Abstract
Epithelial-mesenchymal transition (EMT) is a highly conserved morphogenic process defined by the loss of epithelial characteristics and the acquisition of a mesenchymal phenotype. EMT is associated with increased aggressiveness, invasiveness, and metastatic potential in carcinoma cells. To assess the contribution of extracellular vesicles following EMT, we conducted a proteomic analysis of exosomes released from Madin-Darby canine kidney (MDCK) cells, and MDCK cells transformed with oncogenic H-Ras (21D1 cells). Exosomes are 40–100 nm membranous vesicles originating from the inward budding of late endosomes and multivesicular bodies and are released from cells on fusion of multivesicular bodies with the plasma membrane. Exosomes from MDCK cells (MDCK-Exos) and 21D1 cells (21D1-Exos) were purified from cell culture media using density gradient centrifugation (OptiPrep™), and protein content identified by GeLC-MS/MS proteomic profiling. Both MDCK- and 21D1-Exos populations were morphologically similar by cryo-electron microscopy and contained stereotypical exosome marker proteins such as TSG101, Alix, and CD63. In this study we show that the expression levels of typical EMT hallmark proteins seen in whole cells correlate with those observed in MDCK- and 21D1-Exos, i.e. reduction of characteristic inhibitor of angiogenesis, thrombospondin-1, and epithelial markers E-cadherin, and EpCAM, with a concomitant up-regulation of mesenchymal makers such as vimentin. Further, we reveal that 21D1-Exos are enriched with several proteases (e.g. MMP-1, -14, -19, ADAM-10, and ADAMTS1), and integrins (e.g. ITGB1, ITGA3, and ITGA6) that have been recently implicated in regulating the tumor microenvironment to promote metastatic progression. A salient finding of this study was the unique presence of key transcriptional regulators (e.g. the master transcriptional regulator YBX1) and core splicing complex components (e.g. SF3B1, SF3B3, and SFRS1) in mesenchymal 21D1-Exos. Taken together, our findings reveal that exosomes from Ras-transformed MDCK cells are reprogrammed with factors which may be capable of inducing EMT in recipient cells.
Epithelial-mesenchymal transition (EMT)1 is a cellular process whereby otherwise sessile epithelial cells undergo a shift in plasticity and acquire the ability to disseminate (1–6). Hallmarks of EMT include diminished expression of cell-cell contact and adhesion components (e.g. E-cadherin), diminished expression of cell-matrix components, decreased expression of components involved in cell polarity, elevated expression of proteins involved in cytoskeleton remodelling (e.g. vimentin), and increased expression of various matrix metalloproteinases (7). Established as a central process during the early stages of development (8, 9), EMT also has implications in wound healing, fibrosis and, more recently, cancer progression (10–12). In the latter, EMT is thought to promote metastasis by triggering invasive and anti-apoptotic mechanisms in tumor cells, stimulate the cancer stem cell phenotype, and activate the tumor microenvironment via structural and biochemical modifications (13). Although, crosstalk between numerous intracellular signaling pathways are known to regulate EMT (14), it is now emerging that the EMT process can modulate the tumor microenvironment (15).
The complexity of the tumor microenvironment goes far beyond occupant epithelial cancer cells containing several nonmalignant, albeit genetically altered, heterotypic cell types (e.g. fibroblasts, endothelial cells, and immune cells) (16). Crosstalk is possible, either physically or via secretion of components such as extracellular matrix (ECM) proteins, enzymes, or paracrine signaling molecules such as growth factors and inflammatory cytokines (collectively referred to as the secretome) (17–19). Given that cancer cells at the leading tumor edge can undergo EMT and initiate metastatic lesion formation in response to signals from the microenvironment (11, 20), considerable effort has been directed toward characterizing the tumor secretome (21, 22). To identify extracellular modulators of EMT, which may influence tumor cell state and invasive potential, we have previously analyzed the secretome (soluble-secreted proteins) from Madin-Darby canine kidney (MDCK) and Ras-transformed MDCK (21D1) cells (23, 24). This proteomic-based approach enabled an unbiased global overview of events occurring in the extracellular microenvironment. The expression of components mediating cell-cell and cell-matrix adhesion (collagen XVII, IV, and laminin 5) were attenuated, with concordant up-regulation of proteases and ECM constituents promoting cell motility and invasion (MMP-1, TIMP-1 kallikrein-6, -7, fibronectin, collagen I, fibulin-1, -3, biglycan, decorin, S100A4 and SPARC) (23, 24). It is becoming increasingly clear that in addition to the soluble-secreted cytokines and chemokines that mediate cell communication at primary and secondary tumor sites (25), extracellular membranous vesicles, including exosomes, are important regulators of the tumor microenvironment (19, 26, 27).
Extracellular vesicles (EVs) are capable of enhancing the invasive potential of breast cancer and induce angiogenesis and metastasis in lung cancer (28, 29). In addition, transfer of oncogenic potential to a recipient cell through activation of MAPK and Akt signaling pathways highlights new mechanisms of intercellular communication via EVs in the tumor microenvironment (30, 31). EVs can be categorized by size with apoptotic bodies ranging up to 4000 nm in diameter, shed microvesicles and ectosomes 100–1000 nm, and 40–100 nm exosomes (32, 33). Importantly, exosomes have been associated with modulating the immune response, controlling tumor stroma in the metastatic niche, activating signaling pathways, and transferring genetic and oncogenic information to neighboring cells (32, 34–38). Although many functional activities have been ascribed to exosomes, it should be noted that the majority of sample preparations used for functional studies are heterogeneous in nature containing several EV types including shed microvesicles, exosomes, and apoptotic blebs. As a first step toward characterizing the specific contribution of exosomes to the tumor microenvironment, we report in this study the first protein analysis of highly purified exosomes before and after the EMT process. Comparison of MDCK exosome protein profiles following oncogenic Ras-induced EMT revealed extensive reprogramming in favor of components promoting metastatic niche formation. Additionally, enrichment of transcription and splicing factors known to induce EMT were observed in 21D1 exosomes, suggesting that a recipient cell may undergo EMT following exosome uptake.
EXPERIMENTAL PROCEDURES
Cell Culture and CCM Preparation
MDCK cells (39) and oncogenic H-Ras-transformed MDCK derivative 21D1 cells (23, 24) were routinely cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, NY, USA) supplemented with 10% FCS (Invitrogen), at 37 °C with 10% CO2. MDCK and 21D1 cells were grown to 70% confluence in DMEM containing 10% fetal calf serum (FCS), washed three times with serum-free DMEM, and left to culture in this medium at 37 °C with 10% CO2 for 24 h. Culture medium (CM) from 60 dishes of each cell line (a total of 900 ml from ∼3 × 108 cells) was harvested and centrifuged twice (480 × g 5 min, 2000 × g 10 min) to sediment floating cells and remove cellular debris. CM was centrifuged at 10,000 × g for 30 min to remove shed microvesicles. The resultant supernatant was filtered using a VacuCap® 60 filter unit fitted with a 0.1 μm Supor® Membrane (Pall Life Sciences, Port Washington, NY) and then concentrated to 1 ml concentrated culture medium (CCM) using Amicon® Ultracel-15 centrifugal filter devices with a 5K nominal molecular weight limit (NMWL) (Merck-Millipore, MA).
Exosome Isolation Using OptiPrep™ Density Gradient
Exosomes were isolated as previously described (40). Briefly, to prepare the discontinuous iodixanol gradient, 40% (w/v), 20% (w/v), 10% (w/v) and 5% (w/v) solutions of iodixanol were made by diluting a stock solution of OptiPrep™ (60% (w/v) aqueous iodixanol from Axis-Shield PoC, Norway) with 0.25 m sucrose/10 mm Tris, pH 7.5. The gradient was formed by adding 3 ml of 40% iodixanol solution to a 14 × 89 mm polyallomer tube (Microfuge® Tube, Beckman Coulter), followed by careful layering of 3 ml each of 20 and 10% solutions, and 2 ml of the 5% solution. For each exosome preparation, CCM (1 ml) was overlaid on the gradient, and centrifugation performed at 100,000 × g for 18 h at 4 °C. Twelve individual 1 ml gradient fractions were collected manually (with increasing density). Fractions were diluted with 2 ml PBS and centrifuged at 100,000 × g for 3 h at 4 °C followed by washing with 1 ml PBS, and resuspended in 50 μl PBS. Fractions were monitored for the expression of exosomal markers Alix and TSG101 by Western blotting. To determine the density of each fraction, a control OptiPrep™ gradient containing 1 ml of 0.25 m sucrose/10 mm Tris, pH 7.5 was run in parallel. Fractions were collected as described, serially diluted 1:10,000 with water, and the iodixanol concentration determined by absorbance at 244 nm using a molar extinction coefficient of 320 L g−1cm−1 (41).
Protein Quantitation
The protein content of exosome preparations was estimated by 1D-SDS-PAGE/SYPRO® Ruby protein staining densitometry. This method is reproducible, has a linear quantitation range over three orders of magnitude (42), and is compatible with GeLC-MS/MS (43). Briefly, 5 μl sample aliquots were solubilized in SDS sample buffer (2% (w/v) sodium dodecyl sulfate, 125 mm Tris-HCl, pH 6.8, 12.5% (v/v) glycerol, 0.02% (w/v) bromphenol blue) and loaded into 1 mm, 10-well NuPAGE™ 4–12% (w/v) Bis-Tris Precast gels (Invitrogen). Electrophoresis was performed at 150 V for 1 h in NuPAGE™ 1 × MES running buffer (Invitrogen) using an XCell Surelock™ gel tank (Invitrogen). After electrophoresis, gels were removed from the tank and fixed in 50 ml fixing solution (40% (v/v) methanol, 10% (v/v) acetic acid in water) for 30 min on an orbital shaker and stained with 30 ml SYPRO® Ruby (Invitrogen, NY, USA) for 30 min, followed by destaining twice in 50 ml of 10% (v/v) methanol with 6% (v/v) acetic acid in water for 1 h. Gels were imaged on a Typhoon 9410 variable mode imager (Molecular Dynamics, Sunnyvale, USA), using a green (532 nm) excitation laser and a 610BP30 emission filter at 100 μm resolution. Densitometry quantitation was performed using ImageQuant software (Molecular Dynamics) to determine protein concentration relative to a BenchMark™ Protein Ladder standard of known protein concentration (1.7 μg/μl) (Invitrogen). The yield of purified exosomes was ∼60 μg from 3 × 108 cells for both MDCK- and 21D1-Exos.
Western Blot Analysis
Exosome samples (∼10 μg protein) were prepared for Western blot analysis as previously described (44). Membranes were probed with primary mouse anti-TSG101 (BD Transduction Laboratories; 1:500), mouse anti-Alix (Cell Signaling Technology, Daanvers, MA; 1:1000), mouse anti-H-Ras (Santa Cruz Biotechnology, Santa Cruz, CA; 1:500), mouse anti-E-cadherin (BD Transduction Laboratories; 1:1000), rabbit anti-EpCAM (Abcam, Cambridge, MA; 1:1000), rabbit anti-MMP-1 (Santa Cruz Biotechnology; 1:200), rabbit anti-YB-1(YBX1) (Abcam; 1:500) or mouse anti-vimentin (Merck-Millipore; 1:500), for 1 h in TTBS (50 mm Tris, pH 7, 150 mm NaCl, 0.05% (v/v Tween 20) followed by incubation with the secondary antibody, IRDye 800 goat anti-mouse IgG or IRDye 700 goat anti-rabbit IgG (1:15000, LI-COR Biosciences, Lincoln, NE, USA), for 1 h in darkness. All antibody incubations were carried out using gentle orbital shaking at RT. Western blots were washed three times in TTBS for 10 min after each incubation step and visualized using the Odyssey Infrared Imaging System, version 3.0 (LI-COR Biosciences).
Cryoelectron Microscopy
Purified MDCK-exosomes (MDCK-Exos) and 21D1-exosomes (21D1-Exos) were imaged using cryo-transmission electron microscopy (cryo-(EM)as previously described (39) with slight modifications. Briefly, Aurion Protein-G gold 10 nm (ProSciTech, QLD, Australia) was mixed at a 1:3 ratio with exosomes (2 μg) harvested from OptiPrep™ gradients suspended in PBS buffer and transferred onto glow-discharged C-flat holey carbon grids (ProSciTech). Excess liquid was blotted and grids were plunge-frozen in liquid ethane. Grids were mounted in a Gatan cryoholder (Gatan, Inc., Warrendale, PA, USA) in liquid nitrogen. Images were acquired at 300 kV using a Tecnai G2 F30 (FEI, Eidhoven, NL), in low dose mode.
GeLC-MS/MS
MDCK- and 21D1-Exos (20 μg) were lysed in SDS sample buffer, and proteins separated by SDS-PAGE and visualized by Imperial™ Protein Stain (Thermo Fisher Scientific), according to manufacturer's instructions. Gel lanes were cut into equal slices (33 × 2 mm) using a GridCutter (The Gel Company, San Francisco, CA) and individual gel slices were subjected to in-gel reduction, alkylation and trypsinization (45). Briefly, gel bands were reduced with 10 mm DTT (Calbiochem, San Diego, CA) for 30 min, alkylated for 20 min with 25 mm iodoacetic acid (Fluka, St. Louis, MO), and digested with 150 ng trypsin (Worthington Biochemical Corp, Freehold, NJ) for 4.5 h at 37 °C. Tryptic peptides were extracted with 50 μl 50% (v/v) acetonitrile, 50 mm ammonium bicarbonate, concentrated to ∼10 μl by centrifugal lyophilization and one technical replicate analyzed by LC-MS/MS. RP-HPLC was performed on a nanoAcquity® (C18) 150 × 0.15-mm-internal diameter reversed phase UPLC column (Waters, Milford, MA) using an Agilent 1200 HPLC coupled online to an LTQ-Orbitrap mass spectrometer equipped with a nanoelectrospray ion source (Thermo Fisher Scientific). The column was developed with a linear 60 min gradient with a flow rate of 0.8 μl/min at 45 °C from 0–100% solvent B where solvent A was 0.1% (v/v) aqueous formic acid and solvent B was 0.1% (v/v) aqueous formic acid/60% acetonitrile. Survey MS scans were acquired with the resolution set to a value of 30,000. Real time recalibration was performed using a background ion from ambient air in the C-trap (46). Up to five selected target ions were dynamically excluded from further analysis for 3 min. An additional biological replicate of MDCK- and 21D1-Exos (20 μg) was analyzed on an LTQ-Orbitrap mass spectrometer (supplemental Data) to validate our primary findings. Raw mass spectrometry data is deposited in the PeptideAtlas and can be accessed at http://www.peptideatlas.org/PASS/PASS00225 (47–49).
Database Searching and Protein Identification
Peak lists were extracted using extract-msn as part of Bioworks 3.3.1 (Thermo Fisher Scientific). The parameters used to generate the peak lists were as follows: minimum mass 700, maximum mass 5000, grouping tolerance 0.01 Da, intermediate scans 200, minimum group count 1, 10 peaks minimum and total ion current of 100. Peak lists for each LC-MS/MS run were merged into a single MGF file for Mascot searches. Automatic charge state recognition was used because of the high-resolution survey scan (30,000). MGF files were searched using the Mascot v2.2.01 search algorithm (Matrix Science) against the LudwigNR_Q410 database with a taxonomy filter for human, cow, and dog, comprising 13112897 entries (http://www.ludwig.edu.au/archive/LudwigNR/LudwigNR.pdf). The search parameters consisted of carboxymethylation of cysteine as a fixed modification (+58 Da), NH2-terminal acetylation (+42 Da) and oxidation of methionine (+16 Da) as variable modifications. A peptide mass tolerance of ±20 ppm, #13C defined as 1, fragment ion mass tolerance of ±0.8 Da, and an allowance was made for up to two missed tryptic cleavages. Protein identifications were firstly clustered and analyzed by an in-house developed program MSPro (50). Briefly, peptide identifications were deemed significant if the Ion score was ≥ the Homology score. False-positive protein identifications were estimated by searching MS/MS spectra against the corresponding reverse-sequence (decoy) database (50). MDCK- and 21D1- exosome protein identifications were based on a protein score above the 1% false discovery rate cut-off of 48, and with at least two significant peptides. The BioMart data-mining tool (http://www.ensembl.org/biomart/index.html) was used to obtain Ensembl protein description and gene name as described (51). UniProt (http://www.uniprot.org) and Protein Information Resource (http://pir.georgetown.edu) were used to obtain gene ontology (GO) annotation.
Semiquantitative Label-free Spectral Counting
Significant spectral count fold change ratios (RSC) were determined using a modified formula from a previous serial analysis of gene expression study by Beissbarth et al. (52).
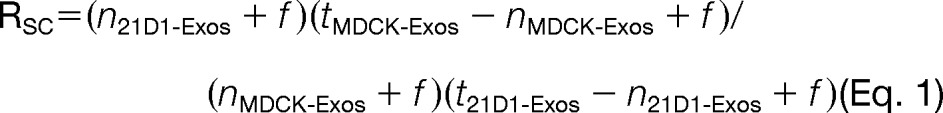 |
where, n is the significant protein spectral count (a peptide spectrum is deemed significant when the Ion score ≥ the Homology score), t is the total number of significant spectra in the sample, and f a correction factor set to 1.25 (53). Total number of spectra was only counted for significant peptides identified (Ion score ≥ Homology score). When Rsc is less than 1, the negative inverse Rsc value was used. The number of significant assigned spectra for each protein was used to determine whether protein abundances between the two categories (MDCK- and 21D1-Exos). For each protein the Fisher's Exact test was applied to significant assigned spectra. The resulting p values were corrected for multiple testing using the Benjamini-Hochberg procedure (54) and computations carried out in R (55).
RESULTS AND DISCUSSION
Exosomes are Released from MDCK Cells Following Oncogenic H-Ras Induced EMT
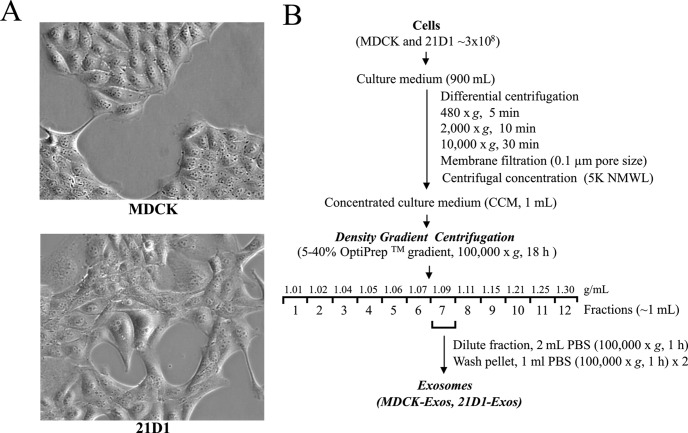
Previously, we established that cultured MDCK cells undergoing oncogenic H-Ras mediated EMT (21D1 cells) secrete protein components that extensively remodel the extracellular microenvironment, e.g. increased expression of ECM proteins, migration factors, and proteases that promote cell motility and invasion (23, 24). MDCK cells exhibit cobblestone-like morphology while 21D1 cells displayed a spindle-shaped mesenchymal phenotype (Fig. 1A). To maintain the mesenchymal phenotype, 21D1 cells require coculture with their own culture medium (supplemental Fig. S1). To isolate exosomes, MDCK and 21D1 cells were cultured to ∼70% confluence, washed with DMEM, and then left to culture in serum-free medium for 24 h. We have previously shown that both cell lines remain greater than 96% viable during this time (23). Culture medium (CM) from ∼3 × 108 cells was harvested, concentrated (CCM) by centrifugal membrane ultrafiltration and crude exosomes were fractionated based upon their buoyant density into 12 fractions using iodixanol density gradient centrifugation (40) as outlined in Fig. 1B. Western blot analysis of these fractions revealed enrichment of exosomes (based on exosome markers Alix/PDCD6IP and TSG101) in a fraction with buoyant density 1.09 g/ml (Fig. 2A, Supplemental Fig. S2). Interestingly, H-Ras was found in both MDCK- and 21D1-Exos, but with much higher levels in 21D1-Exos (Fig. 2A). The existence of H-Ras in the MDCK-Exos suggests that endogenously expressed Ras is implicated in secretory exosomal trafficking; however, it is not clear which form of H-Ras this is (inactive Ras-ADP or active Ras-ATP). Given that v-H-Ras expressed in 21D1 cells is the mutated active form, and that higher levels are observed in 21D1-Exos, it is suggestive of the involvement of the membrane-bound active Ras-ATP form in the secretory 21D1-Exos. This is consistent with the findings regarding K-Ras by Demory Beckler et al., (56). The yield of purified exosomes was ∼60 μg from 3 × 108 cells for both MDCK- and 21D1-Exos. Cryo-EM of purified exosomes revealed a relatively homogenous population of round membranous vesicles 40–100 nm in size, which is in accordance with the typical size reported for exosomes (Fig. 2B) (33).
Fig. 1.
Isolation of exosomes released from MDCK and 21D1 cells. A, Phase contrast images of MDCK cells reveal epithelial cobblestone-like morphology, whereas 21D1 cells display an elongated mesenchymal-like spindle shape. B, Experimental workflow for MDCK and 21D1-Exos isolation.
Fig. 2.
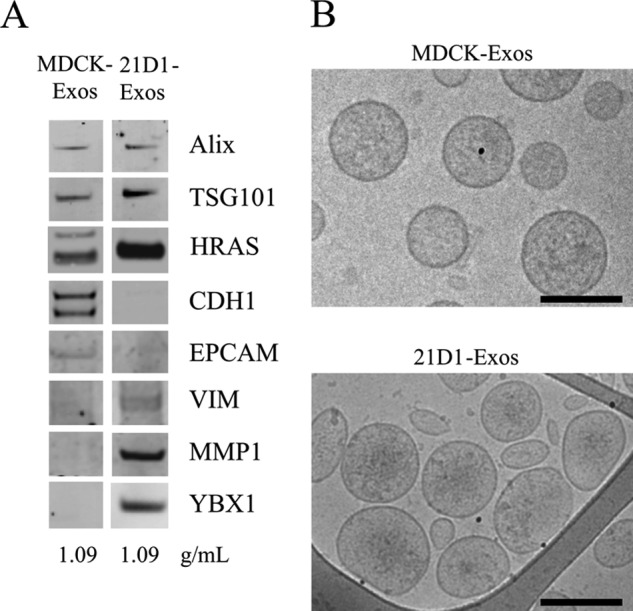
Characterization of MDCK- and 21D1-Exos. A, For Western blotting, exosome preparations (10 μg) were separated by 1D-SDS-PAGE, electrotransferred, and probed with exosome markers Alix and TSG101. Additionally, exosomes were probed with epithelial cell markers CDH1 (E-Cadherin) and EpCAM revealing a down-regulation in 21D1-Exos as compared with MDCK-Exos. HRAS (H-Ras), VIM (vimentin) MMP1 (interstitial collagenase), and YBX1 were significantly enriched in 21D1-Exos. B, MDCK- and 21D1-Exos were imaged using cryo-electron microscopy to reveal textured round vesicles between 40–100 nm. Scale bar, 100 nm.
Proteome Analysis of MDCK- and 21D1-Exos
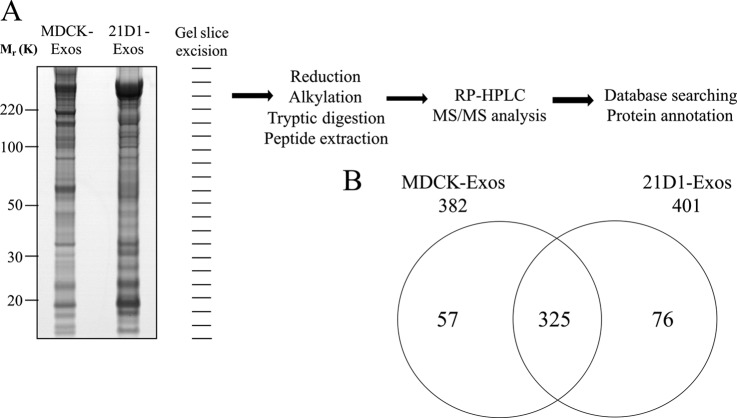
We next compared the protein profiles of MDCK- and 21D1-Exos using GeLC-MS-MS. Protein visualization using ImperialTM Protein Stain indicates significant differences in MDCK- and 21D1-Exos protein profiles following oncogenic H-Ras induced EMT (Fig. 3A). GeLC-MS/MS profiling (45) identified a total of 458 proteins, comprising 382 and 401 in MDCK- and 21D1-Exos, respectively (Fig. 3B and supplemental Tables S1-S3). Of the 325 proteins common to both MDCK- and 21D1-Exos, many are involved in exosome biogenesis (e.g. proteins involved in the endosomal sorting complex required for transport (ESCRT) machinery such as TSG101, VPS28, VPS37B, and ESCRT accessory protein Alix (57)), coordination of intracellular vesicle trafficking (e.g. tetraspanins such as CD63 and CD9 (58, 59), small Rab GTPases such as RAB1B, RAB5A, RAB5B, RAB5C, RAB7A, RAB11A, RAB14, RAB21 (60–62)), and annexins such as ANXA1, ANXA2, ANXA4, ANXA7, ANXA8, ANXA11 (63). 247 of the 325 common proteins (76%) have been reported by other researchers to be present in exosomes released from diverse cell types (see exosomes database ExoCarta containing 13,333 protein entries, Download 4 - release date: 29 May 2012 http://exocarta.org/index.html) (64, 65). Overall, 139 of the 458 MDCK- and 21D1-Exos proteins identified in this study have not been reported in ExoCarta (supplemental Table S4). According to GO subcellular annotation, 28 of these proteins are secreted, 12 cell membrane, 12 membrane, and 4 lipid-anchor proteins. Protein CYR61 (CYR61) is involved in promoting cell proliferation, chemotaxis, angiogenesis and cell adhesion, while protein jagged-1 (JAG1), VEGFR-1 receptor (FLT1), MMP19, and ADAMTS1 involved in angiogenesis and signal transduction. Components involved in cell signaling include AP1M2, CD109, the COP9 signalosome complex protein subunit COPS3, and several tetraspanin proteins, such as TSPAN4 and TSPAN9 were identified. A biological replicate of MDCK- and 21D1-Exos revealed 88% (403/458) similarity in overall protein identifications (supplemental Table S1).
Fig. 3.
Proteomic analysis of exosomes. A, MDCK- and 21D1-Exos proteins were separated by 1D-SDS-PAGE and stained with ImperialTM Protein Stain. Individual gel slices were excised and subjected to in-gel reduction, alkylation, and tryptic digestion. Extracted peptides were separated by reverse phase-high performance liquid chromatography (RP-HPLC) followed by mass spectrometry analysis, database searching and protein annotation. B, A two-way Venn diagram of MDCK- and 21D1-Exos reveals 325 proteins were commonly identified, whereas 57 and 76 proteins were uniquely identified in MDCK- and 21D1-Exos, respectively (supplemental Tables S1-S3).
EMT Hallmark Proteins are Observed in Exosomes Following Oncogenic H-Ras Induced EMT
We next examined whether the pattern of EMT hallmarks typically seen in whole cells (7) are reflected in exosomes released from MDCK cells following H-Ras modulated EMT. For this purpose we used relative spectral count ratios (Rsc) and Western immunoblotting to indicate differential protein expression between samples. Proteins mediating cell-cell contact, cell-matrix contact and cell polarity displayed decreased expression levels in 21D1-Exos (Table I and Fig. 2A), correlating with typical EMT hallmarks seen in whole cells (7). Foremost of these were the adhesive glycoprotein and inhibitor of angiogenesis thrombospondin 1 (THBS1 Rsc -482.8), and the epithelial cell markers E-cadherin (CDH1 Rsc -34.4) and EpCAM (Rsc -16.5). Consistent with these findings were the elevated protein expression levels in 21D1-Exos of vimentin (VIM, Rsc 8.1) and matrix metalloproteins, MMP-1 (Rsc 7.3), MMP-19 (Rsc 11.3) and MMP-14 (Rsc 3.4), typically observed in mesenchymal cells. Confirmatory data for the different abundance levels of CDH1, EpCAM, VIM, and MMP-1 observed in MDCK- and 21D1-Exos was obtained by Western blot analysis (Fig. 2A).
Table I. EMT hallmark proteins identified in MDCK- and 21D1-Exos.
| Categorya | Gene Name | Protein Description | Spectral countsb |
Protein abundance ratioc | |
|---|---|---|---|---|---|
| MDCK-Exos | 21D1-Exos | ||||
| Cell-cell contact | THBS1 | Thrombospondin-1 | 555 | −482.8* | |
| CDH1 | E-cadherin | 41 | −34.4* | ||
| EPCAM | Epithelial cell adhesion molecule | 19 | −16.5* | ||
| Cell-matrix contact | COL12A1 | Collagen alpha-1(XII) chain | 90 | −74.8* | |
| LAMA3 | Laminin subunit alpha-3 | 83 | −69.0* | ||
| LAMB3 | Laminin beta 3 | 56 | −46.7* | ||
| LAMC2 | Laminin-5 gamma 2 | 61 | 1 | −28.2* | |
| COL5A1 | Collagen alpha-1(V) chain | 15 | −13.2* | ||
| LAMC1 | Laminin subunit gamma-1 | 91 | 11 | −7.7* | |
| HSPG2 | Perlecan | 1325 | 211 | −7.3* | |
| LAMB1 | Laminin subunit beta-1 | 89 | 13 | −6.5* | |
| LAMB2 | Laminin subunit beta-2 | 6 | −5.9 | ||
| COL17A1 | Collagen alpha-1(XVII) chain | 9 | 1 | −4.6* | |
| Cell polarity | MUC1 | Endometrial mucin-1 | 24 | −20.5* | |
| CLDN3 | Claudin-3 | 10 | 2 | −3.5 | |
| CLDN4 | Claudin-4 | 12 | 3 | −3.2 | |
| CLDN6 | Claudin-6 | 7 | 5 | −1.3 | |
| Cytoskeleton remodeling | RHOA | Transforming protein RhoA | 2 | 5 | 1.9 |
| VIM | Vimentin | 9 | 8.1* | ||
| Proteases | MMP1 | Interstitial collagenase | 8 | 7.3* | |
| MMP14 | Matrix metalloproteinase-14 | 3 | 3.4 | ||
| MMP19 | Matrix metalloproteinase-19 | 13 | 11.3* | ||
| ADAMTS1 | A disintegrin and metalloproteinase with thrombospondin motifs 1 | 4 | 8 | 1.7 | |
| ADAM10 | ADAM-10 | 12 | 28 | 2.2 | |
b Significant protein spectral counts (SpC) where a peptide spectrum is deemed significant when the Ion score ≥ the Homology score (refer supplemental Table S1).
c Protein abundance ratio (ratio of spectral counts; Rsc) reveals differential protein abundance between MDCK- and 21D1-Exos based on Eq.1. The use of zero spectra is overcome using an arbitrary correction factor (1.25) in Eq. 1. The use of this correction factor allows relative quantitation of all proteins within both normalized datasets to be performed, based upon Old et al. [53]. Positive Rsc values reflect increased protein abundance in 21D1-Exos relative to MDCK-Exos; negative values indicate decreased abundance in 21D1-Exos relative to MDCK-Exos.
* Differential expression with p values <0.05 as reported in supplemental Table S1.
Exosomes Contain Metastatic Niche Factors Following Oncogenic H-Ras-Induced EMT
Melanoma-derived exosomes have been recently implicated in regulating the metastatic microenvironment in sentinel lymph nodes (27) and ‘educating’ circulating bone marrow progenitor cells to promote metastatic progression in vivo (66). In contrast to MDCK-Exos, interrogation of the protein profile of 21D1-Exos revealed increased expression of proteases, annexins, integrins, and other secreted proteins associated with the premetastatic niche formation (19, 67–74), the tumor microenvironment (75) and proteins assisting tissue invasion and metastasis (76, 77) (Table II).
Table II. Exosomal factors involved in metastatic niche formation and metastasis.
| Categorya | Gene Name | Protein description | Spectral countsb |
Protein abundance ratioc | |
|---|---|---|---|---|---|
| MDCK-Exos | 21D1-Exos | ||||
| Proteases | MMP1 | Interstitial collagenase | 8 | 7.3* | |
| MMP14 | Matrix metalloproteinase-14 | 3 | 3.4 | ||
| MMP19 | Matrix metalloproteinase-19 | 13 | 11.3* | ||
| ADAM10 | ADAM-10 | 12 | 28 | 2.2 | |
| ADAMTS1 | A disintegrin and metalloproteinase with thrombospondin motifs 1 | 4 | 8 | 1.7 | |
| Integrins | ITGB1 | Integrin beta-1 | 44 | 191 | 4.3* |
| ITGA3 | Integrin alpha-3 | 18 | 105 | 5.5* | |
| ITGAV | Integrin alpha-V | 6 | 23 | 3.3* | |
| ITGA6 | Integrin alpha-6 | 9 | 8.1* | ||
| Tetraspanins | CD81 | CD81 | 10 | 13 | 1.25 |
| CD82 | CD82 antigen | 2 | 6 | 3.25 | |
| CD151 | CD151 antigen | 4 | 21 | 4.19* | |
| Annexins | ANXA1 | Annexin A1 | 14 | 19 | 1.3 |
| ANXA2 | Annexin A2 | 12 | 24 | 1.9 | |
| ANXA4 | Annexin A4 | 2 | 3 | 1.3 | |
| ANXA7 | Annexin A7 | 2 | 3 | 1.3 | |
| ANXA8 | Annexin A8 | 1 | 4 | 2.3 | |
| ANXA11 | Annexin A11 | 8 | 11 | 1.3 | |
b Significant protein spectral counts (SpC) where a peptide spectrum is deemed significant when the Ion score ≥ the Homology score (refer supplemental Table S1).
c Protein abundance ratio (ratio of spectral counts; Rsc) reveals differential protein abundance between MDCK- and 21D1-Exos based on Eq.1. The use of zero spectra is overcome using an arbitrary correction factor (1.25) in Eq. 1. The use of this correction factor allows relative quantitation of all proteins within both normalized datasets to be performed, based upon Old et al. [53]. Positive Rsc values reflect increased protein abundance in 21D1-Exos relative to MDCK-Exos; negative values indicate decreased abundance in 21D1-Exos relative to MDCK-Exos.
* Differential expression with p values <0.05 as reported in supplemental Table S1.
Proteases
Proteases implicated in metastatic niche preparation and seen highly enriched in 21D1-Exos (relative to MDCK-Exos) include matrix metalloproteinases MMP-1 (Rsc 7.3), MMP-14 (Rsc 3.4), MMP-19 (Rsc 11.3), a disintegrin and metalloproteinase 10 (ADAM10) (Rsc 2.2), and ADAM with thrombospondin motif 1 (ADAMTS1) (Rsc 1.7). The interstitial collagenase MMP-1 is known to assist tumor-induced angiogenesis, tumor invasion, and establishment of metastatic regions at secondary sites (78). Presence of MMP-1 in human colorectal carcinomas correlates with the depth grading of tumor invasion, lymphatic invasion, and lymph node metastasis (79). MMP-14 promotes cell invasion and motility by pericellular ECM degradation, shedding of CD44 (also detected in 21D1-Exos) and syndecan 1, and through activation of ERK (80). Expression of MMP-19 is associated with increased invasion, migratory behavior and early metastasis of melanoma cells (81), and localization of MMP-14 and -19 at the invasive tumor front is characteristic of highly motile invading tumor cells (81, 82). The finding that MMP-14 and -19 are unique to 21D1-Exos and not observed in our previously published MDCK/21D1 secretome analysis (24), may represent a mechanism that allows exosome-bound proteases to traffic and function at distant/metastatic sites. ADAM proteases contain MMP-like catalytic domains (83) and are important mediators of cell surface protein shedding during tumor progression (84). Interestingly, ADAM10 has been shown to be an active vesicle-based protease, cleaving cell adhesion molecule L1 at the cell surface, and subsequently promoting cell migration (85). Given that other substrates of ADAM10 include components of the ECM, epidermal growth factors, chemokines, cytokines, and Notch receptor when bound to its ligands Delta-like 1 or Jagged-1 (also unique to 21D1-Exos, Rsc 4.9) (84), ADAM10 has the ability to extensively modify the tumor microenvironment. Likewise, ADAMTS1 is also capable of degrading various ECM components (86), and increased expression promotes pulmonary metastasis of mammary carcinoma and Lewis lung carcinoma cells (87). ADAMTS1 has also been shown to modulate the metastatic tumor microenvironment by promoting angiogenesis and invasion in osteoclastogenesis (88). These findings suggest that addition to soluble proteases, exosome-associated proteases ADAM10 and ADAMTS1 may also contribute to the EMT process (78) and, additionally, play a role in premetastatic niche formation (19).
Integrins
Integrins represent another class of metastatic niche components that were enriched in 21D1-Exos (Table II). Integrins facilitate cell attachment to surrounding ECM, initiating intracellular signaling cascades that maintain cell survival, proliferation, adhesion, migration and invasion (89). The finding of enriched protein levels of integrins in 21D1-Exos is of particular significance given that a study of ovarian carcinoma identified that collagen-induced activation of integrin receptors caused Ras, Erk and Akt pathway activation (90). In particular, integrins subunits α3, α6, αV and β1, all of which were enriched in 21D1-Exos, have been associated with modulating ECM-induced signaling leading to proliferation, adhesion, migration and invasion of the ovarian cancer cells (90). Moreover, αV mediates latent TGF-β activation, which is required for the maintenance of EMT and tumor cell invasion and dissemination (91). These findings are in accord with our earlier studies showing plasma membrane bound integrins α6β1 and α3β1 were significantly enriched in cell membrane preparations of H-Ras transformed MDCK cells (21D1 cells) when compared with parental MDCK cells (51), further studies are required to ascertain whether these integrins are integral components of the 21D1-Exos membrane.
Tetraspanins
Tetraspanins are characterized by four transmembrane domains, intracellular N- and C termini and two extracellular domains. They are reported to function as scaffolding proteins, which interact with integrins; many tetraspanins have been implicated in tumor progression (92–94). In this study, we observed an enrichment in 21D1-Exos of tetraspanins involved in cancer progression including CD81, CD82, and CD151 (Table II). Interestingly, it has been previously shown that interactions between α6β1 (both integrin components identified in this study) and CD81 may up-regulate cell motility, affecting migration mediated by other integrins (95). Recently, CD81-positive fibroblast-derived exosomes, isolated using differential ultracentrifugation, were reported to regulate breast cancer cell protrusions and motility through Wnt-planar cell polarity signaling (96). Further, CD82 has been implicated in integrin-mediated functions including cell motility and invasiveness (97), while CD151 has been shown to promote cancer cell metastasis via integrins α3β1 and α6β1 (also seen in our study) in vitro (98).
Annexins
Annexins are involved in a diverse array of cellular functions and physiological processes including membrane scaffolding, trafficking and organization of vesicles, exocytosis, endocytosis, and cell migration (99). In this study, we observed increased expression levels of annexins A1, A2, A4, A7, A8, and A11 (Rsc 1.3–2.3) in 21D1-Exos (Table II). In particular, annexin A2 (Rsc 1.9), has been shown to regulate the tumor microenvironment by inducing the remodelling of cytoskeletal structures and actin of breast and colorectal cancer cells (100). siRNA-based experiments have recently demonstrated that annexin A2 is critical in determining the invasive potential of cancer cells, and regulates secretion of pro-angiogenic factors including MMP-14 (101). The precise functional roles played by other annexins during metastatic progression remain to be defined.
Transcriptional Regulators and Splicing Factors are Enriched in Exosomes Following H-Ras-induced EMT
It is well recognized that splicing events and transcription regulation drive critical aspects of EMT-associated phenotypic change (102, 103). For example, the EMT transcription factor twist altered global changes in mRNA splicing in a human mammary epithelial cell line (HMLE cells) resulting in many alternatively spliced genes that are implicated in processes such as cell migration, actin cytoskeletal regulation and cell-cell junction formation, all of which contribute to EMT phenotypic change (102). We report, for the first time, the presence of key transcriptional regulators (e.g. the master transcriptional regulator YBX1) and core splicing complex components in highly-purified exosomes.
Splicing Factors
Recent studies have highlighted an important contribution of alternative splicing to the metastatic cascade, including regulation of EMT at the post-transcriptional level (104, 105). Alternative splicing results in the expression of protein isoforms with distinct structural and functional characteristics, and can even give rise to proteins with opposite properties (106). The involvement of alternative splicing in EMT was first reported in relation to the fibroblast growth factor receptor 2 (FGFR2) (107), and since then, several splicing factors and spliced genes involved in cell migration, actin cytoskeletal regulation and cell-cell junction formation during EMT have been discovered (102, 108, 109). Several splicing factors were identified in 21D1-Exos (Table III) including the splicing regulator protein SRP20 (Rsc 2.7), and SF3B1 (Rsc 8.9) and SF3B3 (Rsc 2.6), which are components of the SF3b complex that interacts with U2 small nuclear ribonucleoprotein (snRNP) complex at the catalytic center of the spliceosome (110). Increased expression levels of splicing factor, arginine/serine-rich 1 (SFRS1/SRSF1) (Rsc 23.2), previously known as (SF2/ASF), in 21D1-Exos is of particular significance given its ability to induce EMT (111). SRSF1 has been shown to regulate the splicing of the tyrosine kinase receptor Ron which is synthesized as a single chain precursor, and is comprised of an extracellular 40 kDa α-subunit and a 145 kDa transmembrane β-subunit (112). SRSF1 promotes the production of ΔRon 165, which is an isoform lacking 49 amino acids in the extracellular β-subunit generated through the skipping of exon 11 (111, 113). ΔRon 165 is unable to undergo proteolytic processing and as a consequence accumulates in the cytoplasm in a constitutively phosphorylated form which induces invasive properties (114). By these means, SRSF1 affects the Ron/ΔRon ratio, which in turn, promotes the morphological and molecular hallmarks of EMT (111). SRSF1 is frequently up-regulated in various human tumors (115). Our finding of the proto-oncogene SRSF1 in H-Ras induced 21D1-Exos may represent a mechanism by which a recipient cell upon uptake of an SRSF1-containing exosome may induce the recipient cell to undergo EMT. Further studies are required to examine this hypothesis.
Table III. Splicing factors and transcription factors enriched in 21D1-Exos.
| Categorya | Gene name | Protein description | Spectral countsb |
Protein abundance ratioc | |
|---|---|---|---|---|---|
| MDCK-Exos | 21D1-Exos | ||||
| Splicing Factors | SF3B1 | Splicing factor 3B subunit 1 | 10 | 8.9* | |
| SF3B3 | Splicing factor 3B subunit 3 | 2 | 2.6 | ||
| SFRS1 | Splicing factor, arginine/serine-rich 1 | 28 | 23.2* | ||
| SRP20 | Serine/arginine-rich splicing factor 3 | 1 | 5 | 2.7 | |
| Transcription Factors | PURA | Transcriptional activator protein Pur-alpha | 2 | 2.6 | |
| NCL | Nucleolin | 28 | 23.2* | ||
| YBX1 | Nuclease-sensitive element-binding protein 1 | 46 | 37.5* | ||
| Other | EHD2 | EH domain-containing protein 2 | 35 | 28.7* | |
b Significant protein spectral counts (SpC) where a peptide spectrum is deemed significant when the Ion score ≥ the Homology score (refer supplemental Table S1).
c Protein abundance ratio (ratio of spectral counts; Rsc) reveals differential protein abundance between MDCK- and 21D1-Exos based on Eq.1. The use of zero spectra is overcome using an arbitrary correction factor (1.25) in Eq. 1. The use of this correction factor allows relative quantitation of all proteins within both normalized datasets to be performed, based upon Old et al. [53]. Positive Rsc values reflect increased protein abundance in 21D1-Exos relative to MDCK-Exos; negative values indicate decreased abundance in 21D1-Exos relative to MDCK-Exos.
* Differential expression with p values <0.05 as reported in supplemental Table S1.
Transcription Factors
A salient finding of this analysis was the identification of Y-box-binding protein (YBX1), a DNA- and RNA- binding protein that has properties of a nucleic acid chaperone (116), in 21D1-Exos (Table III). YBX1 was the most up-regulated protein in exosomes following EMT (Rsc 37.5), and its unique expression in 21D1-Exos was validated by Western blotting (Fig. 2A). YBX1 is known to be involved in almost all DNA- and mRNA-dependent processes including DNA replication and repair, transcription, pre-mRNA splicing, and mRNA translation (116), and is considered to be a master transcriptional regulator. YBX1 can bind RNA to limit protein synthesis, or bind DNA through the Y-box promoter element containing an inverted CCAAT box to either activate or repress transcription (117, 118). YBX1 is known to interact with other DNA binding proteins such as PURα (PURA), also uniquely present in 21D1-Exos (Rsc 2.6) (Table III). PURα regulates cell proliferation through the activation of growth-associated gene transcription (119, 120). MMP-13 expression is also known to be regulated by YBX1 (121), and given that MMP-13 was uniquely identified in MDCK-Exos (Rsc -79.8) it is possible that its diminished expression in 21D1-Exos is because of elevated YBX1 expression. MMP-13, also known as collagenase-3, is an ECM-degrading proteinase (122) that has been reported to be selectively down-regulated in conjunction with MMP-9, by the transcription factor SPDEF during prostate tumor metastasis (123). Given that YBX1 is known to promote an epithelial-mesenchymal transition through translational activation of snail1, it is interesting to hypothesize that 21D1-Exos may also induce EMT via YBX1 in recipient cells (124).
In summary, proteomic profiling of highly-purified exosomes has revealed new insights into the contribution of exosomes to the extracellular microenvironment after oncogenic H-Ras-induced EMT. We show that exosomes released from epithelial MDCK cells undergo extensive reprogramming causing exosome-mediated release of several factors associated with modifying the extracellular tumor microenvironment including proteases, annexins, integrins and secreted ECM components. It is possible that these factors may positively feedback on themselves to maintain the EMT process, or induce neighboring cells to undergo EMT. In addition, our findings reveal for the first time that oncogenic H-Ras transformation induces the packaging and release of mediators associated with nuclear assembly, transcription, splicing, and protein translation. Given that 21D1-Exos contain several features known to induce EMT, it is tempting to speculate that Ras-transformed exosomes are functionally capable of initiating EMT in recipient cells.
Supplementary Material
Footnotes
* This work was supported by the National Health & Medical Research Council (NHMRC) of Australia for program grant #487922 (RJS, JH, DWG), grants #280913 and #433619 (H-JZ), grants #628946 and #400202 (AFH). AFH is also supported by an Australian Research Council (www.arc.gov.au) Future Fellowship (FT100100560). RAM is supported by an Early Career CJ Martin Fellowship #APP1037043, and BMC by an NHMRC Dora Lush Biomedical Postgraduate Scholarship #628959. BJT is supported by The University of Melbourne Research Scholarship. Analysis of proteomic data described in this work was supported using the Australian Proteomics Computational Facility funded by the National Health & Medical Research Council of Australia grant #381413. Electron microscopy was performed at the Advanced Microscopy Facility at the Bio21 Molecular Science and Biotechnology Institute, The University of Melbourne. This work was also supported, in part, by American Recovery and Reinvestment Act funds through National Institutes of Health Grant R01 HG005805 (RLM), the NIGMS Grant 2P50 GM076547 from Center for Systems Biology, the Luxembourg Centre for Systems Biomedicine and the University of Luxembourg, and from the National Science Foundation (MRI Grant 0923536). UK was supported by a fellowship from the German Academic Exchange Service. We thank the NCI of the NIH for support (Grant #1R03CA156667 to RLM).
 This article contains supplemental Figs. S1 and S2 and Tables S1 to S4.
This article contains supplemental Figs. S1 and S2 and Tables S1 to S4.
1 The abbreviations used are:
- EMT
- epithelial-mesenchymal transition
- 21D1
- Ras-transformed MDCK cells
- 21D1-Exos
- Ras-transformed MDCK cell-derived exosomes
- ADAM
- a disintegrin and metalloproteinase
- CM
- culture medium
- CCM
- concentrated culture medium
- DMEM
- Dulbecco's Modified Eagle's Medium
- EM
- electron microscopy
- EVs
- extracellular vesicles
- FCS
- fetal calf serum
- MDCK
- Madin-Darby canine kidney
- MDCK-Exos
- Madin-Darby canine kidney derived exosomes
- MMP
- matrix metalloproteinase
- MVB
- multivesicular body
- NMWL
- nominal molecular weight limit
- Rsc
- relative spectral count fold change ratio
- TEM
- transmission electron microscopy
- YBX1
- Y-box binding protein 1.
REFERENCES
- 1. Hay E. D. (1995) An overview of epithelio-mesenchymal transformation. Acta Anatomica 154, 8–20 [DOI] [PubMed] [Google Scholar]
- 2. Hugo H., Ackland M. L., Blick T., Lawrence M. G., Clements J. A., Williams E. D., Thompson E.W. (2007) Epithelial–mesenchymal and mesenchymal–epithelial transitions in carcinoma progression. J. Cell. Physiol. 213, 374–383 [DOI] [PubMed] [Google Scholar]
- 3. Thiery J. P. (2002) Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2, 442–454 [DOI] [PubMed] [Google Scholar]
- 4. Thiery J. P., Sleeman J. P. (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 7, 131–142 [DOI] [PubMed] [Google Scholar]
- 5. Kalluri R., Weinberg R. A. (2009) The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nieto M. A. (2011) The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu. Rev. Cell Dev. Biol. 27, 347–376 [DOI] [PubMed] [Google Scholar]
- 7. Radisky D. C. (2005) Epithelial-mesenchymal transition. J. Cell Sci. 118, 4325–4326 [DOI] [PubMed] [Google Scholar]
- 8. Greenburg G., Hay E. D. (1982) Epithelia suspended in collagen gels can lose polarity and express characteristics of migrating mesenchymal cells. J. Cell Biol. 95, 333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Trelstad R. L., Hay E. D., Revel J. D. (1967) Cell contact during early morphogenesis in the chick embryo. Develop. Biol. 16, 78–106 [DOI] [PubMed] [Google Scholar]
- 10. Kalluri R., Neilson E. G. (2003) Epithelial-mesenchymal transition and its implications for fibrosis. J. Clin. Invest. 112, 1776–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Berx G., Raspe E., Christofori G., Thiery J. P., Sleeman J. P. (2007) Pre-EMTing metastasis? Recapitulation of morphogenetic processes in cancer. Clin Exp Metastasis 24, 587–597 [DOI] [PubMed] [Google Scholar]
- 12. Thompson E. W., Newgreen D. F., Tarin D. (2005) Carcinoma invasion and metastasis: a role for epithelial-mesenchymal transition? Cancer Res. 65, 5991–5995; discussion 5995 [DOI] [PubMed] [Google Scholar]
- 13. Nistico P., Bissell M. J., Radisky D. C. (2012) Epithelial-mesenchymal transition: general principles and pathological relevance with special emphasis on the role of matrix metalloproteinases. Cold Spring Harb Perspect Biol. 4, pii: a011908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang J., Weinberg R. A. (2008) Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 14, 818–829 [DOI] [PubMed] [Google Scholar]
- 15. Mathias R. A., Gopal S. K., Simpson R. J. (2012) Contribution of cells undergoing epithelial-mesenchymal transition to the tumour microenvironment. J. Proteomics 14, 545–557 [DOI] [PubMed] [Google Scholar]
- 16. Pelham R. J., Rodgers L., Hall I., Lucito R., Nguyen K. C., Navin N., Hicks J., Mu D., Powers S., Wigler M., Botstein D. (2006) Identification of alterations in DNA copy number in host stromal cells during tumor progression. Proc. Natl. Acad. Sci. U.S.A. 103, 19848–19853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang J. D., Nakamura I., Roberts L. R. (2011) The tumor microenvironment in hepatocellular carcinoma: current status and therapeutic targets. Semin Cancer Biol. 21, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bissell M. J., Radisky D. (2001) Putting tumours in context. Nat. Rev. Cancer. 1, 46–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peinado H., Lavotshkin S., Lyden D. (2011) The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Semin. Cancer Biol. 21, 139–146 [DOI] [PubMed] [Google Scholar]
- 20. De Wever O., Pauwels P., De Craene B., Sabbah M., Emami S., Redeuilh G., Gespach C., Bracke M., Berx G. (2008) Molecular and pathological signatures of epithelial-mesenchymal transitions at the cancer invasion front. Histochem. Cell Biol. 130, 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karagiannis G. S., Pavlou M. P., Diamandis E. P. (2010) Cancer secretomics reveal pathophysiological pathways in cancer molecular oncology. Mol. Oncol. 4, 496–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stastna M., Van Eyk J. E. (2012) Secreted proteins as a fundamental source for biomarker discovery. Proteomics 12, 722–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mathias R. A., Chen Y. S., Wang B., Ji H., Kapp E. A., Moritz R. L., Zhu H. J., Simpson R. J. (2010) Extracellular remodelling during oncogenic Ras-induced epithelial-mesenchymal transition facilitates MDCK cell migration. J. Proteome Res. 9, 1007–1019 [DOI] [PubMed] [Google Scholar]
- 24. Mathias R. A., Wang B., Ji H., Kapp E. A., Moritz R. L., Zhu H. J., Simpson R. J. (2009) Secretome-based proteomic profiling of Ras-transformed MDCK cells reveals extracellular modulators of epithelial-mesenchymal transition. J. Proteome Res. 8, 2827–2837 [DOI] [PubMed] [Google Scholar]
- 25. Psaila B., Lyden D. (2009) The metastatic niche: adapting the foreign soil. Nat. Rev. Cancer 9, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ratajczak J., Wysoczynski M., Hayek F., Janowska-Wieczorek A., Ratajczak M. Z. (2006) Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia 20, 1487–1495 [DOI] [PubMed] [Google Scholar]
- 27. Hood J. L., San R. S., Wickline S. A. (2011) Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 71, 3792–3801 [DOI] [PubMed] [Google Scholar]
- 28. Janowska-Wieczorek A., Marquez-Curtis L. A., Wysoczynski M., Ratajczak M. Z. (2006) Enhancing effect of platelet-derived microvesicles on the invasive potential of breast cancer cells. Transfusion 46, 1199–1209 [DOI] [PubMed] [Google Scholar]
- 29. Janowska-Wieczorek A., Wysoczynski M., Kijowski J., Marquez-Curtis L., Machalinski B., Ratajczak J., Ratajczak M. Z. (2005) Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int. J. Cancer 113, 752–760 [DOI] [PubMed] [Google Scholar]
- 30. Al-Nedawi K., Meehan B., Kerbel R. S., Allison A. C., Rak J. (2009) Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc. Natl. Acad. Sci. U.S.A. 106, 3794–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Skog J., Wurdinger T., van Rijn S., Meijer D. H., Gainche L., Sena-Esteves M., Curry W. T., Jr., Carter B. S., Krichevsky A. M., Breakefield X. O. (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee T. H., D'Asti E., Magnus N., Al-Nedawi K., Meehan B., Rak J. (2011) Microvesicles as mediators of intercellular communication in cancer–the emerging science of cellular ‘debris’. Semin. Immunopathol. 33, 455–467 [DOI] [PubMed] [Google Scholar]
- 33. Mathivanan S., Ji H., Simpson R. J. (2010) Exosomes: Extracellular organelles important in intercellular communication. J. Proteomics 73, 1907–1920 [DOI] [PubMed] [Google Scholar]
- 34. Ge R., Tan E., Sharghi-Namini S., Asada H. H. (2012) Exosomes in cancer microenvironment and beyond: have we overlooked these extracellular messengers? Cancer Microenviron. 5, 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hendrix A., Hume A. N. (2011) Exosome signaling in mammary gland development and cancer. Int. J. Dev. Biol. 55, 879–887 [DOI] [PubMed] [Google Scholar]
- 36. Kharaziha P., Ceder S., Li Q., Panaretakis T. (2012) Tumor cell-derived exosomes: A message in a bottle. Biochim. Biophys. Acta 1826, 103–111 [DOI] [PubMed] [Google Scholar]
- 37. Yang C., Robbins P. D. (2011) The roles of tumor-derived exosomes in cancer pathogenesis. Clin. Dev. Immunol. 2011, 842–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Putz U., Howitt J., Doan A., Goh C. P., Low L. H., Silke J., Tan S. S. (2012) The tumor suppressor PTEN is exported in exosomes and has phosphatase activity in recipient cells. Sci. Signal. 5, ra70. [DOI] [PubMed] [Google Scholar]
- 39. Madin S. H., Andriese P. C., Darby N. B. (1957) The in vitro cultivation of tissues of domestic and laboratory animals. Am. J. Vet. Res. 18, 932–941 [PubMed] [Google Scholar]
- 40. Tauro B. J., Greening D. W., Mathias R. A., Ji H., Mathivanan S., Scott A. M., Simpson R. J. (2012) Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 56, 293–304 [DOI] [PubMed] [Google Scholar]
- 41. Schroder M., Schafer R., Friedl P. (1997) Spectrophotometric determination of iodixanol in subcellular fractions of mammalian cells. Anal. Biochem. 244, 174–176 [DOI] [PubMed] [Google Scholar]
- 42. Steinberg T. H., Lauber W. M., Berggren K., Kemper C., Yue S., Patton W. F. (2000) Fluorescence detection of proteins in sodium dodecyl sulfate-polyacrylamide gels using environmentally benign, nonfixative, saline solution. Electrophoresis 21, 497–508 [DOI] [PubMed] [Google Scholar]
- 43. White I. R., Pickford R., Wood J., Skehel J. M., Gangadharan B., Cutler P. (2004) A statistical comparison of silver and SYPRO Ruby staining for proteomic analysis. Electrophoresis 25, 3048–3054 [DOI] [PubMed] [Google Scholar]
- 44. Tauro B. J., Greening D. W., Mathias R. A., Mathivanan S., Ji H., Simpson R. J. (2013) Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol. Cell. Proteomics 12, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Simpson R. J., Connolly L. M., Eddes J. S., Pereira J. J., Moritz R. L., Reid G. E. (2000) Proteomic analysis of the human colon carcinoma cell line (LIM 1215): development of a membrane protein database. Electrophoresis 21, 1707–1732 [DOI] [PubMed] [Google Scholar]
- 46. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 47. Deutsch E. W., Lam H., Aebersold R. (2008) PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Desiere F., Deutsch E. W., King N. L., Nesvizhskii A. I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S. N., Aebersold R. (2006) The PeptideAtlas project. Nucleic Acids Res. 34, D655–D658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Desiere F., Deutsch E. W., Nesvizhskii A. I., Mallick P., King N. L., Eng J. K., Aderem A., Boyle R., Brunner E., Donohoe S., Fausto N., Hafen E., Hood L., Katze M. G., Kennedy K. A., Kregenow F., Lee H., Lin B., Martin D., Ranish J. A., Rawlings D. J., Samelson L. E., Shiio Y., Watts J. D., Wollscheid B., Wright M. E., Yan W., Yang L., Yi E. C., Zhang H., Aebersold R. (2005) Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 6, R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Greening D. W., Glenister K. M., Kapp E. A., Moritz R. L., Sparrow R. L., Lynch G. W., Simpson R. J. (2008) Comparison of human platelet membrane-cytoskeletal proteins with the plasma proteome: Towards understanding the platelet-plasma nexus. Proteomics Clin. Appl. 2, 63–77 [DOI] [PubMed] [Google Scholar]
- 51. Chen Y. S., Mathias R. A., Mathivanan S., Kapp E. A., Moritz R. L., Zhu H. J., Simpson R. J. (2011) Proteomics profiling of Madin-Darby canine kidney plasma membranes reveals Wnt-5a involvement during oncogenic H-Ras/TGF-beta-mediated epithelial-mesenchymal transition. Mol. Cell. Proteomics 10, M110.001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Beissbarth T., Hyde L., Smyth G. K., Job C., Boon W. M., Tan S. S., Scott H. S., Speed T. P. (2004) Statistical modeling of sequencing errors in SAGE libraries. Bioinformatics 20, 1, i31–i39 [DOI] [PubMed] [Google Scholar]
- 53. Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 54. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 [Google Scholar]
- 55. Team R. D.C. R: A language and environment for statistical computing, in R Foundation for Statistical Computing, 2008: Vienna, Austria: p. ISBN 3-900051-900007-900050 [Google Scholar]
- 56. Demory Beckler M., Higginbotham J. N., Franklin J. L., Ham A. J., Halvey P. J., Imasuen I. E., Whitwell C., Li M., Liebler D. C., Coffey R. J. (2013) Proteomic analysis of exosomes from mutant kras colon cancer cells identifies intercellular transfer of mutant KRAS. Mol. Cell. Proteomics 12, 343–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Simpson R. J., Lim J. W., Moritz R. L., Mathivanan S. (2009) Exosomes: proteomic insights and diagnostic potential. Expert Rev. Proteomics 6, 267–283 [DOI] [PubMed] [Google Scholar]
- 58. Nazarenko I., Rana S., Baumann A., McAlear J., Hellwig A., Trendelenburg M., Lochnit G., Preissner K. T., Zoller M. (2010) Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 70, 1668–1678 [DOI] [PubMed] [Google Scholar]
- 59. Rana S., Zoller M. (2011) Exosome target cell selection and the importance of exosomal tetraspanins: a hypothesis. Biochem. Soc. Trans. 39, 559–562 [DOI] [PubMed] [Google Scholar]
- 60. Fukuda M. (2008) Regulation of secretory vesicle traffic by Rab small GTPases. Cell Mol. Life Sci. 65, 2801–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee M. T., Mishra A., Lambright D. G. (2009) Structural mechanisms for regulation of membrane traffic by rab GTPases. Traffic 10, 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 63. Thery C., Ostrowski M., Segura E. (2009) Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 [DOI] [PubMed] [Google Scholar]
- 64. Kalra H., Simpson R. J., Ji H., Aikawa E., Altevogt P., Askenase P., Bond V. C., Borras F. E., Breakefield X., Budnik V., Buzas E., Camussi G., Clayton A., Cocucci E., Falcon-Perez J. M., Gabrielsson S., Gho Y. S., Gupta D., Harsha H. C., Hendrix A., Hill A. F., Inal J. M., Jenster G., Kramer-Albers E. M., Lim S. K., Llorente A., Lotvall J., Marcilla A., Mincheva-Nilsson L., Nazarenko I., Nieuwland R., Nolte-'t Hoen E. N., Pandey A., Patel T., Piper M. G., Pluchino S., Prasad T. S., Rajendran L., Raposo G., Record M., Reid G. E., Sanchez-Madrid F., Schiffelers R. M., Siljander P., Stensballe A., Stoorvogel W., Taylor D., Thery C., Valadi H., van Balkom B. W., Vazquez J., Vidal M., Wauben M. H., Yanez-Mo M., Zoeller M., Mathivanan S. (2012) Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 10, e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mathivanan S., Simpson R. J. (2009) ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000 [DOI] [PubMed] [Google Scholar]
- 66. Peinado H., Aleckovic M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., Garcia-Santos G., Ghajar C., Nitadori-Hoshino A., Hoffman C., Badal K., Garcia B. A., Callahan M. K., Yuan J., Martins V. R., Skog J., Kaplan R. N., Brady M. S., Wolchok J. D., Chapman P. B., Kang Y., Bromberg J., Lyden D. (2012) Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nature Med. 18, 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kaplan R. N., Riba R. D., Zacharoulis S., Bramley A. H., Vincent L., Costa C., MacDonald D. D., Jin D. K., Shido K., Kerns S. A., Zhu Z., Hicklin D., Wu Y., Port J. L., Altorki N., Port E. R., Ruggero D., Shmelkov S. V., Jensen K. K., Rafii S., Lyden D. (2005) VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 438, 820–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sleeman J. P. (2012) The metastatic niche and stromal progression. Cancer Metastasis Rev. 31, 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nguyen D. X., Bos P. D., Massague J. (2009) Metastasis: from dissemination to organ-specific colonization. Nature Rev. Cancer 9, 274–284 [DOI] [PubMed] [Google Scholar]
- 70. Psaila B., Lyden D. (2009) The metastatic niche: adapting the foreign soil. Nature Rev. Cancer 9, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Khamis Z. I., Sahab Z. J., Sang Q. X. (2012) Active roles of tumor stroma in breast cancer metastasis. Int. J. Breast Cancer Epub Feb 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Castellana D., Zobairi F., Martinez M. C., Panaro M. A., Mitolo V., Freyssinet J. M., Kunzelmann C. (2009) Membrane microvesicles as actors in the establishment of a favorable prostatic tumoral niche: a role for activated fibroblasts and CX3CL1-CX3CR1 axis. Cancer Res. 69, 785–793 [DOI] [PubMed] [Google Scholar]
- 73. Jung T., Castellana D., Klingbeil P., Cuesta Hernandez I., Vitacolonna M., Orlicky D. J., Roffler S. R., Brodt P., Zoller M. (2009) CD44v6 dependence of premetastatic niche preparation by exosomes. Neoplasia 11, 1093–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Grange C., Tapparo M., Collino F., Vitillo L., Damasco C., Deregibus M. C., Tetta C., Bussolati B., Camussi G. (2011) Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 71, 5346–5356 [DOI] [PubMed] [Google Scholar]
- 75. Kenny P. A., Lee G. Y., Bissell M. J. (2007) Targeting the tumor microenvironment. Frontiers Biosci. 12, 3468–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kang Y., Siegel P. M., Shu W., Drobnjak M., Kakonen S. M., Cordon-Cardo C., Guise T. A., Massague J. (2003) A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3, 537–549 [DOI] [PubMed] [Google Scholar]
- 77. Nicoloso M. S., Spizzo R., Shimizu M., Rossi S., Calin G. A. (2009) MicroRNAs–the micro steering wheel of tumour metastases. Nat. Rev. Cancer 9, 293–302 [DOI] [PubMed] [Google Scholar]
- 78. Pulukuri S. M., Rao J. S. (2008) Matrix metalloproteinase-1 promotes prostate tumor growth and metastasis. Int. J. Oncol. 32, 757–765 [PMC free article] [PubMed] [Google Scholar]
- 79. Shiozawa J., Ito M., Nakayama T., Nakashima M., Kohno S., Sekine I. (2000) Expression of matrix metalloproteinase-1 in human colorectal carcinoma. Mod. Pathol. 13, 925–933 [DOI] [PubMed] [Google Scholar]
- 80. Itoh Y., Seiki M. (2006) MT1-MMP: a potent modifier of pericellular microenvironment. J. Cell. Physiol. 206, 1–8 [DOI] [PubMed] [Google Scholar]
- 81. Muller M., Beck I. M., Gadesmann J., Karschuk N., Paschen A., Proksch E., Djonov V., Reiss K., Sedlacek R. (2010) MMP19 is upregulated during melanoma progression and increases invasion of melanoma cells. Mod. Pathol. 23, 511–521 [DOI] [PubMed] [Google Scholar]
- 82. Nakahara H., Howard L., Thompson E. W., Sato H., Seiki M., Yeh Y., Chen W. T. (1997) Transmembrane/cytoplasmic domain-mediated membrane type 1-matrix metalloprotease docking to invadopodia is required for cell invasion. Proc. Natl. Acad. Sci. U.S.A. 94, 7959–7964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rocks N., Paulissen G., El Hour M., Quesada F., Crahay C., Gueders M., Foidart J. M., Noel A., Cataldo D. (2008) Emerging roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie 90, 369–379 [DOI] [PubMed] [Google Scholar]
- 84. Murphy G. (2008) The ADAMs: signalling scissors in the tumour microenvironment. Nat. Rev. Cancer 8, 929–941 [DOI] [PubMed] [Google Scholar]
- 85. Gutwein P., Mechtersheimer S., Riedle S., Stoeck A., Gast D., Joumaa S., Zentgraf H., Fogel M., Altevogt D. P. (2003) ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. FASEB J. 17, 292–294 [DOI] [PubMed] [Google Scholar]
- 86. Porter S., Clark I. M., Kevorkian L., Edwards D. R. (2005) The ADAMTS metalloproteinases. Biochem. J. 386(Pt 1), 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Liu Y. J., Xu Y., Yu Q. (2006) Full-length ADAMTS-1 and the ADAMTS-1 fragments display pro- and antimetastatic activity, respectively. Oncogene 25, 2452–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Guise T. A. (2009) Breaking down bone: new insight into site-specific mechanisms of breast cancer osteolysis mediated by metalloproteinases. Genes Dev. 23, 2117–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Guo W., Giancotti F. G. (2004) Integrin signalling during tumour progression. Nat. Rev. Mol. Cell Biol. 5, 816–826 [DOI] [PubMed] [Google Scholar]
- 90. Ahmed N., Riley C., Rice G., Quinn M. (2005) Role of integrin receptors for fibronectin, collagen and laminin in the regulation of ovarian carcinoma functions in response to a matrix microenvironment. Clin. Exp. Metastasis 22, 391–402 [DOI] [PubMed] [Google Scholar]
- 91. Munger J. S., Huang X., Kawakatsu H., Griffiths M. J., Dalton S. L., Wu J., Pittet J. F., Kaminski N., Garat C., Matthay M. A., Rifkin D. B., Sheppard D. (1999) The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell 96, 319–328 [DOI] [PubMed] [Google Scholar]
- 92. Hemler M. E. (2005) Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801–811 [DOI] [PubMed] [Google Scholar]
- 93. Bassani S., Cingolani L. A. (2012) Tetraspanins: Interactions and interplay with integrins. Int J Biochem. Cell Biol. 44, 703–708 [DOI] [PubMed] [Google Scholar]
- 94. Wang H. X., Li Q., Sharma C., Knoblich K., Hemler M. E. (2011) Tetraspanin protein contributions to cancer. Biochem. Soc Trans. 39, 547–552 [DOI] [PubMed] [Google Scholar]
- 95. Domanico S. Z., Pelletier A. J., Havran W. L., Quaranta V. (1997) Integrin alpha 6A beta 1 induces CD81-dependent cell motility without engaging the extracellular matrix migration substrate. Mol. Biol. Cell 8, 2253–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Luga V., Zhang L., Viloria-Petit A. M., Ogunjimi A. A., Inanlou M. R., Chiu E., Buchanan M., Hosein A. N., Basik M., Wrana J. L. (2012) Exosomes Mediate Stromal Mobilization of Autocrine Wnt-PCP Signaling in Breast Cancer Cell Migration. Cell 151, 1542–1556 [DOI] [PubMed] [Google Scholar]
- 97. He B., Liu L., Cook G. A., Grgurevich S., Jennings L. K., Zhang X. A. (2005) Tetraspanin CD82 attenuates cellular morphogenesis through down-regulating integrin alpha6-mediated cell adhesion. J. Biol. Chem. 280, 3346–3354 [DOI] [PubMed] [Google Scholar]
- 98. Fei Y., Wang J., Liu W., Zuo H., Qin J., Wang D., Zeng H., Liu Z. (2012) CD151 promotes cancer cell metastasis via integrins alpha3beta1 and alpha6beta1 in vitro. Mol. Med. Rep. 6, 1226–1230 [DOI] [PubMed] [Google Scholar]
- 99. Gerke V., Creutz C. E., Moss S. E. (2005) Annexins: linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 6, 449–461 [DOI] [PubMed] [Google Scholar]
- 100. Sharma M. R., Koltowski L., Ownbey R. T., Tuszynski G. P., Sharma M. C. (2006) Angiogenesis-associated protein annexin II in breast cancer: selective expression in invasive breast cancer and contribution to tumor invasion and progression. Exp. Mol. Pathol. 81, 146–156 [DOI] [PubMed] [Google Scholar]
- 101. Bao H., Jiang M., Zhu M., Sheng F., Ruan J., Ruan C. (2009) Overexpression of Annexin II affects the proliferation, apoptosis, invasion and production of proangiogenic factors in multiple myeloma. Int. J. Hematol. 90, 177–185 [DOI] [PubMed] [Google Scholar]
- 102. Shapiro I. M., Cheng A. W., Flytzanis N. C., Balsamo M., Condeelis J. S., Oktay M. H., Burge C. B., Gertler F. B. (2011) An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 7, e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Moreno-Bueno G., Portillo F., Cano A. (2008) Transcriptional regulation of cell polarity in EMT and cancer. Oncogene 27, 6958–6969 [DOI] [PubMed] [Google Scholar]
- 104. Brown R. L., Reinke L. M., Damerow M. S., Perez D., Chodosh L. A., Yang J., Cheng C. (2011) CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J. Clin. Invest. 121, 1064–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Warzecha C. C., Jiang P., Amirikian K., Dittmar K. A., Lu H., Shen S., Guo W., Xing Y., Carstens R. P. (2010) An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 29, 3286–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Biamonti G., Bonomi S., Gallo S., Ghigna C. (2012) Making alternative splicing decisions during epithelial-to-mesenchymal transition (EMT). Cell Mol. Life Sci. 69, 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Savagner P., Valles A. M., Jouanneau J., Yamada K. M., Thiery J. P. (1994) Alternative splicing in fibroblast growth factor receptor 2 is associated with induced epithelial-mesenchymal transition in rat bladder carcinoma cells. Mol. Biol. Cell 5, 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wu C. Y., Tsai Y. P., Wu M. Z., Teng S. C., Wu K. J. (2012) Epigenetic reprogramming and post-transcriptional regulation during the epithelial-mesenchymal transition. Trends Genet. 28, 454–463 [DOI] [PubMed] [Google Scholar]
- 109. Warzecha C. C., Carstens R. P. (2012) Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT). Semin Cancer Biol. 22, 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wahl M. C., Will C. L., Luhrmann R. (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718 [DOI] [PubMed] [Google Scholar]
- 111. Ghigna C., Giordano S., Shen H., Benvenuto F., Castiglioni F., Comoglio P. M., Green M. R., Riva S., Biamonti G. (2005) Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 20, 881–890 [DOI] [PubMed] [Google Scholar]
- 112. Lu Y., Yao H. P., Wang M. H. (2007) Multiple variants of the RON receptor tyrosine kinase: biochemical properties, tumorigenic activities, and potential drug targets. Cancer Lett. 257, 157–164 [DOI] [PubMed] [Google Scholar]
- 113. Valacca C., Bonomi S., Buratti E., Pedrotti S., Baralle F. E., Sette C., Ghigna C., Biamonti G. (2010) Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J. Cell Biol. 191, 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Collesi C., Santoro M. M., Gaudino G., Comoglio P. M. (1996) A splicing variant of the RON transcript induces constitutive tyrosine kinase activity and an invasive phenotype. Mol. Cell. Biol. 16, 5518–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Karni R., de Stanchina E., Lowe S. W., Sinha R., Mu D., Krainer A. R. (2007) The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Eliseeva I. A., Kim E. R., Guryanov S. G., Ovchinnikov L. P., Lyabin D. N. (2011) Y-box-binding protein 1 (YB-1) and its functions. Biochemistry 76, 1402–1433 [DOI] [PubMed] [Google Scholar]
- 117. Bader A. G., Vogt P. K. (2004) An essential role for protein synthesis in oncogenic cellular transformation. Oncogene 23, 3145–3150 [DOI] [PubMed] [Google Scholar]
- 118. Kohno K., Izumi H., Uchiumi T., Ashizuka M., Kuwano M. (2003) The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays 25, 691–698 [DOI] [PubMed] [Google Scholar]
- 119. Lasham A., Lindridge E., Rudert F., Onrust R., Watson J. (2000) Regulation of the human fas promoter by YB-1, Puralpha and AP-1 transcription factors. Gene 252, 1–13 [DOI] [PubMed] [Google Scholar]
- 120. Ladomery M., Sommerville J. (1995) A role for Y-box proteins in cell proliferation. Bioessays 17, 9–11 [DOI] [PubMed] [Google Scholar]
- 121. Samuel S., Beifuss K. K., Bernstein L. R. (2007) YB-1 binds to the MMP-13 promoter sequence and represses MMP-13 transactivation via the AP-1 site. Biochim. Biophys. Acta 1769, 525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Knauper V., Lopez-Otin C., Smith B., Knight G., Murphy G. (1996) Biochemical characterization of human collagenase-3. J. Biol. Chem. 271, 1544–1550 [DOI] [PubMed] [Google Scholar]
- 123. Steffan J. J., Koul S., Meacham R. B., Koul H. K. (2012) The transcription factor SPDEF suppresses prostate tumor metastasis. J. Biol. Chem. 287, 29968–29978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Evdokimova V., Tognon C., Ng T., Ruzanov P., Melnyk N., Fink D., Sorokin A., Ovchinnikov L. P., Davicioni E., Triche T. J., Sorensen P. H. (2009) Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell 15, 402–415 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.