Abstract
Laboratory mouse strains carry endogenous copies of the xenotropic mouse leukemia viruses (X-MLVs), named for their inability to infect cells of the laboratory mouse. This resistance to exogenous infection is due to a nonpermissive variant of the XPR1 gammaretrovirus receptor, a resistance that also limits in vivo expression of germ line X-MLV proviruses capable of producing infectious virus. Because laboratory mice vary widely in their proviral contents and in their virus expression patterns, we screened inbred strains for sequence and functional variants of the XPR1 receptor. We also typed inbred strains and wild mouse species for an endogenous provirus, Bxv1, that is capable of producing infectious X-MLV and that also contributes to the generation of pathogenic recombinant MLVs. We identified the active Bxv1 provirus in many common inbred strains and in some Japanese Mus molossinus mice but in none of the other wild mouse species that carry X-MLVs. Our screening for Xpr1 variants identified the permissive Xpr1sxv allele in 7 strains of laboratory mice, including a Bxv1-positive strain, F/St, which is characterized by lifelong X-MLV viremia. Cells from three strains carrying Xpr1sxv, namely, SWR, SJL, and SIM.R, were shown to be infectable by X-MLV and XMRV; these strains carry different alleles at Fv1 and vary in their sensitivities to specific X/P-MLV isolates and XMRV. Several strains with Xpr1sxv lack the active Bxv1 provirus or other endogenous X-MLVs and may provide a useful model system to evaluate the in vivo spread of these gammaretroviruses and their disease potential in their natural host.
Xenotropic mouse gammaretroviruses were first isolated by Levy and Pincus from the NZB strain of laboratory mice (25). The virus was termed xenotropic because of its inability to infect cells of the various inbred mouse strains, although it could infect rat cells. Subsequent studies isolated this virus type from additional strains of inbred and outbred laboratory mice (23) and showed that these viruses are infectious for cells of multiple mammalian species (24, 33).
The various inbred strains of laboratory mice carry up to 15 endogenous proviruses related to infectious xenotropic mouse leukemia viruses (X-MLVs) (8, 34), and some of these germ line X-MLV proviruses, or XMVs, are capable of producing infectious virus or viral proteins (reviewed in reference 40). These inbred mouse strains differ in their abilities to produce infectious X-MLVs because of the different XMVs that they carry and because of the presence of regulatory genes that alter virus expression and spread. Two strains, F/St and NZB, have a “high-virus” phenotype; that is, they are viremic with X-MLVs from an early age, and virus can readily be isolated from cells and tissues of these strains (6, 25, 31). A second and larger set of inbred strains, which includes common strains such as C57BL and BALB/c, carry XMVs that produce virus after chemical or immunological stimulation (1, 9, 27, 38). A third set of strains, including Swiss strain-derived mice and strains such as A and C3H, are not or are rarely capable of producing infectious X-MLV.
Four active XMVs have been identified in the strains that can produce infectious virus. Two of these proviruses are found in NZB mice (6), a third provirus, termed Bxv1, is found in several common inbred strains (8, 18, 19), and the fourth is found along with the Bxv1 provirus in MA/My mice (16). Only one of these proviruses, Bxv1, has been mapped in the mouse genome; it has been mapped to a position on distal chromosome 1 (Chr 1), where it was first located by conventional linkage analysis of chemically inducible virus production (18). The provirus at this locus was subsequently identified by Southern blotting of somatic cell hybrids and recombinant inbred mouse strains (11, 8). The Bxv1 provirus (also termed Xmv43) is present in the sequenced C57BL mouse genome and maps at 179 MB on Chr 1 (12). This provirus is biologically important for two reasons: not only does Bxv1 reliably produce virus in response to specific inducers (18, 19), but it is also involved in virus-induced leukemias. These diseases are associated with the generation of recombinant leukemogenic polytropic MLVs (P-MLVs), and the Bxv1 provirus has been identified as the source of the long terminal repeat sequences of these pathogenic recombinants (11, 41).
The presence of multiple endogenous XMVs in mice that are not susceptible to X-MLV infection can be explained by looking at the wild mouse progenitors of these laboratory strains. The common laboratory strains represent a mosaic of three house mouse species: Mus domesticus, Mus castaneus, and Mus musculus (49). Two of these 3 species carry XMVs (17, 43), and the 3 species carry 3 different alleles of the XPR1 receptor used for entry by X-MLVs, all 3 of which are permissive for X-MLVs (28, 47, 48). In contrast, the XPR1 receptor gene originally cloned from NIH 3T3 cells is not permissive for X-MLV entry (2, 42, 50). This suggests that XMVs were originally acquired by X-MLV-susceptible wild mice and that the receptor variants permissive for this virus were lost in the fancy mouse breeding stocks used to generate laboratory strains.
There are hundreds of strains of laboratory mice. While it is generally recognized that these strains are less genetically diverse than wild Mus species, it is clear that there are distinct lineages of laboratory strains that can be traced to different fancy mouse and wild mouse progenitors. Few of the hundreds of mouse strains have actually been tested for X-MLV susceptibility, and the observations that X-MLV susceptibility is widespread among wild mice and that these inbred strain lineages represent different mosaics of wild mouse species (49) raised the possibility that X-MLV-permissive Xpr1 alleles might have been captured in some of these strains. In this study, we screened inbred strains of laboratory mice for the presence of known and novel variants of the Xpr1 receptor, with the goal of identifying strains susceptible to X-MLV infection. We also identified the inbred strains carrying the inducible Bxv1 provirus and identified the origin of this provirus in the Japanese house mouse species Mus molossinus. The permissive sxv allele of Xpr1 (Xpr1sxv) was identified in 7 inbred strains, including the high-virus, Bxv1-positive F/St strain. We also demonstrate that cells from laboratory mouse strains carrying Xpr1sxv can be efficiently infected with XMRV and X-MLVs.
MATERIALS AND METHODS
Viruses, cells, and virus infectivity.
CAST-X is an X-MLV isolated in our laboratory from the spleen of a CAST/EiJ mouse (46). Cz524 is a novel MLV isolated from the spleen of a CZECHII/EiJ mouse (47). The human xenotropic murine leukemia virus-related virus, XMRV (7), was kindly provided by R. Silverman (Cleveland Clinic, Cleveland, OH). AKR6 X-MLV, CasE#1 (5), Moloney mink cell focus-inducing virus (MoMCF), and Friend MCF (FrMCF) were originally obtained from J. Hartley (NIAID, Bethesda, MD).
Susceptibility to MLVs was tested in NIH 3T3 cells, in Mus dunni cells (20) obtained from M. Lander (NIAID, Bethesda, MD), and in cultured fibroblasts developed from tail biopsy specimens from adult mice (21). Retrovirus stocks carrying the LacZ reporter were generated for the various MLVs by virus infection of GP2-293 cells transfected with pCL-MFG-LacZ (47); the virions produced by these cells represent a mix of infectious virus and particles carrying LacZ. To test for virus susceptibility in a single-round assay, cells were infected with appropriate dilutions of these pseudotype virus stocks in the presence of 4 to 8 μg/ml Polybrene. One day after infection, cells were fixed with 0.4% glutaraldehyde and assayed for β-galactosidase activity using as the substrate 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal, 2 mg/ml; ICN Biomedicals, Aurora, OH). Infectious titers were expressed as the number of foci of blue cells per milliliter of virus.
Genomic DNA and RNA.
DNAs were extracted from mice obtained from several sources. The following mice were obtained from M. Potter (NCI, Bethesda, MD): M. domesticus mice trapped at various sites (Abu Rawash, Egypt; Haven's Farm, Davidsonville, MD; Lewes, DE; J. J. Downs, Ridgely, MD; Watkin's Star, Centreville, MD), M. molossinus mice (MOL/Li), M. castaneus mice (CAST/Ncr, CAS/Li), M. musculus mice (Vejrumbro, Denmark; Skive, Denmark), and M. spretus mice (Spain). Three wild-trapped mice from Lake Casitas, CA, were obtained from S. Rasheed (University of Southern California, Los Angeles). CAST/Rp mice were obtained from R. Elliott (Roswell Park Cancer Institute, Buffalo, NY), and SAMP8 and SAMR1 samples were obtained from R. Carp (New York State Institute for Basic Research in Developmental Disabilities, Staten Island, NY). F/St mice were obtained from H. Morse (NIAID, Bethesda, MD) in the early 1980s; this strain is now extinct. 129-GIX+ mice were obtained from E. Boyse (Memorial Sloan-Kettering Cancer Center, NY). SIM.R and SIM/Ut mice (44) were obtained from A. Axelrad (University of Toronto, Toronto, Canada). AKR/N and NFS/N mice were obtained from the Small Animal Section, NIH (Bethesda, MD). NFS/N-Xpr1sxv congenics were developed in our laboratory and carry the Xpr1 allele from M. domesticus (formerly Mus praetextus), provided by M. Potter. DBA/2J, DBA/1J, C58/J, C57BL/10J, and SWR/J mice were obtained from the Jackson Laboratory (Bar Harbor, ME).
DNAs were also isolated from the cell lines NZB-Q and SC-1, obtained from J. Hartley (NIAID, Bethesda, MD), and from cells of M. domesticus (formerly M. praetextus) obtained from J. Rodgers (Baylor College of Medicine, Houston, TX). Mus fragilicauda DNA was provided by J. Hartley. The Chinese hamster/mouse somatic cell hybrid BE7-2 was produced by the fusion of E36 cells and primary fibroblasts of BALB/c cells, and its mouse chromosome content was determined by examining Giemsa-stained metaphase spreads and by testing for marker loci (14).
Additional DNAs provided by H. Morse and S. Chattopadhyay (NIAID, Bethesda, MD) include Mus abbotti and M. musculus mice trapped in Vejrumbro, Denmark; Brno, Czechoslovakia; Belgrade, Yugoslavia; and Viborg, Denmark. DNA from M. macedonicus was provided by R. Elliott. All other DNAs from common inbred and wild-derived strains were purchased from the Jackson Laboratory (Bar Harbor, ME).
Cloning and sequencing of Xpr1, Fv1, and the Bxv1 insertion site.
The full-length Xpr1 genes were amplified by reverse transcription-PCR (RT-PCR) from cells established from SWR tail biopsy specimens and from the cell line SJLSV, a simian virus 40 (SV40)-transformed line of SJL fibroblasts produced by B. Knowles (Institute of Medical Biology, Singapore). Primers used were 5′-ATGAAGTTCGCCGAGCACCTCTC and 5′-AGTGTTAGCTTCGTCATCTGTGTC. A segment of Xpr1 containing the third and fourth putative extracellular loops (ECL3 and ECL4, respectively) was amplified by RT-PCR from NZB mouse spleen RNA using the forward primer 5′-CGAGTATTTACTGCTCCCTTCC and Ex13R, 5′-CGGAAAACCTCAAGGGGG. Xpr1 exon 13 sequences, containing ECL4, were amplified from the DNA of multiple strains and species using primers Ex13F (5′-GCCTATTACTACTGTGCC) and Ex13R. All PCR products were cloned into pCR2.1-TOPO and sequenced.
PCR primers for the Bxv1 provirus-cell junction fragment and the empty locus at the Bxv1 insertion site were designed based on Mus musculus chromosome 1 clone RP24-65D16 (GenBank accession no. AC115959). The Bxv1 provirus is present in the reverse orientation at coordinates 107533 to 116189. A Bxv1 provirus-cell junction fragment was PCR amplified using primers 5′-AGGAGTAGGAACAGGGACTACAGC (Bxv1ENFP; nucleotide coordinates 108635 to 108612) and 5′-GATCTAGAGTCACCTAGGCAACAAGC (Bxv1FLRP; nucleotide coordinates 106765 to 106740). The empty locus at the Bxv1 insertion site was amplified using primers 5′-CCATGCTAAATACCTCAGACTCCAGTG (Bxv1FLFP; nucleotide coordinates 116359 to 116333) and 5′-GATCTAGAGTCACCTAGGCAACAAGC (Bxv1FLRP; nucleotide coordinates 106765 to 106740) (Fig. 1). Representative PCR products were cloned into pCR2.1-TOPO and sequenced.
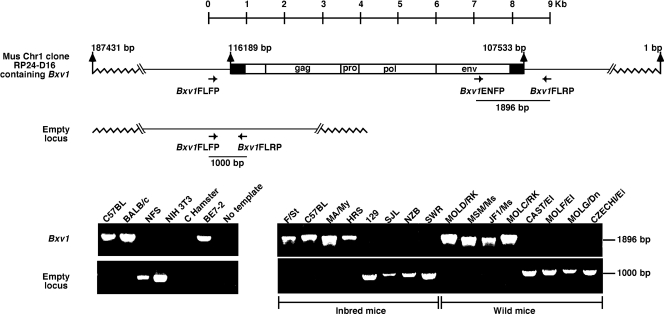
FIG. 1.
Analysis of the Bxv1 provirus in common strains of laboratory mice and wild-derived mouse strains. (Top) Bxv1 provirus integration site and the empty locus on Chr 1 with PCR primers used to amplify these locus alternatives. For clarity, the Bxv1 provirus is diagrammed in the forward orientation, as opposed to its reverse orientation in GenBank accession no. AC115959. (Bottom left) PCR screening of mouse DNAs previously shown to carry Bxv1 (C57BL, BALB/c) or to lack this provirus (NFS, NIH 3T3) and a hamster/mouse somatic cell hybrid, BE7-2, that carries a Chr 1 fragment containing Bxv1. (Bottom right) PCR results for representative common inbred strains and wild-derived mice.
PCR primers for the 3′ half of the Fv1 gammaretrovirus restriction gene, MHRF and GT17a (45), generate fragments of 510 bp for Fv1n and Fv1nr and 1.8 kb for Fv1b. The 510-bp PCR product from SWR/J mice was cloned into pCR2.1-TOPO and sequenced.
Nucleotide sequence accession numbers.
The sequences of the Xpr1 genes for 19 mouse strains (129, AKR, DBA, F/St, FVB, HRS, LP, LT, MA/My, NOD, PWD, RBA, RIIIS, SOD, SWR, YBR, NZB, SIM.R, SJL) were deposited in GenBank under accession no. HQ323662 to HQ323680. Sequences for the empty Chr 1 Bxv1 site in NFS/N and the SWR Fv1 gene were deposited in GenBank under accession no. HQ323681 and HQ323682.
RESULTS
Inbred strain distribution of the inducible XMV locus Bxv1.
We designed primers to identify cell-virus junction fragments diagnostic of the Bxv1 provirus, as well as primers to identify the empty locus (Fig. 1). Primers were initially tested on DNAs from mouse strains previously shown to lack Bxv1, NFS and NIH 3T3, and on DNAs from C57BL and BALB/c, the strains in which Bxv1 was initially discovered (18). We also tested a hamster/mouse somatic cell hybrid, BE 7-2, carrying fragments of two mouse chromosomes, Chr 1 and Chr 15. This hybrid, like mouse cells carrying Bxv1, produces infectious X-MLV following induction (14), and it contains a single XMV provirus (11). The 1.8-kb cell-virus junction fragment was identified only in DNAs from the hybrid cell line and from mice known to carry Bxv1, whereas the 1.0-kb fragment diagnostic of the empty locus was found only in DNAs from mice or cells lacking Bxv1 (Fig. 1), thus confirming that these primers identify the Bxv1 locus. The sequence of the 1.9-kb cell-virus junction fragment produced by Bxv1ENFP and Bxv1FLRP was identical to the corresponding Chr 1 segment in the sequenced C57BL genome (GenBank accession no. AC115959), and the sequence of the Bxv1FLFP/Bxv1FLRP product from the Bxv1-negative NFS/N mouse represents the provirus-free insertion site (GenBank accession no. HQ323681).
We analyzed 50 laboratory mouse strains, most of which fall into several inbred strain categories defined by their related breeding histories (3) (Fig. 2). The majority of the analyzed strains originated from the fancy mouse colonies of breeder Abbie Lathrop and from the early lines developed by William Castle; one set of Lathrop mice provided to William Castle gave rise to many common strains, such as DBA and BALB/c, and additional Lathrop mice produced the C57/C58-related strains. A third set of strains was derived from Swiss mice. Additional strains were developed from stocks in New Zealand, and we also included strains of other or uncertain ancestry in this analysis. We did not test the various strains derived by intercrossing other inbred strains.
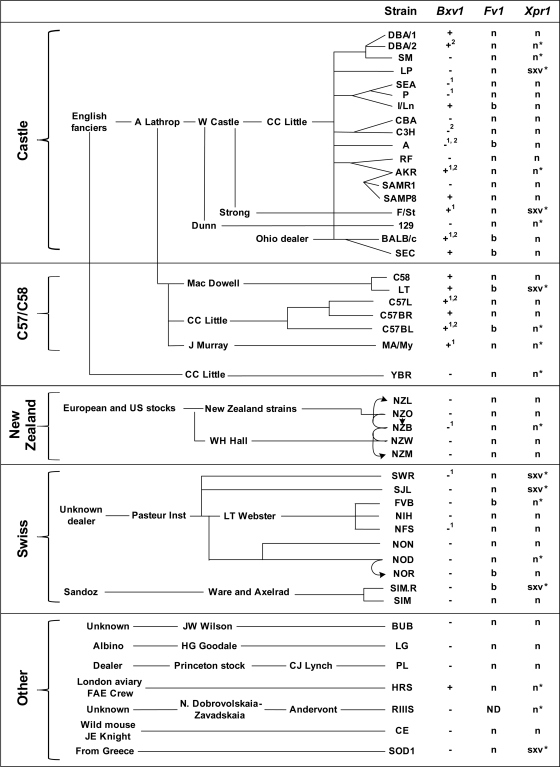
FIG. 2.
Distribution of allelic variants of Bxv1, Fv1, and Xpr1 in inbred strains of the laboratory mouse. The organization of strains into groups based on their related breeding histories is derived from the analysis done by Beck and associates (3). The Bxv1 provirus insertion is identified by + and the empty locus by −; numbers identify strains previously typed for Bxv1 as a Chr 1-linked induction locus (superscript “1”) (18, 19, 51) or by Southern blotting (superscript “2”) (8). Fv1 typing identified the presence of the “n” or “b” restriction fragments that distinguish Fv1n and Fv1nr from Fv1b (45). Identical results were obtained for strain sublines SJL/BmJ and SJL/J, for AKR/N and AKR/J, and for C57BL/6 and C57BL/10. Xpr1 variants confirmed by sequencing are indicated with asterisks; all others were typed by PCR. SAMR1 and SAMP8 are hybrids of AKR and an unknown mouse. NOR was derived from a cross between NOD and C57BLKS/J. NZB was derived from NZO; NZM is a hybrid of NZB and NZW. NZL was developed from NZO with contributions from NZB, C57BL, and 129.
Bxv1 was identified in 17 strains, most of which are Lathrop or Castle strain-derived mice (Fig. 1, 2). Bxv1 was not found in any of the New Zealand or Swiss strain-derived mice. This distribution is consistent with previous analyses that typed 12 strains for the presence or absence of the Chr 1 Bxv1 induction locus (18, 19, 51) and 7 strains for the presence or absence of diagnostic env fragments by Southern blotting (8) (Fig. 2). Previous testing had also identified a Bxv1-like locus in F/St mice, and although the Chr 1 location initially determined for this virus induction locus did not clearly identify it as Bxv1 (31), the present analysis confirms that Bxv1 is present in F/St (Fig. 1).
Wild mouse origin of the Bxv1 provirus.
Laboratory mice are a mosaic of the wild mouse species of house mice, and 3 of the 4 house mouse species (M. molossinus, M. castaneus, M. musculus) carry X-MLV-related sequences and are capable of producing infectious virus (4, 17, 26, 43, 46, 47). It is therefore possible that the Bxv1 provirus originated in one of these wild mouse species. A previous attempt to identify Bxv1 in M. molossinus by screening backcross mice for inducible Chr1-linked proviral loci was unsuccessful because these mice carry too many active proviruses (16). We screened house mouse species and several other Mus species that had not been previously typed for endogenous MLVs for the presence of Bxv1 by PCR (Fig. 1, 3). We identified Bxv1 in 4 wild mouse DNAs, all of which were M. molossinus, although 3 additional breeding lines of M. molossinus mice did not have this provirus.
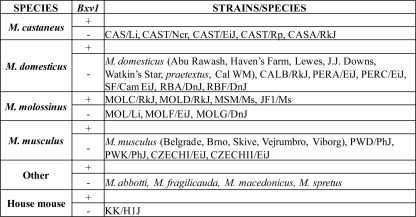
FIG. 3.
Distribution of Bxv1 in wild mouse species and wild-derived inbred strains. DNAs were screened by PCR for the Bxv1 insertion (+) or for the empty locus (−). A total of four wild-trapped California mice, along with California mouse-derived SC-1 cells, were tested.
None of the other house mouse species carries Bxv1. M. molossinus is thought to be a natural hybrid of M. castaneus and M. musculus (52), both of which species carry active XMVs; however, the Bxv1 provirus was not found in any mouse of these 2 species (Fig. 1 and 3). The absence of this provirus in these 2 species and the fact that some, but not all, M. molossinus mice carry Bxv1 indicate that this XMV insertion arose relatively recently in Japanese mice. It is likely that Asian mouse X-MLVs, as well as the ecotropic MLVs carried by these mice (17, 39), were introduced into fancy mouse colonies because of inclusion of Japanese mice in fancy mouse colonies; in fact, Lathrop started her mouse colony with a pair of Japanese waltzing mice (22, 29, 37). It is also probable that the additional active proviruses found in laboratory mouse strains like NZB and MA/My may have been acquired from these Asian mice.
DNAs from M. molossinus and from the common laboratory strains were also typed for alleles of the Fv1 gammaretrovirus restriction gene, a gene that targets the virus capsid and inhibits replication postentry. DNAs were typed using PCR primers that generate products of different sizes for Fv1b (1.8 kb) and for Fv1n or Fv1nr (510 bp) (45). Among the laboratory strains, 28 of the 39 mice typed in this way as Fv1n were found to lack Bxv1, but 6 of the 10 Fv1b mice have Bxv1 (Fig. 2). All M. molossinus DNAs tested (MOLC/RkJ, MOLD/RkJ, MSM/Ms, JF1/Ms, MOLF/EiJ) produced the longer Fv1b-like PCR product, suggesting the possibility that the Bxv1 virus is B tropic. However, M. molossinus cells have not been tested for Fv1-type restriction, and although PCR suggests that this Fv1 resembles the longer Fv1b at its 3′ end, our previous sequence analysis of the M. molossinus Fv1 gene identified Fv1nr codons at key sites in the coding region (GenBank accession no. FJ603569). Furthermore, the Fv1 tropism of the virus produced by Bxv1 has not been determined, and the Bxv1 capsid sequence has novel substitutions at key Fv1 target sites, so its sensitivity to the laboratory mouse Fv1 variants cannot be predicted. It is therefore not yet clear in what way the acquisition of the Bxv1 provirus was influenced by Fv1.
Xpr1 receptor alleles in viremic strains of laboratory mice.
Multiple host factors can influence the expression of proviral loci and the spread of virus in vivo, and the detection of infectious X-MLV or viral proteins depends on what proviruses are present and can also be influenced by genes at or near the major histocompatibility complex (MHC) locus and the Fv1 gene (32, 51). For mice carrying active XMVs, another contributing factor is likely to be the XPR1 receptor used by X-MLVs for entry. Because functional variants of the XPR1 receptor for X-MLVs exist in the various mouse species that contributed to the laboratory mouse genome, we screened inbred strains for variants of Xpr1. Two inbred strains, NZB and F/St, are unusual among laboratory strains in that they constitutively express infectious X-MLV throughout life (6, 31). X-MLVs are readily isolated from lymphoid organs of these mice. NZB carries two active XMVs, and the high-virus expression pattern has been attributed to the constitutive expression of one of those XMVs (6), although the presence of a permissive XPR1 variant could also be a contributing factor. Neither of the XMV loci in NZB loci is Bxv1, as indicated by previous genetic analysis of this strain (16) and by our present study (Fig. 1). To determine if a permissive receptor contributes to the observed high-virus phenotype, we sequenced a segment of the NZB Xpr1 gene encoding the receptor determinants in XPR1 ECL3 and ECL4 (28, 46). This sequencing identified the NZB receptor as the restrictive allele Xpr1n (GenBank accession no. HQ323678), indicating that the viremia observed in these mice has no contribution from receptor-mediated secondary spread.
The second high-virus strain, F/St, produces detectable levels of X-MLV at 1 week of age and high levels from 4 weeks (31). This mouse carries one active XMV that was identified as the Bxv1 provirus (Fig. 1), but this XMV is not associated with viremia in the other strains that carry it. We sequenced exon 13 of F/St Xpr1, which contains ECL4, and found that these mice carry the permissive Xpr1sxv allele (15, 28) (GenBank accession no. HQ323665). While this might suggest an explanation for the F/St high-virus phenotype, analysis of genetic crosses between F/St and other inbred strains have shown that viremia requires Bxv1 and a recessive gene linked to the major histocompatibility locus (51). Xpr1 is closely linked to Bxv1 on Chr 1; its role in viremia cannot be evaluated from the existing data on F/St mice, and this strain has recently become extinct.
Xpr1 variants in other inbred strains.
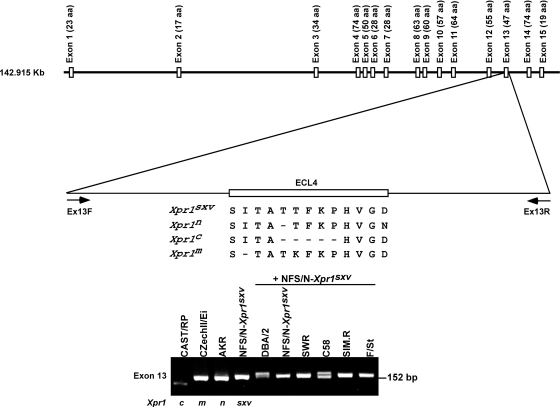
Rather than sequence XPR1 from dozens of mouse strains, we first screened DNAs of laboratory mouse strains by PCR for the presence of the permissive Xpr1sxv allele. There are five functionally distinct Xpr1 alleles in Mus; 4 of these variants are found in the house mouse species and inbred strains, and only 1 of these variants, Xpr1sxv, encodes a fully permissive receptor (15). The other 3 house mouse receptors (Xpr1m, Xpr1n, and Xpr1c) restrict 2 or more of the viruses that rely on XPR1 for entry, and all 3 of these restrictive receptors have deletions in the ECL4 segment of the receptor (48) (Fig. 4). The deletion found in M. castaneus (Xpr1c) removes 5 codons, and the different deletions found in NIH 3T3 (Xpr1n) and M. musculus (Xpr1m) remove single codons. The m, n, and sxv alleles cannot be reliably distinguished by PCR product size differences, but mixtures of DNAs carrying the m, c, and n alleles with Xpr1sxv DNA produce PCR product doublets. Because we were specifically interested in identifying the permissive Xpr1sxv allele in laboratory mouse strains, we did an initial PCR screen by mixing DNAs from each mouse strain with DNA from a mouse carrying wild-mouse Xpr1sxv (Fig. 4).
FIG. 4.
Xpr1 variants in laboratory mouse strains. The exon structure of the Xpr1 gene is indicated at the top. Exon 13 contains the receptor determining the putative ECL4 loop and contains the indicated deletions or substitutions that distinguish the 4 house mouse alleles of Xpr1. aa, amino acids. At the bottom are shown PCR products using exon 13 primers, Ex13F and Ex13R. The first 4 lanes contain DNAs from mice carrying the 4 indicated alleles. The remaining 6 lanes contain DNAs of the indicated strains mixed with DNAs from mice carrying Xprsxv. Doublets are indicative of mice carrying the Xpr1n deletion.
Fifty strains were screened by this method, and 6 strains in addition to F/St were implicated as having Xpr1sxv. Sequencing confirmed the presence of this permissive allele in all 6 strains. Xpr1sxv was found in 3 of the 10 Swiss mouse DNAs and in the 3 mouse strains SOD1, LP, and LT/SvEi. We also sequenced exon 13 in 9 additional strains of interest. This segment of Xpr1 is hypervariable in rodent species (48), and we wanted to identify the presence of any novel polymorphisms due to substitutions that would be missed by the PCR screen for insertions or deletions. The 9 strains selected for sequencing included AKR and HRS, 2 strains that have been used in studies on viral leukemogenesis, and RIIIS, a strain that has acquired an XMV insertion in its Apobec3 gammaretrovirus resistance gene (36). We also sequenced exon 13 in strains expressing high levels of nonecotropic MLV envelope (Env) glycoprotein. For example, DBA mice carry a provirus associated with the Rmcf locus that blocks the entry of viruses that use the Xpr1 receptor (13) and also express high levels of XenCSA and GIX, cell surface antigens related to nonecotropic MLV Env glycoprotein (30). 129-GIX+ mice also express the Env glycoprotein-related GIX antigen. Strains selected for sequencing also included 129, FVB, and NOD, because they are used as human disease models or for genetic manipulations, MA/My, a strain that carries two active XMV proviruses (16), and YBR, a strain produced from a different fancy mouse stock (Fig. 2). No novel sequence variants were identified in these additional mice; all of these strains were found to carry the exon 13 sequence of Xpr1n.
In the last few decades, a number of new inbred strains were derived from wild-trapped mice. These strains are unrelated to the fancy mouse-derived older common inbred strains, and they carry the Xpr1 alleles found in their species of origin. Thus, strains derived from M. molossinus and M. musculus carry Xpr1m and strains derived from M. castaneus carry Xpr1c (48). Two inbred strains derived from wild-trapped M. domesticus mice, PWD and RBA, were found by sequencing to carry Xpr1sxv, as expected (GenBank accession no. HQ323672 and HQ323673).
X-MLV infectivity of cells derived from inbred strains SWR, SJL, and SIM.R.
Three of the strains determined to carry the permissive Xpr1sxv receptor based on initial screening and sequencing of exon 13 were further characterized. We sequenced the entire Xpr1 gene from SJL and SIM.R. The gene in both strains is identical to that originally cloned from M. dunni cells (28). This allele differs from Xpr1n by one insertion, Δ582T, and two substitutions, E500K and N590D.
The presence of a permissive receptor in these mice does not guarantee infectivity, as failure to infect can result from factors such as receptor interference (reviewed in reference 40). Therefore, we examined cells from all three of these strains for X-MLV infectivity. Cultured tail fibroblasts from SIM.R and SWR and an SV40-transformed line of SJL mouse fibroblasts were tested for susceptibility to LacZ retroviral vectors incorporating the Env glycoproteins of 2 mouse X-MLVs and 5 other virus isolates known to use the Xpr1 receptor. These 5 additional viruses included 2 polytropic MLVs (P-MLVs), 2 wild mouse virus isolates (CasE#1 and Cz524), and XMRV, an X-MLV-related virus originally isolated from human patients with prostate cancer (7). Xpr1n restricts all these viruses except the P-MLVs, while Xpr1sxv is permissive for all 7 viruses (47) (Table 1).
TABLE 1.
Susceptibility of mouse cells to LacZ pseudotypes of X/P-MLVs
| Mouse species, cell line, or strain | Xpr1 allele | Fv1 allele | Log10 titera |
||||||
|---|---|---|---|---|---|---|---|---|---|
| X-MLV |
XMRV | Cz524 | CasE#1 | P-MLV |
|||||
| CAST-X | AKR6 | FrMCF | MoMCF | ||||||
| M. dunni | sxv | 0 | 7.1 ± 0.1 | 6.1 ± 0.3 | 5.1 ± 0.3 | 5.9 ± 0.1 | 6.4 ± 0.2 | 5.9 ± 0.5 | 4.6 ± 0.4 |
| NIH 3T3 | n | n | <0 | <0 | <0 | <0 | <0 | 5.5 ± 0.3 | 5.4 ± 0.2 |
| SIM.R | sxv | b | 5.4 ± 0.2 | 5.4 ± 0.5 | 4.1 ± 0.2 | 4.7 ± 0.4 | 6.0 ± 0.8 | 5.4 ± 0.3 | 5.4 ± 0.6 |
| SWR | sxv | n | 4.8 ± 0.2 | 4.0 ± 0.5 | 3.2 ± 1.5 | 4.8 ± 0.3 | 4.2 ± 0.6 | 5.2 ± 0.4 | 4.5 |
| SJLSV | sxv | n | 5.4 ± 0.4 | 3.9 | 2.2 ± 0.4 | 4.5 ± 0.4 | 3.7 ± 0.4 | 4.9 ± 0.3 | 4.8 ± 0.1 |
Titers represent numbers of foci of LacZ-positive cells (log10 in 1.0 ml). Where no standard deviation is given, infectivity was measured only once. <0, no positive cells after infection with undiluted pseudotype virus stock.
The cells from these three inbred mouse strains are susceptible to all viruses (Table 1), confirming that these cells all express a fully permissive XPR1 receptor. Some viruses infected these cells with nearly equal efficiencies, but most of the viruses showed infectivity differences that are consistent with Fv1 restriction. All MLVs are potentially subject to Fv1 restriction, including X-MLVs (35), and mouse-tropic MLVs have been typed as N or B tropic based on their restriction by Fv1b or Fv1n, respectively, or as NB tropic if unaffected by either allele. SIM.R was developed as a congenic strain of Swiss noninbred mice and carries the Fv1b allele from C57BL/6 (44). The presence of Fv1b in the SIM.R cells and Fv1n in SJLSV was also determined by PCR. A segment of Fv1 was cloned from SWR cell DNA and identified as Fv1n by sequencing (GenBank accession no. HQ323682).
Two of the viruses used here are NB tropic, and these two P-MLVs, FrMCF and MoMCF, infected all 5 mouse cells with roughly equal efficiencies (Table 1). The other 5 viruses have unknown Fv1 tropism, but all 5 showed reduced LacZ titers in some or all 3 of the Xpr1sxv laboratory mouse cells relative to titers in M. dunni, which lacks a functional Fv1 gene Fv10 (45). This was somewhat unexpected, as the LacZ pseudotypes used here were produced by virus infection of GP2-293 cells, which carry ecotropic Moloney MLV gag-pol genes and therefore produce NB-tropic MoMLV capsids as well as capsids of the infecting virus. The fact that none of these 5 pseudotypes are NB tropic indicates that tropism is influenced by the infecting virus. Two of those viruses, CasE#1 and AKR6, more efficiently infected SIM.R than they infected SWR or SJL, suggesting that these viruses are restricted by Fv1n and may therefore be B tropic. CAST-X, Cz524, and XMRV are restricted in all 3 of these cells relative to levels of infection in M. dunni cells, although one virus, XMRV, appears somewhat more sensitive to Fv1n than to Fv1b. The restriction of XMRV by Fv1n and Fv1b has been reported previously (10, 47), and these results suggest that CAST-X and Cz524 may have similar Fv1-defined tropisms.
DISCUSSION
The acquisition of endogenous nonecotropic gammaretroviruses, the time course and tissue specificity of their expression, and the horizontal transmission of these viruses to new hosts are governed by various host factors that restrict or enhance replication and spread. While numerous studies have focused on host restriction genes that can inhibit virus replication or host immune system genes that alter in vivo spread, receptor polymorphisms can clearly either facilitate infection or provide an effective host defense against pathogenic and mutagenic viruses. We have identified 5 functional variants of the mouse XPR1 receptor for the xenotropic/polytropic family of gammaretroviruses, 4 of which are capable of restricting infection by multiple members of this family. The distribution of these alleles in wild mice indicates that, not surprisingly, species exposed to infectious virus carry restrictive alleles (48). The first of these alleles to be recognized, Xpr1n, was identified in laboratory mice and was assumed to be representative of all mice; this conclusion, that mice are resistant to infection by xenotropic MLVs, discouraged further analysis of these infectious agents in their natural host. Our findings, that a number of mouse strains carry the permissive allele of the XPR1 receptor and that cells from these mice are as infectable by X-MLVs as cells of the wild mouse species M. dunni, mean that laboratory mouse strains can, in principle, be used to develop mouse models to study expression of endogenous proviruses such as Bxv1 and to describe the course and consequences of exogenous infection by these mouse viruses.
Inbred mouse strains that carry all 5 of the known functional variants of mouse XPR1 are available. Three Eurasian mouse variants (Xpr1c, Xpr1m, Xpr1p) are found in inbred lines recently developed from M. castaneus, M. musculus, M. molossinus, and Mus pahari, but it was surprising to also find that the older common inbred lines carry two variants of Xpr1. One of these variants, Xpr1sxv, is widespread in wild mouse species, while Xpr1n has not been identified in any wild-derived or wild-trapped mouse (48). The X-MLV-restrictive allele, Xpr1n, is carried by the majority of these older strains of laboratory mice, while we identified the X-MLV-permissive allele, Xpr1sxv, in 3 of 10 Swiss mouse strains and in several strains developed from the Lathrop colonies.
Xpr1n is the only one of the 5 Mus Xpr1 alleles to completely restrict X-MLVs, and its origin is unclear. Studies to determine the wild mouse origins of the common inbred strains have concluded that the largest contribution to the inbred mouse genome is from M. domesticus (49). It makes sense for M. domesticus to carry Xpr1n, as these mice lack endogenous XMVs, consistent with Xpr1n restriction of X-MLVs. However, our survey of wild mouse species failed to find Xpr1n in any M. domesticus mouse or in any wild mouse of any species (48). It is of course possible that Xpr1n is present in some unidentified wild mouse populations that contributed to the laboratory mouse genome. Alternatively, it is possible that this allele arose in the fancy mouse colonies that were maintained by hobbyists for centuries (29). These colonies included mice from multiple species that normally do not interbreed in nature, and these hybrids acquired an increased burden of MLV-endogenous retroviruses of different subtypes that are otherwise segregated in the wild mouse species (17). Artificial breeding protocols might also have resulted in the loss of host factors capable of restricting virus in natural populations, like interfering Env genes. Thus, the appearance of a restrictive receptor allele might have provided a survival advantage leading to its selection and near fixation in these fancy mice.
The XPR1 receptor defines the virus subtypes that can infect the host species. XPR1 therefore controls the type of endogenous retroviruses that accumulate in host genomes and also affects expression of these endogenous viruses in the individual animals. Whether the inbred strains carrying Xpr1sxv differ from strains carrying Xpr1n in their expression of endogenous viruses like Bxv1 or whether these strains will permit efficient in vivo replication of exogenous X-MLVs is not yet known but is suggested by the fact that one strain viremic with xenotropic MLV, F/St, carries Bxv1 along with the permissive Xpr1sxv receptor, although other factors, like MHC haplotype (51), clearly influence this unusual phenotype. While F/St is no longer extant, one additional strain identified here, LT/SvEi, carries the sxv receptor as well as Bxv1 and can be examined for X-MLV expression.
The Xpr1sxv strains identified here could be used to develop mouse models for the analysis of X-MLV replication, tissue tropism, and pathogenesis and could also be used to determine whether the different host range subtypes in the X/P-MLV infectious-virus family have different in vivo properties. Previous analyses of the disease process in naturally highly leukemic strains, like AKR, while focused on ecotropic and polytropic MLVs, have recognized that at least one XMV, Bxv1, contributes to leukemogenesis by providing LTR sequences to the pathogenic P-MLV recombinants that arise in these mice (11, 41). These studies can now be expanded to determine how polymorphic variants of the XPR1 receptor affect disease type or latency as well as the type of recombinant viruses that appear. Studies on pathogenesis by exogenous X-MLVs can make use of the Swiss strain-derived mice carrying Xpr1sxv. While SIM.R and SJL have not been characterized for their proviral content, SWR and other Swiss strain-derived mice, like NFS, carry only a single XMV copy (11, 34). The near absence of endogenous XMVs in these X-MLV-susceptible mice should simplify analysis of exogenous-virus infection and pathogenesis. The analysis of X-MLV expression and pathogenesis in such mouse models could have broader relevance, particularly for studies on the origin, pathogenesis, and transspecies transmission of XMRV.
Acknowledgments
This research was supported by the Intramural Research Program of the NIAID, NIH.
We thank Alicia Buckler-White for sequencing.
Footnotes
Published ahead of print on 13 October 2010.
REFERENCES
- 1.Aaronson, S. A., G. J. Todaro, and E. M. Scolnick. 1971. Induction of murine C-type viruses from clonal lines of virus-free BALB/3T3 cells. Science 174:157-159. [DOI] [PubMed] [Google Scholar]
- 2.Battini, J.-L., J. E. J. Rasko, and A. D. Miller. 1999. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. U. S. A. 96:1385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck, J. A., S. Lloyd, M. Hafezparast, M. Lennon-Pierce, J. T. Eppig, M. F. W. Festing, and E. M. C. Fisher. 2000. Genealogies of mouse inbred strains. Nat. Genet. 24:23-25. [DOI] [PubMed] [Google Scholar]
- 4.Chattopadhyay, S. K., M. R. Lander, S. Gupta, E. Rands, and D. R. Lowy. 1981. Origin of mink cytopathic focus-forming (MCF) viruses: comparison with ecotropic and xenotropic murine leukemia virus genomes. Virology 113:465-483. [DOI] [PubMed] [Google Scholar]
- 5.Cloyd, M. W., M. M. Thompson, and J. W. Hartley. 1985. Host range of mink cell focus-inducing viruses. Virology 140:239-248. [DOI] [PubMed] [Google Scholar]
- 6.Datta, S. K., and R. S. Schwartz. 1977. Mendelian segregation of loci controlling xenotropic virus production in NZB crosses. Virology 83:449-452. [DOI] [PubMed] [Google Scholar]
- 7.Dong, B., S. Kim, S. Hong, J. Das Gupta, K. Malathi, E. A. Klein, D. Ganem, J. L. DeRisi, S. A. Chow, and R. H. Silverman. 2007. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc. Natl. Acad. Sci. U. S. A. 104:1655-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frankel, W. N., J. P. Stoye, B. A. Taylor, and J. M. Coffin. 1989. Genetic analysis of endogenous xenotropic murine leukemia viruses: association with two common mouse mutations and the viral restriction locus Fv-1. J. Virol. 63:1763-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenberger, J. S., S. M. Phillips, J. R. Stephenson, and S. A. Aaronson. 1975. Induction of mouse type-C RNA virus by lipopolysaccharide. J. Immunol. 115:317-320. [PubMed] [Google Scholar]
- 10.Groom, H. C. T., M. W. Yap, R. P. Galão, S. J. D. Neil, and K. N. Bishop. 2010. Susceptibility of xenotropic murine leukemia virus-related virus (XMRV) to retroviral restriction factors. Proc. Natl. Acad. Sci. U. S. A. 107:5166-5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoggan, M. D., R. R. O'Neill, and C. A. Kozak. 1986. Nonecotropic murine leukemia viruses in BALB/c and NFS/N mice: characterization of the BALB/c Bxv-1 provirus and the single NFS endogenous xenotrope. J. Virol. 60:980-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jern, P., J. P. Stoye, and J. M. Coffin. 2007. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 3:e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung, Y. T., M. S. Lyu, A. Buckler-White, and C. A. Kozak. 2002. Characterization of a polytropic murine leukemia virus proviral sequence associated with the virus resistance gene Rmcf of DBA/2 mice. J. Virol. 76:8218-8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozak, C. A. 1984. Differential expression of murine leukemia virus loci in chemically induced hybrid cells. J. Virol. 51:876-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozak, C. A. 1985. Susceptibility of wild mouse cells to exogenous infection with xenotropic leukemia viruses: control by a single dominant locus on chromosome 1. J. Virol. 55:690-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak, C. A., J. W. Hartley, and H. C. Morse III. 1984. Laboratory and wild-derived mice with multiple loci for production of xenotropic murine leukemia virus. J. Virol. 51:77-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozak, C. A., and R. R. O'Neill. 1987. Diverse wild mouse origins of xenotropic, mink cell focus-forming, and two types of ecotropic proviral genes. J. Virol. 61:3082-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak, C. A., and W. P. Rowe. 1978. Genetic mapping of xenotropic leukemia virus-inducing loci in two mouse strains. Science 199:1448-1449. [DOI] [PubMed] [Google Scholar]
- 19.Kozak, C. A., and W. P. Rowe. 1980. Genetic mapping of xenotropic murine leukemia virus-inducing loci in five mouse strains. J. Exp. Med. 152:219-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lander, M. R., and S. K. Chattopadhyay. 1984. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ectropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J. Virol. 52:695-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lander, M. R., B. Moll, and W. P. Rowe. 1978. A procedure for culture of cells from mouse tail biopsies: brief communication. J. Natl. Cancer Inst. 60:477-478. [PubMed] [Google Scholar]
- 22.Lathrop, A. E. C., and L. Loeb. 1918. Further investigations on the origin of tumors in mice. V. The tumor rate in hybrid strains. J. Exp. Med. 28:475-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy, J. A. 1973. Xenotropic viruses: murine leukemia viruses associated with NIH Swiss, NZB, and other mouse strains. Science 182:1151-1153. [DOI] [PubMed] [Google Scholar]
- 24.Levy, J. A. 1975. Host range of murine xenotropic virus: replication in avian cells. Nature 253:140-142. [DOI] [PubMed] [Google Scholar]
- 25.Levy, J. A., and T. Pincus. 1970. Demonstration of biological activity of a murine leukemia virus of New Zealand Black mice. Science 170:326-327. [DOI] [PubMed] [Google Scholar]
- 26.Lieber, M., C. Sherr, M. Potter, and G. Todaro. 1975. Isolation of type-c viruses from the Asian feral mouse Mus musculus molossinus. Int. J. Cancer 15:211-220. [DOI] [PubMed] [Google Scholar]
- 27.Lowy, D. R., W. P. Rowe, N. Teich, and J. W. Hartley. 1971. Murine leukemia virus: high-frequency activation in vitro by 5-iododeoxyuridine and 5-bromodeoxyuridine. Science 174:155-156. [DOI] [PubMed] [Google Scholar]
- 28.Marin, M., C. S. Tailor, A. Nouri, S. L. Kozak, and D. Kabat. 1999. Polymorphisms of the cell surface receptor control mouse susceptibilities to xenotropic and polytropic leukemia viruses. J. Virol. 73:9362-9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morse, H. C., III. 1978. Introduction, p. 1-31. In H. C. Morse III (ed.), Origins of inbred mice. Academic Press, New York, NY.
- 30.Morse, H. C., III, T. M. Chused, M. Boehm-Truitt, B. J. Mathieson, S. O. Sharrow, and J. W. Hartley. 1979. XenCSA: cell surface antigens related to the major glycoproteins (gp70) of xenotropic murine leukemia viruses. J. Immunol. 122:443-454. [PubMed] [Google Scholar]
- 31.Morse, H. C., III, C. A. Kozak, R. A. Yetter, and J. W. Hartley. 1982. Unique features of retrovirus expression in F/St mice. J. Virol. 43:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morse, H. C., III, B. A. Taylor, C. A. Kozak, T. M. Chused, S. O. Sharrow, J. W. Hartley, and E. Stockert. 1982. Expression on normal lymphocytes of two cell surface antigens, XenCSA and GIX, related to the major glycoproteins (gp70) of murine leukemia viruses. J. Immunol. 128:2111-2115. [PubMed] [Google Scholar]
- 33.Oie, H. K., E. K. Russell, J. H. Dotson, J. M. Rhoads, and A. F. Gazdar. 1976. Host range properties of murine xenotropic and ecotropic type-C viruses. J. Natl. Cancer Inst. 56:423-426. [DOI] [PubMed] [Google Scholar]
- 34.O'Neill, R. R., A. S. Khan, M. D. Hoggan, J. W. Hartley, M. A. Martin, and R. Repaske. 1986. Specific hybridization probes demonstrate fewer xenotropic than mink cell focus-forming murine leukemia virus env-related sequences in DNAs from inbred laboratory mice. J. Virol. 58:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakai, K., H. Narita, A. Adachi, S. Tsuruta, T. Yorifuji, and A. Ishimoto. 1982. Fv-1 determinants in xenotropic murine leukemia viruses studied with biological assay systems: isolation of xenotropic virus with N-tropic Fv-1 activity in the cryptic form. J. Virol. 42:331-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanville, B., M. A. Dolan, K. Wollenberg, Y. Yan, C. Martin, M. L. Yeung, K. Strebel, A. Buckler-White, and C. A. Kozak. 2010. Adaptive evolution of Mus Apobec3 includes retroviral insertion and positive selection at two clusters of residues flanking the substrate groove. PLoS Pathog. 6:e1000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarz, E. 1942. Origin of the Japanese waltzing mouse. Science 95:46. [DOI] [PubMed] [Google Scholar]
- 38.Sherr, C. J., M. M. Lieber, and G. J. Todaro. 1974. Mixed splenocyte cultures and graft versus host reactions selectively induce an S-tropic murine type C virus. Cell 1:55-58. [Google Scholar]
- 39.Steffen, D. L., S. Bird, and R. A. Weinberg. 1980. Evidence for the Asiatic origin of endogenous AKR-type murine leukemia proviruses. J. Virol. 35:824-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stocking, C., and C. Kozak. 2008. Endogenous retroviruses. Cell. Mol. Life Sci. 65:3383-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoye, J. P., C. Moroni, and J. M. Coffin. 1991. Virological events leading to spontaneous AKR thymomas. J. Virol. 65:1273-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tailor, C. S., A. Nouri, C. G. Lee, C. Kozak, and D. Kabat. 1999. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc. Natl. Acad. Sci. U. S. A. 96:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomonaga, K., and J. M. Coffin. 1998. Structure and distribution of endogenous nonecotropic murine leukemia viruses in wild mice. J. Virol. 72:8289-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ware, L. M., and A. A. Axelrad. 1972. Inherited resistance to N- and B-tropic murine leukemia viruses in vitro: evidence that congenic mouse strains SIM and SIM.R differ at the Fv-1 locus. Virology 50:339-348. [DOI] [PubMed] [Google Scholar]
- 45.Yan, Y., A. Buckler-White, K. Wollenberg, and C. A. Kozak. 2009. Origin, antiviral function and evidence for positive selection of the gammaretrovirus restriction gene Fv1 in the genus Mus. Proc. Natl. Acad. Sci. U. S. A. 106:3259-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan, Y., R. C. Knoper, and C. A. Kozak. 2007. Wild mouse variants of envelope genes of xenotropic/polytropic mouse gammaretroviruses and their XPR1 receptors elucidate receptor determinants of virus entry. J. Virol. 81:10550-10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan, Y., Q. Liu, and C. Kozak. 2009. Six host range variants of the xenotropic/polytropic gammaretroviruses define determinants for entry in the XPR1 cell surface receptor. Retrovirology 6:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan, Y., Q. Liu, K. Wollenburg, C. Martin, A. Buckler-White, and C. A. Kozak. 2010. Evolution of functional and sequence variants of the mammalian XPR1 receptor for mouse xenotropic gammaretroviruses and the human-derived retrovirus XMRV. J. Virol. 84:11970-11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, H., T. A. Bell, G. A. Churchill, and F. Pardo-Manuel de Villena. 2007. On the subspecific origin of the laboratory mouse. Nat. Genet. 39:1100-1107. [DOI] [PubMed] [Google Scholar]
- 50.Yang, Y.-L., L. Guo, S. Xu, C. A. Holland, T. Kitamura, K. Hunter, and J. M. Cunningham. 1999. Receptors for polytropic and xenotropic mouse leukaemia viruses encoded by a single gene at Rmc1. Nat. Genet. 21:216-219. [DOI] [PubMed] [Google Scholar]
- 51.Yetter, R. A., J. W. Hartley, and H. C. Morse III. 1983. H-2-linked regulation of xenotropic murine leukemia virus expression. Proc. Natl. Acad. Sci. U. S. A. 80:505-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yonekawa, H., K. Moriwaki, O. Gotoh, N. Miyashita, Y. Matsushima, L. M. Shi, W. S. Cho, X. L. Zhen, and Y. Tagashira. 1988. Hybrid origin of Japanese mice “Mus musculus molossinus”: evidence from restriction analysis of mitochondrial DNA. Mol. Biol. Evol. 5:63-78. [DOI] [PubMed] [Google Scholar]