Background: Enzymes controlling (de)palmitoylation of ion channels are poorly defined.
Results: Palmitoylation of BK channels by zDHHC22 and zDHHC23 and depalmitoylation by LYPLA1 and LYPLAL1 controls BK channel cell surface expression.
Conclusion: Acyl protein transferases and thioesterases display substrate specificity and control BK channel surface expression.
Significance: Understanding how channels are (de)palmitoylated is essential for defining the role of palmitoylation in ion channel physiology.
Keywords: Ion channels, Membrane transport, Potassium channels, Protein palmitoylation, Signal transduction, APT, Kcnma1, zDHHC
Abstract
Protein palmitoylation is rapidly emerging as an important determinant in the regulation of ion channels, including large conductance calcium-activated potassium (BK) channels. However, the enzymes that control channel palmitoylation are largely unknown. Indeed, although palmitoylation is the only reversible lipid modification of proteins, acyl thioesterases that control ion channel depalmitoylation have not been identified. Here, we demonstrate that palmitoylation of the intracellular S0–S1 loop of BK channels is controlled by two of the 23 mammalian palmitoyl-transferases, zDHHC22 and zDHHC23. Palmitoylation by these acyl transferases is essential for efficient cell surface expression of BK channels. In contrast, depalmitoylation is controlled by the cytosolic thioesterase APT1 (LYPLA1), but not APT2 (LYPLA2). In addition, we identify a splice variant of LYPLAL1, a homolog with ∼30% identity to APT1, that also controls BK channel depalmitoylation. Thus, both palmitoyl acyltransferases and acyl thioesterases display discrete substrate specificity for BK channels. Because depalmitoylated BK channels are retarded in the trans-Golgi network, reversible protein palmitoylation provides a critical checkpoint to regulate exit from the trans-Golgi network and thus control BK channel cell surface expression.
Introduction
Protein palmitoylation, the most common form of S-acylation, resulting from the modification of intracellular cysteine residues by the C16 lipid palmitate via a thioester bond, is emerging as a major post-translational modification to control the activity, trafficking, and function of many proteins, including ion channels (1–5). Indeed, an increasing number of human disorders is associated with dysregulation of protein palmitoylation ranging from cancer to mental retardation (1–5).
To date more than 40 different ion channel subunits have been reported to be palmitoylated (1). Because the thioester bond is labile, protein palmitoylation is the only reversible post-translational lipid modification of proteins, thus providing a potentially highly dynamic mechanism to control ion channel function. Palmitoylation has been implicated in the regulation of multiple stages of the ion channel lifecycle, from channel assembly and trafficking to control of protein-protein interactions and modulation by other signaling pathways (1). However, as for most other palmitoylated proteins, we know very little about the enzymes that palmitoylate or depalmitoylate ion channels. Protein palmitoylation is controlled by a family of ∼23 mammalian protein acyl transferases (zinc finger- and DHHC-domain containing proteins, zDHHC)2 that typically include a conserved cysteine rich domain (CRD) with the DHHC motif being critical for palmitoyltransferase activity (3, 4, 6). Although a limited number of zDHHCs have been reported to control ion channel palmitoylation (7–10) the repertoire of zDHHCs that control palmitoylation for any channel has not been systematically determined. Thus the extent to which zDHHC enzymes display substrate specificity toward ion channel subunits is largely unknown. Furthermore, enzymes that control depalmitoylation of ion channels, and most other proteins, are not defined. Two cytosolic acyl thioesterases (APT1 and APT2, genes LYPLA1 and LYPLA2, respectively) have been identified as palmitoyl thioesterases that depalmitoylate cytosolic cysteine residues of proteins (5, 11–14). However, whether these enzymes control ion channel depalmitoylation and/or display any substrate specificity for ion channels is not known. Addressing these questions is essential to understand the role of this major reversible post-translational modification in controlling ion channels.
We recently demonstrated that large conductance calcium- and voltage-activated potassium (BK) channels are palmitoylated in the intracellular S0–S1 loop (see Fig. 1A), and this regulates cell surface expression of the channel (15). However, the enzymes controlling palmitoylation of the S0–S1 loop and the mechanism by which palmitoylation controls BK channel surface expression is not known. In this study we have undertaken a systematic approach to (i) characterize whether specific zDHHC enzymes control palmitoylation and cell surface trafficking of BK channels, (ii) define whether BK channels are depalmitoylated by candidate cytosolic acyl thioesterases, and (iii) elucidate how reversible protein palmitoylation controls cell surface expression of the channel. We demonstrate that both zDHHCs and thioesterases display substrate specificity for the BK channel. We identify zDHHC22 and zDHHC23 as the palmitoyltransferases and LYPLA1 and LYPLAL1 as acyl thioesterases controlling BK channel palmitoylation. Furthermore, we reveal reversible protein palmitoylation is a critical step in controlling exit of BK channels from the trans-Golgi network (TGN) and thus a major checkpoint for regulating cell surface expression of the channel.
FIGURE 1.
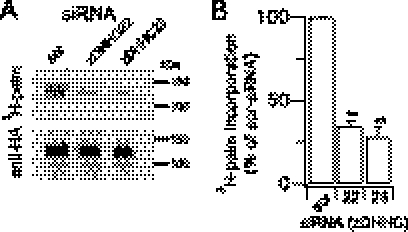
siRNA-based imaging screen reveals zDHHC22 and zDHHC23 as acyl transferases that palmitoylate the S0–S1 loop of BK channels. A, schematic of the pore-forming α-subunit indicating the intracellular S0–S1 loop and the previously identified palmitoylated cysteine residues Cys-53, Cys-54, and Cys-56. B, representative single confocal images of a -YFP fusion protein of the S0–S1 loop in HEK293 cells treated with a scrambled siRNA (scr-siRNA) or siRNAs targeting zDHHC22 or zDHHC23. Scale bars are 5 μm. C, bar graph of membrane expression of the S0–S1-YFP fusion protein, normalized to the scr-siRNA control, after zDHHC knockdown by siRNA. Data are means ± S.E. N >4, n >200. **, p < 0.01 decrease compared with scr-siRNA group, ANOVA with post-hoc Dunnetts test.
EXPERIMENTAL PROCEDURES
Expression Constructs
The generation of the full-length ZERO variant of BK channels (with and without N-terminal (extracellular) FLAG- and C-terminal (intracellular)-HA epitopes, the palmitoylation-deficient mutant C53:54:56A, the S0–S1-YFP fusion construct of the intracellular S0–S1 loop, and the zDHHC constructs have been described previously (10, 15). zDHHC22 was cloned from mouse brain and generated as an HA-tagged fusion protein. LYPLA1 (also known as APT1) as a CFP fusion protein was a generous gift of Prof. G. Schratt (16), and pSUPER vectors for expression of siRNA targeted to LYPLA1 were a generous gift from Prof. H. Waldmann (11). LYPLA2, LYPLAL1, and the in-frame splice variant LYPLAL1Δ66–80 were cloned from HEK293 cells and generated as CFP fusion proteins. Mutation of the “catalytic triad” of both LYPLA1 and LYPLAL1 to generate the catalytically inactive constructs LYPLA1D169A:H203 and LYPLAL1D173A:H211A was performed using QuikChange mutagenesis (Stratagene). The N-terminal HA-tagged GABABR1a(ASRR) mutant receptor subunit was a generous gift from Prof. L. Jan (17). The human HA-tagged PPT1 construct was from Dr. L. Chamberlain. All constructs were fully sequenced on both strands to verify sequence integrity.
Cell Culture, Transfection, RNA Extraction, and qRT-PCR
HEK293 cells were maintained, plated on glass coverslips, and transfected as described before (10, 18). For RNA interference, siRNAs were pre-designed and supplied by Qiagen or Sigma. In addition, for LYPLA1, knockdown was also performed using pSUPER shRNA vector. Knockdown used two independent siRNAs (10–20 nm of each siRNA) for each gene. siRNA transfection was performed using HyperFect (Qiagen) as described (10) or jetPRIME (Polyplus Transfection) according to the manufacturer's protocol. In all imaging and biochemical assays siRNA knockdown was monitored in parallel in each independent experiment. RNA was extracted using a High Pure RNA Isolation Kit (Roche Applied Science), cDNA was synthesized using the Transcriptor High Fidelity cDNA Synthesis Kit (Roche Applied Science), and then quantified in triplicate using the FS Universal SYBR Green Mastermix Rox (Roche Applied Science) on an ABIPrism 7000 real-time PCR machine as described (10). Data were normalized to endogenous β-actin levels (Qiagen primer set AT01680476). All qRT-PCR primers were previously validated with efficiencies calculated to be within 0.1 of the control with knockdown between 70 and 95% for all targets.
Imaging
For S0–S1-loop-YFP fusion experiments, HEK293 cells were fixed and plasma membrane versus intracellular distribution was analyzed as previously described (10). Cell surface labeling of full-length N-terminal FLAG-epitope-tagged BK channels in non-permeabilized cells was performed as described (15, 19) using mouse monoclonal anti-FLAG M2 antibody (Sigma, 1:100) and Alexa-546 (Molecular Probes, 1:1000). Cells were then fixed in 4% paraformaldehyde for 30 min, permeabilized with 0.3% Triton X-100 for 10 min, and blocked with phosphate-buffered saline containing 3% bovine serum albumin plus 0.05% Tween 20 for 1 h. The intracellular C-terminal HA epitope tag was probed with either anti-HA polyclonal rabbit antibody (Zymed Laboratories Inc., 1:500) followed by Alexa-647 (Molecular Probes, 1:1000). Cell surface and total labeling of the HA-tagged GABABR1a(ASRR) mutant receptor subunit was performed as above except that, under non-permeabilized conditions, the HA tag was first probed with anti-HA polyclonal rabbit antibody (Zymed Laboratories Inc., 1:500) followed by Alexa-546 (Molecular Probes, 1:1000) and following permeabilization with anti-HA antibody (1:500) followed by Alexa-647 (1:1000).
Confocal images were acquired on a Zeiss LSM510 laser scanning microscope, using a 63× oil Plan Apochromat (numerical aperture = 1.4) objective lens, in multitracking mode to minimize channel crosstalk. Cell surface expression of full-length channels, or receptor, was determined by quantitative immunofluorescence by calculating the surface to total channel protein ratio using ImageJ as described (15, 19).
For channel co-localization with intracellular compartments co-localization was assayed by co-transfection of the channel with the endoplasmic reticulum (ER) marker plasmid pdsRed-ER (Clontech), or staining for the TGN using mouse anti-TGN38 (BD Bioscience, 1:50) or recycling endosomes labeled using rabbit anti-Rab11 (Invitrogen, 1:50). Confocal images at Nyquist sampling rates were deconvolved using Huygens software (Scientific Volume imaging) and Pearson's correlation coefficient (R) determined using ImageJ (National Institutes of Health) as described (15, 19). An R value of +1 indicates 100% co-localization.
Palmitoylation Assays, Pulldown Assays, and Western Blotting
Transfected HEK293 cells were incubated in DMEM containing 10 mg/ml fatty acid free BSA for 30 min at 37 °C before incubation with 0.5 mCi/ml [3H]palmitic acid (PerkinElmer Life Sciences) for 4 h at 37 °C as described (10, 18). Cells were lysed in 150 mm NaCl, 50 mm Tris-Cl, 1% Triton X-100, pH 8.0 and centrifuged, and channel fusion proteins were captured using magnetic microbeads (μMACSTM epitope tag isolation kits, Miltenyi Biotech). Following extensive washing captured proteins were eluted, separated by SDS-PAGE, transferred to nitrocellulose membranes, dried, and exposed to light-sensitive film at −80 °C using a Kodak Biomax transcreen LE (Amersham Biosciences). The same membrane was then reprobed with a polyclonal HA antibody (Zymed Laboratories Inc., 1:1000).
Statistical Analysis
All data are presented as means ± S.E. with N = number of independent experiments and n = number of individual cells analyzed in imaging assays. Data were analyzed by ANOVA with the post-hoc Dunnett test with significance set at p < 0.05.
RESULTS AND DISCUSSION
S0–S1 Loop of the BK Channel Is Palmitoylated by zDHHC22 and zDHHC23 to Control Surface Expression
We have previously identified three cysteine residues (Cys-53, Cys-54, and Cys-56) within the intracellular S0–S1 loop of BK channels (Fig. 1A) that are palmitoylated and important for controlling cell surface expression of the full-length BK channel (15). Because palmitoylation of the S0–S1 loop targets a -YFP fusion protein of the isolated S0–S1 loop domain (S0–S1-YFP) to the plasma membrane, we exploited this construct to undertake an siRNA-based imaging screen (10) to identify zDHHCs that control palmitoylation of the S0–S1 domain (Fig. 1, B and C). In this assay, knockdown of zDHHCs 22 and 23 (by >70% as determined by qRT-PCR (10)) resulted in a significant reduction of expression of the S0–S1-YFP fusion protein at the plasma membrane (Fig. 1, B and C). Following knockdown, the S0–S1-YFP construct was largely localized in trafficking vesicles, recycling endosomes or lysosomes (data not shown). Paradoxically, knockdown of zDHHC 5, 7, or 17 resulted in a significant increase in localization at the plasma membrane (Fig. 1, B and C). However, in each case this was associated with a significant up-regulation of zDHHC 22 and/or 23 mRNA expression suggesting a level of compensatory feedback. For example, knockdown of DHHC17 resulted in a 2.8 ± 0.2-fold and 1.5 ± 0.1-fold up-regulation of zDHHC 22 and zDHHC 23 mRNA expression, respectively.
We next examined whether overexpression of the cognate zDHHCs would increase surface expression of the S0–S1-YFP construct. Overexpression of zDHHC23 resulted in a significant increase (by 144 ± 5.3% of S0–S1-YFP alone) in cell surface expression of the S0–S1-YFP construct. zDHHC23 predominantly co-localized with markers of the Golgi network (GM130, Pearson's R coefficient of 0.71 ± 0.01, n = 15) and TGN (TGN38, Pearson's R coefficient of 0.38 ± 0.02, n = 15). However, overexpression of zDHHC22 was not well tolerated by HEK293 cells, and thus the effect of overexpressing this palmitoyltransferase could not be reliably determined. In contrast, overexpression of other zDHHCs had no significant effect on surface expression of the S0–S1-YFP construct. For example, overexpression of zDHHC 4 or 17 resulted in surface expression that was 96.8 ± 9.2% and 89.7 ± 6.1% of S0–S1-YFP levels.
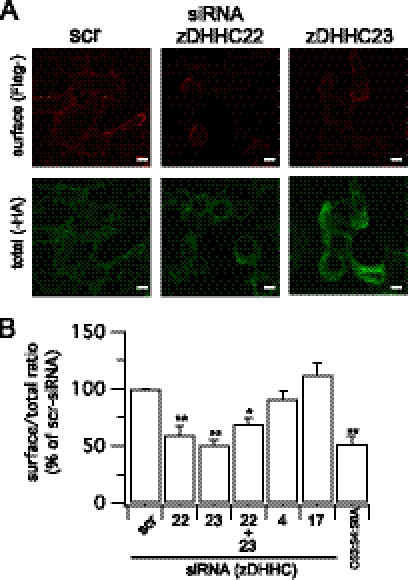
To test whether zDHHCs 22 and 23 play a functional role in controlling palmitoylation and cell surface expression of the full-length BK channel, we exploited the siRNA knockdown strategy in channels with an extracellular FLAG- epitope and intracellular -HA epitope (Fig. 1A) to quantify surface expression. In these assays we used the ZERO splice variant of the BK channel, in which the only sites for protein palmitoylation are within the S0–S1 loop and mutation of the S0–S1 loop palmitoylated cysteines reduces surface expression of the channel (15). Importantly, knockdown of either zDHHC 22 or 23 resulted in a significant reduction in palmitoylation of the full-length BK channel (Fig. 2, A and B), as determined by [3H]palmitate incorporation. In accordance with zDHHCs 22 and 23 being the major zDHHCs controlling palmitoylation of this site knockdown by siRNA also resulted in a significant reduction in cell surface expression of the full-length channel (Fig. 3, A and B). The inhibition of surface expression was not significantly different from that observed with ZERO channels in which all three palmitoylated cysteine residues in the S0–S1 loop were mutated to alanine (C53:54:56A construct). Furthermore, siRNA knockdown of zDHHC 22 or 23 had no significant effect on C53:54:56A surface expression further supporting the major role for these enzymes in controlling S0–S1 loop palmitoylation. Knockdown of zDHHC 22 and 23 together did not have an additive effect suggesting they operate within the same pathway. These data demonstrate that a very restricted subset of zDHHCs regulates palmitoylation of the S0–S1 loop to control surface expression of BK channels. Intriguingly these zDHHCs are distinct from the zDHHCs that control palmitoylation of the Stress regulated exon (STREX) splice variant insert in the C terminus of the BK channel (10) suggesting differential target specificity for zDHHCs even within the same protein.
FIGURE 2.
zDHHC22 and -23 control BK channel palmitoylation. A, representative fluorograph (top panel) of [3H]palmitate incorporation and corresponding Western blot (anti-HA) of the full-length BK channel ZERO variant immunoprecipitated from HEK293 cells following knockdown of the respective zDHHC. B, quantification of [3H]palmitate incorporation from experiments (n = 3) as in A. Data are means ± S.E. **, p < 0.01 decrease compared with scrambled (scr) group, ANOVA with post-hoc Dunnett's test.
FIGURE 3.
zDHHC22 and -23 control cell surface expression of BK channels. A, representative single confocal images of ZERO channel surface (non-permeabilized, Flag-) and total (permeabilized, -HA) labeling following knockdown of the respective zDHHC. Scale bars are 5 μm. B, quantification of the ZERO channel surface (Flag-) to total (-HA) ratio as in A (N >4, n >200) following knockdown of the respective zDHHC or using the palmitoylation-deficient BK channel mutant C53:54:56. Data are means ± S.E. **, p < 0.01 decrease compared with scrambled (scr) group, ANOVA with post-hoc Dunnett's test.
Acyl Protein Thioesterases LYPLA1 and LYPLAL1, but Not LYPLA2, Control BK Channel Depalmitoylation
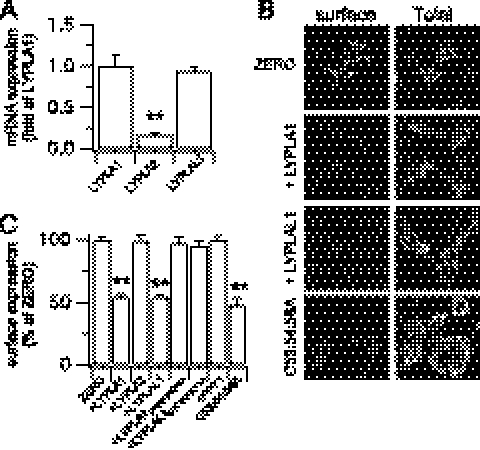
As palmitoylation is the only reversible lipid post-translational modification of proteins, due to the labile thioester linkage, we sought to identify the acyl palmitoyl thioesterases (APTs) that may mediate depalmitoylation of the BK channel S0–S1 loop. Thioesterases controlling depalmitoylation of any ion channel have not been determined however both APT1 (gene: LYPLA1) and APT2 (gene: LYPLA2) are likely candidates for thioesterases that deacylate cytosolic cysteine residues (5, 11–14). In addition, an APT1 homolog (APT1-like thioesterase, gene: LYPLAL1) has been described (5), although whether it functions as an APT is not clear. We first examined the mRNA expression of candidate cytosolic APTs in HEK293 cells using qRT-PCR. These data demonstrate robust mRNA expression for LYPLA1 and the APT1-like thioesterase (LYPLAL1) with significantly lower expression of LYPLA2 (Fig. 4A). We also identified a splice variant of LYPLAL1 that lacks 16 amino acids in the N terminus upstream of the predicted catalytic triad (LYPLAL1Δ66–80, accession: AY341430.1). This variant was expressed at ∼20% of the total LYPLAL1 mRNA transcript level. In agreement with previous studies, overexpression of fluorescently tagged constructs of these proteins revealed predominantly cytosolic distribution in HEK293 cells (12, 20).
FIGURE 4.
Overexpression of acyl thioesterases LYPLA1 and LYPLAL1, but not LYPLA2, reduces BK channel surface expression. A, mRNA expression of acyl thioesterases LYPLA1 and LYAPL2 and the homolog LYPLAL1 in HEK293 cells expressed as a fraction of LYPLA1 mRNA. B, representative single confocal images of full-length ZERO variant BK channels in HEK293 cells co-expressed with CFP- or -CFP-tagged LYPLA1 or LYPLAL1 and the palmitoylation-deficient BK channel mutant C53:54:56A. Surface (Flag-) and total (-HA) channel labeling was determined as in Fig. 2. Scale bars are 5 μm. C, quantitative immunofluorescence analysis of BK channel surface expression under different conditions (N > 5, n > 200/group) expressed as a percentage of the FLAG/HA ratio of ZERO. PPT1 is the lysosomal palmitoyl protein thioesterase 1. LYPLA1D169A:H203 and LYPLAL1D173A:H211A contain mutations in the respective catalytic triad required for thioesterase activity. Data are means ± S.E. **, p < 0.01 compared with ZERO, ANOVA with post-hoc Dunnett's test.
In initial experiments we asked whether knockdown (using siRNA or shRNA constructs) of these APTs increased surface expression of the ZERO variant channels. Even though knockdown of >70% could be achieved, no effects on channel surface expression were observed. We were unable to simultaneously knock down multiple APTs with high efficiency; thus we could not test if the lack of effect of knockdown of a single APT was due to redundancy and/or “basal” APT activity not being a major determinant of BK channel palmitoylation status. We thus implemented an overexpression strategy to define the role of APTs in BK channel depalmitoylation.
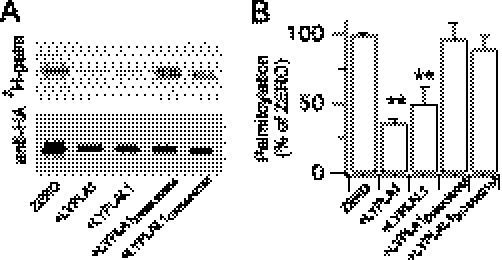
Overexpression of LYPLA1, or LYPLAL1, reduced surface expression of ZERO channels to a similar extent as observed with the palmitoylation-deficient C53:54:56A channels (Fig. 4, B and C). LYPLA1 or LYPLAL1 had no significant effect on total BK channel expression as determined from immunohistochemical (Fig. 4B) or Western blot (Fig. 5) analysis. In contrast, although LYPLA2 was robustly expressed the enzyme had no significant effect on channel surface expression (Fig. 4, B and C). As a further test of APT specificity for BK channel trafficking, overexpression of the lysosomal palmitoyl protein thioesterase PPT1 had no effect on BK channel surface expression (Fig. 4C). Intriguingly, the LYPLAL1Δ66–80 splice variant also had no significant effect (channel surface expression was 98.2 ± 6.3% of control) suggesting that this variant is not functional against BK channels. The splice variant deletion is in the N terminus upstream of the predicted catalytic triad; thus whether the variant is non-functional or has distinct target specificity remains to be determined. Taken together, these data reveal substrate specificity by LYPLA1 and LYPLAL1 for the cysteine residues in the S0–S1 loop of the BK channel.
FIGURE 5.
Acyl thioesterases LYPLA1 and LYPLAL1, but not LYPLA2, depalmitoylate BK channels. A, representative fluorograph (top panel) of [3H]palmitate incorporation and corresponding Western blot (anti-HA) of BK channels co-expressed with LYPLA1, LYPLAL1, or their catalytic triad mutants. B, quantification of [3H]palmitate incorporation from experiments (n = 3) as in A. Data are means ± S.E. **, p < 0.01 compared with ZERO, ANOVA with post-hoc Dunnett's test.
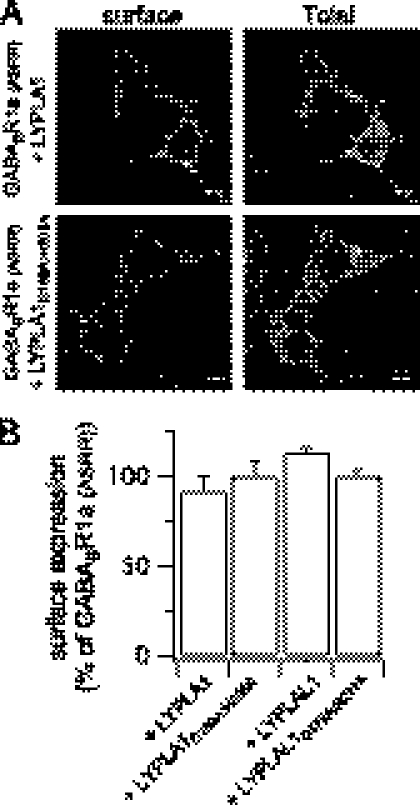
To verify that the effect of APT overexpression was a result of the catalytic activity of the respective enzymes and mediated by depalmitoylation of the BK channel per se we undertook several approaches. Firstly, we mutated key residues in the catalytic triad of LYPLA1 (D169A:H203A mutant) that abolishes thioesterase activity (14). Sequence alignment revealed conservation of the catalytic triad in LYPLAL1 thus we also mutated corresponding residues in LYPLAL1 (D173A:H211A mutant). Both mutant enzymes were robustly expressed, however they had no significant effect on ZERO channel surface expression (Fig. 4, B and C). Secondly, both LYPLA1 and LYPLAL1 overexpression resulted in a significant reduction in [3H]palmitate incorporation into the ZERO variant (Fig. 5, A and B). In contrast, the corresponding catalytic triad mutants D169A:H203A and D173A:H211A had no significant effect (Fig. 5, A and B). Thirdly, we assayed the effect of LYPLA1 and LYPLAL1 overexpression on the palmitoylation-deficient channel mutant C53:54:56A; neither APT had any significant effect on C53:54:56A surface expression. Surface expression of the C53:54:56A mutant channel in the presence of LYPLA1 was 105.3 ± 2.6% (n = 39) and for LYPLAL1 102 ± 4.3% (n = 43) of the C53:54:56A mutant expressed alone. This supports that the effect was dependent upon depalmitoylation of the BK channel per se rather than exerting effects on general trafficking. As a further control to exclude the possibility that overexpression of LYPLA1 or LYPLAL1 may inhibit surface expression of transmembrane proteins through indirect effects, we examined surface trafficking of the γ-aminobutyric acid metabotropic receptor subunit, GABABR1a (17). The GABABR1a subunit is normally retained in the endoplasmic reticulum by an RXRR-dependent ER retrieval and retention mechanism. Mutation of this motif results in a subunit (GABABR1a(ASRR)) that efficiently trafficks to the cell surface (17, 19) (Fig. 6A). Overexpression of LYPLA1 or LYPLAL1 or their catalytic triad mutants had no significant effect on the cell surface expression of the GABABR1a(ASRR) subunit (Fig. 6, A and B). Taken together, these data provide the first functional evidence for APTs that control ion channel depalmitoylation, demonstrate substrate specificity of APTs, and reveal a critical role for reversible protein palmitoylation in controlling BK channel cell surface expression.
FIGURE 6.
LYPLA1 and LYPLAL1 have no effect of GABABR1a(ASRR) cell surface trafficking. A, representative single confocal images of the HA-tagged trafficking competent GABABR1a(ASRR) receptor subunit in HEK293 cells co-expressed with -CFP-tagged LYPLA1, LYPLAL1, or their corresponding catalytic triad mutants LYPLA1D169A:H203 and LYPLAL1D173A:H211A. The extracellular HA tag was labeled under non-permeabilized conditions (surface) and following permeabilization (Total) in the same cells as described under “Experimental Procedures.” Scale bars are 5 μm. B, quantitative immunofluorescence analysis of receptor surface expression under different conditions (N > 3, n > 48/group) expressed as a percentage of the surface expression of GABABR1a(ASRR). Data are means ± S.E.
Channel Depalmitoylation Retards BK Channel Exit from the TGN
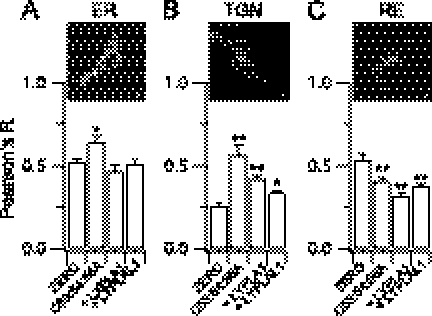
Because the acyl protein thioesterases are predominantly cytosolic proteins (12, 20), whereas the majority of acyltransferases are largely localized at either the ER or Golgi membranes (3, 4, 21), we sought to determine the subcellular location where depalmitoylation of the BK channel is likely to occur to regulate cell surface expression. To address this issue, we undertook quantitative co-localization analysis of the BK channel with markers of the ER, TGN, and recycling endosomes (Fig. 7, A–C, RE). Compared with the ZERO BK channel variant the palmitoylation-deficient mutant C53:54:56A mutant channel displayed a small but significantly increased co-localization with the ER (Fig. 7A). However, this was not recapitulated by co-expressing ZERO with either LYPLA1 or LYPLAL1. This suggests that palmitoylation of BK channels is required for efficient exit from the ER but that the ER is not a major site at which the depalmitoylating acyl thioesterases control channel trafficking. Low co-localization of mutant or wild-type channels with a Golgi marker GM130 precluded quantitative analysis of Golgi co-localization per se. However, depalmitoylation, using either the C53:54:56A mutant or ZERO co-expressed with LYPLA1 or LYPLAL1, resulted in a significantly increased retention of depalmitoylated channels in the TGN (Fig. 7B). Furthermore, this was mirrored by a significant decrease in co-localization of channels with recycling endosomes (Fig. 7C). These data suggest that depalmitoylation at the TGN by LYPLA1 and LYPLAL1 is a critical checkpoint for control of BK channel cell surface expression.
FIGURE 7.
Acyl thioesterases retard BK channels in the TGN. Quantitative co-localization analysis of ZERO variant BK channels co-expressed with LYPLA1 or LYPLAL1 and the C53:54:56A palmitoylation-deficient BK channel with markers of A, the endoplasmic reticulum (ER - pdsRed-ER); B, trans-golgi network (TGN - TGN38); or C, recycling endosomes (RE - rab11). Inset images are representative confocal images indicating distribution of the respective compartment marker, the outline indicates the plasma membrane in the cells stained for the TGN and RE, respectively. Scale bars are 5 μm. Data are expressed as the respective Pearson's correlation coefficient (R) from deconvolved data, where a value of +1 indicates 100% co-localization. Data are means ± S.E., N > 4, n > 48/group. *, p < 0.05; **, p < 0.01 compared with ZERO group, ANOVA with post-hoc Dunnett's test.
Taken together, these data demonstrate that palmitoylation plays a significant role at both the ER and TGN. However, efficient export from the ER requires palmitoylation of the BK channel, but this step is not regulated by acyl thioesterases. In contrast, efficient exit from the TGN is also dependent upon BK channel palmitoylation, and importantly this step may be dynamically controlled by the acyl thioesterases LYPLA1 and LYPLAL1.
This study is the first systematic analysis of the repertoire of acyl transferases and acyl thioesterases that control an ion channel. Indeed, the repertoire of enzymes responsible for controlling palmitoylation of most proteins is not known. Our work lends considerable support to the hypothesis that zDHHC enzymes can display substrate specificity (4, 8, 10, 22–25) in contrast to some reports with peripheral membrane proteins (26). First, we demonstrate that zDHHC22 and zDHHC23 specifically control palmitoylation of the S0–S1 loop of BK channels to control surface trafficking. Second, the zDHHCs that palmitoylate the S0–S1 loop are distinct from zDHHCs 3, 5, 7, 9, and 17 that palmitoylate an alternatively spliced insert Stress regulated exon (STREX) in the C terminus of the BK channel (10, 18). Thus, as suggested for other proteins, zDHHCs may discriminate between different sites of palmitoylation even within the same protein. The molecular basis for this specificity remains a significant challenge to address.
Little is known about the zDHHCs we have identified that control BK channel S0–S1 loop palmitoylation. zDHHC23 (also known as NIDD) assembles with neuronal nitric oxide in presynaptic compartments (27, 28). zDHHC22 has similarities to several of the functional yeast zDHHCs, such as Akr1 (29, 30), which contain a canonical DHHC motif but lack several of the cysteine and histidine residues commonly found in the cysteine rich domain of other mammalian zDHHCs.
Our data also provide the first direct evidence for depalmitoylation of an ion channel and support the hypothesis that acyl thioesterases may also display substrate specificity (5, 12, 31). LYPLA1, but not LYPLA2, regulated BK channel depalmitoylation. In addition, our data reveal that the homolog, LYPLAL1 (previously referred to as acyl protein thioesterase-like 1, APTL1 (5)) also depalmitoylates the BK channel. Although LYPLAL1 has been proposed as a candidate acyl thioesterase, to date evidence to support this has been limited (5, 32). Indeed, although the recent crystal structure of LYPLAL1 reveals a similar fold structure to LYPLA1 with a conserved catalytic triad (32) the crystal structure reveals a shallow active site for LYPLAL1 and in vitro data support preference for short-chain lipid substrates. This would imply LYPLAL1 may have limited function as a palmitoyl thioesterase, and in vitro LYPLAL1 was reported to be unable to depalmitoylate N-Ras (32). Whether the discrepancy between the crystal structure and our observations, that LYPLAL1 depalmitoylated the BK channel in cells, dependent upon a functional catalytic triad, reflects differences in substrate specificity perhaps dependent upon specific protein-protein interactions, or other mechanisms, remains to be resolved. In this regard, although LYPLA1 assembles as a dimer the crystal packing for LYPLAL1 suggests it exists as a monomer (32). Furthermore, we also found that the LYPLAL1Δ66–80 splice variant of LYPLAL1 does not depalmitoylate BK channels, suggesting that it is either non-functional as a thioesterase or splicing controls target specificity.
Importantly, reversible protein palmitoylation regulates surface expression of BK channels by controlling exit from the TGN. zDHHC23 being predominantly localized at the Golgi apparatus and TGN, in agreement with previous studies (3, 4, 21), whereas the thioesterases were predominantly cytosolic (12, 20), supports a role for dynamic regulation of BK channel palmitoylation at the level of the TGN. A challenge for the future is to understand how the cognate zDHHCs and thioesterases identified here are regulated and how this impacts on BK channel trafficking. For example, expression of zDHHC23 is modified in chronic pain states (28), LYPLA1 expression is controlled by microRNAs in neurons (16, 33), and LYPLAL1 expression is up-regulated in obesity (34). Clearly, understanding such mechanisms should provide fundamental new insights into the physiological role of BK channels in a range of systems and disorders.
Acknowledgments
We are grateful to Dr. Trudi Gillespie of the IMPACT imaging facility in the Centre for Integrative Physiology for assistance in confocal imaging and Rhiannon Seymour for assistance with some of the initial S0–S1-YFP measurements. We are grateful to Prof. M. Fukata for the generous gifts of murine DHHC constructs, Prof. G. Schratt for APT1 clones, Prof. H. Waldmann for APT1 shRNA vectors, Prof. L. Jan for the GABABR1a(ASXX) construct, and Dr. L. Chamberlain for the PPT1 construct.
This work was supported by a Wellcome Trust program grant (to M. J. S., H. K., and P. R.).
- zDHHC
- acyl protein palmitoyl transferase gene family
- APT
- acyl protein thioesterase
- BK
- large conductance calcium- and voltage-activated potassium channel
- HEK293
- human embryonic kidney 293
- TGN
- trans-Golgi network
- qRT
- quantitative reverse transcription
- ER
- endoplasmic reticulum
- ANOVA
- analysis of variance.
REFERENCES
- 1. Shipston M. J. (2011) Ion channel regulation by protein palmitoylation. J. Biol. Chem. 286, 8709–8716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Linder M. E., Deschenes R. J. (2007) Palmitoylation. Policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 3. Fukata Y., Fukata M. (2010) Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 11, 161–175 [DOI] [PubMed] [Google Scholar]
- 4. Greaves J., Chamberlain L. H. (2011) DHHC palmitoyl transferases. Substrate interactions and (patho)physiology. Trends Biochem. Sci. 36, 245–253 [DOI] [PubMed] [Google Scholar]
- 5. Zeidman R., Jackson C. S., Magee A. I. (2009) Protein acyl thioesterases. Mol. Membr. Biol. 26, 32–41 [DOI] [PubMed] [Google Scholar]
- 6. Fukata M., Fukata Y., Adesnik H., Nicoll R. A., Bredt D. S. (2004) Identification of PSD-95 palmitoylating enzymes. Neuron 44, 987–996 [DOI] [PubMed] [Google Scholar]
- 7. Rathenberg J., Kittler J. T., Moss S. J. (2004) Palmitoylation regulates the clustering and cell surface stability of GABAA receptors. Mol. Cell Neurosci. 26, 251–257 [DOI] [PubMed] [Google Scholar]
- 8. Hayashi T., Rumbaugh G., Huganir R. L. (2005) Differential regulation of AMPA receptor subunit trafficking by palmitoylation of two distinct sites. Neuron 47, 709–723 [DOI] [PubMed] [Google Scholar]
- 9. Hayashi T., Thomas G. M., Huganir R. L. (2009) Dual palmitoylation of NR2 subunits regulates NMDA receptor trafficking. Neuron 64, 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tian L., McClafferty H., Jeffries O., Shipston M. J. (2010) Multiple palmitoyltransferases are required for palmitoylation-dependent regulation of large conductance calcium- and voltage-activated potassium channels. J. Biol. Chem. 285, 23954–23962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dekker F. J., Rocks O., Vartak N., Menninger S., Hedberg C., Balamurugan R., Wetzel S., Renner S., Gerauer M., Schölermann B., Rusch M., Kramer J. W., Rauh D., Coates G. W., Brunsveld L., Bastiaens P. I., Waldmann H. (2010) Small-molecule inhibition of APT1 affects RAS localization and signalling. Nat. Chem. Biol. 6, 449–456 [DOI] [PubMed] [Google Scholar]
- 12. Tomatis V. M., Trenchi A., Gomez G. A., Daniotti J. L. (2010) Acyl-protein thioesterase 2 catalyzes the deacylation of peripheral membrane-associated GAP-43. PLoS ONE 5, e15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duncan J. A., Gilman A. G. (1998) A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein α subunits and p21(RAS). J. Biol. Chem. 273, 15830–15837 [DOI] [PubMed] [Google Scholar]
- 14. Devedjiev Y., Dauter Z., Kuznetsov S. R., Jones T. L., Derewenda Z. S. (2000) Crystal structure of the human acyl protein thioesterase I from a single X-ray data set to 1.5 A. Structure 8, 1137–1146 [DOI] [PubMed] [Google Scholar]
- 15. Jeffries O., Geiger N., Rowe I. C., Tian L., McClafferty H., Chen L., Bi D., Knaus H. G., Ruth P., Shipston M. J. (2010) Palmitoylation of the S0-S1 linker regulates cell surface expression of voltage- and calcium-activated potassium (BK) channels. J. Biol. Chem. 285, 33307–33314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Siegel G., Obernosterer G., Fiore R., Oehmen M., Bicker S., Christensen M., Khudayberdiev S., Leuschner P. F., Busch C. J., Kane C., Hübel K., Dekker F., Hedberg C., Rengarajan B., Drepper C., Waldmann H., Kauppinen S., Greenberg M. E., Draguhn A., Rehmsmeier M., Martinez J., Schratt G. M. (2009) A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11, 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma D., Zerangue N., Lin Y. F., Collins A., Yu M., Jan Y. N., Jan L. Y. (2001) Role of ER export signals in controlling surface potassium channel numbers. Science 291, 316–319 [DOI] [PubMed] [Google Scholar]
- 18. Tian L., Jeffries O., McClafferty H., Molyvdas A., Rowe I. C., Saleem F., Chen L., Greaves J., Chamberlain L. H., Knaus H. G., Ruth P., Shipston M. J. (2008) Palmitoylation gates phosphorylation-dependent regulation of BK potassium channels. Proc. Natl. Acad. Sci. U.S.A. 105, 21006–21011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen L., Jeffries O., Rowe I. C., Liang Z., Knaus H. G., Ruth P., Shipston M. J. (2010) Membrane trafficking of large conductance calcium-activated potassium channels is regulated by alternative splicing of a transplantable, acidic trafficking motif in the RCK1-RCK2 linker. J. Biol. Chem. 285, 23265–23275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hirano T., Kishi M., Sugimoto H., Taguchi R., Obinata H., Ohshima N., Tatei K., Izumi T. (2009) Thioesterase activity and subcellular localization of acylprotein thioesterase 1/lysophospholipase 1. Biochim. Biophys. Acta 1791, 797–805 [DOI] [PubMed] [Google Scholar]
- 21. Ohno Y., Kihara A., Sano T., Igarashi Y. (2006) Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim. Biophys. Acta 1761, 474–483 [DOI] [PubMed] [Google Scholar]
- 22. Roth A. F., Wan J., Bailey A. O., Sun B., Kuchar J. A., Green W. N., Phinney B. S., Yates J. R., 3rd, Davis N. G. (2006) Global analysis of protein palmitoylation in yeast. Cell 125, 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang K., Sanders S., Singaraja R., Orban P., Cijsouw T., Arstikaitis P., Yanai A., Hayden M. R., El-Husseini A. (2009) Neuronal palmitoyl acyl transferases exhibit distinct substrate specificity. FASEB J. 23, 2605–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsutsumi R., Fukata Y., Noritake J., Iwanaga T., Perez F., Fukata M. (2009) Identification of G protein α subunit-palmitoylating enzyme. Mol. Cell Biol. 29, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Greaves J., Gorleku O. A., Salaun C., Chamberlain L. H. (2010) Palmitoylation of the SNAP25 protein family. Specificity and regulation by DHHC palmitoyl transferases. J. Biol. Chem. 285, 24629–24638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rocks O., Gerauer M., Vartak N., Koch S., Huang Z. P., Pechlivanis M., Kuhlmann J., Brunsveld L., Chandra A., Ellinger B., Waldmann H., Bastiaens P. I. (2010) The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141, 458–471 [DOI] [PubMed] [Google Scholar]
- 27. Saitoh F., Tian Q. B., Okano A., Sakagami H., Kondo H., Suzuki T. (2004) NIDD, a novel DHHC-containing protein, targets neuronal nitric-oxide synthase (nNOS) to the synaptic membrane through a PDZ-dependent interaction and regulates nNOS activity. J. Biol. Chem. 279, 29461–29468 [DOI] [PubMed] [Google Scholar]
- 28. Chen M. L., Cheng C., Lv Q. S., Guo Z. Q., Gao Y., Gao S. F., Li X., Niu S. Q., Shi S. X., Shen A. G. (2007) Altered gene expression of NIDD in dorsal root ganglia and spinal cord of rats with neuropathic or inflammatory pain. J. Mol. Histol. 39, 125–133 [DOI] [PubMed] [Google Scholar]
- 29. Mitchell D. A., Vasudevan A., Linder M. E., Deschenes R. J. (2006) Protein palmitoylation by a family of DHHC protein S-acyltransferases. J. Lipid Res. 47, 1118–1127 [DOI] [PubMed] [Google Scholar]
- 30. Mitchell D. A., Mitchell G., Ling Y., Budde C., Deschenes R. J. (2010) Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J. Biol. Chem. 285, 38104–38114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duncan J. A., Gilman A. G. (2002) Characterization of Saccharomyces cerevisiae acyl-protein thioesterase 1, the enzyme responsible for G protein α subunit deacylation in vivo. J. Biol. Chem. 277, 31740–31752 [DOI] [PubMed] [Google Scholar]
- 32. Bürger M., Zimmermann T. J., Kondoh Y., Stege P., Watanabe N., Osada H., Waldmann H., Vetter I. R. (2012) Crystal structure of the predicted phospholipase LYPLAL1 reveals unexpected functional plasticity despite close relationship to acyl protein thioesterases. J. Lipid Res. 53, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Banerjee S., Neveu P., Kosik K. S. (2009) A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron 64, 871–884 [DOI] [PubMed] [Google Scholar]
- 34. Steinberg G. R., Kemp B. E., Watt M. J. (2007) Adipocyte triglyceride lipase expression in human obesity. Am. J. Physiol. Endocrinol. Metab. 293, E958-E964 [DOI] [PubMed] [Google Scholar]