Background: Specification of terminal fate in trophoblasts derived from human embryonic stem cells is not understood.
Results: Inhibition of activin/nodal signaling triggers extravillous fate, but loss of inhibition causes syncytial fate.
Conclusion: Activin/nodal signaling switches the terminal fate of trophoblasts.
Significance: We provide a model system that allows for targeted derivation of extravillous trophoblasts and syncytiotrophoblasts.
Keywords: Activin, Bone Morphogenetic Protein (BMP), Differentiation, Embryonic Stem Cell, Trophoblast, Activin/Nodal Signaling, BMP Signaling, ELF5, Human Embryonic Stem Cells
Abstract
Human embryonic stem cells (hESCs) have been routinely treated with bone morphogenetic protein and/or inhibitors of activin/nodal signaling to obtain cells that express trophoblast markers. Trophoblasts can terminally differentiate to either extravillous trophoblasts or syncytiotrophoblasts. The signaling pathways that govern the terminal fate of these trophoblasts are not understood. We show that activin/nodal signaling switches the terminal fate of these hESC-derived trophoblasts. Inhibition of activin/nodal signaling leads to formation of extravillous trophoblast, whereas loss of activin/nodal inhibition leads to the formation of syncytiotrophoblasts. Also, the ability of hESCs to form bona fide trophoblasts has been intensely debated. We have examined hESC-derived trophoblasts in the light of stringent criteria that were proposed recently, such as hypomethylation of the ELF5-2b promoter region and down-regulation of HLA class I antigens. We report that trophoblasts that possess these properties can indeed be obtained from hESCs.
Introduction
Trophoblasts have been derived from human embryonic stem cells (hESCs)2 (1–29), embryonic carcinoma cells (30, 31), induced pluripotent stem cells (28, 32), other multipotent cells (33), and mouse embryonic stem cells (mESCs) (34–37). Treatment with bone morphogenetic protein (BMP) and/or inhibitors of the activin/nodal pathway have been used for deriving trophoblasts (1, 6–8, 10, 12–15, 17, 19, 22–29, 33, 38). However, the signaling pathways that guide terminal trophoblast differentiation toward a syncytial fate versus an extravillous fate are poorly understood (39). Trophoblasts of the human placenta are composed of villous cytotrophoblasts (vCTBs), syncytiotrophoblasts (STBs), and extravillous cytotrophoblasts (EVTs) (40). vCTBs are considered as the progenitors of both the STBs and EVTs, although the signaling pathways that regulate the specification of STBs or EVTs are not well known. EVTs are further composed of column cytotrophoblasts and invasive cytotrophoblasts (iCTBs). Column cytotrophoblasts undergo an epithelial-to-mesenchymal transition (EMT) to form iCTBs. ICTBs have never been obtained from hESCs in monolayer cultures, and this has impeded the study of molecular mechanisms that lead to iCTB formation. Some studies have yielded monolayer trophoblast cultures that express markers of iCTBs, such as HLA-G and VE-cadherin, but the formation of mesenchymal cells has not been reported (1, 9–11, 22, 24, 27). These cultures have also shown expression of STB markers, such as β-hCG and syncytin-1. Therefore, the signaling pathways that demarcate an STB fate from an iCTB fate are poorly understood. Recently, inhibition of FGF signaling has been shown to favor a syncytial fate in hESC-derived trophoblasts (26). In primary vCTBs, inhibition of activin/nodal signaling triggers an extravillous fate, whereas activation of activin/nodal signaling inhibits it (41, 42). However, it is not known whether such behavior is conserved in trophoblasts that are derived from hESCs. Therefore, we studied whether activin/nodal signaling has a role in specification of the extravillous fate from hESC-derived trophoblasts. We also studied whether activin/nodal signaling has a role in the specification of the syncytial fate.
Recently, however, severe criticisms have been raised against the possibility of trophoblast differentiation from hESCs (43, 44). mESCs contribute to the placenta at very low frequencies when injected into mouse blastocysts (45). Methylation differences have been found between mESCs and mouse trophoblast stem cells, notably the hypomethylation of Elf5 promoter locus (46). This methylation status is sustained by the epigenetic machinery and is thought to be irreversible (46). Analogously, hESCs are found to be hypermethylated at the ELF5-2b promoter locus, whereas placental vCTBs are hypomethylated at this locus (43, 47). Therefore, hESCs are also thought to be epigenetically restricted from differentiating to trophoblasts (43). Trophoblasts with hypomethylated ELF5-2b promoter locus have never been obtained from hESCs. Also, vCTBs of the placenta down-regulate HLA class I antigens, whereas hESC-derived trophoblasts have not been shown to do so (43). We have attempted to address these criticisms by studying whether hESCs can be differentiated to trophoblasts that have hypomethylated ELF5-2b promoter locus and down-regulate HLA class I antigens.
EXPERIMENTAL PROCEDURES
Cell Culture and Differentiation
H1 and H9 hESCs were cultured on mouse embryonic fibroblasts (MEFs) isolated from embryonic day 13.5 pregnant CD-1 mouse embryos (Charles River, Wilmington, MA) as described previously (48). For feeder-free culture, hESCs were grown on growth factor-reduced MatrigelTM (BD Biosciences) in MEF-conditioned medium (MEF-CM); CM was prepared using previously published protocols (49). Differentiation was carried out by adding SB431542 (25 μm) (Sigma-Aldrich), BMP4 (20 ng/ml) (Invitrogen), and EGF (2.5 ng/ml) (R&D Systems) to H1 or H9 cultures, as specified, in the presence of MEF-CM. Medium was refreshed every day leading up to the passage step, after which medium was refreshed every other day. Enzymatic passaging was carried out by scoring confluent colonies into equally spaced grids of cells using a Pasteur pipette and lifting off the cells using collagenase IV (Invitrogen). For stable isotope labeling by amino acids in cell culture (SILAC), H9 cells were grown in CM without l-lysine and l-arginine but containing the stable isotopes l-[13C6][15N2]lysine and l-[13C6]arginine (Pierce) (SILAC-CM), as described previously (48). Stable isotope-labeled arginine and lysine incorporation of 98.5 and 98.0%, respectively, was achieved. Arginine-to-proline conversion was determined to be ∼5% in our system (50).
RNA Isolation, cDNA Synthesis, and Quantitative PCR
RNA was isolated using TRIzolTM reagent (Invitrogen) using the manufacturer's protocol. For cDNA synthesis, the RNA pellet was dissolved in diethyl pyrocarbonate (Sigma)-treated water, and 15 μg of RNA was heated at 70 °C for 5 min with oligo(dT) 15-mer primers (Integrated DNA Technologies, Coralville, IA). Moloney murine leukemia virus reverse transcriptase (Invitrogen) and dNTP mix (Invitrogen) were added, and the reaction was carried out for 50 min at 42 °C. The reaction mixture was heated to 70 °C for 10 min and further incubated with 0.5 m sodium hydroxide for 30 min at 65 °C. The solution was neutralized with 1 m hydrochloric acid and stored at −20 °C until further use. Quantitative PCRs were carried out using SYBR Green Supermix® (Bio-Rad) in a Mastercycler® ep Realplex system (Eppendorf, Hauppage, NY). The primers used for quantitative PCR analysis are listed in Table 1. GAPDH expression was used for normalization between samples. Analysis of variance of the data was carried out using the SAS software. Quantitative PCR analysis was carried out using biological replicates for H9 and H1 hESCs as specified in the figure legends. The ΔΔCt method was used to determine changes in gene expression (51).
TABLE 1.
List of primers used for quantitative PCR analysis
| Gene | Primer | Sequence |
|---|---|---|
| CDH11 | Forward | AGA GGT CCA ATG TGG GAA CG |
| CDH11 | Reverse | GGT TGT CCT TCG AGG ATA CTG T |
| CDX2 | Forward | GGC AGC CAA GTG AAA ACC AG |
| CDX2 | Reverse | GGT GAT GTA GCG ACT GTA GTG AA |
| CGB | Forward | GAG CTC ACC CCA GCA TCC TAT CAC C |
| CGB | Reverse | GTG GCA TTG ATG GGG CGG CAC |
| CSH1 | Forward | TCC TCA GGA GTA TGT TCG CCA |
| CSH1 | Reverse | GGG TTC CAG GAT TGG TGA CC |
| ELF5 | Forward | GCT GCG ACC AGT ACA AGT TG |
| ELF5 | Reverse | CTG CCT CGA CGA ACT CCT C |
| EOMES | Forward | CAC CGC CAC CAA ACT GAG AT |
| EOMES | Reverse | CGA ACA CAT TGT AGT GGG CAG |
| ERVFRD-1 | Forward | GAA ACA CCA GGG ACA GCT TAT C |
| ERVFRD-1 | Reverse | GAT GGG TCC GAA AAT GGG AGG |
| GAPDH | Forward | CTC CAC GAC GTA CTC AGC G |
| GAPDH | Reverse | TGT TGC CAT CAA TGA CCC CTT |
| HAND1 | Forward | GAG AGC ATT AAC AGC GCA TTC G |
| HAND1 | Reverse | CAC GTC CAT CAG GTA GGC G |
| HLA-G | Forward | CAC GCA CAG ACT GAC AGA ATG |
| HLA-G | Reverse | GCC ATC GTA GGC ATA CTG TTC A |
| HSD3B1 | Forward | TAA CGG GTG GAA TCT GAA AAA CG |
| HSD3B1 | Reverse | CTA GCA GAA AGG AAT CGG CTT C |
| ITGA1 | Forward | CAG CCC CAC ATT TCA AGT CGT |
| ITGA1 | Reverse | ACC TGT GTC TGT TTA GGA CCA |
| ITGA5 | Forward | GCC TGT GGA GTA CAA GTC CTT |
| ITGA5 | Reverse | AAT TCG GGT GAA GTT ATC TGT GG |
| KDR | Forward | GGC CCA ATA ATC AGA GTG GCA |
| KDR | Reverse | CCA GTG TCA TTT CCG ATC ACT TT |
| LMO2 | Forward | GGC CAT CGA AAG GAA GAG CC |
| LMO2 | Reverse | GGC CCA GTT TGT AGT AGA GGC |
| MSI1 | Forward | TAA AGT GCT GGC GCA ATC G |
| MSI1 | Reverse | TCT TCT TCG TTC GAG TCA CCA |
| NES | Forward | CTG CTA CCC TTG AGA CAC CTG |
| NES | Reverse | GGG CTC TGA TCT CTG CAT CTA C |
| OLIG3 | Forward | AGC CGT CTC AAC TCG GTC T |
| OLIG3 | Reverse | CAT GGC TAG GTT CAG GTC GTG |
| PECAM1 | Forward | CCA AGG TGG GAT CGT GAG G |
| PECAM1 | Reverse | TCG GAA GGA TAA AAC GCG GTC |
| PLAU | Forward | GCT TGT CCA AGA GTG CAT GGT |
| PLAU | Reverse | CAG GGC TGG TTC TCG ATG G |
| PSG9 | Forward | AGC TGC CCA TCC CCT ACA T |
| PSG9 | Reverse | TTA CTG CGG AGG CCA CCA TA |
| T | Forward | CTG GGT ACT CCC AAT GGG G |
| T | Reverse | GGT TGG AGA ATT GTT CCG ATG A |
| TBX4 | Forward | TGT TCC CCA GCT ACA AGG TAA |
| TBX4 | Reverse | GCA GGG ACA ATG TCA ATC AGC |
Immunofluorescence
Cells were grown on glass bottom culture dishes (Greiner Bio-one, Monroe, NC) coated with Matrigel. After cytokine treatments as specified, cells were fixed and permeabilized using 1:1 methanol/acetone (BDH Chemicals (London, UK) and Fisher, respectively) and treated with 0.5% Triton X-100 (Acros Organics, Geel, Belgium). Cells were blocked in 1× PBS (Sigma-Aldrich) with 5% BSA (AMRESCO, Solon, OH), 0.1 mg/ml human IgG (Immunoreagents Inc., Raleigh, NC), and 0.3% Triton X-100 and incubated overnight with the primary antibody diluted in blocking buffer. While using goat anti-human antibodies, samples were blocked with 5% normal donkey serum (Immunoreagents Inc.). Rabbit anti-human antibodies for VE-cadherin (Cell Signaling, Danvers, MA) and syncytin (Santa Cruz Biotechnology, Inc.), mouse anti-human antibodies for P63 (4A4, Genetex, Irvine, CA), CK7 (Santa Cruz Biotechnology), CD9 (Millipore, Billerica, MA), β-hCG (Abcam, Cambridge, MA), and HLA-G (4H84, Abcam) were used. Corresponding rabbit, mouse, and goat isotype antibodies for controls were purchased from Cell Signaling, Millipore, Epitomics, Immunoreagents Inc., and Santa Cruz Biotechnology. Secondary antibodies used were Alexa 633-conjugated goat anti-mouse IgG, Alexa 488-conjugated goat anti-rabbit IgG, Alexa 633-conjugated donkey anti-goat IgG, Alexa 488-conjugated donkey anti-mouse IgG, Alexa 488-conjugated donkey anti-rabbit IgG, and Alexa 488-conjugated chicken anti-mouse IgG (Invitrogen) along with DAPI (Invitrogen), as appropriate, and samples were imaged using a Zeiss LSM 710 confocal microscope. Plasma membrane staining in syncytiotrophoblasts was carried out using the CellMask deep red plasma membrane fluorescent dye (Invitrogen).
Flow Cytometry
Cells were dissociated using trypsin/EDTA (Invitrogen), fixed in 4% paraformaldehyde (Thermo Shandon Ltd., Runcorn, UK), and permeabilized in saponin buffer containing 1 mg/ml saponin (Sigma-Aldrich) and 1% BSA in PBS (Sigma-Aldrich). Cells were blocked with 0.1 mg/ml human IgG in saponin buffer for 30 min and incubated with the specified primary antibodies for 1 h at room temperature and subsequently with corresponding secondary antibodies for 1 h. Cells were analyzed using a BD Accuri C6 flow cytometer.
Western Blotting
Cells were lysed in lysis buffer containing 40 mm Tris, 120 mm sodium chloride, 0.5% Triton X-100, 0.3% SDS, Complete® mini protease inhibitors (Roche Applied Science), and phosphatase inhibitor mixtures I and II (Sigma-Aldrich) at pH 7.6. The lysate was stored at −80 °C until use. Polyacrylamide gel electrophoresis and Western blotting were carried out using conventional protocols. Incubations with rabbit antibodies for phospho-SMAD1/5/8 (Cell Signaling), phospho-ERK1/2 (Cell Signaling), and β-ACTIN (Cell Signaling) were carried out using protocols specified by the manufacturer. HRP-conjugated goat anti-rabbit antibodies were purchased from Sigma-Aldrich and Cell Signaling. Chemiluminescence readout was obtained using SuperSignal® West Femto substrate (Pierce).
Zymography Assay
Conditioned medium from differentiated cells was collected and run using a standard SDS-PAGE protocol, except that the sample was not heated prior to loading, and the running gel contained 0.1% gelatin. The zymogram was developed using standard protocols (52). Reagents used included Coomassie Blue R-250 (Pierce), methanol (BDH), acetic acid (Acros Organics), Triton X-100 (Acros Organics), sodium azide (Fisher), Tris base (Fisher), hydrochloric acid (Ricca Chemical Co., Arlington, TX), calcium chloride (Acros Organics), and zinc chloride (Acros Organics).
Invasion Assay
Membrane inserts with 12-μm pores (Millipore) were coated with Geltrex (Invitrogen) and allowed to solidify. Differentiated cells were harvested from cell culture using 0.25% trypsin/EDTA (Sigma-Aldrich), plated on Geltrex, and allowed to invade for 2 days in the presence of MEF-CM, SB431542, and EGF. After 2 days, the upper surface of the membrane was cleaned with a cotton swab to remove Geltrex and cells. Membrane inserts were fixed with methanol (Fisher) and stained with 1% Toludine Blue O (Chem-Impex Internatinal Inc., Wood Dale, IL). Cells on the lower surface of the membrane were imaged.
Immunohistochemistry
Differentiated cells were enzymatically passaged on top of solidified Matrigel and allowed to grow. After the specified number of days, solidified Matrigel was fixed in 70% ethanol and paraffin-embedded. Specimens were processed for immunohistochemistry at the Histology Facility at North Carolina State University using standard procedures.
Bisulfite Sequencing
Cells were washed with PBS (Sigma-Aldrich), and genomic DNA was extracted using the Wizard Genomic DNA kit (Promega). H1 and H9 cells were treated with SB431542 for 12 days. The epithelial core was used for genomic DNA extraction. Bisulfite reaction was carried out using the EpiTect bisulfite kit (Qiagen). The ELF5-2b promoter region was isolated using nested PCR as described previously (2). The primers used were as follows: first PCR forward, GGAAATGATGGATATTGAATTTGA; first PCR reverse, CAATAAAAATAAAAACACCTATAACC; 2nd PCR forward, GAGGTTTTAATATTGGGTTTATAATG; 2nd PCR reverse, ATAAATAACACCTACAAACAAATCC.
The PCR product was cloned into pJET1.2 using the pJET1.2 cloning kit (Thermo Scientific), and 10 clones from each biological sample were sequenced at random.
ELISA
Conditioned medium from STB cultures was collected and centrifuged to clear out cell debris. Samples were stored at −20 °C until use. ELISA for β-hCG was carried out using the ELISA kit (Abcam) using the manufacturer's protocol.
Propidium Iodide Staining
Propidium iodide staining was carried out using a previously published protocol (53). Briefly, cells were harvested and fixed in 70% ethanol. Cells were permeabilized with Triton X-100, and RNA was digested with RNase. DNA was stained using propidium iodide and run on a BD Accuri cytometer.
Subcellular Fractionation
Membrane and cytoplasmic fractions were isolated using previously published protocols (48). The membrane pellets were homogenized in 8 m urea (Fisher) and 50 mm ammonium bicarbonate (BDH Chemicals) and used for mass spectrometric analysis.
Protein Fractionation and In-gel Digestion
The cytosolic, membrane, and nuclear fractions were prepared separately for LC-MS/MS analysis. 25 μg of SILAC-labeled protein sample (day 0, undifferentiated hESCs) was combined with 25 μg of the unlabeled protein sample (day 6 or 12 differentiated cells) and loaded onto a Criterion 10–20% Tris-HCl gel (Bio-Rad). Proteins were separated at 200 V for ∼1 h and visualized with Coomassie Stain (Bio-Rad). Each gel lane was dissected into 12 fractions, reduced with dithiothreitol (DTT), alkylated with iodoacetamide, and digested with trypsin using a protocol adapted from Shevchenko et al. (54). Extracted peptides were dried under vacuum and reconstituted in mobile phase A (98% water, 2% acetonitrile, and 0.2% formic acid) prior to analysis.
LC-MS/MS
An Eksigent 1D+ nano-LC system (Eksigent, Dublin, CA) utilizing a vented column configuration (55) was used for reversed-phase separation of peptides. Magic C18AQ stationary phase (5-μm particle size, 200-Å pore size; Microm BioResources, Auburn, CA) was packed to 15 cm in a 75-μm inner diameter PicoFrit capillary (New Objective, Woburn, MA) for the analytical column and to 5 cm in a 75-μm inner diameter IntegraFrit capillary (New Objective) for the trapping column. LC solvents were purchased from Burdick and Jackson (Muskegon, MI). Mobile phase A contained 98% water, 2% acetonitrile, and 0.2% formic acid, and mobile phase B consisted of 2% water, 98% acetonitrile, and 0.2% formic acid. Flow rate during the gradient was set to 350 nl/min. The gradient was held at 2% B for 5 min, adjusted to 10% B at 7 min, and gradually increased to 50% B over the next 120 min. The gradient was then increased to 95% B and held for 5 min before re-equilibrating at 2% B for 10 min.
All measurements were made using a LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific). A precursor scan with 60,000 resolving power at 400 m/z was performed in the Orbitrap mass analyzer followed by eight data-dependent MS/MS scan events in the ion trap. Collision-induced dissociation was employed with an isolation width of 2 m/z and normalized collision energy of 35% for 30 ms. Unassigned, 1+, and ≥4+ charge states were rejected from MS/MS analysis. Dynamic exclusion was employed and set to 3 min with a repeat count of 1, a repeat duration of 0 s, and an exclusion list size as large as 500. Automatic gain control settings were 8 × 103 and 1 × 106 ions in the ion trap and Orbitrap, respectively.
MS Data Analysis
Peak lists were created from LC-MS/MS. RAW files using MASCOT Distiller (Matrix Science, Boston, MA) and subsequently searched using the MASCOT server (Matrix Science) (56). Search parameters included asparagine and glutamine deamidation, methionine oxidation, [13C6]arginine, and [13C6,15N2]lysine as variable modifications as well as cysteine carbamidomethylation as a fixed modification. Precursor search tolerance was set to ±5 ppm, and product ions were searched with a tolerance of ±0.6 Da. The SwissProt human database was concatenated with reverse sequences and used for searching. ProteoIQ (NuSep, Athens, GA) was used to create protein lists filtered using a 1% false discovery rate and to perform SILAC quantification. Only those proteins that showed a statistically significant change in abundance (p < 0.05) and greater 2-fold change have been reported here; the comprehensive data set is available in supplemental Files 1–3. To evaluate the SILAC data set pertaining to the plasma membrane comparison of Day6 cells versus undifferentiated hESCs, in terms of representation of trophoblast-associated proteins, only one protein from each protein group was considered for analysis so as to avoid redundancy from various isoforms of the same protein.
RESULTS
SB431542 Treatment of hESCs Causes Expression of P63
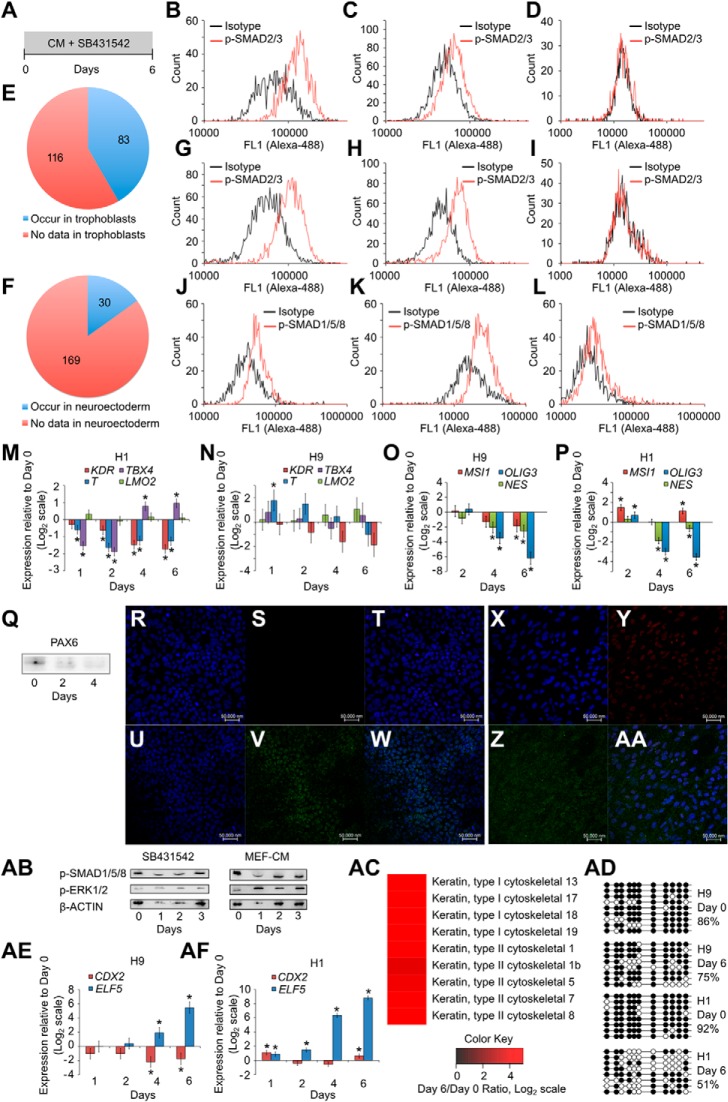
We studied whether SB431542 treatment (Fig. 1A) could lead to formation of trophoblasts. As expected, treatment with SB431542 led to loss of phospho-SMAD2/3 (Fig. 1, B–D and G–I). We labeled undifferentiated H9 cells with the stable isotopes l-[13C6][15N2]lysine and l-[13C6]arginine and isolated the plasma membrane fraction. We compared this with the corresponding plasma membrane fraction of SB431542-treated H9 cells using mass spectrometry to study whether trophoblast differentiation had commenced. We eliminated housekeeping genes from analysis and observed that of 199 up-regulated proteins, 83 had been identified in placental trophoblasts (Fig. 1E and supplemental Table 1). SB431542 treatment has also been reported to cause neural differentiation in serum-free and serum replacer-free protocols. We observed that only 30 of the up-regulated proteins are associated with the neuroectoderm (Fig. 1F), most of which were associated with the adherens and tight junctions (supplemental Table 2), and these proteins are also up-regulated in the trophectoderm. We further observed that phospho-SMAD1/5/8 was present during SB431542 treatment (Fig. 1, J and K). Control cells treated with Noggin and DMH1 along with SB431542 showed decreased phospho-SMAD1/5/8 levels (Fig. 1L). Mesoderm genes T (brachyury) and KDR were down-regulated, and LMO2 was not up-regulated in H1 cells treated with SB431542 (Fig. 1M). Mild up-regulation of TBX4 was seen; however, such behavior was not seen in H9 cells. In H9 cells, whereas T was mildly up-regulated at day 1 and down-regulated thereafter, KDR, TBX4, and LMO2 were not up-regulated (Fig. 1N), showing that mesoderm differentiation did not commence. Also, neural genes MSI1 (musashi 1), OLIG3, and NES (nestin) were down-regulated (Fig. 1O). In H1 cells, however, MSI1 was mildly up-regulated, whereas NES and OLIG3 were down-regulated after 6 days of SB431542 treatment (Fig. 1P). PAX6 was not up-regulated in SB431542-treated H9 cells (Fig. 1Q), showing that neural differentiation did not commence.
FIGURE 1.
SB431542 treatment of hESCs causes expression of P63. A, schematic of protocol showing treatment of hESCs with SB431542 for 6 days. B–D, flow cytometry analysis of H1 cells showing the presence of phospho-SMAD2/3 at day 0 (B), day 1 of treatment with SB431542 (C), and day 6 of treatment with SB431542 (D). Shown is a comparison of membrane fraction from undifferentiated H9 cells and H9 cells treated with SB431542 for 6 days, using SILAC, to show up-regulation of membrane proteins that are found in trophoblasts (E) and in neuroectoderm (F). G–I, flow cytometry analysis of H9 cells showing the presence of phospho-SMAD2/3 at day 0 (G), day 1 of treatment with SB431542 (H), and day 6 of treatment with SB431542 (I). J–L, flow cytometry analysis of H9 cells showing the presence of phospho-SMAD1/5/8 at day 0 (J), day 6 of treatment with SB431542 (K), and day 2 of control treatment with Noggin, DMH1, and SB431542 (L). Shown is expression of T (brachyury), TBX4, LMO2, and KDR after treatment of H1 (M) and H9 cells (N) with SB431542. Shown is expression of MSI1 (musashi 1), NES (nestin), and OLIG3 after treatment of H9 (O) and H1 cells (P) with SB431542. Three biological replicates were used for H9 cells (N and O), and one biological replicate was used for H1 cells (M and P). Error bars, S.E. *, statistically significant changes (p < 0.05). Q, Western blot for PAX6 in H9 cells treated with SB431542. 20 μg protein was loaded in each well. R–W, confocal image showing staining for DAPI (R), P63 (S), and merged (T) in H1 cells at day 0, and DAPI (U), P63 (V), and merged (W) in H1 cells at day 4 of treatment with SB431542. X–AA, confocal image showing staining for DAPI (X), P63 (Y), phospho-SMAD1/5/8 (Z), and merge (AA) in H9 cells at day 4 of treatment with SB431542. Isotype controls did not show staining (data not shown). AB, Western blot showing the presence of phospho-SMAD1/5/8 and phospho-ERK1/2 in both untreated H9 cells and H9 cells treated with SB431542. AC, comparison of cytoplasmic fraction from undifferentiated H9 cells and H9 cells treated with SB431542 for 6 days, using SILAC, to show up-regulation of cytokeratins that are found in trophoblasts. AD, methylation levels of ELF5-2b promoter locus in hESCs and in hESCs treated with SB431542. Data represent DNA sequences sampled randomly from one biological sample. Another biological replicate yielded similar data (data not shown). AE and AF, expression of CDX2 and ELF5 in H9 (AE) and H1 cells (AF) treated with SB431542.
Because P63 and CK-7 have been identified as markers of the trophoblast stem cell state, we studied whether P63 expression was induced by SB431542 treatment. P63 was not seen in undifferentiated H1 cells (Fig. 1, R–T) but was faintly seen after 4 days of SB431542 treatment (Fig. 1, U–W). Similarly, P63 was faintly seen in SB43142-treated H9 cells after 4 days, along with phospho-SMAD1/5/8 (Fig. 1, X–AA). We also confirmed the presence of phospho-SMAD1/5/8 and phoshpo-ERK1/2 in SB431542-treated H9 cells using Western blotting (Fig. 1AB). We labeled undifferentiated H9 cells with the stable isotopes l-[13C6][15N2]lysine and l-[13C6]arginine; isolated the cytoplasmic fraction; and compared this with the corresponding cytoplasmic fraction of SB431542-treated H9 cells using mass spectrometry. We observed up-regulation of cytokeratins of the human trophoblast, such as CK-7, CK-8, and CK-18, in the cytoplasmic fraction (Fig. 1AC). However, we also observed that methylation levels in the ELF5-2b promoter locus only decreased mildly within the span of 6 days (Fig. 1AD). We also observed that whereas ELF5 transcripts were significantly up-regulated in H1 and H9 cells after SB431542 treatment, CDX2 transcripts were mildly down-regulated in H9 cells and mildly up-regulated in H1 cells (Fig. 1, AE–AF). We therefore proceeded to study the effects of prolonged exposure to SB431542 on CDX2 and ELF5 expression and ELF5-2b promoter locus methylation.
HESCs Are Not Epigenetically Restricted from Differentiating to Trophoblasts
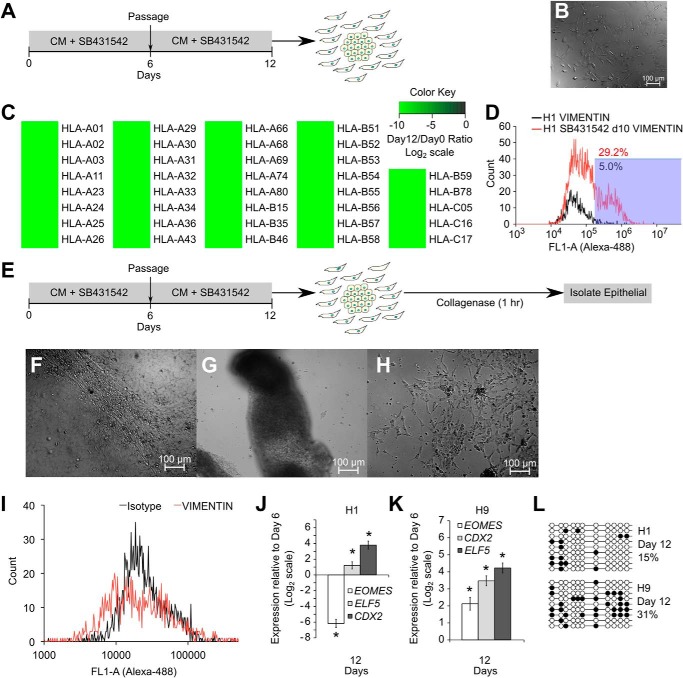
We passaged SB431542-treated cells and continued further treatment with SB431542 (Fig. 2A) and observed the formation of some mesenchymal cells (Fig. 2B). We isolated the plasma membrane fraction from all the cells at day 12 and compared it with the plasma membrane fraction of stable isotope-labeled undifferentiated H9 cells. We observed a complete down-regulation of various HLA class I antigens (Fig. 2C), as expected from trophoblast cells. The occurrence of an epithelial to mesenchymal transition (EMT) was confirmed using vimentin staining (Fig. 2D).
FIGURE 2.
HESCs are not epigenetically restricted from differentiating to trophoblasts. A, protocol for prolonged SB431542 treatment with inclusion of a passage step. B, DIC image showing formation of mesenchymal cells after 12 days of SB431542 treatment of H9 cells. C, comparison of membrane fraction from undifferentiated H9 cells and H9 cells treated with SB431542 for 12 days, using SILAC, to show down-regulation of HLA antigens. D, flow cytometry analysis showing staining for vimentin in cells obtained from 10 days of SB431542 treatment of H9 cells. E, schematic of the protocol for isolating epithelial cells from SB431542-treated cultures using 1-h collagenase treatment. F, DIC image showing epithelial patches in H1 cells treated with SB431542 for 12 days. G and H, DIC image showing these epithelial patches lifting off of the plate after 1-h collagenase treatment (G), whereas mesenchymal cells remain attached to the plate after 1-h collagenase treatment (H). I, flow cytometry analysis showing vimentin staining in isolated epithelial cells. J and K, expression of CDX2, ELF5, and EOMES in these epithelial cells obtained from differentiated H1 (J) and H9 (K) cultures. Three biological replicates were used for both H1 and H9 cells. Error bars, S.E. *, statistically significant changes (p < 0.05). L, methylation levels of ELF5-2b promoter locus in isolated epithelial cells from SB431542 treatment. Data represent DNA sequences sampled randomly from one biological sample. Another biological replicate yielded similar data (data not shown).
To further study trophoblast differentiation, we isolated the epithelial pool of cells at day 12 using a 1-h collagenase IV treatment (Fig. 2E). Before collagenase exposure, epithelial colonies were adhered to the plate (Fig. 2F), but after exposure to collagenase, these colonies lifted off (Fig. 2G). Mesenchymal cells remained adhered to the plate (Fig. 2H). The colonies that lifted off were collected and analyzed for vimentin staining to confirm that they were not mesenchymal (Fig. 2I). These epithelial cells showed substantial up-regulation of CDX2 and ELF5 (Fig. 2, J and K). We also observed hypomethylation of the ELF5-2b promoter region in these cells (Fig. 2L), showing that prolonged SB431542 treatment leads to up-regulation of trophoblast markers CDX2, ELF5, and hypomethylation of the ELF5-2b promoter.
Continued SB431542 Treatment after Passage Leads to iCTB Formation
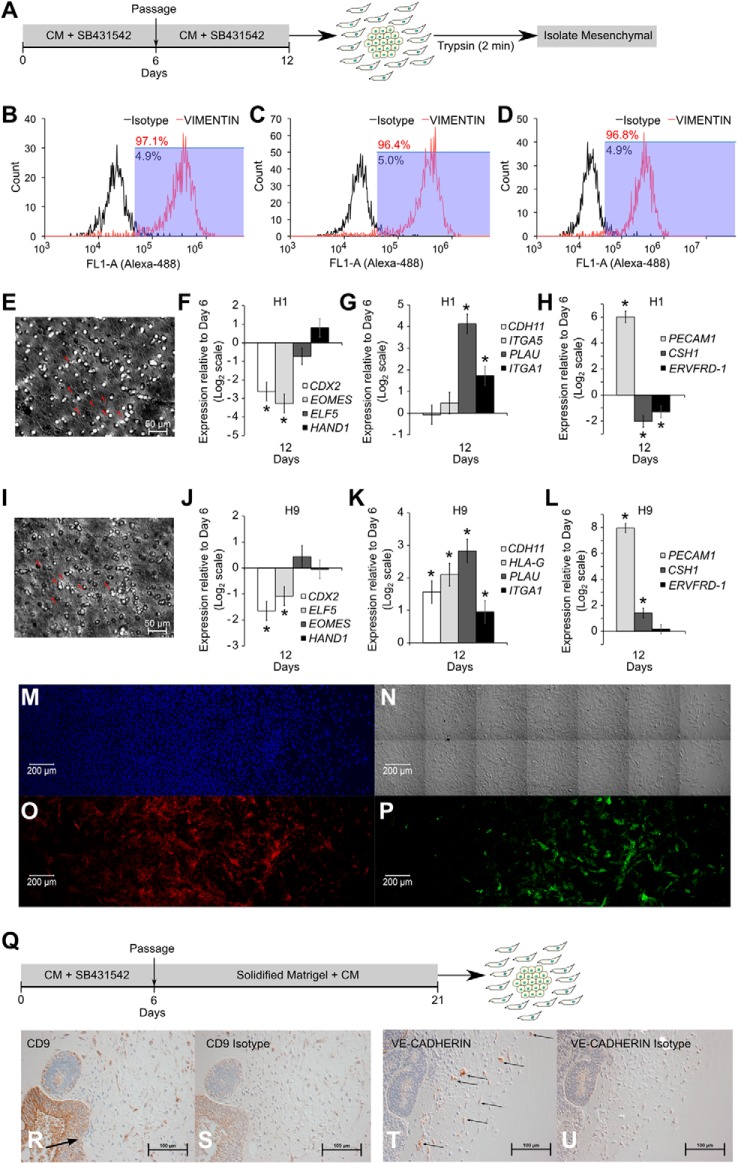
To study whether iCTBs were derived in monolayer cultures, we treated hESCs with SB431542 for 12 days and then isolated the pool of mesenchymal cells with a short trypsin treatment (Fig. 3A). We isolated cells from triplicate cultures and stained with vimentin to confirm that mesenchymal cells had been harvested with >95% purity (Fig. 3, B–D). These mesenchymal cells were capable of invading through solidified Geltrex, showing that these were invasive (Fig. 3, E and I). These mesenchymal cells down-regulated vCTB markers CDX2 and ELF5 but up-regulated iCTB markers PLAU (UPA), ITGA1 (integrin-α1), and PECAM1 and did not up-regulate STB markers CSH1 and ERVFRD-1 (syncytin-2) (Fig. 3, F–H and J–L). More importantly, these cells stained for VE-cadherin and CD9 (Fig. 2, M–P).
FIGURE 3.
Continued SB431542 treatment after passage leads to iCTB formation. A, protocol for SB431542 treatment to form mesenchymal cells and isolation of mesenchymal cells using short trypsin exposure. B–D, mesenchymal cells were isolated from biological triplicate cultures of SB431542-treated H9 cells. Purity of mesenchymal cells was assessed using flow cytometry analysis for vimentin, showing >95% purity. E, an invasion assay of mesenchymal cells obtained from SB431542-treated H9 cells was carried out on Geltrex-coated membrane inserts in duplicate. The image shows the presence of mesenchymal cells on the lower surface of the membrane (arrows indicate nuclei). F–H, expression of CDX2, EOMES, ELF5, HAND1, CDH11, ITGA5, PLAU, IGTA1, PECAM1, CSH1, and ERVFRD-1 (syncytin-2) in H1-derived mesenchymal cells. Three biological replicates were used. Error bars, S.E. *, statistically significant changes (p <0.05). I, invasion assay of mesenchymal cells obtained from SB431542-treated H1 cells was carried out on Geltrex-coated membrane inserts in duplicate. The image shows the presence of mesenchymal cells on the lower surface of the membrane (arrows indicate nuclei). J–L, expression of CDX2, EOMES, ELF5, HAND1, CDH11, HLA-G, PLAU, IGTA1, PECAM1, CSH1, and ERVFRD-1 (syncytin-2) in H9-derived mesenchymal cells. Three biological replicates were used. Error bars, S.E. *, statistically significant changes (p < 0.05). M, confocal image showing staining for DAPI; N, DIC image showing mesenchymal cells; O, staining for VE-cadherin; P, staining for CD9 in H9-derived mesenchymal cells. Q, protocol for obtaining mesenchymal cells from SB431542-treated cells through three-dimensional Matrigel culture. R and S, immunohistochemistry showing staining for CD9 (R) and isotype (S) in H9-derived mesenchymal cells. Arrow, zone of EMT. T and U, immunohistochemistry showing staining for VE-cadherin (indicated by arrows) (T) and isotype (U) in H9-derived mesenchymal cells.
ICTBs have been previously obtained from BMP-treated hESCs through three-dimensional cultures (57). To study whether SB431542-treated hESCs can also give rise to invasive iCTBs in three-dimensional cultures, we passaged SB431542-treated hESCs into solidified Matrigel (Fig. 3Q) and obtained mesenchymal cells that invaded into Matrigel. These cells also co-expressed the iCTB markers, viz. VE-cadherin and CD9 (Fig. 3, R–U). However, staining with VE-cadherin was more sporadic, showing that yield of iCTBs using a three-dimensional differentiation protocol is not homogeneous.
Activation of Activin/Nodal Signaling after Passage Leads to STB Formation
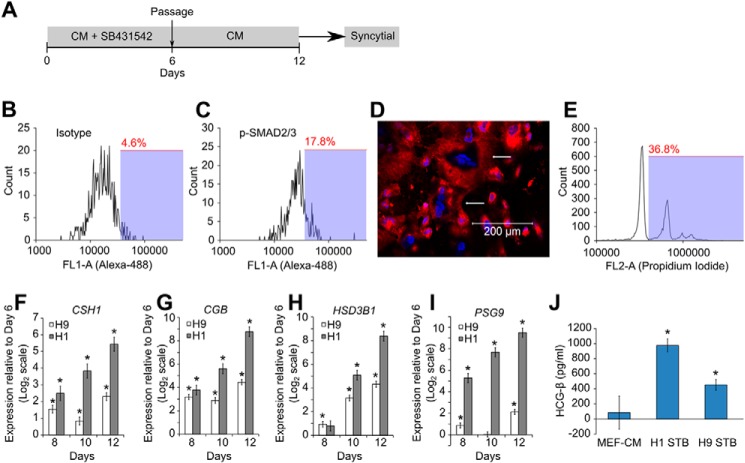
We studied whether inhibition of activin/nodal signaling is necessary for formation of iCTBs by passaging SB431542-treated hESCs into MEF-CM only (Fig. 4A). The presence of phospho-SMAD2/3 staining (Fig. 4, B and C) confirmed that activin/nodal signaling was activated in these cells, whereas phospho-SMAD2/3 staining could not be detected prior to passage (Fig. 1, D and I). Mesenchymal cells were not seen, and instead, syncytial cells were formed, as seen by using a plasma membrane dye (red) and DAPI (blue) (Fig. 4D). Propidium iodide staining and subsequent flow cytometry corroborated the presence of cells with multiple nuclei (Fig. 4E). However, flow cytometry underrepresents the abundance of multinucleated cells because these get fragmented while being handled for analysis, and large cells are filtered and removed prior to flow cytometry. We further observed up-regulation of STB markers CSH1, CGB, HSD3B1, and PSG9 (Fig. 4, F–I). Finally, we collected the conditioned media from these STBs and observed the presence of β-hCG in the media using ELISA (Fig. 4J), showing that STBs capable of hormone production had been obtained.
FIGURE 4.
Activation of activin/nodal signaling after passage leads to STB formation. A, protocol for SB431542 treatment and subsequent activation of activin/nodal signaling to obtain syncytial cells. B and C, H1 cells were treated with SB431542 for 6 days and passaged in MEF-CM for 4 days. Flow cytometry analysis showing staining for isotype control (B) and phospho-SMAD2/3 (C). D, H9 cells were treated with SB431542 for 6 days and passaged into MEF-CM for 6 days. The confocal image shows staining for plasma membrane (red) and nuclei (blue). Arrows, cells with multiple nuclei. E, H1 cells were treated with SB431542 for 6 days and passaged into MEF-CM for 8 days and stained with propidium iodide. Flow cytometry analysis shows multiple peaks corresponding to integer multiples of nuclei in the analyzed cells. F–I, expression of CSH1, CGB, HSD3B1, and PSG9 in H1 and H9 cells exposed to the protocol in A. Three biological replicates were used for H1 and H9 cells. Error bars, S.E. *, statistically significant changes (p < 0.05). J, ELISA showing the amount of β-hCG present in MEF-CM and in media conditioned by STBs from H1 cells and H9 cells. *, statistically significant values (p < 0.05).
Exogenous BMP Treatment Leads to Hypomethylation of ELF5-2b Promoter
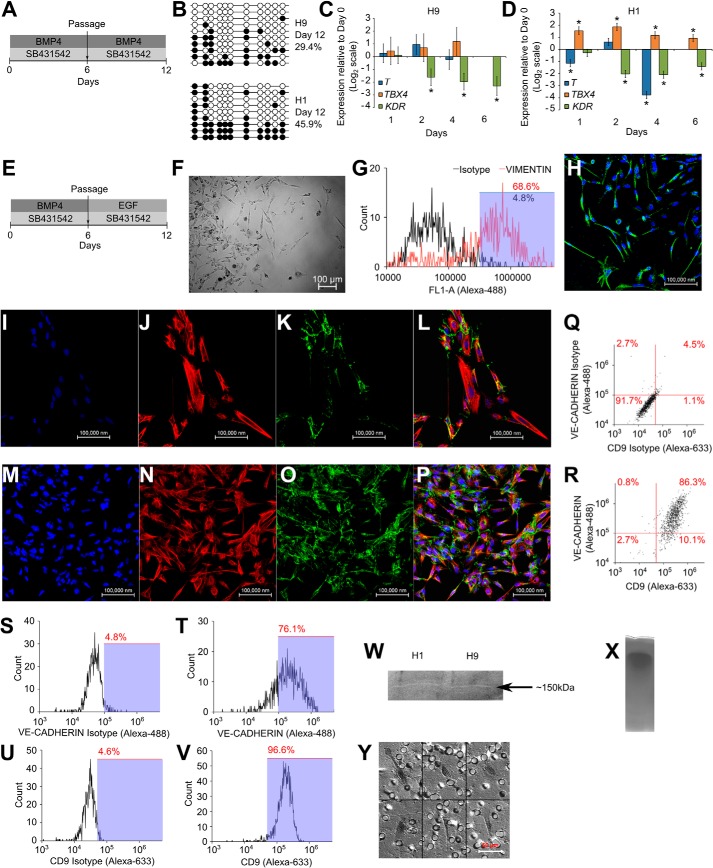
Because endogenous BMP signaling was seen in our previous conditions, we tested whether exogenous BMP treatment also leads to similar effects. We observed that after treatment with BMP4 and SB431542 for 12 days (Fig. 5A), hypomethylation of the ELF5-2b promoter locus was achieved (Fig. 5B). Mesoderm markers were not up-regulated (Fig. 5, C and D). To study if iCTBs could be derived in monolayer cultures using exogenous BMP4, we passaged BMP-treated hESCs in the presence of SB431542 and EGF (Fig. 5E). Mesenchymal cells were observed in culture (Fig. 5F). These cells were harvested with a 2-min trypsin exposure and stained with vimentin to confirm their mesenchymal nature (Fig. 5G). Mesenchymal cells also expressed HLA-G (Fig. 5H). Mesenchymal cells that co-expressed CK-7 and VE-cadherin formed (Fig. 5, I–L and M–P). VE-cadherin and CD9 were widely expressed in the cultures (Fig. 5, Q–V). The conditioned medium from these cultures was able to degrade gelatin in a zymography assay (Fig. 5W), as seen by the presence of a faint clear band at ∼150 kDa. Although the identity of this enzyme could not be discovered using the zymogram alone, it is sufficient to demonstrate the gelatinase activity present in the medium conditioned by these differentiated cells. The control zymogram of MEF-CM did not show any clear band (Fig. 5X). Mesenchymal cells were isolated using trypsin and seeded on Geltrex-coated membrane inserts to study their invasive potential. They were found to invade through Geltrex and reach the lower surface of the membrane (Fig. 5Y), demonstrating their invasiveness. As a control, H1 cells were subjected to the protocol in Fig. 5E, and epithelial cells were isolated using collagenase IV exposure. These cells were dissociated into a single-cell suspension using trypsin and were seeded on Geltrex-coated membrane inserts in duplicates. However, these cells were unable to invade through Geltrex and were not seen on the lower surface of the membrane (data not shown), serving as a control for our invasion assay.
FIGURE 5.
Hypomethylation of ELF5-2b promoter locus and derivation of iCTBs from BMP-treated hESCs. A, schematic of the protocol for prolonged treatment with SB431542 and BMP4, including a passage step, for obtaining cells with hypomethylation of the ELF5-2b promoter locus. B, methylation levels of ELF5-2b promoter locus in hESCs treated with BMP4 and SB431542. Data represent DNA sequences sampled randomly from one biological sample. Another biological replicate yielded similar data (data not shown). C and D, expression of T (brachyury), TBX4, and KDR in differentiated cells. Three biological replicates were used for H9 cells, and one biological replicate was used for H1 cells. Error bars, S.E. *, statistically significant changes (p < 0.05). E, schematic of the protocol for obtaining mesenchymal cells from hESCs treated with SB431542 and BMP4, by passaging them into MEF-CM with EGF and SB431542. F, DIC image showing mesenchymal cells obtained from H9 cultures. G, mesenchymal cells from H9 cultures were harvested using 2-min trypsin exposure. Flow cytometry analysis shows staining for vimentin in harvested mesenchymal cells. H, confocal image showing staining for HLA-G in H9-derived mesenchymal cells. I–L, confocal image showing staining for DAPI (I), CK-7 (J), VE-cadherin (K), and merge (L), in H1-derived mesenchymal cells. M–P, confocal image showing staining for DAPI (M), CK-7 (N), VE-cadherin (O), and merged (P) in H9-derived mesenchymal cells. Q–R, flow cytometry analysis showing co-expression of VE-cadherin and CD9 in H1-derived mesenchymal cells. S and T, flow cytometry analysis showing expression of VE-cadherin in H9-derived mesenchymal cells. U and V, flow cytometry analysis showing expression of CD9 in H9-derived mesenchymal cells. A zymogram shows degradation of gelatin and subsequent clear band at ∼150 kDa by the conditioned medium obtained from differentiated hESC cultures (W), whereas MEF-CM does not show any clear bands on the zymogram (gel shows region from 40 to 260 kDa) (X). Y, invasion assay of mesenchymal cells obtained from H9 cultures, showing the presence of mesenchymal cells on the lower surface of a Geltrex-coated membrane insert.
Activation of Activin/Nodal Signaling after Passage Causes STB Differentiation of Trophoblasts Derived from BMP4 Treatment
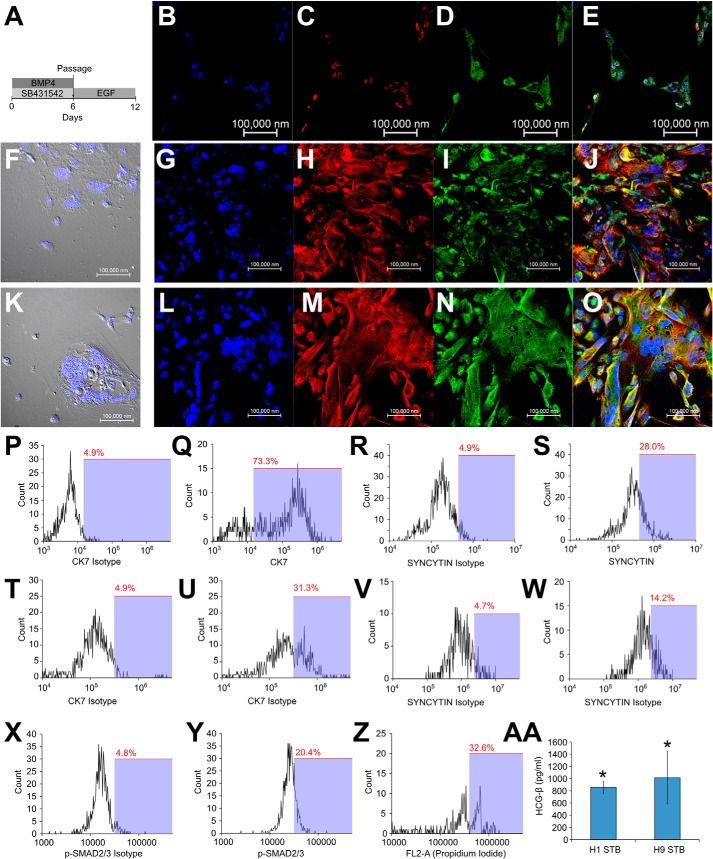
BMP-treated hESCs were passaged into MEF-CM containing EGF (Fig. 6A). These cells co-expressed β-hCG and syncytin-1 (Fig. 6, B–E). These cells were flattened and multinucleate and co-expressed CK-7 and syncytin-1 (Fig. 6, F–O). These cells were removed from the plate using trypsin, fixed using 4% paraformaldehyde, pipetted vigorously to cause fragmentation, and subsequently analyzed using flow cytometry. Expression of CK-7 and synctin-1 increased in most cells of the culture, although the increase in signal from syncytin-1 was lower than that from CK-7 (Fig. 6, P–W). Activation of activin/nodal signaling was confirmed through flow cytometry staining for phospho-SMAD2/3 (Fig. 6, X and Y). Propidium iodide staining and flow cytometry showed peaks corresponding to more than one nucleus (Fig. 6Z). Finally, we collected the conditioned media from these STBs and observed the presence of β-hCG in the media using ELISA (Fig. 6AA), showing that STBs capable of hormone production had been obtained.
FIGURE 6.
Activation of activin/nodal signaling after passage leads to syncytiotrophoblast formation in BMP-treated hESCs. A, schematic of the protocol for obtaining syncytial cells from hESCs treated with SB431542 and BMP4, by passaging them into MEF-CM containing EGF. B–E, confocal image showing staining for DAPI (B), β-hCG (C), syncytin-1 (D), and merge (E) in differentiated H9 cells. F, DIC image showing flattened multinucleate cells in differentiated H9 cultures. G–J, confocal image showing staining for DAPI (G), CK-7 (H), syncytin-1 (I), and merge (J), in differentiated H9 cultures. K, DIC image showing flattened multinucleate cells in differentiated H1 cultures. L–O, confocal image showing staining for DAPI (L), CK-7 (M), syncytin-1 (N), and merge (O) in differentiated H1 cultures. Flow cytometry analysis shows expression of CK-7 (P–Q) and syncytin-1 (R–S) in differentiated H9 cultures. Flow cytometry analysis shows expression of CK-7 (T–U) and syncytin-1 (V–W) in differentiated H1 cultures. X, H1 cells were treated with SB435142 and BMP for 6 days and passaged into MEF-CM containing EGF for 4 days. Y, flow cytometry analysis shows staining for isotype and phospho-SMAD2/3 in these cells. Z, H1 cells were subjected to the protocol in A and stained with propidium iodide. Flow cytometry analysis shows the presence of multiple peaks corresponding to integer multiples of nuclei in analyzed cells. AA, ELISA showing the amount of β-hCG secreted into the medium by STBs derived from H1 and H9 cells. *, statistically significant values (p <0.05).
DISCUSSION
Terminal Differentiation of Trophoblasts Derived from hESCs
Inhibition of FGF signaling promotes a syncytial fate in hESC-derived trophoblasts (26). We show that activin/nodal signaling also governs the terminal differentiation of hESC-derived trophoblasts. Inhibition of activin/nodal signaling leads to iCTB formation, whereas loss of activin/nodal inhibition causes the formation of STBs. Thus, inhibition of activin/nodal signaling is required for iCTB formation, whereas the loss of activin/nodal inhibition is required for STB formation, and activin/nodal signaling switches the terminal fate of hESC-derived trophoblasts from iCTBs toward STBs. Our finding is supported by studies on primary trophoblasts wherein inhibition of activin/nodal signaling triggers extravillous trophoblast formation, but TGFβ inhibits it (41, 42, 58). Also, activin A treatment triggers trophoblast fusion (59). In mouse trophoblast stem cells, activin promotes the acquisition of a labyrinth cell fate (60). Interestingly, the occurrence of EMT upon passage of SB431542-treated hESCs has been reported but in dissimilar culture conditions (61, 62).
We also show that in our protocols, differentiated cultures largely express iCTB markers or STB markers. Therefore, iCTBs and STB can be specifically derived using our protocols for carrying out further studies. For example, we show that iCTBs can be harvested for subsequent study of their invasiveness. Finally, we have used EGF to obtain STBs and iCTBs from BMP-treated hESCs. EGF has been shown to promote the formation of iCTBs and STBs from human placental trophoblasts (63–66). Therefore, our observations are consistent with the behavior of primary human trophoblasts.
We have used new protocols that have been obtained from modifications of previous protocols using BMP4 and/or SB431542 treatment. Importantly, previous protocols have not passaged cells into specific culture conditions supportive of either extravillous differentiation or syncytial differentiation of hESC-derived trophoblasts. However, primary trophoblasts have been routinely isolated and passaged into cultures containing activin/nodal ligands, EGF, etc. (63–66), leading to specific cell fates. Thus, our protocols are analogous to these reported protocols but utilizing hESC-derived trophoblasts.
It must be noted that our protocols do not use completely defined medium conditions because we use MEF-CM that contains undefined factors secreted by MEFs and also from the knock-out serum replacement. However, use of defined media for trophoblast differentiation has yielded significant heterogeneity and formation of non-trophoblastic cells as well (29, 43). The reasons for such differences are beyond the scope of this paper. Therefore, further research needs to be conducted to formulate defined media that may be suitable for such protocols.
The “Epigenetic Barrier” for Trophoblast Differentiation
Both mESCs and hESCs display hypermethylation at the Elf5 promoter locus (43, 46), whereas mouse trophoblast stem cells and human placental villous cytotrophoblasts display hypomethylation at this locus (46, 47), and it was further proposed that hESCs are epigenetically restricted from accessing the trophoblast fate due to irreversible methylation at this locus. We have reported a differentiation protocol, using which we have derived trophoblasts with hypomethylation at the ELF5-2b promoter locus. Therefore, we show that hypermethylation at this locus is not irreversible and that hESCs are not restricted from accessing the trophoblast lineage. Our result is supported by recent studies showing plasticity of DNA methylation in embryonic stem cells (67) and contribution of mESCs toward trophoblasts in cell culture environments as well as upon injection into blastocysts (37, 68).
Bona Fide Trophoblast from hESCs
A stringent set of markers was proposed for assessing trophoblast differentiation of hESCs (44). Many of these markers have never been shown in hESC-derived trophoblasts. We report that upon modification of the previously used differentiation protocols, these markers can be accessed. Such markers include hypomethylation of ELF5-2b promoter locus and down-regulation of HLA class I antigens. Another stringent criterion is whether the hESC-derived trophoblasts can function as bona fide trophoblasts during placental development (44). However, this condition cannot be tested experimentally. Nevertheless, a surrogate criterion is whether the response of hESC-derived trophoblasts toward their signaling environment is similar to those of primary trophoblasts. Inhibition of activin/nodal signaling triggers iCTB formation, but TGFβ suppresses EMT and iCTB formation from placental trophoblasts (41, 42, 69–72). Activin treatment causes fusion in trophoblasts (59). Also, EGF promotes the formation of iCTBs and STBs from human placental trophoblasts (63–66). We report that hESC-derived trophoblasts behave similarly (i.e. both STBs and iCTBs are obtained upon EGF treatment of BMP-treated hESCs). Also, inhibition of activin/nodal signaling leads to EMT and formation of iCTBs, whereas loss of activin/nodal inhibition causes STB formation. Therefore, hESC-derived trophoblasts are capable of forming iCTBs or STBs when exposed to relevant extracellular cues, similar to human placental trophoblasts. Other functional properties of first and third trimester trophoblasts and mouse trophoblasts have been reported, such as their differential response to nodal and activin treatment (58, 73–76). These should also be tested in hESC-derived trophoblasts, although these studies are beyond the scope of this work.
P63 was also recently identified as a marker for the trophoblast stem cell state and was expressed in BMP4-treated hESCs after 4–5 days (27). We show that SB431542-treated hESCs also express P63 after 4 days. We did not observe P63 expression prior to 4 days (data not shown), which is analogous to the behavior of BMP4-treated hESCs. Apart from these critical markers, other trophoblast-associated proteins have been observed in human placental trophoblasts, and these proteins have also been observed in hESC-derived trophoblasts (1, 26, 29, 77). We corroborate these previous studies by showing the presence of trophoblast-associated proteins in hESC-derived trophoblasts, including CK-7, CK-8, CK-18, HLA-G, syncytin, β-hCG, CDX2, ELF5, CD9, VE-cadherin, CSH1, HSD3B1, PSG9, CDH11, PLAU, PECAM1, and the various proteins listed in supplemental Table 1. We further corroborate that these trophoblasts have the properties of invasion through extracellular matrix, gelatinase activity, and the ability to form iCTBs and STBs and lack mesoderm markers.
Role of BMP Signaling in Trophoblast Differentiation
BMP signaling triggers trophoblast differentiation in certain conditions (1, 6–8, 10, 12–15, 19, 22–29), and it has been proposed that BMP signaling is indispensable for trophoblast differentiation (14, 17, 27). We have also observed endogenous BMP signaling while trophoblasts were derived using SB431542 treatment. However, BMP treatment of hESCs also leads to the formation of mesoderm (78, 79), extraembryonic mesoderm (43), extraembryonic endoderm (80, 81), and germ cells (82, 83) in other culture conditions. The fate of BMP-treated hESCs was also shown to depend on the presence of FGF (79). This shows that BMP treatment does not guarantee the formation of trophoblasts. Furthermore, Bmpr1a−/− and Bmpr2−/− knock-out mice do not show trophectoderm defects (84, 85). Mouse two-cell embryos do not develop into blastocysts when exposed to BMP4, although BMP4 induces expression of Cdx2 in the ICM of existing blastocysts (86). On the other hand, human oviductal cells express high levels of Noggin (87) and cause blastocyst formation in mouse embryos (88). Trophoblasts can be derived from mESCs using Noggin (36). Therefore, the exact role of BMP signaling leading to trophoblast (trophectoderm; TE) specification needs to be studied further. Although it is possible that BMP signaling directly up-regulates trophoblast genes, chromatin immunoprecipitation data demonstrating a direct transcriptional role for BMP signaling have not yet been obtained. On the other hand, it is possible that BMP signaling up-regulates trophoblast genes indirectly. In the case of extraembryonic mesoderm differentiation, BMP signaling was shown to trigger CDX2 expression indirectly through brachyury (43). However, others and we have reported that T (brachyury) is not up-regulated in our cultures (26, 29). Nevertheless, it is not clear whether BMP signaling triggers trophoblast gene expression through direct transcriptional control or indirectly. Further studies on the mechanistic role of BMP signaling may help in resolving its exact function in trophoblast differentiation.
Role of FGF Signaling in Trophoblast Differentiation
It has been shown that inhibition of FGF signaling favors a syncytial fate in hESC-derived trophoblasts (26). However, STBs have also been derived from hESCs in the presence of MEF-CM (1, 7), which contains exogenously added FGF2. Indeed, we observe that phospho-ERK1/2 is present in hESCs treated with SB431542. Because reports show that trophoblasts are also obtained in the presence of inhibitors of FGF signaling (26), further studies need to be conducted to study the mechanistic role of FGF signaling in trophoblast differentiation of hESCs.
Role of Activin/Nodal Signaling in Trophoblast Differentiation
It was previously shown that SB431542 treatment leads to trophoblast differentiation (17, 33). However, the study was questioned due to the use of ambiguous markers for characterizing hESC-derived trophoblasts (43). A set of stringent markers was proposed (44), and we have used these markers to establish that trophoblast differentiation occurs when hESCs are treated with SB431542 in MEF-CM. Interestingly, inhibition of activin/nodal signaling also causes neural differentiation but in dissimilar culture conditions (89, 90). We observe that treatment of hESCs with SB431542 in MEF-CM does not induce neural differentiation, and neural genes are down-regulated. We also observe the presence of phospho-SMAD1/5/8 in these cells. Because BMP signaling represses neural differentiation (91), it is likely that endogenous BMP signaling present in our cultures causes suppression of a neural fate.
Our findings are supported by studies on animal blastocysts. Follistatin treatment increases TE formation in bovine blastocysts, whereas follistatin knockdown or activin A treatment decreases TE formation (92, 93). Inhibitors of activin/nodal signaling are expressed in human fallopian tubes (94) and rat oviductal fluid (95, 96), and human oviductal cells cause blastocyst formation in mouse embryos (97–101). Therefore, it is likely that inhibition of activin/nodal signaling has a role in TE formation in embryos.
Interestingly, trophoblasts have also been derived from hESCs without the requirement of adding SB431542 (1, 6–8, 10, 12, 14, 15, 17, 19, 22, 24, 26, 27, 29). In this regard, it was shown that BMP treatment leads to down-regulation of LEFTY-A, LEFTY-B, and NODAL genes (17), which are directly responsive to SMAD2/3 activity (102). Therefore, it is likely that BMP treatment abrogates activin/nodal signaling through cross-talk in hESCs. Interestingly, supplementation with SB431542 accelerates BMP-induced trophoblast differentiation (26). Therefore, the mechanistic role of BMP signaling and activin/nodal inhibition in the specification of trophoblast from hESCs needs to be studied further.
Supplementary Material
This work was supported by National Science Foundation Grant CBET-0966859.

This article contains supplemental Tables 1 and 2 and Files 1–3.
- hESC
- human embryonic stem cell
- mESC
- mouse embryonic stem cell
- BMP
- bone morphogenetic protein
- vCTB
- villous cytotrophoblast
- STB
- syncytiotrophoblast
- EVT
- extravillous cytotrophoblast
- iCTB
- invasive cytotrophoblast
- EMT
- epithelial-to-mesenchymal transition
- MEF
- mouse embryo fibroblast
- MEF-CM
- MEF-conditioned medium
- SILAC
- stable isotope labeling by amino acids in cell culture
- TE
- trophectoderm
- DIC
- differential interference contrast
- β-hCG
- β-human chorionic gonadotropin.
REFERENCES
- 1. Xu R.-H., Chen X., Li D. S., Li R., Addicks G. C., Glennon C., Zwaka T. P., Thomson J. A. (2002) BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 20, 1261–1264 [DOI] [PubMed] [Google Scholar]
- 2. Gerami-Naini B., Dovzhenko O. V., Durning M., Wegner F. H., Thomson J. A., Golos T. G. (2004) Trophoblast differentiation in embryoid bodies derived from human embryonic stem cells. Endocrinology 145, 1517–1524 [DOI] [PubMed] [Google Scholar]
- 3. Matin M. M., Walsh J. R., Gokhale P. J., Draper J. S., Bahrami A. R., Morton I., Moore H. D., Andrews P. W. (2004) Specific knockdown of Oct4 and β2-microglobulin expression by RNA interference in human embryonic stem cells and embryonic carcinoma cells. Stem Cells 22, 659–668 [DOI] [PubMed] [Google Scholar]
- 4. Hay D. C., Sutherland L., Clark J., Burdon T. (2004) Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells 22, 225–235 [DOI] [PubMed] [Google Scholar]
- 5. Hyslop L., Stojkovic M., Armstrong L., Walter T., Stojkovic P., Przyborski S., Herbert M., Murdoch A., Strachan T., Lako M. (2005) Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells 23, 1035–1043 [DOI] [PubMed] [Google Scholar]
- 6. Xu R.-H., Peck R. M., Li D. S., Feng X., Ludwig T., Thomson J. A. (2005) Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat. Methods 2, 185–190 [DOI] [PubMed] [Google Scholar]
- 7. Xu R.-H. (2006) In vitro induction of trophoblast from human embryonic stem cells. Methods Mol. Med. 121, 189–202 [DOI] [PubMed] [Google Scholar]
- 8. Lin G., Martins-Taylor K., Xu R.-H. (2010) Human embryonic stem cell derivation, maintenance, and differentiation to trophoblast. Methods Mol. Biol. 636, 1–24 [DOI] [PubMed] [Google Scholar]
- 9. Harun R., Ruban L., Matin M., Draper J., Jenkins N. M., Liew G. C., Andrews P. W., Li T. C., Laird S. M., Moore H. D. M. (2006) Cytotrophoblast stem cell lines derived from human embryonic stem cells and their capacity to mimic invasive implantation events. Hum. Reprod. 21, 1349–1358 [DOI] [PubMed] [Google Scholar]
- 10. Das P., Ezashi T., Schulz L. C., Westfall S. D., Livingston K. A., Roberts R. M. (2007) Effects of fgf2 and oxygen in the bmp4-driven differentiation of trophoblast from human embryonic stem cells. Stem Cell Res. 1, 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peiffer I., Belhomme D., Barbet R., Haydont V., Zhou Y.-P., Fortunel N. O., Li M., Hatzfeld A., Fabiani J.-N., Hatzfeld J. A. (2007) Simultaneous differentiation of endothelial and trophoblastic cells derived from human embryonic stem cells. Stem Cells Dev. 16, 393–402 [DOI] [PubMed] [Google Scholar]
- 12. Mali P., Ye Z., Hommond H. H., Yu X., Lin J., Chen G., Zou J., Cheng L. (2008) Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells 26, 1998–2005 [DOI] [PubMed] [Google Scholar]
- 13. Yu X., Zou J., Ye Z., Hammond H., Chen G., Tokunaga A., Mali P., Li Y.-M., Civin C., Gaiano N., Cheng L. (2008) Notch signaling activation in human embryonic stem cells is required for embryonic, but not trophoblastic, lineage commitment. Cell Stem Cell 2, 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen G., Ye Z., Yu X., Zou J., Mali P., Brodsky R. A., Cheng L. (2008) Trophoblast differentiation defect in human embryonic stem cells lacking PIG-A and GPI-anchored cell-surface proteins. Cell Stem Cell 2, 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schulz L. C., Ezashi T., Das P., Westfall S. D., Livingston K. A., Roberts R. M. (2008) Human embryonic stem cells as models for trophoblast differentiation. Placenta 29, S10–S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu R.-H., Sampsell-Barron T. L., Gu F., Root S., Peck R. M., Pan G., Yu J., Antosiewicz-Bourget J., Tian S., Stewart R., Thomson J. A. (2008) NANOG is a direct target of TGFβ/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell 3, 196–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu Z., Zhang W., Chen G., Cheng L., Liao J., Jia N., Gao Y., Dai H., Yuan J., Cheng L., Xiao L. (2008) Combinatorial signals of activin/nodal and bone morphogenic protein regulate the early lineage segregation of human embryonic stem cells. J. Biol. Chem. 283, 24991–25002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giakoumopoulos M., Siegfried L. M., Dambaeva S. V., Garthwaite M. A., Glennon M. C., Golos T. G. (2010) Placental-derived mesenchyme influences chorionic gonadotropin and progesterone secretion of human embryonic stem cell-derived trophoblasts. Reprod. Sci. 17, 798–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wolfrum K., Wang Y., Prigione A., Sperling K., Lehrach H., Adjaye J. (2010) The LARGE principle of cellular reprogramming: lost, acquired and retained gene expression in foreskin and amniotic fluid-derived human iPS cells. PloS One 5, e13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frost J. M., Udayashankar R., Moore H. D., Moore G. E. (2010) Telomeric NAP1L4 and OSBPL5 of the KCNQ1 cluster, and the DECORIN gene are not imprinted in human trophoblast stem cells. PLoS One 5, e11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Udayashankar R., Baker D., Tuckerman E., Laird S., Li T. C., Moore H. D. (2011) Characterization of invasive trophoblasts generated from human embryonic stem cells. Hum. Reprod. 26, 398–406 [DOI] [PubMed] [Google Scholar]
- 22. Hoya-Arias R., Tomishima M., Perna F., Voza F., Nimer S. D. (2011) L3MBTL1 deficiency directs the differentiation of human embryonic stem cells toward trophectoderm. Stem Cells Dev. 20, 1889–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Erb T. M., Schneider C., Mucko S. E., Sanfilippo J. S., Lowry N. C., Desai M. N., Mangoubi R. S., Leuba S. H., Sammak P. J. (2011) Paracrine and epigenetic control of trophectoderm differentiation from human embryonic stem cells: the role of bone morphogenic protein 4 and histone deacetylases. Stem Cells Dev. 20, 1601–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marchand M., Horcajadas J. A., Esteban F. J., McElroy S. L., Fisher S. J., Giudice L. C. (2011) Transcriptomic signature of trophoblast differentiation in a human embryonic stem cell model. Biol. Reprod. 84, 1258–1271 [DOI] [PubMed] [Google Scholar]
- 25. Aghajanova L., Shen S., Rojas A. M., Fisher S. J., Irwin J. C., Giudice L. C. (2012) Comparative transcriptome analysis of human trophectoderm and embryonic stem cell-derived trophoblasts reveal key participants in early implantation. Biol. Reprod. 86, 1–21 [DOI] [PubMed] [Google Scholar]
- 26. Sudheer S., Bhushan R., Fauler B., Lehrach H., Adjaye J. (2012) FGF inhibition directs BMP4-mediated differentiation of human embryonic stem cells to syncytiotrophoblast. Stem Cells Dev. 21, 2987–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Y., Moretto-Zita M., Soncin F., Wakeland A., Wolfe L., Leon-Garcia S., Pandian R., Pizzo D., Cui L., Nazor K., Loring J. F., Crum C. P., Laurent L. C., Parast M. M. (2013) BMP4-directed trophoblast differentiation of human embryonic stem cells is mediated through a ΔNp63+ cytotrophoblast stem cell state. Development 140, 3965–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lichtner B., Knaus P., Lehrach H., Adjaye J. (2013) BMP10 as a potent inducer of trophoblast differentiation in human embryonic and induced pluripotent stem cells. Biomaterials 34, 9789–9802 [DOI] [PubMed] [Google Scholar]
- 29. Amita M., Adachi K., Alexenko A. P., Sinha S., Schust D. J., Schulz L. C., Roberts R. M., Ezashi T. (2013) Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc. Natl. Acad. Sci. U.S.A. 110, E1212–E1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kleinsmith L. J., Pierce G. B., Jr. (1964) Multipotentiality of single embryonal carcinoma cells. Cancer Res. 24, 1544–1551 [PubMed] [Google Scholar]
- 31. O'Driscoll C. M., Coulter J. B., Bressler J. P. (2013) Induction of a trophoblast-like phenotype by hydralazine in the p19 embryonic carcinoma cell line. Biochim. Biophys. Acta. 1833, 460–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen Y., Wang K., Chandramouli G. V. R., Knott J. G., Leach R. (2013) Trophoblast lineage cells derived from human induced pluripotent stem cells. Biochem. Biophys. Res. Commun. 436, 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Genbacev O., Donne M., Kapidzic M., Gormley M., Lamb J., Gilmore J., Larocque N., Goldfien G., Zdravkovic T., McMaster M. T., Fisher S. J. (2011) Establishment of human trophoblast progenitor cell lines from the chorion. Stem Cells 29, 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hayashi Y., Furue M. K., Tanaka S., Hirose M., Wakisaka N., Danno H., Ohnuma K., Oeda S., Aihara Y., Shiota K., Ogura A., Ishiura S., Asashima M. (2010) BMP4 induction of trophoblast from mouse embryonic stem cells in defined culture conditions on laminin. In Vitro Cell Dev. Biol. Anim. 46, 416–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He S., Pant D., Schiffmacher A., Meece A., Keefer C. L. (2008) Lymphoid enhancer factor 1-mediated Wnt signaling promotes the initiation of trophoblast lineage differentiation in mouse embryonic stem cells. Stem Cells 26, 842–849 [DOI] [PubMed] [Google Scholar]
- 36. Peng S., Hua J., Cao X., Wang H. (2011) Gelatin induces trophectoderm differentiation of mouse embryonic stem cells. Cell Biol. Int. 35, 587–591 [DOI] [PubMed] [Google Scholar]
- 37. Morgani S. M., Canham M. A., Nichols J., Sharov A. A., Migueles R. P., Ko M. S. H., Brickman J. M. (2013) Totipotent embryonic stem cells arise in ground-state culture conditions. Cell Rep. 3, 1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tiruthani K., Sarkar P., Rao B. (2013) Trophoblast differentiation of human embryonic stem cells. Biotechnol. J. 8, 421–433 [DOI] [PubMed] [Google Scholar]
- 39. Morrish D. W., Dakour J., Li H. (1998) Functional regulation of human trophoblast differentiation. J. Reprod. Immunol. 39, 179–195 [DOI] [PubMed] [Google Scholar]
- 40. Bischof P., Irminger-Finger I. (2005) The human cytotrophoblastic cell, a mononuclear chameleon. Int. J. Biochem. Cell Biol. 37, 1–16 [DOI] [PubMed] [Google Scholar]
- 41. Graham C. H., Lysiak J. J., McCrae K. R., Lala P. K. (1992) Localization of transforming growth factor-β at the human fetal-maternal interface: role in trophoblast growth and differentiation. Biol. Reprod. 46, 561–572 [DOI] [PubMed] [Google Scholar]
- 42. Caniggia I., Taylor C. V., Ritchie J. W., Lye S. J., Letarte M. (1997) Endoglin regulates trophoblast differentiation along the invasive pathway in human placental villous explants. Endocrinology 138, 4977–4988 [DOI] [PubMed] [Google Scholar]
- 43. Bernardo A. S., Faial T., Gardner L., Niakan K. K., Ortmann D., Senner C. E., Callery E. M., Trotter M. W., Hemberger M., Smith J. C., Bardwell L., Moffett A., Pedersen R. A. (2011) BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 9, 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roberts R. M., Loh K. M., Amita M., Bernardo A. S., Adachi K., Alexenko A. P., Schust D. J., Schulz L. C., Telugu B. P. V. L., Ezashi T., Pedersen R. A. (2014) Differentiation of trophoblast cells from human embryonic stem cells: to be or not to be? Reproduction 147, D1–D12 [DOI] [PubMed] [Google Scholar]
- 45. Beddington R. S., Robertson E. J. (1989) An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development 105, 733–737 [DOI] [PubMed] [Google Scholar]
- 46. Ng R. K., Dean W., Dawson C., Lucifero D., Madeja Z., Reik W., Hemberger M. (2008) Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat. Cell Biol. 10, 1280–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hemberger M., Udayashankar R., Tesar P., Moore H., Burton G. J. (2010) ELF5-enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Hum. Mol. Genet. 19, 2456–2467 [DOI] [PubMed] [Google Scholar]
- 48. Collier T. S., Randall S. M., Sarkar P., Rao B. M., Dean R. A., Muddiman D. C. (2011) Comparison of stable-isotope labeling with amino acids in cell culture and spectral counting for relative quantification of protein expression. Rapid Commun. Mass Spectrom. 25, 2524–2532 [DOI] [PubMed] [Google Scholar]
- 49. Xu C., Inokuma M. S., Denham J., Golds K., Kundu P., Gold J. D., Carpenter M. K. (2001) Feeder-free growth of undifferentiated human embryonic stem cells. Nat. Biotechnol. 19, 971–974 [DOI] [PubMed] [Google Scholar]
- 50. Collier T. S., Sarkar P., Rao B., Muddiman D. C. (2010) Quantitative top-down proteomics of SILAC labeled human embryonic stem cells. J. Am. Soc. Mass Spectrom. 21, 879–889 [DOI] [PubMed] [Google Scholar]
- 51. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 52. Birkedal-Hansen H., Yamada S., Windsor J., Pollard A. H., Lyons G., Stetler-Stevenson W., Birkedal-Hansen B. (2008) Matrix metalloproteinases. Curr. Protoc. Cell Biol. 10.1002/0471143030.cb1008s40 [DOI] [PubMed] [Google Scholar]
- 53. Riccardi C., Nicoletti I. (2006) Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat. Protoc. 1, 1458–1461 [DOI] [PubMed] [Google Scholar]
- 54. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 55. Andrews G. L., Shuford C. M., Burnett J. C., Jr., Hawkridge A. M., Muddiman D. C. (2009) Coupling of a vented column with splitless nanoRPLC-ESI-MS for the improved separation and detection of brain natriuretic peptide-32 and its proteolytic peptides. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877, 948–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 57. Telugu B. P., Adachi K., Schlitt J. M., Ezashi T., Schust D. J., Roberts R. M., Schulz L. C. (2013) Comparison of extravillous trophoblast cells derived from human embryonic stem cells and from first trimester human placentas. Placenta 34, 536–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Caniggia I., Lye S. J., Cross J. C. (1997) Activin is a local regulator of human cytotrophoblast cell differentiation. Endocrinology 138, 3976–3986 [DOI] [PubMed] [Google Scholar]
- 59. Gerbaud P., Pidoux G., Guibourdenche J., Pathirage N., Costa J. M., Badet J., Frendo J.-L., Murthi P., Evain-Brion D. (2011) Mesenchymal activin-A overcomes defective human trisomy 21 trophoblast fusion. Endocrinology 152, 5017–5028 [DOI] [PubMed] [Google Scholar]
- 60. Natale D. R. C., Hemberger M., Hughes M., Cross J. C. (2009) Activin promotes differentiation of cultured mouse trophoblast stem cells towards a labyrinth cell fate. Dev. Biol. 335, 120–131 [DOI] [PubMed] [Google Scholar]
- 61. Mahmood A., Harkness L., Schrøder H. D., Abdallah B. M., Kassem M. (2010) Enhanced differentiation of human embryonic stem cells to mesenchymal progenitors by inhibition of TGF-β/activin/nodal signaling using SB-431542. J. Bone Miner. Res. 25, 1216–1233 [DOI] [PubMed] [Google Scholar]
- 62. Chen Y. S., Pelekanos R. A., Ellis R. L., Horne R., Wolvetang E. J., Fisk N. M. (2012) Small molecule mesengenic induction of human induced pluripotent stem cells to generate mesenchymal stem/stromal cells. Stem Cells Transl. Med. 1, 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Morrish D. W., Bhardwaj D., Dabbagh L. K., Marusyk H., Siy O. (1987) Epidermal growth factor induces differentiation and secretion of human chorionic gonadotropin and placental lactogen in normal human placenta. J. Clin. Endocrinol. Metab. 65, 1282–1290 [DOI] [PubMed] [Google Scholar]
- 64. Morrish D. W., Dakour J., Li H., Xiao J., Miller R., Sherburne R., Berdan R. C., Guilbert L. J. (1997) In vitro cultured human term cytotrophoblast: a model for normal primary epithelial cells demonstrating a spontaneous differentiation programme that requires EGF for extensive development of syncytium. Placenta 18, 577–585 [DOI] [PubMed] [Google Scholar]
- 65. Bass K. E., Morrish D., Roth I., Bhardwaj D., Taylor R., Zhou Y., Fisher S. J. (1994) Human cytotrophoblast invasion is up-regulated by epidermal growth factor: evidence that paracrine factors modify this process. Dev. Biol. 164, 550–561 [DOI] [PubMed] [Google Scholar]
- 66. LaMarca H. L., Dash P. R., Vishnuthevan K., Harvey E., Sullivan D. E., Morris C. A., Whitley G. S. J. (2008) Epidermal growth factor-stimulated extravillous cytotrophoblast motility is mediated by the activation of PI3-K, Akt and both p38 and p42/44 mitogen-activated protein kinases. Hum. Reprod. 23, 1733–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hackett J. A., Dietmann S., Murakami K., Down T. A., Leitch H. G., Surani M. A. (2013) Synergistic mechanisms of DNA demethylation during transition to ground-state pluripotency. Stem Cell Reports 1, 518–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Macfarlan T. S., Gifford W. D., Driscoll S., Lettieri K., Rowe H. M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S. L. (2012) Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Caniggia I., Grisaru-Gravnosky S., Kuliszewsky M., Post M., Lye S. J. (1999) Inhibition of TGF-β3 restores the invasive capability of extravillous trophoblasts in preeclamptic pregnancies. J. Clin. Invest. 103, 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Graham C. H. (1997) Effect of transforming growth factor-β on the plasminogen activator system in cultured first trimester human cytotrophoblasts. Placenta 18, 137–143 [DOI] [PubMed] [Google Scholar]
- 71. Graham C. H., Lala P. K. (1991) Mechanism of control of trophoblast invasion in situ. J. Cell Physiol. 148, 228–234 [DOI] [PubMed] [Google Scholar]
- 72. Karmakar S., Das C. (2002) Regulation of trophoblast invasion by IL-1β and TGF-β1. Am. J. Reprod. Immunol. 48, 210–219 [DOI] [PubMed] [Google Scholar]
- 73. Law J., Zhang G., Dragan M., Postovit L.-M., Bhattacharya M. (2014) Nodal signals via β-arrestins and RalGTPases to regulate trophoblast invasion. Cell. Signal. 26, 1935–1942 [DOI] [PubMed] [Google Scholar]
- 74. Nadeem L., Munir S., Fu G., Dunk C., Baczyk D., Caniggia I., Lye S., Peng C. (2011) Nodal signals through activin receptor-like kinase 7 to inhibit trophoblast migration and invasion: implication in the pathogenesis of preeclampsia. Am. J. Pathol. 178, 1177–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bearfield C., Jauniaux E., Groome N., Sargent I. L., Muttukrishna S. (2005) The secretion and effect of inhibin A, activin A and follistatin on first-trimester trophoblasts in vitro. Eur. J. Endocrinol. 152, 909–916 [DOI] [PubMed] [Google Scholar]
- 76. Ma G. T., Soloveva V., Tzeng S. J., Lowe L. A., Pfendler K. C., Iannaccone P. M., Kuehn M. R., Linzer D. I. (2001) Nodal regulates trophoblast differentiation and placental development. Dev. Biol. 236, 124–135 [DOI] [PubMed] [Google Scholar]
- 77. Ezashi T., Telugu B. P. V. L., Roberts R. M. (2012) Model systems for studying trophoblast differentiation from human pluripotent stem cells. Cell Tissue Res. 349, 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang P., Li J., Tan Z., Wang C., Liu T., Chen L., Yong J., Jiang W., Sun X., Du L., Ding M., Deng H. (2008) Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood 111, 1933–1941 [DOI] [PubMed] [Google Scholar]
- 79. Yu P., Pan G., Yu J., Thomson J. A. (2011) FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell 8, 326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pera M. F., Andrade J., Houssami S., Reubinoff B., Trounson A., Stanley E. G., Ward-van Oostwaard D., Mummery C. (2004) Regulation of human embryonic stem cell differentiation by BMP-2 and its antagonist noggin. J. Cell Sci. 117, 1269–1280 [DOI] [PubMed] [Google Scholar]
- 81. Peerani R., Rao B. M., Bauwens C., Yin T., Wood G. A., Nagy A., Kumacheva E., Zandstra P. W. (2007) Niche-mediated control of human embryonic stem cell self-renewal and differentiation. EMBO J. 26, 4744–4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kee K., Gonsalves J. M., Clark A. T., Pera R. A. R. (2006) Bone morphogenetic proteins induce germ cell differentiation from human embryonic stem cells. Stem Cells Dev. 15, 831–837 [DOI] [PubMed] [Google Scholar]
- 83. West F. D., Roche-Rios M. I., Abraham S., Rao R. R., Natrajan M. S., Bacanamwo M., Stice S. L. (2010) KIT ligand and bone morphogenetic protein signaling enhances human embryonic stem cell to germ-like cell differentiation. Hum. Reprod. 25, 168–178 [DOI] [PubMed] [Google Scholar]
- 84. Beppu H., Kawabata M., Hamamoto T., Chytil A., Minowa O., Noda T., Miyazono K. (2000) BMP type II receptor is required for gastrulation and early development of mouse embryos. Dev. Biol. 221, 249–258 [DOI] [PubMed] [Google Scholar]
- 85. Mishina Y., Suzuki A., Ueno N., Behringer R. R. (1995) Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 9, 3027–3037 [DOI] [PubMed] [Google Scholar]
- 86. Home P., Saha B., Ray S., Dutta D., Gunewardena S., Yoo B., Pal A., Vivian J. L., Larson M., Petroff M., Gallagher P. G., Schulz V. P., White K. L., Golos T. G., Behr B., Paul S. (2012) Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc. Natl. Acad. Sci. U.S.A. 109, 7362–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Laurila R., Parkkila S., Isola J., Kallioniemi A., Alarmo E.-L. (2013) The expression patterns of gremlin 1 and noggin in normal adult and tumor tissues. Int. J. Clin. Exp. Pathol. 6, 1400–1408 [PMC free article] [PubMed] [Google Scholar]
- 88. Xu J., Cheung T. M., Chan S. T., Ho P. C., Yeung W. S. (2000) Human oviductal cells reduce the incidence of apoptosis in cocultured mouse embryos. Fertil. Steril. 74, 1215–1219 [DOI] [PubMed] [Google Scholar]
- 89. Chambers S. M., Fasano C. A., Papapetrou E. P., Tomishima M., Sadelain M., Studer L. (2009) Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Patani R., Compston A., Puddifoot C. A., Wyllie D. J. A., Hardingham G. E., Allen N. D., Chandran S. (2009) Activin/Nodal inhibition alone accelerates highly efficient neural conversion from human embryonic stem cells and imposes a caudal positional identity. PLoS One 4, e7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wilson P. A., Hemmati-Brivanlou A. (1995) Induction of epidermis and inhibition of neural fate by Bmp-4. Nature 376, 331–333 [DOI] [PubMed] [Google Scholar]
- 92. Trigal B., Gómez E., Díez C., Caamaño J. N., Martín D., Carrocera S., Muñoz M. (2011) In vitro development of bovine embryos cultured with activin A. Theriogenology 75, 584–588 [DOI] [PubMed] [Google Scholar]
- 93. Lee K.-B., Bettegowda A., Wee G., Ireland J. J., Smith G. W. (2009) Molecular determinants of oocyte competence: potential functional role for maternal (oocyte-derived) follistatin in promoting bovine early embryogenesis. Endocrinology 150, 2463–2471 [DOI] [PubMed] [Google Scholar]
- 94. Refaat B., Ledger W. (2011) The expression of activins, their type II receptors and follistatin in human Fallopian tube during the menstrual cycle and in pseudo-pregnancy. Hum. Reprod. 26, 3346–3354 [DOI] [PubMed] [Google Scholar]
- 95. Argañaraz M. E., Apichela S. A., Miceli D. C. (2012) LEFTY2 expression and localization in rat oviduct during early pregnancy. Zygote 20, 53–60 [DOI] [PubMed] [Google Scholar]
- 96. Argañaraz M. E., Valdecantos P. A., Abate C. M., Miceli D. C. (2007) Rat Lefty2/EBAF gene: isolation and expression analysis in the oviduct during the early pregnancy stage. Genes Genet. Syst. 82, 171–175 [DOI] [PubMed] [Google Scholar]
- 97. Lai Y. M., Wang H. S., Lee C. L., Lee J. D., Huang H. Y., Chang F. H., Lee J. F., Soong Y. K. (1996) Insulin-like growth factor-binding proteins produced by Vero cells, human oviductal cells and human endometrial cells, and the role of insulin-like growth factor-binding protein-3 in mouse embryo co-culture systems. Hum. Reprod. 11, 1281–1286 [DOI] [PubMed] [Google Scholar]
- 98. Salilew-Wondim D., Schellander K., Hoelker M., Tesfaye D. (2012) Oviductal, endometrial and embryonic gene expression patterns as molecular clues for pregnancy establishment. Anim. Reprod. Sci. 134, 9–18 [DOI] [PubMed] [Google Scholar]
- 99. Minami N., Kato H., Inoue Y., Yamada M., Utsumi K., Iritani A. (1994) Nonspecies-specific effects of mouse oviducts on the development of bovine IVM/IVF embryos by a serum free co-culture. Theriogenology 41, 1435–1445 [DOI] [PubMed] [Google Scholar]
- 100. Lloyd R. E., Romar R., Matás C., Gutiérrez-Adán A., Holt W. V., Coy P. (2009) Effects of oviductal fluid on the development, quality, and gene expression of porcine blastocysts produced in vitro. Reproduction 137, 679–687 [DOI] [PubMed] [Google Scholar]
- 101. Yadav P. S., Saini A., Kumar A., Jain G. C. (1998) Effect of oviductal cell co-culture on cleavage and development of goat IVF embryos. Anim. Reprod. Sci. 51, 301–306 [DOI] [PubMed] [Google Scholar]
- 102. Besser D. (2004) Expression of nodal, lefty-a, and lefty-B in undifferentiated human embryonic stem cells requires activation of Smad2/3. J. Biol. Chem. 279, 45076–45084 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.