Abstract
Sequence-specific DNA-binding proteins including transcription factors (TFs) are key determinants of gene regulation and chromatin architecture. Formaldehyde cross-linking and sonication followed by Chromatin ImmunoPrecipitation (X-ChIP) is widely used for profiling of TF binding, but is limited by low resolution and poor specificity and sensitivity. We present a simple protocol that starts with micrococcal nuclease-digested uncross-linked chromatin and is followed by affinity purification of TFs and paired-end sequencing. The resulting ORGANIC (Occupied Regions of Genomes from Affinity-purified Naturally Isolated Chromatin) profiles of Saccharomyces cerevisiae Abf1 and Reb1 provide highly accurate base-pair resolution maps that are not biased toward accessible chromatin, and do not require input normalization. We also demonstrate the high specificity of our method when applied to larger genomes by profiling Drosophila melanogaster GAGA Factor and Pipsqueak. Our results suggest that ORGANIC profiling is a widely applicable high-resolution method for sensitive and specific profiling of direct protein-DNA interactions.
Keywords: chromatin immunoprecipitation, ChIP, native, Abf1, Reb1, GAGA factor, Pipsqueak
Introduction
Sequence-specific DNA-binding proteins reside atop the eukaryotic regulatory hierarchy and functionally interpret signals encoded in the genome to control transcription, modulate chromatin structure, and ultimately shape cellular identity. As a result, comprehensive mapping of genomic loci engaged by regulatory factors is of great interest. Chromatin ImmunoPrecipitation (ChIP) is the most widely used method for profiling genomic targets of DNA-binding proteins. In most ChIP protocols, protein-DNA interactions are fixed by formaldehyde treatment prior to sonication of chromatin and immunoprecipitation of the resulting fragments (X-ChIP)1. After crosslink reversal, immunoprecipitated DNA can be analyzed by microarray hybridization (X-ChIP-chip) or high-throughput sequencing (X-ChIP-Seq)2,3.
Although X-ChIP methods have played a central role in interrogating protein binding genome-wide, they have numerous limitations stemming from crosslinking and sonication4,5 and recent work has uncovered systematic biases in these methodologies6–10. Notably, formaldehyde cross-linking can cause epitope masking and complicate subsequent immunoprecipitation. Formaldehyde also preferentially forms protein-protein crosslinks11, leading to the possible identification of false positive DNA binding events that represent indirect or transient protein-DNA interactions, particularly in highly transcribed regions5,10,12. X-ChIP-Seq resolution is substantially limited by the heterogeneity of fragments resulting from chromatin fragmentation and solubilization by sonication13; however, this limitation is addressed by ChIP-exo, which utilizes exonuclease digestion of crosslinked, sonicated chromatin to achieve single-base resolution13.
ChIP of native chromatin (N-ChIP) is not associated with epitope masking or protein-protein crosslinking and can be used with small amounts of input chromatin5,14. N-ChIP has been applied to histones5 and non-histone proteins, including RNA polymerase II, TFs, and chromatin remodelers15–18. We therefore sought to determine whether N-ChIP could produce high-resolution maps of sequence-specific protein binding sites. We previously demonstrated that micrococcal nuclease (MNase) digestion of native chromatin followed by paired-end sequencing (MNase-Seq) can map both nucleosomal and subnucleosomal particles protecting as little as ~25 bp with single-nucleotide resolution19. This method was recently used in conjunction with N-ChIP to yield ORGANIC (Occupied Regions of Genomes from Affinity-purified Naturally Isolated Chromatin) profiles of chromatin remodeler binding18. Here, we apply ORGANIC profiling to identify binding sites of the structurally distinct Saccharomyces cerevisiae TFs Abf1 and Reb1. With this approach, we identify more Abf1 and Reb1 binding sites than have been previously published and we show high accuracy in the detection of consensus motifs within binding sites. We also apply our method to profile genome-wide binding of Drosophila melanogaster GAGA-binding factor (GAF) and Pipsqueak (Psq), demonstrating the accuracy of ORGANIC maps in more complex eukaryotic genomes.
Results
Robust ORGANIC profiles of Reb1 and Abf1 binding sites
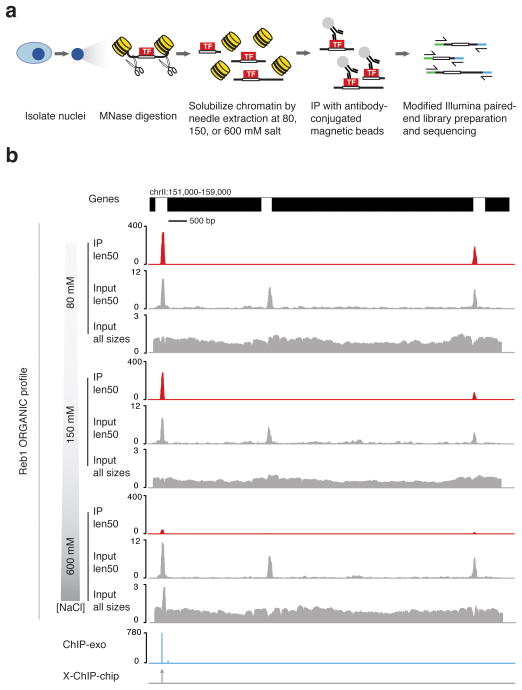
We performed MNase digestion of uncrosslinked intact nuclei from S. cerevisiae strains expressing Reb1-FLAG and Abf1-FLAG, solubilized chromatin by needle extraction, and immunoprecipitated tagged transcription factors at 80, 150, or 600 mM NaCl to obtain different levels of stringency (Fig. 1a and Supplementary Fig. 1). We then prepared TF-bound and input DNAs for paired-end sequencing using a modified library preparation protocol19 (Fig. 1a and Supplementary Fig. 1). Consistent with immunoprecipitation of proteins with small footprints, we found that small fragments were enriched in Reb1 ChIP relative to input (Supplementary Fig. 2a), and we therefore profiled the <100 bp (len50) size class.
Figure 1. Robust identification of Reb1 binding sites on native chromatin.
(a) ORGANIC profiling scheme. (b) Representative example of a region showing ORGANIC profiling ChIP Reb1 occupancy and input tracks for chromatin extracted at 80, 150, and 600mM salt, scaled by normalized counts. The locations and relative occupancies of ChIP-exo calls and location of X-ChIP-chip calls are shown in the lower tracks. Note the different scales in input and IP tracks.
The Reb1 immunoprecipitated (IP) fractions showed sharp peaks over a negligible background relative to the corresponding input chromatin (Fig. 1b). Similar peaks were identified when fragments were not filtered by size (Supplementary Fig. 2). Interestingly, the len50 size class inputs showed strong peaks corresponding to Reb1 binding sites seen in the IP samples, though at a lower level of occupancy. In the input, we observed highly occupied peaks not corresponding to Reb1 binding sites in intergenic regions (len50 tracks, Fig. 1b). With increasing salt concentration, there was a dramatic reduction in both total number and dynamic range of ORGANIC peaks (Fig. 1b), consistent with disruption of relatively weak electrostatic TF-DNA interactions at low affinity sites. Some but not all ORGANIC peaks corresponded to Reb1 binding sites previously identified by ChIP-chip and ChIP-exo (Fig. 1b)13,20. Similar results were obtained for Abf1 when compared to ChIP-chip data (Supplementary Fig. 3). For both Abf1 and Reb1, we observed a high degree of overlap between sites detected at different extents of MNase digestion (Supplementary Figs. 2–4). We conclude that ORGANIC profiling robustly detects both previously published and new Reb1 and Abf1 binding sites.
ORGANIC TF sites have characteristic sequence motifs
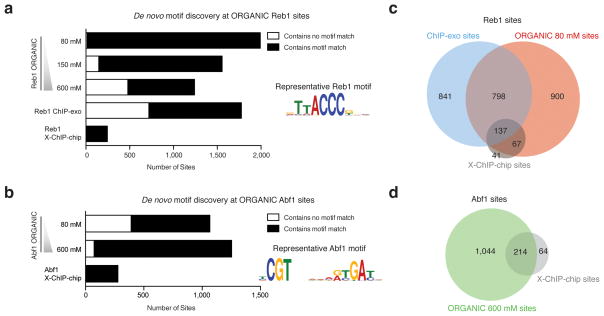
In order to characterize putative Reb1 and Abf1 binding sites, we applied a peak-calling algorithm with a conservative threshold to the len50 ChIP data and asked whether detected peaks were associated with characteristic consensus motifs using the MEME algorithm21. We identified 1,992 ORGANIC peaks in the Reb1 len50 size class 80 mM (low-salt) experiment (Fig. 2a). The low-salt ORGANIC Reb1 sites included 204 (83.3%) ChIP-chip and 935 (52.6%) ChIP-exo sites (Fig. 2b and Supplementary Fig. 5). Low-salt Abf1 ORGANIC peaks included 162 of 278 (58.3%) ChIP-chip peaks, whereas 600 mM (high-salt) Abf1 ORGANIC peaks identified more total sites (1,258), including 214 (of 278) sites also identified by ChIP-chip (Fig. 2b, d). The ORGANIC Reb1 and Abf1 motifs matched those reported in previous studies13,20 (Fig. 2a, b).
Figure 2. ORGANIC TF binding sites have characteristic binding site motifs.
(a, b) Number of sites associated with MEME-discovered Reb1 (a) and Abf1 (b) motifs. Sequence logos displaying representative MEME-derived motifs are included as insets. Note that all of the ChIP-chip TF binding sites have characteristic motifs because of strict motif criteria imposed in determining high-quality binding sites20. (c, d) Venn diagrams show degree of peak overlap between Reb1 (c) and Abf1 (d) datasets. Peaks called for each Abf1 and Reb1 ORGANIC dataset and position-specific log-odds matrices corresponding to MEME-discovered motifs are included (Supplementary Tables 1 and 2).
We characterized the reproducibility of our method by performing pairwise comparisons of positions and occupancies of peaks called using independent biological replicates and from peak sets using varying salt concentrations, and found that datasets were well correlated (R2 = 0.80 – 0.95, Supplementary Fig. 4). Occupancies at Reb1 sites called by both ChIP-exo and ORGANIC profiling were poorly correlated (R2 < 0.05, Supplementary Fig. 5b). We conclude that ORGANIC profiling reproducibly captures a large fraction of previously published Abf1 and Reb1 binding sites, while identifying 2–8 fold more motif-associated sites than other methods.
ORGANIC profiles are highly sensitive and specific
Given the strong sequence specificities of Abf122 and Reb123, we evaluated the accuracy of ORGANIC profiling by using the presence of a MEME-derived motif within a peak region as the ‘gold-standard’ for classifying a peak as a true positive. Strikingly, 99.3% of low-salt Reb1 sites contained the TTACCCG motif (Fig. 2a). The percentage of peaks containing the motif decreased to 61.5% at high salt (Fig. 2a). Whereas virtually all ORGANIC peaks had Reb1 motifs, only 59.6% of all ChIP-exo sites were found to be associated with a Reb1 motif13. In contrast to Reb1 ORGANIC sites, the 1,066 low-salt Abf1 ORGANIC sites contained a smaller percentage of peaks with motifs (63.3%) than peaks called using high-salt extraction (93.7%, Fig. 2b). We estimate a false negative rate of ~0.5% for ORGANIC profiling at Reb1 motifs (see Online Methods).
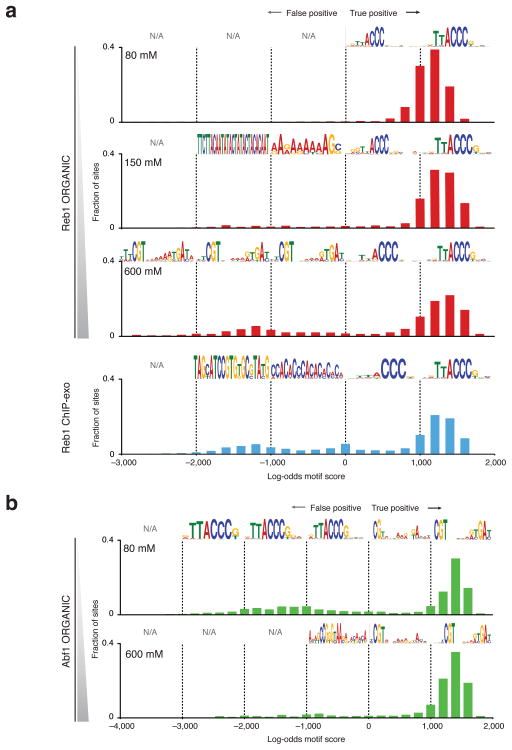
In order to evaluate the specificity of ORGANIC profiling, we determined how well peak sequences matched consensus motifs by scoring peaks using MEME-derived position-specific scoring matrices (PSSMs). Using the Reb1 ORGANIC PSSM, we found a distribution of high motif scores (true positives) with no strongly negative scores at low salt (Fig. 3a). When the salt concentration was increased to 150 mM and 600 mM, we observed a graded reduction in the fraction of true positive Reb1 sites and the appearance of strongly negative scores, giving a bimodal distribution (Fig. 3a). In comparison, ChIP-exo Reb1 calls included a high number of negative calls and showed a motif score distribution similar to the high-salt ORGANIC Reb1 dataset (Fig. 3a bottom panel). A similar trend was obtained using the ChIP-exo-derived PSSM (Supplementary Fig. 6).
Figure 3. High sensitivity and specificity of ORGANIC profiling applied to TF binding sites.
Histograms of motif scores determined using MEME-derived position-specific log-odds scoring matrices are shown for Reb1 (a) and Abf1 (b) binding sites. MEME-ChIP-derived motifs corresponding to each 1,000-unit log-odds motif score cohort are included above each histogram. Bins that contained either too few sites or sequences that did not produce a MEME-ChIP-derived motif are designated ‘N/A.’
Abf1 motif scores were also narrowly distributed (Fig. 3b) and, as expected from the increase in Abf1 motif-containing peaks at high salt, we observed a reduction in false-positive calls and an increase in true-positive calls at 600 mM salt (Fig. 3b). Given the structural differences in the Reb1 and Abf1 DNA-binding domains24,25, it is likely that optimal extraction and ChIP conditions for the proteins differ, explaining the differential specificity of ORGANIC profiles across varying salt concentrations and different DNA binding proteins. The increase in apparent specificity to >90% at higher salt concentrations for Abf1 despite reduction in dynamic range (Supplementary Fig. 3) suggests that experimental parameters can be tailored to the DNA-binding protein of interest.
ORGANIC sites display DNaseI footprints and are conserved
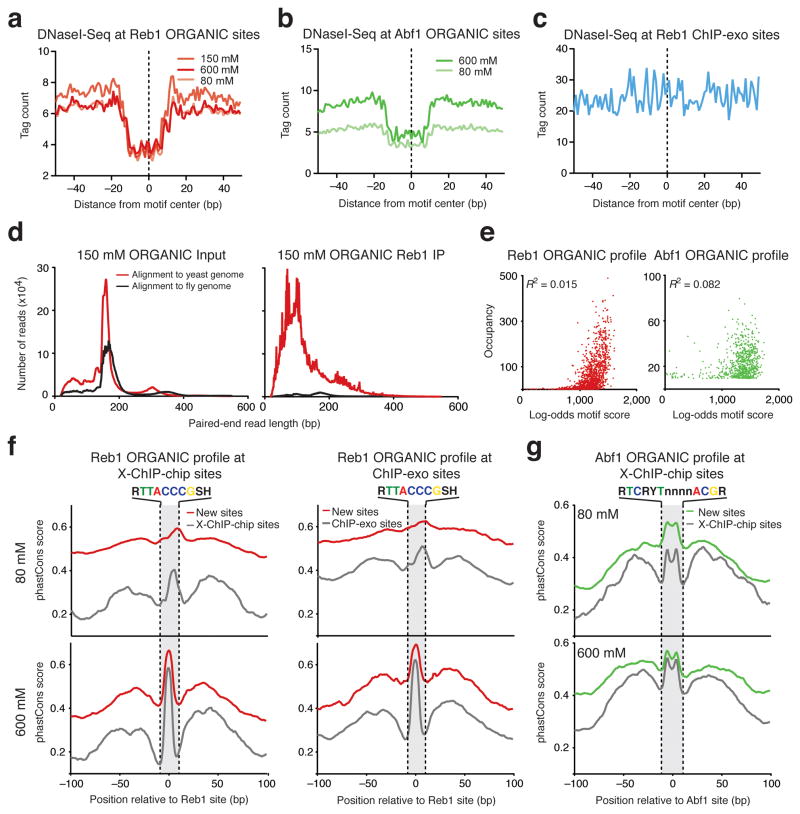
In order to confirm that ORGANIC sites are bound in vivo, we used published S. cerevisiae DNaseI-Seq data26 to ask whether sites detected by ORGANIC profiling are associated with classical footprints indicative of in vivo occupancy26–28. For both Reb1 and Abf1, average DNaseI-Seq profiles at ORGANIC sites showed characteristic footprints (Fig. 4a, b). In contrast, average DNaseI-Seq tag counts at ChIP-exo sites did not show a footprint (Fig. 4c), except at the subset of sites that are also ORGANIC sites (Supplementary Fig. 5). We found that DNaseI footprint depth, which is correlated with in vivo occupancy26,28, corresponds well with Reb1 and Abf1 ORGANIC site occupancies (Supplementary Figs. 7 and 8). These results suggest that ORGANIC sites are qualitatively occupied in vivo and also that relative occupancies determined by ORGANIC profiling are quantitatively correlated with in vivo binding.
Figure 4. ORGANIC sites are stably bound in vivo and are conserved throughout Saccharomyces evolution.
(a–c) DNase I–seq profiles at ORGANIC Reb1 (a), ORGANIC Abf1 (b) and ChIP-exo Reb1 (c) sites. For c, both primary and secondary ChIP-exo sites are shown (see supplementary Fig. 5 for a separate analysis). (d) Paired-end sequencing fragment length distributions of reads from input (left) and IP (right) libraries mapped to fly and yeast genomes in a mixing experiment in which yeast Reb1 ORGANIC profiling was done in the presence of Drosophila nuclei. (e) Correlation between occupancy and log-odds motif score for Reb1 (left) and Abf1 (right) ORGANIC sites. (f,g) Average phastCons scores in 200-bp windows centered at ‘new sites’ (sites detected by ORGANIC but not by other methods) and sites detected by other methods for Reb1 (f) and Abf1 (g).
In order to further exclude the possibility of TF redistribution during chromatin preparation or ChIP, we mixed equal numbers of isolated Drosophila S2 cell nuclei with Reb1-FLAG budding yeast nuclei and performed ORGANIC profiling at 150 mM salt. We expected that, if redistribution occurred, the ~300-fold excess of Drosophila sequences with Reb1 binding sites would be enriched in the ChIP fraction. However, we detected only a background level of Drosophila DNA in the ChIP fraction relative to the input (Fig. 4d). We detected good correlation between Reb1 sites detected in replicates of the mixing experiment and, consistent with stable binding under conditions used in ORGANIC profiling, we found a high level of correlation (R2 = 0.995) between occupancies detected in experiments with mixed and unmixed nuclei (Supplementary Fig. 9a, b). The motif score distribution of Drosophila Reb1 peaks was dominated by negative scores (Supplementary Fig. 9c). These analyses suggest that Reb1 does not shift detectably from yeast to Drosophila chromatin during chromatin preparation and ChIP.
We also considered specificity of identified binding sites by analyzing the correlation between motif score and occupancy of Reb1 and Abf1. At low salt, we observed poor correlation between occupancy and motif score (R2 = 0.02, Fig. 4e). Consistent with a bimodal distribution of motif scores between true positives and false positives (Fig. 3), we observed that low-scoring sites are poorly occupied by the TF, while high-scoring sites show an expected broad distribution of occupancies (Fig. 4e). A similar relationship between occupancy and motif score was observed at high salt and with ChIP-exo Reb1 sites (Supplementary Fig. 10). Because high motif scores are correlated with favorable binding energies29, this analysis also suggests that TFs do not redistribute to thermodynamically favored binding sites during ORGANIC profiling.
We expected that the new Abf1 and Reb1 sites would show evolutionary conservation above background levels because conservation of TF binding sites implies purifying selection30. We plotted phastCons scores31, which represent the probability that a given base is in a conserved region, in windows centered at Reb1 or Abf1 sites (Fig. 4f, g). Interestingly, we observed increased conservation of new sites at motif positions relative to background (Fig. 4f, g and Supplementary Fig. 5). In general, new sites had either higher or comparable conservation scores when compared to sites detected by ChIP-chip or ChIP-exo. Consistent with a role for Abf1 and Reb1 in positioning flanking nucleosomes at a subset of promoters23,32, we detected well-positioned flanking nucleosomes around Reb1 and Abf1 sites (Fig. 5a, b). Since virtually all of the ORGANIC Reb1 sites have a Reb1 binding motif, we asked whether there is a difference in motif strength or TF occupancy that could explain differential phasing of nucleosomes. We ranked ORGANIC sites by nucleosome occupancy and considered the top and bottom 200 sites, which corresponded to sites with relative nucleosome occlusion and depletion, respectively (Fig. 5a, b and Supplementary Fig. 11). We detected no substantial difference in MEME-derived motifs between the two groups (Fig. 5a, b), suggesting that the degree of nucleosome phasing is not associated with motif strength.
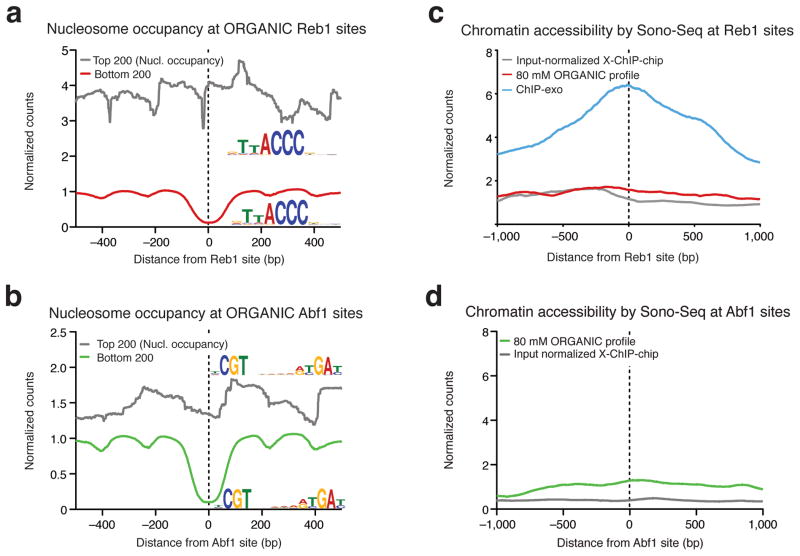
Figure 5. ORGANIC profiling identifies TF binding sites in inaccessible chromatin and does not require input normalization.
(a, b) Nucleosome (Nucl.) occupancy for the top and bottom 200 Reb1 (a) and Abf1 (b) sites ranked by 80 mM ORGANIC occupancy. Motifs discovered by MEME for sites that are in nucleosome-depleted and –occluded DNA are included as insets. (c, d) Average Sono-Seq normalized counts in a 2 kb window centered Reb1 (c) and Abf1 (d) sites. Normalized counts are computed such that the genome-wide average is equal to 1 (see Online Methods).
ORGANIC profiles do not require input normalization
Sequencing formaldehyde cross-linked and sonicated chromatin (Sono-Seq) is known to preferentially recover regions of accessible chromatin9. In order to characterize the observed differences in MNase protection, we performed Sono-Seq and generated profiles of average normalized Sono-Seq counts in 2-kb windows centered at Reb1 binding sites determined by different methods. Strikingly, ChIP-exo Reb1 sites showed enrichment for Sono-Seq reads, whereas there was virtually no enrichment at ORGANIC or ChIP-chip Reb1 sites (Fig. 5c and Supplementary Fig. 5f). Similarly, we detected no enrichment of Sono-Seq reads at ORGANIC or ChIP-chip Abf1 sites (Fig. 5d). Sono-Seq enrichment is consistent with the observation of increased DNase cleavage26 at ChIP-exo sites and comparatively lower cleavage at ORGANIC sites (Fig. 4a–c). We obtained similar results using previously published Sono-Seq data (Supplementary Fig. 12) and using sensitivity to MNase digestion as an independent measure of chromatin accessibility (Supplementary Results). The Reb1 ChIP-chip study was performed using a two-color spotted microarray on which ChIP DNA was co-hybridized with DNA from an un-enriched sample (input)33; this input normalization procedure likely corrected for bias from preference for accessible chromatin, as the input sample was obtained from cross-linked and fragmented DNA. In contrast, input normalization is not performed with ChIP-exo, which likely explains the observed strong preference for binding sites in accessible chromatin. Unlike the Reb1 ChIP-exo map, the ORGANIC accessibility profile is similar to input-normalized ChIP-chip. As the cross-linking and sonication steps of ChIP-exo are essentially the same as those used to obtain accessible chromatin by Sono-Seq9,13, the absence of these steps could account for the insensitivity of ORGANIC profiling to the degree of chromatin accessibility. We conclude that ORGANIC maps do not show chromatin accessibility preferences and therefore do not require input normalization.
Highly specific ORGANIC profiles of Drosophila TFs
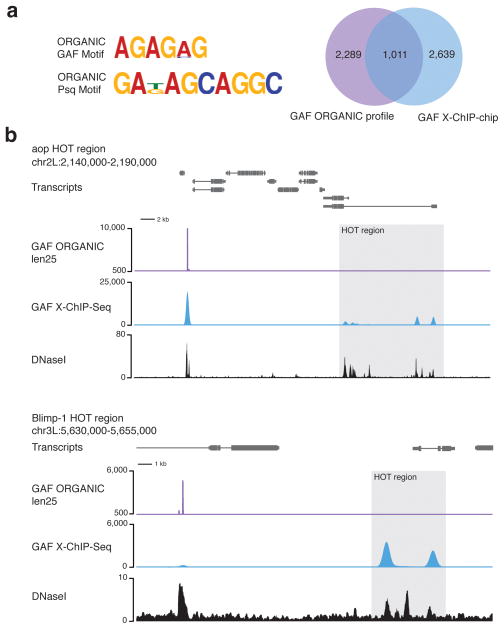
We assessed the applicability of ORGANIC profiling to eukaryotes with larger genomes by mapping GAF and Psq from Drosophila S2 cell active chromatin extracted with 80 mM salt34 (Supplementary Fig. 1). In order to determine whether GAF is lost from the nucleus under native conditions, we summed losses incurred during processing of nuclei during the ORGANIC and modENCODE X-ChIP protocols. In both cases, ~15–20% of total GAF was lost (Supplementary Fig. 13). We observed enrichment for peaks in the len25 (1–50 bp) and len50 size class ChIP fractions relative to input for both GAF and Psq (Supplementary Fig. 14). Enriched peaks were associated with DNaseI hypersensitive sites and were evolutionarily conserved, suggesting they represent bona fide in vivo sites (Supplementary Fig. 15). Consistent with previous work demonstrating that GAF and Psq heterodimerize and act in concert at many loci35, we observed similar genome-wide profiles for GAF and Psq. Using the same peak-calling method and de novo motif analysis approach to characterize yeast TF binding sites, we called 3,300 GAF and 957 Psq sites and recovered expected GAG-repeat containing motifs in 76.5% of GAF and 40% of Psq ORGANIC peaks (Fig. 6a). In contrast, ChIP-chip identified 4,567 GAF sites36, of which only ~5% had characteristic motifs. Therefore, ORGANIC profiling is greater than an order of magnitude more specific than ChIP-chip for factor binding in the Drosophila genome.
Figure 6. ORGANIC profiling of Drosophila GAGA Factor and Pipsqueak DNA-binding proteins.
(a) Representative GAF and Psq motifs found in ORGANIC GAF and Psq sites and Venn diagram showing overlap between GAF X-ChIP-chip (‘all sizes’ size class). (b) Representative TF hotspot-containing regions showing ORGANIC GAF profile and modENCODE X-ChIP-Seq tracks. Hotspots are occupied by GAF in the X-ChIP-Seq tracks, but not in the ORGANIC track. A common peak detected by both X-ChIP-Seq and ORGANIC profiling with high-scoring motifs is to the left of the HOT regions. Len25 GAF and Psq ORGANIC peaks are included (Supplementary Table 1).
In addition to sites with the characteristic GAF motif, GAF is known from X-ChIP-chip data to bind TF ‘hotspots’36,37, which are thought to reflect dynamic, low-affinity binding of multiple transcription factors37. Remarkably, TF hotspots were absent from GAF ORGANIC peak calls (Fig. 6b and Supplementary Fig. 14). When we searched for the GAF consensus motif among TF hotspot regions, only 17.5% displayed such a stringent motif. This lack of robust GAF motifs at TF hotspots can account for the absence of ORGANIC signals at these sites, despite ready detection by X-ChIP. Consistent with the cross-linking of highly transient interactions at commonly used formaldehyde crosslinking times12, we suggest that the dynamic binding of GAF at TF hotspots resulted in trapping of transiently bound GAF by formaldehyde cross-linking to these sites when using X-ChIP. In contrast, ORGANIC profiling detects only sites that are stably bound under native extraction conditions, and so the lack of robust GAF motifs at TF hotspots is further evidence that ORGANIC maps are highly specific for stable, direct TF-DNA interactions.
Discussion
We have shown that ORGANIC profiling identifies direct TF-chromatin interactions at high resolution and with high specificity and sensitivity. De novo motif discovery revealed that the large majority of ORGANIC binding site calls have the expected consensus motif and correspond to DNaseI footprints suggestive of in vivo binding. Our study also demonstrated the flexibility of ORGANIC profiling in mapping genomic binding sites of proteins with structurally distinct DNA-binding domains from different species and showed that the specificity of ORGANIC maps can be modulated by varying salt concentration.
Although native chromatin profiling has been widely used for epigenome mapping in the context of methods like DNaseI-Seq26 and, more recently, the assay for transposase-accessible chromatin using sequencing (ATAC-Seq)38, a potential concern with native chromatin profiling has been that small-footprint DNA-binding proteins that are highly dynamic in vivo could redistribute during chromatin preparation and ChIP, which is a frequently cited rationale for crosslinking protein-DNA interactions with formaldehyde5. However, rearrangement of bound factors is unlikely to occur during ORGANIC profiling for a number of reasons.
First, conditions under which ORGANIC profiling is performed differ substantially from the in vivo state and disfavor rearrangement. MNase digestion is performed at >10-fold lower salt than is present in vivo. Given that salt competes for electrostatic protein-DNA interactions, low salt conditions can functionally fix protein-DNA interactions in a noncovalent manner39. There is some evidence for a role for chromatin remodeling machinery in facilitating the high degree of in vivo TF dynamics40, but given the lack of readily available ATP in the ORGANIC profiling protocol, it is unlikely that these active processes could contribute to TF rearrangement. The cold, dilute conditions under which ORGANIC profiling is performed also render it unlikely that an unbound factor will find its recognition sequence. Under the conditions used for ORGANIC profiling, TF loss during various points of the protocol is comparable to what is observed with X-ChIP, consistent with stable TF binding under ORGANIC profiling conditions41,42.
Second, utilizing MNase to fragment chromatin ensures that any accessible, unbound binding sites will be digested such that they are not available for engagement by free factors. Contrary to the expectation of factor dissociation and rebinding, we showed that there is little change in binding sites detected over a four-fold digestion range. Third, sites identified by ORGANIC profiling are reflective of in vivo occupancy as determined by the classical method of DNaseI footprinting, which is also performed under native, low-salt conditions26. Fourth, inconsistent with a redistribution of factors to the most thermodynamically favorable motifs as inferred by motif strength, ORGANIC profiles show a wide range of occupancies for various motif scores. Fifth, ORGANIC sites are conserved throughout evolution, suggesting a possible functional role for some of these sites. Finally, there is no enrichment for Drosophila sequences in yeast TF ORGANIC profiling when Drosophila S2 nuclei and yeast nuclei are combined prior to MNase digestion, chromatin extraction, and ChIP. The linear correlation between occupancies in yeast-only and yeast/fly-mixed ORGANIC profiles suggests that ORGANIC profiling both qualitatively and quantitatively preserves characteristics of in vivo binding sites. Therefore, ORGANIC profiling captures in vivo, direct TF binding events with high specificity and sensitivity.
Interestingly, the chromatin accessibility bias inherent to some crosslinking and sonication-based methods is corrected by normalizing to input, but such input normalization is not generally done with a sequencing readout, and is not described as an option for ChIP-exo. Moreover, even using X-ChIP-chip normalized by input, some sites in inaccessible chromatin were not detected. In contrast, no accessibility bias was detected using ORGANIC profiling. This lack of bias obviates the need for input normalization, which is impractical for large genomes, where the input library, unlike the ChIP library, must be sequenced at sufficient depth to provide whole-genome coverage.
The high signal-to-noise ratio for ORGANIC profiling means that it is relatively inexpensive to perform even for large genomes. Other advantages include the simple library preparation protocol that requires only a few nanograms of DNA and the precise fragment lengths obtained from paired-end sequencing that can be used to deduce regional features around a binding site by V-plotting19. The successful application of ORGANIC profiling to DNA-binding proteins of all different types, including nucleosomes, RNA polymerases, nucleosome remodelers and sequence-specific DNA binding proteins demonstrates its utility in both large- and small-scale epigenome profiling projects.
Supplementary Material
Supplementary Figure 1. Detailed overview of ORGANIC profiling protocols. (a) S. cerevisiae and (b) D. melanogaster ORGANIC profiling protocols used in this study. References for previously published nuclei extraction procedures, centrifuge specifications, and buffer compositions are included in the Online Methods section. (c) Overview of library preparation protocol used for both yeast and fly ORGANIC DNAs.
Supplementary Figure 2. ORGANIC profiling robustly identifies Reb1 binding sites. (a) Representative paired-end sequencing fragment length distributions for Reb1 ORGANIC input and IP libraries. (b) Representative tracks of Reb1 ORGANIC IP and input tracks at indicated salt concentrations and MNase digestion durations and Sono-Seq profiles (bottom tracks). Note the differences in scale in IP and Input tracks. (c) Average profiles of 2.5′ MNase- and 10′ MNase digestion, 80mM len50 (top panel) and all sizes (bottom panel) Reb1 ORGANIC profiles centered at 10′ MNase, 80mM Reb1 ORGANIC sites.
Supplementary Figure 3. ORGANIC profiling robustly identifies Abf1 binding sites. (a) Representative paired-end sequencing fragment length distributions for Abf1 ORGANIC input and IP libraries. (b) Representative tracks of Abf1 ORGANIC IP and input tracks at indicated salt concentrations and MNase digestion durations and Sono-Seq profiles (bottom tracks). Note the differences in scale in IP and Input tracks. (c) Average profiles of 2.5′ MNase- and 10′ MNase digestion, 80mM len50 (top panel) and all sizes (bottom panel) Abf1 ORGANIC profiles centered at 10′ MNase, 80mM Abf1 ORGANIC sites.
Supplementary Figure 4. Reproducibility of TF binding site and occupancy detection by ORGANIC profiling. (a–g) Pairwise comparisons of Reb1 (a–e) and Abf1 (f–g) datasets. Venn diagrams of sites unique to and shared by each dataset in the indicated comparison (top panels) and correlation plots of occupancies of shared sites (bottom panels). Note that x- and y- axis scales in correlation plots were adjusted to show the majority of data points. R2 values and trend lines are from linear regression.
Supplementary Figure 5. Comparison of Reb1 ORGANIC profiles and ChIP-exo ‘primary’ and ‘secondary’ sites. (a) Overlap between 80 mM, 150 mM, and 600 mM ORGANIC Reb1 sites and ChIP-exo ‘primary’ and ‘secondary’ sites and X-ChIP-chip sites. (b) Correlation between Reb1 occupancies detected by ChIP-exo and ORGANIC profiling at 80 mM, 150 mM, and 600 mM with R2 from linear regression. (c) DNaseI protection (average DNaseI-Seq tag counts) at ChIP-exo ‘primary’ and ‘secondary’ sites. (d) DNaseI-seq tag counts at ChIP-exo primary and secondary sites. Heatmaps are ranked by descending ChIP-exo tag counts and graphs represent average DNaseI tag count for occupancy groups, where each occupancy group represents 20% of total sites detected. Note that the lack of a footprint in the ChIP-exo ‘primary’ site third occupancy group is due to a subset of highly accessible sites located proximal to the yeast rDNA locus. (e) phastCons evolutionary conservation at ChIP-exo sites. (f) Average Sono-Seq normalized counts at ChIP-exo sites.
Supplementary Figure 6. High sensitivity and specificity of Reb1 and Abf1 binding detected by ORGANIC profiling. Motif score histograms of ORGANIC profiling and ChIP-exo sites scored using the ChIP-exo-derived Reb1 position-specific log-odds matrix.
Supplementary Figure 7. Reb1 ORGANIC profile occupancies are correlated with DNaseI-Seq footprint depth. (a, b) DNaseI-Seq at Reb1 ORGANIC (a) and Reb1 ChIP-exo (b) sites. Heatmaps of DNaseI-Seq tag count are ranked by decreasing occupancy and each graph shows the average DNaseI-Seq tag count at the indicated occupancy group, with each occupancy group representing 20% of total sites detected.
Supplementary Figure 8. Abf1 ORGANIC profile occupancies are correlated with DNaseI-Seq footprint depth. DNaseI-Seq at Abf1 ORGANIC sites with heatmaps of DNaseI-Seq tag counts ranked by decreasing occupancy. Each graph shows the average DNaseI-Seq tag count at the indicated occupancy group, with each occupancy group representing 20% of total sites detected.
Supplementary Figure 9. Reb1 does not shift to Drosophila chromatin in mixed yeast/fly ORGANIC profiling. (a) Correlation between occupancies detected by replicates of the yeast/fly mixing experiment. (b) Correlation between occupancies detected by yeast/fly mixed and yeast-only 150 mM ORGANIC profiles. (c) Motif score distribution of peaks called from reads mapping to the Drosophila genome from the yeast/fly-mixed Reb1 ORGANIC profile scored using the Reb1 2.5′ MNase 80 mM ORGANIC PSSM (Supplementary Table 2).
Supplementary Figure 10. Abf1 and Reb1 do not redistribute to thermodynamically favorable binding sites. (a–c) Relationship between TF binding site motif score and occupancy at (a) Abf1 ORGANIC 600 mM, (b) Reb1 ORGANIC 80 and 150 mM, and (c) ChIP-exo Reb1 sites with R2 values from linear regression analysis. Note that the y-axis was rescaled such that most of the data points are visible.
Supplementary Figure 11. Nucleosome occupancy around ORGANIC TF binding sites. (a) Nucleosome occupancy around Reb1 sites rank ordered by ORGANIC profile Reb1 occupancy (left panel) and nucleosome occupancy in a 2 kb window centered at Reb1 sites (right panel). (b) Nucleosome occupancy around Abf1 sites rank ordered by ORGANIC Abf1 occupancy (left panel) and nucleosome occupancy in a 2 kb window (right panel).
Supplementary Figure 12. Sono-Seq chromatin accessibility and Reb1 and Abf1 sites determined using data from Auerbach, et al. (a–c) Sono-Seq (Auerbach, et al.) tag counts at ORGANIC 80, 150, and 600mM, ChIP-exo, and X-ChIP-chip sites (a), at ORGANIC 80 and 600mM and X-ChIP-chip Abf1 sites (b), and at ChIP-exo primary and secondary sites (c).
Supplementary Figure 13. GAF recovery in ORGANIC and modENCODE X-ChIP protocols. (a, b) Western blots of GAF and histone H3 from fractions taken at various stages of the 80 mM ORGANIC (a) and modENCODE X-ChIP (b) protocols (upper panels) and densitometric analysis normalized by fractional volume of total sample loaded (lower panels) with y-axes indicating background-corrected, sample volume-normalized ImageJ densitometry measurements (see Online Methods). Percentages above bars in densitometry measurements represent fraction of GAF or H3 relative to whole cell extracts. Note that the modENCODE X-ChIP pellet could not be completely solubilized and only the soluble fraction was loaded (see Online Methods).
Supplementary Figure 14. ORGANIC profiling robustly identifies Drosophila TF binding sites. (a) Representative tracks of GAF and Psq ORGANIC profiles. (b) Comparison of ORGANIC GAF sites and modENCODE-defined TF hotspots with GAF annotation (see Online Methods).
Supplementary Figure 15. Drosophila TF ORGANIC sites are conserved in evolution and are found in DNaseI hypersensitive regions. (a) Average DNaseI-Seq tag counts and phastCons evolutionary conservation at GAF X-ChIP-chip, X-ChIP-seq, and ORGANIC sites and GAF-bound HOT sites. (b) Average DNaseI-Seq and phastCons evolutionary conservation at Psq ORGANIC sites. (c) Representative tracks of GAGA-motif clusters with specific, discrete GAF binding events detected by ORGANIC profiling. Motif matches were identified by log-odds matrix-based scoring as described in the Online Methods with p < 5e-3.
Supplementary Figure 16. ORGANIC profiling identifies Reb1 and Abf1 binding sites in inaccessible chromatin. (a, b) MNase accessibility at Reb1 (a) and Abf1 (b) sites in 2 kb windows centered at ORGANIC, X-ChIP-chip, and/or ChIP-exo sites.
Supplementary Figure 17. Genomic context analysis of Reb1 and Abf1 ORGANIC and X-ChIP-chip binding sites. (a) Genomic contexts of ORGANIC, X-ChIP-chip and ChIP-exo Reb1 sites. (b) Number of sites falling into intergenic (top), genic (middle), and telomeric (bottom) regions and associated MEME-ChIP-derived motifs (right panel). Telomeric motifs for the 80, 150, and 600mM Reb1 ORGANIC datasets are slightly truncated outside of the core Reb1 consensus (delimited by the gray box). High confidence motifs could not be recovered by MEME-ChIP for X-ChIP-chip genic sites (n = 8) or X-ChIP-chip telomeric sites (n = 0). By chance, 550 Reb1 motifs are expected to occur within genes (see Online Methods). (c) Relative fraction of binding sites in intergenic, genic, telomeric, and ARS (Autonomously Replicating Sequence) annotated regions. (d) Number of sites in each dataset with the indicated annotation and corresponding MEME-ChIP-derived motif. High confidence motifs could not be recovered by MEME-ChIP for ChIP-chip telomeric sites (n = 0) or ChIP-chip ARS sites (n = 2). By chance, 137 Abf1 motifs are expected to occur within genes (see Online Methods).
Reb1: 2.5 min MNase/80mM NaCl/len50, 10 min MNase/80mM NaCl/len50, 10 min MNase/150mM NaCl/len50, 10 min MNase/600mM NaCl/len50; Abf1: 2.5 min MNase/80mM NaCl/len50, 10 min MNase/80mM NaCl/len50, 10 min MNase/600mM NaCl/len50; Psq len25 and GAF len25. Thresholds used for peak calling (see Online Methods) are indicated.
For a given log-odds entry (row, column) in the matrix, row specifies the nucleotide and column the position in the motif.
Acknowledgments
We thank J.G. Henikoff (Fred Hutchinson Cancer Research Center, Seattle, WA, USA) for help with data analysis, members of the Henikoff laboratory and L. Gabrovsek (University of Washington, Seattle, WA, USA) for comments on the manuscript, G. Cavalli (Institut Génétique Humaine, CNRS, Montpellier, France) and C.A. Berg (University of Washington, Seattle, WA, USA) for GAF and Psq antibodies, respectively, and the Drosophila RNAi Screening Center for cells. This work was supported by the Howard Hughes Medical Institute, grant R01 ES020116 from NIH (S.H. and K.A.), the FHCRC Chromosome Metabolism and Cancer Training grant NIH 5T32 CA009657 (G.E.Z), the ERC 7th Framework Program, Marie Curie Actions IOF 300710 (G.A.O), and the Micki & Robert Flowers ARCS Endowment from the Seattle Chapter of the ARCS Foundation (S.K.).
Abbreviations
- TF
Transcription Factor
- ChIP
Chromatin Immunoprecipitation
- X-ChIP-Seq
cross-linking ChIP-Seq
- MNase
Micrococcal Nuclease
- ORGANIC profiling
Occupied Regions of Genomes from Affinity-purified Naturally Isolated Chromatin sequences
- Abf1
ARS Binding Factor 1
- Reb1
rDNA Enhancer Binding Protein 1
- GAF
GAGA Factor
- Psq
Pipsqueak
- PSSM
Position Specific Scoring Matrix
Footnotes
Accession Number
GEO GSE 45672
Author Contributions
S.H. conceived the strategy, S.K., S.H. and G.E.Z. designed the yeast experiments, K.A. and G.A.O. designed and performed the Drosophila experiments, S.K. performed the yeast experiments, S.K. performed the yeast analysis, S.K and G.A.O. performed the Drosophila analysis, and S.K. and S.H. wrote the paper.
Competing Financial Interests
None.
References
- 1.Solomon MJ, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 3.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 4.Zentner GE, Henikoff S. Surveying the epigenomic landscape, one base at a time. Genome Biology. 2012;13:250. doi: 10.1186/gb-2012-13-10-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Neill LP, Turner BM. Immunoprecipitation of native chromatin: NChIP. Methods. 2003;31:76–82. doi: 10.1016/s1046-2023(03)00090-2. [DOI] [PubMed] [Google Scholar]
- 6.Teytelman L, et al. Impact of chromatin structures on DNA processing for genomic analyses. PloS ONE. 2009;4:e6700. doi: 10.1371/journal.pone.0006700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan X, Struhl K. Where does mediator bind in vivo? PloS ONE. 2009;4:e5029. doi: 10.1371/journal.pone.0005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz YB, Kahn TG, Pirrotta V. Characteristic low density and shear sensitivity of cross-linked chromatin containing polycomb complexes. Molecular and Cellular Biology. 2005;25:432–439. doi: 10.1128/MCB.25.1.432-439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auerbach RK, et al. Mapping accessible chromatin regions using Sono-Seq. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14926–14931. doi: 10.1073/pnas.0905443106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teytelman L, Thurtle DM, Rine J, van Oudenaarden A. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proceedings of the National Academy of Sciences of the United States of America; 2013. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson V. Formaldehyde cross-linking for studying nucleosomal dynamics. Methods. 1999;17:125–139. doi: 10.1006/meth.1998.0724. [DOI] [PubMed] [Google Scholar]
- 12.Poorey K, et al. Measuring chromatin interaction dynamics on the second time scale at single-copy genes. Science. 2013;342:369–372. doi: 10.1126/science.1242369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilfillan GD, et al. Limitations and possibilities of low cell number ChIP-seq. BMC Genomics. 2012;13:645. doi: 10.1186/1471-2164-13-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roca H, Franceschi RT. Analysis of transcription factor interactions in osteoblasts using competitive chromatin immunoprecipitation. Nucleic Acids Research. 2008;36:1723–1730. doi: 10.1093/nar/gkn022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teves SS, Henikoff S. Heat shock reduces stalled RNA polymerase II and nucleosome turnover genome-wide. Genes & Development. 2011;25:2387–2397. doi: 10.1101/gad.177675.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Neill LP, Turner BM. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. The EMBO Journal. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zentner GE, Tsukiyama T, Henikoff S. ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies. PLoS Genetics. 2013;9:e1003317. doi: 10.1371/journal.pgen.1003317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S. Epigenome characterization at single base-pair resolution. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18318–18323. doi: 10.1073/pnas.1110731108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacIsaac KD, et al. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings/…. International Conference on Intelligent Systems for Molecular Biology; ISMB. International Conference on Intelligent Systems for Molecular Biology. 1994;2:28–36. [PubMed] [Google Scholar]
- 22.Beinoraviciute-Kellner R, Lipps G, Krauss G. In vitro selection of DNA binding sites for ABF1 protein from Saccharomyces cerevisiae. FEBS Letters. 2005;579:4535–4540. doi: 10.1016/j.febslet.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ju QD, Morrow BE, Warner JR. REB1, a yeast DNA-binding protein with many targets, is essential for growth and bears some resemblance to the oncogene myb. Molecular and Cellular Biology. 1990;10:5226–5234. doi: 10.1128/mcb.10.10.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho G, Kim J, Rho HM, Jung G. Structure-function analysis of the DNA binding domain of Saccharomyces cerevisiae ABF1. Nucleic Acids Research. 1995;23:2980–2987. doi: 10.1093/nar/23.15.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nature Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galas DJ, Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Research. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 30.Blanchette M, Tompa M. Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Research. 2002;12:739–748. doi: 10.1101/gr.6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Research. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganapathi M, et al. Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucleic Acids Research. 2011;39:2032–2044. doi: 10.1093/nar/gkq1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Research. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwendemann A, Lehmann M. Pipsqueak and GAGA factor act in concert as partners at homeotic and many other loci. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12883–12888. doi: 10.1073/pnas.202341499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy S, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moorman C, et al. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006;103:12027–12032. doi: 10.1073/pnas.0605003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature Methods. 2013 doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohman TM, Mascotti DP. Thermodynamics of ligand-nucleic acid interactions. Methods in Enzymology. 1992;212:400–424. doi: 10.1016/0076-6879(92)12026-m. [DOI] [PubMed] [Google Scholar]
- 40.Hager GL, McNally JG, Misteli T. Transcription dynamics. Molecular Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkins RC, Lis JT. GAGA factor binding to DNA via a single trinucleotide sequence element. Nucleic Acids Research. 1998;26:2672–2678. doi: 10.1093/nar/26.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soeller WC, Oh CE, Kornberg TB. Isolation of cDNAs encoding the Drosophila GAGA transcription factor. Molecular and Cellular Biology. 1993;13:7961–7970. doi: 10.1128/mcb.13.12.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Detailed overview of ORGANIC profiling protocols. (a) S. cerevisiae and (b) D. melanogaster ORGANIC profiling protocols used in this study. References for previously published nuclei extraction procedures, centrifuge specifications, and buffer compositions are included in the Online Methods section. (c) Overview of library preparation protocol used for both yeast and fly ORGANIC DNAs.
Supplementary Figure 2. ORGANIC profiling robustly identifies Reb1 binding sites. (a) Representative paired-end sequencing fragment length distributions for Reb1 ORGANIC input and IP libraries. (b) Representative tracks of Reb1 ORGANIC IP and input tracks at indicated salt concentrations and MNase digestion durations and Sono-Seq profiles (bottom tracks). Note the differences in scale in IP and Input tracks. (c) Average profiles of 2.5′ MNase- and 10′ MNase digestion, 80mM len50 (top panel) and all sizes (bottom panel) Reb1 ORGANIC profiles centered at 10′ MNase, 80mM Reb1 ORGANIC sites.
Supplementary Figure 3. ORGANIC profiling robustly identifies Abf1 binding sites. (a) Representative paired-end sequencing fragment length distributions for Abf1 ORGANIC input and IP libraries. (b) Representative tracks of Abf1 ORGANIC IP and input tracks at indicated salt concentrations and MNase digestion durations and Sono-Seq profiles (bottom tracks). Note the differences in scale in IP and Input tracks. (c) Average profiles of 2.5′ MNase- and 10′ MNase digestion, 80mM len50 (top panel) and all sizes (bottom panel) Abf1 ORGANIC profiles centered at 10′ MNase, 80mM Abf1 ORGANIC sites.
Supplementary Figure 4. Reproducibility of TF binding site and occupancy detection by ORGANIC profiling. (a–g) Pairwise comparisons of Reb1 (a–e) and Abf1 (f–g) datasets. Venn diagrams of sites unique to and shared by each dataset in the indicated comparison (top panels) and correlation plots of occupancies of shared sites (bottom panels). Note that x- and y- axis scales in correlation plots were adjusted to show the majority of data points. R2 values and trend lines are from linear regression.
Supplementary Figure 5. Comparison of Reb1 ORGANIC profiles and ChIP-exo ‘primary’ and ‘secondary’ sites. (a) Overlap between 80 mM, 150 mM, and 600 mM ORGANIC Reb1 sites and ChIP-exo ‘primary’ and ‘secondary’ sites and X-ChIP-chip sites. (b) Correlation between Reb1 occupancies detected by ChIP-exo and ORGANIC profiling at 80 mM, 150 mM, and 600 mM with R2 from linear regression. (c) DNaseI protection (average DNaseI-Seq tag counts) at ChIP-exo ‘primary’ and ‘secondary’ sites. (d) DNaseI-seq tag counts at ChIP-exo primary and secondary sites. Heatmaps are ranked by descending ChIP-exo tag counts and graphs represent average DNaseI tag count for occupancy groups, where each occupancy group represents 20% of total sites detected. Note that the lack of a footprint in the ChIP-exo ‘primary’ site third occupancy group is due to a subset of highly accessible sites located proximal to the yeast rDNA locus. (e) phastCons evolutionary conservation at ChIP-exo sites. (f) Average Sono-Seq normalized counts at ChIP-exo sites.
Supplementary Figure 6. High sensitivity and specificity of Reb1 and Abf1 binding detected by ORGANIC profiling. Motif score histograms of ORGANIC profiling and ChIP-exo sites scored using the ChIP-exo-derived Reb1 position-specific log-odds matrix.
Supplementary Figure 7. Reb1 ORGANIC profile occupancies are correlated with DNaseI-Seq footprint depth. (a, b) DNaseI-Seq at Reb1 ORGANIC (a) and Reb1 ChIP-exo (b) sites. Heatmaps of DNaseI-Seq tag count are ranked by decreasing occupancy and each graph shows the average DNaseI-Seq tag count at the indicated occupancy group, with each occupancy group representing 20% of total sites detected.
Supplementary Figure 8. Abf1 ORGANIC profile occupancies are correlated with DNaseI-Seq footprint depth. DNaseI-Seq at Abf1 ORGANIC sites with heatmaps of DNaseI-Seq tag counts ranked by decreasing occupancy. Each graph shows the average DNaseI-Seq tag count at the indicated occupancy group, with each occupancy group representing 20% of total sites detected.
Supplementary Figure 9. Reb1 does not shift to Drosophila chromatin in mixed yeast/fly ORGANIC profiling. (a) Correlation between occupancies detected by replicates of the yeast/fly mixing experiment. (b) Correlation between occupancies detected by yeast/fly mixed and yeast-only 150 mM ORGANIC profiles. (c) Motif score distribution of peaks called from reads mapping to the Drosophila genome from the yeast/fly-mixed Reb1 ORGANIC profile scored using the Reb1 2.5′ MNase 80 mM ORGANIC PSSM (Supplementary Table 2).
Supplementary Figure 10. Abf1 and Reb1 do not redistribute to thermodynamically favorable binding sites. (a–c) Relationship between TF binding site motif score and occupancy at (a) Abf1 ORGANIC 600 mM, (b) Reb1 ORGANIC 80 and 150 mM, and (c) ChIP-exo Reb1 sites with R2 values from linear regression analysis. Note that the y-axis was rescaled such that most of the data points are visible.
Supplementary Figure 11. Nucleosome occupancy around ORGANIC TF binding sites. (a) Nucleosome occupancy around Reb1 sites rank ordered by ORGANIC profile Reb1 occupancy (left panel) and nucleosome occupancy in a 2 kb window centered at Reb1 sites (right panel). (b) Nucleosome occupancy around Abf1 sites rank ordered by ORGANIC Abf1 occupancy (left panel) and nucleosome occupancy in a 2 kb window (right panel).
Supplementary Figure 12. Sono-Seq chromatin accessibility and Reb1 and Abf1 sites determined using data from Auerbach, et al. (a–c) Sono-Seq (Auerbach, et al.) tag counts at ORGANIC 80, 150, and 600mM, ChIP-exo, and X-ChIP-chip sites (a), at ORGANIC 80 and 600mM and X-ChIP-chip Abf1 sites (b), and at ChIP-exo primary and secondary sites (c).
Supplementary Figure 13. GAF recovery in ORGANIC and modENCODE X-ChIP protocols. (a, b) Western blots of GAF and histone H3 from fractions taken at various stages of the 80 mM ORGANIC (a) and modENCODE X-ChIP (b) protocols (upper panels) and densitometric analysis normalized by fractional volume of total sample loaded (lower panels) with y-axes indicating background-corrected, sample volume-normalized ImageJ densitometry measurements (see Online Methods). Percentages above bars in densitometry measurements represent fraction of GAF or H3 relative to whole cell extracts. Note that the modENCODE X-ChIP pellet could not be completely solubilized and only the soluble fraction was loaded (see Online Methods).
Supplementary Figure 14. ORGANIC profiling robustly identifies Drosophila TF binding sites. (a) Representative tracks of GAF and Psq ORGANIC profiles. (b) Comparison of ORGANIC GAF sites and modENCODE-defined TF hotspots with GAF annotation (see Online Methods).
Supplementary Figure 15. Drosophila TF ORGANIC sites are conserved in evolution and are found in DNaseI hypersensitive regions. (a) Average DNaseI-Seq tag counts and phastCons evolutionary conservation at GAF X-ChIP-chip, X-ChIP-seq, and ORGANIC sites and GAF-bound HOT sites. (b) Average DNaseI-Seq and phastCons evolutionary conservation at Psq ORGANIC sites. (c) Representative tracks of GAGA-motif clusters with specific, discrete GAF binding events detected by ORGANIC profiling. Motif matches were identified by log-odds matrix-based scoring as described in the Online Methods with p < 5e-3.
Supplementary Figure 16. ORGANIC profiling identifies Reb1 and Abf1 binding sites in inaccessible chromatin. (a, b) MNase accessibility at Reb1 (a) and Abf1 (b) sites in 2 kb windows centered at ORGANIC, X-ChIP-chip, and/or ChIP-exo sites.
Supplementary Figure 17. Genomic context analysis of Reb1 and Abf1 ORGANIC and X-ChIP-chip binding sites. (a) Genomic contexts of ORGANIC, X-ChIP-chip and ChIP-exo Reb1 sites. (b) Number of sites falling into intergenic (top), genic (middle), and telomeric (bottom) regions and associated MEME-ChIP-derived motifs (right panel). Telomeric motifs for the 80, 150, and 600mM Reb1 ORGANIC datasets are slightly truncated outside of the core Reb1 consensus (delimited by the gray box). High confidence motifs could not be recovered by MEME-ChIP for X-ChIP-chip genic sites (n = 8) or X-ChIP-chip telomeric sites (n = 0). By chance, 550 Reb1 motifs are expected to occur within genes (see Online Methods). (c) Relative fraction of binding sites in intergenic, genic, telomeric, and ARS (Autonomously Replicating Sequence) annotated regions. (d) Number of sites in each dataset with the indicated annotation and corresponding MEME-ChIP-derived motif. High confidence motifs could not be recovered by MEME-ChIP for ChIP-chip telomeric sites (n = 0) or ChIP-chip ARS sites (n = 2). By chance, 137 Abf1 motifs are expected to occur within genes (see Online Methods).
Reb1: 2.5 min MNase/80mM NaCl/len50, 10 min MNase/80mM NaCl/len50, 10 min MNase/150mM NaCl/len50, 10 min MNase/600mM NaCl/len50; Abf1: 2.5 min MNase/80mM NaCl/len50, 10 min MNase/80mM NaCl/len50, 10 min MNase/600mM NaCl/len50; Psq len25 and GAF len25. Thresholds used for peak calling (see Online Methods) are indicated.
For a given log-odds entry (row, column) in the matrix, row specifies the nucleotide and column the position in the motif.