Abstract
The persistence of HIV-Infected cells in individuals on suppressive combination antiretroviral therapy (cART) presents a major barrier for curing HIV infections. HIV integrates its DNA into many sites in the host genome; we identified 2410 integration sites in peripheral blood lymphocytes of five infected individuals on cART. About 40% of the integrations were in clonally expanded cells. Approximately 50% of the infected cells in one patient were from a single clone and some clones persisted for many years. There were multiple independent integrations in several genes, including MKL2 and BACH2; many of these integrations were in clonally expanded cells. Our findings show that HIV integration sites can play a critical role in expansion and persistence of HIV infected cells.
HIV replication is suppressed by combination antiretroviral therapy (cART), but infected cells persist in patients and are a critical obstacle to curing HIV infection (1, 2). Analysis of HIV populations in vivo shows that, after long-term suppressive cART, genetically identical HIV variants emerge (3, 4). The source and mechanisms involved in the emergence of these identical variants are not understood. One possibility is that the identical variants arise from cells that have clonally expanded. Because HIV DNA integrates at many sites in the human genome, the site of integration can be used to identify clonally expanded cells that arose from a single infected progenitor.
Clonal expansion of HIV infected cells inpatients
We analyzed the integration sites in peripheral blood mononuclear cells (PBMCs) or CD4+ T-cells from patients on prolonged cART obtained by negative selection using a previously described technique (5–7). DNA from PBMCs or CD4+ T-cells was randomly sheared to ~400bp fragments and linker-mediated PCR was used to selectively amplify fragments that contained viral/host DNA junctions (8). Both ends of the amplified junction fragments were sequenced on the Illumina platform to detennine the viral/host junctions and the breakpoints in the host DNA. The sequence of the viral/host junction identi fies the exact position and orientation in which the HIV DNA was integrated. The breakpoints in the host DNA can be used to identify the integration sites in clonally expanded cells. If several cells with the same integration site are presert, shearing their DNA will give rise to multiple fragments in which the integration site is the same, but the host DNA breakpoints differ.
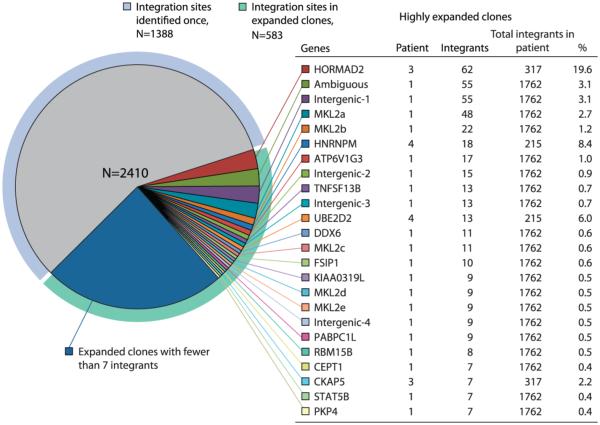
Integration site analysis was performed using PBMCs or CD4+ T-cells from 5 patients (Table S1). A diverse population of viruses was present in each patient either pre-therapy or shortly after cART was initiated; however, after prolonged cART (mean duration of treatment, 11.7 y), identical viral sequences, defined at either the RNA or the DNA level, emerged (Fig. 1, S1). In total 2410 integration sites were mapped; these represented 1632 different integration events. 1388 integration sites (57%) were detected once, and 1022 sites (43%) were associated with more than one host DNA breakpoint, revealing that a large fraction of the infected cells are from expanded clones (Fig. 2, Table S2, Table S3, Fig. S2). We validated the method for identifying expanded clones by making two completely independent libraries from cells from the last time point from patient 1; the same highly expanded clones were identified in both libraries. In some cases, clonal expansion was extensive. For example, in patient 3, the initial analysis found the same site in the HORMAD2 gene in 62 of 317 integrations, but, because of overlaps in the breakpoint analysis, this figure is an underestimate. We also estimated the fraction of the total integration sites that were derived from this expanded clone (8). This analysis implied that approximately 58% of all the HIV infected cells in this patient were derived from a single infected cell.
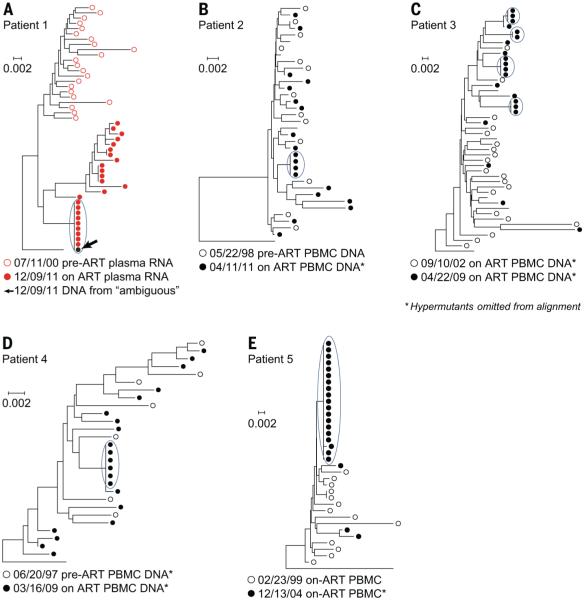
Fig. 1. Long term cART reveals, the presence, in patients, at both the RNA and the DNA level, of HIV genomes that have identical sequences.
(A to E) Single-genome sequences were obtained from plasma virion RNA or from PBMC DNA prior to or shortly after the initiation of cART, and after long-term cART. Sequences were aligned using Clustal W, and neighbor-joining trees were rooted on the consensus sequence of subgroup B HIV (8). Virus populations from pretherapy samples, or samples taken shortly after the initiation of cART, are shown as open circles (red, virion RNA; black, PBMC DNA); populations after prolonged cART, as closed circles (red, virion RNA; black, PBMC DNA). In patient 1, the black dot, marked by the arrow, represents DNA sequences from the provirus in a clone of expanded cells, whose integration site could not be mapped (“ambiguous”, Fig. 2). Short branches and low bootstrap values on major nodes of the trees support a lack of divergence between preor early therapy sequences and populations of identical sequences after prolonged cART (4). To avoid distorting the trees, all hypermutant sequences were removed for the analysis shown in the figure. * Denotes the trees from which G>A hypermutant sequences were removed.
Fig. 2. Distribution of integration sites in the five patients.
A total of 2410 integration sites were obtained from PBMCs or negatively selected CD4+ cells from the five patients. Genes in which we isolated a particular integration site 7 or more times are shown. The table shows which patient harbored each of these expanded clones, how many times the different integrations sites were isolated, and the fraction of the infected cells the expanded clone represents in the patient. The MKL2 clones marked a, b, c, and d correspond to the integration sites marked a, b, c, and d in Fig. 3. One provirus in a highly expanded clone in patient 1 was in a sequence that could not be unambiguously assigned in the human genome (denoted “ambiguous”). Our standard analysis (presented here) underestimated the fraction of the infected cells in patient 3 that had an integration site in HORMAD2; see text and (8).
Among the 5 patients studied, HIV integration sites were found in 985 different genes (Fig. S3). Most of the genes had only a single integrant (659 integrants, 67%); these integrants were not shown to be from clonally expanded cells. The remaining 326 genes (33%) either had a single integrant in a cell that underwent clonal expansion (126 genes) or had multiple integrants (200 genes), some of which (59 genes; 30%) were in cells that underwent clonal expansion (Fig. S3). Approximately 70% (21 of 29) of the genes with multiple integrants in highly expanded clones (listed in Table S4) are known to be directly involved in the regulation of cell growth. HIV proviruses (the integrated form of retroviral DNA is called a provirus) were also found in intergenic regions of the genome, and in sites that could not be uniquely mapped (ambiguous sites); some of the cells with such integrants were also highly expanded (Fig. S2).
Clones of HIV infected cells per sist in patients for more than 11 years
Longitudinal sampling revealed that some of the clones that emerged on cART persisted for many years. Patient 1 had at least 13 different clones that were present in samples taken 11.2 years apart, and 11 other clones were present in samples taken 6.6 years apart (Table 1). A number of these p ersistent clones had integration sites in genes known to be associated with cell growth (STAT5B, PARP8, DDX6), and/or mitosis (PKP4, MAP4). In patient 3, we analyzed a small set of integration sites (47 total sites) from a sample taken before the patient started cART. Most of the integration sites from this pre-cART sample were unique, an observation consistent with the short half-life of infected cells during uncontrolled HIV replication (9). However, one site was from a clonally expanded cell, showing that there is clonal expansion in the absence of cART. Others have reported that there are HIV infected clones that can persist for prolonged periods in patients on cART (10, 11). Our dam show that prolonged persistence of expanded clones is common and is frequently associated with specific integrations in genes involved in controlling cell growth and division. Although there was variation in integration sites among patients, all 5 patients showed evidence of clonal expansion of infected cells, even in the smallest datasets (35 and 46 distinct integration sites, Table S2).
Table 1.
In patient 1, clones persist for many years.
| Detected in year* | ||||||
|---|---|---|---|---|---|---|
| Gene | Integration Site | 0.2 | 4.8 | 11.4 | Orientation† | Gene Description |
| DDX6 | chr11:118636877 | + | + | + | Reverse | DEAD (Asp-Glu-Ala-Asp) box polypep- tide 6 |
| TNFSF13B | chr13:108927255 | + | + | + | Forward | tumor necrosis factor (ligand) superfamily, member 13 |
| PKP4 | chr2:159370503 | + | + | + | Forward | plakophilin 4 |
| RBM15B | chr3:51434782 | + | + | + | Reverse | RNA binding motif protein 15B |
| PARP8 | chr5:50035007 | + | + | + | Reverse | poly (ADP-ribose) polymerase family, member 8 |
| ATP6V1G3 | chr1:198506789 | − | + | + | Forward | ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
| SLC30A7 | chr1:101445575 | − | + | + | Forward | solute carrier family 30 (zinc transporter), member 7 |
| ZCCHC11 | chr1:52956520 | − | + | + | Reverse | zinc finger, CCHC domain containing 11 |
| NUMA1 | chr11:71744577 | − | + | + | Forward | nuclear mitotic apparatus protein 1 |
| CTAGE5 | chr14:39760983 | − | + | + | Reverse | CTAGE family, member 5 |
| PATL2 | chr15:44962782 | − | + | + | Forward | protein associated with topoisomerase II homolog 2 (yeast) |
| P4HB‡ | chr17:79800929 | − | + | + | Reverse | prolyl 4-hydroxylase, beta polypeptide |
| ZC3H4 | chr19:47603701 | − | + | + | Reverse | zinc finger CCCH-type containing 4 |
| COG5 | chr7:107182849 | − | + | + | Reverse | component of oligomeric golgi complex 5 |
| GIMAP6‡ | chr7:150318109 | − | + | + | Reverse | GTPase, IMAP family member 6 |
| ZNF16 | chr8:146170620 | − | + | + | Forward | zinc finger protein 16 |
| R3HDM2‡ | chr12:57731640 | + | − | + | Forward | R3H domain containing 2 (R3HDM2), mRNA.) |
| FSIP1 | chr15:39975096 | + | − | + | Forward | fibrous sheath interacting protein 1 |
| NLRC5 | chr16:57030643 | + | − | + | Forward | NLR family, CARD domain containing 5 |
| STAT5B | chr17:40421711 | + | − | + | Reverse | signal transducer and activator of transcrip- tion 5B |
| PABPC1L | chr20:43561100 | + | − | + | Reverse | poly(A) binding protein, cytoplasmic 1-like |
| MAP4 | chr3:47981132 | + | − | + | Reverse | microtubule-associated protein 4 |
| BTNL3‡ | chr5:180401789 | + | − | + | Forward | butyrophilin-like 3 |
| SQLE | chr8:126012090 | + | − | + | Forward | squalene epoxidase |
| CHD7 | chr8:61594573 | + | + | − | Reverse | chromodomain helicase DNA binding pro- tein 7 |
Relative to start of cART.
The orientation of the provirus relative to the direction of transcription of the gene.
Nearest gene.
Integration in specific genes is associated with the clonal expansion and/or persistence of infected cells
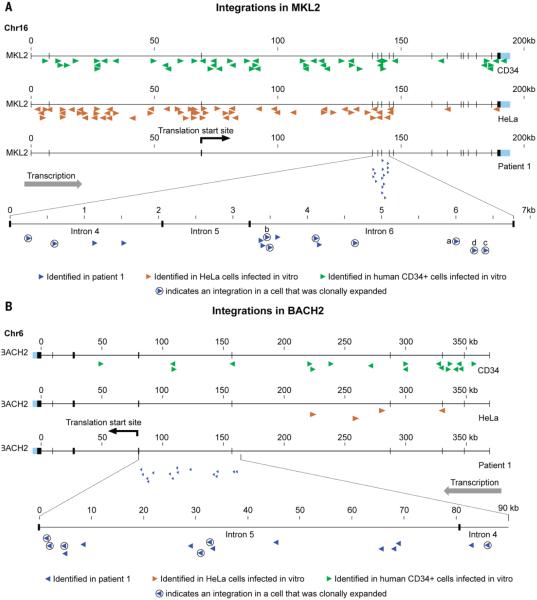
Two genes with remarkable patterns of HIV integration were identified in patient 1 (Fig. 3). In the dataset obtained from CD4+ T-cells after 11.4 year of cART, there were 11 distinct integration sites in intron 6 of MKL2 (intron 6 is ~3.5 kb), more than half of which were in clonally expanded cells; some of these cells were highly expanded (Figs. 2 and 3). There were also 4 nearby integration sites in intron 4 and none in any other part of MKL2 (Fig. 3A). All 15 of these proviruses were integrated in the same transcriptional orientation as the host gene. Thus, about 7% of the infected cells in this patient had proviruses in a region that constitutes a very small fraction (approximately 2×10−6) of the human genome. In the same dataset, there were 15 independent integration sites, also in the same transcriptional orientation as the host gene, in introns 4 and 5 of BACH2 (Fig. 3B); two additional integration sites in BACH2 were identified in earlier samples from this patient.
Fig. 3. Integration sites in the MKL2 and BACH2 genes in patient 1 after 11.4 years of cART.
(A) There were 15 distinct integration sites (blue arrows) in a small region of the MKL2 gene in patient 1. The arrows denote the transcriptional orientation of each provirus. The circled arrows indicate integration sites in clonally expanded cells. The arrows marked a, b, c, and d correspond to the clones marked a, b, c, and d in Fig. 2. (B) There were 15 distinct integration sites in a small region of the BACH2 gene in patient 1 (blue arrows). Some of these integration sites were in clonally expanded cells (circled arrows). Panels A and B also show the HIV integration sites identified in the same two genes in acutely infected HeLa cells (total sites=248,658, brown arrows) and CD34+ cells (total sites=159,484, green arrows).
For comparison, we analyzed two large HIV integration site libraries made from acutely infected HeLa cells (~ 250,000 sites) and human CD34+ hematopoietic stem cells (~ 150,000 sites). The frequencies of HIV integration in MKL2 and BACH2 in cells from patient 1 were much greater than in HeLa or CD34+ cells. In cells from patient 1, integrations in MKL2 were 7% of the total integrations, compared to 0.03% of total integrations in HeLa and CD34+ cells. Similarly, integrations in BACH2 in cells from patient 1 were 1.5% of the total integrations, compared to 0.002% in HeLa cells and 0.01% in CD34+ cells. There was no preference for integrations in specific introns in these genes in HeLa cells or CD34+ cells. Nor was there any indication, in either library, of preferential integration in one orientation in MKL2 or BACH2. Across the entire patient dataset (Table S3), there was a weak, but significant preference for integrants in genes to be in an orientation opposite to the direction in which the gene is transcribed. Of the 1313 integrants that were in genes, 594 were in the same transcriptional orientation as the gene, and 719 were in the opposite orientation (p=0.02, Fisher’s exact test). These findings led us to conclude that the HIV DNA insertions in MKL2 and BACH2 were selected, post integration, because they altered the level of expression of the MKL2 and BACH2 proteins and/or gave rise to the expression of altered forms of the proteins, and that these alterations affected the expansion and survival of the infected cells. In the case of BACH2, all of the integrations were upstream of the initiation site for translation, consistent with the integrations altering the level of expression of BACH2. The integrations in MKL2 were in introns that were between two coding exons and these integrations were more likely to have affected the structure of the protein. Both these mechanisms have been seen with other types of retroviruses and are known to be involved in oncogenic transformation in animals (12).
In all, we found, out of the 985 genes in which there were integrations in the patients, that there were 200 genes that had multiple independent integrations; 59 of these were associated with expanded clones. Table S4 shows the genes that had at least 3 independent integrants; as mentioned earlier, many of these genes have roles in cell growth. In addition, there were integrations in more than one patient in more than 60% (18 of 29) of the genes listed in table S4. For example, a total of 10 independent integrants were found in STAT5B in 4 of the 5 patients, some of these integrants were in expanded clones; however, the proviruses integrated in STAT5B showed no orientation preference. Gene ontology analysis showed that the patient integration sites were enriched for genes in several pathways involved in cell growth. The HeLa and human CD34+ cell datasets (which were similar to each other) were not enriched for genes in these pathways (Fig. S4). This analysis also showed that the patient dataset was related to leukemia and Burkitt’s lymphoma; the HeLa and human CD34+ datasets were not associated with any disease related pathways.
Although, as expected (13), most of the integration sites in the patients were in genes, 21% (509) were in intergenic regions or were in sequences that could not be mapped to a unique location. 44% (226) of the integrants in the intergenic regions, or that could be mapped to a unique location, were in cells that underwent clonal expansion, some of which were highly expanded (Fig. S2, Fig. 2). One of the proviruses in the two most highly expanded clones in patient 1 [each site was identified 55 times, which is an underestimate because of breakpoint overlaps, see (8)], was in an intergenic region; the other could not be mapped to a unique location in the human genome (Fig. 2). The predominant virus in the plasma of patient 1 late in therapy was clonal (Fig. 1, Fig. S1), and was insensitive to a switch in cART (Fig. 1, Fig. S1), suggesting that it was produced by a clone of infected cells, rather than from infection of new cells. More than 1kb of sequence in gag-pro-pol from the RNA genome of this predominant virus exactly matched the sequence of the ambiguously mapped provirus, identifying this provirus as the source of the clonal viral RNA in the plasma (Fig. 1A, black arrow).
Discussion
Our results strongly imply that, in at least some cases, sites of HIV integration play an important role in the expansion and/or persistence of infected cells in patients. This conclusion is particularly strong for the integrations into specific introns of the MKL2 and BACH2 genes. The integrations in MKL2 and BACH2 that were linked to clonal expansion were in internal introns, and in the same transcriptional orientation as the genes in which they are inserted. Even setting aside the fact that that it is extremely unlikely that such a large fraction of the integrations would have occurred in these two small segments of the genome, the probability that all 33 of the integrations we saw in BACH2 and MKL2 in the patients (Table S4) would have been in the same orientation as the genes is approximately 10−10. In prior studies, a limited number of HIV integration sites were identified in patients (14–17), and HIV proviruses were found in intron 6 of MKL2 and intron 5 of BACH2, in the same orientation that the genes are transcribed. However, in the published studies, the integration sites in these genes were not linked to the clonal expansion of the infected cells (Table S5). Both BACH2 and MKL2 are involved in the growth and development of cells, and BACH2 is known to play a key role in T-cell development (18). Both genes (and the MKL2-related gene MKL1, in which there were 4 independent integration sites, some of which were associated with clonally expanded cells in patient 1, Table S4) have been implicated in human cancers (19–21), where they were activated by DNA rearrangements that created gene fusions. The pattern of multiple integrations in MKL2 and BACH2 found in the patients cannot be the result of preferential integration because HIV integration is neither intron specific nor orientation specific (22). Thus, the only plausible explanation for the data that is in accord with the rules for HIV integration is that the cells with the integrations in MKL2 and BACH2 were selected post-integration because the integrations in these genes contributed to the expansion and persistence of the host cells. This interpretation is supported, for BACH2, by a report showing that this gene is a target for retroviral insertional activation in mice infected with murine leukemia virus (23).
Most of our analyses were performed using cells from patients on long-term cART, which blocks the infection of additional cells, but has no effect on cells that have already been infected (9). During untreated HIV infections, approximately 109 cells are infected daily. The vast majority (99%) of the newly infected cells die within 24-48 hours and a substantial proportion of the remaining cells die within 2-4 weeks (24–26). Viremia decreases by 4-5 logs when patients undergo cART; however, the number of cells containing HIV DNA decreases by approximately one log (9), indicating that a substantial fraction (~10%) of the cells that were infected before the initiation of cART persist. Most of these long-lived infected cells contain proviruses that are obviously defective; however, about 12% of the proviruses appear to be functional, although only a small fraction of these apparently functional proviruses can be induced to make virus in ex vivo experiments (27). Cells infected with highly defective or fully latent proviruses that produce little or no viral protein may have a survival advantage relative to cells that produce virions, because cells that express viral proteins are more likely to be lysed by HIV-specific CTL, or be subject to cytopathic effects of the viral proteins. This same logic applies to HIV infected cells that undergo clonal expansion. Although we have not yet shown that clonally expanded cells produce replication competent HIV, we have shown that a highly expanded clone of cells does produce HIV virions in sufficient quantity to cause viremia, which means that the selection against cells that produce viral proteins is not so strong that it prevents extensive clonal expansion of cells that express the viral proteins required to produce virions.
Our data show that many of the infected cells that persist have undergone clonal expansion; these clones were revealed but not created by cART. For some infected cell clones, it is likely that the integration site is only a passive marker of clonal expansions that are driven by another factor or factors, such as antigen stimulation or homeostatic proliferation signals (28). By contrast, we show here that some cells with HIV integration sites in specific genes are strongly selected because these integrations promote the survival and expansion of the infected cells. Although there are obvious similarities in the integration sites seen in the five patients, there is considerable heterogeneity from one patient to another, both in the extent of clonal expansion and in the genes in which proviruses are integrated in the clonally expanded cells (Fig. S5). This complexity highlights the difficulty in attempting to extrapolate, from bulk HIV DNA quantification, the size and nature of the population of HIV proviruses that make up the reservoir that gives rise to HIV rebound following cessation of cART (28).
Our findings have relevance for three important areas: 1) To effectively target HIV persistence with the goal of achieving a cure, it will be important not only to suppress any replication of the virus, but also to block the expansion of infected cells. 2) Although the HIV vectors used in gene therapy have safety features that the parental virus lacks, we now know that, like many other retroviruses, HIV integration can lead to clonal expansion and persistence of infected cells. This discovery suggests that persons treated with HIV-based vectors should be carefully monitored for evidence of clonal expansion of vector-infected cells. 3) We also suggest that it is time to reexamine the question of whether HIV integration can contribute to the development of malignancies. Although there are well-defined cancers in HIV-infected patients that are the result of uncontrolled expression of herpes viruses, there are reports of a small number of lymphomas with HIV proviruses integrated at defined sites; one lymphoma had a provirus integrated in BACH2 (15, 29, 30). Despite these published reports, it is widely believed that HIV DNA is not detectable in most cancers from HIV-infected patients; however, the experiments supporting this belief are not well-documented in the literature. It is possible that prior attempts to detect HIV DNA in cancers examined only a very small portion of the HIV genome; and as such, missed HIV proviruses having large deletions; large deletions are a characteristic of the proviruses that cause murine and avian tumors. Thus, our findings have important implications for designing and implementing strategies to eliminate persistent HIV infection, for the use of lentiviral vectors for gene therapy in human patients, and, possibly, for the origin of some HIV-related malignancies.
Supplementary Material
Acknowledgments
The authors are indebted to the study participants, and to the clinical staff of the NIAID/CCMD clinic who cared for them. We thank C. Lane, H. Malech, H. Imamichi, S. Matsushita, and L. Frenkel for stimulating discussions. We are grateful to J. Meyer and A. Kane for help with the figures, and T. Burdette for help in preparing the manuscript. The data presented in this work is tabulated in the main paper and in the supplementary materials. The integration sites are compiled in Table S3; the data can also be accessed using the NCBI accession number PRJNA241020. Funding for this research was provided with Federal funds from the National Cancer Institute, an NIH Bench to Bedside award (FM), and by funds from the National Cancer Institute under Contract No HSSN261200800001E (XW, LS). JMC was supported by a Research Professorship from the American Cancer Society with additional support from the FM Kirby Foundation and by funding from the National Cancer Institute (Leidos contract 25XS119). JWM was supported by funding from the National Cancer Institute (Leidos contract 25XS119). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References
- 1.Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S, Chomont N, Chun T-W, Churchill M, Di Mascio M, Katlama C, Lafeuillade A, Landay A, Lederman M, Lewin SR, Maldarelli F, Margolis D, Markowitz M, Martinez-Picado J, Mullins JI, Mellors J, Moreno S, O’Doherty U, Palmer S, Penicaud MC, Peterlin M, Poli G, Routy JP, Rouzioux C, Silvestri G, Stevenson M, Telenti A, Van Lint C, Verdin E, Woolfrey A, Zaia J, Barré-Sinoussi F, International AIDS Society Scientific Working Group on HIV Cure Towards an HIV cure: A global scientific strategy. Nat. Rev. Immunol. 2012;12:607–614. doi: 10.1038/nri3262. Medline doi:10.1038/nri3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer S, Josefsson L, Coffin JM. HIV reservoirs and the possibility of a cure for HIV infection. J. Intern. Med. 2011;270:550–560. doi: 10.1111/j.1365-2796.2011.02457.x. Medline doi:10.1111/j.1365-2796.2011.02457.x. [DOI] [PubMed] [Google Scholar]
- 3.Bailey JR, Sedaghat AR, Kieffer T, Brennan T, Lee PK, Wind-Rotolo M, Haggerty CM, Kamireddi AR, Liu Y, Lee J, Persaud D, Gallant JE, Cofrancesco J, Jr., Quinn TC, Wilke CO, Ray SC, Siliciano JD, Nettles RE, Siliciano RF. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 2006;80:6441–6457. doi: 10.1128/JVI.00591-06. Medline doi:10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kearney MF, Spindler J, Shao W, Yu S, Anderson EM, O’Shea A, Rehm C, Poethke C, Kovacs N, Mellors JW, Coffin JM, Maldarelli F. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLOS Pathog. 2014;10:e1004010. doi: 10.1371/journal.ppat.1004010. Medline doi:10.1371/journal.ppat.1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry CC, Gillet NA, Melamed A, Gormley N, Bangham CR, Bushman FD. Estimating abundances of retroviral insertion sites from DNA fragment length data. Bioinformatics. 2012;28:755–762. doi: 10.1093/bioinformatics/bts004. Medline doi:10.1093/bioinformatics/bts004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillet NA, Malani N, Melamed A, Gormley N, Carter R, Bentley D, Berry C, Bushman FD, Taylor GP, Bangham CR. The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood. 2011;117:3113–3122. doi: 10.1182/blood-2010-10-312926. Medline doi:10.1182/blood-2010-10-312926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Ravin SS, Su L, Theobald N, Choi U, Macpherson JL, Poidinger M, Symonds G, Pond SM, Ferris AL, Hughes SH, Malech HL, Wu X. Enhancers Are Major Targets for Murine Leukemia Virus Vector Integration. J. Virol. 2014 doi: 10.1128/JVI.00011-14. 10.1128/JVI.00011-14 doi:10.1128/JVI.00011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Materials and methods are available as supplementary materials on Science Online.
- 9.Coffin J, Swanstrom R. HIV pathogenesis: Dynamics and genetics of viral populations and infected cells. Cold Spring Harb. Perspect. Med. 2013;3:a012526. doi: 10.1101/cshperspect.a012526. Medline doi:10.1101/cshperspect.a012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imamichi H, Natarajan V, Adelsberger JW, Rehm CA, Lempicki RA, Das B, Hazen A, Imamichi T, Lane HC. Lifespan of effector memory CD4+ T cells determined by replication-incompetent integrated HIV-1 provirus. AIDS. 2014;1 doi: 10.1097/QAD.0000000000000223. 10.1097/QAD.0000000000000223 Medline doi:10.1097/QAD.0000000000000223. [DOI] [PubMed] [Google Scholar]
- 11.Josefsson L, von Stockenstrom S, Faria NR, Sinclair E, Bacchetti P, Killian M, Epling L, Tan A, Ho T, Lemey P, Shao W, Hunt PW, Somsouk M, Wylie W, Douek DC, Loeb L, Custer J, Hoh R, Poole L, Deeks SG, Hecht F, Palmer S. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E4987–E4996. doi: 10.1073/pnas.1308313110. Medline doi:10.1073/pnas.1308313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg N, Jolicoeur P. In: Retroviruses. Coffin JM, Hughes SH, Varmus HE, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1997. pp. 475–585. [PubMed] [Google Scholar]
- 13.Schröder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. Medline doi:10.1016/S0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda T, Shibata J, Yoshimura K, Koito A, Matsushita S. Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. J. Infect. Dis. 2007;195:716–725. doi: 10.1086/510915. Medline doi:10.1086/510915. [DOI] [PubMed] [Google Scholar]
- 15.Mack KD, Jin X, Yu S, Wei R, Kapp L, Green C, Herndier B, Abbey NW, Elbaggari A, Liu Y, McGrath MS. HIV insertions within and proximal to host cell genes are a common finding in tissues containing high levels of HIV DNA and macrophage-associated p24 antigen expression. J. Acquir. Immune Defic. Syndr. 2003;33:308–320. doi: 10.1097/00126334-200307010-00004. Medline doi:10.1097/00126334-200307010-00004. [DOI] [PubMed] [Google Scholar]
- 16.Han Y, Lassen K, Monie D, Sedaghat AR, Shimoji S, Liu X, Pierson TC, Margolick JB, Siliciano RF, Siliciano JD. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 2004;78:6122–6133. doi: 10.1128/JVI.78.12.6122-6133.2004. Medline doi:10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katano H, Sato Y, Hoshino S, Tachikawa N, Oka S, Morishita Y, Ishida T, Watanabe T, Rom WN, Mori S, Sata T, Weiden MD, Hoshino Y. Integration of HIV-1 caused STAT3-associated B cell lymphoma in an AIDS patient. Microbes Infect. 2007;9:1581–1589. doi: 10.1016/j.micinf.2007.09.008. Medline doi:10.1016/j.micinf.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu G, Chen J. A genome-wide regulatory network identifies key transcription factors for memory CD8? T-cell development. Nat. Commun. 2013;4:2830. doi: 10.1038/ncomms3830. Medline doi:10.1038/ncomms3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi S, Taki T, Chinen Y, Tsutsumi Y, Ohshiro M, Kobayashi T, Matsumoto Y, Kuroda J, Horiike S, Nishida K, Taniwaki M. Identification of IGHCδ-BACH2 fusion transcripts resulting from cryptic chromosomal rearrangements of 14q32 with 6q15 in aggressive B-cell lymphoma/leukemia. Genes Chromosomes Cancer. 2011;50:207–216. doi: 10.1002/gcc.20845. Medline. [DOI] [PubMed] [Google Scholar]
- 20.Flucke U, Tops BB, de Saint Aubain Somerhausen N, Bras J, Creytens DH, Küsters B, Groenen PJ, Verdijk MA, Suurmeijer AJ, Mentzel T. Presence of C11orf95-MKL2 fusion is a consistent finding in chondroid lipomas: A study of eight cases. Histopathology. 2013;62:925–930. doi: 10.1111/his.12100. Medline doi:10.1111/his.12100. [DOI] [PubMed] [Google Scholar]
- 21.Muehlich S, Hampl V, Khalid S, Singer S, Frank N, Breuhahn K, Gudermann T, Prywes R. The transcriptional coactivators megakaryoblastic leukemia 1/2 mediate the effects of loss of the tumor suppressor deleted in liver cancer 1. Oncogene. 2012;31:3913–3923. doi: 10.1038/onc.2011.560. Medline doi:10.1038/onc 2011 560. [DOI] [PubMed] [Google Scholar]
- 22.Craigie R, Bushman FD. HIV DNA integration. Cold Spring Harb. Perspect. Med. 2012;2:a006890. doi: 10.1101/cshperspect.a006890. Medline doi:10.1101/cshperspect.a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Sørensen AB, Wang B, Wabl M, Nielsen AL, Pedersen FS. Identification of novel Bach2 transcripts and protein isoforms through tagging analysis of retroviral integrations in B-cell lymphomas. BMC Mol. Biol. 2009;10:2. doi: 10.1186/1471-2199-10-2. Medline doi:10.1186/1471-2199-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. Medline doi:10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 25.Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, Saag MS, Shaw GM. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. Medline doi:10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 26.Coffin JM. HIV population dynamics in vivo: Implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. Medline doi:10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 27.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. Medline doi:10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sékaly RP. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 2009;15:893–900. doi: 10.1038/nm.1972. Medline doi:10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiramizu B, Herndier BG, McGrath MS. Identification of a common clonal human immunodeficiency virus integration site in human immunodeficiency virus-associated lymphomas. Cancer Res. 1994;54:2069–2072. Medline. [PubMed] [Google Scholar]
- 30.Herndier BG, Shiramizu BT, Jewett NE, Aldape KD, Reyes GR, McGrath MS. Acquired immunodeficiency syndrome-associated T-cell lymphoma: Evidence for human immunodeficiency virus type 1-associated T-cell transformation. Blood. 1992;79:1768–1774. Medline. [PubMed] [Google Scholar]
- 31.Orenstein JM, Feinberg M, Yoder C, Schrager L, Mican JM, Schwartzentruber DJ, Davey RT, Jr., Walker RE, Falloon J, Kovacs JA, Miller KD, Fox C, Metcalf JA, Masur H, Polis MA. Lymph node architecture preceding and following 6 months of potent antiviral therapy: Follicular hyperplasia persists in parallel with p24 antigen restoration after involution and CD4 cell depletion in an AIDS patient. AIDS. 1999;13:2219–2229. doi: 10.1097/00002030-199911120-00004. Medline doi:10.1097/00002030-199911120-00004. [DOI] [PubMed] [Google Scholar]
- 32.Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RT, Jr., Dewar RL, Metcalf JA, Hammer S, Mellors JW, Coffin JM. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. Medline doi:10 1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. Medline doi:10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 34.Josefsson L, Palmer S, Faria NR, Lemey P, Casazza J, Ambrozak D, Kearney M, Shao W, Kottilil S, Sneller M, Mellors J, Coffin JM, Maldarelli F. Single cell analysis of lymph node tissue from HIV-1 infected patients reveals that the majority of CD4+ T-cells contain one HIV-1 DNA molecule. PLOS Pathog. 2013;9:e1003432. doi: 10.1371/journal.ppat.1003432. Medline doi:10.1371/journal.ppat.1003432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. Medline doi:10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.