Abstract
Mounting evidence suggests a role for innate immunity in the early control of HIV infection, before the induction of adaptive immune responses. Among the early innate immune effector cells, dendritic cells (DCs) respond rapidly following infection aimed at arming the immune system, through the recognition of viral products via pattern recognition receptors. This early response results in the potent induction of a cascade of inflammatory cytokines, intimately involved in directly setting up an antiviral state, and indirectly activating other antiviral cells of the innate immune system. However, epidemiologic data strongly support a role for natural killer (NK) cells as critical innate mediators of antiviral control, through the recognition of virally infected cells through a network of receptors called the killer immunoglobulin-like receptors (KIRs). In this review, the early events in innate immune recognition of HIV, focused on defining the biology underlying KIR-mediated NK-cell control of HIV viral replication, are discussed.
Early events during HIV infection, likely mediated by the innate arm of the immune system, strongly influence disease progression. Natural killer cells, in particular, may contribute to antiviral containment.
Early events following HIV infection determine the course of disease progression in such a way that more robust control of viral replication in acute HIV infection, resulting in lower viral set-point levels, is associated with slower HIV disease progression (Pantaleo et al. 1997). However, reduction in viral replication during acute HIV infection often occurs before the induction of adaptive immune responses such as CD8+ T-cell responses (Alter et al. 2007b), strongly suggesting that the innate immune system, our body’s first line of defense against invading pathogens, may play an early essential role in antiviral control.
THE INNATE IMMUNE SYSTEM
The innate immune system has evolved over millennia to nonspecifically control and clear invading pathogens. Unlike the adaptive arm of the immune system, which uses antigen-specific receptors to recognize foreign antigens, the innate immune system uses an array of pattern recognition receptors to detect patterns associated with bacteria, viruses, and/or parasites. These patterns relate to carbohydrate, protein, or lipid structures that are unique to pathogens, not normally produced in human cells (Murphy et al. 2011). Three classes of pattern recognition receptors have been identified to date, including the (RIG-I)-like receptors (RLRs), the toll-like receptors (TLRs), and the nucleotide oligomerization domain (NOD)-like receptors (NLRs). Activation of different combinations of these receptors, on distinct innate immune cell subsets, results in the induction of distinct inflammatory cues that result in the creation of a nonspecific antiviral environment through the release of cytokines (including interferons [IFNs]) that block viral growth, the activation and recruitment of other immune cells, and the induction of adaptive immune responses.
HIV, like other single-stranded RNA viruses, triggers innate immune receptors, including TLR7 and TLR8, resulting in the potent activation of dendritic cells (DCs) and the release of copious amounts of type 1 IFNs and tumor necrosis factor α (TNF-α), both involved in shutting down viral replication in infected cells while also promoting the activation of the immune response (Diebold et al. 2004; Heil et al. 2004; Beignon et al. 2005). Interestingly, recent data suggest that DCs from females produce higher levels of IFN-α, compared with DCs from age-matched men, on HIV RNA triggering of TLR7/8 (Meier et al. 2009). Given that women show overall lower viral set points than men, it is plausible that enhanced viral control in females may in part relate to this enhanced antiviral innate immune response. The difference in the ability of DCs from women and men to respond to TLR7/8 triggering likely reflects a hormonal sensitization of DCs, specifically promoting TLR-induced IFN-α, but not TNF-α, production in women. However, whether enhanced antiviral control reflects the direct activity of IFN-α alone, or its added effects on activating other innate immune cells (including natural killer [NK] cells), or in the induction of a more potent adaptive immune response is yet to be defined.
In addition to TLR7/8 recognition of HIV, TLR2, TLR4, and TLR9 have been implicated in recognition and modulation of HIV viral replication. Both TLR2 and TLR4 triggering on DCs has been associated with increased and reduced transmission of HIV, respectively, owing to differential induction of type 1 IFNs (Thibault et al. 2009). Furthermore, recent evidence also points to a direct role for gp120 binding to TLR9, resulting in pDC activation, type 1 IFN secretion, and activation of NK cells that may promote early antiviral control (Martinelli et al. 2007). However, the overall role of individual or combined TLR sensing in early recognition and control of HIV has not been fully elucidated.
The early HIV-mediated triggering of DCs, and other TLR expressing innate immune cells, is associated with the induction of a robust cytokine storm (Stacey et al. 2009). This early response is marked by the rapid induction of IFN-α, interleukin-15 (IL-15), and inducible protein-10 (IP-10), followed by a slower increase in proinflammatory factors, associated finally with a sustained increase in immunoregulatory cytokines. Interestingly, the acute cytokine cascade is strikingly more pronounced following HIV infection compared with hepatitis B and C infections. Thus, although the dramatic increase in immunomodulators may be geared toward the priming of a robust immune response against the incoming pathogen, it is plausible that the intensity and magnitude of this cascade may also contribute in part to the observed immunopathology associated with early HIV disease.
INNATE IMMUNE CELLS
An array of cell subsets, all derived from the bone marrow, forms the arsenal of the innate immune system that responds to the acute cytokine cascade, each expressing distinct sets of innate immune receptors, endowing them with a unique capacity to respond to incoming pathogens. These cells include phagocytes (monocytes, macrophages, DCs) primed for antigen clearance, cytolytic cells (NK cells and neutrophils) geared toward the direct destruction of the pathogen or pathogen-infected cells, and professional antigen-presenting cells (DCs) aimed at capturing foreign antigens to present to the adaptive immune response for the induction of immunological memory. These cells persistently patrol peripheral tissues, primed to respond to foreign antigens on receptor engagement without the need for antigen sensitization. Thus, the innate immune response is not only responsible for early pathogen containment, but also plays a central role in shaping the quality of the ensuing adaptive immune response through the release of potent inflammatory cues and the qualitative modulation of DCs.
Among the innate immune cells involved in early antiviral control of HIV, epidemiologic evidence strongly points to a central role for NK cells in antiviral containment. Most convincingly, the coexpression of particular NK-cell receptors (the killer immunoglobulin receptors) in conjunction with their ligands (major histocompatibility complex [MHC] class I alleles) is associated with slower HIV disease progression and early viral control of viremia (Martin et al. 2002, 2007). These data strongly support a role for these cytolytic effector cells early in infection, whereas the adaptive immune response is just developing. However, whether NK cells mediate their antiviral control strictly through cytolytic removal of infected cells or through the editing of particular DC populations resulting in more potent adaptive immune responses is unknown.
NK CELLS
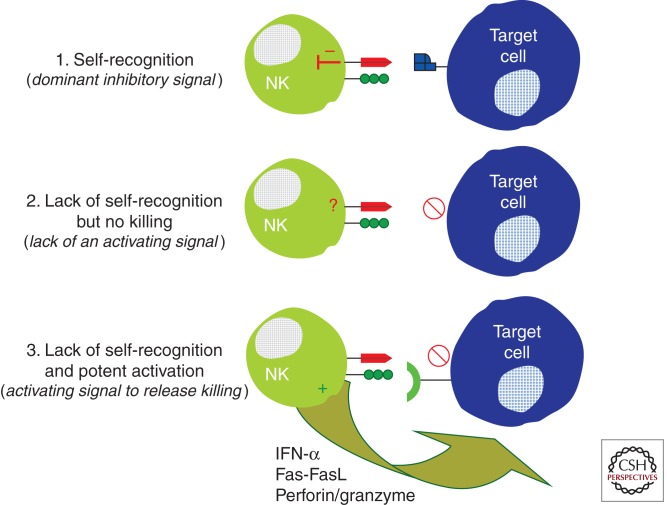
Unlike CD8+ T cells, NK cells are a subset of large granular lymphocytes that do not express an antigen-specific receptor, but rather express a variety of inhibitory and activating receptors on their surface that are involved in sensing changes in their ligands on the surface of the body’s cells (Lanier 1998). As such, these cells are classified as cells of the innate immune system, as they are able to sense viral infection before antigen sensitization. Given that these cells are loaded with cytolytic granules that can cause a great deal of immunopathology, the activation of these cells is under tight regulation by a network of inhibitory and activating self-reactive receptor/ligand interactions. NK cells survey the body for MHC class I expression, using a network of receptors called the killer immunoglobulin-like receptors (KIRs), and are inhibited on interaction with MHC class I. However, lack of engagement of inhibitory receptors alone is not sufficient to activate an NK cell to kill a target cell, but rather an NK cell must receive an additional activating signal through recognition of ligand to induce cytolytic elimination of the target cell (Fig. 1) (Karre et al. 1986; Ljunggren and Karre 1990; Moretta et al. 1993). Alternatively, target cells that up-regulate activating NK receptor ligands to levels that outcompete the dominant inhibitory signals delivered through normal MHC recognition by KIRs can also result in NK cell activation (Cerwenka and Lanier 2001a). Ultimately, NK-cell activation hinges on the delicate balance between inhibition and activation delivered through a variety of NK-cell receptors, including KIRs, that fine-tune their lytic activity. This concept has refined the “missing self” model of NK recognition to include two basic steps: (1) loss of self, which may occur following infection or tumor transformation, as a first signal to alert NK cells that a cell is aberrant, and (2) an activating signal that is required to fully unleash the cytolytic activity of NK cells. Furthermore, over the past decade, accumulating evidence suggests that NK cells may not be as innate as once believed, but that individual NK-cell clones may show some target cell specificity (Malnati et al. 1995; Peruzzi et al. 1996), allowing them to play a critical early role in early antiviral control following infection with HIV.
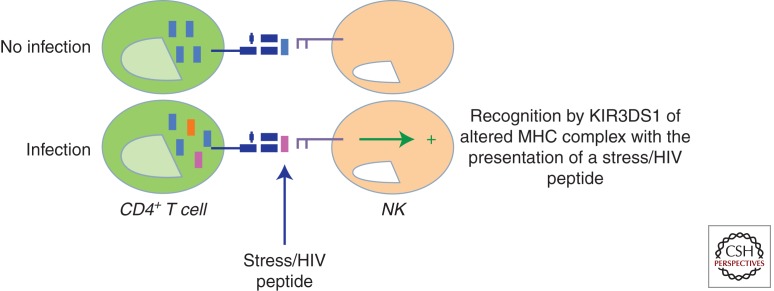
Figure 1.
A model of KIR3DS1+ natural killer (NK)-cell recognition of an HIV-infected target cell. Accumulating evidence suggests that specific amino acid changes in the peptides presented by major histocompatibility complex (MHC) class I can have a profound impact on KIR recognition of peptide/MHC complexes. Along these lines, it is plausible that a viral or stress peptide generated during infection presented by Bw4-80I may alter the affinity of the activating KIR3DS1 receptor expressed on NK cells for its putative ligand, resulting in the potent activation of NK cells and rapid elimination of virally infected cells.
NK CELLS IN HIV
The first immunomodulators in the acute cytokine storm (IFN-α and IL-15) (Stacey et al. 2009) are centrally involved in rapidly arming and activating NK cells following infection (Biron 1999). Thus, as anticipated, NK cells expand rapidly following acute infection, specifically in the acute seronegative window, with a preferential expansion of the cytolytic CD56dim NK-cell subpopulation (Alter et al. 2007b; Alter et al. 2009). However, to compensate for this early burst of innate cytolytic effector cells, HIV has devised multiple strategies to evade NK-cell recognition, indicating that these cells are able to place pressure on the virus.
HIV EVASION OF NK CELLS THROUGH Nef
Viruses have evolved multiple strategies to evade the immune system, including NK-cell recognition, suggesting a role for these cells in the early response to infection (Lodoen and Lanier 2005). Many viruses have specifically evolved strategies to down-regulate MHC class I from the surface of infected cells in an effort to avoid CD8+ T-cell recognition. However, this loss of MHC class I renders infected cells vulnerable to NK-cell-mediated recognition through inhibitory NK-cell receptors. Viruses such as cytomegalovirus (CMV) have evolved a compensatory repertoire of MHC class I homologs aimed at providing inhibitory signals to NK cells (Cerwenka and Lanier 2001b; Arase et al. 2002). Whereas CMV is a large DNA virus that has the opportunity to accommodate multiple genes for the evasion of both innate and adaptive immune responses, HIV is a small RNA virus that encodes only nine genes. Yet a number of studies have shown that HIV uses a single nonstructural gene, Nef, to evade both the innate and adaptive immune response.
Most notably, HIV-1 Nef protein triggers the accelerated endocytosis or retention of MHC class I molecules in the Golgi, resulting in reduced MHC class I expression on the surface of infected cells (Schwartz et al. 1996), thereby preventing recognition by HIV-specific CD8+ T cells. However, reduced MHC class I expression may alert NK cells of a possible infection. Interestingly, Nef may overcome both CD8- and NK-cell-mediated recognition by down-regulating the dominant T-cell receptor ligands HLA-A and -B molecules, while sparing the dominant inhibitory KIR2D ligands, HLA-C (Le Gall et al. 1998; Cohen et al. 1999). However, HLA-A appear to be down-regulated robustly, as compared with HLA-B (Cohen et al. 1999). These data strongly suggest that Nef has evolved a means to spare some KIR ligands, allowing it to strike a balance between T- and NK-cell evasion.
Loss of MHC class I expression is not sufficient to trigger NK-cell destruction of an HIV-infected cell, but requires a second activating signal. Viral infection often results in the up-regulation of the stress-inducible ligands for the activating c-type lectin NK-cell receptor NKG2D (Raulet 2003). These NKG2D-stress ligands, the MHC class I-related chain-A and -B (MIC-A/B) or UL-16 binding proteins-1, -2, and -3 (ULBP-1/2/3), are homologs of MHC class I alleles that are typically expressed following tumor transformation or infection (Raulet 2003). Recent studies reveal that the expression of MIC and ULBP on human tumor cells is sufficient to overcome the inhibitory effects of MHC class I expression (Zhang et al. 2005). To circumvent this activity, the HIV Nef protein has evolved the capacity to prevent the expression of some NKG2D ligands, such as MIC A, ULBP-1, and -2, at the surface of infected cells (Cerboni et al. 2007). It appears then that Nef regulation of host protein expression targets two host defense mechanisms, one involving KIRs and the other NKG2D.
A ROLE FOR KIR IN MODULATING HIV DISEASE PROGRESSION
KIRs can be divided into four groups based on two features: the number of extracellular domains (two domain [2D] or three domain [3D] and the length of the cytoplasmic tail (long [L] or short [S]). The length of the cytoplasmic domains dictates whether the receptor is activating or inhibitory, as long-tail KIRs contain immunoreceptor tyrosine-based inhibition motifs (ITIMs) that deliver strong inhibitory signals, whereas the short cytoplasmic tails associate with molecules that contain immunoreceptor tyrosine-based activation motifs (ITAMs) (Lanier et al. 1998). In addition to differences in gene content, most KIR genes show allelic polymorphism as well (Shilling et al. 2002; Carrington and Norman 2003).
Both epidemiological data and genome-wide association studies (GWASs) have pointed to a central role for particular MHC class I alleles in modulating the rate of disease progression (Carrington and O’Brien 2003; Fellay et al. 2007), the majority of which are encoded by the MHC class 1-B locus. Most of the protective HLA-B alleles express the Bw4 epitope, the primary ligands for KIR3DL1. Given the remarkable homology between alleles of KIR3DL1 and its activating counterpart KIR3DS1, epidemiological studies aimed at defining whether these three-domain KIR had any role in modulating disease progression were tested (Martin et al. 2002, 2007). Interestingly, both the activating and a subset of inhibitory variants of this KIR gene had a profound impact on modulating HIV disease progression in the context of their putative MHC ligands. Furthermore, duplications and deletions within the 3DL1/S1 segment have been observed (Martin et al. 2003), resulting in KIR haplotypes that can have zero or two copies of the KIR3DL1/S1 gene, and increasing doses of KIR3DS1 in the presence of KIR3DL1 and its putative ligand are associated with more robust control of HIV viremia in early disease (K Pelak and DG Goldstein, pers. comm.). These results suggest that NK cells may contribute to control through KIRs through at least two different mechanisms, one modulated by inhibitory receptors and a second mediated by an activating receptor, and that the activating and inhibitory receptors may interact to promote enhanced control of HIV viral replication.
KIR3DS1-MEDIATED CONTROL OF HIV
A number of studies have highlighted the impact of particular KIR/MHC combinations on HIV-1 disease outcome (Martin et al. 2002, 2007; Jennes et al. 2006). Martin et al. showed that subjects that coexpressed the activating KIR3DS1 allele in conjunction with its putative MHC class I ligand, Bw4 alleles with an isoleucine at position 80 of the peptide-binding groove (Bw4-80I) (Barber et al. 1997), progressed significantly more slowly toward AIDS than individuals that do not have this compound genotype (Martin et al. 2002). Although the physical interaction between KIR3DS1 and HLA-Bw4-80I molecules has yet to be shown, this genetic epistasis suggests that this KIR/MHC interaction confers some antiviral signal to NK cells to allow them to control HIV infection more effectively.
Functional data support the interaction between KIR3DS1 and Bw4-80I, as KIR3DS1+ NK cells degranulated more potently in response to HIV-infected Bw4-80I+ CD4+ T cells and suppressed viral replication in a Bw4-80I-dependent manner (Alter et al. 2007a). Additionally, these KIR3DS1+ NK cells expanded robustly following acute HIV infection (Alter et al. 2009), but only in subjects that coexpressed Bw4-80I, further suggesting that KIR3DS1 may receive proliferative signals from its putative ligand early on following infection, allowing NK cells expressing this receptor to expand robustly to help contain early viral replication. Moreover, NK cells derived from individuals that encoded for KIR3DS1 responded more potently to HLA-class I negative target cells than NK cells from KIR3DS1neg subjects (Long et al. 2008). Although KIR3DS1 alone was sufficient to confer elevated NK-cell responsiveness to class I devoid targets, NK-cell responses were strongest among individuals that coexpressed KIR3DS1 and Bw4-80I (Long et al. 2008). Finally, elevated KIR3DS1 transcripts were identified in persistently negative but highly exposed individuals, suggesting that KIR3DS1 may also be involved in protection from infection (Ravet et al. 2007). Taken together, these epidemiological and functional data support a role for KIR3DS1+ NK cells in restricting HIV infection in a “specific” manner in individuals that coexpress its putative ligand Bw4-80I.
Although a physical interaction has yet to be observed between KIR3DS1 and Bw4-80I, epidemiological and functional evidence strongly support that these two molecules are likely to interact either directly or indirectly to activate NK cells during HIV infection. Several potential scenarios may underlie this enigmatic interaction, including the possibility that a viral or stress peptide generated during infection presented by Bw4-80I may alter the affinity of the activating 3DS1 for its putative ligand (Fig. 2). Although data exist demonstrating that amino acid variation within a peptide, particularly at positions 7 and 8, can dramatically alter inhibitory KIR recognition of MHC class I complexes on a target cell, little is known about the particular changes in the MHC class I bound peptide that may alter activating KIR binding and activation. However, recent data now suggest that KIR3DS1 may in fact recognize discrete amino acids within the HIV proteome, as distinct footprints have now been identified that emerge preferentially in individuals that express this activating KIR (G Alter, unpubl.). Like the escape mutations that emerge in CD8+ T-cell-restricted epitopes, it is plausible that KIR-associated footprints may also reflect NK-restricted antiviral pressure. Alternatively, data from the murine model of Ly49p-mediated protection in murine cytomegalovirus (MCMV) infection suggest that instead of a peptide, the activating Ly49p NK-cell receptor interacts with its putative ligand, H2-Dk, only in the presence of a third, undefined protein (Lee et al. 2001). Overall, these data suggest that the affinity of 3DS1 for Bw4-80I may be altered during HIV infection, either by a stress/viral peptide or coactivating protein, resulting in potent NK-cell activation. The attraction of the latter possibility is that it implies nonspecificity of 3DS1 for HIV, which is what is expected for these innate immune receptors.
Figure 2.
A model of two-step NK-cell activation. NK-cell killing of target cells is tightly regulated by a balance of activating and inhibitory signals delivered through the arsenal of NK-cell receptors expressed on the surface of a given NK-cell clone. NK cells survey the body’s cells for normal MHC class-I expression, delivering a potent inhibitory signal to NK cells through inhibitory KIRs (1). Although the missing self-hypothesis states that the loss of MHC class I should trigger NK-cell killing of a target cell, this loss of inhibition is not sufficient to release the cytolytic activity of NK cells (2). Instead, an activating signal (including a stress ligand), to tip the balance toward activation, releases the full cytolytic power of a given NK-cell clone.
3DL1-MEDIATED CONTROL OF HIV
In addition to 3DS1, epidemiological studies later showed that additional inhibitory allotypes of 3DL1 are also associated with slower HIV disease progression (Martin et al. 2007). Distinct 3DL1 allotypes are expressed at variable levels on the surface of NK cells (Yawata et al. 2006), resulting in differing NK-cell functional potencies. Among the 3DL1 allotypes, three subclassifications have been defined: (1) high-expressing alleles that are associated with potent NK-cell effector functions in the presence of MHC-devoid target cells (3DL1*001, *002, *005, *008, *015, and *020), (2) low-expressing alleles that are associated with weaker NK-cell responsiveness to the same target cells (3DL1*005, *007, and *009), and (3) a nonexpressing allotype with unknown functional properties (3DL1*004). Interestingly, 3DL1 alleles expressed at high levels or not expressed at all were associated with slower HIV-1 disease progression, when coexpressed with Bw4-80I alleles (Martin et al. 2007).
Although the role of the nonexpressed 3DL1*004 allele remains an enigma, an explanation has been proposed for the high-expressed 3DL1 alleles. In 2005, a breakthrough was achieved in our understanding of the influence of KIR on NK cell function. In addition to the role of inhibitory KIR in monitoring for normal expression of MHC class I on the surface of cells (“missing self” hypothesis), a series of reports indicated that both Ly49 in mice and KIR in humans regulate NK-cell function by recognition of self-MHC class I providing signals for functional competence of the NK cell during development, a process called “licensing” (Fernandez et al. 2005; Kim et al. 2005; Anfossi et al. 2006; Kim et al. 2008). These studies suggest that NK cells undergo a self-MHC class I-dependent maturation process that delivers a positive signal resulting in the ability of NK cells to distinguish self from autologous target cells that have lost MHC class I (Kim et al. 2005; Anfossi et al. 2006). This model helped explain the fraction of NK cells in the periphery that are hyporesponsive, which are the subgroup of NK cells that lack inhibitory KIR for self, and are not educated to respond against aberrant targets (Anfossi et al. 2006). Additionally, more detailed models termed “arming” or “tuning” helped to refine the licensing model, taking into account the balance between activating MHC-binding receptors that are sometimes expressed in the absence or lower levels of inhibitory self-binding receptors. In these models, the investigators proposed that the presence of a dominant inhibitory signal during development helps to “arm” an NK cell, whereas lack of inhibition and/or excessive activation leads to disarming (Fernandez et al. 2005) or tuning (Salcedo et al. 1998) of NK-cell responsiveness, resulting in the accumulation of a subset of hyporesponsive cells.
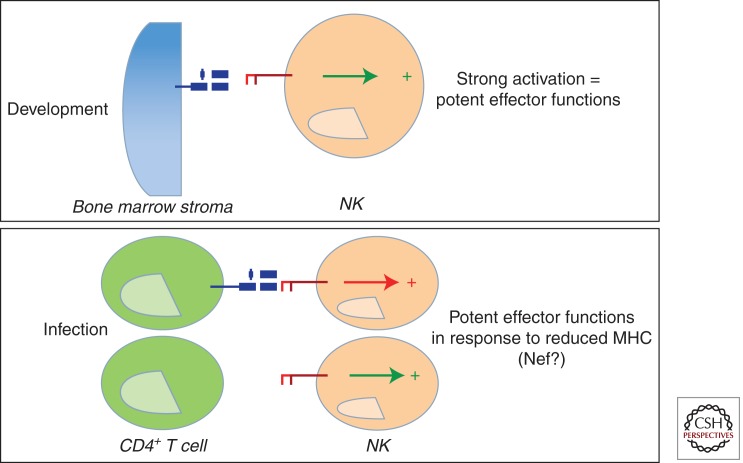
Thus, KIR3DL1 protection may be related to NK-cell education, where higher expression of KIR3DL1 on a developing NK cell in the presence of its ligand may result in the generation of a larger pool of functionally competent cytolytic cells, which on infection may respond more aggressively (Fig. 3). This possibility relates to the “missing self” hypothesis in that cells expressing higher levels of 3DL1 are expected to require a greater number of KIR/MHC interactions to inhibit such a cell, so they may be more sensitive to small losses of MHC class I following infection, responding vigorously to the target.
Figure 3.
A model of KIR3DL1+ NK-cell recognition of an HIV-infected target cell. Given that inhibitory KIRs have been recently implicated in NK-cell education, in such a way that inhibitory KIRs expressed at higher levels are associated with the generation of more functional NK-cell clones, it is possible that the expression of KIR3DL1 at higher levels on a developing NK cell in the presence of its ligand may result in the generation of a larger pool of functionally competent cytolytic cells. These more functionally competent cells may then respond more aggressively on HIV infection to cells that have lost MHC class I ligands, that are down-regulated by the HIV Nef protein.
A POTENTIAL ROLE FOR TWO-DOMAIN KIRs IN CONTROL OF HIV?
GWASs in large cohorts of HIV-infected individuals identified a number of single nucleotide polymorphisms (SNPs) associated with slower HIV disease progression, all of which mapped to a single region of the human genome on chromosome 6 located within the MHC. Among these SNPs, the GWASs confirmed previous epidemiological data demonstrating a protective role for the HLA-B allele B*57 (Carrington and O’Brien 2003), but also identified a number of additional SNPs, including one located 35 kb upstream of HLA-C (Thomas et al. 2009). The protective variant is associated with increased HLA-C expression on the surface of CD3+ T cells (Fellay et al. 2007; Thomas et al. 2009). Interestingly, the protective effect of this SNP could not be assigned to a specific HLA-C allele or phylogenetically related subgroup (Thomas et al. 2009), suggesting a potential non-CD8-dependent protective mechanism. As HLA-C alleles serve as ligands for KIR2D receptors (Vitale et al. 1995; Stewart et al. 2005), several groups have now begun to speculate that this protective effect in HIV infection is NK-cell-dependent through the interaction of KIR2D with its ligand. Based on the NK-cell education models, HLA-C alleles expressed at higher levels on the surface of a cell during development may generate more potent cytolytic NK cells (Kim et al. 2008; Brodin et al. 2009), but this possibility remains to be answered.
KIRs DRIVE VIRAL EVOLUTION
Most recently, efforts to define the mechanism by which NK cells may contribute to HIV viral control have sought to determine whether NK cells may recognize and place pressure on the virus directly in vivo. Historically, the identification of “footprints,” amino acid substitutions in the viral proteome that accumulate specifically in the presence of specific HLA-class I alleles, have been regarded as a marker of CD8+ T-cell pressure (Allen et al. 2000). Likewise, recent data have shown that similar footprints arise in the HIV proteome in the presence of distinct KIR genes (Alter et al. 2011). These data suggest that like T cells, NK cells may also recognize specific regions of the HIV virus, placing pressure on the virus.
How can KIRs see specific regions of the HIV proteome? Several lines of evidence suggest that affinity changes between KIRs and histocompatibility leukocyte antigen (HLA) class I may be induced by the peptide bound in the MHC class I binding groove. Crystal structures of KIR/MHC class I complexes show that KIR interacts with the α1 and α2 helix of MHC class I and makes direct contact with the carboxy-terminal portion of the bound peptide (Boyington et al. 2000; Fan et al. 2001). The impact of the bound peptide on KIR/MHC interactions has further been examined in a number of studies demonstrating that particular amino acid changes in the peptide, particularly at positions 7 or 8, results in the abrogation of inhibition through KIR, resulting in target cell lysis (Correa and Raulet 1995; Malnati et al. 1995; Peruzzi et al. 1996; Rajagopalan and Long 1997; Zappacosta et al. 1997; Fadda et al. 2010). Thus, it is possible that whereas self-peptides bound to MHC class I provide a strong inhibitory signal to the inhibitory KIR, particular viral peptides produced during infection may bind differentially to KIR, whereby decreased binding to an inhibitory KIR may trigger “missing self” NK-cell activation, or increased binding to an activating KIR may activate NK-cell cytotoxicity. This direct KIR-mediated antiviral pressure may drive the virus to incorporate “escape mutations” aimed at evading this form of innate recognition (Alter et al. 2011). However, the overall impact of this specific innate immune response has yet to be defined.
CONCLUSIONS
Over the past two decades, significant advances have been achieved in our basic understanding of the role of innate immunity in the control of viral infections. Moreover, we have come to appreciate that this arm of the immune response may directly contribute to antiviral control but may also play a significant role in modulating the quality of the ensuing adaptive immune response. In the context of HIV infection, mounting epidemiologic data strongly implicate a role for NK cells in antiviral control, underscored by the fact that these innate immune cells expand robustly in response to TLR-induced DC-secreted cytokines and have now been shown to specifically place pressure on HIV in vivo. The failure of recent HIV-1 vaccine trials to induce protective immunity in humans has highlighted our lack of understanding of the correlates of immune protection in HIV-1 infection. Therefore, new therapeutic strategies aimed at harnessing the power of the innate immune response, and particular NK cells, may provide a new approach aimed at enhancing the quality of immune control induced via vaccination.
ACKNOWLEDGMENTS
This project was funded, in whole or in part, by the National Cancer Institute, National Institutes of Health (NIH), contract no. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. This research was also supported, in part, by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Editors: Frederic D. Bushman, Gary J. Nabel, and Ronald Swanstrom
Additional Perspectives on HIV available at www.perspectivesinmedicine.org
REFERENCES
- Allen TM, O’Connor DH, Jing P, Dzuris JL, Mothe BR, Vogel TU, Dunphy E, Liebl ME, Emerson C, Wilson N, et al. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407: 386–390 [DOI] [PubMed] [Google Scholar]
- Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C, et al. 2007a. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med 204: 3027–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter G, Teigen N, Ahern R, Streeck H, Meier A, Rosenberg ES, Altfeld M 2007b. Evolution of innate and adaptive effector cell functions during acute HIV-1 infection. J Infect Dis 195: 1452–1460 [DOI] [PubMed] [Google Scholar]
- Alter G, Rihn S, Walter K, Nolting A, Martin M, Rosenberg ES, Miller JS, Carrington M, Altfeld M 2009. HLA class I subtype-dependent expansion of KIR3DS1+ and KIR3DL1+ NK cells during acute human immunodeficiency virus type 1 infection. J Virol 83: 6798–6805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, Oniangue-Ndza C, Martin M, Li B, Khakoo SI, et al. 2011. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature 476: 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, Breso V, Frassati C, Reviron D, Middleton D, et al. 2006. Human NK cell education by inhibitory receptors for MHC class I. Immunity 25: 331–342 [DOI] [PubMed] [Google Scholar]
- Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296: 1323–1326 [DOI] [PubMed] [Google Scholar]
- Barber LD, Percival L, Arnett KL, Gumperz JE, Chen L, Parham P 1997. Polymorphism in the α 1 helix of the HLA-B heavy chain can have an overriding influence on peptide-binding specificity. J Immunol 158: 1660–1669 [PubMed] [Google Scholar]
- Bashirova AA, Martin MP, McVicar DW, Carrington M 2006. The killer immunoglobulin-like receptor gene cluster: Tuning the genome for defense. Annu Rev Genomics Hum Genet 7: 277–300 [DOI] [PubMed] [Google Scholar]
- Beignon AS, McKenna K, Skoberne M, Manches O, Dasilva I, Kavanagh DG, Larsson M, Gorelick RJ, Lifson JD, Bhardwaj N 2005. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor-viral RNA interactions. J Clin Invest 115: 3265–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron CA 1999. Initial and innate responses to viral infections—Pattern setting in immunity or disease. Curr Opin Microbiol 2: 374–381 [DOI] [PubMed] [Google Scholar]
- Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD 2000. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature 405: 537–543 [DOI] [PubMed] [Google Scholar]
- Brennan J, Mager D, Jefferies W, Takei F 1994. Expression of different members of the Ly-49 gene family defines distinct natural killer cell subsets and cell adhesion properties. J Exp Med 180: 2287–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P, Lakshmikanth T, Johansson S, Karre K, Hoglund P 2009. The strength of inhibitory input during education quantitatively tunes the functional responsiveness of individual natural killer cells. Blood 113: 2434–2441 [DOI] [PubMed] [Google Scholar]
- Carrington M, Norman P 2003. The KIR gene cluster. NCBI, Bethesda, MD [Google Scholar]
- Carrington M, O’Brien SJ 2003. The influence of HLA genotype on AIDS. Annu Rev Med 54: 535–551 [DOI] [PubMed] [Google Scholar]
- Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M 1994. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med 180: 1235–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerboni C, Neri F, Casartelli N, Zingoni A, Cosman D, Rossi P, Santoni A, Doria M 2007. Human immunodeficiency virus 1 Nef protein downmodulates the ligands of the activating receptor NKG2D and inhibits natural killer cell-mediated cytotoxicity. J Gen Virol 88: 242–250 [DOI] [PubMed] [Google Scholar]
- Cerwenka A, Lanier LL 2001a. Ligands for natural killer cell receptors: Redundancy or specificity. Immunol Rev 181: 158–169 [DOI] [PubMed] [Google Scholar]
- Cerwenka A, Lanier LL 2001b. Natural killer cells, viruses and cancer. Nat Rev Immunol 1: 41–49 [DOI] [PubMed] [Google Scholar]
- Ciccone E, Pende D, Viale O, Di Donato C, Tripodi G, Orengo AM, Guardiola J, Moretta A, Moretta L 1992. Evidence of a natural killer (NK) cell repertoire for (allo) antigen recognition: Definition of five distinct NK-determined allospecificities in humans. J Exp Med 175: 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, Baltimore D 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10: 661–671 [DOI] [PubMed] [Google Scholar]
- Correa I, Raulet DH 1995. Binding of diverse peptides to MHC class I molecules inhibits target cell lysis by activated natural killer cells. Immunity 2: 61–71 [DOI] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303: 1529–1531 [DOI] [PubMed] [Google Scholar]
- Dorfman JR, Raulet DH 1996. Major histocompatibility complex genes determine natural killer cell tolerance. Eur J Immunol 26: 151–155 [DOI] [PubMed] [Google Scholar]
- Dorfman JR, Raulet DH 1998. Acquisition of Ly49 receptor expression by developing natural killer cells. J Exp Med 187: 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadda L, Borhis G, Ahmed P, Cheent K, Pageon SV, Cazaly A, Stathopoulos S, Middleton D, Mulder A, Claas FH, et al. 2010. Peptide antagonism as a mechanism for NK cell activation. Proc Natl Acad Sci 107: 10160–10165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan QR, Long EO, Wiley DC 2001. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat Immunol 2: 452–460 [DOI] [PubMed] [Google Scholar]
- Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, et al. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317: 944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH 2005. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood 105: 4416–4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, Vilches C, Parham P 2001. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol 166: 2992–3001 [DOI] [PubMed] [Google Scholar]
- Hanke T, Raulet DH 2001. Cumulative inhibition of NK cells and T cells resulting from engagement of multiple inhibitory Ly49 receptors. J Immunol 166: 3002–3007 [DOI] [PubMed] [Google Scholar]
- Hanke T, Takizawa H, McMahon CW, Busch DH, Pamer EG, Miller JD, Altman JD, Liu Y, Cado D, Lemonnier FA, et al. 1999. Direct assessment of MHC class I binding by seven Ly49 inhibitory NK cell receptors. Immunity 11: 67–77 [DOI] [PubMed] [Google Scholar]
- Hanke T, Takizawa H, Raulet DH 2001. MHC-dependent shaping of the inhibitory Ly49 receptor repertoire on NK cells: Evidence for a regulated sequential model. Eur J Immunol 31: 3370–3379 [DOI] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S 2004. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303: 1526–1529 [DOI] [PubMed] [Google Scholar]
- Held W, Dorfman JR, Wu MF, Raulet DH 1996. Major histocompatibility complex class I-dependent skewing of the natural killer cell Ly49 receptor repertoire. Eur J Immunol 26: 2286–2292 [DOI] [PubMed] [Google Scholar]
- Hsu KC, Liu XR, Selvakumar A, Mickelson E, O’Reilly RJ, Dupont B 2002. Killer Ig-like receptor haplotype analysis by gene content: Evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol 169: 5118–5129 [DOI] [PubMed] [Google Scholar]
- Jennes W, Verheyden S, Demanet C, Adje-Toure CA, Vuylsteke B, Nkengasong JN, Kestens L 2006. Cutting edge: Resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J Immunol 177: 6588–6592 [DOI] [PubMed] [Google Scholar]
- Karre K 2002. NK cells, MHC class I molecules and the missing self. Scand J Immunol 55: 221–228 [DOI] [PubMed] [Google Scholar]
- Karre K, Ljunggren HG, Piontek G, Kiessling R 1986. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature 319: 675–678 [DOI] [PubMed] [Google Scholar]
- Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, French AR, Sunwoo JB, Lemieux S, Hansen TH, et al. 2005. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature 436: 709–713 [DOI] [PubMed] [Google Scholar]
- Kim S, Sunwoo JB, Yang L, Choi T, Song YJ, French AR, Vlahiotis A, Piccirillo JF, Cella M, Colonna M, et al. 2008. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc Natl Acad Sci 105: 3053–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier LL 1998. NK cell receptors. Annu Rev Immunol 16: 359–393 [DOI] [PubMed] [Google Scholar]
- Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH 1998. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature 391: 703–707 [DOI] [PubMed] [Google Scholar]
- Lee SH, Girard S, Macina D, Busa M, Zafer A, Belouchi A, Gros P, Vidal SM 2001. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat Genet 28: 42–45 [DOI] [PubMed] [Google Scholar]
- Le Gall S, Erdtmann L, Benichou S, Berlioz-Torrent C, Liu L, Benarous R, Heard JM, Schwartz O 1998. Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity 8: 483–495 [DOI] [PubMed] [Google Scholar]
- Ljunggren HG, Karre K 1990. In search of the “missing self”: MHC molecules and NK cell recognition. Immunol Today 11: 237–244 [DOI] [PubMed] [Google Scholar]
- Ljunggren HG, Van Kaer L, Ploegh HL, Tonegawa S 1994. Altered natural killer cell repertoire in Tap-1 mutant mice. Proc Natl Acad Sci 91: 6520–6524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodoen MB, Lanier LL 2005. Viral modulation of NK cell immunity. Nat Rev Microbiol 3: 59–69 [DOI] [PubMed] [Google Scholar]
- Long BR, Ndhlovu LC, Oksenberg JR, Lanier LL, Hecht FM, Nixon DF, Barbour JD 2008. Conferral of enhanced natural killer cell function by KIR3DS1 in early human immunodeficiency virus type 1 infection. J Virol 82: 4785–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenaka K, Juji T, Nakayama T, Wyer JR, Gao GF, Maenaka T, Zaccai NR, Kikuchi A, Yabe T, Tokunaga K, et al. 1999. Killer cell immunoglobulin receptors and T cell receptors bind peptide-major histocompatibility complex class I with distinct thermodynamic and kinetic properties. J Biol Chem 274: 28329–28334 [DOI] [PubMed] [Google Scholar]
- Malnati MS, Peruzzi M, Parker KC, Biddison WE, Ciccone E, Moretta A, Long EO 1995. Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science 267: 1016–1018 [DOI] [PubMed] [Google Scholar]
- Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, et al. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet 31: 429–434 [DOI] [PubMed] [Google Scholar]
- Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M 2003. Cutting edge: Expansion of the KIR locus by unequal crossing over. J Immunol 171: 2192–2195 [DOI] [PubMed] [Google Scholar]
- Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, et al. 2007. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 39: 733–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli E, Cicala C, Van Ryk D, Goode DJ, Macleod K, Arthos J, Fauci AS 2007. HIV-1 gp120 inhibits TLR9-mediated activation and IFN-α secretion in plasmacytoid dendritic cells. Proc Natl Acad Sci 104: 3396–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell LD, Wallace A, Middleton D, Curran MD 2002. A common KIR2DS4 deletion variant in the human that predicts a soluble KIR molecule analogous to the KIR1D molecule observed in the rhesus monkey. Tissue Antigens 60: 254–258 [DOI] [PubMed] [Google Scholar]
- Meier A, Chang JJ, Chan ES, Pollard RB, Sidhu HK, Kulkarni S, Wen TF, Lindsay RJ, Orellana L, Mildvan D, et al. 2009. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat Med 15: 955–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moesta AK, Abi-Rached L, Norman PJ, Parham P 2009. Chimpanzees use more varied receptors and ligands than humans for inhibitory killer cell Ig-like receptor recognition of the MHC-C1 and MHC-C2 epitopes. J Immunol 182: 3628–3637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moesta AK, Graef T, Abi-Rached L, Older Aguilar AM, Guethlein LA, Parham P 2010. Humans differ from other hominids in lacking an activating NK cell receptor that recognizes the C1 epitope of MHC class I. J Immunol 185: 4233–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretta A, Vitale M, Bottino C, Orengo AM, Morelli L, Augugliaro R, Barbaresi M, Ciccone E, Moretta L 1993. P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J Exp Med 178: 597–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretta A, Bottino C, Mingari MC, Biassoni R, Moretta L 2002. What is a natural killer cell? Nat Immunol 3: 6–8 [DOI] [PubMed] [Google Scholar]
- Murphy K, Travers P, Walport M 2011. Janeway's immunobiology, 8th ed. Garland Science, New York [Google Scholar]
- Norman PJ, Stephens HA, Verity DH, Chandanayingyong D, Vaughan RW 2001. Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics 52: 195–205 [DOI] [PubMed] [Google Scholar]
- Pantaleo G, Demarest JF, Schacker T, Vaccarezza M, Cohen OJ, Daucher M, Graziosi C, Schnittman SS, Quinn TC, Shaw GM, et al. 1997. The qualitative nature of the primary immune response to HIV infection is a prognosticator of disease progression independent of the initial level of plasma viremia. Proc Natl Acad Sci 94: 254–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peruzzi M, Parker KC, Long EO, Malnati MS 1996. Peptide sequence requirements for the recognition of HLA-B*2705 by specific natural killer cells. J Immunol 157: 3350–3356 [PubMed] [Google Scholar]
- Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330: 1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Long EO 1997. The direct binding of a p58 killer cell inhibitory receptor to human histocompatibility leukocyte antigen (HLA)-Cw4 exhibits peptide selectivity. J Exp Med 185: 1523–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulet DH 2003. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol 3: 781–790 [DOI] [PubMed] [Google Scholar]
- Raulet DH, Held W, Correa I, Dorfman JR, Wu MF, Corral L 1997. Specificity, tolerance and developmental regulation of natural killer cells defined by expression of class I-specific Ly49 receptors. Immunol Rev 155: 41–52 [DOI] [PubMed] [Google Scholar]
- Ravet S, Scott-Algara D, Bonnet E, Tran HK, Tran T, Nguyen N, Truong LX, Theodorou I, Barre-Sinoussi F, Pancino G, et al. 2007. Distinctive NK-cell receptor repertoires sustain high-level constitutive NK-cell activation in HIV-exposed uninfected individuals. Blood 109: 4296–4305 [DOI] [PubMed] [Google Scholar]
- Salcedo M, Andersson M, Lemieux S, Van Kaer L, Chambers BJ, Ljunggren HG 1998. Fine tuning of natural killer cell specificity and maintenance of self tolerance in MHC class I-deficient mice. Eur J Immunol 28: 1315–1321 [DOI] [PubMed] [Google Scholar]
- Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard JM 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat Med 2: 338–342 [DOI] [PubMed] [Google Scholar]
- Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, Parham P 2002. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol 168: 2307–2315 [DOI] [PubMed] [Google Scholar]
- Stacey AR, Norris PJ, Qin L, Haygreen EA, Taylor E, Heitman J, Lebedeva M, DeCamp A, Li D, Grove D, et al. 2009. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol 83: 3719–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CA, Laugier-Anfossi F, Vely F, Saulquin X, Riedmuller J, Tisserant A, Gauthier L, Romagne F, Ferracci G, Arosa FA, et al. 2005. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci 102: 13224–13229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storkus WJ, Alexander J, Payne JA, Cresswell P, Dawson JR 1989a. The α1/α2 domains of class I HLA molecules confer resistance to natural killing. J Immunol 143: 3853–3857 [PubMed] [Google Scholar]
- Storkus WJ, Alexander J, Payne JA, Dawson JR, Cresswell P 1989b. Reversal of natural killing susceptibility in target cells expressing transfected class I HLA genes. Proc Natl Acad Sci 86: 2361–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault S, Fromentin R, Tardif MR, Tremblay MJ 2009. TLR2 and TLR4 triggering exerts contrasting effects with regard to HIV-1 infection of human dendritic cells and subsequent virus transfer to CD4+ T cells. Retrovirology 6: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Apps R, Qi Y, Gao X, Male V, O’hUigin C, O’Connor G, Ge D, Fellay J, Martin JN, et al. 2009. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet 41: 1290–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P 1997. Human diversity in killer cell inhibitory receptor genes. Immunity 7: 753–763 [DOI] [PubMed] [Google Scholar]
- Vales-Gomez M, Reyburn H, Strominger J 2000. Interaction between the human NK receptors and their ligands. Crit Rev Immunol 20: 223–244 [PubMed] [Google Scholar]
- Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, Phillips JH, Lanier LL, Parham P 1997. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity 7: 739–751 [DOI] [PubMed] [Google Scholar]
- Vilches C, Parham P 2002. KIR: Diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol 20: 217–251 [DOI] [PubMed] [Google Scholar]
- Vitale M, Sivori S, Pende D, Moretta L, Moretta A 1995. Coexpression of two functionally independent p58 inhibitory receptors in human natural killer cell clones results in the inability to kill all normal allogeneic target cells. Proc Natl Acad Sci 92: 3536–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wende H, Colonna M, Ziegler A, Volz A 1999. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome 10: 154–160 [DOI] [PubMed] [Google Scholar]
- Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J 2000. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci 97: 4778–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt CS, Dewing C, Sayer DC, Uhrberg M, Parham P, Christiansen FT 1999. Population frequencies and putative haplotypes of the killer cell immunoglobulin-like receptor sequences and evidence for recombination. Transplantation 68: 1784–1789 [DOI] [PubMed] [Google Scholar]
- Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P 2006. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med 203: 633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama WM 2002. The search for the missing “missing-self” receptor on natural killer cells. Scand J Immunol 55: 233–237 [DOI] [PubMed] [Google Scholar]
- Yokoyama WM, Kehn PJ, Cohen DI, Shevach EM 1990. Chromosomal location of the Ly-49 (A1, YE1/48) multigene family. Genetic association with the NK 1.1 antigen. J Immunol 145: 2353–2358 [PubMed] [Google Scholar]
- Yu J, Heller G, Chewning J, Kim S, Yokoyama WM, Hsu KC 2007. Hierarchy of the human natural killer cell response is determined by class and quantity of inhibitory receptors for self-HLA-B and HLA-C ligands. J Immunol 179: 5977–5989 [DOI] [PubMed] [Google Scholar]
- Zappacosta F, Borrego F, Brooks AG, Parker KC, Coligan JE 1997. Peptides isolated from HLA-Cw*0304 confer different degrees of protection from natural killer cell-mediated lysis. Proc Natl Acad Sci 94: 6313–6318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang J, Wei H, Tian Z 2005. Imbalance of NKG2D and its inhibitory counterparts: How does tumor escape from innate immunity? Int Immunopharmacol 5: 1099–1111 [DOI] [PubMed] [Google Scholar]