Abstract
Amyotrophic lateral sclerosis (ALS) is a devastating and universally fatal neurodegenerative disease. Mutations in two related RNA-binding proteins, TDP-43 and FUS, that harbor prion-like domains, cause some forms of ALS. There are at least 213 human proteins harboring RNA recognition motifs, including FUS and TDP-43, raising the possibility that additional RNA-binding proteins might contribute to ALS pathogenesis. We performed a systematic survey of these proteins to find additional candidates similar to TDP-43 and FUS, followed by bioinformatics to predict prion-like domains in a subset of them. We sequenced one of these genes, TAF15, in patients with ALS and identified missense variants, which were absent in a large number of healthy controls. These disease-associated variants of TAF15 caused formation of cytoplasmic foci when expressed in primary cultures of spinal cord neurons. Very similar to TDP-43 and FUS, TAF15 aggregated in vitro and conferred neurodegeneration in Drosophila, with the ALS-linked variants having a more severe effect than wild type. Immunohistochemistry of postmortem spinal cord tissue revealed mislocalization of TAF15 in motor neurons of patients with ALS. We propose that aggregation-prone RNA-binding proteins might contribute very broadly to ALS pathogenesis and the genes identified in our yeast functional screen, coupled with prion-like domain prediction analysis, now provide a powerful resource to facilitate ALS disease gene discovery.
In the future, personalized genome sequencing will become routine, empowering us to define the genetic basis of many human diseases. Currently, however, complete genome sequencing for individuals to discover rare pathogenic mutations is still too costly and time consuming. Thus, more creative approaches are needed to accelerate the discovery of disease genes. Moreover, even once genes are revealed, the need for innovative approaches to elucidate causality remains critical.
ALS, also known as Lou Gehrig's disease, is a devastating adult-onset neurodegenerative disease that attacks upper and lower motor neurons (1). A progressive and ultimately fatal muscle paralysis ensues, usually causing death within 2–5 y of disease onset. ALS is mostly sporadic, but ∼10% of cases are familial. Pathogenic mutations in several genes have been linked to familial and sporadic ALS, including SOD1, TARDBP, FUS/TLS, VAPB, OPTN, VCP, and others (2). Two of these genes, TARDBP (TDP-43) and FUS/TLS (FUS) are notable because they encode related RNA-binding proteins that harbor a prion-like domain (3–6). Moreover, both of these proteins have been identified as components of pathological inclusions in neurons of patients with ALS (7–9). Indeed, an emerging concept suggested by the association of FUS and TDP-43 to ALS is that defects in RNA metabolism might contribute to disease pathogenesis. These observations suggested an intriguing possibility: Could TDP-43 and FUS be just the tip of an iceberg? In other words, could other human RNA-binding proteins with properties similar to those of TDP-43 and FUS also contribute to ALS?
Here we report a simple yeast functional screen, followed by bioinformatics to predict prion-like domains, to identify human proteins with similar properties to TDP-43 and FUS. We then identify mutations in human patients with ALS in one gene from this screen, TAF15, which ranks with the highest prion-like domain score after FUS. Importantly, we show that TAF15 has similar in vitro and in vivo properties to TDP-43 and FUS. Moreover, the ALS-associated TAF15 mutations are more aggregation prone in vitro, have a more severe effect on lifespan than WT when expressed in Drosophila, and increase cytoplasmic mislocalization in mammalian spinal cord neurons. The identification of mutations in an additional RNA-binding protein harboring a prion-like domain further underscores a key role for RNA metabolism defects in ALS and suggests that this class of aggregation-prone RNA-binding proteins might contribute very broadly to ALS and perhaps other related neurodegenerative disorders.
Results
Yeast Screen to Identify RNA-Binding Proteins with Properties Similar to TDP-43 and FUS.
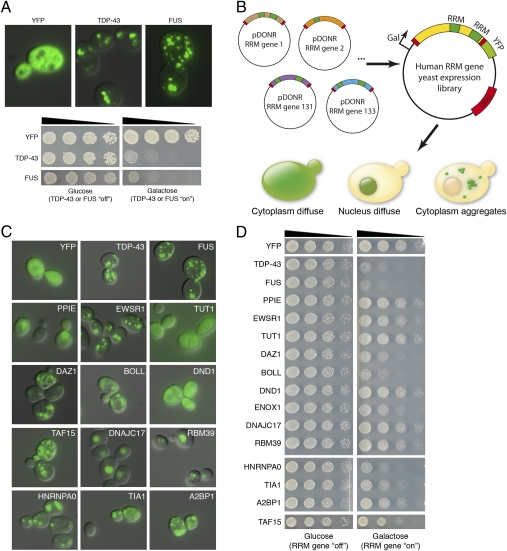
TDP-43 and FUS are both RNA recognition motif (RRM)-containing proteins (RRM proteins) (3) and both form cytoplasmic inclusions and are toxic when expressed in yeast (Fig. 1A) (10–13). Including FUS and TDP-43, there are at least 213 RRM proteins (Protein Families database ID PF00076) present in the human proteome (Dataset S1). This result raised the question of whether other human RRM proteins would show properties like TDP-43 and FUS (e.g., aggregation prone and toxic in yeast). If so, these properties might make these genes top candidates for unique ALS disease genes. We therefore designed a simple yeast functional screen to identify such genes (Fig. 1B). Of the 213 human RRM proteins, we were able to obtain a gene clone for 133. We cloned these 133 different human RRM-containing ORFs into yeast expression vectors as YFP fusion proteins, under the control of a galactose-inducible promoter, and individually transformed them into yeast cells. We have previously found that the addition of the YFP tag to TDP-43 and FUS does not affect the aggregation or toxicity properties (10–12). Fluorescence microscopy was used to determine the localization of each protein (nuclear, cytoplasmic, diffuse, foci; Fig. 1C and Dataset S1) and spotting assays were used to assess toxicity (Fig. 1D and Dataset S1). Some proteins localized to the nucleus (38/133) whereas others were diffusely localized in the cytoplasm (26/133). Interestingly, several others formed multiple foci in the cytoplasm in a pattern strikingly similar to that of FUS and TDP-43 (54/133). Of the proteins that accumulated in the cytoplasm, 38 were also toxic, including FUS and TDP-43 (Table 1). Thus, 38 of 133 human RRM proteins behave like FUS and TDP-43 in yeast cells.
Fig. 1.
A yeast functional screen identifies human RRM proteins with properties similar to FUS and TDP-43. (A) When expressed in yeast, TDP-43 and FUS form multiple cytoplasmic aggregates (Upper) and are toxic (Lower). (B) We designed a yeast functional screen to identify additional human RRM proteins that aggregate and are toxic in yeast. A library of 133 different human ORFs encoding the proteins was cloned into yeast expression vectors as galactose-inducible YFP fusions. We individually transformed each of these plasmids into yeast cells and assessed their effect on aggregation and toxicity by fluorescence microscopy and spotting assays, respectively. (C) Examples of various localization patterns in yeast cells of human RRM proteins. Some proteins were localized diffusely throughout the cytoplasm (TUT1 and DND1) and others were localized diffusely in the nucleus (DNAJC17). Some formed multiple foci in the nucleus (RBM39) and several others resembled FUS and TDP-43, which formed multiple cytoplasmic foci (EWSR1, TAF15, HNRNPA0, and DAZ1). (D) Spotting assays to assess the toxicity of human RRM proteins. Transformants were grown on synthetic media containing either glucose (control, RRM gene ‘‘off’’) or galactose (to induce expression of candidate ORFs, RRM gene ‘‘on’’). Some proteins were very toxic when overexpressed (DAZ1, HNRNPA0, FUS, and TDP-43) whereas others were moderately toxic (EWSR1 and TAF15) and others were not toxic (PPIE and DNAJC17). See Table 1 and Dataset S1 for toxicity and aggregation scores.
Table 1.
Human RRM proteins with similar properties to FUS and TDP-43 when expressed in yeast
| Name | Description | Toxicity score (1–4) | Prion domain score | Prion domain rank |
| BOLL | Boule-like (Drosophila) | 2 | ||
| CPSF6 | Cleavage and polyadenylation-specific factor | 2.5 | ||
| DAZ1 | Deleted in azoospermia 1 | 2.5 | 14 | 143 |
| DAZ2 | Deleted in azoospermia 2 | 3 | 14 | 143 |
| DAZ3 | Deleted in azoospermia 3 | 3.5 | 15 | 136 |
| DAZAP1 | DAZ-associated protein 1 | 2 | 12 | 198 |
| ELAVL1 | Embryonic lethal, abnormal vision-like 1 | 1 | ||
| ELAVL2 | Embryonic lethal, abnormal vision-like 2 | 1 | ||
| ELAVL3 | Embryonic lethal, abnormal vision-like 3 | 2.5 | ||
| ELAVL4 | Embryonic lethal, abnormal vision-like 4 | 1 | ||
| ENOX1 | Ecto-NOX disulfide-thiol exchanger 1 | 2.5 | ||
| EWSR1 | Ewing sarcoma breakpoint region 1 | 3.5 | 32 | 25 |
| FUS | Fusion (involved in malignant liposarcoma) | 1.5 | 38 | 13 |
| G3BP1 | Ras-GTPase–activating protein | 2 | ||
| HNRNPA0 | Heterogeneous nuclear ribonucleoprotein | 1 | 21 | 81 |
| HNRNPA1 | Heterogeneous nuclear ribonucleoprotein | 1 | 28 | 38 |
| HNRNPA2B1 | Heterogeneous nuclear ribonucleoprotein | 1 | 30 | 32 |
| HNRNPM | Heterogeneous nuclear ribonucleoprotein | 3 | ||
| IGF2BP2 | IGF-II mRNA-binding protein 2 | 2.5 | ||
| IGF2BP3 | IGF-II mRNA-binding protein 3 | 2.5 | ||
| MSI2 | Musashi homolog 2 | 2 | ||
| RALYL | RNA-binding protein-like | 2.5 | ||
| RBM12B | RNA-binding motif protein | 3.5 | ||
| RBM14 | RNA-binding motif protein | 2 | 16 | 117 |
| RBM4 | RNA-binding motif protein | 3 | ||
| RBM41 | RNA-binding motif protein | 2.5 | ||
| RBM4B | RNA-binding motif protein | 2.5 | ||
| RBM5 | RNA-binding motif protein | 3 | ||
| RBM9 | RNA-binding motif protein | 3.5 | ||
| RBMS1 | RNA-binding motif, single-stranded interacting protein | 2 | ||
| RBMS2 | RNA-binding motif, single-stranded interacting protein | 2 | ||
| RBPMS | RNA-binding motif, single-stranded interacting protein | 3 | ||
| ROD1 | Regulator of differentiation | 1 | ||
| SNRPA | Small nuclear ribonucleoprotein polypeptide | 2 | ||
| SNRPB2 | Small nuclear ribonucleoprotein polypeptide | 2 | ||
| TAF15 | TATA box binding protein (TBP)-associated factor | 3.5 | 33 | 22 |
| TARDBP | TAR DNA-binding protein (TDP-43) | 1.5 | 27 | 43 |
| TIA1 | Cytotoxic granule-associated RNA binding protein | 2 | 23 | 55 |
Thirty-eight human RRM proteins that formed cytoplasmic aggregates and were toxic when expressed in yeast are shown. Toxicity was scored from 1 (most toxic) to 4 (not toxic). Prion domain score, based on ref. 14, indicates the maximum log-likelihood for prion-like amino acid composition vs. non-prion–like amino acid composition in any 60 consecutive aa window contained in a region parsed as prion-like by the hidden Markov model. No prion score indicates that no region of length ≥60 was parsed as prion-like. Prion domain rank is from 21,873 human proteins.
Prion-Like Domains in TDP-43, FUS, and Several Other RRM Proteins.
To focus this list further, we used a bioinformatics approach. In addition to the RRM domain, FUS and TDP-43 share a glycine-rich domain and a bioinformatics-predicted prion-like domain (4). Like prion domains found in fungal prion proteins (e.g., Sup35, Ure2, and Rnq1), these domains are enriched in uncharged polar amino acids (such as asparagine, glutamine, and tyrosine) and glycine (14). In TDP-43, the predicted prion-like domain overlaps the glycine-rich domain; in FUS, a QGSY-rich region defines the prion-like domain, although there is some overlap with the glycine-rich domain. The prion-like domain is a shared feature that may be important, given the “prionoid” aggregation propensity of many proteins associated with human neurodegenerative disease (15). Moreover, the prion-like domains of TDP-43 and FUS are critical for the aggregation of these proteins (10–13, 16). Remarkably, using an algorithm to score 21,873 human proteins for likelihood of harboring a prion-like domain, FUS and TDP-43 ranked 13th and 43rd, respectively. We therefore interrogated the list of human RRM proteins to identify whether others ranked highly, using the prion domain prediction algorithm (14). Interestingly, 31 of the 213 human RRM proteins ranked in the top 250 (Dataset S1). Among these, FUS and TDP-43 ranked first and 10th, respectively. Of the 38 proteins that were toxic and formed cytoplasmic inclusions in yeast, 13, including FUS and TDP-43, scored highly for a prion-like domain (Table 1). Thus, using the combined yeast screen and prion-like domain analysis, we narrowed the list of RRM proteins by 10-fold (133 human RRM proteins → 38 that aggregate and are toxic in yeast → 13 that also contain a prion domain). We therefore focused on these proteins because they shared similar functional and structural features with FUS and TDP-43: (i) they formed cytoplasmic accumulations, (ii) they were toxic in yeast, and (iii) they contained a predicted prion domain.
Identification of TAF15 Sequence Variants in Patients with ALS.
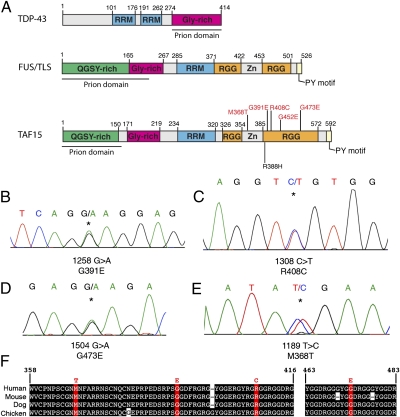
With this list of 13 “FUS- and TDP-43–like” proteins in hand, we sought to test the hypothesis that these additional RRM proteins might contribute to ALS. We gave top priority to TAF15 [RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68 kDa] because it ranked second of 213 human RRM proteins (and 22nd of all 21,873 human proteins) on the basis of the prion domain prediction algorithm (Table 1). Moreover, TAF15 belongs to the same protein family as FUS and is remarkably similar, especially within the RRM, the glycine-rich domain, and the C-terminal RGG domain- and PY-motif–containing region (Fig. 2A). Spotting assays showed that TAF15 expression was also toxic, albeit not as toxic as TDP-43 and FUS (Fig. 1D). Given these commonalities, we proceeded to sequence exons 13–16 of TAF15 (NM_139215), a region analogous to where many FUS mutations are located (3). These exons comprise the RGG- and PY-motif–containing C-terminal domain, which is important for nuclear localization of FUS (17–19). We performed complete sequencing of these exons in 735 individuals diagnosed with ALS and in 1,328 geographically matched healthy population control individuals (see SI Materials and Methods for patient and control demographic information). We found three patient-specific nonsynonymous missense variants (Fig. 2 B–D and Dataset S2), c.1258G > A, p.G391E; c.1308C > T, p.R408C; and c.1504G > A, p.G473E. These variants were found in individuals with ages of onset of sporadic ALS of 67 y, 47 y, and 68 y, respectively, and were all located in highly conserved regions of TAF15 (Fig. 2F). In addition, we identified one nonsynonymous missense variant that was present in both patients and controls (TAF15 c.1249G > A, p.R388H). The presence of this variant in control individuals suggests that it likely represents a benign variant, although functional studies are required to distinguish potentially damaging variants from benign variants (see below). Thus, nonsynonymous missense variations in TAF15 were detected in 4 of 735 North American ALS cases and 1 of 1,328 North American controls. We also identified a fifth variant (M368T; Fig. 2E) in an independent cohort of 351 Swedish patients with ALS and a sixth variant (G452E; Fig. 2A) in a cohort of 176 Australian patients with ALS. Because the TAF15 variants were identified in sporadic ALS cases, familial evidence for segregation with disease was not possible. Notably, however, TARDBP and FUS mutations have also been confirmed in apparent sporadic ALS cases (20). In addition, the parents of the affected individuals were not available to determine whether the mutations occurred de novo or were inherited.
Fig. 2.
Missense mutations in TAF15 in patients with ALS. (A) Comparison of FUS and TAF15 demonstrates similar domain architecture. Both proteins contain a single RRM, a glycine-rich domain, a predicted prion domain, RGG domains, and a C-terminal PY motif, which can function as an NLS (18). Similar domains are also present in TDP-43. The TAF15 variants identified in exons 13–16 in ALS cases are highlighted in red and the variant, R388H, identified in both cases and controls is highlighted in black. (B–E) DNA sequence analysis of TAF15 in North American Caucasian patients with ALS identified three missense variants. (B) A TAF15 mutation in an ALS case: c.1258 G > A, predicted to lead to p.G391E. (C) A TAF15 mutation in an ALS case: c.1308 C > T, predicted to lead to p.R408C. (D) An additional TAF15 variant c.1504G > A, predicted to lead to p.G473E, identified in the ALS cohort from Mayo Clinic. (E) An additional TAF15 variant c.1189T > C, predicted to lead to p.M368T, identified in an ALS cohort from Sweden. (F) Sequence alignment of amino acids 358–416 and 463–483 of TAF15 from diverse vertebrate species indicates that the mutated residues in TAF15 are highly conserved. Identical amino acids have a black background and mutation sites are red.
ALS-Specific TAF15 Variants Form Cytoplasmic Foci in Spinal Cord Neurons.
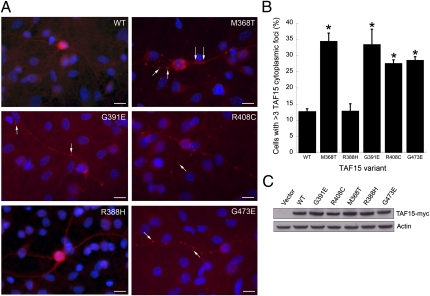
To provide evidence that the specific sequence variants that we found in ALS cases might be pathogenic, we performed an unbiased assessment of all nonsynonymous variants identified in cases and controls. We designed an assay that potentially discriminates functionally deleterious variants from benign variants. ALS-linked mutations in TDP-43 and FUS have been shown to disrupt protein localization, leading to enhanced accumulation of ALS-linked variants in cytoplasmic structures (8, 9, 18, 21–23). Given this common feature, we assessed the effects of the TAF15 variants (Fig. 2A) on subcellular localization. We transfected myc-tagged WT or the mutant forms of TAF15 into spinal cord neurons cultured from rat embryos (Fig. 3A). The transfection efficiency in these primary neuronal cultures was too low to quantify protein expression levels; however, immunoblotting of HEK293T cells transfected with these same constructs confirmed that the WT and variant constructs are expressed at similar levels (Fig. 3C). WT or mutant TAF15 expression under these conditions did not result in toxicity; however, we did observe significant effects of the variants on localization.
Fig. 3.
A functional assay to distinguish potentially damaging TAF15 variants from benign variants. We performed an unbiased assessment of all TAF15 missense variants identified from sequencing of ALS cases (M368T, G391E, R408C, and G473E) and controls (R388H). (A) Primary rat embryonic neuron cultures were transfected with myc-tagged WT or mutant TAF15 and stained with α-myc (red). Transfection of WT TAF15 or the R388H variant found in controls results in localization within the nucleus and cytoplasm of neurons in a diffuse pattern. In contrast, the ALS-linked mutant forms of TAF15 showed a striking accumulation of cytoplasmic foci (arrows) in dendrites and axons. (Scale bar, 20 μm.) (B) Quantitation of transfected WT or mutant TAF15 that accumulates in cytoplasmic puncta. Four of four TAF15 variants found in ALS cases (M368T, G391E, R408C, and G473E) showed cytoplasmic puncta formation whereas the one variant found in both cases and controls (R388H) behaved like WT. *P < 0.01 (cytoplasmic puncta formation of TAF15 variants compared with WT, Student's t test). Error bars show mean ± SEM. (C) Because the transfection efficiency in the primary rat spinal cord neuron cultures was not high enough to detect TAF15 overexpression by immunoblot, we transfected the same constructs of WT and TAF15 mutants into HEK293T cells. Immunoblotting with α-myc antibodies was used to detect myc-tagged TAF15 and α-actin antibody was used as a loading control. In contrast to the primary neuronal cultures, we did not observe a significant difference in aggregation between WT TAF15 and the variants in HEK293T cells.
The transfected WT and mutant TAF15 proteins were restricted to the nucleus in ∼40% of cells and present in both nucleus and cytoplasm in ∼60% of cells. The TAF15 mutations did not affect nuclear localization per se, but rather promoted the formation of cytoplasmic foci. WT TAF15 and the R388H variant, which was present in both patients and controls, localized to the nucleus and cytoplasm, in a diffuse pattern (Fig. 3 A and B). In contrast, all three of the ALS-linked mutant forms of TAF15 (G391E, R408C, and G473E) found in the North American patients with ALS showed a striking accumulation of TAF15 in cytoplasmic foci in dendrites and axons (Fig. 3 A and B). Thus, all of the TAF15 variants that we found in patients with ALS were deleterious; that is, they caused mislocalization of the protein to cytoplasmic foci within neurons, whereas the one variant found in controls was benign and behaved like WT. This unbiased assessment of all variants provides strong support for the variants that we found in ALS cases as being risk factors for ALS (3 damaging TAF15 variants of 735 ALS cases vs. 0 damaging TAF15 variants of 1,328 controls, P = 0.0451, Fisher's exact test). The fourth ALS-specific variant (M368T; Fig. 2E), found in an independent cohort of 351 Swedish patients with ALS, also affected localization in this assay (Fig. 3 A and B), but we do not include this variant in the above statistical assessment because we did not sequence an equivalent number of matched controls. The fifth ALS-specific variant (G452E; Fig. 2A), in 1 Australian patient with ALS of a cohort of 176 patients with ALS, was not present in 72 sequenced Australian controls or in our collection of 1,328 sequenced North American controls. We have not yet tested the effects of this variant on TAF15 localization in our functional assay. Unlike several of the ALS-linked FUS mutations, which are located in the conserved PY motif at the extreme C terminus of the protein and have strong effects on nuclear localization (18), the TAF15 variants that we have identified are not located in TAF15's PY motif (Fig. 2A), possibly explaining why they do not affect nuclear localization per se but seem to promote the accumulation of TAF15 in cytoplasmic foci. As additional variants in TAF15 are identified, this functional assay will be a powerful tool for assessing their potential pathogenicity.
TAF15 Is Inherently Aggregation Prone.
These genetic and functional studies highlight a potential role for TAF15 in ALS pathogenesis. We next sought further functional evidence that TAF15 has properties similar to TDP-43 and FUS. We asked, does TAF15 spontaneously aggregate in vitro like TDP-43 and FUS (11, 12) and does TAF15 confer neurodegeneration when expressed in the nervous system, like TDP-43 and FUS (24–31)?
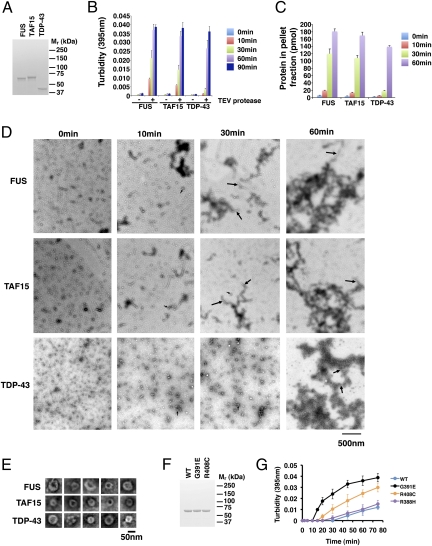
We purified bacterially expressed GST-tagged TAF15 as a soluble protein under native conditions, as previously described for TDP-43 and FUS (11, 12). TAF15, TDP-43, and FUS were all competent in RNA binding, suggesting that the RRM domains were correctly folded (11, 12). Upon addition of tobacco etch virus (TEV) protease to specifically remove the N-terminal GST tag and SDS-PAGE to assess purity and expected molecular weight (Fig. 4A), we found that TAF15 rapidly aggregated at 25 °C with gentle agitation (Fig. 4 B and C). Turbidity and sedimentation analysis revealed that TAF15 aggregated with kinetics similar to FUS and slightly more rapidly than TDP-43, assessed by turbidity (Fig. 4B). If TEV protease was omitted, then little aggregation occurred (Fig. 4B). Electron microscopy studies established that TAF15 rapidly accessed oligomeric forms (Fig. 4D), which would frequently adopt a pore-like conformation [Fig. 4D (small arrows) and E], similar to those formed by TDP-43 and FUS (Fig. 4 D and E) (11, 12), which are reminiscent of pathological oligomers formed by α-synuclein and amyloid-β (32). Furthermore, TAF15 also assembled into linear polymers with a cross-sectional diameter of ∼15–20 nm (Fig. 4D, large arrows) that increased in length over time and would often become tangled into large masses by 60 min (Fig. 4D). In general, the morphology of TAF15 aggregates was more similar to FUS (12) than to TDP-43 (11), which over this time frame formed shorter polymers that would clump together to form large masses (Fig. 4D) (11). Importantly, a human RRM protein, DND1, which did not aggregate and was not toxic in yeast (Fig. 1 C and D), remained soluble and did not aggregate in this in vitro assay, providing evidence that in vitro aggregation is not a property shared by all RRM proteins. Thus, similar to TDP-43 and FUS, and concordant with the yeast data, TAF15 is an inherently aggregation-prone protein.
Fig. 4.
TAF15 is an aggregation-prone protein like TDP-43 and FUS. (A) Following TEV protease cleavage to remove the N-terminal GST tag, FUS, TAF15, and TDP-43 proteins were processed for SDS-PAGE and Coomassie stained to confirm purity and expected molecular weight. (B) GST-TDP-43, GST-FUS, or GST-TAF15 (3 μM) were incubated in the presence or absence of TEV protease at 25 °C for 0–90 min with agitation. Note that very little aggregation occurs in the absence of TEV protease. The extent of aggregation was determined by turbidity. Values represent means ± SEM (n = 3). (C) GST-TDP-43, GST-FUS, or GST-TAF15 (3 μM) were incubated in the presence of TEV protease at 25 °C for 0–60 min. At the indicated times, reactions were processed for sedimentation analysis. Pellet and supernatant fractions were resolved by SDS-PAGE and stained with Coomassie Brilliant Blue. The amount of protein in the pellet fraction was determined by densitometry in comparison with known quantities of the appropriate protein. Values represent means ± SEM (n = 3). A human RRM protein, DND1, which did not aggregate and was not toxic in yeast (Fig. 1 C and D), was also soluble and did not form aggregates in this assay. (D) GST-TDP-43, GST-FUS, or GST-TAF15 (3 μM) were incubated in the presence of TEV protease at 25 °C for 0–60 min. At various times, reactions were processed for EM. Small arrows denote small pore-shaped oligomers and large arrows denote linear polymers. (Scale bar, 500 nm.) (E) Gallery of TDP-43, FUS, and TAF15 oligomers formed during aggregation reactions. (Scale bar, 50 nm.) (F) Following TEV protease cleavage to remove the N-terminal GST tag, TAF15 wild-type (WT), G391E, and R408C proteins were processed for SDS-PAGE and Coomassie stained to confirm purity and expected molecular weight. (G) ALS-linked TAF15 variants G391E and R408C displayed accelerated aggregation kinetics whereas the R388H variant found in both cases and controls aggregated with similar kinetics to WT TAF15.
It is likely that sophisticated cellular proteostasis mechanisms, not recapitulated with this in vitro assay, mitigate rapid TAF15 aggregation in vivo. However, age-associated decline in these quality control measures, along with environmental triggers (for example, injury or exposure to toxins), might allow TAF15 to aggregate in disease. In vitro aggregation assays similar to these have been tremendously powerful tools in defining basic mechanisms underpinning TDP-43 and FUS aggregation (11, 12) as well as the aggregation events in Parkinson's disease and Alzheimer's disease (33–35).
ALS-Specific Variants Accelerate TAF15 Aggregation in Vitro.
Next, we tested two of the ALS-linked mutant forms of TAF15 (G391E and R408C) as well as the variant that we also found in controls (R388H) in this aggregation assay (Fig. 4F). We noticed that if we omitted agitation during the in vitro aggregation reaction, we were able to slow down the aggregation process, allowing us to detect potential differences in aggregation between WT and mutant TAF15 proteins. R388H aggregated with similar kinetics to WT TAF15 (Fig. 4G). By contrast, the two ALS-linked mutants, G391E and R408C, aggregated with more rapid kinetics (Fig. 4G). This increased aggregation propensity might help explain why ALS-linked mutant TAF15 is more prone to accumulate in cytoplasmic inclusions in spinal cord neurons (Fig. 3) and supports the hypothesis that these are likely pathogenic variants.
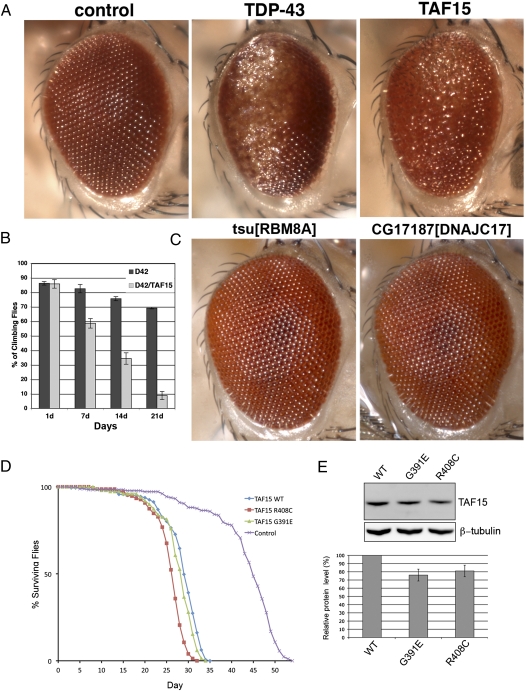
TAF15 Up-Regulation Causes Neurodegeneration in Drosophila and ALS-Linked Mutants Have More Severe Effect on Lifespan.
To extend these findings from in vitro to in vivo, and analyze the effects of TAF15 in the nervous system, we used Drosophila. We and others have previously shown that directing TDP-43 or FUS expression to the fly nervous system causes neurodegeneration (24–28, 30, 31). A series of transgenic lines were generated that expressed WT and mutant human TAF15. Directing expression of TAF15 to the eye of the fly caused degeneration of the structure (Fig. 5A) and led to progressive loss of motility when directed to motor neurons (Fig. 5B). As for the in vitro aggregation assay (Fig. 4), the effect was specific because two other unrelated Drosophila RRM proteins, the human counterparts of which did not aggregate and were not toxic in yeast, did not confer neurodegeneration when up-regulated in Drosophila (Fig. 5C). Thus, TAF15 possesses activity sufficient to confer neurodegeneration in the nervous system, in a manner similar to that of TDP-43 and FUS. TAF15 also induced a markedly shortened lifespan when expressed in the nervous system (Fig. 5D). Notably, expression of ALS-linked TAF15 variants G391E and R408C, inserted at the same genomic location to ensure similar protein expression (Fig. 5E), had a more severe effect, resulting in an even shorter lifespan than that of flies expressing WT TAF15 (Fig. 5D), supporting the notion that these missense variants in TAF15 are potentially deleterious.
Fig. 5.
TAF15 confers neurodegeneration and dysfunction in Drosophila. (A) Toxicity of RRM proteins in the eye. TAF15 causes degeneration and disruption of the retinal structure, akin to TDP-43 (also see ref. 24). Control is driver line alone, gmr-GAL4/+. TDP-43 is gmr-GAL4/UAS-TDP-43-YFP. TAF15 is gmr-GAL4/UAS-TAF15 (grown at 29 °C). (B) Progressive loss of climbing behavior upon expression of TAF15 selectively in motor neurons using the motor neuron-specific D42-GAL4 driver. (C) Up-regulation of two other RRM proteins does not cause neurodegeneration in Drosophila. As a specificity control for the neurodegenerative phenotype conferred by up-regulation of TDP-43 and TAF15 in Drosophila, we tested the effects of up-regulating the fly homologs of two other human RRM proteins in the eye. Up-regulation of tsu and CG17187, which are Drosophila homologs of human genes RBM8A and DNAJC17, respectively, did not cause neurodegeneration. (D) Expression of TAF15 in the nervous system reduces lifespan (blue, compared with normal in purple). Up-regulation of TAF15 variants G391E and R408C causes more rapid death (red and green, compared with WT TAF15 in blue). (E) Immunoblot showing TAF15 WT and mutant expression levels in transgenic flies. β-Tubulin levels were used as loading control.
TAF15 Is Mislocalized in Motor Neurons of Patients with ALS.
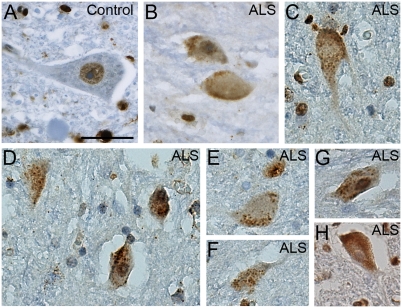
We next performed immunohistochemistry on human postmortem spinal cord tissue to determine whether TAF15 was present in motor neurons and whether its localization was affected in ALS, as for TDP-43 and FUS. TAF15 was robustly expressed in spinal cord motor neurons and localized to the nucleus, as for TDP-43 and FUS (Fig. 6A). We next examined TAF15 localization in autopsy tissue from three sporadic ALS cases (without mutations in TAF15), all with confirmed TDP-43 pathology. Immunostaining of spinal cord sections from these patients with ALS revealed significant cytoplasmic staining in a punctate granular pattern (Fig. 6 B–H), which did not colocalize with TDP-43 inclusions. In contrast to TDP-43 (7), and similar to FUS (8, 9), we did not observe significant nuclear clearing of TAF15. The strong punctate granular localization for TAF15 was not seen in any of the control cases that we examined (n = 3). Because these sporadic ALS cases that we examined also contained prominent TDP-43 cytoplasmic inclusions, which did not colocalize with TAF15, future studies will be required to extend these initial findings and to determine whether TAF15 mislocalization in disease is an early initiator or rather a later consequence of motor neuron degeneration. Thus, TAF15 is expressed in a disease-relevant cell type and can be mislocalized to the cytoplasm in ALS, further supporting the notion that TAF15 can contribute to pathogenesis. Notably, a recent study has also observed TAF15 mislocalization in neurons and glial cells of patients with frontotemporal lobar degeneration (FTLD)-FUS (36).
Fig. 6.
TAF15 accumulates in the cytoplasm in some sporadic spinal cord neurons of patients with ALS. (A) Immunohistochemistry for TAF15 in a postmortem control spinal cord neuron indicates nuclear localization, whereas, in addition to nuclear localization, TAF15 accumulated in cytoplasmic puncta of spinal cord neurons of patients with ALS (B–H). (Scale bar, 25 μm.)
Discussion
In an effort to streamline the identification of new ALS genetic risk factors, we devised a simple yeast functional screen to define additional RNA-binding proteins with properties shared by the known ALS disease genes FUS and TDP-43. This screen resulted in the enrichment of 38 proteins that behave like FUS and TDP-43 in yeast (cytoplasmic inclusions and toxicity), 13 of which contain a predicted prion-like domain (Table 1). Indeed, the combination of yeast screen and prion prediction algorithm enabled us to significantly focus our list of candidate genes ∼10-fold. As evidence of the usefulness of this approach to define genes with a role in ALS, we identified patient-specific missense variants in one of these genes, TAF15, in five unrelated patients with ALS (three variants from our initial cohort of North American Caucasian patients with ALS, a fourth variant from a cohort of Swedish patients with ALS, and a fifth variant from a cohort of Australian patients with ALS). Further, we provide in vitro and in vivo evidence that TAF15 has functional properties similar to those of TDP-43 and FUS: It is intrinsically aggregation prone and can confer neurodegeneration in Drosophila and the ALS-linked variants can increase aggregation in vitro, decrease lifespan in Drosophila, and alter protein subcellular localization in spinal cord neurons. Although familial segregation could not be assessed, the absence of the variants in a large number of healthy controls, the shared structural evidence with known ALS genes, and functional in vitro and in vivo data strongly support the notion that these variants in TAF15 represent pathogenic disease mutations for ALS.
Future studies will be required to determine the relative contribution of TAF15 variants to ALS risk compared with known genetic risk factors such as TDP-43, FUS, SOD1, and others. Our initial analyses with TAF15 in patients with ALS and control populations, as well as recent studies by Ticozzi and colleagues with TAF15 and the related gene EWSR1 (37), suggest that if indeed TAF15 mutations contribute to ALS, they will likely be rarer than FUS and TDP-43 mutations. However, as for all complicated human diseases there will very likely be common genetic contributors as well as rare genetic risk factors. For ALS, we propose that there may be a delicate balance in RNA processing within motor neurons such that slight perturbations from any one of several different aggregation-prone RNA-binding proteins could lead to neurodegeneration. An interesting additional concept that emerges from our findings is that perhaps variants in multiple RNA-binding proteins could synergize with each other to contribute to ALS. There are likely to be some variants that are extremely damaging and thus fully penetrant and aggressive on their own. Case in point: P525L and R495X mutations in FUS lead to relatively severe ALS clinical phenotypes and very early age of disease onset (18, 23), whereas other FUS mutations are less severe (e.g., R521G) (18). This result seems to be due to the effect of the mutations on FUS nuclear localization, with variants having the strongest effect on nuclear localization resulting in the earliest age of onset of ALS (18). Those aggressive FUS variants might sit at one end of a spectrum, with weaker variants at the other. Perhaps then the accumulation of multiple weaker variants in two, three, or more different aggregation-prone RNA-binding proteins (e.g., the top candidates listed in Table 1) might be required to tip the balance in RNA metabolism toward ALS. Future studies will be required to test this hypothesis and to better resolve the complexities of the ALS genetic landscape.
These findings predict that additional aggregation-prone RRM or other RNA-binding proteins, like TAF15, FUS, and TDP-43, could contribute to ALS. Notably, the prion-like domain algorithm ranked FUS and TAF15 first and second of 213 RRM proteins, respectively, and ranked TDP-43 10th. We suggest that genes ranked third through ninth should now be given top priority for genetic analysis in populations of patients with ALS, especially EWSR1, which ranked third and is a close relative of both FUS and TAF15 (38). In addition to ALS, these candidates should also be examined in related clinico-pathological disorders including FTLD and inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia (IBMPFD). For example, mutations in ALS genes TARDBP and FUS have been identified in patients with FTLD and mutations in an IBMPFD gene, VCP, have been identified in patients with ALS (39–41).
Next-generation sequencing and exon capture approaches will eventually become routine in personalized medicine (42–44) and promise to identify all genetic contributors to ALS. Meanwhile, the list of ALS candidate genes that we provide here (Table 1 and Dataset S1), generated by the combination of the yeast functional screen and prion-like domain prediction, will be a powerful resource to jumpstart efforts to identify new genetic risk factors for ALS and spur innovative new diagnostic and therapeutic approaches.
Materials and Methods
Plasmids, Yeast Strains, and Media.
Strain and plasmid construction is detailed in SI Materials and Methods.
Human RNA-Binding Protein Plasmid Library.
We constructed a library of yeast expression plasmids containing 133 unique human RRM-containing ORFs, as detailed in SI Materials and Methods.
Yeast Transformation and Spotting Assays.
Yeast transformation and spotting assays were performed as described in ref. 10.
Microscopy.
Fluorescence microscopy was performed as described in SI Materials and Methods.
Prion-Prediction Algorithm.
Putative prion domains were predicted using a hidden Markov model as described in SI Materials and Methods.
Sequencing TAF15 in Patients with ALS and Controls.
ALS patient and control population demographics and TAF15 mutational analysis procedures are described in SI Materials and Methods.
TAF15 Protein Purification and in Vitro Aggregation Assays.
TDP-43 and FUS were purified as described (11, 12). WT and mutant TAF15 proteins were expressed and purified from Escherichia coli as GST-tagged proteins and in vitro aggregation assays were performed as described in SI Materials and Methods.
Drosophila Experiments.
Transgenic flies expressing human TAF15 were generated by standard techniques using the pUAST vector and analyzed as described in SI Materials and Methods.
Rat Primary Neuron Transfection and Immunofluorescence.
Primary neurons were isolated from rat embryos, cultured, and transfected as described in SI Materials and Methods.
Statistical Analysis.
Two-tailed Fisher's exact tests were used to evaluate genetic association between TAF15 sequence variants and ALS. We tested all TAF15 nonsynonymous missense variants detected in cases and controls in an assay that potentially discriminates functionally deleterious variants from benign variants (e.g., the subcellular localization assay reported in Fig. 3), then classified the variants as deleterious or benign on the basis of their properties in this assay, and then performed a statistical comparison of the frequency of functionally deleterious variants in cases vs. controls.
Immunohistochemistry.
Immunohistochemistry to detect TAF15 localization in postmortem spinal cord tissue was performed as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the patients and their families for their dedication and for their invaluable contributions to this research. We thank the Packard Center for ALS Research at Johns Hopkins for their generosity and collaborative spirit. We thank contributors, including the Alzheimer's Disease Centers who collected samples used in this study. We thank C. Cecere for assistance in sample collection. We thank Rose Li, Phoebe Leboy, and Marisa Bartolomei for helpful suggestions on the manuscript. This work was supported by National Institutes of Health Director's New Innovator Awards 1DP2OD004417 (to A.D.G.) and 1DP2OD002177-01 (to J.S.); National Institutes of Health Grants 1R01NS065317 (to A.D.G.), 5R21NS067354-02 (to J.S.), AG17586 (to V.V.D., J.Q.T., and R.G.), AG10124 (to V.V.D. and J.Q.T.), P01-AG-09215 (to N.M.B.), NS056070 and NS072561 (to Z.M.), T32-AG00255 [to F.I. and V.M.-Y.L. (program director)], R01 AG26251-03A1 (to R.R.), R01 NS065782 (to R.R.), and P50 AG16574 (to R.R.); the University of Pennsylvania Institute on Aging and Alzheimer's Disease Core Center Pilot Grant Program (AG10124) (to V.V.D.); the ALS Association (R.R); a grant from the Packard Center for ALS Research at Johns Hopkins (to A.D.G. and J.S.); and an Ellison Medical Foundation New Scholar in Aging Award (to J.S.). A.D.G. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts, and a Rita Allen Scholar, supported by the Rita Allen Foundation. J.Q.T. is the William Maul Measey–Truman G. Schnabel, Jr., Professor of Geriatric Medicine and Gerontology. N.M.B. is an Investigator of the Howard Hughes Medical Institute. Samples from the National Cell Repository for Alzheimer's Disease, which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging, were used in this study. In Australia, the work was supported by the National Health and Medical Research Council of Australia (1004670 and 511941) and a Peter Stearne grant from the Motor Neurone Disease Research Institute of Australia. This research was conducted while J.C. was an Ellison Medical Foundation/AFAR Postdoctoral Fellow.
Footnotes
Conflict of interest statement: A.D.G. is an inventor on patents and patent applications that have been licensed to FoldRx Pharmaceuticals.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109434108/-/DCSupplemental.
References
- 1.Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: Deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 2.Van Damme P, Robberecht W. Recent advances in motor neuron disease. Curr Opin Neurol. 2009;22:486–492. doi: 10.1097/WCO.0b013e32832ffbe3. [DOI] [PubMed] [Google Scholar]
- 3.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: The FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cushman M, Johnson BS, King OD, Gitler AD, Shorter J. Prion-like disorders: Blurring the divide between transmissibility and infectivity. J Cell Sci. 2010;123:1191–1201. doi: 10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Udan M, Baloh RH. Implications of the prion-related Q/N domains in TDP-43 and FUS. Prion. 2011;5:1–5. doi: 10.4161/pri.5.1.14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gitler AD, Shorter J. RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion. 2011;5:179–187. doi: 10.4161/pri.5.3.17230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 8.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 9.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci USA. 2008;105:6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson BS, et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Z, et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ju S, et al. A yeast model of FUS/TLS-dependent cytotoxicity. PLoS Biol. 2011;9:e1001052. doi: 10.1371/journal.pbio.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguzzi A, Rajendran L. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron. 2009;64:783–790. doi: 10.1016/j.neuron.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 16.Furukawa Y, Kaneko K, Watanabe S, Yamanaka K, Nukina N. A seeding reaction recapitulates intracellular formation of Sarkosyl-insoluble transactivation response element (TAR) DNA-binding protein-43 inclusions. J Biol Chem. 2011;286:18664–18672. doi: 10.1074/jbc.M111.231209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Araya N, et al. Cooperative interaction of EWS with CREB-binding protein selectively activates hepatocyte nuclear factor 4-mediated transcription. J Biol Chem. 2003;278:5427–5432. doi: 10.1074/jbc.M210234200. [DOI] [PubMed] [Google Scholar]
- 18.Dormann D, et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee BJ, et al. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: Emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19(R1):R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barmada SJ, et al. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabashi E, et al. Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum Mol Genet. 2010;19:671–683. doi: 10.1093/hmg/ddp534. [DOI] [PubMed] [Google Scholar]
- 23.Bosco DA, et al. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet. 2010;19:4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elden AC, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanson KA, Kim SH, Wassarman DA, Tibbetts RS. Ubiquilin modifies TDP-43 toxicity in a Drosophila model of amyotrophic lateral sclerosis (ALS) J Biol Chem. 2010;285:11068–11072. doi: 10.1074/jbc.C109.078527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, et al. A Drosophila model for TDP-43 proteinopathy. Proc Natl Acad Sci USA. 2010;107:3169–3174. doi: 10.1073/pnas.0913602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y, Ferris J, Gao FB. Frontotemporal dementia and amyotrophic lateral sclerosis-associated disease protein TDP-43 promotes dendritic branching. Mol Brain. 2009;2:30. doi: 10.1186/1756-6606-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritson GP, et al. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J Neurosci. 2010;30:7729–7739. doi: 10.1523/JNEUROSCI.5894-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Estes PS, et al. Wild-type and A315T mutant TDP-43 exert differential neurotoxicity in a Drosophila model of ALS. Hum Mol Genet. 2011;20:2308–2321. doi: 10.1093/hmg/ddr124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanson NA, Jr, et al. A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum Mol Genet. 2011;20:2510–2523. doi: 10.1093/hmg/ddr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang JW, Brent JR, Tomlinson A, Shneider NA, McCabe BD. The ALS-associated proteins FUS and TDP-43 function together to affect Drosophila locomotion and life span. J Clin Invest. 2011;121:4118–4126. doi: 10.1172/JCI57883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Neurodegenerative disease: Amyloid pores from pathogenic mutations. Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 33.Conway KA, Harper JD, Lansbury PT. Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat Med. 1998;4:1318–1320. doi: 10.1038/3311. [DOI] [PubMed] [Google Scholar]
- 34.Conway KA, et al. Acceleration of oligomerization, not fibrillization, is a shared property of both alpha-synuclein mutations linked to early-onset Parkinson's disease: Implications for pathogenesis and therapy. Proc Natl Acad Sci USA. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lashuel HA, et al. Mixtures of wild-type and a pathogenic (E22G) form of Abeta40 in vitro accumulate protofibrils, including amyloid pores. J Mol Biol. 2003;332:795–808. doi: 10.1016/s0022-2836(03)00927-6. [DOI] [PubMed] [Google Scholar]
- 36.Neumann M, et al. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain. 2011;134:2595–2609. doi: 10.1093/brain/awr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ticozzi N, et al. Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:285–290. doi: 10.1002/ajmg.b.31158. [DOI] [PubMed] [Google Scholar]
- 38.Tan AY, Manley JL. The TET family of proteins: Functions and roles in disease. J Mol Cell Biol. 2009;1:82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Broustal O, et al. French clinical and genetic research network on FTD/FTD-MND FUS mutations in frontotemporal lobar degeneration with amyotrophic lateral sclerosis. J Alzheimers Dis. 2010;22:765–769. [PubMed] [Google Scholar]
- 40.Gitcho MA, et al. TARDBP 3′-UTR variant in autopsy-confirmed frontotemporal lobar degeneration with TDP-43 proteinopathy. Acta Neuropathol. 2009;118:633–645. doi: 10.1007/s00401-009-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson JO, et al. ITALSGEN Consortium Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biesecker LG. Exome sequencing makes medical genomics a reality. Nat Genet. 2010;42:13–14. doi: 10.1038/ng0110-13. [DOI] [PubMed] [Google Scholar]
- 43.Ng SB, et al. Exome sequencing identifies the cause of a Mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.