Abstract
The pathogenesis of systemic lupus erythematosus (SLE) is multi-factorial, and the interferon regulatory factors (IRFs) play an important role. Autoantibodies formed in SLE target nuclear antigens, and immune complexes formed by these antibodies contain nucleic acid. These immune complexes can activate anti-viral pattern-recognition receptors (PRRs), resulting in the downstream activation of interferon regulatory factors (IRFs), which can induce type I interferon and other inflammatory mediators. Genetic variations in IRFs have been associated with susceptibility to SLE, and current evidence supports the idea that these polymorphisms are gain-of-function in humans. Recent studies suggest that these genetic variations contribute to the break in humoral tolerance that allows for nucleic acid binding autoantibodies, and that the same polymorphisms also augment type I IFN production in the presence of these autoantibody immune complexes, forming a feed-forward loop. In this review, we will outline major features of the PRR/IRF systems and describe the role of the IRFs in human SLE pathogenesis.
Introduction
Systemic lupus erythematosus (SLE) is a chronic complex multisystem autoimmune disease that often arises during early adulthood, resulting in great morbidity. Clinical symptomology varies between individuals and within individuals over time, as inflammation often affects multiple organs and tissues. Almost any organ system can be involved, and sites commonly impacted by disease include the joints, skin, lungs, neurological system, kidneys, and hematological systems 1. Antibodies reactive to nucleic acids and nuclear proteins are prominent in lupus subjects and most individuals have circulating antibodies to numerous autoantigens. Antinuclear antibodies found in the disease can be reactive to nucleosomes, double-strand DNA, Ro/SSA, Smith, small nuclear ribonucleoproteins, and La/SSB, with up to 90% of patients having reactivity against nucleosomes 1. Disease is thought to result from a break in immune tolerance to self-proteins, including DNA and other nuclear proteins, with both genetic and environmental factors contributing to disease susceptibility and symptomology.
Though the etiology of SLE is unknown, many individuals with SLE show defects in their ability to clear apoptotic cells and cellular debris, suggesting that an excess of nucleic material contributes to autoreactivity to nuclear antigens 2-10. Reactivity to nuclear material leads to the formation of circulating immune complexes that serve as a stimulus for the production of type I interferon by plasmacytoid dendritic cells (PDCs) 11-14. Immune complexes are internalized in PDCs via surface expressed Fc receptors and traffic to endosomal compartments where the nucleic acid contained within them binds to toll like receptors (TLRs) that recognize RNA (TLR7, TLR8) and DNA (TLR9). TLR7, TLR8, and TLR9 triggering activates members of interferon regulatory factor (IRFs) family of transcription factors including IRF1, IRF3, IRF5, IRF7, and IRF9, that are positive regulators of type I interferons (IFN-1) 15,16-21. Uptake of immune complexes is thought to trigger IFN-1 production by PDCs, and elevated levels of type I IFNs are detectable in the blood of roughly half of SLE individuals 22-26. This concept is supported by the fact that the strongest predictor of high type I IFN levels in human SLE patients is the presence of antibodies targeting DNA and RNA-binding proteins 27. Interestingly, these autoantibodies are not sufficient for high serum type I IFN 28, suggesting that there are also disease-associated factors that modify the association between nucleic acid ICs and high IFN in SLE. One likely factor is intrinsic over-activity within the TLR signaling system. Human genetic variants in IRF5, IRF7, and IRF8 have been implicated in SLE susceptibility 29-36, suggesting that gain-of-function polymorphisms in these molecules augment TLR signaling. Many studies have placed the IRF family as central mediators in human SLE 37. In this review, we will summarize the TLR/IRF/type I IFN pathways and the role of this system in human SLE pathogenesis.
Type I IFN in SLE
Type I IFNs are classically involved in viral defense, and induce activation of antigen presenting cells and increased expression of MHC and co-stimulatory molecules. Type I IFN has been implicated as a primary pathogenic factor in human SLE by various lines of evidence 38. High type I IFN levels are clustered in SLE families as a heritable trait 39, 40, and this clustering supporting heritability is not observed with other SLE-associated cytokines such as tumor necrosis factor alpha 41, 42. Many of the genetic factors associated with SLE function within the type I IFN pathway 43, with clear over-representation of the IRF family as noted above (three of the nine IRF family members have genetic variants associated with SLE). When SLE-risk polymorphisms in IFN pathway genes have been investigated functionally in humans, they have been associated with increased type I IFN pathway signaling 44-46, supporting the idea that gain-of-function variants in the type I IFN pathway underlie human SLE 47, 48. Additionally, human subjects given recombinant human type I IFN in the form of IFN-α2 as a treatment for malignancy or chronic viral infection have developed de novo SLE 49, which has generally resolved when the IFN-α was discontinued 50. These data all support a causal role for type I IFN in human SLE. There are two major categories of type I interferons, alpha and beta, with the majority of circulating IFN activity in SLE attributable to the IFN-α isoforms 39, 51, 52. A prominent IFN gene signature, comprised of genes strongly activated by type I IFNs (interferon stimulated genes (ISGs)), is present in circulating peripheral blood mononuclear cells (PBMC) in a sub group of SLE patients and is associated with the presence of anti-dsDNA antibodies, elevated IFN levels, and worse disease activity 23, 24, 51, 53. Type I IFNs also induce members of the STAT and IRF transcription factor families, including IRF5 and IRF7, and which can then influence the expression of extended gene networks 54.
Type I IFNs (IFN-α and IFN-β) bind to a single receptor; a heterodimer (IFNAR) consisting of IFNAR1 and IFNAR2 chains. Additionally, IFN-β has the capacity to bind to and trigger signaling via the IFNAR1 chain in the absence of the IFNAR2 chain 55. IFNAR signaling activates the receptor-associated kinases Janus kinase I (JAKI) and tyrosine kinase 2 (TYK2) that then phosphorylate STAT1 and STAT2 (pSTAT1 and pSTAT2) 56. Heterodimers of pSTAT1 and pSTAT2 translocate to the nucleus and conjoin with interferon regulatory factor 9 (IRF9) to form IFN-stimulated gene factor 3 (ISGF3) 56, 57. ISGF3 binds to DNA-sequences called IFN-stimulated response elements (ISREs; consensus sequence TTTCNNTTTC) present in the promoters of many ISGs inducing their transcription. Most cells are competent to express low levels of early or immediate IFN-1 genes, including IFN-α4 and IFN-β, which then bind to the IFN receptor, an autocrine or paracrine manner, to activate the JAK-STAT pathway leading to ISGF3 formation 58. IRF1, IRF3, and IRF5 have each been shown to play essential roles in different in vitro assays in the induction of this early IFN response 58. ISGF3 induces the expression of IRF7 which is required for the induction of high levels of late or delayed type I IFNs by plasmacytoid dendritic cells (PDCs) downstream of multiple PRR pathways 58-60. Thus the following sequence of events is thought to occur in SLE, leading to a positive feedback loop. Nucleic acids bind to cytoplasmic or transmembrane expressed PRRs, resulting in the induction of early type I IFNs, which bind to the type I IFN receptor. This primes the cells by inducing IRFs such as IRF5, IRF7, IRF9, as well as STAT family members, and any further triggering of PRRs in the cells leads to induction of high levels of late IFN-1s. Multiple IRFs, including IRF3, IRF5, and IRF7 are integral to the pattern-recognition receptors (PRRs) response and the IFN response within this cycle 61.
Pattern Recognition Receptors
Pattern recognition receptors (PRRs) were initially characterized as sensing receptors that recognized pathogen associated molecular patterns (PAMPs) associated with fungi, bacteria, flagella, and viruses, and are now understood to also recognize damage associated molecular patterns (DAMPs) released by damaged cells and tissues 62. PRRs may be broadly subdivided into membrane bound types that include the toll like receptors (TLRs) and c-type lectin receptors (CLRs); cytoplasmic sensors that include NOD like receptors (NLRs), pyrin, and HIN domain-containing members (PYHIN), RIG-I-like receptors (RLRs), and additional cytoplasmic and nuclear nucleic acid sensing receptors 63. Several PRRs that recognize nucleic acids (cytosolic RLRs, cytosolic DNA sensors, and several members of the TLRs) are of particular interest in understanding the pathogenesis of SLE in that nucleic acids (and the proteins that bind nucleic acids, nucleosomal proteins) are targets of the autoreactive response in SLE patients, and triggering these PRRs induces type I IFN production 64. Events that lead to the initial break in tolerance and reactivity to nuclear matter in SLE and also to the ongoing reactivity to nucleic acid in lupus patients are thought to likely involve multiple types of nucleic acid sensing PRRs that signal through members of the IRF family 65.
Nucleic Acid Sensing TLRs
Nucleic acid sensing TLRs are expressed within endosomes and include TLR3 and TLR7-9 66. TLR3 binds dsRNA and synthetic dsRNA analogues such as poly I:C. TLR7 and TLR8 recognize single strand viral RNA, self RNA bound in immune complexes, and imidazoquiloline derivatives such as resiquimod (R848). Though TLR7 and TLR8 both bind ssRNA, TLR7 preferentially binds ssRNA rich in GU while TLR8 is activated with ssRNA rich in AU. TLR9 binds dsDNA and oligonucleotides that have a high content of unmethylated CpG motifs resembling bacterial DNA. The CpG motifs have been thought to be key PAMP structures associated TLR9 sensing, however mammalian DNA low in CpG motifs can also potently activate TLR9 when efficiently translocated to endosomes 67. Some studies indicate that the key structure in DNA associated with TLR9 recognition appears to be the 2′ deoxyribose phosphate backbone 67-69. TLR3 signals through the adaptor TRIF, which associates with TRAF3. TRAF3 associates with TBK1 and IKK-ε which phosphorylates IRF3 leading to IFN-β production. TLR7-9 signal through the adapter MyD88 and IRAK4 which complex with TRAF6, TRAF3, osteopontin and the kinases IRAK1 and IKK-α that phosphorylate IRF7 in PDCs and IRF5 in macrophages and myeloid dendritic cells (MDCs).
Cytosolic Pattern Recognition Receptors
The two main cytosolic RNA sensing PRRs that sense intracellular RNA are the RLRs, retinoic acid-inducible gene 1 (RIG-1) and melanoma differentiation factor 5 (MDA5) 70. RIG-1 recognizes single-stranded RNA (sRNA) that contains a 5′-triphosphate moiety and short double stranded RNA (dsRNA) while MDA5 is efficiently activated by long dsRNA 71. Both sensors signal through adapter mitochondrial antiviral signaling (MAVS; also known as IPS-1/CARDIF/VISA) that leads to activation of the kinases, TANK-binding kinase 1 (TBK1) and inducible kinase IκB kinase (IKK-i; also known as IKK-ε) that then phosphorylate IRF3 and IRF7 which induce IFN-1 genes 72, 73. IRF5 also appears able to activate the early IFN-1 response downstream of MAVS independent of IRF3 and IRF7 20, 74. Importantly, gain of function mutations in MDA5 lead to a lupus like disease in mice 75. Furthermore, polymorphisms in or near IFIH1, the gene encoding MDA5, leads to increased susceptibility to lupus and patients with anti-dsDNA antibodies have an heightened IFN gene signature that is associated with increased responsiveness to type I IFNs 46, 76. Additionally, loss-of-function variants in MAVS result in lower type I IFN levels in SLE patients 77, further supporting the importance of the cytosolic PRR pathways in human SLE.
DNA sensing PRRs are less well understood than are the RNA sensors but several candidate sensors have been identified and include DNA-dependent activator of interferon regulator factors (DAI, also known as ZBP1), DEAD box polypeptide 41 (DDX41), interferon-γ inducible protein 16 (IFI16), cyclic guanosine monophosphate-adenosine monophosphate synthase (cGAS), meiotic recombination homolog 11a (MRE11), and absent in melanoma (Aim2) 78-90. Signaling through DAI, DDX41, IFI16, MRE11, and cGAS leads to IRF3-dependent IFN production while NLRs and Aim2 signaling leads to inflammasome activation. In addition to the sensors listed above, cytosolic double stranded DNA (poly dA-dT) is detected by RNA polymerase III which transcribes the dsDNA into poly (A-U) RNA that results in IFN production by the RIG-1/MAVS pathway described above 91.
IFI16, DDX41, cGAS, MRE11, and likely DAI, all induce type I IFN expression in a stimulator of IFN genes (STING) dependent manner 89, 92. Importantly, DNA-bound cGAS synthesizes cyclic GMP-AMP which binds STING and acts as intermediary between cGAS and STING and possibly for other DNA sensors as well 82, 83. STING (also known as MITA/MPYS/ERIS) provides a scaffold for bringing IRF3 in close proximity to tank binding kinase (TBK1) which phosphorylates IRF3 leading to its activation and downstream activation of type I IFN gene expression. Triggering of these sensors by poorly cleared cytoplasmic DNA appears to be linked to lupus type diseases in humans and in mice in studies involving mutations in the exonuclease Trex1. Trex1 digests single-stranded DNA in the cytoplasm arising from the retrotranscription of endogenous retroelements 93, 94. In humans, rare loss of function mutations in Trex1 cause Aicardi-Goutieres syndrome (AGS) and chilblain lupus, while other more common variants are associated with SLE 95-99. TREX1 deficient mice show increased levels of cytoplasmic DNA, develop myocarditis and lupus-like symptoms including multi-organ inflammation and renal IgG deposition that are dependent on host expression of IRF3 and associated with anti-nuclear antibodies and an IFN gene signature 94, 100. An IFN gene signature is still detectable in heart tissue of Trex−/−Rag2−/− mice suggesting that an increased type I IFN response precedes the autoreactive adaptive immune response in these mice 94, 100.
Interferon Regulatory Factors
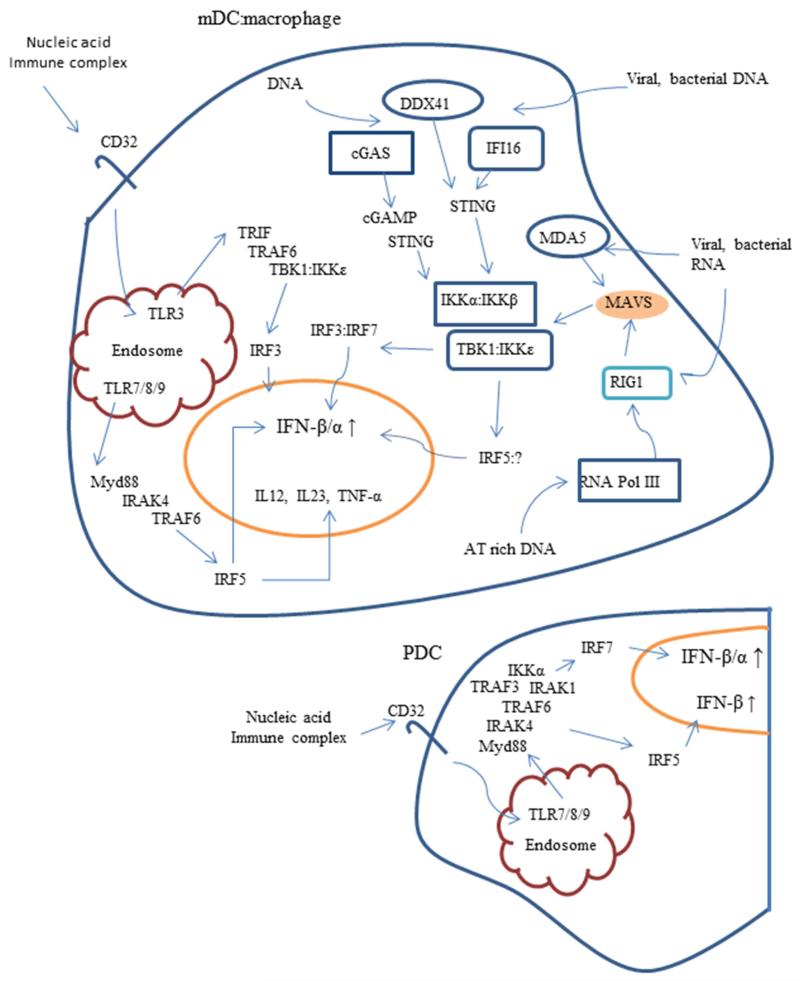
Interferon regulatory factors (IRFs) comprise a family of transcription factors that include nine members, IRF1, IRF2, IRF3, IRF4 (also known as LSIRE, PIP or ICSAT), IRF5, IRF6, IRF7, IRF8 (also known as ICSBP), and IRF9 (also known as ISGFγ) in humans and mice (reviewed in 65, 101). IRFs play prominent roles in the signaling cascades of pattern-recognition receptors (PRRs) such as TLRs that induce type I IFNs, cytokine secretion, cellular apoptosis, and cell activation and differentiation. In 1998, X-ray crystallographic studies showed that the N-terminal domain of IRF1, the first IRF family member discovered bound to a sequence 5′-GAAA-3′ within the promoter region of IFNβ. The N-terminal region of IRF1 forms a helix-turn-helix structure that is common among DNA-binding proteins and is referred to as the N-terminal DNA binding domain (NDB) 102. Further studies on other IRF family members defined the NDB has a region approximately 120 amino acids shared among all IRFs family members that binds to the core recognition sequence, GAAANNGAAAG/CT/C, where N denotes any nucleotide 103, 104. IRF members, except for IRF1 and IRF2, also contain a C-terminal IRF association domain (IAD). The IAD allows for homodimeric interactions or heterodimeric binding to other IRF family members. Some IRF members have also been shown to interact with members of the ETS, STAT, and AP1 family members via binding to this region. A conserved auto inhibitory region blocks IAD binding which is relieved by phosphorylation of key residues within this region in several IRF family members (including IRF3, IRF4, IRF5, and IRF7) 105-108. IRF family members regulate the expression of numerous genes, including the type I IFNs (IFN-β and IFN-α) and genes induced by type I IFNs (IFN-stimulated genes, ISGs), via different signaling pathways in cell specific manners. A diagram outlining interactions between PRRs and IRFs is shown in Figure 1.
Figure 1.
Pattern Recognition Receptor IFN-α Response to Nucleic Acids MDCs/Macrophages and PDCs. In MDCs and macrophages, nucleic acid receptors are detected by RNA and DNA sensing receptors in the cytoplasm and by Toll like receptors (TLR3, TLR7, TLR8, and TLR9) that detect nucleic acids and signal within endolysosomes. The cytosolic DNA sensors, cGAS, DDX41, and IFI16 signal through STING to activate the kinase TBK1 that then phosphorylates IRF3 that then leads to IFN-1 production. cGAS, once activated induces synthesis of cGAMP which binds to and activates STING. The two cytosolic RNA receptors, MDA5 and RIG1, signal through MAVS to activate the kinases TBK1 and IKKε that then phosphorylate IRF3 and IRF7 respectively, and also IRF5 under certain conditions. TLR3, TLR7 and TLR8 sense RNA and TLR9 sensed DNA when imported to endolysosomes by absorptive receptors, such as Fc receptors expressed by dendritic cells and macrophages. TLR3 signals through TRIF and TRAF6 to activate IRF3 while TLR7, TLR8, and TLR9 signal through Myd88 to activate IRF5. PDCs have the capacity to produce large amounts of IFN-α when activated through the TLR7 and TLR9 receptors by signaling through IRF7. TLR triggering in these cells also seems able to activate IRF5 leading to production of IFN-β.
Genetic polymorphisms in several molecules of the nucleic acid sensing PRR pathways have been associated with SLE susceptibility. As noted above, three members of the IRF family have been implicated as genetic risk factors for in SLE, and these will be dealt with in detail in later sections. Additionally, genetic polymorphisms in other genes involved in PRR pathways have been reported, and these are summarized in Table 1. These PRR pathway gene polymorphisms include a variant of the RNA sensor, IFIH1, tagged by rs1990760 that shows genetic association for the presence of anti-dsDNA antibodies and also for lower levels of type I IFN levels in individuals with anti-dsDNA antibodies (i.e. dsDNA antibody positive and IFN low)34, 109. Though these individuals have lower IFN levels, the IFIH1 variant also associates with increased cellular response to type I IFNs as measured by IFN-induced gene expression in peripheral blood mononuclear cells, suggesting an effect on sensitivity to IFN signaling109. A STAT4 variant shows a similar association with low IFN levels and increased cellular response to type I IFNs in SLE patients as does the IFIH1 variant, and these two genetic effects are independent46109. SLE-risk genes not directly associated with PRR signaling pathways may also influence the type I IFN pathway in SLE patients. For example, methyl CpG binding protein 2 (MECP2) represses gene transcription by affecting DNA methylation and histone acetylation, but may also augment gene expression by recruiting CREB1 to chromatin. An SLE-risk polymorphism in the MECP2 gene is associated with altered transcript levels in PBMC of lupus patients 110-112. This variant appears to modulate gene expression differently than the non-risk MECP2 variant and its presence is linked to increased expression of a wide range of genes, many of which are induced by type I IFNs111, 112. Collectively, these studies emphasize the importance of PRR pathways in human SLE pathogenesis, and ways in which different genetic polymorphisms may be tuning the PRR and IFN pathways in SLE patients. Studies of the IRF family of transcription factors in humans provide further support for this idea, and we will discuss the SLE-associated IRFs in detail next.
Table 1.
Proteins involved in IRF signaling pathways that have genetic variants linked to SLE susceptibility
| Protein | Gene | Name | IRF | Function/V ariant Specific Effects |
|---|---|---|---|---|
| TNFAIP3 (A20) |
TNFAIP3 | tumor necrosis factor alpha inducible protein 3 |
3, 4, 5 | ubiquitin-modifying enzyme that limits NF-κB signaling following TNF-α, TLR, IL-1R, and NOD2 signaling; risk variant associated with reduced transcript and protein levels113, 114 |
| ABIN1 | tnipl | A20-binding inhibitor of NF-κB 1 (also TNFAIP3 interacting protein 1) |
3, 4, 5 | facilitates IKKλ, ubiquitination by recruiting A20 to IKKγ (NEMO); limits NF-κB signaling; two independent risk loci linked to lower TNIP1 transcripts and ABIN1 protein levels31, 115 |
| IRAK1 | irakl | interleukin-1 receptor related kinase-1 | 5, 7 | essential kinase for IRF5 and IRF7 activation in the TLR7/8/9 signaling pathways;19, 116 variant linked to increased NF-κB signaling112 |
| TREX1 | trex1 | three prime repair exonuclease | 3, 5, 7, | cytosolic 3′–5′ DNA exonuclease’ (DNAse III) risk variant = loss of function;95, 99 variant may lead to increased levels of cytosolic nucleic acid |
| STAT4 | stat4 | signal transducer and activator of transcription-4 |
risk variant associated with increased response to type I IFN and with ischaemic cerebro- vascular events and antibodies to phospholipids and dsDNA;117-120 |
|
| IFIH1 (MDA5) |
ifihl | IFN induced with helicase C domain 1 |
3, 5, 7 | cytosolic RNA sensor; variant associates with an increased response to type I IFN in SLE patients that have dsDNA antibody titers34, 46, 121 |
| PTPN22 | ptpn22 | protein tyrosine phosphatase nonreceptor 22 |
3, 7 | interacts with TRAF3 to augment type I IFN response, risk variant associates with increased circulating levels of type I IFNs31, 122, 123 |
| MAVS | mavs | mitochondrial anti-viral signaling | 3, 5, 7 | signals from RIG1 and from MDA5; loss of function variant associated with reduced protein levels of IFN-α and the lack of antibody titers to RNA associated proteins109, 124 |
IRF5
IRF5 is a key regulator of numerous biological functions that contribute to the pathogenesis of SLE. Early studies using IRF deficient mice showed a definite role for IRF5 in TLR-mediated production of pro-inflammatory cytokines including IL-6, TNF-α, and IL-12 60, 125. High expression levels of IRF5 are associated with macrophage polarization and conversion from M2 to M1 macrophages 126. In vitro studies of virally infected or TLR7 triggered human cells showed that IRF5 plays a role in induction of type I IFNs and that IRF5 homodimers or heterodimers formed between IRF5 and other IRF members bind to promoter regions of specific IFN genes and selectively induced the transcription of different IFN gene members including IFN-β 127. In vivo, IRF5 contributes to IFN production in virally infected mice in an MAVS dependent manner 20. IRF5 also influences B cell responses by regulating transcription of B-lymphocyte-induced maturation protein (Blimp-1) crucial for B cell maturation and plasma cell formation, promoting B cell switching to the pathogenic IgG2a isotype, and possibly through induction of increased circulating levels of Blys 128. Finally, IRF5 appears to be strongly proapoptotic and promotes DNA damage-induced apoptosis 129-131.
Several genetic polymorphisms in IRF5 are associated with SLE susceptibility, and with increased circulating type I IFN levels 132, 133. The IRF5 gene structure has 4 non-coding first exons (in sequence order of exon 1d, 1a, 1b, and 1c) that are alternatively used and 8 coding exons (exons 2 – 9). Each first exon has its own promoter that directs transcription of the IRF5 gene to yield more than 14 different mRNA isoforms expressed in cell specific manners, resulting in significant complexity in the IRF5 gene transcriptome 134. The genetic epidemiology of IRF5 in SLE is similarly complex, as there are multiple polymorphisms which occur together in combinations on various haplotypes that result in graded degrees of SLE risk. Functional polymorphisms on IRF5 haplotypes include a promoter insertion/deletion, a splice site variation that allows exon 1b to be spliced in, two coding change variations in exon 6, and a 3′ UTR polymorphism that results in alternate polyadenylation 135-138 (Table 2). These functional genetic variations come together on chromosomes in particular combinations (haplotypes), and various combinations are associated with risk of SLE the formation of anti-Ro, anti-La, and anti-dsDNA autoantibodies 30 (Table 3). Interestingly, when type I IFN levels are examined in the context of IRF5 genotype, elevations in type I IFN related to IRF5 genotype are only observed in patients who have one of these associated autoantibodies 30. This suggests a model in which Gene + Autoantibody = High IFN, and would fit an induced model of function for SLE-associated variants in IRF5, in which the autoantibodies provide a chronic endogenous stimulus which along with IRF5 variants is sufficient to result in dysregulation of circulating IFN levels 37. When case-control genetic data for IRF5 are re-analyzed with respect to autoantibodies, >70% of the genetic risk of SLE related to IRF5 is carried within these two serologic subgroups of patients, which compose approximately half of the SLE population 30. And it is notable that this gene is one of the strongest overall case-control genetic associations in SLE, despite this strong subgroup effect. While this subset effect could relate to the effect of the gene-antibody interaction upon IFN levels, IRF5 variants could also predispose to autoantibody production, and this hypothesis could fit the data as well. TLRs are important in B cell differentiation, and it is plausible that IRFs could augment TLR signaling in B cells resulting in an increased propensity to autoantibody production. In a study that explores this question in humans, Cherian et al studied IRF5 genotypes in a group of subjects who had SLE-associated autoantibodies but were otherwise healthy to assess whether IRF5 was associated with serologic immunity in the absence of SLE disease and high IFN 139. They found that IRF5 haplotypes were enriched in serologically positive healthy individuals to the same degree as SLE patients, suggesting that IRF5 is involved in serologic autoimmunity in humans 139. This supports a model in which the SLE-associated variants in IRF5 predispose to autoantibodies that can bind nucleic acid, and then these same autoantibodies form immune complexes which trigger the TLR system in innate immune cells, resulting in over-production of IFN in the setting of the same genetic variants. This would be an example of the same variant functioning in the TLR system in two different immune cell types, with a synergistic or feed-forward effect in which the product of the hyperactive TLR system in B cells provides a chronic TLR stimulus to innate immune cells with a similarly hyperactive TLR system. The complexity of IRF5 genetics and the functional effects described thus far is impressive, and these are summarized in Table 2.
Table 2.
IRF5 variants linked to susceptibility to SLE
| SNP or variant | gene effect | disease impact | functional impact |
|---|---|---|---|
| promoter CGGGG/- insertion | CGGGG insertion 64 bp upstream exon 1a that creates a Sp1 binding site; in LD with rs10954213 and rs2004640135 |
increased risk of SLE MS, IBD, and Crohn’s |
increased mRNA and protein levels |
| rs2004640 | novel site that allows exon1b splicing to exon 2 with additional IRF5 mRNA isoforms137 |
increased risk | increased protein levels and altered IRF5 isoforms134 |
| 30 bp in-frame insertion in exon 6 |
occurs within the PEST domain | risk and protective haplotypes136 |
affects protein stability; alters binding of TRIM21 to IRF5140 |
| rs10954213 | altered polyadenylation signal;137 shortened poly A tail and greater mRNA stability |
increased risk | increase mRNA levels |
Table 3.
Major IRF5 haplotypes in European ancestry, showing the combinations of functional genetic polymorphisms present and disease associations in SLE
| Promoter indel |
Splice Site Variation |
Exon 6 indel | Alternate poly-a site |
Disease associations |
|---|---|---|---|---|
| + | + | + | + | Risk of disease, anti-Ro and anti-dsDNA antibodies, high IFN |
| + | + | − | + | Disease risk in seropositive individuals, anti-dsDNA antibodies, high IFN |
| − | + | + | − | Anti-La antibodies |
| − | − | − | + | Protective haplotype |
| − | − | + | − | Protective haplotype |
means indel present, or novel splice site or alternate poly-A site present
IRF7
IRF7 functions downstream of the endosomal TLRs via the MyD88 adaptor protein, and is activated by phosphorylation 141. In addition to phosphorylation, ubiquitination likely plays a role in IRF7 regulation as well 141. IRF7 is a major transcription factor involved in type I IFN production in plasmacytoid dendritic cells, and can cooperate with IRF3 to induce robust IFN production following viral infection 17. Genetic variations in IRF7 have been associated with SLE susceptibility 142, 143. The genetic polymorphisms in IRF7 that are associated with SLE are also associated with anti-dsDNA and anti-RNA-binding protein antibodies 144, similar to the above situation with IRF5. In a similar parallel fashion, genetic variations in IRF7 are associated with increased type I IFN only in the presence of these specific SLE-associated autoantibodies. These data further support an induced model of IRFs impacting circulating IFN levels, in which the autoantibody immune complexes presumably act as a chronic endogenous stimulus. This parallel relationship between IRF5 and IRF7 in human SLE strengthens the confidence in this mechanism as a common pathogenic route in SLE. In the case of both IRF5 and IRF7, these associations between genotype and cytokine phenotype extended across difference ancestral backgrounds.
IRF8
IRF8 is another member of the IRF family which can be activated downstream of PRRs. IRF8 differs in that it does not interact directly with MyD88, but is important to signals downstream of TLR9, as IRF8 deficiency greatly reduces TLR9-induced cytokine production by murine dendritic cells 145. In humans, IRF8 deficiency results in an immunodeficiency characterized by the loss of monocytes and dendritic cells 146, supporting a role for IRF8 in monocyte and dendritic cell development. IRF8 is involved in transcriptional responses to type I IFNs, and can induce transcription of inflammatory cytokines 147. Genetic variations in IRF8 have been associated with susceptibility to both SLE and multiple sclerosis (MS), although the specific variants associated with each disease differ somewhat 35, 148. Type I IFNs play opposite roles in MS and SLE, while IFN-α is elevated and thought to be causal in SLE, type I IFN levels are lower in MS patients than in healthy controls 149, 150, and recombinant human IFN-β is commonly used as a therapeutic in multiple sclerosis. While both SLE and MS are considered autoimmune diseases, it seems likely that stark differences exist between the two conditions in the regulation of type I IFNs 151. Given this background, it is interesting that genetic variations in IRF8 are associated with both of these autoimmune conditions. Type I IFN levels were not different in SLE patients in the context of the SLE-associated genetic polymorphisms 36. Interestingly, the MS-associated genetic variant was associated with decreased type I IFN levels in both SLE and MS, and was associated with anti-dsDNA antibodies and increased IRF8 in B cells in the SLE patients 36. These data suggest that the MS-associated allele of IRF8 contributes to autoimmunity in the setting of low type I IFN levels, and may play a role in humoral autoimmunity given the antibody and B-cell associations. It is possible that the SLE-associated allele exerts a different influence on the immune system which was not assessed in the study referenced above.
Conclusions
These studies illustrate how deeply interwoven the IRF family of transcription factors is within human SLE. IRFs provide a link between the autoantibodies formed in SLE and the characteristic activation of the type I IFN pathway observed in the disease. In fact, the studies also support involvement of the IRFs in the initial break in humoral tolerance, as in each case the IRF variant are associated with specific autoantibodies. The fact that SLE-associated variants in IRF5 are associated with these antibodies in the absence of disease 139, and that the SLE-associated variant of IRF8 alters IRF8 expression specifically in B cells support the concept that IRFs play a primary role in humoral autoimmunity in addition to aberrant type I IFN production in SLE. This suggests a feed-forward loop, in which the IRF variations predispose to autoantibody formation in SLE, possibly via effects in B cells. Then immune complexes made from these autoantibodies provide a chronic stimulus to PRRs, and the same gain-of-function genetic variations result in increased type I IFN production. This provides a fascinating paradigm in human autoimmune disease pathogenesis, that the same gain-of-function polymorphisms could play multiple roles across different immune cell types, with activation of the TLR/IRF pathway in one cell producing a chronic stimulus to that same pathway in another cell type. This type of feed-forward model could explain the particular relevance of the IRF family of transcription factors in human SLE.
Acknowledgements
Funding Sources: TB Niewold – Research grants from the NIH (AR060861, AI083790, AI071651), Rheumatology Research Foundation, the Mayo Clinic Foundation, and the Lupus Foundation of Minnesota.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts of interest and have read the journal of Translational Research authorship agreement. The manuscript was prepared by and approved by both authors.
References
- 1.Goldblatt F, O’Neill SG. Clinical aspects of autoimmune rheumatic diseases. Lancet. 2013;382:797–808. doi: 10.1016/S0140-6736(13)61499-3. [DOI] [PubMed] [Google Scholar]
- 2.Perniok A, Wedekind F, Herrmann M, Specker C, Schneider M. High levels of circulating early apoptic peripheral blood mononuclear cells in systemic lupus erythematosus. Lupus. 1998;7:113–118. doi: 10.1191/096120398678919804. [DOI] [PubMed] [Google Scholar]
- 3.Poon IK, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol. 2014;14:166–180. doi: 10.1038/nri3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casciola-Rosen LA, Anhalt G, Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med. 1994;179:1317–1330. doi: 10.1084/jem.179.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mevorach D, Mascarenhas JO, Gershov D, Elkon KB. Complement-dependent clearance of apoptotic cells by human macrophages. J Exp Med. 1998;188:2313–2320. doi: 10.1084/jem.188.12.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korb LC, Ahearn JM. C1q binds directly and specifically to surface blebs of apoptotic human keratinocytes: complement deficiency and systemic lupus erythematosus revisited. J Immunol. 1997;158:4525–4528. [PubMed] [Google Scholar]
- 7.Baumann I, Kolowos W, Voll RE, et al. Impaired uptake of apoptotic cells into tingible body macrophages in germinal centers of patients with systemic lupus erythematosus. Arthritis Rheum. 2002;46:191–201. doi: 10.1002/1529-0131(200201)46:1<191::AID-ART10027>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 8.Botto M, Dell’Agnola C, Bygrave AE, et al. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat Genet. 1998;19:56–59. doi: 10.1038/ng0598-56. [DOI] [PubMed] [Google Scholar]
- 9.Wermeling F, Chen Y, Pikkarainen T, et al. Class A scavenger receptors regulate tolerance against apoptotic cells, and autoantibodies against these receptors are predictive of systemic lupus. J Exp Med. 2007;204:2259–2265. doi: 10.1084/jem.20070600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen XW, Shen Y, Sun CY, Wu FX, Chen Y, Yang CD. Anti-class a scavenger receptor autoantibodies from systemic lupus erythematosus patients impair phagocytic clearance of apoptotic cells by macrophages in vitro. Arthritis Res Ther. 2011;13:R9. doi: 10.1186/ar3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Means TK, Latz E, Hayashi F, Murali MR, Golenbock DT, Luster AD. Human lupus autoantibody-DNA complexes activate DCs through cooperation of CD32 and TLR9. J Clin Invest. 2005;115:407–417. doi: 10.1172/JCI23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrat FJ, Meeker T, Gregorio J, et al. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med. 2005;202:1131–1139. doi: 10.1084/jem.20050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lovgren T, Eloranta ML, Bave U, Alm GV, Ronnblom L. Induction of interferon-alpha production in plasmacytoid dendritic cells by immune complexes containing nucleic acid released by necrotic or late apoptotic cells and lupus IgG. Arthritis Rheum. 2004;50:1861–1872. doi: 10.1002/art.20254. [DOI] [PubMed] [Google Scholar]
- 14.Kelly KM, Zhuang H, Nacionales DC, et al. “Endogenous adjuvant” activity of the RNA components of lupus autoantigens Sm/RNP and Ro 60. Arthritis Rheum. 2006;54:1557–1567. doi: 10.1002/art.21819. [DOI] [PubMed] [Google Scholar]
- 15.Taniguchi T, Lamphier MS, Tanaka N. IRF-1: the transcription factor linking the interferon response and oncogenesis. Biochim Biophys Acta. 1997;1333:M9–17. doi: 10.1016/s0304-419x(97)00014-0. [DOI] [PubMed] [Google Scholar]
- 16.Nakaya T, Sato M, Hata N, et al. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem Biophys Res Commun. 2001;283:1150–1156. doi: 10.1006/bbrc.2001.4913. [DOI] [PubMed] [Google Scholar]
- 17.Sato M, Suemori H, Hata N, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 18.Paun A, Reinert JT, Jiang Z, et al. Functional characterization of murine interferon regulatory factor 5 (IRF-5) and its role in the innate antiviral response. J Biol Chem. 2008;283:14295–14308. doi: 10.1074/jbc.M800501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoenemeyer A, Barnes BJ, Mancl ME, et al. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J Biol Chem. 2005;280:17005–17012. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- 20.Lazear HM, Lancaster A, Wilkins C, et al. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 2013;9:e1003118. doi: 10.1371/journal.ppat.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai P, Cao H, Merghoub T, et al. Myxoma virus induces type I interferon production in murine plasmacytoid dendritic cells via a TLR9/MyD88-, IRF5/IRF7-, and IFNAR-dependent pathway. J Virol. 2011;85:10814–10825. doi: 10.1128/JVI.00104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hooks JJ, Moutsopoulos HM, Geis SA, Stahl NI, Decker JL, Notkins AL. Immune interferon in the circulation of patients with autoimmune disease. N Engl J Med. 1979;301:5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- 23.Baechler EC, Batliwalla FM, Karypis G, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett L, Palucka AK, Arce E, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hua J, Kirou K, Lee C, Crow MK. Functional assay of type I interferon in systemic lupus erythematosus plasma and association with anti-RNA binding protein autoantibodies. Arthritis Rheum. 2006;54:1906–1916. doi: 10.1002/art.21890. [DOI] [PubMed] [Google Scholar]
- 26.Niewold TB, Hua J, Lehman TJ, Harley JB, Crow MK. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8:492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weckerle CE, Franek BS, Kelly JA, et al. Network analysis of associations between serum interferon-alpha activity, autoantibodies, and clinical features in systemic lupus erythematosus. Arthritis Rheum. 2011;63:1044–1053. doi: 10.1002/art.30187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niewold TB, Rivera TL, Buyon JP, Crow MK. Serum type I interferon activity is dependent on maternal diagnosis in anti-SSA/Ro-positive mothers of children with neonatal lupus. Arthritis Rheum. 2008;58:541–546. doi: 10.1002/art.23191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niewold TB, Kelly JA, Flesch MH, Espinoza LR, Harley JB, Crow MK. Association of the IRF5 risk haplotype with high serum interferon-alpha activity in systemic lupus erythematosus patients. Arthritis Rheum. 2008;58:2481–2487. doi: 10.1002/art.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niewold TB, Kelly JA, Kariuki SN, et al. IRF5 haplotypes demonstrate diverse serological associations which predict serum interferon alpha activity and explain the majority of the genetic association with systemic lupus erythematosus. Ann Rheum Dis. 2012;71:463–468. doi: 10.1136/annrheumdis-2011-200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harley JB, Alarcon-Riquelme ME, Criswell LA, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salloum R, Franek BS, Kariuki SN, et al. Genetic variation at the IRF7/PHRF1 locus is associated with autoantibody profile and serum interferon-alpha activity in lupus patients. Arthritis Rheum. 2010;62:553–561. doi: 10.1002/art.27182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu Q, Zhao J, Qian X, et al. Association of a functional IRF7 variant with systemic lupus erythematosus. Arthritis Rheum. 2011;63:749–754. doi: 10.1002/art.30193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gateva V, Sandling JK, Hom G, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lessard CJ, Adrianto I, Ice JA, et al. Identification of IRF8, TMEM39A, and IKZF3-ZPBP2 as susceptibility loci for systemic lupus erythematosus in a large-scale multiracial replication study. Am J Hum Genet. 2012;90:648–660. doi: 10.1016/j.ajhg.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chrabot BS, Kariuki SN, Zervou MI, et al. Genetic Variation near IRF8 is Associated with Serologic and Cytokine Profiles in Systemic Lupus Erythematosus and Multiple Sclerosis. Genes Immun. 2013 doi: 10.1038/gene.2013.42. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salloum R, Niewold TB. Interferon regulatory factors in human lupus pathogenesis. Transl Res. 2011;157:326–331. doi: 10.1016/j.trsl.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niewold TB. Interferon alpha as a primary pathogenic factor in human lupus. Journal of interferon & cytokine research: the official journal of the International Society for Interferon and Cytokine Research. 2011;31:887–892. doi: 10.1089/jir.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niewold TB, Hua J, Lehman TJ, Harley JB, Crow MK. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8:492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niewold TB, Adler JE, Glenn SB, Lehman TJ, Harley JB, Crow MK. Age- and sex-related patterns of serum interferon-alpha activity in lupus families. Arthritis Rheum. 2008;58:2113–2119. doi: 10.1002/art.23619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangale D, Kariuki SN, Chrabot BS, et al. Familial aggregation of high tumor necrosis factor alpha levels in systemic lupus erythematosus. Clin Dev Immunol. 2013;2013:267430. doi: 10.1155/2013/267430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weckerle CE, Mangale D, Franek BS, et al. Large-scale analysis of tumor necrosis factor alpha levels in systemic lupus erythematosus. Arthritis Rheum. 2012;64:2947–2952. doi: 10.1002/art.34483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghodke-Puranik Y, Niewold TB. Genetics of the type I interferon pathway in systemic lupus erythematosus. Int J Clin Rheumtol. 2013;8 doi: 10.2217/ijr.13.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kariuki SN, Niewold TB. Genetic regulation of serum cytokines in systemic lupus erythematosus. Transl Res. 2010;155:109–117. doi: 10.1016/j.trsl.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agik S, Franek BS, Kumar AA, et al. The autoimmune disease risk allele of UBE2L3 in African American patients with systemic lupus erythematosus: a recessive effect upon subphenotypes. J Rheumatol. 2012;39:73–78. doi: 10.3899/jrheum.110590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson T, Kariuki SN, Franek BS, et al. Autoimmune disease risk variant of IFIH1 is associated with increased sensitivity to IFN-alpha and serologic autoimmunity in lupus patients. J Immunol. 2011;187:1298–1303. doi: 10.4049/jimmunol.1100857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kariuki SN, Franek BS, Kumar AA, et al. Trait-stratified genome-wide association study identifies novel and diverse genetic associations with serologic and cytokine phenotypes in systemic lupus erythematosus. Arthritis Res Ther. 2010;12:R151. doi: 10.1186/ar3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koldobskaya Y, Ko K, Kumar AA, et al. Gene-expression-guided selection of candidate loci and molecular phenotype analyses enhance genetic discovery in systemic lupus erythematosus. Clin Dev Immunol. 2012;2012:682018. doi: 10.1155/2012/682018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ronnblom LE, Alm GV, Oberg KE. Possible induction of systemic lupus erythematosus by interferon-alpha treatment in a patient with a malignant carcinoid tumour. J Intern Med. 1990;227:207–210. doi: 10.1111/j.1365-2796.1990.tb00144.x. [DOI] [PubMed] [Google Scholar]
- 50.Niewold TB, Swedler WI. Systemic lupus erythematosus arising during interferon-alpha therapy for cryoglobulinemic vasculitis associated with hepatitis C. Clin Rheumatol. 2005;24:178–181. doi: 10.1007/s10067-004-1024-2. [DOI] [PubMed] [Google Scholar]
- 51.Kirou KA, Lee C, George S, et al. Coordinate overexpression of interferon-alpha-induced genes in systemic lupus erythematosus. Arthritis Rheum. 2004;50:3958–3967. doi: 10.1002/art.20798. [DOI] [PubMed] [Google Scholar]
- 52.Niewold TB, Clark DN, Salloum R, Poole BD. Interferon alpha in systemic lupus erythematosus. J Biomed Biotechnol. 2010;2010:948364. doi: 10.1155/2010/948364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kariuki SN, Franek BS, Kumar AA, et al. Trait-stratified genome-wide association study identifies novel and diverse genetic associations with serologic and cytokine phenotypes in systemic lupus erythematosus. Arthritis Res Ther. 2010;12:R151. doi: 10.1186/ar3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiao Y, Giannopoulou EG, Chan CH, et al. Synergistic activation of inflammatory cytokine genes by interferon-gamma-induced chromatin remodeling and toll-like receptor signaling. Immunity. 2013;39:454–469. doi: 10.1016/j.immuni.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Weerd NA, Vivian JP, Nguyen TK, et al. Structural basis of a unique interferon-beta signaling axis mediated via the receptor IFNAR1. Nat Immunol. 2013;14:901–907. doi: 10.1038/ni.2667. [DOI] [PubMed] [Google Scholar]
- 56.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stark GR, Darnell JE., Jr. The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. Embo J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441:106–110. doi: 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 60.Honda K, Yanai H, Negishi H, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 61.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 62.Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008;8:279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Broz P, Monack DM. Newly described pattern recognition receptors team up against intracellular pathogens. Nat Rev Immunol. 2013;13:551–565. doi: 10.1038/nri3479. [DOI] [PubMed] [Google Scholar]
- 64.Shrivastav M, Niewold TB. Nucleic Acid Sensors and Type I Interferon Production in Systemic Lupus Erythematosus. Frontiers in immunology. 2013;4:319. doi: 10.3389/fimmu.2013.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 66.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 67.Yasuda K, Yu P, Kirschning CJ, et al. Endosomal translocation of vertebrate DNA activates dendritic cells via TLR9-dependent and -independent pathways. J Immunol. 2005;174:6129–6136. doi: 10.4049/jimmunol.174.10.6129. [DOI] [PubMed] [Google Scholar]
- 68.Yasuda K, Rutz M, Schlatter B, et al. CpG motif-independent activation of TLR9 upon endosomal translocation of “natural” phosphodiester DNA. Eur J Immunol. 2006;36:431–436. doi: 10.1002/eji.200535210. [DOI] [PubMed] [Google Scholar]
- 69.Haas T, Metzger J, Schmitz F, et al. The DNA sugar backbone 2′ deoxyribose determines toll-like receptor 9 activation. Immunity. 2008;28:315–323. doi: 10.1016/j.immuni.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 70.Schlee M, Roth A, Hornung V, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kato H, Takeuchi O, Mikamo-Satoh E, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matsui K, Kumagai Y, Kato H, et al. Cutting edge: Role of TANK-binding kinase 1 and inducible IkappaB kinase in IFN responses against viruses in innate immune cells. J Immunol. 2006;177:5785–5789. doi: 10.4049/jimmunol.177.9.5785. [DOI] [PubMed] [Google Scholar]
- 73.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takaki H, Honda K, Atarashi K, et al. MAVS-dependent IRF3/7 bypass of interferon beta-induction restricts the response to measles infection in CD150Tg mouse bone marrow-derived dendritic cells. Mol Immunol. 2014;57:100–110. doi: 10.1016/j.molimm.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 75.Funabiki M, Kato H, Miyachi Y, et al. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity. 2014;40:199–212. doi: 10.1016/j.immuni.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 76.Molineros JE, Maiti AK, Sun C, et al. Admixture mapping in lupus identifies multiple functional variants within IFIH1 associated with apoptosis, inflammation, and autoantibody production. PLoS Genet. 2013;9:e1003222. doi: 10.1371/journal.pgen.1003222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pothlichet J, Niewold TB, Vitour D, Solhonne B, Crow MK, Si-Tahar M. A loss-of-function variant of the antiviral molecule MAVS is associated with a subset of systemic lupus patients. EMBO Mol Med. 2011;3:142–152. doi: 10.1002/emmm.201000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parvatiyar K, Zhang Z, Teles RM, et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Unterholzner L, Keating SE, Baran M, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A. 2012;109:E3008–3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu J, Sun L, Chen X, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kondo T, Kobayashi J, Saitoh T, et al. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc Natl Acad Sci U S A. 2013;110:2969–2974. doi: 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hornung V, Ablasser A, Charrel-Dennis M, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burckstummer T, Baumann C, Bluml S, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 87.Roberts TL, Idris A, Dunn JA, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 88.Fernandes-Alnemri T, Yu JW, Juliana C, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takaoka A, Wang Z, Choi MK, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 90.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lindahl T, Gally JA, Edelman GM. Properties of deoxyribonuclease 3 from mammalian tissues. J Biol Chem. 1969;244:5014–5019. [PubMed] [Google Scholar]
- 94.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Crow YJ, Hayward BE, Parmar R, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 96.Lee-Kirsch MA, Chowdhury D, Harvey S, et al. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. J Mol Med (Berl) 2007;85:531–537. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- 97.Lee-Kirsch MA, Gong M, Chowdhury D, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 98.de Vries B, Steup-Beekman GM, Haan J, et al. TREX1 gene variant in neuropsychiatric systemic lupus erythematosus. Ann Rheum Dis. 2010;69:1886–1887. doi: 10.1136/ard.2009.114157. [DOI] [PubMed] [Google Scholar]
- 99.Namjou B, Kothari PH, Kelly JA, et al. Evaluation of the TREX1 gene in a large multi-ancestral lupus cohort. Genes Immun. 2011;12:270–279. doi: 10.1038/gene.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gall A, Treuting P, Elkon KB, et al. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhao GN, Jiang DS, Li H. Interferon regulatory factors: at the crossroads of immunity, metabolism, and disease. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbadis.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 102.Escalante CR, Yie J, Thanos D, Aggarwal AK. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature. 1998;391:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- 103.Kusumoto M, Fujii Y, Tsukuda Y, et al. Crystallographic Characterization of the DNA-Binding Domain of Interferon Regulatory Factor-2 Complexed with DNA. J Struct Biol. 1998;121:363–366. doi: 10.1006/jsbi.1998.3970. [DOI] [PubMed] [Google Scholar]
- 104.Fujii Y, Shimizu T, Kusumoto M, Kyogoku Y, Taniguchi T, Hakoshima T. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. Embo J. 1999;18:5028–5041. doi: 10.1093/emboj/18.18.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen W, Lam SS, Srinath H, et al. Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5. Nat Struct Mol Biol. 2008;15:1213–1220. doi: 10.1038/nsmb.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Barnes BJ, Kellum MJ, Field AE, Pitha PM. Multiple regulatory domains of IRF-5 control activation, cellular localization, and induction of chemokines that mediate recruitment of T lymphocytes. Mol Cell Biol. 2002;22:5721–5740. doi: 10.1128/MCB.22.16.5721-5740.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lin R, Mamane Y, Hiscott J. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol Cell Biol. 1999;19:2465–2474. doi: 10.1128/mcb.19.4.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin R, Mamane Y, Hiscott J. Multiple regulatory domains control IRF-7 activity in response to virus infection. J Biol Chem. 2000;275:34320–34327. doi: 10.1074/jbc.M002814200. [DOI] [PubMed] [Google Scholar]
- 109.Pothlichet J, Niewold TB, Vitour D, Solhonne B, Crow MK, Si-Tahar M. A loss-of-function variant of the antiviral molecule MAVS is associated with a subset of systemic lupus patients. EMBO Mol Med. 2011;3:142–152. doi: 10.1002/emmm.201000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sawalha AH, Webb R, Han S, et al. Common variants within MECP2 confer risk of systemic lupus erythematosus. PLoS One. 2008;3:e1727. doi: 10.1371/journal.pone.0001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Webb R, Wren JD, Jeffries M, et al. Variants within MECP2, a key transcription regulator, are associated with increased susceptibility to lupus and differential gene expression in patients with systemic lupus erythematosus. Arthritis Rheum. 2009;60:1076–1084. doi: 10.1002/art.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kaufman KM, Zhao J, Kelly JA, et al. Fine mapping of Xq28: both MECP2 and IRAK1 contribute to risk for systemic lupus erythematosus in multiple ancestral groups. Ann Rheum Dis. 2013;72:437–444. doi: 10.1136/annrheumdis-2012-201851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Graham RR, Cotsapas C, Davies L, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Adrianto I, Wen F, Templeton A, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat Genet. 2011;43:253–258. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Adrianto I, Wang S, Wiley GB, et al. Association of two independent functional risk haplotypes in TNIP1 with systemic lupus erythematosus. Arthritis Rheum. 2012;64:3695–3705. doi: 10.1002/art.34642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Uematsu S, Sato S, Yamamoto M, et al. Interleukin-1 receptor-associated kinase-1 plays an essential role for Toll-like receptor (TLR)7- and TLR9-mediated interferon-{alpha} induction. J Exp Med. 2005;201:915–923. doi: 10.1084/jem.20042372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Namjou B, Sestak AL, Armstrong DL, et al. High-density genotyping of STAT4 reveals multiple haplotypic associations with systemic lupus erythematosus in different racial groups. Arthritis Rheum. 2009;60:1085–1095. doi: 10.1002/art.24387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kariuki SN, Kirou KA, MacDermott EJ, Barillas-Arias L, Crow MK, Niewold TB. Cutting edge: autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-alpha in lupus patients in vivo. J Immunol. 2009;182:34–38. doi: 10.4049/jimmunol.182.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Svenungsson E, Gustafsson J, Leonard D, et al. A STAT4 risk allele is associated with ischaemic cerebrovascular events and anti-phospholipid antibodies in systemic lupus erythematosus. Ann Rheum Dis. 2010;69:834–840. doi: 10.1136/ard.2009.115535. [DOI] [PubMed] [Google Scholar]
- 120.Chung SA, Taylor KE, Graham RR, et al. Differential genetic associations for systemic lupus erythematosus based on anti-dsDNA autoantibody production. PLoS Genet. 2011;7:e1001323. doi: 10.1371/journal.pgen.1001323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ferreira RC, Pan-Hammarstrom Q, Graham RR, et al. Association of IFIH1 and other autoimmunity risk alleles with selective IgA deficiency. Nat Genet. 2010;42:777–780. doi: 10.1038/ng.644. [DOI] [PubMed] [Google Scholar]
- 122.Kyogoku C, Langefeld CD, Ortmann WA, et al. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet. 2004;75:504–507. doi: 10.1086/423790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kariuki SN, Crow MK, Niewold TB. The PTPN22 C1858T polymorphism is associated with skewing of cytokine profiles toward high interferon-alpha activity and low tumor necrosis factor alpha levels in patients with lupus. Arthritis Rheum. 2008;58:2818–2823. doi: 10.1002/art.23728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 125.Takaoka A, Yanai H, Kondo S, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 126.Krausgruber T, Blazek K, Smallie T, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011;12:231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 127.Barnes BJ, Richards J, Mancl M, Hanash S, Beretta L, Pitha PM. Global and distinct targets of IRF-5 and IRF-7 during innate response to viral infection. J Biol Chem. 2004;279:45194–45207. doi: 10.1074/jbc.M400726200. [DOI] [PubMed] [Google Scholar]
- 128.Yasuda K, Watkins AA, Kochar GS, et al. Interferon Regulatory Factor-5 Deficiency Ameliorates Disease Severity in the MRL/lpr Mouse Model of Lupus in the Absence of a Mutation in DOCK2. PLoS One. 2014;9:e103478. doi: 10.1371/journal.pone.0103478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Couzinet A, Tamura K, Chen HM, et al. A cell-type-specific requirement for IFN regulatory factor 5 (IRF5) in Fas-induced apoptosis. Proc Natl Acad Sci U S A. 2008;105:2556–2561. doi: 10.1073/pnas.0712295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lien C, Fang CM, Huso D, Livak F, Lu R, Pitha PM. Critical role of IRF-5 in regulation of B-cell differentiation. Proc Natl Acad Sci U S A. 2010;107:4664–4668. doi: 10.1073/pnas.0911193107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fang CM, Roy S, Nielsen E, et al. Unique contribution of IRF-5-Ikaros axis to the B-cell IgG2a response. Genes Immun. 2012;13:421–430. doi: 10.1038/gene.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cham CM, Ko K, Niewold TB. Interferon regulatory factor 5 in the pathogenesis of systemic lupus erythematosus. Clin Dev Immunol. 2012;2012:780436. doi: 10.1155/2012/780436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lazzari E, Jefferies CA. IRF5-mediated signaling and implications for SLE. Clin Immunol. 2014;153:343–352. doi: 10.1016/j.clim.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 134.Mancl ME, Hu G, Sangster-Guity N, et al. Two discrete promoters regulate the alternatively spliced human interferon regulatory factor-5 isoforms. Multiple isoforms with distinct cell type-specific expression, localization, regulation, and function. J Biol Chem. 2005;280:21078–21090. doi: 10.1074/jbc.M500543200. [DOI] [PubMed] [Google Scholar]
- 135.Sigurdsson S, Goring HH, Kristjansdottir G, et al. Comprehensive evaluation of the genetic variants of interferon regulatory factor 5 (IRF5) reveals a novel 5 bp length polymorphism as strong risk factor for systemic lupus erythematosus. Hum Mol Genet. 2008;17:872–881. doi: 10.1093/hmg/ddm359. [DOI] [PubMed] [Google Scholar]
- 136.Graham RR, Kyogoku C, Sigurdsson S, et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci U S A. 2007;104:6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Graham RR, Kozyrev SV, Baechler EC, et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 138.Sigurdsson S, Nordmark G, Goring HH, et al. Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. Am J Hum Genet. 2005;76:528–537. doi: 10.1086/428480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cherian TS, Kariuki SN, Franek BS, Buyon JP, Clancy RM, Niewold TB. Brief Report: IRF5 systemic lupus erythematosus risk haplotype is associated with asymptomatic serologic autoimmunity and progression to clinical autoimmunity in mothers of children with neonatal lupus. Arthritis Rheum. 2012;64:3383–3387. doi: 10.1002/art.34571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lazzari E, Korczeniewska J, Ni Gabhann J, Smith S, Barnes BJ, Jefferies CA. TRIpartite Motif 21 (TRIM21) Differentially Regulates the Stability of Interferon Regulatory Factor 5 (IRF5) Isoforms. PLoS One. 2014;9:e103609. doi: 10.1371/journal.pone.0103609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ning S, Pagano JS, Barber GN. IRF7: activation, regulation, modification and function. Genes Immun. 2011;12:399–414. doi: 10.1038/gene.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Harley JB, Alarcon-Riquelme ME, Criswell LA, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fu Q, Zhao J, Qian X, et al. Association of a functional IRF7 variant with systemic lupus erythematosus. Arthritis Rheum. 2011;63:749–754. doi: 10.1002/art.30193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Salloum R, Franek BS, Kariuki SN, et al. Genetic variation at the IRF7/PHRF1 locus is associated with autoantibody profile and serum interferon-alpha activity in lupus patients. Arthritis Rheum. 2010;62:553–561. doi: 10.1002/art.27182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tsujimura H, Tamura T, Kong HJ, et al. Toll-like receptor 9 signaling activates NF-kappaB through IFN regulatory factor-8/IFN consensus sequence binding protein in dendritic cells. J Immunol. 2004;172:6820–6827. doi: 10.4049/jimmunol.172.11.6820. [DOI] [PubMed] [Google Scholar]
- 146.Hambleton S, Salem S, Bustamante J, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365:127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 148.De Jager PL, Jia X, Wang J, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Feng X, Reder NP, Yanamandala M, et al. Type I interferon signature is high in lupus and neuromyelitis optica but low in multiple sclerosis. J Neurol Sci. 2012;313:48–53. doi: 10.1016/j.jns.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Feng X, Han D, Kilaru BK, Franek BS, Niewold TB, Reder AT. Inhibition of interferon-beta responses in multiple sclerosis immune cells associated with high-dose statins. Arch Neurol. 2012;69:1303–1309. doi: 10.1001/archneurol.2012.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Reder AT, Feng X. Aberrant Type I Interferon Regulation in Autoimmunity: Opposite Directions in MS and SLE, Shaped by Evolution and Body Ecology. Front Immunol. 2013;4:281. doi: 10.3389/fimmu.2013.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]