Abstract
Despite triggering strong immune responses, Epstein-Barr virus (EBV) has colonized more than 90% of the adult human population. Successful persistence of EBV depends on the establishment of a balance between host immune responses and viral immune evasion. Here we have extended our studies on the EBV-encoded BILF1 protein, which was recently identified as an immunoevasin that functions by enhancing degradation of major histocompatibility complex class I (MHC-I) antigens via lysosomes. We now demonstrate that disruption of the EKT signaling motif of BILF1 by a K122A mutation impairs the ability of BILF1 to enhance endocytosis of surface MHC-I molecules, while subsequent lysosomal degradation was impaired by deletion of the 21-residue C-terminal tail of BILF1. Furthermore, we identified another mechanism of BILF1 immunomodulation: it targets newly synthesized MHC-I/peptide complexes en route to the cell surface. Importantly, although the diversion of MHC-I on the exocytic pathway caused a relatively modest reduction in cell surface MHC-I, presentation of endogenously processed target peptides to immune CD8+ effector T cells was reduced by around 65%. The immune-modulating functions of BILF1 in the context of the whole virus were confirmed in cells lytically infected with a recombinant EBV in which BILF1 was deleted. This study therefore extends our initial observations on BILF1 to show that this immunoevasin can target MHC-I antigen presentation via both the exocytic and endocytic trafficking pathways. The results also emphasize the merits of including functional T cell recognition assays to gain a more complete picture of immunoevasin effects on the antigen presentation pathway.
For viruses to establish a persistent infection, they need to have mechanisms for evading the host immune responses. A passive form of evasion involves latency, where viral antigens are silenced and the infected cells are therefore invisible to immune responses. In addition, active mechanisms of immune evasion are frequently evident during the productive stage of the virus life cycle. For viruses to be successful, a delicate virus-host balance needs to be established to ensure survival and transmission of the virus while minimizing morbidity.
Epstein-Barr virus (EBV) is a prime example of a successful persistent virus, having coevolved with its human host over millions of years to colonize more than 90% of the adult population worldwide (28). EBV is a gammaherpesvirus type 1 that replicates in permissive cells in the oropharynx and persists as a latent infection in long-lived memory B lymphocytes. That EBV is usually carried as an asymptomatic infection is remarkable, considering it is a potent growth-transforming agent for resting B lymphocytes in vitro and is, in some patients, associated with lymphoproliferative disease or malignant tumors of lymphoid or epithelial cell origin (28, 37). The importance of host T cell surveillance for preventing EBV pathogenesis is well-illustrated by the increased incidence of potentially fatal lymphoproliferative lesions in patients receiving immunosuppressive therapy following organ transplants, which can be reversed by infusion of EBV-specific immune T cells (13, 30). These lymphoproliferative lesions are comprised of EBV-transformed B cells, which are phenotypically similar to lymphoblastoid cell lines (LCLs). LCLs are easily established following experimental infection of resting B cells with EBV in vitro, and they express a limited number of transformation-associated genes that are targets of the immune responses (15, 33, 36).
While the pathogenic potential of EBV is normally kept in check by T cell responses, it is evident that these responses do not succeed in eliminating the virus completely from its host. In healthy infected hosts, EBV-transformed B cells are rarely detectable, since the virus reverts to an immunologically silent latent infection in resting memory B cells (28, 36). Nevertheless, infectious virus is regularly detected in oropharyngeal secretions (16, 22, 39). This implies that during the lytic virus productive cycle EBV has effective immune evasion mechanisms. Indeed, a number of immunoevasins targeting the MHC-I and MHC-II antigen presentation pathways have recently been identified (27, 34). EBV genes reported to modulate MHC-I antigen presentation include vIL10, BGLF5, BNLF2a, and BILF1, each of which acts at a different point along the presentation pathway (14, 32, 34, 40, 41). The most recent of these immunoevasin genes to be identified, BILF1, encodes a viral G protein-coupled receptor (vGPCR) that was previously shown to physically associate with MHC-I molecules, to enhance internalization of MHC-I from the cell surface, and to target MHC-I for degradation via lysosomes (41).
The present study was initiated to characterize the molecular mechanisms by which the BILF1 protein effects its immunomodulatory functions. The experiments revealed an unexpected complexity and led to the identification of an additional functional component that promotes diversion of newly synthesized MHC-I molecules away from the normal trafficking pathway. Biochemical analyses of MHC-I molecule expression and localization can reveal modest or marked differences that do not necessarily reflect larger qualitative differences in the relevant MHC-I/peptide complexes expressed at the cell surface. To address this, we also employed functional T cell assays with specific CD8+ effectors to investigate the contribution of the different mechanisms of BILF1 to the availability of endogenously processed antigen on target cells. The results showed that the separate effects of BILF1 on both the exocytic and endocytic pathways of MHC-I trafficking caused marked impairment of T cell recognition.
MATERIALS AND METHODS
Plasmids, retroviral expression vectors, and small interfering RNA (siRNA).
The N-terminal hemagglutinin (HA)-tagged wild-type (wt) BILF1 and a K122A mutant BILF1 have been described previously (41). The C-terminal deletion mutant was generated by PCR amplification of the BILF1 gene with an introduced stop codon mutation. HA-tagged wt BILF1 and all BILF1 mutants were cloned into the Retro-XTM universal packaging system (Clontech Laboratories, Inc.) to prepare retroviruses. This system is based on the PQCXIH vector, which contains a hygromycin resistance gene. The expression plasmid pCDNA3-HABILF1-GFP and control empty vector pCDNA3-IRES-nlsGFP have been described previously (41), as have the pCEP4-SM plasmid, containing the EBV BSLF2/BMLF1 spliced gene, and p509, containing the EBV BZLF1 gene (42).
Mutant recombinant EBV construction and generation of a virus producer cell line.
Wild-type (clone 2089) and BZLF1-negative (ΔBZLF1) recombinant EBV bacterial artificial chromosomes (BACs) have been previously described (12). The EBV BILF1-negative (ΔBILF1) mutant was constructed (see Fig. S1 in the supplemental material) by replacing the complete BILF1 gene (coordinates 151706 to 152641 of EBV strain B95.8; accession number NC_007605) with the kanamycin resistance gene by homologous recombination with a linear PCR fragment as described previously (8, 21). The kanamycin resistance gene from pCP15 was amplified using primers BILF1-Kan1, CAGGCCTGTGTGTCAGTTTGCAGGGCCATCCTCGCACTCAACCAGTCACGACGTTGTAAAACGAC, and BILF1-Kan2, TTTGCTGCAGACACCACCCAGTCTGGCTCTGACCAGCAAGAACAGCTATGACCATGATTACGCC, resulting in a linear fragment containing the kanamycin resistance gene flanked by 40-bp stretches of homology (underlined) to sequences next to the BILF1 gene. This fragment was transformed into electrocompetent Escherichia coli DH10B cells carrying EBV BAC p2089 as described previously (9, 21). Recombinant clones were selected with kanamycin and analyzed for BAC DNA integrity by restriction enzyme cleavage. Plasmid DNA from a successfully recombined clone was prepared (Nucleobond; Machery-Nagel) and transfected into HEK293 cells by lipofection (Metafectene; Biontex). Cells were kept under hygromycin selection (100 μg/ml) for 3 weeks, and outgrowing green fluoresceint protein (GFP)-positive cell clones were tested for virus production after transfection of BZLF1 and gp110 (BALF4) expression plasmids.
Recombinant EBV strains and generation of LCLs.
Stable 293 cell clones carrying the EBV BACs were selected by hygromycin (100 μg/ml) and induced to produce virus by transfection with BZLF1 and gp110 plasmids by using Lipofectamine 20000 (Invitrogen). Virus supernatants were harvested 3 days posttransfection, filtered through a 0.8-μm-pore-size filter, and stored at 4°C until used to infect transformed laboratory donor peripheral blood B lymphocytes to generate LCLs. LCLs were generated using wt EBV (clone 2089) and ΔBZLF1 and ΔBILF1 recombinant EBVs.
Cells and transfections.
LCLs were maintained in RPMI 1640 supplemented with10% fetal calf serum (FCS). EBV-specific CD8+ cytotoxic T cells were grown in 10% FCS in RPMI 1640 medium supplemented with 30% supernatant from the interleukin-2 (IL-2)-producing MLA 144 cell line (26) and 50 U/ml recombinant IL-2, as described elsewhere (25).
The HEK293 epithelial cell line (American Type Culture Collection) and derived cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS and penicillin-streptomycin antibiotics. HEK293 lines transduced by PQCXIH-based retroviruses were selected with 400 μg/ml hygromycin. Transient transfection of HEK293 with plasmid DNA was routinely performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Retroviral infection.
For the production of replication-defective recombinant retrovirus, the packaging cell line (GP2-293) was transfected with retroviral vectors and PVSV-G plasmid. Supernatants containing the retrovirus particles were harvested 72 h after transfection and filtered through a 0.45-μm-pore-size, low-protein-binding filter. To generate stable cell lines by transduction with retrovirus, HEK293 cells were infected with 1 ml retrovirus supernatants with Polybrene added to a final concentration of 4 μg/ml. The stable cell lines were selected with 400 μg/ml hygromycin.
Antibodies.
The murine monoclonal antibodies (MAbs) used to detect human MHC class I were W6/32 (3), which recognizes native β2 microglobulin-associated MHC-I complexes (HLA-A, -B, and -C alleles), and HC10 (35), which recognizes free HLA class I heavy chains. For flow cytometry experiments, phycoerythrin (PE)-conjugated MAb to HLA-A, -B, and -C (MCA81PE; clone W6/32) was purchased from AbD Serotec. Goat antibodies to calregulin (sc6467) were purchased from Santa Cruz Biotechnology. The 3F10 rat MAb directed against the influenza virus-derived HA tag was purchased from Roche Diagnostics. The rabbit serum with anti-BMLF1 (EB2) antibodies (6) was a kind gift of A. Sergeant, Lyon, France. The BZ.1 murine MAb specific for the EBV BZLF1-encoded protein was generated by our investigators.
Flow cytometry analysis of cell surface MHC molecules.
Cell surface expression of MHC-I on viable cells was determined by staining with PE-labeled W6/32 antibodies or PE-labeled isotype control MAb and detection on a Beckman Coulter XL flow cytometer. The data were analyzed using FlowJo software (Tree Star).
To assay the kinetics of internalization of surface MHC-I, HEK293 cells were incubated for 60 min on ice with saturating amounts of W6/32 MAb, then washed three times in phosphate-buffered saline (PBS) and replaced in warm culture medium to incubate at 37°C for the length of time indicated below in Results. To terminate MHC/antibody complex internalization, cells were rapidly cooled to 0°C. Finally, the MAb-bound surface MHC-I molecules were stained at 0°C with PE-conjugated goat anti-mouse IgG2a antibody (AbD Serotec), and cells were analyzed by flow cytometry.
To assay the kinetics of appearance of surface MHC-I molecules, HEK293 cells were incubated with saturating amounts of W6/32 MAb, then incubated at 37°C for different lengths of time as described for the internalization assay (see above). After cooling to 0°C to prevent further appearance of unblocked surface MHC-I molecules, the cells were stained with PE-conjugated W6/32 MAb and analyzed by flow cytometry.
To assay the kinetics of recycling of surface MHC-I molecules, HEK293 cells were incubated for 60 min on ice with saturating amounts of W6/32 MAb, then washed three times in PBS, replaced in warm culture medium, and incubated at 37°C for 30 min. The cells were then washed with PBS, and remaining surface-bound W6/32 MAb was stripped by gently resuspending the cells in pH 3.1 citrate/phosphate buffer (0.131 M citric acid, 0.066 M Na2HPO4) for 1 min on ice before neutralization by addition of excess standard medium. Stripped target cells were then washed twice with PBS, resuspended in warm medium, and incubated at 37°C for the length of time indicated below in Results. To terminate further recycling of MHC-I molecules, cells were rapidly cooled to 0°C. Finally, recycled MHC-I molecules were stained with PE-conjugated goat anti-mouse IgG2a antibody (Serotec), and the cells were analyzed by flow cytometry.
Flow cytometric analysis of intracellular staining for lytic cycle antigens.
The percentages of cells spontaneously reactivating into the lytic cycle in LCLs were measured by intracellular staining for BZLF1. Cells were first fixed using 100 μl of Ebiosciences intracellular (IC) fixative (catalog number 00-8222-49) for 1 h on ice, followed by permeabilization through the addition of 100 μl Triton X-100 (final concentration, 0.2%) and a further 30 min of incubation on ice. After extensive washing with PBS, cells were incubated with 1 μg/ml of either MAb BZ.1 (anti-BZLF1) or with an IgG1 isotype control MAb for 1 h at 37°C. Cells were washed twice in PBS and then incubated with 1:50-diluted R-phycoerythrin-conjugated goat anti-mouse IgG1 antibody (catalog number STAR132PE; AbD Serotec) for 1 h at 37°C. Following further washes, cells were analyzed on a Beckman Coulter XL flow cytometer, and the data were processed using FlowJo software (Tree Star).
For comparison of surface MHC-I expression in lytic and latent 293/EBV cells, the 293/wt EBV cells and 293/ΔBILF1 cells were induced to the EBV lytic cycle by transfecting BZLF1 plasmids. At 24 h postinduction, the surface HLA class I and intracellular EBV gp110 antigens were detected simultaneously. First, viable 293 cells were stained with 1:15-diluted allophycocyanin-conjugated anti-human HLA-A, -B, or -C anitbody (catalog number 311410; Biolegend) for 30 min on ice. Cells were then washed extensively in PBS and fixed and permeabilized as described above, followed by incubation for 1 h at 37°C with 1 μg/ml of L2 MAb (late antigen gp110) or with isotype control MAb. After washing in PBS, cells were incubated for 1 h with 1:20-diluted peridinin chlorophyll protein-conjugated goat anti-mouse IgG1 antibody. Cells were analyzed by flow cytometry as described above.
Western blotting.
For Western blotting, total cell lysates were denatured in reducing sample buffer (final concentrations, 2% SDS, 72.5 mM Tris-HCl [pH 6.8], 10% glycerol, 0.2 M sodium 2-mercaptoethane sulfonate, 0.002% bromophenol blue) and then sonicated and heated to 100°C for 5 min. Solubilized proteins equivalent to 1 × 105 cells/20-μl sample were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 4-to-12% acrylamide gradient bis-Tris NuPage minigels with morpholinepropanesulfonic acid running buffer (Invitrogen).
Following electroblotting to polyvinylidene difluoride membranes (Invitrogen) and blocking with I-Block (Tropix; Applied Biosystems) in PBS with 0.1% Tween 20 detergent, specific proteins were detected by incubating the membranes with primary antibodies at 4°C overnight. The purified mouse MAb HC10 and the goat antibody to calregulin were used at 1 μg/ml, the rat anti-HA MAb was used at 50 ng/ml, and the anti-BMLF1 rabbit serum was used at 1/6,000. Primary antibodies specifically bound to blotted proteins were detected by incubation for 30 min with appropriate alkaline phosphatase-conjugated secondary antibodies, developed using a CDP-Star detection kit (Tropix; Applied Biosystems), and exposed to autoradiographic film.
T cell function assays.
The effector T cell clone GLC, specific for BMLF1 and restricted through HLA-A2, was generated as described elsewhere (25). Targets for the GLC clone were generated by cotransfection of HLA-A2-type HEK293 cells with a BMLF1 expression plasmid and wt BILF1 or different BILF1 mutants' expression plasmids. At 24 h posttransfection, recognition of target cells by the effector T cells was determined by enzyme-linked immunosorbent assay (ELISA) of gamma interferon (IFN-γ) release using a standard protocol described elsewhere (19). Briefly, 104 effector T cells were incubated for 18 h at 37°C in V-bottom microtest plate wells with 105 target cells, then the supernatants were harvested for quantitation of IFN-γ by ELISA (Endogen) in accordance with the manufacturer's recommended protocol. Specificity control targets included HLA-matched and -mismatched EBV-transformed LCLs, empty vector-transfected HEK293 cells, and empty vector-transfected HEK293 cells pulsed with the GLCTLVAML (GLC) synthetic peptide.
For the peptide disappearance assay, HEK293 cells were pulsed with synthetic GLC peptide on ice for 1 h, then washed extensively with PBS, replaced in warm culture medium, and incubated at 37°C for different periods of time. At each time point, the cells were washed twice with PBS and fixed with 1% paraformaldehyde (PFA) in PBS at room temperature for 10 min. The PFA was subsequently quenched with 0.2 M glycine in PBS. The fixed, quenched, and washed peptide-pulsed cells were cocultured with the CD8+ effector GLC T cell clone for a further 18 h, and the supernatants were tested for the release of IFN-γ as a measure of T cell recognition. All results are expressed in terms of IFN-γ release (in pg/ml), and error bars indicate standard deviations of triplicate cultures.
For the EBV lytic antigen CD8+ T cell against LCLs recognition assay, 104 effector T cells were incubated for 18 h at 37°C in V-bottom microtest plate wells with 2 × 105 target LCLs, and then the supernatants were harvested for quantitation of IFN-γ by ELISA (Endogen) in accordance with the manufacturer's recommended protocol.
NF-κB reporter assays.
HEK293 cells were seeded at 2 × 105 cells/well in 24-well plates at 24 h prior to transfection with a constitutively expressed Renilla luciferase reporter construct (phRL-TK; Promega) for normalizing transfection efficiency, and 3 enh-ConA (an NF-κB-dependent luciferase reporter construct in which transcription of the firefly luciferase gene is driven by three NF-κB binding sites [1]), together with vectors for wt BILF or BILF1 mutants or empty vector DNA. Transfection was performed with Lipofectamine 2000 (Invitrogen) and DNA mix in Opti-MEM, prepared and used according to the supplier's instructions. Cell extracts were generated after 48 h using cell culture lysis buffer (Promega), and extracts were assayed for firefly luciferase and Renilla luciferase activities by using the dual-luciferase reporter assay system (Promega).
RESULTS
BILF1 contributes to the downregulation of surface MHC-I in the EBV lytic cycle.
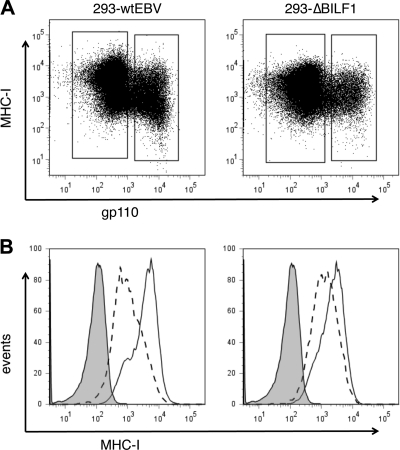
BILF1 as an immune evasion gene causing downregulation of surface MHC-I expression was originally demonstrated using a reductionist approach by expressing the BILF1 gene in isolation. We therefore sought first to confirm the contribution of BILF1 in the context of the whole viral genome. To this end, a recombinant EBV lacking BILF1 was generated (EBV ΔBILF1), and stable 293 cell clones carrying the EBV wt BAC and EBV ΔBILF1 BAC were generated by transfection and hygromycin selection. The EBV lytic cycle was then synchronously induced in these cells by transient transfection of a BZLF1-expressing plasmid. These induced cultures were analyzed by two-color immunoflourescence staining for EBV gp110 (BALF4) late lytic cycle antigen and cell surface MHC-I. As shown in Fig. 1, surface MHC-I levels in the 293/EBV wt cells were reduced by about 80% in the lytically infected subpopulation expressing gp110. In contrast, in the 293/ΔBILF1 cells, the surface MHC-I expression in the lytically infected subpopulation was reduced by less than 50%. This result is consistent with the hypothesis that BILF1 cooperates with other EBV immune evasion genes to reduce the expression of MHC-I at the cell surface.
FIG. 1.
Surface HLA class I expression in wt 293 cells and ΔBILF1 293 cells expressing late lytic antigens. 293 epithelial cells carrying recombinant wt EBV or ΔBILF1 EBV were induced into the EBV lytic cycle by transfecting a BZLF1 expression plasmid. At 24 h postinduction, these two cell lines were stained for surface HLA class I and intracellular gp110 and then analyzed by flow cytometry. (A) Two-color analysis of surface MHC-I and intracellular viral gp110 expression, showing the gates used to analyze latent (gp110-negative) and lytic (gp110-positive) populations. (B) Histograms of surface MHC-I expression on latent cells (solid line histogram) and lytic cells (dotted line histogram). The shaded histogram shows the isotype control staining.
The EKT signaling motif and a C-terminal domain of BILF1 cooperate for degradation of MHC-I.
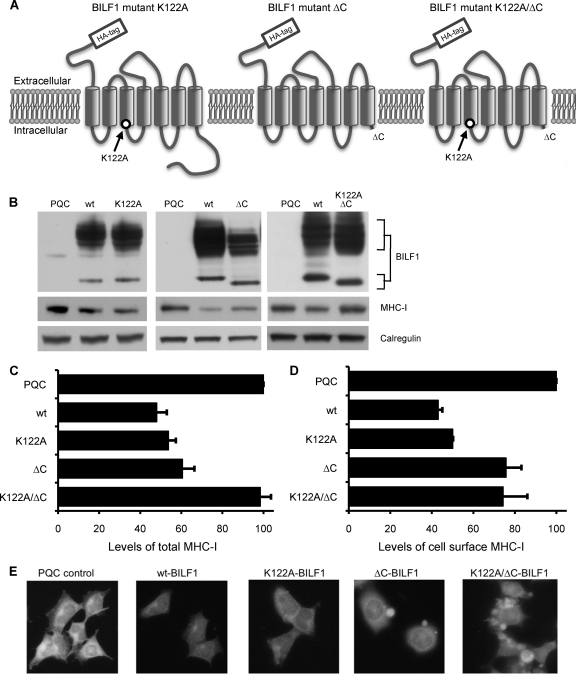
To identify the functional domain(s) of BILF1 responsible for the diversion of MHC-I for degradation and the impairment of T cell recognition, a series of BILF1 mutants were made. Three mutants proved to be informative for the current study: K122A mutant BILF1, in which the lysine at codon 122 is replaced with an alanine, disrupting a DRY-like EKT motif (31, 38) necessary for NF-κB signaling (41); ΔC mutant BILF1, from which the last 21 C-terminal amino acids are deleted; the K122A/ΔC double mutant BILF1 (see Fig. 2A, below). An N-terminal HA tag sequence was engineered into wt BILF1 and each of the mutants, which were then cloned into the PQCXIH retrovirus vector to generate the recombinant retroviruses used to transduce HEK293 cells. Stable expressing cell lines were selected with hygromycin.
FIG. 2.
The EKT signaling motif and a C-terminal domain of BILF1 cooperate for degradation of MHC-I. (A) Schematic representation of the mutated BILF1 proteins, showing the seven transmembrane helices, the location of the K122A mutation in the DRY-like EKT motif, and the truncation of the C terminus. (B) HEK293 cells stably transduced with control (PQC), wt BILF1, K122A mutant BILF1, ΔC mutant BILF1, or K122A/ΔC mutant BILF1 retroviruses were analyzed by Western blotting. Total cell lysates from 105 cells were separated by SDS-PAGE and analyzed by Western blotting with MAbs specific for BILF1 (3F10; anti-HA tag), MHC-I (HC10), or with polyclonal antibodies to calregulin as a loading control. (C) Histogram showing the mean results of quantification of Western blotting results by densitometry from three independent experiments. The densities of the HC10 bands were normalized relative to their own calregulin loading control. All results are expressed as amounts of total MHC-I expression as a percentage of the expression in PQC-293 cells, and error bars indicate standard deviations of triplicate experiments. (D) Histogram showing the mean results of quantification of surface MHC-I expression by flow cytometry. Viable cells of the same panel of lines as shown in panel C were stained with PE-labeled W6/32 MAb and analyzed by flow cytometry. All results are expressed as the amount of surface MHC-I staining as a percentage of the staining in PQC-293 cells, and error bars indicate standard deviations of triplicate experiments. (E) Immunofluorescence staining with W6/32 MAb to HLA class I complexes in fixed cell cultures of HEK293 cells transduced with control retrovirus (PQC control) or retroviruses expressing wt BILF1, K122A BILF1, ΔC BILF1, or K122A/ΔC BILF1.
Total cell lysates of these transduced cell lines showed comparable expression levels of wt and mutant BILF1 proteins when analyzed by SDS-PAGE and immunoblotting, while the levels of MHC-I were affected by the expression of BILF1, as shown by the representative immunoblotting experiments with whole-cell lysates in Fig. 2 B. Densitometry was performed on immunoblots from three separate experiments to quantitate expression of total MHC-I; consistent with previous observations (41), wt BILF1 caused a 52% reduction in the expression of total MHC-I relative to control (PQC) transduced HEK293 cells (Fig. 2C). In addition, expression of K122A mutant BILF1 similarly reduced the level of total MHC-I by 46%, and expression of the ΔC mutant BILF1 was associated with a 40% reduction (Fig. 2C). In marked contrast, cells transduced with virus expressing the K122A/ΔC double mutant BILF1 showed levels of total MHC-I that did not differ significantly from the levels seen in control (PQC) transduced cells (Fig. 2C). The results thus far described (Fig. 2) suggest that the C-terminal domain of BILF1 contains a determinant that cooperates with the EKT signaling function of BILF1 to target MHC class I for degradation.
However, when the effects on levels of cell surface MHC-I expression were examined by flow cytometry of W6/32 MAb-stained cells, a different pattern of results was obtained. While cells transduced with the wt BILF1, K122A mutant, or ΔC mutant BILF1 showed a reduction in cell surface expression that reflected the reduction in total MHC-I expression, the KI22A/ΔC double mutant BILF1 showed an approximately 25% reduction in cell surface MHC-I expression, although there was no significant change in total cellular MHC-I expression (cf. Fig. 2C and D). However, we did see a pronounced accumulation of intracellular MHC-I in K122A/ΔC mutant BILF1-expressing cells, as revealed by immunofluorescence staining of fixed and permeabilized cells (Fig. 2E).
The EKT signaling motif, but not the C terminus of BILF1, is necessary for enhanced endocytosis of surface MHC-I.
The disconnection between the levels of total and cell surface MHC-I expression with the K122A/ΔC mutant BILF1 hinted at a greater mechanistic complexity for the effect of BILF1 on MHC-I antigen presentation than was previously envisaged. We therefore included this panel of mutants in a reexamination of key known phenotypes of BILF1.
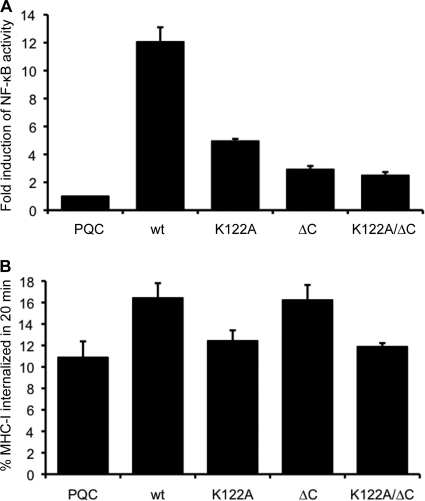
First we examined the signaling functions of the mutants in a reporter assay by cotransfecting an NF-κB reporter plasmid with or without wt or mutant BILF1 expression vectors (Fig. 3 A). The wt BILF1 constitutively activated NF-κB signaling by approximately 12-fold above background, and disruption of the EKT motif in the K122A mutant BILF1 reduced this activation by almost 70%. In addition, these experiments showed for the first time an even greater impairment of NF-κB activation in the ΔC and K122A/ΔC mutants.
FIG. 3.
Signaling functions of BILF1 mutants and the requirement of the EKT signaling motif for enhanced endocytosis of surface MHC-I. (A) Control (PQC), wt BILF1, K122A BILF1, ΔC BILF1, or K122A/ΔC BILF1 expression plasmids were transfected into HEK293 cells together with an NF-κB reporter plasmid and a Renilla luciferase reporter construct. The degree of NF-κB activation was measured by detection of luciferase activity. The results are the means ± standard deviations for three independent experiments which were themselves performed in triplicate. (B) Assay for the rate of internalization of cell surface MHC-I complexes. HEK293 cells stably transduced with different recombinant retroviruses were incubated at 0°C with saturating concentrations of W6/32 MAb to MHC-I and then washed and incubated at 37°C for 20 min. The viable cells were then stained with PE-conjugated goat anti-mouse IgG antibody and analyzed by flow cytometry. The mean fluorescence intensities of staining were averaged for triplicate samples. The histogram shows the percentages of internalized MHC-I in 20 min. The results are the means of three independent experiments, and error bars indicate standard deviations of three experiments.
Next, we examined the ability of these mutants to mediate the previously reported ability of wt BILF1 to enhance endocytosis of surface MHC-I. As before, we used an internalization assay in which cell surface MHC-I was first labeled with W6/32 MAb at 0°C, and the amount remaining on the cell surface after 20 min of incubation at 37°C was determined by detection with PE-conjugated anti-mouse IgG and analyzed by flow cytometry. The pooled results from three independent experiments are shown in Fig. 3B. While wt BILF1 caused an enhanced rate of MHC-I internalization (16.4 ± 1.4% versus the control, 10.9 ± 1.5%, in 20 min), the K122A mutant almost completely abrogated this effect. The rate of MHC-I internalization in K122A mutant BILF1-expressing cells (12.4 ± 1.0% in 20 min) was not significantly different from that in the control transduced cells. In contrast, the ΔC mutant BILF1 retained the ability to enhance surface MHC-I internalization (16.2 ± 1.4%). Similar to the K122A mutant, the K122A/ΔC double mutant BILF1 showed a rate of internalization of surface MHC-I (11.9 ± 0.3%) that was indistinguishable from control cells.
The data in Fig. 3 show that the EKT signaling motif of BILF1 is essential for the ability of BILF1 to enhance internalization of surface MHC-I, while the C terminus is dispensable. This cannot be explained simply in terms of the NK-κB signaling properties of BILF1.
The K122A/ΔC double mutant BILF1 selectively reduces the rate of appearance of surface MHC-I.
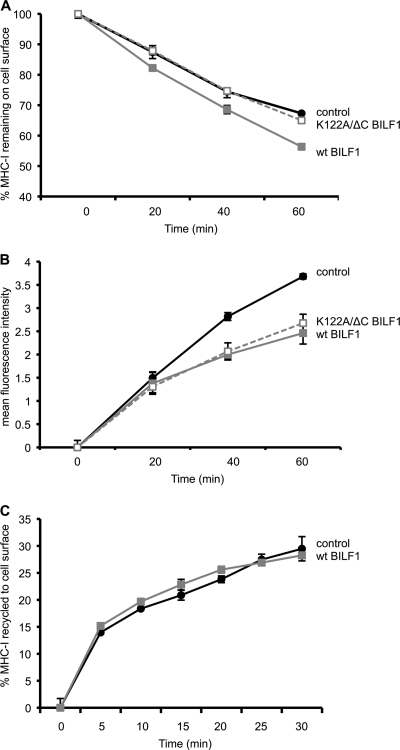
Since the K122A/ΔC double mutant BILF1 is completely defective with regard to enhancing surface MHC-I internalization (Fig. 3B), we questioned how it achieves the small but reproducible reduction in surface MHC-I levels shown in Fig. 2D. We therefore examined the phenotype of the K122A/ΔC mutant in more detail, by comparing the rate of internalization and the rate of appearance of MHC-I at the cell surface. Figure 4 A shows a representative internalization assay in which the percentage of MHC-I remaining on the cell surface was measured over a 60-min time course. As expected from the data shown in Fig. 3B, cells expressing wt BILF1 showed a higher rate of internalization than control cells, while K122A/ΔC double mutant BILF1 expressing cells showed the same rate of internalization as the control cells (Fig. 4A). In contrast, when we used a modification of the assay that measured the rate of appearance of surface MHC-I, the K122A/ΔC double mutant BILF1 behaved exactly as did wt BILF1 in reducing the rate of appearance of surface MHC-I (Fig. 4B).
FIG. 4.
The K122A/ΔC double mutant BILF1 selectively reduces the rate of appearance of surface MHC-I. Internalization, appearance, and recycling assays were performed on HEK293 cells stably transduced with PQC control, wt BILF1, or K122A/ΔC mutant BILF1 retroviruses. (A) Internalization assay. As for Fig. 2B, cells were treated with saturating concentrations of W6/32 MAb to MHC-I and then washed and incubated at 37°C for up to 60 min. At the indicated times, viable cells were stained with PE-conjugated goat anti-mouse IgG antibody and analyzed by flow cytometry. The mean fluorescence intensities of staining were averaged for triplicate samples and then normalized to the time zero samples; error bars indicating standard deviations of triplicate samples are shown for all samples, although the errors were often smaller than the symbols used in the graphs and are therefore not always visible. (B) Appearance assay. Cells were incubated at 0°C with saturating concentrations of W6/32 MAb to MHC-I molecules and then washed and incubated at 37°C for different periods of time. The appearance of new MHC-I molecules was assayed by staining with PE-conjugated W6/32 antibody. The mean fluorescence intensities of staining were averaged for triplicate samples; error bars indicate standard deviations of triplicate samples. (C) Recycling assay. Cells were incubated at 0°C with saturating concentrations of W6/32 MAb to MHC-I and then washed and incubated at 37°C for 30 min before stripping the remaining surface-bound W6/32 MAb and resuming incubation at 37°C for the indicated periods of time. The cells were then stained with PE-conjugated goat anti-mouse IgG antibody and analyzed by flow cytometry. The mean fluorescence intensities of staining were averaged for triplicate samples and are expressed as the percentage of the amount of MHC-I internalized in the initial 30-min incubation; error bars indicate standard deviations of triplicate samples.
We previously attributed the effect of wt BILF1 on the rate of appearance of surface MHC-I to a secondary and delayed effect of the BILF1-mediated enhanced internalization (41). However, these new results with the K122A/ΔC double mutant BILF1 (Fig. 4A and B) are inconsistent with that interpretation, as K122A/ΔC double mutant BILF1 reduced the rate of appearance of surface MHC-I under conditions when this mutant did not enhance internalization. Since the assay shown in Fig. 4B measures the appearance of both recycled and newly synthesized MHC-I molecules, we therefore devised a modification of the assay to measure the amount of MHC-I recycling to the cell surface after internalization.
In this recycling assay, the surface MHC-I was first saturated with W6/32 MAb and then allowed to internalize for 30 min at 37°C before acid stripping away all MAb remaining on the surface. The reappearance of internalized MHC-I/MAb complexes was then determined at serial time points by staining the surface with PE-conjugated anti-mouse IgG antibodies. In such assays, we found that BILF1 had no effect on the rate of recycling of internalized MHC-I molecules (Fig. 4C). Notably, 15% of the internalized MHC-I was recycled to the surface within 5 min; thereafter, the rate of recycling was slower, such that the following 15% recycled portion took more than 25 min. This suggests that the initial MHC-I to arrive at the surface in the appearance assay (Fig. 4B) will be derived predominantly from the recycling pool and, thereafter, newly synthesized MHC-I molecules arriving from the endoplasmic reticulum (ER)/Golgi compartments make a greater contribution to the MHC-I molecules appearing at the cell surface.
Together, the results in Fig. 4 showed that wt BILF1 both enhances the rate of internalization of surface MHC-I molecules and reduces the rate of appearance of newly synthesized MHC-I molecules at the cell surface. The K122A/ΔC double mutant BILF1 only reduces the rate of appearance of newly synthesized MHC-I. Thus, the EKT signaling motif is necessary for enhanced internalization, and the C terminus is necessary for subsequent enhanced degradation of MHC-I (Fig. 2 and 3). Neither the EKT motif nor the C-terminal domain is required for diverting newly synthesized MHC-I on the exocytic pathway (Fig. 4B).
BILF1-mediated enhanced endocytosis of MHC-I from the cell surface is important for antigen presentation to CD8+ T cells.
As BILF1 can act on both the exocytic and endocytic trafficking pathways for MHC-I, we next addressed which of these functions contributed to evading recognition by immune CD8+ T cells.
To address this question with regard to the endocytic pathway, we first pulsed control 293, wt BILF1 293, and K122A/ΔC BILF1 293 cells with synthetic GLCTLVAML peptide before fixing the cells and using them as targets in T cell assays with the HLA-A2-restricted GLC CD8+ T cell clone. As expected, the results reflected the levels of MHC-I molecules on the surface of the respective cell lines (cf. Fig. 5A and 2D). Thus, T cell recognition of peptide-pulsed cells expressing wt BILF1 was reduced by 61% compared to the recognition of peptide-pulsed control cells, and recognition of peptide-pulsed cells expressing K122A/ΔC double mutant BILF1 was 31% lower than the recognition of peptide-pulsed control cells (Fig. 5A).
FIG. 5.
BILF1-mediated enhanced endocytosis of MHC-I from cell surface modulates antigen presentation to CD8+ T cells. HEK293 cells stably transduced with control (PQC), wt BILF1, or K122A/ΔC mutant BILF1 recombinant retroviral vectors were pulsed with synthetic GLCTLVAML peptide and used as targets in T cell assays. (A) Peptide-pulsed cells were extensively washed to remove unbound peptide, and replicate aliquots of cells were fixed with 1% PFA. Following coculture of the fixed peptide-pulsed cells with GLC effector CD8+ T cells for 18 h, the supernatants were tested for the release of IFN-γ as a measure of T cell recognition. All results are expressed as IFN-γ release (in pg/ml), and error bars indicate standard deviations of triplicate cultures. (B) Peptide-pulsed cells (PQC 293 controls, wild-type BILF1-293, and K122A/ΔC BILF1-293) were extensively washed to remove unbound peptide and then incubated at 37°C for up to 3 h. At the indicated periods of time, triplicate aliquots of peptide-pulsed cells were fixed with 1% PFA and were cocultured with GLC effector CD8+ T cells for a further 18 h. The supernatants were tested for the release of IFN-γ as a measure of T cell recognition. All results are expressed as IFN-γ release as a percentage of that observed at time zero. Error bars indicate standard deviations of triplicate cultures.
Next, viable peptide-pulsed cells were incubated at 37°C for up to 3 h before harvesting and fixing for use as targets in T cell assays. In these experiments, the peptide-pulsed control 293 cells rapidly internalized peptide/MHC-I complexes from the cell surface and became less potent targets for stimulating the effector T cells to produce IFN-γ; thus, about 45% of T cell recognition was lost within the first hour (Fig. 5B). The peptide-pulsed wt BILF1 293 cells showed an even higher rate of peptide/MHC-I complex internalization, showing an almost 80% loss of T cell recognition within 1 h of incubation (Fig. 5B). In contrast, the peptide-pulsed K122A/ΔC mutant BILF1 293 cells showed loss of surface peptide/MHC-I complexes with kinetics and magnitude indistinguishable from control 293 cells (Fig. 5B), consistent with the inability of this double mutant to enhance endocytosis (Fig. 4A). These results demonstrate a role for BILF1-mediated enhanced endocytosis of MHC-I complexes in modulating antigen presentation to immune effector T cells.
Diversion of MHC-I on the exocytic pathway by BILF1 affects antigen presentation to CD8+ T cells.
We next asked to what extent the effect of BILF1 on the MHC-I exocytic trafficking pathway might influence antigen presentation for functional CD8+ T cell recognition. To address this question, HEK293 cells were cotransfected with BMLF1 target antigen plasmid together with plasmid vectors for wt BILF1 or the different BILF1 mutants. These target cells were then examined for antigen presentation to effector GLC CD8+ T cells specific for a BMLF1-derived peptide (GLCTLVAML). Following coculture of transfected target cells with effector CD8+ T cells, T cell recognition was measured by ELISA for release of IFN-γ from the effector cells.
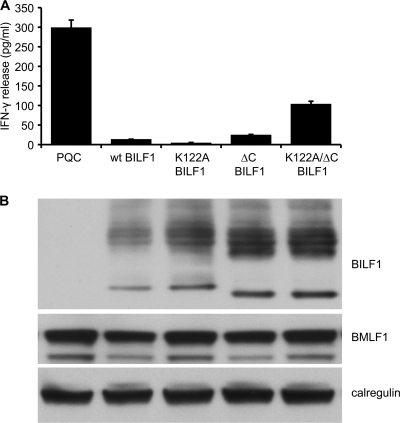
The representative results in Fig. 6A show good recognition of cells transfected with BMLF1 alone, which was inhibited by more than 90% when wt BILF1 was coexpressed. Expression of the K122A mutant BILF1 or ΔC mutant BILF1 resulted in similarly efficient inhibition of CD8+ T cell recognition. Notably, expression of the K122A/ΔC double mutant BILF1 resulted in an up-to-10-times-greater CD8+ T cell recognition than when wt BILF1 or K122A mutant BILF1 or ΔC mutant BILF1 were expressed. Nevertheless, the CD8+ T cell recognition in the K122A/ΔC double mutant BILF1 transfectant was typically only about 35% of that seen in the control transfectant lacking any BILF1. Immunoblotting of target cells showed that expression of transfected BMLF1 target protein was comparable in the different transfected targets (Fig. 6B). Similar results were obtained in three separate experiments. In addition, experiments using MelJuSo target cells with RAK CD8+ effector T cells specific for a BZLF1-derived peptide (41) gave the same pattern of results with the panel of BILF1 mutants (data not shown).
FIG. 6.
Diversion of MHC-I from the exocytic pathway by BILF1 modulates antigen presentation to CD8+ T cells. (A) HEK293 cells were cotransfected with a BMLF1 expression vector together with control (PQC), wt BILF1, K122A BILF1, ΔC BILF1, or K122A/ΔC BILF1 expression plasmids. At 24 h posttransfection, the cells were cocultured with GLC CD8+ effector T cells (specific for a BMLF1-derived peptide) for a further 18 h, and the supernatants were tested for the release of IFN-γ as a measure of T cell recognition. All results are expressed as IFN-γ release (in pg/ml), and error bars indicate standard deviations of triplicate cultures. (B) Total cell lysates were generated from aliquots of the above target cell transfections and were analyzed by Western blotting using antibodies specific for HA-tagged BILF1(3F10; anti-HA tag), BMLF1, or calregulin as a loading control.
As the K122A/ΔC double mutant BILF1 selectively disrupts trafficking of newly synthesized MHC-I to the cell surface and still inhibits CD8+ T cell recognition by 65%, these results imply that diversion of MHC-I on the exocytic pathway by BILF1 substantially affects antigen presentation to CD8+ T cells.
Together, the results in Fig. 5 and 6 demonstrate that the mechanism of immune evasion of BILF1 includes both enhanced internalization of existing surface peptide/MHC-I complexes and diversion of newly synthesized peptide/MHC-I complexes. From Fig. 5 and 6, it can be estimated that the effect of BILF1 on the exocytic pathway is responsible for about 70% of the immune evasion property of wt BILF1 in this model.
Deletion of BILF1 confers an increase in recognition by EBV lytic antigen-specific CD8+ T cells.
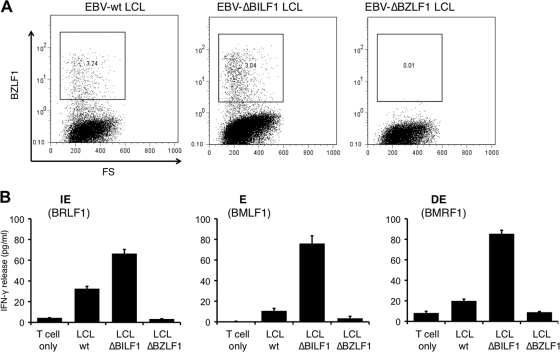
Having used a reductionist approach in epithelial cells to elucidate the broad mechanisms by which BILF1 modulates MHC-I expression and thereby recognition of antigen by immune CD8+ effector cells, we returned to the question of whether BILF1 contributes to immune evasion in the context of the whole virus during the lytic cycle in B cells. A panel of LCLs from an HLA-A2-matched donor was derived by transformation of peripheral blood B cells with either recombinant EBV wt, EBV ΔBILF1, or EBV ΔBZLF1. As LCLs normally show variable numbers of cells spontaneously entering the lytic cycle, we generated sets of LCLs in parallel from the same donor bleeds and monitored levels of the lytic cycle by intracellular staining for the immediate-early (IE) gene product BZLF1. Lines which had similar levels of spontaneous lytic cycle were selected for T cell recognition assays. In the representative experiment shown, the EBV wt and EBV ΔBILF1 LCLs both contained around 3% BZLF1+ cells, while the control EBV ΔBZLF1 LCLs were defective for lytic cycle entry (Fig. 7A). These lines were used as targets in T cell assays with HLA-A2-restricted, EBV lytic antigen-specific CD8+ T cells: the YVL effectors specific for a peptide which comes from the IE lytic antigen BRLF1; GLC effectors specific for the early (E) lytic antigen BMLF1; TLD specific for the delayed early (DE) lytic antigen BMRF1. As shown in Fig. 7B, the ΔBILF1 LCLs were recognized more efficiently than the wild-type LCLs by each of the three effector CD8+ T cell clones.
FIG. 7.
A role for BILF1 in modulating T cell recognition during the lytic cycle in B cells. (A) The proportions of LCLs spontaneously reactivating into the lytic cycle in EBV ΔBILF1 and EBV wt LCLs were assessed by intracellular BZLF1 staining and analysis by flow cytometry. (B) CD8+ T cell recognition of wt, ΔBILF1, and ΔBZLF1 LCLs using three HLA-A*0201-restricted CD8+ effector clones specific for IE, E, or DE lytic cycle antigens. Effector clone YVL was specific for BRLF1; clone GLC was specific for BMLF; clone TLD was specific for BMRF1. T cell recognition of the HLA-A*0201-matched LCL targets was measured in an IFN-γ ELISA. All results are expressed as IFN-γ release (in pg/ml), and error bars indicate standard deviations of triplicate cultures.
Together, the results in Fig. 1 and 7 show that, in the context of whole EBV, BILF1 not only contributes to the downregulation of surface MHC-I in the EBV lytic cycle but also contributes to the impairment of CD8+ T cell recognition of EBV lytic cycle antigens in B cells.
DISCUSSION
We previously reported that the BILF1 lytic cycle virus protein can downregulate the expression of surface MHC-I molecules and that one element of the molecular mechanism involved enhanced internalization of MHC-I from the cell surface and subsequent degradation via the lysosomal compartment (41). In this paper we show that an additional mechanism involves diversion of newly synthesized MHC-I molecules en route to the cell surface. The effects of BILF1 on both the exocytic pathway and the endocytic pathway contribute to impaired antigen presentation of MHC-I/peptide targets to CD8+ T cells. Importantly, we also demonstrated by using cells lytically infected with a recombinant EBV in which BILF1 was deleted that the immune-modulating functions of BILF1 operated in the context of the whole virus in both epithelial cells and B cells.
The complexity of the molecular mechanisms of BILF1 first became apparent in this study when we investigated the relationship between the NF-κB signaling properties of mutant BILF1 proteins and their ability to modulate MHC-I expression. The K122A mutant BILF1, which disrupts the EKT DRY-like signaling motif (24, 31, 38) and has impaired signaling functions (20, 41), was previously shown to retain the ability to reduce the steady-state levels of surface MHC-I (41) (Fig. 2D). Surprisingly, we found that this signaling-impaired mutant had substantially lost the ability to enhance internalization of MHC-I molecules (Fig. 3). Furthermore, a deleted mutant of BILF1, ΔC, which was similarly impaired for NF-κB signaling, retained the enhanced MHC-I internalization function (Fig. 3). These observations implied that the EKT motif is necessary for the enhanced MHC-I internalization but that the mechanism is apparently independent of NF-κB signaling.
How, therefore, does the EKT motif affect BILF1-induced enhanced internalization of MHC-I molecules? One possibility is that the EKT motif and the C terminus of BILF1 might trigger qualitatively different NF-κB activities, similarly to the two functional domains of EBV-encoded the LMP1 transmembrane protein that selectively activate the canonical and noncanonical NF-κB pathways (2, 11, 23). Alternatively, an NF-κB-independent signaling pathway activated by the BILF1 EKT motif might be involved. BILF1 has multiple constitutive signaling functions that are mediated through activation of Gαi proteins (4, 24), including activation of NF-κB, modulation of CRE-mediated signaling, inhibition of RNA-dependent PKR, and possibly others. Therefore, while the finer molecular details remain to be determined, our observations remain consistent with the interpretation that enhanced internalization of cell surface MHC-I is a signaling-dependent function of BILF1.
With regard to how BILF1 targets MHC-I molecules for lysosomal degradation, the results with the BILF1 mutants (Fig. 2C, D, and E) suggest a critical role for the C-terminal domain in cooperation with the EKT motif for this function. These observations are consistent with previous studies in other models, which have reported that sorting of transmembrane proteins to endosomes and lysosomes is normally mediated by signals present within the cytosolic domains of the proteins (5). However, as BILF1 does not contain any typical tyrosine-based sorting signals (e.g., NPXY or YXXØ) or dileucine-based signals (e.g., [DE]XXXL[LI] or DXXLL), it is difficult to predict what sorting signals may be involved. Various adapter proteins have been implicated in the regulation of membrane protein trafficking (29). In the coimmunoprecipitation experiments, we can demonstrate a physical association between BILF1 and the AP-1 adapter protein (see Fig. S2A in the supplemental material). However, in siRNA knockdown experiments, we were unable to demonstrate a role for AP-1, AP-2, phosphofurin acidic cluster sorting protein 1 (PACS-1), or clathrin in BILF1-mediated modulation of MHC-I (data not shown, but see Fig. S2B and C in the supplemental material). This contrasts with observations on HIV nef, which was one of the first viral proteins reported to disrupt antigen presentation in the secretory pathway (17). MHC-I binds to HIV-Nef, which binds to PACS-1, which binds AP1 and clathrin; these interactions bring MHC-I into an endosome-trans-Golgi network recycling loop and keep it off the plasma membrane (10). Determination of which membrane trafficking proteins are involved in BILF1 functions will require further work that was outside the scope of the present study.
One important new finding from the present study is that, in addition to inducing more rapid turnover of MHC-I from the cell surface, BILF1 impacts the transport of MHC-I to the plasma membrane. Although we observed a marked difference in the appearance of MHC-I at the cell surface between BILF1-expressing and control cells at later time points in the appearance assay (Fig. 4B), we previously interpreted this result as a secondary effect of the enhanced internalization (41). However, an analysis of the properties of the K122A/ΔC double mutant BILF1, in which enhanced internalization and targeted degradation are completely abolished, leads to the conclusion that the delayed effects in the appearance assay must actually reflect a marked effect of BILF1 in diverting MHC-I molecules away from the normal secretory trafficking pathway. As already discussed, the precise molecular mechanisms by which BILF1 modulates MHC-I trafficking are unknown, but we do know that BILF1 can physically associate with endo H enzyme-sensitive MHC-I molecules in the ER and with endo H-resistant MHC-I at the cell surface (41) (see Fig. S3 in the supplemental material).
Although the interference with MHC-I secretory pathway by BILF1 causes a relatively modest 20 to 25% reduction in the steady-state levels of MHC-I molecules at the cell surface, as revealed by the flow cytometry results with K122A/ΔC double mutant BILF1 in Fig. 2D, the qualitative effect is more pronounced. Thus, the K122A/ΔC double mutant BILF1 inhibits CD8+ T cell recognition of endogenously processed antigen by around 65% (Fig. 6). That CD8+ T cell recognition is affected disproportionately to the effect of BILF1 on the levels of MHC-I molecules at the cell surface may be explained by the fact that the peptide/MHC-I complexes targeted by the CD8+ T cell effectors in these experiments are mostly derived from newly synthesized proteins. The surface MHC-I molecules presenting newly generated peptides represent only a fraction of the surface MHC-I molecules, most of which will be recycled and/or arrived at the cell surface before the target antigen was first expressed. These experiments emphasize the limitation of only measuring levels of cell surface MHC-I molecules and extrapolating the results to draw conclusions with regard to antigen presentation.
Lemmermann et al. (18) showed that the immune evasion proteins from murine cytomegalovirus preferentially affect cell surface display of recently generated peptide presentation complexes. These immunoevasins only slowly downmodulate preexisting cell surface MHC-I/peptide complexes, which is a passive mechanism due to constitutive turnover in the absence of resupply. This raises the question of whether BILF1 might similarly be acting predominantly by inhibiting newly synthesized MHC-I/peptide complexes. However, when we used specific CD8+ effector T cells as an indicator of the internalization of preexisting peptide complexes from the cell surface, it was clear that the expression of BILF1 resulted in a marked reduction of T cell recognition (Fig. 5).
Therefore, we can conclude that the BILF1 immunoevasin can efficiently impair recognition of both newly synthesized MHC-I/peptide complexes by targeting the secretory pathway and also of preexisting MHC-I/peptide complexes by targeting an endocytosis/degradation pathway. This is particularly relevant for the role of BILF1 as an immunoevasin in the lytic cycle.
EBV has now been demonstrated to target the MHC-I antigen-processing pathway at the level of translation through BGLF5 (32, 42), peptide transport through BNLF2a (14), and by targeting mature MHC-I molecules for degradation through BILF1 (41). By using cells carrying a BNLF2a-deficient recombinant EBV, Croft et al. (7) showed that BNLF2a impaired antigen presentation of IE and E cycle antigens, but not of late viral targets. This observation can probably be explained by the fact that the BNLF2a protein is rapidly expressed at the onset of the EBV lytic cycle, but only transiently. In contrast, the kinetics and persistence of BILF1 expression through the lytic cycle allow BILF1 to potentially also affect late lytic cycle antigen presentation. Indeed, this prediction is supported by our results in Fig. 7, when we used a BILF1-deleted recombinant EBV in the same experimental model described by Croft et al. Furthermore, the results in Fig. 7 suggest that BILF1 can fulfill the immune evasion function in the EBV lytic cycle through targeting preexisting and newly synthesized MHC/peptide complexes. Since IE antigens such as BRLF1 will generate MHC-I/peptide complexes prior to the appearance of BILF1 (7), the enhanced recognition of ΔBILF1 LCLs by BRLF1-specific T cells implicates a role for the effect of BILF1 on the endocytic pathway to modulate presentation of preexisting MHC-I/peptide complexes. However, it is likely that the effect of BILF1 on the exocytic trafficking pathway for newly synthesized MHC-I/peptide complexes is the reason why BILF1 more efficiently impairs recognition of E and DE antigens, which are expressed coincident with or subsequent to BILF1 expression.
As with other successful persistent viruses, therefore, EBV has evolved multiple complex mechanisms for immunomodulation. Through targeting of at least four elements of the MHC-I antigen processing pathway, EBV immunoevasins complement and synergize with each other to achieve an appropriate level of immune modulation at particular points in the virus life cycle.
Supplementary Material
Acknowledgments
We thank Emmanuel Wiertz (Utrecht, Netherlands) for critical and helpful discussions during this study and Alan Rickinson and Caroline Rowe for their critical reading of the manuscript.
This work was supported by grants from the Wellcome Trust and the Medical Research Council.
Footnotes
Published ahead of print on 1 December 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Arenzana-Seisdedos, F., et al. 1993. Phosphatidylcholine hydrolysis activates NF-kappa B and increases human immunodeficiency virus replication in human monocytes and T lymphocytes. J. Virol. 67:6596-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson, P. G., H. J. Coope, M. Rowe, and S. C. Ley. 2003. Latent membrane protein 1 of Epstein-Barr virus stimulates processing of NF-kappa B2 p100 to p52. J. Biol. Chem. 278:51134-51142. [DOI] [PubMed] [Google Scholar]
- 3.Barnstable, C. J., et al. 1978. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell 14:9-20. [DOI] [PubMed] [Google Scholar]
- 4.Beisser, P. S., et al. 2005. The Epstein-Barr virus BILF1 gene encodes a G protein-coupled receptor that inhibits phosphorylation of RNA-dependent protein kinase. J. Virol. 79:441-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonifacino, J. S., and L. M. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395-447. [DOI] [PubMed] [Google Scholar]
- 6.Buisson, M., et al. 1989. The Epstein-Barr virus (EBV) early protein EB2 is a posttranscriptional activator expressed under the control of EBV transcription factors EB1 and R. J. Virol. 63:5276-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Croft, N. P., et al. 2009. Stage-specific inhibition of MHC class I presentation by the Epstein-Barr virus BNLF2a protein during virus lytic cycle. PLoS Pathog. 5:e1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. U. S. A. 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doms, R. W., and D. Trono. 2000. The plasma membrane as a combat zone in the HIV battlefield. Genes Dev. 14:2677-2688. [DOI] [PubMed] [Google Scholar]
- 11.Eliopoulos, A. G., et al. 2003. Epstein-Barr virus-encoded latent infection membrane protein 1 regulates the processing of p100 NF-κB2 to p52 via an IKKγ/NEMO-independent signalling pathway. Oncogene 22:7557-7569. [DOI] [PubMed] [Google Scholar]
- 12.Feederle, R., et al. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haque, T., et al. 2002. Treatment of Epstein-Barr virus-positive post-transplantation lymphoproliferative disease with partly HLA-matched allogeneic cytotoxic T cells. Lancet 360:436-442. [DOI] [PubMed] [Google Scholar]
- 14.Hislop, A. D., et al. 2007. A CD8+ T cell immune evasion protein specific to Epstein-Barr virus and its close relatives in Old World primates. J. Exp. Med. 204:1863-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hislop, A. D., G. S. Taylor, D. Sauce, and A. B. Rickinson. 2007. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus. Annu. Rev. Immunol. 25:587-617. [DOI] [PubMed] [Google Scholar]
- 16.Hudnall, S. D., et al. 2005. Distribution and phenotype of Epstein-Barr virus-infected cells in human pharyngeal tonsils. Mod. Pathol. 18:519-527. [DOI] [PubMed] [Google Scholar]
- 17.Kasper, M. R., et al. 2005. HIV-1 Nef disrupts antigen presentation early in the secretory pathway. J. Biol. Chem. 280:12840-12848. [DOI] [PubMed] [Google Scholar]
- 18.Lemmermann, N. A. W., et al. 2010. Immune evasion proteins of murine cytomegalovirus preferentially affect cell surface display of recently generated peptide presentation complexes. J. Virol. 84:1221-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long, H. M., et al. 2005. CD4+ T-cell responses to Epstein-Barr virus (EBV) latent-cycle antigens and the recognition of EBV-transformed lymphoblastoid cell lines. J. Virol. 79:4896-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyngaa, R., et al. 2010. Cell transformation mediated by the Epstein-Barr virus G protein-coupled receptor BILF1 is dependent on constitutive signaling. Oncogene 29:4388-4398. [DOI] [PubMed] [Google Scholar]
- 21.Neuhierl, B., and H. J. Delecluse. 2005. Molecular genetics of DNA viruses: recombinant virus technology. Methods Mol. Biol. 292:353-370. [DOI] [PubMed] [Google Scholar]
- 22.Niedobitek, G., A. Agathanggelou, N. Steven, and L. S. Young. 2000. Epstein-Barr virus (EBV) in infectious mononucleosis: detection of the virus in tonsillar B lymphocytes but not in desquamated oropharyngeal epithelial cells. Mol. Pathol. 53:37-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paine, E., R. I. Scheinman, A. S. Baldwin, Jr., and N. Raab-Traub. 1995. Expression of LMP1 in epithelial cells leads to the activation of a select subset of NF-kappa B/Rel family proteins. J. Virol. 69:4572-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulsen, S. J., M. M. Rosenkilde, J. Eugen-Olsen, and T. N. Kledal. 2005. Epstein-Barr virus-encoded BILF1 is a constitutively active G protein-coupled receptor. J. Virol. 79:536-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pudney, V. A., A. M. Leese, A. B. Rickinson, and A. D. Hislop. 2005. CD8+ immunodominance among Epstein-Barr virus lytic cycle antigens directly reflects the efficiency of antigen presentation in lytically infected cells. J. Exp. Med. 201:349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rabin, H., et al. 1981. Spontaneous release of a factor with properties of T cell growth factor from a continuous line of primate tumor T cells. J. Immunol. 127:1852-1856. [PubMed] [Google Scholar]
- 27.Ressing, M. E., et al. 2008. Epstein-Barr virus evasion of CD8+ and CD4+ T cell immunity via concerted actions of multiple gene products. Semin. Cancer Biol. 18:397-408. [DOI] [PubMed] [Google Scholar]
- 28.Rickinson, A. B., and E. Kieff. 2007. Epstein-Barr virus, p. 2655-2700. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott, Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 29.Robinson, M. S. 2004. Adaptable adaptors for coated vesicles. Trends Cell Biol. 14:167-174. [DOI] [PubMed] [Google Scholar]
- 30.Rooney, C. M., et al. 1995. Use of gene-modified virus-specific T lymphocytes to control Epstein-Barr virus-related lymphoproliferation. Lancet 345:9-13. [DOI] [PubMed] [Google Scholar]
- 31.Rosenkilde, M. M., M. J. Smit, and M. Waldhoer. 2008. Structure, function and physiological consequences of virally encoded chemokine seven transmembrane receptors. Br. J. Pharmacol. 153(Suppl. 1):S154-S166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rowe, M., et al. 2007. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. U. S. A. 104:3366-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rowe, M., G. L. Kelly, A. I. Bell, and A. B. Rickinson. 2009. Burkitt's lymphoma: the Rosetta Stone deciphering Epstein-Barr virus biology. Semin. Cancer Biol. 19:377-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rowe, M., and J. Zuo. 2010. Immune responses to Epstein-Barr virus: molecular interactions in the virus evasion of CD8+ T cell immunity. Microbes Infect. 12:173-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stam, N. J., H. Spits, and H. L. Ploegh. 1986. Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J. Immunol. 137:2299-2306. [PubMed] [Google Scholar]
- 36.Thorley-Lawson, D. A. 2001. Epstein-Barr virus: exploiting the immune system. Nat. Rev. Immunol. 1:75-82. [DOI] [PubMed] [Google Scholar]
- 37.Thorley-Lawson, D. A., and A. Gross. 2004. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 350:1328-1337. [DOI] [PubMed] [Google Scholar]
- 38.Wess, J. 1998. Molecular basis of receptor/G-protein-coupling selectivity. Pharmacol. Ther. 80:231-264. [DOI] [PubMed] [Google Scholar]
- 39.Yao, Q. Y., A. B. Rickinson, J. S. Gaston, and M. A. Epstein. 1985. In vitro analysis of the Epstein-Barr virus: host balance in long-term renal allograft recipients. Int. J. Cancer 35:43-49. [DOI] [PubMed] [Google Scholar]
- 40.Zeidler, R., et al. 1997. Downregulation of TAP1 in B lymphocytes by cellular and Epstein-Barr virus-encoded interleukin-10. Blood 90:2390-2397. [PubMed] [Google Scholar]
- 41.Zuo, J., et al. 2009. The Epstein-Barr virus G-protein-coupled receptor contributes to immune evasion by targeting MHC class I molecules for degradation. PLoS Pathog. 5:e1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuo, J., et al. 2008. The DNase of gammaherpesviruses impairs recognition by virus-specific CD8+ T cells through an additional host shutoff function. J. Virol. 82:2385-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.