Abstract
DNA double-strand breaks resulting from normal cellular processes including replication and exogenous sources such as ionizing radiation pose a serious risk to genome stability, and cells have evolved different mechanisms for their efficient repair. The two major pathways involved in the repair of double-strand breaks in eukaryotic cells are non-homologous end joining and homologous recombination. Numerous factors affect the decision to repair a double-strand break via these pathways, and accumulating evidence suggests these major repair pathways both cooperate and compete with each other at double-strand break sites to facilitate efficient repair and promote genomic integrity.
Keywords: Double-strand break, DNA repair, Non-homologous end joining, Homologous recombination, Single-strand annealing
1. Introduction
DNA damage constantly occurs in cells as a result of both environmental and endogenous insults. Double-strand breaks (DSBs), which may arise during the normal course of DNA replication and as a result of exposure to DNA damaging agents, are considered one of the most cytotoxic forms of DNA damage [1]. The ability to accurately repair these breaks is essential for faithful propagation of genetic information. Deficiencies in DSB repair can lead to mutations and chromosomal rearrangements that ultimately may result in genomic instability and tumorigenesis. Therefore, cells have evolved effective mechanisms for the accurate and timely repair of DSBs in DNA.
Two major pathways are involved in the repair of DSBs in eukaryotic cells: non-homologous end joining (NHEJ) and homologous recombination (HR) [2,3]. NHEJ is an efficient pathway that functions throughout the cell cycle and involves the ligation of DNA ends with minimal processing at the site of end joining, while HR, occurring specifically in late S and G2 phases of the cell cycle, utilizes an undamaged homologous sequence as a repair template, preferably the sister chromatid, and is considered a more precise method for repairing DSBs in DNA. This review focuses on the collaboration and competition of the two major pathways of DSB repair in mammalian cells, with an emphasis on factors affecting the decision to repair breaks via HR or NHEJ.
2. Mechanisms of homologous recombination
HR is initiated by resection of DNA ends at the DSB site to yield 3′-single-stranded DNA (ssDNA) overhangs which are capable of invading duplex DNA containing a homologous sequence [4]. Studies in Saccharomyces cerevisiae suggest that the MRX complex, encoded by MRE11, RAD50 and XRS2 (the ortholog of NBS1 in mammalian cells), together with the Sae2 protein, is required for the initial end processing step of HR. More extensive processing involves the 5′–3′ exonuclease Exo1 or the combined helicase/nuclease activities of Sgs1/Dna2 [5,6]. The functional counterpart of Sae2 in vertebrate cells is CtIP [7]. The protein product of the breast cancer susceptibility gene BRCA1 interacts with both MRN and CtIP, and genetic and physical evidence suggests that BRCA1 may be involved in end resection [8–10], although its exact role remains uncertain. There is, however, data supporting a role for mammalian counterparts of Exo1 and Sgs1 in end resection: human EXO1 can resect DNA ends in vitro, and its activity is stimulated by Bloom’s syndrome protein (BLM), the Sgs1 ortholog [11]. DNA ends resected by EXO1 and BLM are utilized in subsequent strand exchange reactions.
The 3′-single-stranded DNA overhang generated during end resection is bound by replication protein A (RPA), which is required for the subsequent recruitment of checkpoint and HR proteins such as RAD51 [12]. RAD51, a homolog of the bacterial RecA protein, is a DNA-dependent ATPase that forms nucleoprotein filaments with DNA. In mammalian cells, RAD51 is recruited to DSBs by the protein product of the breast cancer susceptibility gene BRCA2 [13]. BRCA2 is a large (410kD) protein that binds RAD51 through interactions with a series of eight short conserved repeats termed BRC repeats [14,15]. Recent biochemical analysis has shown that one or more BRC repeats stimulate RAD51 nucleoprotein filament formation on ssDNA in the presence of ATP [16,17]. Moreover, structural studies have demonstrated that BRCA2 itself binds to ssDNA [18].
Both BRCA1 and BRCA2 mutant cells are defective for HR [19,20]. BRCA2 appears to interact with BRCA1 via the BRCA2 “partner” PALB2; mutations in PALB2 that disrupt binding to either BRCA1 or BRCA2, as well as clinically relevant mutations in BRCA1 which abrogate binding to PALB2, result in decreased levels of HR [21–24]. Additionally, BRCA2 forms a complex with DSS1, a conserved 70 amino acid protein required for DNA damage-induced RAD51 foci formation, and presumably HR, in mammalian cells [18,25,26]. In yeast, BRCA2 (as well as BRCA1 and PALB2) is not present; therefore, other proteins such as Rad52 assist in loading Rad51 onto ssDNA [12].
Once recruited to the DSB, RAD51 catalyzes strand exchange during which ssDNA invades homologous duplex DNA forming a displacement loop (D-loop). Recently solved crystal structures of Escherichia coli RecA-ssDNA and RecA-heteroduplex filaments have shed new light on how RAD51 may facilitate strand exchange [27]: RecA-bound ssDNA is stretched globally but maintains a B-DNA-like conformation locally in base triplets; this unusual structure favors Watson–Crick type base pairing during homology sampling with the complementary strand in a destabilized donor duplex DNA.
Once formed, the D-loop has multiple fates [12]. In the primary pathway in mitotic cells, termed synthesis-dependent strand annealing, the 3′ end in the D-loop is extended by repair synthesis, and then the newly synthesized DNA strand dissociates to anneal to the other DNA end to complete the reaction. If the second DNA end is “captured” by the D-loop, a double Holliday junction forms that can potentially be resolved by several different proteins, including in humans GEN1 and SLX1/SLX4 [4,28]. As double Holliday junction resolution can occur in different ways, crossover and non-crossover products are possible. While crossovers play an important role in facilitating chromosome segregation during meiotic recombination [29], crossovers occurring during mitotic recombination may have serious deleterious effects, including loss of heterozygosity [3]. Proteins that disrupt D-loops or “dissolve” Holliday junctions (such as BLM) suppress mitotic crossovers, thereby decreasing the risk of genomic instability [30].
3. Repair by single-strand annealing
Another repair pathway involving sequence homology, but distinct from HR, is single-strand annealing (SSA) [31]. SSA can occur following end resection if sequence repeats exist on both sides of the DSB. The complementary single strands formed at the repeats then anneal and flaps formed from the annealing reaction are trimmed off, resulting in a loss of sequence between the repeats. Compared to HR, SSA is therefore more mutagenic because it involves loss of genetic information. Proteins identified to promote SSA in mammalian cells and yeast include RAD52 (annealing) and ERCC1 and Rad1/Rad10 (flap endonuclease) [8,32,33].
4. Repair by non-homologous end joining
NHEJ proteins were initially identified through their requirement for resistance to ionizing radiation and V(D)J recombination in the immune system [2]. The first protein in this pathway to bind DNA ends is the Ku70/80 heterodimer (Ku). In mammalian cells, Ku interacts with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and together they may act to synapse the two DNA ends to be repaired [34]. DNA ends are joined by the DNA ligase IV/XRCC4 complex [35]. XRCC4 does not possess any known enzymatic activity but acts as a scaffold that forms interactions with both Ku and DNA and both stabilizes and stimulates the ligase activity of DNA ligase IV [36].
NHEJ can join DNA ends with a number of different structures. As a result, this pathway makes use of a number of processing steps that may include cleavage and gap filling prior to ligation. The nuclease Artemis is recruited to a DSB site by its interactions with DNA-PKcs, and the Artemis/DNA-PKcs complex is able to cleave a variety of damaged DNA overhangs [37]. Cleavage of DNA ends may result in gaps in the DNA that need to be filled in by polymerases involved in NHEJ. Members of the PolX family include polymerases μ and λ, which interact with the Ku:DNA complex via BRCT domains [38], although data in yeast suggests that other polymerase families can substitute [39]. The modification of DNA ends prior to joining by these processing steps can lead to deletions and insertions accounting for the more error-prone nature of NHEJ compared to HR.
The pathway thus described is considered to be the “canonical” NHEJ pathway. However, almost from their initial characterization, cell mutants for the canonical NHEJ factors have been recognized to join DSBs with good efficiency in certain contexts, for example, endonuclease-generated DSBs in plasmid and chromosomal DNA [40,41] and immune system-generated DSBs in mice [42,43]. NHEJ in the absence of the canonical factors is termed alternative NHEJ (alt-NHEJ), but whether this is a distinct pathway is uncertain, although a number of reports have suggested the involvement of a number of other factors (see e.g., [44]). Canonical NHEJ and alt-NHEJ seem to differ by the amount of microhomology at the site of joining. Small sequence microhomologies may help to align broken strands of DNA, but whereas microhomology at breakpoint junctions can occur in canonical NHEJ at frequencies expected by chance, longer microhomologies are over-represented in junctions arising from alt-NHEJ [45].
5. Regulation of repair pathway choice
DSB repair pathway choice is regulated by several factors, including the nature of the lesion and cell cycle phase. Programmed DSBs are channeled into specific repair pathways, e.g., DSBs generated during V(D)J recombination by the RAG proteins are repaired by NHEJ [46], whereas DSBs generated during meiosis by the Spo11 protein are repaired by HR [29]. Several factors may enforce pathway choice for programmed DSBs. For example, Spo11 forms a covalent linkage with DNA and is cleaved off of DNA by the MRX/Sae2 proteins [47]. Moreover, in mouse Ku has been reported to be down regulated early in meiotic prophase, which would presumably lessen NHEJ [48]. Finally, DSBs arising during DNA replication may typically be one ended, requiring HR for repair, as NHEJ of two one-ended DSBs would give rise to translocations [49].
More generally, cell cycle phase is a primary determinant in restricting HR, whereas NHEJ operates throughout the cell cycle [50]. The restriction in HR to the S/G2 phases of the cell cycle makes sense from the standpoint that the primary repair template in mammalian cells is the sister chromatid, which is not present in G1 cells. By contrast, in yeast diploid cells are able to efficiently use the homolog for DSB repair [51,52]. Why might the homolog be used efficiently in yeast for repair but not in mammalian cells? A simple explanation may be that the chance for random collision between homologs is greater in the much smaller yeast nucleus than it is in the mammalian nucleus [53]. That proximity matters is supported by efficient interhomolog recombination in Drosophila, where homologs are actually paired [54].
Cell cycle phase also plays a more active role in regulating HR in that end resection is promoted by cyclin-dependent protein kinases (CDKs). In yeast, CDK activity is required for efficient end resection of DSBs, and hence, HR, during S/G2 [55,56]. A key target of CDK is Sae2, which is phosphorylated at serine 267 [57]. Cells expressing a non-phosphorylatable Sae2 protein have similar phenotypes to a sae2 null mutant, including delayed HR; conversely, cells expressing the phospho-mimicking sae2-S267E protein undergo some end resection even in the absence of CDK activity, while demonstrating accelerated HR. Interestingly, the limited homology between Sae2 and CtIP includes Sae2 Ser267 and the CtIP equivalent Thr847, and abrogation of phosphorylation at CtIP Thr847 impairs end resection, as measured by RPA phosphorylation [58].
CDK also phosphorylates another site on CtIP, Ser327, which has been reported to promote CtIP/BRCA1 interaction during S/G2 phases [9,59,60]. Furthermore, CDK-dependent phosphorylation of BRCA2 at Ser3291 interferes with the ability of Rad51 to interact with BRCA2 C-terminus; interestingly, phosphorylation peaks during M phase, suggesting a link between disassembly of Rad51 filaments and mitotic entry [61–63].
6. Collaboration between NHEJ and HR
There is significant evidence that HR and NHEJ collaborate to enhance overall DNA repair and safeguard genomic integrity. Both HR and NHEJ are essential DNA damage repair pathways in mice, as single knockouts for multiple components of either pathway die during embryogenesis (e.g., HR: Rad51, BRCA2, and XRCC2; NHEJ: DNA ligase IV and XRCC4) [64]. A notable example of the requirement for both pathways for genomic integrity is in the developing brain, where differential effects are seen with loss of either pathway: apoptosis arises during early proliferative stages from disruption of HR (likely from unrepaired DNA damage arising during replication) and during post-mitotic stages from disruption of NHEJ (where HR would not be possible) [65].
Further evidence of the collaboration between pathways comes from double mutant analysis of HR and NHEJ components. These studies suggest that concomitant loss of a protein involved in HR and a protein involved in NHEJ results in a more severe phenotype than would be expected from loss of either single pathway, with significant evidence of genomic instability [66,67]. Mice with mutations in both the HR component Rad54 and the NHEJ protein Ku80, both of which are viable as single mutants, show decreased survival, extreme sensitivity to even low doses of ionizing radiation, and, at the cellular level, increased accumulation of unrepaired DSBs, as measured by γ-H2AX foci, even in the absence of DNA damage [67]. Similarly, embryonic fibroblasts deficient for both Rad54 and the DNA ligase IV also exhibit high levels of spontaneous DNA damage and accumulate a substantial number of chromatid breaks [66]. Chicken cells mutated for Rad54 and Ku70 or DNA-PKcs also show extreme radiosensitivity [68,69]. The substantially increased DNA damage in these double mutant combinations implies cooperation between HR and NHEJ, even during the same phase of the cell cycle. One suggestion is that when HR is reduced one-ended DSBs arising during replication convert into two-ended DSBs by fork convergence, providing a substrate for NHEJ [67].
7. Competition between HR, SSA, and NHEJ
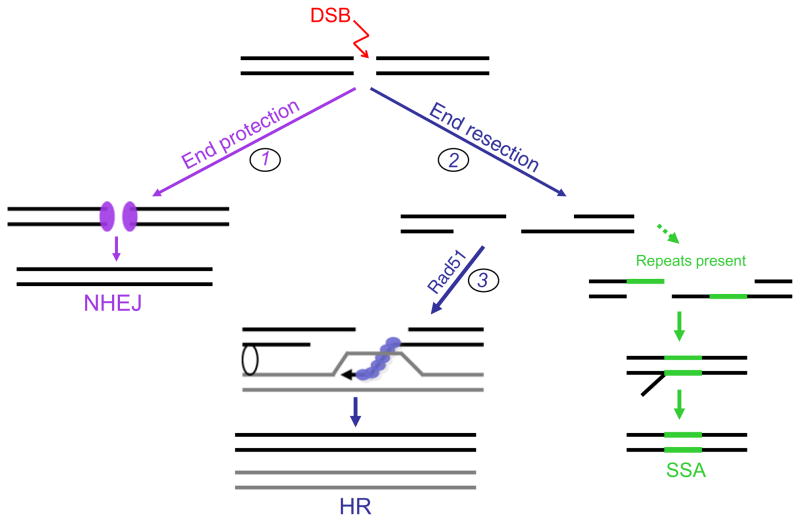
While collaboration between HR and NHEJ is necessary for the maintenance of genomic integrity, competition between DSB repair pathways is also evident. This competition is most apparent in mutants that affect either end resection (Fig. 1, 1 and 2) or the ability of a resected end to be channeled into HR (Fig. 1, 3). Thus, NHEJ mutants that have enhanced end resection (e.g., Ku) have increased HR and SSA, while mutants with decreased end resection (e.g., Sae2/CtIP) have increased NHEJ.
Fig. 1.
Competition between double-strand break repair pathways. DNA double-strand breaks (DSBs) are repaired by two distinct pathways, homologous recombination (HR) and non-homologous end joining (NHEJ). HR is initiated by 5′ to 3′ end resection, forming a 3′ single-stranded tail onto which Rad51 assembles. The Rad51 nucleoprotein filament allows single-strand DNA invasion into a homologous duplex, typically the sister chromatid, to initiate repair synthesis. The newly synthesized strand is then displaced to anneal to the other DNA end (not shown) to complete the HR reaction. When a DSB and subsequent end resection occurs at sequence repeats (green lines), an alternative pathway, single-strand annealing (SSA), can take place. The complementary single strands at the repeats can anneal, giving rise to a copy number variant, in this case a product with a single copy of the repeat and a deletion of the intervening sequence. NHEJ involves the joining of DNA ends with no or little homology (microhomology). In this pathway, the Ku heterodimer binds to DNA ends, protecting them from end resection. A number of processing factors are subsequently recruited (not shown), which allows a variety of end structures to be joined. Mutational analysis has demonstrated that factors involved in HR and NHEJ likely directly “compete” at steps indicated by the numbers: (1) loss of canonical NHEJ factors (Ku, DNA ligase IV/XRCC4) leads to increased end resection and hence HR and SSA. (2) End resection mutants (e.g., Sae2) have increased NHEJ. (3) Disruption of Rad51 filament formation allows DNA ends to be channeled into SSA.
7.1. NHEJ mutants with enhanced end resection
An enhanced rate of end resection has been demonstrated in a yeast Ku mutant using direct molecular analysis [70]; since Ku binds DNA ends, it presumably physically blocks access of the end resection machinery. More recently, the homolog of DNA ligase IV/XRCC4 in yeast, Dnl4-Lif1, has also been shown to have an inhibitory effect on end resection, possibly by stabilizing Ku at DNA ends, although the effect is somewhat less pronounced than with Ku [71].
Consistent with an increase in the end resected intermediate, mutation of Ku or DNA ligase IV/XRCC4 in mammalian cells leads to increased HR, with mutation of Ku showing a more profound effect [49]. Loss of DNA-PKcs, which does not have an ortholog in yeast, also increases HR [49,72], although as yet a role for DNA-PKcs in inhibiting end resection has not been shown. As with mammalian cells, chicken cells mutated for Ku or DNA-PKcs also show increased HR compared with wild-type cells [69]. Notably, the increase of HR in NHEJ mutants occurs whether the DSB is introduced by I-SceI endonuclease, which generates a 3′ overhang, or the RAG recombinase, which generates a hairpin capped end [49,73]. SSA, which has an end-resected intermediate like HR, is also increased with loss of Ku or DNA ligase IV/XRCC4 [8,74].
The finding that Ku can suppress repair through other pathways has implications for other NHEJ mutants. Ku and DNA ligase IV/ XRCC4 mutants share many phenotypes; however, DNA ligase IV/ XRCC4 mice are not viable whereas Ku mutant mice are viable, although they have reduced vigor [67,75]. Intriguingly, the embryonic lethality of DNA ligase IV-deficient mice can be rescued by deletion of Ku, suggesting that Ku blocks access to other repair pathways even in other NHEJ mutants [76]. (But see also discussion in [76].) Moreover, in chicken cells, Ku mutation actually leads to increased resistance to ionizing radiation during late S/G2 both in otherwise wild-type cells as well as in DNA-PKcs mutant cells [69]; this has been interpreted as Ku interference of HR during these phases of the cell cycle.
In addition to canonical NHEJ factors, the DNA damage response factor 53BP1 has been suggested to play an important role in NHEJ [77,78], and reports have linked 53BP1 disruption to increased HR [77,79]. Notably, 53BP1 loss restores viability to BRCA1 null cells [80] and BRCA1 mutant mice [81]. Mechanistic studies have provided evidence that 53BP1 loss restores HR levels in BRCA1-deficient cells [80,82], possibly by unleashing end resection at DSBs from inhibition by 53BP1 [82]. Interestingly, 53BP1 loss does not rescue the viability of BRCA2 null cells [82]; these results are consistent with genetic studies that place BRCA1 upstream of BRCA2 in HR, such that BRCA2 mutants are deficient at a step after end resection (see below). These studies point to the important consequences of the interplay of HR and NHEJ, which may have implications for human tumorigenesis [80,83].
7.2. End resection mutants
In yeast, resected DNA ends appear to be less prone to repair via NHEJ [84], consistent with the observation that HR mutants like BRCA2, which have resected intermediates, do not show an apparent increase in NHEJ [8]. Thus, interference of end resection would be predicted to increase NHEJ levels. Consistent with this, both sae2 null and sae2-S267A mutants show increased frequencies of NHEJ in addition to delayed HR [57]. Vertebrate cells deficient for CtIP have also been reported to show increased NHEJ [44,60], although the role of CDK phosphorylation in modulating its activity is as yet unresolved [85].
7.3. HR mutants with resected DNA ends
Once a resected DNA end has been formed, it becomes a substrate for Rad51 filament formation. Several HR mutants have been identified in which Rad51 filament formation is disrupted, including BRCA2, RAD54, RAD51 paralogs, and ATP binding mutants of RAD51 itself, and in each of these mutants, SSA is increased in model substrates [8,86–88]. For example, with the RAD51-K133A mutant, the ratio of HR to SSA is shifted 93-fold (22-fold decrease in HR and 4.2-fold increase in SSA) [8]. In endogenous genomic sequences, homologous repeats are not as likely to be present close by and with identical or nearly identical sequence (see [89,90] for effects of repeat heterology and distance on SSA). Nonetheless, the tremendous shift towards this mutagenic pathway likely contributes to the mutation load found in these various HR mutants.
8. HR vs. NHEJ pathway choice and the Fanconi anemia pathway
More recent analysis has implicated Ku in regulating pathway choice in the repair of other DNA lesions, in particular those caused by crosslinking agents like cisplatin or mitomycin C. Cells derived from Fanconi anemia patients are extremely sensitive to crosslinking agents due to mutations in any number of FANC genes [91]. Like BRCA1 mutant cells, FANC mutant cells show small defects in both HR and SSA in the repair of a DSB [92], as well as increased radial chromosomes, apparently from aberrant NHEJ.
In chicken cells, loss of Ku in FANCC mutant cells greatly relieves their cisplatin sensitivity, possibly because HR is increased in the Ku/FANCC cells compared with FANCC single mutant cells [93]. A separate study found that Ku mutation also relieves the sensitivity of human patient derived cell lines (FANCC or FANCD2 mutant) to mitomycin C [94]. These findings have potentially profound implications for possible therapies for Fanconi anemia patients.
Conflicting results were obtained in these two studies with other NHEJ mutants. In chicken cells, neither DNA ligase IV nor DNA-PKcs deficiency relieves cisplatin sensitivity of FANCC mutant cells; in fact, DNA ligase IV deficiency further sensitizes the FANCC cells to cisplatin, consistent with additive repair defects rather than a suppression of repair defects found with Ku deficiency [93]. By contrast, in patient derived cells, loss of DNA ligase IV or DNA-PKcs, like Ku, relieves the sensitivity of FANC mutant cells to mitomycin C, and in worms, loss of DNA ligase IV relieves the sensitivity of FANCD2 mutants to cisplatin [94]. What accounts for the difference between the two reports is uncertain, e.g., whether the interplay between repair pathways differs in some respect between humans and worms compared with chickens. However, it is notable that in DSB repair in mouse cells, all three canonical NHEJ factors suppress HR, but Ku has the most profound suppression [49], especially for DSBs induced by the RAG recombinase [73].
9. Summary
DNA damage is constantly occurring in eukaryotic cells, posing an ongoing threat to genomic stability. Consequently, eukaryotic cells have evolved multiple pathways for the efficient repair of DNA damage like DSBs. The coordinated actions of components of different repair pathways at DSBs that ultimately navigate repair pathway choice play a critical role in the ability of cells to repair DNA lesions in a timely and accurate manner. These safeguards ensure proper development while preventing tumorigenesis.
Acknowledgments
The authors thank past and present members of the laboratory for discussion. Work in the author’s laboratory is supported by the National Institutes of Health grants P01CA94060 (M.J.) and R01GM54668 (M.J.).
References
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 3.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mimitou EP, Symington LS. Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci. 2009;34:264–272. doi: 10.1016/j.tibs.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 10.Schlegel BP, Jodelka FM, Nunez R. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 2006;66:5181–5189. doi: 10.1158/0008-5472.CAN-05-3209. [DOI] [PubMed] [Google Scholar]
- 11.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci USA. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 13.Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 14.Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 15.Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51–BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 16.Shivji MK, Mukund SR, Rajendra E, Chen S, Short JM, Savill J, Klenerman D, Venkitaraman AR. The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single-versus double-stranded DNA to stimulate strand exchange. Proc Natl Acad Sci USA. 2009;106:13254–13259. doi: 10.1073/pnas.0906208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, Kowalczykowski SC. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang H, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 19.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 20.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 21.Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 22.Zhang F, Fan Q, Ren K, Andreassen PR. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res. 2009;7:1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sy SM, Huen MS, Zhu Y, Chen J. PALB2 regulates recombinational repair through chromatin association and oligomerization. J Biol Chem. 2009;284:18302–18310. doi: 10.1074/jbc.M109.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci USA. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marston NJ, Richards WJ, Hughes D, Bertwistle D, Marshall CJ, Ashworth A. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol. 1999;19:4633–4642. doi: 10.1128/mcb.19.7.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gudmundsdottir K, Lord CJ, Witt E, Tutt AN, Ashworth A. DSS1 is required for RAD51 focus formation and genomic stability in mammalian cells. EMBO Rep. 2004;5:989–993. doi: 10.1038/sj.embor.7400255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 28.Klein HL, Symington LS. Breaking up just got easier to do. Cell. 2009;138:20–22. doi: 10.1016/j.cell.2009.06.039. [DOI] [PubMed] [Google Scholar]
- 29.Cole F, Keeney S, Jasin M. Evolutionary conservation of meiotic DSB proteins: more than just Spo11. Genes Dev. 2010;24:1201–1207. doi: 10.1101/gad.1944710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 31.Lin F-L, Sperle K, Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: Role for DNA ends in the recombination process. Mol Cell Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fishman-Lobell J, Haber JE. Removal of nonhomologous DNA ends in double-strand break recombination: The role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 34.DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Critchlow SE, Jackson SP. DNA end-joining: from yeast to man. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 36.Grawunder U, Zimmer D, Kulesza P, Lieber MR. Requirement for an interaction of XRCC4 with DNA ligase IV for wild-type V(D)J recombination and DNA double-strand break repair in vivo. J Biol Chem. 1998;273:24708–24714. doi: 10.1074/jbc.273.38.24708. [DOI] [PubMed] [Google Scholar]
- 37.Ma Y, Schwarz K, Lieber MR. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 2005;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 38.Ma Y, et al. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 39.Wilson TE, Lieber MR. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J Biol Chem. 1999;274:23599–23609. doi: 10.1074/jbc.274.33.23599. [DOI] [PubMed] [Google Scholar]
- 40.Liang F, Jasin M. Ku80 deficient cells exhibit excess degradation of extrachromosomal DNA. J Biol Chem. 1996;271:14405–14411. doi: 10.1074/jbc.271.24.14405. [DOI] [PubMed] [Google Scholar]
- 41.Delacote F, Han M, Stamato TD, Jasin M, Lopez BS. An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 2002;30:3454–3463. doi: 10.1093/nar/gkf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferguson DO, Alt FW. DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene. 2001;20:5572–5579. doi: 10.1038/sj.onc.1204767. [DOI] [PubMed] [Google Scholar]
- 43.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 44.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- 47.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goedecke W, Eijpe M, Offenberg HH, van Aalderen M, Heyting C. Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat Genet. 1999;23:194–198. doi: 10.1038/13821. [DOI] [PubMed] [Google Scholar]
- 49.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fabre F. Induced intragenic recombination in yeast can occur during the G1 mitotic phase. Nature. 1978;272:795–798. doi: 10.1038/272795a0. [DOI] [PubMed] [Google Scholar]
- 52.Bressan DA, Baxter BK, Petrini JH. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stark JM, Jasin M. Extensive Loss of Heterozygosity is suppressed during homologous repair of chromosomal breaks. Mol Cell Biol. 2003;23:733–743. doi: 10.1128/MCB.23.2.733-743.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fung JC, Marshall WF, Dernburg A, Agard DA, Sedat JW. Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J Cell Biol. 1998;141:5–20. doi: 10.1083/jcb.141.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 62.Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 63.Shivji MK, Davies OR, Savill JM, Bates DL, Pellegrini L, Venkitaraman AR. A region of human BRCA2 containing multiple BRC repeats promotes RAD51-mediated strand exchange. Nucleic Acids Res. 2006;34:4000–4011. doi: 10.1093/nar/gkl505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pierce AJ, Stark JM, Araujo FD, Moynahan ME, Berwick M, Jasin M. Double-strand breaks and tumorigenesis. Trends Cell Biol. 2001;11:S52–S59. doi: 10.1016/s0962-8924(01)02149-3. [DOI] [PubMed] [Google Scholar]
- 65.Orii KE, Lee Y, Kondo N, McKinnon PJ. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc Natl Acad Sci USA. 2006;103:10017–10022. doi: 10.1073/pnas.0602436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mills KD, Ferguson DO, Essers J, Eckersdorff M, Kanaar R, Alt FW. Rad54 and DNA Ligase IV cooperate to maintain mammalian chromatid stability. Genes Dev. 2004;18:1283–1292. doi: 10.1101/gad.1204304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Couedel C, et al. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev. 2004;18:1293–1304. doi: 10.1101/gad.1209204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takata M, et al. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fukushima T, et al. Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S-G2 phase DNA double-strand break repair. J Biol Chem. 2001;276:44413–44418. doi: 10.1074/jbc.M106295200. [DOI] [PubMed] [Google Scholar]
- 70.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y, et al. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol. 2007;14:639–646. doi: 10.1038/nsmb1261. [DOI] [PubMed] [Google Scholar]
- 72.Allen C, Kurimasa A, Brenneman MA, Chen DJ, Nickoloff JA. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc Natl Acad Sci USA. 2002;99:3758–3763. doi: 10.1073/pnas.052545899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weinstock DM, Jasin M. Alternative pathways for the repair of RAG-induced DNA breaks. Mol Cell Biol. 2006;26:131–139. doi: 10.1128/MCB.26.1.131-139.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bennardo N, Gunn A, Cheng A, Hasty P, Stark JM. Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet. 2009;5:e1000683. doi: 10.1371/journal.pgen.1000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P. Deletion of Ku86 causes early onset of senescence in mice. Proc Natl Acad Sci USA. 1999;96:10770–10775. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karanjawala ZE, Adachi N, Irvine RA, Oh EK, Shibata D, Schwarz K, Hsieh CL, Lieber MR. The embryonic lethality in DNA ligase IV-deficient mice is rescued by deletion of Ku: implications for unifying the heterogeneous phenotypes of NHEJ mutants. DNA Repair (Amst) 2002;1:1017–1026. doi: 10.1016/s1568-7864(02)00151-9. [DOI] [PubMed] [Google Scholar]
- 77.Nakamura K, Sakai W, Kawamoto T, Bree RT, Lowndes NF, Takeda S, Taniguchi Y. Genetic dissection of vertebrate 53BP1: a major role in non-homologous end joining of DNA double strand breaks. DNA Repair (Amst) 2006;5:741–749. doi: 10.1016/j.dnarep.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 78.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xie A, et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell. 2007;28:1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cao L, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell. 2009;35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kass EM, Moynahan ME, Jasin M. Loss of 53BP1 is a gain for BRCA1 mutant cells. Cancer Cell. 2010;17:423–425. doi: 10.1016/j.ccr.2010.04.021. [DOI] [PubMed] [Google Scholar]
- 84.Frank-Vaillant M, Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol Cell. 2002;10:1189–1199. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- 85.Nakamura K, et al. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 2010;6:e1000828. doi: 10.1371/journal.pgen.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dronkert ML, Beverloo HB, Johnson RD, Hoeijmakers JH, Jasin M, Kanaar R. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol Cell Biol. 2000;20:3147–3156. doi: 10.1128/mcb.20.9.3147-3156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tutt A, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sugawara N, Goldfarb T, Studamire B, Alani E, Haber JE. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc Natl Acad Sci USA. 2004;101:9315–9320. doi: 10.1073/pnas.0305749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Elliott B, Richardson C, Jasin M. Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol Cell. 2005;17:885–894. doi: 10.1016/j.molcel.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 91.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 92.Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D’Andrea AD, Wang ZQ, Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci USA. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329:219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 94.Adamo A, et al. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]