Abstract
Actin is a key player for nuclear structure and function regulating both chromosome organization and gene activity. In the cell nucleus actin interacts with many different proteins. Among these proteins several studies have identified classical nuclear factors involved in chromatin structure and function, transcription and RNA processing as well as proteins that are normally involved in controlling the actin cytoskeleton. These discoveries have raised the possibility that nuclear actin performs its multi task activities through tight interactions with different sets of proteins. This high degree of promiscuity in the spectrum of protein-to-protein interactions correlates well with the conformational plasticity of actin and the ability to undergo regulated changes in its polymerization states. Several of the factors involved in controlling head-to-tail actin polymerization have been shown to be in the nucleus where they seem to regulate gene activity. By focusing on the multiple tasks performed by actin and actin-binding proteins, possible models of how actin dynamics controls the different phases of the RNA polymerase II transcription cycle are being identified.
Keywords: Nuclear actin, nuclear structure and function, RNA polymerase, transcription, actin polymerization
Introduction
In recent years, several studies have underscored the importance of actin in the eukaryotic cell nucleus, permanently sweeping away decades of skepticism in the field. Actin participates in chromatin remodeling and histone modifications.1,2 Actin is involved in different phases of gene transcription by all three eukaryotic RNA polymerases. Actin is co-transcriptionally assembled into nascent RNPs, actin is potentially involved in RNA processing and actin is a component of ribosomes.3-9 In addition, nuclear actin interacts with certain transcription factors and determines their subcellular localization.10-12 Finally, recent work points to a primary role for actin even in nuclear reprogramming.13 Taken altogether, these observations indicate that actin functions as an essential player in gene regulation at multiple layers.
A most puzzling aspect is why and how actin participates in so many nuclear functions. Nuclear actin is part of different multiprotein complexes. Therefore, a fascinating possibility is that through specific interactions with different nuclear factors, actin connects molecular machines with nuclear compartments and this is precisely how actin would end up being critical for many steps within the gene expression pathway. These considerations are also valid at gene level where in principle actin is a primary candidate to mediate the crosstalk between different transcriptional phases and RNA processing events.
Another important aspect for which our present knowledge still lags behind, concerns the polymerization state of nuclear actin. In the cytoplasm regulated actin polymerization drives motile processes. Whether nuclear actin polymerizes and above all how, is not fully understood. This is also a rather controversial point, partly due to the difficulty to visualize nuclear actin filaments by microscopy methods. Conformational differences and specific posttranslational modifications in the nuclear vs. the cytoplasmic actin population may be responsible for differences in the polymerization propensities.14-16 However, the fraction of nuclear actin is biochemically undistinguishable from the cytoplasmic counterpart. The same non-muscle actin isoforms, β-actin and γ-actin, are present both in the nucleus and in the cytoplasm. So, even though we currently lack detailed mechanistic insights, nuclear actin probably maintains the same ability as the cytoplasmic one to undergo changes in its polymerization state. Early reports already supported this view.17,18 More recently, studies based on the use of fluorescence recovery after photobleaching (FRAP) techniques have tracked down nuclear actin subpopulations with different mobilities,19 suggesting the presence of both monomeric (G) actin and polymeric (F) actin in the nucleus. Furthermore, in the past years many proteins which are known to regulate actin polymerization in the cytoplasm have also been discovered in the cell nucleus.3,5,20 For some of these proteins, we even know that there is an involvement in basal nuclear functions. Therefore, altogether, the assumptions that (1) regulated changes in nuclear actin polymerization states are likely to occur and (2) they are important for specific nuclear events are entirely plausible.
This review focuses on the fundamental role of actin in transcription, and the idea that regulated changes in the state of actin polymerization help to mediate transitions between different stages of transcription.
Evidence for Actin in Transcription Initiation
Two milestone studies provided the first circumstantial evidence on the potential role of actin in transcription.21,22 Biochemically, actin was co-purified with the RNA polymerase II machinery and shown to be required for the initial phases of mRNA synthesis.21 In parallel, injections of anti-actin antibodies or actin binding proteins such as gelsolin into nuclei of living oocytes from the amphibian Pleurodeles waltlii led to a transcriptional block and collapse of the fine structures of lampbrush chromosomes. These results were taken to suggest a primary role for acin on the active gene.22 However, both studies generated skepticism and as consequence, mechanistic studies on nuclear actin in gene transcription lagged behind for years.
Many studies have confirmed and built on the above discoveries. Actin co-immunoprecipitated with the largest RNA polymerase I subunit from nucleolar protein extracts, occupying the entire rDNA transcription unit.23-26 In abortive transcription initiation assays where the formation of the ACU trimer by the mouse RNA polymerase I enzyme is monitored, antibodies to actin inhibited RNA synthesis.24 Since actin was found to occupy rRNA gene promoter, an interpretation of these results is that actin has a key role in the pre-initiation and initiation phases of RNA polymerase I transcription.26 Actin was also localized at RNA polymerase II transcription sites in an RNA-dependent manner.27 Biochemical evidence demonstrated association of actin with pre-initiation complex (PIC) at inducible and constitutively expressed promoters of protein coding genes.28,29 Actin association with the PIC complex is functionally important as actin depletion prevented PIC assembly and inhibited in vitro transcription initiation. Importantly, exogenously added actin stimulated RNA polymerase II transcription in vitro,28 which supports earlier work on the fundamental role of actin as transcription factor.21,22 Concomitantly, actin was also found as genuine component of a highly purified fraction of the active RNA polymerase III enzyme.30 Similarly to RNA polymerase II, the association of actin with RNA polymerase III is functional since depletion of actin from the RNA polymerase III preparation inhibited transcription from a U6 snRNA gene promoter.30 In the same study, specific interactions were identified between actin and the RNA polymerase III subunits RPABC2 and RPABC3.30 Since these subunits are shared by all three RNA polymerases,31 these observations support a common association mode for actin with the basal transcription machinery independent of the type of RNA polymerase. The above studies, at least in vitro, support a specific role for the β-actin isoform in transcription, as opposed to other actin isoforms present in non-muscle cells. Remarkably, the importance of β-actin has been confirmed in vivo.32 In a recent study, transcriptional profiling on mouse embryonic fibroblasts obtained from an embryonic-lethal β-actin knockout mouse revealed that the lack of β-actin has an impact on genetic reprogramming but does not affect cell motility.32
Further mechanistic insights are needed. However these findings support an essential role for β-actin in gene activity during the initial phases of transcription by all three eukaryotic RNA polymerases.
Actin in Transcription Elongation
Actin also appears to function along the entire transcription unit and therefore, in transcription elongation. This was first shown in the dipteran insect Chironomus tentans.27 At the fourth instar larval stage, C. tentans has got large salivary glands with a characteristic monolayer of saddle-shaped cells; each has four polytene chromosomes that can be isolated with intact morphology.33 In situ histochemistry with an actin antibody revealed actin at all transcription sites, including the exceptionally large Balbiani ring (BR) transcription puffs on chromosome four.27,34 Actin was not detected at transcription sites after RNase treatment,27 suggesting actin occupies transcription sites in an RNA-dependent manner. This gave rise to the idea that actin is part of the nascent RNP complex. Immunoelectronmicroscopy on cryosections of intact salivary glands confirmed actin association with the RNP still coupled to chromatin axis and showed that actin remains incorporated in mature RNPs all the way to the cytosol.27 A similar scenario was discovered in mammals. The nuclear distribution of actin is partly sensitive to in vivo RNase treatment, actin occupies RNA polymerase II gene promoters and coding sequences as revealed by chromatin immunoprecipitation methods, and actin is a component of pre-mRNP/mRNPs.29,35 The straightforward interpretation of these findings is that actin is important along the active gene, presumably to facilitate the elongation of nascent pre-mRNA molecules. Why actin remains incorporated in mature RNPs is presently unknown.
Several reports support a role for actin in commitment and maintenance of transcription elongation. The positive elongation factor P-TEFb, required by the RNA polymerase II for escape from pausing, is recruited via polymerase-associated actin, a mechanism that leads to hyperphosphorylation of the CTD and commitment to elongation (Fig. 1).36 To find out how actin actually functions in transcription elongation, nuclear RNP preparations from both C. tentans and mammalian cells were screened by DNase I affinity chromatography to pull-down actin and potential actin-binding proteins.35,37,38 These experiments led to the identification of a subset of heterogeneous nuclear ribonucleoproteins (hnRNPs) which is in complex with actin. Within these hnRNP proteins, specific interactions of actin with the C. tentans hrp65–2, homolog to the vertebrate transcriptional co-activators PSF-NonO (polypyrimidine-tract-binding-protein-associated splicing factor-non-Pou-domain octamer-binding protein/p54nrb), and with the mammalian hnRNP U/SAF-A (Scaffold Attachment Factor-A) were found to be required for transcription elongation in living cells.37,38 The actin-hrp65 and actin-hnRNP U interactions are highly conserved occurring through a novel, yet uncharacterized actin binding motif located in the C-termini. These actin-hnRNP interactions promote recruitment of histone acetyl transferases (HATs).29,39 In mammals, the actin-hnRNP U interaction facilitates recruitment of the HAT PCAF which leads to H3K9 acetylation along the gene. What places this mechanism in the context of transcription elongation is the fact that the actin-hnRNP U interaction does not seem to take place at gene promoter. It rather occurs immediately after promoter clearance when the heptapeptide repeats of the C-terminal domain (CTD) of the RNA polymerase II are hyperphosphorylated on Ser2 and Ser5 (Fig. 1).29 Therefore, it seems that the primary outcome of this actin-based mechanism is to provide an open chromatin configuration which allows passage of the elongating polymerase to scan the gene and elongate the nascent transcript (Fig. 2). We define this open chromatin configuration required for transcription elongation as permissive chromatin.
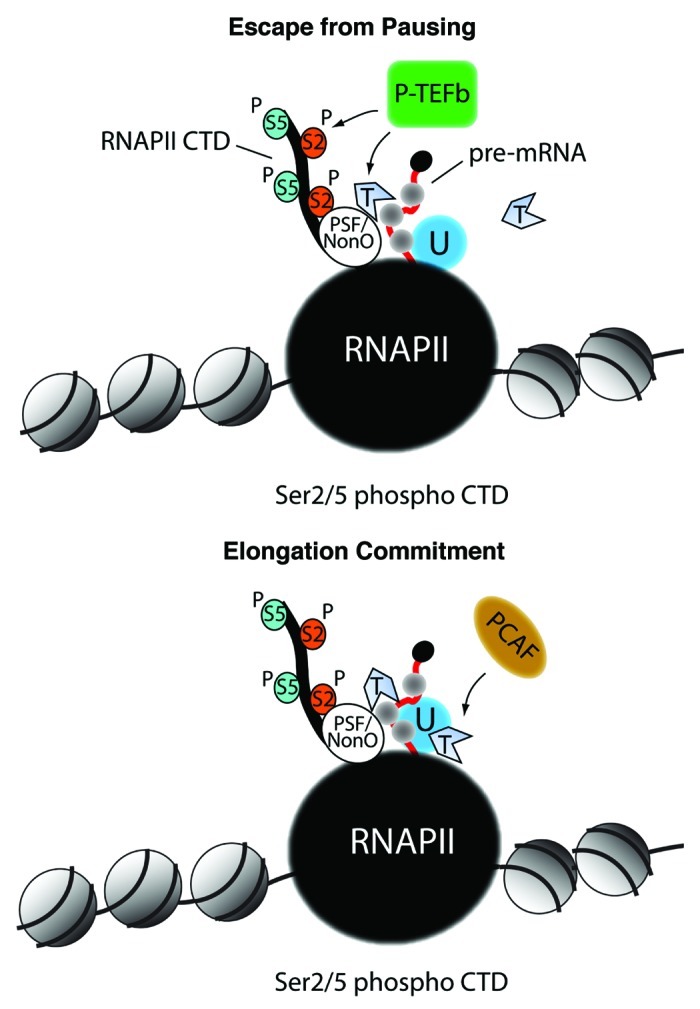
Figure 1. The possible contribution of actin in RNA polymerase II transcription activation. Top panel, monomeric actin interacts with the PSF/NonO complex to recruit the positive elongation factor P-TEFb with the subunit cdk9. This in turn leads to Ser2 phosphorylation within the heptapeptide repeats of the RNA polymerase II CTD. This mechanism promotes RNA polymerase II CTD escape from pausing. Bottom panel, the CTD associated-actin interacts with hnRNP U and this mechanism commits the hyperphosphorylated RNA polymerase II to transcription elongation through recruitment of the HAT PCAF. RNAPII, RNA polymerase II; U, hnRNP U; T, ATP-actin; P-S2, phosphorylated Ser2; P-S5, phosphorylated Ser5.
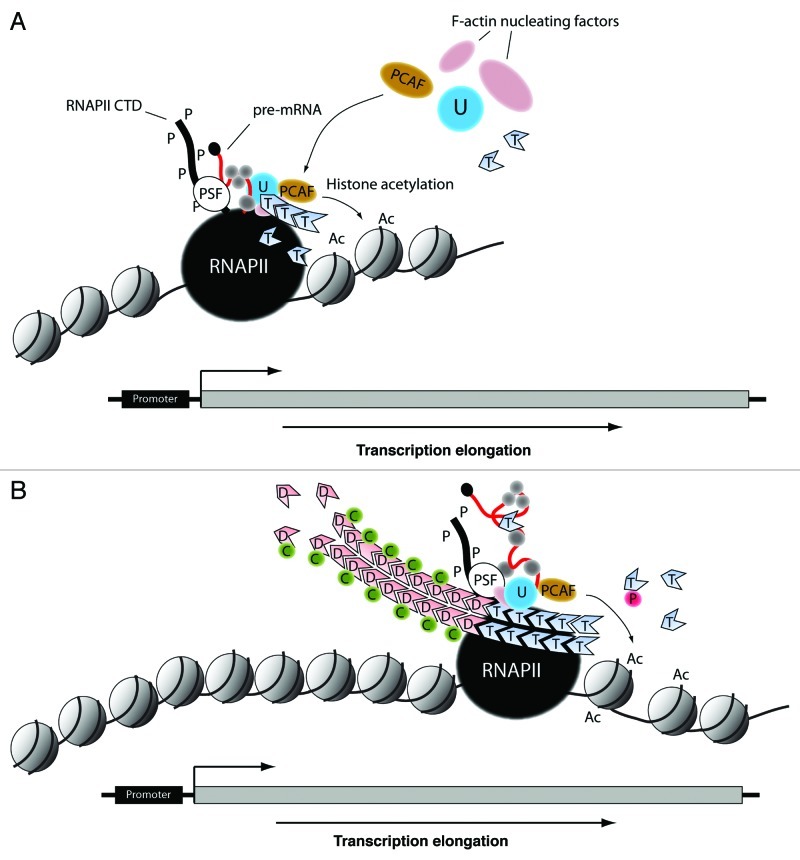
Figure 2. Hypothetical models depicting actin-based mechanisms during transcription elongation, with respect to a eukaryotic gene. (A) After commitment to elongation, as part of the PSF/NonO complex, actin interacts with hnRNP U to recruit the HAT PCAF. Concomitantly, actin polymers are nucleated through F-actin nucleating factors such as N-WASP and ARP2/3 that are also part of the same multiprotein complex together with PSF/NonO. This mechanism leads to H3K9 acetylation at the exit of the gene promoter, it generates an open chromatin configuration and favors passage of the elongating polymerase through the nucleosome barrier to start transcribing the gene. (B) During transcription elongation the polymerase-associated actin undergoes head-to-tail polymerization. This mechanism is controlled by the F-actin severing activity of cofilin that accompanies the elongating polymerase to maintain a pool of polymerization competent actin. Co-transcriptional actin polymerization occurs throughout the entire length of the transcribed gene and contributes to provide directionality to the elongating polymerase through maintenance of permissive chromatin. RNAPII, RNA polymerase II; U, hnRNP U; T, ATP-actin; P-S2, phosphorylated Ser2; P-S5, phosphorylated Ser5; Ac, acetylated histone; D, ADP-actin; T, ATP-actin; C, cofilin; P, profilin.
Based on the above considerations, one would expect that actin is preferentially coupled to transcriptionally active genes. Several observations support this hypothesis. First, as already mentioned, actin association with chromatin is RNA-dependent both in C. tentans and in mammals.27,29 Furthermore, actin does not occupy untranslated regions (UTRs) and flanking regions of RNA polymerase II genes,40 its nuclear distribution depends on the transcriptional activity of the cell41 and own unpublished evidence indicates that actin occupancy is considerably reduced in intergenic regions separating consecutive rDNA transcription units. Whether actin association with the gene is transcription dependent or not still remains controversial due to a study where chromatin immunoprecipitations showed actin binding to chromatin isolated from actinomycin D-treated cells and thus, transcriptionally silent.42 Chromatin immunoprecipitations rely on different chromatin preparations and actin antibodies.27,29,40-42 In any case, the idea that actin preferentially occupies transcriptionally active chromatin is corroborated by an independent recent work where in Drosophila, chromatin was systematically mapped to five principal types and actin exclusively associates with transcriptionally competent euchromatin.43 We conclude that during the RNA polymerase II transcription cycle actin is directly involved in PIC formation, escape from pausing and transcription elongation (Fig. 3).
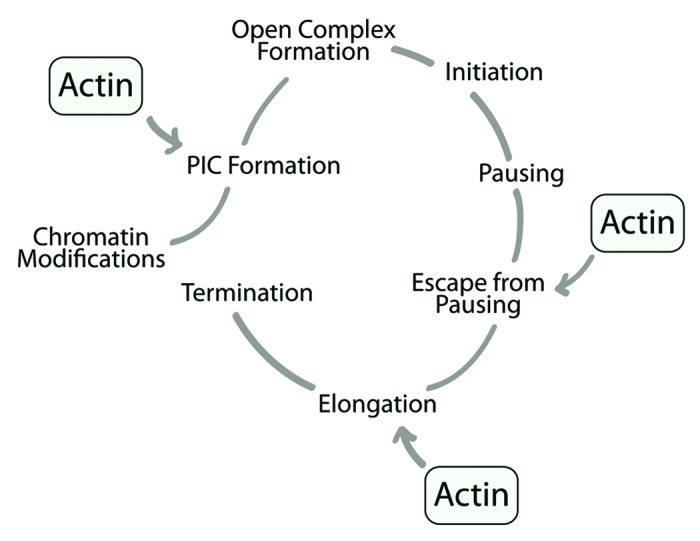
Figure 3. A schematic diagram summarizing the RNA polymerase II transcription cycle. Actin is known to be directly involved in PIC formation, escape from pausing and transcription elongation.
The hypothesis that actin-based mechanisms are required to generate permissive chromatin for transcription elongation unleashes many fundamental questions. One of the most intriguing aspects concerns the possibility that actin cooperates with myosin. Nuclear myosin 1c (NM1), its canonical form and a recently identified third isoform,44-47 myosin Va and Vb,48,49 myosin VI,50 myosin 16b51 and myosin 18b52,53 are all motor proteins which have been convincingly localized to the cell nucleus. So far NM1c is the best characterized from a mechanistic point of view. NM1c cooperates with actin in both initiation and elongation phases during RNA polymerase I transcription.4,54 In vitro NM1c is involved in transcription44 but we do not know whether actomyosin-based mechanisms are conserved in RNA polymerase II transcription elongation.
Another fundamental question concerns the polymerization state of actin.5,55,56 In the following sections focus will be on emerging evidence that actin polymerization is likely to occur along active gene and that it is a prime requirement for transcription elongation.
Actin Polymerization at the Gene Level
Evidence for actin polymerization in the nucleus has been discussed in an insightful review by Gieni and Hendzel (2009).57 Here it is worth adding that nuclear actin filaments have been observed in both non-physiologic58-60 and physiologic conditions.61-63 Furthermore, even though the nuclear actin rich filaments are not exactly the same as the canonical cytoplasmic filaments,63 their stability depends on actin polymerization. They connect different nuclear compartments63 and thus, they are also likely to have an impact on the structure of the cell nucleus. Finally, actin polymerization also seems to be implicated in nuclear function as it is needed for transcription-dependent long range chromosome loci movement toward Cajal bodies,64 a potential mechanism for the formation of neighborhoods where active genes are readily transcribed.65,66
A most challenging question that has not been discussed until now is whether dynamic actin polymerization accompanies the RNA polymerase along active gene to facilitate transcription elongation. When going back to the C. tentans model system, immunoelectron microscopy on cryosections of isolated polytene chromosomes or salivary gland cells did not reveal any sign of actin labeled filamentous structures connected to transcription sites or ribonucleoprotein particles.27 However these studies did not preclude the possibility that there is polymerized actin coupled to the transcription unit. In fact an interesting scenario emerged where during transcription there might be a rapid and dynamic equilibrium between monomeric G-actin and oligomeric/polymeric forms that in principle could locally coexist.67
Several arguments provide indirect evidence for the hypothesis outlined above. First, the use of drugs targeting different polymerization states of actin has proven effective in RNA polymerase I transcription assays,42 where the F-actin stabilizing drug jasplakinolide led to increased transcription rates. Furthermore, in the same study the use of actin mutants that stabilize F-actin stimulated rRNA synthesis. Some of these experiements were performed in vitro and therefore they represent snapshots of the transcription reaction. However, they pointed to the possibility that actin polymerization is necessary for the transcriptional process. Similarly in the case of RNA polymerase II transcription, an interesting study demonstrated the requirement for actin polymerization in retinoic acid (RA)-induced HoxB gene transcription. Induction of the HoxB genes was specifically prevented by inhibition of actin polymerization with cytochalasin D, a drug that caps F-actin. This specific inhibition also prevented RA-induced recruitment of elongating RNA polymerase II, β-actin and the accessory factors p54Nrb and PSF.68
The bulk of evidence that during transcription regulated nuclear actin polymerization is likely to take place probably comes from the discovery that the nucleus hosts several actin-binding proteins that are known to control actin polymerization.3 In the cytoplasm these proteins target both filamentous F-actin and monomeric G-actin and they are involved in multiple aspects of actin polymerization, including de novo formation of actin filaments, stabilization or branching of growing filaments as well as crosslinking, capping and severing actin filaments.69,70 For most of these proteins it is not known how they affect the state of actin polymerization in the context of specific nuclear functions. However, there are some exceptions. The F-actin nucleating factor N-WASP (Neuronal Wiskott-Aldrich syndrome protein), a member of the WASP family of proteins is in a large complex with PSF-NonO, nuclear actin and RNA polymerase II. This association seems to be functional since together with the ARP2/3 complex, N-WASP is implicated in RNA polymerase II transcription71,72 and N-WASP is recruited to RA-induced HoxB genes in a manner that is dependent on actin polymerization.68 Likewise the small actin binding proteins actin depolymerizing factor ADF/cofilin and profilin have been recently shown to be important for transcription of both rRNA and protein coding genes.40,42,73 These observations indicate that the RNA polymerase machinery is equipped with some of the proteins that control actin polymerization.
Co-transcriptional Actin Polymerization: Lessons from the Cytoplasm
Many cellular movements such as cell migration, adhesion, cytokinesis, endocytosis and axonal growth are based on polarized turnover of actin filaments. The growing actin polymer is dynamically maintained by subunit addition to the plus (“barbed”) end, and removal from the minus (“pointed”) end, a process altogether termed “treadmilling”.74 Disassembly is primarily mediated by ADF/cofilin which interacts with the ADP-actin rich pointed ends of the actin polymer, mediates severing and generates a local pool of ADP-actin monomers. Following ADP to ATP nucleotide exchange, profilin mediates addition of ATP-actin monomers to the barbed end of the polymer.75
Here, the most intriguing question is whether such an exquisite regulatory mechanism is conserved in the cell nucleus and whether it could also be occurring along active genes. Profilin and cofilin are known to be relatively abundant in the nucleus.76,77 Profilin and cofilin shuttle between cytoplasm and nucleus. Cofilin serves as adaptor for active Ran-dependent nuclear import of the cofilactin complex via importin 9, whereas profilin is an adaptor for the nuclear export of actin through exportin 6.78,79 Above all, nuclear import of actin supports transcription (Fig. 4).79 Therefore profilin and cofilin are also important for gene activity. In C. tentans polytene chromosomes, profilin localizes to active transcription units and appears to be required for ongoing transcription, since a transcriptional block with actinomycin D or DRB leads to a global release of profilin from the transcription sites.73 These findings confirm and extend the importance of profilin in mammalian gene expression.77,80 In the nucleus, profilin was found to partly co-localize with snRNP-core proteins, Cajal bodies and gems,77,80 which altogether suggest a complex role(s) for profilin at different layers, in both transcription and pre-mRNA processing.
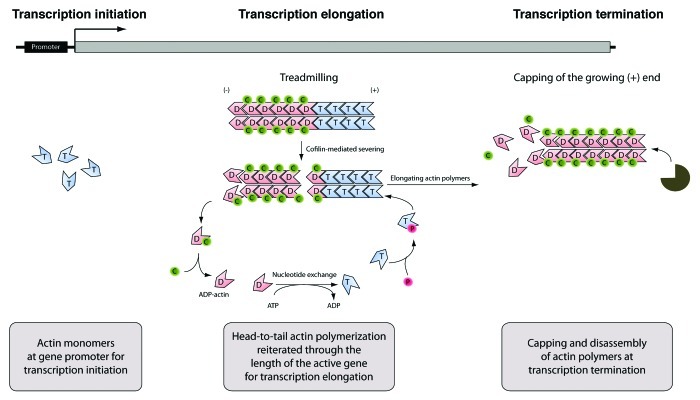
Figure 4. The actin nucleocytoplasmic transport cycle. Actin is actively imported into the nucleus in complex with cofilin. Ran-dependent nuclear import requires the import factor importin 9 which interacts with actin-associated cofilin. In the nucleus actin is excluded from heterochromatin and it is mostly found to be coupled to euchromatin and consistently, actin import supports transcription. From the cell nucleus actin is exported in complex with profilin. Active nuclear export of the profilin-actin complex is regulated by exportin 6 which specifically targets profilin. By controlling the nucleocytoplasmic shuttling of actin, cofilin and profilin tightly regulate transcription and gene activity.
Cofilin was recently found to occupy protein coding genes in a rather selective manner. Cofilin localizes to exonic sequences - both promoter proximal, medial and distal - being excluded from gene promoter, UTRs and flanking regions. In living cells cofilin gene knockdown induced drastic drops in transcriptional levels. Under the same conditions exonic sequences were found to be devoid of both actin and elongating RNA polymerase II, exhibiting decreased levels of H3K9 acetylation.40 Based on these results and the finding that cofilin import correlates with transcription rates,79,81 it is conceivable that cofilin has a primary role in transcription activation. In the nucleus, cofilin seems to be functionally coupled to actin. We know that cofilin gene knockdown induces accumulation of nuclear F-actin foci. In addition, cofilin binds to the polymerase-associated actin.40 Given that in vitro a role for cofilin in RNA polymerase I transcription was also observed,42 an interpretation of these findings is that during transcription elongation cofilin de facto functions by targeting actin.
RNA Polymerase II Transcription and Changes in Actin Polymerization: A Working Model
The RNA polymerase II transcription cycle is a complex multistep process82 and some of these steps require actin (see Fig. 3). A challenging task which is the focus of the present section is to place the potential mechanisms of regulated actin polymerization in the context of the RNA polymerase II transcription cycle.
Gene activators bind to the chromatin and facilitate recruitment of chromatin remodelers and histone modifying enzymes. This mechanism clears the gene promoter and allows for cooperative binding of general transcription factors to specialized elements. As mentioned earlier, there is evidence that at this point PIC formation requires an actin-based mechanism.28 At this stage actin is likely to be in a monomeric G-actin form. Once the PIC has been assembled, transcription initiation formally begins. After local DNA unwinding, an open complex is generated. At this point the RNA polymerase II, presumably still in complex with G-actin is liberated from the interactions with promoter-associated transcription factors, the RNA polymerase II leaves the promoter and synthesizes a short (20–50 bp) transcript. The RNA polymerase II machinery then pauses. In mammals this pausing is mediated by negative elongation factors. Transcription elongation begins when the conserved heptapeptide repeats of the RNA polymerase II C-terminal domain (CTD) are phosphorylated at Ser-5. Subsequently, escape from pausing is mediated by P-TEFb, a Cdk9/cyclin T1 heterodimer which is recruited to the polymerase machinery and phosphorylates the conserved Ser 2 of the RNA polymerase II CTD repeats (see Fig. 1). P-TEFb recruitment is mediated by the direct interaction of monomeric G-actin with the catalytic subunit Cdk9 within the elongation complex.36 Cdk9 binds to G-actin through the conserved Thr186 in the T-loop. Importantly, the cdk9/G-actin interaction promotes cdk9-mediated phosphorylation of the CTD and stimulates RNA polymerase II transcription elongation. Therefore, altogether these observations support the idea that PIC assembly and escape from pausing require monomeric actin.
As mentioned earlier, immediately after promoter escape, actin remains associated with the hyperphosphorylated RNA polymerase II CTD in a manner that is dependent on the nascent transcript.29 It is after promoter clearance that actin interacts with hnRNP U to recruit the HAT PCAF for H3K9 acetylation (Fig. 1),29 a mechanism required to maintain efficient transcription elongation. At this stage, we propose that actin polymers begin to be nucleated by F-actin nucleators (Fig. 2). Immediately after, our working model is that a treadmilling regime responsible for head-to-tail actin polymerization closely associated with the elongating polymerase is established and it is maintained through the combined action of cofilin and profilin (Fig. 2). In support of this view, it has already been mentioned that cofilin is required for RNA polymerase II transcription elongation, cofilin targets the polymerase-associated actin and cofilin promotes gene occupancies of actin and elongating polymerase II.40 Similarly, in the C. tentans polytene chromosomes profilin is involved in transcription elongation.73 The suggested head-to-tail actin polymerization may then facilitate processivity and directionality of the polymerase by promoting chromatin alterations that grant the elongation complex license to pass through the nucleosome barrier to transcribe the gene (Fig. 2).
On a more speculative basis, since cofilin does not occupy untranslated regions and flanking regions of active genes, it is suggested that actin polymerization ceases upon termination when the RNA polymerase II complex is removed from the DNA and the RNA is released in the form of a ribonucleoprotein complex.
Future Directions
The treadmilling hypothesis during transcription elongation is very challenging and raises many questions, some of which are highlighted below.
One of the major issues concerns the way actin polymers are initially nucleated outside the gene promoter, concomitantly with hyperphosphorylation of the polymerase CTD. One possibility is that nucleating factors concentrate at gene promoter to kick off filament formation and remain behind at gene promoter as soon as the polymerase becomes committed to elongation. An alternative to this line of thinking is that nucleators of actin polymers travel with the elongating polymerase along the gene. What factors match these properties, at least in part, is difficult to tell at this stage. The F-actin nucleating factor N-WASP which is part of the same complex with nuclear actin, RNA polymerase II and the PSF-NonO complex is a prime candidate as nucleator of nuclear actin polymers in the transcription context,71 possibly synergizing with the ARP2/3 complex.72 However to date there is no causal correlation between occupancies of N-WASP, ARP2/3 complex with actin and elongating polymerase along the entire transcription unit.
In the nucleus, there are other potential F-actin nucleating factors that may have an impact on the transcription process. One candidate F-actin nucleating factor is JMY (a transcriptional coactivator of p53), which is actively imported into the nucleus83 and is known to activate Arp2/3 complexes and mediate F-actin assembly in the cytoplasm.84 Other candidates include diaphanous-related formins [e.g., mDia2 (ref. 85); multidomain proteins that regulate nucleation of actin filaments and barbed-end elongation in the cytoplasm (ref. 86), by recruiting profilin-actin complexes (ref. 87)], or novel factors potentially including RNA polymerase complex components (e.g., hnRNP U).
In any case, if actin polymerization accompanies the elongating polymerase, one would also expect specialized machinery terminating the polymerization event when the polymerase stops transcribing. In the cytoplasm, to control actin filament growth the barbed ends are capped by gelsolin-like proteins, which are also suggested to work as transcriptional co-activators.88 CapG, one of the family members, represents an example of gelsolin-like proteins being actively imported in the cell nucleus and to localize also in the nucleolus.89,90 Furthermore a large fraction of CapG freely diffuses but there is a smaller fraction of about 12% that appears to be slowed down in diffusion by 100-fold. These findings and evidence that active transcription loops of lampbrush chromosomes collapse upon injections of gelsolin in the oocytes nuclei of Pleurodeles waltlii,22 are consistent with a capping event that leads to disassembly of actin polymers within the transcription unit. Further experiments are required to support this hypothesis, but this mechanism may be required for transcription termination when actin polymerization must come to a halt.
Concluding Remarks
In summary during the RNA polymerase II transcription cycle, a key role for actin has been demonstrated for PIC formation and in the post-initiation phases to facilitate escape from pausing and elongation (Fig. 3). However, we would like to propose that regulated actin polymerization is not a requirement for transcription initiation but it affects the later phases of gene transcription. We hypothesize that regulated actin polymerization is needed for the transition from initiation to elongation and to maintain productive transcription elongation (see Figure 5). These mechanisms appear to be isoform-specific for β-actin as the other non-muscle form, γ-actin, has not been implicated in gene expression regulation in vivo.32 Whether these mechanisms support only gene activation and not gene repression is a fascinating question for future work. As mentioned earlier a recent β-actin knockout mouse model demonstrated that in vivo, the primary task of β-actin is not to promote cell motility but it is required for gene activity.32 Remarkably, in this study the authors found that the lack of β-actin leads to both activation and repression of different sets of genes. This finding is compatible with a dual role for β-actin as activator and repressor of gene transcription. How this dual function is exerted in the same nuclear context is a matter of speculation. An attractive possibility is that it is precisely the control of head-to-tail actin polymerization along a gene that contributes to set the rules as to whether the gene is activated or repressed via an actin-based mechanism. Changes in the rate of actin depolymerization from the pointed ends may ultimately affect the spatial distribution of the polymerase machinery and lead either to a stall or to higher polymerase processivity. In this scenario, cofilin becomes a central player in the regulation of gene activity.
Figure 5. Speculative model on the dynamic actin polymerization during transcription. In the initial phases of transcription actin is in a monomeric form. This actin fraction contributes to PIC assembly and to facilitate transcription initiation. Upon commitment of the polymerase enzyme to elongation, actin polymerization accompanies the elongation process in a treadmilling regime which is controlled by cofilin and profilin. This mechanism is reiterated throughout the entire length of the transcribed gene. For termination we speculate that a yet unidentified mechanism at the anchorage point of polymeric actin leads to disassembly of actin polymers and contributes to transcription termination. D, ADP-actin; T, ATP-actin; C, cofilin; P, profilin.
The above ideas are partly speculative but they rely on our current knowledge of actin in gene transcription and the huge amount of mechanistic details available from the cytoplasmic actin field. To be able to further analyze how actin polymerization is regulated and facilitates polymerase mediated transcription, a way to go is to apply quantitative and high throughput analyses that will identify the repertoire of nuclear actin-associated proteins and correlate their gene occupancies with the polymerase machinery. In this complexity, as already mentioned, how the nuclear myosin species talk with the elongating RNA polymerase II will be an important question to address in the future along with the corresponding signaling pathways. At this stage there are no mechanistic insights. Furthermore, we have limited clues as to the significance of having multiple myosin species in the nucleus. We leave these considerations for future discussion. In any case, since there is a clear correlation between nuclear myosin and active transcription sites,23,41 it is not difficult to imagine that a myosin motor that requires a degree of actin polymerization is also involved in RNA polymerase II-mediated transcription elongation.
Disclosure of Potential Conflicts of Interest
The author declares that to the best of his knowledge there are no conflicts of interest associated with the present manuscript.
Acknowledgments
This work is supported by grants from the Swedish Research Council (Vetenskapsrådet) and the Swedish Cancer Society (Cancerfonden).
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/22798
References
- 1.Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem. 2002;71:755–81. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- 2.Farrants AK. Chromatin remodelling and actin organisation. FEBS Lett. 2008;582:2041–50. doi: 10.1016/j.febslet.2008.04.032. [DOI] [PubMed] [Google Scholar]
- 3.Percipalle P. The long journey of actin and actin-associated proteins from genes to polysomes. Cell Mol Life Sci. 2009;66:2151–65. doi: 10.1007/s00018-009-0012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visa N, Percipalle P. Nuclear functions of actin. Cold Spring Harb Perspect Biol. 2010;2:a000620. doi: 10.1101/cshperspect.a000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Lanerolle P, Serebryannyy L. Nuclear actin and myosins: life without filaments. Nat Cell Biol. 2011;13:1282–8. doi: 10.1038/ncb2364. [DOI] [PubMed] [Google Scholar]
- 6.Skarp KP, Vartiainen MK. Actin on DNA-an ancient and dynamic relationship. Cytoskeleton (Hoboken) 2010;67:487–95. doi: 10.1002/cm.20464. [DOI] [PubMed] [Google Scholar]
- 7.Saitoh N, Spahr CS, Patterson SD, Bubulya P, Neuwald AF, Spector DL. Proteomic analysis of interchromatin granule clusters. Mol Biol Cell. 2004;15:3876–90. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Obrdlik A, Louvet E, Kukalev A, Naschekin D, Kiseleva E, Fahrenkrog B, et al. Nuclear myosin 1 is in complex with mature rRNA transcripts and associates with the nuclear pore basket. FASEB J. 2010;24:146–57. doi: 10.1096/fj.09-135863. [DOI] [PubMed] [Google Scholar]
- 9.Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, et al. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods. 2007;4:951–6. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- 10.Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–52. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- 11.Miralles F, Visa N. Actin in transcription and transcription regulation. Curr Opin Cell Biol. 2006;18:261–6. doi: 10.1016/j.ceb.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–96. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Miyamoto K, Pasque V, Jullien J, Gurdon JB. Nuclear actin polymerization is required for transcriptional reprogramming of Oct4 by oocytes. Genes Dev. 2011;25:946–58. doi: 10.1101/gad.615211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonsior SM, Platz S, Buchmeier S, Scheer U, Jockusch BM, Hinssen H. Conformational difference between nuclear and cytoplasmic actin as detected by a monoclonal antibody. J Cell Sci. 1999;112:797–809. doi: 10.1242/jcs.112.6.797. [DOI] [PubMed] [Google Scholar]
- 15.Schoenenberger CA, Buchmeier S, Boerries M, Sütterlin R, Aebi U, Jockusch BM. Conformation-specific antibodies reveal distinct actin structures in the nucleus and the cytoplasm. J Struct Biol. 2005;152:157–68. doi: 10.1016/j.jsb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann WA, Arduini A, Nicol SM, Camacho CJ, Lessard JL, Fuller-Pace FV, et al. SUMOylation of nuclear actin. J Cell Biol. 2009;186:193–200. doi: 10.1083/jcb.200905016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark TG, Merriam RW. Diffusible and bound actin nuclei of Xenopus laevis oocytes. Cell. 1977;12:883–91. doi: 10.1016/0092-8674(77)90152-0. [DOI] [PubMed] [Google Scholar]
- 18.Clark TG, Rosenbaum JL. An actin filament matrix in hand-isolated nuclei of X. laevis oocytes. Cell. 1979;18:1101–8. doi: 10.1016/0092-8674(79)90223-X. [DOI] [PubMed] [Google Scholar]
- 19.McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ. Nucleoplasmic β-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol. 2006;172:541–52. doi: 10.1083/jcb.200507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic ‘network of networks’. Nat Rev Mol Cell Biol. 2011;12:695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- 21.Egly JM, Miyamoto NG, Moncollin V, Chambon P. Is actin a transcription initiation factor for RNA polymerase B? EMBO J. 1984;3:2363–71. doi: 10.1002/j.1460-2075.1984.tb02141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheer U, Hinssen H, Franke WW, Jockusch BM. Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell. 1984;39:111–22. doi: 10.1016/0092-8674(84)90196-X. [DOI] [PubMed] [Google Scholar]
- 23.Fomproix N, Percipalle P. An actin-myosin complex on actively transcribing genes. Exp Cell Res. 2004;294:140–8. doi: 10.1016/j.yexcr.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 24.Philimonenko VV, Zhao J, Iben S, Dingová H, Kyselá K, Kahle M, et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol. 2004;6:1165–72. doi: 10.1038/ncb1190. [DOI] [PubMed] [Google Scholar]
- 25.Percipalle P, Fomproix N, Cavellán E, Voit R, Reimer G, Krüger T, et al. The chromatin remodelling complex WSTF-SNF2h interacts with nuclear myosin 1 and has a role in RNA polymerase I transcription. EMBO Rep. 2006;7:525–30. doi: 10.1038/sj.embor.7400657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grummt I. Actin and myosin as transcription factors. Curr Opin Genet Dev. 2006;16:191–6. doi: 10.1016/j.gde.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Percipalle P, Zhao J, Pope B, Weeds A, Lindberg U, Daneholt B. Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J Cell Biol. 2001;153:229–36. doi: 10.1083/jcb.153.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol. 2004;6:1094–101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- 29.Obrdlik A, Kukalev A, Louvet E, Farrants AK, Caputo L, Percipalle P. The histone acetyltransferase PCAF associates with actin and hnRNP U for RNA polymerase II transcription. Mol Cell Biol. 2008;28:6342–57. doi: 10.1128/MCB.00766-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu P, Wu S, Hernandez N. A role for beta-actin in RNA polymerase III transcription. Genes Dev. 2004;18:3010–5. doi: 10.1101/gad.1250804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 32.Tondeleir D, Lambrechts A, Müller M, Jonckheere V, Doll T, Vandamme D, et al. Cells lacking β-actin are genetically reprogrammed and maintain conditional migratory capacity. Mol Cell Proteomics. 2012;11:255–71. doi: 10.1074/mcp.M111.015099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiesler E, Visa N. Intranuclear pre-mRNA trafficking in an insect model system. Prog Mol Subcell Biol. 2004;35:99–118. doi: 10.1007/978-3-540-74266-1_5. [DOI] [PubMed] [Google Scholar]
- 34.Daneholt B. Assembly and transport of a premessenger RNP particle. Proc Natl Acad Sci U S A. 2001;98:7012–7. doi: 10.1073/pnas.111145498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Percipalle P, Jonsson A, Nashchekin D, Karlsson C, Bergman T, Guialis A, et al. Nuclear actin is associated with a specific subset of hnRNP A/B-type proteins. Nucleic Acids Res. 2002;30:1725–34. doi: 10.1093/nar/30.8.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi T, Tang W, Wang L, Zhai L, Guo L, Zeng X. G-actin participates in RNA polymerase II-dependent transcription elongation by recruiting positive transcription elongation factor b (P-TEFb) J Biol Chem. 2011;286:15171–81. doi: 10.1074/jbc.M110.184374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Percipalle P, Fomproix N, Kylberg K, Miralles F, Bjorkroth B, Daneholt B, et al. An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II. Proc Natl Acad Sci U S A. 2003;100:6475–80. doi: 10.1073/pnas.1131933100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kukalev A, Nord Y, Palmberg C, Bergman T, Percipalle P. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat Struct Mol Biol. 2005;12:238–44. doi: 10.1038/nsmb904. [DOI] [PubMed] [Google Scholar]
- 39.Sjölinder M, Björk P, Söderberg E, Sabri N, Farrants AK, Visa N. The growing pre-mRNA recruits actin and chromatin-modifying factors to transcriptionally active genes. Genes Dev. 2005;19:1871–84. doi: 10.1101/gad.339405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Obrdlik A, Percipalle P. The F-actin severing protein cofilin-1 is required for RNA polymerase II transcription elongation. Nucleus. 2011;2:72–9. doi: 10.4161/nucl.2.1.14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kyselá K, Philimonenko AA, Philimonenko VV, Janácek J, Kahle M, Hozák P. Nuclear distribution of actin and myosin I depends on transcriptional activity of the cell. Histochem Cell Biol. 2005;124:347–58. doi: 10.1007/s00418-005-0042-8. [DOI] [PubMed] [Google Scholar]
- 42.Ye J, Zhao J, Hoffmann-Rohrer U, Grummt I. Nuclear myosin I acts in concert with polymeric actin to drive RNA polymerase I transcription. Genes Dev. 2008;22:322–30. doi: 10.1101/gad.455908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–24. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nowak G, Pestic-Dragovich L, Hozák P, Philimonenko A, Simerly C, Schatten G, et al. Evidence for the presence of myosin I in the nucleus. J Biol Chem. 1997;272:17176–81. doi: 10.1074/jbc.272.27.17176. [DOI] [PubMed] [Google Scholar]
- 45.Pestic-Dragovich L, Stojiljkovic L, Philimonenko AA, Nowak G, Ke Y, Settlage RE, et al. A myosin I isoform in the nucleus. Science. 2000;290:337–41. doi: 10.1126/science.290.5490.337. [DOI] [PubMed] [Google Scholar]
- 46.Dzijak R, Yildirim S, Kahle M, Novák P, Hnilicová J, Venit T, et al. Specific nuclear localizing sequence directs two myosin isoforms to the cell nucleus in calmodulin-sensitive manner. PLoS One. 2012;7:e30529. doi: 10.1371/journal.pone.0030529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ihnatovych I, Migocka-Patrzalek M, Dukh M, Hofmann WA. Identification and characterization of a novel myosin Ic isoform that localizes to the nucleus. Cytoskeleton (Hoboken) 2012;69:555–65. doi: 10.1002/cm.21040. [DOI] [PubMed] [Google Scholar]
- 48.Pranchevicius MC, Baqui MM, Ishikawa-Ankerhold HC, Lourenço EV, Leão RM, Banzi SR, et al. Myosin Va phosphorylated on Ser1650 is found in nuclear speckles and redistributes to nucleoli upon inhibition of transcription. Cell Motil Cytoskeleton. 2008;65:441–56. doi: 10.1002/cm.20269. [DOI] [PubMed] [Google Scholar]
- 49.Lindsay AJ, McCaffrey MW. Myosin Vb localises to nucleoli and associates with the RNA polymerase I transcription complex. Cell Motil Cytoskeleton. 2009;66:1057–72. doi: 10.1002/cm.20408. [DOI] [PubMed] [Google Scholar]
- 50.Vreugde S, Ferrai C, Miluzio A, Hauben E, Marchisio PC, Crippa MP, et al. Nuclear myosin VI enhances RNA polymerase II-dependent transcription. Mol Cell. 2006;23:749–55. doi: 10.1016/j.molcel.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 51.Cameron RS, Liu C, Mixon AS, Pihkala JP, Rahn RJ, Cameron PL. Myosin16b: The COOH-tail region directs localization to the nucleus and overexpression delays S-phase progression. Cell Motil Cytoskeleton. 2007;64:19–48. doi: 10.1002/cm.20162. [DOI] [PubMed] [Google Scholar]
- 52.Salamon M, Millino C, Raffaello A, Mongillo M, Sandri C, Bean C, et al. Human MYO18B, a novel unconventional myosin heavy chain expressed in striated muscles moves into the myonuclei upon differentiation. J Mol Biol. 2003;326:137–49. doi: 10.1016/S0022-2836(02)01335-9. [DOI] [PubMed] [Google Scholar]
- 53.Ajima R, Akazawa H, Kodama M, Takeshita F, Otsuka A, Kohno T, et al. Deficiency of Myo18B in mice results in embryonic lethality with cardiac myofibrillar aberrations. Genes Cells. 2008;13:987–99. doi: 10.1111/j.1365-2443.2008.01226.x. [DOI] [PubMed] [Google Scholar]
- 54.Percipalle P, Farrants AK. Chromatin remodelling and transcription: be-WICHed by nuclear myosin 1. Curr Opin Cell Biol. 2006;18:267–74. doi: 10.1016/j.ceb.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 55.Pederson T, Aebi U. Nuclear actin extends, with no contraction in sight. Mol Biol Cell. 2005;16:5055–60. doi: 10.1091/mbc.E05-07-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pederson T. As functional nuclear actin comes into view, is it globular, filamentous, or both? J Cell Biol. 2008;180:1061–4. doi: 10.1083/jcb.200709082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gieni RS, Hendzel MJ. Actin dynamics and functions in the interphase nucleus: moving toward an understanding of nuclear polymeric actin. Biochem Cell Biol. 2009;87:283–306. doi: 10.1139/O08-133. [DOI] [PubMed] [Google Scholar]
- 58.Fukui Y. Intranuclear actin bundles induced by dimethyl sulfoxide in interphase nucleus of Dictyostelium. J Cell Biol. 1978;76:146–57. doi: 10.1083/jcb.76.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wieland T, Faulstich H. Amatoxins, phallotoxins, phallolysin and antamanide: the biological active components of poisonous Amanita mushrooms. Crit Rev Biochem. 1978;5:185–260. doi: 10.3109/10409237809149870. [DOI] [PubMed] [Google Scholar]
- 60.Sanger JW, Sanger JM, Kreis TE, Jockusch BM. Reversible translocation of cytoplasmic actin into the nucleus caused by dimethyl sulfoxide. Proc Natl Acad Sci U S A. 1980;77:5268–72. doi: 10.1073/pnas.77.9.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bohnsack MT, Stüven T, Kuhn C, Cordes VC, Görlich D. A selective block of nuclear actin export stabilizes the giant nuclei of Xenopus oocytes. Nat Cell Biol. 2006;8:257–63. doi: 10.1038/ncb1357. [DOI] [PubMed] [Google Scholar]
- 62.Gall JG. Exporting actin. Nat Cell Biol. 2006;8:205–7. doi: 10.1038/ncb0306-205. [DOI] [PubMed] [Google Scholar]
- 63.Kiseleva E, Drummond SP, Goldberg MW, Rutherford SA, Allen TD, Wilson KL. Actin- and protein-4.1-containing filaments link nuclear pore complexes to subnuclear organelles in Xenopus oocyte nuclei. J Cell Sci. 2004;117:2481–90. doi: 10.1242/jcs.01098. [DOI] [PubMed] [Google Scholar]
- 64.Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, et al. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol. 2007;179:1095–103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumaran RI, Thakar R, Spector DL. Chromatin dynamics and gene positioning. Cell. 2008;132:929–34. doi: 10.1016/j.cell.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malyavantham KS, Bhattacharya S, Berezney R. The architecture of functional neighborhoods within the mammalian cell nucleus. Adv Enzyme Regul. 2010;50:126–34. doi: 10.1016/j.advenzreg.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vieu E, Hernandez N. Actin’s latest act: polymerizing to facilitate transcription? Nat Cell Biol. 2006;8:650–1. doi: 10.1038/ncb0706-650. [DOI] [PubMed] [Google Scholar]
- 68.Ferrai C, Naum-Onganía G, Longobardi E, Palazzolo M, Disanza A, Diaz VM, et al. Induction of HoxB transcription by retinoic acid requires actin polymerization. Mol Biol Cell. 2009;20:3543–51. doi: 10.1091/mbc.E09-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carlier MF. Control of actin dynamics. Curr Opin Cell Biol. 1998;10:45–51. doi: 10.1016/S0955-0674(98)80085-9. [DOI] [PubMed] [Google Scholar]
- 70.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–76. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 71.Wu X, Yoo Y, Okuhama NN, Tucker PW, Liu G, Guan JL. Regulation of RNA-polymerase-II-dependent transcription by N-WASP and its nuclear-binding partners. Nat Cell Biol. 2006;8:756–63. doi: 10.1038/ncb1433. [DOI] [PubMed] [Google Scholar]
- 72.Yoo Y, Wu X, Guan JL. A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J Biol Chem. 2007;282:7616–23. doi: 10.1074/jbc.M607596200. [DOI] [PubMed] [Google Scholar]
- 73.Söderberg E, Hessle V, von Euler A, Visa N. Profilin is associated with transcriptionally active genes. Nucleus. 2012;3:290–9. doi: 10.4161/nucl.20327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bugyi B, Carlier M-F. Control of actin filament treadmilling in cell motility. Annu Rev Biophys. 2010;39:449–70. doi: 10.1146/annurev-biophys-051309-103849. [DOI] [PubMed] [Google Scholar]
- 75.Paavilainen VO, Bertling E, Falck S, Lappalainen P. Regulation of cytoskeletal dynamics by actin-monomer-binding proteins. Trends Cell Biol. 2004;14:386–94. doi: 10.1016/j.tcb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 76.Pendleton A, Pope B, Weeds A, Koffer A. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J Biol Chem. 2003;278:14394–400. doi: 10.1074/jbc.M206393200. [DOI] [PubMed] [Google Scholar]
- 77.Skare P, Kreivi JP, Bergström A, Karlsson R. Profilin I colocalizes with speckles and Cajal bodies: a possible role in pre-mRNA splicing. Exp Cell Res. 2003;286:12–21. doi: 10.1016/S0014-4827(03)00102-2. [DOI] [PubMed] [Google Scholar]
- 78.Stüven T, Hartmann E, Görlich D. Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J. 2003;22:5928–40. doi: 10.1093/emboj/cdg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dopie J, Skarp KP, Rajakylä EK, Tanhuanpää K, Vartiainen MK. Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci U S A. 2012;109:E544–52. doi: 10.1073/pnas.1118880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giesemann T, Rathke-Hartlieb S, Rothkegel M, Bartsch JW, Buchmeier S, Jockusch BM, et al. A role for polyproline motifs in the spinal muscular atrophy protein SMN. Profilins bind to and colocalize with smn in nuclear gems. J Biol Chem. 1999;274:37908–14. doi: 10.1074/jbc.274.53.37908. [DOI] [PubMed] [Google Scholar]
- 81.Huet G, Skarp K-P, Vartiainen MK. Nuclear actin levels as an important transcriptional switch. Transcr. 2012;3:1–5. doi: 10.4161/trns.21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–92. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zuchero JB, Belin B, Mullins RD. Actin binding to WH2 domains regulates nuclear import of the multifunctional actin regulator JMY. Mol Biol Cell. 2012;23:853–63. doi: 10.1091/mbc.E11-12-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–9. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miki T, Okawa K, Sekimoto T, Yoneda Y, Watanabe S, Ishizaki T, et al. mDia2 shuttles between the nucleus and the cytoplasm through the importin-alpha/beta- and CRM1-mediated nuclear transport mechanism. J Biol Chem. 2009;284:5753–62. doi: 10.1074/jbc.M806191200. [DOI] [PubMed] [Google Scholar]
- 86.Dominguez R. Structural insights into de novo actin polymerization. Curr Opin Struct Biol. 2010;20:217–25. doi: 10.1016/j.sbi.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–35. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 88.Archer SK, Claudianos C, Campbell HD. Evolution of the gelsolin family of actin-binding proteins as novel transcriptional coactivators. Bioessays. 2005;27:388–96. doi: 10.1002/bies.20200. [DOI] [PubMed] [Google Scholar]
- 89.Van Impe K, Hubert T, De Corte V, Vanloo B, Boucherie C, Vandekerckhove J, et al. A new role for nuclear transport factor 2 and Ran: nuclear import of CapG. Traffic. 2008;9:695–707. doi: 10.1111/j.1600-0854.2008.00720.x. [DOI] [PubMed] [Google Scholar]
- 90.Hubert T, Van Impe K, Vandekerckhove J, Gettemans J. The F-actin filament capping protein CapG is a bona fide nucleolar protein. Biochem Biophys Res Commun. 2008;377:699–704. doi: 10.1016/j.bbrc.2008.10.048. [DOI] [PubMed] [Google Scholar]