Abstract
Aurora kinases are a family of mitotic kinases that play important roles in the tumorigenesis of a variety of cancers including pancreatic cancer. A number of Aurora kinase inhibitors (AKIs) are currently being tested in preclinical and clinical settings as anti-cancer therapies. However, the antitumor activity of AKIs in clinical trials has been modest. In order to improve the antitumor activity of AKIs in pancreatic cancer, we utilized a kinome focused RNAi screen to identify genes that, when silenced, would sensitize pancreatic cancer cells to AKI treatment. A total of 17 kinase genes were identified and confirmed as positive hits. One of the hits was the platelet-derived growth factor receptor, alpha polypeptide (PDGFRA), which has been shown to be overexpressed in pancreatic cancer cells and tumor tissues. Imatinib, a PDGFR inhibitor, significantly enhanced the anti-proliferative effect of ZM447439, an Aurora B specific inhibitor, and PHA-739358, a pan-Aurora kinase inhibitor. Further studies showed that imatinib augmented the induction of G2/M cell cycle arrest and apoptosis by PHA-739358. These findings indicate that PDGFRA is a potential mediator of AKI sensitivity in pancreatic cancer cells.
Introduction
Due to the lack of early diagnosis and effective therapeutic modalities, pancreatic cancer remains a devastating disease with a five-year survival of less than 5% (1). Gemcitabine, a nucleoside analog which was approved for the treatment of patients with locally advanced or metastatic pancreatic cancer, only has moderate therapeutic effects with an average median survival of 6 months. The FDA approved erlotinib plus gemcitabine combination treatment for locally advanced, inoperable or metastatic pancreatic cancer only demonstrated a moderate survival benefit in a Phase III study (median 6.24 months vs. 5.91 months) (2). Most recently, a Phase I/II clinical trial showed promising activity of the gemcitabine plus nab-paclitaxel combination in patients with advanced pancreatic cancer (3). This regimen is currently being evaluated in a randomized Phase III trial. In addition, the FOLFIRINOX (5-FU/leucovorin, irinotecan, and oxaliplatin) regimen was shown to have improved survival compared to gemcitabine alone in a Phase III trial, albeit, with more toxicity (4). To further improve the treatment outcome and increase the survival rate of pancreatic cancer patients, better tumor markers for diagnosis and new therapeutics are urgently needed.
Aurora kinases are serine-threonine kinases that play important, yet distinct, roles in mitosis (5, 6). There are three Aurora kinases, Aurora A, B, and C in mammals. Since its identification in the late 1990’s (7, 8), the human Aurora A kinase gene has been reported to be overexpressed and/or amplified in many malignant diseases including breast, colon, bladder, ovarian, melanoma, and pancreatic cancers (9, 10). Deregulation of Aurora A and Aurora B has been linked to advanced tumor stages and poor prognosis of patients (reviewed in (9)). Aurora A is shown to be oncogenic and play an important role in cancer initiation and progression (11). Although the role of Aurora B in tumorigenesis is less clear, many studies support an association between Aurora B and malignant transformation (11, 12). In pancreatic cancer, we and others have shown that both Aurora A and Aurora B kinases are highly expressed in tumor tissues and the Aurora A gene is amplified in tumor cells (13-15). In recent years, several small-molecule Aurora kinase inhibitors have been developed and shown to exhibit antitumor activity in both pancreatic cancer cell lines and xenograft models (16, 17). A number of Aurora kinase inhibitors including VX680 (MK-0457) (18, 19), AZD1152 (20, 21), MLN8237 (22, 23), PHA-739358 (24, 25), either have been or are currently in Phase I/II clinical development. Although some of the AKIs have shown evidence of clinical activity, the overall patient response has been modest. For instance, the clinical activity of PHA-739358, a pan-Aurora kinase inhibitor with a dominant Aurora B kinase inhibition related cellular phenotype (24), has largely been consistent with cytostatic effects, with the best response so far being stable disease in about 23.7% of evaluable patients (26). Recently, a phase I study of PHA-739358 in patients with advanced solid tumors showed that one patient with refractory small cell lung cancer had an objective response lasting 23 weeks (27). Although the reason for the modest clinical activity of AKIs could be multifaceted, one of the most plausible possibilities is that patient tumors may harbor additional genetic changes (i.e. context of vulnerability) that may affect the sensitivity of tumor cells to AKI therapies. For example, it has been shown that Aurora A protects ovarian cancer cells from cisplatin-induced apoptosis by activating the Akt pathway in p53 wildtype cells (28). This indicates that cisplatin might increase the activity of AKIs in p53 wildtype cells and combining inhibitors of the Akt pathway and AKIs might be synergistic. We hypothesize that similar contexts of vulnerability might also exist in pancreatic cancer cells. By identifying such contexts of vulnerability we will be able to develop either new biomarkers for selecting patient populations for AKI therapies or new AKI-based combination therapies that increase patient response. With the advances in genome-based techniques, particularly in the area of high-throughput RNAi screening, it is possible to carry out systematic searches for the context of vulnerabilities for individual targeted therapies.
As kinases are major control points in cellular signaling and are considered to be highly druggable, the kinome has been the target of large scale functional genomics with RNAi screens and of drug discovery efforts, especially in cancer therapeutics (29). The aim of this study was to identify kinases that, when inhibited, sensitize pancreatic cancer cells to the treatment of AKIs. To achieve this aim, we carried out a screen using the Human Validated Kinase siRNA Set from Qiagen in combination with an Aurora kinase inhibitor (AKI-1) previously reported by Lampson et al (30) in pancreatic cells. Positive hits were further subjected to confirmation/validation studies using multiple AKIs in multiple pancreatic cell lines. Using this approach we identified a list of 17 genes that, when silenced by siRNA oligonucleotides, sensitize pancreatic cancer cells to the treatment of AKIs. These genes present potential new targets against which agents that enhance the antitumor activity of AKIs can be developed.
Materials and Methods
Chemicals and Reagents
VX-680 (MK-0457), sorafenib, and imatinib were purchased from ChemieTek, LLC (Indianapolis, IN). ZM447439 was purchased from Tocris Bioscience (Ellisville, MI). Aurora kinase inhibitor-1 (AKI-1) and MP235 were synthesized in our lab (16, 31). PHA-739358 was purchased from Selleck Chemicals (Houston, TX). Etopside was purchased from Sigma-Aldrich (St. Louis, MO). The chemical structures of the Aurora kinase inhibitors used in this study are shown in Supplementary Figure S1.
The Human Validated Kinase siRNA Set (HVKS) V2 was purchased from Qiagen (Valencia, CA). This siRNA library contains two validated siRNA oligonucleotides for each of 588 kinase and kinase related genes (a total of 1,176 siRNA oligonucleotides; see www.qiagen.com). Additional siRNA oligonucleotides targeting individual genes or negative (non-targeting) siRNA oligonucleotides were also purchased from Qiagen. The siRNA oligonucleotides were dissolved in RNase free water at 10μM stock concentration and stored at −80°C until use.
Cell culture
BxPC-3, Mia PaCa-2, AsPC-1, CFPAC-1, PANC-1 and SU 86.86 pancreatic cancer cell lines were purchased from American Type Tissue Culture Collection (ATCC, Manassas, VA) and cultured in RPMI 1640 supplemented with 10% (v/v) fetal bovine serum, 100 units/ml penicillin, and 100μg/ml streptomycin (Invitrogen, Carlsbad, CA).
Cell line identities were verified by STR profiling (32) using the AmpFISTR Identifiler PCR amplification kit (Applied Biosystems, Foster City, CA). This method simultaneously amplifies 15 STR loci and Amelogenin in a single tube, using 5 dyes, 6-FAM™, JOE™, NED™, PET™, and LIZ™ which are then separated on a 3100 Genetic Analyzer (Applied Biosystems). GeneMapper ID v3.2. Software was used for analysis (Applied Biosystems). AmpFISTR control DNA and the AmpFISTR allelic ladder were run concurrently. Results were compared to published STR sequences from the ATCC. The STR profiling is repeated once a cell line has been passaged more than 6 months after previous STR profiling.
Optimization of transfection conditions for HT-siRNA screen
To find the most optimal transfection reagent and conditions for pancreatic cancer cells, we first tested a panel of transfection reagents with two siRNA oligonucleotides, a non-silencing negative control siRNA (Qiagen) and a positive control siRNA (UBB1, Qiagen) (33) in a panel of pancreatic cancer cell lines, including AsPC-1, BxPC-3, CFPAC-1, Mia PaCA-2, PANC-1, and SU86.86. The panel of transfection reagents includes Lipofectamine 2000 (Invitrogen), Lipofectamine RANiMax (Invitrogen), siLentFect (Bio-Rad, Hercules, CA), Oligofectamine (Invitrogen). The siRNA (9 ng in 2μl) was first printed onto solid white 384-well plates using a Biomek FX liquid handling system (Beckman Coulter, Fullerton, CA). The transfection reagents were diluted in OptiMEM (Invitrogen) at five different ratios (1:2, 1:3, 1:5, 1:7, and 1:8) from 200 nl/well. The final volumes of the transfection reagents tested were therefore 100, 66.7, 40, 28.6, and 25 nl/well. Diluted transfection reagents (20μl) were added to the 384-well plates containing siRNA oligonucleotides and were allowed to complex for 20 minutes. Equal volume of cells was added in growth media resulting in 1,000-1,200 cells per well depending on growth characteristics of the cell lines (determined in separate experiments). The cells were then incubated in a CO2 incubator at 37°C for 96 hours at which point 25μl of CellTiter-Glo® reagent (Promega, Madison, WI) was added to each well to determine cell viability. The luminescence intensities were obtained for each plate using an Analyst GT microplate reader (Molecular Devices, Sunnyvale, CA). Percent viability values were calculated by comparing the intensity units from each treatment condition with that of the untreated controls. The transfection reagent and conditions that give the highest difference in cell viability between the Non-silencing siRNA (negative control purchased from Qiagen) and the lethal siRNA (positive control) were then chosen for the subsequent HT RNAi screening in combination with AKIs.
Selection of cell lines and AKIs for HT-siRNA screening
To select a cell line and an AKI that would maximize our chances of finding siRNA hits that are specific to Aurora kinase inhibition, we first evaluated three different AKIs in a panel of pancreatic cancer cells, including AsPC-1, BxPC-3, CFPAC-1, Mia PaCa-2, PANC-1, and SU86.86, using the same growth and assay conditions as those for the siRNA transfection. The three AKIs were VX-680 (34), MP235(35, 36), and AKI-1 (30). All three AKIs have been shown to inhibit Aurora kinases in cell-free assays with nM IC50s and induce phenotypes in cancer cells that are consistent with the inhibition of Aurora kinases (30, 34, 36). The cells were treated with varying doses of AKIs. Cell viability was determined 96 hours after adding the drug using CellTiter-Glo® Assay. The cell line that provided the most consistent dose response results with a modest sensitivity crossing all the AKIs tested was selected as the screening cell line (BxPC3). The AKI showing relatively smooth dose response curves with modest activity crossing all cell lines tested would be selected as the screening compound (AKI-1).
High-throughput RNAi screening
The siRNA library was printed onto 384-well cell culture plates (Corning, Lowell, MA) at 2μl/well (the final concentration of siRNA in the screening assay was 16nM). A set of control siRNA oligonucleotides including GFP siRNA, All Star Negative Control siRNA, Non-silencing siRNA, and the UBB1 positive control siRNA (all purchased from Qiagen) were also included. Each of library siRNA sequences was printed in duplicates and the control siRNAs were printed in quadruplicates. A set of siRNA buffer only wells were also printed for inclusion of negative controls such as buffer and transfection reagent only controls. For each compound concentration one set of the plates printed with siRNA library were used. The assay procedure is shown in Supplementary Figure S2. Briefly, on Day 1, 20μl of SilentFect diluted in serum free medium was added onto the pre-printed siRNA library plates and incubated for 30 minutes at room temperature. 20μl of BxPC-3 cells (1,200 cells/well) were then added to each well of the plates. Following an overnight (16 hours) incubation in a CO2 incubator, 5μl of the Aurora kinase inhibitor, AKI-1, at appropriate concentrations was added into each well of the plates. To ensure the quality of the positive hits to be identified, we designed a screening scheme with five different AKI-1 drug concentrations, EC10, EC20, EC30, EC50 (concentrations required to achieve 10%, 20%, 30% and 50% of the maximal growth inhibitory effect, respectively), as well as a vehicle control, all of which were calculated based on the non-regression curve fitting equations of the dose-response curves using the Prism 5 software (GraphPad Inc., La Jolla, CA) in the screening cell line. The zero, EC10 and EC30 concentration sets were performed in duplicate to ensure the screening quality (this makes a total of nine screening sets of the siRNA library). After adding drugs, the cells were further incubated for 96 hours in a CO2 incubator. On Day 6, the viability of cells in each well was measured by the CellTiter-Glo® Luminescent Cell Viability Assay (Promega) as per manufacturer’s instructions.
Hit selection
Hit selection was based on the detection of changes between two drug-dose response curves (DDRC) generated from individual siRNA and the negative siRNA control. In general, the following steps were involved: data normalization, filtration of toxic siRNA and outliers, IC50 calculation, and ranking. The data normalization was done by fixing the DDRC of plate median and shifting the sample DDRC so that both curves have identical origins at drug dose 0 (siRNA only control). Such normalization allowed us to calculate the size of enclosed area formed by two DDRCs (sample and plate median). Toxic siRNA oligonucleotides (percentage cell survival for siRNA only control is less than 50%) were removed from further analysis. EC50s and EC30s (drug concentration required to achieve 50% and 30% of the maximal cell growth inhibitory effect) for plate median and each siRNA were calculated by fitting the data to a sigmoid dose-response model using nonlinear regression with the Matlab software (2007a, The MathWorks Inc). The EC30 and EC50 shift between sample DDRC and the DDRC of plate median was then used to rank the siRNA. For the validation experiments, since more potential sensitizer hits were tested, we used a negative siRNA control (Non-silencing siRNA from Qiagen) as a reference instead of plate median in data normalization.
Confirmation screening
From primary screening, we identified kinase genes targeted by siRNA that mediate sensitivity of AKI-1 in the BxPC-3 cell line. To exclude the possibility of siRNA with biological off-target effects, we performed a confirmation screen using four siRNA sequences per gene in combination with AKI-1 in the BxPC-3 cell line and defined confirmed hits as those kinases whose inhibition was synthetically lethal with AKIs in pancreatic cancer cells with concordant results from two or more unique siRNAs.
Drug combination treatment
Cells were seeded at 2,000 cells/well in 96-well plates and allowed to grow overnight. On the second day, a serial dilution of the Aurora kinase inhibitors (ZM447439 or PHA-739358) combined with fixed concentrations of the second drug (imatinib or sarofenib) as indicated in the figures was added to cells and incubated for 96 hours. At the end of drug incubation, cell viability was determined using the SRB (Sulforhodamine B) assay.
Sulforhodamine B (SRB) assay
After drug treatment, culture media were removed from the 96-well plate and the cells were fixed by adding 65 μl of 10% trichloroacetic acid (TCA) solutions and incubating for 30 minutes at 4°C. Cells were then rinsed five times with deionized water and stained with 0.04% SRB solution (40 μl/well) for 30 minutes at room temperature. Cells were then washed five times with 1% acetic acid to remove unbound dye, and left to air dry. The bound SRB dye was then solubilized by adding 50 mM Tris-base solution (100 μl/well), and plates were incubated at room temperature for 40 minutes with shaking. Plates were finally read at OD 564 nm using a BioTek plate reader (BioTek, Winooski, VT). Cell viability was calculated by dividing the average of the reading number for the drug treated wells by the average of the reading number for vehicle treated wells. The IC50 values (concentration required to achieve 50% growth inhibition) were determined using the Prism 5 software (GraphPad Software).
Cell cycle analysis using flow cytometry
Cells were seeded in T-25 tissue culture flasks (6×105 cells per flask) (Corning, Lowell, MA) and grown overnight before drug treatment. For cell cycle analysis, AsPC-1 cells were treated with PHA-739358 (3μM), imatinib (15μM), or PHA-739358 (3μM) plus imatinib (15μM) for 24, 48, and 72 hours. The drug-treated cells and untreated control samples were harvested by trypsinization and stained with propidium iodide (Sigma-Aldrich, St. Louis, Mo) in a modified Krishan buffer for 1 hour at 4°C. The propidium iodide-stained samples were then analyzed with a FACSCalibur Flow Cytometer (BD Immunocytometry Systems, San Jose, CA). Histograms were analyzed for cell cycle compartments, and the percentage of cells at each phase of the cell cycle was calculated using CellQuest Pro Software (BD Immunocytometry Systems).
Caspase 3/7 activity based apoptosis assay
Cells were seeded in 6-well plates (0.3 × 106 cells/well) and incubated for 24 hours at 37°C to allow attachment. Then cells were treated with various concentrations of drugs as indicated in the figure legends. Culture media were collected at 72 hours after drug treatment. After washing with Phosphate Buffer Saline (PBS) solution, the cells were detached by trypsinization and combined with the culture media for each sample. The cell suspension was pelleted by centrifugation at 1,000 rpm for 5 min. 200 μl of NP40 lysis buffer (10 mM Tris-Cl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, 0.5% NP40) was then added into the cell pellet and mixed by pipetting and incubated on ice for at least 30 min. The lysed cell mixture was then spun down at 13,000 g for 10 min to remove cell debris. Protein concentrations were determined using the BCA protein assay kit (Pierce, Rockford, IL). Caspase 3/7 activity was measured using the Caspase-Glo® 3/7 Assay kit (Promega) according to the manufacture instructions. Briefly, an equal volume (100 μl) of Caspase-Glo® 3/7 reagent was added to each cell lysate sample (100 μl) in a 96-well assay plate with a final assay volume of 200 μl. Samples were incubated at room temperature for one hour (protected from light) with shaking, and the luminescence of each sample is measured using a Veritas™ Microplate Luminometer (Turner BioSystems, Sunnyvale, CA). The Caspase 3/7 activity was normalized to the amount of total protein contained in the cell lysate as determined by the BCA protein assay (Thermo Scientific, Rockford, MI).
Western blotting analysis
The cells were treated with PHA-739358 (3μM), imatinib (15μM), or PHA-739358 (3μM) plus imatinib (15μM), for 72 hours and then harvested by trypsinization. The cell lysates were prepared as described for the Caspase 3/7 activity assay. Cell lysates containing equal amount of protein (20 μg) were resolved on 4-12% SDS-PAGE (polyacrylamide gel electrophoresis) gels. The separated proteins were transferred to nitrocellulose membranes. Membranes were then probed with primary antibodies against Phospho-PDGFRA, Bcl-xL, Bcl-2, PI3K, Phospho-PI3K, ERK, Phospho-ERK and β-actin (All from Cell Signaling Technology, Inc., Danvers, MA). β-actin was included to serve as a protein loading control. The bound primary antibodies were detected using peroxidase-conjugated secondary antibodies (Cell Signaling) and chemiluminescence by the ImmobilonTM Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA) according to manufacturer’s instructions. The luminescent signal of the membrane was then detected by photographic film.
RESULTS
Optimization of conditions for HT-siRNA screening
Selection of cell lines and AKIs
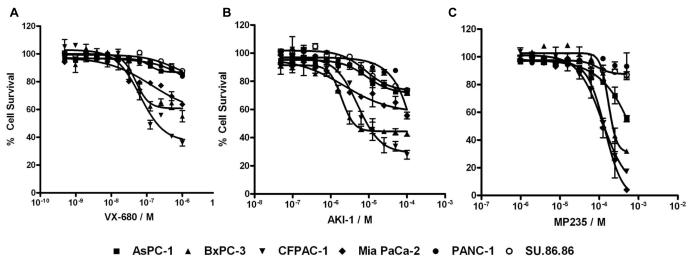
To select an AKI that would maximize our chances of finding siRNA hits that are specific to Aurora kinase inhibition, we first evaluated 3 different AKIs, VX-680, MP235, and AKI-1, in a panel of pancreatic cancer cells, including AsPC-1, BxPC-3, CFPAC-1, Mia PaCa-2, PANC-1 and SU86.86, using the same growth and assay conditions as described in material and methods. As shown in Figure 1, the three AKIs showed different levels of cell growth inhibition in pancreatic cancer cell lines. VX-680 was the most potent with EC50s (concentration required to achieve 50% of the maximal cell growth inhibition) below 100 nM; AKI-1 had modest EC50s (low μM); and MP235 was the least potent with EC50s over 100 μM. Although the reason for the different cellular potency of the AKIs is potentially complex, we believed that AKI-1 would be a good compound for HT-siRNA due to its modest activity and relatively smooth dose response curves in the cell lines. BxPC-3 is one of the cell lines that gave the most consistent dose responses to all three AKIs and its sensitivity to the AKIs is modest among the cell lines (Fig. 1). We therefore decided to carry out the HT-siRNA screen with AKI-1 in the BxPC-3 cell line.
Figure 1. Inhibition of pancreatic cancer cell growth by AKIs.
AsPC-1, BxPC-3, CFPAC-1, Mia PaCa-2, PANC-1, and SU86.86 pancreatic cancer cells were treated with a serial dilution of VX-680 (A), AKI-1 (B), or MP235 (C). Cell viability was measured at 96 hours after drug treatment by CellTiter Glo® Assay.
Optimization of transfection conditions
Efficient delivery of siRNA into cells is critical to the success of a HT-RNAi screen. To find the best transfection reagent and conditions for pancreatic cancer cells, we first tested a panel of 4 transfection reagents (Lipofectamine 2000), RNAiMax, SilentFect, and Oligofectamine) with two siRNA oligonucleotides, a negative control siRNA control (Non-silencing siRNA, Qiagen) and a positive control siRNA (UBB1, Qiagen) which was found to be lethal in all cell lines tested (data not shown). Among the 4 transfection reagents, SilentFect (SLF) showed the most consistent highly transfection efficiency across different pancreatic cancer cell lines (data not shown). The transfection conditions were further optimized by evaluating the transfection efficiency at different SLF dilutions. The optimal SLF dilutions for 6 pancreatic cancer cell lines are shown in Supplementary Figure S3A. For BxPC-3 cells, the optimal transfection reagent is SLF with a dilution rate at 1:5 (SLF to Serum free medium, Fig. S3B).
Identification of siRNAs sensitizing pancreatic cancer cells to AKIs
We first performed an RNAi screen with the Human Validated Kinase Set (HVKS) siRNA library from Qiagen (two siRNA sequences/gene), in combination with AKI-1 in the BxPC-3 cell line. The screen was performed in duplicates. From this initial screen, a total of 172 siRNAs targeting 152 different kinase or kinase related genes showed greater than 1.5 fold decrease in the EC50 or EC30 of the AKI-1 dose response curves compared to the plate median and were selected as positive hits. We then obtained four different siRNA sequences for each of the 152 gene hits and performed a confirmation screen using the same procedure as the initial screen. A total of 17 different kinase genes were confirmed to have at least 2 out of 4 siRNA oligonucleotides to show greater than 1.5 fold decrease in EC50 or EC30 values. Table 1 lists those 17 genes and the drug dose response curves in the presence of the positive siRNAs are shown in Supplementary Figure S4. Many of the 17 gene hits have been previously reported to be involved in tumorigenesis or progression of various tumor types including pancreatic cancer. For instance, PDGFRA (Platelet-derived growth factor receptor alpha) has been shown to be overexpressed in human pancreatic cancer and PDGFR inhibitors such as imatinib reduce the growth and metastasis of pancreatic tumors in mouse xenograft models (37-39). Our analysis of DNA microarray gene expression profiling datasets of pancreatic normal and cancerous tissues deposited in the oncomine database (www.oncomine.com) also showed overexpression of PDGFRA in pancreatic tumor tissues (Supplementary Fig. S5).
Table 1.
Confirmed gene hits from the HT-RNAi screen
| HUGO Symbol | # of siRNAs | Entrez ID | Description |
|---|---|---|---|
| BMPR2 | 3 | 659 | bone morphogenetic protein receptor, type II |
| CSNK1A1 | 3 | 1452 | casein kinase 1, alpha 1 |
| LIMK2 | 2 | 3985 | LIM domain kinase 2 |
| MAP3K10 | 2 | 4294 | mitogen-activated protein kinase kinase kinase 10 |
| MET | 2 | 4233 | met proto-oncogene (hepatocyte growth factor receptor) |
| NEK2 | 2 | 4751 | NIMA (never in mitosis gene a)-related kinase 2 |
| NME2 | 2 | 4831 | non-metastatic cells 2, protein (NM23B) |
| PAK4 | 2 | 10298 | p21(CDKN1A)-activated kinase 4 |
| PAK7 | 2 | 57144 | p21(CDKN1A)-activated kinase 7 |
| PDGFRA | 2 | 5156 | platelet-derived growth factor receptor, alpha polypeptide |
| PIM2 | 2 | 11040 | pim-2 oncogene |
| PINK1 | 3 | 65018 | PTEN induced putative kinase 1 |
| PIP5K2B | 2 | 8396 | phosphatidylinositol-4-phosphate 5-kinase, type II, beta |
| PTK2B | 2 | 2185 | PTK2B protein tyrosine kinase 2 beta |
| STK32B | 2 | 55351 | serine/threonine-protein kinase 32B |
| STK39 | 2 | 27347 | serine threonine kinase 39 |
| TESK2 | 2 | 10420 | testis-specific kinase 2 |
Inhibition of PDGFRA by small molecule inhibitors sensitizes pancreatic cancer cells to AKIs
To further validate PDGFRA as a sensitizing target for AKIs in pancreatic cancer, we examined the anti-proliferation activity of combination treatment of PDGFR inhibitors (imatinib and sorafenib) and different AKIs.
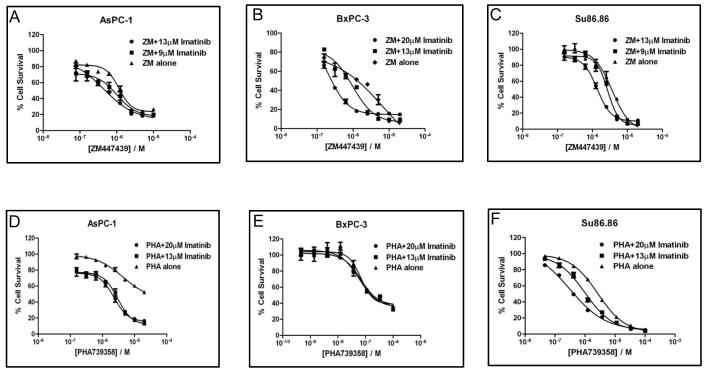
Various concentrations of imatinib combined with a serial dilution of two AKIs (ZM447439 and PHA-739358) were first evaluated in three pancreatic cancer cell lines (AsPC-1, BxPC-3, and SU.86.86). As shown in Figure 2, addition of 9 or 13μM of imatinib to ZM447439 (a selective Aurora B inhibitor) resulted in a left shift of the dose response curves in all 3 cell lines (Fig. 2). Imatinib at 13 μM reduced the IC50 values of ZM447439 by 2 and 3 fold in the AsPC-1 and SU.86.86 cell lines, respectively (Fig. 2A and 2C). Addition of imatinib to PHA-739358 (a pan-Aurora kinase inhibitor) also increased the sensitivity of two of the cell lines. Imatinib (20 μM) reduced the IC50 of PHA-739358 by 2 fold in AsPC-1 and 9 fold in SU.86.86 (Fig. 2D and 2F). In addition to the IC50 decrease in the AsPC-1 cell line, this combination enhanced the cytotoxicity effect at the higher concentration of PHA-739358 (Fig. 2D). Table 2 summarizes the IC50 values of the AKIs in combination with imatinib after normalization with the imatinib only treatment (assuming the Bliss independence criterion (40)) and their ratios to the IC50 values of AKI only treatments in the three pancreatic cancer cell lines. A ratio of less than 1 indicates a synergistic interaction between the AKIs and imatinib at the concentrations tested.
Figure 2. Imatinib sensitizes pancreatic cancer cells to the treatment of ZM447439 and PHA-739358.
AsPC-1, BxPC-3, and SU86.86 cells were treated with a serial dilution of ZM447439 (A~C) or PHA-739358 (D~F) in combination with different fixed concentrations of imatinib. Each of the drug dose response curves was normalized to the percentage cell survival of imatinib only treatment at the indicated concentration based on the Bliss independence drug interaction model. ZM: ZM447439; PHA: PHA-739358
Table 2.
IC50 of AKIs in combination with imatinib in pancreatic cancer
| cell lines | Treatments | AKI IC50 (μM)* | IC50 ratio to AKI alone |
|---|---|---|---|
| BxPC-3 | PHA Alone | 0.07 | -- |
| PHA+Imatinib (20 μM) | 0.07 | 1.00 | |
| PHA+Imatinib (13 μM) | 0.06 | 0.88 | |
|
| |||
| ZM Alone | 10.59 | N/A | |
| ZM+Imatinib (20 μM) | 1.81 | 0.17 | |
| ZM+Imatinib (13 μM) | 2.92 | 0.28 | |
|
| |||
| AsPC-1 | PHA Alone | 4.49 | -- |
| PHA+Imatinib (20 μM) | 1.83 | 0.41 | |
| PHA+Imatinib (13 μM) | 1.95 | 0.43 | |
|
| |||
| ZM Alone | 12.00 | -- | |
| ZM+Imatinib (13 μM) | 4.46 | 0.37 | |
| ZM+Imatinib (9 μM) | 5.31 | 0.44 | |
|
| |||
| Su86.86 | PHA Alone | 2.66 | -- |
| PHA+Imatinib (20 μM) | 0.31 | 0.12 | |
| PHA+Imatinib (13 μM) | 0.99 | 0.37 | |
|
| |||
| ZM Alone | 3.32 | -- | |
| ZM+Imatinib (13 μM) | 1.30 | 0.39 | |
| ZM+Imatinib (9 μM) | 2.32 | 0.70 | |
IC50 values for the combination treatments were normalized to imatinib only treatment. PHA: PHA-739358; ZM: ZM447439.
Since imatinib is known to inhibit other kinases besides PDGFR, to further confirm that the synergism observed is specific to PDGFR inhibition we tested another known small molecule inhibitor of PDGFR, sorafenib. Similar to imatinib, sorafenib (2 μM and 4 μM) caused a left shift of PHA-739358 dose response curves in AsPC-1 and SU.86.86 cell lines but not in BxPC-3 (Supplementary Fig. S6).
Effects of imatinib and PHA-739358 combination on cell cycle progression
Since Aurora kinase inhibition has been shown to induce cell cycle arrest we examined the effects of the combination treatment of imatinib and PHA-739358 on cell cycle progression in AsPC-1 cells. As expected, PHA-739358 alone induced significant G2/M arrest and polyploidy. PHA-739358 (3 μM) significantly increased the G2/M population from 19.37% to 30.56% and the population of polyploidy cells from 5.80% to 15.61% within 24 hours (Table 3). Imatinib (15 μM) does not affect the cell cycle distribution of at 24 hours. However, the combination treatment of both drugs resulted in further induction of G2/M arrest (45.72% of total cell population) compared to PHA-739358 alone (30.56%)). Similar synergistic effect was observed at both 48 and 72 hour time points where the combination treatment significantly increased G2/M arrest when compared to either drug alone (Table 3). Interestingly, the addition of imatinib to PHA-739358 reduced the polyploidy (>4N DNA content) population induced by PHA-739358 at all 3 time points (Table 3). For instance, at the 24 hour time point, the cell population with >4N DNA increased from 5.8% in untreated control and 5.6% in imatinib only treatment to 15.6% in PHA-739358 only treatment, and reduced back to 5.4% in the imatinib plus PHA-739358 combination treatment.
Table 3.
Induction of G2/M arrest by the combination treatment of PHA-739358 and imatinib in AsPC-1 pancreatic cancer cells
| Treatment | Cell cycle | 24hr | 48hr | 72hr |
|---|---|---|---|---|
| Untreated control |
G0-G1(%) | 52.1 | 60.7 | 72.7 |
| S(%) | 21.0 | 12.8 | 9.1 | |
| G2-M(%) | 19.4 | 19.8 | 12.9 | |
| Polyploidy(%) | 5.8 | 4.8 | 3.8 | |
| Debris(%) | 0.3 | 0.3 | 0.4 | |
|
| ||||
| PHA alone | G0-G1(%) | 33.6 | 55.4 | 41.1 |
| S(%) | 19.1 | 14.4 | 12.0 | |
| G2-M(%) | 30.6 | 22.7 | 24.7 | |
| Polyploidy(%) | 15.6 | 22.4 | 16.9 | |
| Debris(%) | 0.9 | 0.2 | 3.0 | |
|
| ||||
| Imatinib alone | G0-G1(%) | 57.5 | 31.4 | 68.0 |
| S(%) | 22.1 | 11.3 | 11.5 | |
| G2-M(%) | 12.8 | 31.7 | 15.3 | |
| Polyploidy(%) | 5.6 | 4.1 | 2.9 | |
| Debris(%) | 0.4 | 1.3 | 0.4 | |
|
| ||||
| PHA + Imatinib | G0-G1(%) | 34.5 | 31.4 | 31.2 |
| S(%) | 12.3 | 8.5 | 8.6 | |
| G2-M(%) | 45.7 | 48.8 | 48.3 | |
| Polyploidy(%) | 5.4 | 9.8 | 10.4 | |
| Debris(%) | 0.2 | 0.2 | 0.2 | |
Induction of apoptotic cell death by the combination treatment of imatinib and PHA-739358 in pancreatic cancer cells
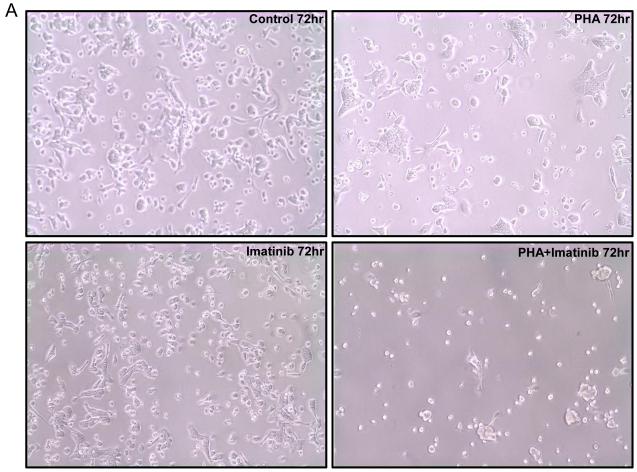
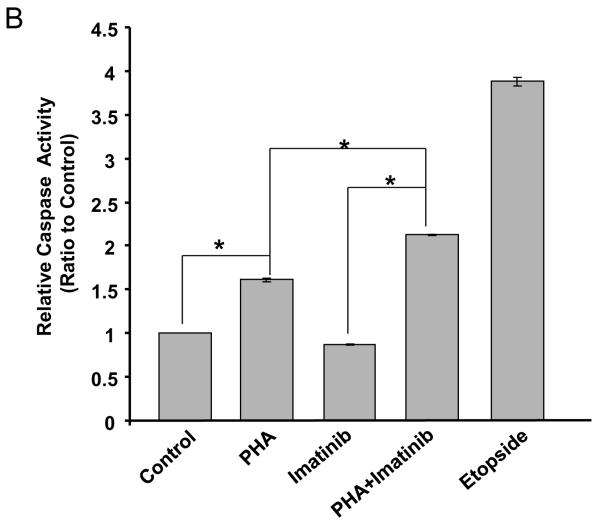
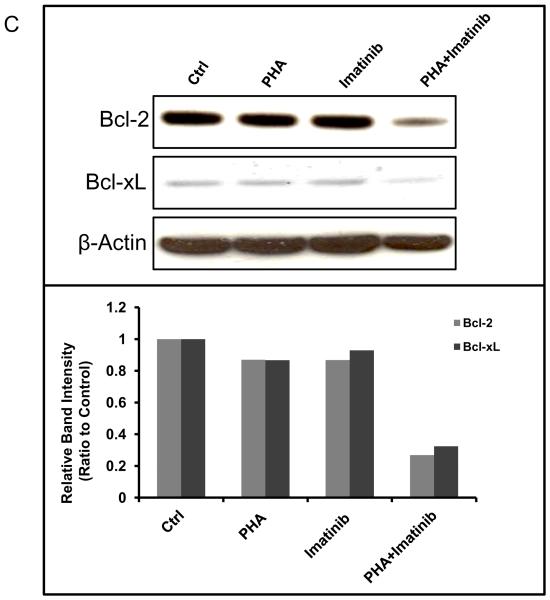
Consistent with its inhibitory activity against both Aurora A and B, PHA-739358 (3 μM) as a single agent reduced the proliferation of AsPC-1 cells and increased the formation of multinucleated cells (top right panel in Fig. 3A). Imatinib (15μM), as a single agent, did not significantly affect the growth of AsPC-1 cells (Bottom left panel in Fig. 3A). However, combination treatment of PHA-739358 and imatinib induced dramatic cell death (Bottom right panel in Fig. 3A). Caspase 3/7 activity assays indicated that PHA-739358 (3 μM) alone significantly induced apoptosis at 72 hours compared to vehicle control (P < 0.001) whereas imatinib (15 μM) did not (Fig. 3B). When the two drugs were combined, the induction of apoptosis further increased significantly when compared to PHA-739358 only treatment (P = 0.00003, Fig. 3B), indicating that PHA-739358 and imatinib act synergistically in inducing apoptosis. Furthermore, combination of another AKI, ZM447439, and imatinib also showed a significant increase in the induction of caspase activity in comparison to either drug alone in the BxPC-3 cell line (Supplementary Fig. S7). To explore the mechanism of action of the increased apoptotic effect of the combination treatment, the expression of the two anti-apoptotic proteins, Bcl-2 and Bcl-xL, were examined by Western blotting. As shown in Figure 3C, treatment with either PHA-739358 or imatinib alone did not significantly affect the level of either protein whereas the combination treatment reduced the expression of Bcl-2 and Bcl-xL by 73% and 68%, respectively, compared to the untreated control, indicating that the increased anti-apoptotic effect of the combination treatment might be a result of the synergistic down-regulation of Bcl-2 and Bcl-xL expression by the two drugs.
Figure 3. Increased apoptotic cell death induced by the combination treatment of PHA-739358 and imatinib.
AsPC-1 cells were treated with PHA-739358 (PHA) (3μM), imatinib (15 μM), or combination of PHA and imatinib for 72 hours and then subjected to evaluation of cell morphology changes by microscope (A), Caspase activity levels by the Caspase 3/7 Glo® assay (B), and Bcl2 and Bcl-xL expression levels by Western blotting (C). Etopside (100μM) was used as a positive control for the Caspase 3/7 assay in B. The intensities of the Western blotting bands (Top panel in C) were quantified using the ImageJ software (63) and each band was normalized with their corresponding β-Actin loading controls (Bottom panel in C). * indicates significant differences between the two treatments (P<0.001).
Combination treatment of imatinib and AKIs decreased the phosphorylation of PI3K but not ERK
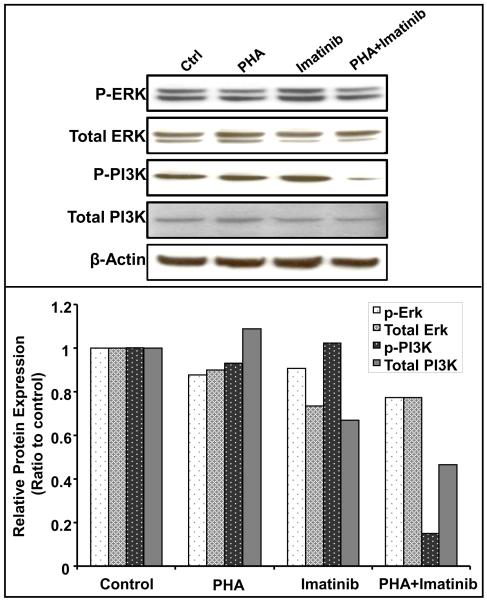
Two major effector pathways of PDGF/PDGFR signaling are the Ras/Erk pathway and the PI3K/Akt pathway. To investigate the effect of combination treatment of imatinib and AKI on these two pathways, we examined the phosphorylation of PI3K and Erk1/2 upon the drug treatment. As shown in Fig. 4, AsPC-1 cells treated with single agent PHA-739358 (3μM) or imatinib (15μM) did not significantly affect the phosphorylation of either Erk1/2 or PI3K. However, combination treatment of PHA-739358 and imatinib resulted in decreased phosphorylation of PI3K but not the ERK1/2 kinases. Similarly, combination of ZM447439 and imatinib resulted in a significant decrease of PI3K phosphorylation level, but not the phosphorylation of Erk kinase in the BxPC-3 cell line (Supplementary Fig. S8). These results suggest that AKIs and imatinib might act synergistically in inhibiting the PI3K/Akt induced cell survival in pancreatic cancer cells.
Figure 4. Combination of PHA-739358 and imatinib inhibits the phosphorylation of PI3K but not ERK.
AsPC-1 cells were treated with PHA-739358 (PHA) (3μM), imatinib (15 μM), or the combination of PHA and imatinib. Cells were harvested 72 hours after drug treatment and 20 μg of whole cell lysates were used in Western blotting detection of the proteins (Top panel). The intensities of the bands were quantified using the ImageJ software and normalized with their corresponding β-Actin loading controls (Bottom panel)
Discussion
Over the past decade, more than a dozen of small molecule Aurora kinase inhibitors have been developed and entered into clinical studies. Many of these inhibitors were reported to show impressive in vitro and in vivo activities in a variety of tumor types including colon, breast, ovarian and pancreatic cancers (41-43). Phase I and early phase II results reported for some of the AKIs are promising with stable disease observed in about 20% of the patients. Hence, combining with other agents might be needed to further enhance the efficacy of AKIs (44). In this study, we utilized high throughput RNAi (HT-RNAi) screening to identify genes that can potentiate AKI response in pancreatic cancer cells. Using HT-RNAi screening as a tool to identify drug sensitizing targets has gained wide attraction in recent years (45-50). However, the majority of those screens use one or two drug concentrations in combination with RNAi. Since the synergism between siRNA and drug is often drug concentration dependent, using only one or two drug concentrations could miss a significant number of potential positive hits. In our study we used 5-dose serial dilutions of the drugs, which allowed us to generate drug dose response curves for comparison of growth inhibitory effects. This approach not only significantly reduces the impact of experimental variations amongst different drug concentrations but also provides activity data on the combination of RNAi and multiple drug concentration, therefore, reducing false positive and negative rates.
Among the 17 kinase gene targets we identified, some are involved in cell cycle regulation. For instance, NEK2 is a centrosomal resident protein that regulates centrosome separation and mitotic spindle assembly. Overexpression of NEK2 has been shown to cause centrosome missegregation and aneuploidy (51). Both NEK2 and Aurora A kinase have been reported to interact with protein phosphatase 1 (PP1) and regulate cell cycle progression (52, 53). Another gene hit, the c-Met oncogene, is known for signaling the invasive growth of tumor cells. Recently, overexpression of c-Met is shown to induce centrosome amplification and chromosomal instability (CIN) via the PI3K-Akt pathway in a p53-dependent manner (54). In pancreatic cancer, we and others have shown that c-Met is overexpressed in cancer cells and tumor tissues (55-57). Besides c-Met and PDGFRA, a number of the other gene targets have also been associated with pancreatic cancer. For instance, BMPR2 is reported to be overexpressed by 8 fold in pancreatic cancer tissues in comparison to normal pancreas (58). Knockdown of LIMK2 expression is shown to reduce the invasiveness and metastatic capabilities of pancreatic cancer cells in a zebrafish xenograft metastasis assay (59). The p21-activating kinase 4 (PAK4) gene is amplified in pancreatic tumors and is shown to promote the motility and invasion of pancreatic ductal carcinoma cells (60, 61). Although the mechanisms of the synergistic effect between the knockdown of these genes and the AKIs remain to be investigated, it is possible that the signaling pathways involving these genes may crosstalk with one or more of Aurora kinases and act in augmentation to promote pancreatic cancer progression and/or metastasis. Molecules that modulate the activity/expression of these gene targets may hence enhance the antitumor activity of AKIs. In this study, we demonstrated that the multi-targeted kinase inhibitor (PDGFR, ABL, and c-Kit), imatinib, synergize with AKIs in inhibiting pancreatic cancer cell growth. It has been reported that imatinib treatment reduced the level of phosphorylated PDGFRA in a pancreatic cancer mouse xenograft model (38). We also observed the inhibition of PDGFRA autophosphorylation by imatinib in AsPC-1 pancreatic cancer cell line (data not shown). Furthermore, a second PDGFR inhibitor, sorafenib, also showed synergistic effect in combination with the pan-Aurora kinase inhibitor PHA-739358 in pancreatic cancer cells. These results further support the conclusion that PDGFR inhibition can sensitize pancreatic cancer cells to the treatment of Aurora kinase inhibitors. However, further studies are needed to test whether or not the inhibition of other cellular targets of imatinib and sorafenib (e.g. ABL and RAF) also contributes to the synergism. Although our study was performed in pancreatic cancer cells, considering the fact that both Aurora kinases and PDGFR have been implicated in multiple tumor types, it is plausible that agents targeting these kinases may also show synergist effects in other cancer types. In fact, a recent study reported that the combination of PHA-739358 and sorafenib showed significantly increased antitumor activity compared to single drug treatments in a mouse xenograft model for hepatocellular carcinoma (62).
PHA-739358 is among the few AKIs that have entered Phase II clinical trials for patients with solid tumors (www.clinicaltrials.gov). In vitro studies have shown that PHA-739358 causes a failure of cell division, resulting in polyploidy and reduction in viability (27). In agreement with these results, our study shows PHA-739358 induces G2/M arrest and polyploidy (Fig. 3), and inhibited proliferation in pancreatic cancer cell lines (Fig. 2). We further showed that imatinib and sorafenib could sensitize pancreatic cancer cells to the treatment of PHA-739358 (Fig. 2). Imatinib further enhances the G2/M arrest and apoptosis induced by PHA-739358 (Table 3). Such synergistic effect is potentially mediated through inhibition of PI3K activation but not ERK activation (Fig. 4).
In conclusion, this is the first report describing the use of kinome-wide siRNA library to functionally screen for sensitizer targets of AKIs in pancreatic cancer cells. The findings from this study further demonstrated the power of high-throughput RNAi screening identifying sensitizers for existing therapeutic agents. The genes identified from this study present new opportunities for the development of rational combination regimens that include Aurora kinase inhibitors.
Supplementary Material
Supplementary Figure S1. Chemical structures of the Aurora kinase inhibitors used in this study.
Supplementary Figure S2. Experimental scheme for high-throughput RNAi screening
Supplementary Figure S3. Optimization of transfection conditions for pancreatic cancer cell lines. A) Optimal dilution ratios for SLF in pancreatic cancer cell lines; B) representative of the transfection optimization experiment for SLF in the BxPC-3 cell line.
Supplementary Figure S4. Drug dose response curves of confirmed siRNA hits. Curves for siRNA hits targeting the same gene are plotted in the same graph along with the curve for the non-silencing control siRNA. The curves were normalized to the siRNA only controls.
Supplementary Figure S5. PDGFRA mRNA levels in pancreatic tumor and normal pancreas tissues from DNA microarray datasets in Oncomine. A) Whiskers plots of PDGFRA expression in normal pancreas (N=39) and pancreatic ductal adenocarcinoma tissues (N=39) from the dataset deposited by Badea and colleagues (Badea et al., Hepatogastroenterology, 2008, 55:2016-27.); The median expression level of PDGFRA in the cancer samples is 2.9 fold higher than that of normal pancreas tissues (p value < 0.0001). B) Whiskers plots of PDGFRA expression in normal pancreas (N=12) and pancreatic ductal adenocarcinoma tissues (N=12) from the dataset deposited by lacobuzio-Donahue and colleagues (Lacobuzio-Donahue CA, et al., Am J Pathol, 2003 162:1151-62). The median expression level of PDGFRA in the cancer samples is 3.0 fold higher than that of normal pancreas tissues (P value < 0.005).
Supplementary Figure S6. Combination treatment of sorafenib and PHA-739358 in pancreatic cancer cell lines. The drug dose response curves in three pancreatic cancer cell lines AsPC-1 (A), BxPC-3 (B), and SU.86.86 (C) were determined by treating the cells with a serial dilution of PHA-739358 (PHA) in combination with different fixed concentrations of sorafenib. The drug dose response curves were normalized to the sorafenib only treatment based on the Bliss independence drug interaction model.
Supplementary Figure S7. Increased apoptotic cell death induced by the combination treatment of ZM447439 and imatinib. BxPC-3 cells were treated with ZM447439 (ZM) (2 μM), imatinib (15 μM), or combination of ZM447439 and imatinib for 72 hours and then subjected to the caspase activity measurement using the Caspase 3/7 Glo® assay. Etopside (100μM) was included as a positive control for the Caspase 3/7 assay. * indicates significant differences between the two treatments (P<0.001).
Supplementary Figure S8. Combination of ZM447439 and imatinib inhibits the phosphorylation of PI3K. BxPC-3 cells were treated with ZM447439 (ZM) (2μM), imatinib (15 μM), or the combination of ZM447439 and imatinib. Cells were harvested 72 hours after drug treatment and 20 μg of whole cell lysates were used in Western blotting detection of the proteins (Top panel). The intensities of the corresponding bands in the Western blot were quantified using ImageJ (Bottom panel).
Acknowledgements
We would like to thank Steve Warner and Stanley Nwokenkwo for their technical help with the transfection optimization experiments. This work was supported by NIH/NCI grants CA095031 and CA109552.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, Au HJ, Murawa P, Walde D, Wolff RA, Campos D, Lim R, Ding K, Clark G, Voskoglou-Nomikos T, Ptasynski M, Parulekar W. Erlotinib Plus Gemcitabine Compared With Gemcitabine Alone in Patients With Advanced Pancreatic Cancer: A Phase III Trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 3.Von Hoff DD, Ramanathan R, Borad M, Laheru D, Smith L, Wood T, Korn R, Desai N, Trieu V, Iglesias J, Zhang H, Soo-shiong P, Shi T, Rajeshkumar NV, Maitra A, Hidalgo M. Gemcitabine plus nab-paclitaxel Is an active regimen in patients with advanced pancreatic cancer: A Phase I/II trial. J. Clin. Oncol. 2011 doi: 10.1200/JCO.2011.36.5742. Published online on October 3, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, Adenis A, Raoul JL, Gourgou-Bourgade S, de la Fouchardiere C, Bennouna J, Bachet JB, Khemissa-Akouz F, Pere-Verge D, Delbaldo C, Assenat E, Chauffert B, Michel P, Montoto-Grillot C, Ducreux M. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 5.Warner SL, Bearss DJ, Han H, Von Hoff DD. Targeting Aurora-2 Kinase in Cancer. Mol Cancer Ther. 2003;2:589–595. [PubMed] [Google Scholar]
- 6.Carmena M, Earnshaw WC. The cellular geography of Aurora kinases. Nat Rev Mol Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 7.Sen S, Zhou H, White R. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene. 1997;14:2195–2200. doi: 10.1038/sj.onc.1201065. [DOI] [PubMed] [Google Scholar]
- 8.Bischoff J, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, Chan C, Novotny M, Slamon D, Plowman G. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loka W, Kleinb RQ, Saifa MW. Aurora kinase inhibitors as anti-cancer therapy. Anti-Cancer Drugs. 2010 doi: 10.1097/CAD.0b013e3283350dd1. Ahead of print. [DOI] [PubMed] [Google Scholar]
- 10.Pirker C, Lotsch D, Spiegl-Kreinecker S, Jantscher F, Sutterluty H, Micksche M, Grusch M, Berger W. Response of experimental malignant melanoma models to the pan-Aurora kinase inhibitor VE-465. Exp Dermatol. 2010;19:1040–1047. doi: 10.1111/j.1600-0625.2010.01182.x. [DOI] [PubMed] [Google Scholar]
- 11.Kanda A, Kawai H, Suto S, Kitajima S, Sato S, Takata T, Tatsuka M. Aurora-B//AIM-1 kinase activity is involved in Ras-mediated cell transformation. Oncogene. 2005;24:7266–7272. doi: 10.1038/sj.onc.1208884. [DOI] [PubMed] [Google Scholar]
- 12.Meraldi P, Honda R, A.Nigg E. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rojanala S, Han H, Munoz RM, Browne W, Nagle R, Von Hoff DD, Bearss DJ. The mitotic serine threonine kinase, Aurora-2, is a potential target for drug development in human pancreatic cancer. Mol Cancer Ther. 2004;3:451–457. [PubMed] [Google Scholar]
- 14.Han H, Bearss DJ, Browne LW, Calaluce R, Nagle RB, Von Hoff DD. Identification of Differentially Expressed Genes in Pancreatic Cancer Cells Using cDNA Microarray. Cancer Res. 2002;62:2890–2896. [PubMed] [Google Scholar]
- 15.Li D, Zhu J, Firozi PF, Abbruzzese JL, Evans DB, Cleary K, Friess H, Sen S. Overexpression of oncogenic STK15/BTAK/Aurora A kinase in human pancreatic cancer. Clin Cancer Res. 2003;9:991–997. [PubMed] [Google Scholar]
- 16.Warner SL, Bashyam S, Vankayalapati H, Bearss DJ, Han H, Von Hoff DD, Hurley LH. Identification of a lead small-molecule inhibitor of the Aurora kinases using a structure-assisted, fragment-based approach. Mol Cancer Ther. 2006;5:1764–1773. doi: 10.1158/1535-7163.MCT-05-0524. [DOI] [PubMed] [Google Scholar]
- 17.Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA, Demur C, Hercend T, Diu-Hercend A, Su M, Golec JMC, Miller KM. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 18.Rubin EH, Shapiro GI, Stein MN, Watson P, Bergstrom D, Xiao A, Clark JB, Freedman SJ, Eder JP. A Phase I clinical and pharmacokinetic (PK) trial of the aurora kinase (AK) inhibitor MK-0457 in cancer patients. J Clin Oncol. 2006;24(18S) Abstract 3009. [Google Scholar]
- 19.Bebbington D, Binch H, Charrier JD, Everitt S, Fraysse D, Golec J, Kay D, Knegtel R, Mak C, Mazzei F, Miller A, Mortimore M, O’Donnell M, Patel S, Pierard F, Pinder J, Pollard J, Ramaya S, Robinson D, Rutherford A, Studley J, Westcott J. The discovery of the potent aurora inhibitor MK-0457 (VX-680) Bioorg Med Chem Lett. 2009;19:3586–3592. doi: 10.1016/j.bmcl.2009.04.136. [DOI] [PubMed] [Google Scholar]
- 20.Pluim D, Beijnen JH, Schellens JH, van Tellingen O. Simultaneous determination of AZD1152 (prodrug) and AZD1152-hydroxyquinazoline pyrazol anilide by reversed phase liquid chromatography. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:3549–3555. doi: 10.1016/j.jchromb.2009.08.035. [DOI] [PubMed] [Google Scholar]
- 21.Wilkinson RW, Odedra R, Heaton SP, Wedge SR, Keen NJ, Crafter C, Foster JR, Brady MC, Bigley A, Brown E, Byth KF, Barrass NC, Mundt KE, Foote KM, Heron NM, Jung FH, Mortlock AA, Boyle FT, Green S. AZD1152. a selective inhibitor of Aurora B kinase, inhibits human tumor xenograft growth by inducing apoptosis, Clin Cancer Res. 2007;13:3682–3688. doi: 10.1158/1078-0432.CCR-06-2979. [DOI] [PubMed] [Google Scholar]
- 22.Gorgun G, Calabrese E, Hideshima T, Ecsedy J, Perrone G, Mani M, Ikeda H, Bianchi G, Hu Y, Cirstea D, Santo L, Tai YT, Nahar S, Zheng M, Bandi M, Carrasco RD, Raje N, Munshi N, Richardson P, Anderson KC. A novel Aurora-A kinase inhibitor MLN8237 induces cytotoxicity and cell-cycle arrest in multiple myeloma. Blood. 115:5202–5213. doi: 10.1182/blood-2009-12-259523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maris JM, Morton CL, Gorlick R, Kolb EA, Lock R, Carol H, Keir ST, Reynolds CP, Kang MH, Wu J, Smith MA, Houghton PJ. Initial testing of the aurora kinase A inhibitor MLN8237 by the Pediatric Preclinical Testing Program (PPTP) Pediatr Blood Cancer. 55:26–34. doi: 10.1002/pbc.22430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpinelli P, Ceruti R, Giorgini ML, Cappella P, Gianellini L, Croci V, Degrassi A, Texido G, Rocchetti M, Vianello P, Rusconi L, Storici P, Zugnoni P, Arrigoni C, Soncini C, Alli C, Patton V, Marsiglio A, Ballinari D, Pesenti E, Fancelli D, Moll J. PHA-739358, a potent inhibitor of Aurora kinases with a selective target inhibition profile relevant to cancer. Mol Cancer Ther. 2007;6:3158–3168. doi: 10.1158/1535-7163.MCT-07-0444. [DOI] [PubMed] [Google Scholar]
- 25.Cohen RB, Jones SF, Aggarwal C, von Mehren M, Cheng J, Spigel DR, Greco FA, Mariani M, Rocchetti M, Ceruti R, Comis S, Laffranchi B, Moll J, Burris HA. A phase I dose-escalation study of danusertib (PHA-739358) administered as a 24-hour infusion with and without granulocyte colony-stimulating factor in a 14-day cycle in patients with advanced solid tumors. Clin Cancer Res. 2009;15:6694–6701. doi: 10.1158/1078-0432.CCR-09-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steeghs N, Eskens FALM, Gelderblom H, Verweij J, Nortier JWR, Ouwerkerk J, van Noort C, Mariani M, Spinelli R, Carpinelli P, Laffranchi B, de Jonge MJA. Phase I Pharmacokinetic and Pharmacodynamic Study of the Aurora Kinase Inhibitor Danusertib in Patients With Advanced or Metastatic Solid Tumors. J Clin Oncol. 2009;27:5094–5101. doi: 10.1200/JCO.2008.21.6655. [DOI] [PubMed] [Google Scholar]
- 27.Benten D, Keller G, Quaas A, Schrader J, Gontarewicz A, Balabanov S, Braig M, Wege H, Moll J, Lohse AW, Brummendorf TH. Aurora kinase inhibitor PHA-739358 suppresses growth of hepatocellular carcinoma in vitro and in a xenograft mouse model. Neoplasia. 2009;11:934–944. doi: 10.1593/neo.09664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, He L, Kruk P, Nicosia SV, Cheng JQ. Aurora-A induces cell survival and chemoresistance by activation of Akt through a p53-dependent manner in ovarian cancer cells. International Journal of Cancer. 2006;119:2304–2312. doi: 10.1002/ijc.22154. [DOI] [PubMed] [Google Scholar]
- 29.Workman P. Drugging the cancer kinome: progress and challenges in developing personalized molecular cancer therapeutics. Cold Spring Harb Symp Quant Biol. 2005;70:499–515. doi: 10.1101/sqb.2005.70.020. [DOI] [PubMed] [Google Scholar]
- 30.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 31.Lampson M, Renduchitala K, Khodjakov A, Kapoor T. Correcting improper chromosome–spindle attachments during cell division. Nature Cell Biology. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 32.Collins PJ, Hennessy LK, Leibelt CS, Roby RK, Reeder DJ, Foxall PA. Developmental validation of a single-tube amplification of the 13 CODIS STR loci, D2S1338, D19S433, and amelogenin: the AmpFlSTR Identifiler PCR Amplification Kit. J Forensic Sci. 2004;49:1265–1277. [PubMed] [Google Scholar]
- 33.Tiedemann RE, Zhu YX, Schmidt J, Yin H, Shi C-X, Que Q, Basu G, Azorsa D, Perkins LM, Braggio E, Fonseca R, Bergsagel PL, Mousses S, Stewart AK. Kinome-wide RNAi studies in human multiple myeloma identify vulnerable kinase targets, including a lymphoid-restricted kinase, GRK6. Blood. 115:1594–1604. doi: 10.1182/blood-2009-09-243980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA, Demur C, Hercend T, Diu-Hercend A, Su M, Golec JM, Miller KM. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 35.Carvajal RD, Tse A, Schwartz GK. Aurora kinases: new targets for cancer therapy. Clin Cancer Res. 2006;12:6869–6875. doi: 10.1158/1078-0432.CCR-06-1405. [DOI] [PubMed] [Google Scholar]
- 36.Warner SL, Bashyam S, Vankayalapati H, Bearss DJ, Han H, Mahadevan D, Von Hoff DD, Hurley LH. Identification of a lead small-molecule inhibitor of the Aurora kinases using a structure-assisted, fragment-based approach. Mol Cancer Ther. 2006;5:1764–1773. doi: 10.1158/1535-7163.MCT-05-0524. [DOI] [PubMed] [Google Scholar]
- 37.Ebert M, Yokoyama M, Friess H, Kobrin MS, Buchler MW, Korc M. Induction of platelet-derived growth factor A and B chains and over-expression of their receptors in human pancreatic cancer. Int J Cancer. 1995;62:529–535. doi: 10.1002/ijc.2910620507. [DOI] [PubMed] [Google Scholar]
- 38.Hwang RF, Yokoi K, Bucana CD, Tsan R, Killion JJ, Evans DB, Fidler IJ. Inhibition of platelet-derived growth factor receptor phosphorylation by STI571 (Gleevec) reduces growth and metastasis of human pancreatic carcinoma in an orthotopic nude mouse model. Clin Cancer Res. 2003;9:6534–6544. [PubMed] [Google Scholar]
- 39.Baker CH, Trevino JG, Summy JM, Zhang F, Caron A, Nesbit M, Gallick GE, Fidler IJ. Inhibition of PDGFR phosphorylation and Src and Akt activity by GN963 leads to therapy of human pancreatic cancer growing orthotopically in nude mice. Int J Oncol. 2006;29:125–138. [PubMed] [Google Scholar]
- 40.Greco WR, Bravo G, Parsons JC. The search for synergy: a critical review from a response surface perspective. Pharmacol Rev. 1995;47:331–385. [PubMed] [Google Scholar]
- 41.Green MR, Woolery JE, Mahadevan D. Update on Aurora Kinase Targeted Therapeutics in Oncology. Expert Opin Drug Discov. 2011;6:291–307. doi: 10.1517/17460441.2011.555395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheung CH, Coumar MS, Chang JY, Hsieh HP. Aurora kinase inhibitor patents and agents in clinical testing: an update (2009 - 10) This article is an update to aurora kinase inhibitors review, which appeared in: Expert Opin. Ther. Patents 2009, 19, 1-36 and Expert Opin. Investig. Drugs 2009, 18, 1-20. Expert Opin Ther Pat. 2011;21:857–884. doi: 10.1517/13543776.2011.574614. [DOI] [PubMed] [Google Scholar]
- 43.Keen N, Taylor S. Aurora-kinase inhibitors as anticancer agents. Nat Rev Cancer. 2004;4:927–936. doi: 10.1038/nrc1502. [DOI] [PubMed] [Google Scholar]
- 44.Boss DS, Beijnen JH, Schellens JH. Clinical experience with aurora kinase inhibitors: a review. Oncologist. 2009;14:780–793. doi: 10.1634/theoncologist.2009-0019. [DOI] [PubMed] [Google Scholar]
- 45.Bartz SR, Zhang Z, Burchard J, Imakura M, Martin M, Palmieri A, Needham R, Guo J, Gordon M, Chung N, Warrener P, Jackson AL, Carleton M, Oatley M, Locco L, Santini F, Smith T, Kunapuli P, Ferrer M, Strulovici B, Friend SH, Linsley PS. Small interfering RNA screens reveal enhanced cisplatin cytotoxicity in tumor cells having both BRCA network and TP53 disruptions. Mol Cell Biol. 2006;26:9377–9386. doi: 10.1128/MCB.01229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Azorsa DO, Gonzales IM, Basu GD, Choudhary A, Arora S, Bisanz KM, Kiefer JA, Henderson MC, Trent JM, Von Hoff DD, Mousses S. Synthetic lethal RNAi screening identifies sensitizing targets for gemcitabine therapy in pancreatic cancer. J Transl Med. 2009;7:43. doi: 10.1186/1479-5876-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diep CH, Munoz RM, Choudhary A, Von Hoff DD, Han H. Synergistic effect between erlotinib and MEK inhibitors in KRAS wild-type human pancreatic cancer cells. Clin Cancer Res. 2011;17:2744–2756. doi: 10.1158/1078-0432.CCR-10-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramaswamy S. Rational design of cancer-drug combinations. N Engl J Med. 2007;357:299–300. doi: 10.1056/NEJMcibr072593. [DOI] [PubMed] [Google Scholar]
- 49.Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, Minna JD, Michnoff C, Hao W, Roth MG, Xie XJ, White MA. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 50.Giroux V, Iovanna J, Dagorn JC. Probing the human kinome for kinases involved in pancreatic cancer cell survival and gemcitabine resistance. Faseb J. 2006;20:1982–1991. doi: 10.1096/fj.06-6239com. [DOI] [PubMed] [Google Scholar]
- 51.Hayward DG, Fry AM. Nek2 kinase in chromosome instability and cancer. Cancer Lett. 2006;237:155–166. doi: 10.1016/j.canlet.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 52.Katayama H, Zhou H, Li Q, Tatsuka M, Sen S. Interaction and feedback regulation between STK15/BTAK/Aurora-A kinase and protein phosphatase 1 through mitotic cell division cycle. J Biol Chem. 2001;276:46219–46224. doi: 10.1074/jbc.M107540200. [DOI] [PubMed] [Google Scholar]
- 53.Meraldi P, Nigg EA. Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. J Cell Sci. 2001;114:3749–3757. doi: 10.1242/jcs.114.20.3749. [DOI] [PubMed] [Google Scholar]
- 54.Nam HJ, Chae S, Jang SH, Cho H, Lee JH. The PI3K-Akt mediates oncogenic Met-induced centrosome amplification and chromosome instability. Carcinogenesis. 2010;31:1531–1540. doi: 10.1093/carcin/bgq133. [DOI] [PubMed] [Google Scholar]
- 55.Kiehne K, Herzig KH, Folsch UR. c-met expression in pancreatic cancer and effects of hepatocyte growth factor on pancreatic cancer cell growth. Pancreas. 1997;15:35–40. doi: 10.1097/00006676-199707000-00005. [DOI] [PubMed] [Google Scholar]
- 56.Han H, Bearss DJ, Browne LW, Calaluce R, Nagle RB, Von Hoff DD. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 2002;62:2890–2896. [PubMed] [Google Scholar]
- 57.Ebert M, Yokoyama M, Friess H, Buchler MW, Korc M. Coexpression of the c-met proto-oncogene and hepatocyte growth factor in human pancreatic cancer. Cancer Res. 1994;54:5775–5778. [PubMed] [Google Scholar]
- 58.Kleeff J, Maruyama H, Ishiwata T, Sawhney H, Friess H, Buchler MW, Korc M. Bone morphogenetic protein 2 exerts diverse effects on cell growth in vitro and is expressed in human pancreatic cancer in vivo. Gastroenterology. 1999;116:1202–1216. doi: 10.1016/s0016-5085(99)70024-7. [DOI] [PubMed] [Google Scholar]
- 59.Vlecken DH, Bagowski CP. LIMK1 and LIMK2 are important for metastatic behavior and tumor cell-induced angiogenesis of pancreatic cancer cells. Zebrafish. 2009;6:433–439. doi: 10.1089/zeb.2009.0602. [DOI] [PubMed] [Google Scholar]
- 60.Chen S, Auletta T, Dovirak O, Hutter C, Kuntz K, El-ftesi S, Kendall J, Han H, Von Hoff DD, Ashfaq R, Maitra A, Iacobuzio-Donahue CA, Hruban RH, Lucito R. Copy number alterations in pancreatic cancer identify recurrent PAK4 amplification. Cancer Biol Ther. 2008;7:1793–1802. doi: 10.4161/cbt.7.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kimmelman AC, Hezel AF, Aguirre AJ, Zheng H, Paik JH, Ying H, Chu GC, Zhang JX, Sahin E, Yeo G, Ponugoti A, Nabioullin R, Deroo S, Yang S, Wang X, McGrath JP, Protopopova M, Ivanova E, Zhang J, Feng B, Tsao MS, Redston M, Protopopov A, Xiao Y, Futreal PA, Hahn WC, Klimstra DS, Chin L, DePinho RA. Genomic alterations link Rho family of GTPases to the highly invasive phenotype of pancreas cancer. Proc Natl Acad Sci U S A. 2008;105:19372–19377. doi: 10.1073/pnas.0809966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benten D, Keller G, Quaas A, Schrader J, Gontarewicz A, Balabanov S, Braig M, Wege H, Moll J, Lohse AW, Brummendorf TH. Aurora kinase inhibitor PHA-739358 suppresses growth of hepatocellular carcinoma in vitro and in a xenograft mouse model. Neoplasia. 2009;11:934–944. doi: 10.1593/neo.09664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Chemical structures of the Aurora kinase inhibitors used in this study.
Supplementary Figure S2. Experimental scheme for high-throughput RNAi screening
Supplementary Figure S3. Optimization of transfection conditions for pancreatic cancer cell lines. A) Optimal dilution ratios for SLF in pancreatic cancer cell lines; B) representative of the transfection optimization experiment for SLF in the BxPC-3 cell line.
Supplementary Figure S4. Drug dose response curves of confirmed siRNA hits. Curves for siRNA hits targeting the same gene are plotted in the same graph along with the curve for the non-silencing control siRNA. The curves were normalized to the siRNA only controls.
Supplementary Figure S5. PDGFRA mRNA levels in pancreatic tumor and normal pancreas tissues from DNA microarray datasets in Oncomine. A) Whiskers plots of PDGFRA expression in normal pancreas (N=39) and pancreatic ductal adenocarcinoma tissues (N=39) from the dataset deposited by Badea and colleagues (Badea et al., Hepatogastroenterology, 2008, 55:2016-27.); The median expression level of PDGFRA in the cancer samples is 2.9 fold higher than that of normal pancreas tissues (p value < 0.0001). B) Whiskers plots of PDGFRA expression in normal pancreas (N=12) and pancreatic ductal adenocarcinoma tissues (N=12) from the dataset deposited by lacobuzio-Donahue and colleagues (Lacobuzio-Donahue CA, et al., Am J Pathol, 2003 162:1151-62). The median expression level of PDGFRA in the cancer samples is 3.0 fold higher than that of normal pancreas tissues (P value < 0.005).
Supplementary Figure S6. Combination treatment of sorafenib and PHA-739358 in pancreatic cancer cell lines. The drug dose response curves in three pancreatic cancer cell lines AsPC-1 (A), BxPC-3 (B), and SU.86.86 (C) were determined by treating the cells with a serial dilution of PHA-739358 (PHA) in combination with different fixed concentrations of sorafenib. The drug dose response curves were normalized to the sorafenib only treatment based on the Bliss independence drug interaction model.
Supplementary Figure S7. Increased apoptotic cell death induced by the combination treatment of ZM447439 and imatinib. BxPC-3 cells were treated with ZM447439 (ZM) (2 μM), imatinib (15 μM), or combination of ZM447439 and imatinib for 72 hours and then subjected to the caspase activity measurement using the Caspase 3/7 Glo® assay. Etopside (100μM) was included as a positive control for the Caspase 3/7 assay. * indicates significant differences between the two treatments (P<0.001).
Supplementary Figure S8. Combination of ZM447439 and imatinib inhibits the phosphorylation of PI3K. BxPC-3 cells were treated with ZM447439 (ZM) (2μM), imatinib (15 μM), or the combination of ZM447439 and imatinib. Cells were harvested 72 hours after drug treatment and 20 μg of whole cell lysates were used in Western blotting detection of the proteins (Top panel). The intensities of the corresponding bands in the Western blot were quantified using ImageJ (Bottom panel).