Abstract
The NLR protein, NLRC5 is an important regulator of MHC class I gene expression, however, the role of NLRC5 in other innate immune responses is less well defined. In the present study, we report that NLRC5 binds RIG-I and that this interaction is critical for robust antiviral responses against influenza virus. Overexpression of NLRC5 in the human lung epithelial cell line, A549, and normal human bronchial epithelial cells resulted in impaired replication of influenza virus A/Puerto Rico/8/34 virus (PR8) and enhanced IFN-β expression. Influenza virus leads to induction of IFN-β that drives RIG-I and NLRC5 expression in host cells. Our results suggest that NLRC5 extends and stabilizes influenza virus induced RIG-I expression and delays expression of the viral inhibitor protein NS1. We show that NS1 binds to NLRC5 to suppress its function. Interaction domain mapping revealed that NLRC5 interacts with RIG-I via its N-terminal death domain and that NLRC5 enhanced antiviral activity in an leucine-rich repeat domain independent manner. Taken together, our findings identify a novel role for NLRC5 in RIG-I-mediated antiviral host responses against influenza virus infection, distinguished from the role of NLRC5 in MHC class I gene regulation.
Keywords: Influenza, Interferon, NLRC5, NS1, RIG-I Antiviral
Introduction
The innate immune system relies on pathogen sensors to provide defense against invading pathogens. To detect diverse pathogen-associated molecular patterns (PAMPs), several classes of pattern-recognition receptors (PRRs) have evolved in mammals including RIG-I-like receptors, Toll-like receptors (TLRs), and nucleotide-binding domain and leucine-rich repeat containing receptors (NLRs) [1–3]. Recognition and binding of PRRs to their cognate ligands often result in conformational changes triggering downstream innate immune signaling [4].
In recent years, there has been marked progress in our understanding of NLRs as critical regulators of innate and adaptive immune responses. Emerging evidence suggests that NLR family members play crucial roles in antiviral responses [5, 6]. NLR proteins have a typical tripartite structure: a C-terminal LRR domain that, in most cases, is associated with PAMP sensing, a centrally located nucleotide-triphosphatase (NTPase) (NACHT - domain present in NAIP, CIITA, HET-E, and TP1) domain responsible for self-oligomerization, and an N-terminal effector domain that mediates protein–protein interactions for initiating downstream signaling [5]. Activation of these receptors induces the production of proinflammatory cytokines, in many cases by the formation of high molecular weight complexes called inflammasomes that lead to caspase-1 activation. However, some of the family members such as NLRX1 play a role in modulating innate immune responses at the level of mitochondria [7–9], whereas class II, major histocompatibility complex (MHC), transactivator (CIITA) and NLRC5 act as transcriptional enhancers for MHC gene expression [10–14]. To date, only a few NLRs have been extensively characterized and the precise role of their function(s) as innate immune receptors remains fragmentary. NLRC5 shares the common NLR architecture but differs from the other NLR members in having an unusual (i.e. non-PYD non-CARD) death domain (DD) fold N-terminal effector domain [13, 15]. Moreover, it possesses the longest LRR domain of all known human NLRs [13, 16]. Only recently NLRC5 has been experimentally characterized and shown to contribute to MHC class I gene regulation [11, 12, 17, 18]. Additionally, there is evidence to suggest that NLRC5 plays a role in the regulation of the inflammasome signaling pathway, the NF-κB pathway [19], and antiviral innate immune responses [13, 16, 20]. A role of NLRC5 in antiviral responses was first reported by Kuenzel et al. [16] as well as by our group [13] for polyinosinic:polycytidylic acid (polyI:C), cytomegalovirus (CMV), and Sendai virus induced interferon (IFN) responses in human cells. A negative role for NLRC5 in antiviral response was reported. A study by Cui et al. [20] showed that NLRC5 can prevent IFN induction by directly binding to cytoplasmic receptors such as RIG-I and MDA5, thereby blocking their binding to mitochondrial antiviral signaling protein and subsequent downstream signaling. Surprisingly, IFN and cytokines responses of macrophages and DCs stimulated with Newcastle disease virus, HSV-1, or polyI:C were not significantly different between cells derived from NLRC5-deficient and WT animals [21]. Sequence comparison studies suggest that human and mouse NLRC5 share 64% amino acid sequence identity [20]. While a general role for NLRC5 in regulating host innate immune responses has yet to be established, discrepancies in these reports need more independent studies to confirm if NLRC5 performs similar functions in response to viral infections in both humans and mice. In the current study, we investigated the role of NLRC5 in influenza A virus infection in human respiratory epithelial cells.
Despite current prevention and treatment strategies, influenza virus infection remains a major threat to public health and accounts for 250,000–500,000 deaths globally each year [21]. The ability of influenza viruses to undergo frequent genetic changes increases the risk of emergence of drug-resistant strains with epidemic or pandemic potential (http://www.who.int/csr/disease/swineflu/notes/h1n1_antiviral_resistance_20090708/en/index.html) [22–24]. Recently, the evolutionarily conserved innate immune receptors of the RIG-I-like receptor family have been shown to play a critical role in protection against single- and double-stranded RNA viruses [25]. It has been reported that ligand-induced activation of RIG-I can inhibit influenza virus replication irrespective of subtypes, drug-sensitivity status, or virulence [26–30] as well as Ebola virus replication [31]. In the present work, we report that the NLR family member NLRC5 enhances the RIG-I-dependent antiviral response against influenza virus (PR8) infection in epithelial cells.
Results
NLRC5 expression impairs influenza A virus replication in respiratory epithelial cells
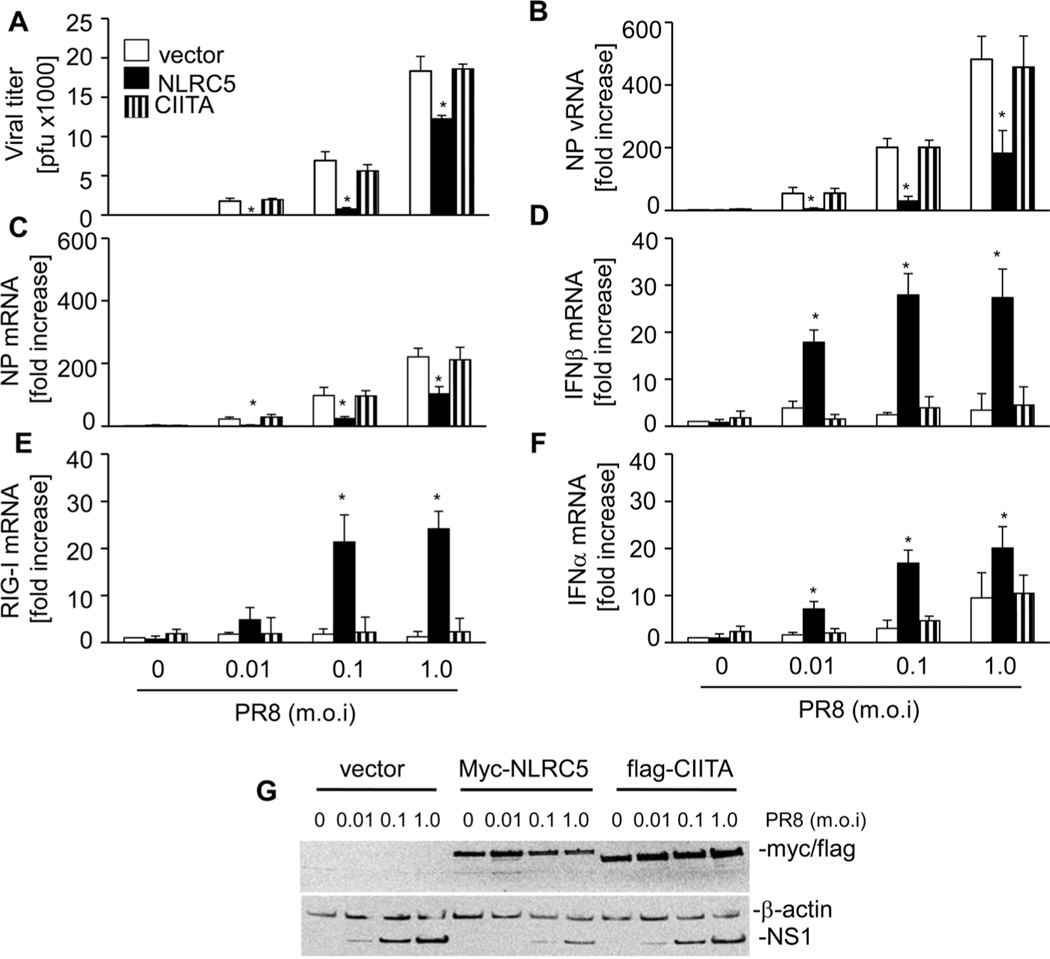
To evaluate the role of NLRC5 in influenza virus infection, we transfected the lung epithelial cell line A549 with myc-NLRC5 expression or human flag-CIITA expression vector as A549 cells express only very low levels of NLRC5 (Fig. 1). After 24 h, cells were infected with different doses of PR8 (multiplicity of infection (MOI) 0.01, 0.1, and 1.0) for a further 24 h, which resulted in a dose-dependent increase in NS1 expression (Fig. 1G). NLRC5 overexpression significantly reduced PR8 replication compared with cells transfected with the empty vector at all infection doses used as determined by viral plaque assays (Fig. 1A). Interestingly, we observed a more robust inhibition of PR8 replication at lower MOI (0.01 and 0.1) as compared with an MOI of 1.0 in NLRC5-transfected cells. In contrast, overexpression of CIITA, an NLRC5-related NLR protein, did not inhibit virus replication as viral titers were comparable to that of control vector transfected cells (Fig. 1A). Accordingly, we observed a significant decrease in nucleoprotein (NP) vRNA and NP mRNA copy numbers in infected A549 that had been transfected with NLRC5 expression vector as compared with control vector or CIITA expression vector transfected A549 cells (Fig. 1B and C). Accordingly, NS1 levels were strongly reduced in NLRC5-expressing cells (Fig. 1G). Host defense against viral infection is associated with type I IFN induction. Interestingly, NLRC5 overexpression (Fig. 1G) enhanced mRNA expression of IFN-β in A549 cells infected with PR8 (Fig. 1D). Of note, we also observed a significant increase in RIG-I expression in NLRC5-transfected cells (Fig. 1E), which we and others recently identified as an important factor to restrict influenza replication in human cells [26–30]. Increase in IFN-β and RIG-I in NLRC5-transfected cells was observed only in response to PR8 infection, which reached a maximum at 0.1 MOI infection dose. This was also observed for IFN-α, mRNA expression, another type I IFN (Fig. 1F). By contrast, PR8-induced expression of IFN-β, RIG-I, and IFN-α was not changed in A549 cells expressing CIITA (Fig. 1G).
Figure 1.
NLRC5 overexpression inhibits influenza virus PR8 replication and induces RIG-I and IFN-β expression in A549 cells. A549 cells were transfected with 2 μg of vector alone, myc-NLRC5 or flag-CIITA expression vector. Twenty-four hours post-transfection, the cells were infected with PR8 virus (MOI 0, 0.01, 0.1, and 1.0) for 24 h. (A) Supernatants were tested for viral titers by plaque assay using MDCK cells. (B–F) The expression of (B) NP vRNA, (C) NP mRNA, (D) IFN-β mRNA, (E) RIG-I mRNA, and (F) IFN-α was analyzed by real-time RT-PCR, relative to β-actin. Data shown are mean + SD of three samples per group, pooled from three independent experiments carried out in duplicate. (F) Expression of myc-NLRC5 and NS1 was analyzed by immunoblotting and the immunoblot shown is from one single experiment representative of three independent experiments. β-Actin was used as a loading control. ANOVA was performed to compare vector control versus myc-NLRC5 or flag-CIITA-transfected A549 cells and p values <0.05 are indicated with an asterisk.
We carried out similar studies in normal human bronchial epithelial (NHBE) cells. As shown in Supporting Information Figure 1, NHBE cells transfected with NLRC5 expression vector showed significant reduction in viral titer as compared with vector control upon PR8 infection (MOI 1.0) (Supporting Information Fig. 1A). This NLRC5-mediated inhibition of PR8 replication in NHBE cells was consistent with a reduction in NP vRNA (Supporting Information Fig. 1B). NLRC5 expression also induced IFN-β (Supporting Information Fig. 1D) and RIG-I mRNA expression (Supporting Information Fig. 1C) in PR8-infected NHBE cells. NLRs have also been reported to upregulate expression of many proinflammatory cytokines including CCL5 (RANTES) [32, 33]. Previously, we have shown that NLRC5 affects RANTES and IFN-β secretion in PMA-differentiated THP-1 cells and primary human dermal fibroblasts following Sendai virus infection and polyI:C stimulation [13]. Analysis of IFN-β and RANTES in NHBE culture supernatants by ELISA indicates that NLRC5 overexpression also resulted in increased IFN-β and RANTES levels upon PR8 infection (Supporting Information Fig. 1E and F). These observations were further supported by immunohistochemistry where we observed a significant reduction in NP-positive NHBE cells, transfected with NLRC5 expression vector (Supporting Information Fig. 1G and H).
Contrary to our findings in A549 and NHBE cells, overexpression of NLRC5 in HEK293T cells did not affect PR8 replication irrespective of the virus dose and viral titers (Supporting Information Fig. 2A and B). Immunoblot analysis of NS1 expression confirmed no change in NS1 expression level in vector- and NLRC5-transfected HEK293T cells (Supporting Information Fig. 2A). Furthermore, IFN-β promoter activity was not significantly different in NLRC5-transfected cells following PR8 infection compared with cells transfected with the empty vector (Supporting Information Fig. 2C).
These results suggest that NLRC5 plays a role in the induction of antiviral innate immune responses against influenza virus infection and that NLRC5 contributes to the restriction of influenza replication in human respiratory epithelial cells, but not in HEK293T cells.
Viral NS1 counteracts NLRC5, RIG-I, and IFN-β expression
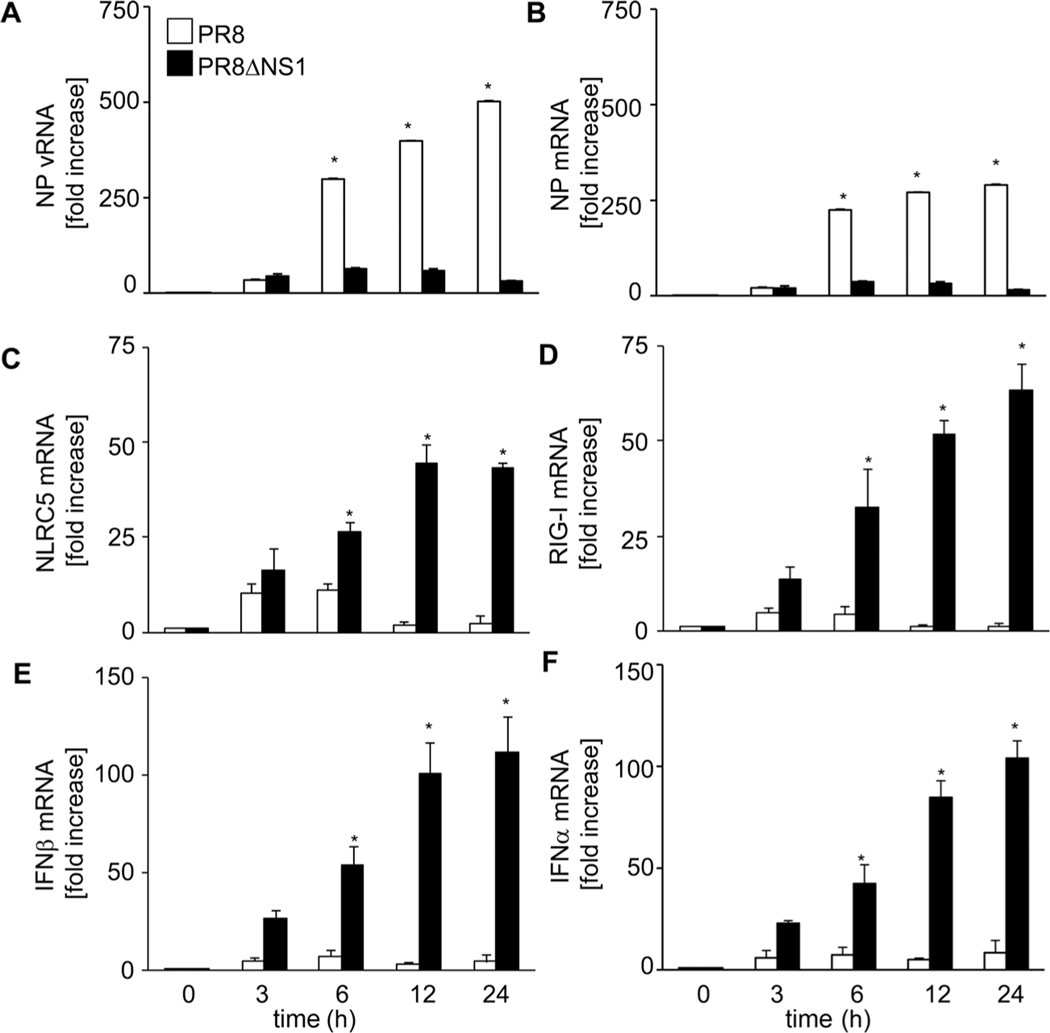
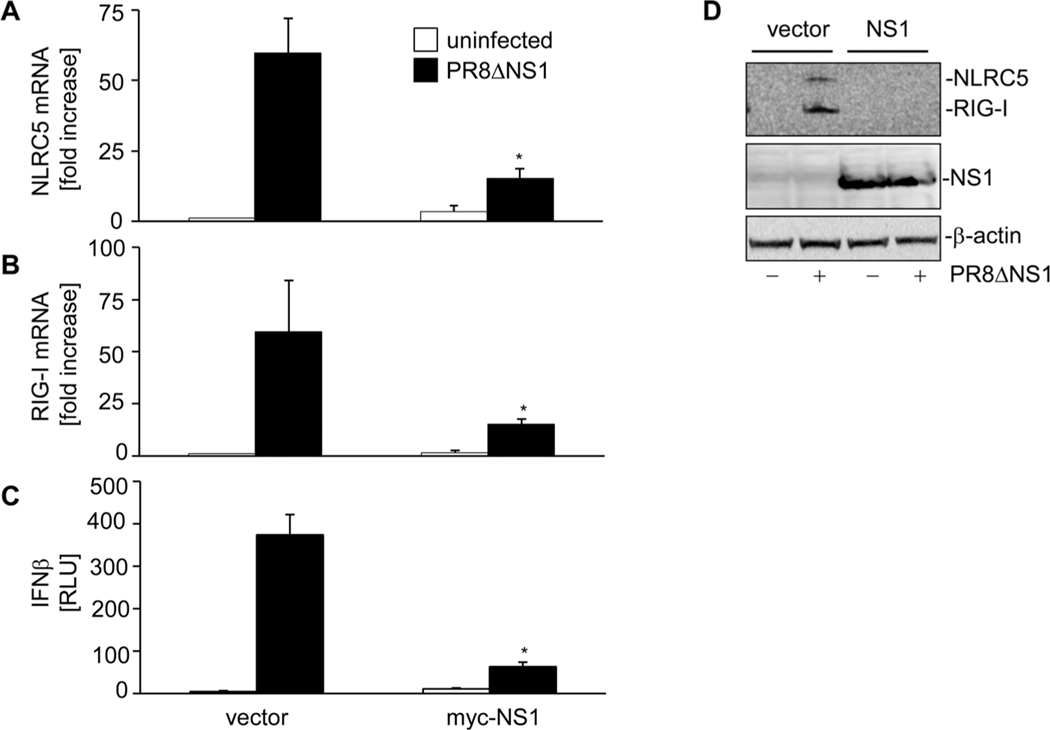
NLRC5 expression has been shown to be induced upon viral infection in different human cell types [13, 16, 19]. Therefore, we investigated the level of endogenous NLRC5 mRNA in A549 cells in response to PR8 infection. The kinetic of virus infection was studied by quantitative RT-PCR of NP vRNA and NP mRNA, which showed a time-dependent increase in their copy numbers (Fig. 2A and B). Further, quantitative RT-PCR studies showed that PR8 infection induced NLRC5 mRNA expression in A549 cells within 3 h of infection (Fig. 2C). However, NLRC5 expression declined thereafter and reached nearly basal levels by 12 h of infection. Similar kinetics were observed for RIG-I mRNA expression (Fig. 2D). We observed only a marginal increase in IFN-β expression at 6 h following PR8 infection (Fig. 2E). Expression of many PRRs has been shown to be induced by viral PAMPs as well as cytokines [34] but viruses have devised potent strategies to counteract PRR expression, recognition, activation, and subsequent cytokine production. Influenza NS1 has been shown to suppress RIG-I activation and subsequent IFN-β induction [30, 35–38]. To determine the effect of NS1 on virus-induced NLRC5 expression PR8ΔNS1, a mutant of influenza virus PR8 that lacks functional NS1 (PR8ΔNS1) was used. Unlike WT PR8 virus, infection with PR8ΔNS1 resulted in a strong IFN-β expression in a time-dependent manner (Fig. 2E), a concordant increase in RIG-I (Fig. 2D) and NLRC5 mRNA (Fig. 2C) levels was also observed. Very low PR8ΔNS1 NP vRNA and NP mRNA expression levels confirmed replication deficiency of PR8ΔNS1 in A549 cells (Fig. 2A and B). To provide further evidence that NS1 suppresses NLRC5 induction and antiviral response, we complemented viral NS1 expression by transfecting a myc-NS1 expression vector into A549 cells prior to infection with PR8ΔNS1 (Fig. 3D). Real-time RT-PCR revealed that expression of NS1 in the host cells significantly (p < 0.05) inhibited NLRC5 (Fig. 3A) and RIG-1 mRNA (Fig. 3B) and protein (Fig. 3D) expression upon viral infection. LUC reporter assays of the same cells revealed reduced IFN-β promoter activity (Fig. 3C) in response to PR8ΔNS1 compared with cells not expressing myc-NS1.
Figure 2.
Viral NS1 counteracts endogenous NLRC5, RIG-I, and IFN-β expression. A549 cells were infected with PR8 or PR8ΔNS1 (MOI 1.0) for 0, 3, 6, 12, and 24 h and the expression of (A) NP vRNA, (B) NP mRNA, (C) NLRC5 mRNA, (D) RIG-I mRNA, (E) IFN-β mRNA, and (F) IFN-α was analyzed by real-time RT-PCR, relative to β-actin. Data shown are mean + SD of three samples per group, pooled from three independent experiments carried out in duplicate. ANOVA was performed to compare PR8-infected versus PR8ΔNS1-infected A549 cells and p values <0.05 are indicated with an asterisk.
Figure 3.
NS1 complementation inhibits PR8ΔNS1-induced NLRC5. A549 cells were cotransfected with 2 μg of vector or myc-NS1 expression vector and IFN-β promoter LUC reporter using lipofectamine 2000. Twenty-four hours post-transfection, these cells were infected with PR8ΔNS1 for another 24 h. (A, B) Cells were harvested and mRNA expression of (A) NLRC5 and (B) RIG-I was determined by real-time RT-PCR, relative to β-actin. (C) IFN-β induction was assayed by LUC reporter assay. (D) NLRC5, RIG-I, and myc-NS1 expression was analyzed by immunoblotting. The immunoblot shown is from one single experiment representative of three independent experiments. β-Actin was used as a loading control. Data shown are mean + SD of three samples per group, pooled from three independent experiments carried out in duplicate. ANOVA was performed to compare vector control versus myc-NS1-transfected A549 cells and p values <0.05 are indicated with an asterisk.
Influenza virus mediated induction of NLRC5 and type-I IFN requires RIG-I
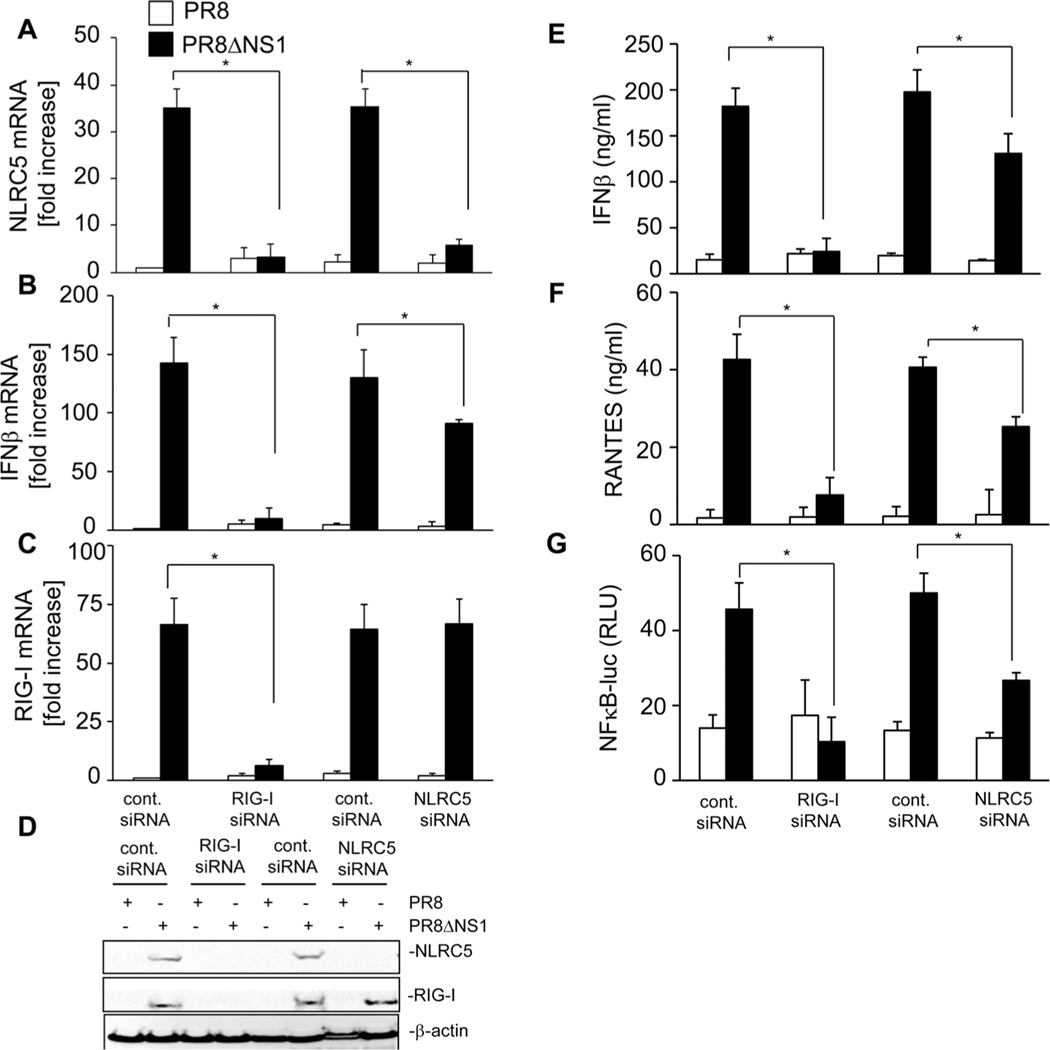
RIG-I has a well-established role in inhibiting influenza virus replication [26, 28, 36–38]. We observed a significant increase in RIG-I expression in response to NLRC5 overexpression in A549 as well as in NHBE cells upon influenza virus infection (Fig. 1E and Supporting Information Fig. 1C). Furthermore, infection with PR8ΔNS1 induced RIG-I expression (Fig. 2D). This likely is mediated by the IFN pathway as RIG-I is well known to be IFN inducible [39]. To delineate the specific role of NLRC5 and RIG-I in the antiviral response, we applied a knockdown approach using gene-specific siRNAs in A549 cells. A549 cells were transfected with NLRC5 or RIG-I siRNA for 24 h, and subsequently infected with PR8 or PR8ΔNS1 for another 24 h. Knockdown efficiency of NLRC5 and RIG-I was determined by quantitative RT-PCR (Fig. 4A and C) as well as by Western blot (Fig. 4D). Knockdown of RIG-I expression completely abrogated the induction of NLRC5 and IFN-β mRNA (Fig. 4A, B, and D). Analysis of RIG-I expression showed that NLRC5 knockdown did not affect RIG-I mRNA or protein expression as compared with control siRNA treated A549 cells infected with PR8ΔNS1 (Fig. 4C and D). In contrast, reduction of NLRC5 mRNA levels partially, but significantly (p < 0.03), inhibited influenza virus induced IFN-β (Fig. 4B), as shown recently for Sendai virus [13] and CMV [16] in other cell types. In any case, control siRNA did not alter the expression level of NLRC5 (Fig. 4A and D) or RIG-I (Fig. 4C and D). Release of IFN-β and RANTES also was fully blunted in RIG-I-specific siRNA-treated cells and partially reduced in cells that received NLRC5-specific siRNA (Fig. 4E and F). The RIG-I-dependent antiviral pathway also involves NFκB activation [40], therefore, we investigated NFκB activation using NFκB gene reporter LUC assays. As shown in Figure 4G, infection with PR8ΔNS1, but not PR8, resulted in increased NFκB promoter activity. Knocking down RIG-I completely inhibited NFκB activation, while NLRC5 knockdown resulted in partial but significant (p < 0.05) reduction in NFκB promoter activation. To substantiate these results, we further tested two independent sets of control and NLRC5 specific siRNAs. As shown in Supporting Information Figure 3A, both of these NLRC5-targeting siRNAs, but not the control siRNAs, effectively reduced PR8ΔNS1-induced NLRC5 protein expression below the detection limit. In contrast, RIG-I protein expression was not affected by the control or NLRC5 siRNAs (Supporting Information Fig. 3A). IFN-β promoter LUC reporter assays confirmed that the NLRC5-specific siRNAs but not the control siRNAs efficiently reduced PR8ΔNS1-induced IFN-β activation (Supporting Information Fig. 3B).
Figure 4.
NS1ΔPR8 virus induces NLRC5, IFN-β, RANTES expression, and NFκB activation in a RIG-I-dependent manner. The expression of endogenous NLRC5 or RIG-I was silenced using gene-specific NLRC5 or RIG-I siRNA in A549 cells followed by infection with PR8 or PR8ΔNS1 (MOI 1.0). Cells were also cotransfected with NFκB promoter LUC reporter using lipofectamine 2000. (A–C) Cells were harvested 24 h postinfection to assess the expression of (A) NLRC5 mRNA, (B) IFN-β mRNA, and (C) RIG-I mRNA, relative to β-actin by real-time RT-PCR. (D) Cells were analyzed for endogenous RIG-I, NLRC5, and β-actin (loading control) protein expression by immunoblotting and the immunoblot shown is from one single experiment representative of three independent experiments. Cell supernatants were assayed for (E) IFN-β and (F) RANTES by ELISA. (G) NFκB activation was measured by LUC reporter assay. Data shown are mean + SD of three samples per group, pooled from three independent experiments carried out in duplicate. ANOVA was performed to compare PR8-infected versus PR8ΔNS1-infected A549 cells and p values <0.05 are indicated with an asterisk.
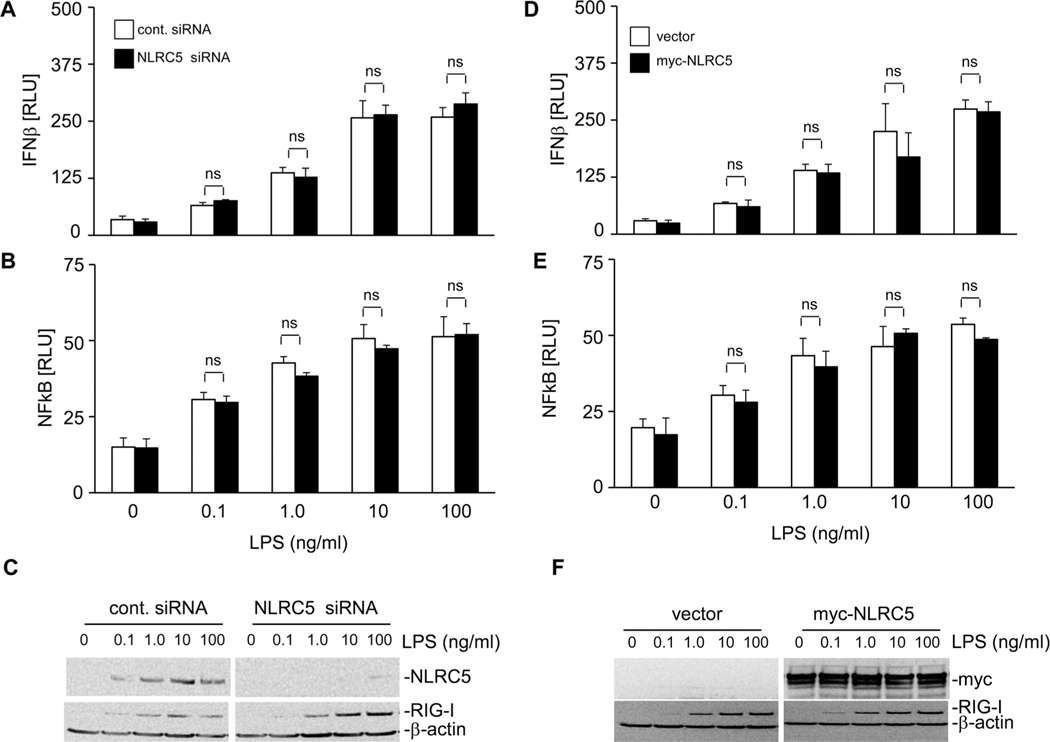
Results of the above studies suggest that NLRC5 is involved in RIG-I-dependent antiviral responses against influenza virus. Influenza virus is a natural ligand for RIG-I that induces its activation [41, 42]. In order to establish the specificity of this response, we carried out studies to investigate if NLRC5 influences other innate immune responses induced by other PAMPs such as LPS. A549 cells either transfected with NLRC5 siRNA or NLRC5 expression vector and/or IFN-β or NFκB promoter LUC reporter plasmids were treated with PBS or LPS (0.1, 1.0, 10, or 100 ng/mL) for 24 h. As shown in Figure 5, LPS treatment resulted in a dose-dependent increase in IFN-β and NFκB promoter activation in A549 cells. NLRC5 knockdown strongly reduced LPS-induced NLRC5 protein levels (Fig. 5C). However, this neither significantly altered IFN-β nor NFκB promoter activation compared with cells treated with control siRNA (Fig. 5A and B), and this trend was consistent with all the doses of LPS used in the study. Similarly, overexpression of NLRC5 in A549 cells followed by LPS treatment also did not affect IFN-β or NFκB promoter activation (Fig. 5D and E). Figure 5F shows NLRC5 and RIG-I protein expression in these cells. These results provide additional support that NLRC5 is involved specifically in RIG-I-mediated antiviral responses in A549 cells.
Figure 5.
LPS-induced IFN-β induction and NFκB activation remain unchanged in the presence or absence of NLRC5. (A–C) The expression of endogenous NLRC5 in A549 cells was silenced using gene-specific NLRC5 siRNA. (D–F) Alternatively, A549 cells were transfected with 2 μg of vector alone or myc-NLRC5 expression vector. (A–F) Cells were also cotransfected with IFN-β promoter or NFκB promoter LUC reporter using lipofectamine 2000 and treated with indicated dose of LPS. (A–F) Cells were harvested 24 h post-LPS treatment to assess (A and D) IFN-β induction and (B and E) NFκB activation by LUC reporter assay. (C and F) Cells were analyzed for myc-NLRC5, RIG-I, and β-actin (loading control) protein expression by immunoblotting and the immunoblot shown is from one single experiment representative of two independent experiments. Data are shown as mean + SD of three samples per group, pooled from three independent experiments carried out in duplicate. ANOVA was performed to compare control siRNA versus NLRC5 siRNA treated A549 cells or vector versus NLRC5-transfected A549 cells and p values <0.05 are indicated with an asterisk; ns: not significant.
We further extended these studies to primary mouse normal bronchial and tracheal epithelial cells (Supporting Information Fig. 4). To this end, we knocked down mouse NLRC5 using mouse gene specific NLRC5 siRNA and infected these cells with PR8 or PR8ΔNS1 for 6 h. We observed a strong NLRC5 and RIG-I expression in response to both PR8 and PR8ΔNS1 at this time point (Supporting Information Fig. 4A). NLRC5 siRNA effectively reduced NLRC5 expression without affecting RIG-I expression (Supporting Information Fig. 4A). Analysis of IFN-β and RANTES in culture supernatants 24 h postinfection by ELISA showed that NLRC5 knockdown significantly reduced IFN-β and RANTES level (Supporting Information Fig. 4B and C). These findings confirm that our results obtained in cell lines are transferrable to primary epithelial cells.
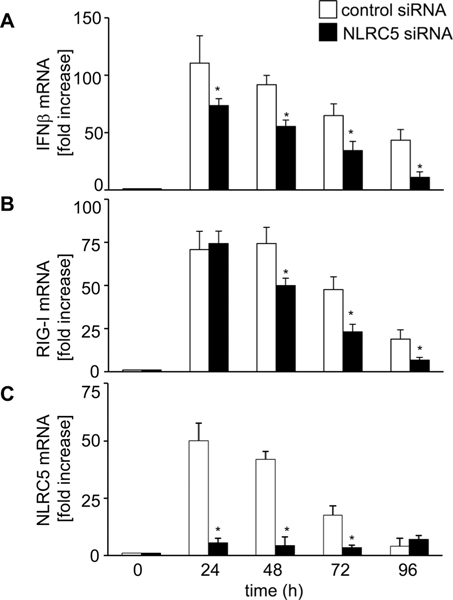
To characterize the role of NLRC5 in antiviral response in more detail, we analyzed the expression level of RIG-I and IFN-β in infected cells over time. A549 cells treated with control or NLRC5-specific siRNA were infected with PR8ΔNS1 for the indicated time and IFN-β, RIG-I, and NLRC5 mRNA expression was measured by quantitative RT-PCR (Fig. 6). NLRC5 knockdown resulted in a significant reduction of IFN-β expression at all time points (p < 0.03, 0.01, 0.01, and 0.008) (Fig. 6A). Interestingly, RIG-I mRNA expression was not different in control siRNA versus NLRC5 siRNA treated cells at 24 h postinfection (see also Fig. 4C), however was significantly lower in the absence of NLRC5 starting from 48 h postinfection (p < 0.04, 0.04, and 0.03) (Fig. 6B). In any case, NLRC5 mRNA was efficiently reduced by the siRNA treatment (Fig. 6C). These results strongly suggest that while NLRC5 expression is RIG-I dependent, NLRC5 contributes to mount a robust IFN-β induction in response to influenza infection. The type I IFN, IFN-β, is pivotal for the antiviral host defense. It is involved in the transcriptional activation of many PRRs including NLRC5 [11, 16], a large number of so-called IFN-stimulated genes as well as its own production. To examine whether NLRC5 expression is driven directly by IFN-β also upon influenza infection, we treated A549 with recombinant human IFN-β and analyzed the expression of IFN-β, NLRC5, and RIG-I mRNA. As shown in Supporting Information Figure 5A, recombinant IFN-β treatment not only induced the expression of IFN-β, but also the expression of NLRC5 and RIG-I. Dependency of NLRC5 expression on RIG-I-induced IFN-β was further confirmed in studies using neutralizing IFN-α/β antibodies, which prevented PR8ΔNS1-induced expression of RIG-I, NLRC5, and IFN-β (Supporting Information Fig. 5B).
Figure 6.
NLRC5 is required for robust IFN-β and RIG-I expression. A549 cells transfected with control siRNA or NLRC5 siRNA were infected with NS1-del PR8 (MOI 1.0). (A–C) Cells were harvested 0, 24, 48, 72, and 96 h postinfection and analyzed for (A) IFN-β, (B) RIG-I, and (C) NLRC5 mRNA expression, relative to β-actin by real-time RT-PCR. Data are shown as mean + SD of three samples per group, pooled from three independent experiments carried out in duplicate. ANOVA was performed to compare control siRNA versus NLRC5 siRNA treated A549 cells and p values <0.05 are indicated with an asterisk.
Taken together, these results confirmed previous findings, showing that NLRC5 mRNA is regulated by type I IFNs and suggest that it is induced upon activation of RIG-I by influenza virus. More interestingly, we revealed that NLRC5 positively affects RIG-I signaling and that NLRC5 is required to maintain a long-lasting antiviral response toward influenza virus in human host cells.
The N-terminal and NACHT domains of NLRC5 are critical for its antiviral function
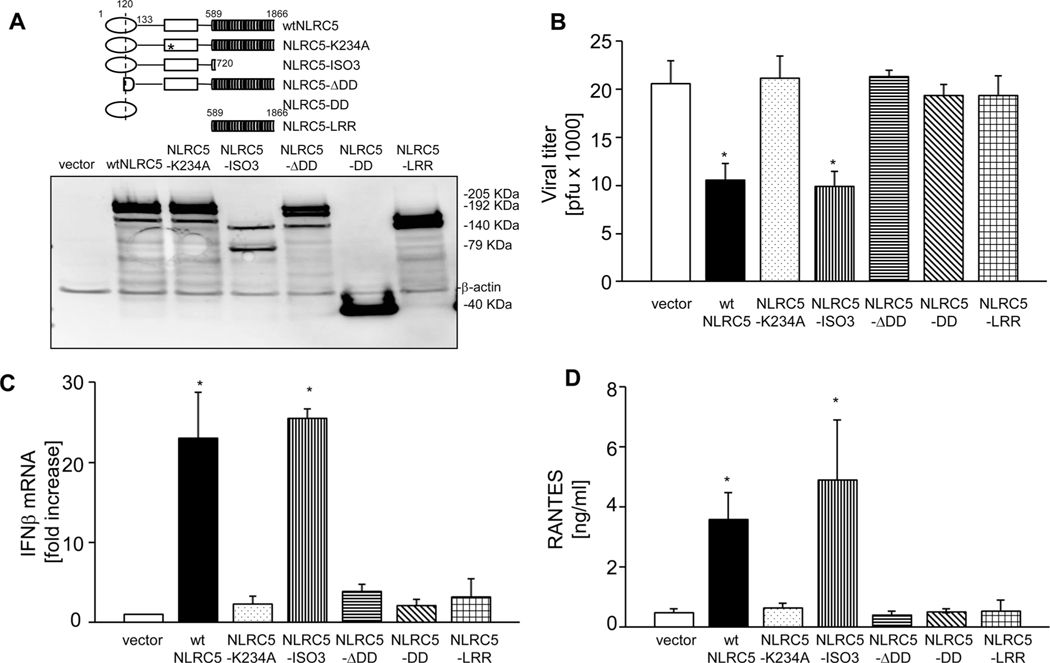
Next, we examined, which domains of NLRC5 contribute to the antiviral activity. To this end, myc-tagged wtNLRC5 and deletion mutants thereof [17] were expressed in A549 cells (Fig. 7A). As expected, transfection of A549 cells with NLRC5 significantly reduced influenza viral titers following infection with PR8 (Fig. 7B). By contrast, overexpression of a Walker A mutant of NLRC5 (NLRC5-K234A) or of a deletion construct that lacks the N-terminal domain (NLRC5-ΔDD), contain only the N-terminal death-fold domain (NLRC5-DD) or of the LRR domain of NLRC5 (NLRC5-LRR), all did not significantly affect viral titers (Fig. 7B). However, overexpression of isoform 3 of NLRC5 (NLRC5-ISO3), which lacks the LRR domain [13], inhibited PR8 replication to a similar extent as wtNLRC5 (Fig. 7B). These results indicate that NLRC5-mediated restriction of influenza virus replication is dependent on a functional ATPase domain and the presence of N-terminal domain of NLRC5. Consistent with these results, only wtNLRC5 and isoform 3 but not NLRC5-K234A, NLRC5-ΔDD, NLRC5-DD, or NLRC5-LRR induced IFN-β mRNA expression upon infection (Fig. 7C). Further, increased levels of RANTES were detectable in the supernatants of A549 cells transfected with wtNLRC5 and NLRC5-ISO3 but not in the supernatants from cells transfected with the other NLRC5 constructs (Fig. 7D).
Figure 7.
The NLRC5 death domain and nucleotide-binding domain is critical for NLRC5-mediated antiviral function. A549 cells were transfected with vector alone or with myc-tagged wtNLRC5, NLRC5-K234A, NLRC5-ISO3, NLRC5-ΔDD, NLRC5-DD, or LRR domain of NLRC5 and subsequently infected with PR8 (MOI 1.0) for 24 h. (A) The upper panel shows the schematic representation of the NLRC5 constructs used and the lower panel shows expression of NLRC5 constructs in the cells by immunoblotting. β-Actin was used as a loading control. (B, C) Supernatants were collected and cells were harvested to determine (B) viral titers by plaque assay and (C) IFN-β mRNA expression, relative to β-actin by real-time RT-PCR. (D) Secretion of CCL5 (RANTES) in cell supernatants was measured by ELISA. Data are shown as mean + SD of three samples per group, pooled from three independent experiments carried out in duplicate. ANOVA was performed to compare control vector versus myc-NLRC5 expression vectors transfected A549 cells and p values <0.05 are indicated with an asterisk.
Taken together, these data suggest that the DD and NACHT ATPase domain are sufficient for NLRC5-mediated antiviral responses toward influenza. Of note, a construct comprising only these domains is unable to promote MHC class I expression [17], functionally separating the role of NLRC5 in RIG-I-mediated antiviral immunity from its contribution to MHC-I-dependent antigen presentation.
NLRC5 forms a complex with RIG-I and influenza virus protein NS1
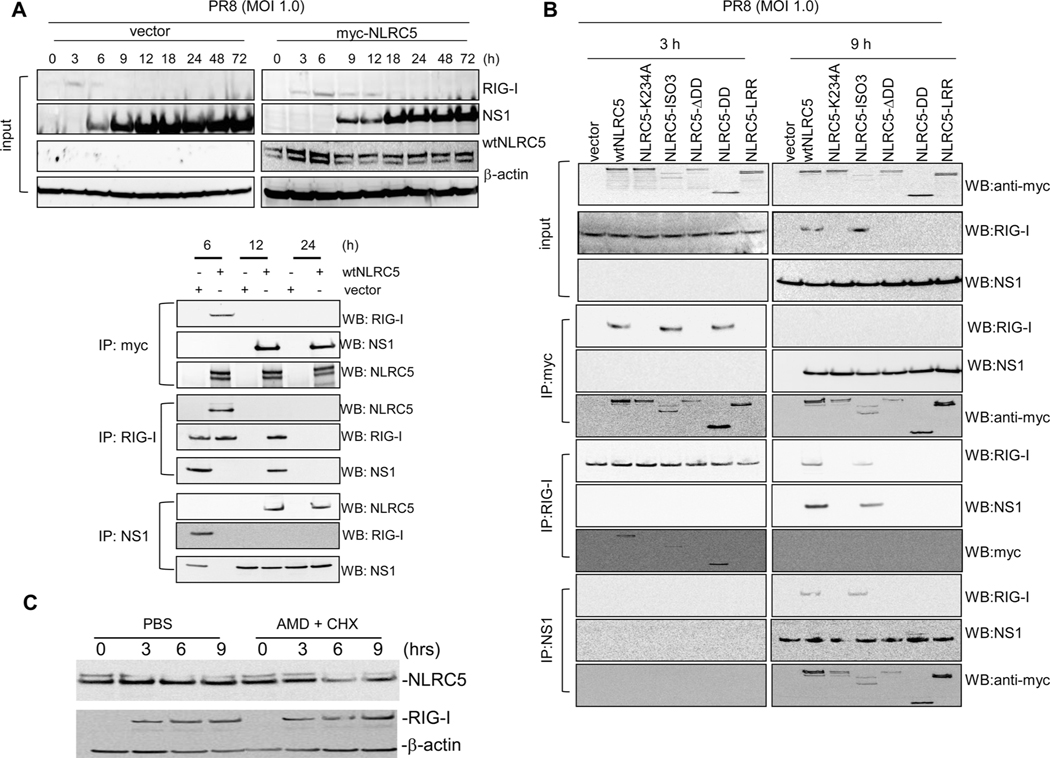
Viruses can suppress host innate immune responses. Influenza NS1 has been shown to interact directly with RIG-I and other PRRs to dampen host innate immune activation [28, 30, 35, 38, 43–46]. Our observation that the antiviral activity of NLRC5 is dependent on RIG-I prompted us to investigate if NLRC5 might act in a physical complex with RIG-I. To this end, we carried out coimmunoprecipitation studies. A549 cells, transfected either with vector or myc-tagged wtNLRC5, were infected with PR8 24 h post-transfection. Cells were harvested at the indicated time postinfection and protein expression of RIG-I, myc-NLRC5, and viral NS1 was determined by immunoblotting (Fig. 8A). In vector-transfected A549 cells, RIG-I protein expression was detected at 3 h post-PR8 infection and rapidly declined starting 6 h postinfection, and was undetectable after 9 h. Consistent with our quantitative RT-PCR data (Fig. 1E), NLRC5 overexpression increased and prolonged RIG-I expression that was still detectable up to 12 h p.i. Notably, NLRC5 expression also delayed the expression of NS1 in PR8-infected A549 cells. In vector-transfected cells, NS1 was detectable starting at 6 h following PR8 infection, whereas in NLRC5-transfected A549 cells, NS1 appeared only after 9 h postinfection and its expression appeared to be lower than in vector-transfected A549 cells, consistent with the lower viral titer found in these cells (Fig. 8A). To examine if NLRC5 formed a complex with endogenous RIG-I or viral NS1, coimmunoprecipitations were conducted at 6, 12, and 24 h postinfection using anti-myc, anti-RIG-I, and anti-NS1 antibodies to precipitate myc-NLRC5, RIG-I, and NS1, respectively. Precipitation of myc-NLRC5 copurified RIG-I at 6 h post-PR8 infection but not at the later time points. Notably, NS1 bound to NLRC5 at 12 and 24 h postinfection but not at 6 h, suggesting that NLRC5 can form a complex with NS1 and RIG-I upon infection at the time when these proteins are most highly expressed (Fig. 8A). Precipitation of RIG-I copurified NLRC5 at 6 h postinfection. Notably, at this time point, NS1 was absent in A549 cells (Fig. 8A). NS1 coprecipitated with RIG-I at 12 h post-infection, however, we did not detect NLRC5 in coimmunoprecipitate at this time point despite its presence in the cells (Fig. 8A). These data indicate that while RIG-I is capable of interacting with NLRC5, NS1 can prevent this interaction. Precipitation of NS1 using NS1-specific antibodies copurified RIG-I in vector-transfected cells 6 h postinfection. At this time point, NS1 was absent in NLRC5 transfected cells (Fig. 8A). NS1 copurified NLRC5 at 12 and 24 h postinfection suggesting NS1 can bind to NLRC5 regardless of the presence of absence of RIG-I (Fig. 8A).
Figure 8.
NLRC5 stabilizes RIG-I. (A) A549 cells transfected with vector alone or myc-wtNLRC5 were infected with PR8 (MOI 1.0) for 0, 3, 6, 9, 12, 18, 24, 48, and 72 h and harvested for NLRC5, RIG-I, and NS1 expression and coimmunoprecipitation assay. β-Actin was used as a loading control. Cell lysates from 6, 12, and 24 h were immunoprecipitated with anti-myc, anti-NS1, or anti-RIG-I antibodies and immunoprecipitates were analyzed for the presence of RIG-I, NS1, and NLRC5 by immunoblotting. (B) To map the domain responsible for NLRC5 interaction with RIG-I and NS1, A549 cells were transfected for 24 h with myc-vector alone or with myc-tagged wtNLRC5, NLRC5-K234A, NLRC5-ISO3, NLRC5-ΔDD, NLRC5-DD, or NLRC5-LRR mutants and then infected with PR8 (MOI 1.0) for 3 or 9 h. Cell lysates were immunoprecipitated with anti-myc, anti-NS1, or anti-RIG-I antibodies and immunoprecipitates were analyzed for the presence of RIG-I, NS1, and NLRC5 by immunoblotting. β-Actin was used as a loading control. The input for the immunoblot was about 5% of the total cell lysate. (C) A549 cells transfected with wtNLRC5 were infected with PR8 (MOI 1.0) in the presence or absence of actinomycin D (5 μg/mL)/cyclohexamide (20 μg/mL) combination. Cell lysates were analyzed for RIG-I and NLRC5 expression at 0, 3, 6, and 9 h postinfection by immunoblotting. Data shown are from one single experiment representative of two independent experiments.
Our domain mapping studies revealed that the antiviral activity of NLRC5 depended on the DD and NACHT domain. We next investigated if the N-terminal DD and the central NACHT domain also participate in the above observed interaction with RIG-I and NS1. To this end, we overexpressed myc-tagged wtNLRC5 or the NLRC5 constructs NLRC5-K234A, NLRC5-ISO3, NLRC5-ΔDD, NLRC5-DD, or NLRC5-LRR domain in A549 cells and subsequently infected these cells with PR8 influenza virus. Cell lysates were harvested at 3 and 9 h postinfection and were analyzed for the expression of NLRC5, RIG-I, and NS1 by immunoblotting (Fig. 8B). These time points were chosen due to the high abundance of RIG-I, myc-NLRC5, and/or NS1 at these times. RIG-I expression was well detectable at 3 h post-PR8 infection irrespective of the expression of the NLRC5 constructs, whereas NS1 was not detectable at that this time. As expected RIG-I was not detectable at 9 h postinfection in mock-treated cells, however it was readily detected in cells expressing wtNLRC5 or the ISO3 construct (Fig. 8B). In any case, NS1 was detectable at 9 h postinfection (Fig. 8B). Immunoprecipitation of NLRC5 showed that wtNLRC5 and NLRC5-ISO3 but also the N-terminal DD of NLRC5 interacted with RIG-I at 3 h post-PR8 infection. However, at 9 h postinfection, we did not detect RIG-I in any coimmunoprecipitation, although RIG-I was clearly present in cells expressing wtNLRC5 or ISO3 (Fig. 8B). Interestingly, NS1 coimmunoprecipitated with all the NLRC5 constructs 9 h postinfection, however not in control immunoprecipitations in both directions (Fig. 8B). Immunoprecipitation of RIG-I copurified wtNLRC5, NLRC5-ISO3, and NLRC5-DD. Notably, NLRC5-DD did not induce antiviral effects (Fig. 7). Immunoprecipitation of NS1 copurified RIG-I in wtNLRC5 and NLRC5-ISO3 expressing A549 cells at 9 h postinfection when RIG-I was present only in wtNLRC5 or NLRC5-ISO3 expressing A549 cells (Fig. 8B).
NLRC5 overexpression enhanced RIG-I expression in PR8-infected cells, and RIG-I was detectable up to 12 h by immunoblotting. In contrast, in vector-transfected cells, PR8-induced RIG-I was detectable only up to 6 h postinfection (Fig. 8A). This could be a result of enhanced transcription and/or translation of RIG-I or decreased RIG-I degradation. To understand the role of NLRC5 in enhanced RIG-I expression, we treated NLRC5-transfected A549 cells with actinomycin D and cyclohexamide to block transcription and translation, and subsequently infected these with PR8 (MOI 1.0). Cells were harvested at 0, 3, 6, and 9 h postinfection to analyze expression of RIG-I by immunoblotting. These data show that treatment with actinomycin D and cyclohexamide did not reduce the steady-state levels of RIG-I suggesting that NLRC5 affects the half-life of the RIG-I protein (Fig. 8C).
In summary, our results suggest that NLRC5 extends and stabilizes influenza virus induced RIG-I expression and delays NS1 expression. NS1 counteracts NLRC5-mediated enhancement of RIG-I activity by competing for binding to RIG-I and/or by interacting with NLRC5 thereby preventing the binding of NLRC5 to RIG-I. RIG-I interaction with NLRC5 required the NLRC5 DD, which was sufficient for binding, though a Walker A mutant of NLRC5 failed to interact with RIG-I despite the presence of DD.
Discussion
Members of NLR protein family are known for their primary role in mediating inflammatory response upon pathogenic insult. Recently, several members of this family including NLRC5 have been proposed to function as critical regulators of antiviral responses [5, 13, 16, 20, 47, 48]. NLRC5 has been shown to contribute to type I IFN signaling and MHC class I gene expression [10–12, 17, 18, 49–52], both pathways that are pivotal for the host to combat viral infection. NLRC5 is predominantly expressed in cells of the lymphoid lineage, in spleen, thymus, BM, and LNs, and, in addition, in mucosal epithelial surfaces such as the lung, small intestine, colon, and the uterus that are in direct contact with pathogens [13, 16, 19]. Several isoforms of NLRC5 have been reported, which differ in the composition of the LRR domain [13]. The presence and expression of these isoforms are tissue and cell-type specific, however, their functional roles remain elusive. NLRC5 has been reported to be a cytosolic protein [13, 16, 19, 53] that can shuttle to the nucleus, which is consistent with the function of NLRC5 as a transcriptional enhancer and its contribution to cytosolic type I IFN pathways.
Abundant expression of NLRC5 in lung tissue and immune cells [20] prompted us to investigate its role in influenza virus infection. We observed a partial but significant reduction in viral titers as well as increased expression of IFN-β and RIG-I expression in human respiratory epithelial cell lines overexpressing NLRC5, suggesting an involvement of NLRC5 in the antiviral innate immune response. These results confirm previous findings, showing that Sendai virus and polyI:C-mediated type I IFN responses are partially dependent on NLRC5 in different human cells [13] and that both CMV- or polyI:C-induced type I IFN and proinflammatory cytokines are reduced in NLRC5 knockdown fibroblasts compared with control cells [16]. In contrast, NLRC5 was also reported to negatively regulate antiviral signaling and type I IFN production by interacting with IKKα/β, and RIG-I and MDA5 [20]. The reasons for these discrepant findings are unclear. However, different cell types have been used in these studies with different outcomes. A study by Kumar et al. suggests that NLRC5 deficiency does not influence cytokine induction by virus and bacterial infection in BM-derived cells in NLRC5 KO mice [21]. Also, Tong and colleagues found little effect of the NLRC5 status on PAMP stimulation of BM-derived cells, however they reported increased IFN-γ and IL-6 in mouse embryonic fibroblast derived from NLRC5 KO mice [47]. This might suggest that NLRC5-mediated host responses vary greatly among different cell types and in response to diverse stimuli. Accordingly, we could not detect endogenous NLRC5 in HEK293T cells even upon PR8 or PR8ΔNS1 infection. Moreover, NLRC5 overexpression did not result in enhanced antiviral effect against influenza in these cells, which is consistent with our previous study that showed that NLRC5 does not positively affect type I IFN responses in HEK293T cells [13]. This might not come as a surprise as influenza virus primarily infects respiratory epithelial cells; and HEK293T cells are not a natural target for influenza virus infection and thus may not have evolved to response to flu the same way as the lung epithelial cells do.
It is well established that influenza viruses activate RIG-I [26, 28, 54]. Here, we confirmed interaction of NLRC5 with RIG-I and expanded on this finding by revealing a novel role of human NLRC5 in antiviral response toward influenza infection. NLRC5 overexpression significantly reduced PR8 replication and enhanced antiviral effects in both A549 and NHBE cells. A specific role of NLRC5 in innate immune responses toward influenza virus was confirmed by overexpressing an unrelated NLR family protein CIITA in A549 cells, which did not reduce PR8 replication or enhanced RIG-I or type I IFN induction. NLRC5 was neither necessary nor sufficient for viral induction of RIG-I expression, however, NLRC5 knockdown significantly reduced IFN-β induction. Conversely, RIG-I knockdown completely abrogated virus-induced NLRC5 expression as well as type I IFN. Importantly, we confirmed the contribution of NLRC5 to influenza induced innate immune responses in primary mouse normal bronchial and tracheal epithelial cells, showing that our results in cell lines were not affected by the transformed state of these cells. Moreover, NLRC5 did not affect LPS-induced responses in A549 cells suggesting that the effect of NLRC5 on influenza virus induced IFN responses is rather specific.
We and others have reported that initial induction of IFN-β by viral PAMPs can strengthen antiviral response by increasing PRRs expression and subsequent IFN-β production in an autocrine and paracrine manner [26, 55, 56]. In fact, this is crucial for the antiviral response in early stages of infection, where viral PAMPs might be limiting to induce strong antiviral response [42]. This generally accepted hypothesis is also supported by our findings that treatment with IFN-β induced an antiviral response, whereas neutralizing antibodies against IFN-α/β abolished the antiviral effect. The final outcome of host defense against pathogens depends on the strength and duration of the antiviral response. Our studies indicate that NLRC5 expression was required for a robust RIG-I-dependent type I IFN response and inhibition of virus replication, in particular, at later time points postinfection.
Human NLRC5 is the largest protein among the human NLR family members and consists of an N-terminal domain that comprises a predicted DD. The predicted NACHT domain is located between position N-terminal and C-terminal domains. The predicted C-terminal LRR region consists of 713 amino acid residues and is largest among NLR members [13]. We and others have identified that these domains contribute to NLRC5 subcellular localization and are involved in its transit through nucleus [17, 53]. In order to determine the role of these domains in antiviral signaling, we used a series of NLRC5 mutants and truncated versions as described in materials and methods section. Results indicated that the DD along with the NACHT domain (referred to as NLRC5 isoform 3 (NLRC5-ISO3)) were sufficient for the NLRC5 function in antiviral responses. By contrast, a point mutation in the conserved lysine (K234) of the Walker A motif of NLRC5 (NLRC5-K234A) resulted in loss of NLRC5 function. Of note, NLRC5 isoform 3 (NLRC5-ISO3), which lacks the entire LRR domain, was found to be equally potent as wtNLRC5. In contrast, NLRC5-ΔDD, a deletion mutant that lacks DD or the DD alone and the LRR domain alone, failed to induce an antiviral response. This showed that the DD and NACHT domain are needed, whereas the LRR domain of NLRC5 is dispensable for the antiviral effect of NLRC5 described here. We and others have recently shown that both the DD and LRR domain are critical for NLRC5-dependent MHC class I activation [12, 17]. Notably, NLRC5 isoform 3 is unable to activate MHC class I expression despite its competence to shuttle to the nucleus [17]. In contrast, this isoform was sufficient to enhance the influenza-induced type I IFN response. Our results provide evidence that the functions of NLRC5 in MHC class I gene expression and in the regulation of type-I IFN responses are mediated through different domains in NLRC5. Whether cytoplasmic availability or nuclear localization of NLRC5 is the limiting factor for its antiviral function is not known and needs further investigation.
Based on our findings that human NLRC5 contributes to influenza-induced RIG-I activation, we wanted to explore if NLRC5 might form a complex with RIG-I and which domains of NLRC5 are involved in such an interaction. We used an in vitro influenza virus infection model to study the interaction of NLRC5 with endogenous RIG-I and viral NS1 protein. NLRC5 did interact with RIG-I and the N-terminal DD of NLRC5 appeared to be involved in this interaction. Consistent with a loss of function, the Walker A mutant K234A of NLRC5 did not pull down RIG-I despite the presence of the DD. It is not clear if NLRC5 undergoes structural rearrangements needed for protein–protein interaction upon activation like other PRRs. If so, a functional ATPase NACHT domain might be required to achieve this. RIG-I interaction with NLRC5 has also been shown by Cui et al. [20], however, these authors identified a negative regulation of antiviral response by NLRC5. Discrepancies between Cui et al. and our findings in the functional outcome of the NLRC5 and RIG-I interaction might be due to different cell types or experimental conditions, and certainly requires more independent investigations. We provide evidence that NLRC5 can stabilize the RIG-I protein upon influenza virus infection to enhance RIG-I signaling, suggesting that at least in human epithelial cells, which have not been analyzed by Cui and co-workers, NLRC5 positively contributes to RIG-I-dependent antiviral responses
To evade host antiviral response, pathogens have evolved various strategies to escape the innate immune system. We [35] and others [28, 30, 57] have reported that influenza virus NS1 protein can target innate immune receptor RIG-I and its signaling components to suppress host innate immune defenses. In the present study, we found that while influenza virus PR8 only modestly induced NLRC5 expression and IFN-β, PR8ΔNS1, which lacks the NS1 protein, induced robust expression of NLRC5 as well as IFN-β. Ectopic expression of NS1 in human cells not only inhibited type I IFN but also significantly decreased NLRC5 and RIG-I mRNA levels. It is known that NS1 protein directly interacts with RIG-I by coimmunoprecipitation studies [28]. Our data cannot discriminate if NS1 interacts with NLRC5 directly, if it targets an NLRC5/RIG-I complex, or if it sequesters NLRC5 protein thereby preventing its interaction with RIG-I. However, in human cells NS1 coimmunoprecipitated with all of the tested NLRC5 constructs, supporting an interaction of NS1 with multiple domains of NLRC5. Moreover, the presence of RIG-I was not needed for its interaction.
In summary, we show that during early influenza virus infection, NLRC5 binds to RIG-I to form a complex in an LRR-independent manner. This interaction stabilizes RIG-I and enhances downstream type I IFN responses. Viral NS1 interferes with the NLRC5/RIG-I complex formation to inhibit antiviral signaling. This might be due to direct interaction with NLRC5 and/or RIG-I. NS1 interaction with NLRC5 may also sequester NLRC5 thereby preventing formation of RIG-I/NLRC5 complex and activation of antiviral defenses. Of note, this is the first report of a role for NLRC5 in antiviral immunity that is functionally distinguishable from the well-established function of NLRC5 in MHC class I gene expression, which, in contrast to the antiviral activity of NLRC5, depends on the LRRs of NLRC5 [17]. Differential expression of isoforms of NLRC5 that lack part or all of the LRRs might, therefore, relate to different antiviral activities of the corresponding cell types.
Further studies will help to determine the structural and functional role of the domains involved in the NLRC5/RIG-I/NS1 complex formation during viral infection and to gain insights into developing novel targets for next-generation antiviral agents.
Materials and methods
Plasmids and reagents
Myc-tagged NLRC5 full-length (NP 115582; flag-NLRC5 or myc-NLRC5) and mutants (NLRC5-K234A, NLRC5-ISO3, NLRC5-ΔDD, NLRC5-DD, and NLRC5-LRR) are described previously [13]. Flag-tagged CIITA was a kind gift from Victor Steimle, Sherbrook University, Canada. The myc-NS1 expression vector used in this study has been described previously [35]. NFκB and IFN LUC reporter plasmid was obtained from Dr. Rongtuan Lin, McGill University, Canada. Anti-RIG-I and anti-NS1 antibodies were purchased from Santa Cruz Biotechnology (CA, USA). Actinomycin D, cyclohexamide, anti-flag, anti-myc, and anti-β-actin antibodies were purchased from Sigma Aldrich (St. Louis, USA). Anti-CIITA and anti-IFN-α/β neutralizing antibodies were purchased from Millipore (MA, USA). Anti-NLRC5 monoclonal antibody 3H8 was described previously [13]. The anti-influenza A virus NP antibody and laboratory strain A/Puerto Rico/8/34 [(PR8); H1N1] were obtained from the Influenza Reagents Repository, Centers for Disease Control and Prevention (CDC), Atlanta, USA; the NS1-del mutant (PR8ΔNS1) was provided by Adolfo García-Sastre, Mount Sinai School of Medicine, NY, USA. Human and mouse IFN-β and CCL5 ELISA kits were from R&D system Inc. (MN, USA).
Cell cultures and virus infection
Human lung epithelial cell line A549, HEK293T (ATCC, VA, USA), and NHBE cells (Lonza, Switzerland) were maintained as described [13, 26]. Mouse normal bronchial and tracheal epithelial cells were purchased from (CHI Scientific, MA, USA) and maintained as per manufacturer instructions. 106 cells in six-well plates were transfected with the empty vector or NLRC5 expression vectors for 24 h. Cells were then infected with PR8 or PR8ΔNS1 virus at an MOI of 1.0 or 1.0 focal forming units with trypsin supplement as described previously [26]. Twenty-four hours postinfection, cells were harvested for RNA and protein analysis and cell-culture supernatants were collected and stored at −80°C for determination of viral titer by plaque assay as described previously using MDCK cells [26]. Three independent experiments were performed at different times with each treatment carried out in duplicate cultures.
siRNA knockdown studies
Gene-specific siRNAs to silence NLRC5 [13] or RIG-I [26] in A549 or NHBE or mouse normal bronchial and tracheal epithelial cells were purchased from Thermo Fisher Scientific (Lafayette, CO, USA), and were used as described in the manufacturer’s protocol. Briefly, cells were plated at a density of 106/well in a six-well plate and transfected with 75 nM each of the gene-specific siRNA 24 h prior to infection with PR8 or PR8-NS1-del virus. After 24 h, transfected cells were harvested and analyzed for NLRC5, RIG-I, IFN-β, or NS1 expression by real-time PCR or Western blot.
Real-time RT-PCR
Total RNA was isolated from cells using the RNAeasy kit (Qiagen, Valencia, CA, USA) and real-time RT-PCR was conducted using a Stratagene Mx3005P Q-PCR machine for mRNA expression of NLRC5, RIG-I, IFN-β, IFN-α NP vRNA, RANTES, and β-actin. For each sample, 2 μg of RNA was reverse-transcribed using Superscript II Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Parallel reactions without reverse transcriptase were included as negative controls. Reverse transcription reactions were analyzed using syber green Q-PCR reagents (Sigma Aldrich, St. Louis, USA). PCR conditions were as follows: 95°C for 15 s, annealing at 57 °C for 30 s, and extension at 72 °C for 30 s for a total of 45 cycles. The threshold cycle number for cDNA was normalized to that of βactin mRNA, and the resulting value was converted to a linear scale. Data from three independent experiments were used for analysis. Primer sets used for these studies have been described previously [13, 26, 28].
Immunoblotting and coimmunoprecipitation
Cells plated in 100-mm tissues culture plates were harvested in RIPA buffer (Sigma Aldrich), and cell lysates were incubated with primary antibody overnight at 4°C followed by incubation with protein A Dynabeads (Invitrogen) for 2 h. The beads were washed three times with PBS, suspended in Laemmli buffer (62.5 mM Tris-HCl pH 6.8, 25% glycerol, 2% SDS, 0.01% Bromophenol blue), boiled for 10 min, and centrifuged at 12 000 × g for 10 min at 4°C. Supernatants were collected and analyzed by Western blotting as described previously [26].
LUC assay
A549 or NHBE cells were cotransfected with IFN-β or NFκB promoter LUC reporter plasmid and harvested at indicated time points for LUC activity using dual LUC assay kit (Promega, WI, USA) as per manufacturer’s instructions. Mean and SDs were calculated from duplicate or triplicate cultures and are representative of at least three independent experiments.
Confocal microscopy
A549 or NHBE cells were cultured on collagen-coated glass cover slips and transfected with vector or wtNLRC5 using lipofectamine as described previously [28] followed by PR8 infection. Cells on the cover slips were washed with cold PBS and fixed with 4% paraformaldehyde for 10 min. Cells were permeabilized in 0.1% Triton X-100 in PBS and blocked with 1% BSA and 5% normal goat serum in PBS. Following blocking, cover slips were incubated overnight with anti-myc mouse monoclonal antibodies (1:1000 dilution), and anti-influenza A virus NP mouse monoclonal antibody (1:2 500). Alexa 488 conjugated anti-mouse IgG (LifeTechnologies, USA) was used as the secondary antibody. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells weremounted on slides with Prolong Antifade Mounting Media (Molecular Probes). Images were examined using an LSM 710 inverted confocal microscope (Zeiss, Oberkochen, Germany). For calculating NP-positive cells, five independent ×20 image fields with approximately 100 cells/field from each condition were evaluated. The total number of NP-positive cells was divided by the total number of DAPI positive cells for each field to estimate the percent of NP positive cells. Data are represented as the percentage ± SD of five ×20 fields from each experimental condition.
Statistical analysis
To determine the statistical significance, we used analysis of variance (ANOVA) using GraphPad PRISM 5 and a value of- p ≤ 0.05 was considered significant when compared with respective controls.
Supplementary Material
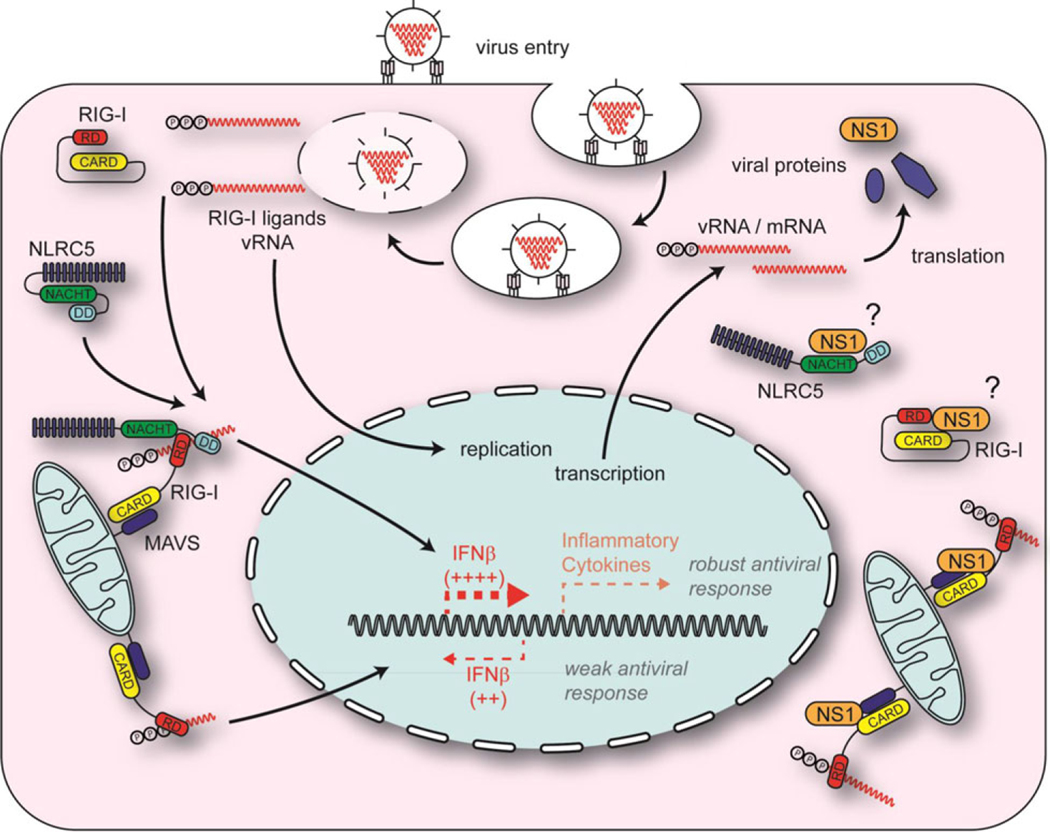
Figure 9.
Schematic representation of the role of NLRC5 in influenza virus infection. NLRC5 induces a RIG-I-dependent robust antiviral response against influenza virus and induced type I IFN. Type I IFN upregulated NLRC5, RIG-I expression, and subsequently its own production (left). Absence of NLRC5 results in a weaker type-I IFN response. NS1 can suppress the NLRC5-mediated antiviral response by interacting with the RIG-I/NLRC5 complex or by sequestering NLRC5 and preventing its interaction with RIG-I (top right). While 5′PPP-RNA is a natural ligand for RIG-I, it is not known if NLRC5 binds to viral RNA or undergoes conformational changes (bottom left and right). (Protein–protein interactions shown in the figure do not represent the specific domains responsible for binding.)
Acknowledgments:
This work was supported by the German Research Foundation (DFG) grants SFB670 and KU1945/2-1 to T.A.K. A.N. acknowledges support by the Koeln Fortune Program/Faculty of Medicine, University of Cologne.
Disclaimer:
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of Centers for Disease Control and Prevention.
Abbreviations:
- DD
death domain
- NHBE
normal human bronchial epithelial
- NLR
nucleotide-binding domain and leucine-rich repeat containing receptor
- NP
nucleoprotein
Footnotes
Conflict of interest: The authors declare no financial or commercial conflict of interest.
References
- 1.Akira S, Uematsu S. and Takeuchi O, Pathogen recognition and innate immunity. Cell 2006. 124: 783–801. [DOI] [PubMed] [Google Scholar]
- 2.Kawai T. and Akira S, TLR signaling. Cell Death Differ. 2006. 13: 816–825. [DOI] [PubMed] [Google Scholar]
- 3.Ranjan P, Bowzard JB, Schwerzmann JW, Jeisy-Scott V, Fujita T. and Sambhara S, Cytoplasmic nucleic acid sensors in antiviral immunity. Trends Mol. Med 2009. 15: 359–368. [DOI] [PubMed] [Google Scholar]
- 4.Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M. et al. , Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell 2008. 29: 428–440. [DOI] [PubMed] [Google Scholar]
- 5.Lamkanfi M. and Kanneganti TD, Regulation of immune pathways by the NOD-like receptor NLRC5. Immunobiology 2012. 217: 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y. and Shao F, NLRC5: a NOD-like receptor protein with many faces in immune regulation. Cell Res. 2012. 22: 1099–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanneganti TD, Central roles of NLRs and inflammasomes in viral infection. Nat. Rev. Immunol 2010. 10: 688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanneganti TD, Lamkanfi M. and Nunez G, Intracellular NOD-like receptors in host defense and disease. Immunity 2007. 27: 549–559. [DOI] [PubMed] [Google Scholar]
- 9.Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA, Gris D. et al. , NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity 2011. 34: 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi KS and van den Elsen PJ, NLRC5: a key regulator of MHC class I-dependent immune responses. Nat. Rev. Immunol 2012. 12: 813–820. [DOI] [PubMed] [Google Scholar]
- 11.Staehli F, Ludigs K, Heinz LX, Seguin-Estevez Q, Ferrero I, Braun M, Schroder K. et al. , NLRC5 deficiency selectively impairs MHC class I-dependent lymphocyte killing by cytotoxic T cells. J. Immunol 2012. 188: 3820–3828. [DOI] [PubMed] [Google Scholar]
- 12.Meissner TB, Li A, Biswas A, Lee KH, Liu YJ, Bayir E, Iliopoulos D. et al. , NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc. Natl. Acad. Sci. USA 2010. 107: 13794–13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neerincx A, Lautz K, Menning M, Kremmer E, Zigrino P, Hosel M, Buning H. et al. , A role for the human nucleotide-binding domain, leucine-rich repeat-containing family member NLRC5 in antiviral responses. J. Biol. Chem 2010. 285: 26223–26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou H. and Glimcher LH, Human MHC class II gene transcription directed by the carboxyl terminus of CIITA, one of the defective genes in type II MHC combined immune deficiency. Immunity 1995. 2: 545–553. [DOI] [PubMed] [Google Scholar]
- 15.Gutte PG, Jurt S, Grutter MG and Zerbe O, Unusual structural features revealed by the solution NMR structure of the NLRC5 caspase recruitment domain. Biochemistry 2014. 53: 3106–3117. [DOI] [PubMed] [Google Scholar]
- 16.Kuenzel S, Till A, Winkler M, Hasler R, Lipinski S, Jung S, Grotzinger J. et al. , The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J. Immunol 2010. 184: 1990–2000. [DOI] [PubMed] [Google Scholar]
- 17.Neerincx A, Rodriguez GM, Steimle V. and Kufer TA, NLRC5 controls basal MHC class I gene expression in an MHC enhanceosome-dependent manner. J. Immunol 2012. 188: 4940–4950. [DOI] [PubMed] [Google Scholar]
- 18.Biswas A, Meissner TB, Kawai T. and Kobayashi KS, Cutting edge: impaired MHC class I expression in mice deficient for Nlrc5/class I transactivator. J. Immunol 2012. 189: 516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benko S, Magalhaes JG, Philpott DJ and Girardin SE, NLRC5 limits the activation of inflammatory pathways. J. Immunol 2010. 185: 1681–1691. [DOI] [PubMed] [Google Scholar]
- 20.Cui J, Zhu L, Xia X, Wang HY, Legras X, Hong J, Ji J. et al. , NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell 2010. 141: 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar H, Pandey S, Zou J, Kumagai Y, Takahashi K, Akira S. and Kawai T, NLRC5 deficiency does not influence cytokine induction by virus and bacteria infections. J. Immunol 2011. 186: 994–1000. [DOI] [PubMed] [Google Scholar]
- 22.Fiore AE, Shay DK, Broder K, Iskander JK, Uyeki TM, Mootrey G, Bresee JS et al. , Prevention and control of influenza: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2008. MMWR Recomm. Rep 2008. 57: 1–60. [PubMed] [Google Scholar]
- 23.Deyde VM, Xu X, Bright RA, Shaw M, Smith CB, Zhang Y, Shu Y. et al. , Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J. Infect. Dis 2007. 196: 249–257. [DOI] [PubMed] [Google Scholar]
- 24.He G, Qiao J, Dong C, He C, Zhao L. and Tian Y, Amantadine-resistance among H5N1 avian influenza viruses isolated in Northern China. Antiviral Res. 2008. 77: 72–76. [DOI] [PubMed] [Google Scholar]
- 25.Ireton RC and Gale M Jr., RIG-I like receptors in antiviral immunity and therapeutic applications. Viruses 2011. 3: 906–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranjan P, Jayashankar L, Deyde V, Zeng H, Davis WG, Pearce MB, Bowzard JB et al. , 5’PPP-RNA induced RIG-I activation inhibits drug-resistant avian H5N1 as well as 1918 and 2009 pandemic influenza virus replication. Virol. J 2010. 7: 102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Opitz B, Rejaibi A, Dauber B, Eckhard J, Vinzing M, Schmeck B, Hippenstiel S. et al. , IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol. 2007. 9: 930–938. [DOI] [PubMed] [Google Scholar]
- 28.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F. and Reis e Sousa C, RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science 2006. 314: 997–1001. [DOI] [PubMed] [Google Scholar]
- 29.Le Goffic R, Pothlichet J, Vitour D, Fujita T, Meurs E, Chignard M. and Si-Tahar M, Cutting edge: influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J. Immunol 2007. 178: 3368–3372. [DOI] [PubMed] [Google Scholar]
- 30.Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M Jr. and Garcia-Sastre A, Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol 2007. 81: 514–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spiropoulou CF, Ranjan P, Pearce MB, Sealy TK, Albarino CG, Gangappa S, Fujita T. et al. , RIG-I activation inhibits ebolavirus replication. Virology 2009. 392: 11–15. [DOI] [PubMed] [Google Scholar]
- 32.Scott MJ, Chen C, Sun Q. and Billiar TR, Hepatocytes express functional NOD1 and NOD2 receptors: a role for NOD1 in hepatocyte CC and CXC chemokine production. J. Hepatol 2010. 53: 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu F, Maniar A, Diaz MQ, Chapoval AI and Medvedev AE, Activation of cytokine-producing and antitumor activities of natural killer cells and macrophages by engagement of Toll-like and NOD-like receptors. Innate Immun. 2011. 17: 375–387. [DOI] [PubMed] [Google Scholar]
- 34.Kumar H, Kawai T. and Akira S, Pathogen recognition by the innate immune system. Int. Rev. Immunol 2011. 30: 16–34. [DOI] [PubMed] [Google Scholar]
- 35.Guo Z, Chen LM, Zeng H, Gomez JA, Plowden J, Fujita T, Katz JM et al. , NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am. J. Respir. Cell Mol. Biol 2007. 36: 263–269. [DOI] [PubMed] [Google Scholar]
- 36.Chakravarthy KV, Bonoiu AC, Davis WG, Ranjan P, Ding H, Hu R, Bowzard JB et al. , Gold nanorod delivery of an ssRNA immune activator inhibits pandemic H1N1 influenza viral replication. Proc. Natl. Acad. Sci. USA 2010. 107: 10172–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N. et al. , RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 2010. 140: 397–408. [DOI] [PubMed] [Google Scholar]
- 38.Wolff T, Zielecki F, Abt M, Voss D, Semmler I. and Matthaei M, Sabotage of antiviral signaling and effectors by influenza viruses. Biol. Chem 2008. 389: 1299–1305. [DOI] [PubMed] [Google Scholar]
- 39.Sheikh F, Dickensheets H, Gamero AM, Vogel SN and Donnelly RP, An essential role for IFN-beta in the induction of IFN-stimulated gene expression by LPS in macrophages. J. Leukoc. Biol 2014. 96: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoboua F, Martel A, Duval A, Mukawera E. and Grandvaux N, Respiratory syncytial virus-mediated NF-kappa B p65 phosphorylation at serine 536 is dependent on RIG-I, TRAF6, and IKK beta. J. Virol 2010. 84: 7267–7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber M, Gawanbacht A, Habjan M, Rang A, Borner C, Schmidt AM, Veitinger S. et al. , Incoming RNA virus nucleocapsids containing a 5’-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe 2013. 13: 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowzard JB, Ranjan P. and Sambhara S, RIG-I goes beyond naked recognition. Cell Host Microbe 2013. 13: 247–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Min JY, Krug RM and Sen GC, Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 2006. 349: 13–21. [DOI] [PubMed] [Google Scholar]
- 44.Min JY, Li S, Sen GC and Krug RM, A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 2007. 363: 236–243. [DOI] [PubMed] [Google Scholar]
- 45.Tan SL and Katze MG, Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J. Interferon Cytokine Res 1998. 18: 757–766. [DOI] [PubMed] [Google Scholar]
- 46.Hale BG, Randall RE, Ortin J. and Jackson D, The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol 2008. 89: 2359–2376. [DOI] [PubMed] [Google Scholar]
- 47.Tong Y, Cui J, Li Q, Zou J, Wang HY and Wang RF, Enhanced TLR-induced NF-kappaB signaling and type I interferon responses in NLRC5 deficient mice. Cell Res. 2012. 22: 822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lian L, Ciraci C, Chang G, Hu J. and Lamont SJ, NLRC5 knockdown in chicken macrophages alters response to LPS and poly (I:C) stimulation. BMC Vet. Res 2012. 8: 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao Y. and Qian Y, Expression regulation and function of NLRC5. Protein Cell 2013. 4: 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robbins GR, Truax AD, Davis BK, Zhang L, Brickey WJ and Ting JP, Regulation of class I major histocompatibility complex (MHC) by nucleotide-binding domain, leucine-rich repeat-containing (NLR) proteins. J. Biol. Chem 2012. 287: 24294–24303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao Y, Wang Y, Chen F, Huang Y, Zhu S, Leng Q, Wang H. et al. , NLRC5 regulates MHC class I antigen presentation in host defense against intracellular pathogens. Cell Res. 2012. 22: 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meissner TB, Liu YJ, Lee KH, Li A, Biswas A, van Eggermond MC, van den Elsen PJ et al. , NLRC5 cooperates with the RFX transcription factor complex to induce MHC class I gene expression. J. Immunol 2012. 188: 4951–4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meissner TB, Li A, Liu YJ, Gagnon E. and Kobayashi KS, The nucleotide-binding domain of NLRC5 is critical for nuclear import and transactivation activity. Biochem. Biophys. Res. Commun 2012. 418: 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barber MR, Aldridge JR Jr., Webster RG and Magor KE, Association of RIG-I with innate immunity of ducks to influenza. Proc. Natl. Acad. Sci. USA 2010. 107: 5913–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu J, Lou D, Carow B, Winerdal ME, Rottenberg M, Wikstrom AC, Norstedt G. et al. , LPS regulates SOCS2 transcription in a type I interferon dependent autocrine-paracrine loop. PLoS One 2012. 7: e30166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gautier G, Humbert M, Deauvieau F, Scuiller M, Hiscott J, Bates EE, Trinchieri G. et al. , A type I interferon autocrine-paracrine loop is involved in Toll-like receptor-induced interleukin-12p70 secretion by dendritic cells. J. Exp. Med 2005. 201: 1435–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pachler K. and Vlasak R, Influenza C virus NS1 protein counteracts RIG-I-mediated IFN signalling. Virol. J 2011. 8: 48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.