Abstract
The adult CNS contains an abundant population of oligodendrocyte precursor cells (NG2+ cells) that generate oligodendrocytes and repair myelin, but how these ubiquitous progenitors maintain their density is unknown. Here we generated NG2-mEGFP mice and used in vivo two-photon imaging to study their dynamics in the adult brain. Time-lapse imaging revealed that NG2+ cells in the cortex are highly dynamic; they survey their local environment with motile filopodia, extend growth cones, and continuously migrate. They maintain unique territories through self-avoidance, and NG2+ cell loss through death, differentiation, or ablation triggered rapid migration and proliferation of adjacent cells to restore their density. NG2+ cells recruited to sites of focal CNS injury were similarly replaced by a proliferative burst surrounding the injury site. Thus, homeostatic control of NG2+ cell density through a balance of active growth and self-repulsion ensures that these progenitors are available to replace oligodendrocytes and participate in tissue repair.
Introduction
Homeostatic control of cell density is an essential feature of tissue and organ maintenance, allowing cell replacement and regeneration to offset cell loss resulting from injury, disease or age-dependent degeneration1, 2. Tight control over cell proliferation is especially critical in the adult central nervous system (CNS), which has a limited capacity to accommodate growth due to its complex cellular architecture and its encasement in bone. In contrast to neurons, which apart from restricted populations in the hippocampus and olfactory bulb are not replaced even in the context of injury and disease3, many glial cells exhibit a remarkable capacity for self-renewal4, 5. However, it is not known how the density and distribution of different classes of glial cells are maintained in the adult CNS.
Glial progenitor cells that express the chondroitin sulfate proteoglycan NG2, termed NG2+ cells (or oligodendrocyte precursor cells), comprise the majority of proliferating cells in the adult CNS6. During development these glial cells migrate from germinal zones, proliferate, and differentiate into myelinating oligodendrocytes7-9. Although myelinated tracts are formed early in life, NG2+ cells are retained throughout the adult CNS, where they are organized in a grid-like or tiled manner, with individual cells occupying non-overlapping domains10. In vivo genetic fate tracing studies indicate that NG2+ cells continue to differentiate into oligodendrocytes in adults7, 11-13, and are rapidly mobilized to replace oligodendrocytes in animal models of acute and chronic demyelination4, 14, 15, suggesting that they play a key role in both normal oligodendrocyte homeostasis and regeneration of myelin. Although continual renewal of these progenitors is likely to be crucial for efficient oligodendrogenesis, the mechanisms that control their uniform distribution and high density in the adult CNS remain unknown, in part, because their dynamics have not been examined in the intact adult CNS9, 16, 17.
NG2+ cell proliferation is enhanced following demyelination15, traumatic injury to the CNS18, and in chronic neurodegenerative disease7, 19; however, the relationship between proliferation of these progenitors and the generation of new oligodendrocytes remains uncertain20. Moreover, uncontrolled growth of these progenitors leads to tumor formation21, and recent studies suggest that NG2+ cells are likely to be a cell of origin for certain forms of glioma22, 23, highlighting the importance of understanding how the proliferation of these cells is controlled in vivo.
To address these questions, we developed a line of transgenic mice that express a membrane anchored form of EGFP under control of the NG2 (Cspg4) promoter (NG2-mEGFP-H mice) and performed in vivo two-photon imaging of NG2+ cells in the mouse somatosensory cortex. We find that NG2+ cells are highly dynamic in the adult brain; they extend motile filopodia, reorganize their processes, and continuously move through the parenchyma. Although their position is not fixed, NG2+ cells maintain independent domains through self-repulsion, and loss of cells through death, differentiation, or experimental ablation triggers rapid migration and proliferation of adjacent NG2+ cells to preserve their density. Long-term imaging revealed that NG2+ cells directly differentiate into oligodendrocytes without proliferation, indicating that division of these progenitors is a homeostatic response to cell removal, rather than the generation of oligodendrocytes through asymmetric division. Although adult NG2+ cells can serve as oligodendrocyte progenitors, they also migrated to sites of focal injury to help form a glial scar and were similarly replaced through proliferation of neighboring NG2+ cells. By balancing active growth with self-repulsion, NG2+ cells maintain a constant density in the CNS, ensuring that they are available to participate in regeneration and repair of the CNS throughout life.
Results
Adult NG2+ cells extend processes with dynamic filopodia
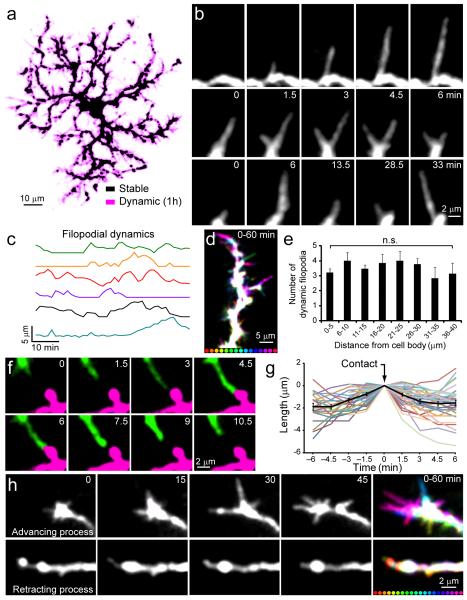
To define the behavior of NG2+ cells in the adult brain, we generated BAC transgenic mice that express a membrane anchored form of EGFP (ref. 24) under control of the NG2 promoter (NG2-mEGFP-H mice) (Supplementary Fig. 1), which allowed visualization of the full extent of their complex morphologies (Supplementary Fig. 2). In vivo two-photon imaging through a cranial window revealed that fluorescent NG2+ cells were distributed in a highly ordered manner throughout the upper layers of the cortex in these mice (Supplementary Movie 1), with cells occupying non-overlapping domains. NG2+ cells extended highly ramified processes studded with numerous filopodia-like protrusions into the surrounding neuropil (Fig. 1a). In time-lapse recordings, these thin protrusions were highly dynamic, continually extending, retracting, branching and altering their trajectory (Fig. 1a-c, Supplementary Movies 2,3) with an average speed of 0.3 ± 0.01 μm/min (185 protrusions on 10 cells in 5 mice). Motile filopodia were present along the length of each process, and even extended from the soma (Fig. 1a,d,e), indicating that NG2+ cells actively survey their local environment. This dynamic behavior is similar to that exhibited by microglial cells25, 26, although NG2+ cell filopodia lacked the bulbous endings characteristic of microglial processes and moved more slowly (microglia process speed = 1.47 ± 0.1 μm/min; from ref 26).
Fig. 1.
NG2+ cells extend dynamic filopodia and exhibit self-repulsion in the adult cortex. a, Maximum-intensity projection of an individual NG2+ cell during 1 hour time-lapse imaging. Stable regions are shown in black and dynamic regions are shown in magenta. b, Individual NG2+ cell filopodia extend (top), branch (middle), and alter their trajectory (bottom) in minutes. c, Plots showing the change in length over time of six NG2+ cell filopodia. d, Maximum-intensity projection of one NG2+ cell process, color-coded for time (intervals below = 4 min). Stable areas are represented in white. e, Graph showing the distribution of dynamic filopodia along NG2+ cell processes (n = 13 branches on 7 cells in 5 mice; P > 0.05 one-way ANOVA with Tukey post-hoc test). Error bars show s.e.m. f, In vivo time-lapse images of two pseudo-colored NG2+ cell processes, showing that contact leads to filopodial retraction. g, Plot of the change in length of NG2+ cell filopodia before and after contacting another NG2+ cell processes (n = 35 filopodia on 10 cells in 5 mice). Average change in length for all processes is shown in black. h, In vivo time-lapse imaging of advancing (top) and retracting (bottom) NG2+ cell processes. Montages at far right (0 – 60 min) are maximum-intensity projection images showing two NG2+ cell processes color-coded for time (intervals below = 3 min). Stable areas are represented in white. Note the presence of highly motile filopodia at the tip of the advancing process and the absence of filopodia on the retracting process.
Despite the active growth of NG2+ cell processes, these cells maintained a radial morphology and contacts between processes were rarely observed. Previous studies indicate that contact mediated inhibition can help establish a radial morphology and control cell spacing through homotypic repulsion27, 28. In time-lapse imaging, growing filopodia of NG2+ cells always retracted when they contacted a process of the same or an adjacent NG2+ cell (Fig. 1f,g; Supplementary Movie 3), indicating that these progenitors engage in continual self-avoidance through contact mediated inhibition.
Previous studies have shown that removal of the skull and implantation of a chronic cranial window can induce reactive changes in glial cells and enhance the dynamics of dendritic spines on neurons29. To determine if the dynamic behavior of NG2+ cells was a reactive response to injury, we prepared NG2-mEGFP-H mice with thinned-skull cranial windows30 and performed time-lapse imaging. NG2+ cells imaged through the skull had complex morphologies and highly dynamic filopodia were observed along their processes, features comparable to those observed in mice implanted with chronic cranial windows (Supplementary Fig. 3; Supplementary Movie 4). Moreover, mice implanted with chronic cranial windows did not show reactive changes in microglia or increases in the number of proliferating cells relative to controls when in vivo imaging was initiated (Supplementary Figure 4). These results indicate that the extension of dynamic filopodia from NG2+ cell processes is a normal feature of these progenitors in the uninjured brain, rather than a reactive response to cranial window implantation.
NG2+ cells migrate continuously through the adult cortex
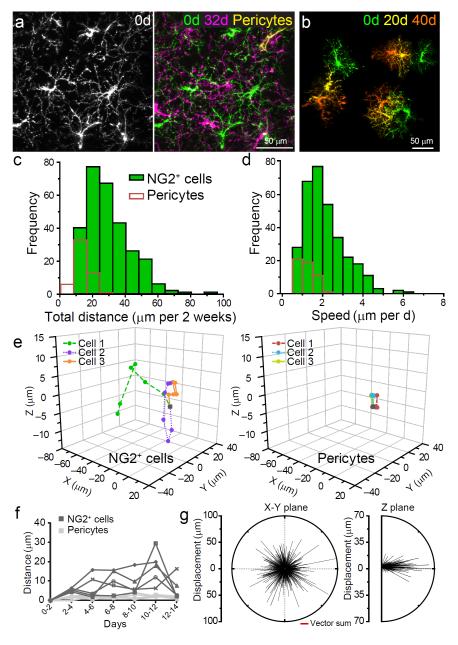
The tips of advancing NG2+ cell processes displayed numerous motile filopodia, reminiscent of neuronal growth cones, whereas retracting processes lacked filopodia (Fig. 1h), suggesting that these dynamics may enable cell migration. To determine if process motility is accompanied by somatic translocation, we re-imaged NG2+ cells in the same volume of tissue one month later. Although the position of vascular-associated pericytes in these regions was unchanged, the distribution of NG2+ cell somata and the orientation of their processes were dramatically altered, such that it was not possible to unambiguously identify the same NG2+ cells from the previous imaging session (Fig. 2a).
Fig. 2.
NG2+ cells continually change their position in the adult cortex. a, Image of NG2+ cells in one region of cortex on day 0 (left panel) and 32 days later (Right panel = montage of two time points). Stable pericytes are shown in yellow. b, Montage showing the change in morphology and position of three NG2+ cells over 40 days. c, Histogram showing the distances that NG2+ cells (green bars) and perivascular cells (red bars) moved over two weeks (318 NG2+ cells, 53 perivascular cells in 5 mice; P < 0.05 Mann-Whitney test). d, Histogram of the speed that NG2+ cells and pericytes moved over two weeks (318 NG2+ cells, 53 perivascular cells in 5 mice; P < 0.05 Mann-Whitney test). e, Three-dimensional graphs showing the movements of three NG2+ cells (left) and three perivascular cells (right) in the somatosensory cortex over a two week period. f, Graph showing the displacement of somata over time for five NG2+ cells and five pericytes. g, Vector plots of the direction and displacement of 905 NG2+ cell movements in the XY plane (left; P = 0.578 Moore-Rayleigh test) and the Z plane (right) for cells > 90 μm below the pia mater.
More frequent imaging revealed that NG2+ cells continually reoriented their processes and moved an average distance of 2.1 ± 0.1 μm per day (318 NG2+ cells in 5 mice) (Fig. 2b-d; Supplementary Movies 5,6), in contrast to pericytes that remained largely stationary (Fig. 2e,f). Individual NG2+ cells exhibited heterogeneous behaviors, with some cells moving at a continuous rate during the two-week imaging period, while others interrupted periods of relative stability with large movements of > 30 μm over a period of a few days (Fig. 2f). Long-range cues guide the migration of NG2+ cells during development31, raising the possibility that similar gradients influence their movement in the adult CNS. However, analysis of the trajectories of all NG2+ cells in a 0.06 mm3 cortical volume revealed that there was no directional bias in their movement (Fig. 2g), suggesting that their behavior is controlled by local interactions rather than large-scale gradients of attractive or repulsive cues.
Constant turnover of NG2+ cells in the adult brain
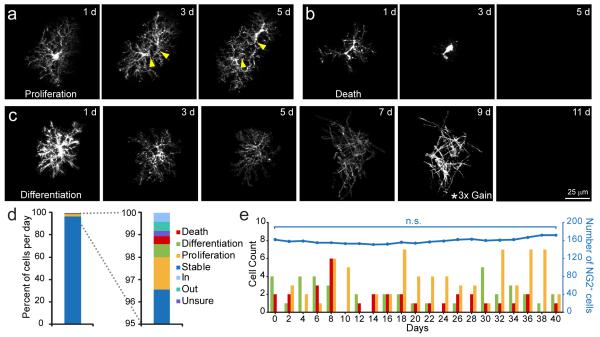
In addition to the constant reorganization of NG2+ cell position in the cortex, there were dynamic changes in the overall population, due to the proliferation, differentiation, and death of individual NG2+ cells (Fig. 3a-c). Cell death was characterized by hypertrophy, retraction of processes and abrupt disappearance of the cell (Supplementary Fig. 5), whereas differentiation into oligodendrocytes was accompanied by a gradual decrease in fluorescence, transformation of their highly ramified processes to long, unbranched processes, and expression of myelin proteins (Supplementary Fig. 6). NG2+ cells did not differentiate into astrocytes or neurons in this region, providing further evidence that these glial cells serve as lineage restricted progenitors in the normal CNS7; however, if EGFP is degraded more rapidly during transdifferentiation, adoption of these alternate fates would be difficult to assess using this approach.
Fig. 3.
NG2+ cell density is maintained despite proliferation, differentiation, and death. a-c, Sequential images from in vivo time lapse recordings illustrating individual NG2+ cells undergoing division a, death b, and differentiation c. The image intensity at 9 days in c was increased 3x to illustrate the transition to an oligodendrocyte morphology. EGFP intensity decreased with differentiation due to down regulation of the NG2 promoter10. d, Graph illustrating the proportion of NG2+ cells engaged in different behaviors on each day (1118 NG2+ cells in 5 mice). e, Combined plot showing the number of NG2+ cells undergoing proliferation, differentiation, and death (colored bars) and the total NG2+ cell density (blue line) in a 0.06 mm3 volume of adult somatosensory cortex over a 40 day period.
In contrast to the segregation of principal neurons into distinct cortical lamina, NG2+ cells were evenly distributed among cortical layers, and the core behaviors (proliferation, differentiation, and death) were not biased to a particular lamina (Supplementary Fig. 7). On each day, 3 % of NG2+ cells were engaged in these dynamic behaviors (Fig. 3d), which expanded to include 36 % of the population over two weeks. Notably, cell loss through death or differentiation (1.2 ± 0.1 %/day) was balanced by cell addition through proliferation (1.5 ± 0.1 %/day, P = 0.09), allowing the density of NG2+ cells to remain constant (Fig. 3e), indicating that this progenitor pool is constantly reorganized by the continual loss and addition of new cells.
NG2+ cell density is maintained through local proliferation
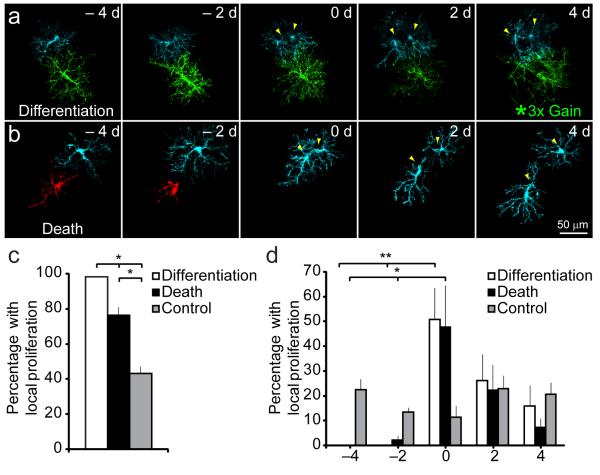
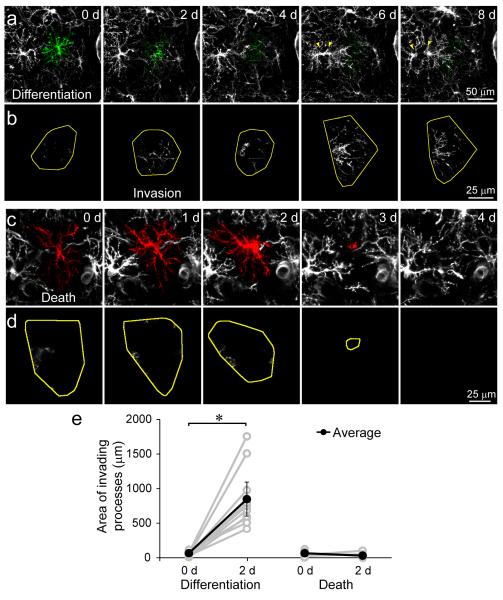
If homeostatic control of NG2+ cell density is governed by local interactions, then differentiation or death should be associated with division of a neighboring cell. We observed that 98 % of differentiating and 76 % of dying NG2+ cells were associated with proliferation of a neighboring NG2+ cell (< 50 μm away), whereas only 43 % of stable NG2+ cells were associated with a local proliferation (P < 0.005 for both death and differentiation) (Fig. 4a-c). Moreover, the probability of local proliferation in instances of death or differentiation was highest at the time of cell loss, but did not vary with time for control NG2+ cells (Fig. 4d). Neighboring NG2+ cells rapidly invaded the territory of differentiating cells (Fig. 5a,b,e), suggesting that the homotypic repulsive cues that prevent overgrowth are down regulated with differentiation. In contrast, dying NG2+ cells maintained distinct, though progressively smaller, territories until removal (Fig. 5c,d,e). The gradual decrease in the territory of dying cells (Supplementary Fig. 5), occasionally resulted in repositioning of neighboring NG2+ cells without cell proliferation, accounting for the somewhat lower incidence of neighbor proliferation in cases of death.
Fig. 4.
NG2+ cell density is maintained through local proliferation. a-b, Images from time-lapse images showing that NG2+ cell differentiation (green cell) a or death (red cell) b is associated with proliferation (yellow arrowheads) of a neighboring NG2+ cell (cyan). The intensity for the differentiating cell at day 4 in b has been increased 3x to highlight the morphological change. c, Graph showing the percent of differentiating, dying, and stable NG2+ cells that were associated with proliferation of a neighboring NG2+ cell (72 differentiating, 54 dying, 187 stable NG2+ cells in 5 mice; *P < 0.005 one-way ANOVA with Tukey post-hoc test). d, Graph showing the time-course of local proliferation relative to the onset of cell loss through death or differentiation (*P < 0.03, **P < 0.005, one-way ANOVA with Tukey post-hoc test). Error bars show s.e.m.
Fig. 5.
Neighboring NG2+ cell processes invade the territory of differentiating but not dying NG2+ cells. a, In vivo time-lapse images of an individual NG2+ cell (pseudo-colored green) that differentiated into an oligodendrocyte during the 8 day imaging period. Note that differentiation resulted in processes extension and proliferation (yellow arrowheads) of the neighboring NG2+ cell to the left of the differentiating cell. b, Montage showing the territory of the differentiating cell (yellow) overlaid with the processes of neighboring NG2+ cells (white) that entered the territory of the differentiating cell. Note that there was extensive invasion of cell territory very early in the differentiation process. c, In vivo time-lapse images of an individual NG2+ cell (pseudo-colored red) that died during the 4 day imaging period. d, Montage showing the territory of the dying cell (yellow) overlaid with the processes of neighboring NG2+ cells (white) that entered the territory of the dying cell. e, Graph showing the area of processes that invaded into the territory of differentiating or dying NG2+ cells. (10 differentiating, 10 dying NG2+ cells in 5 mice; *P < 0.0005 paired two-tailed Student’s t test). Error bars show s.e.m.
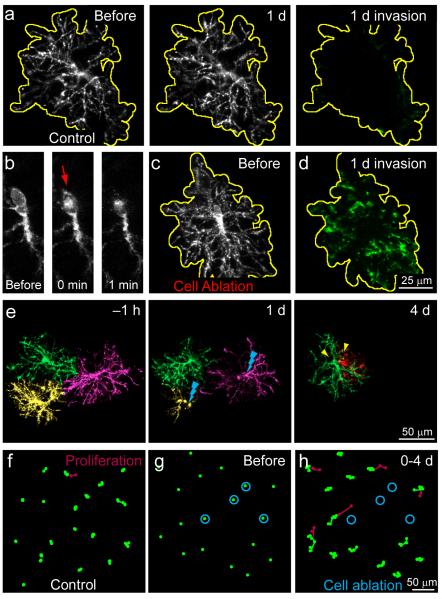
To further assess whether cell loss is sufficient to induce local proliferation, we ablated individual NG2+ cells in vivo using focal laser illumination and examined the response of neighboring NG2+ cells. Following cell removal, neighboring NG2+ cells reoriented their processes and invaded the former territory of the ablated cell, providing further evidence that NG2+ cell domains are maintained by self-avoidance (Fig. 6a-d; Percent territory invasion: Control, 6 ± 1 %, n = 10 cells; Cell ablation, 52 ± 10 %, n = 12 cells; *P < 0.001, Student’s t test). Moreover, removal of several NG2+ cells within the imaging field triggered reorientation, cell migration, and proliferation of NG2+ cells in the vicinity, resulting in rapid restoration of their density (Fig. 6e-h; Number of proliferating cells: Control 1 ± 0.4 cells, Cell ablation 5 ± 0.3 cells; volume analyzed = 0.02 mm3; from 4 control and 3 ablation mice; *P < 0.001, Student’s t test). These data indicate that NG2+ cells maintain a constant density though a local homeostatic mechanism, in which cell loss results in release from growth inhibition and rapid replacement through division of a neighboring cell.
Fig. 6.
NG2+ cell ablation triggers territory invasion and division of a neighboring NG2+ cell. a, In vivo time-lapse images of an individual NG2+ cell over 1 day. The territory of the cell is outlined in yellow. Processes of neighboring NG2+ cells have been pseudo-colored green. Note the lack of territory invasion over 1 day (far right, Invasion). b, Single z-plane images of an individual NG2+ cell before and after brief exposure of the cell body to the focused laser beam to induce ablation. Red arrow highlights the increase in auto-fluorescence of the nucleus after illumination. c, Maximum intensity projection image of the NG2+ cell shown in f prior to ablation. The territory of the cell is outlined in yellow. d, Montage of the same volume of tissue in c one day following cell ablation. Processes of neighboring NG2+ cells have been pseudo-colored green. e, In vivo time-lapse images of three neighboring NG2+ cells (pseudo-colored green, yellow and magenta) from a thinned-skull preparation. Two NG2+ cells (shown in yellow and magenta) were removed by laser-mediated ablation on day 1. On day 4, a neighboring NG2+ cell (green) divided (yellow arrowheads). f, Map of the soma position of NG2+ cells over 4 days in control. Newly generated NG2+ cells are represented in red. g, Snapshot of NG2+ cells distribution before cell ablation. Cells to be targeted for ablation are circled in blue. h, Map of the soma position of NG2+ cells over 4 days following ablation of 4 NG2+ cells (shown in g). Newly generated NG2+ cells are represented in red.
NG2+ cells directly differentiate into oligodendrocytes
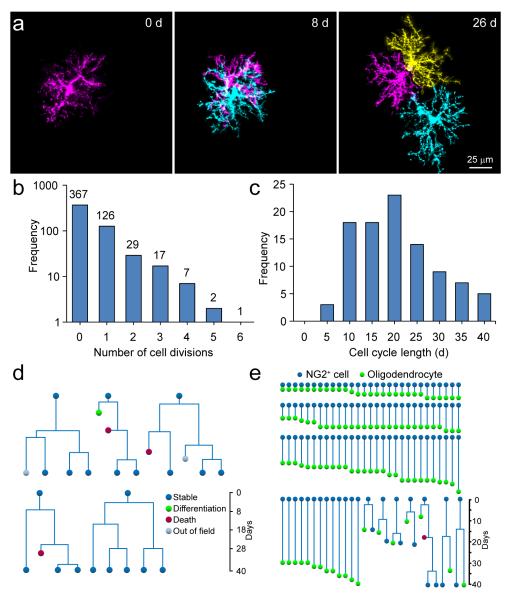
NG2+ cells in the adult CNS can be generated from SVZ progenitors32, which may have a greater capacity for proliferation and differentiation than cells with a longer residence time. To determine if repopulation is accomplished by a subset of highly proliferative, self-renewing NG2+ cells33, we followed the behavior of proliferating NG2+ cells by repeatedly imaging the same cortical volume for many weeks. During this period, the majority of proliferating NG2+ cells divided only once, and for cells that divided multiple times, cell cycle length was variable (average cell cycle: 18.5 ± 1 days, n = 97 cells) (Fig. 7a-c), as expected if extrinsic factors (i.e. death/differentiation of a neighbor) determine when NG2+ cells divide. Indeed, cells that underwent multiple divisions were located in regions where there was substantial turnover/differentiation. Moreover, lineage trees of highly proliferative cells revealed instances where both sister cells underwent multiple divisions, as well as instances where one sister cell either died or differentiated and the other remained stable (Fig. 7d). These findings, and the consistent proliferation of immediate neighbors following cell loss, suggest that homeostasis is achieved by endowing all NG2+ cells with the capacity to divide7, rather than by seeding the brain with a subset of highly proliferative cells.
Fig. 7.
NG2+ cells directly differentiate into oligodendrocytes without asymmetric division. a, Sequential images from in vivo time-lapse recordings of one NG2+ cell that divided twice over 26 days. Newly generated cells are shown in cyan and yellow. b, Histogram of the distribution of proliferation frequencies of NG2+ cells in the somatosensory cortex (828 NG2+ cells in 3 mice imaged for 40 days). c, Histogram of NG2+ cell cycle length (97 NG2+ cell divisions in 3 mice). d, Lineage trees of highly proliferative NG2+ cells, illustrating the range of fates of sister cells. e, Lineage trees of NG2+ cells that differentiated into oligodendrocytes (107 NG2+ cells in 5 mice).
New neurons are formed in the CNS through symmetric or asymmetric division of progenitors34; however, it is unclear whether oligodendrocytes are formed through similar mechanisms or by direct differentiation of NG2+ cells20. Lineage tracing through in vivo imaging revealed that differentiation was rarely preceded by cell division (7/107 differentiation events) (Fig. 7e), indicating that oligodendrocytes in the adult CNS are formed primarily through direct differentiation of these progenitors. Thus, proliferation of NG2+ cells in vivo reflects a homeostatic response to cell depletion rather than oligodendrogenesis.
NG2+ cells participate in glial scar formation
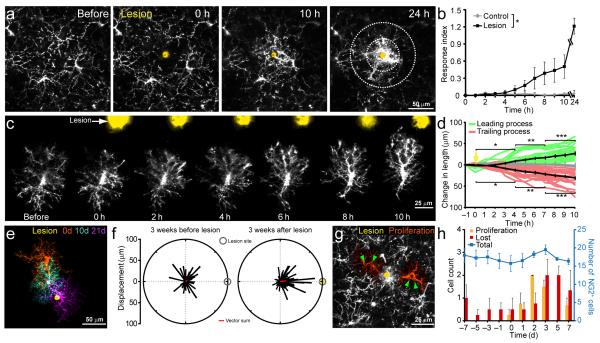
NG2+ cell proliferation is often enhanced after injury and the NG2 proteoglycan accumulates at lesion sites where it can inhibit axon outgrowth18, 35. However, the reason for their enhanced proliferation is unknown, and the participation of NG2+ cells in scar formation has not been established, as NG2 also is expressed by macrophages and pericytes in injured tissue36, 37. To assess the behavior of NG2+ glial cells to CNS injury, we induced focal laser lesions25 in the cortex of adult NG2-mEGFP-H mice. Immediately following injury, NG2+ cells adjacent to the lesion extended processes towards the lesion site and eventually surrounded the site of injury (Fig. 8a-d, Supplementary Movie 7). Although their response to focal injury was similar to microglia25, NG2+ cells extended their processes more slowly than microglia to the lesion site (Fig. 8b). As a consequence, scars acquired a layered structure consisting of microglial cell processes surrounded by NG2+ cell processes (Supplementary Fig. 8). Reorientation and extension of NG2+ cell processes after injury was followed by migration of these cells to the lesion site within several weeks, and eventual removal of these cells as the scar resolved (Fig. 8e,f and Supplementary Movie 8). As predicted by their homeostatic behavior to cell loss, depletion of NG2+ cells from the area surrounding the lesion was accompanied by proliferation of neighboring NG2+ cells (Fig. 8g), with the number of proliferating cells matching the number lost (before lesion, 18 ± 2 NG2+ cells; after lesion, 16 ± 1 NG2+ cells, n = 4 mice, P = 0.5) (Fig. 8h). These findings demonstrate that NG2+ cells contribute to glial scar formation, and provide an explanation for the enhanced proliferation of NG2+ cells that occurs following many types of CNS trauma.
Fig. 8.
NG2+ cells surround areas of CNS damage and proliferate to maintain their density. a, Time-lapse imaging of NG2+ cell responses to focal laser injury (site of lesion shown in yellow). b, Graph showing accumulation of EGFP+ NG2+ cell processes within 75 μm of the lesion over time. Concentric circles on the 24hr time-point in a, highlight the areas measured to determine Response index. (n = 5 mice each condition; *P < 0.05 two-way ANOVA). c, Sequential images showing the response of a NG2+ cell to a focal laser injury (yellow spot). d, Quantification of extension/retraction of leading and trailing processes of NG2+ cells relative to the lesion (n = 20 branches on 10 cells in 4 mice; *P < 0.05, **P < 0.005, ***P < 0.0005 one-way ANOVA with Tukey post-hoc test). e, Montage of three images of one NG2+ cell collected on different days, showing migration of this cell towards the lesion (yellow). f, Vector plots showing the direction and displacement of NG2+ cells within 75 μm of the lesion site three weeks before and after lesion induction (73 cells in 4 mice; before, P = 0.073; after, P = 1.02 × 10−9 Moore-Rayleigh test). Line direction represents the angle of the displacement relative to the lesion site. Red line is the vector sum of all displacements. g, Maximum-intensity projection of 15 μm above and below a focal laser injury (yellow), two days after lesion induction. Two proliferating NG2+ cells (green arrowheads) adjacent to the lesion site are highlighted in orange. h, Combined plot showing the number of NG2+ cells undergoing proliferation and death/differentiation (combined) and the total NG2+ cell density (blue line) within 75 μm of the lesion. Focal laser injury was induced on day 0 (n = 4 mice). Error bars show s.e.m.
Discussion
NG2+ glial cells comprise the most abundant population of proliferative cells in the adult CNS. These progenitors retain the capacity to differentiate into oligodendrocytes and contribute to regeneration of myelin following injury and disease. Homeostatic control of their density and distribution allows not only efficient replacement of oligodendrocytes, but also ensures that cell growth does not proceed unchecked and result in tumor formation. To determine how NG2+ cell density is maintained in the adult CNS, we developed mice that express membrane anchored EGFP in NG2+ cells (NG2-mEGFP-H mice) and examined the behavior of these ubiquitous progenitors in the somatosensory cortex of adult mice using in vivo two-photon imaging. Here, we report that NG2+ cells are highly dynamic in the resting brain; they actively survey their local environment with motile filopodia and continually migrate through the parenchyma, maintaining exclusive territories through self-repulsion. NG2+ cells removed from the population by differentiation or death are rapidly replaced through proliferation of an immediate neighbor (Supplementary Fig. 9). By balancing active growth with self-repulsion, NG2+ cells ensure that proliferation is coupled both spatially and temporally to cell loss, preserving a high density of these progenitors throughout the CNS to efficiently generate oligodendrocytes and participate in tissue repair.
NG2+ glial cells are distributed in grid-like or tiled manner in the adult CNS, with individual cells occupying non-overlapping domains. As this grid is constantly reorganized, mechanisms must exist to actively limit the growth of these progenitors. In the mammalian retina, the mosaic spacing of neurons is regulated by homotypic repulsive interactions through transmembrane receptors (e.g. MEGFs, DSCAM)27, 38. Similar repulsive interactions are likely to play a key role in both establishing and maintaining the density of NG2+ cells, as selective ablation of individual NG2+ cells was sufficient to trigger rapid invasion of the territory of the removed cell, and contact between NG2+ cell processes was always followed by retraction (see Figs. 1, 6). Homotypic repulsion would also provide the means to establish and maintain the radial orientation of their processes28. Alternatively, proliferation of these progenitors may be triggered by enhanced access to mitogens, such as platelet-derived growth factor (PDGF), as overexpression of PDGF increases the number of oligodendrocyte progenitors in the developing spinal cord39 and induces tumors from these cells in adults40. However, oligodendrocyte progenitors exhibit a density-dependent reduction in proliferation in saturating PDGF in vitro41, suggesting that direct interactions can limit their growth. Although guidance cues, such as netrin and semaphorins have been shown to influence the dispersion and differentiation of oligodendrocyte progenitors during development31, the molecules that maintain their density in the adult CNS remain to be identified. As NG2+ cell processes overlap extensively with other glial cells that exhibit tiling, such as astrocytes and microglia, the molecular pathways that control the distribution and density of different glial cell types are likely to be distinct.
Long-term time-lapse imaging of NG2+ cells in the cortex revealed that the entire network of NG2+ cells is constantly reorganized, due to the continual loss of cells and their intrinsic pressure to actively survey their local environment. The migration of individual cells was induced by local events – the differentiation, death, or displacement of neighboring cells – rather than large scale attractive or repulsive gradients (see Fig. 2d). In some instances, multiple cells within the imaged volume died or differentiated, creating larger voids that resulted in greater reorganization through migration of nearby NG2+ cells. However, large scale migration of individual cells through the parenchyma was not observed, and maintenance of the population was achieved through local proliferation of neighboring cells, rather than through a subpopulation of highly migratory, proliferative NG2+ cells (see Fig. 7d), consistent with the profound self-avoidance exhibited by these cells.
Dynamic filopodia were distributed along the length of NG2+ cells processes and even projected from their somata. This constant exploration of the local environment by NG2+ cells may allow them to not only assess their density, but also the viability of oligodendrocytes and the state of myelination of nearby axons4. If local interactions are required to stimulate oligodendrogenesis, it would explain why a high density of these progenitors is maintained throughout the adult CNS. In multiple sclerosis (MS), NG2+ cell density is dramatically reduced in many chronically demyelinated white matter lesions42. This impairment of NG2+ cell homeostasis may prevent detection of demyelinated axons and limit oligodendrocyte regeneration, raising the possibility that focal restoration of NG2+ cell numbers in chronic lesions may be therapeutically beneficial in MS.
NG2+ cells form functional synapses with glutamatergic neurons in all regions of the CNS that have been examined43-45. The high motility of NG2+ cell processes and constant movement of these cells through the parenchyma contrast with the remarkable stability of glutamatergic axons in the cortex observed through similar in vivo imaging, which apart from the refinement of some boutons, do not change their position over several months46. The extent of NG2+ cell movement suggests that synaptic connections between axons and NG2+ cells are continually remodeled on a time scale of days to weeks. The transient nature of these synapses may explain why NG2+ cells in the same brain region exhibit widely varying levels of synaptic connectivity45. Although the role of this rapid form of neuron-glial cell communication has not been established in vivo, glutamatergic signaling influences the proliferation and differentiation of these progenitors in vitro47. As NG2+ cells occupy non-overlapping domains, the continual reorganization of their processes may enable these progenitors to sample the activity of a greater proportion of axons to enable learning-induced changes in myelination48, and ensure that oligodendrocytes that degenerate as a result of injury, disease, or normal aging are rapidly replaced.
In vivo genetic fate tracing studies indicate that NG2+ cells continue to generate oligodendrocytes in the adult CNS, although less frequently than during early postnatal life7, 11-13. In accordance with these findings, time lapse imaging revealed that NG2+ cells in the cortex of mice 5 months old continue to differentiate into oligodendrocytes at a low rate (see Fig. 3). In vitro studies indicate that oligodendrocytes are generated through asymmetric division of these progenitors20; however, the steps leading to oligodendrogenesis in vivo have remained uncertain. Through long-term imaging we discovered that the majority of NG2+ cells in the adult cortex directly differentiate into oligodendrocytes without undergoing asymmetric division. Thus, proliferation of NG2+ cells in vivo is not a direct reflection of oligodendrogenesis, but rather a homeostatic response to replace progenitors that have differentiated or died.
This analysis of NG2+ cells focused exclusively on layers 1 and 2/3 of the cortex, due the limitations of two-photon imaging at greater depths. Although each cortical layer has a distinct cytoarchitecture, NG2+ cell behavior was similar through these upper layers of cortex (see Supplemental Fig. 7), consistent with the ability of these cells to proliferate and generate new oligodendrocytes throughout the CNS7, 13. Although not considered a traditional white matter region, layer 1 of the cortex contains a relatively high density of myelinated axons that course horizontally through the dendritic tufts of pyramidal neurons. Nevertheless, it is possible that NG2+ cell behaviors may differ between gray and white matter in the adult CNS, particularly if these regions vary with regard to cell turnover or oligodendrogenesis. Development of new approaches for high-resolution imaging in deep brain structures such as the corpus callosum, or long-term imaging of dorsal column white matter in the spinal cord, will be required to examine NG2+ cell behavior in adult white matter.
Injury to the CNS, whether induced acutely or through chronic disease, is often associated with gliosis, characterized by enhanced proliferation and accumulation of glia around lesions to form glial scars. This recruitment of glia to sites of trauma is thought to play a crucial role in limiting further injury and promoting repair. In addition to astrocytes and microglia/macrophages, cells that express the NG2 proteoglycan accumulate at lesions, with their numbers peaking one week following injury then declining slowly over an extended period37, 49. Determining the source of NG2 has been problematic, because NG2 is also expressed by pericytes and a subset of microglia/macrophages following injury36, 37, and can be released into the extracellular space through enzymatic cleavage by matrix metalloproteinases (MMPs)50. Time-lapse imaging of individual NG2+ cells revealed that these progenitors are attracted to the site of injury in a manner similar to microglia25; they reorient and extend their processes toward the lesion and eventually migrate to the peri-lesion area to participate in the formation of the scar. NG2+ cells recruited to the lesion no longer exhibit self-repulsion, suggesting that injury may transform these cells into distinct scar forming cells with unique characteristics. Accordingly, loss of cells to the scar triggered homeostatic proliferation of surrounding NG2+ cells to restore their density. Although NG2+ cells began to reorient their processes within minutes of injury, they arrived at the lesion several hours later than microglia (Extension rate: NG2+ cells: 2.7 ± 0.4 μm/hour, see Fig. 8; microglia: 1.25 ± 0.06 μm/min, from ref. 25). The sequential appearance of these different types of glial cells at the lesion suggests that they play distinct roles in the response to acute injury. Long-term time-lapse imaging revealed that accumulation of NG2+ cell processes reached its peak one week following injury induction (see Supplementary movie 8) and were eventually removed as the scar resolved. The participation of NG2+ cells in the formation and resolution of glial scars, suggests that that these ubiquitous progenitors not only serve as a reservoir to generate oligodendrocytes, but also as a surveillance network to detect CNS injury and promote tissue repair.
Methods
Generation of NG2-mEGFP transgenic mice
A mouse bacterial artificial chromosome (BAC) clone containing the NG2 (Cspg4) gene (RP23-309G21) was purchased from BACPAC Resources Center and modified by homologous recombination51. To target EGFP to the plasma membrane, the first 26 amino acids of Lck, a Src family tyrosine kinase which contains two palmitoylation domains and a myristoylation domain (gift from Drs. M. Dailey and S.Green, University of Iowa)24, was fused to the N-terminus of EGFP in pEGFP-N1 (Clontech). A cDNA encoding this membrane anchored EGFP (mEGFP) and subsequent rabbit β-globin polyA signal were placed at the translational initiator (ATG) of the NG2 (Cspg4) gene with two flanking homology arms (500 bps for each arm). The modified BAC was linearized by NotI digestion, injected into the pronucleus of mouse embryos in the JHU Transgenic Core, and implanted into pseudopregnant females. Of the three lines of NG2-mEGFP mice generated, two lines were selected for further studies.
In vivo two-photon microscopy
All experiments were performed in strict accordance with protocols approved by the Animal Care and use Committee at Johns Hopkins University. To prepare cranial windows, NG2-mEGFP-H mice (2–3 months old) were anesthetized by interperitoneal injection of ketamine (100 mg/kg b.w.) / xylazine (10 mg/kg b.w.); body temperature was maintained at 37°C with a thermostat-controlled heating plate. The skin over the right cerebral hemisphere was retracted and the skull cleaned. A metal plate with a center hole was attached to the skull with dental cement (C&B-Metabond) to allow for head stabilization. A ~400 μm diameter region of skull over somatosensory cortex (−0.5 to −2 mm post bregma and 1 to 3.5 mm lateral) was either thinned (~20 μm thickness) or removed using a high-speed dental drill30, 52. For cranial windows, a piece of cover glass (VWR, No. 1) was placed within the craniotomy and sealed with dental cement. In vivo imaging sessions began immediately after surgery (thinned-skull preparation) or after a minimum of 3 weeks (chronic cranial window). Although craniotomy can induce gliosis29, these small cranial windows did not induce reactive changes in microglial cells or NG2+ cells 3 weeks following implantation (Supplementary Fig. 13). Mice were anesthetized with isoflurane (1.0–1.5%; mixed with 0.5 liters/min O2) during imaging. Images were collected using a Zeiss LSM 710 microscope equipped with a GaAsP detector using a mode-locked Ti:sapphire laser (Coherent Ultra II) tuned to 920 nm. The average power at the sample during imaging was <50 mW. Vascular landmarks (EGFP+ pericytes) were utilized to identify the same imaging area on subsequent days.
Two-photon laser-induced lesions and individual cell ablation
Small lesions were induced in cortical gray matter by illuminating with the Ti:sapphire laser for several seconds (780 nm; ~150 mW, 1–3 s)25. This protocol induced a ~ 20 μm area of damage visible as an autofluorescent sphere. To ablate individual NG2+ cells in mice with thinned-skull windows, cell somata were subjected to repeated, short-duration laser irradiation (780 nm; ~150 mW, 100-500 ms, 1–6 repetitions)53. Successful ablations were preceded by slight swelling of the soma that was followed by cell disappearance within 1–8 hours. Laser power, pulse duration, and pulse repetition were varied depending on the depth of the targeted cell and thickness of the overlying thinned-skull.
Image processing and analysis
Image stacks and times series were analyzed using Fiji/ImageJ54. All analysis was performed on unprocessed images. For presentation in figures, image brightness and contrast levels were adjusted for clarity. Filopodial images were additionally processed with a Gaussian filter (radius = 2 pixels) to remove detector noise. For pseudo-color display of individual cells, the 3D cell volume was defined plane-by-plane for each time point and a custom Fji/ImageJ script was used to segment and/or colorize the cell.
Filopodial analysis
Filopodia on NG2+ cell processes (≥ 50 μm in length) were analyzed frame-by-frame using a custom Fiji/ImageJ script to quantify filopodial length, motility, density, and lifetime. Filopodia were considered dynamic if the length varied more than 2 μm during the imaging period. For analysis of filopodial density in fixed tissue, protrusions with a length at least twice the width and a minimum length of 2 μm were classified as filopodia.
Cell tracking
NG2+ cells were tracked in 3D using custom Fiji/ImageJ scripts by defining soma position at each time-point, recording XYZ coordinates and categorizing “cellular behavior” (e.g. proliferation, differentiation, etc.). NG2+ cells were classified as proliferating if, 1) two cells appeared in the same location where there was one cell on the previous time-point, 2) the somata were separated by less than 50 μm 3) the processes of recently divided cells were oriented in opposing directions, and 4) the somata lacked major cell processes at the point closest to the sister cell. The 3D volume around each proliferating cell was examined to exclude neighboring NG2+ cells that migrated into the field. NG2+ cells were classified as differentiating if, 1) cells exhibited a progressive decrease in EGFP fluorescence, 2) cells changed from having radial, highly branched processes, to long, unbranched processes characteristic of oligodendrocytes, and 3) the processes of neighboring NG2+ cells invaded the territory of the differentiating cell. NG2+ cells were classified as dying if cells exhibited a, 1) shortening and/or blebbing of processes, 2) reduction in territory size, 3) increase in brightness of the cell soma, and 4) complete disappearance of EGFP fluorescence in < 3 days without movement from the imaging area. NG2+ cells that lost EGFP fluorescence but could not be definitively identified as differentiated or dying were classified as “Unsure.”
NG2+ cell territory invasion
To calculate territory invasion following death or differentiation, a convex hull was computed plane-by-plane for each time-point around the differentiating/dying cell, the cell masked and EGFP+ processes contained within the convex hull autothresholded using the Fiji/ImageJ IsoData algorithm. A maximum projection of 3D volume defined by the convex hull was generated and the area of processes for each time-point determined. To calculate territory invasion following NG2+ cell ablation, the 3D volume of the ablated cell was merged with the image of the following time-point to identify “invading” processes. For control cells, the cell in the second time-point was masked and the region of the 3D volume of the cell in the first time-point was overlaid onto the second time-point. A maximum projection of 3D volume of the invading processes was autothresholded using the Fiji/ImageJ IsoData algorithm and the extent of territory invasion calculated by normalizing the area occupied by “invading” processes at the last time-point to the area occupied by the cell at the first time-point.
Analysis of local proliferation
NG2+ cells that underwent proliferation, differentiation, or death were identified during the imaging period. For each differentiating cell, the time-point at which the cell displayed a premyelinating oligodendrocyte morphology55, and neighboring cells had begun invading that cell’s territory, was marked as day 0. For each cell that died, the time-point that the cell displayed altered morphology (increased brightness of cell soma and reduced territory size), immediately preceding disappearance, was marked as day 0. After identification of a NG2+ cell that underwent differentiation/death, immediately adjacent NG2+ cells were examined for cell division preceding and following the event. The day of proliferation was marked as the time-point at which the two sister cells had undergone cytokinesis and had no adjoining processes. Cells were excluded from analysis if the entire differentiation/death process did not occur during the imaging period or if not all adjacent cells were located within the field of view. For controls, we analyzed cells that did not undergo proliferation, differentiation, or cell death for the entire imaging period and did not have more than 1 cell division within 100 μm.
Analysis of the response to focal injury
To follow appearance of NG2+ cells at lesions, EGFP+ processes entering a 75 μm zone surrounding lesion sites (area X) from a larger 150 μm zone (area Y) were measured as a function of time (Fig. 4a)25. To calculate the area surrounding the lesion occupied by NG2+ cell processes, images were autothresholded using the Fiji/ImageJ IsoData algorithm and the number of pixels was assessed. The change in pixel number over time (Rx(t)) compared to the initial value of pixels (R (0)) represented NG2+ x cell processes accumulation. To control for variations in the number NG2+ cells surrounding the lesion site in different experiments, accumulation of NG2+ cell processes was calculated relative to the number of processes in the outer area Y immediately following induction of the lesion (Ry(0)). The NG2+ cell response index at any time-point, (R(t)), is given by the equation:
| (1) |
To quantify response speed, we measured the length of NG2+ cell processes within 150 μm of the lesion site over time. To determine the leading and trailing processes of NG2+ cells, the coordinate system was rotated so that the y-axis passed through the lesion site at the top and the soma of the cell was located at the origin (lesion site = 90° from the cell of interest). The angle at which processes extended from the soma determined if they were on the leading edge (0–180°) or trailing edge (181–360°) of the NG2+ cell.
To determine the magnitude and direction of NG2+ cell movements with respect to the lesion site, the coordinate system was rotated such that the x-axis passed through the lesion site at the right and the start point of the cell was located at the origin (lesion site = 0° from the start point). Next, the angle θ between the displacement vector of NG2+ cell movement and the x-axis (displacement vector between the start point and the lesion site) was determined using:
| (2) |
where ai is the displacement vector for the cell, i and aL is the displacement vector between the start point of the cell and the lesion site, respectively, ai • aL is the dot product between the two vectors, and . The sign of the angle was determined by calculating the determinant of the two vectors using:
| (3) |
where the Cartesian coordinates of the lesion site, the start position, and the final position of the NG2+ cell of interest are defined as a, b, c respectively.
Immunohistochemistry
Mice were administered an overdose of anesthesia (pentobarbital; 100 mg/kg, i.p.) perfused transcardially with 4% formaldehyde (PFA in 0.1M phosphate buffer [pH 7.4]), their brains extracted and postfixed in 4% formaldehyde for 4 hours or overnight at 4°C, then cryoprotected in a 30% sucrose solution (in PBS, pH 7.4) at 4°C for up to 36 hours. Brains were frozen in TissueTek, sectioned (coronal, bregma 0.2 to –1.9 mm) at 30–50 μm thick, and incubated free-floating for 1–2 hr at room temperature (RT) in a blocking solution (5% normal donkey serum, 0.3% Triton X-100 in PBS, pH 7.4). Sections were incubated with primary antibodies (Supplementary Table 1) suspended in blocking solution overnight at 4°C on an orbital shaker. For Olig2 immunostaining, brain sections were incubated in LAB solution (Polysciences) for 10 min prior to blocking. For BrdU immunostaining, sections were treated with 2N HCl at 37°C for 30 min., then neutralized with 0.1 M sodium borate buffer (pH 8.5) prior to incubation with primary antibodies. After washing in blocking solution, sections were incubated with fluorescently conjugated secondary antibodies (Supplementary Table 2) for two hours at RT then mounted on slides with Aqua Poly/Mount (Polysciences). Images were acquired using either an epifluorescence microscope (Zeiss Axio-imager M1) and Axiovision software (Zeiss) or a confocal laser-scanning microscope (Zeiss LSM 510 Meta). For cell proliferation analysis, mice were provided with BrdU-containing drinking water (1 mg/ml supplemented with 1% sucrose) and received two injections of BrdU (50 mg/kg, i.p.) per day, at least 8 hr apart, for one week prior to perfusion. To analyze the effects of cranial window implantation, burr holes were drilled <1 mm outside the crainiotomy area and 1 μl of dextran-conjugated rhodamine was injected into the brain to label the position of the cranial window just prior to sacrifice.
Statistical analysis
Statistical analyses were performed in OriginPro (OriginLab), Excel (Microsoft), or Fiji/ImageJ. Sample sizes were chosen according to the standard practice in the field. Normality of the data was tested with the Shapiro-Wilk test. Significance was determined using the Mann-Whitney test, unpaired or paired two-tailed Student’s t test, or one-way ANOVA with Tukey post-hoc test, as noted. All direction statistics were performed using the modification by Moore to the Rayleigh’s test56, implemented in Chemotaxis and Migration Tool (ImageJ plug-in, Ibidi). All data are presented as mean ± standard error of the mean (s.e.m.).
Supplementary Material
Acknowledgments
We thank M. Pucak, N. Ye, and T. Lee for technical assistance, B. Cudmore (Johns Hopkins University) and S. Wang (Princeton University) for advice on cranial window implantation, W-B Gan (New York University) for advice on preparing thinned skull windows, and members of the Bergles laboratory for discussions. E.G.H was supported by a NRSA grant from the NIH (F32NS076098). Funding was provided by grants from the NIH (NS051509, NS050274) and the Brain Science Institute at Johns Hopkins University.
Footnotes
Contributions: E.G.H., M.F., S.H.K., and D.E.B. designed experiments; E.G.H. designed, executed, and analyzed the experiments described in the figures, movies, and text. M.F. made seminal observations of NG2+ cell dynamics and their response to laser-induced lesions in thinned skull preparations, and generated data for Supplementary Fig. 8. S.H.K. generated and characterized the NG2-mEGFP-H and NG2-mEGFP-L mouse lines and created Supplementary Fig. 1. E.G.H. and D.E.B. wrote the manuscript.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Biteau B, Hochmuth CE, Jasper H. Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell. 2011;9:402–411. doi: 10.1016/j.stem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simons BD, Clevers H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 2011;145:851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 3.Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 4.Franklin RJ, Gilson JM, Blakemore WF. Local recruitment of remyelinating cells in the repair of demyelination in the central nervous system. J Neurosci Res. 1997;50:337–344. doi: 10.1002/(SICI)1097-4547(19971015)50:2<337::AID-JNR21>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 5.Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci. 2007;10:1538–1543. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- 6.Dawson MR, Polito A, Levine JM, Reynolds R. NG2-expressing glial progenitor cells: an abundant and widespread population of cycling cells in the adult rat CNS. Mol Cell Neurosci. 2003;24:476–488. doi: 10.1016/s1044-7431(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 7.Kang SH, Fukaya M, Yang JK, Rothstein JD, Bergles DE. NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron. 2010;68:668–681. doi: 10.1016/j.neuron.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kessaris N, et al. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirby BB, et al. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat Neurosci. 2006;9:1506–1511. doi: 10.1038/nn1803. [DOI] [PubMed] [Google Scholar]
- 10.Nishiyama A, Komitova M, Suzuki R, Zhu X. Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat Rev Neurosci. 2009;10:9–22. doi: 10.1038/nrn2495. [DOI] [PubMed] [Google Scholar]
- 11.Rivers LE, et al. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat Neurosci. 2008;11:1392–1401. doi: 10.1038/nn.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimou L, Simon C, Kirchhoff F, Takebayashi H, Gotz M. Progeny of Olig2-expressing progenitors in the gray and white matter of the adult mouse cerebral cortex. J Neurosci. 2008;28:10434–10442. doi: 10.1523/JNEUROSCI.2831-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young KM, et al. Oligodendrocyte Dynamics in the Healthy Adult CNS: Evidence for Myelin Remodeling. Neuron. 2013;77:873–885. doi: 10.1016/j.neuron.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tripathi RB, Rivers LE, Young KM, Jamen F, Richardson WD. NG2 glia generate new oligodendrocytes but few astrocytes in a murine experimental autoimmune encephalomyelitis model of demyelinating disease. J Neurosci. 2011;30:16383–16390. doi: 10.1523/JNEUROSCI.3411-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine JM, Reynolds R. Activation and proliferation of endogenous oligodendrocyte precursor cells during ethidium bromide-induced demyelination. Exp Neurol. 1999;160:333–347. doi: 10.1006/exnr.1999.7224. [DOI] [PubMed] [Google Scholar]
- 16.Haber M, Vautrin S, Fry EJ, Murai KK. Subtype-specific oligodendrocyte dynamics in organotypic culture. Glia. 2009;57:1000–1013. doi: 10.1002/glia.20824. [DOI] [PubMed] [Google Scholar]
- 17.Haberlandt C, et al. Gray matter NG2 cells display multiple Ca2+-signaling pathways and highly motile processes. PLoS One. 2011;6:e17575. doi: 10.1371/journal.pone.0017575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McTigue DM, Wei P, Stokes BT. Proliferation of NG2-positive cells and altered oligodendrocyte numbers in the contused rat spinal cord. J Neurosci. 2001;21:3392–3400. doi: 10.1523/JNEUROSCI.21-10-03392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magnus T, et al. Adult glial precursor proliferation in mutant SOD1G93A mice. Glia. 2008;56:200–208. doi: 10.1002/glia.20604. [DOI] [PubMed] [Google Scholar]
- 20.Sugiarto S, et al. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell. 2011;20:328–340. doi: 10.1016/j.ccr.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivkovic S, Canoll P, Goldman JE. Constitutive EGFR signaling in oligodendrocyte progenitors leads to diffuse hyperplasia in postnatal white matter. J Neurosci. 2008;28:914–922. doi: 10.1523/JNEUROSCI.4327-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Persson AI, et al. Non-stem cell origin for oligodendroglioma. Cancer Cell. 18:669–682. doi: 10.1016/j.ccr.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benediktsson AM, Schachtele SJ, Green SH, Dailey ME. Ballistic labeling and dynamic imaging of astrocytes in organotypic hippocampal slice cultures. J Neurosci Methods. 2005;141:41–53. doi: 10.1016/j.jneumeth.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 25.Davalos D, et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- 26.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 27.Kay JN, Chu MW, Sanes JR. MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature. 2012;483:465–469. doi: 10.1038/nature10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. 2012;488:517–521. doi: 10.1038/nature11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu HT, Pan F, Yang G, Gan WB. Choice of cranial window type for in vivo imaging affects dendritic spine turnover in the cortex. Nat Neurosci. 2007;10:549–551. doi: 10.1038/nn1883. [DOI] [PubMed] [Google Scholar]
- 30.Yang G, Pan F, Parkhurst CN, Grutzendler J, Gan WB. Thinned-skull cranial window technique for long-term imaging of the cortex in live mice. Nat Protoc. 2010;5:201–208. doi: 10.1038/nprot.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spassky N, et al. Directional guidance of oligodendroglial migration by class 3 semaphorins and netrin-1. J Neurosci. 2002;22:5992–6004. doi: 10.1523/JNEUROSCI.22-14-05992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menn B, et al. Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci. 2006;26:7907–7918. doi: 10.1523/JNEUROSCI.1299-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Psachoulia K, Jamen F, Young KM, Richardson WD. Cell cycle dynamics of NG2 cells in the postnatal and ageing brain. Neuron Glia Biol. 2009;5:57–67. doi: 10.1017/S1740925X09990354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Busch SA, Silver J. The role of extracellular matrix in CNS regeneration. Curr Opin Neurobiol. 2007;17:120–127. doi: 10.1016/j.conb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Goritz C, et al. A pericyte origin of spinal cord scar tissue. Science. 2011;333:238–242. doi: 10.1126/science.1203165. [DOI] [PubMed] [Google Scholar]
- 37.Jones LL, Yamaguchi Y, Stallcup WB, Tuszynski MH. NG2 is a major chondroitin sulfate proteoglycan produced after spinal cord injury and is expressed by macrophages and oligodendrocyte progenitors. J Neurosci. 2002;22:2792–2803. doi: 10.1523/JNEUROSCI.22-07-02792.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuerst PG, Koizumi A, Masland RH, Burgess RW. Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature. 2008;451:470–474. doi: 10.1038/nature06514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calver AR, et al. Oligodendrocyte population dynamics and the role of PDGF in vivo. Neuron. 1998;20:869–882. doi: 10.1016/s0896-6273(00)80469-9. [DOI] [PubMed] [Google Scholar]
- 40.Assanah MC, et al. PDGF stimulates the massive expansion of glial progenitors in the neonatal forebrain. Glia. 2009;57:1835–1847. doi: 10.1002/glia.20895. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H, Miller RH. Density-dependent feedback inhibition of oligodendrocyte precursor expansion. J Neurosci. 1996;16:6886–6895. doi: 10.1523/JNEUROSCI.16-21-06886.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang A, Nishiyama A, Peterson J, Prineas J, Trapp BD. NG2-positive oligodendrocyte progenitor cells in adult human brain and multiple sclerosis lesions. J Neurosci. 2000;20:6404–6412. doi: 10.1523/JNEUROSCI.20-17-06404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chittajallu R, Aguirre A, Gallo V. NG2-positive cells in the mouse white and grey matter display distinct physiological properties. J Physiol. 2004;561:109–122. doi: 10.1113/jphysiol.2004.074252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergles DE, Roberts JD, Somogyi P, Jahr CE. Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature. 2000;405:187–191. doi: 10.1038/35012083. [DOI] [PubMed] [Google Scholar]
- 45.De Biase LM, Nishiyama A, Bergles DE. Excitability and synaptic communication within the oligodendrocyte lineage. J Neurosci. 2010;30:3600–3611. doi: 10.1523/JNEUROSCI.6000-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Paola V, et al. Cell type-specific structural plasticity of axonal branches and boutons in the adult neocortex. Neuron. 2006;49:861–875. doi: 10.1016/j.neuron.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 47.Gallo V, et al. Oligodendrocyte progenitor cell proliferation and lineage progression are regulated by glutamate receptor-mediated K+ channel block. J Neurosci. 1996;16:2659–2670. doi: 10.1523/JNEUROSCI.16-08-02659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bengtsson SL, et al. Extensive piano practicing has regionally specific effects on white matter development. Nat Neurosci. 2005;8:1148–1150. doi: 10.1038/nn1516. [DOI] [PubMed] [Google Scholar]
- 49.Levine JM. Increased expression of the NG2 chondroitin-sulfate proteoglycan after brain injury. J Neurosci. 1994;14:4716–4730. doi: 10.1523/JNEUROSCI.14-08-04716.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larsen PH, Wells JE, Stallcup WB, Opdenakker G, Yong VW. Matrix metalloproteinase-9 facilitates remyelination in part by processing the inhibitory NG2 proteoglycan. J Neurosci. 2003;23:11127–11135. doi: 10.1523/JNEUROSCI.23-35-11127.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- 52.Holtmaat A, et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chung SH, Mazur E. Femtosecond laser ablation of neurons in C. elegans for behavioral studies. Applied Physics a-Materials Science & Processing. 2009;96:335–341. [Google Scholar]
- 54.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapp BD, Nishiyama A, Cheng D, Macklin W. Differentiation and death of premyelinating oligodendrocytes in developing rodent brain. J Cell Biol. 1997;137:459–468. doi: 10.1083/jcb.137.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moore BR. A Modification of the Rayleigh Test for Vector Data. Biometrika. 1980;67:175–180. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.