Abstract
The small GTPase Cdc42 is a key regulator of polarity, but little is known in mammals about its spatial regulation and the relevance of spatial Cdc42 pools for polarity. Here, we report the identification of a GM130-RasGRF complex as a regulator of Cdc42 at the Golgi. Silencing GM130 results in RasGRF-dependent inhibition of the Golgi pool of Cdc42, but does not affect Cdc42 at the cell surface. Furthermore, active Cdc42 at the Golgi is important to sustain asymmetric front-rear Cdc42-GTP distribution in directionally-migrating cells. Concurrently to Cdc42 inhibition, silencing GM130 also results in RasGRF-dependent Ras-ERK pathway activation. Moreover, depletion of GM130 is sufficient to induce E-cadherin down-regulation, indicative of a loss in cell polarity and epithelial identity. Accordingly, GM130 expression is frequently lost in colorectal and breast cancer patients. These findings establish a previously unrecognized role for the GM130-RasGRF-Cdc42 connection in regulating polarity and tumourigenesis.
Introduction
Cell polarity is a highly orchestrated multi-step cellular process that regulates a plethora of biological functions of relevance for cell migration, wound healing and cancer. Among the regulators of polarity, the small GTPase Cdc42 is known to play a central role 1. Given the emerging role of polarity in tumourigenesis 2–4, it is now important to study the regulation of Cdc42 in time and space. A large body of evidence exists on Cdc42 signalling at or from the plasma membrane, but in comparison only little is known about Cdc42 signalling from endomembranes such as the Golgi apparatus, where this Rho family GTPase has been detected previously 5, 6. It currently remains unclear whether Cdc42 at the Golgi is regulated, what factors contribute to its activation and whether this pool is of any relevance for cell polarity. Endomembranes are increasingly recognized as sites where cellular signals are either initiated or modulated 7, thus supporting the notion that the Golgi pool of Cdc42 might be biologically relevant. Furthermore, the Golgi apparatus is known to play a role in directional migration and polarity 8, but despite wide acceptance there is surprisingly little mechanistic understanding of the role of this organelle in directional motility and cell polarity. Therefore, investigating whether a Golgi protein regulates spatial Cdc42 signalling might provide mechanistic insight into the role of the Golgi in polarity.
RasGRF family Guanine Nucleotide Exchange Factors are well-known regulators of the small GTPase Ras 9, 10. In addition, RasGRFs were shown to mediate functional crosstalk between Ras signaling and Cdc42 11, 12. In this respect, it has been recently demonstrated that RasGRF binds to and inhibits Cdc42, thereby regulating cellular motility, transformation and invasion 13.
In the current work, we hypothesized that the Golgi matrix protein GM130 might regulate the activity of the Golgi pool of Cdc42. Using fluorescent reporters, we show that this is indeed the case. We identify, through siRNA screening of a guanine nucleotide exchange factor (GEF) library, candidate GEFs that contribute to the regulation of Cdc42 specifically at the Golgi, but surprisingly none of these is involved in regulating the GM130-Cdc42 axis. We go on to identify RasGRF2 as a novel interaction partner for GM130 and demonstrate that this interaction is pivotal for the regulation of both Ras and Cdc42. Loss of GM130 releases RasGRF, allowing it to inhibit Cdc42 and activate Ras, leading to alterations in cell polarity and hyperactivity of the Ras-ERK pathway. Accordingly, we show that GM130 is frequently downregulated in cancer.
Results
The Golgi pool of Cdc42 controls cell polarity
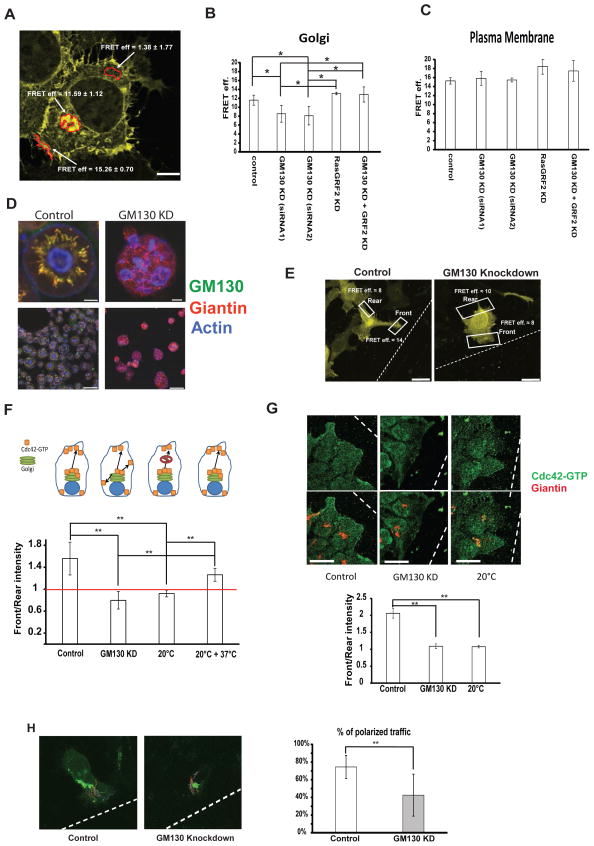
To study spatial Cdc42 activation patterns we used a Cdc42 Raichu probe 14, 15. For all our measurements we used a probe containing the C-terminal domain of Cdc42 15 (Cdc42-EM hereafter). This C-terminal domain targets the FRET reporter to different membranes including the plasma membrane and various cellular endomembranes such as the Golgi and endosomes (Supplementary Fig 1A) localizing as reported for endogenous Cdc425. Cdc42-EM appeared in a juxtanuclear pool colocalizing with GM130, indicating its presence at the Golgi complex (Fig. S1B), thus allowing us to monitor Cdc42 activation at this cellular location where Cdc42 is active 6 (Fig. 1 A). Measuring FRET outside the Golgi-area or the plasma membrane did not reveal any specific signal, thus supporting the specificity of our FRET measurements (Fig. 1A). Silencing GM130 reduced the total cellular pool of Cdc42-GTP 16 (Fig. S1C), and reduced substantially the activity of Cdc42 at the Golgi (Fig. 1B). However, GM130 depletion did not affect the activity of this reporter at the plasma membrane indicating that it did not affect Cdc42 activation therein (Fig. 1C). Silencing GM130 inhibited 2D cell migration (Fig S1E) indicating that alteration of spatial Cdc42 signalling is relevant for directed cell motility. Moreover, the recruitment of aPKC to the leading edge of directionally migrating cells was also reduced (Supplementary Fig. 1F). To test for effects on polarity in a 3D cell culture model, we used Caco-2 cells, which are known to spontaneously polarize when plated in a 3D matrix, forming cysts with a single lumen. Previous work showed that alteration of Cdc42 activity (and thus of polarity) leads to the formation of multi-lumen (aberrant) cysts 17, 18. Depletion of GM130 in Caco-2 cells led to an increase in the number of aberrant cysts, which is indicative of a polarity defect (Fig. 1D). These results suggested that regulation of spatial Cdc42 activity by GM130 was relevant for cell polarization in 2D and 3D cell culture. In contrast, GM130 depletion did not affect Golgi morphology (Supplementary Fig 2), ER-Golgi trafficking, Golgi-to-plasma membrane trafficking or coatomer recruitment to the Golgi (Supplementary Fig. 3A–C). Furthermore, the capability of the Golgi to nucleate microtubules was also unaffected (Supplementary Fig. 3D), in agreement with previous findings 19. Thus, the indirect impact of these processes on polarity could be excluded 8.
Figure 1. GM130 spatially regulates Cdc42 activity to control polarity.
(A) One representative HEK293 cell transfected with the Cdc42-EM FRET reporter is shown. Areas where FRET was measured are marked in red and average FRET values ± standard deviation calculated from 5 independent experiments in control-transfected cells are shown. Scale bar = 25 μm. (B, C) HEK293 cells were transfected with the indicated siRNA. After 48h, cells were transfected with the Cdc42-EM FRET reporter. After additional 24 h, cells were fixed and FRET at the Golgi region (B) or at the plasma membrane (C). Averages of at least 3 independent experiments ± standard deviations (at least 15 cells per condition in every experiment) are shown on the graph. Asterisks indicate statistically significant differences tested using ANOVA with Newman-Keuls multiple comparison test (*p<0.05). (D) Caco-2 cells were transfected with the indicated siRNA. Cells were grown in Geltrex® matrix for four days and cysts were stained for Giantin (red) and GM130 (green) using immunofluorescence and for F-actin (blue) using fluorescent phalloidin. A magnification of a single cyst is displayed in the upper row and an overview of several cysts in shown in the lower row. Scale bar = 7.5 μm for the magnified images (upper images) and 75 μm for the overview images. (E) HeLa cells were transfected with the indicated siRNA. After 48 h, cells were transfected with a plasmid encoding the Cdc42-EM Raichu probe. After 24 h, a wound was generated using a 10 μl pipette tip and cells were allowed to migrate and polarize for approximately 6 h at 37°C followed by fixation. Representative images of cells expressing the Raichu probe that are facing the wound are shown. FRET was measured on the plasma membrane part facing the wound (indicated as “Front”) and on the opposing end of the cell (indicated as “Rear”). Scale bar is 10 μm. (F) Graphic representation of FRET measurements from three independent experiments of HeLa cells expressing the indicated Raichu probe and that that were transfected with the indicated siRNA 72 h prior to the experimental measurements. Above the graph a schematic of the experimental setup: control cells have Cdc42-GTP at the Golgi and deliver it to the leading edge in a polarized way. GM130 knockdown cells have less Cdc42-GTP at the Golgi and lose the ability to polarize their traffic towards the leading edge. Blocking post-Golgi traffic by incubation at 20°C abolishes front-rear asymmetry and re-warming the cells to 37°C re-established asymmetry. White bars represent Front/Rear ratios of FRET values. Results are means ± SD from four independent experiments. Asterisks indicate statistically significant differences tested using ANOVA with Newman-Keuls multiple comparison test (** p<0.01). (G) HeLa cells were transfected with siRNA. After 72 hours, a wound was made with a 10 μl tip and the cells were allowed to migrate for 6 hours. Cells were then fixed and immunostained for Giantin and Cdc42-GTP. Intensities of Cdc42-GTP were measured and the Front/Rear ratio is plotted as a bar graph. Results are means ± SD of 3 independent experiments. Asterisks indicate statistically significant differences tested using ANOVA with Newman-Keuls multiple comparison test (** p<0.01). Scale bar = 25 μm. (H) HeLa cells were transfected with the indicated siRNA. After 48 h, cells were transfected with the plasmid encoding for VSVG-RUSH-GFP. After further 24 h, a wound was made with a 10 μl tip and cells were allowed to migrate at 37°C for approximately 6 hours followed by live video microscopy. VSVG-RUSH-GFP was released from the ER by addition of biotin and after 25 min VSVG carriers were tracked using ImageJ. Individual tracks are displayed on the representative images. Results are means ± SD from three independent experiments. Asterisks indicate statistically significant differences tested using Student’s T-test (** p<0.01).
Directed trafficking causes asymmetric distribution of Cdc42
To obtain direct evidence for the role of the GM130-regulated Golgi pool of active Cdc42, we measured Cdc42 activity at the front and the rear of directionally migrating cells using the Cdc42-EM probe. Strikingly, Cdc42 activity exhibited an asymmetric distribution, being more active at the front compared to the rear of the cell (Fig. 1E, F). Silencing GM130 reduced the amount of activated Cdc42 at the leading edge, thereby breaking the asymmetric distribution of active Cdc42 (Fig. 1E, F). These results were confirmed by immunofluorescence staining of endogenous Cdc42-GTP (Fig. 1G). Sustained accumulation of active Cdc42 at the leading edge could be the consequence of an increased local activation. Alternatively, the Golgi might deliver Cdc42-GTP to the leading edge. If the latter mechanism was true, then blocking trafficking from the Golgi ought to diminish the amount of Cdc42-GTP at the leading edge. To investigate this point, directionally migrating cells in a wound closure assay were incubated at 20°C for one hour, which blocks exit from the trans-Golgi 20. Measuring FRET reporter activity or staining for Cdc42-GTP revealed that blocking post-Golgi trafficking diminished the presence of GTP-Cdc42 at the cell front, thereby abolishing Cdc42-GTP asymmetry (Fig. 1E&F). Importantly, re-incubation of the cells at 37°C for one hour re-established Cdc42-GTP asymmetry with higher levels at the leading edge compared to the rear (Fig. 1F). The loss of Cdc42-GTP asymmetry in GM130 knockdown cells was likely due to abrogation of directed trafficking. Live cell imaging of GFP-VSVG tracks forming from the Golgi showed that while carriers formed readily in GM130-depleted cells, they were non-directed, contrarily to tracks in control cells, which mostly trafficked towards the leading edge (Fig. 1H). Of note, the directionality of post-Golgi trafficking was affected, whereas the global rate of post-Golgi trafficking was not (Supplementary Fig. 3B). Since, both GDP- and GTP-restricted Cdc42 is known to block post-Golgi trafficking 21 we conclude that GM130 depletion does not affect the GTP/GDP cycling of Cdc42. Taken together, the Golgi pool of active Cdc42 is relevant for cell polarization.
The GM130-Cdc42 crosstalk does not involve GEFs
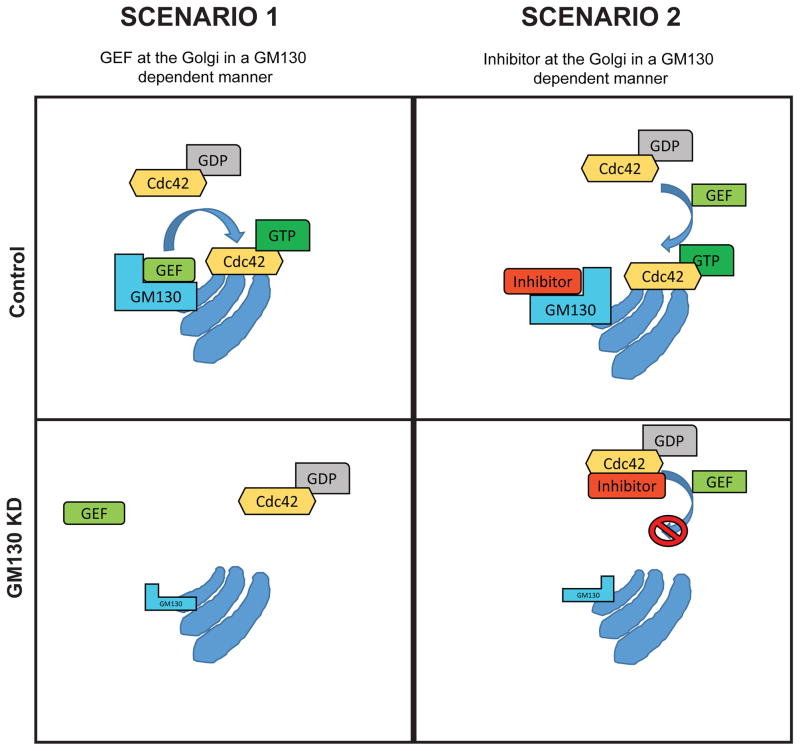
Next we asked how the Cdc42 activity at the Golgi was regulated by GM130. One possibility is that GM130 recruited a Cdc42 GEF leading to local activation of this GTPase. If true, then this GEF must interact with GM130 and localize to the Golgi in a GM130-dependent manner (schematic in Fig. 2).
Figure 2. Models for the GM130-Cdc42 crosstalk.
Schematic representation of two different models. On the left-hand side: GM130 acts as a scaffold for a Cdc42-GEF at the Golgi. In this case, a GEF should be located at the Golgi in a GM130 dependent manner. Upon GM130 depletion, the GEF is lost from the Golgi and subsequently, Cdc42-GTP doesn’t accumulate at the organelle. In the second scenario (right-hand side), GM130 binds to an inhibitor of Cdc42, protecting the small GTPase from inactivation. If GM130 is lost, the inhibitor will bind to Cdc42 in places other than the Golgi, thereby preventing a GEF mediated activation of Cdc42. In this way, Cdc42-GTP will not be accumulated at the Golgi.
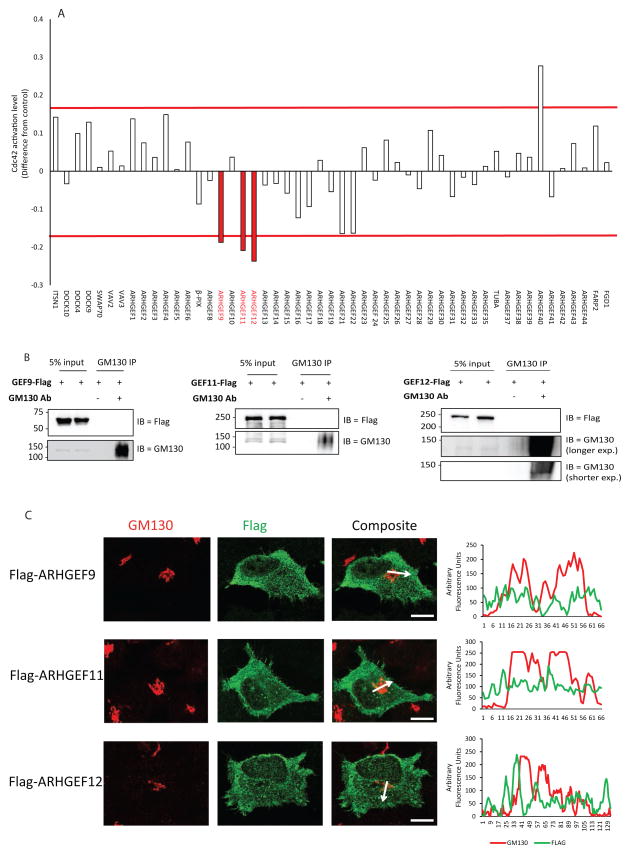
To identify GEFs that contribute to activate Cdc42 at the Golgi, we screened a library of 51 siRNAs against Rho family GEFs for effects on the Golgi pool of Cdc42 using the Cdc42-EM FRET probe. We focused primarily on the knockdowns that affected Cdc42 activity at the Golgi in a manner more conspicuous than GM130 depletion. While several GEF knockdowns decreased the FRET values at the Golgi, only three had an effect that was comparable to the effect of GM130, namely ARHGEF9, 11 and 12 (Fig. 3A). Depletion of GEF11 and 12 also resulted in reduced total cellular Cdc42-GTP (Supplementary Fig. 4B) supporting these FRET measurements. Depletion of any of these GEFs also resulted in an inhibition of directed migration as determined in a wound-healing assay (Supplementary Fig. 4C). In addition, polarization of the Golgi to the leading edge of migrating cells was also affected (Supplementary Fig. 4D). However, none of these GEFs interacted with GM130 in co-immunoprecipitation experiments, nor did they localize to the Golgi (Fig. 3B, C). We also failed to detect any evidence for an involvement of the GEF Tuba, as it did not localize to the Golgi (Fig. S5A, B), nor did it interact with GM130 (Supplementary Fig. 5C, D) contrarily to previous findings 16, and in agreement with others who failed to detect this GEF at the Golgi 22, 23. In addition, neither Tuba depletion (Fig 3A, Fig S5E) nor its overexpression (Supplementary Fig. 5F) changed the activation of Cdc42 at the Golgi. Of note, none of the GEFs tested localizes to the Golgi (Oliver Rocks, personal communication). Therefore, the GM130-Cdc42 crosstalk was not mediated via recruitment of a GEF driving local activation of Cdc42.
Figure 3. Screening for GEFs that regulate the Golgi pool of Cdc42.
(A) HEK293 cells were transfected with control or with the indicated siRNA. After 48h, cells were transfected with the plasmid encoding for the Cdc42-EM-FRET probe. After a further 24h the cells were fixed and mounted. FRET was measured as described in the Material and Methods section and FRET values are displayed as fold of control and are averages from the measurements of at least 30 cells per condition. Red bars represent CI95 intervals (B) HeLa cells were transfected with plasmids encoding the indicated GEF. 24 h later, cells were lysed and the lysates were subjected to immunoprecipitation either with a GM130 antibody (+) or with protein G sepharose beads alone. “5% input” indicates lanes where 5% of the total material that was used in the IP is loaded. The immunoprecipitated material was eluted and subjected to SDS-PAGE followed by immunoblotting against the indicated proteins. (C) HeLa cells were plated on coverslips, then transfected with plasmids encoding the specified GEF. 24 h later, cells were fixed and immunostained for GM130 and Flag. Intensity plot corresponding to the region marked by a white arrow are shown on the right. Scale bar is 10 μm.
The GM130-Cdc42 crosstalk involves RasGRF
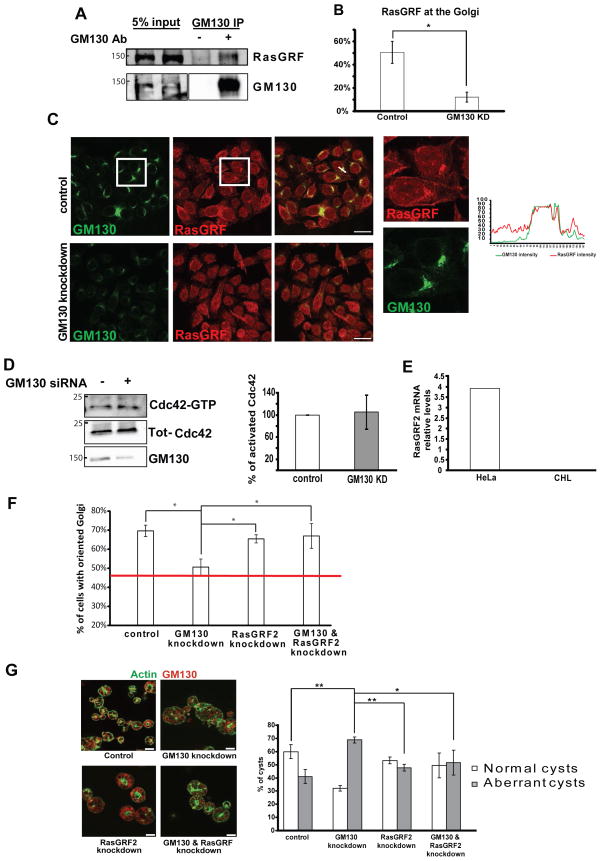
Another explanation for the GM130-Cdc42 connection could be that GM130 was preventing the function of some Cdc42 inhibitor (schematic in Fig. 2). One possible candidate was the GEF RasGRF2, the RasGRF isoform expressed in epithelial cells, which has been recently shown to act as an inhibitor of Cdc42 13. A marked association of endogenous GM130 and RasGRF2 could be detected in co-immunoprecipitation experiments (Fig. 4A). Moreover, in about 50% of the cells, RasGRF2 co-localized with GM130 at the juxtanuclear region, consistent with Golgi complex localization (Fig. 4B, C). RasGRF in addition also exhibited a localization pattern reminiscent of the ER, which is in agreement with previous work 24. Noticeably, upon GM130 depletion, RasGRF2 juxtanuclear localization dropped strongly to less than 10% of the cells (Fig. 4B, C). Moreover we found that the interaction between these proteins was mediated by the C-terminal domain of GM130 and the Cdc25 domain of RasGRF2 (Supplementary Fig. 6A& B). These data suggested that GM130 is involved in localizing RasGRF to the Golgi complex.
Figure 4. The effect of GM130 on Cdc42 is dependent on RasGRF.
(A) HeLa cells were lysed and the lysate was subjected to immunoprecipitation either with a GM130 antibody (+) or with protein G sepharose beads alone. “5% input” indicates lanes where 5% of the total material that was used in the IP is loaded. The immunoprecipitated material was eluted and subjected to SDS-PAGE followed by immunoblotting against the indicated protein. (B) Quantification of cells with RasGRF at the Golgi. (C) HeLa cells grown on glass cover slips were fixed followed by immunofluorescence staining of RasGRF and GM130. Images were acquired using a confocal laser-scanning microscope. The magnifications correspond to the areas inside the white squares. Scale bar is 25 μm. An intensity plot corresponding to the region marked by a white arrow is shown. (D) CHL cells were depleted of GM130 by siRNA transfection. After 72 h, cells were lysed and GST-PAK1 was added to the lysate followed by pulldown with GSH-sepharose. The eluate was subjected to SDS-PAGE followed by immunoblotting against Cdc42 (Cdc42-GTP). 5% of the original lysate were subjected to SDS-PAGE followed by immunoblotting against Cdc42 (Tot-Cdc42) and against GM130. Bar graph in the right panel represents a quantification of the amount active Cdc42 (GTP-Cdc42) to total Cdc42. Results are means ± SD from three independent experiments. (E) RT-PCR of RasGRF2 comparing HeLa and CHL cells. (F) Cells were transfected with the indicated siRNA. After 72 h, a wound was made using a 10μl pipette tip and cells were allowed to migrate and polarize for approximately 6 h at 37°C followed by fixation and immunofluorescence staining of Giantin to label the Golgi. The Golgi was counted as oriented towards the wound if its major mass was located in a 120° angle facing the wound edge. The red line shows the average orientation of the Golgi immediately after wounding. Results are means ± SD from three independent experiments. Asterisk indicates statistically significant differences using ANOVA with Newman-Keuls multiple comparison test (*p<0.05). (G) Caco-2 cells were transfected with siRNA against GM130, or RasGRF2, or with both siRNA. Cells were grown in Matrigel® for four days to allow cyst formation. Afterwards, cysts were stained for GM130 using immunofluorescence and for F-actin using fluorescent phalloidin as described in “Materials and Methods”. Cysts with a single lumen were counted as normal, while cysts with multiple or no lumen were counted as aberrant. The percentage of normal and aberrant cysts in each condition was counted from three independent experiments which are displayed in the bar graph in the lower panel. Results are means ± SD. Asterisk indicates statistically significant differences using ANOVA with Newman-Keuls multiple comparison test (*p<0.05; **p<0.01). Scale bar is 7.5 μm.
Overexpression of RasGRF1 or 2 blocked activation of Cdc42 at the Golgi as determined using the Cdc42-EM probe, in a manner dependent on the GEFs DH domain (Fig. S6C). Importantly, co-knockdown of GM130 and RasGRF2 restored Cdc42 activation levels at the Golgi (Fig. 1A). The involvement of RasGRF in the GM130-Cdc42 crosstalk was further supported by the finding that GM130 depletion did not affect the levels of GTP-Cdc42 in CHL cells which naturally lack all RasGRF isoforms (Fig. 4D). Therefore, we conclude that RasGRF is an essential mediator in the GM130-Cdc42 crosstalk.
Regulation of the GM130-RasGRF complex
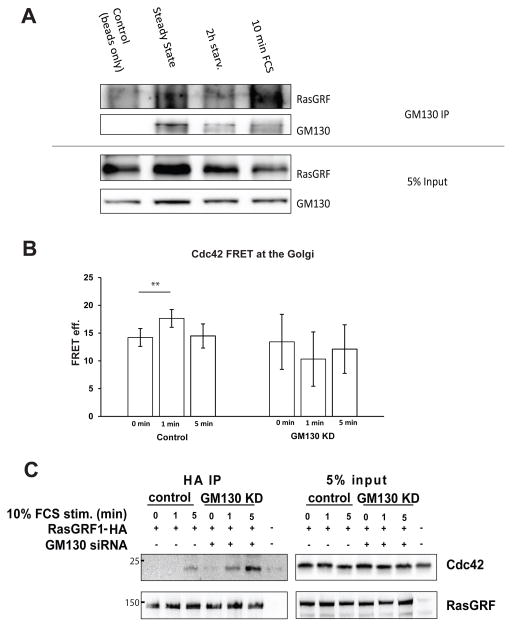
We hypothesized that GM130 could sequester RasGRF, thereby preventing it from binding and inhibiting Cdc42. Thus, under conditions that activate Cdc42, GM130 would sequester RasGRF to prevent it from inhibiting Cdc42. To test this hypothesis, we determined whether the GM130-RasGRF complex is affected by serum-starvation, a condition under which Cdc42 is not activated. Under these conditions, GM130 did not co-immunoprecipitate with RasGRF (Fig. 5A), contrary to cells in steady-state cultured under complete conditions (Fig. 3). Treatement with 10% FCS for 10 min resulted in re-formation of the GM130-RasGRF complex (Fig. 5A). If RasGRF were free under serum-starved conditions, then we would expect the Golgi pool of Cdc42 to be unresponsive to GM130 depletion. Cdc42 at the Golgi responded to serum treatment in control siRNA transfected cells, while GM130 depleted cells did not respond to this treatment (Fig. 5B). Of note, the FRET values at the Golgi were similar in control and GM130 silenced serum-starved cells, further supporting the notion that this pool becomes GM130-independent under serum-starved conditions (Fig. 5B).
Figure 5. The GM130-RasGRF complex is regulated by serum.
(A) MCF7 cells were grown to confluency. Then, cells were serum-starved for 2 h followed by stimulation with 10% FCS for the indicated time points. Cell lysates were subjected to immunoprecipitation against GM130. The immunoprecipitate was subjected to SDS-PAGE followed by immunoblotting against the indicated proteins. (B) HeLa cells were transfected with control or GM130 siRNA 72h prior to the experiment. After 48 h, cells were transfected with the plasmid encoding for the Cdc42 Raichu probe. Cells were serum-starved for 2 h followed by stimulation with 10% FCS for 0,1 and 5 min and FRET was measured as indicated in “Materials and Methods. Results are means ± SD from three independent experiments. Asterisks indicate statistically significant differences tested using ANOVA with Newman-Keuls multiple comparison test (*p<0.05). (C) HeLa cells were transfected with the indicated siRNA. After 48 h, cells were transfected with empty vector or with a plasmid encoding HA-tagged RasGRF1. After 24 h, cells were serum-starved for 2 h followed by stimulation with 10% FCS for the indicated time points. Cell lysates were subjected to immunoprecipitation against HA. The immunoprecipitate was subjected to SDS-PAGE followed by immunoblotting against the indicated proteins.
If GM130 acts to protect Cdc42 from inhibition by RasGRF, then depletion of GM130 ought to increase the association between RasGRF and Cdc42. Co-immunoprecipitation experiments demonstrated that this was the case: the amount of RasGRF in association with Cdc42 increased in GM130 knockdown cells compared to control cells (Fig. 5C).
Finally, we tested whether RasGRF is relevant for the GM130-Cdc42 crosstalk in the context of polarity. This was the case, as co-depletion of RasGRF2 and GM130 rescued the polarity defects imposed by GM130 depletion (Fig. 3F&G).
The GM130-RasGRF complex controls Ras activity
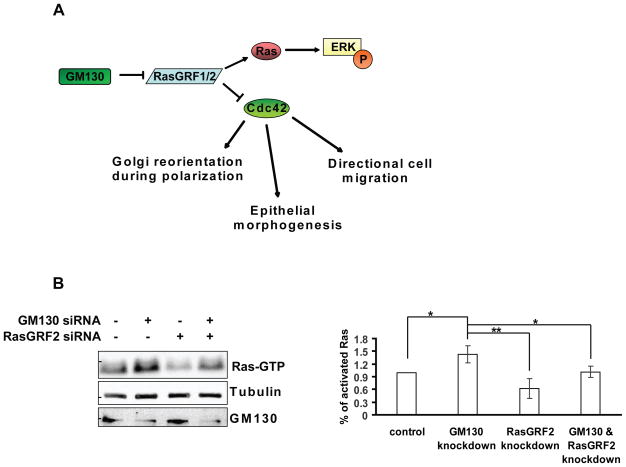
Our data indicated that GM130 acts as a repressor of RasGRF. Consistent with this model (Fig. 6A), knockdown of GM130 would release RasGRF, allowing it to inhibit Cdc42. However, RasGRF is also a well-known activator of Ras. Thus, in agreement with our model, loss of GM130 should also result in an increase of Ras activation. To test this prediction, we performed a knockdown of GM130 and measured Ras activation using a pulldown assay. Depletion of GM130 increased the levels of GTP-loaded Ras and co-knockdown of RasGRF abolished this effect (Fig. 6B), indicating that the increase in Ras activity was dependent on RasGRF. We also tested whether signalling downstream of Ras was modulated at the level of ERK1/2 in response to mitogen treatment. Knockdown of GM130 increased ERK1/2 activation in response to serum stimulation and this effect was again dependent on RasGRF (Supplementary Fig. 7A). To test whether the effect on ERK1/2 signaling is biologically relevant, we depleted GM130 in PC12 cells. These cells are a standard model for ERK-dependent cell fate decisions 25. Higher and more sustained ERK1/2 signalling increases the likelihood that PC12 cells shift from proliferation to differentiation, which is evident by the emergence of neurites. Depletion of GM130 resulted in elevated and sustained ERK1/2 activation (Supplementary Fig. 8A) and accordingly the percentage of cells that underwent differentiation was significantly increased (Supplementary Fig. 8B&C). Thus, the effects of GM130 on Ras signalling are relevant for ERK-dependent cell fate decisions.
Figure 6. GM130 depletion increases Ras activity.
(A) Working model: GM130 acts as a suppressor of RasGRF activity, thereby modulating the activities of Ras and Cdc42. According to this model, the knockdown of GM130 results in increased Ras activation and decreased Cdc42 activation. (B) HepG2 cells were transfected with the indicated siRNAs. After 72 h, cells were lysed and Ras pull-down was performed using GST-RBD. The eluate form the beads was subjected to SDS-PAGE followed by immunoblotting with a pan-Ras antibody (Ras-GTP). 5% of the original lysate was subjected to SDS-PAGE followed by immunoblotting against tubulin and GM130. Bar graph represents a quantification of the amount active Ras (Ras-GTP) normalized to tubulin. Results are means ± SD from three independent experiments. Asterisks indicate statistically significant differences tested using ANOVA with Newman-Keuls multiple comparison test (*p<0.05; ** p<0.01).
Expression of GM130 in colon cancer
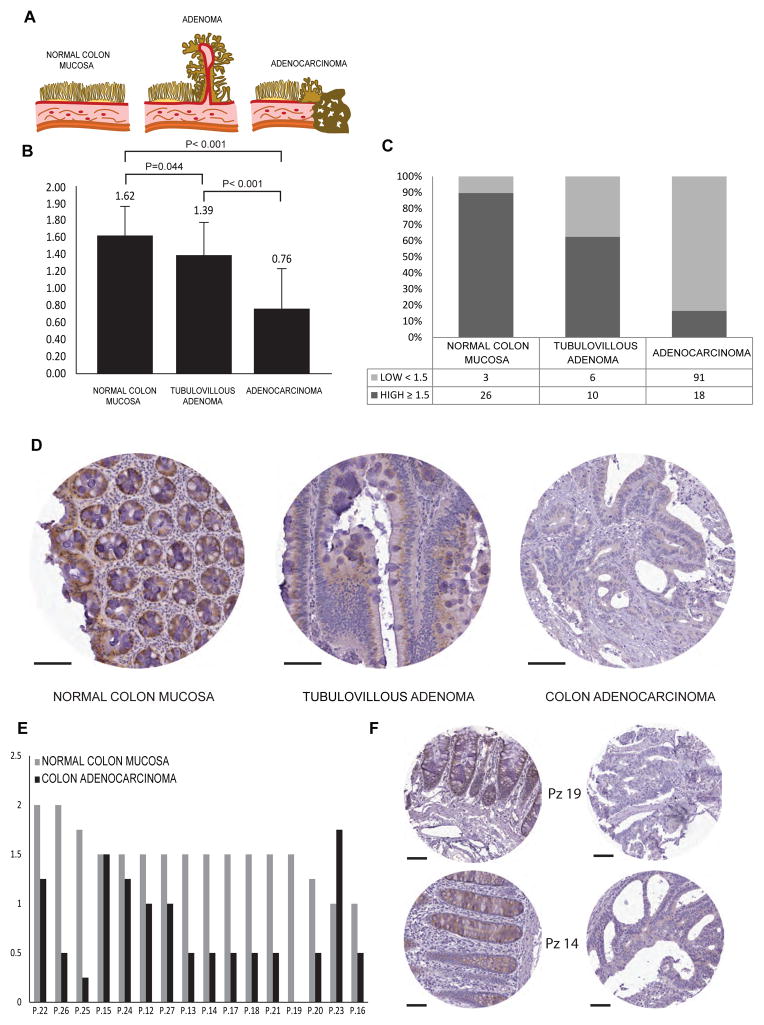
Loss of polarity is a frequent feature of epithelial tumours and is associated with dysplasia and increased metastatic potential 3, 4. Our results indicated that GM130 regulates polarity by modulating the Golgi pool of active Cdc42 and, in addition, Ras activity. Therefore, we asked whether GM130 might be associated with the malignant phenotype. To investigate this possible connection, we used two strategies. First, we analysed GM130 expression in a colon cancer progression TMA comparing normal colon mucosa (29 cases), adenoma (16 cases), and adenocarcinoma (109 cases) (Fig 7A). While the expression of GM130 only differed marginally in normal colon and adenoma, the expression in carcinoma was reduced markedly (Fig. 7B–D). We next extended this analysis in samples from 16 patients, present on TMA, with matched normal and cancer tissues. We compared the scored intensity of GM130 expression and found that in 14 out of 16 individual patients the expression of GM130 was reduced in tumours compared to the matched normal tissue (Fig. 7E&F). The down-regulation of GM130 in cancer was further confirmed by an analysis of 11 matched normal and cancer colon tissues from a different source (Supplementary Fig. 9). In this case, the intensity of the GM130 staining was measured and healthy tissues were compared with the matching cancerous tissues. In agreement with the previous results, GM130 was down-regulated in the cancerous tissue in 9 out of 11 patients (Supplementary Fig. 9A–C).
Figure 7. Dysregulation of GM130 in human colon cancer.
GM130 expression was measured by IHC on a Colon Progression TMA. (A) Graphic representation of the neoplastic evolution of colon carcinoma from the normal colon mucosa through the adenoma. (B) Bar graphs depicting the average expression level (± SD) of GM130 in Normal Colon Mucosa, Colon Tubulovillous Adenomas and Colon Adenocarcinomas or (C) the percentage of low and high GM130-expressing samples when using a cut off score value of 1.5 (differences are significant as assessed by Pearson χ2 analysis P < 0.001). (D) Representative images of immunohistochemical staining for GM130. (E) Bar graphs depict the expression score of GM130 in 16 patients for which matched normal and tumor tissue were available on the TMAs. GM130 is downregulated in colon cancer with respect to the normal colon mucosa in 14 out of 16 patients. (F) Representative core images of normal and tumor tissue from patient 14 and 19. Scale bar, 100 μm
In support for a role of GM130 loss in tumour progression, we found that knockdown of GM130 in Caco-2 cells led to a loss of E-cadherin expression (Supplementary Fig. 9D), a phenotype that is often considered to be linked to tumour progression and a loss of epithelial identity and is in line with the notion that loss of polarity is a typical phenomenon in EMT 26. We could show that the effect of GM130 on E-Cadherin was not cell line specific, as the depletion of GM130 also caused loss of E-Cadherin in another epithelial cell line (Supplementary Fig. 9D).
Discussion
A large body of research on directional migration and cellular polarization has focused on dissecting signaling events that originate from and are regulated by proteins at the plasma membrane. However, comparatively little is known about how polarity is regulated by signaling from endomembrane compartments like the Golgi apparatus. This organelle was shown to orient towards the leading edge in directionally migrating cells 27 and disrupting Golgi structural integrity resulted in a block of directional motility 8. Our aim was to explore the functional relevance of the Golgi localized pool of Cdc42 5. Overall, our data establish for the first time a role for active Cdc42 at the Golgi in the control of cell polarity and that the complex formed by GM130 and RasGRF is critical in this process. Of note, depletion of GM130 did not disrupt Golgi integrity or membrane trafficking, which is in agreement with others 28, 29. Different findings by others 30–32 can be attributed to the use of different cell lines, different siRNA concentrations and transfection reagents. We interpret our data to mean that the Golgi might supply the leading edge of migrating cells with active Cdc42. An alternative explanation could be that the Golgi is supplying the leading edge with a GEF for Cdc42, and recent work has already speculated about such a possibility 33. However, such a model cannot explain our data showing that co-depletion of RasGRF2 and GM130 rescues Cdc42 activation and cell polarity, which strongly implies that the presence of active Cdc42 at the Golgi is important for polarization. Thus, we consider it more likely that active Cdc42 is delivered from the Golgi to the leading edge. The Golgi could provide a permissive environment for accumulating active Cdc42 by being free or poor in GAPs. In fact, no Rho family GAPs were have been found at the Golgi, except for small amounts of ARHGAP21 34 (Oliver Rocks, personal communication).
Recent computational modelling approaches tried to elucidate the contribution of polarized trafficking to polarization in yeast 35–37. While trafficking appeared to be an important component, endocytic removal of Cdc42 from the leading edge, its lateral diffusion and ‘off’ kinetics from membranes were also found to be a key factor. This is in line with published work where directional polarity was found to be dependent on Arf6 33 a GTPase known to regulate endocytosis 38.
Altogether, our findings establish for the first time a connection between endomembrane Cdc42 activity and cell polarity and allow us to propose a role for this regulation in neoplasia. The GM130-RasGRF-Cdc42 axis appears as a potentially valuable new therapeutic target and future work will elucidate whether the loss of GM130 observed in tumours is associated with a change in the invasive or metastatic potential of cancer cells.
Methods
Cell Culture and transfection
HeLa, HEK293, HepG2, MCF7 cells were purchased from ATCC and were cultured in Dulbecco’s modified Eagles medium (DMEM) supplemented with 10% FCS and 100 U/ml penicillin/streptomycin. RPE-1 cells were grown in DMEM/F12 supplemented with 10% FCS and 100U/ml penicillin/streptomycin. MDA-MB-231 were cultured in RPMI supplemented with 10% FCS and 100 U/ml penicillin/streptomycin. PC12 cells were cultured in RPMI supplemented with 10% HS, 5% FCS and 100U/ml penicillin/streptomycin. Caco-2 cells were cultured in minimal essential medium supplemented with 20% FCS and 100 U/ml penicillin/streptomycin. To generate stable cell lines, cells were transduced with lentiviruses (produced according to the protocols of Trono’s lab) and transduced cells were either selected with antibiotics or sorted via FACS. Transfection of siRNA was performed using HiPerFect (Qiagen) or Lipofectamine2000 (Invitrogen) according to the manufacturer’s instructions. The siRNAs used for the GEF screening were purchased from Thermo Fisher Scientific. Control siRNA was purchased from Thermo Fisher Scientific (ON_TARGETplus Control pool – Cat. Nr. D-001810-10-20). The siRNAs against GM130 were from Thermo Fisher Scientific (ON-TARGETplus smart pool – Cat. Nr. L-017282-00 – indicated in the text as siRNA1) and from QIAGEN (SI04235147 - indicated in the text as siRNA2 – and SI00429191, indicated in the text as siRNA3). The siRNA against RasGRF2 was from Qiagen (SI04329941). The siRNA against TUBA was from QIAGEN (SI04220237 – used in Fig 1B). Plasmid transfection was performed using Fugene6 (Promega) according to the manufacturer’s instructions. Cells were typically transfected 24 h prior to the experiment. PC12 cells were transfected with FugeneHD according to the manufacturer’s instructions.
Plasmids
Cdc42-GFP-wt (nr. 12599), Cdc42-GFP-TN (nr. 12601), Cdc42-GFP-QL (nr. 12600) were from Addgene (Klaus Hahn’s Laboratory6). RasGRF1-HA, RasGRF2-FLAG, RasGRF1-ΔPH-DH, RasGRF1ΔCdc25, RasGRF2-ΔDH and RasGRF2-ΔCdc25 were described previously13. myc-GM130-ΔN690 was cloned using pcDNA 3.1 myc-His as backbone pL-Venus-Cdc42 was cloned using a modified version of pLVTHM as backbone. RAICHU-Cdc42-EM and RAICHU-Cdc42-PM were kind gifts from Dr. Michiyuki Matsuda (Kyoto University, Japan). The RUSH-MannII-GFP expressing HeLa cells, as well as the VSVG-GFP-RUSH plasmid were a kind gift from Dr. Franck Perez39. Temperature-sensitive VSVGts045-GFP was a kind gift from Dr. Kai Simons. Flag tag ARHGEF9, 11 and 12 were a gift from Dr. Oliver Rocks. Lentiviral packaging plasmids were bought from Addgene (psPAX2 nr. 12260, pMD2.G nr. 12259 and pLVTHM nr. 12247, from Didier Trono’s laboratory). GM130 GIPZ shRNA was bought from Thermo Scientific (Clone ID: V3LHS_313361). GM130 rat shRNAs (RSH050521 nr 1 to 4) and Control shRNAs (MSH029723-CH1) were bought from Genecopoeia. pmTurquoise2-Golgi was from Addgene (nr. 36205 40).
Pull-down experiments
To pull down active Cdc42 cells were lysed in buffer (25mM HEPES pH7.5, 150mM Nacl, 1% Nonidet-P-40, 10mM MgCl2). Purified GST-tagged effector domain of PAK1 immobilized on GSH-sepharose beads was added to the lysate that was pre-cleared by centrifugation at 21,000xg for 5 min at 4°C. After 1 h incubation at 4 °C, beads were washed twice followed by boiling in sample buffer to elute bound material.
To precipitate active Ras, the same procedure was used with the exception the purified GST-tagged Raf1-RBD immobilized to GSH-sepharose beads were added to the lysate.
ERK1/2 activation assays
250,000 HeLa or HEK293 cells were plated in 35mm dishes and transfected with siRNA at the same time (HiPerFect from Qiagen). After 72h cells were serum-starved for at least 2 h. Cells were subsequently stimulated with 10% FCS followed by lysis in ice cold MAPK buffer (150mM NaCl 20mM TRIS-HCl pH7.5, 1% Triton X-100) supplemented with protease and phosphatase inhibitors. The amount of phosphorylated ERK1/2 was determined by immunoblotting.
PC12 differentiation
PC12 cells were plated at low density on Collagen-coated coverslips. 24 h later, cells were transfected with plasmids encoding either control shRNA or GM130 shRNA. Cells were then serum starved overnight. Cells were then stimulated with 10% NGF and 1% HS for 48h, then coverslips were stained and fixed. Cells with neurites as long as the body length were scored as ‘differentiated’. Due to the presence of GFP in the plasmid encoding for the shRNAs, we were able to count only cells effectively transfected. To measure P-ERK intensity, cells were stimulated as described above for 0, 30 or 90 minutes and then stained.
Antibodies and immunofluorescent labels
Rabbit monoclonal anti-Cdc42 (dilution 1:300 – Cat. Nr. 2466), rabbit polyclonal ERK1/2 (dilution 1:1000 – Cat. Nr. 4695) and rabbit polyclonal against phospho-ERK1/2 (dilution 1:1000 – Cat. Nr. 9101) were purchased from Cell Signalling. Mouse monoclonal GTP-Cdc42 was purchased from NewEast Biosciences (dilution 1:50 - Cat. Nr. 26905). To stain for GTP-Cdc42, Image-iT, FX Signal Enhancer from Invitrogen was used according to manufacturer’s instructions. Mouse monoclonal anti-GM130 antibody was from BD-Biosciences (IF dilution: 1:1000 – WB dilution 1:250 – Cat. Nr. 610823). Rabbit monoclonal anti-GM130 was from cell signalling (dilution 1:500 – Cat. Nr. 12480). Rabbit monoclonal anti-RasGRF (WB dilution 1:200 – IF dilution 1:50 – Cat. Nr. Sc-863) and Mouse monoclonal E-Cadherin antibody (dilution 1:500 – Cat. Nr. 21791) were purchased from Santa-Cruz. Rabbit polyclonal anti-Giantin antibody was from Covance (dilution 1:1000 – Cat. Nr. PRB-114C). Monoclonal ANTI-FLAG® M2-Peroxidase (HRP) was from Sigma (dilution 1:2000 – Cat. Nr. A8592). Mouse monoclonal anti-HA 12CA5 antibody was from Roche (WB dilution 1:4000 – IF dilution 1:500 – Cat. Nr. 11583816001). Mouse monoclonal anti-γ-tubulin (dilution 1:1000 – Cat. Nr. Ab11317), Rabbit polyclonal anti TGN46 (dilution 1:1000 – Cat. Nr. Ab50595) were from Abcam, Mouse monoclonal anti-GFP (dilution 1:1000 – Cat. Nr. 11814460001) was from Roche. AlexaFluor647 Phalloidin (Cat. Nr. A22287) and AlexaFluor488 concanavalin (Cat. Nr. C11252) and AlexaFluor647HPA (Cat. Nr. L32454) were from Invitrogen. For immunofluorescence staining cells were grown on glass coverslips. To stain for RasGRF, cells were fixed on ice with 4% formaldehyde for 10 min followed by permeabilization with PBS containing 0.05% triton X100 for 3 min at RT. To stain for β-COP cells were fixed with methanol-acetone (50/50) for 2 min on ice. For all other stainings, cells were fixed with 4% paraformaldehyde for 15 min at RT followed by permeabilization with PBS containing 3% BSA and 0.2% triton X100.
Microtubule nucleation assay
RPE-1 cells were plated on coverslips and transfected with siRNAs using HIPerFect. After 72 h, 0.1 mM HEPES pH 7.35 was added to the cells and the cells were then transferred on ice for 40 minutes. Then cells were put back at room temperature and fixed after 0, 3 and 4 minutes at room temperature in fixation/permeabilization buffer (10mM Citric Acid, 0.05% Tween 20, pH 6.0, PFA 2% in buffer CB, 0,025% glutaraldheide, 0,2% Tx-100). Immunostaining was then performed using antibodies against GM130, TGN46 and Tubulin.
Caco2 Cysts
Caco-2 cells were plated in 35mm dishes. 24 h later they were transfected with siRNAs using Lipofectamine 2000. After an additional 24 h cells were trypsinized and an additional siRNA transfection using Lipofectamine 2000 was performed. Cells (6×104) were re-suspended in 60% DMEM + 40% Matrigel (previously thawed overnight on ice). 100ul of this solution was plated into 8-wells chambers (BD-biosciences), incubated at 37°C for 30 minutes and, once solid, Matrigel was covered with 400ul of DMEM. Media was changed every other day. On the 4th day of culture cysts were fixed with 300ul of paraformaldehyde (4%) 30 min at RT, washed with PBS, permeabilized with 0.5% triton X-100 for 10 min and washed with PBSG (PBS containing 7.5mg/ml glycine). Non-specific staining was blocked with 300ul of blocking buffer (7.7mM NaN3, 0.1% BSA, 0.2% Triton X-100, 0.05% Tween20) and afterwards the 8-wells chambers were incubated overnight with the primary antibodies diluted in PBS containing 3% BSA. On the next day, cells were washed with blocking buffer and secondary antibodies + fluorescent phalloidin were added for 2 h at RT. Cells were washed again with blocking buffer and the plastic support was removed. A glass coverslip was put on the top of the matrigel and the samples were imaged.
Confocal microscopy image analysis and FRET
Microscopy was performed on a LeicaSP5 confocal laser-scanning microscope equipped with a Ludin chamber. Most imaging was performed using the 63X objective (1.4 NA). Images were converted into TIFF files and analysed using ImageJ. FRET was performed using the donor-recovery after acceptor photobleaching method using the FRET-AB wizard of the Leica software. Acceptor (YFP) fluorescence was bleached at 100% laser intensity assuring a minimal extent of bleaching by 75%. To analyse FRET at the Golgi, we chose a region in the juxtanuclear region where the reporter accumulated. To test whether we could recognize the Golgi correctly when looking at the reporter, we initially performed experiments where we colocalized the reporter with Golgi markers and found that we could always recognize the Golgi accurately. To measure FRET at the plasma membrane, we chose random regions at the plasma membrane of cells grown up to 80% confluency. Briefly, cells were divided into 4 regions (quadrants of a Cartesian Coordinates Plane) and FRET was measured every time in one different region (the first cell was measured in the first quadrant, the next cell in the second quadrant, and so on). The only limitation we imposed was to avoid regions overlapping with neighbor transfected cells. ImageJ was used to process the images. Briefly, the process “subtract background” was used on every image. If needed to improve visibility, brightness and contrast of the images were corrected (linearly) using the “adjust” menu of ImageJ. Images coming from the same experiment were all modified the same way to leave unaltered the information. Merged images were created with the appropriate function of ImageJ, after correcting the single channel images as described above.
GEF screening
FRET measurements at the Golgi were performed in HEK293 cells expressing the Cdc42-EM probe. We calculated the CI95 of GM130 knockdown vs. control cells according to:
Any value below the CI95 (red line in Fig. 2) was considered a hit. Our previous measurements of control and GM130 knockdown cells were used to quantitate the CI95 with the formula described above. 30–45 cells were measured for each condition from two-three independent experiments. The rationale for measuring at least 30 cells was based on our calculation that 18 data points (i.e. cells) are necessary to distinguish a population that differs from the control by 80% of the GM130 effect.
FRET of directional migrating cells
HeLa cells were plated on glass coverslips and transfected with siRNA (HIPerfect). After 48 h, cells were transfected with plasmids encoding Cdc42-EM or Cdc42-PM FRET probes (Fugene6). After 72 h, a wound was made using a 200μl pipette tip and cells were allowed to migrate at 37°C for 6 hours. Then cells were either fixed or incubated for 1 h at 20°C and subsequently fixed. To measure FRET, we selected the region of the cells directly facing the wound and named it “front”. For regions designated as “rear” we chose regions from the side of the cell opposing the wound. Up to 3 different regions were measured in the “rear” and averaged. We did not detect any dependency on where we measured as long as the plasma membrane was opposing the wound. We avoided measuring FRET in regions overlapping with neighbor transfected cells.
Video acquisition of VSVG-GFP vesicles
HeLa cells were plated on glass coverslips and transfected with siRNA (HIPerfect). After 48 h, cells were transfected with plasmids encoding for the VSVG-GFP-RUSH (Fugene6). After 72 h, a wound was made using a 200μl pipette tip and cells were allowed to migrate at 37°C for 4 hours. Then imaging was started and biotin added to release the VSVG construct from the ER. Images were acquired every 1.5 sec.
Golgi orientation, cell migration and aPKC recruitment assay
For Golgi orientation assays, cells were plated on glass coverslips. After 72 h, a wound was made manually using a 200μl pipette tip. Cells were stained with antibodies anti γ-tubulin and anti-giantin immediately after wounding and after 6 hours. For assays measuring aPKC recruitment to the leading edge, cells were fixed 1 h after wounding.
For migration assays, cells were transfected with siRNA, and trypsinized after 48 h and plated into Ibidi® chambers placed on glass cover slips. This insert consists of two chambers separated by a 600 μm wall. Removal of the insert generates a wound with a defined width of 600 μm. 12000 cells were plated into each chamber. After 6h, the IBiDi insert was removed and cells were allowed to migrate at 37°C for 24 h. Then, cells were fixed and stained with concanavalin-alexa 488.
Coimmunoprecipitations
1×106 cells were plated in 10cm2 cell culture dishes. After 24 h, cells were transfected. After an additional 24 h, cells were lysed in IP buffer (20mM HEPES pH 7.3, 200mM KCl, 0,5% Triton X-100), then centrifuged for 15 min at 20’000xg. The supernatant was incubated with protein G sepharose beads coupled to the appropriate antibodies. For anti-HA immunoprecipitation commercial anti-HA beads (Sigma) were used. After 2 h incubation at 4°C, beads were recovered by centrifugation, washed and the bound material was eluted by boiling in sample buffer.
RUSH assay to study ER-Golgi trafficking
The RUSH assay was performed as described previously 39. Briefly, 250,000 HeLa cells stably expressing RUSH-ManII-GFP were plated in 35mm cell culture dishes and transfected with siRNA (using HiPerFect from Qiagen). After 72 h, cells were either fixed directly or were treated with 40 μM of biotin for 1 h followed by fixation in 4% paraformaldehyde at RT.
RT-PCR
Total RNA was isolated from cells by using the Qiagen RNeasy kit. cDNA was prepared from 100ng total RNA by using the high capacity cDNA reverse transcription kit (Applied Biosystems) following manufacturers’ instructions. Q-PCR was performed by using Fast SYBR green PCR MasterMix (Applied Biosystems). RT-PCR were run on 7900HT Fast RT-PCR system (Applied Biosystems). Expression of each gene was normalized to the expression of ubiquitin and β-2-microglobulin. Specific primers for RasGRF2 were described in Jacinto et al. 41. Primers for TUBA, ARHGEF9, ARHGEF11 and ARHGEF12 were designed using PrimerExpress®.
Immunohistochemistry
Three-μm slides cut from three different tissue microarray block containing a total of 229 different formalin-fixed, paraffin-embedded samples from colorectal cancers (n=133), tubulovillous adenomas (n=27) and normal colonic mucosa (n=69) were used (duplicate cores for each samples). The project was approved by the ethical committee of the IEO. Samples were rehydrated through xylene and graded alcohols. Antigen retrieval was accomplished using 1 mM EDTA, 0.05% Tween. After peroxidase block with 3% H2O2 for 5 min, and block with 2% goat serum in PBS for 1 h, samples were incubated with primary antibodies (Mouse monoclonal anti-GM130 antibody, BD-Biosciences) in a 1:50 dilution for 60’ min at RT in 2% goat serum. Immunocomplexes were visualized by the EnVision™+ HRP Rabbit (DAKO, K4011), and acquired with the Aperio ScanScope system. Duplicate cores for every sample were evaluated and then the scores were averaged and considered as weak expression (average score 0.5 – 1), moderate expression (average score 1.5 – 2), strong expression (average score 2.5 – 3), or as negative if they scored 0. IHC data for GM130 expression were available only for 29 normal colonic mucosa, 16 tubulovillous adenoma and 109 colorectal cancer samples, since in some cases, individual cores detached from the slides during the manipulations.
11 matched colon cancer samples were analysed at the University of Regensburg according to the following procedure: two-μm slides cut from a tissue microarray block containing 60 different formalin-fixed, paraffin-embedded samples from colorectal cancers (n=20) and matched normal tissues (n=40) were used. Immunohistochemistry of GM130 was carried out in a BenchMark ULTRA (VENTANA) autostainer according to the manufacturer’s instructions. Briefly, retrieval was done using a citrate buffer (pH 6.0) for 44 min. The anti-GM130 antibody was used in a 1:100 dilution and incubated for 32 min. The slides were detected using the ULTRA View DAB detection kit (VENTANA). Negative controls without primary antibody were included in each experiment.
Statistical analysis
The data were analysed using Student T Test, Chi Square test or ANOVA test for multiple comparison in GraphPad Prism. All IF images are representative of at least 3 independent experiments. Western blots evaluations and IF evaluations are averages of at least 3 independent experiments. For TMA analysis differences between experimental groups were examined for statistical significance using the nonparametric comparisons for each pair using Wilcoxon Method (Figure 7B) or Pearson Chi Square test (Figure 7C). Where applicable, data are expressed as average ± SD. TMA data analysis were performed using JMP 10.0 statistical software (SAS Institute, Inc).
Supplementary Material
Acknowledgments
HF is supported by the German Science Foundation (DFG), by the Biotechnology Institute Thurgau, and by the Young Scholar Fund of the University of Konstanz. FB acknowledges support by a fellowship from the Company of Biologists. We thank Beate Reiler for excellent technical assistance on staining the tissue samples. We thank Elisabeth Genot and Paolo Ciufici (University of Bordeaux) for help with the GTP-Cdc42 staining. PPDF acknowledges support by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC IG 14404), MIUR (the Italian Ministry of University and Scientific Research), the Italian Ministry of Health, the European Research Council (Mammastem Project) and The Monzino Foundation.
Footnotes
Author contributions
FB performed experiments, analysed experiments and wrote the manuscript
HF conceived the study, analysed experiments and wrote the manuscript
PC provided valuable reagents and wrote the manuscript
IGM provided valuable reagents and wrote the manuscript
WD performed and analysed experiments on GM130 expression in colorectal cancer
SC performed and analysed experiments on GM130 expression in colorectal cancer and wrote the manuscript
GB performed and analysed experiments on GM130 expression in colorectal cancer
PPDF supervised the analysis of GM130 expression in tumours and wrote the manuscript
ML provided reagents and wrote the manuscript
Competing financial interests
The authors declare that no competing financial interests exist.
References
- 1.Etienne-Manneville S. Cdc42 - the centre of polarity. J Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- 2.Iden S, et al. Tumor Type-Dependent Function of the Par3 Polarity Protein in Skin Tumorigenesis. Cancer Cell. 2012;22:389–403. doi: 10.1016/j.ccr.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 3.McCaffrey Luke M, Montalbano J, Mihai C, Macara Ian G. Loss of the Par3 Polarity Protein Promotes Breast Tumorigenesis and Metastasis. Cancer cell. 2012;22:601–614. doi: 10.1016/j.ccr.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue B, Krishnamurthy K, Allred DC, Muthuswamy SK. Loss of Par3 promotes breast cancer metastasis by compromising cell–cell cohesion. Nat Cell Biol. 2012;15:189–200. doi: 10.1038/ncb2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erickson JW, Zhang C-j, Kahn RA, Evans T, Cerione RA. Mammalian Cdc42 Is a Brefeldin A-sensitive Component of the Golgi Apparatus. Journal of Biological Chemistry. 1996;271:26850–26854. doi: 10.1074/jbc.271.43.26850. [DOI] [PubMed] [Google Scholar]
- 6.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of Endogenous Cdc42 Visualized in Living Cells. Science. 2004;305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 7.Farhan H, Rabouille C. Signalling to and from the secretory pathway. J Cell Sci. 2011;124:669. doi: 10.1242/jcs.076455. [DOI] [PubMed] [Google Scholar]
- 8.Yadav S, Puri S, Linstedt AD. A Primary Role for Golgi Positioning in Directed Secretion, Cell Polarity, and Wound Healing. Mol Biol Cell. 2009;20:1728–1736. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farnsworth CL, et al. Calcium activation of Ras mediated by neuronal exchange factor Ras-GRF. Nature. 1995;376:524–527. doi: 10.1038/376524a0. [DOI] [PubMed] [Google Scholar]
- 10.Fernández-Medarde A, Santos E. The RasGrf family of mammalian guanine nucleotide exchange factors. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2011;1815:170. doi: 10.1016/j.bbcan.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Arozarena I, et al. The Rho Family GTPase Cdc42 Regulates the Activation of Ras/MAP Kinase by the Exchange Factor Ras-GRF. J Biol Chem. 2000;275:26441–26448. doi: 10.1074/jbc.M002992200. [DOI] [PubMed] [Google Scholar]
- 12.Arozarena I, Matallanas D, Crespo P. Maintenance of Cdc42 GDP-bound State by Rho-GDI Inhibits MAP Kinase Activation by the Exchange Factor Ras-GRF: EVIDENCE FOR Ras-GRF FUNCTION BEING INHIBITED BY Cdc42-GDP BUT UNAFFECTED BY Cdc42-GTP. J Biol Chem. 2001;276:21878–21884. doi: 10.1074/jbc.M011383200. [DOI] [PubMed] [Google Scholar]
- 13.Calvo F, et al. RasGRF suppresses Cdc42-mediated tumour cell movement, cytoskeletal dynamics and transformation. Nat Cell Biol. 2011;13:819–826. doi: 10.1038/ncb2271. [DOI] [PubMed] [Google Scholar]
- 14.Itoh RE, et al. Activation of Rac and Cdc42 Video Imaged by Fluorescent Resonance Energy Transfer-Based Single-Molecule Probes in the Membrane of Living Cells. Mol Cell Biol. 2002;22:6582–6591. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshizaki H, et al. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. The Journal of Cell Biology. 2003;162:223–232. doi: 10.1083/jcb.200212049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kodani A, Kristensen I, Huang L, Sütterlin C. GM130-dependent Control of Cdc42 Activity at the Golgi Regulates Centrosome Organization. Mol Biol Cell. 2009;20:1192–1200. doi: 10.1091/mbc.E08-08-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durgan J, Kaji N, Jin D, Hall A. Par6B and Atypical PKC Regulate Mitotic Spindle Orientation during Epithelial Morphogenesis. Journal of Biological Chemistry. 2011;286:12461–12474. doi: 10.1074/jbc.M110.174235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaffe AB, Kaji N, Durgan J, Hall A. Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. J Cell Biol. 2008;183:625–633. doi: 10.1083/jcb.200807121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oddoux S, et al. Microtubules that form the stationary lattice of muscle fibers are dynamic and nucleated at Golgi elements. The Journal of Cell Biology. 2013;203:205–213. doi: 10.1083/jcb.201304063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matlin KS, Simons K. Reduced temperature prevents transfer of a membrane glycoprotein to the cell surface but does not prevent terminal glycosylation. Cell. 1983;34:233–243. doi: 10.1016/0092-8674(83)90154-x. [DOI] [PubMed] [Google Scholar]
- 21.Wang B, Wylie FG, Teasdale RD, Stow JL. Polarized trafficking of E-cadherin is regulated by Rac1 and Cdc42 in Madin-Darby canine kidney cells. American Journal of Physiology - Cell Physiology. 2005;288:C1411–C1419. doi: 10.1152/ajpcell.00533.2004. [DOI] [PubMed] [Google Scholar]
- 22.Kovacs EM, Makar RS, Gertler FB. Tuba stimulates intracellular N-WASP-dependent actin assembly. J Cell Sci. 2006;119:2715–2726. doi: 10.1242/jcs.03005. [DOI] [PubMed] [Google Scholar]
- 23.Salazar MA, et al. Tuba, a Novel Protein Containing Bin/Amphiphysin/Rvs and Dbl Homology Domains, Links Dynamin to Regulation of the Actin Cytoskeleton. J Biol Chem. 2003;278:49031–49043. doi: 10.1074/jbc.M308104200. [DOI] [PubMed] [Google Scholar]
- 24.Arozarena I, et al. Activation of H-Ras in the Endoplasmic Reticulum by the RasGRF Family Guanine Nucleotide Exchange Factors. Mol Cell Biol. 2004;24:1516–1530. doi: 10.1128/MCB.24.4.1516-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teng K, Greene L. Cultured PC12 cells: a model for neuronal function and differentiation. In: Celis JE, editor. Cell Biology: A Laboratory Handbook. Vol. 218. San Diego: Academic Press; 1994. p. 224. [Google Scholar]
- 26.Nieto MA. Epithelial Plasticity: A Common Theme in Embryonic and Cancer Cells. Science. 2013;342 doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- 27.Kupfer A, Louvard D, Singer SJ. Polarization of the Golgi apparatus and the microtubule-organizing center in cultured fibroblasts at the edge of an experimental wound. PNAS. 1982;79:2603–2607. doi: 10.1073/pnas.79.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kodani A, Sütterlin C. The Golgi Protein GM130 Regulates Centrosome Morphology and Function. Mol Biol Cell. 2008;19:745–753. doi: 10.1091/mbc.E07-08-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson JC, et al. Genome-wide RNAi screening identifies human proteins with a regulatory function in the early secretory pathway. Nat Cell Biol. 2012;14:764–774. doi: 10.1038/ncb2510. [DOI] [PubMed] [Google Scholar]
- 30.Diao A, Frost L, Morohashi Y, Lowe M. Coordination of Golgin Tethering and SNARE Assembly: GM130 BINDS SYNTAXIN 5 IN A p115-REGULATED MANNER. Journal of Biological Chemistry. 2008;283:6957–6967. doi: 10.1074/jbc.M708401200. [DOI] [PubMed] [Google Scholar]
- 31.Marra P, et al. The Biogenesis of the Golgi Ribbon: The Roles of Membrane Input from the ER and of GM130. Molecular Biology of the Cell. 2007;18:1595–1608. doi: 10.1091/mbc.E06-10-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- 33.Osmani N, Peglion F, Chavrier P, Etienne-Manneville S. Cdc42 localization and cell polarity depend on membrane traffic. J Cell Biol. 2010;191:1261–1269. doi: 10.1083/jcb.201003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubois T, et al. Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp2/3 complex and F-actin dynamics. Nat Cell Biol. 2005;7:353–364. doi: 10.1038/ncb1244. [DOI] [PubMed] [Google Scholar]
- 35.Falkenberg CV, Loew LM. Computational Analysis of Rho GTPase Cycling. PLoS Comput Biol. 2013;9:e1002831. doi: 10.1371/journal.pcbi.1002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozubowski L, et al. Symmetry-Breaking Polarization Driven by a Cdc42p GEF-PAK Complex. Current Biology. 2008;18:1719–1726. doi: 10.1016/j.cub.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Layton AT, et al. Modeling Vesicle Traffic Reveals Unexpected Consequences for Cdc42p-Mediated Polarity Establishment. Current Biology. 2011;21:184–194. doi: 10.1016/j.cub.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 39.Boncompain G, et al. Synchronization of secretory protein traffic in populations of cells. Nat Meth. 2012;9:493–498. doi: 10.1038/nmeth.1928. [DOI] [PubMed] [Google Scholar]
- 40.Goedhart J, et al. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93% Nat Commun. 2012;3 doi: 10.1038/ncomms1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacinto FV, Ballestar E, Ropero S, Esteller M. Discovery of Epigenetically Silenced Genes by Methylated DNA Immunoprecipitation in Colon Cancer Cells. Cancer Research. 2007;67:11481–11486. doi: 10.1158/0008-5472.CAN-07-2687. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.