabeta K., Georgel P., Janssen E., Du X., Hoebe K., Crozat K., Mudd S., Shamel L., Sovath S., Goode J., Alexopoulou L., Flavell R. A., and Beutler B. (2004) Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc. Natl. Acad. Sci. U.S.A. 101, 3516–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krug A., French A. R., Barchet W., Fischer J. A., Dzionek A., Pingel J. T., Orihuela M. M., Akira S., Yokoyama W. M., and Colonna M. (2004) TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity 21, 107–119 [DOI] [PubMed] [Google Scholar]
- 93.Delale T., Paquin A., Asselin-Paturel C., Dalod M., Brizard G., Bates E. E., Kastner P., Chan S., Akira S., Vicari A., Biron C. A., Trinchieri G., and Brière F. (2005) MyD88-dependent and -independent murine cytomegalovirus sensing for IFN-α release and initiation of immune responses in vivo. J. Immunol. 175, 6723–6732 [DOI] [PubMed] [Google Scholar]
- 94.van Gent M., Griffin B. D., Berkhoff E. G., van Leeuwen D., Boer I. G., Buisson M., Hartgers F. C., Burmeister W. P., Wiertz E. J., and Ressing M. E. (2011) EBV lytic-phase protein BGLF5 contributes to TLR9 downregulation during productive infection. J. Immunol. 186, 1694–1702 [DOI] [PubMed] [Google Scholar]
- 95.Furr S. R., Chauhan V. S., Moerdyk-Schauwecker M. J., and Marriott I. (2011) A role for DNA-dependent activator of interferon regulatory factor in the recognition of herpes simplex virus type 1 by glial cells. J. Neuroinflammation 8, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rasmussen S. B., Sørensen L. N., Malmgaard L., Ank N., Baines J. D., Chen Z. J., and Paludan S. R. (2007) Type I interferon production during herpes simplex virus infection is controlled by cell-type-specific viral recognition through Toll-like receptor 9, the mitochondrial antiviral signaling protein pathway, and novel recognition systems. J. Virol. 81, 13315–13324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang J. P., Bowen G. N., Zhou S., Cerny A., Zacharia A., Knipe D. M., Finberg R. W., and Kurt-Jones E. A. (2012) Role of specific innate immune responses in herpes simplex virus infection of the central nervous system. J. Virol. 86, 2273–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nour A. M., Reichelt M., Ku C. C., Ho M. Y., Heineman T. C., and Arvin A. M. (2011) Varicella-zoster virus infection triggers formation of an interleukin-1β (IL-1β)-processing inflammasome complex. J. Biol. Chem. 286, 17921–17933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yu H. R., Huang H. C., Kuo H. C., Sheen J. M., Ou C. Y., Hsu T. Y., and Yang K. D. (2011) IFN-α production by human mononuclear cells infected with varicella-zoster virus through TLR9-dependent and -independent pathways. Cell. Mol. Immunol. 8, 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hasan U. A., Bates E., Takeshita F., Biliato A., Accardi R., Bouvard V., Mansour M., Vincent I., Gissmann L., Iftner T., Sideri M., Stubenrauch F., and Tommasino M. (2007) TLR9 expression and function is abolished by the cervical cancer-associated human papillomavirus type 16. J. Immunol. 178, 3186–3197 [DOI] [PubMed] [Google Scholar]