This study demonstrates that the tomato APETALA2a (AP2a) transcription factor modulates fruit ripening by negatively regulating ethylene biosynthesis and signaling. Various ripening regulators are shown to act upstream of AP2a. Gene expression analysis reveals that AP2a is involved in chloroplast to chromoplast transition.
Abstract
Fruit ripening in tomato (Solanum lycopersicum) requires the coordination of both developmental cues as well as the plant hormone ethylene. Although the role of ethylene in mediating climacteric ripening has been established, knowledge regarding the developmental regulators that modulate the involvement of ethylene in tomato fruit ripening is still lacking. Here, we show that the tomato APETALA2a (AP2a) transcription factor regulates fruit ripening via regulation of ethylene biosynthesis and signaling. RNA interference (RNAi)-mediated repression of AP2a resulted in alterations in fruit shape, orange ripe fruits, and altered carotenoid accumulation. Microarray expression analyses of the ripe AP2 RNAi fruits showed altered expression of genes involved in various metabolic pathways, such as the phenylpropanoid and carotenoid pathways, as well as in hormone synthesis and perception. Genes involved in chromoplast differentiation and other ripening-associated processes were also differentially expressed, but softening and ethylene biosynthesis occurred in the transgenic plants. Ripening regulators RIPENING-INHIBITOR, NON-RIPENING, and COLORLESS NON-RIPENING (CNR) function upstream of AP2a and positively regulate its expression. In the pericarp of AP2 RNAi fruits, mRNA levels of CNR were elevated, indicating that AP2a and CNR are part of a negative feedback loop in the regulation of ripening. Moreover, we demonstrated that CNR binds to the promoter of AP2a in vitro.
INTRODUCTION
Tomato (Solanum lycopersicum) is the primary model for climacteric fruit ripening for a combination of scientific and agricultural reasons. Its fruit plays an important role in the human diet and provides health benefits as a source of vitamins, minerals, and antioxidants (phenolics, folate, lycopene, and β-carotene) (Toor et al., 2005; Carrari and Fernie, 2006; Fraser et al., 2009). Fruit ripening is a complex, genetically programmed process that culminates in dramatic changes in color, texture, flavor, and aroma of the fruit flesh (reviewed in Alexander and Grierson, 2002; Carrari and Fernie, 2006). During tomato fruit ripening, free amino acids increase dramatically and their abundance changes differentially (Boggio et al., 2000; Sorrequieta et al., 2010). Ripening of climacteric fruits is characterized by an autocatalytic increase in respiration and ethylene biosynthesis just prior to the initiation of ripening. Ethylene biosynthesis occurs via a pathway (reviewed in Bleecker and Kende, 2000; Alexander and Grierson, 2002; Lin et al., 2009) involving two key biosynthetic enzymes: 1-aminocyclopropane-1-carboxylate (ACC) synthase (ACS), which converts S-adenosyl-l-methionine (SAM) into ACC (Sato and Theologis, 1989), and ACC oxidase (ACO), which further converts ACC to ethylene. Unraveling the regulation of these gene activities and the ethylene signaling pathway is important to understanding the processes of ripening, senescence, abscission, and response to stress.
Several genes that regulate tomato ripening through ethylene signal transduction have been identified. These are, among others, encoding two ethylene receptors, Never-ripe (Wilkinson et al., 1995; Yen et al., 1995) and ETHYLENE RESPONSE6 (ETR6) (Kevany et al., 2007), and Green-ripe (Gr), a gene encoding a protein of unknown function (Barry et al., 2005; Barry and Giovannoni, 2006). Ripening-associated transcription factors have been found to regulate, either directly or indirectly, the biosynthesis of ethylene. For example, two transcription factors, the MADS box protein RIPENING-INHIBITOR (RIN) (Vrebalov et al., 2002) and COLORLESS NON-RIPENING (CNR), a SQUAMOSA promoter binding protein (SBP) (Manning et al., 2006), were proposed to function early in the transcriptional activation cascade regulating ripening-related processes, including ethylene production and/or signaling. Mutations in both loci cannot be rescued by exogenous application of ethylene. RIN was shown to bind to the promoter of ACS2 (Ito et al., 2008) as does another MADS box protein, TAGL1, which was subsequently proposed to regulate ripening through regulation of ACS2 (Itkin et al., 2009; Vrebalov et al., 2009). Another transcription factor, the homeobox protein HB-1, was also recently reported to regulate fruit ripening and ACO1 expression (Lin et al., 2008). Unraveling both ethylene-dependent and -independent transcriptional networks that regulate fruit ripening is crucial for the understanding of this complex process. Here, we describe a member of another class of genes, an APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF).
AP2 is the founding member of the AP2/ERF superfamily, a family of transcription factors unique to plants. Together with the ethylene-responsive element binding protein and RAV subfamilies, the AP2 subfamily forms the AP/ERF superfamily, which plays important roles in diverse processes, such as development, ethylene response, and pathogen resistance. It is defined by the conserved 68–amino acid DNA binding AP2 domain, first described by Jofuku et al. (1994) for the AP2 protein from Arabidopsis thaliana. The AP2 subfamily is distinguished from the ethylene-responsive element binding protein and RAV subfamilies by the occurrence of two AP2 domain copies in each protein, while the proteins in the other two subfamilies have only one copy.
The archetypal AP2 from Arabidopsis (Koornneef et al., 1980; Jofuku et al., 1994) is characterized by mutants that show modifications of the outer two floral organ whorls. In weak ap2 mutants, sepals are converted to cauline leaf-like structures with stigmatic papillae at their tips, and petals show incomplete conversion into stamens (Bowman et al., 1989). In strong ap2 mutants, sepals are converted into ovule-bearing carpels and petals are absent (Bowman et al., 1991). AP2 is also involved in seed development, as shown by alterations in the seed coat epidermis in ap2 mutants (Jofuku et al., 1994) and regulation of seed size (Jofuku et al., 2005; Ohto et al., 2005). It has been shown that AP2 and its closest homologs are targets of miR172, which downregulates these target genes by a translational inhibition mechanism rather than by RNA cleavage (Aukerman and Sakai, 2003). The targets of miR172 were shown to regulate flowering time genes from Arabidopsis as well. Plants lacking TARGET OF EAT1 (TOE1) and TOE2 are early flowering, whereas plants overexpressing TOE1, TOE2, SCHLAFMUTZE (SMZ), and SCHNARCHZAPFEN (SNZ) flower late (Aukerman and Sakai, 2003; Schmid et al., 2003; Jung et al., 2007). AP2 homologs that share similarities in gene structure and function with AP2 have been isolated from numerous species. The putative Petunia hybrida ortholog, AP2A, is capable of complementing ap2 mutants of Arabidopsis. In Petunia, however, knockout mutations of AP2A do not affect floral organ development, suggesting that AP2 function is redundant in this species (Maes et al., 2001). Similarly, in Antirrhinum majus, two close homologs have been identified, LIPLESS1 (LIP1) and (LIP2), both of which need to be inactivated to get an ap2-like phenotype (Keck et al., 2003).
For tomato, no ap2-like mutants have been identified so far. In this study, using a transgenic approach that involves targeted RNA interference (RNAi) repression, we show that the closest tomato homolog of AP2, AP2a, plays a critical role in fruit ripening. This gene was also recently described as a negative regulator of ripening and of ethylene production (Chung et al., 2010). We found that tomato AP2-like genes also function in floral development, similar to Arabidopsis AP2, possibly in a redundant manner. In addition, by transcriptome profiling, we identified many direct and indirect target genes of AP2a, among which components of the ethylene and auxin signaling pathways and genes involved in the differentiation of chloroplasts into chromoplasts. Significant changes in key primary metabolites and carotenoids were observed as a result of AP2a downregulation. Our data show that AP2a has positive ripening regulatory functions besides its negative regulatory function in ethylene synthesis. Moreover, we show that the ripening regulators RIN, NON-RIPENING (NOR), and CNR as well as ethylene positively regulate AP2a expression in either a direct or indirect manner, while AP2a in turn negatively regulates CNR expression, thus positioning AP2a in a ripening regulatory network that includes a negative feedback loop.
RESULTS
AP2 Homologs in Tomato
To gain insight into the molecular regulation of tomato fruit development, we selected potentially important transcription factor genes for suppression in transgenic tomato using an RNAi strategy. Comparison of EST frequencies in various tomato EST libraries indicated that the AP2 homolog represented by TC162117 is highly expressed in the pericarp of developing tomato fruit. Publicly available microarray expression data show that the level of mRNA represented by this contig increases steadily during tomato fruit development, to a level at day 48 that was 2.5 times the level at 7 d postanthesis (http://ted.bti.cornell.edu/cgi-bin/array/unigene.cgi, SGN-U213383). This observation and the demonstrated important regulatory role of AP2-like transcription factors in developmental processes indicated that the AP2 homolog might play an important role in tomato fruit development as well.
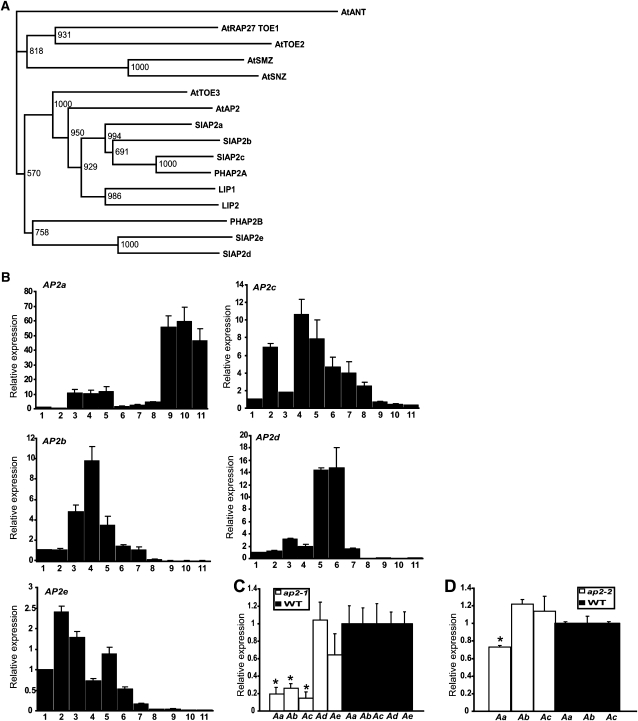
We mined the Solanaceae Genomics Network (SGN) tomato Unigene database for other unigenes with homology to Arabidopsis AP2 and, where necessary, extended cDNA sequences by 5′ and 3′ rapid amplification of cDNA ends to obtain sequences covering the entire open reading frame of each gene. At completion, we had identified five distinct tomato cDNAs encoding AP2 homologs, including the one represented by the tentative consensus mentioned above. Figure 1A (see Supplemental Data Set 1 online) shows a phylogenetic tree of the encoded proteins together with AP2 homologs of Arabidopsis, Petunia, and Antirrhinum, as far as available. One of these is the previously identified AP2 from tomato represented by Unigenes SGN-U579591, 580201, and 593325, which we here call AP2a (S. lycopersicum AP2a). For four other genes, full-length cDNA sequences were obtained by 5′ and 3′ rapid amplification of cDNA ends from available Unigene sequences. Two tomato proteins, which are homologous to the previously identified AP2A from Petunia, were designated AP2b (represented by SGN-U580135 and 589866) and AP2c (represented by SGN-U580203 and 579384). These three proteins are identical to AP2a, b, and c, respectively, published recently by Chung et al. (2010). The remaining two tomato AP2-like proteins are more distantly related to Arabidopsis AP2, are more similar to the P. hybrida AP2B (Maes et al., 2001), and were designated AP2d (represented by SGN-U563871) and AP2e (represented by SGN-U585439 and 585539). A more elaborate phylogenetic three including these five proteins with AP2 subfamily proteins from diverse species is shown in Supplemental Figure 1 online (see Supplemental Data Set 2 online). This analysis firmly places tomato AP2a, b, and c proteins together with Arabidopsis AP2 in a subclade of the AP2 subgroup of the AP2 subfamily of transcription factors as defined by Shigyo et al. (2006), indicating that these three proteins are putative tomato orthologs of AP2. Tomato AP2d and AP2e, with P. hybrida AP2B, are more similar to Arabidopsis TOE1 and TOE2. Several other tomato AP2 subfamily proteins, or fragments thereof, were identified from the SGN Unigene database and were included in the analysis. An alignment of the tomato proteins with Arabidopsis AP2, TOE2, and AINTEGUMENTA (ANT) is shown in Supplemental Figure 2A online. The phylogenetic position of AP2d and AP2e is less clear than for the other three proteins (as shown by the low bootstrap values on the major nodes). However, all five proteins belong to the euAP2 lineage, as they all contain a putative miR172 target site and lack the characteristic ANT lineage motifs, such as the 10–amino acid and 1–amino acid insertions in AP2 domain repeats 1 and 2, respectively (Kim and Nam, 2006).
Figure 1.
Phylogenetic Analysis of AP2a and Its Homologs and Comparison of Their Expression Pattern in Different Wild-Type Tissues and at Various Fruit Developmental Stages.
(A) Protein phylogenetic tree of tomato AP2s and selected homologs indicate that five AP2 paralogs exist in tomato. Numbers at tree nodes indicate bootstrap values from a total of 1000 trials.
(B) Relative expression profiles of AP2a, AP2b, AP2c, AP2d, and AP2e in different cv Moneymaker tissues obtained by quantitative RT-PCR. 1, Seedlings; 2, leaves; 3, roots; 4, flowers; 5, 5-mm fruits; 6, 1-cm fruits; 7, early green fruits; 8, mature green fruits, 9, breaker; 10, turning; 11-Br+7, 7 d after breaker stage, red fruits. Values represent means of two biological replicates, and vertical bars represent se of the means.
(C) Relative expression profiles of Aa, AP2a; Ab, AP2b; Ac, AP2c; Ad, AP2d; and Ae, AP2e in sepals of AP2i-1 transgenic lines or the wild type (WT). Values represent means of two biological replicates, and vertical bars represent sd. The wild-type expression data are normalized to 1. Asterisks indicate P value < 0.05 (t test) when comparing data for each measurement between the AP2i and wild-type plants.
(D) Relative expression profiles of Aa, AP2a; Ab, AP2b; and Ac, AP2c in the sepals of AP2i-2 transgenic lines or the wild type. Values represent means of two biological replicates, and vertical bars represent sd. The wild-type expression data are normalized to 1. Asterisk indicates P value < 0.05 (t test) when comparing data for each measurement between the AP2i and wild-type plants.
AP2a Expression Increases during Fruit Ripening
To better understand possible functions as well as to identify possible functional redundancy through overlapping expression patterns for the tomato AP2 genes, we determined, by quantitative RT-PCR (qRT-PCR), the expression levels of the five AP2-like genes in tomato seedlings, leaves, roots, and flowers at anthesis, flowers at 2 d postanthesis (Fruit I), and fruit at different developmental stages. All expression levels as related to the expression of the β-actin gene and normalized to the first sample (seedling) are shown in Figure 1B. From this figure it is apparent that of the three genes most related to Arabidopsis AP2, AP2b, and AP2c have similar expression patterns, with highest expression in flowers and some expression in roots, leaves, and early stage fruits, but no significant expression beyond the mature green stage. By contrast, AP2a is expressed at a relatively low level in flowers and early fruit stages but is strongly upregulated between the mature green and breaker stages and is highly expressed up to the red ripe stage. AP2d and AP2e are differentially expressed, with AP2d being preferentially expressed in very early fruit development and AP2e being expressed particularly in vegetative tissues, but also in flowers. All homologs were expressed in seeds, with AP2d and AP2e being the highest expressed (see Supplemental Figure 3A online). Thus, the five identified tomato AP2 paralogs are differentially expressed in vegetative tissues, flowers, and developing fruit. The differential expression of the three closest homologs of AP2, with only AP2a being expressed in ripening fruit and AP2b and AP2c both being expressed in flowers, suggests that extensive gene duplication and subsequent subfunctionalization of these AP2 homologs through differential expression have occurred in the tomato lineage since its divergence from the Arabidopsis lineage in evolution. If this is indeed the case, the genes that, like Arabidopsis AP2, are predominantly expressed in flowers may function in floral organ identity determination, as AP2 does, while AP2a may have acquired a novel function more specific for fleshy fruit development.
Phenotype of AP2a RNAi Lines
We constructed an RNAi vector from a 468-bp cDNA fragment (underlined in Supplemental Figure 2B online) of AP2a. The resulting construct, designated AP2i-1, contains two copies of a sequence encoding most of the two AP2 domains (see Supplemental Figure 2B online, bold) in an inverted repeat orientation under the transcriptional control of the cauliflower mosaic virus 35S promoter. Transgenic tomato cv Moneymaker plants harboring the AP2i-1 construct were generated to examine the effect of the suppressed AP2a expression on development.
Out of 12 analyzed diploid transformants, three lines with AP2i-1 exhibited severe alterations both in flower development and in fruit development and ripening, and three showed a milder phenotype. Figure 2 and Supplemental Figures 3B to 3E online show representative phenotypes of flowers and fruit of AP2i-1 lines. The sepals were greatly increased in size, both in length and in width (see Supplemental Figures 3B and 3C online). Sepals were fused along the margins and curled at the tip, forming a bell shape that completely enclosed the flower, leaving the petals invisible (Figure 2A; see Supplemental Figure 3C online). As the fruits developed, the sepals continued to increase in size (cf. the length and width of sepals in flowers and on fruit for wild-type and transgenic lines in Supplemental Figures 3B and 3C online). Because all five tomato AP2 paralogs are expressed in flowers, they might be functionally redundant in that organ; therefore, the flower phenotype may be the result of downregulation of one or more of the other AP2 homologs in addition to AP2a. To test this hypothesis, we performed qRT-PCR to compare the expression of the five AP2 homologs in AP2i-1 and wild-type sepals (Figure 1C). We found that not only AP2a but also its closest homologs, AP2b and AP2c, were downregulated in sepals, while the expression of the other two members of the family, AP2d and AP2e, was not affected.
Figure 2.
Ripening Effects of Decreased AP2a Expression.
(A) Typical example of the ripening phenotype of AP2i-1 RNAi lines. Fully developed fruit (equivalent to Br + 7, 7 d after breaker stage) are orange in color, never achieving the full red color of the wild-type ripe tomatoes at the same stage.
(B) Different stages of fruit ripening: (from left to right) mature green (~40 d postanthesis), turning, pink and red ripe (Br + 7) for wild-type (WT) and AP2i-1 RNAi lines. Fruits from independent transformant lines are shown (5, 8, 10, and 11, respectively), which are delayed in color development, never developing full red color.
(C) Ripening wild-type fruits have an even, homogenous color and a smooth, intact surface during the orange and red stages of ripening. The color of the ripening AP2i-1 RNAi lines at the Br + 7 stage is not homogenous, with sectors displaying different colors. The fruit does not develop the red color but remains orange, even when the tissue has characteristics of being fully ripe or overripe (i.e., soft and mushy). The surface of the fruit splits open (arrows and in [A]).
(D) Compared with the wild type (left), the fruit from AP2i-1 lines (right) at the mature green stage has a distorted shape, exhibiting strong indentations resulting in less round fruit. Additionally, the surface of the fruit is bumpy (arrows).
(E) Wild-type (left) and AP2i-2-15 RNAi fruits (ap2-2-15) at Br + 7, showing ripening defects and uneven pigmentation.
Additionally, the morphology of the fruit was altered in some of the AP2i-1 lines (Figures 2B to 2D). A wild-type fruit (Figure 2D) is round, with a smooth, consistent epidermal surface. Fruits of the AP2i-1-8 line, on the other hand, were not round but had large indentations delineating the location of each locular cavity. Another common feature of these fruits was the raised bumpy areas that were present intermittently on the epidermal surface of the fruit (Figure 2D, arrowheads). When the seeds from these AP2i-1-8 fruits were closely examined, we found that seed morphology was also affected. In AP2i-1 lines, a variety of aberrant seed shapes was observed (see Supplemental Figures 4A to 4E online). Moreover, seed coat morphology, specifically the size and shape of the surface hairs, was strongly affected. In tomato, the seed coat hairs are the cell wall remains of the outer epidermal cells. Their size and shape varies between cultivars and cv Moneymaker has laterally banded hairs as shown in Supplemental Figures 4B and 4E online (Chakrabarti et al., 2003).
AP2a Is Involved in Fruit Ripening
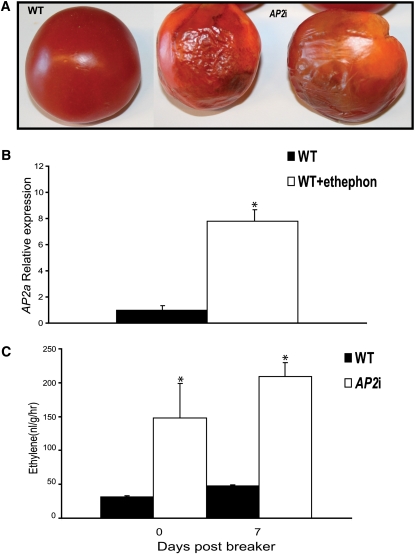
In addition to the alterations observed in the early development of the fruit, AP2i-1 lines exhibited distinct alterations in the ripening process (Figure 2). Ripe AP2i-1 fruits are shown in Figure 2A. All fruits had ripened, yet never turned from orange to homogenous red. The fruit phenotype for wild-type and AP2i-1 lines at various stages of ripening is shown in Figure 2B (several fruits of the same truss, representing different ripening stages, are shown). In the wild type, fruit ripening (Figure 2B, top row, left to right) begins with mature full-sized green fruit. In the next stage, the breaker (Br) stage, a visible color change just begins to occur. By the turning stage, the entire fruit has a consistent orange color. In the final stage (breaker + 7 d) of ripening, the fruit has a deep red color and starts to soften. The differences in ripening between the AP2i-1 and wild-type fruits became apparent at the turning stage. At breaker + 7 d, the fruit retained its orange color, even though other aspects of the ripening process, such as tissue softening, had begun. In Figure 2C, fruits from the wild type and two independent AP2i lines, AP2i-1-8 and AP2i-1-10, at turning (top) and breaker + 7 stage (bottom) are shown. Wild-type fruit at the turning stage was a homogenous orange. However, the color of fruit from AP2i-1 lines was patchy with sectors of different shades of yellow and orange. At the breaker + 7 d stage, wild-type fruit was homogenous red, whereas AP2i-1 lines still displayed sectors of different colors and did not turn red. The internal fruit anatomy at breaker + 7 d is shown in Supplemental Figure 4F online. The inside of wild-type fruit was red throughout. The locular cavities were filled with seeds and jelly, resulting in a juicy appearance. While AP2i-1-10 line fruits were more similar to the wild type in texture, fruits from line AP2i-1-8 appeared drier and the fruit appeared to be crumbly, with the internal structure collapsing (see Supplemental Figure 4F online). Line AP2i-1-8 showed the most severe phenotype. Additionally, the peel of AP2i-1-8 fruits was frequently cracked (Figure 2C, arrows). We noted that the AP2i-1-8 fruits deteriorated faster than the wild-type fruits (Figure 3A), and this could explain the observed peel cracks in the fruits shown in Figure 2C.
Figure 3.
AP2 RNAi Fruits Senescenced Faster Than the Wild-Type Fruits and Produced More Ethylene.
(A) Comparison of wild-type (WT) and AP2i-1 fruits 14 d after harvesting at stage Br + 7 d.
(B) Relative expression profiles of AP2a in mature green fruits, treated or not with ethephon. Values represent means of two biological replicates, and vertical bars represent sd. The wild-type expression data are normalized to 1. Asterisks indicate a significant P value < 0.05 (t test) when comparing data for each measurement between the treated and nontreated wild-type samples.
(C) Fruits from Br and Br + 7 stages were sealed in airtight vials, and 2.5 mL of gas was sampled after 35 min. Values represent means of at least four fruits, and vertical bars represent sd. Asterisks indicate a significant P value < 0.05 (t test).
[See online article for color version of this figure.]
Because the AP2i-1 construct targets AP2a as well as AP2b and AP2c, and although the latter two show no expression in ripening fruits, we designed another RNAi fragment named AP2i-2 (highlighted in gray in Supplemental Figure 2B online), which excludes the conserved AP2 domains. In agreement with the proposed role of AP2a in fruit ripening, fruits from three out of 15 transformant lines containing this construct showed the same ripening defects (Figure 2E), without the sepal or seed phenotypes observed in the AP2i-1 lines. This supports the proposed function of AP2a in fruit ripening, while flower- and seed-specific functions may be shared with, or are exclusively determined by, AP2b and AP2c. To confirm this, we performed qRT-PCR to compare the expression of the five AP2 homologs in AP2i-2 and wild-type sepals (Figure 1D). We found that AP2a was downregulated in the sepals of AP2i-2, but, in contrast with AP2i-1 plants, neither of its closest homologs, AP2b or AP2c, was downregulated. To confirm downregulation of AP2a in fruits of both types of RNAi lines, we performed protein gel blot analysis using AP2a-specific peptide antiserum. In contrast with the situation in wild-type fruits, AP2a was not detected in the transgenic fruits, in both breaker stage (Br) and in 7 d after breaker stage (Br+7) fruits of AP2i-1 or AP2i-2 lines (see Supplemental Figure 3F online).
Fruits of line AP2i-1-8 were used for more detailed characterization of the ripening-specific function of AP2a. In the progeny of this line, the described phenotypes all cosegregated with the transgene construct as determined by PCR.
AP2a Expression Is Stimulated by Ethylene, and Its Repression Resulted in Fruits Defective in Ripening but Producing Higher Levels of Ethylene
To achieve full ripening, tomato fruits require synthesis, perception, and signal transduction of the plant hormone ethylene (Alexander and Grierson, 2002; Kevany et al., 2007). As ripening progresses, the expression of many genes is initiated or upregulated (reviewed in Alexander and Grierson, 2002). We observed that the expression of AP2a is increased during ripening. To distinguish if this regulation is ethylene dependent or ethylene independent, we incubated wild-type tomato fruits at the mature green stage with the ethylene precursor ethephon. After 6 h of induction, we observed an 8-fold upregulation of AP2a expression (Figure 3B).
To investigate if a high level of ethylene production in the AP2i fruits may be responsible for the observed early fruit senescence (Figure 3A), we measured the ethylene production in fruits during ripening. As shown in Figure 3C, AP2i-1-8 fruits produced 4- to 6-fold more ethylene at breaker and breaker + 7 stages than wild-type fruits at the same stage. Thus, AP2a appears to function as a negative regulator of ethylene synthesis in tomato fruits by a negative feedback loop.
AP2a Downregulation Alters Fruit Carotenoid Levels
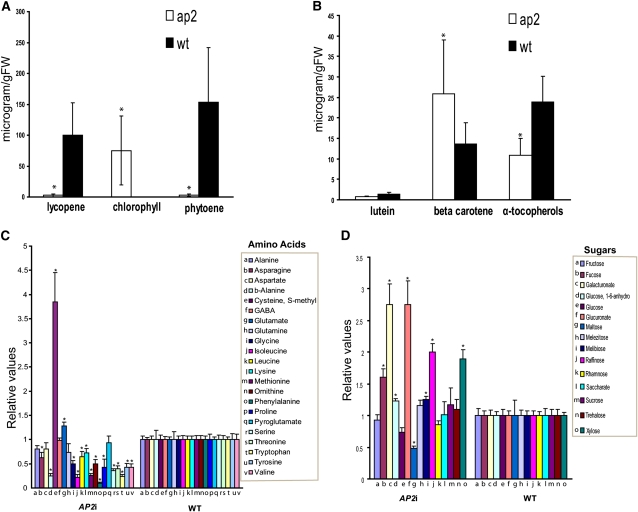
The green-to-red color change typical of ripening tomato fruits is largely due to chlorophyll degradation and accumulation of carotenoids, including β-carotene and lycopene (reviewed in Giovannoni, 2001). Lycopene, which confers the red color to ripe tomatoes, accounts for 70 to 90% of the total carotenoids present (Burns et al., 2003; Alba et al., 2005). To examine the underlying causes of the color differences observed between wild-type and AP2i ripe fruits in more detail, biochemical analysis of carotenoid (β-carotene, all-trans lycopene, lutein, and their precursor phytoene) levels was performed on wild-type and AP2a silenced fruits at a similar developmental stage (Figure 4). The AP2i fruits had decreased levels of both phytoene and lycopene. By contrast, an increase in the β-carotene content was found, accounting for the orange color of the ripe AP2i fruits. The α-tocopherol content was also slightly decreased in these fruits. Elevated levels of chlorophyll were observed in the AP2i fruits compared with the wild-type fruits 7 d after the breaker stage (Figure 4A). No difference in the lutein content was observed between the wild-type and transgenic fruits.
Figure 4.
Metabolic Comparison of the AP2i-1 and Wild-Type Fruits.
(A) and (B) Comparison of the chlorophyll, carotenoid, and α-tocopherol levels (microgram/g fresh weight [FW]) in wild-type (wt) and transgenic AP2i-1 tomatoes at stage Br + 7. Biological replicates (11 fruits per sample) were performed in triplicate, and the data are presented as means ± sd. Student’s t test was used to determine significant differences between wild-type and transgenic fruits P < 0.01 (asterisks).
(C) and (D) Comparison of the primary metabolites (amino acid and sugar) levels in wild-type and AP2i-1 tomatoes (six biological replicates) at stage Br + 7. Wild-type values are normalized to one. Vertical bars represent se of the means. Asterisks show statistically significant changes according to Student’s t test (P < 0.05).
[See online article for color version of this figure.]
Gene Expression Analysis Identifies Genes Regulated by AP2a during Ripening
To identify genes that might be directly or indirectly regulated by AP2a in tomato, we compared gene expression between transgenic knockdown plants in the T1 generation, in which the transgene cosegregated correctly with the phenotype, and their wild-type siblings. For this purpose, fruit RNA from three plants derived from transformant line AP2i-1-8 and from three plants of wild-type siblings were hybridized to the Syngenta Tomato Affymetrix GeneChip. Analysis of differentially expressed genes revealed 1093 genes with significant differential expression at a P value cutoff of 0.001 (false discovery rate Q-value < 0.002), and genes with a >2-fold difference in expression were identified. After removal of apparent redundancies in the probe set, 527 genes were found to be expressed at significantly lower levels (2log≤−1), and 523 genes at higher levels (2log≥1) in the transgenic fruits. A full list of significantly differentially expressed genes can be found in Supplemental Data Set 3 online. A shortlist of differentially regulated genes, discussed below, is shown in Table 1.
Table 1.
Gene Expression Changes for Selected Ripening-Related Genes in AP2 RNAi Ripe (Breaker +7 d) Fruits
| SGN Unigene | Possible Function | Fold Change RNAi/Wild Type | Annotation | Reference |
| SGN-U581293 | – | −49 | HSP21/pTOM11 (chloroplast) | Alba et al. (2005) |
| SGN-U578203 | – | −27 | Putative HSP20 | Alba et al. (2005) |
| SGN-U566498 | Brassinosteroid synthesis | −25 | DWARF | Ozaki et al. (2010) |
| SGN-U578017 | Metabolic intermediate biosynthesis | −24 | HMG-CoA reductase 2 | Rodriguez-Concepcion and Gruissem (1999) |
| SGN-U572041 | Lipid metabolism; oxylipin biosynthesis | −14 | LOXC | Alba et al. (2005); Eriksson et al. (2004) |
| SGN-U584210 | – | −11 | HSP70 (chloroplast) | Alba et al. (2005) |
| SGN-U579266 | – | −6.7 | Putative heat shock protein | Alba et al. (2005) |
| SGN-U573533 | Hormone metabolism, auxin induced | −6.7 | GH3.3 (indole-3-acetic acid amido synthetase) | Ozaki et al. (2010) |
| SGN-U572734 | Ripening chlorophyll degradation | −6.6 | SGR1 (stay green protein) | Barry et al. (2008) |
| SGN-U563074 | Cell wall metabolism | −6.3 | PME1.9 (pectin methylesterase) | Eriksson et al. (2004) |
| SGN-U581241 | Brassinosteroid synthesis | −5.2 | DWF1 (DIMINUTO) | Ozaki et al. (2010) |
| SGN-U576599 | Cell wall metabolism | −4.9 | EXP3 | Eriksson et al. (2004) |
| SGN-U584987 | Carotenoid biosynthesis | −3.5 | CrtISO (carotenoid isomerase) | Isaacson et al. (2002) |
| SGN-U582628 | Chlorophyll degradation | −3.3 | ACD2; RCCR (red chlorophyll catabolite reductase) | – |
| SGN-U578028 | Lipid metabolism | −3.1 | LOXB | Eriksson et al. (2004) |
| SGN-U568611 | Carotenoid biosynthesis | −2.8 | CrtR-b2 | Bramley (2002) |
| SGN-U580564 | Auxin metabolism | −2.8 | SAUR family (auxin responsive) | – |
| SGN-U569639 | Auxin responses | −2.6 | ARF4/DR12 | Jones et al. (2002) |
| SGN-U582562 | Carotenoid desaturation | −2.4 | PTOX (plastid terminal oxidase) | Josse et al. (2000) |
| SGN-U565008 | – | −2.3 | FtsH-like protein Pftf | – |
| SGN-U576134 | – | −2.2 | ORANGE, DNAJ-like protein | – |
| SGN-U593894 | Carotenoid biosynthesis | −2.3 | PDS1 (phytoene desaturase) | – |
| SGN-U583842 | Ethylene biosynthesis | 2.2 | ACS6 | Barry et al. (2000) |
| SGN-U580893 | Ethylene biosynthesis | 2.3 | SAM3 | – |
| SGN-U576603 | Pathogen and insect response | 2.5 | MPKA1 | – |
| SGN-U571201 | Cell wall metabolism | 2.6 | MANS2 | – |
| SGN-U577086 | Ethylene signaling | 3.0 | ETR5 | Kevany et al. (2007) |
| SGN-U580563 | Cell wall metabolism | 3.2 | Polygalacturonase | – |
| SGN-U564043 | Protein phosphorylation | 3.9 | MKK4 | Alba et al. (2005) |
| SGN-U569421 | Carotenoid biosynthesis | 3.9 | ZEP1 (zeaxanthin epoxidase) | Thompson et al. (2000) |
| SGN-U579176 | Cell wall metabolism | 4.0 | MAN3 | – |
| SGN-U563352 | Ethylene signaling | 4.2 | MBF1b/ER24 | Zegzouti et al. (1999) |
| SGN-U571269 | Auxin signaling | 5.3 | MONOPTEROS-like protein | – |
| ? | Auxin signaling | 5.4 | GH3/ IAA synthetase | – |
| SGN-U572472 | Auxin metabolism | 6.6 | Putative SAUR family protein | – |
| SGN-U578110 | Protein degradation | 6.8 | EBF1 | Guo and Ecker (2003) |
| SGN-U579235 | Biotic stress response | 7.0 | PR-P2 | – |
| SGN-U579104 | Ethylene biosynthesis | 7.1 | LeACO4 | Nakatsuka et al. (1998) |
| SGN-U569953 | Amino acids metabolism | 8.2 | BCAT1 | Maloney et al. (2010) |
| SGN-U581694 | Ethylene signaling | 9.2 | ETR6 | Kevany et al. (2007) |
| SGN-U580736 | Flavonoid synthesis | 12 | Phe ammonia lyase | – |
| SGN-U565756 | Regulation of transcription | 12 | AIM1 (MYB TF) | Abuqamar et al. (2009) |
| SGN-U570620 | Cell wall metabolism | 14 | CEL1 (endo-1,4-β-glucanase) | – |
| SGN-U585765 | Ethylene signaling | 15 | EIL2 | Tieman et al. (2001) |
| SGN-U578033 | Biotic stress response | 79 | PepEST (esterase, pathogen, wounding and jasmonate-induced) | – |
| SGN-U580143 | Defense response | 85 | PR6/PR1b1 | – |
Strikingly, the extreme ends of the spectrum of AP2a-regulated genes, as shown in Table 1, are made up predominantly of genes involved in heat shock responses (downregulated in RNAi fruits, 7- to 50-fold) and by genes involved in ethylene synthesis and signal transduction, as well as ethylene-(co-)regulated pathogen and wounding response genes (upregulated in RNAi plants 7- to 58-fold). Although heat shock–responsive genes have not been described in association with fruit ripening, inspection of publicly available gene expression data derived from Alba et al. (2005) and Ozaki et al. (2010) shows that many of these are upregulated from the breaker stage onwards, indicating that AP2a positively regulates these genes during ripening. Pathogenesis-related proteins that are also ethylene induced in other tissues are not known to be regulated during ripening, and the significant higher expression of these genes in fruits of RNAi plants may represent misregulation of their expression as a result of AP2a downregulation, higher ethylene production, or both. Several genes encoding pathogenesis-related proteins were identified as being enriched in the Cnr mutant fruit mRNA in a differential cDNA screen against wild-type fruit, suggesting that these genes are also negatively regulated by CNR (Eriksson et al., 2004). Several genes that were previously characterized as being ripening induced (Alba et al., 2005) had altered expression levels in AP2 RNAi fruits, but with opposite effects. LIPOXYGENASE C (LOXC), PECTIN METHYLESTERASE 1.9, EXPANSIN3 (EXP3), and LOXB were all downregulated compared with the wild-type fruits. On the other hand, MITOGEN-ACTIVATED PROTEIN KINASE1 (MPKA1), MITOGEN-ACTIVATED PROTEIN KINASE KINASE4 (MKK4), and MULTIPROTEIN BRIDGING FACTOR1 (MBF1), which are all induced during ripening as well, were upregulated in the RNAi fruits compared with wild-type fruits (Table 1). These observations suggest that AP2a may have activating and suppressing effects on genes that are normally induced during ripening.
During tomato fruit development, significant changes occur in the cell wall components and in the expression of polysaccharide-degrading enzymes (reviewed in Carrari and Fernie, 2006). We found several of the genes encoding these enzymes to be differentially regulated in the AP2 RNAi fruits. A polygalacturonase gene (PG) is significantly upregulated compared with the wild type, as are genes encoding endo-1,3-1,4-β-d-gluconase (CEL1), mannosidase (MAN3), and mannan synthase (MANS2). A PECTIN METHYLESTERASE1 (PE1) gene and the gene encoding expansin precursor EXP3, both found to be downregulated in the Cnr mutant, were also downregulated in AP2 RNAi fruits (Table 1). These data suggest that AP2a and CNR regulate a number of cell wall–related genes in a partially overlapping manner.
In agreement with the observed higher ethylene production in AP2 RNAi plants (Figure 3C), we found genes involved in ethylene synthesis, specifically SAM3, ACO4, and ACS6, to be upregulated in the transgenic fruits, suggesting that, during ripening in wild-type fruits, AP2a represses ethylene production via these genes. Genes involved in ethylene signal transduction that were found to be significantly upregulated in the transgenic fruits are EIN3-LIKE2 (EIL2), ETR5, ETR6, and EIN3 BINDING F-BOX1 (EBF1).
In AP2i plants, several carotenoid biosynthesis genes, PHYTOENE DESATURASE (PDS), CAROTENE-β-HYDROXYLASE (CtR-b2), and CAROTENE ISOMERASE (CtrISO), were significantly downregulated (Table 1). Expression of the other carotenoid synthesis genes was detected but not significantly altered. Additionally, we found ZEP1, which encodes zeaxanthin epoxidase and is involved in abscisic acid synthesis, to be upregulated (4-fold) in the transgenic fruits. These results suggest that AP2a positively regulates carotenoid synthesis during ripening through activation of specific genes in the biosynthetic pathway.
The expression of several key transcripts related to biosynthetic genes involved in phenylpropanoid and flavonoid formation was also changed in the AP2 RNAi fruits (Table 1; see Supplemental Data Set 3 online).
We found several genes that are possibly involved in hormone synthesis to be downregulated in the transgenic fruits. Among these are two genes that are involved in brassinosteroid synthesis: the D (dwarf) gene and a DWARF1/DIMINUTO-like gene (DWF1). These results suggest that AP2a positively regulates brassinosteroid synthesis during ripening. Furthermore, we observed that the auxin response factor ARF4 gene was downregulated. Upregulation of a MONOPTEROS/ARF5 homolog and of two homologs of early auxin-responsive genes, gh3, and a SAUR family protein homolog and downregulation of another gh3 homolog and SAUR family homolog suggest that auxin responses are perturbed in these plants.
Finally, we found the expression of a number of genes implicated in chromoplast differentiation to be significantly downregulated. These were the STAY GREEN PROTEIN1 (SGR1) gene, RCCR (encoding red chlorophyll catabolite reductase), a tomato homolog of the cauliflower (Brassica oleracea) ORANGE gene (OR), and a homolog of a target of the latter, a tomato homolog of the pepper PFTF gene.
AP2a Silenced Fruits Show Increased Accumulation of Sugars and Putrescine and Reduced Free Amino Acid Levels
To identify differentially regulated metabolic pathways in AP2i-1-8 plants, we analyzed our microarray data using the Plant MetGenMAP database (Joung et al., 2009). Among the pathways identified, we found an overrepresentation of primary metabolic processes (see Supplemental Table 1 online). Correspondingly, gas chromatography–mass spectrometry (GC-MS) profiles (Lisec et al., 2006) revealed that the AP2a silenced fruits displayed substantial changes in the level of primary metabolites (Figures 4C and 4D; see Supplemental Table 2 online). To visually compare alterations in sectors of metabolism and interactions between metabolite levels in the AP2i fruits and the wild type, the fold changes in metabolite levels were painted onto biochemical pathway displays (see Supplemental Figure 5 online). The majority of the amino acids (15 out of 22) were present at significantly lower levels in the AP2i fruits than in the wild type, and the most dramatic reductions were in β-Ala, Ile, Met, Phe, and Trp. The observed reduced levels of Ile and Leu (branched-chain amino acids) are in agreement with the higher expression of BRANCHED-CHAIN AMINOTRANSFERASE1 (BCAT1; 8.6-fold), which was shown to be involved in the degradation of these two amino acids (Maloney et al., 2010). Similarly, nine of the 15 organic acids measured displayed significant changes. However, these were predominantly increased in the AP2i line, with the exception of the tricarboxylic acid cycle intermediates citrate, fumarate and succinate, which were decreased. The increase in benzoate, dehydroascorbate, glycerate, glycolate, nicotinate, and nonanate was relatively mild. The fatty acids dodeconate and octodeconate were also present at mildly, yet significantly, higher levels in the AP2i line. Of the 15 sugars we detected, seven (Fuc, galacturonate, glucose-1,6-anhydro-glucuronate, melibiose, raffinose, and Xyl) were present at higher levels in the AP2i line, whereas maltose was present at lower levels. With the exception of maltose and raffinose, these sugars and sugar derivatives are generally associated with cell wall or ascorbate metabolism. It is interesting to note that the content of the major sugars of the fruit were consistent between genotypes. The content of sugar alcohols in the genotypes was somewhat variable, with an increase in glycerol content but a decrease in inositol in the AP2i line. By contrast, inositol 3-phosphate levels were higher in the AP2i line, whereas all other sugar phosphates were invariant. Of the six miscellaneous compounds we measured, five were present at significantly different levels between the genotypes, with gluconate-1,5-lactone, phosphorate, putrescine, 3-hydroxy-pyridine, and 3-caffeoyl quinate being elevated in the AP2i line. Putrescine was elevated to levels that were 12-fold higher than those observed in the wild type.
Regulatory Interactions between AP2a and Other Ripening Regulators
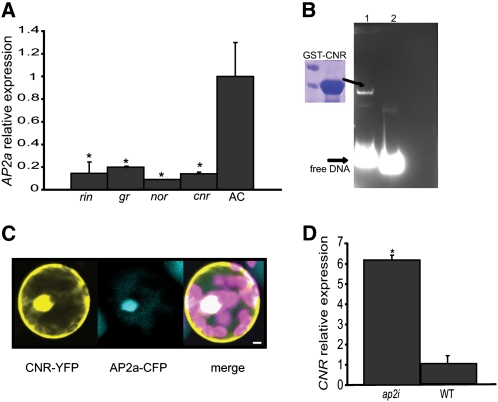
Several transcription factors are positive regulators of tomato fruit ripening. Although AP2i fruits failed to complete ripening, they did soften in a similar way as wild-type fruits, in contrast with, for example, Cnr mutant fruits (Thompson et al., 1999). The Cnr tomato mutant has a colorless, nonripening phenotype and mealy pericarp (Eriksson et al., 2004; Manning et al., 2006). CNR and other ripening regulators, like RIN and TAGL1, were found to have ethylene-dependent and ethylene-independent functions (Giovannoni, 2007; Itkin et al., 2009; Vrebalov et al., 2009). We investigated AP2a expression in nor (Tigchelaar et al., 1973), rin, Gr, and Cnr mutant backgrounds (Figure 5A). The expression of AP2a appeared to be reduced in all four mutants. Our results suggest that, although AP2a appears at first sight to be a negative regulator of ripening through inhibition of ethylene production, it is itself positively regulated by essential regulators of ripening, like CNR, RIN, and NOR (Barry et al., 2005; Giovannoni, 2007; Cantu et al., 2009), and its expression is ethylene dependent, as shown by its downregulation in the Gr mutant. Next, to test whether the CNR protein could bind directly to the AP2a promoter, we expressed the full-length CNR protein tagged with glutathione S-transferase (GST) (GST-CNR) in Escherichia coli (Figure 5B. Two AP2a promoter regions were selected, one containing the predicted (GTAC) core binding motif of the plant-specific SBP domain (Birkenbihl et al., 2005) and the other one not containing the binding site, to be used as a negative control. To confirm that CNR binds specifically to this core element, we performed an electrophoretic mobility shift assay using the two selected promoter elements (Figure 5B). Our data showed that CNR can bind specifically to the promoter element of AP2a in vitro and positively regulates its expression in vivo. Furthermore, CNR and AP2a were found to be colocalized in the nucleus of transfected Arabidopsis protoplast cells (Figure 5C).
Figure 5.
Relative Expression of AP2a in Fruits from Different Ripening-Defective Mutant Backgrounds.
CNR protein binds directly to the promoter element of AP2a, and the two proteins colocalized in the nucleus of plant cells.
(A) Relative expression profile of AP2a in wild-type cv WT-AC (Ailsa Craig), nor, rin, Gr, and Cnr fruits at Br +7 d, obtained by qRT-PCR. Values represent means of two biological replicates, and vertical bars represent sd. The wild-type expression data are normalized to 1. Asterisks indicate P value < 0.05 (t test).
(B) SDS gel (left) showing GST-purified CNR protein used for the gel retardation assay. Main panel, gel retardation experiments with biotinylated DNA probes and GST-CNR purified protein. 1, AP2a promoter element containing the SBP true binding site; 2, AP2a promoter element, which does not contain the conserved SBP binding motif, used as a negative control.
(C) Confocal scanning laser microscopy images of an Arabidopsis protoplast cotransfected with both CNR-YFP and AP2a-CFP constructs. Bar = 5 μm.
(D) Upregulation of CNR in the pericarp of Br stage of the AP2i fruit compared with the wild-type cv Moneymaker (WT). The relative expression levels were obtained by qRT-PCR. Values represent the means of two biological replicates, and vertical bars represent sd.
[See online article for color version of this figure.]
The ripening regulatory transcription factors NOR, CNR, and RIN activate the expression of the AP2a transcription factor, which in turn acts partially as a negative regulator of ripening through modulation of ethylene signaling. Our microarray analysis was performed with RNA from AP2i whole fruits and yielded only a few transcription factors that were significantly regulated (Table 1; see Supplemental Data Set 3 online). Therefore, we decided to investigate in greater detail the expression of CNR, RIN, and TAGL1 specifically in the pericarp of the AP2i fruits by qRT-PCR. Although no significant change in the expression of RIN or TAGL1 was observed (see Supplemental Figure 6 online), CNR mRNA levels were significantly higher in the pericarp of AP2i fruits (Figure 5D). Taking into account our previous observation that AP2a is positively regulated by CNR, these data suggest that AP2 in the pericarp of wild-type fruits suppresses the expression of CNR by a negative feedback loop.
DISCUSSION
In this article, we characterized a putative tomato ortholog of AP2, here named AP2a. We studied the function of the AP2a transcription factor by analyzing transgenic plants silenced for the AP2a gene. A phenotype in tomato fruit ripening was observed, which is similar to that described by Chung et al. (2010). The function of the other tomato AP2 homologs was apparent from the use of another RNAi construct that targets the conserved AP2 domains, which resulted in additional floral phenotypes. In agreement with Chung et al. (2010), we found that AP2b and AP2c are not significantly expressed in ripening fruit, thus showing that, also in plants containing the less specific AP2i-1 construct, the fruit ripening phenotype is due to downregulation of AP2a. Transcriptomic and metabolic analysis of the AP2i-silenced fruits described in this study indicate that the AP2a transcription factor plays important roles during tomato fruit development in ripening-related metabolism, in ethylene biosynthesis and signaling pathways, and in the differentiation of chromoplasts. Although, using qRT-PCR, we found a decrease in the mRNA of target gene(s) in the sepals of the RNAi lines, we did not detect such a decrease in ripe fruits, where AP2a expression is highest. This observation was supported by the microarray analysis, which also did not detect a significant decrease in AP2a mRNA in transgenic fruits. Nonetheless, at the protein level, AP2a was downregulated in fruits of both types of RNAi plants. These results point to a relatively inefficient cleavage of the mRNA and, hence, a relatively inefficient degradation of the endogenous mRNA. In Arabidopsis, AP2 protein expression is regulated by a microRNA, miR172, which acts as a translational repressor (Aukerman and Sakai, 2003; Chen, 2004). It was shown that cleavage of the endogenous AP2 mRNA occurs also but is normally not detectable because of feedback regulation of AP2 transcription by the AP2 protein (Schwab et al., 2005). It was suggested that although perfect or near-perfect complementarity between miRNA and target sequence is usually thought to cause the miRNA to enter the RNAi pathway (Rhoades et al., 2002), and cleavage products of the target mRNA can be detected by PCR in many cases, it is possible that translational inhibition may be more common than appreciated so far (Aukerman and Sakai, 2003).
AP2b, AP2c, or Both, Possibly with AP2a, Function in Floral Organ Development
The flower phenotype of the tomato AP2i-1 lines is reminiscent of some of the aspects of weaker ap2 phenotypes in Arabidopsis as well as of that in Antirrhinum lip1 lip2 double mutants (Bowman et al., 1989; Keck et al., 2003). The Arabidopsis A-class genes, APETALA1 (AP1) and AP2, confer sepal identity in the first floral whorl. While MADS-MC (Macrocalyx) is a likely ortholog of AP1 (Vrebalov et al., 2002), a gene or genes with an A-class function similar to that of AP2 has not yet been found in tomato. Here, we found that sepals of AP2i-1 lines are elongated and show distinct leaf-like characteristics, such as a thinner blade in which venation can be easily observed. However, this sepal phenotype was not observed in the transgenic plants expressing the AP2i-2 construct for silencing, suggesting that there may be additional tomato AP2 genes that are either solely responsible for this sepal phenotype or functionally redundant with each other or with AP2a for the floral development function, similar to the redundancy observed in Antirrhinum AP2 genes. We identified five homologs of the Arabidopsis AP2 gene in tomato and showed that the observed sepal phenotype in the AP2i-1 lines is correlated with downregulation of AP2a and its closest homologs, AP2b and AP2c. Unlike in strong Arabidopsis ap2 mutants (Bowman et al., 1989, 1991) and in Antirrhinum lip1 lip2 mutants (Keck et al., 2003), we observed no changes in the other floral whorls. However, similar to Arabidopsis ap2 mutants, suppression of tomato AP2 homologs also affected seed shape and seed coat morphology. The AP2a suppression could cause defects in the development of the seed testa, specifically in the outer epidermal layer as in Arabidopsis ap2 mutants (Jofuku et al., 1994; Leon-Kloosterziel et al., 1994), and may well explain the aberrant seed hair morphology that we observed in the AP2i-1 lines.
SlAP2a Functions in Fruit Development
Our data demonstrate that AP2a plays a role in fruit development. The effect of AP2 suppression on early fruit development is a bumpy appearance of the green fruits, which is conceivably caused by a defect in the normal or coordinated expansion of the pericarp. Ripening fruits had a number of anatomical differences from wild-type fruits, such as, to a varying degree, a smaller volume of jelly, a dry and crumbly appearance (observed in one line only) of the pericarp, and cracking of the skin, indicating a defect in the expansion or elasticity of the epidermis. Differences in the ripening process were apparent from the differences in color development between wild-type and AP2i lines. Orange fruits of tomato AP2i lines had a blotchy appearance with light colored or white sectors and failed to turn completely red. Biochemical analysis of the major colored compounds of the tomato fruit, the carotenoids, indicated that AP2i fruits are affected in phytoene and lycopene production, explaining their failure to turn fully red. The reduction in color development correlates with the reduced expression of PDS, PTOX, and CRTISO in the AP2i fruits, since they encode proteins involved in the biosynthesis of the red all-trans lycopene (for review, see Bramley, 2002). PDS, PTOX, and CRTISO expression in tomato fruits increases dramatically during tomato fruit ripening (Josse et al., 2000; Isaacson et al., 2002), as does expression of AP2a. Although reduction of the expression of CrtR-b2 was also detected, no difference in lutein accumulation was observed in the AP2i transgenic fruits (CrtR-b converts α-carotene to lutein together with CrtR-e; Bramley, 2002). Two genes involved in the biosynthesis of flavonoids, FLAVONOL SYNTHASE (FLS) and CHALCONE SYNTHASE2 (CHS2) (Table 1), were also found to be downregulated in AP2i fruits, and the accumulation of tocopherols was reduced when compared with the wild-type fruits.
Carotenoids, flavonoids, and tocopherols are all bioactive compounds with potent antioxidant properties. Carotenoids can be further metabolized to abscisic acid in plants and to vitamin A in animal cells (reviewed in Cunningham and Gantt, 1998). In this study, we demonstrated that AP2a positively regulates the synthesis of several carotenoids and of tocopherol during tomato fruit ripening. One of the enzymes dramatically downregulated (24-fold) in the AP2i fruits is 3-hydroxy-3methylglutaryl-CoA (HMGR) reductase 2 (Table 1). The HMGR reaction makes mevalonate, a necessary component in the synthesis of all isoprene-containing compounds, such as sterols, carotenoids, phytoalexins, and other growth hormones (Rodriguez-Concepcion and Gruissem, 1999). The expression of HMG2 was shown to increase strongly during ripening and in parallel with the accumulation of lycopene (Rodriguez-Concepcion and Gruissem, 1999).
AP2a Regulates the Expression of Genes Involved in Chromoplast Differentiation
Differentiation of chloroplasts to chromoplasts is an integral part of tomato fruit ripening, and chromoplasts are the site of lycopene production and accumulation. Chromoplast differentiation involves extensive changes in plastid ultrastructure, accompanied by similarly extensive changes in gene expression (for review, see Egea et al., 2010). A large number of genes that change expression during chromoplast differentiation or have a function in the process have been identified through transcriptome profiling and by natural mutations in genes affected in the process. Several of these are downregulated in our AP2 RNAi plants, indicating that AP2a may be a major regulator of the process. These genes are SGR1, as well as a gene encoding a predicted putative RCCR (red chlorophyll catabolite reductase), which is involved in chlorophyll degradation (Barry et al., 2008), and a tomato homolog of the cauliflower OR gene, which encodes a plastidic DnaJ-like protein. The mutation or in cauliflower induces the production of chromoplasts in the normally white cauliflower curd, causing it to turn orange due to the accumulation of carotenoids (Lu et al., 2006). In AP2i fruits, a tomato homolog of the pepper (Capsicum annuum) Pftf gene, which encodes an FtsH-like protein, the homolog in cauliflower of which is positively regulated by Or (Lu et al., 2006), is downregulated. Apart from the Or gene homolog, many other heat shock protein-encoding genes are strongly downregulated in the AP2i plants, several of which were identified as being plastid localized. DnaJ-like cochaperones interact with chaperones, such as Hsp70 (which is also downregulated in AP2i plants) and are involved in functions such as protein folding, assembly/disassembly, and translocation, which may be required in higher amounts during chromoplast differentiation. The other chloroplast-localized heat shock protein homologs may have similar functions. As downregulation of AP2a likely interferes with this process, the downregulation of the genes encoding the chloroplast-localized heat shock protein homologs in AP2i plants would follow from this.
AP2a Is a Negative Regulator of Ethylene Production
Climacteric fruits such as tomato are distinguished from nonclimacteric fruits by their increased respiration and ethylene biosynthesis rates during ripening. Ethylene is formed from Met via S-adenosyl-l-methionine (AdoMet) and the cyclic nonprotein amino acid ACC. ACC is formed from AdoMet by the action of ACS and the conversion of ACC to ethylene is performed by ACO (Kende, 1989). Biochemical evidence suggests that ethylene production may be influenced or regulated by interactions between its biosynthesis and other metabolic pathways (reviewed in Carrari and Fernie, 2006). For example S-adenosylmethionine is the substrate for both the polyamine pathways and ethylene. SAM, one of the enzymes in the ethylene biosynthesis pathway that is also required for the biosynthesis of polyamines, was found to be upregulated in the AP2i transgenic fruits. High levels of putrescine, which together with S-adenosylmethionine form spermidine in the polyamine biosynthetic pathway, were observed. The lower levels of Orn (a precursor of putrescine biosynthesis) found in the AP2i fruits correlate with higher levels of putrescine. The lower levels of Met measured in the AP2a silenced fruits could be attributed to the higher levels of ethylene production, since these fruits produced up to 5-fold more ethylene than the wild-type fruits. This observation suggests that the AP2a transcription factor is a negative regulator of ethylene biosynthesis in ripening fruits. While ethylene action is associated with senescence and ripening, polyamines, on the other hand, have been considered to be senescence inhibitors. Interestingly, in the Sl AP2i fruits, both ethylene and putrescine levels are elevated. In general, putrescine is the predominant polyamine during tomato ripening, and its levels are high at the immature-green stage and decline throughout the ripening process (Rastogi and Davies, 1991), which coincides with the accumulation of AP2a transcript. Together, our data suggest that AP2a is a negative regulator of both ethylene and putrescine synthesis in wild-type fruits. Interestingly, we show that AP2a expression is induced by ethylene, suggesting that AP2a indirectly regulates ethylene biosynthesis by a negative feedback loop mechanism.
Ethylene biosynthesis is tightly regulated, and two systems of ethylene regulation have been proposed to operate in climacteric plants (Barry et al., 2000). System 1 functions during normal vegetative growth, is ethylene autoinhibitory, and is responsible for producing basal ethylene levels that are detected in all tissues, including those of nonclimacteric fruits. System 2 operates during the ripening of climacteric fruit. It has been proposed that tomato ACS1 and ACS6 are involved in the production of system 1 ethylene in green fruits (Barry et al., 2000). System 1 continues until the fruit ripening initiates, when ACS1 expression increases and ACS2, ACS4, ACO2, and ACO4 are induced as a result of positive feedback regulation (Nakatsuka et al., 1998; Barry et al., 2000). We found that ACS6, but not the other ACS genes, together with ACO4 and two of the ethylene receptor genes, ETR5 and ETR6, as well as the transcription factor gene EIL2 are induced in the AP2i fruits, suggesting that AP2a is a negative regulator of their expression. A model was proposed for tomato fruit in which preclimacteric system 1 ethylene is mediated via constitutively expressed ACS1 and ACS3 and negatively regulated through a feedback loop involving ACS6 (Nakatsuka et al., 1998). Our data suggest that AP2a might be a regulator of the switch between the two ethylene systems (1 and 2) through negative regulation of ACS6 expression. The other genes upregulated in AP2i fruits, the ETR6 receptor gene and ACO4 (Nakatsuka et al., 1998; Kevany et al., 2007), were shown to be regulated by elevated ethylene levels, which could be the reason for their higher expression in the AP2i fruits, as opposed to regulation by AP2a directly. Since the expression of both ETR5 and EIL2 is not ethylene dependant (Tieman et al., 2001; Kevany et al., 2007), their expression could be under direct regulation of AP2a. Finally, we also found tomato EBF1, a homolog of Arabidopsis EBF1, to be upregulated in AP2i fruits. EBF1 is involved in targeting EIN3 for degradation and thus modulates the ethylene response (Guo and Ecker, 2003). The two tomato homologs, EBF1 and EBF2, are regulated by ethylene and auxin, and their silencing causes a constitutive ethylene response, plant senescence, and early fruit ripening (Yang et al., 2010).
In addition to ripening, ethylene is also known to be involved in other processes, such as pathogen and wounding responses, leaf senescence, and the abiotic and biotic stress response (for review, see Alexander and Grierson, 2002). In AP2i fruits, a large group of genes encoding pathogenesis-related proteins (PR proteins and chitinases), as well as one key regulator of the biotic stress response, the MYB transcription factor gene AIM1 (Abuqamar et al., 2009), are upregulated. Many of these genes are also likely upregulated in Cnr mutant fruits (Eriksson et al., 2004). Our data suggest that in wild-type fruits, AP2a acts not only as repressor of ethylene biosynthesis, but also as a repressor of ethylene-induced pathogen defense and wounding responses.
In comparison with ethylene, very little is known about the role of other hormones in fruit ripening. The role of auxins has been extensively investigated in other fruits, such as strawberry (Fragaria × ananassa) (Harpster et al., 1998) and grape (Vitis vinifera) (Davies et al., 1997). Our transcriptomics data show a high level of repression of GH3, which is involved in maintaining auxin homeostasis by conjugating excess indole-3-acetic acid to amino acids (Staswick et al., 2005). In addition to GH3, ARF4, a transcription factor gene involved in auxin signaling, was suppressed. ARF4 (also known as DEVELOPMENTALLY REGULATED12 [DR12]; Jones et al., 2002) is developmentally regulated, and its expression increases in fruits during ripening and decreases after ethylene treatment of immature fruits. Knockdown of DR12 in transgenic tomato plants caused blotchy fruit ripening, similar to the defects observed here for AP2 RNAi plants, suggesting that at least part of the effects of AP2a downregulation is caused by deregulation of ARF4. Genes involved in the biosynthesis of brassinosteroid hormones, like DWARF (Nomura et al., 2005) and the homolog of Arabidopsis DWF1/DIMINUTO (Klahre et al., 1998), were downregulated in the AP2i fruits. Previously, it was shown by treatment of tomato pericarp discs with brassinosteroids that these could accelerate ripening and increase ethylene production (Vidya Vardhini and Rao, 2002). Although brassinosteroids recently have been shown to play a role in early fruit development in cucumber (Cucumis sativus) (Fu et al., 2008) and in fruit ripening in grape (Symons et al., 2006), their function in tomato fruit development or ripening remains to be determined.
Effect of AP2a Downregulation on Fruit Metabolism
From the gene ontology analysis of our transcriptomics data, it is apparent that most of the metabolic pathways that are significantly regulated are related to primary metabolism. In agreement with these data are the changes in the levels of accumulation of different primary metabolites that were measured in the AP2i fruits compared with the wild-type ripe fruits. We found that AP2a positively regulates the accumulation of Leu and Ile, probably through regulation of BCAT1. BCAT1 transcripts were shown to accumulate at the highest level in ripening fruits, and BCAT1 is likely the primary enzyme involved in the recycling of branched-chain amino acids generated by protein degradation. This statement is in agreement with data obtained in a recent study of the tomato BCAT gene family (Maloney et al., 2010).
Taken together, our metabolic and transcriptomics data indicate that the downregulation of AP2a transcription factor has a major effect on cell wall metabolism, which is associated with fruit softening and shelf life and the biosynthesis of secondary metabolites and their primary metabolite precursors, as well as decreased flux through the tricarboxylic acid cycle.
Additionally, the downregulation of LOXC may well affect volatile production in AP2 RNAi plants during ripening. The chromoplast-located TomloxC protein has been shown to be involved in the production of flavor volatiles hexanal, hexenal, and hexenol from linoleic and linolenic acid, which may be produced through thylakoid membrane disruption at the transition from chloroplasts to chromoplasts (Chen et al., 2004).
AP2a Participates in a Transcriptional Regulatory Network That Regulates Tomato Fruit Ripening
Unlike Arabidopsis AP2, which is mostly expressed in flowers and functions in floral organ identity, tomato AP2a has acquired a novel function in fleshy fruit development. Some of the other tomato AP2 paralogs may still be involved in floral organ identity as A-class homeotic genes, like AP2 in Arabidopsis (Bowman et al., 1991; Coen and Meyerowitz, 1991); however, further research is needed to unravel the function of these other tomato genes.
The AP2a transcription factor appeared to have dual functions in tomato fruit ripening as a positive and negative regulator of different processes occurring during ripening. An interesting observation was that AP2a most probably functions downstream of known major positive regulators of fruit ripening, such as NOR, RIN, and CNR. At least in the case of CNR, this regulation is likely to be direct, since CNR is able to bind directly to a promoter element of AP2a. On the other hand, in the fruit pericarp, AP2a regulates the expression of CNR in a negative manner. In both the Cnr (Eriksson et al., 2004) mutant and in AP2i transgenic fruits, several ripening-associated genes are downregulated, such as the carotene synthesis pathway genes, LOXB and LOXC, pectin methylesterase, and EXP3, indicating that AP2a has positive ripening regulatory functions besides its negative regulatory function in ethylene synthesis. Further research, such as the identification of direct and indirect targets of the key transcription factors, will likely contribute to further mapping of the transcriptional network that regulates fruit ripening, a process in which AP2a plays an essential function.
METHODS
Greenhouse/Growth Conditions
Transgenic plants were transferred to soil and grown in the greenhouse at ambient temperatures (>20°C) under natural light supplemented with artificial sodium lights, according to a 16-h-light/8-h-dark cycle. Standard greenhouse culture conditions, with regular fertilizer application, were used.
Construction of RNAi Construct and Transgenic Tomato Plants
The sequences, and associated names, available on the Tomato Functional Genomics Database and The Institute for Genomic Research (TIGR) websites, are always being updated; however, old names remain linked to updated information. Therefore, the original names first notated for this research are kept for the article. The primers used (see Supplemental Table 3 online) for the RNAi construct were designed based on the sequence of EST311591 (GenBank AW442195; http://www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=tomato). RT-PCR was performed with red ripe fruit RNA, using an annealing temperature of 54°C, and the products were cloned into pENTR/D-TOPO (Invitrogen) (resulting plasmid designated pARC131 and CZN098 for specific RNAi fragment) and subcloned into pHellsgate8 (designated pARC139 and CZN096, respectively) (Helliwell et al., 2002). pARC139 and CZN096 were transformed into electrocompetent Agrobacterium tumefaciens strain EHA105, followed by transformation into Solanum lycopersicum cv Moneymaker and cv M82, respectively, as described by van Roekel et al. (1993). Tissue culture-generated nontransgenic Moneymaker and M82 plants were used for wild-type controls. Sixteen independent transgenic AP2i-1 and 14 independent transgenic AP2i-2 RNAi lines were generated and analyzed for ploidy level by flow cytometry as described elsewhere (de Laat et al., 1987), and polyploid plants were removed from the population.
Plasmid Constructions
AP2a and CNR were amplified from the cDNA made from mixed fruit stages using the primer pairs described in Supplemental Table 4 online. The full-length open reading frames of AP2a and CNR were inserted into the pENTR-D TOPO vector following the Gateway protocol (Invitrogen). All plasmids were controlled by sequence analyses (DETT sequence kit; Amersham). Finally, expression vectors for the colocalization analysis were obtained by LR reaction following the Gateway protocol. For the C-terminal yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP) fusions, CZN576 and CZN575 destination vectors, respectively, were used. For constructing CZN575, the open reading frame from sCFP3A was amplified from pSCFP3A-C1 (Kremers et al., 2006) and was cloned into pGD120 (Immink et al., 2002). The obtained product was digested with XbaI, which is located just before the start codon of sCFP3A, and the overhangs were blunted with Klenow. Subsequently, a Gateway conversion cassette (Invitrogen) was ligated into this vector, resulting in the destination vector CZN575, containing the expression cassette pCaMV35S:Gateway-sCFP3A:NOS terminator. For the CZN576 vector, the open reading frame from sYFP2 was amplified from pSYFP2-C1 (Kremers et al., 2006), and the remaining cloning strategy was identical to that used for the CZN575 vector.
For the N-terminal GST fusion, the full-length CNR PCR fragment was obtained with the primers listed in Supplemental Table 3 online. The purified PCR fragment was digested with EcoRI and NotI and ligated into pGEX4T1. The resulting construct was verified by sequencing and transformed into Escherichia coli Arctic Express strain (Stratagene). The protein expression and GST purification were performed as described before (Karlova et al., 2009).
For the electrophoretic mobility shift assay or gel retardation assay, the two AP2a promoter regions chosen were PCR amplified with the primers listed in Supplemental Table 3 online, purified, and ligated into the pGEMT vector (Promega).
Ethephon Treatment of Tomato Fruit
Ethephon (Acros Organics) treatment of intact mature green fruits involved a 1-min dip in 50 mM fresh aqueous ethephon solution for 6 h.
Gel Retardation Assays
The binding reactions for the gel retardation assays were performed as described by Winter et al. (2002). Fifty nanograms of in vitro GST-CNR purified protein were used per binding reaction. PCR fragments from the above described promoter fragment construct were biotinylated, and 40 nM of probe was used per lane. After native polyacrylamide gel electrophoresis, the DNA was blotted to positively charged nylon membranes (Hybond N+; Amersham), and signal was detected using the chemiluminescent nucleic acid detection module (Peirce).
Images
Micrographs of seeds were taken on a Zeiss STEMI SV8 microscope. Seed hairs were processed and viewed using a JEOL JSM-6330F field emission electron scanning microscope as described elsewhere (Angenent et al., 1995).
Fluorescence Microscopy in Living Cells
Arabidopsis thaliana leaf protoplasts were transfected as described by Aker et al. (2006). Plasmid DNA (15 μg) was used and the protoplasts were incubated overnight at 25°C before imaging. Images were acquired using an upright confocal laser scanning microscope (Leica SPE DM5500) with a 10 × 0.30 CS ACS APO (air) lens, using the LAS AF 1.8.2 software (Leica).
Quantitative Real-Time PCR
RNA isolation from fruits or pericarp of the fruits was performed with the InviTrap Spin Plant RNA kit (Invitek) according to the manufacturer’s protocol and from all other tissues by the Trizol method (Sigma-Aldrich), except from seeds, where a modified method was used. Seeds were ground in a 5:5:1 mixture of buffer (180 mM Tris pH 8.2, 90 mM LiCl, 4.5 mM EDTA, 1% SDS, and 0.1% β-mercaptoethanol), phenol, and chloroform. After phase separation by centrifugation, the watery phase was reextracted with 1 mL phenol/chloroform (1:1) and subsequently with chloroform. The extracted RNA was precipitated with one-third volume of 8 M LiCl overnight at −20°C. Precipitated RNA was centrifuged, washed with 70% ethanol, dried, and dissolved in RNase-free water.
DNaseI-treated RNA was reverse transcribed (Applied Biosystems), and 1 μL of cDNA solution was used for the real-time PCR analysis. Real-time qPCR was performed with iQ SYBR Green Supermix (Bio-Rad) using the iCycler iQ5 system (Bio-Rad) and gene-specific primers (see Supplemental Table 4 online). Two independent biological replicates were analyzed per sample (or two to three fruits from the same plant were mixed together per sample). Relative quantification of specific mRNA levels was performed using the cycle threshold (Ct) 2–Δ(ΔCt) method (Software IQ5 2.0; Livak and Schmittgen, 2001). Expression values were normalized using the housekeeping gene actin or CAC and TIP41 in case of sepals, petals, and seeds (Expósito-Rodriguez et al., 2008) from tomato.
Ethylene Measurements
Fruits at breaker or breaker +7 d stages were harvested and placed in open 500-mL jars for 2 h. Jars were then sealed and incubated at room temperature for 35 min, and 1.5 mL of headspace gas was injected into a Focus GS gas chromatograph (Thermo-Electron) equipped with a flame ionization detector. Samples were compared with reagent grade ethylene standards of known concentration and normalized for fruit weight.
Carotenoid Analysis
Pooled fruits from independent plants were harvested and homogenized. Aliquots from these samples were taken separately for extraction and analysis of primary and secondary metabolite content by HPLC coupled with photodiode array detection and MS. The extraction, HPLC separation, photodiode array detection, and quantification of carotenes, carotenoids, and xanthophylls were previously described (Fraser et al., 2000). In this study, 0.5 to 1 g of frozen fruit tissue was extracted with chloroform and methanol (2.5:1 by volume), mixed, and incubated for 20 min on ice. Then, 1 volume of deionized water was added. A partition was formed after mixing by centrifugation. The organic hypophase was removed, and the aqueous phase was reextracted with chloroform (2.5 times by volume). HPLC separations were performed using a C30 reverse-phase column (250 × 4.6 mm) purchased from YMC. The mobile phases used were methanol (A), water/methanol (20/80 by volume) containing 0.2% ammonium acetate (B), and tert-methyl butyl ether (C). The gradient used was 95% A:5% B, isocratically for 12 min and then stepped to 80% A:5% B:15%C, from which a linear gradient to 30% A:5% B:65% C over 30 min was performed. A Waters Alliance model 2695 injection and solvent delivery system was used. Detection was performed continuously from 220 to 700 nm with an online photodiode array detector (Waters 966). Identification was performed by cochromatography and comparison of spectral properties with authentic standards and reference spectra. Annotated chromatograms of this HPLC system are provided elsewhere (Fraser et al., 2007). Quantitative determination of carotenoids was performed by comparison with dose–response curves (0.2 to 1.0 mg) constructed from authentic standards. Purchasing, preparation, and characterization of authentic standards are as described previously (Fraser et al., 2000).
GC-MS Analysis
Metabolite extraction, derivatization, GC-MS analysis, and data processing were performed as described previously (Roessner et al., 2001; Lisec et al., 2006). Metabolites were identified in comparison to database entries of authentic standards (Kopka et al., 2005; Schauer et al., 2005).
Peptide Antibody Generation
For polyclonal AP2a antibodies, the peptide CRTQNNGFHHYFMRP was generated and used for rabbit immunization by Genosphere Biotechnologies.
Protein Gel Blot Analysis
Plant material (tomato fruits) was ground in liquid nitrogen, and 0.2 g was dissolved in sample buffer (100 mM Tris-HCl, pH 6.8, 4% SDS, 0.2% bromphenol blue, 20% glycerol, and 100 mM DTT) and boiled for 5 min. Protein gel blotting was performed as described before (Karlova et al., 2006). Proteins were detected with anti-AP2 antibodies (2000× diluted).
Phylogenetic Analysis
Protein homology searches in the National Center for Biotechnology Information (NCBI) GenBank protein database were done using the BLASTP algorithm (Altschul et al., 1997). Homology searches in the TIGR Tomato Gene Index were done with the TBLASTN algorithm. Multiple protein alignments of the full-length proteins were produced with the ClustalW program of the DNAstar Megalign software package (DNASTAR). Phylogenetic trees were produced with the ClustalX package (Thompson et al., 1997). Phylogenetic reconstruction was obtained by the neighbor-joining method (Saitou and Nei, 1987) together with bootstrap analysis using 1000 replicates. Kimura correction for multiple substitutions was applied (Kimura and Takahata, 1983). The tree was visualized using the TreeView package (Page, 1996).
Microarray Analysis
Transcript profiling was performed on fruits of wild-type tomato and AP2-1 RNAi mutant lines, at the breaker +7 d stage (U.S. standards for grades of fresh tomatoes, 1991). RNA was isolated with the Trizol method (Sigma-Aldrich) and hybridized to the Syngenta Tomato Affymetrix GeneChip. Three biological replicates and two technical replicates were analyzed from either unmodified tomato variety Moneymaker or from an AP2-1 RNAi line. Twelve experiments (slides) were analyzed. Data were analyzed using the GeneData Expressionist suite of tools. All 12 experiments passed quality controls and were used for analysis. The Bioconductor RMA algorithm was used for background subtraction according to a Bayesian model using the PM Features Only option ('bgversion = 2' in the BioConductor routine) for quantile normalization of the arrays (Bolstad et al., 2003) and for condensing probes into probe regions (hereafter referred to as genes) (Irizarry et al., 2003).
A Welch t test was performed to identify genes with significant differences in expression level between the two data sets. P values were adjusted for false discovery rate 0.05, as described by Benjamini and Hochberg (1995).
Accession Numbers
Sequence data from this article can be found in Swiss-Prot or Protein Bank/NCBI databases under the following accession numbers: Arabidopsis ANT protein, Q38914; At RAP27/TOE1, Q9SK03; At TOE2, Q9LVG2; At SMZ, Q6PV68; At SNZ, Q6PV67; At TOE3, Q9FH95; At AP2, AAC13770; At WRI1, NP_191000; At AIL1, Q1PFE1; At AIL5, Q6PQQ3; At AIL6, Q52QU2; At AIL7, Q6J9N8; At BBM, Q6PQQ4; At PUCHI, NP_197357; At PLT1, Q5YGP8; At PLT2, Q5YGP7; Tomato AP2a, ACD62792, Sl AP2c, HQ586951, Sl AP2b, HQ586952, Sl AP2d, HQ586953; Sl AP2e, HQ586954; Petunia PHAP2A, AAD39439, and PHAP2B, AAD39440; Antirrhinum LIP1, AAO52746 and LIP2, AAO52747; Malus domestica AHAP2, AAL57045; Zea mays GLOSSY15, AAC49567; IDS1, NP_001104904. The accession numbers of genes used in the microarray analysis are provided in Table 1 and Supplemental Data Set 3 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Tree of the AP2 Subfamily, with the AP2 and ANT Subgroups Indicated.
Supplemental Figure 2. Protein Sequence Alignment and Open Reading Frame Nucleotide Sequence of AP2a and Its Homologs.
Supplemental Figure 3. Decreased Expression of AP2a Protein Results in Alterations in Sepal Morphology.
Supplemental Figure 4. Decreased Expression of AP2a Protein Results in Alterations in Fruit and Seed Morphology.
Supplemental Figure 5. Schematic Representation of the Metabolic Changes Occurring in the AP2i-1 Silenced Ripe Fruits Compared with the Wild Type.
Supplemental Figure 6. Relative Expression of RIN and TAGL1 in Wild-Type and AP2i-1 Breaker Fruits.
Supplemental Table 1. Pathways Significantly Altered in AP2i Fruits.
Supplemental Table 2. Metabolite Levels in Wild-Type and AP2i Transgenic Fruits.
Supplemental Table 3. Oligonucleotides Used in This Study.
Supplemental Data Set 1. FASTA Format File of the Alignment Used for Creating Figure 1A.
Supplemental Data Set 2. FASTA Format File of the Alignment Used for Creating Supplemental Figure 1.
Supplemental Data Set 3. Gene Expression Changes in AP2 RNAi Ripe Fruits.
Supplementary Material
Acknowledgments
We thank Jurriaan Mes, Marco Busscher, and Michiel Lammers (Plant Research International) for technical assistance. We thank Graham Seymour (University of Nottingham, UK) for kindly providing the Cnr mutant seeds and the C.M. Rick Tomato Genetics Resource Center (Davis, CA) for other mutant seeds. The project was financially supported by the European Commission Framework 6 project “EU-SOL” (Contract FOOD-CT-2006-016214; J.B.-L., G.C.A., and R.A.D.) and by the Dutch Organization for Scientific Research (Nederlandse Organisatie voor Wetenschappelijk Onderzoek) in the framework of the ERA-NET Plant Genomics program project “TomQML” (R.K.).
References
- Abuqamar S., Luo H., Laluk K., Mickelbart M.V., Mengiste T. (2009). Crosstalk between biotic and abiotic stress responses in tomato is mediated by the AIM1 transcription factor. Plant J. 58: 347–360 [DOI] [PubMed] [Google Scholar]
- Aker J., Borst J.W., Karlova R., de Vries S. (2006). The Arabidopsis thaliana AAA protein CDC48A interacts in vivo with the somatic embryogenesis receptor-like kinase 1 receptor at the plasma membrane. J. Struct. Biol. 156: 62–71 [DOI] [PubMed] [Google Scholar]
- Alba R., Payton P., Fei Z.J., McQuinn R., Debbie P., Martin G.B., Tanksley S.D., Giovannoni J.J. (2005). Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17: 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander L., Grierson D. (2002). Ethylene biosynthesis and action in tomato: A model for climacteric fruit ripening. J. Exp. Bot. 53: 2039–2055 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent G.C., Franken J., Busscher M., van Dijken A., van Went J.L., Dons H.J., van Tunen A.J. (1995). A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell 7: 1569–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C.S., Giovannoni J.J. (2006). Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proc. Natl. Acad. Sci. USA 103: 7923–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C.S., Llop-Tous M.I., Grierson D. (2000). The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 123: 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C.S., McQuinn R.P., Chung M.Y., Besuden A., Giovannoni J.J. (2008). Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 147: 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C.S., McQuinn R.P., Thompson A.J., Seymour G.B., Grierson D., Giovannoni J.J. (2005). Ethylene insensitivity conferred by the Green-ripe and Never-ripe 2 ripening mutants of tomato. Plant Physiol. 138: 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Roy. Stat. Soc. Ser. B 57: 289–300 [Google Scholar]
- Birkenbihl R.P., Jach G., Saedler H., Huijser P. (2005). Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 352: 585–596 [DOI] [PubMed] [Google Scholar]
- Bleecker A.B., Kende H. (2000). Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16: 1–18 [DOI] [PubMed] [Google Scholar]
- Boggio S.B., Palatnik J.F., Heldt H.W., Valle E.M. (2000). Changes in amino acid composition and nitrogen metabolizing enzymes in ripening fruits of Lycopersicon esculentum Mill. Plant Sci. 159: 125–133 [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Åstrand M., Speed T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20 [DOI] [PubMed] [Google Scholar]
- Bramley P.M. (2002). Regulation of carotenoid formation during tomato fruit ripening and development. J. Exp. Bot. 53: 2107–2113 [DOI] [PubMed] [Google Scholar]
- Burns J., Fraser P.D., Bramley P.M. (2003). Identification and quantification of carotenoids, tocopherols and chlorophylls in commonly consumed fruits and vegetables. Phytochemistry 62: 939–947 [DOI] [PubMed] [Google Scholar]
- Cantu D., Blanco-Ulate B., Yang L., Labavitch J.M., Bennett A.B., Powell A.L.T. (2009). Ripening-regulated susceptibility of tomato fruit to Botrytis cinerea requires NOR but not RIN or ethylene. Plant Physiol. 150: 1434–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F., Fernie A.R. (2006). Metabolic regulation underlying tomato fruit development. J. Exp. Bot. 57: 1883–1897 [DOI] [PubMed] [Google Scholar]
- Chakrabarti A.K., Mukherjee S., Maiti G., Kabi M. (2003). SEM studies of seed and seed coat structures in cultivars of Lycopersicon esculentum Mill. J. Veg. Crop. Prod. 9: 75–85 [Google Scholar]
- Chen G., Hackett R., Walker D., Taylor A., Lin Z., Grierson D. (2004). Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol. 136: 2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M.Y., Vrebalov J., Alba R., Lee J., McQuinn R., Chung J.D., Klein P., Giovannoni J. (2010). A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 64: 936–947 [DOI] [PubMed] [Google Scholar]
- Coen E.S., Meyerowitz E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Cunningham F.X., Gantt E. (1998). Genes and enzymes of carotenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 557–583 [DOI] [PubMed] [Google Scholar]
- Davies C., Boss P.K., Robinson S.P. (1997). Treatment of grape berries, a nonclimacteric fruit with a synthetic auxin, retards ripening and alters the expression of developmentally regulated genes. Plant Physiol. 115: 1155–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat A.M.M., Gohde W., Vogelzang M.J.D.C. (1987). Determination of ploidy of single plants and plant-populations by flow-cytometry. Plant Breed. 99: 303–307 [Google Scholar]
- Egea I., Barsan C., Bian W., Purgatto E., Latché A., Chervin C., Bouzayen M., Pech J.C. (2010). Chromoplast differentiation: Current status and perspectives. Plant Cell Physiol. 51: 1601–1611 [DOI] [PubMed] [Google Scholar]
- Eriksson E.M., Bovy A., Manning K., Harrison L., Andrews J., De Silva J., Tucker G.A., Seymour G.B. (2004). Effect of the Colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol. 136: 4184–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expósito-Rodríguez M., Borges A.A., Borges-Pérez A., Pérez J.A. (2008). Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P.D., Enfissi E.M., Bramley P.M. (2009). Genetic engineering of carotenoid formation in tomato fruit and the potential application of systems and synthetic biology approaches. Arch. Biochem. Biophys. 483: 196–204 [DOI] [PubMed] [Google Scholar]
- Fraser P.D., Enfissi E.M., Halket J.M., Truesdale M.R., Yu D., Gerrish C., Bramley P.M. (2007). Manipulation of phytoene levels in tomato fruit: Effects on isoprenoids, plastids, and intermediary metabolism. Plant Cell 19: 3194–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P.D., Pinto M.E., Holloway D.E., Bramley P.M. (2000). Technical advance: Application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J. 24: 551–558 [DOI] [PubMed] [Google Scholar]
- Fu F.Q., Mao W.H., Shi K., Zhou Y.H., Asami T., Yu J.Q. (2008). A role of brassinosteroids in early fruit development in cucumber. J. Exp. Bot. 59: 2299–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J. (2001). Molecular biology of fruit maturation and ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 725–749 [DOI] [PubMed] [Google Scholar]
- Giovannoni J.J. (2007). Fruit ripening mutants yield insights into ripening control. Curr. Opin. Plant Biol. 10: 283–289 [DOI] [PubMed] [Google Scholar]
- Guo H.W., Ecker J.R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Harpster M.H., Brummell D.A., Dunsmuir P. (1998). Expression analysis of a ripening-specific, auxin-repressed endo-1, 4-beta-glucanase gene in strawberry. Plant Physiol. 118: 1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C.A., Varsha Wesley S., Wielopolska A.J., Waterhouse P.M. (2002). High-throughput vectors for efficient gene silencing in plants. Funct. Plant Biol. 29: 1217–1225 [DOI] [PubMed] [Google Scholar]
- Immink R.G.H., Gadella T.W., Jr, Ferrario S., Busscher M., Angenent G.C. (2002). Analysis of MADS box protein-protein interactions in living plant cells. Proc. Natl. Acad. Sci. USA 99: 2416–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Isaacson T., Ronen G., Zamir D., Hirschberg J. (2002). Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of beta-carotene and xanthophylls in plants. Plant Cell 14: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M., Seybold H., Breitel D., Rogachev I., Meir S., Aharoni A. (2009). TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Ito Y., Kitagawa M., Ihashi N., Yabe K., Kimbara J., Yasuda J., Ito H., Inakuma T., Hiroi S., Kasumi T. (2008). DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 55: 212–223 [DOI] [PubMed] [Google Scholar]
- Jofuku K.D., den Boer B.G.W., Van Montagu M., Okamuro J.K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku K.D., Omidyar P.K., Gee Z., Okamuro J.K. (2005). Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. USA 102: 3117–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B., Frasse P., Olmos E., Zegzouti H., Li Z.G., Latché A., Pech J.C., Bouzayen M. (2002). Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant J. 32: 603–613 [DOI] [PubMed] [Google Scholar]
- Josse E.M., Simkin A.J., Gaffé J., Labouré A.M., Kuntz M., Carol P. (2000). A plastid terminal oxidase associated with carotenoid desaturation during chromoplast differentiation. Plant Physiol. 123: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J.G., Corbett A.M., Fellman S.M., Tieman D.M., Klee H.J., Giovannoni J.J., Fei Z. (2009). Plant MetGenMAP: an integrative analysis system for plant systems biology. Plant Physiol. 151: 1758–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.H., Seo Y.H., Seo P.J., Reyes J.L., Yun J., Chua N.H., Park C.M. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R., Boeren S., Russinova E., Aker J., Vervoort J., de Vries S. (2006). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R., Boeren S., van Dongen W., Kwaaitaal M., Aker J., Vervoort J., de Vries S. (2009). Identification of in vitro phosphorylation sites in the Arabidopsis thaliana somatic embryogenesis receptor-like kinases. Proteomics 9: 368–379 [DOI] [PubMed] [Google Scholar]
- Keck E., McSteen P., Carpenter R., Coen E. (2003). Separation of genetic functions controlling organ identity in flowers. EMBO J. 22: 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H. (1989). Enzymes of ethylene biosynthesis. Plant Physiol. 91: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevany B.M., Tieman D.M., Taylor M.G., Cin V.D., Klee H.J. (2007). Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J. 51: 458–467 [DOI] [PubMed] [Google Scholar]
- Kim V.N., Nam J.W. (2006). Genomics of microRNA. Trends Genet. 22: 165–173 [DOI] [PubMed] [Google Scholar]
- Kimura M., Takahata N. (1983). Selective constraint in protein polymorphism: Study of the effectively neutral mutation model by using an improved pseudosampling method. Proc. Natl. Acad. Sci. USA 80: 1048–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U., Noguchi T., Fujioka S., Takatsuto S., Yokota T., Nomura T., Yoshida S., Chua N.H. (1998). The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 10: 1677–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., de Bruine J.H., Goettsch P. (1980). A provisional map of chromosome 4 of Arabidopsis. Arabidopsis Inf. Serv. 17: 11–18 [Google Scholar]
- Kopka J., et al. (2005). GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kremers G.J., Goedhart J., van Munster E.B., Gadella T.W., Jr (2006). Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 45: 6570–6580 [DOI] [PubMed] [Google Scholar]
- Leon-Kloosterziel K.M., Keijzer C.J., Koornneef M. (1994). A seed shape mutant of Arabidopsis that is affected in integument development. Plant Cell 6: 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Hong Y., Yin M., Li C., Zhang K., Grierson D. (2008). A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 55: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Zhong S., Grierson D. (2009). Recent advances in ethylene research. J. Exp. Bot. 60: 3311–3336 [DOI] [PubMed] [Google Scholar]
- Lisec J., Schauer N., Kopka J., Willmitzer L., Fernie A.R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu S., et al. (2006). The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. Plant Cell 18: 3594–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes T., Van de Steene N., Zethof J., Karimi M., D’Hauw M., Mares G., Van Montagu M., Gerats T. (2001). Petunia Ap2-like genes and their role in flower and seed development. Plant Cell 13: 229–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney G.S., Kochevenko A., Tieman D.M., Tohge T., Krieger U., Zamir D., Taylor M.G., Fernie A.R., Klee H.J. (2010). Characterization of the branched-chain amino acid aminotransferase enzyme family in tomato. Plant Physiol. 153: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K., Tör M., Poole M., Hong Y., Thompson A.J., King G.J., Giovannoni J.J., Seymour G.B. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Nakatsuka A., Murachi S., Okunishi H., Shiomi S., Nakano R., Kubo Y., Inaba A. (1998). Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiol. 118: 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Kushiro T., Yokota T., Kamiya Y., Bishop G.J., Yamaguchi S. (2005). The last reaction producing brassinolide is catalyzed by cytochrome P-450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. J. Biol. Chem. 280: 17873–17879 [DOI] [PubMed] [Google Scholar]
- Ohto M.A., Fischer R.L., Goldberg R.B., Nakamura K., Harada J.J. (2005). Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 102: 3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki S., et al. (2010). Coexpression analysis of tomato genes and experimental verification of coordinated expression of genes found in a functionally enriched coexpression module. DNA Res. 17: 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R.D. (1996). TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Rastogi R., Davies P.J. (1991). Polyamine metabolism in ripening tomato fruit: II. Polyamine metabolism and synthesis in relation to enhanced putrescine content and storage life of a/c tomato fruit. Plant Physiol. 95: 41–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. (2002). Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M., Gruissem W. (1999). Arachidonic acid alters tomato HMG expression and fruit growth and induces 3-hydroxy-3-methylglutaryl coenzyme A reductase-independent lycopene accumulation. Plant Physiol. 119: 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U., Luedemann A., Brust D., Fiehn O., Linke T., Willmitzer L., Fernie A. (2001). Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sato T., Theologis A. (1989). Cloning the mRNA encoding 1-aminocyclopropane-1-carboxylate synthase, the key enzyme for ethylene biosynthesis in plants. Proc. Natl. Acad. Sci. USA 86: 6621–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N., Steinhauser D., Strelkov S., Schomburg D., Allison G., Moritz T., Lundgren K., Roessner-Tunali U., Forbes M.G., Willmitzer L., Fernie A.R., Kopka J. (2005). GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Schmid M., Uhlenhaut N.H., Godard F., Demar M., Bressan R., Weigel D., Lohmann J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012 [DOI] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Shigyo M., Hasebe M., Ito M. (2006). Molecular evolution of the AP2 subfamily. Gene 366: 256–265 [DOI] [PubMed] [Google Scholar]
- Sorrequieta A., Ferraro G., Boggio S.B., Valle E.M. (2010). Free amino acid production during tomato fruit ripening: a focus on L-glutamate. Amino Acids 38: 1523–1532 [DOI] [PubMed] [Google Scholar]
- Staswick P.E., Serban B., Rowe M., Tiryaki I., Maldonado M.T., Maldonado M.C., Suza W. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons G.M., Davies C., Shavrukov Y., Dry I.B., Reid J.B., Thomas M.R. (2006). Grapes on steroids. Brassinosteroids are involved in grape berry ripening. Plant Physiol. 140: 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.J., Jackson A.C., Parker R.A., Morpeth D.R., Burbidge A., Taylor I.B. (2000). Abscisic acid biosynthesis in tomato: Regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol. Biol. 42: 833–845 [DOI] [PubMed] [Google Scholar]
- Thompson A.J., Tor M., Barry C.S., Vrebalov J., Orfila C., Jarvis M.C., Giovannoni J.J., Grierson D., Seymour G.B. (1999). Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. Plant Physiol. 120: 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D.M., Ciardi J.A., Taylor M.G., Klee H.J. (2001). Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J. 26: 47–58 [DOI] [PubMed] [Google Scholar]
- Tigchelaar E.C., Tomes M.L., Kerr E.A., Barman R.J. (1973). A new fruit ripening mutant, non-ripening (nor). Rep. Tomato Genet. Coop. 23: 33–34 [Google Scholar]
- Toor R.K., Lister C.E., Savage G.P. (2005). Antioxidant activities of New Zealand-grown tomatoes. Int. J. Food Sci. Nutr. 56: 597–605 [DOI] [PubMed] [Google Scholar]
- van Roekel J., Damm B., Melchers L., Hoekema A. (1993). Factors influencing transformation frequency of tomato (Lycopersicon esculentum). Plant Cell Rep. 12: 644–647 [DOI] [PubMed] [Google Scholar]
- Vidya Vardhini B., Rao S.S. (2002). Acceleration of ripening of tomato pericarp discs by brassinosteroids. Phytochemistry 61: 843–847 [DOI] [PubMed] [Google Scholar]
- Vrebalov J., Pan I.L., Arroyo A.J.M., McQuinn R., Chung M., Poole M., Rose J., Seymour G., Grandillo S., Giovannoni J., Irish V.F. (2009). Fleshy fruit expansion and ripening are regulated by the Tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J., Ruezinsky D., Padmanabhan V., White R., Medrano D., Drake R., Schuch W., Giovannoni J. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wilkinson J.Q., Lanahan M.B., Yen H.C., Giovannoni J.J., Klee H.J. (1995). An ethylene-inducible component of signal transduction encoded by never-ripe. Science 270: 1807–1809 [DOI] [PubMed] [Google Scholar]
- Winter K.U., Weiser C., Kaufmann K., Bohne A., Kirchner C., Kanno A., Saedler H., Theissen G. (2002). Evolution of class B floral homeotic proteins: obligate heterodimerization originated from homodimerization. Mol. Biol. Evol. 19: 587–596 [DOI] [PubMed] [Google Scholar]
- Yang Y., Wu Y., Pirrello J., Regad F., Bouzayen M., Deng W., Li Z. (2010). Silencing Sl-EBF1 and Sl-EBF2 expression causes constitutive ethylene response phenotype, accelerated plant senescence, and fruit ripening in tomato. J. Exp. Bot. 61: 697–708 [DOI] [PubMed] [Google Scholar]
- Yen H.C., Lee S., Tanksley S.D., Lanahan M.B., Klee H.J., Giovannoni J.J. (1995). The tomato Never-ripe locus regulates ethylene-inducible gene expression and is linked to a homolog of the Arabidopsis ETR1 gene. Plant Physiol. 107: 1343–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegzouti H., Jones B., Frasse P., Marty C., Maitre B., Latch A., Pech J.C., Bouzayen M. (1999). Ethylene-regulated gene expression in tomato fruit: Characterization of novel ethylene-responsive and ripening-related genes isolated by differential display. Plant J. 18: 589–600 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.