Abstract
MicroRNAs (miRNAs) are small (~22 nucleotide) noncoding RNAs that play pivotal roles in regulation of gene expression. The value of miRNAs as circulating biomarkers is now broadly recognized; such tissue-specific biomarkers can be used to monitor tissue injury and several pathophysiological conditions in organs. In addition, miRNA profiles of normal organs and tissues are important for obtaining a better understanding of the source of modulated miRNAs in blood and how those modulations reflect various physiological and toxicological conditions. This work was aimed at creating an miRNA atlas in rats, as part of a collaborative effort with the Toxicogenomics Informatics Project in Japan (TGP2). We analyzed genome-wide miRNA profiles of 55 different organs and tissues obtained from normal male rats using miRNA arrays. The work presented herein represents a comprehensive dataset derived from normal samples profiled in a single study. Here we present the whole dataset with miRNA profiles of multiple organs, as well as precise information on experimental procedures and organ-specific miRNAs identified in this dataset.
Background & Summary
MicroRNAs (miRNAs) are small (~ 22 nucleotide) noncoding RNAs that play pivotal roles in regulation of gene expression; miRNAs bind via complementary base-pairing to target transcripts to repress translation or promote mRNA degradation. Major miRNAs are highly conserved across species, allowing translation of biomarkers to the clinic. The machinery for miRNA-mediated gene regulation is conserved from plants to humans, and miRNAs are encoded by their respective genes. As with other regulatory molecules, miRNAs are frequently subject to changes in expression level due to a large range of physiological processes, such as development, immune responses, metabolism and diseases, as well as toxicological outcomes1. Mounting evidence indicates that miRNAs are frequently overexpressed or downregulated as a result of cancer, obesity, diabetes, inflammation, neurological disorders, cardiovascular diseases or autoimmune diseases2–7,2–7,2–7,2–7,2–7,2–7. Previous studies have shown that distinct miRNA signatures can be assigned to particular organs and tumor types8. For example, miR-21 is ubiquitously expressed and upregulated in various types of cancer, including lymphoma, lung, prostate and colorectal cancers among others9. Furthermore, tumor cells have been shown to release tumor-specific miRNAs into systemic circulation. Because of the fundamental roles played by miRNAs in cellular functions, the potential for miRNAs as novel therapeutic targets is now widely recognized. Several candidate therapeutic miRNAs have progressed into clinical and preclinical development; for example, antisense miR-122 is being developed as a treatment for hepatitis C virus, miR-208/499 for chronic heart failure, miR-195 for myocardial infarction and miR-34 and let-7 for cancer10,11. However, serious obstacles obstructing the development of miRNA-based therapies remain, including lack of tissue specificity and risks of systemic toxicity. Characterizing miRNA expression profiles of normal organs and tissues may improve our understanding of the etiology of diseased organs and of organ- and tissue-specificity of miRNAs, and the success rate of development of new miRNA-based therapeutics.
Additionally, miRNAs have the potential to be useful biomarkers for monitoring physiological conditions and tissue injury. Upon tissue injury, miRNAs are released into systemic circulation or other body fluids; upon release, these miRNAs can be detected in small-volume samples via specific and sensitive quantitative real-time PCR. The precise mechanisms underlying the remarkable stability of miRNAs in the RNase-rich environment of blood are not well understood; nevertheless, miRNAs persist and are remarkably stable in blood; and these miRNAs can persist in an encapsulated state in exosomes or in protein complexes with carriers such as Argonaute 2 and Nucleophosmin or the High Density Lipoprotein12. Moreover, several studies have successfully identified circulating miRNA-based biomarkers; for example, Mitchell et al.13 found that miR-141 was highly elevated in serum from patients with prostate cancer and hypothesized that this miRNA may be useful as a diagnostic marker. Similarly, Wang et al.14 found that miR-122a, which is specifically expressed in the liver, was circulating systemically in mice with acetaminophen- (APAP-) induced hepatotoxicity; the authors reported that miR-122a and miR-192 were detected in plasma as early as the point when elevated alanine aminotransferase activity was found evident. Laterza et al.15 also reported that plasma miRNA measurements could be useful for monitoring tissue injury in the liver, muscle and brain. Even in clinical research, plasma miR-122a was significantly elevated in APAP-induced hepatitis patients16. These reports clearly indicate that circulating organ-specific miRNAs can serve as useful biomarkers for tissue injury and disease status in various organs. The organ-specificity of circulating miRNAs, or in other words, the detailed tissue distribution of miRNAs, is important information to be used in obtaining biomarkers for organ toxicity, although desirable properties of biomarkers vary with intended use. In addition, miRNA profiles of normal organs and tissues are important for gaining a better understanding of the source of modulated miRNAs in blood and in gaining an understanding of how those modulations reflect various physiological and toxicological conditions.
To date, organ-specific miRNA profiles have been reported for several animal species, including human, mouse and rat17–20,17–20,17–20,17–20. However, the amount of data currently available is still inadequate, since there are only limited datasets with profiles in a limited number of organs. The main objectives of our data analysis were (i) to establish the validity of this dataset and (ii) to demonstrate organ-specific miRNAs identified in this dataset. In this report, we present a large-scale reference dataset constituting genome-wide miRNA profiles for 55 normal rat organs or tissues. The validity of this dataset was confirmed by comprehensive statistical analysis. We ultimately identified several organ-specific miRNAs in rats. These organ-specific miRNAs can potentially be used as biomarkers for identifying the origin of metastatic tumors and for monitoring toxicity in targeted organs. Furthermore, establishing combinations of organ-specific miRNA measurements may be a novel biomarker for monitoring simultaneous impairment in multiple organs. Our data also support the hypothesis that specificity of miRNA expression is conserved among different species, since the majority of organ-specific miRNAs identified in this rat study were also confirmed as organ-specific in humans. The value of miRNAs as novel therapeutic targets and circulating biomarkers to monitor tissue injury and several pathophysiological conditions in organs is now broadly recognized. In the course of development of miRNAs for practical use as targeted therapeutics and biomarkers, there is no doubt about the importance of open access large-scale datasets for a free miRNA atlas for normal organs/tissues and comparative data analysis. The work presented herein represents a comprehensive dataset derived from normal samples profiled in a single study. We believe that our dataset will be of particular value to both basic and translational scientists in biological and biomedical sciences, especially for novel target discovery and biomarker identification.
Methods
Animal experiments
For each miRNA microarray experiment, 9-week-old male Sprague-Dawley rats were obtained from Charles River Japan, Inc. (Kanagawa, Japan). After a 7-day quarantine and acclimatization period, 10-week-old animals were used (N=6). The animals were individually housed in stainless-steel cages in an animal room set to the following conditions: 12 h (7:00–19:00) light phase; ventilation rate, 12/h; temperature, 20 °C–26 °C; and relative humidity, 35–75%. Each animal had free access to water and pellet diet (CRF-1, sterilized by radiation, Oriental Yeast Co., Ltd., Tokyo, Japan). Each animal was anesthetized with ether. The animals were divided into 2 groups to compare the effects of peripheral blood cells in organs; 3 animals were perfused with saline to remove blood from all organs, while the remaining 3 animals that were not perfused were subjected to collection of only the heart (atrium and interventricular septum), kidney, liver and lung. All organs evaluated in this study are listed in Table 1 (available online only). Experimental protocols were reviewed and approved by the Ethics Review Committee for Animal Experimentation of the National Institute of Health Sciences.
Table 1. List of organs/tissues evaluated in this study.
| Organ ID | Organ name | Organ ID | Organ name |
|---|---|---|---|
| 1 | Liver* | 29 | Seminal vesicle |
| 2 | Kidney† | 30 | Prostate_ventral lobe |
| 3 | Lung | 31 | Prostate_others |
| 4 | Heart_cardiac atrium | 32 | Testis |
| 5 | Heart_interventricular septum | 33 | Epididymis |
| 6 | Skin | 34 | Eyeball |
| 7 | Lymph node_cervical | 35 | Harderian gland |
| 8 | Lymph node_mesenteric | 36 | Optic nerve |
| 9 | Sublingual gland | 37 | Cerebrum_cerebral cortex‡ |
| 10 | Mandibular gland | 38 | Cerebrum_hippocampus‡ |
| 11 | Parotid gland | 39 | Cerebrum_thalamus‡ |
| 12 | Thymus | 40 | Cerebellum |
| 13 | Trachea | 41 | Medulla oblongata |
| 14 | Thyroid gland | 42 | Pituitary gland |
| 15 | Tongue | 43 | Spinal cord_cervical |
| 16 | Esophagus | 44 | Spinal cord_pectoral |
| 17 | Stomach_anterior | 45 | Spinal cord_pars lumbalis |
| 18 | Stomach_glandular | 46 | Femoris muscle |
| 19 | Duodenum§ | 47 | Gastrocnemial muscle |
| 20 | Jejunum§ | 48 | Soleus muscle |
| 21 | Ileum§ | 49 | Sciatic nerve |
| 22 | Cecum§ | 50 | Interseptum |
| 23 | Colon§ | 51 | Bone marrow_femur |
| 24 | Rectum§ | 52 | Aorta_ abdominal |
| 25 | Pancreas | 53 | Vein_ abdominal |
| 26 | Spleen | 54 | White adipose tissue |
| 27 | Adrenal gland | 55 | Brown adipose tissue |
| 28 | Urinary bladder |
*Left lateral lobe.
†Whole kidney containing cortex, medulla, and papilla.
‡Each cerebrum region, including the cortex, hippocampus and thalamus, was macroscopically collected from the sagittal section of the cerebrum.
§Scrapped mucosal epithelial tissue.
RNA extraction and miRNA microarray analysis
RNA was prepared using the miRNeasy kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions. RNA was quantified using a DU-7400 spectrophotometer (Beckman Coulter, Brea, CA) and quality was monitored with the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Cyanine-3 (Cy-3) labeled RNA was prepared from 0.1 μg RNA using the miRNA Complete Labeling and Hyb Kit (Agilent Technologies), according to the manufacturer’s instructions. The entire volume of Cy3-labeled RNA was incubated at 100 °C for 5 min in a 45 μl reaction volume containing 1x Agilent blocking agent, Hybridization Spike-In and Hi-RPM Hybridization Buffer. Upon completion of this incubation, samples were hybridized to an 8×15 k customized Agilent Rat miRNA microarray containing both miRBase 15.0 and 16.0 probes for 20 h at 55 °C on a rotating rack in an Agilent hybridization oven. After hybridization, microarrays were washed for 5 min at room temperature with GE Wash Buffer 1 (Agilent Technologies) and then for 5 min at 37 °C with GE Wash buffer 2 (Agilent Technologies); slides were then dried immediately. Immediately after this washing and drying step, slides were scanned using an Agilent DNA Microarray Scanner (G2565AA) at the following settings: one-color scan for 8×16 k array slides, scan area, 61×21.6 mm; scan resolution, 5 μm; dye channel, green; and extended dynamic range scan mode (Hi=100%, Lo=5%).
Microarray data analysis
Expressionist analysis software Ver. 7.6 (Genedata AG, Basel, Switzerland) was used for normalization, principal component analysis (PCA) and hierarchical clustering. First, all signal intensities were scaled to the 75th percentile of the median of the dynamic target value for each array using the central tendency normalization method. Next, the following two statistical parameters were calculated for each probe: (i) P-value of the Shapiro-Wilk test with Bonferroni correction (R software) and (ii) maximal signal intensity. Probes were then filtered by a P-value of <0.05 and a maximal signal intensity of >100. To identify organ-specific miRNAs, expression profiles of filtered probes were further analyzed by model-based clustering using the ‘mclust’ R package (http://cran.r-project.org/web/packages/mclust/index.html). For this analysis, a histogram was created to determine the distribution of expression values among different organs for each gene. For multiple distributions, the presence of an isolated peak was considered attributable to organ-specific expression of miRNAs. Genes with isolated peaks consisting of less than 37 microarrays (20% of all microarrays) in the histogram were selected as candidate miRNAs that were specifically expressed in one or a few organs.
Data Records
Microarray data are available in the NCBI Gene Expression Omnibus (GEO), accession GSE52754 (Data Citation 1). This accession contains matrix files of normalized miRNA expression data used in this report and raw data files generated from Agilent microarray systems as.gz files.
Technical Validation
miRNA microarray experiments
In this study, microarray experiments were performed according to the manufacturer’s instructions, and the quality of each experiment was assessed in QC reports generated from the Agilent microarray systems.
Possible effects of peripheral blood cell-derived miRNAs
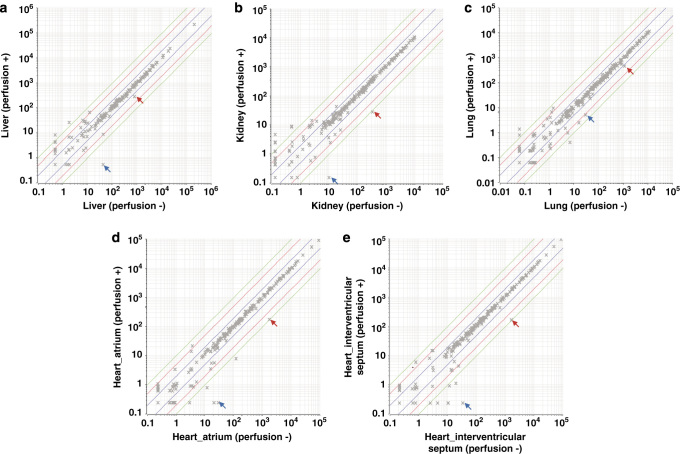
To evaluate the possible effects of peripheral blood cell-derived miRNAs on the miRNA expression profiles of organs, two different sampling conditions, with or without perfusion, were compared to assess effects on miRNA expression profiles in four organs (heart, kidney, liver and lung). As shown in Figure 1, blood-cell–specific miRNAs (e.g., miR-451 and miR-144)21 were clearly evident in nonperfused samples, but not in perfused samples. On the other hand, for each organ, miRNA profiles for perfused samples were highly correlated with those for nonperfused samples (R2>0.995). These data support the hypothesis that there are no distinct differences in miRNA profiles between these major organs and peripheral blood cells with some exceptions, such as miR-451 and miR-144, and that expression profiles for the majority of miRNAs were not markedly affected by remaining peripheral blood. Moreover, our data showed that miR-451 and miR-144 were specifically expressed in hematopoietic tissues e.g., spleen and bone marrow (refer to the original dataset for details). Therefore, microarray data obtained from nonperfused samples were analyzed in combination with those from perfused samples when generating the final comprehensive dataset.
Figure 1.
Effect of circulating blood on miRNA expression in organs. Arrows indicate blood specific miRNAs; red arrows: miR-451, blue arrows: miR-144. Horizontal axis: Perfusion (-), vertical axis: Perfusion (+). Correlation coefficient (R 2 ); (a) Liver: 0.9998, (b) Kidney: 0.9960, (c) Lung: 0.9956, (d) Heart, atrium: 0.9988, (e) Heart, interventricular septum: 0.9994.
Principal component analysis (PCA)
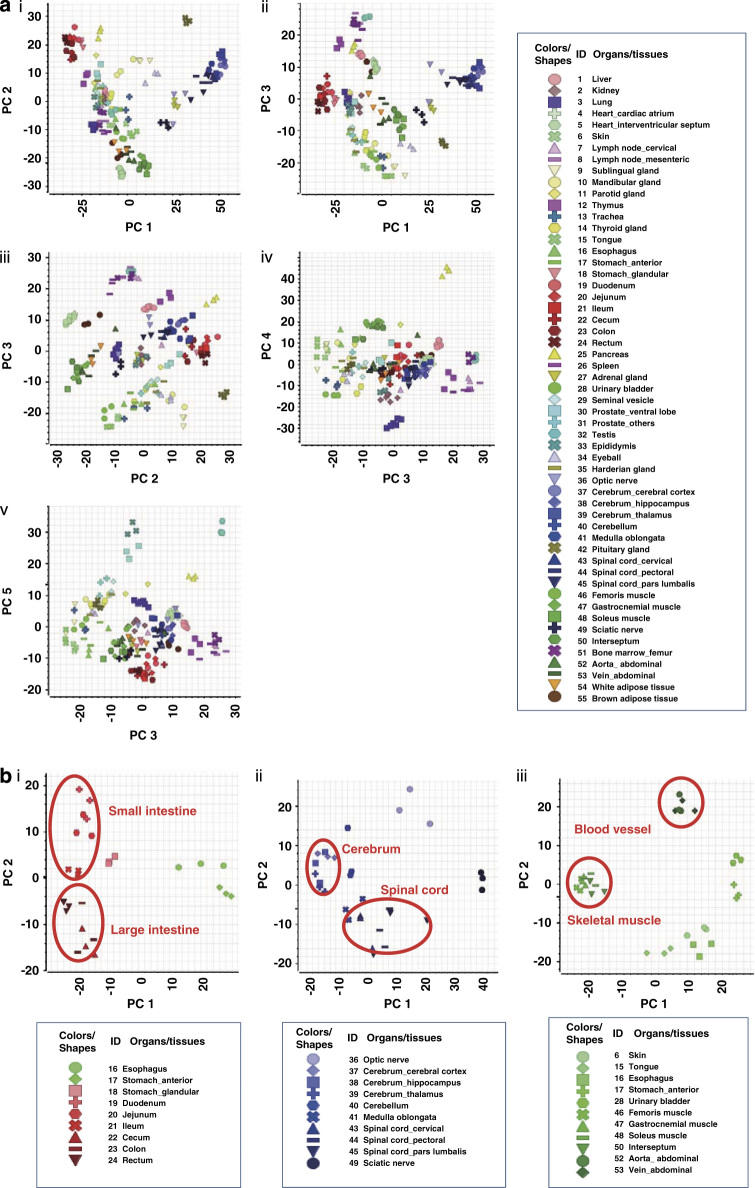
PCA was performed to comprehensively compare miRNA expression profiles among all organs in the entire experimental dataset (Figure 2a). Along the principal component (PC) 1 axis, which accounts for 29.1% of all variation, the clusters of signals from nervous system (brain, spinal cord, optic nerve and sciatic nerve; shown in blue colors), pituitary gland and adrenal gland samples were clearly separated from intestine samples (shown in red), whereas along the PC2 axis (11.4%), muscle (shown in green) samples were clearly separated from nervous system and intestine samples. Along the PC3 axis (8.1%), the clusters of signals from lymph and hematopoietic tissue (shown in pink) samples were clearly separated from those of other samples. Similarly, along the PC4 axis (6.7%) and PC5 axis (5.0%), the clusters of signals from pancreas and lung samples, and those from reproductive organ samples, were clearly separated from others. A tendency towards densely clustered signals from triplicate samples of the same organ was observed in PCA results. Additional information and eigenvalues of PC1 to PC5 axes are summarized in Tables 2 and 3 (available online only).
Figure 2.
Principal component analysis (PCA). (a) PCA for all organ miRNAs. Spot colors represent broad types of organs; blue: nervous system, red: intestines, green: smooth or skeletal muscles, pink: lymphoid or hematopoietic system, light blue: reproductive system. Eigenvalues for each component: 29.1% (PC1), 11.4% (PC2); 8.1% (PC3), 6.7% (PC4), 5.0% (PC5). (b) PCA for digestive organs, nervous system or muscles. Eigenvalues for each component: (i) 37.2% (PC1), 10.8% (PC2); (ii) 35.7% (PC1), 13.2% (PC2); (iii) 31.0% (PC1), 16.4% (PC2).
Table 2. Individual sample IDs and additional information.
| Sample ID * | Rat number | Tissue † | Perfused? y/n | Organ type category |
|---|---|---|---|---|
| GSM1275420 | 1 | No.1_liver | n | others |
| GSM1275421 | 1 | No.1_kidney | n | others |
| GSM1275422 | 1 | No.1_lung | n | others |
| GSM1275423 | 1 | No.1_heart_atrium | n | smooth or skeletal muscles |
| GSM1275424 | 1 | No.1_heart_interventricular septum | n | smooth or skeletal muscles |
| GSM1275425 | 2 | No.2_liver | n | others |
| GSM1275426 | 2 | No.2_kidney | n | others |
| GSM1275427 | 2 | No.2_lung | n | others |
| GSM1275428 | 2 | No.2_heart_atrium | n | smooth or skeletal muscles |
| GSM1275429 | 2 | No.2_heart_interventricular septum | n | smooth or skeletal muscles |
| GSM1275430 | 3 | No.3_liver | n | others |
| GSM1275431 | 3 | No.3_kidney | n | others |
| GSM1275432 | 3 | No.3_lung | n | others |
| GSM1275433 | 3 | No.3_heart_atrium | n | smooth or skeletal muscles |
| GSM1275434 | 3 | No.3_heart_interventricular septum | n | smooth or skeletal muscles |
| GSM1275435 | 4 | No.4_liver | y | others |
| GSM1275436 | 4 | No.4_kidney | y | others |
| GSM1275437 | 4 | No.4_lung | y | others |
| GSM1275438 | 4 | No.4_heart_atrium | y | smooth or skeletal muscles |
| GSM1275439 | 4 | No.4_heart_interventricular septum | y | smooth or skeletal muscles |
| GSM1275440 | 4 | No.4_skin | y | smooth or skeletal muscles |
| GSM1275441 | 4 | No.4_lymph node—cervical | y | lymphoid or hematopoietic system |
| GSM1275442 | 4 | No.4_lymph node—mesenteric | y | lymphoid or hematopoietic system |
| GSM1275443 | 4 | No.4_sublingual gland | y | gland |
| GSM1275444 | 4 | No.4_mandibular gland | y | gland |
| GSM1275445 | 4 | No.4_parotid gland | y | gland |
| GSM1275446 | 4 | No.4_thymus | y | lymphoid or hematopoietic system |
| GSM1275447 | 4 | No.4_trachea | y | others |
| GSM1275448 | 4 | No.4_thyroid | y | gland |
| GSM1275449 | 4 | No.4_tongue | y | smooth or skeletal muscles |
| GSM1275450 | 4 | No.4_esophagus | y | smooth or skeletal muscles |
| GSM1275451 | 4 | No.4_stomach_anterior | y | smooth or skeletal muscles |
| GSM1275452 | 4 | No.4_stomach_glandular | y | smooth or skeletal muscles |
| GSM1275453 | 4 | No.4_duodenum | y | intestines (mucosal epithelial tissue) |
| GSM1275454 | 4 | No.4_jejunum | y | intestines (mucosal epithelial tissue) |
| GSM1275455 | 4 | No.4_ileum | y | intestines (mucosal epithelial tissue) |
| GSM1275456 | 4 | No.4_cecum | y | intestines (mucosal epithelial tissue) |
| GSM1275457 | 4 | No.4_colon | y | intestines (mucosal epithelial tissue) |
| GSM1275458 | 4 | No.4_rectum | y | intestines (mucosal epithelial tissue) |
| GSM1275459 | 4 | No.4_pancreas | y | gland |
| GSM1275460 | 4 | No.4_spleen | y | lymphoid or hematopoietic system |
| GSM1275461 | 4 | No.4_adrenal gland | y | gland |
| GSM1275462 | 4 | No.4_bladder | y | smooth or skeletal muscles |
| GSM1275463 | 4 | No.4_seminal vesicle | y | reproductive system |
| GSM1275464 | 4 | No.4_prostate_ventral | y | reproductive system |
| GSM1275465 | 5 | No.5_liver | y | others |
| GSM1275466 | 5 | No.5_kidney | y | others |
| GSM1275467 | 5 | No.5_lung | y | others |
| GSM1275468 | 5 | No.5_heart_atrium | y | smooth or skeletal muscles |
| GSM1275469 | 5 | No.5_heart_interventricular septum | y | smooth or skeletal muscles |
| GSM1275470 | 5 | No.5_skin | y | smooth or skeletal muscles |
| GSM1275471 | 5 | No.5_lymph node—cervical | y | lymphoid or hematopoietic system |
| GSM1275472 | 5 | No.5_lymph node—mesenteric | y | lymphoid or hematopoietic system |
| GSM1275473 | 5 | No.5_sublingual gland | y | gland |
| GSM1275474 | 5 | No.5_mandibular gland | y | gland |
| GSM1275475 | 5 | No.5_parotid gland | y | gland |
| GSM1275476 | 5 | No.5_thymus | y | lymphoid or hematopoietic system |
| GSM1275477 | 5 | No.5_trachea | y | others |
| GSM1275478 | 5 | No.5_thyroid | y | gland |
| GSM1275479 | 5 | No.5_tongue | y | smooth or skeletal muscles |
| GSM1275480 | 5 | No.5_esophagus | y | smooth or skeletal muscles |
| GSM1275481 | 5 | No.5_stomach_anterior | y | smooth or skeletal muscles |
| GSM1275482 | 5 | No.5_stomach_glandular | y | smooth or skeletal muscles |
| GSM1275483 | 5 | No.5_duodenum | y | intestines (mucosal epithelial tissue) |
| GSM1275484 | 5 | No.5_jejunum | y | intestines (mucosal epithelial tissue) |
| GSM1275485 | 5 | No.5_ileum | y | intestines (mucosal epithelial tissue) |
| GSM1275486 | 5 | No.5_cecum | y | intestines (mucosal epithelial tissue) |
| GSM1275487 | 5 | No.5_colon | y | intestines (mucosal epithelial tissue) |
| GSM1275488 | 5 | No.5_rectum | y | intestines (mucosal epithelial tissue) |
| GSM1275489 | 5 | No.5_pancreas | y | gland |
| GSM1275490 | 5 | No.5_spleen | y | lymphoid or hematopoietic system |
| GSM1275491 | 5 | No.5_adrenal gland | y | gland |
| GSM1275492 | 5 | No.5_bladder | y | smooth or skeletal muscles |
| GSM1275493 | 5 | No.5_seminal vesicle | y | reproductive system |
| GSM1275494 | 5 | No.5_prostate_ventral | y | reproductive system |
| GSM1275495 | 5 | No.5_prostate_other | y | reproductive system |
| GSM1275496 | 6 | No.6_liver | y | others |
| GSM1275497 | 6 | No.6_kidney | y | others |
| GSM1275498 | 6 | No.6_lung | y | others |
| GSM1275499 | 6 | No.6_heart_atrium | y | smooth or skeletal muscles |
| GSM1275500 | 6 | No.6_heart_interventricular septum | y | smooth or skeletal muscles |
| GSM1275501 | 6 | No.6_skin | y | smooth or skeletal muscles |
| GSM1275502 | 6 | No.6_lymph node—cervical | y | lymphoid or hematopoietic system |
| GSM1275503 | 6 | No.6_lymph node—mesenteric | y | lymphoid or hematopoietic system |
| GSM1275504 | 6 | No.6_sublingual gland | y | gland |
| GSM1275505 | 6 | No.6_mandibular gland | y | gland |
| GSM1275506 | 6 | No.6_parotid gland | y | gland |
| GSM1275507 | 6 | No.6_thymus | y | lymphoid or hematopoietic system |
| GSM1275508 | 6 | No.6_trachea | y | others |
| GSM1275509 | 6 | No.6_thyroid | y | gland |
| GSM1275510 | 6 | No.6_tongue | y | smooth or skeletal muscles |
| GSM1275511 | 6 | No.6_esophagus | y | smooth or skeletal muscles |
| GSM1275512 | 6 | No.6_stomach_anterior | y | smooth or skeletal muscles |
| GSM1275513 | 6 | No.6_stomach_glandular | y | smooth or skeletal muscles |
| GSM1275514 | 6 | No.6_duodenum | y | intestines (mucosal epithelial tissue) |
| GSM1275515 | 6 | No.6_jejunum | y | intestines (mucosal epithelial tissue) |
| GSM1275516 | 6 | No.6_ileum | y | intestines (mucosal epithelial tissue) |
| GSM1275517 | 6 | No.6_cecum | y | intestines (mucosal epithelial tissue) |
| GSM1275518 | 6 | No.6_colon | y | intestines (mucosal epithelial tissue) |
| GSM1275519 | 6 | No.6_rectum | y | intestines (mucosal epithelial tissue) |
| GSM1275520 | 6 | No.6_pancreas | y | gland |
| GSM1275521 | 6 | No.6_spleen | y | lymphoid or hematopoietic system |
| GSM1275522 | 6 | No.6_adrenal gland | y | gland |
| GSM1275523 | 6 | No.6_bladder | y | smooth or skeletal muscles |
| GSM1275524 | 6 | No.6_seminal vesicle | y | reproductive system |
| GSM1275525 | 6 | No.6_prostate_ventral | y | reproductive system |
| GSM1275526 | 4 | No.4_prostate_other | y | reproductive system |
| GSM1275527 | 4 | No.4_testicle | y | reproductive system |
| GSM1275528 | 4 | No.4_epididymis | y | reproductive system |
| GSM1275529 | 4 | No.4_eyeball | y | nervous system |
| GSM1275530 | 4 | No.4_Harderian gland | y | gland |
| GSM1275531 | 4 | No.4_optic nerve | y | nervous system |
| GSM1275532 | 4 | No.4_cerebrum_cerebral cortex | y | nervous system |
| GSM1275533 | 4 | No.4_cerebrum_hippocampus | y | nervous system |
| GSM1275534 | 4 | No.4_cerebrum_Thalamus | y | nervous system |
| GSM1275535 | 4 | No.4_cerebellum | y | nervous system |
| GSM1275536 | 4 | No.4_medulla oblongata | y | nervous system |
| GSM1275537 | 4 | No.4_Pituitary gland | y | gland |
| GSM1275538 | 4 | No.4_spinal cord_cervical | y | nervous system |
| GSM1275539 | 4 | No.4_spinal cord_pectoral | y | nervous system |
| GSM1275540 | 4 | No.4_spinal cord_pars lumbalis | y | nervous system |
| GSM1275541 | 4 | No.4_femoris muscle | y | smooth or skeletal muscles |
| GSM1275542 | 4 | No.4_gastrocnemial muscle | y | smooth or skeletal muscles |
| GSM1275543 | 4 | No.4_musculus soleus | y | smooth or skeletal muscles |
| GSM1275544 | 4 | No.4_ischial nerve | y | nervous system |
| GSM1275545 | 4 | No.4_interseptum | y | smooth or skeletal muscles |
| GSM1275546 | 4 | No.4_bone marrow | y | lymphoid or hematopoietic system |
| GSM1275547 | 4 | No.4_aorta | y | smooth or skeletal muscles |
| GSM1275548 | 4 | No.4_vein | y | smooth or skeletal muscles |
| GSM1275549 | 4 | No.4_white adipose tissue | y | others |
| GSM1275550 | 4 | No.4_brown adipose tissue | y | others |
| GSM1275551 | 5 | No.5_testicle | y | reproductive system |
| GSM1275552 | 5 | No.5_epididymis | y | reproductive system |
| GSM1275553 | 5 | No.5_eyeball | y | nervous system |
| GSM1275554 | 5 | No.5_Harderian gland | y | gland |
| GSM1275555 | 5 | No.5_optic nerve | y | nervous system |
| GSM1275556 | 5 | No.5_cerebrum_cerebral cortex | y | nervous system |
| GSM1275557 | 5 | No.5_cerebrum_hippocampus | y | nervous system |
| GSM1275558 | 5 | No.5_cerebrum_Thalamus | y | nervous system |
| GSM1275559 | 5 | No.5_cerebellum | y | nervous system |
| GSM1275560 | 5 | No.5_medulla oblongata | y | nervous system |
| GSM1275561 | 5 | No.5_Pituitary gland | y | gland |
| GSM1275562 | 5 | No.5_spinal cord_cervical | y | nervous system |
| GSM1275563 | 5 | No.5_spinal cord_pectoral | y | nervous system |
| GSM1275564 | 5 | No.5_spinal cord_pars lumbalis | y | nervous system |
| GSM1275565 | 5 | No.5_femoris muscle | y | smooth or skeletal muscles |
| GSM1275566 | 5 | No.5_gastrocnemial muscle | y | smooth or skeletal muscles |
| GSM1275567 | 5 | No.5_musculus soleus | y | smooth or skeletal muscles |
| GSM1275568 | 5 | No.5_ischial nerve | y | nervous system |
| GSM1275569 | 5 | No.5_interseptum | y | smooth or skeletal muscles |
| GSM1275570 | 5 | No.5_bone marrow | y | lymphoid or hematopoietic system |
| GSM1275571 | 5 | No.5_aorta | y | smooth or skeletal muscles |
| GSM1275572 | 5 | No.5_vein | y | smooth or skeletal muscles |
| GSM1275573 | 5 | No.5_white adipose tissue | y | others |
| GSM1275574 | 5 | No.5_brown adipose tissue | y | others |
| GSM1275575 | 6 | No.6_prostate_other | y | reproductive system |
| GSM1275576 | 6 | No.6_testicle | y | reproductive system |
| GSM1275577 | 6 | No.6_epididymis | y | reproductive system |
| GSM1275578 | 6 | No.6_eyeball | y | nervous system |
| GSM1275579 | 6 | No.6_Harderian gland | y | gland |
| GSM1275580 | 6 | No.6_optic nerve | y | nervous system |
| GSM1275581 | 6 | No.6_cerebrum_cerebral cortex | y | nervous system |
| GSM1275582 | 6 | No.6_cerebrum_hippocampus | y | nervous system |
| GSM1275583 | 6 | No.6_cerebrum_Thalamus | y | nervous system |
| GSM1275584 | 6 | No.6_cerebellum | y | nervous system |
| GSM1275585 | 6 | No.6_medulla oblongata | y | nervous system |
| GSM1275586 | 6 | No.6_Pituitary gland | y | gland |
| GSM1275587 | 6 | No.6_spinal cord_cervical | y | nervous system |
| GSM1275588 | 6 | No.6_spinal cord_pectoral | y | nervous system |
| GSM1275589 | 6 | No.6_spinal cord_pars lumbalis | y | nervous system |
| GSM1275590 | 6 | No.6_femoris muscle | y | smooth or skeletal muscles |
| GSM1275591 | 6 | No.6_gastrocnemial muscle | y | smooth or skeletal muscles |
| GSM1275592 | 6 | No.6_musculus soleus | y | smooth or skeletal muscles |
| GSM1275593 | 6 | No.6_ischial nerve | y | nervous system |
| GSM1275594 | 6 | No.6_interseptum | y | smooth or skeletal muscles |
| GSM1275595 | 6 | No.6_bone marrow | y | lymphoid or hematopoietic system |
| GSM1275596 | 6 | No.6_aorta | y | smooth or skeletal muscles |
| GSM1275597 | 6 | No.6_vein | y | smooth or skeletal muscles |
| GSM1275598 | 6 | No.6_white adipose tissue | y | others |
| GSM1275599 | 6 | No.6_brown adipose tissue | y | others |
*Individual sample IDs created by GEO.
†Resistered tissue names in GSE52754.
Table 3. Eigen values and vectors of PCAs in Figure 1 .
| Probe ID | Eigenvalue 1: 29.1% | Eigenvalue 2: 11.4% | Eigenvalue 3: 8.1% | Eigenvalue 4: 6.7% | Eigenvalue 5: 5.0% |
|---|---|---|---|---|---|
| rno-let-7a | 0.001 | −0.024 | −0.008 | −0.013 | 0.003 |
| rno-let-7a-1* | 0.004 | −0.015 | −0.026 | −0.029 | −0.035 |
| rno-let-7b | −0.002 | −0.018 | −0.013 | −0.002 | 0.004 |
| rno-let-7b* | −0.025 | 0.013 | 0.013 | 0.040 | 0.015 |
| rno-let-7c | 0.002 | −0.020 | −0.017 | −0.007 | 0.009 |
| rno-let-7c-1* | 0.023 | −0.008 | −0.014 | −0.006 | 0.030 |
| rno-let-7d | 0.002 | −0.027 | −0.005 | −0.014 | −0.010 |
| rno-let-7d* | 0.008 | −0.036 | 0.001 | −0.027 | −0.060 |
| rno-let-7e | 0.023 | −0.034 | −0.020 | −0.044 | 0.001 |
| rno-let-7f | −0.002 | −0.023 | −0.005 | −0.018 | −0.005 |
| rno-let-7i | 0.001 | −0.027 | −0.008 | −0.014 | −0.004 |
| rno-let-7i* | −0.015 | −0.004 | 0.008 | 0.035 | 0.014 |
| rno-miR-1 | 0.011 | −0.204 | −0.135 | 0.113 | −0.015 |
| rno-miR-1* | −0.023 | 0.015 | 0.019 | 0.053 | 0.002 |
| rno-miR-100 | 0.026 | −0.056 | −0.018 | −0.018 | 0.033 |
| rno-miR-100* | −0.012 | −0.006 | 0.011 | 0.037 | 0.014 |
| rno-miR-101a | −0.006 | −0.016 | −0.008 | −0.003 | 0.008 |
| rno-miR-101a* | −0.014 | −0.006 | 0.007 | 0.037 | 0.015 |
| rno-miR-101b | −0.007 | −0.012 | 0.011 | −0.025 | 0.001 |
| rno-miR-103 | 0.007 | −0.014 | −0.003 | −0.054 | −0.030 |
| rno-miR-105 | −0.015 | −0.007 | 0.007 | 0.041 | 0.013 |
| rno-miR-106b | −0.022 | −0.011 | 0.012 | −0.030 | −0.026 |
| rno-miR-106b* | −0.026 | −0.007 | 0.041 | 0.012 | 0.002 |
| rno-miR-107 | 0.004 | −0.016 | −0.002 | −0.043 | −0.026 |
| rno-miR-10a-3p | −0.036 | 0.000 | −0.016 | −0.035 | −0.018 |
| rno-miR-10a-5p | −0.043 | −0.055 | 0.000 | −0.027 | −0.038 |
| rno-miR-10b | −0.045 | −0.071 | −0.039 | 0.007 | −0.024 |
| rno-miR-10b* | −0.006 | −0.027 | 0.019 | −0.009 | −0.021 |
| rno-miR-1188-3p | −0.019 | 0.004 | 0.014 | 0.038 | 0.012 |
| rno-miR-1188-5p | −0.011 | 0.015 | 0.018 | 0.068 | 0.031 |
| rno-miR-122 | −0.029 | −0.009 | 0.044 | 0.041 | 0.019 |
| rno-miR-122* | −0.020 | −0.004 | 0.037 | 0.061 | 0.016 |
| rno-miR-1224 | −0.028 | 0.016 | 0.027 | 0.078 | 0.011 |
| rno-miR-124 | 0.131 | 0.069 | 0.066 | −0.009 | 0.016 |
| rno-miR-124* | 0.035 | 0.025 | 0.025 | 0.020 | 0.013 |
| rno-miR-1249 | −0.062 | 0.040 | 0.063 | 0.080 | −0.063 |
| rno-miR-125a-3p | −0.009 | 0.024 | 0.022 | 0.012 | −0.046 |
| rno-miR-125a-5p | 0.023 | −0.039 | −0.015 | −0.054 | −0.001 |
| rno-miR-125b* | 0.037 | 0.002 | −0.002 | 0.004 | 0.020 |
| rno-miR-125b-5p | 0.027 | −0.044 | −0.023 | −0.009 | 0.038 |
| rno-miR-126 | −0.003 | −0.069 | 0.005 | −0.019 | −0.006 |
| rno-miR-126* | 0.004 | −0.086 | 0.004 | −0.047 | −0.028 |
| rno-miR-127 | 0.113 | −0.023 | −0.097 | 0.063 | 0.012 |
| rno-miR-127* | 0.079 | 0.033 | −0.025 | 0.039 | −0.016 |
| rno-miR-128 | 0.046 | −0.019 | 0.021 | −0.028 | −0.041 |
| rno-miR-128-2* | −0.004 | 0.002 | 0.021 | 0.032 | 0.011 |
| rno-miR-129 | 0.102 | 0.067 | 0.046 | 0.007 | 0.034 |
| rno-miR-129-1* | 0.068 | 0.058 | 0.025 | −0.002 | 0.023 |
| rno-miR-129-2* | 0.116 | 0.079 | 0.036 | −0.030 | 0.017 |
| rno-miR-130a | −0.005 | −0.040 | −0.009 | −0.041 | −0.010 |
| rno-miR-130b | −0.077 | 0.074 | 0.044 | −0.061 | −0.095 |
| rno-miR-130b* | −0.043 | 0.037 | 0.033 | 0.023 | −0.050 |
| rno-miR-132 | 0.093 | 0.074 | 0.025 | −0.029 | −0.011 |
| rno-miR-132* | −0.006 | 0.000 | 0.012 | 0.036 | 0.016 |
| rno-miR-133a | 0.001 | −0.172 | −0.096 | 0.115 | −0.009 |
| rno-miR-133a* | −0.009 | −0.171 | −0.092 | 0.157 | 0.000 |
| rno-miR-133b | 0.013 | −0.151 | −0.130 | 0.063 | −0.005 |
| rno-miR-134 | 0.002 | 0.038 | 0.003 | 0.085 | 0.018 |
| rno-miR-134* | −0.006 | 0.027 | 0.001 | 0.036 | −0.011 |
| rno-miR-135a | 0.070 | 0.073 | −0.033 | −0.032 | 0.090 |
| rno-miR-135a* | 0.004 | 0.017 | 0.007 | 0.035 | 0.044 |
| rno-miR-135b | 0.075 | 0.046 | −0.019 | −0.012 | 0.046 |
| rno-miR-136 | 0.100 | 0.000 | −0.099 | 0.052 | −0.015 |
| rno-miR-136* | 0.101 | 0.015 | −0.081 | 0.058 | −0.037 |
| rno-miR-137 | 0.094 | 0.058 | 0.032 | 0.003 | −0.014 |
| rno-miR-137* | 0.013 | 0.011 | 0.024 | 0.030 | 0.014 |
| rno-miR-138 | 0.090 | 0.053 | 0.071 | −0.012 | −0.009 |
| rno-miR-138-1* | −0.011 | −0.005 | 0.011 | 0.037 | 0.013 |
| rno-miR-138-2* | 0.045 | 0.024 | 0.035 | 0.021 | −0.001 |
| rno-miR-139-3p | −0.017 | −0.001 | −0.004 | 0.067 | −0.001 |
| rno-miR-139-5p | 0.020 | −0.035 | 0.020 | −0.044 | −0.093 |
| rno-miR-140 | −0.003 | −0.030 | −0.002 | −0.047 | −0.035 |
| rno-miR-140* | −0.006 | −0.028 | 0.001 | −0.037 | −0.033 |
| rno-miR-141 | −0.128 | 0.143 | −0.149 | −0.046 | 0.068 |
| rno-miR-141* | −0.012 | 0.000 | 0.001 | 0.038 | 0.016 |
| rno-miR-142-3p | −0.046 | −0.021 | 0.050 | −0.054 | −0.056 |
| rno-miR-142-5p | −0.090 | −0.019 | 0.060 | −0.085 | −0.101 |
| rno-miR-143 | −0.024 | −0.054 | −0.039 | −0.022 | 0.013 |
| rno-miR-143* | −0.033 | −0.038 | −0.036 | −0.024 | 0.037 |
| rno-miR-144 | −0.027 | −0.069 | 0.078 | 0.000 | 0.002 |
| rno-miR-144* | −0.019 | −0.042 | 0.051 | −0.001 | 0.006 |
| rno-miR-145 | −0.023 | −0.072 | −0.055 | −0.023 | 0.021 |
| rno-miR-145* | −0.016 | −0.026 | −0.014 | 0.023 | 0.008 |
| rno-miR-146a | −0.024 | −0.038 | 0.008 | −0.015 | −0.016 |
| rno-miR-146b | 0.009 | −0.019 | −0.026 | −0.051 | −0.021 |
| rno-miR-147 | −0.041 | 0.037 | 0.015 | 0.034 | −0.052 |
| rno-miR-148b-3p | −0.002 | −0.007 | −0.009 | −0.041 | −0.005 |
| rno-miR-148b-5p | −0.011 | 0.004 | 0.003 | 0.015 | 0.015 |
| rno-miR-150 | 0.002 | −0.065 | 0.038 | −0.049 | −0.046 |
| rno-miR-150* | −0.060 | 0.035 | 0.056 | 0.051 | −0.101 |
| rno-miR-151 | 0.003 | −0.032 | −0.011 | −0.042 | −0.009 |
| rno-miR-151* | −0.001 | −0.008 | 0.011 | −0.009 | 0.013 |
| rno-miR-152 | −0.022 | −0.029 | −0.045 | 0.012 | 0.016 |
| rno-miR-153 | 0.066 | 0.090 | 0.011 | −0.014 | −0.015 |
| rno-miR-154 | 0.099 | −0.001 | −0.104 | 0.033 | −0.042 |
| rno-miR-154* | 0.069 | 0.058 | −0.012 | 0.018 | 0.007 |
| rno-miR-15b | −0.027 | −0.012 | 0.015 | −0.026 | −0.014 |
| rno-miR-15b* | −0.037 | 0.018 | 0.042 | −0.012 | −0.032 |
| rno-miR-16 | −0.015 | −0.024 | 0.011 | −0.022 | −0.021 |
| rno-miR-17-1-3p | −0.039 | 0.003 | 0.057 | −0.005 | −0.032 |
| rno-miR-17-5p | −0.031 | −0.010 | 0.022 | −0.038 | −0.038 |
| rno-miR-181a | 0.012 | −0.013 | 0.027 | −0.016 | −0.010 |
| rno-miR-181a-1* | 0.040 | −0.038 | 0.053 | −0.067 | −0.034 |
| rno-miR-181b | 0.022 | −0.016 | 0.019 | −0.016 | −0.024 |
| rno-miR-181b-1* | −0.018 | −0.003 | 0.022 | 0.032 | 0.009 |
| rno-miR-181c | 0.006 | −0.019 | −0.009 | −0.053 | −0.032 |
| rno-miR-181d | 0.005 | −0.007 | 0.005 | −0.022 | −0.022 |
| rno-miR-182 | −0.042 | 0.081 | −0.127 | −0.031 | 0.026 |
| rno-miR-183 | −0.040 | 0.115 | −0.158 | −0.072 | 0.032 |
| rno-miR-183* | −0.024 | 0.033 | −0.019 | 0.032 | −0.010 |
| rno-miR-184 | −0.013 | −0.016 | 0.006 | 0.010 | 0.030 |
| rno-miR-185 | 0.012 | 0.000 | 0.003 | −0.018 | −0.017 |
| rno-miR-186 | −0.005 | −0.009 | 0.008 | −0.002 | −0.010 |
| rno-miR-187* | −0.015 | −0.001 | 0.011 | 0.044 | 0.016 |
| rno-miR-188 | −0.075 | 0.065 | 0.050 | 0.085 | −0.067 |
| rno-miR-188* | −0.026 | −0.008 | 0.055 | 0.010 | −0.009 |
| rno-miR-18a | −0.071 | 0.025 | 0.034 | −0.049 | −0.082 |
| rno-miR-190 | 0.004 | 0.002 | −0.003 | 0.032 | −0.022 |
| rno-miR-190* | −0.007 | 0.003 | 0.008 | 0.038 | −0.003 |
| rno-miR-191 | −0.020 | −0.007 | 0.032 | 0.022 | −0.001 |
| rno-miR-191* | −0.007 | −0.013 | 0.028 | −0.027 | −0.041 |
| rno-miR-192 | −0.045 | 0.062 | 0.023 | −0.045 | −0.133 |
| rno-miR-192* | −0.067 | 0.073 | 0.025 | 0.025 | −0.092 |
| rno-miR-193 | −0.036 | −0.042 | −0.027 | 0.023 | −0.027 |
| rno-miR-193* | −0.024 | −0.065 | −0.039 | 0.035 | −0.050 |
| rno-miR-194 | −0.045 | 0.095 | 0.039 | −0.052 | −0.181 |
| rno-miR-194* | −0.054 | 0.056 | 0.018 | 0.026 | −0.070 |
| rno-miR-1949 | −0.055 | −0.039 | 0.009 | 0.001 | −0.054 |
| rno-miR-195 | −0.002 | −0.063 | −0.018 | −0.022 | −0.008 |
| rno-miR-196a | −0.032 | −0.021 | −0.046 | 0.027 | −0.081 |
| rno-miR-196b | −0.044 | −0.026 | −0.032 | 0.017 | −0.061 |
| rno-miR-196c | −0.039 | −0.004 | −0.044 | 0.033 | −0.088 |
| rno-miR-196c* | −0.051 | 0.060 | 0.011 | 0.036 | −0.091 |
| rno-miR-199a-3p | −0.027 | −0.069 | −0.048 | −0.001 | −0.005 |
| rno-miR-199a-5p | −0.041 | −0.080 | −0.053 | −0.009 | −0.015 |
| rno-miR-19a | −0.056 | 0.008 | 0.035 | −0.042 | −0.033 |
| rno-miR-19b | −0.039 | 0.002 | 0.028 | −0.009 | −0.008 |
| rno-miR-19b-1* | −0.025 | −0.011 | 0.058 | 0.011 | −0.006 |
| rno-miR-200a | −0.130 | 0.157 | −0.197 | −0.039 | 0.081 |
| rno-miR-200a* | −0.052 | 0.065 | −0.025 | 0.028 | 0.000 |
| rno-miR-200b | −0.134 | 0.158 | −0.206 | −0.040 | 0.082 |
| rno-miR-200b* | −0.082 | 0.099 | −0.090 | 0.006 | 0.067 |
| rno-miR-200c | −0.098 | 0.118 | −0.161 | −0.018 | 0.090 |
| rno-miR-200c_v15.0 | −0.134 | 0.121 | −0.135 | −0.055 | 0.111 |
| rno-miR-201 | −0.028 | −0.016 | 0.031 | −0.003 | 0.103 |
| rno-miR-201* | −0.017 | −0.008 | 0.022 | 0.034 | 0.065 |
| rno-miR-201_v15.0 | −0.026 | −0.014 | 0.028 | 0.006 | 0.094 |
| rno-miR-202 | −0.014 | −0.007 | 0.013 | 0.038 | 0.022 |
| rno-miR-202* | −0.029 | 0.034 | 0.027 | 0.071 | 0.021 |
| rno-miR-202_v15.0 | −0.015 | −0.008 | 0.025 | 0.040 | 0.046 |
| rno-miR-203 | −0.090 | 0.046 | −0.139 | −0.034 | −0.071 |
| rno-miR-204 | 0.039 | 0.005 | −0.051 | −0.061 | 0.029 |
| rno-miR-204* | 0.026 | 0.015 | 0.011 | 0.018 | 0.017 |
| rno-miR-205 | −0.080 | 0.042 | −0.210 | −0.041 | 0.051 |
| rno-miR-205_v15.0 | −0.088 | 0.037 | −0.216 | −0.035 | 0.032 |
| rno-miR-206 | 0.020 | −0.080 | −0.104 | 0.189 | −0.035 |
| rno-miR-207 | −0.015 | −0.007 | 0.015 | 0.036 | 0.013 |
| rno-miR-207_v15.0 | −0.015 | −0.007 | 0.015 | 0.036 | 0.013 |
| rno-miR-208 | −0.016 | −0.053 | 0.035 | 0.043 | 0.031 |
| rno-miR-208* | −0.017 | −0.052 | 0.030 | 0.040 | 0.034 |
| rno-miR-208_v15.0 | −0.017 | −0.071 | 0.046 | 0.045 | 0.037 |
| rno-miR-208b-3p | −0.013 | −0.011 | 0.007 | 0.041 | 0.012 |
| rno-miR-20a | −0.031 | −0.013 | 0.025 | −0.039 | −0.037 |
| rno-miR-20a* | −0.054 | 0.027 | 0.045 | −0.023 | −0.041 |
| rno-miR-20b-3p | −0.016 | −0.006 | 0.018 | 0.032 | 0.008 |
| rno-miR-20b-5p | −0.031 | −0.014 | 0.026 | −0.037 | −0.037 |
| rno-miR-20b-5p_v15.0 | −0.026 | −0.003 | 0.081 | 0.035 | −0.002 |
| rno-miR-21 | −0.038 | −0.001 | −0.006 | −0.017 | −0.007 |
| rno-miR-21* | −0.033 | 0.019 | 0.020 | 0.028 | −0.032 |
| rno-miR-210 | −0.003 | −0.021 | −0.003 | −0.007 | −0.012 |
| rno-miR-211 | 0.023 | 0.009 | 0.012 | −0.041 | 0.064 |
| rno-miR-211* | −0.064 | 0.065 | 0.023 | 0.075 | −0.077 |
| rno-miR-212 | 0.014 | 0.082 | 0.040 | 0.065 | −0.064 |
| rno-miR-212* | 0.007 | 0.015 | 0.011 | 0.033 | 0.022 |
| rno-miR-214 | −0.035 | −0.077 | −0.048 | 0.006 | −0.008 |
| rno-miR-215 | −0.076 | 0.096 | 0.012 | 0.046 | −0.133 |
| rno-miR-215_v15.0 | −0.082 | 0.103 | 0.014 | 0.047 | −0.146 |
| rno-miR-216a | −0.011 | 0.025 | 0.017 | 0.085 | 0.044 |
| rno-miR-216b-5p | −0.011 | 0.017 | 0.012 | 0.071 | 0.039 |
| rno-miR-217 | −0.012 | 0.019 | 0.016 | 0.081 | 0.043 |
| rno-miR-217_v15.0 | −0.016 | 0.007 | 0.019 | 0.071 | 0.033 |
| rno-miR-218 | 0.067 | −0.055 | −0.041 | −0.072 | 0.057 |
| rno-miR-219-2-3p | 0.096 | 0.049 | 0.068 | 0.003 | −0.002 |
| rno-miR-219-5p | 0.053 | 0.092 | 0.062 | −0.087 | −0.096 |
| rno-miR-22 | −0.005 | −0.049 | −0.014 | 0.017 | −0.002 |
| rno-miR-22* | 0.006 | −0.054 | −0.038 | −0.012 | −0.028 |
| rno-miR-221 | 0.000 | −0.017 | −0.025 | −0.055 | 0.019 |
| rno-miR-221* | −0.004 | 0.001 | 0.008 | 0.020 | 0.023 |
| rno-miR-222 | 0.013 | 0.025 | −0.036 | −0.088 | 0.047 |
| rno-miR-223 | −0.025 | −0.026 | 0.013 | 0.000 | −0.018 |
| rno-miR-224 | −0.012 | −0.004 | −0.005 | 0.036 | 0.016 |
| rno-miR-224* | −0.011 | −0.003 | −0.001 | 0.040 | 0.016 |
| rno-miR-224_v15.0 | −0.019 | −0.110 | −0.102 | 0.015 | 0.055 |
| rno-miR-23a | −0.011 | −0.046 | −0.028 | −0.007 | −0.020 |
| rno-miR-23a* | −0.027 | 0.009 | 0.001 | 0.048 | −0.013 |
| rno-miR-23b | 0.001 | −0.029 | −0.033 | −0.008 | −0.008 |
| rno-miR-24 | −0.006 | −0.038 | −0.026 | −0.007 | −0.015 |
| rno-miR-24-2* | 0.002 | −0.061 | −0.037 | −0.037 | −0.045 |
| rno-miR-25 | −0.019 | −0.012 | 0.013 | −0.004 | −0.006 |
| rno-miR-26a | −0.003 | −0.028 | −0.005 | −0.011 | −0.006 |
| rno-miR-26b | −0.012 | −0.024 | −0.010 | −0.017 | −0.008 |
| rno-miR-26b* | −0.026 | −0.014 | 0.020 | −0.017 | −0.006 |
| rno-miR-27a | −0.017 | −0.040 | −0.023 | −0.005 | −0.032 |
| rno-miR-27b | −0.005 | −0.024 | −0.026 | 0.004 | −0.011 |
| rno-miR-28 | −0.021 | −0.037 | −0.019 | −0.025 | −0.027 |
| rno-miR-28* | −0.016 | −0.011 | 0.003 | 0.031 | 0.012 |
| rno-miR-290 | −0.076 | −0.011 | 0.025 | 0.054 | −0.011 |
| rno-miR-290_v15.0 | −0.043 | 0.028 | 0.050 | 0.061 | −0.003 |
| rno-miR-291a-5p | −0.021 | 0.008 | 0.011 | 0.074 | 0.032 |
| rno-miR-292-5p | −0.031 | 0.025 | 0.016 | 0.063 | −0.012 |
| rno-miR-294 | −0.015 | −0.006 | 0.007 | 0.043 | 0.015 |
| rno-miR-295 | −0.017 | −0.003 | 0.012 | 0.038 | 0.008 |
| rno-miR-295_v15.0 | −0.017 | −0.003 | 0.011 | 0.038 | 0.008 |
| rno-miR-296* | 0.019 | 0.035 | 0.084 | 0.046 | −0.016 |
| rno-miR-298 | 0.006 | 0.043 | 0.039 | 0.042 | 0.007 |
| rno-miR-2985 | −0.015 | −0.006 | 0.011 | 0.037 | 0.013 |
| rno-miR-299 | 0.074 | −0.029 | −0.122 | 0.070 | −0.017 |
| rno-miR-299* | −0.011 | 0.001 | 0.002 | 0.037 | 0.018 |
| rno-miR-29a | 0.002 | −0.012 | 0.002 | −0.010 | 0.005 |
| rno-miR-29a* | 0.002 | −0.019 | −0.018 | −0.040 | −0.008 |
| rno-miR-29b | 0.003 | 0.009 | 0.005 | −0.029 | 0.005 |
| rno-miR-29b-1* | −0.015 | −0.007 | 0.010 | 0.034 | 0.017 |
| rno-miR-29b-2* | 0.011 | 0.014 | 0.015 | 0.031 | 0.014 |
| rno-miR-29c | 0.004 | −0.007 | −0.012 | −0.011 | 0.008 |
| rno-miR-29c* | 0.021 | −0.019 | −0.042 | −0.023 | 0.004 |
| rno-miR-300-3p | 0.102 | −0.001 | −0.102 | 0.050 | −0.048 |
| rno-miR-300-5p | 0.057 | 0.043 | 0.031 | 0.024 | 0.006 |
| rno-miR-301a | 0.000 | −0.008 | 0.007 | −0.054 | −0.029 |
| rno-miR-301b | −0.032 | 0.031 | 0.024 | 0.028 | −0.034 |
| rno-miR-3085 | −0.015 | −0.006 | 0.013 | 0.038 | 0.013 |
| rno-miR-30a | −0.006 | −0.039 | −0.010 | 0.011 | 0.008 |
| rno-miR-30a* | 0.003 | −0.053 | −0.021 | −0.018 | −0.002 |
| rno-miR-30b-3p | −0.025 | −0.021 | 0.010 | −0.002 | −0.011 |
| rno-miR-30b-5p | −0.013 | −0.024 | −0.009 | −0.004 | 0.008 |
| rno-miR-30c | −0.002 | −0.035 | −0.006 | −0.006 | −0.002 |
| rno-miR-30c-1* | −0.006 | −0.016 | 0.033 | −0.002 | −0.012 |
| rno-miR-30c-2* | 0.006 | −0.037 | −0.023 | 0.017 | 0.012 |
| rno-miR-30d | −0.009 | −0.026 | 0.003 | 0.012 | 0.005 |
| rno-miR-30d* | −0.024 | −0.006 | 0.017 | −0.015 | −0.010 |
| rno-miR-30e | −0.014 | −0.025 | 0.010 | 0.005 | −0.015 |
| rno-miR-30e* | 0.001 | −0.043 | −0.001 | −0.020 | −0.020 |
| rno-miR-31 | −0.018 | 0.084 | −0.078 | −0.047 | −0.028 |
| rno-miR-31* | −0.043 | 0.055 | −0.066 | 0.011 | −0.044 |
| rno-miR-32 | −0.055 | 0.040 | 0.035 | −0.049 | −0.061 |
| rno-miR-32* | −0.021 | 0.003 | 0.021 | 0.058 | 0.018 |
| rno-miR-320 | −0.003 | −0.009 | −0.021 | 0.007 | −0.024 |
| rno-miR-322 | 0.002 | −0.082 | −0.009 | −0.033 | 0.003 |
| rno-miR-322* | 0.033 | −0.125 | −0.009 | −0.034 | 0.020 |
| rno-miR-323 | 0.091 | 0.058 | 0.028 | 0.004 | 0.001 |
| rno-miR-324-3p | −0.004 | −0.002 | 0.002 | 0.013 | 0.007 |
| rno-miR-324-5p | 0.014 | −0.012 | −0.012 | −0.047 | −0.013 |
| rno-miR-325-3p | 0.092 | 0.057 | 0.030 | 0.003 | 0.003 |
| rno-miR-325-5p | 0.044 | 0.036 | 0.023 | 0.019 | 0.016 |
| rno-miR-326 | 0.022 | 0.025 | 0.009 | −0.103 | −0.048 |
| rno-miR-326* | −0.015 | −0.002 | 0.015 | 0.053 | 0.023 |
| rno-miR-327 | −0.079 | 0.063 | 0.018 | 0.066 | −0.091 |
| rno-miR-328a | 0.023 | −0.023 | 0.002 | −0.041 | −0.018 |
| rno-miR-328a* | −0.050 | 0.046 | 0.027 | 0.102 | −0.081 |
| rno-miR-328b-3p | 0.012 | 0.010 | 0.029 | 0.028 | 0.008 |
| rno-miR-329 | 0.111 | −0.007 | −0.108 | 0.042 | −0.037 |
| rno-miR-33 | −0.014 | 0.027 | 0.019 | −0.046 | −0.064 |
| rno-miR-33* | 0.008 | 0.046 | 0.038 | −0.032 | −0.048 |
| rno-miR-330 | 0.019 | 0.013 | 0.026 | 0.028 | 0.010 |
| rno-miR-330* | 0.067 | 0.036 | 0.030 | −0.026 | 0.010 |
| rno-miR-331 | 0.016 | −0.017 | −0.007 | −0.035 | −0.011 |
| rno-miR-331* | −0.025 | −0.004 | 0.051 | −0.016 | 0.004 |
| rno-miR-335 | 0.068 | −0.079 | −0.002 | −0.020 | −0.038 |
| rno-miR-337 | 0.081 | −0.012 | −0.092 | 0.066 | −0.045 |
| rno-miR-337* | 0.070 | 0.003 | −0.050 | 0.067 | −0.029 |
| rno-miR-338 | 0.060 | −0.037 | 0.008 | −0.037 | −0.022 |
| rno-miR-338* | 0.052 | 0.020 | 0.039 | 0.018 | −0.006 |
| rno-miR-339-3p | −0.013 | −0.010 | 0.021 | 0.002 | 0.004 |
| rno-miR-339-5p | −0.014 | −0.008 | 0.012 | 0.036 | 0.013 |
| rno-miR-340-3p | 0.032 | −0.011 | 0.014 | −0.083 | 0.016 |
| rno-miR-340-5p | 0.028 | 0.060 | 0.046 | −0.109 | 0.011 |
| rno-miR-341 | 0.095 | 0.041 | −0.047 | 0.064 | −0.033 |
| rno-miR-342-3p | 0.014 | −0.016 | 0.036 | −0.075 | −0.016 |
| rno-miR-342-5p | 0.021 | 0.019 | 0.081 | −0.079 | 0.027 |
| rno-miR-344a-3p | 0.083 | 0.056 | 0.036 | 0.009 | 0.008 |
| rno-miR-344b-2-3p | 0.019 | 0.020 | 0.022 | 0.029 | 0.019 |
| rno-miR-344b-5p | 0.023 | 0.026 | 0.020 | 0.029 | 0.021 |
| rno-miR-345-3p | −0.024 | 0.016 | 0.018 | 0.072 | 0.006 |
| rno-miR-345-5p | −0.003 | −0.017 | 0.000 | 0.006 | 0.002 |
| rno-miR-346 | 0.029 | 0.019 | 0.032 | 0.026 | 0.008 |
| rno-miR-347 | −0.045 | 0.011 | 0.015 | 0.039 | −0.034 |
| rno-miR-34a | 0.023 | −0.036 | 0.024 | −0.007 | 0.027 |
| rno-miR-34a* | 0.022 | 0.000 | 0.039 | −0.008 | 0.021 |
| rno-miR-34b | 0.032 | −0.013 | 0.057 | −0.110 | 0.122 |
| rno-miR-34b* | −0.020 | −0.014 | 0.020 | −0.016 | 0.106 |
| rno-miR-34b_v15.0 | 0.034 | 0.006 | 0.061 | −0.121 | 0.129 |
| rno-miR-34c | 0.041 | −0.003 | 0.066 | −0.109 | 0.144 |
| rno-miR-34c* | −0.018 | −0.013 | 0.017 | −0.003 | 0.066 |
| rno-miR-350 | 0.034 | −0.053 | 0.059 | 0.003 | −0.004 |
| rno-miR-351 | −0.012 | −0.016 | 0.006 | 0.011 | 0.013 |
| rno-miR-351* | −0.024 | 0.002 | 0.042 | 0.039 | 0.025 |
| rno-miR-352 | 0.005 | −0.035 | −0.014 | −0.035 | −0.021 |
| rno-miR-3541 | −0.015 | −0.007 | 0.016 | 0.035 | 0.012 |
| rno-miR-3544 | −0.032 | 0.016 | 0.022 | 0.048 | 0.003 |
| rno-miR-3545-3p | −0.019 | −0.032 | 0.008 | 0.017 | −0.003 |
| rno-miR-3546 | −0.018 | 0.004 | 0.021 | 0.081 | 0.035 |
| rno-miR-3547 | −0.032 | 0.012 | 0.020 | 0.052 | −0.001 |
| rno-miR-3549 | −0.037 | 0.040 | 0.036 | 0.087 | −0.012 |
| rno-miR-3550 | −0.015 | −0.007 | 0.015 | 0.036 | 0.013 |
| rno-miR-3551-3p | −0.016 | −0.004 | 0.009 | 0.043 | 0.029 |
| rno-miR-3554 | −0.015 | −0.004 | 0.012 | 0.046 | 0.019 |
| rno-miR-3557-3p | −0.027 | 0.010 | 0.011 | 0.081 | −0.004 |
| rno-miR-3558-3p | −0.029 | 0.018 | 0.011 | −0.029 | 0.081 |
| rno-miR-3559-5p | −0.064 | 0.046 | 0.017 | −0.067 | −0.063 |
| rno-miR-3562 | −0.029 | 0.020 | 0.010 | 0.093 | −0.011 |
| rno-miR-3563-3p | 0.001 | 0.007 | −0.027 | 0.041 | 0.008 |
| rno-miR-3563-5p | −0.009 | −0.001 | −0.001 | 0.037 | 0.013 |
| rno-miR-3564 | −0.059 | 0.061 | 0.006 | 0.087 | −0.089 |
| rno-miR-3568 | −0.015 | −0.005 | 0.011 | 0.038 | 0.013 |
| rno-miR-3572 | −0.015 | −0.007 | 0.014 | 0.036 | 0.013 |
| rno-miR-3573-3p | −0.017 | −0.002 | 0.014 | 0.039 | 0.010 |
| rno-miR-3573-5p | −0.016 | −0.005 | 0.011 | 0.039 | 0.012 |
| rno-miR-3580-3p | −0.032 | −0.012 | 0.082 | −0.014 | 0.080 |
| rno-miR-3582 | −0.031 | 0.019 | 0.035 | 0.058 | 0.013 |
| rno-miR-3584-3p | −0.015 | −0.007 | 0.015 | 0.036 | 0.013 |
| rno-miR-3584-5p | −0.052 | 0.033 | −0.006 | 0.084 | 0.027 |
| rno-miR-3585-5p | −0.033 | −0.039 | 0.041 | −0.004 | 0.116 |
| rno-miR-3588 | 0.003 | −0.013 | 0.029 | −0.001 | 0.057 |
| rno-miR-3593-3p | −0.079 | 0.049 | 0.044 | 0.067 | −0.074 |
| rno-miR-3593-5p | −0.024 | −0.006 | 0.060 | 0.001 | −0.008 |
| rno-miR-3594-5p | −0.021 | 0.005 | 0.013 | 0.042 | −0.005 |
| rno-miR-361 | 0.008 | −0.015 | 0.013 | −0.040 | −0.032 |
| rno-miR-361* | 0.006 | 0.004 | 0.035 | 0.025 | 0.001 |
| rno-miR-362 | −0.041 | −0.008 | −0.016 | 0.001 | −0.107 |
| rno-miR-362* | −0.022 | −0.025 | −0.010 | −0.018 | −0.047 |
| rno-miR-363 | −0.037 | −0.012 | 0.086 | −0.060 | 0.002 |
| rno-miR-363_v15.0 | −0.043 | 0.002 | 0.081 | −0.088 | −0.012 |
| rno-miR-365 | −0.013 | −0.046 | −0.027 | 0.013 | −0.009 |
| rno-miR-369-3p | 0.048 | 0.040 | 0.016 | 0.023 | 0.007 |
| rno-miR-369-5p | 0.105 | 0.011 | −0.071 | 0.058 | −0.040 |
| rno-miR-370 | 0.032 | 0.055 | 0.010 | 0.116 | −0.036 |
| rno-miR-370_v15.0 | −0.013 | 0.011 | 0.015 | 0.073 | 0.033 |
| rno-miR-374 | 0.003 | −0.024 | −0.023 | −0.061 | −0.032 |
| rno-miR-375 | −0.070 | 0.162 | −0.088 | −0.002 | 0.099 |
| rno-miR-376a | 0.108 | 0.031 | −0.067 | 0.005 | −0.027 |
| rno-miR-376a* | 0.044 | 0.038 | 0.014 | 0.025 | 0.010 |
| rno-miR-376b-3p | 0.090 | 0.067 | −0.020 | 0.002 | 0.006 |
| rno-miR-376b-5p | 0.078 | 0.021 | −0.048 | 0.042 | −0.021 |
| rno-miR-376c | 0.074 | −0.006 | −0.090 | 0.057 | −0.042 |
| rno-miR-377 | −0.002 | 0.009 | −0.006 | 0.037 | 0.014 |
| rno-miR-377_v15.0 | 0.039 | 0.035 | −0.012 | 0.033 | 0.007 |
| rno-miR-378 | −0.029 | −0.040 | −0.014 | 0.044 | −0.057 |
| rno-miR-378* | −0.031 | −0.072 | −0.030 | 0.031 | −0.097 |
| rno-miR-379 | 0.103 | −0.007 | −0.104 | 0.039 | −0.021 |
| rno-miR-379* | 0.075 | 0.024 | −0.022 | 0.029 | −0.021 |
| rno-miR-380 | 0.005 | 0.006 | 0.015 | 0.035 | 0.012 |
| rno-miR-380* | 0.082 | 0.053 | 0.027 | 0.008 | −0.005 |
| rno-miR-381 | 0.096 | −0.003 | −0.091 | 0.070 | −0.050 |
| rno-miR-381* | 0.029 | 0.008 | −0.009 | 0.052 | −0.004 |
| rno-miR-381_v15.0 | −0.011 | 0.000 | 0.003 | 0.037 | 0.018 |
| rno-miR-382 | 0.102 | 0.011 | −0.074 | 0.041 | −0.040 |
| rno-miR-382* | 0.071 | 0.053 | 0.007 | 0.018 | −0.001 |
| rno-miR-383 | 0.072 | 0.053 | 0.037 | 0.008 | 0.005 |
| rno-miR-383_v15.0 | 0.065 | 0.051 | 0.043 | 0.020 | 0.006 |
| rno-miR-384-3p | 0.057 | 0.042 | 0.028 | 0.014 | 0.014 |
| rno-miR-384-5p | 0.112 | 0.065 | 0.023 | −0.013 | 0.009 |
| rno-miR-409-3p | −0.012 | −0.002 | 0.011 | 0.036 | 0.013 |
| rno-miR-409-5p | 0.079 | 0.032 | −0.016 | 0.047 | −0.021 |
| rno-miR-409-5p_v15.0 | 0.065 | 0.040 | 0.005 | 0.032 | −0.004 |
| rno-miR-410 | 0.111 | 0.022 | −0.046 | 0.057 | −0.035 |
| rno-miR-411 | 0.090 | 0.005 | −0.086 | 0.056 | −0.040 |
| rno-miR-411* | 0.102 | −0.009 | −0.104 | 0.041 | −0.037 |
| rno-miR-412* | −0.003 | 0.001 | 0.016 | 0.035 | 0.016 |
| rno-miR-421* | 0.046 | 0.015 | −0.003 | −0.075 | −0.004 |
| rno-miR-423 | −0.017 | −0.012 | 0.023 | 0.028 | 0.011 |
| rno-miR-423* | −0.016 | −0.004 | 0.016 | 0.009 | −0.016 |
| rno-miR-425 | −0.015 | −0.003 | 0.020 | −0.049 | −0.029 |
| rno-miR-425* | −0.015 | −0.008 | 0.017 | 0.034 | 0.011 |
| rno-miR-429 | −0.122 | 0.146 | −0.190 | −0.041 | 0.071 |
| rno-miR-431 | 0.075 | 0.031 | −0.044 | 0.061 | −0.019 |
| rno-miR-433 | 0.101 | 0.049 | 0.013 | 0.027 | −0.016 |
| rno-miR-433* | 0.067 | 0.041 | 0.027 | 0.020 | −0.004 |
| rno-miR-434 | 0.121 | −0.021 | −0.105 | 0.054 | 0.012 |
| rno-miR-434* | 0.087 | 0.036 | −0.026 | 0.037 | −0.017 |
| rno-miR-448 | 0.014 | 0.010 | 0.023 | 0.028 | 0.010 |
| rno-miR-448* | 0.012 | 0.010 | 0.023 | 0.030 | 0.011 |
| rno-miR-448_v15.0 | −0.010 | −0.004 | 0.011 | 0.036 | 0.015 |
| rno-miR-449a | −0.045 | −0.003 | 0.055 | −0.057 | 0.073 |
| rno-miR-449c-5p | −0.017 | −0.010 | 0.005 | 0.017 | 0.021 |
| rno-miR-450a | 0.019 | −0.103 | −0.002 | −0.036 | 0.031 |
| rno-miR-450a* | −0.011 | −0.007 | 0.009 | 0.037 | 0.011 |
| rno-miR-450a_v15.0 | −0.004 | −0.119 | −0.008 | −0.025 | −0.001 |
| rno-miR-451 | −0.034 | −0.131 | 0.032 | −0.025 | −0.055 |
| rno-miR-455 | −0.017 | −0.018 | 0.004 | −0.015 | −0.003 |
| rno-miR-455* | 0.021 | −0.045 | −0.007 | −0.068 | −0.059 |
| rno-miR-463 | −0.021 | −0.007 | 0.030 | 0.029 | 0.099 |
| rno-miR-463* | −0.015 | −0.008 | 0.017 | 0.037 | 0.036 |
| rno-miR-465 | −0.015 | −0.005 | 0.011 | 0.038 | 0.013 |
| rno-miR-465* | −0.016 | −0.008 | 0.022 | 0.037 | 0.047 |
| rno-miR-466b | −0.033 | 0.018 | 0.024 | 0.043 | −0.001 |
| rno-miR-466b-1* | −0.015 | −0.005 | 0.011 | 0.043 | −0.011 |
| rno-miR-466b-2* | −0.012 | −0.010 | 0.075 | 0.001 | −0.031 |
| rno-miR-466c* | −0.018 | −0.006 | 0.008 | 0.040 | 0.001 |
| rno-miR-471 | −0.023 | −0.004 | 0.030 | 0.029 | 0.092 |
| rno-miR-471* | −0.021 | −0.007 | 0.029 | 0.030 | 0.094 |
| rno-miR-483 | 0.008 | −0.020 | 0.024 | 0.076 | 0.003 |
| rno-miR-483* | −0.056 | 0.054 | 0.009 | 0.101 | −0.121 |
| rno-miR-484 | −0.001 | −0.015 | −0.004 | −0.034 | −0.077 |
| rno-miR-485 | 0.073 | 0.051 | 0.014 | 0.019 | 0.001 |
| rno-miR-487b | 0.107 | 0.008 | −0.069 | 0.054 | −0.047 |
| rno-miR-487b_v15.0 | 0.070 | 0.056 | 0.013 | 0.027 | −0.024 |
| rno-miR-488 | 0.050 | 0.037 | 0.034 | 0.015 | 0.021 |
| rno-miR-489 | −0.008 | −0.001 | 0.012 | 0.029 | 0.018 |
| rno-miR-490 | 0.026 | −0.036 | 0.028 | 0.008 | 0.033 |
| rno-miR-490* | 0.030 | −0.013 | 0.030 | 0.020 | 0.028 |
| rno-miR-493 | −0.013 | 0.014 | 0.008 | 0.072 | 0.035 |
| rno-miR-493* | −0.010 | 0.000 | −0.004 | 0.040 | 0.016 |
| rno-miR-494 | −0.025 | 0.015 | 0.011 | 0.077 | 0.022 |
| rno-miR-494_v15.0 | −0.028 | 0.016 | 0.021 | 0.081 | 0.012 |
| rno-miR-495 | 0.109 | 0.025 | −0.054 | 0.054 | −0.033 |
| rno-miR-496 | 0.049 | 0.032 | 0.028 | 0.021 | 0.001 |
| rno-miR-496_v15.0 | 0.077 | 0.048 | 0.015 | 0.012 | −0.007 |
| rno-miR-497 | −0.005 | −0.055 | −0.010 | −0.007 | −0.001 |
| rno-miR-499 | 0.017 | −0.101 | −0.048 | −0.017 | 0.010 |
| rno-miR-500 | −0.014 | −0.009 | 0.004 | 0.007 | −0.006 |
| rno-miR-501* | −0.015 | −0.004 | 0.012 | 0.045 | 0.019 |
| rno-miR-503 | −0.003 | −0.117 | −0.011 | −0.002 | −0.019 |
| rno-miR-505 | 0.002 | −0.015 | 0.013 | −0.051 | −0.036 |
| rno-miR-505* | 0.029 | 0.010 | 0.027 | −0.052 | −0.009 |
| rno-miR-511* | −0.040 | −0.108 | 0.010 | 0.011 | −0.004 |
| rno-miR-532-3p | −0.005 | −0.027 | −0.008 | −0.023 | −0.040 |
| rno-miR-532-5p | −0.014 | −0.030 | 0.000 | −0.013 | −0.051 |
| rno-miR-539 | 0.082 | 0.009 | −0.054 | 0.060 | −0.024 |
| rno-miR-540 | −0.011 | 0.000 | 0.005 | 0.037 | 0.019 |
| rno-miR-540* | −0.012 | −0.002 | 0.007 | 0.037 | 0.018 |
| rno-miR-540_v15.0 | −0.005 | 0.008 | 0.001 | 0.036 | 0.022 |
| rno-miR-541 | 0.075 | 0.031 | −0.023 | 0.042 | −0.018 |
| rno-miR-541* | −0.006 | 0.005 | −0.008 | 0.040 | 0.014 |
| rno-miR-542-3p | 0.027 | −0.114 | 0.017 | −0.037 | 0.010 |
| rno-miR-542-5p | 0.040 | −0.119 | −0.002 | −0.026 | 0.015 |
| rno-miR-543* | 0.092 | 0.052 | 0.006 | 0.019 | −0.007 |
| rno-miR-543_v15.0 | 0.033 | 0.027 | 0.020 | 0.025 | 0.009 |
| rno-miR-547 | −0.034 | −0.033 | 0.037 | −0.007 | 0.121 |
| rno-miR-547* | −0.016 | −0.008 | 0.025 | 0.039 | 0.052 |
| rno-miR-547_v15.0 | −0.018 | −0.009 | 0.024 | 0.033 | 0.074 |
| rno-miR-551b | 0.091 | 0.061 | 0.037 | −0.042 | 0.023 |
| rno-miR-582 | 0.010 | 0.075 | −0.050 | −0.067 | −0.048 |
| rno-miR-582* | 0.007 | 0.060 | 0.027 | 0.048 | −0.014 |
| rno-miR-592 | 0.038 | 0.031 | 0.025 | 0.022 | 0.014 |
| rno-miR-598-3p | 0.082 | 0.039 | −0.022 | −0.080 | 0.004 |
| rno-miR-652 | −0.009 | −0.009 | 0.005 | −0.023 | 0.008 |
| rno-miR-652* | −0.034 | 0.005 | 0.009 | 0.053 | 0.004 |
| rno-miR-653 | −0.007 | 0.001 | 0.012 | 0.024 | 0.018 |
| rno-miR-664 | 0.000 | −0.008 | 0.007 | 0.010 | 0.007 |
| rno-miR-664-1* | −0.041 | 0.036 | 0.012 | 0.037 | −0.053 |
| rno-miR-664-2* | 0.016 | 0.012 | 0.018 | 0.035 | 0.008 |
| rno-miR-665 | 0.019 | 0.023 | 0.013 | 0.029 | 0.016 |
| rno-miR-665_v15.0 | −0.004 | 0.011 | −0.001 | 0.039 | 0.016 |
| rno-miR-666 | −0.017 | 0.007 | 0.023 | 0.080 | 0.036 |
| rno-miR-666_v15.0 | −0.018 | 0.005 | 0.020 | 0.069 | 0.026 |
| rno-miR-667 | 0.052 | 0.060 | 0.014 | 0.031 | −0.056 |
| rno-miR-667* | −0.020 | −0.001 | 0.005 | 0.046 | −0.003 |
| rno-miR-667_v15.0 | 0.029 | 0.023 | 0.029 | 0.024 | 0.011 |
| rno-miR-668 | −0.015 | −0.007 | 0.014 | 0.036 | 0.013 |
| rno-miR-671 | −0.015 | −0.007 | 0.014 | 0.036 | 0.013 |
| rno-miR-672 | 0.026 | 0.071 | −0.038 | −0.056 | 0.178 |
| rno-miR-674-3p | 0.013 | −0.023 | −0.006 | −0.035 | −0.059 |
| rno-miR-674-5p | 0.019 | −0.004 | 0.032 | −0.033 | −0.010 |
| rno-miR-675* | −0.006 | −0.063 | −0.055 | 0.111 | −0.029 |
| rno-miR-678 | −0.024 | 0.011 | 0.016 | 0.080 | 0.011 |
| rno-miR-702-3p | −0.014 | −0.006 | 0.011 | 0.036 | 0.013 |
| rno-miR-708 | −0.014 | −0.007 | 0.008 | 0.040 | 0.014 |
| rno-miR-711 | −0.017 | −0.003 | 0.011 | 0.038 | 0.008 |
| rno-miR-741-3p | −0.026 | −0.002 | 0.033 | 0.029 | 0.118 |
| rno-miR-742 | −0.026 | −0.001 | 0.024 | 0.033 | 0.130 |
| rno-miR-742* | −0.023 | −0.002 | 0.025 | 0.034 | 0.112 |
| rno-miR-743a | −0.025 | −0.001 | 0.023 | 0.033 | 0.124 |
| rno-miR-743a* | −0.019 | −0.007 | 0.023 | 0.034 | 0.087 |
| rno-miR-743b | −0.027 | −0.001 | 0.026 | 0.032 | 0.147 |
| rno-miR-743b* | −0.021 | −0.005 | 0.024 | 0.036 | 0.102 |
| rno-miR-758 | 0.070 | 0.050 | 0.033 | 0.014 | 0.004 |
| rno-miR-760-3p | 0.000 | 0.018 | 0.072 | 0.035 | −0.017 |
| rno-miR-760-5p | −0.021 | 0.010 | 0.017 | 0.074 | 0.023 |
| rno-miR-764* | 0.001 | 0.004 | 0.015 | 0.036 | 0.012 |
| rno-miR-764_v15.0 | 0.016 | 0.012 | 0.025 | 0.028 | 0.010 |
| rno-miR-770* | 0.050 | 0.054 | 0.038 | 0.061 | 0.022 |
| rno-miR-7a | 0.007 | 0.051 | 0.038 | −0.017 | −0.006 |
| rno-miR-7a-1* | 0.000 | 0.014 | −0.001 | −0.032 | −0.016 |
| rno-miR-7a-2* | 0.024 | 0.032 | 0.014 | 0.030 | 0.026 |
| rno-miR-7b | 0.047 | 0.100 | 0.068 | −0.032 | −0.033 |
| rno-miR-802 | −0.030 | 0.019 | 0.026 | 0.055 | −0.018 |
| rno-miR-802_v15.0 | −0.038 | 0.041 | 0.030 | 0.059 | −0.037 |
| rno-miR-871* | −0.021 | −0.007 | 0.029 | 0.030 | 0.096 |
| rno-miR-871_v15.0 | −0.017 | −0.008 | 0.021 | 0.033 | 0.066 |
| rno-miR-872 | −0.009 | −0.011 | 0.006 | −0.041 | −0.013 |
| rno-miR-872* | −0.011 | −0.001 | 0.025 | −0.025 | −0.015 |
| rno-miR-873 | 0.043 | 0.034 | 0.045 | 0.010 | 0.005 |
| rno-miR-874 | 0.017 | 0.032 | 0.059 | 0.008 | 0.026 |
| rno-miR-877 | −0.042 | 0.023 | 0.056 | 0.044 | −0.031 |
| rno-miR-878 | −0.015 | −0.008 | 0.016 | 0.037 | 0.036 |
| rno-miR-880 | −0.022 | −0.007 | 0.028 | 0.029 | 0.097 |
| rno-miR-881 | −0.015 | −0.008 | 0.022 | 0.039 | 0.039 |
| rno-miR-881* | −0.018 | −0.009 | 0.024 | 0.033 | 0.075 |
| rno-miR-881_v15.0 | −0.020 | −0.010 | 0.029 | 0.030 | 0.098 |
| rno-miR-883 | −0.026 | −0.001 | 0.025 | 0.034 | 0.133 |
| rno-miR-883* | −0.023 | 0.003 | 0.034 | 0.060 | 0.123 |
| rno-miR-9 | 0.109 | 0.026 | 0.025 | −0.053 | 0.091 |
| rno-miR-9* | 0.117 | 0.044 | 0.028 | −0.050 | 0.101 |
| rno-miR-92a | −0.029 | −0.011 | 0.036 | −0.003 | −0.015 |
| rno-miR-92a-2* | −0.019 | −0.009 | 0.033 | 0.029 | 0.003 |
| rno-miR-92b | 0.025 | 0.035 | 0.035 | 0.026 | 0.006 |
| rno-miR-93 | −0.018 | −0.014 | 0.010 | −0.040 | −0.033 |
| rno-miR-96 | −0.060 | 0.132 | −0.147 | −0.079 | 0.024 |
| rno-miR-96* | −0.012 | −0.004 | 0.003 | 0.035 | 0.019 |
| rno-miR-98 | 0.007 | −0.015 | −0.007 | −0.024 | −0.014 |
| rno-miR-99a | 0.019 | −0.051 | −0.021 | 0.005 | 0.055 |
| rno-miR-99a* | 0.033 | −0.071 | −0.050 | −0.010 | 0.053 |
| rno-miR-99b | 0.024 | −0.044 | −0.022 | −0.056 | 0.004 |
| rno-miR-99b* | 0.016 | 0.010 | −0.002 | 0.021 | 0.006 |
Furthermore, miRNA microarray data obtained from digestive organs, the nervous system and skeletal/smooth muscles were extracted from the whole dataset and subjected to system-specific PCA (Figure 2b). In the intestinal-specific subanalysis, clusters of signals from upper digestive organ (esophagus and stomach) samples were clearly separated from those of lower digestive organs (small and large intestine) along PC1 (37.2%), whereas along the PC2 axis (10.8%), those from small and large intestine samples were further divided into each organ by functional anatomy of the digestive system. In the nervous system subanalysis, a clear separation of clusters of signals were observed along both PC1 (35.7%) and PC2 (13.2%); those from cerebrum samples were clearly separated from ischial nerve samples along PC1, and those from the spinal cord were separated from optic nerve samples along PC2 (13.2%). Skeletal/smooth muscles were separated into four clusters: skeletal muscles, smooth muscles (blood vessel), smooth muscles (bladder and stomach) and others (esophagus, tongue and skin) (31.0% for PC1 and 16.4% for PC2). Overall, PCA successfully showed the similarity in gene expression profiles among functionally similar organs. These results provided evidence that the microarray data were of high enough quality to assess similarities and differences among miRNA profiles from different organs and tissues. In addition, the global miRNA expression profiles for these 55 normal organs included high-quality, reliable data that reflected the biological function of each organ.
Organ specific miRNA selection
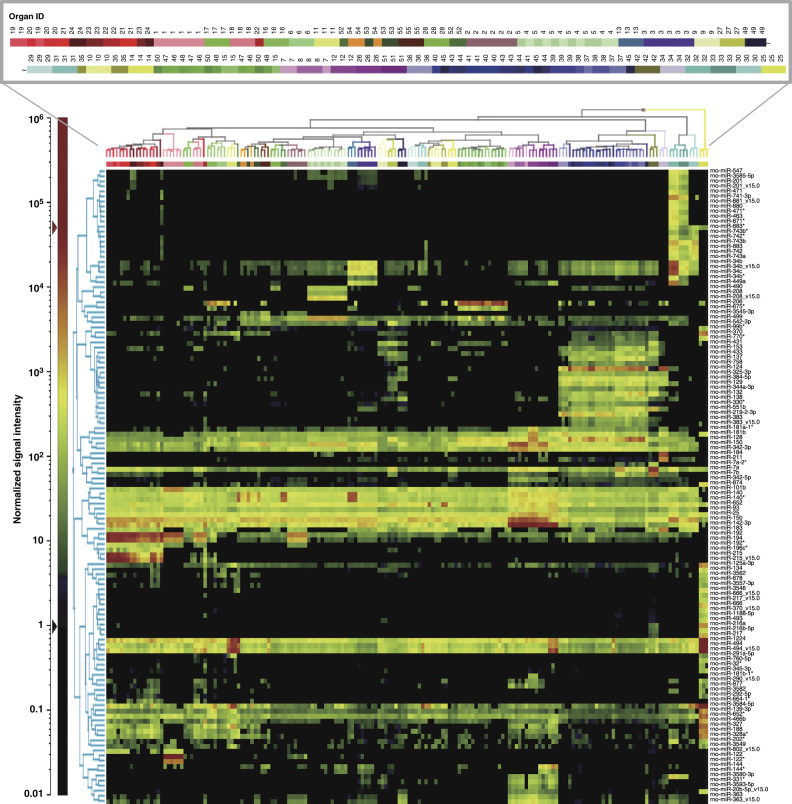
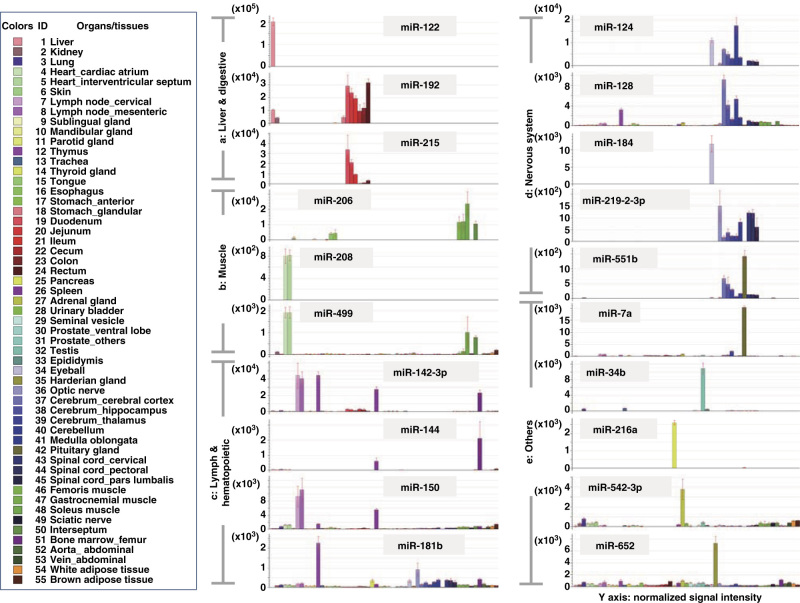
To identify organ-specific miRNAs, all probes with any detectable signal were further filtered by a statistical analysis with criteria described in the Methods section; 296 probes met these criteria and were further analyzed using model-based clustering. In the model-based clustering, 128 probes were identified as miRNAs that were specifically expressed in one or more organs; further details on the statistical parameters calculated are available in Table 4 (available online only). Figure 3 shows a 2-dimensional hierarchical clustering diagram of groups of organs; the clustering was based on the expression profiles of the 128 filtered probes. Triplicate samples of the same organ were densely clustered; moreover, functionally similar organs were closely clustered to each other. Among these clusters, several miRNAs that were specifically expressed in specific organ(s) were successfully identified. Expression profiles of several representative organ-specific miRNAs are summarized (Figure 4 and Table 5 (available online only)). As shown in Table 5, more than half of these miRNAs were expressed in the corresponding human organs/tissues22–31,22–31,22–31,22–31,22–31,22–31,22–31,22–31,22–31,22–31.
Table 4. Summary of statistical parameters of miRNAs calculated in this study.
| Probe ID | Min | 25% | Med | 75% | Max | P value of Shapiro-Wilk test with Bonferroni correction | Minimum cluster size by model-based clustering |
|---|---|---|---|---|---|---|---|
| rno-let-7a | 1046.42 | 6845.00 | 10232.27 | 13716.02 | 31554.59 | 8.52E-03 | NaN |
| rno-let-7a-1* | 0.17 | 6.19 | 9.34 | 12.50 | 32.73 | 1.08E-13 | NaN |
| rno-let-7b | 1989.44 | 4032.61 | 5945.58 | 8531.73 | 19369.52 | 1 | NaN |
| rno-let-7b* | 0.05 | 3.49 | 9.43 | 18.50 | 110.78 | 1.00E-08 | NaN |
| rno-let-7c | 1934.18 | 5813.99 | 8433.22 | 11790.22 | 28792.33 | 1.00E+00 | NaN |
| rno-let-7c-1* | 0.04 | 0.15 | 0.27 | 2.35 | 14.76 | 1.02E-07 | NaN |
| rno-let-7d | 218.80 | 2090.72 | 3021.18 | 4325.45 | 9045.13 | 3.86E-04 | NaN |
| rno-let-7d* | 0.15 | 6.63 | 11.02 | 17.19 | 51.65 | 1.71E-10 | NaN |
| rno-let-7e | 1.05 | 538.17 | 1224.57 | 1897.35 | 5184.52 | 4.55E-09 | NaN |
| rno-let-7e* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-let-7f | 590.94 | 6021.56 | 9451.42 | 12156.47 | 22912.60 | 1.21E-07 | NaN |
| rno-let-7f-1* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-let-7f-2* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-let-7i | 432.57 | 1570.24 | 3054.18 | 4569.44 | 8432.51 | 1.98E-03 | NaN |
| rno-let-7i* | 0.04 | 0.10 | 0.15 | 0.25 | 9.35 | 5.45E-04 | NaN |
| rno-miR-1 | 0.04 | 0.59 | 28.12 | 1022.22 | 158531.73 | 3.63E-04 | NaN |
| rno-miR-1* | 0.04 | 0.13 | 0.27 | 3.76 | 84.42 | 2.15E-07 | NaN |
| rno-miR-100 | 0.32 | 147.83 | 330.78 | 609.36 | 2208.20 | 8.61E-08 | NaN |
| rno-miR-100* | 0.04 | 0.10 | 0.15 | 0.25 | 2.49 | 1.00E+00 | NaN |
| rno-miR-101a | 92.73 | 216.13 | 364.40 | 521.12 | 1626.91 | 1.00E+00 | NaN |
| rno-miR-101a* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-101b | 0.69 | 132.64 | 170.04 | 238.46 | 3695.66 | 1.03E-14 | 6 |
| rno-miR-101b* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-103 | 0.66 | 592.23 | 856.15 | 1066.14 | 1943.70 | 2.45E-20 | NaN |
| rno-miR-103-1* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-103-2* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-105 | 0.04 | 0.10 | 0.15 | 0.25 | 6.54 | 1.05E-02 | NaN |
| rno-miR-106b | 1.05 | 189.31 | 292.88 | 433.68 | 2364.88 | 4.44E-07 | NaN |
| rno-miR-106b* | 0.04 | 0.11 | 0.17 | 0.30 | 28.20 | 4.10E-10 | NaN |
| rno-miR-107 | 0.69 | 758.82 | 1049.45 | 1375.37 | 6350.00 | 2.12E-18 | NaN |
| rno-miR-107* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-10a-3p | 0.04 | 0.14 | 0.26 | 3.28 | 31.83 | 4.68E-09 | NaN |
| rno-miR-10a-5p | 0.08 | 107.73 | 813.28 | 1755.77 | 5844.59 | 1.32E-07 | NaN |
| rno-miR-10b | 0.04 | 67.78 | 405.06 | 1718.67 | 5033.55 | 1.89E-10 | NaN |
| rno-miR-10b* | 0.04 | 0.14 | 0.26 | 3.97 | 33.39 | 1.66E-08 | NaN |
| rno-miR-1188-3p | 0.05 | 6.21 | 14.55 | 25.71 | 119.77 | 1.56E-11 | NaN |
| rno-miR-1188-5p | 0.04 | 0.11 | 0.16 | 0.26 | 201.28 | 3.29E-15 | 8 |
| rno-miR-1193-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-1193-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-122 | 0.04 | 0.10 | 0.17 | 0.27 | 237948.69 | 1.83E-19 | 6 |
| rno-miR-122* | 0.04 | 0.10 | 0.15 | 0.25 | 6544.33 | 2.58E-20 | 6 |
| rno-miR-1224 | 75.01 | 218.56 | 418.63 | 792.43 | 752897.90 | 4.84E-12 | 6 |
| rno-miR-124 | 0.04 | 0.14 | 0.24 | 0.66 | 21435.00 | 1.80E-15 | 30 |
| rno-miR-124* | 0.04 | 0.13 | 0.20 | 0.32 | 35.59 | 7.23E-12 | NaN |
| rno-miR-1249 | 0.04 | 0.12 | 0.25 | 14.28 | 413.90 | 5.82E-10 | NaN |
| rno-miR-125a-3p | 0.10 | 3.58 | 7.98 | 13.73 | 3683.65 | 6.08E-07 | 5 |
| rno-miR-125a-5p | 0.66 | 150.66 | 369.09 | 587.12 | 1078.25 | 1.32E-11 | NaN |
| rno-miR-125b* | 0.06 | 0.16 | 0.28 | 3.45 | 19.28 | 1.69E-08 | NaN |
| rno-miR-125b-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-125b-5p | 80.47 | 2268.48 | 4729.35 | 7736.27 | 33595.29 | 4.84E-06 | NaN |
| rno-miR-126 | 95.16 | 533.36 | 1111.75 | 3040.94 | 10867.25 | 1.00E+00 | NaN |
| rno-miR-126* | 0.12 | 30.75 | 64.82 | 185.69 | 729.14 | 4.18E-10 | NaN |
| rno-miR-127 | 0.06 | 0.33 | 35.30 | 270.18 | 2092.46 | 1.01E-07 | NaN |
| rno-miR-127* | 0.06 | 0.15 | 0.27 | 9.22 | 74.43 | 1.27E-11 | NaN |
| rno-miR-128 | 0.66 | 61.76 | 110.78 | 493.04 | 10166.81 | 2.70E-06 | 12 |
| rno-miR-128-1* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-128-2* | 0.04 | 0.11 | 0.17 | 0.27 | 9.24 | 1.31E-09 | NaN |
| rno-miR-129 | 0.04 | 0.14 | 0.23 | 0.64 | 794.15 | 1.55E-14 | 33 |
| rno-miR-129-1* | 0.06 | 7.83 | 13.37 | 31.37 | 2391.57 | 1.34E-05 | NaN |
| rno-miR-129-2* | 0.06 | 0.18 | 0.33 | 26.50 | 2838.37 | 5.43E-11 | NaN |
| rno-miR-130a | 1.05 | 422.65 | 754.01 | 1435.88 | 3143.45 | 1.50E-08 | NaN |
| rno-miR-130a* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-130b | 0.04 | 0.16 | 0.31 | 16.33 | 1214.23 | 2.34E-09 | NaN |
| rno-miR-130b* | 0.04 | 0.11 | 0.19 | 0.39 | 41.61 | 1.20E-10 | NaN |
| rno-miR-132 | 0.06 | 0.17 | 0.27 | 8.36 | 1255.41 | 5.05E-12 | 30 |
| rno-miR-132* | 0.04 | 0.11 | 0.16 | 0.26 | 6.34 | 5.66E-06 | NaN |
| rno-miR-133a | 0.04 | 0.16 | 4.03 | 59.24 | 5011.82 | 3.26E-07 | NaN |
| rno-miR-133a* | 0.04 | 0.12 | 0.23 | 37.46 | 5935.05 | 1.58E-11 | NaN |
| rno-miR-133b | 0.18 | 33.24 | 188.85 | 2606.57 | 191928.27 | 0.023667643 | NaN |
| rno-miR-133b* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-134 | 0.04 | 0.12 | 0.20 | 0.34 | 478.02 | 6.77E-13 | 31 |
| rno-miR-134* | 0.04 | 0.12 | 0.19 | 0.30 | 11.72 | 3.23E-11 | NaN |
| rno-miR-135a | 0.06 | 0.16 | 0.29 | 29.68 | 189.61 | 2.69E-11 | NaN |
| rno-miR-135a* | 0.04 | 0.12 | 0.18 | 0.29 | 31.84 | 6.93E-10 | NaN |
| rno-miR-135b | 0.06 | 0.15 | 0.27 | 21.71 | 277.53 | 3.49E-12 | NaN |
| rno-miR-135b* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-136 | 0.06 | 0.25 | 21.91 | 121.71 | 650.14 | 3.47E-08 | NaN |
| rno-miR-136* | 0.06 | 0.18 | 0.39 | 50.11 | 296.84 | 2.19E-10 | NaN |
| rno-miR-137 | 0.06 | 0.14 | 0.23 | 0.61 | 627.29 | 4.13E-14 | 27 |
| rno-miR-137* | 0.04 | 0.12 | 0.17 | 0.27 | 8.60 | 4.73E-10 | NaN |
| rno-miR-138 | 0.06 | 0.16 | 0.27 | 9.34 | 302.15 | 2.48E-12 | 27 |
| rno-miR-138-1* | 0.04 | 0.10 | 0.15 | 0.25 | 1.61 | 1 | NaN |
| rno-miR-138-2* | 0.04 | 0.13 | 0.20 | 0.33 | 18.75 | 9.68E-13 | NaN |
| rno-miR-139-3p | 0.08 | 10.92 | 19.65 | 28.99 | 5126.95 | 5.10E-10 | 8 |
| rno-miR-139-5p | 0.04 | 6.03 | 15.07 | 28.62 | 311.08 | 5.65E-09 | NaN |
| rno-miR-140 | 0.66 | 204.35 | 269.11 | 353.18 | 10030.81 | 2.24E-14 | 8 |
| rno-miR-140* | 0.66 | 140.24 | 207.53 | 291.22 | 11001.85 | 2.46E-12 | 8 |
| rno-miR-141 | 0.04 | 0.20 | 60.09 | 901.04 | 8798.65 | 8.83E-10 | NaN |
| rno-miR-141* | 0.04 | 0.10 | 0.16 | 0.25 | 5.62 | 2.71E-05 | NaN |
| rno-miR-142-3p | 0.69 | 138.15 | 336.86 | 746.27 | 53948.66 | 1.09E-05 | 15 |
| rno-miR-142-5p | 0.04 | 0.25 | 20.19 | 70.80 | 5412.80 | 1.47E-05 | NaN |
| rno-miR-143 | 84.82 | 1078.62 | 3009.63 | 6557.55 | 83261.41 | 1 | NaN |
| rno-miR-143* | 0.04 | 0.13 | 0.22 | 0.64 | 97.33 | 2.40E-11 | NaN |
| rno-miR-144 | 0.04 | 0.12 | 0.20 | 0.64 | 3290.75 | 1.84E-12 | 6 |
| rno-miR-144* | 0.04 | 0.12 | 0.17 | 0.31 | 186.03 | 9.82E-14 | 22 |
| rno-miR-145 | 0.05 | 19.09 | 66.42 | 129.77 | 3396.35 | 7.12E-05 | NaN |
| rno-miR-145* | 0.04 | 0.11 | 0.17 | 0.27 | 42.00 | 5.64E-14 | NaN |
| rno-miR-146a | 30.98 | 227.01 | 423.47 | 791.65 | 3351.58 | 0.19575736 | NaN |
| rno-miR-146a* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-146b | 0.49 | 62.61 | 93.85 | 132.40 | 537.37 | 4.71E-15 | NaN |
| rno-miR-146b* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-147 | 0.04 | 0.11 | 0.17 | 0.30 | 32.55 | 2.17E-12 | NaN |
| rno-miR-148b-3p | 0.66 | 122.74 | 161.12 | 212.00 | 465.38 | 1.27E-18 | NaN |
| rno-miR-148b-5p | 0.04 | 0.12 | 0.18 | 0.28 | 4.78 | 4.06E-06 | NaN |
| rno-miR-150 | 0.66 | 105.59 | 216.92 | 582.12 | 14086.78 | 3.94E-04 | 9 |
| rno-miR-150* | 0.04 | 0.15 | 0.37 | 18.79 | 495.74 | 1.23E-07 | NaN |
| rno-miR-151 | 0.66 | 427.76 | 565.99 | 801.16 | 1599.04 | 1.46E-18 | NaN |
| rno-miR-151* | 0.04 | 0.13 | 0.21 | 0.43 | 8.31 | 1.26E-07 | NaN |
| rno-miR-152 | 2.87 | 68.16 | 135.46 | 224.96 | 1138.08 | 4.77E-02 | NaN |
| rno-miR-152* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-153 | 0.06 | 0.16 | 0.27 | 10.36 | 242.15 | 3.54E-11 | 18 |
| rno-miR-153* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-154 | 0.06 | 0.23 | 10.01 | 67.91 | 453.98 | 2.33E-09 | NaN |
| rno-miR-154* | 0.06 | 0.14 | 0.23 | 0.64 | 268.89 | 4.88E-13 | NaN |
| rno-miR-15b | 32.42 | 689.50 | 946.72 | 1473.16 | 13923.56 | 0.001740291 | 15 |
| rno-miR-15b* | 0.04 | 0.13 | 0.25 | 1.05 | 25.81 | 1.48E-07 | NaN |
| rno-miR-16 | 175.81 | 2288.05 | 3249.48 | 4493.43 | 16073.99 | 7.06E-04 | NaN |
| rno-miR-16* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-17-1-3p | 0.04 | 0.12 | 0.20 | 0.39 | 31.27 | 1.28E-09 | NaN |
| rno-miR-17-2-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-17-5p | 0.66 | 43.54 | 75.97 | 130.46 | 1399.39 | 0.009362903 | NaN |
| rno-miR-181a | 102.77 | 301.09 | 544.85 | 938.27 | 17649.29 | 0.08351972 | NaN |
| rno-miR-181a-1* | 0.05 | 0.34 | 10.53 | 29.86 | 1086.48 | 2.93E-06 | 3 |
| rno-miR-181a-2* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-181b | 0.33 | 49.48 | 93.90 | 144.48 | 2639.77 | 1.88E-10 | 5 |
| rno-miR-181b-1* | 0.04 | 0.10 | 0.15 | 0.25 | 104.73 | 6.56E-15 | 3 |
| rno-miR-181b-2* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-181c | 0.33 | 51.45 | 64.06 | 93.04 | 419.34 | 1.17E-15 | NaN |
| rno-miR-181c* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-181d | 0.28 | 29.56 | 40.93 | 54.59 | 264.35 | 7.58E-14 | NaN |
| rno-miR-181d* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-182 | 0.04 | 0.22 | 7.37 | 28.07 | 405.23 | 2.45E-07 | NaN |
| rno-miR-183 | 0.04 | 0.25 | 54.17 | 151.77 | 4643.37 | 1.23E-09 | 3 |
| rno-miR-183* | 0.04 | 0.11 | 0.20 | 0.33 | 50.48 | 6.73E-11 | NaN |
| rno-miR-184 | 0.04 | 0.11 | 0.17 | 0.28 | 14536.44 | 3.84E-17 | 15 |
| rno-miR-185 | 0.53 | 80.36 | 118.70 | 206.17 | 545.17 | 9.99E-11 | NaN |
| rno-miR-185* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-186 | 74.90 | 119.75 | 153.84 | 208.21 | 385.44 | 1.00E+00 | NaN |
| rno-miR-186* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-187 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-187* | 0.04 | 0.10 | 0.15 | 0.25 | 32.01 | 3.91E-11 | NaN |
| rno-miR-188 | 0.04 | 0.14 | 0.30 | 30.56 | 4285.67 | 1.18E-08 | 4 |
| rno-miR-188* | 0.04 | 0.11 | 0.17 | 0.28 | 95.27 | 1.09E-14 | NaN |
| rno-miR-18a | 0.05 | 0.27 | 6.44 | 28.29 | 665.88 | 1.01E-04 | NaN |
| rno-miR-18a* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-190 | 0.05 | 0.13 | 0.22 | 0.56 | 29.40 | 5.69E-10 | NaN |
| rno-miR-190* | 0.04 | 0.11 | 0.17 | 0.28 | 11.36 | 1.44E-10 | NaN |
| rno-miR-190b | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-190b* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-191 | 0.04 | 0.10 | 0.17 | 0.27 | 12.93 | 3.73E-10 | NaN |
| rno-miR-191* | 0.22 | 8.84 | 13.77 | 20.62 | 72.24 | 2.32E-12 | NaN |
| rno-miR-1912-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-1912-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-192 | 0.22 | 16.65 | 31.09 | 53.94 | 38627.58 | 1.33E-10 | 33 |
| rno-miR-192* | 0.04 | 0.10 | 0.20 | 0.34 | 371.98 | 1.25E-14 | 33 |
| rno-miR-193 | 0.08 | 51.76 | 111.89 | 206.11 | 8566.40 | 1.73E-04 | NaN |
| rno-miR-193* | 0.04 | 0.13 | 0.27 | 6.03 | 219.24 | 1.86E-08 | NaN |
| rno-miR-194 | 0.08 | 0.35 | 9.57 | 17.46 | 39188.27 | 2.97E-07 | 13 |
| rno-miR-194* | 0.04 | 0.10 | 0.20 | 0.34 | 82.50 | 2.96E-13 | NaN |
| rno-miR-1949 | 0.05 | 20.69 | 57.74 | 147.83 | 1029.60 | 1.72E-08 | NaN |
| rno-miR-195 | 1.05 | 327.86 | 776.76 | 1827.52 | 6119.92 | 4.34E-05 | NaN |
| rno-miR-195* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-196a | 0.04 | 0.14 | 0.27 | 61.40 | 890.40 | 2.17E-11 | NaN |
| rno-miR-196a* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-196b | 0.04 | 0.14 | 0.30 | 131.12 | 1359.48 | 1.56E-10 | NaN |
| rno-miR-196b* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-196c | 0.04 | 0.14 | 0.27 | 22.64 | 861.60 | 2.72E-09 | NaN |
| rno-miR-196c* | 0.04 | 0.11 | 0.19 | 0.31 | 405.68 | 8.47E-16 | 22 |
| rno-miR-199a-3p | 13.15 | 355.53 | 974.47 | 2118.58 | 9731.91 | 2.26E-02 | NaN |
| rno-miR-199a-5p | 0.04 | 85.46 | 215.36 | 436.09 | 3267.33 | 2.79E-10 | NaN |
| rno-miR-19a | 0.08 | 23.81 | 52.30 | 177.81 | 1593.88 | 1.24E-03 | NaN |
| rno-miR-19a* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-19b | 41.12 | 308.66 | 550.75 | 1267.64 | 7230.97 | 1 | NaN |
| rno-miR-19b-1* | 0.04 | 0.10 | 0.17 | 0.28 | 39.53 | 1.77E-13 | NaN |
| rno-miR-19b-2* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-200a | 0.04 | 0.21 | 219.88 | 2433.63 | 24103.62 | 3.47E-10 | NaN |
| rno-miR-200a* | 0.04 | 0.11 | 0.21 | 0.61 | 66.60 | 6.01E-12 | NaN |
| rno-miR-200b | 0.04 | 0.21 | 324.56 | 3718.40 | 40718.36 | 2.73E-10 | NaN |
| rno-miR-200b* | 0.04 | 0.15 | 0.35 | 47.52 | 408.48 | 3.62E-09 | NaN |
| rno-miR-200c | 0.04 | 0.17 | 4.11 | 238.36 | 1678.72 | 5.31E-10 | NaN |
| rno-miR-200c* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-200c_v15.0 | 0.04 | 0.30 | 268.55 | 4255.80 | 22974.50 | 2.91E-08 | NaN |
| rno-miR-201 | 0.04 | 0.11 | 0.17 | 0.28 | 435.09 | 3.43E-15 | 16 |
| rno-miR-201* | 0.04 | 0.10 | 0.16 | 0.27 | 34.83 | 6.54E-12 | NaN |
| rno-miR-201_v15.0 | 0.04 | 0.11 | 0.17 | 0.28 | 288.72 | 2.79E-14 | 16 |
| rno-miR-202 | 0.04 | 0.10 | 0.15 | 0.25 | 5.64 | 0.02992306 | NaN |
| rno-miR-202* | 0.04 | 0.13 | 0.26 | 7.74 | 316.00 | 4.91E-09 | 7 |
| rno-miR-202_v15.0 | 0.04 | 0.10 | 0.15 | 0.25 | 71.39 | 1.97E-13 | NaN |
| rno-miR-203 | 0.04 | 0.32 | 38.31 | 263.16 | 16980.79 | 5.63E-04 | NaN |
| rno-miR-203* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-204 | 0.22 | 71.23 | 196.05 | 545.70 | 16085.60 | 5.68E-05 | NaN |
| rno-miR-204* | 0.04 | 0.13 | 0.21 | 0.34 | 41.27 | 2.17E-10 | NaN |
| rno-miR-205 | 0.04 | 0.22 | 11.18 | 278.55 | 2563.11 | 1.05E-07 | NaN |
| rno-miR-205_v15.0 | 0.04 | 0.23 | 21.09 | 401.87 | 6690.92 | 5.51E-07 | NaN |
| rno-miR-206 | 0.04 | 0.12 | 0.23 | 8.49 | 28471.33 | 2.52E-13 | 19 |
| rno-miR-206* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-207 | 0.04 | 0.10 | 0.15 | 0.25 | 24.12 | 8.93E-07 | NaN |
| rno-miR-207_v15.0 | 0.04 | 0.10 | 0.15 | 0.25 | 16.73 | 1.10E-05 | NaN |
| rno-miR-208 | 0.04 | 0.10 | 0.15 | 0.27 | 101.96 | 1.75E-16 | 12 |
| rno-miR-208* | 0.04 | 0.10 | 0.15 | 0.27 | 85.59 | 4.71E-16 | NaN |
| rno-miR-208_v15.0 | 0.04 | 0.10 | 0.15 | 0.27 | 939.09 | 1.21E-18 | 12 |
| rno-miR-208b-3p | 0.04 | 0.10 | 0.15 | 0.25 | 53.09 | 6.48E-09 | NaN |
| rno-miR-208b-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-20a | 0.66 | 152.50 | 272.69 | 524.16 | 4352.14 | 2.59E-06 | NaN |
| rno-miR-20a* | 0.04 | 0.16 | 0.33 | 4.81 | 64.68 | 1.10E-05 | NaN |
| rno-miR-20b-3p | 0.04 | 0.10 | 0.16 | 0.26 | 6.98 | 5.95E-07 | NaN |
| rno-miR-20b-5p | 0.66 | 81.30 | 145.67 | 272.59 | 2663.03 | 1.40E-04 | NaN |
| rno-miR-20b-5p_v15.0 | 0.04 | 0.12 | 0.18 | 0.31 | 323.96 | 1.04E-13 | 24 |
| rno-miR-21 | 373.94 | 2398.23 | 5423.12 | 13183.28 | 81002.91 | 1 | NaN |
| rno-miR-21* | 0.04 | 0.11 | 0.18 | 0.30 | 28.43 | 1.38E-10 | NaN |
| rno-miR-210 | 0.23 | 31.30 | 60.01 | 124.81 | 449.33 | 5.74E-11 | NaN |
| rno-miR-210* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-211 | 0.07 | 0.17 | 0.29 | 3.64 | 3010.04 | 8.88E-10 | 3 |
| rno-miR-211* | 0.04 | 0.16 | 0.50 | 27.16 | 865.84 | 1.87E-07 | NaN |
| rno-miR-212 | 0.06 | 0.25 | 8.51 | 21.78 | 243.00 | 5.49E-08 | NaN |
| rno-miR-212* | 0.04 | 0.11 | 0.17 | 0.27 | 7.29 | 2.12E-10 | NaN |
| rno-miR-214 | 0.06 | 89.62 | 201.93 | 441.91 | 3191.01 | 1.83E-09 | NaN |
| rno-miR-214* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-215 | 0.04 | 0.10 | 0.19 | 0.32 | 50159.24 | 2.32E-17 | 17 |
| rno-miR-215_v15.0 | 0.04 | 0.10 | 0.19 | 0.33 | 83077.24 | 3.33E-17 | 17 |
| rno-miR-216a | 0.04 | 0.11 | 0.16 | 0.26 | 2713.17 | 2.88E-18 | 7 |
| rno-miR-216a* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-216b-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-216b-5p | 0.04 | 0.11 | 0.16 | 0.25 | 317.72 | 1.03E-16 | 6 |
| rno-miR-217 | 0.04 | 0.11 | 0.16 | 0.25 | 1731.97 | 3.17E-18 | 5 |
| rno-miR-217* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-217_v15.0 | 0.04 | 0.10 | 0.15 | 0.25 | 263.55 | 8.28E-17 | 3 |
| rno-miR-218 | 0.09 | 34.91 | 72.44 | 239.79 | 1898.88 | 2.47E-10 | NaN |
| rno-miR-218-1* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-218-2* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-219-1-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-219-2-3p | 0.04 | 0.13 | 0.20 | 0.34 | 1914.99 | 7.96E-17 | 27 |
| rno-miR-219-5p | 0.08 | 0.30 | 12.75 | 38.46 | 2717.50 | 1.69E-05 | NaN |
| rno-miR-22 | 843.43 | 2160.93 | 4330.57 | 9292.37 | 32360.32 | 3.67E-03 | NaN |
| rno-miR-22* | 0.11 | 31.42 | 48.56 | 104.38 | 342.60 | 3.44E-12 | NaN |
| rno-miR-220_v15.0 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-221 | 0.27 | 75.38 | 134.42 | 228.59 | 954.62 | 1.21E-11 | NaN |
| rno-miR-221* | 0.05 | 0.11 | 0.17 | 0.27 | 10.91 | 4.55E-10 | NaN |
| rno-miR-222 | 0.05 | 0.63 | 19.31 | 48.50 | 282.61 | 2.98E-09 | NaN |
| rno-miR-222* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-223 | 23.93 | 187.66 | 299.18 | 467.34 | 5118.20 | 0.50673044 | NaN |
| rno-miR-223* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-224 | 0.04 | 0.11 | 0.17 | 0.27 | 8.53 | 1.57E-08 | NaN |
| rno-miR-224* | 0.04 | 0.10 | 0.16 | 0.26 | 7.23 | 2.50E-06 | NaN |
| rno-miR-224_v15.0 | 0.04 | 0.15 | 2.06 | 50.02 | 316.76 | 2.65E-09 | NaN |
| rno-miR-23a | 518.56 | 2297.45 | 4144.09 | 10485.93 | 22126.41 | 2.84E-02 | NaN |
| rno-miR-23a* | 0.04 | 0.11 | 0.18 | 0.30 | 20.47 | 6.75E-10 | NaN |
| rno-miR-23b | 199.38 | 2860.93 | 4452.26 | 7512.34 | 31341.74 | 0.39951342 | NaN |
| rno-miR-23b* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-24 | 338.79 | 1692.48 | 2632.61 | 5498.91 | 17408.16 | 1 | NaN |
| rno-miR-24-1* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-24-2* | 0.22 | 11.68 | 23.87 | 60.70 | 222.18 | 4.96E-09 | NaN |
| rno-miR-25 | 55.71 | 282.96 | 364.47 | 431.25 | 2319.58 | 1.76E-05 | 15 |
| rno-miR-25* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-26a | 903.59 | 3240.30 | 5775.75 | 7505.45 | 15300.83 | 0.022537716 | NaN |
| rno-miR-26a* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-26b | 257.77 | 1580.83 | 2622.57 | 3702.03 | 7388.00 | 1.27E-04 | NaN |
| rno-miR-26b* | 0.04 | 0.13 | 0.21 | 0.38 | 8.68 | 9.15E-08 | NaN |
| rno-miR-27a | 241.69 | 1051.70 | 2023.35 | 4124.49 | 9816.97 | 3.44E-02 | NaN |
| rno-miR-27a* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-27b | 365.23 | 1234.89 | 1993.22 | 3007.14 | 10523.20 | 1.00E+00 | NaN |
| rno-miR-27b* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-28 | 0.66 | 81.12 | 152.59 | 231.23 | 773.62 | 5.23E-10 | NaN |
| rno-miR-28* | 0.04 | 0.11 | 0.17 | 0.27 | 12.45 | 1.41E-06 | NaN |
| rno-miR-290 | 0.04 | 8.40 | 26.93 | 58.80 | 504.09 | 4.67E-11 | NaN |
| rno-miR-290_v15.0 | 0.04 | 0.11 | 0.19 | 0.38 | 222.56 | 1.52E-12 | 34 |
| rno-miR-291a-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-291a-5p | 0.04 | 0.10 | 0.15 | 0.26 | 315.16 | 1.37E-15 | 9 |
| rno-miR-291b | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-292-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-292-5p | 0.04 | 0.10 | 0.17 | 0.28 | 124.35 | 1.43E-12 | 16 |
| rno-miR-293 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-293* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-293_v15.0 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-294 | 0.04 | 0.10 | 0.15 | 0.25 | 7.42 | 2.86E-04 | NaN |
| rno-miR-295 | 0.04 | 0.10 | 0.15 | 0.25 | 11.27 | 6.85E-06 | NaN |
| rno-miR-295* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-295_v15.0 | 0.04 | 0.10 | 0.15 | 0.25 | 10.05 | 1.21E-05 | NaN |
| rno-miR-296 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-296* | 0.06 | 0.27 | 6.26 | 12.46 | 275.70 | 1.49E-06 | NaN |
| rno-miR-2964 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-297 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-298 | 0.04 | 0.16 | 0.35 | 3.77 | 50.88 | 1.32E-06 | NaN |
| rno-miR-298* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-2985 | 14.52 | 36.11 | 57.58 | 92.83 | 334.21 | 1 | NaN |
| rno-miR-299 | 0.06 | 0.22 | 8.07 | 35.75 | 580.29 | 5.12E-08 | NaN |
| rno-miR-299* | 0.04 | 0.11 | 0.16 | 0.25 | 3.07 | 9.76E-03 | NaN |
| rno-miR-29a | 1385.37 | 2855.73 | 4038.49 | 5158.04 | 10243.52 | 1.00E+00 | NaN |
| rno-miR-29a* | 0.06 | 3.80 | 6.14 | 8.75 | 18.16 | 3.41E-12 | NaN |
| rno-miR-29b | 564.99 | 1824.15 | 3005.00 | 4352.34 | 8235.58 | 0.032288026 | NaN |
| rno-miR-29b-1* | 0.04 | 0.10 | 0.15 | 0.25 | 1.21 | 1 | NaN |
| rno-miR-29b-2* | 0.04 | 0.12 | 0.18 | 0.28 | 2.23 | 3.68E-06 | NaN |
| rno-miR-29c | 279.41 | 1177.41 | 1809.01 | 2455.62 | 4203.73 | 0.003115777 | NaN |
| rno-miR-29c* | 0.18 | 33.60 | 69.00 | 111.90 | 213.56 | 5.27E-15 | NaN |
| rno-miR-300-3p | 0.06 | 0.23 | 9.86 | 98.29 | 382.83 | 4.95E-09 | NaN |
| rno-miR-300-5p | 0.06 | 0.14 | 0.21 | 0.38 | 24.50 | 2.47E-13 | NaN |
| rno-miR-301a | 0.64 | 76.14 | 106.91 | 138.00 | 502.36 | 1.37E-14 | NaN |
| rno-miR-301a* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-301b | 0.04 | 0.11 | 0.19 | 0.31 | 31.81 | 5.67E-11 | NaN |
| rno-miR-301b* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3065-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3065-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3074 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3085 | 12.49 | 34.09 | 55.18 | 91.03 | 365.92 | 1 | NaN |
| rno-miR-30a | 241.54 | 533.22 | 992.92 | 2045.90 | 4579.74 | 2.48E-03 | NaN |
| rno-miR-30a* | 0.66 | 74.42 | 123.14 | 262.67 | 1047.27 | 6.97E-08 | NaN |
| rno-miR-30b-3p | 0.05 | 4.34 | 8.75 | 14.23 | 43.12 | 2.56E-11 | NaN |
| rno-miR-30b-5p | 117.83 | 636.72 | 948.75 | 1587.02 | 3418.97 | 0.28570926 | NaN |
| rno-miR-30c | 185.38 | 750.98 | 1126.84 | 1926.20 | 6611.22 | 8.33E-01 | NaN |
| rno-miR-30c-1* | 0.08 | 2.58 | 5.19 | 8.80 | 117.18 | 3.43E-04 | NaN |
| rno-miR-30c-2* | 0.11 | 13.16 | 22.62 | 50.80 | 109.04 | 4.15E-12 | NaN |
| rno-miR-30d | 205.44 | 412.59 | 599.76 | 913.21 | 2167.96 | 1.00E+00 | NaN |
| rno-miR-30d* | 0.04 | 0.13 | 0.24 | 1.65 | 12.27 | 1.46E-07 | NaN |
| rno-miR-30e | 195.39 | 376.31 | 612.28 | 1142.90 | 3760.12 | 1.01E-01 | NaN |
| rno-miR-30e* | 0.66 | 114.11 | 153.97 | 247.02 | 973.51 | 2.41E-13 | NaN |
| rno-miR-31 | 0.07 | 0.28 | 20.69 | 99.53 | 1923.98 | 1.32E-06 | NaN |
| rno-miR-31* | 0.05 | 0.17 | 0.35 | 19.20 | 485.65 | 2.97E-08 | NaN |
| rno-miR-3120 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-32 | 0.04 | 0.16 | 0.35 | 7.33 | 65.21 | 4.94E-07 | NaN |
| rno-miR-32* | 0.04 | 0.10 | 0.16 | 0.25 | 111.41 | 8.65E-15 | 6 |
| rno-miR-320 | 0.22 | 38.41 | 54.77 | 82.60 | 326.09 | 2.30E-13 | NaN |
| rno-miR-320* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-322 | 0.66 | 96.72 | 204.54 | 559.02 | 4215.70 | 6.98E-03 | NaN |
| rno-miR-322* | 0.05 | 0.46 | 13.59 | 47.27 | 424.21 | 5.56E-08 | NaN |
| rno-miR-323 | 0.06 | 0.14 | 0.23 | 0.61 | 145.83 | 4.96E-14 | NaN |
| rno-miR-323* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-324-3p | 97.09 | 153.53 | 190.21 | 228.06 | 567.56 | 0.9403239 | NaN |
| rno-miR-324-5p | 0.53 | 84.47 | 111.33 | 172.01 | 479.90 | 1.01E-15 | NaN |
| rno-miR-325-3p | 0.04 | 0.14 | 0.23 | 0.56 | 145.62 | 1.36E-14 | 33 |
| rno-miR-325-5p | 0.04 | 0.13 | 0.21 | 0.37 | 11.82 | 3.27E-11 | NaN |
| rno-miR-326 | 0.13 | 15.71 | 32.45 | 58.49 | 253.35 | 5.62E-10 | NaN |
| rno-miR-326* | 0.04 | 0.10 | 0.15 | 0.25 | 62.22 | 2.56E-12 | NaN |
| rno-miR-327 | 0.04 | 0.17 | 0.36 | 47.16 | 8657.61 | 5.35E-08 | 5 |
| rno-miR-328a | 0.66 | 58.14 | 95.08 | 157.00 | 571.77 | 3.52E-11 | NaN |
| rno-miR-328a* | 0.05 | 0.19 | 4.68 | 25.25 | 6184.15 | 7.92E-06 | 3 |
| rno-miR-328b-3p | 0.04 | 0.12 | 0.17 | 0.28 | 22.52 | 5.67E-10 | NaN |
| rno-miR-329 | 0.06 | 0.23 | 10.54 | 154.81 | 569.60 | 2.72E-09 | NaN |
| rno-miR-329* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-33 | 0.19 | 9.19 | 13.67 | 28.96 | 218.33 | 2.37E-07 | NaN |
| rno-miR-33* | 0.08 | 0.19 | 0.33 | 3.43 | 17.94 | 4.43E-08 | NaN |
| rno-miR-330 | 0.04 | 0.12 | 0.19 | 0.30 | 4.33 | 1.75E-08 | NaN |
| rno-miR-330* | 0.07 | 0.15 | 0.25 | 2.24 | 104.32 | 2.27E-12 | 26 |
| rno-miR-331 | 0.66 | 77.60 | 106.87 | 168.48 | 439.70 | 8.64E-15 | NaN |
| rno-miR-331* | 0.04 | 0.12 | 0.19 | 0.36 | 255.54 | 1.10E-13 | 29 |
| rno-miR-335 | 0.09 | 7.13 | 58.89 | 148.06 | 2165.22 | 1.19E-08 | NaN |
| rno-miR-336 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-336* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-337 | 0.06 | 0.19 | 0.50 | 22.14 | 193.43 | 2.32E-09 | NaN |
| rno-miR-337* | 0.06 | 0.15 | 0.27 | 10.81 | 101.89 | 7.08E-11 | NaN |
| rno-miR-338 | 0.12 | 60.04 | 121.47 | 387.67 | 9238.49 | 8.28E-07 | NaN |
| rno-miR-338* | 0.04 | 0.13 | 0.21 | 0.38 | 31.56 | 1.34E-12 | NaN |
| rno-miR-339-3p | 0.04 | 0.12 | 0.21 | 0.38 | 9.32 | 1.05E-07 | NaN |
| rno-miR-339-5p | 0.04 | 0.10 | 0.15 | 0.25 | 2.24 | 1.00E+00 | NaN |
| rno-miR-340-3p | 0.13 | 9.41 | 18.28 | 30.28 | 125.05 | 1.27E-11 | NaN |
| rno-miR-340-5p | 0.13 | 0.28 | 16.95 | 41.46 | 104.19 | 7.08E-11 | NaN |
| rno-miR-341 | 0.06 | 0.17 | 0.32 | 26.55 | 301.99 | 1.88E-10 | NaN |
| rno-miR-342-3p | 0.32 | 84.38 | 129.62 | 318.07 | 3113.84 | 1.90E-08 | 12 |
| rno-miR-342-5p | 0.07 | 0.20 | 0.53 | 5.99 | 107.70 | 3.94E-07 | 12 |
| rno-miR-343 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-344a | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-344a-3p | 0.04 | 0.14 | 0.22 | 0.49 | 172.03 | 2.61E-14 | 35 |
| rno-miR-344a-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-344b-2-3p | 0.04 | 0.12 | 0.17 | 0.28 | 18.80 | 2.40E-12 | NaN |
| rno-miR-344b-5p | 0.04 | 0.12 | 0.17 | 0.28 | 39.41 | 8.40E-14 | NaN |
| rno-miR-345-3p | 0.04 | 0.11 | 0.17 | 0.27 | 178.55 | 3.62E-13 | 16 |
| rno-miR-345-5p | 37.05 | 113.77 | 154.58 | 207.68 | 483.57 | 1.00E+00 | NaN |
| rno-miR-346 | 0.04 | 0.13 | 0.19 | 0.31 | 8.74 | 4.32E-10 | NaN |
| rno-miR-347 | 0.05 | 4.71 | 13.69 | 23.74 | 284.87 | 2.47E-08 | NaN |
| rno-miR-349 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-34a | 30.20 | 215.22 | 441.72 | 915.14 | 5711.36 | 1 | NaN |
| rno-miR-34a* | 0.04 | 0.13 | 0.20 | 0.34 | 29.61 | 1.44E-12 | NaN |
| rno-miR-34b | 0.09 | 0.30 | 9.52 | 25.97 | 12120.03 | 2.57E-05 | 16 |
| rno-miR-34b* | 0.04 | 0.12 | 0.18 | 0.29 | 526.82 | 4.11E-15 | 16 |
| rno-miR-34b_v15.0 | 0.09 | 0.27 | 9.26 | 36.52 | 18265.09 | 6.74E-06 | 15 |
| rno-miR-34c | 0.09 | 0.21 | 6.94 | 24.07 | 13443.18 | 7.45E-07 | 15 |
| rno-miR-34c* | 0.04 | 0.12 | 0.17 | 0.28 | 52.00 | 1.74E-12 | NaN |
| rno-miR-350 | 0.08 | 0.25 | 3.36 | 7.25 | 14.86 | 2.88E-09 | NaN |
| rno-miR-351 | 0.04 | 0.11 | 0.17 | 0.27 | 48.60 | 1.77E-11 | NaN |
| rno-miR-351* | 0.04 | 0.10 | 0.16 | 0.27 | 40.56 | 1.90E-13 | NaN |
| rno-miR-352 | 0.66 | 108.90 | 154.39 | 230.93 | 662.42 | 8.53E-15 | NaN |
| rno-miR-3541 | 0.04 | 0.10 | 0.15 | 0.25 | 78.79 | 6.81E-10 | NaN |
| rno-miR-3542 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3543 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3544 | 0.04 | 0.12 | 0.21 | 2.24 | 136.81 | 6.04E-10 | NaN |
| rno-miR-3545-3p | 0.04 | 0.10 | 0.18 | 0.30 | 124.42 | 3.07E-14 | 18 |
| rno-miR-3545-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3546 | 0.04 | 0.10 | 0.15 | 0.25 | 360.25 | 1.81E-17 | 5 |
| rno-miR-3547 | 0.04 | 0.15 | 2.01 | 8.62 | 179.58 | 1.15E-07 | NaN |
| rno-miR-3548 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3549 | 0.04 | 0.13 | 0.29 | 11.77 | 293.89 | 2.55E-08 | 6 |
| rno-miR-3550 | 0.04 | 0.10 | 0.15 | 0.25 | 24.91 | 7.21E-07 | NaN |
| rno-miR-3551-3p | 0.04 | 0.10 | 0.15 | 0.25 | 47.13 | 2.17E-11 | NaN |
| rno-miR-3551-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3552 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3553 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3554 | 0.04 | 0.10 | 0.15 | 0.25 | 35.86 | 4.18E-08 | NaN |
| rno-miR-3555 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3556a | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3556b | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3557-3p | 0.04 | 0.10 | 0.17 | 0.27 | 115.08 | 3.94E-14 | 16 |
| rno-miR-3557-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3558-3p | 0.04 | 0.15 | 0.35 | 33.13 | 536.17 | 7.91E-09 | NaN |
| rno-miR-3558-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3559-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3559-5p | 0.04 | 0.24 | 8.80 | 35.48 | 218.12 | 3.02E-07 | NaN |
| rno-miR-3560 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3561-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3561-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3562 | 0.04 | 0.11 | 0.19 | 0.34 | 171.89 | 1.37E-12 | 33 |
| rno-miR-3563-3p | 0.04 | 0.11 | 0.17 | 0.28 | 31.40 | 8.41E-14 | NaN |
| rno-miR-3563-5p | 0.04 | 0.11 | 0.17 | 0.27 | 4.68 | 1.68E-05 | NaN |
| rno-miR-3564 | 0.04 | 0.11 | 0.21 | 0.49 | 694.83 | 1.89E-13 | 37 |
| rno-miR-3565 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3566 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3567 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3568 | 0.04 | 0.10 | 0.15 | 0.25 | 5.97 | 2.05E-02 | NaN |
| rno-miR-3569 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3570 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3571 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3572 | 0.04 | 0.10 | 0.15 | 0.25 | 7.48 | 0.003819167 | NaN |
| rno-miR-3573-3p | 6.44 | 16.68 | 26.78 | 42.66 | 262.95 | 1 | NaN |
| rno-miR-3573-5p | 0.04 | 0.10 | 0.15 | 0.25 | 20.68 | 2.58E-06 | NaN |
| rno-miR-3574 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3575 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3576 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3577 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3578 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3579 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3580-3p | 0.04 | 0.12 | 0.19 | 0.34 | 1014.44 | 1.90E-14 | 28 |
| rno-miR-3580-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3581 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3582 | 0.04 | 0.10 | 0.17 | 0.28 | 107.42 | 4.72E-14 | 16 |
| rno-miR-3583-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3583-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3584-3p | 0.04 | 0.10 | 0.15 | 0.25 | 25.37 | 6.39E-07 | NaN |
| rno-miR-3584-5p | 0.05 | 18.94 | 45.51 | 170.77 | 167704.17 | 6.16E-04 | 17 |
| rno-miR-3585-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3585-5p | 0.04 | 0.11 | 0.18 | 0.38 | 498.76 | 1.18E-12 | 31 |
| rno-miR-3586-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3586-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3587 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3588 | 0.08 | 34.71 | 94.50 | 161.39 | 1283.51 | 2.39E-12 | NaN |
| rno-miR-3589 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3590-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3590-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3591 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3592 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3593-3p | 0.04 | 0.13 | 0.31 | 29.77 | 809.20 | 1.68E-09 | NaN |
| rno-miR-3593-5p | 0.04 | 0.11 | 0.17 | 0.30 | 132.07 | 2.72E-14 | 20 |
| rno-miR-3594-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3594-5p | 0.04 | 0.10 | 0.16 | 0.26 | 37.19 | 1.53E-11 | NaN |
| rno-miR-3595 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3596a | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3596b | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3596c | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3596d | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-3597-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-3597-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-361 | 0.66 | 92.90 | 129.77 | 160.59 | 517.80 | 1.99E-14 | NaN |
| rno-miR-361* | 0.04 | 0.12 | 0.17 | 0.28 | 7.86 | 2.17E-09 | NaN |
| rno-miR-362 | 0.04 | 0.27 | 4.90 | 12.77 | 46.66 | 4.36E-07 | NaN |
| rno-miR-362* | 0.28 | 25.38 | 45.62 | 75.92 | 180.15 | 8.85E-11 | NaN |
| rno-miR-363 | 0.04 | 0.13 | 0.25 | 5.11 | 373.45 | 3.50E-10 | 18 |
| rno-miR-363* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-363_v15.0 | 0.04 | 0.15 | 0.30 | 13.32 | 759.81 | 1.92E-08 | 19 |
| rno-miR-365 | 45.52 | 142.17 | 393.00 | 631.97 | 7466.02 | 1.00E+00 | NaN |
| rno-miR-365* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-369-3p | 0.06 | 0.13 | 0.21 | 0.34 | 30.60 | 3.15E-13 | NaN |
| rno-miR-369-5p | 0.06 | 0.18 | 0.39 | 46.37 | 238.33 | 2.53E-10 | NaN |
| rno-miR-370 | 0.06 | 0.16 | 0.30 | 11.38 | 6400.64 | 2.71E-09 | 3 |
| rno-miR-370* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-370_v15.0 | 0.04 | 0.11 | 0.16 | 0.26 | 425.72 | 3.95E-16 | 3 |
| rno-miR-374 | 0.12 | 3.35 | 6.66 | 10.15 | 42.00 | 2.00E-09 | NaN |
| rno-miR-374* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-375 | 0.04 | 0.17 | 0.57 | 435.36 | 5547.82 | 4.68E-10 | NaN |
| rno-miR-375* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-376a | 0.06 | 0.21 | 0.54 | 37.42 | 535.99 | 5.48E-09 | NaN |
| rno-miR-376a* | 0.06 | 0.13 | 0.21 | 0.38 | 14.63 | 1.33E-11 | NaN |
| rno-miR-376b-3p | 0.06 | 0.15 | 0.25 | 12.15 | 338.83 | 1.56E-12 | NaN |
| rno-miR-376b-5p | 0.06 | 0.17 | 0.32 | 9.17 | 65.89 | 2.31E-09 | NaN |
| rno-miR-376c | 0.06 | 0.19 | 0.39 | 19.64 | 286.94 | 8.13E-09 | NaN |
| rno-miR-376c* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-377 | 0.05 | 0.11 | 0.17 | 0.27 | 14.26 | 9.50E-12 | NaN |
| rno-miR-377* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-377_v15.0 | 0.06 | 0.13 | 0.21 | 0.39 | 62.78 | 2.41E-12 | NaN |
| rno-miR-378 | 28.04 | 157.43 | 388.76 | 1103.04 | 12683.83 | 1.00E+00 | NaN |
| rno-miR-378* | 0.06 | 22.50 | 60.38 | 211.05 | 1932.92 | 3.46E-05 | NaN |
| rno-miR-379 | 0.06 | 0.24 | 11.27 | 103.85 | 449.41 | 2.43E-09 | NaN |
| rno-miR-379* | 0.06 | 0.16 | 0.28 | 6.91 | 37.43 | 1.11E-10 | NaN |
| rno-miR-380 | 0.04 | 0.12 | 0.17 | 0.27 | 66.00 | 6.69E-09 | NaN |
| rno-miR-380* | 0.06 | 0.14 | 0.23 | 0.53 | 139.53 | 4.20E-14 | 37 |
| rno-miR-381 | 0.06 | 0.18 | 0.35 | 50.14 | 385.12 | 4.22E-11 | NaN |
| rno-miR-381* | 0.06 | 0.13 | 0.21 | 0.39 | 25.86 | 9.19E-11 | NaN |
| rno-miR-381_v15.0 | 0.04 | 0.11 | 0.16 | 0.25 | 1.94 | 1.12E-01 | NaN |
| rno-miR-382 | 0.06 | 0.20 | 0.57 | 29.66 | 229.16 | 2.96E-09 | NaN |
| rno-miR-382* | 0.06 | 0.14 | 0.23 | 0.54 | 79.77 | 1.12E-13 | NaN |
| rno-miR-383 | 0.04 | 0.14 | 0.22 | 0.39 | 186.17 | 1.51E-14 | 32 |
| rno-miR-383* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-383_v15.0 | 0.04 | 0.13 | 0.21 | 0.37 | 128.08 | 1.93E-14 | 31 |
| rno-miR-384-3p | 0.04 | 0.13 | 0.22 | 0.39 | 30.03 | 1.75E-12 | NaN |
| rno-miR-384-5p | 0.06 | 0.15 | 0.25 | 8.71 | 703.42 | 4.20E-14 | 33 |
| rno-miR-409-3p | 11.14 | 32.35 | 48.29 | 76.19 | 283.30 | 1.00E+00 | NaN |
| rno-miR-409-5p | 0.06 | 0.15 | 0.25 | 11.91 | 111.01 | 1.74E-12 | NaN |
| rno-miR-409-5p_v15.0 | 0.06 | 0.14 | 0.23 | 0.66 | 43.44 | 5.91E-12 | NaN |
| rno-miR-410 | 0.06 | 0.17 | 0.30 | 47.07 | 403.41 | 3.88E-11 | NaN |
| rno-miR-410* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-411 | 0.06 | 0.18 | 0.37 | 40.19 | 150.32 | 1.46E-10 | NaN |
| rno-miR-411* | 0.06 | 0.22 | 7.05 | 106.44 | 329.22 | 3.37E-10 | NaN |
| rno-miR-412 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-412* | 0.04 | 0.11 | 0.16 | 0.26 | 3.90 | 2.99E-06 | NaN |
| rno-miR-421 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-421* | 0.10 | 0.22 | 0.54 | 9.87 | 29.73 | 2.98E-10 | NaN |
| rno-miR-423 | 0.04 | 0.11 | 0.16 | 0.26 | 40.50 | 1.18E-10 | NaN |
| rno-miR-423* | 0.38 | 17.65 | 27.47 | 39.71 | 298.32 | 1.28E-08 | NaN |
| rno-miR-425 | 0.64 | 43.33 | 68.48 | 108.07 | 440.43 | 8.54E-11 | NaN |
| rno-miR-425* | 0.04 | 0.10 | 0.15 | 0.25 | 18.44 | 3.79E-06 | NaN |
| rno-miR-429 | 0.04 | 0.21 | 123.53 | 1486.43 | 10686.49 | 1.94E-10 | NaN |
| rno-miR-431 | 0.06 | 0.15 | 0.27 | 11.74 | 300.11 | 1.54E-10 | 10 |
| rno-miR-433 | 0.06 | 0.14 | 0.25 | 8.71 | 249.28 | 2.37E-13 | 32 |
| rno-miR-433* | 0.06 | 0.14 | 0.22 | 0.49 | 44.11 | 1.90E-13 | NaN |
| rno-miR-434 | 0.06 | 0.27 | 35.25 | 337.29 | 1430.38 | 1.32E-08 | NaN |
| rno-miR-434* | 0.06 | 0.15 | 0.27 | 12.25 | 93.55 | 1.25E-11 | NaN |
| rno-miR-448 | 0.04 | 0.12 | 0.17 | 0.28 | 15.59 | 3.59E-10 | NaN |
| rno-miR-448* | 0.04 | 0.12 | 0.17 | 0.27 | 7.93 | 2.58E-10 | NaN |
| rno-miR-448_v15.0 | 0.04 | 0.10 | 0.15 | 0.25 | 7.45 | 0.001404246 | NaN |
| rno-miR-449a | 0.04 | 0.13 | 0.24 | 0.88 | 5573.39 | 3.00E-12 | 15 |
| rno-miR-449a* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-449c-3p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-449c-5p | 0.04 | 0.11 | 0.17 | 0.27 | 4.92 | 5.17E-05 | NaN |
| rno-miR-450a | 0.12 | 14.61 | 38.98 | 96.74 | 1039.39 | 1.31E-09 | NaN |
| rno-miR-450a* | 0.04 | 0.10 | 0.15 | 0.25 | 10.05 | 5.28E-07 | NaN |
| rno-miR-450a_v15.0 | 0.04 | 0.17 | 0.67 | 21.04 | 255.90 | 8.70E-08 | NaN |
| rno-miR-451 | 0.04 | 8.96 | 65.30 | 276.08 | 36495.07 | 7.24E-02 | NaN |
| rno-miR-451* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-455 | 0.04 | 0.12 | 0.20 | 0.34 | 92.19 | 9.07E-13 | NaN |
| rno-miR-455* | 0.12 | 11.45 | 26.24 | 57.54 | 1000.44 | 1.26E-09 | NaN |
| rno-miR-463 | 0.04 | 0.10 | 0.16 | 0.27 | 785.95 | 9.62E-18 | 7 |
| rno-miR-463* | 0.04 | 0.10 | 0.15 | 0.25 | 12.26 | 9.11E-08 | NaN |
| rno-miR-465 | 0.04 | 0.10 | 0.15 | 0.25 | 5.71 | 0.028226383 | NaN |
| rno-miR-465* | 0.04 | 0.10 | 0.15 | 0.26 | 25.14 | 6.67E-11 | NaN |
| rno-miR-466b | 0.05 | 6.11 | 13.57 | 23.81 | 2351.78 | 8.53E-07 | 9 |
| rno-miR-466b-1* | 7.99 | 35.75 | 63.40 | 118.92 | 440.03 | 1.00E+00 | NaN |
| rno-miR-466b-2* | 0.04 | 0.21 | 3.86 | 17.00 | 199.69 | 5.04E-07 | NaN |
| rno-miR-466c | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-466c* | 0.04 | 0.10 | 0.16 | 0.26 | 17.99 | 8.57E-11 | NaN |
| rno-miR-466d | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-471 | 0.04 | 0.10 | 0.16 | 0.27 | 761.81 | 2.91E-17 | 8 |
| rno-miR-471* | 0.04 | 0.10 | 0.16 | 0.27 | 661.55 | 2.90E-17 | 7 |
| rno-miR-483 | 0.04 | 3.19 | 10.82 | 19.88 | 152.51 | 1.43E-09 | NaN |
| rno-miR-483* | 0.05 | 0.20 | 4.16 | 26.93 | 2233.57 | 1.32E-04 | NaN |
| rno-miR-484 | 0.10 | 9.28 | 14.34 | 17.90 | 86.14 | 9.32E-14 | NaN |
| rno-miR-485 | 0.06 | 0.14 | 0.23 | 0.61 | 47.93 | 3.87E-13 | NaN |
| rno-miR-485* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-487b | 0.06 | 0.19 | 0.50 | 47.18 | 393.01 | 1.04E-09 | NaN |
| rno-miR-487b* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-487b_v15.0 | 0.06 | 0.15 | 0.25 | 5.11 | 65.72 | 2.52E-11 | NaN |
| rno-miR-488 | 0.04 | 0.13 | 0.21 | 0.37 | 19.70 | 1.75E-12 | NaN |
| rno-miR-488* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-489 | 0.04 | 0.11 | 0.16 | 0.27 | 5.70 | 3.44E-08 | NaN |
| rno-miR-489* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-490 | 0.04 | 0.13 | 0.22 | 1.41 | 162.94 | 3.22E-12 | 21 |
| rno-miR-490* | 0.04 | 0.14 | 0.23 | 5.55 | 85.26 | 1.42E-11 | NaN |
| rno-miR-493 | 0.04 | 0.11 | 0.16 | 0.26 | 362.36 | 3.50E-16 | 7 |
| rno-miR-493* | 0.04 | 0.11 | 0.16 | 0.26 | 7.96 | 3.40E-08 | NaN |
| rno-miR-494 | 17.53 | 83.14 | 134.14 | 246.16 | 60962.48 | 1.38E-10 | 11 |
| rno-miR-494* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-494_v15.0 | 22.42 | 117.53 | 205.23 | 352.73 | 170953.80 | 3.27E-11 | 10 |
| rno-miR-495 | 0.06 | 0.17 | 0.32 | 38.19 | 304.66 | 9.24E-11 | NaN |
| rno-miR-496 | 0.06 | 0.13 | 0.21 | 0.36 | 15.30 | 1.43E-12 | NaN |
| rno-miR-496* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-496_v15.0 | 0.06 | 0.14 | 0.23 | 0.64 | 52.92 | 2.24E-13 | NaN |
| rno-miR-497 | 69.62 | 295.98 | 658.38 | 1389.89 | 4721.54 | 4.95E-01 | NaN |
| rno-miR-497* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-499 | 0.11 | 8.57 | 17.70 | 43.21 | 2232.83 | 4.08E-05 | 18 |
| rno-miR-499* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-500 | 30.07 | 114.63 | 159.30 | 231.00 | 1074.50 | 2.69E-01 | NaN |
| rno-miR-500* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-501 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-501* | 0.04 | 0.10 | 0.15 | 0.25 | 37.84 | 3.05E-08 | NaN |
| rno-miR-501_v15.0 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-503 | 0.04 | 0.15 | 0.45 | 16.94 | 71.56 | 1.01E-08 | NaN |
| rno-miR-503* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-504 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-505 | 0.24 | 8.82 | 12.64 | 18.64 | 36.73 | 3.38E-14 | NaN |
| rno-miR-505* | 0.07 | 0.19 | 0.33 | 6.97 | 13.07 | 1.27E-10 | NaN |
| rno-miR-511 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-511* | 0.04 | 0.27 | 18.10 | 36.97 | 164.31 | 1.17E-09 | NaN |
| rno-miR-511_v15.0 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-532-3p | 0.30 | 11.76 | 18.01 | 25.98 | 104.57 | 4.05E-11 | NaN |
| rno-miR-532-5p | 0.38 | 24.22 | 46.55 | 63.28 | 220.53 | 8.85E-09 | NaN |
| rno-miR-539 | 0.06 | 0.17 | 0.35 | 13.34 | 68.50 | 2.92E-09 | NaN |
| rno-miR-539* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-540 | 0.04 | 0.10 | 0.15 | 0.25 | 4.26 | 1.09E-03 | NaN |
| rno-miR-540* | 0.04 | 0.10 | 0.15 | 0.25 | 1.15 | 1.00E+00 | NaN |
| rno-miR-540_v15.0 | 0.04 | 0.11 | 0.17 | 0.27 | 6.52 | 1.18E-06 | NaN |
| rno-miR-541 | 0.06 | 0.15 | 0.25 | 9.14 | 67.13 | 6.64E-12 | NaN |
| rno-miR-541* | 0.04 | 0.11 | 0.17 | 0.27 | 15.19 | 1.94E-11 | NaN |
| rno-miR-542-3p | 0.05 | 0.32 | 9.15 | 29.89 | 439.16 | 3.04E-07 | 3 |
| rno-miR-542-5p | 0.05 | 0.27 | 10.09 | 42.04 | 332.95 | 3.51E-08 | NaN |
| rno-miR-543 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-543* | 0.06 | 0.15 | 0.25 | 6.97 | 123.49 | 6.60E-13 | NaN |
| rno-miR-543_v15.0 | 0.06 | 0.13 | 0.21 | 0.34 | 6.68 | 2.43E-09 | NaN |
| rno-miR-544 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-544* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-544_v15.0 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-547 | 0.04 | 0.11 | 0.18 | 0.36 | 590.02 | 2.20E-13 | 6 |
| rno-miR-547* | 0.04 | 0.10 | 0.15 | 0.26 | 53.27 | 2.66E-13 | NaN |
| rno-miR-547_v15.0 | 0.04 | 0.10 | 0.16 | 0.27 | 83.96 | 3.92E-14 | NaN |
| rno-miR-551b | 0.04 | 0.15 | 0.24 | 0.72 | 1606.61 | 7.81E-14 | 24 |
| rno-miR-551b* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-568 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-582 | 0.07 | 10.02 | 29.92 | 62.51 | 292.17 | 6.17E-11 | NaN |
| rno-miR-582* | 0.06 | 0.18 | 2.43 | 9.86 | 76.63 | 1.60E-07 | NaN |
| rno-miR-592 | 0.04 | 0.13 | 0.20 | 0.32 | 28.42 | 1.44E-12 | NaN |
| rno-miR-598-3p | 0.09 | 0.23 | 0.75 | 31.30 | 549.92 | 2.96E-08 | NaN |
| rno-miR-598-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-615 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-628 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-632 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-652 | 120.14 | 223.45 | 322.41 | 550.64 | 8315.28 | 6.36E-07 | 3 |
| rno-miR-652* | 7.64 | 43.83 | 77.36 | 135.38 | 8521.49 | 5.71E-05 | 3 |
| rno-miR-653 | 0.04 | 0.11 | 0.17 | 0.27 | 12.47 | 1.83E-11 | NaN |
| rno-miR-653* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-653_v15.0 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-664 | 10.92 | 29.60 | 38.83 | 53.96 | 202.78 | 1 | NaN |
| rno-miR-664-1* | 0.04 | 0.10 | 0.17 | 0.28 | 110.07 | 1.77E-14 | 17 |
| rno-miR-664-2* | 0.04 | 0.12 | 0.18 | 0.29 | 15.86 | 4.76E-10 | NaN |
| rno-miR-665 | 0.06 | 0.12 | 0.18 | 0.29 | 11.78 | 8.61E-11 | NaN |
| rno-miR-665_v15.0 | 0.04 | 0.11 | 0.17 | 0.27 | 17.09 | 4.11E-08 | NaN |
| rno-miR-666 | 0.04 | 0.10 | 0.15 | 0.25 | 1619.00 | 1.03E-18 | 3 |
| rno-miR-666* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-666_v15.0 | 0.04 | 0.10 | 0.15 | 0.25 | 219.96 | 6.46E-16 | 4 |
| rno-miR-667 | 0.06 | 0.17 | 0.28 | 9.34 | 51.51 | 8.64E-11 | NaN |
| rno-miR-667* | 0.04 | 0.10 | 0.16 | 0.26 | 56.34 | 3.07E-12 | NaN |
| rno-miR-667_v15.0 | 0.04 | 0.13 | 0.20 | 0.32 | 5.22 | 9.24E-10 | NaN |
| rno-miR-668 | 0.04 | 0.10 | 0.15 | 0.25 | 12.40 | 9.27E-05 | NaN |
| rno-miR-671 | 0.04 | 0.10 | 0.15 | 0.25 | 8.22 | 1.90E-03 | NaN |
| rno-miR-672 | 0.09 | 0.21 | 8.84 | 64.83 | 947.30 | 1.03E-08 | NaN |
| rno-miR-672* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-673 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-673* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-674-3p | 0.08 | 7.30 | 11.78 | 18.83 | 33.72 | 3.66E-13 | NaN |
| rno-miR-674-5p | 0.05 | 0.17 | 0.28 | 2.79 | 8.47 | 1.32E-08 | NaN |
| rno-miR-675 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-675* | 0.04 | 0.11 | 0.17 | 0.31 | 330.99 | 5.69E-15 | 25 |
| rno-miR-678 | 0.04 | 0.10 | 0.17 | 0.27 | 152.95 | 5.19E-15 | 11 |
| rno-miR-702-3p | 13.14 | 31.60 | 52.15 | 82.56 | 282.00 | 1 | NaN |
| rno-miR-702-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-708 | 0.04 | 0.10 | 0.15 | 0.25 | 3.01 | 1.00E+00 | NaN |
| rno-miR-708* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-711 | 0.04 | 0.10 | 0.15 | 0.25 | 9.44 | 4.45E-05 | NaN |
| rno-miR-741-3p | 0.04 | 0.10 | 0.16 | 0.27 | 3094.37 | 1.18E-18 | 9 |
| rno-miR-741-5p | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-742 | 0.04 | 0.10 | 0.16 | 0.27 | 795.81 | 9.51E-18 | 11 |
| rno-miR-742* | 0.04 | 0.10 | 0.16 | 0.27 | 332.55 | 5.09E-17 | 10 |
| rno-miR-743a | 0.04 | 0.10 | 0.16 | 0.27 | 454.12 | 2.23E-17 | 11 |
| rno-miR-743a* | 0.04 | 0.10 | 0.16 | 0.27 | 90.13 | 6.48E-15 | NaN |
| rno-miR-743b | 0.04 | 0.10 | 0.16 | 0.27 | 1633.21 | 1.48E-18 | 11 |
| rno-miR-743b* | 0.04 | 0.10 | 0.16 | 0.27 | 165.57 | 2.79E-16 | 9 |
| rno-miR-758 | 0.06 | 0.14 | 0.22 | 0.39 | 105.29 | 3.60E-14 | 31 |
| rno-miR-758* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-759 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-760-3p | 0.05 | 0.14 | 0.24 | 1.71 | 98.91 | 1.45E-09 | NaN |
| rno-miR-760-5p | 0.04 | 0.10 | 0.16 | 0.26 | 191.93 | 8.64E-15 | 8 |
| rno-miR-761 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-764 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-764* | 0.04 | 0.11 | 0.17 | 0.27 | 4.38 | 1.35E-06 | NaN |
| rno-miR-764_v15.0 | 0.04 | 0.12 | 0.18 | 0.28 | 7.53 | 1.30E-09 | NaN |
| rno-miR-770 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-770* | 0.06 | 0.14 | 0.21 | 0.41 | 439.91 | 3.73E-13 | 36 |
| rno-miR-7a | 17.69 | 40.39 | 73.38 | 254.21 | 21015.92 | 2.57E-07 | 3 |
| rno-miR-7a-1* | 0.22 | 9.06 | 13.98 | 25.05 | 84.92 | 1.17E-08 | NaN |
| rno-miR-7a-2* | 0.04 | 0.12 | 0.17 | 0.28 | 177.53 | 4.30E-15 | 12 |
| rno-miR-7b | 0.04 | 0.19 | 0.41 | 14.70 | 5325.66 | 6.82E-08 | 6 |
| rno-miR-802 | 0.04 | 0.10 | 0.16 | 0.27 | 49.84 | 2.36E-15 | NaN |
| rno-miR-802* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-802_v15.0 | 0.04 | 0.11 | 0.17 | 0.28 | 298.16 | 1.17E-16 | 18 |
| rno-miR-871 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-871* | 0.04 | 0.10 | 0.16 | 0.27 | 603.88 | 2.07E-17 | 6 |
| rno-miR-871_v15.0 | 0.04 | 0.10 | 0.16 | 0.27 | 22.48 | 3.02E-12 | NaN |
| rno-miR-872 | 0.64 | 47.14 | 63.84 | 83.29 | 253.45 | 9.13E-16 | NaN |
| rno-miR-872* | 0.05 | 0.14 | 0.24 | 0.66 | 10.50 | 1.70E-07 | NaN |
| rno-miR-873 | 0.04 | 0.13 | 0.21 | 0.39 | 45.10 | 7.13E-12 | NaN |
| rno-miR-873* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-874 | 0.04 | 0.21 | 1.34 | 4.93 | 832.60 | 2.62E-07 | 4 |
| rno-miR-874* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-875 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-876 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-877 | 0.04 | 0.11 | 0.19 | 0.34 | 187.38 | 1.22E-12 | 30 |
| rno-miR-878 | 0.04 | 0.10 | 0.15 | 0.25 | 11.70 | 1.63E-07 | NaN |
| rno-miR-879 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-879* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1.00E+00 | NaN |
| rno-miR-880 | 0.04 | 0.10 | 0.16 | 0.27 | 520.57 | 1.34E-17 | 7 |
| rno-miR-880* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-881 | 0.04 | 0.10 | 0.15 | 0.25 | 19.00 | 6.71E-10 | NaN |
| rno-miR-881* | 0.04 | 0.10 | 0.16 | 0.27 | 59.57 | 2.69E-14 | NaN |
| rno-miR-881_v15.0 | 0.04 | 0.10 | 0.16 | 0.27 | 480.61 | 1.21E-17 | 6 |
| rno-miR-883 | 0.04 | 0.10 | 0.16 | 0.27 | 1019.83 | 6.28E-18 | 11 |
| rno-miR-883* | 0.04 | 0.10 | 0.16 | 0.27 | 344.36 | 4.11E-17 | 12 |
| rno-miR-9 | 0.06 | 0.19 | 0.67 | 45.69 | 5173.33 | 1.90E-08 | NaN |
| rno-miR-9* | 0.06 | 0.18 | 0.41 | 55.45 | 8290.03 | 5.73E-10 | NaN |
| rno-miR-92a | 30.51 | 270.04 | 447.36 | 805.31 | 6072.39 | 1 | NaN |
| rno-miR-92a-1* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-92a-2* | 0.04 | 0.10 | 0.16 | 0.27 | 19.81 | 9.73E-12 | NaN |
| rno-miR-92b | 0.04 | 0.15 | 0.28 | 3.70 | 62.60 | 3.00E-07 | NaN |
| rno-miR-92b* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-93 | 0.66 | 76.99 | 122.35 | 185.60 | 1312.71 | 4.93E-11 | 6 |
| rno-miR-93* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-935 | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-96 | 0.04 | 0.28 | 111.69 | 447.51 | 9114.44 | 9.25E-09 | NaN |
| rno-miR-96* | 0.04 | 0.10 | 0.16 | 0.25 | 7.75 | 3.77E-07 | NaN |
| rno-miR-98 | 1.05 | 251.33 | 342.31 | 409.05 | 750.08 | 3.82E-17 | NaN |
| rno-miR-98* | 0.04 | 0.10 | 0.15 | 0.25 | 1.05 | 1 | NaN |
| rno-miR-99a | 15.96 | 919.18 | 1485.49 | 2515.58 | 5746.90 | 1.94E-10 | NaN |
| rno-miR-99a* | 0.07 | 7.03 | 16.18 | 27.46 | 52.84 | 1.18E-12 | NaN |
| rno-miR-99b | 0.14 | 54.23 | 118.53 | 208.20 | 823.34 | 4.90E-11 | NaN |
| rno-miR-99b* | 0.05 | 0.17 | 0.27 | 2.31 | 181.48 | 1.44E-08 | 3 |
| rno-miR-218a | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| rno-miR-218a-1* | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| rno-miR-218a-2* | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| rno-miR-218b | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
Figure 3.
Two-dimensional hierarchical clustering analysis based on the expression profiles of the 128 filtered probes. Horizontal: individual organs, vertical: miRNAs. In this analysis, expression profiles for the 128 probes filtered by model-based clustering were used for clustering.
Figure 4.
Expression profiles of organ-specific miRNAs identified in this study. Horizontal: individual organs, vertical: normalized signal intensity. Each bar represents mean±S.D. (Organ ID 1–5: N=6, organ ID 6–55: N=3).
Table 5. Expression profiles of organ-specific miRNAs.
| miRNAs |
Signal intensity across all organs
|
Highly-expressed organ(s)/tissue(s) | Human expression report | ||||
|---|---|---|---|---|---|---|---|
| Min. | 25% | Med. | 75% | Max. | |||
| rno-miR-7a | 17.69 | 40.39 | 73.38 | 254.21 | 21,016 | Pituitary gland, cerebrum_thalamus, adrenal gland, lymph node_cervical, lymph node_mesenteric | Bottoni, A. et al. (22) |
| rno-miR-34b | 0.09 | 0.30 | 9.52 | 25.97 | 12,120 | Testis, trachea, lung, epididymis | Liang, Y. (23) |
| rno-miR-122 | 0.04 | 0.10 | 0.17 | 0.27 | 237,949 | Liver | Starkey Lewis, P. J. et al. (16) |
| rno-miR-124 | 0.04 | 0.14 | 0.24 | 0.66 | 21,435 | Cerebellum, eyeball, cerebrum_cerebral cortex, cerebrum_hippocampus, medulla oblongata | Smirnova, L. et al. (24) |
| rno-miR-128 | 0.66 | 61.76 | 110.78 | 493.04 | 10,167 | Cerebrum_cerebral cortex, cerebellum, cerebrum_hippocampus, thymus, medulla oblongata | Smirnova, L. et al. (24) |
| rno-miR-142-3p | 0.69 | 138.15 | 336.86 | 746.27 | 53,949 | Lymph node_cervical, thymus, lymph node_mesenteric, spleen, bone marrow | No report |
| rno-miR-144 | 0.04 | 0.12 | 0.20 | 0.64 | 3,291 | Bone marrow, spleen | No report |
| rno-miR-150 | 0.66 | 105.59 | 216.92 | 582.12 | 14,087 | Lymph node_mesenteric, lymph node_cervical, spleen, thymus, brown adipose tissue | No report |
| rno-miR-181b | 0.33 | 49.48 | 93.90 | 144.48 | 2,640 | Thymus, optic nerve, bone marrow, medulla oblongata, spinal cord_cervical | No report |
| rno-miR-184 | 0.04 | 0.11 | 0.17 | 0.28 | 14,536 | Eyeball | Tian, L. et al. (25) |
| rno-miR-192 | 0.22 | 16.65 | 31.09 | 53.94 | 38,628 | Intenstines (duodenum, jejunum, ileum, cecum, colon, rectum), stomach_glandular, liver, kidney | Jenkins, R. H. et al. (26) |
| rno-miR-206 | 0.04 | 0.12 | 0.23 | 8.49 | 28,471 | Skeletal muscles (musculus soleus, femoris muscle, gastrocnemial muscle), interseptum, esophagus, tongue | Nielsen, S. et al. (27) |
| rno-miR-208_v15.0 | 0.04 | 0.10 | 0.15 | 0.27 | 939 | Heart_interventricular septum, heart_atrium | van Rooij, E. et al. (28) |
| rno-miR-215 | 0.04 | 0.10 | 0.19 | 0.32 | 50,159 | Duodenum, jejunum, ileum, rectum, colon | Sharbati, S. et al. (29) |
| rno-miR-216a | 0.04 | 0.11 | 0.16 | 0.26 | 2,713 | Pancreas | Szafranska, A. E. et al. (30) |
| rno-miR-219-2-3p | 0.04 | 0.13 | 0.20 | 0.34 | 1,915 | Optic nerve, spinal cord_cervical, spinal cord_pectoral, medulla oblongata, spinal cord_pars lumbalis | No report |
| rno-miR-499 | 0.11 | 8.57 | 17.70 | 43.21 | 2,233 | Heart_interventricular septum, heart_atrium, interseptum, musculus soleus | Sluijter, J. P. et al. (31) |
| rno-miR-542-3p | 0.05 | 0.32 | 9.15 | 29.89 | 439 | Adrenal gland | No report |
| rno-miR-551b | 0.04 | 0.15 | 0.24 | 0.72 | 1,607 | Pituitary gland, cerebrum, medulla oblongata, spinal cord | No report |
| rno-miR-652 | 120.14 | 223.45 | 322.41 | 550.64 | 8,315 | Harderian gland, bone marrow, adrenal gland, rectum, lung | No report |
Technical validity and limitation of microarray analysis
Microarray technology has become a crucial tool for large-scale and high-throughput measurement. Non-specific hybridization or cross-hybridization is a common concern when interpreting microarrays, particularly with closely related gene family members having highly similar sequences. However, in such cases cross-hybridization is unavoidable since designing appropriate, highly specific, probes is not easy. Advances in microarray technology have successfully reduced, but not completely eliminated, cross-hybridization between the same subfamilies of miRNAs32. Therefore, attention should be paid to interpreting data from genes with highly similar sequences particularly in a microarray dataset similar to the present data. Recently, RNA-sequencing (RNA-seq) technology has emerged as a powerful tool for transcriptomics. This new technology is expected to be a useful tool for miRNA expression analysis.
Usage Notes
Since a large amount of numerical data is produced with microarray analysis, sophisticated bioinformatics approaches are required to analyze and interpret the results. A great number of statistical algorithms for filtering differentially expressed genes are available through the BioConductor project website. In addition, there are several software packages available that combine visualization with statistical analysis, such as Genedata Expressionist (Genedata), GeneSpring (Agilent Technology), Spotfire DecisionSite (Spotfire), TIBCO Spotfire (TIBCO Software Inc.) and ArrayTrack (Tong et al.,33).
Additional information
Tables 1, 2, 3, 4, 5 are only available in the online version of this paper.
How to cite this article: Minami, K. et al. miRNA expression atlas in male rat. Sci. Data 1:140005 doi: 10.1038/sdata.2014.5 (2014).
Supplementary Material
Acknowledgments
This work was conducted as part of a collaborative effort with the Toxicogenomics Informatics Project in Japan (TGP2; starting from 2007 and ending in 2012), which includes the National Institute of Biomedical Innovation (NIBIO), the National Institute of Health Science (NIHS) and several Japanese pharmaceutical companies. This work was supported in part by H14-001-Toxico and H19-001-Toxico grants from the Japanese Ministry of Health, Labour and Welfare.
Footnotes
The authors declare no competing financial interests.
Data Citations
- Minami K., Uehara T. 2013. Gene Expression Omnibus. GSE52754
References
- Stefani G. & Slack F. J. Small noncoding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9, 219–230 (2008). [DOI] [PubMed] [Google Scholar]
- Farazi T. A., Hoell J. I., Morozov P. & Tuschl T. MicroRNAs in human cancer. Adv. Exp. Med. Biol. 774, 1–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S. D. et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 13, 434–446 (2011). [DOI] [PubMed] [Google Scholar]
- Lu T. X., Munitz A. & Rothenberg M. E. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 182, 4994–5002 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P. T., Wang W. X. & Rajeev B. W. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 18, 130–138 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbich C., Kuehbacher A. & Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc. Res. 79, 581–588 (2008). [DOI] [PubMed] [Google Scholar]
- Stanczyk J. et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 58, 1001–1009 (2008). [DOI] [PubMed] [Google Scholar]
- Lu J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838 (2005). [DOI] [PubMed] [Google Scholar]
- Jazbutyte V. & Thum T. MicroRNA-21: from cancer to cardiovascular disease. Curr. Drug Targets 11, 926–935 (2010). [DOI] [PubMed] [Google Scholar]
- Janssen H. L. et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368, 1685–1694 (2013). [DOI] [PubMed] [Google Scholar]
- Srivastava A., Suy S., Collins S. P. & Kumar D. Circulating MicroRNA as biomarkers: An update in prostate cancer. Mol. Cell. Pharmacol. 3, 115–124 (2011). [PMC free article] [PubMed] [Google Scholar]
- Cortez M. A. et al. MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 8, 467–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. S. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 105, 10513–10518 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA 106, 4402–4407 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laterza O. F. et al. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin. Chem. 55, 1977–1983 (2009). [DOI] [PubMed] [Google Scholar]
- Starkey Lewis P. J. et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology 54, 1767–1776 (2011). [DOI] [PubMed] [Google Scholar]
- Miska E. A. et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 5, R68 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P. et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Identification of rat lung-specific microRNAs by micoRNA microarray: valuable discoveries for the facilitation of lung research. BMC Genomics 8, 29 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y. J. et al. Identification and target prediction of miRNAs specifically expressed in rat neural tissue. BMC Genomics 10, 214 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore L. C. et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc. Natl. Acad. Sci. USA 105, 3333–3338 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottoni A. et al. Identification of differentially expressed microRNAs by microarray: a possible role for microRNA genes in pituitary adenomas. J. Cell. Physiol. 210, 370–377 (2007). [DOI] [PubMed] [Google Scholar]
- Liang Y. An expression meta-analysis of predicted microRNA targets identifies a diagnostic signature for lung cancer. BMC Med. Genomics 1, 61 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova L. et al. Regulation of miRNA expression during neural cell specification. Eur. J. Neurosci. 21, 1469–1477 (2005). [DOI] [PubMed] [Google Scholar]
- Tian L., Huang K., DuHadaway J. B., Prendergast G. C. & Stambolian D. Genomic profiling of miRNAs in two human lens cell lines. Curr. Eye Res. 35, 812–818 (2010). [DOI] [PubMed] [Google Scholar]
- Jenkins R. H., Martin J., Phillips A. O., Bowen T. & Fraser D. J. Transforming growth factor β1 represses proximal tubular cell microRNA-192 expression through decreased hepatocyte nuclear factor DNA binding. Biochem. J. 443, 407–416 (2012). [DOI] [PubMed] [Google Scholar]
- Nielsen S. et al. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. J. Physiol. 588, 4029–4037 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E. et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579 (2007). [DOI] [PubMed] [Google Scholar]
- Sharbati S. et al. Deciphering the porcine intestinal microRNA transcriptome. BMC Genomics 11, 275 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranska A. E. et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene 26, 4442–4452 (2007). [DOI] [PubMed] [Google Scholar]
- Sluijter J. P. et al. MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler. Thromb. Vasc. Biol. 30, 859–868 (2010). [DOI] [PubMed] [Google Scholar]
- Wang H., Ach R. A. & Curry B. Direct and sensitive miRNA profiling from low-input total RNA. RNA 13, 151–159 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W. et al. ArrayTrack—supporting toxicogenomic research at the U.S. Food and Drug Administration National Center for Toxicological Research. Environ. Health Perspect. 111, 1819–1826 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Minami K., Uehara T. 2013. Gene Expression Omnibus. GSE52754