Abstract
Rhabdomyosarcoma (RMS) is a pediatric tumor that arises from muscle precursor cells. RMS cells express several markers of early myogenic differentiation, but they fail to complete both differentiation program and cell cycle arrest, resulting in uncontrolled proliferation and incomplete myogenesis. Previous studies showed that EZH2, which is involved in both differentiation and cancer progression, is overexpressed in RMS, but a functional binding between its expression and its functional role in tumor formation or progression has not yet been demonstrated. We hypothesized that EZH2 is a key regulator of muscular differentiation program in RMS cells.
In this study, we demonstrated that EZH2 directly binds muscle specific genes in RD cells. Silencing of EZH2 promotes the recruitment of a multiprotein complex at muscle-specific promoters, their transcriptional activation and protein expression. Moreover, we demonstrated that EZH2 is directly involved in transcriptional repression of MyoD, the main factor promoting myogenesis. EZH2 ablation induces MyoD activation the recovery of its binding on muscle-specific genes.
Keywords: PRC complex, EZH2, MyoD, rhabdomyosarcoma, myogenesis
Introduction
Rhabdomyosarcoma (RMS) is one of the most common pediatric tumors that arise from muscle precursor cells. RMS cells fail to complete both cell cycle arrest and differentiation program, resulting in uncontrolled proliferation and incomplete myogenesis.1
RMS is defined as a small, round, blue cell tumor expressing several markers of early myogenic differentiation to varying degrees, which discriminates it from other soft tissues or bone marrow; however, RMS displays limited terminal differentiated cells and does not form myotubes or functional muscle units.2-4
An important regulatory role in the development of skeletal muscle tissue is performed by a family of muscle-restricted bHLH proteins. After myogenesis activation, these proteins bind sequence-specific DNA elements, named E-box (CANNTG), which are located in both enhancer and promoter sequences of muscle-specific genes and induce their transcriptional activation.5 The most important member of the bHLH family is MyoD, which has a crucial role in the regulation of more than 300 muscle-specific genes.6
MyoD-DNA binding is achieved by heterodimerization with other non-myogenic bHLH proteins, such as the products of the E2A gene (E12, E47).5 At muscle-specific genes promoters, MyoD heterodimeres form a multiprotein complex with Brg1, pTEFIIb and acetyltransferases p300 and PCAF. This complex induces histone acetylation, changes in nucleosomal conformation and RNA polymerase II (Pol II) C terminus (CTD) phosphorylation, promoting gene expression.7,8
In Rhabdomyosarcoma, MyoD is not able to activate transcription of muscle-specific genes, despite that it is still able to bind some of its protein partners and DNA.2,9 Therefore, it is crucial to clarify the molecular mechanism of MyoD inhibition in RMS in order to find new therapeutic agents able to induce the reversion of tumor phenotype.
In recent years, it has been shown that histone post-translational modifications are involved in numerous cellular processes, including oncogenesis.10-12 In particular, altered acetylation and methylation patterns of histones H3 and H4 are associated with various cancer types. Moreover, specific epigenetic patterns enable the distinction of several tumor subtypes.13,14
Polycomb group proteins (PcG) are regulators of epigenetic gene silencing whose activity is essential to ensure the establishment and maintenance of the correct transcriptome during development.12 PcG-mediated gene expression silencing, which involves the tri-methylation of lysine 27 on histone 3 (H3K27me3),15,16 is required for the timely expression of genes involved in stem cell fate and lineage commitment17-20 and for mammalian X-inactivation and imprinting.21,22 Enhancer of zeste homolog 2 (EZH2) is the catalytic subunit of the Polycomb repressive complex 2 (PRC2), which is activated by other PCR2-subunits as embryonic ectoderm development (EED), suppressor of zeste 12 homolog (SUZ12) and histone binding protein RbAp46.23-25
EZH2 has a widespread role in early mouse development,26,27 in contrast to its limited post-embryonic expression.28,29 In adult tissues, EZH2 is specifically expressed in undifferentiated cell progenitors, such as hematopoietic cells of the pro-B lymphocyte lineage.30
Interestingly, EZH2 is overexpressed in various tumors, and its expression is related to advanced stages of diseases and poor prognosis.25
For what concerns muscle differentiation, it has been shown that PRC2 binds and represses numerous MyoD target genes in embryonic stem cells by establishing tri-methylation of H3K27.18 Furthermore, PRC2 is able to bind and silence several MyoD target genes in undifferentiated skeletal myoblasts, despite MyoD expression. After the commitment of myogenesis, PRC2 binding and H3K27me3 presence at MyoD loci are lost, inducing muscle gene expression and differentiation.31,32
Interestingly, recent studies showed that transcript and protein levels of EZH2 are consistently higher in RMS compared with normal myoblasts.33 These data suggest that PRC2 complex plays a key role in the maintenance of the undifferentiated state of muscle cell precursors and in RMS.
In this paper, we studied the functional role of EZH2 in RMS. Our studies confirm that EZH2 is overexpressed in RMS cell lines. Moreover, our results indicate that EZH2-knockdown RMS cells show the reactivation of muscle-specific genes and a partial recovery of myocite phenotype. Finally, we demonstrated that EZH2 is directly involved in both MyoD expression and binding at muscle-specific genes in RMS cells.
Results
RD cells fail to complete the differentiation program and show high levels of EZH2
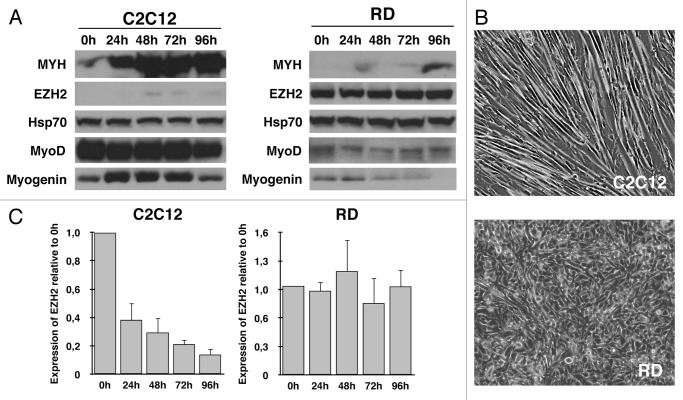
In order to confirm the data showed in previous studies, we analyzed expressions of MyoD and two muscle-specific genes, myogenin and myosin heavy chain (Myh), in normal myoblasts (C2C12) and in RMS cells (RD) in both proliferative and differentiation conditions. Protein expression was examined by immunoblot. As shown in Figure 1A, MyoD dependent-genes myogenin and Myh exhibit significantly lower protein levels in RD, according to previous studies. Interestingly, expression of MyoD, the principal activator of myogenesis, shows very low levels if compared with normal myoblasts. In agreement with this result, after 96 h in differentiation medium (DM), RD cells fail cell cycle arrest and do not form myotubes (Fig. 1B).
Figure 1. RD cells fail to complete the differentiation program and show high levels of EZH2. Myoblasts (C2C12) and RMS cells (RD) were grown to 90% and induced to differentiate by serum withdrawal and addition of 2% horse serum (DM) for 96 h. Pellets were collected each 24 h. (A) Immunoblot analysis reveals the expression of EZH2 in concurrence with decreased levels of MyoD, Myogenin and Myh expression. HSP70 was used as loading control. (B) C2C12 and RD phenotype after 96 h of differentiation induction. C2C12 cells form myotubes, whereas no phenotypic changes can be seen in RD. (C) EZH2 transcription at various times after differentiation induction. Although in differentiation conditions, RD cells maintain EZH2 expression.
Recent studies showed that expression of EZH2, the catalytic subunit of PRC2 complex, frequently elevated in several tumors,25 is overexpressed in RMS cell lines and samples.33 In order to confirm these data, we analyzed gene and protein expression in RD after myogenesis induction.
According to previous studies, protein levels of EZH2 were considerably higher in proliferating RMS cells, and its expression persist in differentiation conditions; contrariwise, normal myoblasts express low levels of EZH2 in all stages of differentiation process (Fig. 1A). Moreover, transcript analysis showed lack of transcriptional repression of EZH2 in RD cells, after differentiation induction, compared with C2C12 (Fig. 1C).
Previous studies demonstrated that EZH2 promotes a displacement of MyoD from its target genes in proliferating myoblasts up to muscle differentiation triggering. We hypothesized a similar mechanism in RMS, where a deregulated PRC2 complex would prevent MyoD binding at muscle specific genes. Nevertheless, this result indicates that the inhibition of myogenesis triggering is accounted for by a downregulation in MyoD expression.
EZH2 binds MyoD, myogenin and Myh promoter in RMS cells
The PcG proteins represent a global gene silencing system that plays a crucial role in organisms development by contributing to the maintenance of a stemness potential.34 Furthermore, EZH2 levels are abnormally elevated in cancer tissues vs. corresponding normal ones.25
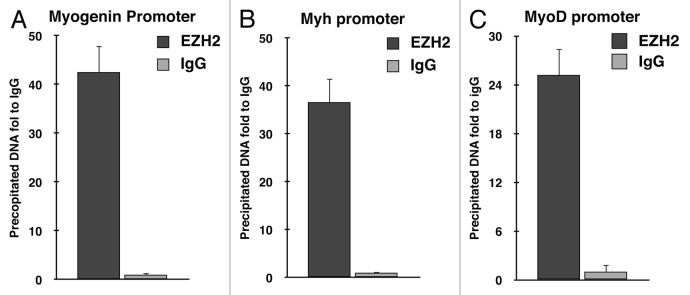
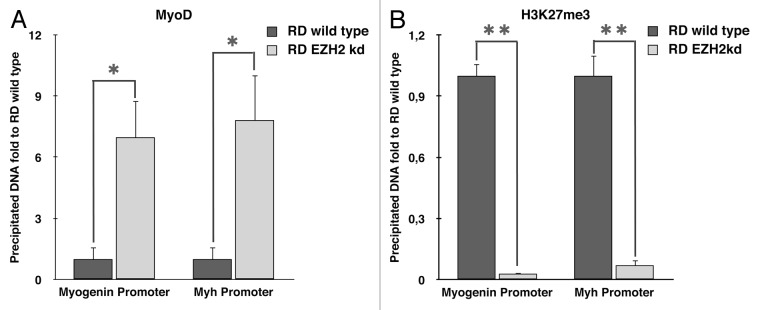
In order to clarify the role of EZH2 in RMS formation, we investigated whether EZH2 is able to bind muscle-specific promoters in RD cells. Chromatin immunoprecipitation (ChIP) was performed using anti-EZH2 antibody. Enrichment of Myogenin and Myh promoter regions in immunoprecipitated RD DNA was evaluated by real-time PCR. EZH2 was detected in both promoter regions analyzed (Fig. 2A and B).
Figure 2. EZH2 binds muscle-specific gene promoters in RD cells. Chromatin immunoprecipitation revealed that EZH2 binds muscle-specific gene promoters in RD cells. The chromatin-immunoprecipitated DNA was amplified by real-time PCR using specific primers for myogenin (A), Myh (B) and MyoD (C) promoter regions. Input DNA was used as loading reference and normal IgG control as calibrator.
Furthermore, ChIP assay with anti-EZH2 antibody was performed in order to investigate a putative role of PRC2 in the inhibition of MyoD expression. Surprisingly, our results demonstrated that EZH2 is able to bind MyoD promoter, differently from current literature (Fig. 2C).
EZH2 inhibits the expression of genes involved in skeletal muscle differentiation in RMS
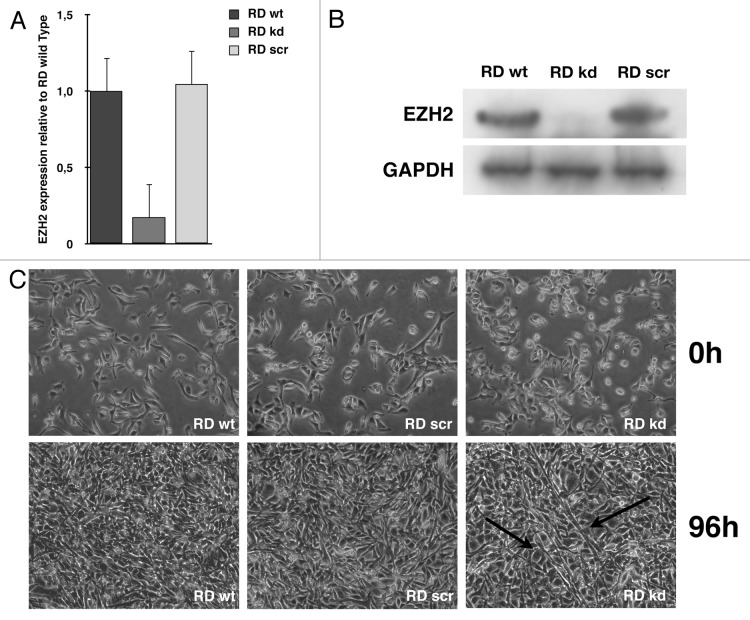
In order to evaluate the functional role of EZH2 in RMS cells, a stable EZH2 knockdown RD cell line was generated using a vector-based shRNA. EZH2 knockdown RD cells showed a specific transcript and protein reduction of 80% in MyoD compared with normal and shRNA scrambled-infected cells (Fig. 3A and B).
Figure 3. EZH2-knockdown RD cells show partial reversion of tumor phenotype. Real-time PCR (A) and immunoblotting (B) show a decrease of EZH2 levels of 80% in EZH2 knockdown RD (kd) compared with RD wild type (wt) and scramble (scr). (C) RD wild type, RD scramble and RD EZH2-knockdown morphology in GM and after 96 h in DM. RD EZH2 knockdown show the formation of several myotube-like cells. Black arrows indicate myotube-like morphology.
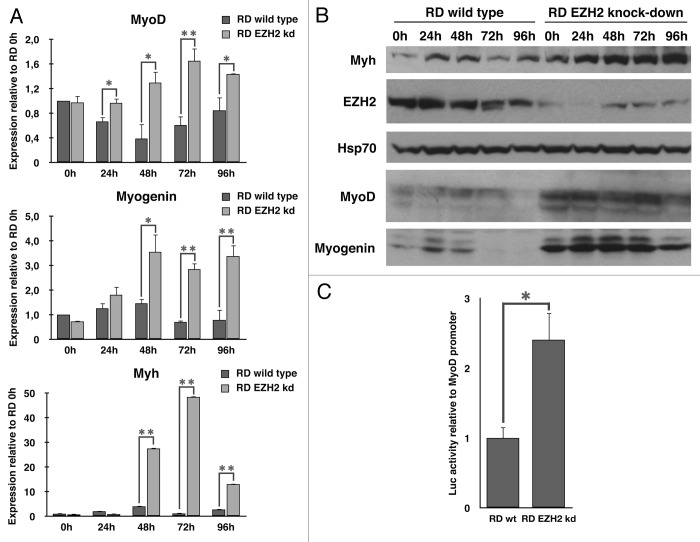
Therefore, all cell lines were grown in growth medium (GM) or induced to differentiate, replacing GM with differentiating medium (DM) for various time periods. As shown in Figure 3C, after 96 h in DM, EZH2 knockdown cells showed a partial recovery of the differentiated phenotype compared with wild type, and some myotube can be detected. In order to assess whether the reactivation of the myogenic program overlaps with a resumption of the expression of differentiation markers, transcripts and proteins levels were assessed every 24 h after induction of differentiation in both wild type and EZH2 knockdown cells. As expected, differentiation induction in the absence of EZH2 resulted in a significant increase of muscle specific factors (Fig. 4A and B). In order to confirm the role of EZH2 in downregulating muscle-specific genes, and, in particular, MyoD, wild type and EZH2 knockdown cells were transiently transfected with MyoD promoter luciferase reporter and induced to differentiate for 72h. EZH2 knockdown cells showed a greater MyoD promoter activation following differentiation promotion, confirming the data previously obtained (Fig. 4C). The partial reactivation of muscle-specific genes, in particular MyoD, strongly supports the role of EZH2 in myogenesis control and, if deregulated, in RMS formation.
Figure 4. EZH2 inhibits the expression of skeletal muscle-specific genes. Differentiation induction in absence of EZH2 results in a significant increase of muscle-specific genes. (A) Real-time qPCR was performed using cDNA from RD wild type and EZH2 knockdown at several stage of differentiation (24h-96 h). Reported data were normalized to GAPDH levels. EZH2-kd RD cells showed increase of muscle specific gene transcription following differentiation induction. (B) Immunoblot analysis reveals that protein levels of muscle specific genes increase in EZH2-knockdown RD cells following differentiation induction. (C) Luciferase assay shows that EZH2-knockdown cells have a greater MyoD promoter activation following differentiation promotion. The assay was conducted on RD wild type and RD EZH2-knockdown cells transfected with MyoD-luc reporter and RL-TK plasmid as calibrator. Transfected cells were cultured in DM for 72 h. Luciferase activity was normalized to TK-directed Renilla expression. The results are expressed in arbitrary units relative to the activity of the basic luciferase vector control. Student’s t-test was used to evaluate statistical significance: * p < 0.05, ** p < 0.01. Values were obtained by the average of at least three independent experiments.
Downregulation of EZH2 restores MyoD binding and complex activation to muscle-specific promoters
As discussed in the introduction, in normal myoblasts EZH2 is able to silence muscle-specific promoters. In order to investigate whether silencing of EZH2 would allow a restoration of MyoD binding to its target genes, we performed a ChIP assay using anti-MyoD antibody. As previously explained, cells were growth in GM and induced to differentiate in DM for 72 h. Enrichment of myogenin and Myh promoter regions in immunoprecipitated DNA was evaluated by real-time PCR. As expected, MyoD is unable to bind muscle-specific genes in RMS cells, whereas its binding is restored in EZH2 knockdown RD cells (Fig. 5A).
Figure 5. EZH2 ablation restores MyoD binding on muscle-specific genes. ChIP assays, performed in RD wild type and EZH2 knockdown 72 h following differentiation induction, show (A) the recovery of MyoD binding at myogenin and Myh regulatory regions in absence of EZH2, (B) a strong decrease of H3K27me3 enrichment at myogenin and Myh promoters. Student’s t-test was used to evaluate statistical significance: * p < 0.05, ** p < 0.01. Values were obtained by the average of at least three independent experiments.
ChIP assay was performed evenly with anti-H3K27me3 antibody that confirmed a decrease of try-methylation of H3K27 in EZH2 knockdown cells, demonstrating the inhibition of EZH2 activity in knockdown cells (Fig. 5B).
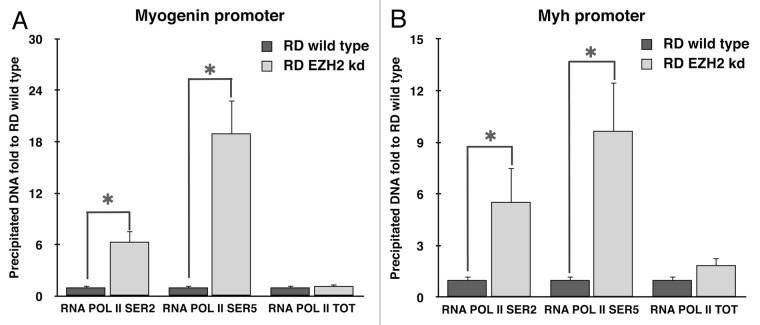
MyoD is able to recruit numerous factors in order to activate transcription of its target genes, such as acetyltransferases p300 and PCAF, and pTEFIIb, which promotes transcriptional activation. In particular, pTEFIIb complex, by phosphorylation of RNApolII CTD, converts the pre-initiation form of RNApolII into the active form. Furthermore, recent studies demonstrated that UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. The removal of the UTX-dependent-repressive H3K27me3 mark requires RNA PolII elongation form.35
Therefore, we analyzed the phosphorylation of RNApolII CTD in RD wild type and EZH2 knockdown after 72 h of myogenesis induction. ChIP assays were performed using specific antibodies for phosphorylated forms of RNAPolII at Serine 2 and Serine 5. As control, chromatin immunoprecipitation using total RNApolII antibody was added.
As shown in Figure 6A and B, Ser2 and Ser5 phosphorylation of RNA Pol II at Myogenin and Myh promoters were significantly increased in the absence of EZH2. In contrast, RNA Pol II occupancy at the promoter regions analyzed did not show significant differences.
Figure 6. EZH2 ablation restores the phosphorylation of RNA polymerase II on muscle-specific genes promoting transcriptional activation. Silencing of EZH2 promotes the recruitment of transcriptional activation complex at MyoD-dependent genes promoting phosphorylations of RNA PolII CTD. Chromatin immunoprecipitation assay was performed 72 h after induction of myogenesis. Samples were analyzed with real-time PCR using specific primers for the myogenin and Myh promoter regions (A and B, respectively). Input DNA was used as reference and normal IgG immunoprecipitated DNA as calibrator. The data were compared with RD wild type precipitated DNA. Student’s t-test was used to evaluate statistical significance. * p < 0.05, ** p < 0.01. Values were obtained by the average of at least three experiments.
These data demonstrated the crucial role of EZH2 in muscle-specific gene inhibition and in RMS progression.
Discussions
Soft tissue sarcomas (STSs) are a group of cancers that originate from cellular aberrations occurring during the differentiation of mesenchymal stem cells (MSCs).36Rhabdomyosarcoma (RMS), which accounts for almost 50% of all STS and is most common in children,1 arises from skeletal muscle precursor cells.
The 5-year survival rate after therapy is strictly related to risk factors; usually, if the disease recurs despite intensive initial therapy, rarely it is cured.4,37 Clarifying the mechanisms leading to deregulation of the muscle differentiation process and tumor formation is therefore essential in order to find new targets and new therapeutic strategies able to increase the survival chance and reduce side effects of traditional therapies.
Epigenetic alterations in cancer cells are recognized as important aspects of tumor biology. Unlike genetic alterations, epigenetic modifications can be reversed with specific inhibitors or enzymes. Indeed, epigenetically silenced tumor suppressor genes can be reactivated to induce apoptosis or senescence. This peculiarity makes epigenetic signatures an interesting target for therapeutic intervention in cancer.38-40
PcG proteins are repressor of transcription. Genome-wide studies showed that in murine ESCs17 and in human embryonic fibroblast,41 Polycomb complexes transcriptionally modulate numerous development regulators and differentiation-related genes. Two main complexes have been discovered: the polycomb repressive complex (PRC) 1 and 2. PRC2 is responsible for tri-methylation of lysine-27 of histone H3, which is the fundamental feature of PcG-mediated repression. H3K27me3 recruits PRC1 which is viewed as a direct executor of silencing.34 PRC1 catalyzes the monoubiquitylation of lysine 119 at histone H2A (H2AK119ub), which is involved in chromatin compaction and is viewed as a direct executor of silencing. Silencing mechanisms PRC1-independent are discovered. For example Lysine 26 of Histone H1 (H1K26) can be methylated by PRC2 in vitro42; methylated H1K26 can recruits heterocromatin binding protein 1 (HP1) to chromatin influencing chromatin structure.43
Our studies focused on the role of PRC2 in RMS carcinogenesis. In particular, we analyzed the function of EZH2, the catalytic subunit of PRC2, on muscle-specific gene transcription.25
Recent studies showed that EZH2 is overexpressed in some RMS cell lines,33 but a functional binding between its expression and its functional role on tumor formation or progression has not yet been demonstrated.
In undifferentiated myoblasts, EZH2 is expressed and detected at genomic loci of repressed muscle-specific genes.31 Following myogenesis activation, EZH2 expression decreases, and PRC2 complex dissociates from gene promoters, and muscle-specific genes are activated. On this basis, we hypothesized that a similar mechanism would occur in RMS, where deregulation of EZH2 expression leads to lack of differentiation.
We confirmed previous studies demonstrating that EZH2 is strongly expressed also in differentiation conditions in RMS cells compared with normal myoblasts (Fig. 1A). This expression pattern is associated with the maintenance of muscle-specific gene repression (Fig. 4). Interestingly, we showed also decreased protein expression of MyoD, the key regulator of skeletal myogenesis (Fig. 4B). This result represents a new important finding that allowed us to hypothesize a different role of EZH2 in RMS. Indeed, in normal myoblasts, MyoD is expressed, and EZH2 displaces it from target genes; in RMS, our results let us assume that EZH2 inhibits MyoD expression.
This hypothesis is supported by ChIP assays that showed the binding of EZH2 at MyoD promoter but also a direct binding at muscle-specific genes, similar to what occurs in normal muscle differentiation (Fig. 2).
These results allow us to hypothesize a double role of PRC2 in the inhibition of muscle-specific gene activation. On the one hand, the presence of EZH2 in RMS cells prevents binding of MyoD at muscle-specific genes. On the other hand, we showed that EZH2 binds MyoD promoter in RD cells, which lets us hypothesize its functional role in the repression of MyoD transcription.
In order to confirm the repressive role of PRC2 on muscle-specific genes, we generated an EZH2-knockdown RMS cell line. As showed in Figure 3C, EZH2 knockdown RD cells showed a partial recovery of muscle phenotype with some myotubes. Moreover, gene and protein expression analysis showed that downregulation of EZH2 increased the expression of MyoD and muscle-specific genes (Fig. 4).
Afterwards, we postulated that ablation of EZH2 would induce the restoration of MyoD binding at muscle-specific promoters and their activation. ChIP assays confirmed this hypothesis, indeed, which corresponds also to a decrease in H3K27 trimethylation (Fig. 5).
Recent studies demonstrated that in differentiating myoblasts, H3K27me3 is removed by a demethylase, UTX, and replaced with H3-Lys4 trimethylation (H3K4me3), which is involved in transcriptional activation.44-47 Removal of H3K27me3 from muscle-specific genes occurs through a two-step process. Initially, Six4 targets UTX to muscle-specific genes, resulting in the demethylation of regulatory regions upstream the transcription start site. Subsequently, UTX needs elongating RNA PolII to demethylate coding regions and allow transcription. Elongating RNA PolII corresponds to its phosphorylated form, which is established by pTEFIIb complex.35,48
Since MyoD recruits p300, PCAF, SWI/SNF and pTEFII on muscle-specific gene promoters to induce gene expression,8 we assumed that the recovery of MyoD binding could restore the entire complex, leading to phosphorylation of RNA PolII, demethylation of H3K27me3 and transcriptional activation. CTD phosphorylation at Ser2 and Ser5 at Myogenin and Myh genes promoters was evaluated by ChIP. As expected, CTD phosphorylation was significantly increased in the EZH2 knockdown RD cells compared with wild type demonstrating a recovery of MyoD functions in absence of EZH2 (Fig. 6).
These studies significantly contribute to a better understanding of the mechanisms triggering RMS carcinogenesis.
Materials and Methods
Cell culture and differentiation
Myoblasts (C2C12) were grown in DMEM supplemented with 20% FBS 1% L-glutamine and antibiotics (growth medium, GM). RMC cells (RD) were grown in DMEM supplemented with 10% FBS, 1% L-glutamine and antibiotics. The differentiation was induced by serum withdrawal in presence of 2% horse serum (differentiation medium, DM). All cell lines were obtained from ATCC. Pellets were collected every 24h for 96h.
Lentivirus production and infection
Knockdown cells were generated as described in www.broadinstitute.org/genome_bio/trc/publicProtocols.html. Briefly 293FT-packaging cells (Invitrogen) were seed at 1.5 × 105 cells/ml (6 ml per plate) in low-antibiotic growth media (DMEM + 10% FBS) in 6 cm tissue culture plates.
Packaging cells were transfected with 3μg of Hairpin-pLKO.1 vector, shRNA for EZH2 (SHCLNG-NM_004456- Sigma-Aldrich) or scrambled shRNA (Addgene plasmid 17920), 0.9 μg of packaging plasmid psPAX2 (Addgene plasmid 12260), 0.1 μg of envelope plasmid pMD2G, (Addgene plasmid 12259) using 24 μl of FuGene HD (Promega). The media containing virus were filtered with 0.45 μm filter. Polyclonal populations of EZH2-knockdown and scrambled RD cells were generated by infection with 1 MOI (multiplicity of infectious) of shRNA lentiviral particles. After 3 d, the cells were selected with 2 μg /ml of puromycin for 1 week.
Plasmids, transient transfections and luciferase assay
The plasmid MyoD promoter-luc reporter was constructed as follow. RD cells DNA was extracted by DNeasy blood and tissue kit (Qiagen) following the manufacture’s protocol. PCR was performed with specific primers for MyoD promoter: For: 5′ -GGGTGGCTAGCAGGACTTTGCTGCCTCTGCCA-3′; Rev: 5′- GCAAGATCTCGATATAGCGGATGGCGTTGC-3′. The amplified product was cloned in NheI and BglII site of the pGL3 basic vector (Promega). The correct sequence of the construct was confirmed by sequencing.
Transient transfections were performed using FuGene HD (Promega). 2 μg of total DNA diluted in 100 μl Opti-MEM (CellGro) was incubated with 8 μl of FuGene HD for 20 min to let the transfection complex form. The transfection complex was added in a ratio 1:16 to the volume of the incubation medium of the cells (6 μl of transfection complex was added to 100 μl of complete medium in 96 wells plate).
Dual luciferase reporter assay (Promega) was used to measure the firefly luciferase and renilla luciferase activities within the transfected cells. Luciferase assay was conducted with MyoD promoter-luc reporter and RL-TK renilla. Both RD and EZH2-knockdown transfected cells were cultured in DM for 72 h. Luciferase activity was normalized to TK-directed Renilla expression, in a ratio 1:20 respect to firefly luciferase vector, to control the variability in transfection efficiencies. The assay was performed with Sirius Luminometer (Berthold detection systems).
The results are expressed in arbitrary units relative to the activity of the basic luciferase vector in RD wild type.
Total RNA extraction, cDNA synthesis and real-time PCR
Total RNA was extracted using High Pure RNA Isolation Kit (Roche). Retrotranscription was performed with 1 μg of RNA and random primers, using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems), following the manufacture’s protocol. Real-time PCR was performed using FastStart Universal SYBR Green Master (ROX) (Roche) and primers 250 nM. Accumulation of fluorescent products was monitored using an Applied Biosystem 7,300 system (Applied Biosystems). Each data point was obtained from at least three independent experiments. Transcripts for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as a reference. To ensure specific PCR amplification, every real-time PCR run was followed by a dissociation phase analysis (denaturation curve) and by gel electrophoresis. ΔΔCT method was used to calculate relative changes in gene expression; primer efficiency was calculated for every target using 5 × 10-fold serial dilutions of PCR products.
The primers used in Real-time PCR are listed below: EZH2: For: 5′-GCGGGACGAAGAATAATCATGG -3′, Rev: 5′-CCAAAATTTTCTGACGATTGGAAC-3′; GAPDH: For: 5′-GAAGGTGAAGGTCGGAGT-3′, Rev: 5′-CATGGGTGGAATCATATTGGA-3′; MyoD: For: 5′-GACGGCATGATGGACTACAGC-3′, Rev: 5′-GGAGATGCGCTCCACGATGC-3′; Myogenin: For: 5′-CCTGCTCAGCTCCCTCAACC-3′, Rev: 5′-AGGGTCAGCCGTGAGCAGATG-3′; MYH: For: 5′-GGAGGAGGCTGATGAACAAGC-3′, Rev: 5′ CTGGCTCACTCTTCACTCTCG-3′.
Immunoblotting
Cells were lysed in lysis buffer (20 mM TRIS-HCl pH 8; 137 mM NaCl; 10% glycerol 1% Nonidet P-40; 2 mM EDTA; Protease Inhibitor Cocktails).
The protein concentration was determined by Bradford assay (Biorad), following the manufacturer’s instructions and by using BSA as a standard.
The protein extract (50 μg) was resolved in 8% -10% SDS/PAA gel and transferred to a nitrocellulose membrane. The protein levels were detected with the followed antibody anti-MyoD, anti-Myogenin, anti-Myh (Santa Cruz Biotechnology), anti EZH2 (Invitrogen). Equal loading was controlled with anti-GAPDH and anti-Hsp70 (Santa Cruz Biotechnology).
Anti-mouse, rabbit (1:10000) peroxidase conjugated (Pierce) and ECL detection system (PerkinElmer) were used for detection.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (XChIP) assays were performed using the ChIP-IT Express Enzymatic kit (Active Motif) according to the protocol provided by manufacturer. Briefly, cells were inducted to differentiate for 72 h in 15 cm plates and fixed with 1% formaldehyde for 5 min at room temperature. The nuclei of cells were enzymatically treated for 10 min at 37°C in digestion buffer. Ten μg of sheared chromatins were used for immunoprecipitation with normal rabbit/mouse IgG (negative controls) or anti-EZH2 (Millipore), anti-H3K27me3 (Active Motif), anti-MyoD, anti-RNApolII, (Santa Cruz Biotechnology), anti-phospho RNA polII (S2) and anti- phospho RNA polII (S5) (Bethyl) at 4°C with rotation overnight. DNA from ChIP was purified and analyzed by real-time PCR.
Real-time PCR was performed using the Applied Biosystem 7300 System with a FastStart Universal SYBR Green Master (ROX), (Roche). Each data point was obtained from three independent experiments. Input DNA (1/5 of total chromatin used for the IP reactions) was used as reference and the background signal as calibrator. Reported data represent real-time PCR amplification values obtained with the formula: 2 [ ( Ct background − Ct Input background) − ( Ct Ab − Ct Input Ab ) ] where Ct is the threshold cycle, background is the normal rabbit IgG immunoprecipitated sample, and Ab is the EZH2, H3K27me3, MyoD, RNA polII, phospho RNA polymerases immmunoprecipitated sample.
To ensure specific PCR amplification, every real-time PCR run was followed by a dissociation phase analysis (denaturation curve).
The primers used in real-time PCR are listed below: MyoD for: GTTCCTATTGGCCTCGGACTC, rev: CTGTCCGGCCTGATTTGTGGT; Myogenin for: GAGAGGGGAATGTGTCCTCC, rev: CTTCTTTCTCCCGCATGGCC; MYH: for: CTCGTCAGAAGCCGATTCTAC, rev: CCAACTTCTCACCTGAGAGTC;
Acknowledgments
This work was supported by grants from POR FSE 2007–2013 “Regione Autonoma della Sardegna,” and from “Fondazione Banco di Sardegna” (Rif. Vs Prot. 469/2011.271).
Glossary
Abbreviations:
- RMS
Rhabdomyosarcoma
- H3K27me3
tri-methylation of Lysine 27 on Histone H3
- CTD
C-terminal domain
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/22025
References
- 1.Merlino G, Helman LJ. Rhabdomyosarcoma--working out the pathways. Oncogene. 1999;18:5340–8. doi: 10.1038/sj.onc.1203038. [DOI] [PubMed] [Google Scholar]
- 2.Otten AD, Firpo EJ, Gerber AN, Brody LL, Roberts JM, Tapscott SJ. Inactivation of MyoD-mediated expression of p21 in tumor cell lines. Cell Growth Differ. 1997;8:1151–60. [PubMed] [Google Scholar]
- 3.Sebire NJ, Malone M. Myogenin and MyoD1 expression in paediatric rhabdomyosarcomas. J Clin Pathol. 2003;56:412–6. doi: 10.1136/jcp.56.6.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saab R, Spunt SL, Skapek SX. Chapter 7 - Myogenesis and Rhabdomyosarcoma: The Jekyll and Hyde of Skeletal Muscle. 1sted. Elsevier Inc; 2011. [DOI] [PubMed] [Google Scholar]
- 5.Puri PL, Sartorelli V. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J Cell Physiol. 2000;185:155–73. doi: 10.1002/1097-4652(200011)185:2<155::AID-JCP1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 6.Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell. 2002;9:587–600. doi: 10.1016/S1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- 7.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet. 2004;36:738–43. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- 8.Giacinti C, Bagella L, Puri PL, Giordano A, Simone C. MyoD recruits the cdk9/cyclin T2 complex on myogenic-genes regulatory regions. J Cell Physiol. 2006;206:807–13. doi: 10.1002/jcp.20523. [DOI] [PubMed] [Google Scholar]
- 9.Knudsen ES, Pazzagli C, Born TL, Bertolaet BL, Knudsen KE, Arden KC, et al. Elevated cyclins and cyclin-dependent kinase activity in the rhabdomyosarcoma cell line RD. Cancer Res. 1998;58:2042–9. [PubMed] [Google Scholar]
- 10.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 11.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome--components and functional correlates. Genes Dev. 2006;20:3215–31. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 12.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–56. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 13.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 14.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–6. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 15.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–96. doi: 10.1016/S0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 16.Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/S0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 17.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 18.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–36. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol. 2008;20:201–7. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–5. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 22.Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, et al. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36:1296–300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- 23.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 24.Pasini D, Bracken AP, Helin K. Polycomb group proteins in cell cycle progression and cancer. Cell Cycle. 2004;3:396–400. doi: 10.4161/cc.3.4.773. [DOI] [PubMed] [Google Scholar]
- 25.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–9. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 26.O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–6. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erhardt S, Lyko F, Ainscough JF-X, Surani MA, Paro R. Polycomb-group proteins are involved in silencing processes caused by a transgenic element from the murine imprinted H19/Igf2 region in Drosophila. Dev Genes Evol. 2003;213:336–44. doi: 10.1007/s00427-003-0331-y. [DOI] [PubMed] [Google Scholar]
- 28.Hobert O, Sures I, Ciossek T, Fuchs M, Ullrich A. Isolation and developmental expression analysis of Enx-1, a novel mouse Polycomb group gene. Mech Dev. 1996;55:171–84. doi: 10.1016/0925-4773(96)00499-6. [DOI] [PubMed] [Google Scholar]
- 29.Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, et al. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 1997;16:3219–32. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su I-H, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4:124–31. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 31.Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–38. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell. 2009;36:61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciarapica R, Russo G, Verginelli F, Raimondi L, Donfrancesco A, Rota R, et al. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle. 2009;8:172–5. doi: 10.4161/cc.8.1.7292. [DOI] [PubMed] [Google Scholar]
- 34.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 35.Seenundun S, Rampalli S, Liu Q-C, Aziz A, Palii C, Hong S, et al. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 2010;29:1401–11. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siddiqi S, Mills J, Matushansky I. Epigenetic remodeling of chromatin architecture: exploring tumor differentiation therapies in mesenchymal stem cells and sarcomas. Curr Stem Cell Res Ther. 2010;5:63–73. doi: 10.2174/157488810790442859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pappo AS, Anderson JR, Crist WM, Wharam MD, Breitfeld PP, Hawkins D, et al. Survival after relapse in children and adolescents with rhabdomyosarcoma: A report from the Intergroup Rhabdomyosarcoma Study Group. J Clin Oncol. 1999;17:3487–93. doi: 10.1200/JCO.1999.17.11.3487. [DOI] [PubMed] [Google Scholar]
- 38.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–63. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 39.Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005;97:1498–506. doi: 10.1093/jnci/dji311. [DOI] [PubMed] [Google Scholar]
- 40.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 41.Bracken AP, Helin K. Epigenetics and genetics: Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:1–12. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 42.Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14:183–93. doi: 10.1016/S1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- 43.Daujat S, Zeissler U, Waldmann T, Happel N, Schneider R. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J Biol Chem. 2005;280:38090–5. doi: 10.1074/jbc.C500229200. [DOI] [PubMed] [Google Scholar]
- 44.Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–4. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 45.Hong S, Cho Y-W, Yu L-R, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18439–44. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–94. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 47.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–50. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 48.Ni Z, Saunders A, Fuda NJ, Yao J, Suarez J-R, Webb WW, et al. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol Cell Biol. 2008;28:1161–70. doi: 10.1128/MCB.01859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]