Abstract
Inflammation is a multifaceted defense response of immune system against infection. Chronic inflammation has been implicated as an imminent threat for major human malignancies and is directly linked to various steps involved in tumorigenesis. Inflammatory cytokines, interleukins, interferons, transforming growth factors, chemokines, and adhesion molecules have been associated with chronic inflammation. Numerous cytokines are reported to be aberrantly regulated by different epigenetic mechanisms like DNA methylation and histone modifications in tumor tissues, contributing to pathogenesis of tumor in multiple ways. Some of these cytokines also work as epigenetic regulators of other crucial genes in tumor biology, either directly or indirectly. Such regulations are reported in lung, breast, cervical, gastric, colorectal, pancreatic, prostate, and head and neck cancers. Epigenetics of inflammatory mediators in cancer is currently subject of extensive research. These investigations may help in understanding cancer biology and to develop effective therapeutic strategies. The purpose of this paper is to have a brief view of the aberrant regulation of inflammatory cytokines in human malignancies.
1. Introduction
Inflammation is a complex defense response of immune system, attempted to neutralize an insult and reestablish normal tissue structure and function [1]. Inflammation is characterized by redness, swelling, and pain and sometimes failure of function. It is mainly mediated and regulated by inflammatory cytokines. Proinflammatory cytokines are concerned with the upgrading of inflammatory reactions while anti-inflammatory cytokines dampen the proinflammatory cytokine response. Chronic inflammation has been associated with different diseases, such as cancer, diabetes, cardiovascular disorders, pulmonary, and neurological diseases [2–6].
Chronic inflammation is now well acknowledged as a threat feature for major types of cancer [7–11]. About 25% of all cancers are connected to chronic inflammation, which is linked to various stages of tumorigenesis including cellular transformation, tumor progression, endurance, propagation, invasion, angiogenesis, and metastasis [12–14]. Proinflammatory cytokines like chemokines, adhesion molecules, and inflammatory enzymes are known to cause chronic inflammation. Several proinflammatory genes, for example, tumor necrosis factor (TNF) and members of its superfamily, IL-1a, IL-1b, IL-6, IL-8, IL-18, chemokines, VEGF, MMP-9, 5-LOX, and COX-2, play critical role in the control of apoptosis, angiogenesis, proliferation, invasion, and metastasis. Overexpression of transcription factors like NF-κB, which becomes constitutively active in most tumors, is principally responsible for the expression of these genes [15–17].
The survival and growth of the initiated cells are prerequisite for tumor development. Many inflammatory mediators such as interleukins, eicosanoids, and chemokines are able to motivate the propagation of both normal and cancer cells [18]. Clinical and epidemiological studies have suggested a strong connection between cancer, inflammation, and chronic infection [12, 19–22]. Proinflammatory mediators can contribute to tumor promotion and progression when produced intolerably and persistently because of events like aberrant epigenetic changes [12, 18, 23, 24].
Epigenetic processes are direct heritable changes in gene expression without changes in DNA sequence [25]. Epigenetic gene silencing is the transcriptional repression of specific genes throughout growth and cellular differentiation [26, 27]. The active or silent gene states are controlled by the processes of addition or removal of chemical modifications in the chromatin [28, 29]. These modifications include DNA methylation and a variety of posttranslational histone modifications (acetylation, methylation, phosphorylation, etc.). These modifications are recognized by protein complexes that decide the fate of gene expression [30]. In mammals, DNA methylation mainly involves the attachment of methyl groups (–CH3) to cytosine residues present at the CpG sites. Hypermethylation of promoter regions of genes is typically associated with transcriptional silencing while hypomethylation facilitates gene expression.
A number of reviews have been published on diverse aspects of cancer epigenetics, inflammation and the relationship between the chronic inflammation and cancer. The current review is focused on epigenetic regulation of different inflammatory cytokines involved in various human malignances with emphasis on aberrant methylation.
2. Basic Concept of Histone Modifications and DNA Methylation
Epigenetic regulation is the mechanism by which chromatin undergoes multiple types of alterations including histone modifications and DNA methylation. These modifications are important for chromatin remodeling and accessibility of transcriptional machinery to facilitate and regulate the transfer of genetic information.
Histone modifications (like acetylation and methylation, etc.) are important regulators of gene transcription. These can activate transcription by acetylation and are depending on the level of histone methylation and repress or inactivate transcription [31]. These modifications have not only the ability to regulate the binding of effector molecules essential to DNA processes including transcription, repair, and replication, but also the ability to regulate higher order chromatin structure and stability [32]. Therefore, it is not surprising that many chromatin-modifying enzymes are deranged during malignant transformation.
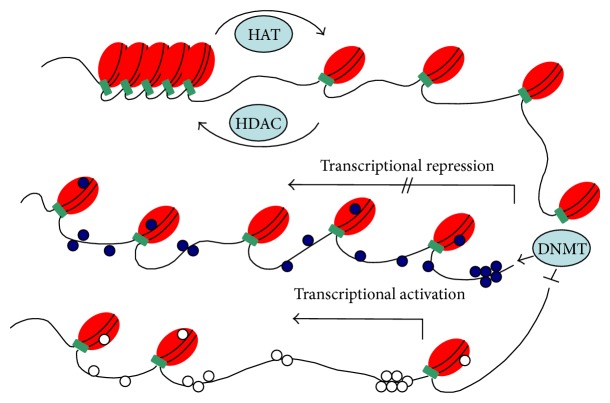
Histone acetylation of the lysine residues is regulated by enzymes that catalyze the addition or removal of acetyl group at numerous positions in histones. Histone acetyltransferases (HATs) and histone deacetyltransferases (HDATs) are the enzymes that acetylate or deacetylate histones, respectively, and thus activate or suppress the transcriptional activity [33]. However, exact mechanism of this gene regulation by histone acetylation is not well understood. It is believed that acetylation process neutralizes positively charged histones and reduces their interaction with the negatively charged DNA thus loosening the chromatin structure (Figure 1). Histone methyltransferases (HMTs) are the most recently discovered histone-modifying enzymes that regulate gene expression by histone methylation at specific sites [34]. These enzymes catalyze the transfer of methyl groups to lysine and arginine residues of histones (H3 and H4). Depending upon the number of methyl groups transferred and the histone residues involved, transcription of a gene can be suppressed or activated [35].
Figure 1.
Overview of different epigenetic mechanisms involved in the regulation of chromatin (heterochromatin and euchromatin). Histone acetylation catalyzed by histone acetylase (HAT) that slightly unwraps the DNA from histones and promotes gene expression. Histone deacetylation is catalyzed by histone deacetylase (HADC), which tightens the DNA and histone core together and represses transcription. CpG islands are mostly located in the regulatory regions of genome. DNA methyltransferase (DNMT) adds methyl group to the CpG site, which is the main epigenetic mechanism reported for transcriptional repression.
DNA methylation is the most widely studied mechanism of epigenetic modifications meant to regulate gene expression. It involves covalent attachment of a methyl group to the cytosine residue by the DNA methyltransferase enzymes (DNMTs) [36]. In DNA methylation, cytosine is converted to methyl-cytosine at CpG (cytosine-phosphate-guanine) site. These CpG sites can span at an area of more than 200 bp (called CpG islands), are present in about 70% of human gene promoters, and are important modulators of gene transcription (Figure 1) [37–39]. The CpG islands are methylated on the inactive female X-chromosome and in certain other conditions like tissue specific and age-related genes [40]. However, in normal cells most of the CpG islands are unmethylated. The aberrant methylation of the CpG islands especially in the promoter region of tumor suppressor genes are responsible for inducing cancer and related human diseases [41].
3. Cytokines as Epigenetic Regulators in Cancer
Along with being epigenetically regulated in malignancies, cytokines also affect the regulation of other genes. Few studies report certain cytokines as epigenetic regulators of other genes having role in cancer initiation and progression.
TGF-β is an inflammatory cytokine having role in organ development, cellular differentiation apoptosis, and fibrosis [42]. It regulates the expression of CD133 (gene symbol: PROM1) in cancer stem cells (CSCs) by DNA methylation (Figure 2) [43]. CD133 is used extensively as a stem cell marker for the identification of normal and cancer cells especially in liver [44]. The high expression of CD133 by promoter demethylation adds to the resistance of tumor cells to chemotherapy and apoptosis. TGFB1 itself and its receptors (TGFBR1 and TGFGR2) are aberrantly regulated by promoter methylation in various cancers including gastric, breast, lung, and ovarian. Downregulation of TGFB1 has also been linked with the paclitaxel resistance, a mitotic inhibitor used in cancer chemotherapy [45–51].
Figure 2.
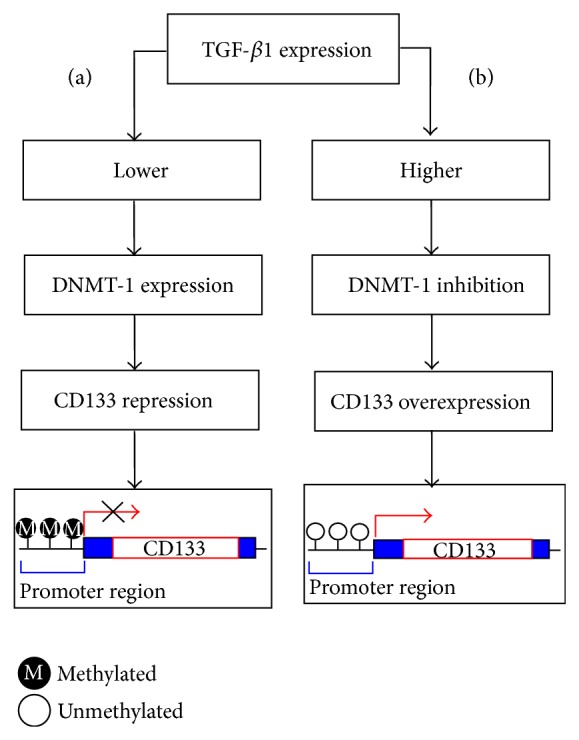
A schematic presentation elaborating epigenetic regulation of CD133 by TGF-β1 mediated by DNA methyltransferase (DNMT-1) in Huh 7 cell (HCC cell line). (a) Low expression of TGF-β1 does not affect DNMT-1 and thus methylation of CD133 promoter is maintained. (b) Elevated expression of TGF-β1 inhibits DNMT-1 which results in demethylation of CD133 promoter resulting in the initiation of transcription.
Suppressor of cytokine signaling 1 (SOCS1), a negative regulator of cytokine signaling and suppressor of inflammation related diseases, is also regulated epigenetically. SOCS1 promoter hypermethylation is one of the best-categorized epigenetic changes in macrophages and hepatocellular carcinoma [52–54]. A recent study provides evidence that loss of SOCS1 expression inside tumor cells via promoter hypermethylation is strongly associated with overproduction of inflammatory cytokines like TNF-α and IL-6. [55].
Inflammatory cytokines such as CXCL1/GROα exert cancer-promoting activities by increasing tumor angiogenesis. CXCL1/GROα decreases the expression of extracellular matrix and plasma protein, fibulin-1D (gene symbol: FBLN1) in prostate cancer cells [56].
In highly invasive castration-resistant prostate cancer (CRPC), CXCL1/GROα signaling results in nuclear factor-kappa B (NF-κB) activation that interacts with histone deacetylase 1 (HDAC1) and form gene-silencing complex. This complex represses the expression of fibulin-1D by deacetylation of histones H3 and H4 on the NF-κB-binding site of the fibulin-1D promoter [56].
4. The Interplay between Epigenetically Regulated Cytokines and Cancer
Epigenetic changes can alter the pattern of normal gene expression thus resulting in pathological conditions including cancer. Aberrant epigenetic events are common in cancer [57]. These changes typically result in silencing of tumor suppressor genes or activation of tumor inducing genes. The transcriptional silencing usually involves hypermethylation of gene promoters while overexpression is a result of DNA hypomethylation. Various inflammatory cytokines are regulated aberrantly in human cancer via epigenetic mechanisms [58–60].
Although epigenetic regulation, especially aberrant DNA methylation, is considered a common mechanism in tumorigenesis, there are only few studies regarding the role of epigenetic regulations of cytokines in malignancies. It is evident that inflammatory cytokines and chemokines, for example, tumor necrosis factor-alpha (TNF-α), IL-1 and IL-6, and interferon gamma (IFN-γ), which can be produced by the tumor cells and/or tumor-associated leukocytes and platelets, may add directly to the development of malignancy. Cytokines can also mediate the activities of immune cells in the fight against malignant cells [61].
4.1. Chemokines Hypermethylation in Cancer
Human chemokine superfamily is composed of more than 50 small secreted proteins. Recent evidences show that chemokines and their receptors also play a critical role in neoplastic transformations, cancer progression, and angiogenesis, in addition to their role in development and inflammatory responses [62]. CXCL14 also known as BRAK is a member of chemokine family. It acts as a chemoattractant and serves immunity by stimulating trafficking of natural killer cells to the sites of inflammation or malignancy [63]. Several parallel studies show that CXCL14 mRNA and protein are universally expressed in normal tissues but are absent in tumor cell lines and primary tumors [64, 65]. CXCL14 was found to be transcriptionally inactivated by promoter CpG hypermethylation in human prostate cancer, in vitro demethylation of the promoter in PC3 cells reexpressed the chemokine [66]. Aberrant methylation of CpG islands in promoter region and the first exon of the gene was associated with its downregulation in gastric cancer [67]. CXCL14 is also known to control colorectal cancer by inhibiting migration and invasion by suppressing NF-κB signaling. Apart from gastric cancer CXCL14 is silenced in colorectal cancer by frequent methylation of the promoter region [68]. Another member of the same family, CXCL12, and its receptor CXCR4 are also associated with tumorigenesis. Interaction between chemokines CXCL12 and CXCR4 promotes cellular adhesion, survival, proliferation, and migration. Its upregulation is reported in skin, lung, pancreas, brain, and breast cancer, while, in pancreatic cancer and melanoma, CXCR4 is downregulated by promoter aberrant methylation [69–71]. Breast cancer cells, with upregulated CXCR4 genes, have been found engrossed to CXCL12 expressing cells in the lymph nodes, liver, and lungs, thus causing metastasis of disconnected tumor cells [72]. Epigenetic mechanisms like demethylation of CXCR4 and hypermethylation of CXCL12 and ESR1 are a characteristic of tumor stage, size, metastasis, and poor overall survival [73]. Knowing the methylation status of both these genes can serve as a biomarker for diagnosis and prognosis in breast cancer [74].
4.2. Epigenetic Regulation of Interleukins in Cancer
Many proinflammatory cytokines including interleukins are frequently claimed to be epigenetically regulated in cancers, especially lung cancer [75–77]. DNA methylation at the promoters of IL-1B, IL-6, and IL-8 genes and their consequential downregulation is reported in lung cancer [59]. Among them, IL-1B promoter has the highest methylation status. In addition, IL-23, a member of the IL-6 superfamily that plays a key role in cancer, is also epigenetically modified in malignancies. It promotes inflammatory responses in the defense against pathogenic infection and upregulation of angiogenic factors and MMPs [78]. Recently, interleukin-23 (IL-23) is claimed to be epigenetically regulated in non-small-cell lung cancer (NSCLC) by both histone acetylation and DNA methylation [79]. IL-23A together with its receptor IL-23R mediates inflammatory pathways; the receptor is also regulated by epigenetic mechanisms in lung cancer [80].
Another member of the same family, IL-12 is known to inhibit directly the growth of human lung adenocarcinoma [81]. It holds a powerful antitumor potential, due to activation of combined immune stimulatory and antiangiogenic mechanisms [82–84]. IL-12 facilitates cytotoxic natural killer cells and induces the production of IFN-γ from NK and T cells. In addition, IL-12 downregulates the production of the proangiogenic factors VEGF and FGF-2 [85–88].
The IL-12 receptor (R) contains two subunits IL-12Rb1 and IL-12Rb2 [89]. IL12RB2 gene encodes IL-12R chain essential for the IL-12 signal transduction [82, 90]. Epigenetic silencing of IL12RB2 is a recurrent event in human lung cancers. Aberrant methylation of this gene sounds like a useful forecaster of long-standing result for adenocarcinoma of lung [91]. IL12RB2 methylation is also reported to be a more frequent in the patients suffering from both chronic obstructive pulmonary diseases (COPD) and non-small-cell lung cancer (NSCLC) [92].
4.3. Epigenetic Regulation of Interferons in Cancer
Interferons are fighters against viral invaders. IFN-γ is a pleiotropic cytokine secreted by type-1 helper (Th1) T cells, cytotoxic T cells, and stimulated natural killer. Production of IFN-γ is related to the induction of reaction in T lymphocytes, which contributes to enhancement of an immune response against malignant cells. IFN-γ (gene symbol: IFNG) stimulates antitumor immune activity by inhibiting cell proliferation and sensitizes tumor cells to apoptosis [93, 94]. Downregulation of IFNG mediated by hypermethylation has been observed in lung and cervical cancer [95, 96]. Human papillomavirus (HPV) is now a well-known risk factor involved in the progression of cervical cancer targeting keratinocytes which produces IFN-κ [97]. A recent study reported that IFN-κ is suppressed in the presence of E6, a HPV protein, signifying the involvement of E6 in IFN-κ de novo methylation followed by transcriptional silencing [98].
5. Epigenetic Targeting Agents in Cancer Therapeutics
Epigenetic therapy is emerging as an exciting, novel approach to treat a variety of diseases, particularly cancer. This therapy consists of using DNA methylation inhibitors and HADC inhibitors for the reversal of the epigenetic aberrations inside the diseased genome. The reversal of aberrant gene methylation is more considerable to reversal of gene mutations or deletions. Epigenetic drugs, whether demethylating agent or HDAC inhibitor, target aberrantly heterochromatic regions, leading to reactivation of tumor suppressor genes and/or other genes that are vital for the normal cells [99]. There are two classes of DNA methylation inhibitors: nucleoside analogues and nonnucleoside analogues. DNA methylation inhibitors include 5-azacytidine (5-Aza-CR), zebularine, 5-aza-2′-deoxycytidine (5-Aza-CdR), and 5-fluoro-2′ deoxycytidine (5-F-CdR), and so forth. 5-Aza-CR and 5-Aza-CdR have been widely studied for the treatment of hematological diseases. 5-Aza-CR is approved by FDA for the treatment of myelodysplasia in 2004 [100]. HDAC inhibitors include short chain fatty acids, hydroxamic acids, cyclic tetrapeptides and benzamides, each of them possesses different functional groups.
Many histone acetylases and deacetylases have been identified but their specific inhibitors still need to be investigated. These agents inhibit histone deacetylase enzymes, so histones remain acetylated and tailed by changes in cellular processes that were malfunctioning in malignant cells. Unlike chemotherapeutic drugs, demethylating agents do not mark cells for instant death. The cells are left to proliferate and reactivate the aberrantly hypermethylated genes. Inhibition of DNA methylation and consequently reactivation of genes, including apoptotic genes and cell-cycle regulators, finally lead the transformed cell to death and cell-cycle arrest [101]. Many investigators have combined DNA-methylation inhibitors with HDAC inhibitors and have shown synergistic tumour-cell-growth inhibition and gene reexpression [102, 103]. It might be advantageous to combine both drugs for the treatment of solid tumors because treatment with DNA-methylation inhibitors alone in solid tumors is not sufficient [103]. Epigenetic targeting of different misregulated genes inside human malignancies is currently in phases I, II, and III clinical trials, to restore them back.
Considering the great potential of epigenetic therapy, there is a hope that in near future it will be possible to target the aberrant regulation of inflammatory cytokines including interleukins, interferons, and chemokines that are facilitating the malignancies. Complete understanding of these epigenetic modifiers and their specific target cytokine will make development of the most effective therapies possible, not only to treat but also to prevent cancer.
6. Conclusion
The literature cited in this review demonstrates that aberrant epigenetic regulation of diverse members of inflammatory cytokines inside different cancers is linked to tumor initiation, endurance, invasion, and progression in one way or the other. Both under- and overexpression of these cytokines are dependent on their epigenetic regulations. Epigenetically regulated cytokines mediate the expression of tumor-associated genes and manipulate their biological role in cancer. Chemokines and interleukins appear to be actively regulated in lung cancer. Targeting the reversal of aberrantly regulated cytokines could be a good potential target for cancer therapy and advanced research in this field could enable us to develop more efficient methods for cancer treatment.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Hirahara K., Poholek A., Vahedi G., et al. Mechanisms underlying helper T-cell plasticity: implications for immune-mediated disease. Journal of Allergy and Clinical Immunology. 2013;131(5):1276–1287. doi: 10.1016/j.jaci.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aggarwal B. B. Nuclear factor-κB: the enemy within. Cancer Cell. 2004;6(3):203–208. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Mason J. C., Libby P. Cardiovascular disease in patients with chronic inflammation: mechanisms underlying premature cardiovascular events in rheumatologic conditions. European Heart Journal. 2014 doi: 10.1093/eurheartj/ehu403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pugliese S. C., Poth J. M., Fini M. A., Olschewski A., El Kasmi K. C., Stenmark K. R. The role of inflammation in hypoxic pulmonary hypertension: from cellular mechanisms to clinical phenotypes. The American Journal of Physiology—Lung Cellular and Molecular Physiology. 2015;308(3):L229–L252. doi: 10.1152/ajplung.00238.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singhal G., Jaehne E. J., Corrigan F., Toben C., Baune B. T. Inflammasomes in neuroinflammation and changes in brain function: a focused review. Frontiers in Neuroscience. 2014;8, article 315 doi: 10.3389/fnins.2014.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Felice F. G., Ferreira S. T. Inflammation, defective insulin signaling, and mitochondrial dysfunction as common molecular denominators connecting type 2 diabetes to Alzheimer disease. Diabetes. 2014;63(7):2262–2272. doi: 10.2337/db13-1954. [DOI] [PubMed] [Google Scholar]
- 7.Peek R. M., Jr., Crabtree J. E. Helicobacter infection and gastric neoplasia. The Journal of Pathology. 2006;208(2):233–248. doi: 10.1002/path.1868. [DOI] [PubMed] [Google Scholar]
- 8.Garcea G., Dennison A. R., Steward W. P., Berry D. P. Role of inflammation in pancreatic carcinogenesis and the implications for future therapy. Pancreatology. 2005;5(6):514–529. doi: 10.1159/000087493. [DOI] [PubMed] [Google Scholar]
- 9.Murphy S. J., Anderson L. A., Johnston B. T., et al. Have patients with esophagitis got an increased risk of adenocarcinoma? Results from a population-based study. World Journal of Gastroenterology. 2005;11(46):7290–7295. doi: 10.3748/wjg.v11.i46.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vagefi P. A., Longo W. E. Colorectal cancer in patients with inflammatory bowel disease. Clinical Colorectal Cancer. 2005;4(5):313–319. doi: 10.3816/CCC.2005.n.003. [DOI] [PubMed] [Google Scholar]
- 11.Nelson W. G., de Marzo A. M., DeWeese T. L., et al. The role of inflammation in the pathogenesis of prostate cancer. The Journal of Urology. 2004;172(5):S6–S12. doi: 10.1097/01.ju.0000142058.99614.ff. [DOI] [PubMed] [Google Scholar]
- 12.Coussens L. M., Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mantovani A. Cancer: inflammation by remote control. Nature. 2005;435(7043):752–753. doi: 10.1038/435752a. [DOI] [PubMed] [Google Scholar]
- 14.Vendramini-Costa D. B., Carvalho J. E. Molecular link mechanisms between inflammation and cancer. Current Pharmaceutical Design. 2012;18(26):3831–3852. doi: 10.2174/138161212802083707. [DOI] [PubMed] [Google Scholar]
- 15.Dong J., Jimi E., Zeiss C., Hayden M. S., Ghosh S. Constitutively active NF-kappaB triggers systemic TNFalpha-dependent inflammation and localized TNFalpha-independent inflammatory disease. Genes and Development. 2010;24(16):1709–1717. doi: 10.1101/gad.1958410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bharti A. C., Shishodia S., Reuben J. M., et al. Nuclear factor-κB and STAT3 are constitutively active in CD138+ cells derived from multiple myeloma patients, and suppression of these transcription factors leads to apoptosis. Blood. 2004;103(8):3175–3184. doi: 10.1182/blood-2003-06-2151. [DOI] [PubMed] [Google Scholar]
- 17.Nakshatri H., Bhat-Nakshatri P., Martin D. A., Goulet R. J., Jr., Sledge G. W., Jr. Constitutive activation of NF-kappaB during progression of breast cancer to hormone-independent growth. Molecular and Cellular Biology. 1997;17(7):3629–3639. doi: 10.1128/mcb.17.7.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balkwill F., Mantovani A. Inflammation and cancer: back to Virchow? The Lancet. 2001;357(9255):539–545. doi: 10.1016/s0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 19.Shacter E., Weitzman S. A. Chronic inflammation and cancer. Oncology. 2002;16(2):217–230. [PubMed] [Google Scholar]
- 20.Fox J. G., Wang T. C. Inflammation, atrophy, and gastric cancer. The Journal of Clinical Investigation. 2007;117(1):60–69. doi: 10.1172/jci30111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobrovolskaia M. A., Kozlov S. V. Inflammation and cancer: When NF-κB amalgamates the perilous partnership. Current Cancer Drug Targets. 2005;5(5):325–344. doi: 10.2174/1568009054629645. [DOI] [PubMed] [Google Scholar]
- 22.Hussain S. P., Hofseth L. J., Harris C. C. Radical causes of cancer. Nature Reviews Cancer. 2003;3(4):276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Baruch A. Inflammation-associated immune suppression in cancer: the roles played by cytokines, chemokines and additional mediators. Seminars in Cancer Biology. 2006;16(1):38–52. doi: 10.1016/j.semcancer.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Smyth M. J., Cretney E., Kershaw M. H., Hayakawa Y. Cytokines in cancer immunity and immunotherapy. Immunological Reviews. 2004;202(1):275–293. doi: 10.1111/j.0105-2896.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- 25.Holliday R. Epigenetics: a historical overview. Epigenetics. 2006;1(2):76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
- 26.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nature Biotechnology. 2010;28(10):1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- 27.Smith Z. D., Meissner A. DNA methylation: roles in mammalian development. Nature Reviews Genetics. 2013;14(3):204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 28.Bannister A. J., Kouzarides T. Regulation of chromatin by histone modifications. Cell Research. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohli R. M., Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothbart S. B., Strahl B. D. Interpreting the language of histone and DNA modifications. Biochimica et Biophysica Acta—Gene Regulatory Mechanisms. 2014;1839(8):627–643. doi: 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morillon A., Karabetsou N., Nair A., Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Molecular Cell. 2005;18(6):723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Eberharter A., Becker P. B. Histone acetylation: a switch between repressive and permissive chromatin. EMBO Reports. 2002;3(3):224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rea S., Eisenhaber F., O'Carroll D., et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 35.Kouzarides T. Histone methylation in transcriptional control. Current Opinion in Genetics and Development. 2002;12(2):198–209. doi: 10.1016/S0959-437X(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 36.Mann M. R., Bartolomei M. S. Epigenetic reprogramming in the mammalian embryo: struggle of the clones. Genome Biology. 2002;3(2):1003.1–1003.4. doi: 10.1186/gb-2002-3-2-reviews1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 38.Lister R., Pelizzola M., Dowen R. H., et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saxonov S., Berg P., Brutlag D. L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(5):1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohandas T., Sparkes R. S., Shapiro L. J. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981;211(4480):393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- 41.Clark S. J., Melki J. DNA methylation and gene silencing in cancer: which is the guilty party? Oncogene. 2002;21(35):5380–5387. doi: 10.1038/sj.onc.1205598. [DOI] [PubMed] [Google Scholar]
- 42.Kajdaniuk D., Marek B., Borgiel-Marek H., Kos-Kudła B. Transforming growth factor β1 (TGFβ1) in physiology and pathology. Endokrynologia Polska. 2013;64(5):384–396. doi: 10.5603/ep.2013.0022. [DOI] [PubMed] [Google Scholar]
- 43.You H., Ding W., Rountree C. B. Epigenetic regulation of cancer stem cell marker CD133 by transforming growth factor-β . Hepatology. 2010;51(5):1635–1644. doi: 10.1002/hep.23544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding W., Mouzaki M., You H., et al. CD133+ liver cancer stem cells from methionine adenosyl transferase 1A-deficient mice demonstrate resistance to transforming growth factor (TGF)-beta-induced apoptosis. Hepatology. 2009;49(4):1277–1286. doi: 10.1002/hep.22743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang N., Zhang H., Yao Q., Wang Y., Dai S., Yang X. TGFBI promoter hypermethylation correlating with paclitaxel chemoresistance in ovarian cancer. Journal of Experimental and Clinical Cancer Research. 2012;31(1, article 6) doi: 10.1186/1756-9966-31-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah J. N., Shao G., Hei T. K., Zhao Y. Methylation screening of the TGFBI promoter in human lung and prostate cancer by methylation-specific PCR. BMC Cancer. 2008;8, article 284 doi: 10.1186/1471-2407-8-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y.-Q., Li Y.-M., Li X., et al. Hypermethylation of TGF-β1 gene promoter in gastric cancer. World Journal of Gastroenterology. 2013;19(33):5557–5564. doi: 10.3748/wjg.v19.i33.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shipitsin M., Campbell L. L., Argani P., et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11(3):259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 49.Shin H. K., Bang Y.-J., Im Y.-H., et al. Transcriptional repression of the transforming growth factor-beta type I receptor gene by DNA methylation results in the development of TGF-beta resistance in human gastric cancer. Oncogene. 1999;18(51):7280–7286. doi: 10.1038/sj.onc.1203146. [DOI] [PubMed] [Google Scholar]
- 50.Kim S.-J., Im Y.-H., Markowitz S. D., Bang Y.-J. Molecular mechanisms of inactivation of TGF-β receptors during carcinogenesis. Cytokine and Growth Factor Reviews. 2000;11(1-2):159–168. doi: 10.1016/s1359-6101(99)00039-8. [DOI] [PubMed] [Google Scholar]
- 51.Irigoyen M., Pajares M. J., Agorreta J., et al. TGFBI expression is associated with a better response to chemotherapy in NSCLC. Molecular Cancer. 2010;9, article 130 doi: 10.1186/1476-4598-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herath N. I., Leggett B. A., MacDonald G. A. Review of genetic and epigenetic alterations in hepatocarcinogenesis. Journal of Gastroenterology and Hepatology. 2006;21(1, part 1):15–21. doi: 10.1111/j.1440-1746.2005.04043.x. [DOI] [PubMed] [Google Scholar]
- 53.Zhou Y., Zhang X., Klibanski A. Genetic and epigenetic mutations of tumor suppressive genes in sporadic pituitary adenoma. Molecular and Cellular Endocrinology. 2014;386(1-2):16–33. doi: 10.1016/j.mce.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amodio N., Bellizzi D., Leotta M., et al. miR-29b induces SOCS-1 expression by promoter demethylation and negatively regulates migration of multiple myeloma and endothelial cells. Cell Cycle. 2013;12(23):3650–3662. doi: 10.4161/cc.26585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng C., Huang C., Ma T.-T., et al. SOCS1 hypermethylation mediated by DNMT1 is associated with lipopolysaccharide-induced inflammatory cytokines in macrophages. Toxicology Letters. 2014;225(3):488–497. doi: 10.1016/j.toxlet.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 56.Kuo P.-L., Shen K.-H., Hung S.-H., Hsu Y.-L. CXCL1/GROα increases cell migration and invasion of prostate cancer by decreasing fibulin-1 expression through NF-kappaB/HDAC1 epigenetic regulation. Carcinogenesis. 2012;33(12):2477–2487. doi: 10.1093/carcin/bgs299.bgs299 [DOI] [PubMed] [Google Scholar]
- 57.You J. S., Jones P. A. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22(1):9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenzweig J. M., Glenn J. D., Calabresi P. A., Whartenby K. A. KLF4 modulates expression of IL-6 in dendritic cells via both promoter activation and epigenetic modification. Journal of Biological Chemistry. 2013;288(33):23868–23874. doi: 10.1074/jbc.M113.479576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tekpli X., Landvik N. E., Anmarkud K. H., Skaug V., Haugen A., Zienolddiny S. DNA methylation at promoter regions of interleukin 1B, interleukin 6, and interleukin 8 in non-small cell lung cancer. Cancer Immunology, Immunotherapy. 2013;62(2):337–345. doi: 10.1007/s00262-012-1340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen X., He Z., Li H., et al. Distinct functional patterns of gene promoter hypomethylation and hypermethylation in cancer genomes. PLoS ONE. 2012;7(9) doi: 10.1371/journal.pone.0044822.e44822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mocellin S., Wang E., Marincola F. M. Cytokines and immune response in the tumor microenvironment. Journal of Immunotherapy. 2001;24(5):392–407. doi: 10.1097/00002371-200109000-00002. [DOI] [PubMed] [Google Scholar]
- 62.Singh R. K., Sudhakar A., Lokeshwar B. L. Role of chemokines and chemokine receptors in prostate cancer development and progression. Journal of Cancer Science & Therapy. 2010;2(4):89–94. doi: 10.4172/1948-5956.1000030. [DOI] [PubMed] [Google Scholar]
- 63.Starnes T., Rasila K. K., Robertson M. J., et al. The chemokine CXCL14 (BRAK) stimulates activated NK cell migration: implications for the downregulation of CXCL14 in malignancy. Experimental Hematology. 2006;34(8):1101–1105. doi: 10.1016/j.exphem.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 64.Shellenberger T. D., Wang M., Gujrati M., et al. BRAK/CXCL14 is a potent inhibitor of angiogenesis and a chemotactic factor for immature dendritic cells. Cancer Research. 2004;64(22):8262–8270. doi: 10.1158/0008-5472.CAN-04-2056. [DOI] [PubMed] [Google Scholar]
- 65.Shurin G. V., Ferris R., Tourkova I. L., et al. Loss of new chemokine CXCL14 in tumor tissue is associated with low infiltration by Dendritic Cells (DC), while restoration of human CXCL14 expression in tumor cells causes attraction of DC both in vitro and in vivo. The Journal of Immunology. 2005;174(9):5490–5498. doi: 10.4049/jimmunol.174.9.5490. [DOI] [PubMed] [Google Scholar]
- 66.Song E. Y., Shurin M. R., Tourkova I. L., Gutkin D. W., Shurin G. V. Epigenetic mechanisms of promigratory chemokine CXCL14 regulation in human prostate cancer cells. Cancer Research. 2010;70(11):4394–4401. doi: 10.1158/0008-5472.CAN-10-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu C., Lin F., Zhu G., et al. Abnormal hypermethylation of promoter region downregulates chemokine CXC ligand 14 expression in gastric cancer. International Journal of Oncology. 2013;43(5):1487–1494. doi: 10.3892/ijo.2013.2078. [DOI] [PubMed] [Google Scholar]
- 68.Cao B., Yang Y., Pan Y., et al. Epigenetic silencing of CXCL14 induced colorectal cancer migration and invasion. Discovery Medicine. 2013;16(88):137–147. [PMC free article] [PubMed] [Google Scholar]
- 69.Sato N., Matsubayashi H., Fukushima N., Goggins M. The chemokine receptor CXCR4 is regulated by DNA methylation in pancreatic cancer. Cancer Biology & Therapy. 2005;4(1):70–76. doi: 10.4161/cbt.4.1.1378. [DOI] [PubMed] [Google Scholar]
- 70.Mori T., Kim J., Yamano T., et al. Epigenetic up-regulation of C-C chemokine receptor 7 and C-X-C chemokine receptor 4 expression in melanoma cells. Cancer Research. 2005;65(5):1800–1807. doi: 10.1158/0008-5472.can-04-3531. [DOI] [PubMed] [Google Scholar]
- 71.Lippitz B. E. Cytokine patterns in patients with cancer: a systematic review. The Lancet Oncology. 2013;14(6):e218–e228. doi: 10.1016/s1470-2045(12)70582-x. [DOI] [PubMed] [Google Scholar]
- 72.Müller A., Homey B., Soto H., et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 73.Ramos E. A. S., Camargo A. A., Braun K., et al. Simultaneous CXCL12 and ESR1 CpG island hypermethylation correlates with poor prognosis in sporadic breast cancer. BMC Cancer. 2010;10(1, article 23) doi: 10.1186/1471-2407-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramos E. A. S., Grochoski M., Braun-Prado K., et al. Epigenetic changes of CXCR4 and its ligand CXCL12 as prognostic factors for sporadic breast cancer. PLoS ONE. 2011;6(12) doi: 10.1371/journal.pone.0029461.e29461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baird A.-M., Gray S. G., O'Byrne K. J. Epigenetics underpinning the regulation of the CXC (ELR+) chemokines in non-small cell lung cancer. PLoS ONE. 2011;6(1) doi: 10.1371/journal.pone.0014593.e14593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baird A.-M., Gray S. G., O'Byrne K. J. IL-20 is epigenetically regulated in NSCLC and down regulates the expression of VEGF. European Journal of Cancer. 2011;47(12):1908–1918. doi: 10.1016/j.ejca.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 77.Cathcart M.-C., Gray S. G., Baird A.-M., et al. Prostacyclin synthase expression and epigenetic regulation in nonsmall cell lung cancer. Cancer. 2011;117(22):5121–5132. doi: 10.1002/cncr.26168. [DOI] [PubMed] [Google Scholar]
- 78.Langowski J. L., Zhang X., Wu L., et al. IL-23 promotes tumour incidence and growth. Nature. 2006;442(7101):461–465. doi: 10.1038/nature04808. [DOI] [PubMed] [Google Scholar]
- 79.Baird A.-M., Leonard J., Naicker K. M., Kilmartin L., O'Byrne K. J., Gray S. G. IL-23 is pro-proliferative, epigenetically regulated and modulated by chemotherapy in non-small cell lung cancer. Lung Cancer. 2013;79(1):83–90. doi: 10.1016/j.lungcan.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 80.Baird A.-M., Dockry É., Daly A., et al. IL-23R is epigenetically regulated and modulated by chemotherapy in non-small cell lung cancer. Frontiers in Oncology. 2013;3, article 162 doi: 10.3389/fonc.2013.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Airoldi I., Di Carlo E., Cocco C., et al. IL-12 can target human lung adenocarcinoma cells and normal bronchial epithelial cells surrounding tumor lesions. PLoS ONE. 2009;4(7) doi: 10.1371/journal.pone.0006119.e6119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Airoldi I., Di Carlo E., Banelli B., et al. The IL-12Rbeta2 gene functions as a tumor suppressor in human B cell malignacies. The Journal of Clinical Investigation. 2004;113(11):1651–1659. doi: 10.1172/jci200420303. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 83.Brunda M. J., Luistro L., Warrier R. R., et al. Antitumor and antimetastatic activity of interleukin 12 against murine tumors. The Journal of Experimental Medicine. 1993;178(4):1223–1230. doi: 10.1084/jem.178.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Colombo M. P., Trinchieri G. Interleukin-12 in anti-tumor immunity and immunotherapy. Cytokine & Growth Factor Reviews. 2002;13(2):155–168. doi: 10.1016/s1359-6101(01)00032-6. [DOI] [PubMed] [Google Scholar]
- 85.Dias S., Boyd R., Balkwill F. IL-12 regulates VEGF and MMPs in a murine breast cancer model. International Journal of Cancer. 1998;78(3):361–365. doi: 10.1002/(sici)1097-0215(19981029)78:3x003C;361::aid-ijc17x0003e;3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 86.Sgadari C., Angiolillo A. L., Tosato G. Inhibition of angiogenesis by interleukin-12 is mediated by the interferon-inducible protein 10. Blood. 1996;87(9):3877–3882. [PubMed] [Google Scholar]
- 87.Voest E. E., Kenyon B. M., O'Reilly M. S., Truitt G., D'Amato R. J., Folkman J. Inhibition of angiogenesis in vivo by interleukin 12. Journal of the National Cancer Institute. 1995;87(8):581–586. doi: 10.1093/jnci/87.8.581. [DOI] [PubMed] [Google Scholar]
- 88.Duda D. G., Sunamura M., Lozonschi L., et al. Direct in vitro evidence and in vivo analysis of the antiangiogenesis effects of interleukin 12. Cancer Research. 2000;60(4):1111–1116. [PubMed] [Google Scholar]
- 89.Presky D. H., Yang H., Minetti L. J., et al. A functional interleukin 12 receptor complex is composed of two β-type cytokine receptor subunits. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(24):14002–14007. doi: 10.1073/pnas.93.24.14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Airoldi I., Cocco C., di Carlo E., et al. Methylation of the IL-12Rbeta2 gene as novel tumor escape mechanism for pediatric B-acute lymphoblastic leukemia cells. Cancer Research. 2006;66(8):3978–3980. doi: 10.1158/0008-5472.can-05-4418. [DOI] [PubMed] [Google Scholar]
- 91.Suzuki M., Iizasa T., Nakajima T., et al. Aberrant methylation of IL-12Rβ2 gene in lung adenocarcinoma cells is associated with unfavorable prognosis. Annals of Surgical Oncology. 2007;14(9):2636–2642. doi: 10.1245/s10434-006-9310-7. [DOI] [PubMed] [Google Scholar]
- 92.Suzuki M., Wada H., Yoshino M., et al. Molecular characterization of chronic obstructive pulmonary disease-related non-small cell lung cancer through aberrant methylation and alterations of EGFR signaling. Annals of Surgical Oncology. 2010;17(3):878–888. doi: 10.1245/s10434-009-0739-3. [DOI] [PubMed] [Google Scholar]
- 93.Miller C. H. T., Maher S. G., Young H. A. Clinical use of interferon-γ . Annals of the New York Academy of Sciences. 2009;1182:69–79. doi: 10.1111/j.1749-6632.2009.05069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zaidi M. R., Merlino G. The two faces of interferon-γ in cancer. Clinical Cancer Research. 2011;17(19):6118–6124. doi: 10.1158/1078-0432.ccr-11-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma D., Jiang C., Hu X., et al. Methylation patterns of the IFN-gamma gene in cervical cancer tissues. Scientific Reports. 2014;4, article 6331 doi: 10.1038/srep06331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang F., Xu J., Zhu Q., et al. Downregulation of IFNG in CD4+ T cells in lung cancer through hypermethylation: a possible mechanism of tumor-induced immunosuppression. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0079064.e79064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hausen H. Z. Papillomaviruses and cancer: from basic studies to clinical application. Nature Reviews Cancer. 2002;2(5):342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 98.Rincon-Orozco B., Halec G., Rosenberger S., et al. Epigenetic silencing of interferon-κ in human papillomavirus type 16-positive cells. Cancer Research. 2009;69(22):8718–8725. doi: 10.1158/0008-5472.can-09-0550. [DOI] [PubMed] [Google Scholar]
- 99.Yoo C. B., Cheng J. C., Jones P. A. Zebularine: a new drug for epigenetic therapy. Biochemical Society Transactions. 2004;32(6):910–912. doi: 10.1042/bst0320910. [DOI] [PubMed] [Google Scholar]
- 100.Kaminskas E., Farrell A., Abraham S., et al. Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clinical Cancer Research. 2005;11(10):3604–3608. doi: 10.1158/1078-0432.ccr-04-2135. [DOI] [PubMed] [Google Scholar]
- 101.Zhang J., Zhong Q. Histone deacetylase inhibitors and cell death. Cellular and Molecular Life Sciences. 2014;71(20):3885–3901. doi: 10.1007/s00018-014-1656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cameron E. E., Bachman K. E., Myöhänen S., Herman J. G., Baylin S. B. Synergy of demethylation and histone deacetylase inhibition in the re- expression of genes silenced in cancer. Nature Genetics. 1999;21(1):103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 103.Rudek M. A., Zhao M., He P., et al. Pharmacokinetics of 5-azacitidine administered with phenylbutyrate in patients with refractory solid tumors or hematologic malignancies. Journal of Clinical Oncology. 2005;23(17):3906–3911. doi: 10.1200/jco.2005.07.450. [DOI] [PubMed] [Google Scholar]