Abstract
The signaling pathways in tick salivary glands that control “sialo-secretome” secretion at the tick-host interface remain elusive. The complex processes by which tick sialo-secretome is exocytosed from the salivary gland cells and manipulates host hemostatic responses are essential for successful prolonged blood feeding. Exocytosis of the sialo-secretome in the salivary glands requires a core of Soluble N-ethylmaleimide-sensitive fusion attachment proteins (SNAPs) and their receptor proteins (SNAREs). SNAREs have been identified as the key components in regulating the secretion of the sialo-secretome in the salivary gland cells. In this study, we investigated the functional role of two Amblyomma maculatum SNARE complex proteins, AmNSF and AmSNAP-25, in the tick salivary glands during extended blood-feeding on the vertebrate host. The qRT-PCR analysis exhibited cyclic regulation of AmNSF transcript, increasing nearly four-fold around 48 hour post infestation in the salivary glands. Similarly, AmSNAP-25 transcript followed cyclic regulation, increasing three-fold around 72 hour post infestation in the midguts. Immunolocalization of AmNSF showed the presence of NSF in secretory granule containing cells of Acini II and Acini III in the unfed stage, with widespread localization in the partially fed glands. Knockdown of AmNSF and AmSNAP-25 transcript resulted in death, impaired feeding on the host, and lack of engorgement in both experimental groups at eleven days post infestation, 13 mg for dsRNA-AmNSF, 191 mg for dsRNA-AmSNAP-25, and 383 mg for control ticks. Depletion also led to important morphological changes in the collapse of the Golgi apparatus in the salivary gland cells. Our results imply a functional significance of AmNSF and AMSNAP-25 in the prolonged tick feeding, and survival on the host. Further characterization of the factors that regulate exocytosis will lead to novel approaches to prevent tick-borne diseases.
1. Introduction
Exocytosis is mediated by SNAREs, Soluble NSF Attachment Protein Receptors. SNAREs are small molecules that are mostly found in the plasma membrane and are classified in one of two ways; one, as target (t) or vesicle (v) SNAREs, or alternatively as arginine (R) or glutamine (Q) containing SNAREs which is determined by the presence of the residue found in the SNARE domain of these molecules (Fasshauer et al. 1998). Syntaxin and SNAP-25, synaptosomal associated protein of 25 kDa, comprise the t-SNARE classification, and synaptobrevin and VAMPs, vessical associated membrane proteins, comprise the v-SNAREs. The SNARE complex contains a syntaxin, SNAP-25, and VAMP molecule; these three proteins come together in a zipper like method, which begins at the N-termini and moves towards the C-termini, fusing the vesicle membrane with the plasma membrane of the cell. Once exocytosis is complete, and the vesicle contents have been deposited into the cell, two molecules aid in the disassembly and recycling of the components of the SNARE complex: α SNAP and NSF, N-ethylmaleimide sensitive fusion protein (Zhao et al. 2007). In this study, we focused on two SNARE molecules, AmSNAP-25 and AmNSF as the functional significance of these two molecules in obtaining a bloodmeal, to date, has yet to be investigated in any tick species.
The regulated fusion of vesicles with the plasma membrane in neural and endocrine systems requires a core SNARE complex (synaptobrevin, Syntaxin, SNAP-25). This complex is proposed to function in vesicle targeting, docking and fusion. For instance, in PC12 cells, RNA interference on SNAP-25 greatly reduced the level of catecholamine secretion (Cahill et al. 2006). Additionally, a temperature sensitive SNAP-25 mutant in Drosophila exhibits decreased neurotransmitter release at 37°C in comparison to the control (Rao et al. 2001). Interestingly, SNAP-25 homologues have been identified in other tissues of various organisms, suggesting that SNARE mediated exocytosis is highly conserved throughout tissues. Most notably are SNAP-24 found in Drosophila and SNAP-23, also called syndet, in both humans and mice (Ravichandran et al. 1996, Wang et al. 1997, Niemeyer and Schwarz 2000). NSF is a protein required for constitutive and Ca++-regulated neuro-secretion (Sollner et al. 1993a, Sollner et al. 1993b, Rothman and Warren 1994). In Drosophila, the two NSF genes, dNSF1 and dNSF2, are involved in either neuronal or non-neuronal secretion, respectively (Boulianne and Trimble 1995). Expression of dNSF1 is seen in the nervous system of adults while dNSF2 is seen during larval development in secretory tissues such as the ring gland and salivary glands (Boulianne and Trimble 1995, Golby et al. 2001). The functional significance of SNARE proteins in non-neuronal arthropod systems remains an enigma.
Ticks are blood sucking arthropods that are found in nearly every region of the world and are second only to mosquitoes in their public health importance (Soneshine 1991). Ticks transmit a variety of pathogens to both humans and veterinary species, such as, Anaplasmosis, Ehrlichiosis, Heartwater, Lyme disease, Rocky Mountain spotted fever, and Spotted Fever Rickettsiosis. The Gulf Coast tick, Amblyomma maculatum, is a three host tick; the larvae and nymphs feed on small rodents while the adults feed on large mammals. These ticks are found along the Atlantic and Gulf Coast regions with inland stretches as far as Oklahoma. A. maculatum is a known carrier of rickettsial species including Rickettsia parkeri, R. africae, and R. slovaca. These pathogens pass from the midguts of the tick to the salivary glands and then into the host via the tick saliva; the mechanism by which these pathogens move is through exocytosis.
Here we use an in vivo gene silencing technique, RNA interference (RNAi), to test the functional significance of AmSNAP-25 and AmNSF during prolonged tick feeding on the host. We see that depletion of both AmSNAP-25 and AmNSF in the Gulf Coast tick significantly impeded tick feeding and ultimate survival on the host.
Results
Transcriptional Expression of AmNSF and AmSNAP-25
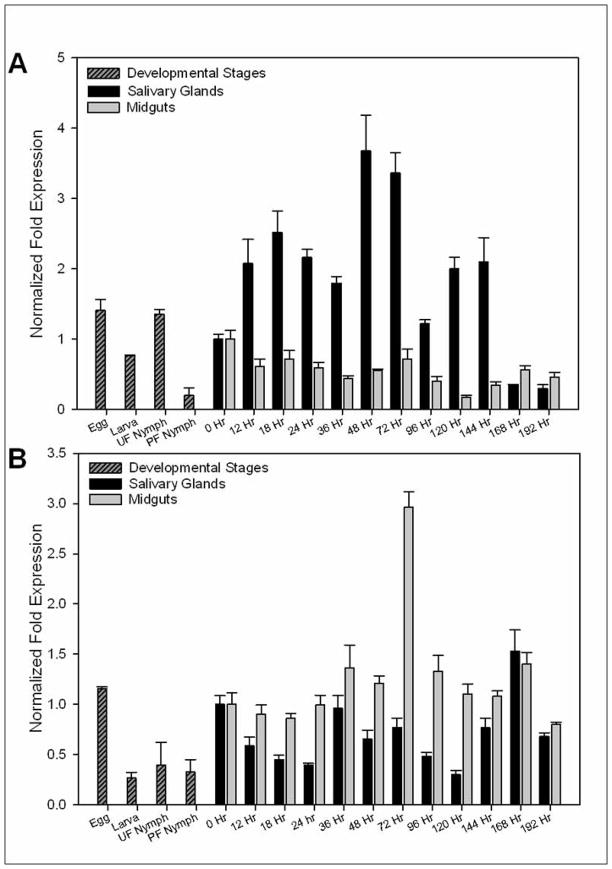
Since NSF and SNAP-25 are involved in exocytosis, we first examined the relative transcript level of both genes during the developmental stages of A. maculatum. We found that AmNSF was most vital during embryonic development, 1.41 fold, and during the unfed nymphal stage, 1.35 fold, as the relative gene expression was equivalent, or higher, to that of the unfed salivary glands, 1.0 fold (Fig. 1A). We observed both an increases and decreases in AmNSF transcript levels in the salivary glands of adult female A. maculatum throughout the blood meal. During the slow feeding stage, from 0 hour to approximately 96 hour, there are two points where AmNSF transcript is significantly increased in abundance; first, at 18 hour there is a 2.5 fold increase compared to unfed ticks and then a 3.5 fold increase at 48 hour (Fig. 1A). Interestingly, as the tick is entering the fast feeding stage, around 120 hours post infestation there is an increase in transcriptional gene expression, approximately 2 fold higher than unfed (Fig. 1A). The transcript level of AmNSF in the midguts exhibits reduction in the relative transcript abundance at the beginning of the blood meal; there are some slight variances in the relative transcript level of AmNSF in the midgut, but none that ever rise above the unfed stage (Fig. 1A). This cyclic regulation of AmNSF is consistent with the theory that the SNARE partners are recycled after completion of the vesicle fusion event.
Figure 1.
AmSNAP-25 transcript is slightly higher during embryonic development, 1.15 fold, when compared to adult unfed salivary gland, 1.0 fold, but is lower remaining nearly constant, during the subsequent life stages, i.e., larvae, 0.26 fold, unfed nymph, 0.39 fold, and partially fed nymph, 0.32 fold (Fig. 1B). The transcriptional expression of AmSNAP-25 in the salivary glands appears to follow a trend of cyclic regulation, with decreases and increases seen throughout the entire blood meal; this is consistent with the thought that SNARE molecules are recycled after each vesicle fusion event. AmSNAP-25 in the midguts, however, was constitutively expressed until 72 hours post infestation where the transcript abundance increased to 2.96 fold compared to unfed midguts.
Endogenous protein expression and Immunolocalization of AmNSF
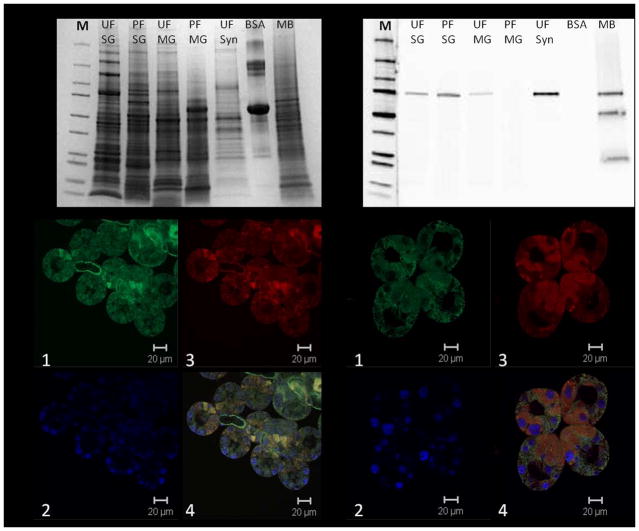
After examining the relative transcript abundance of AmNSF and AmSNAP-25, we next focused on the endogenous protein expression of these molecules (Fig. 2). However, we were unable to find a commercial SNAP-25 antibody which adequately cross reacted with our tissues homogenates. The abundance of AmNSF increased in partially fed salivary glands in comparison to the unfed salivary glands, which is seen by the prominent band at approximately 75 kDa (Fig. 2B). Surprisingly, none of the NSF protein was detected in the partially fed midguts and only a faint band was detected in the unfed midguts (Fig. 2B). Additionally, a prominent band is seen in the unfed synganglia extract even though the protein concentration was significantly lower than that of the salivary glands or midguts, 5ug of proteins from synganglia extract compared to 20ug of salivary glands or midguts extract; this is consistent with previous knowledge linking SNARE proteins to neuronal secretion/synaptic transmission. The three bands seen in the mouse brain extract, which served as a positive control, is a common occurrence.
Figure 2.
Next we localized endogenous AmNSF in unfed and partially blood-fed salivary glands. We first examined the localization pattern of AmNSF in the unfed salivary glands. AmNSF was stained distinctly in both acini II and III, and also in the salivary ducts (Fig. 2C). Upon further examination, AmNSF was also seen localized in the two cell types connecting to the salivary duct (Fig. 2C). This clear localization may indicate that these two cells of the acini are the first to begin secretion and, therefore, explain why AmNSF is highly expressed here. In the partially fed A. maculatum salivary glands ubiquitous expression of AmNSF can be seen (Fig. 2D), indicating an activation of the cellular machinery within the cells of the acini.
In vivo Gene Silencing of AmNSF and AmSNAP-25
In order to reveal the importance of AmNSF and AmSNAP-25 we implored the technique of RNA interference (RNAi) by injected 800 ng of dsRNA for either gene into separate experimental groups of unfed, adult, female A. maculatum ticks. The observed 99.9% transcript depletion of AmNSF and AmSNAP-25 in the respective dsRNA treated salivary glands indicates the successful silencing of these genes through RNAi (Fig. 3A). Depletion of AmNSF resulted in a 70% mortality rate (Fig. 3D) as well as impeded attachment to the host. Additionally, it was noticed during dissections that ticks treated with dsRNA-NSF had midguts that appeared disintegrated and would completely fall apart when touched (data not shown). Ticks treated with dsRNA-SNAP-25 showed no difference in attachment from the control.
Figure 3.
Fitness and Fecundity of RNAi Ticks
The engorged body weight of partially fed female A. maculatum did show marked differences between the mock control and both experimental groups (Fig. 3B, D). Ticks injected with dsRNA-NSF had a lower average body weight 11 days post infestation compared to that of the control ticks (12.83 mg ± 1.5 vs. 382.23 mg ± 87.6, p<0.05). Likewise, dsRNA-SNAP-25 treated ticks weighed less at 11 days post infestation when compared to the control ticks (91.01 mg ± 22.9 vs. 382.23 mg ± 87.6, p<0.05). None of the dsRNA-NSF ticks engorged, however the final engorgement weight of dsRNA-SNAP-25 was lower than that of the control ticks (145.7 mg ± 46.6 vs. 638.2 mg ± 33.7, p<0.05).
A. maculatum ticks that were injected with dsRNA-SNAP-25 showed a reduction in average egg mass in comparison to control ticks average egg mass size (58.2 mg ± 35.75 vs. 451.4 mg ± 28.7, p<0.05) (Fig. 3C, D). Additionally, dsRNA-SNAP-25 treated ticks laid eggs which were dark brown in colorization, compared to the light brown, tan color of eggs from control ticks (Fig 3C). Finally, no larvae emerged from the dsRNA-SNAP-25 eggs while the eggs from control ticks exhibited a normal larval hatching rate (data not shown). None of the dsRNA-NSF ticks reached a sufficient weight to be examined for effects on oviposition.
Compensatory Action of SNARE partners
We hypothesized that the depletion of AmNSF or AmSNAP-25 would affect the gene expression of interacting molecules that form the SNARE complex, and therefore examined the transcriptional level of other SNARE proteins (Fig. 3A) identified from the pyrosequencing of the cDNA library generated from A. maculatum salivary glands (Karim et al. 2011). The transcript level of Syntaxin 16 exhibited a decrease in transcript level in the salivary glands of ticks treated with both dsRNA-NSF and dsRNA-SNAP-25 in comparison to the mock control. However, the other SNARE molecules, Syntaxin 1a, Syntaxin 17, Syntaxin 18, VAMP 1/2, and VAMP7 all show no significant changes in the transcript level in salivary glands in dsRNA-SNAP-25 treated ticks, except Syntaxin 8 transcription level which was decreased in the dsRNA-SNAP-25 ticks. In ticks treated with dsRNA-NSF, Syntaxin 18 and VAMP7 both exhibited an increase in the relative transcript abundance in the salivary glands when compared to the control, while the remaining SNARE molecules, Syntaxin 1a, Syntaxin 8, Syntaxin 17, and VAMP 1/2 do not exhibit any differential expression.
Immunolocalization of AmNSF in RNAi salivary glands
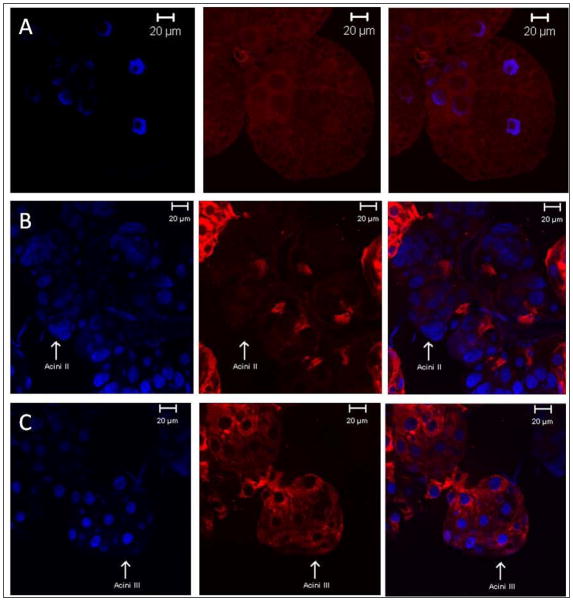
Next, we examined the immunolocalization of AmNSF in partially blood fed control versus the knockdown, RNAi, ticks. The immunofluorescence of AmNSF in salivary glands from 6 day fed control ticks (Fig. 4A) showed that AmNSF is ubiquitously expressed throughout the salivary acini cells. However, in salivary glands from 6 day fed ticks treated with dsRNA-NSF there is significant de-localization of AmNSF (Fig. 4B,C). The strongest effect was seen in acini II glands (Fig. 4B) where there was minimal detection of AmNSF with slight protein localization towards the salivary gland duct. Likewise, acini III glands (Fig. 4C) exhibit a diffuse localization with perinuclear and ductal localization of AmNSF.
Figure 4.
Transmission Electron microscopy
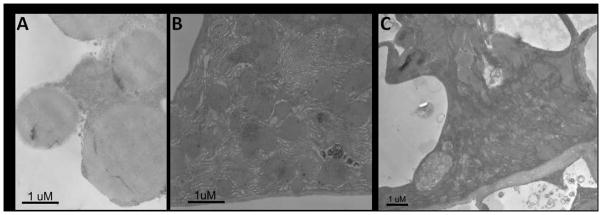
Ultrastructural analysis of the AmNSF and AmSNAP-25 knockdown salivary glands revealed a clear and corresponding phenotype. In contrast to the inclusion bodies comprising the secretory vesicles filled with proteins in normal cells (Fig. 5A), there were no detectable secretory vesicles in cells after knockdown of AmNSF and AmSNAP-25. Instead, the area of salivary gland cells exhibited striking collapse of the cells (Fig. 5B,C). The effect of AmNSF and AmSNAP-25 on Golgi structure was more pronounced in partially blood-fed salivary gland samples, and these Golgi structures were seen to be dispersed. In the control salivary glands, the cell structures (Golgi apparatus and secretory vesicles) appear as a highly organized structure in which stacks of flattened cisternae are tightly attached to each other. By contrast, in AmNSF and AmSNAP-25 depleted salivary gland cells the Golgi stacks were considerably smaller, collapsed, disorganized and had an altered morphology.
Figure 5.
Discussion
The Gulf-Coast tick (Amblyomma maculatum) has been implicated as a vector for many pathogens of veterinarian and human health importance, namely Rickettsia parkeri and Ehrlichia ruminantium (Parker et al. 1939, Uilenberg 1982, Walker and Olwage 1987, Paddock et al. 2004). In order to prevent the tick-borne infections, a basic understanding of the sialo-secretome release is crucial. Biochemical studies on secretagogue induced saliva revealed that tick saliva contains numerous bioactive proteins (Sauer et al. 1995, Sauer et al. 2000, Ribeiro and Francischetti 2003, Francischetti et al. 2009). We have previously shown that certain SNARE proteins are essential for saliva secretion through Ca2+-regulated exocytosis (Karim et al. 2002, Karim et al. 2004). SNARE proteins have become a major point of interest in membrane trafficking in the secretory cells due to their role in exocytosis (Hay and Scheller 1997). A high degree of conservation exists among intracellular SNARE complex proteins involved in vesicle transport in invertebrate and vertebrate systems. Eukaryotic cells contain several SNARE proteins involved in multiple steps in the secretory pathway, and their main functions are to transport proteins and lipids through organelles en route to their final destinations, such as the plasma membrane, the extracellular medium and endosomal/lysosomal compartments. Our previous studies have identified 21 SNARE proteins in the salivary glands of A. maculatum (Karim et al. 2011). To date, most studies concerning tick SNAREs have focused around vesicle associated molecules, therefore, in this study, we investigated the functional significance of two key non-vesicular SNARE proteins: N-ethylmaleimide sensitive fusion protein (NSF), and synaptosomal associated protein of 25 kDa (SNAP-25).
Quantitative RT-PCR analysis demonstrated that AmNSF and AmSNAP-25 were expressed in all developmental stages (Fig. 1). Our results showed that both AmNSF and AmSNAP-25 exhibit cyclic experssion at the transcript level throughout the blood meal of A. maculatum (Fig. 1). The cyclic nature of these SNARE molecules is consistent with previous reports showing SNARE proteins are recycled after a membrane fusion event, allowing for one SNARE molecule to mediate multiple exocytotic events (Hong 2005). The induced expression of AmNSF in salivary glands facilitates the recycling of the secretory vesicles and release of key saliva factors involved in tick attachment and suppressing the host immune system. Up-regulation of AmSNAP-25 in the tick midgut enables the release of digestive enzymes. It is likely that the wide distribution of AmNSF and AmSNAP-25 in egg, larval, unfed and partially nymphal ticks facilitates the release of saliva, as well as neuropeptides/neurotransmitters from synganglion, digestive enzymes from midgut, secretion of vitellogenin from the fat body, accumulation of vitellogenin in the hemolymph, and phagocytosis of oocytes (Soneshine 1991). AmNSF and AmSNAP-25 proteins are the core SNAREs that are essential to mediate most intracellular vesicle fusion events in the tick tissues. Western blot analysis showed the presence of AmNSF in tick synganglia (Fig. 2). It may be possible that AmNSF and AmSNAP-25 are expressed in other tissue types and play a vital role in the protein secretion. For example, neurotransmitters, such as dopamine, which stimulates fluid secretion, are also released by exocytosis (Maritz-Olivier et al. 2005).
The results from confocal scanning microscopy of AmNSF in the salivary glands of unfed versus partially blood fed A. maculatum support an essential role in vesicular trafficking (Fig. 2C–D). In the unfed salivary glands, highly localized staining was observed in Acinus II cell types. A diversity of cell types exists in salivary glands of Amblyomma maculatum and Acinus II contains 14–15 granular cells of 6 types, a, b, c1–c4 and interstitial epithelial cells surrounded a central lumen which opens via a cuticular valve into a lobular duct (Binnington 1978). All cells in unfed salivary glands contained secretory vesicles (granules) (Fig. 2C–D). Acinus III contains 16–15 cells of three types d, e and f in female salivary glands (Binnington 1978). Cell d is adjacent to the valvular duct and filled with secretory vesicles. Although, AmNSF possessed a prominent intracellular pool in the unfed salivary glands, it is challenging to identify specific cell types from confocal microscopy as tick salivary glands are a very complex tissue. As the blood meal begins, a more ubiquitous expression is seen suggesting the process of vesicular trafficking from the acini (Fig. 2-B). Despite the immunolocalization of AmNSF, a detailed study of intracellular distribution of AmNSF and AmSNAP-25 is needed in tick salivary cell types. Future immunolocalization work coupled with specific cell organelle and SNARE partner antibodies may help to more distinctly describe specific location of SNARE proteins in the cells of the salivary glands.
The present study is the first to examine the functional role of AmNSF and AmSNAP-25 in tick salivary glands on the organismal level. The qRT-PCR analysis demonstrated the depletion of AmNSF and AmSNAP-25 mRNA in tick salivary glands 6 days post attachment (Fig. 3A). The knockdown of both AmNSF and AmSNAP-25 resulted in significant phenotypic changes, indicative of a pivotal role for both of these molecules in successful blood feeding. The knockdown of AmNSF impaired tick blood feeding and engorgement 11 days post infestation and 70% of the ticks died on the host (Fig. 3E). The disruption of AmSNAP25 also impeded weight gain 11 days post infestation on the host. The transcript level of both AmNSF and AmSNAP-25 showed a significant reduction of 99.9% in the salivary glands of RNAi ticks (Fig. 3A). The ticks obtained were not big enough to extract saliva to investigate protein secretion in salivary glands. The salivary gland samples obtained were insufficient in size or volume and concentration to perform western blot. The AmNSF gene disruption exhibited a more significant lethal phenotype in that they had an average engorged body weight of only 13 mg as compared to 383 mg of the mock control. Furthermore, the midguts appeared disintegrated, and ticks were unable to transition into oviposition. Ticks injected with dsRNA-SNAP-25 also showed a reduction in body weight but were able to oviposit. However, the eggs which resulted from the dsRNA-SNAP-25 treated ticks were reduced in number and average weight, approximately 58 mg, and were noticeably darker in color than that of the control which were healthy in appearance with a light tan colorization and produced a normal number of eggs, with an average egg mass weight of approximately 450 mg. Furthermore, the eggs which resulted from the control ticks had a normal larvae hatching rate while none of the dsRNA-SNAP-25 treated ticks developed past the egg. It is possible that the more significant lethal phenotype observed in the AmNSF knockdown ticks resulted from, not only the exocytotic mechanism of this molecule (i.e. the disassembly of the SNARE complex), but its actions in other cellular processes, such as binding to membrane receptors (AMPA and GABA) (Zhao et al. 2007). Interestingly, NSF has been shown to bind to the dopaminergic receptor, with the most specificity seen with D1 and D5 (Heydorn et al. 2004). Tick salivary secretion has been shown to be directly mediated by dopamine in other ixodid species and the presence of a D1 dopamine receptor has been found in A. americanum (Kaufman 1976, Schmidt et al. 1981, Lindsay and Kaufman 1986, McSwain et al. 1992, Sauer et al. 2000, Bowman and Sauer 2004). It is also important to note that the focus of this study is on the action of exocytosis in the salivary glands but the depletion of NSF transcript most likely affected other tissues within the tick, for example the synganglia, midguts, or the reproduction system. The AmNSF and AmSNAP-25 gene disruption in tick salivary glands was assumed to lead to the failure of membrane fusion that facilitates blood feeding. Concurrently, it can be speculated that secretory vesicles may accumulate on the plasma membrane and Golgi apparatus exhibited collapsed structure in salivary glands (Fig. 4–5). Based on confocal and TEM results, this may be construed by the essential functional significance of AmNSF and AmSNAP-25 in transport of pharmacologicaly active molecules from the salivary Acinus cells into the host. Depletion of AmNSF and AmSNAP-25 may have blocked the exocytosis of a battery of enzymes involved in the digestion of the blood meal in the midgut. It could also be possible that ticks were unable to digest the blood meal in order to synthesize the variety of molecules involved in tick physiology. Lack of essential components required for the processing of the blood-meal caused the death or impaired engorgement in the tick (Karim and Adamson 2012). Other SNAREs have been silenced in Amblyomma americanum, Ixodes scapularis and Haemaphysalis longicornis (Karim et al. 2004, Karim et al. 2005, Gong et al. 2009). The results suggest a vital role of Synaptobrevin, Syntaxin, nSec1, and HIYkt6 in tick feeding and disruption of either gene may lead to the impaired blood feeding and ultimate death on the host.
It has previously been suggested that SNARE molecules exhibit a compensatory action to overcome any detrimental effect (Bethani et al. 2009). To examine this, we looked at the relative transcript abundance of select SNARE molecules through qRT-PCR. Based on the results, Syntaxin 16 exhibited down-regulation of transcript level in AmNSF and AmSNAP-25 knockdown salivary glands in comparison to that of the control; furthermore, Syntaxin 8 appears to be down regulated at the transcript level in the salivary glands of dsSNAP-25 knockdown ticks. None of the SNARE molecules were up-regulated in the AmSNAP-25 knockdown tissues; however, in the AmNSF knockdown salivary glands, Syntaxin 17 and VAMP 7 both showed up-regulation at the transcript level in salivary glands. As more information becomes available about exocytotic mechanism in the tick salivary glands, it may be possible to test whether the same SNARE complex proteins regulate the release of proteins from different cell types in Acinus II and III during both slow and fast feeding phases.
Overall, our results indicate a functional significance of AmNSF and AmSNAP-25 in tick survival and engorgement, and potentially oviposit and embryogenesis. SNARE proteins are believed to mediate exocytotic events in all secretory pathways, such as the secretion of salivary proteins from the tick salivary glands. As pathogen transmission from the tick to the host most likely utilizes this route of saliva secretion, examining the effects of AmSNARE knockdowns will allow for a better understanding of the exact machinery and proteins involved in this process. Gaining insight into the role of AmSNARE molecules in the exocytosis of the secretory granules found in Acini II and Acini III will aid in elucidating a mechanism to block tick salivary protein secretion and prevention of pathogen transmission.
Experimental Procedures
Unless otherwise indicated, the protocols followed standard procedures (Sambrook and Russell 1999), and all of the experiments were performed at room temperature (25 ± 1°C). All water used was of 18-megaohm quality, produced by a MilliQ apparatus (Millipore Corp., Bedford, MA). Commercial antibodies to human NSF (mouse: monoclonal) and human SNAP-25 (goat: polyclonal) were obtained from Santa Cruz Biotechnology, Inc (Santa Cruz, California, USA). Alexa546-rabbit anti-goat, and Alexa546-donkey anti-mouse, Taq DNA polymerase, reverse transcriptase and polymerase chain reaction (PCR), plasmid DNA purification kits were purchased from Invitrogen (Carlsbad, CA, USA) and Qiagen (Valencia, CA, USA).
Tick Rearing
The Gulf coast ticks, Amblyomma maculatum, were purchased from Tick Rearing Facility at Oklahoma State University, and maintained according to the methods of Patrick and Hair (Patrick and Hair 1975). Tick examined in this experiment were unfed and Adult ticks were fed on sheep specifically for this study and all studies with animals were performed in accordance with an approved protocol. Prior to infestation on the host, all unfed ticks were kept at approximately 28°C, with 90% relative humidity, for a 14 hour light and 10 hour dark photoperiod. All use of animals for this research was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources Commission of Life Sciences, National Research Council (http://www.nal.usda.gov/awic/pubs/noawicpubs/careuse.htm). The protocols were approved by the University of Southern Mississippi Institutional Animal Care and Use Committee (IACUC). The approved protocols are on the file in the University of Southern Mississippi’s Office of Research Integrity (IACUC# 10042001).
Tick Tissue Dissection
Tick tissues were dissected in ice-cold 100 mM 3-(N-Morpholino)-propanesulfonic acid (MOPS) buffer containing 20 mM ethylene glycol bis-(b-aminoethyl ether)-N, N, N′, N′-tetraacetic acid (EGTA), pH 6.8. Once removed, glands were gently washed in the same ice-cold buffer. Once dissected, the tissues were either immediately used or stored at −70°C in RNALater (Invitrogen, Carlsbad, CA, USA).
RNA Isolation, cDNA synthesis, and RT-PCR
RNA was isolated from salivary glands and midguts of unfed and fed ticks by using illustra RNAspin Mini (GE Healthcare, Amersham Place, Little Chalfont, Buckinghamshire, United Kingdom). Briefly, the total RNA was eluted into nuclease free water and the concentration of total RNA was determined using the Nanodrop spectrophotometer and stored at −80°C (Karim et al. 2011, Karim et al. 2012). The total RNA was reverse transcribed using Moloney Murine Leukemia Virus (MMLV) reverse transcriptase according to manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). Gene specific primers were designed to amplify cDNA fragments from A. maculatum tissues. All primer sequences used in this study are listed in Table 1. PCR amplification was performed using a program of 94 °C for 1 min, 29 cycles of 94 °C for 1 min, 49 °C for 1 min, and 72 °C for 1 min, followed by 72 °C for 8 min. PCR products were analyzed on a 2% agarose gel stained with ethidium bromide, and visualized using GelDoc system (Bio-Rad). PCR products were cleaned using Qiagen PCR cleaning kit (Qaigen, CA, USA) and submitted to Eurofins MWG Operon for sequencing using the gene specific primers. The resulting sequences were used to search the DNA and protein databases (www.ncbi.nih.gov/blast).
Table 1.
Summary of the gene specific primers used in this study
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Size (bp) | |
|---|---|---|---|---|
| Am β-Actin | Quantitative | TGGCTCCTTCCACCATGAAGATCA | TAGAAGCACTTGCGGTGCACAATG | 177 |
| Am NSF | Quantitative | GATGTGCGGCCGTATGTGTTTGAT | TTGGAAACTGCTGCACAAAGTCCG | 145 |
| Am SNAP-25 | Quantitative | TCCAGTGTCTTCTCAAGCTCCGTT | TCAAGAGCTACTTTCGTGGTGGCA | 118 |
| Am Syntaxin1a | Quantitative | TGAGCTTGAAGACCTTATGGCCGA | CTGCGTTTGCTCCAACTGCTCAAT | 103 |
| Am Syntaxin 8 | Quantitative | TGCTAAACAAAGGGCGGGAAATGG | ACCACCTTCACTTACAGCCACACT | 122 |
| Am Syntaxin 16 | Quantitative | TTGCAAGAGGCTTCAGTCCAGAGA | TGTTGATCTCTCGTTCCCGCACTT | 182 |
| Am Syntaxin 17 | Quantitative | TGGGCGCTGGATTCTTAGGTTACA | AACTGAGTGACTCGTCTGGTGCTT | 135 |
| Am Syntaxin 18 | Quantitative | AAGGCACCTCATGAGACAGAAGGA | ATCACCACATTCTCATCCCAGCCT | 134 |
| Am VAMP 1/2 | Quantitative | AGGGTCAATACTGTGAACAGGCCA | ACTGATCCTCTTTGGAGCACGTCA | 182 |
| Am VAMP 7 | Quantitative | ACTTCTCGGAGGTCACCGAACAAA | ACTCACTTGGAACCTGCCCTGAAT | 197 |
| Am NSF | dsRNA | ATGGGCAGAGCTTTCAGTCAGTCA | TCAGGTGGAACACTCTCGATGCAA | 533 |
| Am SNAP-25 | dsRNA | TCAAGAGCTACTTTCGTGGTGGCA | TGAGGATCCGTTTGATCTGGCTGT | 465 |
qRT-PCR
Approximately 1500 ng of total RNA was reverse transcribed using Moloney Murine Leukemia Virus (MMLV) reverse transcriptase according to manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). The cDNA used for qRT-PCR gene expression analysis was approximately 25ng/μl (Karim et al. 2012). First strand cDNA was used to measure mRNA levels through qRT-PCR. For quantification with BIORAD CFX96 Real Time System, the Maxima SYBR Green qPCR Master Mix (Fermentas) was used according to manufacturers recommendations; approximately 50 ng of cDNA and gene specific primers (150 nM) were used for each reaction mixture. The C1000 Thermal Cycler was used to control the thermal treatment which was 10 minutes at 95 °C, followed by 35 cycles of 15 seconds at 95 °C, 30 seconds at 60 °C, and 30 seconds at 72 °C. All protocols for qRT-PCR experiments are in line with MIQE quidelines (Bustin et al. 2009) All primer sequences used in this study are listed in Table 1; AmActin was used as the reference gene, which was proven to be the most stable and accurate internal control from (Browning et al. 2013). All Fold Expression values were calculated by the BIORAD CFX Manager Software using the 2ΔΔCt Method.
Protein Extraction
Proteins were partially purified from salivary gland, midgut, and synganglia tissues in 1x PBS treated with protease inhibitor by crushing tissue with pestles and then subjected to sonication (50 duty cycle, 3–4 output, two 3 second pulses). Homogenates were then centrifuged at 5000 × g for 10 minutes at 4°C and the supernatant was collected. Protein concentrations were estimated by the Bradford method (Bradford 1976).
SDS-PAGE and Western Blotting
Protein extracts for salivary glands and midguts (20 ug) and for synganglia (5 ug), were separated on a 4–20% SDS-PAGE and transferred onto a nitrocellulose membrane in a Transblot cell (Bio-Rad, Hercules, California, USA) following the manufacturer’s instructions. The transfer buffer was comprised of 25 mM Tris HCl, 192 mM glycine in 20% methanol. Nonspecific protein binding sites were blocked with a 5% skim milk solution and the membranes were incubated with an NSF antibody (Santa Cruz Biotechnology, Santa Cruz, California, USA) at a dilution of 1:500. The antigen-antibody complexes were visualized with horseradish peroxidase-conjugated anti-mouse IgG (KPL, Gaithersburg, Maryland, USA) at a dilution of 1:20,000 and detected with SuperSignal chemiluminescent substrate (Thermo Scientific, Pierce, Rockford, Illinois, USA) using Bio-Rad ChemiDoc XRS.
Synthesis of dsRNA, Tick Injections, and Feeding
PCR products of N-ethylmaleimide sensitive fusion protein (NSF) and Synaptosomal associated protein 25 kDa (SNAP-25) were joined to the Block-iT T7 TOPO linker. The TOPO linking reaction was used to produce sense and anti-sense linear DNA templates using gene specific and T7 PCR primers in two separate PCR reactions. The sense and anti-sense DNA templates were used to create sense and anti-sense transcripts using BLOCK-iT RNA TOPO transcription kit (Invitrogen, Carlsbad, CA, USA). The dsRNA was analyzed to verify size through agarose gel electrophoresis. Subsequently, unfed females were injected with 1 μl of NSF dsRNA, SNAP-25 dsRNA, or irrelevant GFP dsRNA using a 31-gauge needle. Once injected with dsRNA or buffer, ticks were maintained at 37 °C overnight under high humidity to monitor tick survival. Surviving ticks were exposed to naïve-sheep and allowed to blood feed to repletion; un-injected males were co-infested with all experimental groups to aid females in feeding. Ticks were pulled 6 days and 11 days post infestation. Their feeding success was determined by total engorged weight, survival, and egg lying (Karim and Adamson 2012).
Confocal scanning microscopy
For immunolocalization, dissected salivary glands from unfed and partially fed females were stored in 4% paraformaldehyde at 4°C. Dissected glands were rinsed in PBS for 30 minutes then soaked in 4% formaldehyde in PBS for 30 minutes at room temperature then washed in PBS. Glands were permeabilized in 0.5% tritonX-100 for 30 minutes at room temperature and washed in PBS. Next, glands were incubated in 3% BSA for 1 hour at room temperature and excess BSA was washed away with PBS. Glands were soaked in the primary antibody (1:100) in 3% BSA and incubated at 4°C for overnight (Santa Cruz Biotechnology, Santa Cruz, California, USA). Glands were washed in PBS. Salivary glands were incubated in Alexa Flour-546 anti-mouse secondary antibody (1:100) in 3% BSA for 1 hour at room temperature in the dark. Salivary glands were incubated in Alexa Flour – 488 phalloidin (1:100) in 3% BSA for 1 hour at room temperature in the dark (Invitrogen, Eugene, Oregon, USA). Excess antibody and stain was rinsed away with three PBS washes. Glands were incubated in DAPI for 5 minutes and thoroughly washed with PBS at room temperature. Glands were mounted on glass slides and viewed under a Leica TCS 4D confocal microscope.
TEM
Tissues were transferred to 2%formaldehyde/0.2% picric acid in 0.1M sodium phosphate buffer, pH 7.2 solution and stored at 4°C. Tissues were washed in sodium cacodylate buffer, pH7.0 and postfixed in 1% osmium tetraoxide in 0.1M sodium cacodylate buffer, pH 7.0 for 45 minutes at room temperature. Tissues were washed in distilled water and then dehydrated in ethanol (50%–100%) washes and acetone (100%) washes. The tissues were then embedded in Spurr’s Epoxy. 100 μm thick sections were cut from the block using the MT-9 ultramicrotome and stained with Uracil Acetate and Lead Citrate for light microscopy. Sections were observed using a Zeiss photomicroscope and photographed on Panatomic X film (Kodak).
Statistical Analysis
All data are expressed as ± SEM. Statistical significance was determined by Student’s t test; differences in multiple comparisons among different experimental groups were determined by analysis of variance using the Tukey test. Statistical analysis of qRT-PCR data was done using the REST 2009 software (Pfaffl et al. 2002).
Acknowledgments
We sincerely thank Drs. Ken Curry and Steve Adamson for useful discussions and gratefully acknowledge Baobin Kang and Brandon Drescher for technical assistance in microscopy. This work was supported by grants from the American Heart Association award (09SDG2280207), USDA National Institute of Food and Agriculture award (2007-35607-20200), and US Department of State award (PGA-P21049) to SK. The core-facility is supported by the Mississippi INBRE funded by grants from the National Center for Research Resources (5P20RR016476-11) and the National Institute of General Medical Sciences (8 P20 GM103476-11) from the National Institutes of Health.
Abbreviations
- SNARE
Soluble NSF Attachment Protein Receptor
- NSF
N-ethylmaleimide sensitive fusion protein
- SNAP-25
Synaptosomal Associated Protein of 25 kDa
- AmNSF
A. maculatum NSF
- AmSNAP-25
A. maculatum SNAP-25
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
References
- Bethani I, Werner A, Kadian C, Geumann U, Jahn R, Rizzoli SO. Endosomal fusion upon SNARE knockdown is maintained by residual SNARE activity and enhanced docking. Traffic. 2009;10:1543–59. doi: 10.1111/j.1600-0854.2009.00959.x. [DOI] [PubMed] [Google Scholar]
- Binnington KC. Sequential changes in salivary gland structure during attachment and feeding of the cattle tick, Boophilus microplus. Int J Parasitol. 1978;8:97–115. doi: 10.1016/0020-7519(78)90004-8. [DOI] [PubMed] [Google Scholar]
- Boulianne GL, Trimble WS. Identification of a second homolog of N-ethylmaleimide-sensitive fusion protein that is expressed in the nervous system and secretory tissues of Drosophila. Proc Natl Acad Sci U S A. 1995;92:7095–9. doi: 10.1073/pnas.92.15.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman AS, Sauer JR. Tick salivary glands: function, physiology and future. Parasitology. 2004;129(Suppl):S67–81. doi: 10.1017/s0031182004006468. [DOI] [PubMed] [Google Scholar]
- Bradford MM. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Browning R, Adamson SW, Karim S. Choice of a Stable Set of Reference Genes for qRT-PCR Analysis in Amblyomma maculatum (Acari: Ixodidae) Journal of Medical Entomology. 2013;50 doi: 10.1603/me12123. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Cahill AL, Herring BE, Fox AP. Stable silencing of SNAP-25 in PC12 cells by RNA interference. BMC Neurosci. 2006;7:9. doi: 10.1186/1471-2202-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q-and R-SNAREs. Proc Natl Acad Sci U S A. 1998;95:15781–6. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francischetti IM, Sa-Nunes A, Mans BJ, Santos IM, Ribeiro JM. The role of saliva in tick feeding. Front Biosci. 2009;14:2051–88. doi: 10.2741/3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golby JA, Tolar LA, Pallanck L. Partitioning of N-ethylmaleimide-sensitive fusion (NSF) protein functionin Drosophila melanogaster: dNSF1 is required in the nervous system, and dNSF2 is required in mesoderm. Genetics. 2001;158:265–78. doi: 10.1093/genetics/158.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H, Umemiya R, Zhou J, Liao M, Zhang H, Jia H, Nishikawa Y, Xuan X, Fujisaki K. Blocking the secretion of saliva by silencing the HlYkt6 gene in the tick Haemaphysalis longicornis. Insect Biochem Mol Biol. 2009;39:372–81. doi: 10.1016/j.ibmb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Hay JC, Scheller RH. SNAREs and NSF in targeted membrane fusion. Curr Opin Cell Biol. 1997;9:505–12. doi: 10.1016/s0955-0674(97)80026-9. [DOI] [PubMed] [Google Scholar]
- Heydorn A, Sondergaard BP, Hadrup N, Holst B, Haft CR, Schwartz TW. Distinct in vitro interaction pattern of dopamine receptor subtypes with adaptor proteins involved in post-endocytotic receptor targeting. FEBS Lett. 2004;556:276–80. doi: 10.1016/s0014-5793(03)01431-5. [DOI] [PubMed] [Google Scholar]
- Hong W. SNAREs and traffic. Biochim Biophys Acta. 2005;1744:493–517. [PubMed] [Google Scholar]
- Karim S, Adamson SW. RNA Interference in ticks: a function genomics tool. In: Jockusch EL, editor. Small RNAs: Their Diversity, Roles, and Practical Uses. Elsevier, Storrs; 2012. [Google Scholar]
- Karim S, Singh P, Ribeiro JM. A Deep Insight into the Sialotranscriptome of the Gulf Coast Tick, Amblyomma maculatum. PLoS One. 2011;6:e28525. doi: 10.1371/journal.pone.0028525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S, Browning R, Ali A, Truhett R. Laboratory infected Ehrlichia chaffeensis female adult Amblyomma americanum salivary glands reveal differential gene expression. Journal of Medical Entomology. 2012 doi: 10.1603/me11214. [DOI] [PubMed] [Google Scholar]
- Karim S, V, Ramakrishnan G, Tucker JS, Essenberg RC, Sauer JR. Amblyomma americanum salivary gland homolog of nSec1 is essential for saliva protein secretion. Biochem Biophys Res Commun. 2004;324:1256–63. doi: 10.1016/j.bbrc.2004.09.189. [DOI] [PubMed] [Google Scholar]
- Karim S, Miller NJ, Valenzuela J, Sauer JR, Mather TN. RNAi-mediated gene silencing to assess the role of synaptobrevin and cystatin in tick blood feeding. Biochem Biophys Res Commun. 2005;334:1336–42. doi: 10.1016/j.bbrc.2005.07.036. [DOI] [PubMed] [Google Scholar]
- Karim S, Essenberg RC, Dillwith JW, Tucker JS, Bowman AS, Sauer JR. Identification of SNARE and cell trafficking regulatory proteins in the salivary glands of the lone star tick, Amblyomma americanum (L.) Insect Biochem Mol Biol. 2002;32:1711–21. doi: 10.1016/s0965-1748(02)00111-x. [DOI] [PubMed] [Google Scholar]
- Kaufman W. The influence of various factors on fluid secretion by in vitro salivary glands of ixodid Ticks. J Exp Biol. 1976;64:727–42. doi: 10.1242/jeb.64.3.727. [DOI] [PubMed] [Google Scholar]
- Lindsay PJ, Kaufman WR. Potentiation of salivary fluid secretion in ixodid ticks: a new receptor system for gamma-aminobutyric acid. Can J Physiol Pharmacol. 1986;64:1119–26. doi: 10.1139/y86-191. [DOI] [PubMed] [Google Scholar]
- Maritz-Olivier C, Louw AI, Neitz AW. Similar mechanisms regulate protein exocytosis from the salivary glands of ixodid and argasid ticks. J Insect Physiol. 2005;51:1390–6. doi: 10.1016/j.jinsphys.2005.08.012. [DOI] [PubMed] [Google Scholar]
- McSwain JL, Essenberg RC, Sauer JR. Oral secretion elicited by effectors of signal transduction pathways in the salivary glands of Amblyomma americanum (Acari: Ixodidae) J Med Entomol. 1992;29:41–8. doi: 10.1093/jmedent/29.1.41. [DOI] [PubMed] [Google Scholar]
- Niemeyer BA, Schwarz TL. SNAP-24, a Drosophila SNAP-25 homologue on granule membranes, is a putative mediator of secretion and granule-granule fusion in salivary glands. J Cell Sci. 2000;113(Pt 22):4055–64. doi: 10.1242/jcs.113.22.4055. [DOI] [PubMed] [Google Scholar]
- Paddock CD, Sumner JW, Comer JA, Zaki SR, Goldsmith CS, Goddard J, McLellan SL, Tamminga CL, Ohl CA. Rickettsia parkeri: a newly recognized cause of spotted fever rickettsiosis in the United States. Clin Infect Dis. 2004;38:805–11. doi: 10.1086/381894. [DOI] [PubMed] [Google Scholar]
- Parker RR, Kohls GM, Cox GW, Davis GE. Observations of an infectious agent from Amblyomma maculatum. Public Health Reports. 1939;54:1482–1484. [Google Scholar]
- Patrick CD, Hair JA. Laboratory rearing procedures and equipment for multi-host ticks (Acarina: Ixodidae) J Med Entomol. 1975;12:389–90. doi: 10.1093/jmedent/12.3.389. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Stewart BA, Rivlin PK, Vilinsky I, Watson BO, Lang C, Boulianne G, Salpeter MM, Deitcher DL. Two distinct effects on neurotransmission in a temperature-sensitive SNAP-25 mutant. EMBO J. 2001;20:6761–71. doi: 10.1093/emboj/20.23.6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran V, Chawla A, Roche PA. Identification of a novel syntaxin-and synaptobrevin/VAMP-binding protein, SNAP-23, expressed in non-neuronal tissues. J Biol Chem. 1996;271:13300–3. doi: 10.1074/jbc.271.23.13300. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, I, Francischetti M. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Warren G. Implications of the SNARE hypothesis for intracellular membrane topology and dynamics. Curr Biol. 1994;4:220–33. doi: 10.1016/s0960-9822(00)00051-8. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular Cloning A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1999. [Google Scholar]
- Sauer JR, Essenberg RC, Bowman AS. Salivary glands in ixodid ticks: control and mechanism of secretion. J Insect Physiol. 2000;46:1069–1078. doi: 10.1016/s0022-1910(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Sauer JR, McSwain JL, Bowman AS, Essenberg RC. Tick salivary gland physiology. Annu Rev Entomol. 1995;40:245–67. doi: 10.1146/annurev.en.40.010195.001333. [DOI] [PubMed] [Google Scholar]
- Schmidt SP, Essenberg RC, Sauer JR. Evidence for a D1 dopamine receptor in the salivary glands of Amblyomma americanum (L.) J Cyclic Nucleotide Res. 1981;7:375–84. [PubMed] [Google Scholar]
- Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993a;75:409–18. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993b;362:318–24. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Soneshine D. Biology ofTicks. Oxford University Press; New York: 1991. [Google Scholar]
- Uilenberg G. Experimental transmission of Cowdria ruminantium by the Gulf coast tick Amblyomma maculatum: danger of introducing heartwater and benign African theileriasis onto the American mainland. Am J Vet Res. 1982;43:1279–82. [PubMed] [Google Scholar]
- Walker JB, Olwage A. The tick vectors of Cowdria ruminantium (Ixodoidea, Ixodidae, genus Amblyomma) and their distribution. Onderstepoort J Vet Res. 1987;54:353–79. [PubMed] [Google Scholar]
- Wang G, Witkin JW, Hao G, Bankaitis VA, Scherer PE, Baldini G. Syndet is a novel SNAP-25 related protein expressed in many tissues. J Cell Sci. 1997;110(Pt 4):505–13. doi: 10.1242/jcs.110.4.505. [DOI] [PubMed] [Google Scholar]
- Zhao C, Slevin JT, Whiteheart SW. Cellular functions of NSF: not just SNAPs and SNAREs. FEBS Lett. 2007;581:2140–9. doi: 10.1016/j.febslet.2007.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]