SUMMARY
Growth–defense tradeoffs are thought to occur in plants due to resource limitations to optimize plant fitness. Hormone crosstalk appears to be the primary means for plant modulation of growth and defense. Understanding the molecular processes governing plant prioritization and diversion of resources towards growth or defense may enable genetic tailoring of plants to harness this natural plasticity for optimization of both growth and defense under variable environmental conditions.
Key words: plant immunity, plant hormone, salicylic acid, jasmonate, PAMP, plant growth.
Abstract
Growth–defense tradeoffs are thought to occur in plants due to resource restrictions, which demand prioritization towards either growth or defense, depending on external and internal factors. These tradeoffs have profound implications in agriculture and natural ecosystems, as both processes are vital for plant survival, reproduction, and, ultimately, plant fitness. While many of the molecular mechanisms underlying growth and defense tradeoffs remain to be elucidated, hormone crosstalk has emerged as a major player in regulating tradeoffs needed to achieve a balance. In this review, we cover recent advances in understanding growth–defense tradeoffs in plants as well as what is known regarding the underlying molecular mechanisms. Specifically, we address evidence supporting the growth–defense tradeoff concept, as well as known interactions between defense signaling and growth signaling. Understanding the molecular basis of these tradeoffs in plants should provide a foundation for the development of breeding strategies that optimize the growth–defense balance to maximize crop yield to meet rising global food and biofuel demands.
INTRODUCTION
While the deployment of defense mechanisms is imperative for plant survival, defense activation generally comes at the expense of plant growth (Figure 1). The ‘growth–defense tradeoff’ phenomenon was first observed in forestry studies of plant–insect interactions, and is based on the assumption that plants possess a limited pool of resources that can be invested either in growth or in defense (Coley et al., 1985; Simms and Rausher, 1987; Herms and Mattson, 1992). As plants must both grow and defend in order to survive and reproduce, growth–defense tradeoffs have important ecological, agricultural, and economic consequences. In nature, plants live in diverse and complex environments in which they constantly encounter a variety of pathogens and insect herbivores with a wide array of life styles and infection strategies. In adaptation to such natural conditions, plants have evolved sophisticated mechanisms to balance growth and defense (Herms and Mattson, 1992; Baldwin, 2001; Walling, 2009). However, in agricultural settings, crops have been bred for centuries to maximize growth-related traits resulting in a loss of genetic diversity that often compromises defense (Strange and Scott, 2005). Understanding the molecular mechanisms used by plants to balance growth and defense can enrich plant breeding and engineering strategies for selection of elite genetic traits that will maximize plant fitness.
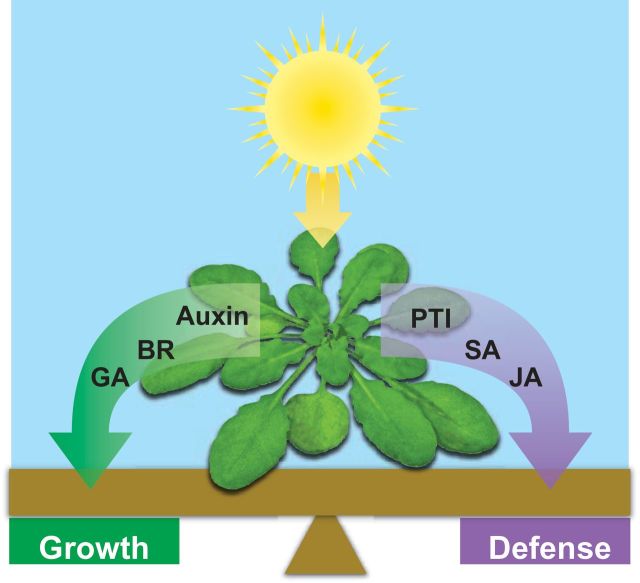
Figure 1.
A diagram depicting the concept of growth-defense tradeoffs.
Plants use photosynthesis to convert light energy into chemical energy in the form of carbohydrates. These resources are then allocated towards growth or defense, depending on the presence or absence of specific stresses. This process is facilitated by hormone crosstalk and is referred to as the growth–defense tradeoff. BR, brassinosteroid; GA, gibberellin; PTI, pathogen-associated-molecular-pattern-triggered immunity; SA, salicylic acid; JA, jasmonates.
In this review, we discuss the evidence supporting the concept of growth–defense tradeoffs in plants as well as the recent advances in deciphering the molecular mechanisms underlying their occurrence. As numerous studies have implicated hormone crosstalk as having a fundamental role in fine-tuning the growth–defense process, we provide brief descriptions of each defense and growth signaling pathway to introduce key players, and then discuss relevant hormone crosstalk. Due to space constraints, we focus our discussion on tradeoffs between defenses mediated by pathogen-associated-molecular-pattern (PAMP)-triggered immunity (PTI), salicylic acid (SA), and jasmonate (JA) versus growth mediated by auxin, brassinosteroids (BR), and gibberellins (GA), for which most progress has been made (Figure 1). Readers are referred to several recent reviews related to this topic, including discussions of the roles of ethylene (ET) and cytokinins (Bari and Jones, 2009; Robert-Seilaniantz et al., 2011a; Naseem and Dandekar, 2012; Bartoli et al., 2013; De Vleesschauwer et al., 2013; Denance et al., 2013; Yang et al., 2013). We conclude with a summary of concepts that may be drawn from current knowledge as well as several key areas where further research is needed.
DEFENSE SIGNALING
The ability to perceive and mount a rapid response to pathogen attack is critical for plant survival. Plants have evolved a sophisticated immune system that is initiated upon detection of highly conserved PAMPs by membrane-associated pattern recognition receptors (PRRs), which leads to activation of PTI (Boller and Felix, 2009; Monaghan and Zipfel, 2012). While PTI is believed to provide sufficient defense against non-pathogenic microbes, pathogens have developed the ability to secrete virulence effectors into the plant cell to suppress PTI and promote disease (Boller and He, 2009; Dou and Zhou, 2012; Xin and He, 2013). Plants have evolved resistance (R) genes to recognize these effectors and activate a much stronger immune response, effector-triggered immunity (ETI), which often results in a type of programmed cell death response known as the hypersensitive response (HR) in pathogen-infected tissue (Chisholm et al., 2006; Jones and Dangl, 2006; Bent and Mackey, 2007; Caplan et al., 2008). ETI may also trigger secondary immune responses in distal, uninfected tissues and lead to so-called systemic acquired resistance (SAR) (Grant and Lamb, 2006; Fu and Dong, 2013).
Plant hormones are small organic molecules that are required by plants in low concentrations and regulate growth, development, reproduction, and immune responses. Changes in environmental signals—both abiotic and biotic—induce changes in the quantity and composition of these signal molecules to facilitate appropriate plant responses (Kazan and Manners, 2009; Santner and Estelle, 2009; Robert-Seilaniantz et al., 2011a; Denance et al., 2013). Plant defense hormones such as SA, JA, and ET play important roles in the precise regulation of plant immune responses both locally and systemically to coordinate plant defense against different types of pathogens and in different parts of the plant (Erb et al., 2012; Pieterse et al., 2012; Wasternack, 2013). SA signaling is primarily induced by and involved in defense against biotrophic pathogens, whereas JA signaling is primarily induced by and involved in defense against insect herbivores and, in conjunction with ET, against necrotrophic pathogens (Thomma et al., 1998; Glazebrook, 2005). SA and JA signaling pathways are generally antagonistic to each other (Pieterse et al., 2012). For example, elevated SA signaling in response to biotrophic pathogens is often correlated with reduced JA signaling and decreased resistance to necrotrophic pathogens (Spoel et al., 2007). The following sections provide brief summaries of PTI, SA, and JA signaling pathways relevant to this review.
PAMP-Triggered Immunity
As mentioned above, PTI is triggered following detection of PAMPs by PRRs (Monaghan and Zipfel, 2012). The best characterized PRRs are leucine-rich repeat receptor kinases (LRR–RKs) consisting of an extracellular LRR domain, which can vary in the number of repeats and is directly involved in ligand perception, a transmembrane domain, and an intracellular kinase domain (Nicaise et al., 2009). FLAGELLIN SENSING 2 (FLS2) and ELONGATION FACTOR-TU RECEPTOR (EFR) are LRR–RKs that recognize bacterial flagellin and bacterial EF-Tu, respectively (Gomez-Gomez and Boller, 2000; Zipfel et al., 2004, 2006; Sun et al., 2013). Upon ligand perception, both FLS2 and EFR rapidly recruit a LRR–RK, BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1), resulting in their transphosphorylation (Chinchilla et al., 2007; Heese et al., 2007; Schulze et al., 2010; Roux et al., 2011). Treatment with flg22, a bioactive 22-amino acid peptide derived from bacterial flagellin, activates the FLS2/BAK1 co-receptor complex and triggers a phosphorylation cascade, including the phosphorylation and displacement of BOTRYTIS-INDUCED KINASE 1 (BIK1) from the FLS2 complex to promote the immune response (Lu et al., 2010; Lin et al., 2014). Initial PTI responses occur within minutes to hours following PAMP perception, and include elevation of reactive oxygen species (ROS), calcium influx, activation of calcium/calmodulin-dependent kinase and mitogen-activated protein kinase signaling cascades, and transcriptional reprogramming (Boller and Felix, 2009; Dodds and Rathjen, 2010). PTI-associated transcriptional reprogramming is facilitated in part by the WRKY family of transcription factors, members of which are involved in both positive and negative regulation of PTI (Thilmony et al., 2006; Pandey and Somssich, 2009; Rushton et al., 2010). Later responses attributed to PTI activation include deposition of callose at the cell wall near the site of pathogen infection and seedling growth inhibition (Gomez-Gomez et al., 1999; Boller and Felix, 2009).
Salicylic Acid
SA is a phenolic hormone shown to affect many plant processes including growth, development, senescence, and stress responses (Vlot et al., 2009; Rivas-San Vicente and Plasencia, 2011). It is primarily recognized for its role in local defense induced against biotrophic and hemi-biotrophic pathogens and in the establishment of SAR (Fu and Dong, 2013). After years of searching, two recent studies have proposed NONEXPRESSOR OF PATHOGENESIS RELATED PROTEINS 1 (NPR1) and its paralogs, NPR3 and NPR4, to act as SA receptors (Attaran and He, 2012; Fu et al., 2012; Wu et al., 2012). Multiple genetic screens led to the identification of NPR1, which is a key regulator of SA signaling (Cao et al., 1994; Delaney et al., 1995; Cao et al., 1997; Shah et al., 1997). Under non-induced conditions, NPR1 proteins oligomerize in the cytoplasm (Mou et al., 2003). SA accumulation in response to pathogen detection triggers the release of NPR1 monomers, which then translocate to the nucleus and activate defense gene expression (Kinkema et al., 2000; Mou et al., 2003; Tada et al., 2008). NPR1 regulates gene expression through physical interaction with TGA transcription factors, which bind to promoters of PATHOGENESIS RELATED (PR) genes to activate expression in the presence of SA and repress expression in the absence of SA (Zhang et al., 2003b; Fu and Dong, 2013). PR genes encode small proteins, some of which have been shown to possess antimicrobial or antifungal properties in vitro (van Loon et al., 2006). Of the many PR genes identified, PR1, PR2, and PR5 have been shown to be induced by SA and have long been used as markers of SA signaling (Fu and Dong, 2013). Other genes identified as direct targets of NPR1 include WRKY transcription factors and components required for the synthesis and secretion of PR proteins (Wang et al., 2006). WRKYs are involved in both NPR1-dependent and NPR1-independent SA signaling and, as in the case of PTI, include both positive and negative regulators of SA-mediated defense (Yu et al., 2001; Wang et al., 2006; Rushton et al., 2010; Fu and Dong, 2013).
Jasmonate
JAs are a group of lipid-derived hormones that regulate plant defense against necrotrophic pathogens and insect herbivores (Pieterse et al., 2012) and also affect several other physiological processes including abiotic stress responses, reproductive development, and primary and secondary metabolism (Wasternack, 2007; Browse, 2009). Jasmonoyl isoleucine (JA-Ile) is perceived by a co-receptor complex consisting of the F-box protein CORONATINE INESENSTIVE 1 (COI1) and the JASMONATE ZIM DOMAIN (JAZ) family of transcription repressors (Sheard et al., 2010). COI1 is required for almost all known JA-dependent responses (Feys et al., 1994; Xie et al., 1998; Browse, 2009). The JAZ-family proteins repress JA signaling by directly binding to the MYC family of transcription factors required for the expression of JA-responsive genes (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). Under normal growth conditions where JA-Ile levels are low, JAZ proteins recruit co-repressors, TOPLESS (TPL) or TPL-related proteins, either directly through their ETHYLENE RESPONSE FACTOR-ASSOCIATED AMPHIFILIC REPRESSION (EAR) motifs or indirectly through NOVEL INTERACTOR of JAZ (NINJA) protein to suppress MYC activities (Pauwels et al., 2010; Shyu et al., 2012). It was recently shown that physical association of JAZ proteins with MYC2 is required for the nuclear localization of JAZ repressors (Withers et al., 2012); however, the mechanism for JAZ repression of MYC activity is not clearly understood. Upon wounding or pathogen attack, JA-Ile is rapidly synthesized in both local and distal tissues (Staswick and Tiryaki, 2004; Fonseca et al., 2009). An increasing concentration of JA-Ile promotes physical interaction between COI1 and JAZ proteins, which leads to ubiquitination and subsequent degradation of JAZs through the 26S proteasome, thereby relieving the repression on MYC transcription factors and initiating the expression of JA-responsive genes (Chini et al., 2007; Thines et al., 2007; Katsir et al., 2008).
GROWTH-PROMOTING HORMONE SIGNALING
Plant growth and development are coordinately regulated by a complement of hormones in order to optimize growth and reproduction (Depuydt and Hardtke, 2011). Growth hormones implicated in growth–defense tradeoffs are auxin, BRs, GAs, and cytokinins. As excellent reviews have been written on each of these hormones (Santner and Estelle, 2009; Sun, 2011; Zhao and Li, 2012), we will briefly describe what is known regarding the main signaling components for the three growth hormones relevant to this review.
Auxin
Auxins regulate many fundamental aspects of plant growth and development including stem and petiole elongation and root architecture in response to light, temperature, and gravity (Santner and Estelle, 2009; Vanneste and Friml, 2009; Kieffer et al., 2010; Kazan, 2013). Biosynthesis of indole-3-acetic acid (IAA), one of the primary auxins studied, occurs primarily in young leaves via multiple biosynthetic pathways and IAA is transported throughout the plant (Woodward and Bartel, 2005; Normanly, 2010). Once synthesized, accumulation of free IAA is regulated by GH3 proteins, which conjugate IAA with amino acids to yield metabolites for storage (IAA-alanine and IAA-leucine) or oxidation and degradation (IAA-aspartate and IAA-glutamic acid) (Ljung et al., 2002; Staswick et al., 2005; Ludwig-Muller, 2011). When auxin levels are low, auxin response genes are actively repressed by heterodimerization of the AUX/IAA family of transcriptional repressor proteins with the AUXIN RESPONSIVE FACTORS (ARF) family of transcription factors (Ulmasov et al., 1999; Tiwari et al., 2001; Liscum and Reed, 2002; Tiwari et al., 2004). The F-box proteins, TRANSPORT INHIBITOR RESISTANT 1 (TIR1) and AUXIN SIGNALING F-BOX (AFB), are substrate-recognition components of an SKP–Cullin–F-box (SCF) E3 ubiquitin ligase complex, SCFTIR1/AFB (Gray et al., 1999, 2001). When auxin concentration reaches a threshold in the cell, auxin directly facilitates SCFTIR1/AFB binding to AUX/IAA proteins, resulting in the ubiquitination and degradation of AUX/IAA repressors via the 26S proteasome thereby derepressing ARF-dependent transcription of auxin-regulated genes (Kepinski and Leyser, 2004; Dharmasiri et al., 2005; Kepinski and Leyser, 2005). Auxin-regulated genes include the AUX/IAA and GH3 gene families (Hagen et al., 1984; Abel et al., 1994), expression of which forms part of a feedback mechanism to reset auxin signaling homeostasis.
Brassinosteroids
BRs are polyhydroxylated steroid phytohormones that influence diverse developmental processes from seed germination to plant senescence (Gruszka, 2013; Hao et al., 2013; Fariduddin et al., 2014). Plants that are insensitive to or deficient in BR signaling have severely stunted growth and are male infertile, whereas exogenous application of BR has a positive impact on the quality and quantity of crop yield (Khripach et al., 2000; Gruszka, 2013; Hao et al., 2013; Fariduddin et al., 2014). In the absence of BR, the glycogen-synthase-kinase-3-like kinase BRASSINOSTEROID INSENSITIVE 2 (BIN2) phosphorylates two nuclear-localized transcription factors, BRI1-EMS-SUPPRESSOR 1 (BES1) and BRASSINAZOLE-RESISTANT 1 (BZR1), to block activation of BR-responsive genes (He et al., 2002; Wang et al., 2002; Yin et al., 2002; He et al., 2005; Yin et al., 2005; Vert and Chory, 2006). The presence of BR stabilizes the BRASSINOSTEROID INSENSITIVE 1 (BRI1)/BAK1 co-receptor complex, causing activation of their respective kinase domains and subsequent transphosphorylation (Li and Chory, 1997; Li et al., 2002; Nam and Li, 2002; Wang and Chory, 2006; Hothorn et al., 2011; She et al., 2011). The resulting cascade of phosphorylation events leads to the phosphorylation and inactivation of BIN2 kinase and the dephosphorylation and activation of BES1 and BZR1 to promote the expression of BR-regulated genes (Mora-Garcia et al., 2004; Tang et al., 2011).
Gibberellins
GAs are tetracyclic diterpene acids that control seed development and germination, vegetative growth, and flower initiation and development (Sun, 2011). GA induces gene expression by relieving the repression of a family of transcriptional repressors known as DELLA proteins (Peng et al., 1997; Silverstone et al., 2001). In the absence of bioactive GAs, DELLAs bind to and inactivate PHYTOCHROME INTERACTING FACTORS (PIFs), a group of bHLH-family transcription factors (Sun, 2011). The presence of a growth signal stimulates the biosynthesis of GA, which is perceived by GA INSENSTIVE DWARF 1 (GID1) through direct binding. This leads to a conformational change of GID1, facilitating its binding to DELLA proteins (Murase et al., 2008). The formation of the GID1–DELLA complex enhances the interaction between DELLA and the F-box protein SLEEPY 1 in the SCFSLY1 E3 ubiquitin ligase complex, which results in DELLA ubiquitination and degradation that relieves PIF repression and promotes GA-mediated gene expression and growth (McGinnis et al., 2003; Dill et al., 2004; Achard et al., 2007; Sun et al., 2011).
IN DEFENSE OF THE GROWTH TRADEOFF
Implementation of defense imposes a substantial demand for resources, which has been suggested to reduce growth. This negative impact on growth could result from diminished photosynthesis, which would decrease the overall pool of energy reserves, and/or from a diversion of resources away from growth and towards defense. As deficiencies in defense capabilities can result in pathogen-induced decimation of a plant population, a balance must be achieved between growth and defense to optimize plant fitness.
Finding Balance to Optimize Fitness
Fitness costs associated with defense have been clearly demonstrated (Heil and Baldwin, 2002; Tian et al., 2003; Heidel et al., 2004; Zavala et al., 2004; Kempel et al., 2011; Meldau et al., 2012). For example, silencing components in JA-mediated defense signaling was shown to alleviate fitness costs observed in wild-type plants (Meldau et al., 2012). In the case of constitutive defense responses, reduced fitness may be due in part to unnecessary diversion of energy reserves away from growth in the absence of stress. Benzothiadiazole (BTH) is a synthetic analog of SA used commercially to enhance disease resistance by inducing SAR in crops (Gorlach et al., 1996; Lawton et al., 1996). Application of BTH to wheat was observed to negatively impact fitness in the absence of pathogens (Heil et al., 2000) and to increase fitness in the presence of the biotrophic fungus powdery mildew (Gorlach et al., 1996). Another fitness cost attributed to constitutive defense is the inability of the plant to respond appropriately to environmental conditions that limit energy production. In support of this, enhanced susceptibility to the hemi-biotrophic pathogen Pseudomonas syringae (P. syringae) and the necrotrophic pathogen Botrytis cinerea observed in shade-grown plants has been attributed to the need to prioritize growth under these light-restrictive conditions (Cerrudo et al., 2012; de Wit et al., 2013; Ballaré, 2014).
While it is easy to understand the costs associated with constitutive defense, fitness is also compromised in the absence of defense. Loss of NPR1-dependent, SA-mediated defense was shown to reduce the fitness of field-grown plants (Heidel et al., 2004), whereas overexpression of NPR1 was shown to enhance resistance to biotrophic and hemi-biotrophic pathogens without adversely affecting growth or fitness (Cao et al., 1998; Heidel et al., 2004). This is most likely due to the fact that SA signaling is not constitutively active but rather primed for quicker response to pathogen detection in these plants (Cao et al., 1998). Together, these studies indicate that approaches used to achieve an enhanced primed state can ameliorate the fitness costs associated with constitutive defense, while optimizing the fitness benefits of rapid defense induction upon pathogen detection. They also emphasize the point that increased growth is not equivalent to enhanced fitness. Rather, plant fitness is optimized when growth and defense are appropriately prioritized in response to both environmental and developmental cues (Valverde et al., 2003; Heidel et al., 2004).
Impacts on Photosynthesis
Pathogen/herbivore activity that results in damage to photosynthetic machinery, loss of photosynthetic tissue, and/or disruption of the vasculature affecting water and sugar transport has been shown to negatively impact photosynthesis (Aldea et al., 2005; Zou et al., 2005; Gutsche et al., 2009; Nabity et al., 2009; Kerchev et al., 2012). In addition, pathogen/herbivore attack has been shown to suppress components of photosynthesis at the levels of gene expression and of protein abundance (Zou et al., 2005; Jung et al., 2007; Denoux et al., 2008; Ishiga et al., 2009; Bilgin et al., 2010; Sugano et al., 2010; Chen et al., 2011b; Gohre et al., 2012; Guo et al., 2012; Borges et al., 2013). The negative impact of defense on photosynthesis has been best demonstrated in response to JA treatment, which results in a reduction of components essential for light harvesting and carbon fixation (Wierstra and Kloppstech, 2000; Chen et al., 2011b; Shan et al., 2011; Guo et al., 2012) as well as a substantial decrease in photosynthetic activities and chlorophyll contents in Arabidopsis (Jung, 2004). supporting the need for energy acquisition to enable the defense response.
However, down-regulation of photosynthetic genes following defense activation does not always correlate with changes in protein profiles (Gohre et al., 2012), leading to the hypothesis that the stability of most photosynthetic proteins allows for a temporary halt at the transcriptional level without a significant impact on photosynthesis itself. This appears to be supported by some studies using chlorophyll fluorescence to measure photosynthetic rates following infection with biotrophic, hemi-biotrophic, or necrotrophic pathogens. A similar spatial pattern has been reported for each pathogen type where inhibition of photosynthesis is confined to infected cells and is offset by elevated photosynthesis in the surrounding cells whereas no impact is observed in distal, uninfected tissues (Chou et al., 2000; Berger et al., 2004; Bonfig et al., 2006; Berger et al., 2007). Also, proteomic and biochemical analyses of resistant and susceptible plants have shown that the ability to maintain photosynthesis during infection is a vital element of defense (Gutsche et al., 2009; Zhang et al., 2013). Furthermore, loss of RuBPCase activase (RCA), which has a critical role in carbon fixation, has been shown to diminish JA-mediated defenses (Mitra and Baldwin, 2014). Together, these studies indicate that the ability to appropriately maintain photosynthesis is crucial for defense. Whether or not the observed effects on photosynthesis are a programmed part of the defense response or merely a by-product remains to be determined.
Resource Diversion
In support of the growth–defense tradeoff theory, diversion of plant resources has been shown to occur at all levels, including machinery involved in transcription, translation, and protein secretion from cells as well as prioritization of carbon and nitrogen towards production of defense compounds. Transcriptomic and proteomic studies have demonstrated transcriptional reprogramming and altered protein profiles upon pathogen/herbivore detection to promote defense at the expense of growth (Wang et al., 2006; Jung et al., 2007; Denoux et al., 2008; Bilgin et al., 2010; Sugano et al., 2010; Chen et al., 2011b; Gohre et al., 2012; Guo et al., 2012; Borges et al., 2013). Production and secretion of proteins with specific defensive properties, such as PR proteins, place a significant demand on the protein folding and secretory systems, which have also been shown to be required for defense (Wang et al., 2005; Kwon et al., 2008; Pajerowska-Mukhtar et al., 2012). Allocation of resources involved in protein folding and secretion towards defense has been proposed to be regulated in part by TL1 BINDING TRANSCRIPTION FACTOR 1 (TBF1) (Pajerowska-Mukhtar et al., 2012). Many TBF1-regulated genes encode ER resident proteins involved in protein folding and secretion, and loss of TBF1 was shown to compromise the unfolded protein response as well as to impair PTI and SAR (Pajerowska-Mukhtar et al., 2012). Furthermore, tbf1 knockout mutants were shown to exhibit partial suppression of growth inhibition associated with defense activation, and transcriptional profiling of these mutants showed a general promotion of growth-related genes and repression of defense-related genes (Pajerowska-Mukhtar et al., 2012).
Studies using radiolabeled carbon or nitrogen have shown that pathogen/herbivore detection alters the normal metabolic flux to enable the incorporation of these resources into defense-related compounds (Engelsdorf et al., 2013; Ullmann-Zeunert et al., 2013). Reallocation of labeled nitrogen from ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) into nicotine and phenolamide compounds following simulated herbivory was shown to rely on a functional JA pathway (Ullmann-Zeunert et al., 2013). Carbon availability has been shown to be important for SA-regulated defense, as starch-free mutants, which have a general reduction in carbohydrates compared with wild-type plants, showed a delayed production of SA-regulated defense compounds resulting in increased susceptibility to the hemi-biotrophic pathogen Colletotrichum higginsianum (Engelsdorf et al., 2013). Carbohydrates are produced in photosynthetic ‘source’ tissues and transported in the form of sucrose to non-photosynthetic ‘sink’ tissues (Roitsch and Gonzalez, 2004). Upon pathogen infection in the leaves, this process is disrupted by up-regulation of cell wall invertases, which cleave sucrose into glucose and fructose thereby preventing sucrose export from infected cells (Sturm, 1999; Roitsch and Gonzalez, 2004; Swarbrick et al., 2006; Kocal et al., 2008). Transgenic suppression of cell wall invertase activity results in elevated sucrose-to-hexose ratios accompanied by reduced and delayed callose deposition and inhibition of PR gene expression following pathogen infection (Essmann et al., 2008; Kocal et al., 2008), whereas ectopic expression of a yeast cell wall invertase has been shown to activate defense responses in tobacco (Herbers et al., 1996). Comparison of resistant and susceptible barley interactions with the biotrophic fungal pathogen Blumeria graminis revealed a more robust activation of cell wall invertase in the resistant interaction resulting in accumulation of hexose sugars localized to regions of actively defending cells (Swarbrick et al., 2006). In addition, a recent study has shown cell wall invertase activity to be a possible virulence target of the biotrophic pathogen Xanthomonas campestris pv. vesicatoria, to promote disease in pepper (Sonnewald et al., 2012), providing further evidence supporting a role for cell wall invertases in redirecting carbon resources to enable plant defense.
Together, these studies begin to reveal some of the regulatory mechanisms underlying resource reallocation to mediate the growth–defense tradeoff in plants. Along with the co-opting of energy reserves and cellular machinery to produce compounds necessary for defense, transcriptional reprogramming induced by defense activation is often accompanied by repression of growth hormone signaling as a fundamental aspect of growth–defense tradeoffs. In the following sections, we discuss current knowledge regarding crosstalk between defense signaling and growth hormones.
PAMP-TRIGGERED IMMUNITY-MEDIATED DEFENSE VERSUS GROWTH
One of the most noticeable physiological consequences of prolonged or constitutively active PTI is growth inhibition, which is observed upon treatment of a plant with a PAMP (Gomez-Gomez et al., 1999; Zipfel et al., 2006). As discussed in the previous section, there is mounting evidence to support a mechanism whereby resources normally allocated towards growth are diverted to support defense and, as discussed below, hormonal crosstalk appears to play a major role in regulating the tradeoff between growth and PTI-mediated defense.
PAMP-Triggered Immunity Crosstalk with Auxin
Auxin has long been implicated in suppressing plant defense due to the fact that many pathogens, including P. syringae and Agrobacterium tumefaciens, can directly synthesize auxin or manipulate auxin synthesis and signaling in plants to promote disease (Yamada, 1993; Glickmann et al., 1998; O’Donnell et al., 2003; Chen et al., 2007; Kidd et al., 2011). Microorganisms primarily synthesize IAA from tryptophan and, in some cases, the genes encoding the enzymes required for this process are located on a pathogen virulence plasmid (Yamada, 1993). Analysis of plant transcriptional reprogramming following some pathogen infections has shown a general derepression of the auxin pathway including promotion of auxin biosynthetic genes and repression of AUX/IAA genes resulting in enhanced plant susceptibility (O’Donnell et al., 2003; Thilmony et al., 2006). Furthermore, virulence of the bacterial hemi-biotrophic pathogen P. syringae pv. tomato DC3000 (Pto DC3000) can be enhanced by treatment with synthetic auxins prior to pathogen inoculation (Navarro et al., 2006; Chen et al., 2007).
To combat the effects of pathogen produced or induced auxin to promote disease, plants actively suppress auxin signaling during defense (Navarro et al., 2004). Following flg22-treatment, wild-type Arabidopsis plants show a reduction in both transcript and protein levels of the auxin F-box receptors, resulting in stabilization of AUX/IAA proteins and repression of auxin-responsive genes (Navarro et al., 2006). This suppression is partially due to the activity of the microRNA miR393 (Figure 2), which is induced by flg22 and directly targets and cleaves TIR1, AFB2, and AFB3 transcripts (Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Navarro et al., 2006). However, additional mechanisms such as transcriptional repression must also contribute to PTI inhibition of auxin signaling, as partial reduction in transcript levels is still observed in the DICER LIKE 1 (DCL1) mutant, dcl1-9, which is required for miR393 function (Navarro et al., 2006). Also, the AFB1 transcript is partially resistant to miR393 activity, and shows reduced transcript levels in both wild-type and dcl1-9 mutant plants (Navarro et al., 2006).
Figure 2.
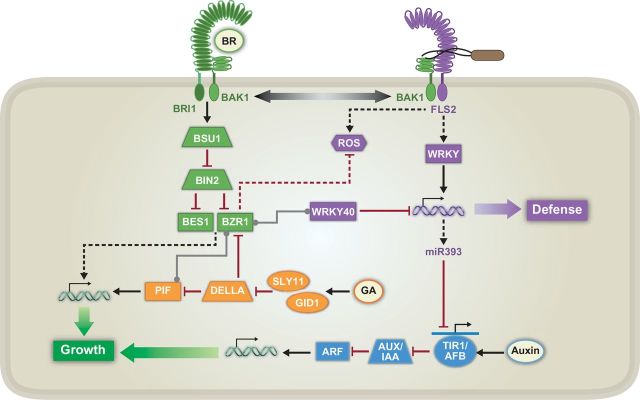
Known Signaling Contributing to Growth–Defense Tradeoffs between PTI-Mediated Defense and Auxin-, Brassinosteroid (BR)-, and Gibberellin (GA)-Mediated Growth.
Black arrows and red, blunted lines represent positive and negative regulation, respectively. Double helices with arrows represent global transcriptional reprogramming, and gray lines with dots at both ends indicate protein–protein interactions. Solid lines indicate a known connection between two components, whereas dashed lines indicate unknown connections or missing steps between two components. The solid blue line with an arrow represents expression of TIR1/AFB genes, the transcripts of which are targeted by miR393. FLS2, FLAGELLIN SENSING 2; ROS, reactive oxygen species; WRKY, WRKY DNA-BINDING PROTEIN; miR393, microRNA 393; TIR1, TRANSPORT INHIBITOR RESPONSE 1; AFB, AUXIN SIGNALING F-BOX; AUX/IAA, AUXIN-INDUCIBLE/INDOLE-3-ACETIC ACID INDUCIBLE; ARF, AUXIN RESPONSE FACTOR; BAK1, BRI1-ASSOCIATED RECEPTOR KINASE 1; BRI1, BRASSINOSTEROID INSENSITIVE 1; BSU1, BRI1 SUPPRESSOR 1; BIN2, BRASSINOSTEROID INSENSITIVE 2; BES1, BRI1-EMS-SUPPRESSOR 1; BZR1, BRASSINAZOLE-RESISTANT 1; SLY1, SLEEPY 1; GID1, GA INSENSITIVE DWARF 1A; DELLA, repressor protein; PIF, PHYTOCHROME INTERACTING FACTOR.
Suppression of auxin signaling has been shown to be biologically relevant to PTI, as overexpression of miR393 enhances resistance to virulent pathogens and overexpression of AFB1 increases susceptibility relative to that observed in wild-type plants, as measured by bacterial growth (Navarro et al., 2006). One study has shown that pathogen manipulation of auxin metabolism to generate higher levels of IAA-aspartate (IAA-Asp) promotes disease by positively regulating the expression of bacterial virulence genes rather than by directly suppressing PTI (González-Lamothe et al., 2012). This was shown to require the GH3.2 enzyme, as gh3.2 knockout plants exhibited reduced susceptibility to Pto DC3000 (González-Lamothe et al., 2012). However, Mutka et al., (2013) were unable to reproduce these results, making the role of GH3.2 in this process unclear. If GH3.2 is involved, it cannot fully account for auxin-induced susceptibility because gh3.2 knockout plants crossed with plants overexpressing the auxin biosynthetic gene, YUCCA 1, retained enhanced susceptibility (Mutka et al., 2013). Therefore, while there is much evidence to implicate auxin in promoting plant disease, the exact mechanism underlying this phenomenon remains unclear.
PAMP-Triggered Immunity Crosstalk with Brassinosteroids and Gibberellins
Unlike the mutually antagonistic interactions observed between PTI and auxin-mediated growth, negative crosstalk between PTI and BR-mediated growth is unidirectional (Albrecht et al., 2012; Belkhadir et al., 2012). Elevation of BR signaling in Arabidopsis using either transgenic modifications (Jaillais et al., 2011; Belkhadir et al., 2012) or exogenous application of BR (Albrecht et al., 2012) results in inhibition of flg22-mediated protection against Pto DC3000. Conversely, treatment with brassinazole, which inhibits BR biosynthesis, elevates ROS production in response to PAMP treatment (Lozano-Durán et al., 2013), indicating that endogenous levels of BR are sufficient to suppress PTI. Due to the association of BAK1 with both FLS2 and BRI1 receptors, it was hypothesized that FLS2 and BRI1 competition for BAK1 might facilitate BR-mediated suppression of PTI-mediated defense (Figure 2). However, while overexpression of BRI1 was shown to inhibit PTI responses in a BAK1-dependent manner (Belkhadir et al., 2012), neither exogenous BR nor expression of a hyperactive form of BRI1, BRI1sud1, were shown to affect FLS2–BAK1 complex formation, transphosphorylation, or phosphorylation of downstream targets (Albrecht et al., 2012; Lozano-Durán et al., 2013).
A recent study by Lozano-Duran et al. (2013) has shown that constitutively active BZR1, but not BES1, is sufficient to block PAMP-triggered ROS burst, gene expression, and seedling growth inhibition (Figure 2). BZR1 inhibition of PTI appears to be mediated through its downstream targets, which include transcription factors known to promote BR responses and/or block defense (Lozano-Durán et al., 2013; Malinovsky et al., 2014). For example, a group of WRKY transcription factors known to negatively regulate PTI were identified as BR-induced BZR1 targets (Lozano-Durán et al., 2013). Of these, WRKY40 was shown to have a role in suppression of PAMP-induced ROS production and seedling growth inhibition. It is possible that BZR1 and WRKY40 act together to suppress PTI, as co-immunoprecipitation experiments indicated that these two proteins physically interact (Figure 2), and analysis of publicly available gene expression data revealed that all WRKY40-regulated genes are also targets of BZR1 (Lozano-Durán et al., 2013). Another example of a BZR1 target involved in PTI suppression is HBI1, which encodes a bHLH transcription factor shown to promote BR-regulated cell elongation by inducing the expression of expansin genes (Bai et al., 2012a). In addition to enhanced growth phenotypes, overexpression of HBI1 was shown to suppress PAMP-induced ROS and seedling growth inhibition downstream of FLS2–BAK1 complex formation (Malinovsky et al., 2014). While the mechanism for HBI1-mediated suppression of PAMP-responses is not known, identification of specific defense gene targets of BZR1-regulated transcription factors, including the WRKYs identified by Lozano-Duran et al. (2013), begins to shed light on the molecular mechanism behind BR suppression of PTI-mediated defense.
GA suppression of PAMP-induced seedling growth inhibition most likely occurs through promotion of BR signaling. BR- and GA-mediated signaling pathways work additively or synergistically to promote growth in response to environmental and developmental cues (Jaillais and Vert, 2012; Lilley et al., 2013). This cooperative relationship is facilitated in part by the formation of a BZR1/PIF4 heterodimer (Figure 2), which binds to the promoters of some 2,000 shared target genes to promote growth (Oh et al., 2012). DELLA proteins have been shown to inhibit both BZR1 and PIF4 proteins and may also target the BZR1/PIF4 heterodimer (De Lucas et al., 2008; Bai et al., 2012b; Gallego-Bartolome et al., 2012). While exogenous application of GA did not affect PAMP-induced seedling growth inhibition, chemical inhibition of GA synthesis completely blocked the effect of BR on seedling growth inhibition, and GA treatment in combination with BR resulted in an additive effect on PAMP-induced seedling growth inhibition (Lozano-Durán et al., 2013). This is most likely due to the effect of GA on DELLA stabilization. In the absence of GA, increased DELLA stabilization would result in BZR1 inhibition and loss of BR-mediated seedling growth inhibition (Figure 2). In support of this, flg22-induced stabilization of DELLA proteins has been shown to be a mechanism for PTI inhibition of GA-mediated growth (Navarro et al., 2008).
SALICYLIC ACID-MEDIATED DEFENSE VERSUS GROWTH
Suppression of growth by SA is best illustrated by constitutive defense mutants, which typically have a dwarf plant phenotype due in part to elevated SA accumulation or signaling (Clarke et al., 2000; Zhang et al., 2003a); however, as these mutants may be perturbed in cellular processes other than SA defense, it is difficult to ascertain how SA itself is directly contributing to growth suppression in these plants. To demonstrate the effect of SA on plant growth, experiments employing chemical inducers or genetic manipulation to alter SA accumulation or perception have been used. Cold temperature-induced growth reduction in Arabidopsis has been shown to be due to endogenous elevation of SA as it was lost in plants compromised in SA accumulation (Scott et al., 2004). Also, repeated application of BTH reduced plant biomass in a reproducible and dose-dependent manner that was correlated with induction of SA-mediated defense responses (Canet et al., 2010a). Mutants isolated in a screen based on resistance to BTH-induced growth inhibition were compromised in SA-mediated disease resistance and were primarily identified as non-functional alleles of NPR1 (Canet et al., 2010b). The mechanisms for SA-induced suppression of growth are most likely mediated by crosstalk with growth hormone signaling pathways, as discussed in the following sections.
Salicylic Acid Crosstalk with Auxin
One of the primary ways SA has been shown to inhibit growth is by suppression of auxin signaling (Figure 3). A microarray study revealed that a number of auxin-responsive genes were affected by BTH treatment, namely 21 genes encoding proteins involved in auxin reception, import and export and signaling were down-regulated and two genes encoding GH3 enzymes were up-regulated (Wang et al., 2006, 2007). As GH3 enzymes are responsible for regulating auxin homeostasis by conjugating IAA with different amino acids (Staswick et al., 2005), the transcriptional profile indicates a general BTH-dependent repression of auxin homeostasis and signaling. A follow-up study confirmed this by investigating the effect of SA on auxin levels, uptake, sensitivity, and signaling (Wang et al., 2007). It was shown that SA does not affect auxin synthesis, but instead represses the expression of the TIR1/ABF F-box genes (Figure 3), resulting in stabilization of AUX/IAA repressor proteins to decrease auxin signaling (Wang et al., 2007).
Figure 3.
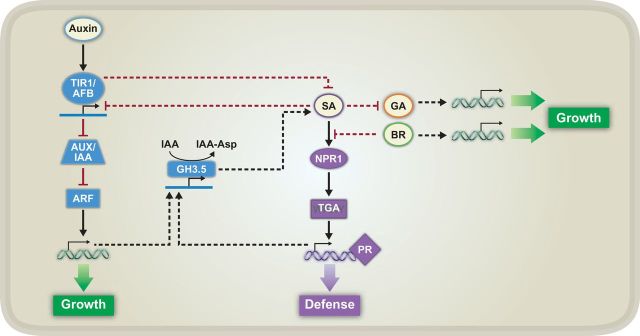
Known Signaling Contributing to Growth–Defense Tradeoffs between Salicylic Acid (SA)-Mediated Defense and Auxin-, Brassinosteroid (BR)-, and Gibberellin (GA)-Mediated Growth.
As in Figure 2, black arrows and red, blunted lines represent positive and negative regulation, respectively. Double helices with arrows represent global transcriptional reprogramming, and solid lines associated with arrows represent expression of TIR1/AFB and GH3.5 genes. Solid lines indicate a known connection between two components, whereas dashed lines indicate unknown connections or missing steps in between two components. NPR1, NONEXPRESSOR OF PR GENES 1; TGA, TGACG SEQUENCE-SPECIFIC BINDING PROTEIN; PR, PATHOGENESIS RELATED; IAA, INDOLE-3-ACETIC ACID; Asp, aspartate; TIR1, TRANSPORT INHIBITOR RESPONSE 1; AFB, AUXIN SIGNALING F-BOX; AUX/IAA, AUXIN-INDUCIBLE/IAA INDUCIBLE; ARF, AUXIN RESPONSE FACTOR.
One of the two GH3 genes identified in the microarray study encodes GH3.5 (Wang et al., 2006, 2007), which conjugates IAA with Asp (Staswick et al., 2005). The gh3.5 knockout mutants were shown to be compromised in SAR while overexpression lines exhibited a dwarf phenotype, accumulated higher levels of SA, had elevated expression of PR1, and increased resistance to Pto DC3000 (Park et al., 2007; Zhang et al., 2007, 2008). IAA-Asp is an inactive form of auxin that is targeted for metabolism (Ostin et al., 1998; Ljung et al., 2002); therefore, it would seem logical to infer that GH3.5 directly facilitates the growth–defense tradeoff between SA and auxin by simultaneously elevating SA levels and reducing active IAA levels. However, the dwarf phenotype observed in several GH3.5 overexpression lines did not always correlate with a reduction in free IAA (Park et al., 2007; Zhang et al., 2007). As GH3.5 expression is also induced by IAA to regulate its homeostasis (Hagen et al., 1984; Hagen and Guilfoyle, 2002), it is possible for GH3.5 to inhibit the auxin pathway directly by conjugating IAA and also indirectly by promoting SA biosynthesis and signaling, which then acts to block auxin responses (Figure 3).
SA-mediated defense has also been shown to be affected by auxin, as transgenic overexpression of the AFB1 gene, which enhances auxin signaling, led to a reduction in pathogen-induced SA biosynthesis relative to wild-type plants (Figure 3) (Robert-Seilaniantz et al., 2011b). However, transgenic overexpression of the YUCCA 1 gene showed that elevation of auxin levels alone can promote plant disease without affecting SA levels or signaling (Mutka et al., 2013). Auxin positively regulates expansins, which are involved in cell wall loosening, to promote growth (Cosgrove, 2005; Ding et al., 2008), and the ability of Xanthomonas oryzae pv. oryzae to induce expansins in rice was shown to be important in determining the outcome of the plant–pathogen interaction (Ding et al., 2008). Together, these studies indicate a dual function for auxin in direct interference with SA-mediated defense and in positive regulation of physiological changes that aid pathogen proliferation in the plant.
Salicylic Acid Crosstalk with Brassinosteroids and Gibberellins
There is much less known regarding the relationships between SA-mediated defense and BR- and GA-mediated growth. BR treatment was shown to block BTH-mediated resistance in rice, indicating suppression of SA signaling (De Vleesschauwer et al., 2012). Based on analysis of mutant plants affected in SA production or NPR1-mediated signaling, it was concluded that this antagonism occurs downstream of SA biosynthesis and upstream of NPR1 signaling (Figure 3), but the mechanism for this suppression is unknown (De Vleesschauwer et al., 2012). A recent study showed that down-regulation of the gene encoding the hydroxycinnamoyl CoA (HCT) enzyme resulted in stunted plant growth that was directly correlated with lignin reduction and endogenous SA elevation (Gallego-Giraldo et al., 2011a). These same plants were also shown to be impaired in both GA accumulation and perception (Gallego-Giraldo et al., 2011a, 2011b). Crosses between HCT RNAi plants and plants defective in SA biosynthesis, accumulation, or perception by NPR1 revealed that loss of SA production and accumulation, but not NPR1-dependent SA perception, was responsible for growth suppression in these plants (Gallego-Giraldo et al., 2011a). Loss of SA accumulation was also shown to restore gene induction and growth enhancement in response to exogenous GA, implicating SA in repression of GA signaling and growth (Gallego-Giraldo et al., 2011a). As mentioned previously, BZR1 is directly targeted and suppressed by the DELLA family of growth-suppressing proteins (Gallego-Bartolome et al., 2012). SA-mediated suppression of GA would most likely result in increased DELLA stability, which may lead to suppression of BR-mediated signaling. Further studies are needed to both establish a molecular mechanism for SA-inhibition of GA signaling and to determine whether this suppression of GA results in loss or reduction in BR signaling.
JASMONATE-MEDIATED DEFENSE VERSUS GROWTH
It has long been known that activation of JA signaling by applying JA into the growth medium results in growth inhibition (Staswick et al., 1992). Correlated with growth inhibition, JA suppresses mitosis, arrests the cell cycle in G1 prior to the S transition, and delays the switch from the mitotic cell cycle to the endoreduplication cycle (Zhang and Turner, 2008; Noir et al., 2013). Transcriptomic analysis further confirmed that JA activates several critical regulators of endoreduplication and affects the expression of key determinants of DNA replication (Noir et al., 2013). As in the case of PTI and SA-mediated defense, the effects of JA on growth appear to be mediated by crosstalk with growth hormone signaling.
Jasmonate Crosstalk with Auxin and Brassinosteroids
The auxin signaling pathway has been implicated in JA-induced growth inhibition in Arabidopsis (Figure 4) (Wasternack and Hause, 2013). JA not only suppresses the expression of the auxin efflux carrier PINFORMED 2 (PIN2), but also inhibits PIN2 endocytosis and membrane accumulation (Sun et al., 2011). Consequently, the normal auxin distribution in roots is disrupted after JA treatment (Sun et al., 2011). Moreover, MYC2 has been shown to negatively regulate the expression of PLETHORA (PLT1 and PLT2) transcription factors (Chen et al., 2011a), which are important regulators of auxin-mediated root stem cell development and auxin biosynthesis in roots (Figure 4) (Pinon et al., 2013). Taken together, it is postulated that JA changes the spatial and temporal distribution pattern of auxin in plants to suppress normal plant growth mediated by auxin. However, JA was also shown to increase auxin biosynthesis by inducing ANTHRANILATE SYNTHASE (ASA1 and ASB1) and YUCCA (YUC8 and YUC9) gene expression in certain plant tissues (Sun et al., 2009; Hentrich et al., 2013), and JA-induced auxin biosynthesis and lateral root formation were impaired in yuc knockout mutants (Hentrich et al., 2013). Conversely, auxin has been shown to induce expression of JAZ1, suggesting that auxin may suppress JA signaling through JAZ1 (Figure 4) (Grunewald et al., 2009). These latter studies illustrate the complexity of the interaction between JA and auxin signaling pathways.
Figure 4.
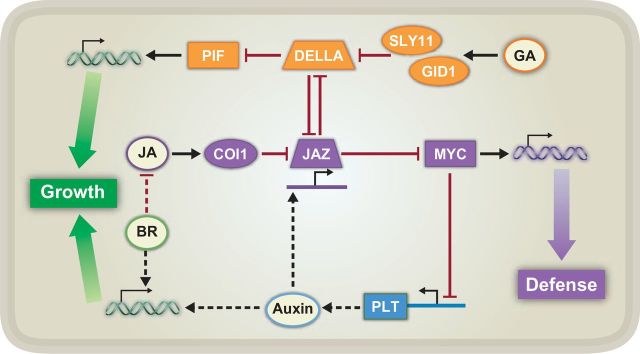
Known Signaling Contributing to Growth–Defense Tradeoffs between Jasmonate (JA)-Mediated Defense and Auxin-, Brassinosteroid (BR)-, and Gibberellin (GA)-Mediated Growth.
As in Figures 2 and 3, black arrows and red, blunted lines represent positive and negative regulation, respectively. Double helices with arrows represent global transcriptional reprogramming, and solid lines with arrows represent expression of JAZ and PLT genes. Solid lines indicate a known connection between two components, whereas dashed lines indicate unknown connections or missing steps in between two components. COI1, CORONATINE INSENSITIVE 1; JAZ, JASMONATE ZIM DOMAIN; MYC, transcription factor; SLY1, SLEEPY 1; GID1, GA INSENSITIVE DWARF 1A; DELLA, repressor protein; PIF, PHYTOCHROME INTERACTING FACTOR; PLT, PLETHORA.
BR signaling has also been implicated in antagonizing JA-induced growth suppression (Figure 4) (Wasternack, 2013). The first indication of a connection between JA-mediated defense and BR-mediated growth was the identification of a partially suppressing coi1 (psc1) mutant, which carries a mutation in a key enzyme involved in BR biosynthesis, DWARF 4 (DWF4) (Ren et al., 2009). In Arabidopsis, the psc1 mutation partially suppresses the loss of JA-induced growth inhibition in the coi1 mutant background (Ren et al., 2009) and displays increased JA-induced growth inhibition in the wild-type background (Huang et al., 2010). The negative impact of BR signaling on JA signaling has also been demonstrated in tomato, where BR was shown to antagonize several JA-dependent traits including trichome density and allelochemical content (Campos et al., 2009). Unlike in Arabidopsis, BR appears to act upstream of COI1 in tomato since loss of BR synthesis cannot suppress the tomato coi1 mutation (Figure 4) (Campos et al., 2009). However, BR has also been shown to have positive effects on some JA-mediated traits, as JA-induced anthocyanin accumulation is reduced both in BR-biosynthetic mutants and a BR signaling mutant (Peng et al., 2011; Song et al., 2011). Thus, as in the case of the JA-auxin interaction, the crosstalk between JA and BR appears to be complicated.
Jasmonate Crosstalk with Gibberellins
A wave of recent studies has shown an important role for JA–GA signaling crosstalk in regulating the growth–defense tradeoff (Figure 4) (Hou et al., 2010; Wild et al., 2012; Yang et al., 2012; Heinrich et al., 2013). In Nicotiana attenuata, elevated JA has a negative effect on GA biosynthesis in stems resulting in growth inhibition (Heinrich et al., 2013). In several Arabidopsis mutants in which the DELLA transcriptional repressors are stabilized, MYC2-dependent JA-responsive genes are hypersensitive to JA treatment resulting in increased growth inhibition (Hou et al., 2010). In addition, overexpression of a DELLA protein, RGA LIKE 3 (RGL3), which reduces GA-mediated growth, increases MYC2-dependent gene expression; whereas rgl3 mutation reduces MYC2-dependent gene expression (Wild et al., 2012). MYC2 has also been shown to positively regulate RGL3 by directly binding to the promoter of this gene, creating a positive feedback loop in JA signaling (Wild et al., 2012). Consistently with GA antagonism of JA signaling, DELLA repressor proteins have been shown to be positive regulators of JA-mediated disease resistance against necrotrophic pathogens, as JA-mediated defense is compromised in DELLA loss-of-function mutants and is enhanced by overexpression of RGL3 (Navarro et al., 2008; Wild et al., 2012).
Direct physical interaction between JAZ and DELLA repressor proteins has been shown to be crucial for the JA–GA crosstalk in regulating growth and defense (Figure 4) (Hou et al., 2010; Wild et al., 2012; Yang et al., 2012). JAZ proteins interact with the GRAS domain of DELLA proteins, which is important for the interaction between DELLAs and growth-promoting PIF transcription factors (De Lucas et al., 2008). JAZ binding to DELLA proteins was shown to block the interaction between DELLAs and PIFs, thereby relieving the inhibition of DELLAs on PIFs and promoting GA-dependent growth in Arabidopsis (Yang et al., 2012). Accordingly, Arabidopsis coi1 mutants, JAZ overexpression lines, and COI1-silenced rice plants show enhanced growth, whereas Arabidopsis della mutants and PIF overexpression lines are compromised in JA-induced growth inhibition (Yang et al., 2012). These results suggest that, in response to pathogen or herbivore attack, degradation of JAZ proteins makes more DELLA proteins available for interaction with and inhibition of PIF transcription factors as part of a mechanism to inhibit growth (Figure 4) (Yang et al., 2012; Kazan and Manners, 2013). Conversely, GA has also been demonstrated to have a positive effect on some JA-mediated traits such as sesquiterpene synthase gene expression (Hong et al., 2012). The RGA DELLA protein can interact with and repress MYC2 activity resulting in inhibition of JA-mediated terpene biosynthesis; in this case, GA-mediated degradation of DELLAs promotes a specific JA-mediated trait (Hong et al., 2012). Together, these findings suggest that interactions between JA and GA signaling pathways can occur at multiple levels and in different directions, illustrating the dynamic nature of JA–GA crosstalk in regulating the growth–defense tradeoff.
CONCLUSIONS AND FUTURE PERSPECTIVES
Pathogen and herbivore-induced damage is known to reduce plant yield, causing substantial economic losses (Smedegaardpetersen and Stolen, 1981; Oerke, 2006). However, simply breeding plants to have constitutively active defense is not a viable solution, as there are known fitness costs associated with the induction of defense responses (Heil and Baldwin, 2002; Tian et al., 2003; Heidel et al., 2004; Kempel et al., 2011), as well as conditions under which growth must be prioritized in spite of pathogen or herbivore attack (Lozano-Durán et al., 2013; Ballaré, 2014). Plants have evolved mechanisms, such as hormone crosstalk, to optimize fitness in response to the dynamic environments in which they live. A critical step in harnessing this process for the improvement of crop performance is the identification of molecular targets responsible for implementing resource reallocation to facilitate prioritization of growth or defense.
Studies reviewed here and elsewhere have revealed a web of interconnected hormone signaling networks that enable fine-tuning of plant responses to environmental and developmental cues (Bari and Jones, 2009; Robert-Seilaniantz et al., 2011a; Naseem and Dandekar, 2012; Bartoli et al., 2013; De Vleesschauwer et al., 2013; Denance et al., 2013; Hao et al., 2013; Yang et al., 2013). However, it can be challenging to compare and integrate data collected using different experimental parameters, namely plant growth conditions and/or age. Untangling this web is also constrained by the tools and methods available. For instance, while ‘omic’ methods have enabled global visualization of changes in gene expression and protein profiles to some extent, the snapshots they provide are incapable of capturing the full range of dynamic temporal and spatial processes of growth–defense interactions. Also, tools currently available to isolate or amplify certain effects, such as the use of exogenous application of elicitors/hormones and stable genetic manipulation, may result in the identification of interactions that do not exist in nature or fail to identify those that do (Heil and Baldwin, 2002). Other issues include the limitations of using whole seedlings or tissues to investigate changes occurring on a sub-organismal scale, and the relatively few studies conducted to investigate the effects of multiple or variable stresses on growth–defense interactions.
Therefore, while the use of simple laboratory conditions is essential for establishing foundational knowledge of individual signaling pathways, it will also be necessary in the future to design experiments that more accurately reflect natural environments—fluctuating conditions, exposure to multiple stresses, and field studies—to identify network interactions and to test putative molecular mechanisms. As technology advances, the ability to observe plant growth and plant–pathogen/herbivore interactions at a cellular level and in a spatiotemporal manner will provide valuable insight towards elucidating the timing and subcellular localization of molecular interactions as well as to distinguish between local and global effects on plant growth and defense. Understanding the specific molecular interactions that facilitate these tradeoffs will provide powerful tools to genetically tailor plants that optimize this balance to maximize crop yield in fluctuating environmental conditions.
FUNDING
Research in the authors’ laboratories and preparation of this review article were supported by grants from the US Department of Energy (the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science ; DE–FG02-91ER20021), the National Institutes of Health (R01AI068718 and R01AI060761), and the Gordon and Betty Moore Foundation (GBMF3037) to S.Y.H., and the National Science Foundation (MCB-0919100), to B.M. B.H. is supported by the Michigan State University Enrichment Fellowship.
ACKNOWLEDGMENTS
We thank Andre Velasquez and Lori Imboden for critically reading and commenting on the manuscript and Marlene Cameron for her graphic design expertise. No conflict of interest declared.
REFERENCES
- Abel S., Oeller P.W., Theologis A. (1994). Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl Acad. Sci. U S A. 91, 326–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Liao L.L., Jiang C.F., Desnos T., Bartlett J., Fu X.D., Harberd N.P. (2007). DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143, 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C., Boutrot F., Segonzac C., Schwessinger B., Gimenez-Ibanez S., Chinchilla D., Rathjen J.P., de Vries S.C., Zipfel C. (2012). Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl Acad. Sci. U S A. 109, 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldea M., Hamilton J.G., Resti J.P., Zangerl A.R., Berenbaum M.R., DeLucia E.H. (2005). Indirect effects of insect herbivory on leaf gas exchange in soybean. Plant Cell Environ. 28, 402–411 [Google Scholar]
- Attaran E., He S.Y. (2012). The long-sought-after salicylic acid receptors. Mol. Plant. 5, 971–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Fan M., Oh E., Wang Z.Y. (2012a). A triple helix–loop–helix/basic helix–loop–helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis . Plant Cell. 24, 4917–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012b). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis . Nat. Cell Biol. 14, 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin I.T. (2001). An ecologically motivated analysis of plant–herbivore interactions in native tobacco. Plant Physiol. 127, 1449–1458 [PMC free article] [PubMed] [Google Scholar]
- Ballaré C.L. (2014). Light regulation of plant defense. Annu. Rev. Plant Biol. 65, 335–363 [DOI] [PubMed] [Google Scholar]
- Bari R., Jones J.D.G. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488 [DOI] [PubMed] [Google Scholar]
- Bartoli C.G., Casalongué C.A., Simontacchi M., Marquez-Garcia B., Foyer C.H. (2013). Interactions between hormone and redox signalling pathways in the control of growth and cross tolerance to stress. Environ. Exp. Bot. 94, 73–88 [Google Scholar]
- Belkhadir Y., Jaillais Y., Epple P., Balsemão-Pires E., Dangl J.L., Chory J. (2012). Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl Acad. Sci. U S A. 109, 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A.F., Mackey D. (2007). Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436 [DOI] [PubMed] [Google Scholar]
- Berger S., Benediktyova Z., Matous K., Bonfig K., Mueller M.J., Nedbal L., Roitsch T. (2007). Visualization of dynamics of plant-pathogen interaction by novel combination of chlorophyll fluorescence imaging and statistical analysis: differential effects of virulent and avirulent strains of P. syringae and of oxylipins on A. thaliana . J. Exp. Bot. 58, 797–806 [DOI] [PubMed] [Google Scholar]
- Berger S., Papadopoulos M., Schreiber U., Kaiser W., Roitsch T. (2004). Complex regulation of gene expression, photosynthesis and sugar levels by pathogen infection in tomato. Physiol. Plant. 122, 419–428 [Google Scholar]
- Bilgin D.D., Zavala J.A., Zhu J., Clough S.J., Ort D.R., DeLucia E.H. (2010). Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ. 33, 1597–1613 [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406 [DOI] [PubMed] [Google Scholar]
- Boller T., He S.Y. (2009). Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 324, 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfig K.B., Schreiber U., Gabler A., Roitsch T., Berger S. (2006). Infection with virulent and avirulent P. syringae strains differentially affects photosynthesis and sink metabolism in Arabidopsis leaves. Planta. 225, 1–12 [DOI] [PubMed] [Google Scholar]
- Borges L.L., Santana F.A., Castro I.S.L., Arruda K.M.A., Ramos H.J.D., Moreira M.A., de Barros E.G. (2013). Differentially expressed proteins during an incompatible interaction between common bean and the fungus Pseudocercospora griseola . Mol. Breed. 32, 933–942 [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205 [DOI] [PubMed] [Google Scholar]
- Campos M.L., de Almeida M., Rossi M.L., Martinelli A.P., Litholdo C.G., Figueira A., Rampelotti-Ferreira F.T., Vendramim J.D., Benedito V.A., Peres L.E.P. (2009). Brassinosteroids interact negatively with jasmonates in the formation of anti-herbivory traits in tomato. J. Exp. Bot. 60, 4346–4360 [DOI] [PubMed] [Google Scholar]
- Canet J.V., Dobon A., Ibanez F., Perales L., Tornero P. (2010a). Resistance and biomass in Arabidopsis: a new model for salicylic acid perception. Plant Biotechnol. J. 8, 126–141 [DOI] [PubMed] [Google Scholar]
- Canet J.V., Dobon A., Roig A., Tornero P. (2010b). Structure-function analysis of npr1 alleles in Arabidopsis reveals a role for its paralogs in the perception of salicylic acid. Plant Cell Environ. 33, 1911–1922 [DOI] [PubMed] [Google Scholar]
- Cao H., Bowling S.A., Gordon A.S., Dong X.N. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired-resistance. Plant Cell. 6, 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J.D., Volko S., Dong X.N. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 88, 57–63 [DOI] [PubMed] [Google Scholar]
- Cao H., Li X., Dong X.N. (1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl Acad. Sci. U S A. 95, 6531–6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan J., Padmanabhan M., Dinesh-Kumar S.P. (2008). Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe. 3, 126–135 [DOI] [PubMed] [Google Scholar]
- Cerrudo I., Keller M.M., Cargnel M.D., Demkura P.V., de Wit M., Patitucci M.S., Pierik R., Pieterse C.M.J., Ballare C.L. (2012). Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1–JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol. 158, 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Sun J.Q., Zhai Q.Z., Zhou W.K., Qi L.L., Xu L., Wang B., Chen R., Jiang H.L., Qi J., et al. (2011a). The basic helix–loop–helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis . Plant Cell. 23, 3335–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.Z., Pang Q.Y., Dai S.J., Wang Y., Chen S.X., Yan X.F. (2011b). Proteomic identification of differentially expressed proteins in Arabidopsis in response to methyl jasmonate. J. Plant Physiol. 168, 995–1008 [DOI] [PubMed] [Google Scholar]
- Chen Z.Y., Agnew J.L., Cohen J.D., He P., Shan L.B., Sheen J., Kunkel B.N. (2007). Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc. Natl Acad. Sci. U S A. 104, 20131–20136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nurnberger T., Jones J.D.G., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 448, 497–U412 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernandez G., Adie B., Chico J.M., Lorenzo O., Garcia-Casado G., Lopez-Vidriero I., Lozano F.M., Ponce M.R., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 448, 666–U664 [DOI] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host–microbe interactions: shaping the evolution of the plant immune response. Cell. 124, 803–814 [DOI] [PubMed] [Google Scholar]
- Chou H.M., Bundock N., Rolfe S.A., Scholes J.D. (2000). Infection of Arabidopsis thaliana leaves with Albugo candida (white blister rust) causes a reprogramming of host metabolism. Mol. Plant Pathol. 1, 99–113 [DOI] [PubMed] [Google Scholar]
- Clarke J.D., Volko S.M., Ledford H., Ausubel F.M., Dong X.N. (2000). Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis . Plant Cell. 12, 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley P.D., Bryant J.P., Chapin F.S. (1985). Resource availability and plant antiherbivore defense. Science. 230, 895–899 [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861 [DOI] [PubMed] [Google Scholar]
- De Lucas M., Daviere J.M., Rodriguez-Falcon M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blazquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature. 451, 480–U411 [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D., Gheysen G., Höfte M. (2013). Hormone defense networking in rice: tales from a different world. Trends Plant Sci. 18, 555–565 [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D., Van Buyten E., Satoh K., Balidion J., Mauleon R., Choi I.R., Vera-Cruz C., Kikuchi S., Hofte M. (2012). Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 158, 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit M., Spoel S.H., Sanchez-Perez G.F., Gommers C.M.M., Pieterse C.M.J., Voesenek L.A.C.J., Pierik R. (2013). Perception of low red:far-red ratio compromises both salicylic acid- and jasmonic acid-dependent pathogen defences in Arabidopsis . Plant J. 75, 90–103 [DOI] [PubMed] [Google Scholar]
- Delaney T.P., Friedrich L., Ryals J.A. (1995). Arabidopsis signal-transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl Acad. Sci. U S A. 92, 6602–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denance N., Sanchez-Vallet A., Goffner D., Molina A. (2013). Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4, 10.3389/fpls.2013.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoux C., Galletti R., Mammarella N., Gopalan S., Werck D., De Lorenzo G., Ferrari S., Ausubel F.M., Dewdney J. (2008). Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant. 1, 423–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S., Hardtke C.S. (2011). Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 21, R365–R373 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005). The F-box protein TIR1 is an auxin receptor. Nature. 435, 441–445 [DOI] [PubMed] [Google Scholar]
- Dill A., Thomas S.G., Hu J.H., Steber C.M., Sun T.P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell. 16, 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X.H., Cao Y.L., Huang L.L., Zhao J., Xu C.G., Li X.H., Wang S.P. (2008). Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell. 20, 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548 [DOI] [PubMed] [Google Scholar]
- Dou D.L., Zhou J.M. (2012). Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe. 12, 484–495 [DOI] [PubMed] [Google Scholar]
- Engelsdorf T., Horst R.J., Proels R., Proeschel M., Dietz F., Hueckelhoven R., Voll L.M. (2013). Reduced carbohydrate availability enhances the susceptibility of Arabidopsis toward Colletotrichum higginsianum . Plant Physiol. 162, 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M., Meldau S., Howe G.A. (2012). Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 17, 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann J., Schmitz-Thom I., Schoen H., Sonnewald S., Weis E., Scharte J. (2008). RNA interference-mediated repression of cell wall invertase impairs defense in source leaves of tobacco. Plant Physiol. 147, 1288–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fariduddin Q., Yusuf M., Ahmad I., Ahmad A. (2014). Brassinosteroids and their role in response of plants to abiotic stresses. Biol. Plant. 58, 9–17 [Google Scholar]
- Feys B.J.F., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male-sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell. 6, 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350 [DOI] [PubMed] [Google Scholar]
- Fu Z.Q., Dong X.N. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863 [DOI] [PubMed] [Google Scholar]
- Fu Z.Q., Yan S.P., Saleh A., Wang W., Ruble J., Oka N., Mohan R., Spoel S.H., Tada Y., Zheng N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature. 486, 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolome J., Minguet E.G., Grau-Enguix F., Abbas M., Locascio A., Thomas S.G., Alabadi D., Blazquez M.A. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis . Proc. Natl Acad. Sci. U S A. 109, 13446–13451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Giraldo L., Escamilla-Trevino L., Jackson L.A., Dixon R.A. (2011a). Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc. Natl Acad. Sci. U S A. 108, 20814–20819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Giraldo L., Jikumaru Y., Kamiya Y., Tang Y.H., Dixon R.A. (2011b). Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol. 190, 627–639 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227 [DOI] [PubMed] [Google Scholar]
- Glickmann E., Gardan L., Jacquet S., Hussain S., Elasri M., Petit A., Dessaux Y. (1998). Auxin production is a common feature of most pathovars of Pseudomonas syringae . Mol. Plant Microbe Interact. 11, 156–162 [DOI] [PubMed] [Google Scholar]
- Gohre V., Jones A.M.E., Sklenar J., Robatzek S., Weber A.P.M. (2012). Molecular crosstalk between PAMP-triggered immunity and photosynthesis. Mol. Plant Microbe Interact. 25, 1083–1092 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis . Mol. Cell. 5, 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L., Felix G., Boller T. (1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana . Plant J. 18, 277–284 [DOI] [PubMed] [Google Scholar]
- González-Lamothe R., El Oirdi M., Brisson N., Bouarab K. (2012). The conjugated auxin indole-3-acetic acid–aspartic acid promotes plant disease development. Plant Cell. 24, 762–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlach J., Volrath S., KnaufBeiter G., Hengy G., Beckhove U., Kogel K.H., Oostendorp M., Staub T., Ward E., Kessmann H., et al. (1996). Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell. 8, 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M., Lamb C. (2006). Systemic immunity. Curr. Opin. Plant Biol. 9, 414–420 [DOI] [PubMed] [Google Scholar]
- Gray W.M., del Pozo J.C., Walker L., Hobbie L., Risseeuw E., Banks T., Crosby W.L., Yang M., Ma H., Estelle M. (1999). Identification of an SCF ubiquitin–ligase complex required for auxin response in Arabidopsis thaliana . Genes Dev. 13, 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., Kepinski S., Rouse D., Leyser O., Estelle M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature. 414, 271–276 [DOI] [PubMed] [Google Scholar]
- Malinovsky F.G., Batoux M., Schwessinger B., Youn J.H., Stransfeld L., Win J., Kim S.-K., Zipfel C. (2014). Antagonistic regulation of growth and immunity by the Arabidopsis basic helix-loop-helix transcription factor HOMOLOG OF BRASSINOSTEROID ENHANCED EXPRESSION2 INTERACTING WITH INCREASED LEAF INCLINATION1 BINDING bHLH1. Plant Physiol. 164, 1443–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., Vanholme B., Pauwels L., Plovie E., Inze D., Gheysen G., Goossens A. (2009). Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 10, 923–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruszka D. (2013). The brassinosteroid signaling pathway: new key players and interconnections with other signaling networks crucial for plant development and stress tolerance. Int. J. Mol. Sci. 14, 8740–8774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Pang Q.Y., Wang L.H., Yu P., Li N., Yan X.F. (2012). Proteomic identification of MYC2-dependent jasmonate-regulated proteins in Arabidopsis thaliana . Proteome Sci. 10, 10.1186/1477-5956-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsche A.R., Heng-Moss T.M., Higley L.G., Sarath G., Mornhinweg D.W. (2009). Physiological responses of resistant and susceptible barley, Hordeum vulgare to the Russian wheat aphid, Diurpahis noxia (Mordvilko). Arthropod–Plant Interact. 3, 233–240 [Google Scholar]
- Hagen G., Guilfoyle T. (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385 [PubMed] [Google Scholar]
- Hagen G., Kleinschmidt A., Guilfoyle T. (1984). Auxin-regulated gene-expression in intact soybean hypocotyl and excised hypocotyl sections. Planta. 162, 147–153 [DOI] [PubMed] [Google Scholar]
- Hao J.J., Yin Y.H., Fei S.Z. (2013). Brassinosteroid signaling network: implications on yield and stress tolerance. Plant Cell Rep. 32, 1017–1030 [DOI] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S.L., Gendron N., Sun C.Q., Wang Z.Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 307, 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Yang Y.L., Li J.M., Wang Z.Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis . Proc. Natl Acad. Sci. U S A. 99, 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M.E., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl Acad. Sci. U S A. 104, 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel A.J., Clarke J.D., Antonovics J., Dong X.N. (2004). Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana . Genetics. 168, 2197–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M., Baldwin I.T. (2002). Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci. 7, 61–67 [DOI] [PubMed] [Google Scholar]
- Heil M., Hilpert A., Kaiser W., Linsenmair K.E. (2000). Reduced growth and seed set following chemical induction of pathogen defence: does systemic acquired resistance (SAR) incur allocation costs? J. Ecol. 88, 645–654 [Google Scholar]
- Heinrich M., Hettenhausen C., Lange T., Wunsche H., Fang J.J., Baldwin I.T., Wu J.Q. (2013). High levels of jasmonic acid antagonize the biosynthesis of gibberellins and inhibit the growth of Nicotiana attenuata stems. Plant J. 73, 591–606 [DOI] [PubMed] [Google Scholar]
- Hentrich M., Bottcher C., Duchting P., Cheng Y.F., Zhao Y.D., Berkowitz O., Masle J., Medina J., Pollmann S. (2013). The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 74, 626–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbers K., Meuwly P., Frommer W.B., Metraux J.P., Sonnewald U. (1996). Systemic acquired resistance mediated by the ectopic expression of invertase: possible hexose sensing in the secretory pathway. Plant Cell. 8, 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herms D.A., Mattson W.J. (1992). The dilemma of plants: to grow or defend. Q. Rev. Biol. 67, 283–335 [Google Scholar]
- Hong G.J., Xue X.Y., Mao Y.B., Wang L.J., Chen X.Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell. 24, 2635–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M., Belkhadir Y., Dreux M., Dabi T., Noel J.P., Wilson I.A., Chory J. (2011). Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature. 474, 467–U490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X.L., Lee L.Y.C., Xia K.F., Yen Y.Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell. 19, 884–894 [DOI] [PubMed] [Google Scholar]
- Huang Y., Han C., Peng W., Peng Z., Xiong X., Zhu Q., Gao B., Xie D., Ren C. (2010). Brassinosteroid negatively regulates jasmonate inhibition of root growth in Arabidopsis . Plant Signal. Behav. 5, 140–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga Y., Uppalapati S.R., Ishiga T., Elavarthi S., Martin B., Bender C.L. (2009). The phytotoxin coronatine induces light-dependent reactive oxygen species in tomato seedlings. New Phytol. 181, 147–160 [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Vert G. (2012). Brassinosteroids, gibberellins and light-mediated signalling are the three-way controls of plant sprouting. Nat. Cell Biol. 14, 788–790 [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Belkhadir Y., Balsemao-Pires E., Dangl J.L., Chory J. (2011). Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc. Natl Acad. Sci. U S A. 108, 8503–8507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature. 444, 323–329 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 14, 787–799 [DOI] [PubMed] [Google Scholar]
- Jung C., Lyou S.H., Yeu S., Kim M.A., Rhee S., Kim M., Lee J.S., Do Choi Y., Cheong J.J. (2007). Microarray-based screening of jasmonate-responsive genes in Arabidopsis thaliana . Plant Cell Rep. 26, 1053–1063 [DOI] [PubMed] [Google Scholar]
- Jung S. (2004). Effect of chlorophyll reduction in Arabidopsis thaliana by methyl jasmonate or norflurazon on antioxidant systems. Plant Physiol. Biochem. 42, 225–231 [DOI] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl Acad. Sci. U S A. 105, 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. (2013). Auxin and the integration of environmental signals into plant root development. Ann. Bot. 112, 1655–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2009). Linking development to defense: auxin in plant–pathogen interactions. Trends Plant Sci. 14, 373–382 [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2013). MYC2: the master in action. Mol. Plant. 6, 686–703 [DOI] [PubMed] [Google Scholar]
- Kempel A., Schadler M., Chrobock T., Fischer M., van Kleunen M. (2011). Tradeoffs associated with constitutive and induced plant resistance against herbivory. Proc. Natl Acad. Sci. U S A. 108, 5685–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2004). Auxin-induced SCFTIR1–Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc. Natl Acad. Sci. U S A. 101, 12381–12386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 435, 446–451 [DOI] [PubMed] [Google Scholar]
- Kerchev P.I., Fenton B., Foyer C.H., Hancock R.D. (2012). Plant responses to insect herbivory: interactions between photosynthesis, reactive oxygen species and hormonal signalling pathways. Plant Cell Environ. 35, 441–453 [DOI] [PubMed] [Google Scholar]
- Khripach V., Zhabinskii V., De Groot A. (2000). Twenty years of brassinosteroids: steroidal plant hormones warrant better crops for the XXI century. Ann. Bot. 86, 441–447 [Google Scholar]
- Kidd B.N., Kadoo N.Y., Dombrecht B., Tekeoglu M., Gardiner D.M., Thatcher L.F., Aitken E.A.B., Schenk P.M., Manners J.M., Kazan K. (2011). Auxin signaling and transport promote susceptibility to the root-infecting fungal pathogen Fusarium oxysporum in Arabidopsis . Mol. Plant Microbe Interact. 24, 733–748 [DOI] [PubMed] [Google Scholar]
- Kieffer M., Neve J., Kepinski S. (2010). Defining auxin response contexts in plant development. Curr. Opin. Plant Biol. 13, 12–20 [DOI] [PubMed] [Google Scholar]
- Kinkema M., Fan W.H., Dong X.N. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell. 12, 2339–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocal N., Sonnewald U., Sonnewald S. (2008). Cell wall-bound invertase limits sucrose export and is involved in symptom development and inhibition of photosynthesis during compatible interaction between tomato and Xanthomonas campestris pv vesicatoria . Plant Physiol. 148, 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C., Neu C., Pajonk S., Yun H.S., Lipka U., Humphry M., Bau S., Straus M., Kwaaitaal M., Rampelt H., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature. 451, 835–U810 [DOI] [PubMed] [Google Scholar]
- Lawton K.A., Friedrich L., Hunt M., Weymann K., Delaney T., Kessmann H., Staub T., Ryals J. (1996). Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 10, 71–82 [DOI] [PubMed] [Google Scholar]
- Li J., Wen J.Q., Lease K.A., Doke J.T., Tax F.E., Walker J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 110, 213–222 [DOI] [PubMed] [Google Scholar]
- Li J.M., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 90, 929–938 [DOI] [PubMed] [Google Scholar]
- Lilley J.L.S., Gan Y.B., Graham I.A., Nemhauser J.L. (2013). The effects of DELLAs on growth change with developmental stage and brassinosteroid levels. Plant J. 76, 165–173 [DOI] [PubMed] [Google Scholar]
- Lin W.W., Li B., Lu D., Chen S., Zhu N., He P., Shan L. (2014). Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc. Natl Acad. Sci. U S A. 111, 3632–3637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E., Reed J.W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400 [PubMed] [Google Scholar]
- Ljung K., Hull A.K., Kowalczyk M., Marchant A., Celenza J., Cohen J.D., Sandberg G. (2002). Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana . Plant Mol. Biol. 50, 309–332 [DOI] [PubMed] [Google Scholar]
- Lozano-Durán R., Macho A.P., Boutrot F., Segonzac C., Somssich I.E., Zipfel C., Nürnberger T. (2013). The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. eLIFE. 2, e00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D.P., Wu S.J., Gao X.Q., Zhang Y.L., Shan L.B., He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl Acad. Sci. U S A. 107, 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Muller J. (2011). Auxin conjugates: their role for plant development and in the evolution of land plants. J. Exp. Bot. 62, 1757–1773 [DOI] [PubMed] [Google Scholar]
- McGinnis K.M., Thomas S.G., Soule J.D., Strader L.C., Zale J.M., Sun T.P., Steber C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell. 15, 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldau S., Ullman-Zeunert L., Govind G., Bartram S., Baldwin I.T. (2012). MAPK-dependent JA and SA signalling in Nicotiana attenuata affects plant growth and fitness during competition with conspecifics. BMC Plant Biol. 12, 10.1186/1471-2229-12-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S., Baldwin I.T. (2014). RuBPCase activase (RCA) mediates growth–defense trade-offs: silencing RCA redirects jasmonic acid (JA) flux from JA-isoleucine to methyl jasmonate (MeJA) to attenuate induced defense responses in Nicotiana attenuata . New Phytol. 201, 1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J., Zipfel C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357 [DOI] [PubMed] [Google Scholar]
- Mora-Garcia S., Vert G., Yin Y.H., Cano-Delgado A., Cheong H., Chory J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis . Genes Dev. 18, 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z., Fan W.H., Dong X.N. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 113, 935–944 [DOI] [PubMed] [Google Scholar]
- Murase K., Hirano Y., Sun T.P., Hakoshima T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 456, 459–U415 [DOI] [PubMed] [Google Scholar]
- Mutka A.M., Fawley S., Tsao T., Kunkel B.N. (2013). Auxin promotes susceptibility to Pseudomonas syringae via a mechanism independent of suppression of salicylic acid-mediated defenses. Plant J. 74, 746–754 [DOI] [PubMed] [Google Scholar]
- Nabity P.D., Zavala J.A., DeLucia E.H. (2009). Indirect suppression of photosynthesis on individual leaves by arthropod herbivory. Ann. Bot. 103, 655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K.H., Li J.M. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 110, 203–212 [DOI] [PubMed] [Google Scholar]
- Naseem M., Dandekar T. (2012). The role of auxin-cytokinin antagonism in plant–pathogen interactions. PLoS Pathog. 8, e1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L., Bari R., Achard P., Lison P., Nemri A., Harberd N.P., Jones J.D.G. (2008). DELLAs control plant immune responses by modulating the balance and salicylic acid signaling. Curr. Biol. 18, 650–655 [DOI] [PubMed] [Google Scholar]
- Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J.D.G. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 312, 436–439 [DOI] [PubMed] [Google Scholar]
- Navarro L., Zipfel C., Rowland O., Keller I., Robatzek S., Boller T., Jones J.D.G. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135, 1113–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise V., Roux M., Zipfel C. (2009). Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol. 150, 1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noir S., Bomer M., Takahashi N., Ishida T., Tsui T.L., Balbi V., Shanahan H., Sugimoto K., Devoto A. (2013). Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol. 161, 1930–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J. (2010). Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb. Perspect. Biol. 2, a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell P.J., Schmelz E.A., Moussatche P., Lund S.T., Jones J.B., Klee H.J. (2003). Susceptible to intolerance: a range of hormonal actions in a susceptible Arabidopsis pathogen response. Plant J. 33, 245–257 [DOI] [PubMed] [Google Scholar]
- Oerke E.C. (2006). Crop losses to pests. J. Agric. Sci. 144, 31–43 [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–U864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostin A., Kowalyczk M., Bhalerao R.P., Sandberg G. (1998). Metabolism of indole-3-acetic acid in Arabidopsis . Plant Physiol. 118, 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar K.M., Wang W., Tada Y., Oka N., Tucker Chandra L., Fonseca J.P., Dong X. (2012). The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr. Biol. 22, 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S.P., Somssich I.E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Park J.Y., Kim Y.S., Staswick P.E., Jeon J., Yun J., Kim S.Y., Kim J., Lee Y.H., Park C.M. (2007). GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis . J. Biol. Chem. 282, 10036–10046 [DOI] [PubMed] [Google Scholar]
- Pauwels L., Barbero G.F., Geerinck J., Tilleman S., Grunewald W., Perez A.C., Chico J.M., Vanden Bossche R., Sewell J., Gil E., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 464, 788–U169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J.R., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P., Harberd N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z.H., Han C.Y., Yuan L.B., Zhang K., Huang H.M., Ren C.M. (2011). Brassinosteroid enhances jasmonate-induced anthocyanin accumulation in Arabidopsis seedlings. J. Integr. Plant Biol. 53, 632–640 [DOI] [PubMed] [Google Scholar]
- Pieterse C.M.J., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S.C.M. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521 [DOI] [PubMed] [Google Scholar]
- Pinon V., Prasad K., Grigg S.P., Sanchez-Perez G.F., Scheres B. (2013). Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis . Proc. Natl Acad. Sci. U S A. 110, 1107–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C.M., Han C.Y., Peng W., Huang Y., Peng Z.H., Xiong X.Y., Zhu Q., Gao B.D., Xie D.X. (2009). A leaky mutation in DWARF4 reveals an antagonistic role of brassinosteroid in the inhibition of root growth by jasmonate in Arabidopsis . Plant Physiol. 151, 1412–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-San Vicente M., Plasencia J. (2011). Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62, 3321–3338 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J.D.G. (2011a). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., MacLean D., Jikumaru Y., Hill L., Yamaguchi S., Kamiya Y., Jones J.D.G. (2011b). The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant J. 67, 218–231 [DOI] [PubMed] [Google Scholar]
- Roitsch T., Gonzalez M.C. (2004). Function and regulation of plant invertases: sweet sensations. Trends Plant Sci. 9, 606–613 [DOI] [PubMed] [Google Scholar]
- Roux M., Schwessinger B., Albrecht C., Chinchilla D., Jones A., Holton N., Malinovsky F.G., Tor M., de Vries S., Zipfel C. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell. 23, 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton P.J., Somssich I.E., Ringler P., Shen Q.J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258 [DOI] [PubMed] [Google Scholar]
- Santner A., Estelle M. (2009). Recent advances and emerging trends in plant hormone signalling. Nature. 459, 1071–1078 [DOI] [PubMed] [Google Scholar]
- Schulze B., Mentzel T., Jehle A.K., Mueller K., Beeler S., Boller T., Felix G., Chinchilla D. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285, 9444–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I.M., Clarke S.M., Wood J.E., Mur L.A.J. (2004). Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis . Plant Physiol. 135, 1040–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J., Tsui F., Klessig D.F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant Microbe Interact. 10, 69–78 [DOI] [PubMed] [Google Scholar]
- Shan X.Y., Wang J.X., Chua L.L., Jiang D.A., Peng W., Xie D.X. (2011). The role of Arabidopsis rubisco activase in jasmonate-induced leaf senescence. Plant Physiol. 155, 751–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J., Han Z.F., Kim T.W., Wang J.J., Cheng W., Chang J.B., Shi S.A., Wang J.W., Yang M.J., Wang Z.Y., et al. (2011). Structural insight into brassinosteroid perception by BRI1. Nature. 474, 472–U496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L.B., Tan X., Mao H.B., Withers J., Ben-Nissan G., Hinds T.R., Kobayashi Y., Hsu F.F., Sharon M., Browse J., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 468, 400–U301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu C., Figueroa P., DePew C.L., Cooke T.F., Sheard L.B., Moreno J.E., Katsir L., Zheng N., Browse J., Howe G.A. (2012). JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis . Plant Cell. 24, 536–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Jung H.S., Dill A., Kawaide H., Kamiya Y., Sun T.P. (2001). Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis . Plant Cell. 13, 1555–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms E.L., Rausher M.D. (1987). Costs and benefits of plant-resistance to herbivory. Am. Nat. 130, 570–581 [Google Scholar]
- Smedegaardpetersen V., Stolen O. (1981). Effect of energy-requiring defense reactions on yield and grain quality in a powdery mildew-resistant barley cultivar. Phytopathology. 71, 396–399 [Google Scholar]
- Song S.S., Qi T.C., Huang H., Ren Q.C., Wu D.W., Chang C.Q., Peng W., Liu Y.L., Peng J.R., Xie D.X. (2011). The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis . Plant Cell. 23, 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnewald S., Priller J.P.R., Schuster J., Glickmann E., Hajirezaei M.-R., Siebig S., Mudgett M.B., Sonnewald U. (2012). Regulation of cell wall-bound invertase in pepper leaves by Xanthomonas campestris pv. vesicatoria type three effectors. PLoS One. 7, e51763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., Johnson J.S., Dong X. (2007). Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl Acad. Sci. U S A. 104, 18842–18847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Tiryaki I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis . Plant Cell. 16, 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Serban B., Rowe M., Tiryaki I., Maldonado M.T., Maldonado M.C., Suza W. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 17, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Su W.P., Howell S.H. (1992). Methyl jasmonate inhibition of root-growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl Acad. Sci. U S A. 89, 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange R.N., Scott P.R. (2005). Plant disease: a threat to global food security. Annu. Rev. Phytopathol. 43, 83–116 [DOI] [PubMed] [Google Scholar]
- Sturm A. (1999). Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 121, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S., Jiang C.J., Miyazawa S.I., Masumoto C., Yazawa K., Hayashi N., Shimono M., Nakayama A., Miyao M., Takatsuji H. (2010). Role of OsNPR1 in rice defense program as revealed by genomewide expression analysis. Plant Mol. Biol. 74, 549–562 [DOI] [PubMed] [Google Scholar]
- Sun J.Q., Chen Q., Qi L.L., Jiang H.L., Li S.Y., Xu Y.X., Liu F., Zhou W.K., Pan J.W., Li X.G., et al. (2011). Jasmonate modulates endocytosis and plasma membrane accumulation of the Arabidopsis PIN2 protein. New Phytol. 191, 360–375 [DOI] [PubMed] [Google Scholar]
- Sun J.Q., Xu Y.X., Ye S.Q., Jiang H.L., Chen Q., Liu F., Zhou W.K., Chen R., Li X.G., Tietz O., et al. (2009). Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell. 21, 1495–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.-p. (2011). The molecular mechanism and evolution of the GA–GID1–DELLA signaling module in plants. Curr. Biol. 21, R338–R345 [DOI] [PubMed] [Google Scholar]
- Sun Y., Li L., Macho A.P., Han Z., Hu Z., Zipfel C., Zhou J.-m., Chai J. (2013). Structural basis for flg22-induced activation of the Arabidopsis FLS2–BAK1 immune complex. Science. 342, 624–628. [DOI] [PubMed] [Google Scholar]
- Sunkar R., Zhu J.K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis . Plant Cell. 16, 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbrick P.J., Schulze-Lefert P., Scholes J.D. (2006). Metabolic consequences of susceptibility and resistance (race-specific and broad-spectrum) in barley leaves challenged with powdery mildew. Plant Cell Environ. 29, 1061–1076 [DOI] [PubMed] [Google Scholar]
- Tada Y., Spoel S.H., Pajerowska-Mukhtar K., Mou Z.L., Song J.Q., Wang C., Zuo J.R., Dong X.N. (2008). Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science. 321, 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.Q., Yuan M., Wang R.J., Yang Y.H., Wang C.M., Oses-Prieto J.A., Kim T.W., Zhou H.W., Deng Z.P., Gampala S.S., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13, 124–U149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thilmony R., Underwood W., He S.Y. (2006). Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 46, 34–53 [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G.H., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCFC°11 complex during jasmonate signalling. Nature. 448, 661–U662 [DOI] [PubMed] [Google Scholar]
- Thomma B., Eggermont K., Penninckx I., Mauch-Mani B., Vogelsang R., Cammue B.P.A., Broekaert W.F. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl Acad. Sci. U S A. 95, 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Traw M.B., Chen J.Q., Kreitman M., Bergelson J. (2003). Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana . Nature. 423, 74–77 [DOI] [PubMed] [Google Scholar]
- Tiwari S.B., Hagen G., Guilfoyle T.J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell. 16, 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.B., Wang X.J., Hagen G., Guilfoyle T.J. (2001). AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell. 13, 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann-Zeunert L., Stanton M.A., Wielsch N., Bartram S., Hummert C., Svatos A., Baldwin I.T., Groten K. (2013). Quantification of growth–defense trade-offs in a common currency: nitrogen required for phenolamide biosynthesis is not derived from ribulose-1,5-bisphosphate carboxylase/oxygenase turnover. Plant J. 75, 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. (1999). Activation and repression of transcription by auxin-response factors. Proc. Natl Acad. Sci. U S A. 96, 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde P.L., Fornoni J., Nunez-Farfan J. (2003). Evolutionary ecology of Datura stramonium: equal plant fitness benefits of growth and resistance against herbivory. J. Evol. Biol. 16, 127–137 [DOI] [PubMed] [Google Scholar]
- van Loon L.C., Rep M., Pieterse C.M.J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162 [DOI] [PubMed] [Google Scholar]
- Vanneste S., Friml J. (2009). Auxin: a trigger for change in plant development. Cell. 136, 1005–1016 [DOI] [PubMed] [Google Scholar]
- Vert G., Chory J. (2006). Downstream nuclear events in brassinosteroid signalling. Nature. 441, 96–100 [DOI] [PubMed] [Google Scholar]
- Vlot A.C., Dempsey D.A., Klessig D.F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206 [DOI] [PubMed] [Google Scholar]
- Walling L.L. (2009). Adaptive defense responses to pathogens and insects. In Advances in Botanical Research: Plant Innate Immunity, Loon L.C.V, ed. (London: Academic Press; ), pp. 551–612 [Google Scholar]
- Wang D., Amornsiripanitch N., Dong X.N. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathogens. 2, 1042–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Pajerowska-Mukhtar K., Culler A.H., Dong X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17, 1784–1790 [DOI] [PubMed] [Google Scholar]
- Wang D., Weaver N.D., Kesarwani M., Dong X.N. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science. 308, 1036–1040 [DOI] [PubMed] [Google Scholar]
- Wang X.L., Chory J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 313, 1118–1122 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J.X., Chen M., Vafeados D., Yang Y.L., Fujioka S., Yoshida S., Asami T., et al. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell. 2, 505–513 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007). Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100, 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C. (2013). Action of jasmonates in plant stress responses and development: applied aspects. Biotechnol. Adv. 32, 31–39 [DOI] [PubMed] [Google Scholar]
- Wasternack C., Hause B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development: an update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierstra I., Kloppstech K. (2000). Differential effects of methyl jasmonate on the expression of the early light-inducible proteins and other light-regulated genes in barley. Plant Physiol. 124, 833–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild M., Daviere J.M., Cheminant S., Regnault T., Baumberger N., Heintz D., Baltz R., Genschik P., Achard P. (2012). The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell. 24, 3307–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers J., Yao J., Mecey C., Howe G.A., Melotto M., He S.Y. (2012). Transcription factor-dependent nuclear localization of a transcriptional repressor in jasmonate hormone signaling. Proc. Natl Acad. Sci. U S A. 109, 20148–20153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A.W., Bartel B. (2005). Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhang D., Chu J.Y., Boyle P., Wang Y., Brindle I.D., De Luca V., Després C. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Reports. 1, 639–647 [DOI] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 280, 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xin X.F., He S.Y. (2013). Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51, 473–498 [DOI] [PubMed] [Google Scholar]
- Yamada T. (1993). The role of auxin in plant-disease development. Annu. Rev. Phytopathol. 31, 253–273 [DOI] [PubMed] [Google Scholar]
- Yan Y.X., Stolz S., Chetelat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 19, 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.L., Yang Y.N., He Z.H. (2013). Roles of plant hormones and their interplay in rice immunity. Mol. Plant. 6, 675–685 [DOI] [PubMed] [Google Scholar]
- Yang D.L., Yao J., Mei C.S., Tong X.H., Zeng L.J., Li Q., Xiao L.T., Sun T.P., Li J.G., Deng X.W., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl Acad. Sci. U S A. 109, E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y.H., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis . Cell. 120, 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y.H., Wang Z.Y., Mora-Garcia S., Li J.M., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 109, 181–191 [DOI] [PubMed] [Google Scholar]
- Yu D.Q., Chen C.H., Chen Z.X. (2001). Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell. 13, 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala J.A., Patankar A.G., Gase K., Baldwin I.T. (2004). Constitutive and inducible trypsin proteinase inhibitor production incurs large fitness costs in Nicotiana attenuata . Proc. Natl Acad. Sci. U S A. 101, 1607–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.H., Fu J.M., Hiromasa Y., Pan H.Y., Bai G.H. (2013). Differentially expressed proteins associated with fusarium head blight resistance in wheat. PLoS One. 8, e82079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Turner J.G. (2008). Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS One. 3, e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.L., Goritschnig S., Dong X.N., Li X. (2003a). A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1 . Plant Cell. 15, 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.L., Tessaro M.J., Lassner M., Li X. (2003b). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell. 15, 2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wang M., Li Z., Li Q., He Z. (2008). Arabidopsis GH3.5 regulates salicylic acid-dependent and both NPR1-dependent and independent defense responses. Plant Signal. Behav. 3, 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.Q., Li Q., Li Z.M., Staswick P.E., Wang M.Y., Zhu Y., He Z.H. (2007). Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis–Pseudomonas syringae interaction. Plant Physiol. 145, 450–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B.L., Li J. (2012). Regulation of brassinosteroid biosynthesis and inactivation. J. Integr. Plant Biol. 54, 746–759 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D.G., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 125, 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D.G., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 428, 764–767 [DOI] [PubMed] [Google Scholar]
- Zou J.J., Rodriguez-Zas S., Aldea M., Li M., Zhu J., Gonzalez D.O., Vodkin L.O., DeLucia E., Clough S.J. (2005). Expression profiling soybean response to Pseudomonas syringae reveals new defense-related genes and rapid HR-specific downregulation of photosynthesis. Mol. Plant Microbe Interact. 18, 1161–1174 [DOI] [PubMed] [Google Scholar]