Abstract
Long-range regulatory interactions play an important role in shaping gene-expression programs. However, the genomic features that organize these activities are still poorly characterized. We conducted a large operational analysis to chart the distribution of gene regulatory activities along the mouse genome, using hundreds of insertions of a regulatory sensor. We found that enhancers distribute their activities along broad regions and not in a gene-centric manner, defining large regulatory domains. Remarkably, these domains correlate strongly with the recently described TADs, which partition the genome into distinct self-interacting blocks. Different features, including specific repeats and CTCF-binding sites, correlate with the transition zones separating regulatory domains, and may help to further organize promiscuously distributed regulatory influences within large domains. These findings support a model of genomic organization where TADs confine regulatory activities to specific but large regulatory domains, contributing to the establishment of specific gene expression profiles.
Specificity of gene expression is key to tissue function and identity, and is in great part determined at the transcriptional level. Promoters, located proximally to transcriptional start sites, play an essential role in initiating gene expression, but their activity greatly depends on the action of more distal regulatory elements. Among these, enhancers remain the best characterized (Bulger and Groudine 2011; Ong and Corces 2011). They often show a specific chromatin signature, which has helped the annotation of enhancers active in human or mouse cell lines and tissues (Heintzman et al. 2009; Creyghton et al. 2010; Rada-Iglesias et al. 2011; The ENCODE Project Consortium 2012; Shen et al. 2012). Remarkably, a substantial fraction of these putative enhancers was found hundreds of kilobases from the nearest gene. Yet, how their activity is allocated to their target gene(s) across such large distances and sometimes several genes, remains unclear.
The evidence to date indicates that enhancer–promoter communication can be influenced by specific regulatory elements, which may contribute to enhancer–promoter interactions positively (promoter architecture [Ohtsuki et al. 1998], tethering elements [Calhoun and Levine 2003]), or negatively (e.g., insulators/enhancer blockers [for review, see Gaszner and Felsenfeld 2006]). Also, the regulatory interactions between promoters and associated remote enhancers are usually associated with physical proximity (Bulger and Groudine 2011; for review, see Bickmore and van Steensel 2013). Several protein complexes, including CTCF, cohesin, and Mediator, have been proposed to have a role in organizing these regulatory contacts (Parelho et al. 2008; Wendt et al. 2008; Hadjur et al. 2009; Kagey et al. 2010). Furthermore, recent studies suggest that the genome is organized into relatively cell-type invariant topological domains (TADs) characterized by preferential self-contacts (Dixon et al. 2012; Hou et al. 2012; Nora et al. 2012; Sexton et al. 2012). Genes located in the same TAD show greater expression correlation than genes located in distinct ones (Nora et al. 2012), and TAD borders are enriched for factors implicated as insulator elements (Dixon et al. 2012; Hou et al. 2012). These observations suggested that TADs may form a backbone for tissue-specific regulatory interactions (Bickmore and van Steensel 2013; Gibcus and Dekker 2013; Nora et al. 2013).
However, despite a growing body of knowledge on individual regulatory elements and the three-dimensional organization of the genome, the “rules of engagement” (Splinter and de Laat 2011) that determine the activity of remote cis-acting elements on the surrounding genes remain elusive, in part because a direct assessment of how such activities are distributed in the genomic environment is still lacking. Scrutinizing endogenous gene activity offers only a partial view of this distribution, since expression of endogenous genes also depends on their different promoters, and distinct post-transcriptional regulation. Furthermore, it does not provide information about the gene deserts that constitute ∼25% of mammalian genomes and where many functionally important regulatory elements lie (Nobrega et al. 2003; Ovcharenko et al. 2004).
We have recently developed an efficient in vivo transposition system that allows the rapid production of mice with a single-copy insertion of a regulatory sensor (Ruf et al. 2011). This regulatory sensor consists of a lacZ reporter gene driven by a weak promoter, and has minimal effect—if any—on the expression of the surrounding genes. The sensor is carried in a Sleeping Beauty transposon that integrates almost randomly in the genome (Horie et al. 2003; Liu et al. 2005). Importantly, the sensor is “naïve”: It has not been subjected to the evolutionary selection that has shaped endogenous genes to favor or avoid regulatory influences. Therefore, a comparison of the sensor's expression pattern at different insertions provides a simple and direct readout for the regulatory input acting on those positions.
By analyzing the activity of the regulatory sensor in mouse embryos at several hundred insertion sites, we identified multiple factors that impinge on the action of cis-regulatory elements. We confirm circumstantial evidence that the influence of enhancers extends over several hundred kilobases, irrespective of the position of the normal target genes. By comparing adjacent insertions, we identified large regulatory domains as well as transition zones. We find that regulatory domains are included within TADs, and accordingly depleted in features associated with insulating activities, whereas TAD boundaries correspond to regulatory transitions. Thus, the influence of enhancers appears limited to the topological domain they are part of, providing direct support to the notion that TADs correspond to a functional subdivision of the genome into regions where otherwise promiscuous regulatory influences are confined.
Results
Our initial analysis of about 160 insertions of a regulatory sensor had suggested that tissue-specific regulatory activities are pervasively present throughout the genome (Ruf et al. 2011). From this, we inferred that with an increased number and density of insertions, one could map—in the natural genomic context—the intervals that were responsive to enhancers, and thus investigate the regulatory architecture of the mouse genome in greater detail (Fig. 1A).
Figure 1.
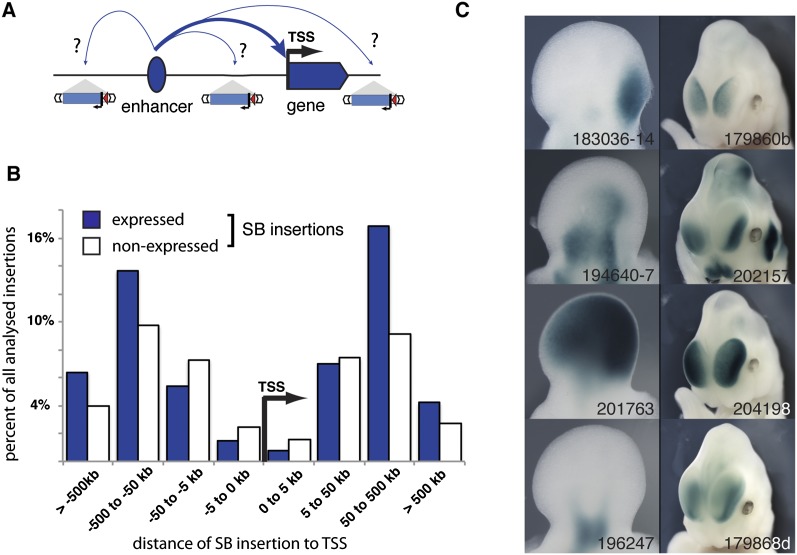
Mapping the distribution of regulatory activities along the genome. (A) Insertion of a regulatory sensor (drawing of sensor) at different distances from an enhancer (blue oval) and different positions relative to its target gene (blue arrow) can report on the domains of action of the enhancer. (B) A total of 734 insertions were characterized for expression in mid-gestation mouse embryos. About 55% of these reported regulatory activities. Insertions with (blue) or without (white) expression were broadly distributed around endogenous gene transcriptional start sites (distribution made with GREAT) (McLean et al. 2010). (C) Examples of the diverse expression patterns obtained in E11.5 embryonic forelimb (left) or forebrain (right). Numbers refer to insertion identifiers used in the TRACER database (Chen et al. 2013).
For this purpose, we generated and mapped more than 1000 new integration sites using extensive transposition from single-copy starting points. We characterized a substantial subset of these (747 insertions) for lacZ expression (Supplemental Table 1; online TRACER database) (Chen et al. 2013). These insertions are distributed throughout the genome, with a subset clustered in domains of up to 1 Mb, owing to the local hopping of the Sleeping Beauty transposon (Horie et al. 2003). Overall, most insertions are localized far from endogenous transcriptional start sites (TSS) (Fig. 1B), with two-thirds more than 50 kb from the nearest TSS. Consistent with our first analysis (Ruf et al. 2011), ∼55% of all insertions showed expression, with the vast majority expressed in a tissue-restricted manner (Symmons and Spitz 2013). Insertions far from TSS are more likely to show expression than the ones next to TSS (Fig. 1B), probably due to local competition with endogenous gene promoters (Ruf et al. 2011). Importantly, independent insertions show extremely diverse expression patterns (Fig. 1C): Only nearby insertions or insertions around paralogous genes yielded expression patterns that were not clearly distinguishable.
Binding of the enhancer-associated protein EP300 is predictive of tissue-specific expression over large distances
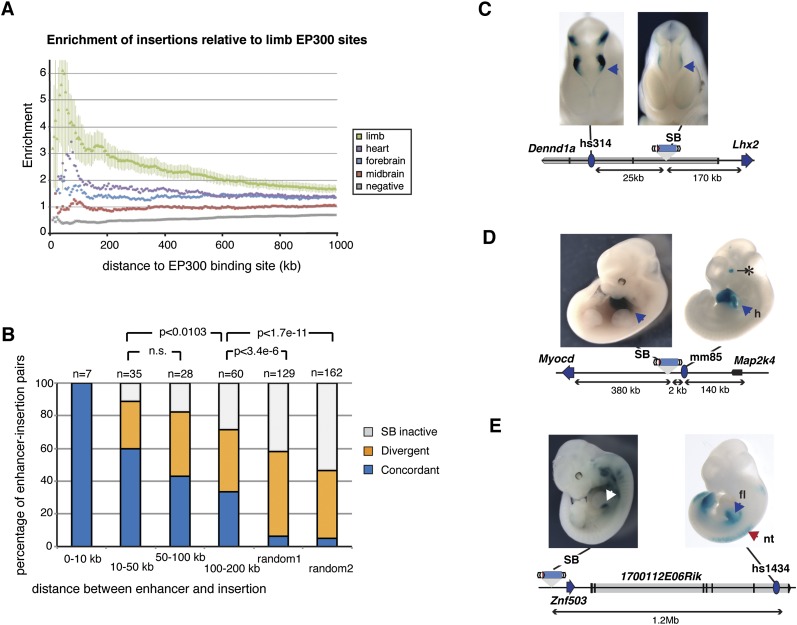
We first sought to obtain deeper insight into the range of action of enhancers. For this, we investigated whether insertions were more likely to show expression in a given tissue if located in the proximity of an active enhancer, and—if so—how far we could detect such enrichment. We compiled a list of 670 nonredundant viewpoints, annotated with a simple controlled vocabulary (Chen et al. 2013; Supplemental Table 2). Since binding of the coactivator protein EP300 has been reported to demarcate active enhancers in a given tissue (Visel et al. 2009), we used EP300 binding sites from E11.5 mouse embryonic forebrain, midbrain, limb, and heart (Visel et al. 2009; Blow et al. 2010). We identified pairs of sensor insertion/EP300 binding sites, and classified them as concordant, when the sensor was expressed in the tissue where the EP300 binding site was detected. By calculating the enrichment of such concordant pairs compared with a set of random insertion/EP300 site pairs, we revealed significant enrichment for concordant pairs up to a distance of 200 kb (Fig. 2A; Supplemental Fig. 1). When EP300 binding sites were paired with silent insertions or insertions expressed in different tissues, enrichment was weaker or not found. This analysis implied that enhancers—represented here by EP300-bound regions—exert their effects across large distances. A similar typical range has been observed when comparing the distance between enhancer regions and the TSS of endogenous genes (Blow et al. 2010; Chepelev et al. 2012; Li et al. 2012).
Figure 2.
Expression of the regulator sensor is correlated with surrounding enhancers up to large distances. (A) Enrichment of insertions showing LacZ activity in a given tissue relative to limb EP300 binding sites. Enrichment for insertions with expression in the limb (green) compared with random insertions is calculated at increasing distances from the nearest EP300 site (x-axis). Error bars represent one standard deviation from the mean. Enrichments of insertions with activity in other tissues but not in the limb (heart: purple; forebrain: blue; midbrain: red) or with no LacZ activity (gray) are also displayed. Results for EP300 sites from other tissues are shown in Supplemental Figure 1. (B) Comparison of enhancer and sensor activity. Different groups were considered, according to the relative distance between the insertions and the enhancers (number of insertion–enhancer pairs indicated above each bar). The two random data sets are described in the Methods section. The proportions of concordant enhancer–insertion pairs in different groups were compared using Fisher's exact test. (C–E) Examples of concordant enhancer–insertion pairs. The different loci are schematized (enhancer: blue oval with VISTA reference; sensor: drawing of transposon; endogenous genes: arrows or gray bars with black exons), with putative target genes of enhancers indicated by labeling the gene the same color as the enhancer. Photos of representative embryos of the in vivo enhancer assays are from the Vista Enhancer Browser (Visel et al. 2007). (C) The sensor reported the activity of an intronic diencephalon/midbrain enhancer, which likely contributes to the regulation of the distant Lhx2 gene (Gray et al. 2004). (D) Heart-specific expression of the sensor when inserted adjacent to a heart-specific enhancer, possibly regulating the adjacent Myocd gene (Wang et al. 2001). The eye expression shown on the representative transgenic embryo (*) is ectopic. (E) The sensor, inserted next to Znf503 (McGlinn et al. 2008), showed expression in the posterior forelimb, which overlapped with the activity of a distant enhancer (fl, blue/white arrow). The enhancer was also active in the neural tube, but the sensor was not expressed in that region.
Enhancer activity is not gene-centric and broadly distributed
To refine this analysis, we made use of enhancers for which transgenic activity had been previously documented in E11.5 embryos, mostly from the Vista Enhancer database (Visel et al. 2007). We carefully compared the spatial activity of these enhancers with that of the regulatory sensor when inserted in their vicinity (Fig. 2B–E). Given the highly specific patterns of the different insertions and enhancers, this offered much greater confidence in a direct relationship between matching patterns. We defined pairs as “concordant” when the domain of enhancer activity was included in the one shown by the regulatory sensor; as “divergent” or “inactive,” when the regulatory sensor was expressed elsewhere or not expressed (Supplemental Table 3). As shown in Figure 2B, all insertions positioned in close proximity (<10 kb) of an enhancer showed expression patterns highly concordant with its autonomous activity. This ratio decreased with increasing distance, but even at 100–200 kb, a third of insertions–enhancer pairs were concordant, compared with <7% for random associations of enhancers and insertions (Fig. 2B) (Fisher's exact test, P < 3.305 × 10−6). Expression of our regulatory sensor was not dependent on its orientation (Supplemental Fig. 2).
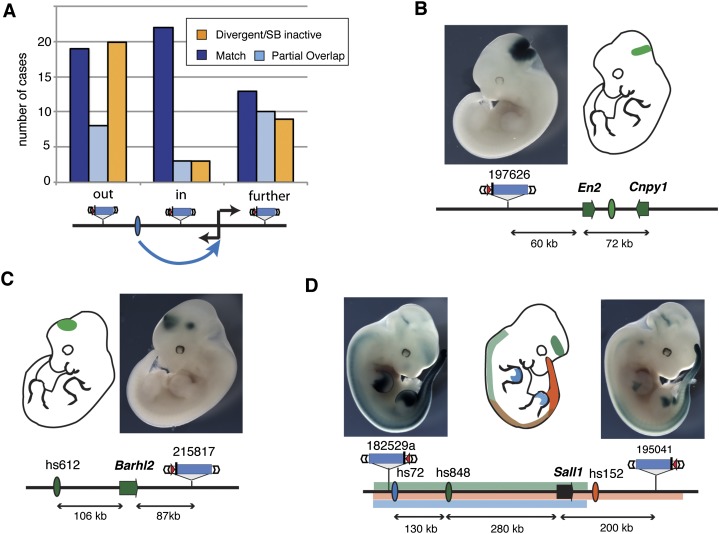
We then examined whether enhancers distributed their activities preferentially toward their endogenous target gene. For this purpose, we considered a subset of enhancers for which we could confidently assign a—putative—target gene, using in situ gene expression data from the literature. We categorized these 107 enhancer-gene-insertion triplets (corresponding to 33 genomic loci) based on the relative order of their components, and determined whether the inserted sensor showed complete, partial, or no overlapping expression with the enhancer/gene pair (Fig. 3; Supplemental Table 4). Insertions located between an enhancer and its target gene showed overlap with the reported enhancer activity more frequently than insertions located either opposite to the gene or beyond the transcriptional start site (Fisher's exact test, P = 0.001457 and P = 0.04238, respectively). However, about half of the insertions located opposite the target gene or beyond its promoter showed an expression pattern corresponding to the associated enhancer (Fig. 3), indicating that enhancer activity is not exclusive to the promoter of their target gene. Furthermore, given the relatively small sample size, it is possible that the weak directionality of enhancer activity that we observed is due to the different distances associated with the different categories.
Figure 3.
Non-gene-centric enhancer activities are detected across large distances. (A) The number of insertions that correctly (blue) or partially (light blue) reported the activity of a neighboring tissue-specific enhancer, or showed a different activity (orange). Insertions were grouped depending on their position relative to the enhancer/target gene, as shown below the chart. (B–D) Examples of expression detected with the regulatory sensor (photos) in non-gene-centered situations. Gene (arrows) and enhancer (ovals) activities are color-coded and shown on the embryo outline. (B) An insertion between the En2 and Cnpy1 genes matches their expression at the mid/hindbrain junction (Jukkola et al. 2006), as well as the activity of an enhancer on the far side of En2 (see also Supplemental Fig. 8) (C) Barhl2 expression in the midbrain and diencephalon requires remote enhancers (Saba et al. 2005), and a diencephalon enhancer (hs612) (Visel et al. 2007) is present upstream of this gene. Enhancer activity extends to a downstream insertion. (D) Sall1 gene expression is controlled by multiple enhancers spread in the two surrounding gene deserts, and insertions flanking the gene display overlapping expression patterns.
Overall, these multiple analyses show that enhancers act on their environment broadly and in a largely undiscriminating manner.
Extended enhancer activity results in large regulatory domains
Consistent with these findings, we noted that adjacent insertions frequently had similar expression, even when hundreds of kilobases apart. Large domains of co-regulation have been reported before, usually around developmental genes (Spitz et al. 2003; Zuniga et al. 2004; Kikuta et al. 2007), but with our extensive survey of the mouse genome, we could expand this list and refine the extent of such domains. Importantly, since the regulatory sensor is driven by the same promoter at each insertion site, changes in expression are not due to differently responsive promoters, but really outline the limits of enhancer action. We cannot formally exclude that co-expression of the sensor at different positions results from activation by different enhancers with overlapping activities, like those discovered around developmental genes (Carvajal et al. 2001; Uchikawa et al. 2003; Hong et al. 2008; Marinić et al. 2013; Visel et al. 2013). But individual enhancers usually have distinct spatial activities (Visel et al. 2007) and therefore, the spatially restricted expression patterns that we use to define co-expression provide confidence for genuine co-regulation. Supporting this further, insertions into loci where enhancers have been meticulously mapped allowed us to assess that they respond to the same enhancer(s) at multiple distant positions (e.g., Shh, Supplemental Fig. 3; Hoxd, Supplemental Fig. 4).
We found 311 chromosomal intervals defined by the presence of two or more insertions within <2 Mb. We assigned these intervals to different categories, depending on the expression patterns of the insertions (Fig. 4A–C; Supplemental Figs. 3–5; Supplemental Table 5). We defined regulatory domains (RDs; 46 regions; size from 3.8 kb to 2.1 Mb; median size: 359 kb; total combined length: 22.3 Mb) as the largest possible interval containing multiple insertions with shared expression. This is a conservative definition, and RDs likely extend beyond the insertions that define them. RDs are found on all chromosomes, except possibly Y, where we did not obtain insertions with expression. Within RDs, we occasionally saw quantitative variation or no detectable expression of the sensor at different positions. However, we never observed insertions with divergent expression between two insertions with the same activity.
Figure 4.
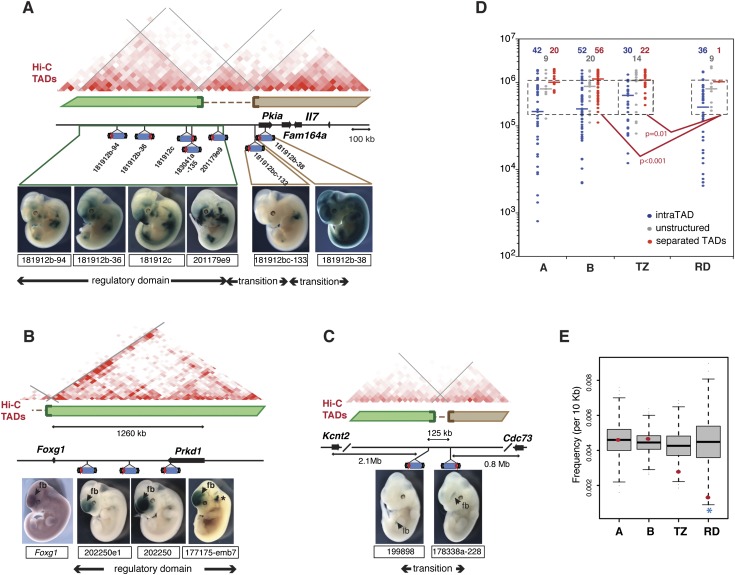
Extended domains of co-regulation correlate with the subdivision of the genome into topological domains. (A–C) Outlines of loci, with genes displayed as arrows and insertions as drawings of the transposon. Regulatory domains and transition zones are labeled, TADs (identified by Hi-C in mouse ES cells) are indicated by green and brown bars and unstructured regions by dashed lines. Hi-C interaction frequencies are represented as a two-dimensional heat map (from Dixon et al. 2012). (A) Multiple insertions in the chr3:7.3–8.3M interval outlined an extended regulatory domain characterized by shared expression in the facial and trunk mesenchyme, and in neural crest derivatives. This domain extends into the adjacent unstructured region (insertion 201179e9), but two telomeric insertions, located in a different TAD, showed different patterns (the proximal limb expression of 181912bc-133 is anterior, whereas insertions in the flanking RD have a more medial expression), defining two transition zones. (B) Multiple insertions in the vicinity of the Foxg1 gene display the typical forebrain (fb) expression of the gene (adapted from Chen et al. 2013, with permission from Elsevier © 2013). Expression in the ear (*) is due to another insertion also present in 177175-emb7. The regulatory domain defined by the gene and the insertions is contained within a single TAD. A more detailed version of this panel is shown in Supplemental Figure 5E. (C) Two insertions in a gene desert between Kcnt2 and Cdc73 are divergently expressed in the limb bud (lb) and forebrain (fb), delineating a transition zone. This coincides with the respective insertions being located in different TADs. (D) Size distribution (y-axis) and relationship with TADs (color-coded) of functionally defined intervals. Only a single regulatory domain (RD) overlaps a TAD boundary. Random permutation of regions 200 kb–1 Mb in length (boxed area), where the size distribution of the different functional categories is not statistically different (Kolmogorov-Smirnov test, P = 0.8560 for RD vs. TZ + A + B; P = 0.9240 for RD vs. TZ), showed that RDs are significantly underrepresented in the “separated TADs” category. (E) Unlike control regions (classes A, B) and transition zones (TZ), RDs show depletion in topological boundaries compared with equally sized, randomly distributed fragments. Gray box-plots represent the results of randomization; red dots, the position of the real data. The depletion is statistically significant (P = 0.009), as indicated by the blue star.
Next, we defined transition zones (TZs) as regions separating insertions with distinct expression profiles (66 intervals, ranging from 14 kb to 1.9 Mb; median size: 734 kb; total combined length: 53.8 Mb). Other types of intervals (class A: two or more insertions without detectable expression; class B: one insertion with expression and one with no expression) were also annotated. B regions may be a type of transition, but we considered them separately, since several local features may lead to inactivity of the sensor (Ruf et al. 2011).
Regulatory domains are included in topologically associating domains
Next, we looked to see whether our operational subdivision of the genome matched any structural features of the genome. To this end, we compared RDs with the largely cell-invariant self-associating “topologically associating domains” (TADs) identified by Hi-C (Dixon et al. 2012). We found that the great majority of RDs (78%) were contained within one TAD (Fig. 4D). The remaining ones extended into flanking unstructured regions (for which Hi-C data did not highlight specific compartmentalization). In only one case, two adjacent insertions with partially overlapping—but not identical—patterns of expression were found in distinct adjacent TADs. Similarly, insertions mirroring the expression patterns of adjacent but remote genes were also found within the same TAD (Supplemental Fig. 5C–E). In contrast, one third of TZs, including relatively short regions in gene-deserts, were located in different TADs (Fig. 4C; Supplemental Fig. 6). Since, on average, RDs were smaller than TZs, we performed several analyses to verify the significance of this different distribution relative to TADs. We randomly permutated RD, TZ, A, and B regions, and found it was significant (P < 0.05) that RDs almost never overlapped two or more TADs, unless we considered only extreme size ranges (below 400 kb; above 1.5 Mb). In particular, for intervals between 200 kb and 1 Mb, a range where the size distribution of the RD, TZ, A, and B regions was not significantly different, the altered distribution of RDs and TZs relative to TADs was highly significant (Fig. 4D). We also calculated the density of TAD ends for the real intervals and for 1000 random distributions of intervals of the same size in the genome, and found that RDs, but not the other categories, showed a significant depletion of TAD ends (Fig. 4E).
Distribution of regulatory activities and insulators
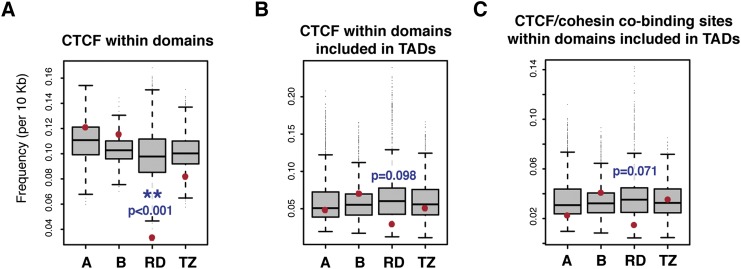
Next, we compared how insulators or elements with enhancer-blocking activities are distributed between the four types of operationally defined domains. We considered CTCF sites, SINE B1 and SINE B2 elements, as well as the TSS of protein-coding and noncoding genes from the RefSeq collection (Supplemental Table 6). All four elements were clearly depleted from RDs (Fig. 5A; Supplemental Fig. 7), whereas type A and B control regions never showed depletion, and TZs showed no, or far less depletion. This depletion was largely maintained, even when we considered potentially confounding factors, such as the clustering of CTCF sites and TSS (Kim et al. 2007; Shen et al. 2012) or the overlap of CTCF sites and SINE B2 elements (Schmidt et al. 2012) (Supplemental Fig. 7). The proportion of RDs containing at least one cell-invariant CTCF site was also lower than for TZ (39% compared with 70%), consistent with the proposed contribution of this protein in organizing regulatory interactions (Phillips and Corces 2009). However, since TAD boundaries are enriched for the different elements associated with insulator activity (Dixon et al. 2012), we also restricted our analysis to intervals located within TADs. In this case, we still found lower than expected density of TSS and SINE B2. However, the depletion of CTCF binding sites and SINE B1 X35S in RDs was no longer statistically significant (Fig. 5B; Supplemental Fig. 7), suggesting that it correlated with the topological, rather than regulatory subdivision of the genome.
Figure 5.
Depletion of CTCF and cohesin in regulatory domains reflects the topological segmentation of the genome. Tissue-invariant CTCF sites (bound in more than nine tissues) are significantly depleted (but not absent) in regulatory domains when considering all functionally defined regions (A). However, the statistical significance of this depletion becomes marginal if the comparison is limited to intervals included in TADs, and not compared against the overall genome (B). Constitutive cohesin complex-binding sites (data not shown), or CTCF/cohesin co-occupied regions (C) also showed only a slightly reduced density in RDs, when compared with the part of the genome that is included in TADs.
As CTCF (Phillips and Corces 2009), together with cohesin complexes (Hadjur et al. 2009; Kagey et al. 2010; Merkenschlager and Odom 2013), has been proposed to have a major role in organizing regulatory interactions we examined their relationship to RDs in more detail, using available data sets (Supplemental Table 6). In agreement with previous analyses, we found a high degree of overlap between CTCF and cohesin-binding across mouse cell-types and tissues (Parelho et al. 2008; Wendt et al. 2008; Remeseiro et al. 2012). In several cases, cohesin and CTCF-bound regions lie within regulatory TZs, often also corresponding to TAD boundaries, an observation consistent with their proposed role (Phillips-Cremins et al. 2013).
However, even if slightly less abundant in RDs (Fig. 5C), many CTCF and cohesin binding sites are also interspersed within RDs, between co-expressed genes, insertions, and their associated enhancers (Fig. 6; Supplemental Figs. 4, 5, 8). We noted that RDs can also be further subdivided in more “specialized” ones, each characterized by expression specificities additional to the ones defining the RD (Fig. 5C; Supplemental Fig. 5A,E). Some of these subdivisions, with the current resolution offered by the available insertions, might be outlined by the presence of CTCF/cohesin sites (Fig. 5). However, by and large, it appeared difficult to match the mere distribution of the CTCF/cohesin sites with the distribution of regulatory activities, highlighting that, within TADs, the binding of cohesin/CTCF, defined by chromatin-immunoprecipitation, may not be a sufficient indicator of regulatory boundaries.
Figure 6.
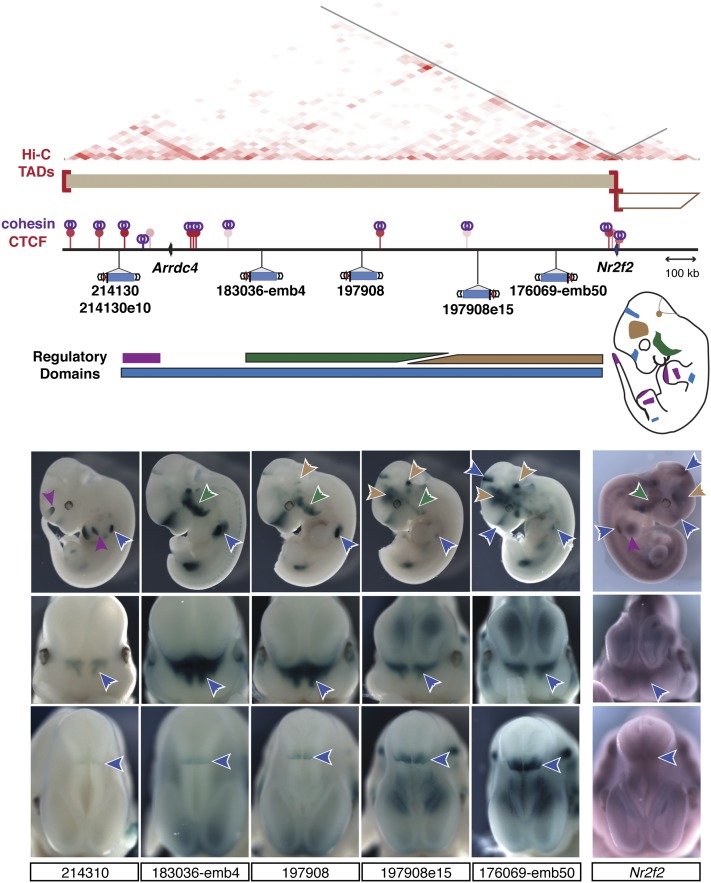
CTCF and cohesin sites are interspersed in regulatory domains. Schematic representation of the large topological/regulatory domain on chr7:75.5M–77.8M. The two genes (Arrdc4 and Nr2f2) are represented as arrows. The corresponding TAD is represented by a two-dimensional heat map (Dixon et al. 2012). Several constitutive CTCF sites (red lollipop, color intensity proportional to cell invariance), largely co-bound by cohesin (purple rings), are interspersed in this interval. Insertions spread across almost 2 Mb showed highly overlapping patterns in the proximal limb (blue arrow, top), face (blue arrow, middle), and at the midbrain/diencephalon boundary (blue arrow, bottom), forming a large regulatory domain. This large domain can be subdivided into smaller tissue-specific landscapes (green, purple, and brown) based on expression patterns displayed by only a subset of the insertions and quantitative differences in LacZ staining intensity. These different regulatory influences overlap with Nr2f2 expression, detected by whole-mount in situ hybridization. In situ hybridization with Arrdc4 probes did not reveal specific expression in E11.5 embryos. Embryos 183036-emb4 and 176069-emb50 were described previously (Ruf et al. 2011).
Discussion
In vertebrates, since many critical enhancers lie at considerable distances from the genes they influence, ensuring and controlling proper interactions between regulatory elements and promoters is essential (Bulger and Groudine 2011; Splinter and de Laat 2011; Williamson et al. 2011). Our mechanistic understanding of this process is derived from studies carried out on a limited number of model loci (Gaszner and Felsenfeld 2006; Simonis et al. 2006; Vernimmen et al. 2007; Jhunjhunwala et al. 2008; Amano et al. 2009; Montavon et al. 2011; Tena et al. 2011; Marinić et al. 2013). Here, we expanded these by probing the regulatory landscape of the entire mouse genome with a transposable, naïve lacZ sensor. This large-scale exploration of the genomic regulatory architecture identified—in an operational manner—the widespread presence of large regulatory domains (RDs), within which the sensor displayed highly similar expression patterns at multiple distant positions. These RDs largely overlapped with TADs, the sub-megabase-sized self-interacting intervals defined by chromosomal conformation capture analysis (Dixon et al. 2012; Nora et al. 2012). Insertions located in adjacent TADs almost systematically reported distinct regulatory activities, providing direct support for the suggested role of TADs as the basic building blocs of genomic regulatory architecture (Dixon et al. 2012; Hou et al. 2012; Nora et al. 2012; Gibcus and Dekker 2013). RDs are usually gene poor and shared several other features with TADs, such as a depletion of various elements (SINE B2 repeats, constitutive CTCF-binding sites).
Our operational approach also shed some light on the organization of these domains. Firstly, despite the overall inclusion of regulatory domains into TADs, the positions of the relative transitions were not always exactly superimposed, similarly to chromatin domains, which do not exactly correlate with TADs (Hou et al. 2012). These differences may partially arise from the low resolution of Hi-C, which can locate topological transitions with only limited precision (20 kb). Furthermore, topological transitions may not necessarily constitute absolute barriers, but act more like dampers (Andrey et al. 2013).
Within TADs, we found that enhancer activities are broadly distributed, and not targeted to specific regions (i.e., proximity of gene promoters or TAD borders). From their discovery, enhancers have been shown to act irrespectively of their orientation (Banerji et al. 1981). Genome-wide studies have further shown that enhancers can be found both 3′ and 5′ of their endogenous target genes (The ENCODE Project Consortium 2012; Li et al. 2012; Shen et al. 2012). Our data further stresses that orientation independence is an intrinsic property of enhancers in their normal context: in addition to controlling of target genes, enhancers generally act pervasively throughout their regulatory domains.
This broad distribution of enhancer activities along large domains has several implications. It may account for the transcriptional “ripple” effect observed upon growth-factor stimulation (Ebisuya et al. 2008) and bystander gene activation (Spitz et al. 2003; Cajiao et al. 2004; Zuniga et al. 2004). Furthermore, widespread enhancer activities may have been evolutionarily advantageous for genes brought into a new neighborhood by chromosomal rearrangements (Cande et al. 2009; De et al. 2009) or retrotransposition (Vinckenbosch et al. 2006), or for emerging lncRNAs (Ponting et al. 2009; Kutter et al. 2012), facilitating the acquisition of new expression domains.
Constraining enhancer activity through the formation of distinct topological compartments may contribute to gene regulation in two important ways. It may help specify gene-enhancer interactions, and restrict ectopic “enhancer adoption” to accidental disruption of existing topologies, for example, through chromosomal rearrangements (Kokubu et al. 2003; Spitz et al. 2003; Niedermaier et al. 2005; Gostissa et al. 2009; Kantaputra et al. 2010; Marinić et al. 2013). Alternatively, it may help integrate the activity of multiple regulatory elements spread along large intervals (Carvajal et al. 2001; Uchikawa et al. 2003; Montavon et al. 2011; Li et al. 2012; Sanyal et al. 2012; Shen et al. 2012; Delpretti et al. 2013; Marinić et al. 2013; Visel et al. 2013) into the coherent regulatory units that have been described as regulatory archipelagos, holo-enhancers or chromatin-hubs (Palstra et al. 2003; Montavon et al. 2011; Marinić et al. 2013). Our observation that in intact endogenous loci, randomly inserted sensors report the complete integrated output provided to normal target genes (and do not decompose it into the individual activities of the closest enhancers) (e.g., Hoxd, Supplemental Fig. 4; Foxg1, Fig. 4B; Twist1, Ruf et al. 2011; Birnbaum et al. 2012), further supports the notion that regulatory control is exerted by coordinated, and not individual action, of enhancers. It indicates that this integration is orchestrated at the level of the regulatory domain, not at endogenous gene promoters (Marinić et al. 2013).
Along a given regulatory domain, the expression level detected by the sensor can vary quantitatively and reveal subdomains, including few positions apparently refractory to activation. We speculate that these “cold-spots” within otherwise permissive domains may shield genes from the influence of surrounding enhancers (Marinić et al. 2013), adding to other mechanisms of specificity such as core promoter sequences (Ohtsuki et al. 1998). This fine-scale organization of activities may be due to the positions of the corresponding enhancers, to local epigenetic modifications (Akhtar et al. 2013; Arnold et al. 2013), or to different tissue-specific three-dimensional organization within domains. Indeed, restructuring of physical interactions within TADs has been shown during lineage-commitment (Phillips-Cremins et al. 2013) and in different cell-types (Dixon et al. 2012). We suggest that such changes in local interactions may correlate with the different subregulatory domains that we observed within TADs. It will be an exciting avenue to explore what proteins (Hadjur et al. 2009; Kagey et al. 2010; Remeseiro et al. 2012; Aragon et al. 2013; Merkenschlager and Odom 2013) and nuclear substructures (Bickmore and van Steensel 2013; Gibcus and Dekker 2013; Meuleman et al. 2013) may be involved. Even though we found frequent co-occurrence of CTCF-bound regions with transitions between regulatory domains, our current analyses also highlight the difficulty of inferring the structure of these domains from the mere presence of CTCF/cohesin binding sites detected by chromatin-immunoprecipitation. This reinforces the growing perception that CTCF may have a versatile role (Handoko et al. 2011; Sanyal et al. 2012; Phillips-Cremins et al. 2013), perhaps different within topological domains and at their borders, and that analyzing these activities in the context of a 3D genome will be essential (Handoko et al. 2011; DeMare et al. 2013). Our data also emphasizes the need of direct functional approaches, as the one developed here, to map regulatory interactions and to compare them with physical conformations or chromatin maps. The confrontation of these approaches will be essential to understand the mechanistic basis and principles underlying the organization of the genome. Ultimately, such an understanding will be crucial to predict whether and how variants (such as structural variants) affect topological and regulatory organization, and consequently influence gene expression and phenotype (Weischenfeldt et al. 2013).
Methods
Mouse lines and embryos with SBlac insertions
Insertion sites of the transposon were generated, mapped, and lacZ expression analysis was performed as described previously (Ruf et al. 2011). Information on insertions is found in Supplemental Table 1 and details are on the TRACER website (tracerdatabase.embl.de) (Chen et al. 2013). Whole-mount in situ hybridizations were carried out as described in Ruf et al. (2011). Templates for mRNA antisense probe synthesis were described in Spitz et al. (2003) (Hoxd13), Ruf et al. (2011) (Sall1), or produced by PCR on mouse cDNA using the following primers (Foxg1_2F: GCCAAGCTGGCCTTTAAGC; Foxg1_2R: ATTCTCCCACATTGCACCTC; Nr2f2_3Sp6: CATTTAGGTGACACTATAGCCACATGGGCTACATCAGAC; Nr2f2_5: GGGCGGAGGAACCTGAGCTACAC; Hand2_F: AGGACTCAGAGCATCAACAGC; Hand2-R: AGCGGATGCTCAAAGGTG).
Mouse experiments were conducted in accordance with the principles and guidelines in place at the European Molecular Biology Laboratory, as defined and overseen by its Institutional Animal Care and Use Committee, in accordance with the European Convention 18/3/1986 and Directives 86/609/EEC and 2010/63/EU.
Genomic data sets and resources used for the study
Genomic data sets were obtained from public sources and are summarized in Supplemental Table 6.
SB-EP300 comparison
We considered SB insertions with expression in heart, limb, midbrain, or forebrain, where EP300 binding data has been generated (Visel et al. 2009; Blow et al. 2010), and merged insertions with the same expression pattern <5 kb apart into one (Supplemental Table 2). In a given tissue, we calculated the enrichment of insertions with expression in that tissue, compared with the same number of randomly selected insertions with no expression in that tissue (200 randomizations). Enrichment was calculated for increasing distances (with steps of 5000 bp from 0 to 1 Mb, provided more than five insertions were found within). To test whether this enrichment was specific to the relevant tissue, we repeated the analysis using insertions expressed in other tissues or not expressed. For this, we removed EP300 sites if they were within 10 kb of a EP300 site in the first tissue considered, to reduce the possible confounding signal arising from clusters of different tissue-specific EP300 sites (Visel et al. 2009).
Comparison of insertions, autonomous enhancer activity, and endogenous genes
We complemented the set of enhancers active at E11.5 from the VISTA Enhancer Browser (Visel et al. 2007) with additional enhancers from the literature. We visually inspected expression overlap between enhancer–insertion pairs (where insertions were within 200 kb of these enhancers; Supplemental Table 3), and annotated pairs as concordant, discordant, or inactive. As random control data sets (Supplemental Table 3), we first performed random permutation of the positions of the insertions and enhancers, and extracted new adjacent pairs. Second, we randomly drew insertion—enhancer pairs and compared their expression patterns.
For the enhancer-regulatory sensor-gene triplets, we compared the expression of characterized enhancers to reported in situ hybridization gene expression of either the first two flanking genes or genes <2 Mb away using gene expression resources (Supplemental Table 6). We considered sensor insertions lying within the same genomic interval, and annotated their expression as concordant, partially overlapping or divergent (no or different expression) with the enhancer-gene pair. These enhancer-gene-insertion triplets were then categorized depending on the position of the insertion relative to the enhancer and gene TSS. For neighboring insertions that showed the same annotation/relative position for a given enhancer, we kept only one triplet. Informative triplets are listed in Supplemental Table 4.
Defining regions with characteristic expression patterns
We functionally defined different classes of genomic intervals by comparing the expression of adjacent insertions <2 Mb apart. Intervals where both insertions showed no expression were annotated as class A. Class B was formed by pairs where one insertion was expressed whereas the other was not. Intervals defined by two insertions with discordant (nonoverlapping) expression patterns formed transition zones. Intervals where insertions showed overlapping lacZ expression were grouped as regulatory domains (RDs). To define a RD, the observed spatial pattern had to be identical, not only correspond to the same broad anatomical domain: Patterns as in Figure 1B are considered divergent. In cases where multiple insertions within a 2-Mb window were concordantly expressed, we considered the most centromeric and telomeric insertions as the boundaries of the RD, but only if no insertions in between were discordantly expressed. Thus, we merged adjacent RDs with the same activity together, sometimes including small B regions. Similarly, we determined the minimal extent of transition zones as the smallest region flanked by divergently expressed insertions (i.e., where none of the expression patterns overlapped), even if they were disrupted by insertions with no expression. We did not further group insertion pairs, where one or both insertions are not expressed, except when three or more consecutive insertions showed no expression, and one pair was <10 kb apart. The extended annotated regions are listed in Supplemental Table 5.
Comparison to topological domains and other genomic landmarks
We compared the occurrence of certain features in our functionally defined regions to a random model. To estimate the significance (P-value) of the frequency of certain features (e.g., topological boundaries, CTCF sites, SINE B2 elements) for each category (i.e., all A, B, RD, and TZ domains, or those found intra-TAD only), we applied bootstrap following recommendations from Phipson and Smyth (2010). For details, see Supplemental Note 1. For TAD boundaries we also performed random permutation, as described in the text and Supplemental Note 1. For the analysis we used the data sets described in Supplemental Table 6.
Data access
All insertions generated for this study are documented in the TRACER database (Chen et al. 2013) (http://tracerdatabase.embl.de; or http://www.ebi.ac.uk/panda-srv/tracer/index.php).
Supplementary Material
Acknowledgments
We thank Angel-Carlos Roman for SINEB1-X35S coordinates. We thank the members of the EMBL Laboratory Animal Resources Facility for animal welfare and husbandry, especially Silke Feller and Michaela Wesch. We thank Chao-Kung Chen for kindly keeping the TRACER database up and running. For computational analysis we received invaluable support from Charles Girardot and the EMBL Genome Biology Computational Support Group. We also thank members of the Spitz laboratory and colleagues from EMBL for helpful discussion and sharing reagents, and Laura Panavaite for providing the in situ data for Hand2. O.S. and V.V.U. were supported by PhD fellowships from the Louis-Jeantet Foundation and Jeff Schell Darwin Trust, respectively. T.T. was supported successively by postdoctoral fellowships awarded by the Uehara Memorial Foundation and the Japan Society for the Promotion of Science. W.S. was supported by an EMBL Interdisciplinary Postdoc (EIPOD) fellowship under Marie Curie Actions COFUND. This work was supported by the European Molecular Biology Laboratory, the European Commission-FP7 (grant Health 223210/CISSTEM), and the Human Frontier Science Program (grant RGY0081/2008-C) (F.S.).
Author contributions: F.S. and O.S. designed the experiments. O.S., V.V.U., T.T., S.R., S.N., and W.S. performed the experiments. O.S., L.E., and F.S. analyzed the data. O.S. and F.S. wrote the manuscript with input from the other authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.163519.113.
References
- Akhtar W, de Jong J, Pindyurin AV, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels LFA, van Lohuizen M, van Steensel B 2013. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell 154: 914–927 [DOI] [PubMed] [Google Scholar]
- Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T 2009. Chromosomal dynamics at the Shh locus: Limb bud-specific differential regulation of competence and active transcription. Dev Cell 16: 47–57 [DOI] [PubMed] [Google Scholar]
- Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D 2013. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340: 1234167. [DOI] [PubMed] [Google Scholar]
- Aragon L, Martinez-Perez E, Merkenschlager M 2013. Condensin, cohesin and the control of chromatin states. Curr Opin Genet Dev 23: 204–211 [DOI] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Boryń ŁM, Rath M, Stark A 2013. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339: 1074–1077 [DOI] [PubMed] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W 1981. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell 27: 299–308 [DOI] [PubMed] [Google Scholar]
- Bickmore WA, van Steensel B 2013. Genome architecture: Domain organization of interphase chromosomes. Cell 152: 1270–1284 [DOI] [PubMed] [Google Scholar]
- Birnbaum RY, Clowney EJ, Agamy O, Kim MJ, Zhao J, Yamanaka T, Pappalardo Z, Clarke SL, Wenger AM, Nguyen L, et al. 2012. Coding exons function as tissue-specific enhancers of nearby genes. Genome Res 22: 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2010. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet 42: 806–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajiao I, Zhang A, Yoo EJ, Cooke NE, Liebhaber SA 2004. Bystander gene activation by a locus control region. EMBO J 23: 3854–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VC, Levine M 2003. Long-range enhancer-promoter interactions in the Scr-Antp interval of the Drosophila Antennapedia complex. Proc Natl Acad Sci 100: 9878–9883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cande JD, Chopra VS, Levine M 2009. Evolving enhancer-promoter interactions within the tinman complex of the flour beetle, Tribolium castaneum. Development 136: 3153–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal JJ, Cox D, Summerbell D, Rigby PW 2001. A BAC transgenic analysis of the Mrf4/Myf5 locus reveals interdigitated elements that control activation and maintenance of gene expression during muscle development. Development 128: 1857–1868 [DOI] [PubMed] [Google Scholar]
- Chen C-K, Symmons O, Uslu VV, Tsujimura T, Ruf S, Smedley D, Spitz F 2013. TRACER: A resource to study the regulatory architecture of the mouse genome. BMC Genomics 14: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K 2012. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res 22: 490–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Teichmann SA, Babu MM 2009. The impact of genomic neighborhood on the evolution of human and chimpanzee transcriptome. Genome Res 19: 785–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpretti S, Montavon T, Leleu M, Joye E, Tzika A, Milinkovitch M, Duboule D 2013. Multiple enhancers regulate Hoxd genes and the Hotdog LncRNA during cecum budding. Cell Reports 5: 137–150 [DOI] [PubMed] [Google Scholar]
- DeMare LE, Leng J, Cotney J, Reilly SK, Yin J, Sarro R, Noonan JP 2013. The genomic landscape of cohesin-associated chromatin interactions. Genome Res 23: 1224–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisuya M, Yamamoto T, Nakajima M, Nishida E 2008. Ripples from neighbouring transcription. Nat Cell Biol 10: 1106–1113 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G 2006. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7: 703–713 [DOI] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J 2013. The hierarchy of the 3D genome. Mol Cell 49: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostissa M, Yan CT, Bianco JM, Cogné M, Pinaud E, Alt FW 2009. Long-range oncogenic activation of Igh-c-myc translocations by the Igh 3′ regulatory region. Nature 462: 803–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, Yuk D-I, Tsung EF, Cai Z, et al. 2004. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science 306: 2255–2257 [DOI] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M 2009. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CWH, Ye C, Ping JLH, Mulawadi F, et al. 2011. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 43: 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J-W, Hendrix DA, Levine MS 2008. Shadow enhancers as a source of evolutionary novelty. Science 321: 1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie K, Yusa K, Yae K, Odajima J, Fischer SEJ, Keng VW, Hayakawa T, Mizuno S, Kondoh G, Ijiri T, et al. 2003. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol Cell Biol 23: 9189–9207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Li L, Qin ZS, Corces VG 2012. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell 48: 471–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJM, Grosveld FG, Knoch TA, Murre C 2008. The 3D structure of the immunoglobulin heavy-chain locus: Implications for long-range genomic interactions. Cell 133: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukkola T, Lahti L, Naserke T, Wurst W, Partanen J 2006. FGF regulated gene-expression and neuronal differentiation in the developing midbrain–hindbrain region. Dev Biol 297: 141–157 [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantaputra PN, Klopocki E, Hennig BP, Praphanphoj V, Le Caignec C, Isidor B, Kwee ML, Shears DJ, Mundlos S 2010. Mesomelic dysplasia Kantaputra type is associated with duplications of the HOXD locus on chromosome 2q. Eur J Hum Genet 18: 1310–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engström PG, Fredman D, Akalin A, Caccamo M, Sealy I, Howe K, et al. 2007. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res 17: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubu C, Wilm B, Kokubu T, Wahl M, Rodrigo I, Sakai N, Santagati F, Hayashizaki Y, Suzuki M, Yamamura K-I, et al. 2003. Undulated short-tail deletion mutation in the mouse ablates Pax1 and leads to ectopic activation of neighboring Nkx2-2 in domains that normally express Pax1. Genetics 165: 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, Ponting CP, Odom DT, Marques AC 2012. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet 8: e1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. 2012. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148: 84–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Geurts AM, Yae K, Srinivasan AR, Fahrenkrug SC, Largaespada DA, Takeda J, Horie K, Olson WK, Hackett PB 2005. Target-site preferences of Sleeping Beauty transposons. J Mol Biol 346: 161–173 [DOI] [PubMed] [Google Scholar]
- Marinić M, Aktas T, Ruf S, Spitz F 2013. An integrated holo-enhancer unit defines tissue and gene specificity of the Fgf8 regulatory landscape. Dev Cell 24: 530–542 [DOI] [PubMed] [Google Scholar]
- McGlinn E, Richman JM, Metzis V, Town L, Butterfield NC, Wainwright BJ, Wicking C 2008. Expression of the NET family member Zfp503 is regulated by hedgehog and BMP signaling in the limb. Dev Dyn 237: 1172–1182 [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, Odom DT 2013. CTCF and cohesin: Linking gene regulatory elements with their targets. Cell 152: 1285–1297 [DOI] [PubMed] [Google Scholar]
- Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B 2013. Constitutive nuclear lamina–genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res 23: 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D 2011. A regulatory archipelago controls hox genes transcription in digits. Cell 147: 1132–1145 [DOI] [PubMed] [Google Scholar]
- Niedermaier M, Schwabe GC, Fees S, Helmrich A, Brieske N, Seemann P, Hecht J, Seitz V, Stricker S, Leschik G, et al. 2005. An inversion involving the mouse Shh locus results in brachydactyly through dysregulation of Shh expression. J Clin Invest 115: 900–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM 2003. Scanning human gene deserts for long-range enhancers. Science 302: 413. [DOI] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Dekker J, Heard E 2013. Segmental folding of chromosomes: A basis for structural and regulatory chromosomal neighborhoods? Bioessays 35: 818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S, Levine M, Cai HN 1998. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev 12: 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C-T, Corces VG 2011. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat Rev Genet 12: 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Nobrega MA, Hardison RC, Miller W, Stubbs L 2004. Evolution and functional classification of vertebrate gene deserts. Genome Res 15: 137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra R-J, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W 2003. The β-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35: 190–194 [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG 2009. CTCF: master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, Ong C-T, Hookway TA, Guo C, Sun Y, et al. 2013. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153: 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Smyth GK 2010. Permutation P-values should never be zero: Calculating exact P-values when permutations are randomly drawn. Stat Appl Genet Mol Biol 9 doi: 10.2202/1544-6115.1585 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W 2009. Evolution and functions of long noncoding RNAs. Cell 136: 629–641 [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Gómez-López G, Pisano DG, Losada A 2012. A unique role of cohesin-SA1 in gene regulation and development. EMBO J 31: 2090–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S, Symmons O, Uslu VV, Dolle D, Hot C, Ettwiller L, Spitz F 2011. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat Genet 43: 379–386 [DOI] [PubMed] [Google Scholar]
- Saba R, Johnson JE, Saito T 2005. Commissural neuron identity is specified by a homeodomain protein, Mbh1, that is directly downstream of Math1. Development 132: 2147–2155 [DOI] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J 2012. The long-range interaction landscape of gene promoters. Nature 489: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Wilson MD, Ballester B, Goncalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT 2012. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148: 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472 [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38: 1348–1354 [DOI] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D 2003. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113: 405–417 [DOI] [PubMed] [Google Scholar]
- Splinter E, de Laat W 2011. The complex transcription regulatory landscape of our genome: Control in three dimensions. EMBO J 30: 4345–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons O, Spitz F 2013. From remote enhancers to gene regulation: Charting the genome's regulatory landscapes. Philos Trans R Soc Lond B Biol Sci 368: 20120358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena JJ, Alonso ME, de la Calle-Mustienes E, Splinter E, de Laat W, Manzanares M, Gómez-Skarmeta JL 2011. An evolutionarily conserved three-dimensional structure in the vertebrate Irx clusters facilitates enhancer sharing and coregulation. Nat Commun 2: 310. [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H 2003. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev Cell 4: 509–519 [DOI] [PubMed] [Google Scholar]
- Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR 2007. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J 26: 2041–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinckenbosch N, Dupanloup I, Kaessmann H 2006. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci 103: 3220–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA 2007. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res 35: D88–D92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. 2013. A high-resolution enhancer atlas of the developing telencephalon. Cell 152: 895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN 2001. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105: 851–862 [DOI] [PubMed] [Google Scholar]
- Weischenfeldt J, Symmons O, Spitz F, Korbel JO 2013. Phenotypic impact of genomic structural variation: Insights from and for human disease. Nat Rev Genet 14: 125–138 [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- Williamson I, Hill RE, Bickmore WA 2011. Enhancers: From developmental genetics to the genetics of common human disease. Dev Cell 21: 17–19 [DOI] [PubMed] [Google Scholar]
- Zuniga A, Michos O, Spitz F, Haramis A-PG, Panman L, Galli A, Vintersten K, Klasen C, Mansfield W, Kuc S, et al. 2004. Mouse limb deformity mutations disrupt a global control region within the large regulatory landscape required for Gremlin expression. Genes Dev 18: 1553–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.