Abstract
Lung cancer cell lines have made a substantial contribution to lung cancer translational research and biomedical discovery. A systematic approach to initiating and characterizing cell lines from small cell and non–small cell lung carcinomas has led to the current collection of more than 200 lung cancer cell lines, a number that exceeds those for other common epithelial cancers combined. The ready availability and widespread dissemination of the lines to investigators worldwide have resulted in more than 9000 citations, including multiple examples of important biomedical discoveries. The high (but not perfect) genomic similarities between lung cancer cell lines and the lung tumor type from which they were derived provide evidence of the relevance of their use. However, major problems including misidentification or cell line contamination remain. Ongoing studies and new approaches are expected to reveal the full potential of the lung cancer cell line panel.
The current scale of biomedical cancer research requires an extensive source of human tumor materials. It would appear that resected human tumors could satisfy such needs. However, increasingly stringent requirements from institutional review boards, governmental requirements for protection of patients’ privacy rights, restrictions on the international exchange of biological reagents, for example, severely limit the availability and distribution of human tumors. Thus, animal and in vitro models have been developed for experimental studies and are being widely used. We have recently written another review article on the pros and cons of using lung cancer cells lines (1) that focuses on the use of cell lines to investigate the hallmarks of cancer (2) and on the development of in vitro culture systems to study multistage pathogenesis. In this review, we focused on the utility of cell lines that have been derived from human lung tumors for the study of lung cancer.
Methods
In writing this review, we followed the “Methodologic guidelines for review papers” (3). We performed a search of the National Library of Medicine Medline (http://www.nlm.nih.gov/pubs/) database from within the EndNote program (Thomson Reuters, Carlsbad, CA) that used the following Medical Search Heading terms: 1) “lung neoplasms,” 2) “cell line, tumor,” and 3) “humans.” On June 10, 2010, the search yielded 9741 citations. Because most of the cell lines cited in this article were established by the authors, we used our knowledge of the literature and our own experiences to select relevant articles that illustrate the major purpose of this review article—the utility of cell lines that have been derived from human lung tumors for the study of lung cancer. The selected references and the topics that they illustrate are not meant to be a comprehensive review of all of the literature but only to serve as representative examples. Portable Document Format (PDF) files of approximately 300 articles were obtained, and the contents of the subset that were selected as being most relevant were summarized and used as illustrative examples. The Wellcome Trust Sanger Institute (www.sanger.ac.uk) maintains public databases of genomic alterations in cancers and cell lines. We searched the Sanger databases for the numbers and types of human cancer cell lines that they had accumulated. Finally, we examined the Catalogue of Cell Cultures and Hybridomas from the American Type Culture Collection (ATCC, Manassas, VA) (http://www.atcc.org/culturesandproducts/cellbiology/celllinesandhybridomas/tabid/169/default.aspx) for the number and origin of currently available human lung cancer cell lines.
We also reviewed the literature on human cell line contamination, its recognition, and its prevention from the pioneer observations by Nelson-Rees and colleagues (4,5) about HeLa cell contamination of supposedly independently derived human cultures until June 2010. We used our database of cell line DNA “fingerprints” to construct a database containing the reference information for correct identification of our lung cancer cell lines as well as those established by others that we have studied (Supplementary Table 1, available online).
A Brief History of Human Lung Culture—the National Cancer Institute and Hamon Cancer Center Series
We have reviewed the early history of lung cancer culture and of our experience previously (6,7). Lung cancer cells were successfully cultured approximately 25 years after the establishment of HeLa, the first human tumor to be propagated on a long-term basis (8). In 1975, John Minna was appointed the head of the (then) National Cancer Institute (NCI)-Veterans Administration Medical Oncology Branch, an NCI intramural branch that was located at the Veterans Administration Medical Center (Washington, DC) and that had as its clinical research focus the development of lung cancer therapeutics. At this location, we initiated a program to establish lung cancer cell lines as tools for discovery of basic and translational biology of lung cancer. In 1981, the NCI relocated this intramural branch to the National Naval Medical Center (Bethesda, MD) as the NCI-Navy Medical Oncology Branch, where these efforts continued. Several key components allowed these studies to be successful, including intramural NCI funding; the large numbers of lung cancer patients coming to these branches for novel clinical trials who provided biopsy specimens as a source of fresh tumor material; and dedicated NCI, Veterans Administration, and Navy senior staff and fellows who were committed to this multidisciplinary endeavor. Specialists included medical, radiation, and thoracic surgical oncologists, pulmonary physicians, diagnostic radiologists, and of course pathologists. We would especially like to acknowledge the invaluable contributions of Paul Bunn, Daniel Ihde, Mary Matthews, Martin Cohen, Desmond Carney, Bruce Johnson, James Mulshine, Harvey Pass, and Eli Glastein for the clinical investigations and of James Battey, Edward Sausville, Frederick Kaye, Marion Nau, Frank Cuttitta, and James Mitchell for the associated laboratory studies. Finally, there was a team of dedicated laboratory research associates or technicians who derived and maintained these cell lines, including Herbert Oie, Edward Russell, Harvey Sims, and Sylvia Stephenson.
In 1991, Minna and Gazdar left the NCI for positions at the UT Southwestern Medical Center, Dallas, Texas. Establishment of cell cultures continued at this site, with the emphasis on non–small cell lung cancer (NSCLC) lines that had defined molecular characteristics. The lines established at the NCI have the prefix “NCI-” and the lines established at UT Southwestern Medical Center have the prefix “HCC” (Hamon Cancer Center). Although not the subject of this review, the same effort yielded a number of other important human tumor–derived lines that were also obtained from patients coming to the NCI or UT Southwestern Medical Center for clinical studies. These cell lines include NCI-H78 and NCI-H102 T-cell lymphoma–derived lines that were of great importance in the identification of human T-lymphotrophic virus 1 and HIV, NCI-H929 myeloma cells, various colorectal carcinoma lines, and the large HCC series of breast cancer cell lines (9–14). To date, more than 250 lung cancer lines have been established, and more than 200 have been deposited with the ATCC. Currently, the ATCC catalog lists 173 human lung cancer cell lines of which 134 (78%) are from the NCI or HCC series. Although the NCI series were not the first lung cancer lines established, the large numbers and histological origin of lines and their ease of availability (from those who established the lines or the ATCC) led to their wide dissemination to scientific investigators worldwide. A perusal of the literature available from PubMed led to the conclusion that as much as 70%–80% of the literature from North America or Europe with results from lung cancer lines used one or more cell lines from the NCI or HCC series (A. F. Gazdar, unpublished data). Because the full and correct nomenclature of cell lines were not always cited in reports, these percentages are only rough approximations. Because small cell lung cancer (SCLC) tumors are seldom surgically resected, laboratory studies are limited by lack of material, with only small tissue samples from occasional biopsy examinations, malignant aspirates, and rare malignant effusions being available for investigational use. Thus, almost all of our knowledge about the biology and molecular pathogenesis of SCLC has evolved from studies of continuous cell cultures, with somewhat less from xenografts and from formalin-fixed paraffin-embedded tissues. SCLC was first successfully cultured in Japan in 1971 (15) and later cultured in the United States (16,17). A major step forward occurred when we realized that SCLC lacked the ability to adhere to culture dishes (a characteristic that is also manifested by most epithelial tumor lines) and that the SCLC tumor cells grew in vitro as floating cell aggregates or “spheroids.” Another major step forward was suggested to us by the work of Sato and his colleagues (18,19) on the use of chemically defined medium for culturing mammalian cells. Through laborious testing of defined medium and many growth factors, we identified the combination of hydrocortisone, insulin, transferrin, estradiol, and selenium (“HITES” medium) for the selective outgrowth and maintenance of SCLC cells (20,21). Later, this defined medium approach was applied to NSCLC cells and led to the identification of ACL4 medium for the selective propagation of NSCLC lines, particularly those from adenocarcinomas (21). The SCLC cultures retained the cytological characteristics of SCLC cells and expressed the entire program of neuroendocrine differentiation that characterizes SCLC tumors (17,22). Characterization of a large number of SCLC lines revealed a subset of “variant” lines that had lacked the characteristic cytological features of SCLC and had partially lost neuroendocrine differentiation (17,22). Many of the variant lines also demonstrated amplification of a member of the MYC family of oncogenes. Later, many other facets of SCLC biology were gleaned from studies of cell lines, including the role of the basic helix-loop-helix transcription factor human achaete–scute homologue-1 and the role of the Notch signaling pathway in neuroendocrine differentiation (21) and growth (23,24) of SCLC.
Later, as clinical protocols for the therapy of NSCLC tumors were implemented, our attention switched to the culture of specific histological types of NSCLC. The NSCLC cell line A549 was established in 1976 (25) and has been very widely studied since then. The culture of NSCLC cell lines requires a different approach than that of SCLC cell lines. Both primary and metastatic tumor materials are frequently available, but NSCLC is a heterogeneous disease that has three major histological types and multiple histological subtypes (26). Although cells from metastatic tumors, especially from malignant effusions, were relatively easy to culture, cell cultures from primary cancers and small biopsy specimens could also be established at relatively high success rates (21,27). Although all major histological types of NSCLC could be cultured, assay conditions favored adenocarcinoma because of identification of defined medium for the selective culture of this type. The lack of a similar defined medium for culture of squamous lung cancers resulted in a bias favoring culture of adenocarcinomas (20,21).
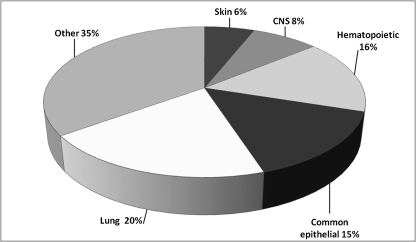
These efforts have resulted in the establishment of more than 200 lung cancer cell lines, of which perhaps 150 are well characterized and widely distributed. These 200 cell lines far exceed the number of cell lines available from the other common human epithelial tumor types. The Wellcome Trust Sanger Institute has accumulated data on almost all cancer cell lines that are readily available to the scientific community. We were able to download data from 784 cell lines, of which 154 (20%) were lung cancer cell lines (Figure 1). By contrast, the total number of breast, colorectal, prostate, gastric, and liver cancer lines was 121 (15%). Thus, of the six most common tumors occurring worldwide, the number of lung cancer lines exceeded the sum of the cell lines from the other five epithelial cancer types. In our laboratory, we maintain fully defined stocks and use approximately 140 lines (100 NSCLC lines and 40 SCLC lines) for routine studies (28).
Figure 1.
Distribution of human tumor cell lines in the Sanger database (www.sanger.ac.uk). Of the 784 cell lines for which data were available, 154 were of lung origin. The common epithelial category group includes breast, colorectal, prostate, gastric, and hepatic cancers. The “other” group includes all other types of human cancers listed in the database. CNS = central nervous system.
Cell Lines as Experimental Systems to Study Cancer Biology and for Translational Research
Initially, it was believed that tumor cells lost their differentiated properties during cell culture. However, it was later shown that this “dedifferentiation” was the result of stromal cell overgrowth and that “true” tumor cell cultures often retained their differentiated properties (29). Thus, cultured tumor cells accurately represent tumor cells in vivo without the complex in vivo environment and are basically populations of pure tumor cells without admixed stromal or inflammatory cells (29). The lack of stromal and inflammatory cells provides both advantages and disadvantages. Their absence results in a pure tumor cell population, greatly aiding tumor cell characterization; however, stromal (and inflammatory) cells play crucial roles in tumor formation, differentiation, growth, and localized and metastatic spread and are essential for angiogenesis. Stromal components may mask the mutations that are present in tumor cells and make detection of gene deletions and accurate estimates of gene copy number difficult. In addition, cell lines are capable of infinite replication and so can provide a limitless source of materials that can be dispersed to laboratories worldwide to allow scientists to directly compare their results from identical study materials. Thus, investigators have a wealth of materials that they can use to study tumor gene functions and interactions, including studies of “driver” or “passenger” genetic changes.
The Crucial Role of Cell Lines in Elucidating the Molecular and Translational Biology of Lung Cancer
The cancer genome is amazingly complex, as manifested by a detailed analysis of lung cancer cell lines (30,31). Cell lines have played crucial roles in the identification and characterization of driver mutations (Table 1). Every single important driver mutation present in lung cancer tumors is also represented in the large bank of lung cancer cell lines that are available for investigation, providing crucial, and in some cases essential, resources for the study of lung cancer pathogenesis.
The relevance of cell lines for biomedical studies is dependent on how closely they resemble the tumors from which they were derived. We have demonstrated previously (32) that the genomic drift during culture life is not as great as commonly believed. During the preparation for this review, we compared the regions of frequent gain and loss in lung adenocarcinoma tumors and cell lines (Figure 2, B) as determined by comparative genomic hybridization (36,37). As shown in Figure 2, many of the regions of frequent genomic gains and losses known to occur in tumors are represented in the cell lines. Many of these sites of gains or losses are the locations of genes known to be important in the pathogenesis of lung cancers. However, in general, the frequencies of gains or losses in cell lines are greater than in the corresponding type of tumor, which may reflect 1) preferential culture of tumors containing copy number changes at locations of crucial oncogenes and tumor suppressor genes, 2) contamination of malignant cells by nonmalignant stromal cells in tumors, or 3) enhanced frequencies of genomic instability reflecting the short doubling times of cultured cells.
Figure 2.
Regions of frequent amplification and deletion in lung adenocarcinoma cell lines and tumors. A) Alteration frequency. Thirty-three lung adenocarcinoma cell lines and 169 tumor samples were analyzed by use of a whole-genome tiling path array for comparative genomic hybridization analysis (33). Genomic imbalances were identified by use of aCGH-Smooth (34), and the frequency of alteration across all samples in each group was determined as described previously (35). The resulting alteration frequencies are displayed across the entire human genome with amplifications on the left and deletions on the right. For each chromosome, the shaded portion represents the p arm. Chromosome location is depicted on the left of the histograms with the p and q arms separated by differential shading (gray and white, respectively). The genomic similarity between the cell lines and tumors is evident by the close association of the plots. B) Commonly reported sites of amplification and deletion that are frequently altered (>15% of samples) in both cell line and tumor samples. Frequency values for chromosome bands were generated as described above. Known oncogenes and tumor suppressor genes that are located in each region are shown.
The role of tumor cell lines in understanding the molecular biology of lung cancer and the ability to translate these findings to clinical applications would have been severely hampered and delayed without the availability of these cell lines (see below). However, for many decades, a debate has been going on as to the relevance of cell lines and in vitro models derived from them for the study of cancer and translational biology. An excellent review of this complex and multifaceted subject has been published recently (38). In general, cell lines maintain the expression of the “hallmarks of cancer” (2), with the exception of angiogenesis (which requires the presence of stromal tissues). Acquisition of the hallmarks of cancer results in genomic instability, with the appearance of numerous genetic and epigenetic changes that characterize the cancer genome (30). They include driver mutations essential for the appearance and maintenance of the malignant phenotype, as well as many passenger mutations that contribute little or nothing. Lung cancer cell lines have contributed greatly to sorting out the driver mutations from the passenger mutations by their use in functionality tests after genetic manipulations (eg, reexpression of tumor suppressor genes or knockdown of candidate oncogenes), which are difficult if not impossible to perform in tumor tissues. Thus, these cell lines have contributed to the identification of TP53 mutations in lung cancer and to our understanding of the relationship between copy number gains, mutations, and mutant allele–specific imbalance in cancers (28,39,40). They also led to the finding that uniparental disomy frequently accompanied mutations of oncogenes, including KRAS and EGFR in lung and other cancers (40).
In 1982, shortly after the establishment of panels of SCLC lines, a specific cytogenetic change—the deletion of much of the short arm of chromosome 3 (41)—was present at very high frequency. Related changes were subsequently found in NSCLC lines (42). Later, molecular techniques that used polymorphic markers identified at least three regions of loss in chromosome 3p. Multiple putative tumor suppressor genes have been identified in these regions, including RASSF1A, FUS1, and FHIT. Some of these genes, especially RASSF1A, may act as tumor suppressor genes in multiple tumor types. Thus, the initial cytogenetic observations of SCLC lines have led to extensive studies of many tumor types. Another early observation with SCLC lines was the finding of MYC amplification in a subset of SCLC lines (43) that had variant features, including atypical morphology and partial loss of neuroendocrine properties (44). The observations in cell lines also led to the identification of the LMYC gene, to an understanding of the role of NMYC (45), and to the finding that amplification of MYC family members is a frequent occurrence in SCLC tumors (46), with MYC overexpression in most lung cancers.
Determining sites of frequent allelic loss in lung cancer cell lines, initially by use of relatively simple techniques (such as a panel of polymorphic markers) and later by use of global high-density comparative genomic hybridization or single-nucleotide polymorphism arrays, has identified many sites of recurrent gain or loss in the lung cancer genome (47–49). As shown in Figure 2, sites of gain or loss have considerable overlap between cell lines and tumors, although the frequencies in cell lines usually exceed those of tumors. Lung cancer cell lines were used to demonstrate an important role for RB in the pathogenesis of SCLC (50). Although both RB and CDKN2A act as cell cycle checkpoints, the latter is inactivated in many tumors, whereas inactivating point mutations in RB are largely limited to SCLC and bladder cancers. The concept that there was a mutually exclusive RB–cyclin–CDKN2A tumor suppressor pathway, which could be inactivated by mutational or epigenetic alterations in a wide range of human cancers, was initiated by a study of lung cancer cell lines (51,52). The gene LKB1 (also known as STK11) is frequently mutated and inactivated in NSCLC cells (53). This gene is located at a site of frequent loss in lung cancer cell lines that was identified years ago (47). Determining the sites of frequent gain in cell lines has identified many genes of interest for lung cancer pathogenesis (35,48). Of particular interest was the finding that the gene TITF1, which is a master transcription factor for peripheral airway differentiation and frequently amplified in lung cancer tumors and cell lines (Figure 2), functioned as a lineage-dependent oncogene (ie, lineage-specific genes that function as oncogenes) (36,54).
A relatively new finding in lung cancer with major clinical and biological implications has been the discovery of activating mutations in the EGFR kinase domain. Cell line studies were an integral part of an initial report (55), demonstrating the relationship between mutations and sensitivity to tyrosine kinase inhibitors. Cell lines have made important contributions to virtually all of the subsequent important biological characterization of the EGFR mutations, including intrinsic and acquired resistance (56,57) and the cancer cell’s “addiction” to the product of activating oncogenes, such as EGFR (58).
Global Approaches That Use Cell Lines
Until recently, identification of specific genetic, epigenetic, and expression changes in tumor cells required laborious examination at the single-gene level. With the advent of very high-throughput DNA sequencing (59) and whole-genome platforms for querying the transcriptome, the methylome, microRNAs, and copy number changes, the landscape of the cancer genome is rapidly evolving (30). Although currently limited to a modest number of studies, these approaches have been used in SCLC and NSCLC cell lines to identify copy number changes (36,37,48,49,60), to discover novel methylated genes (61), for mutation profiling (62), to identify molecular signatures of oncogene addiction (39) and the effect of tobacco smoke on pathway analyses (63), and to predict drug sensitivity and investigate its mechanisms (64,65).
Pathway Analysis and Prediction of Drug Sensitivity
Genome-wide profiling of gene expression and identification of copy number alterations have confirmed the major differences between the two primary subdivisions of lung cancers, SCLC and NSCLC (35). This study identified more than 100 genes that are differentially expressed in SCLC and NSCLC cell lines and demonstrated the different cell cycle pathways involved in their pathogenesis. In SCLC cells, increased expression of MRP5, activation of Wnt pathway inhibitors, and increased expression of p38 mitogen–activating protein kinase–activating genes occur; and in NSCLC cells, decreased expression of CDKN2A and increased expression of MAPK9 and EGFR have been described. This information highlights the need for differential molecular target selection in the treatment of lung cancers.
Use of gene expression signatures for signaling pathway analysis has been suggested as an improved method for rational therapeutic drug selection (2). We have performed a detailed study of the EGFR signaling pathway in a large panel of lung cancer cell lines (with and without acquired EGFR mutations) and correlated the findings with response to tyrosine kinase inhibitors (28). Mutations in seven pathway-related genes (EGFR, HER2, HER3, HER4, KRAS, BRAF, and PIK3CA) were detected at frequencies similar to those described in resected tumors. The resistance or sensitivity to tyrosine kinase inhibitors of all cell lines, without exception, could be explained by their molecular signatures. It is of great interest that cell lines NCI-H1795 and NCI-H820 had dual EGFR mutations including an activating mutation and the resistance-associated T790M mutation and were derived from tumors before EGFR tyrosine kinase inhibitors were developed, indicating that the mutation probably is present in a small percentage of tumor cells at diagnosis and is preferentially selected by targeted therapy. Our studies as a group confirmed and extended the clinical observations that 1) EGFR mutations and copy number gains of EGFR and HER2 were independent factors related to tyrosine kinase inhibitor sensitivity, in descending order of importance, and 2) KRAS mutations were associated with increased in vitro resistance. For all 45 cell lines tested, a genetic basis for sensitivity or resistance to tyrosine kinase inhibitor therapy was evident. Thus, after decades in culture, the driver mutations, secondary resistance-associated mutations and copy number gains, and their patterns of tyrosine kinase inhibitor response are retained in cell lines.
The NCI60 Tumor Cell Line Panel for Anticancer Drug Screening
In the late 1980s, the failure of mouse screening model systems to identify useful anticancer compounds led to a fundamental change in testing from the mouse model systems to a cell line–based panel. The NCI assembled a panel of 60 human cancer cell lines (from the approximately 1500 lines then currently available) that represented nine human cancer types for the screening of drugs for anticancer activities (66). The panel includes eight NSCLC cell lines, four of them from the NCI series. However, because most of the classic SCLC lines are nonadherent (making them technically more difficult to use in screening systems), two adherent SCLC lines from another source were selected. These lines lacked neurosecretory granules and other neuroendocrine properties and thus appeared unsuitable for inclusion, and they have been assigned to the “additional cell lines” category. Because the panel has been widely distributed and used (it has been tested for sensitivities to >100 000 compounds), the panel members have received considerable scrutiny as to their provenances (67) and for suitable representation of their respective cancer types. A massive study was undertaken, directly comparing the global expression patterns of panel members with their respective primary tumor types (68). Most cell lines tested (51 [86%] of the 59 lines) represented their presumed tumor types, including six of the NSCLC lines. One of the two discordant lines had its subtype attribution corrected, although the original subtype had been the subject of discrepancy. Thus, most members of the panel had retained their characteristic cell type expression patterns after many passages and culture years.
Pleural Mesotheliomas
Pleural mesotheliomas are asbestos exposure-related tumors that arise from the mesothelial cells of the pleura. Consequently, they are not, in the strict sense, lung cancers. However, because of their close anatomical relationship to the lung, we briefly discuss mesothelioma cell lines. Although continuous mesothelioma lines were reported more than 34 years ago (69), the NCI group, under the leadership of Harvey Pass, undertook a systemic approach to culture such cells starting in 1990 (70). A similar approach was undertaken by other NCI scientists (71). Mesothelial cells are biphasic, and cell lines recapitulate the two major morphological forms of mesothelioma, namely epitheloid and sarcomatoid (72). These cell lines have been used to identify and to study oncogenes, tumor suppressor genes, tumor biomarkers, global gene expression, and methylation profiles and to identify targets for novel therapies.
NF2 is a tumor suppressor gene in pleural mesothelioma whose product, merlin, provides regulated linkage between membrane-associated proteins and the actin cytoskeleton (73) and inhibits cell proliferation (74). Inactivating mutations in NF2 were first demonstrated to be frequent in mesothelioma (but not in lung cancer) cell lines (75). Cell lines were also used to demonstrate that merlin exerts its antiproliferative effect by repression of p21-activated kinase-induced cyclin D1 expression (74). Osteopontin (the product of the SPP1 gene) is an inflammatory cytokine that is used as a diagnostic marker in patients with asbestos-induced malignant mesothelioma. Mesothelioma cell lines have played an important role in understanding the role of osteopontin and its isoforms in mesothelioma biology (76). Mesothelioma lines have also proved important for the testing of targeted and conventional therapies and as models for gene therapy (77). An unexpected complication (and possible biohazard) of xenotransplantation has been contamination of the tumor cells with murine xenotropic retrovirus, especially contamination of SCLC cells, including the widely distributed NCI-N417 cell line [A. F. Gazdar and Y.-A. Zhang, UT Southwestern Medical Center, unpublished data and (78,79)].
Xenografts From Lung Cancer Cell Lines
The major limitations of tumor cell lines are their lack of stromal, vascular, and inflammatory components (1). These deficiencies can be partially overcome by xenograft transplantation (of tumor fragments or cell line suspensions) into immunocompromised rodents, usually mice (80,81). However, xenografts also have certain limitations, including the presence of stromal components of heterologous origin that support the human tumor cells. In addition, most human xenografts, even from highly metastatic tumors such as SCLC, fail to metastasize. Attempts to develop metastatic xenografts have included inoculation via alternate routes, including intravenous, intracranial, intraperitoneal, subrenal capsule, or orthotopic sites. By use of various techniques, metastatic models from xenografted human lung cancer cell lines have been developed (82–84) and intracranial inoculation of SCLC results in a model for leptomeningeal spread (85). By combining reporter systems that tag specific cellular processes, tumor growth, metastatic spread, and molecular events can now be studied in vivo (86,87). Cell lines are ideal models for such studies because bioluminescence reporter genes can readily be inserted in vivo before xenografting and then are easily monitored.
Among the many uses of xenografts generated with human cell lines are determination of tumorigenicity and histological appearance, the identification of cancer stem cells, and the study of metabolic functions and gene interactions. Perhaps the most widely used translational application of xenograft tumors has been the in vivo testing of conventional or targeted therapies (88–90). A recent example exemplifies how cell lines could be used to identify and to test novel therapeutic compounds (64). Integrated genomic profiling was used to demonstrate that the genomes of a large panel of human NSCLC cell lines were highly representative of those of primary NSCLC tumors and that mutations conferred enhanced heat shock protein 90 dependency; this finding was then validated in mice with KRAS-driven lung adenocarcinoma because when these mice were treated with an heat shock protein 90 inhibitor, their tumors regressed. Thus, genomically annotated collections of cell lines may help translate cancer genomics information into clinical practice by defining critical pathway dependencies that are amenable to therapeutic inhibition.
The Problems of Cell Line Contamination With Mycoplasma, Xenotropic Viruses, or Other Cell Lines
Contamination of long-term cultured cells represents a major problem and has cast doubt on the results of many published medical reports (see below). Two major forms of contamination account for most of the problems—mycoplasma contamination and cell line contamination or mistaken attribution (see below). Mycoplasma contamination is common, and the usual sources are human organisms from contamination from the oral cavity or bovine strains from inadequately tested or treated serum used for cell culture. Cross-culture contamination during laboratory handling is also common. Various tests are routinely used to detect mycoplasma, including polymerase chain reaction–based, fluorescence, and enzymatic assays (91). Although investigators have used several methods for eradication (91), all methods usually result in temporary (but often long-term) suppression rather than permanent cure and so provide a false sense of security. Mycoplasma contamination may result in ragged cell appearance or slow growth, but often it has no visible effects. The possibility exists that results of genomic or functional assays may be influenced by mycoplasma contamination and the difficulties of true eradication require constant surveillance and, in our opinion, the contaminated cultures should be discarded or isolated. Contamination of lung and other cell cultures with murine xenotropic retrovirus strains has already been discussed. Contamination with other cells and misidentification of cell lines is discussed below.
Screening for Cross-Contamination of Tumor Cell Lines and the Importance of Cell Line Provenance
Successful long-term culture of human cells was achieved less than 60 years ago when George Gey established the HeLa cell line from an African American woman with cervical cancer (8). Within 25 years, human cell culture entered a renaissance, with the successful culture of many types of cancers. However, the robustly growing HeLa cells had been distributed to numerous laboratories worldwide, and in the 1970s, Nelson-Rees et al. (5) and others (8) noted that many human lines of diverse origins and sources contained isoenzyme and cytogenetic characteristics of HeLa cells (8). The obvious conclusion was that HeLa cells had somehow contaminated many (perhaps the majority) of human cell lines reported to be of independent origin. For several years, many scientists, cell bank repositories, and journal editors refused to accept this conclusion, further confounding and propagating the problem. However, the introduction of molecular methods for DNA fingerprinting has removed all reasonable doubt that cell line contamination is an enormous problem in scientific investigations and that approximately 20% of cell cultures show evidence of intra- or interspecies contamination (8). The devastating effects of contamination were recently highlighted when an analysis of 40 thyroid cell lines revealed that more than one-third showed evidence of redundancy or misidentification, effectively nullifying much of the literature regarding in vitro studies on thyroid cancer (92,93) and (indirectly) questioning the relevance of many funded grants that relied on cultures with incorrect provenances. More recently, three of the most commonly used esophageal adenocarcinoma cell lines were found to have been misidentified and were actually derived from other forms of human cancers, including lung cancer, resulting in doubts about the relevance of approximately 100 published reports and of clinical trials and patents that were based on the reports (94). With cell line contamination noted more than 30 years ago and molecular “fingerprinting” methods available for a decade, it is difficult to understand why these important problems were not identified and corrected earlier. A recent study (95) that fingerprinted the NCI60 panel of cell lines confirmed previous reports that several sets of cell lines had common fingerprints. Fortunately, all lung cancer cell lines in that panel proved to be from individual isolates.
We as well as other centers doing large-scale culturing invariably encounter the occasional contaminant. The key to minimizing the problem is to identify the offending line through a surveillance program. The use of short tandem repeat analysis of specific loci in the human genome has become the standard for contamination detection (96). An inexpensive, easy to use, and reliable kit, the PowerPlex 1.2 System (http://www.promega.com/applications/cellularanalysis/cellauthentication.htm), is available from Promega Corporation (Madison, WI) that combines analysis of nine loci (including amelogenin for sex identification of the donor). The ATCC (http://www.atcc.org/CulturesandProducts/CellBiology/STRProfileDatabase/tabid/174/Default.aspx) and our center maintain databases of the DNA fingerprints of several hundred cell lines, providing evidence of their independent origin from human tumors. Such databases can also be used to identify the true origin of contaminated lines. Elimination (or minimization) of the enormous contamination problem requires several steps: 1) awareness of the problem; 2) a constant ongoing surveillance program; 3) obtaining cell lines from the original source or a responsible cell bank rather than from a convenient source or investigator who provides materials of untested provenance; 4) large publicly available databases of the fingerprints of all human cell lines that are used in scientific research and can be used to validate the origin of a cell line; 5) editors of journals and granting authorities that require evidence of the true provenance of cell lines that are used in reports submitted for publication or in grant applications submitted to funding agencies, respectively (97,98). Fortunately, the Journal has included the following statement in its instructions to authors: “Authors of provisionally accepted manuscripts that use cell lines should state the methods used to authenticate any cell lines used in their studies and should give the date of the last authentication.” As our contribution toward attributing the correct provenance to lung cancer cell lines, we include the Promega fingerprint results of nearly 900 cell lines (40% of which are from the lung) in Supplementary Table 1 (available online). These data include reference fingerprints from the ATCC as well as from our laboratory. We hope that these fingerprints will be regarded as the gold standard for identification and authentication of the NCI and HCC series of lung cancer cell lines and that widespread use of this information will greatly diminish the use of contaminated cell lines in the future.
Table 1.
The role of cell lines in some important discoveries regarding lung cancer pathogenesis*
| Discovery or finding | Reference |
| Deletion of chromosome 3p and identification of inactivated suppressor genes | (102–105) |
| Frequent MYC family amplification in SCLC | (42,106) |
| Discovery of LMYC gene | (44) |
| Mutations and inactivation of the RB gene in SCLC | (49) |
| The spectrum of TP53 gene mutations in lung cancer | (107) |
| Identification of the RB–CDKN2 pathway in lung cancer | (50) |
| Frequent silencing of RASSF1A by methylation in lung cancers | (108,109) |
| BRAF mutations in lung and other tumors | (110,111) |
| Resistance mechanisms in EGFR mutant cells | (112–114) |
| Identification of sites of frequent DNA amplification in lung cancers | (47) |
| PIK3CA mutations and amplifications in squamous cell cancers | (115) |
| Identification of TITF1 as a lineage-specific oncogene for lung adenocarcinoma | (36,54) |
| EGFR mutations in NSCLC and response to tyrosine kinase inhibitors | (28) |
| Role of microRNAs in lung cancer and regulation of RAS and EGFR protein | (116–118) |
| Expression signature of KRAS “addiction” | (39) |
| Frequent mutant allele-specific imbalance and uniparental disomy of oncogenes in NSCLCs | (28,40) |
This table is not meant to be a comprehensive review of all important discoveries in which cell lines were involved but to highlight some relevant findings. NSCLC = non–small cell lung cancer; SCLC = small cell lung cancer.
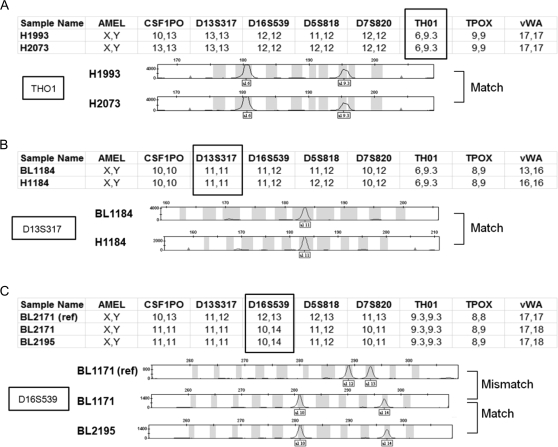
Reference fingerprints are determined as follows: All ATCC fingerprints are considered gold standards. Cell lines that were obtained from sources other than the ATCC are considered gold standards when they have been fingerprinted at least twice from two different sources and produced consistent fingerprint results. Cell lines whose identity is ambiguous are excluded from this list. Any new fingerprint is compared with these references and a probable match is called when at least seven of the nine markers are identical (Figure 3). So far, of the 1800 fingerprint assays that resulted in a correct cell line match, 70% showed complete nine of the nine marker alignment, 21% showed alignment of eight of the nine markers, and 9% showed alignment of seven of the nine markers. Individual markers have comparable frequencies of mismatch (4% on average). These marker mismatches are presumably caused by a low frequency of unfaithful replication of short tandem repeats that may occur during extended cell culture. Alternatively, they could be caused by polymerase chain reaction artifacts. Interestingly, the amelogenin marker for sex identification of the donor shows a 53% mistyping of cell lines from male donors (in both reference and nonreference fingerprints) compared with a baseline 4% mistyping of cell lines from female donors. This result indicates that the Y chromosome may be lost at high frequency in cancer cell lines (or tumors) from male donors, and therefore, the presence or absence of the male-specific form of amelogenin (AMELY) in tumor cells may not be a reliable test for identification of cell lines from male donors. Overall, approximately 10% of the cell line samples (from internal and external sources) that we have fingerprinted have been incorrectly identified. Institutions that do not frequently monitor for contamination may have a higher proportion of such lines in their collections. Although lung cancer cell lines have not avoided the contamination problem, claims of incorrect attribution that have been based solely on sex chromosome analyses (99) cannot be accepted without further evidence. Thus, we retain amelogenin as a marker for cell line identification, whether or not the reference standard results match the sex of the cell line donor.
Figure 3.
Verification of the identity of lung cancer cell lines. Representative electropherograms from PowerPlex fingerprint analyses are shown that verify the identity of cell lines from different sources. For sample analysis, the allele sizes of the nine polymorphic markers analyzed are displayed below the electropherograms. The electropherogram results of one marker (boxed and also identified to the left) are displayed. When at least seven of the nine markers matched, the cell lines are assumed to be identical. A) Match of cell lines H1993 and H2073. Both cell lines were isolated from the same patient and, thus, this represents a match. B) Match of B lymphoblastoid cell line BL1184 and non–small cell lung cancer cell line H1184. Both cell lines were isolated from the same patient. C) Incorrect attribution. BL1171 cells (middle) match BL2195 cells as a result of cell line cross-contamination or mislabeling. Note that full designations of the cell lines include the prefix NCI (eg, NCI-BL1171).
A listing of known contaminated human cell cultures has recently been published and is a useful starting source for identification of such contaminations (100). The report also discusses DNA barcoding, a taxonomic method that uses amplification of a segment of mitochondrial DNA for species identification.
Global Approaches That Use Cell Lines
Until recently, identification of individual genetic, epigenetic, and expression changes in tumor cells required laborious examination at the single-gene level. With the advent of very high-throughput DNA sequencing (59) and whole-genome platforms for querying the transcriptome, the methylome, microRNAs, and copy number changes, the landscape of the cancer genome is rapidly being elucidated (30). Although currently limited to a modest number of studies, these approaches have been used in SCLC and NSCLC cell lines to identify copy number changes (36,37,48,49,60), to discover novel methylated genes (61), to do mutation profiling (62), to identify molecular signatures of oncogene addiction (39) and the effects of tobacco smoke on pathway analyses (63), to predict drug sensitivity, and to investigate its mechanisms (64,65).
The power of such a global approach to studying the human genome was demonstrated recently by the resequencing of an SCLC cell line, NCI-H209, and its corresponding B lymphoblastoid culture (31,101). A total of 132 somatic substitutions in coding exons were identified, many of which bore evidence of being induced by tobacco carcinogens. In addition, there was evidence of the imprint of a novel and more general form of expression-linked DNA repair through which mutation frequency is reduced on both strands of highly expressed genes.
The Use of Cell Lines by the Biomedical Community
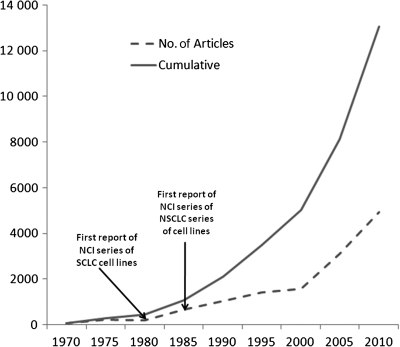
The ultimate evidence of the value of cell lines is how widely used they are by the biomedical community. We used the medical search heading terms as described above to search the PubMed database and also to search within the EndNote program because this technique yielded results that were more focused, more restricted, and more relevant, than the direct search of PubMed. As of June 2010, a total of more than 9700 citations were available for human lung cancer cell lines (Figure 4). With approximately 1000 new citations being added per year, the number of new citations for human lung cancer cell lines has been rising exponentially. Lung cancer cell lines have been widely distributed to the scientific community, which in turn has used them in a highly productive manner, and their utilization continues to increase.
Figure 4.
Number of citations for human lung cancer cell lines. Data were obtained from a search of the PubMed database from within the EndNote program by use of the medical search heading terms “lung neoplasms”; “cell line, tumor”; and “humans.” The cumulative number of citations and the number of citations per 5 years are shown. The results are from a search performed in June 2010. NCI = National Cancer Institute; NSCLC = non–small cell lung cancer; SCLC = small cell lung cancer.
Future Directions
Although studies of cancer lines have made major contributions to our understanding of lung cancer biology and pathogenesis and to translational research, much remains to be done concerning several issues. 1) The lack of sufficient numbers of lung cancer cell lines that have been isolated from tumors with certain histological types, especially squamous cell carcinomas and lines from lifetime never-smokers, needs to be addressed. 2) There needs to be universal agreement among investigators and journal editors that accurate cell line authentication is an absolute prerequisite for publication. 3) A comprehensive panel of fully characterized and certified lines from repositories needs to be made readily available worldwide at a reasonable cost. Currently, many countries place limitations on the international importing or exporting of lines. Such practices have prevented lines with specific geographic associated genetic changes (such as EGFR mutations) from being freely available to scientists outside Asia. Of the 10 cell lines carrying EGFR mutations that are currently available in the United States (28), only one originated in East Asia and that line has a resistant mycoplasma infection. Repositories should offer not only living or cryopreserved cultures but also cell products, such as nucleic acids and cell pellets for proteomic research. The already high and increasing cost of cultures from major repositories is limiting the use of these cultures in many laboratories, resulting in distribution from secondary unauthorized sources, which contributes to the contamination problem. Funding should be made available from agencies to reduce the costs of cell line materials. Awarded grants should contain adequate funding so that the investigators can obtain cell line samples from reputable major repositories for study, instead of relying on third-party sources. 4) The complete growth factor requirements for all lung cancer types should be determined so that all cultures can be maintained in fully defined medium. 5) Methods need to be available to study the multistage pathogenesis of lung cancer in vitro and in vivo, including cell lines from preinvasive lesions and other in vitro model systems. 6) Improved techniques should be developed to culture slow-growing well-differentiated tumors, including those with low malignant potential. The development of such techniques may also permit rapid culture of most lung cancers, leading to selection of individualized therapies and other translational applications. 7) Improved systems and methods need to be developed for the in vitro study of tumor cell interactions with stroma and the immune system and also for angiogenesis. Such techniques may permit cell culture systems to more closely model in vivo tumor growth, invasiveness, and metastatic potential. 8) Methods for the identification, isolation, and manipulation of the stem cell component of cultured tumor cells are needed. 9) Complete characterization at a global level, and for critical individual genes at an individual gene level, is needed to be completed for a comprehensive number and variety of cell lines. Such studies should include the resequencing of the complete genome or a large panel of cell lines. 10) Public databases need to be developed to house the vast data from the new global approaches that are currently available and expected in the future. Such databases would need to be updated periodically. 11) Methods need to be developed so that the information available from multiple platforms and tools can be seamlessly integrated so that investigators without specialized training or knowledge can query the integrated data. Some of these issues are currently being addressed, and so we expect that some shortcomings will be at least partially corrected in the not too distant future.
Concluding Remarks
Cell cultures offer many intrinsic advantages, and some disadvantages, for the investigation of lung cancer pathogenesis and for the identification and testing of novel therapeutic approaches. Some of the advantages are unique for cell lines, enhancing their value. The high numbers and varieties of lung cancer cultures (more than the total number of cultures from other common epithelial cell lines combined) and their ready availability to the scientific community have resulted in a very large body of literature (approximately 9000 citations), covering virtually every aspect of lung cancer research. In particular, the contributions to tumor suppressor and oncogene discovery and understanding have been noteworthy. Cell lines have proved useful in elucidating the important signaling pathways in lung cancers and for therapeutic applications resulting from these studies. Many of the NCI and HCC series of lung cancer lines have been licensed to pharmacological and other biomedical companies, demonstrating their usefulness for therapeutic and other translational applications.
A major and universal problem with the use of cell cultures remains contamination and wrongful attribution. Minimization of the problem requires constant vigilance, acquisition of defined cell lines from recognized repositories or trusted sources, and the requirement that journals publish only research articles that use fully defined cell lines. To aid the scientific community with this problem, we are supplying a large DNA fingerprint database of lung cancer and other cultures in Supplementary Table 1 (available online). We hope that widespread usage of this gold standard will result both in increased awareness of the problem and in greater use of identification tests.
Despite the large number of lung cell lines available and their widespread contributions to a variety of biomedical applications, much remains to be accomplished until the full potential of lung cancer cell lines and their applications can be achieved. These goals include availability of panels of fully defined lines from reputable sources at reasonable costs and public databases of the vast amounts of data that have been and are being generated by the increasing use of global approaches.
Supplementary Data
Supplementary data can be found at http://www.jnci.oxfordjournals.org/.
Funding
Specialized Program of Research Excellence in Lung Cancer (P50CA70907); Early Detection Research Network, National Cancer Institute (Bethesda, MD) (U01CA084971); Canary Foundation (Palo Alto, CA).
Supplementary Material
Footnotes
The authors had full responsibility for the design of the study; the collection, the analysis, and interpretation of the data; the decision to submit the manuscript for publication; and the writing of the manuscript.
Drs Gazdar and Minna receive royalties from the National Cancer Institute and in the future may receive royalties from UT Southwestern from the licensing of cell lines, according to the policies of these two institutions.
References
- 1.Gazdar AF, Gao B, Minna JD. Lung cancer cell lines: useless artifacts or invaluable tools for medical science? Lung Cancer. 2010;68(3):309–318. doi: 10.1016/j.lungcan.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Weed D. Methodologic guidelines for review papers. J Natl Cancer Inst. 1997;89(1):6–7. [Google Scholar]
- 4.Lucey BP, Nelson-Rees WA, Hutchins GM. Henrietta Lacks, HeLa cells, and cell culture contamination. Arch Pathol Lab Med. 2009;133(9):1463–1467. doi: 10.5858/133.9.1463. [DOI] [PubMed] [Google Scholar]
- 5.Nelson-Rees WA, Zhdanov VM, Hawthorne PK, Flandermeyer RR. HeLa-like marker chromosomes and type-A variant glucose-6-phosphate dehydrogenase isoenzyme in human cell cultures producing Mason-Pfizer monkey virus-like particles. J Natl Cancer Inst. 1974;53(3):751–757. doi: 10.1093/jnci/53.3.751. [DOI] [PubMed] [Google Scholar]
- 6.Gazdar A. Cell culture of lung cancers. In: Hay RJ, Park J-G, Gazdar A, editors. Atlas of Human Tumor Cell Lines. New York: Academic Press; 1994. [Google Scholar]
- 7.Gazdar AF, Minna JD. NCI series of cell lines: an historical perspective. J Cell Biochem Suppl. 1996;suppl 24:1–11. doi: 10.1002/jcb.240630502. [DOI] [PubMed] [Google Scholar]
- 8.Masters JR. HeLa cells 50 years on: the good, the bad and the ugly. Nat Rev Cancer. 2002;2(4):315–319. doi: 10.1038/nrc775. [DOI] [PubMed] [Google Scholar]
- 9.Rubinstein E. The untold story of HUT78. Science. 1990;248(4962):1499–1507. doi: 10.1126/science.2193399. [DOI] [PubMed] [Google Scholar]
- 10.Baylin SB, Abeloff MD, Goodwin G, Carney DN, Gazdar AF. Activities of L-dopa decarboxylase and diamine oxidase (histaminase) in human lung cancers and decarboxylase as a marker for small (oat) cell cancer in cell culture. Cancer Res. 1980;40(6):1990–1994. [PubMed] [Google Scholar]
- 11.Gazdar AF, Oie HK, Kirsch IR, Hollis GF. Establishment and characterization of a human plasma cell myeloma culture having a rearranged cellular myc proto-oncogene. Blood. 1986;67(6):1542–1549. [PubMed] [Google Scholar]
- 12.Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci U S A. 1980;77(12):7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mann DL, O'Brien SJ, Gilbert DA, et al. Origin of the HIV-susceptible human CD4+ cell line H9. AIDS Res Hum Retroviruses. 1989;5(3):253–255. doi: 10.1089/aid.1989.5.253. [DOI] [PubMed] [Google Scholar]
- 14.Gazdar AF, Kurvari V, Virmani A, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int J Cancer. 1998;78(6):766–774. doi: 10.1002/(sici)1097-0215(19981209)78:6<766::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 15.Oboshi S, Tsugawa S, Seido T, Shimosato Y, Koide T. A new floating cell line derived from human pulmonary carcinoma of oat cell type. Gann. 1971;62(6):505–514. [PubMed] [Google Scholar]
- 16.Pettengill OS, Faulkner CS, Wurster-Hill DH, et al. Isolation and characterization of a hormone-producing cell line from human small cell anaplastic carcinoma of the lung. J Natl Cancer Inst. 1977;58(3):511–518. doi: 10.1093/jnci/58.3.511. [DOI] [PubMed] [Google Scholar]
- 17.Gazdar AF, Carney DN, Russell EK, et al. Establishment of continuous, clonable cultures of small-cell carcinoma of lung which have amine precursor uptake and decarboxylation cell properties. Cancer Res. 1980;40(10):3502–3507. [PubMed] [Google Scholar]
- 18.Barnes D, van der Bosch J, Masui H, Miyazaki K, Sato G. The culture of human tumor cells in serum-free medium. Methods Enzymol. 1981;79(pt 8):368–391. doi: 10.1016/s0076-6879(81)79050-5. [DOI] [PubMed] [Google Scholar]
- 19.Bottenstein J, Hayashi I, Hutchings S, et al. The growth of cells in serum-free hormone-supplemented media. Methods Enzymol. 1979;58:94–109. doi: 10.1016/s0076-6879(79)58127-0. [DOI] [PubMed] [Google Scholar]
- 20.Simms E, Gazdar AF, Abrams PG, Minna JD. Growth of human small cell (oat cell) carcinoma of the lung in serum-free growth factor-supplemented medium. Cancer Res. 1980;40(12):4356–4363. [PubMed] [Google Scholar]
- 21.Oie HK, Russell EK, Carney DN, Gazdar AF. Cell culture methods for the establishment of the NCI series of lung cancer cell lines. J Cell Biochem. 1996;suppl 24:24–31. doi: 10.1002/jcb.240630504. [DOI] [PubMed] [Google Scholar]
- 22.Carney DN, Gazdar AF, Bepler G, et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res. 1985;45(6):2913–2923. [PubMed] [Google Scholar]
- 23.Jiang T, Collins BJ, Jin N, et al. Achaete-scute complex homologue 1 regulates tumor-initiating capacity in human small cell lung cancer. Cancer Res. 2009;69(3):845–854. doi: 10.1158/0008-5472.CAN-08-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sriuranpong V, Borges MW, Ravi RK, et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res. 2001;61(7):3200–3205. [PubMed] [Google Scholar]
- 25.Lieber M, Smith B, Szakal A, Nelson-Rees W, Todaro G. A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int J Cancer. 1976;17(1):62–70. doi: 10.1002/ijc.2910170110. [DOI] [PubMed] [Google Scholar]
- 26.Brambilla E, Travis WD, Colby TV, Corrin B, Shimosato Y. The new World Health Organization classification of lung tumours. Eur Respir J. 2001;18(6):1059–1068. doi: 10.1183/09031936.01.00275301. [DOI] [PubMed] [Google Scholar]
- 27.Oie HK, Gazdar AF. Initiation of cell cultures from lung tumor biopsies. In: Pass HI, Mitchell JB, Johnson dH, Turrisi AT, Minna JD, editors. Lung Cancer: Principles and Practice. 2nd ed. Philadelphia, PA: Lippincott, Williams and Wilkins; 2000. pp. 337–346. [Google Scholar]
- 28.Gandhi J, Zhang J, Xie Y, et al. Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS One. 2009;4(2) doi: 10.1371/journal.pone.0004576. e4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato G. Tissue culture: the unrealized potential. Cytotechnology. 2008;57(2):111–114. doi: 10.1007/s10616-007-9109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wistuba, Bryant D, Behrens C, et al. Comparison of features of human lung cancer cell lines and their corresponding tumors. Clin Cancer Res. 1999;5(5):991–1000. [PubMed] [Google Scholar]
- 33.Ishkanian AS, Malloff CA, Watson SK, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004;36(3):299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 34.Jong K, Marchiori E, Meijer G, Vaart AV, Ylstra B. Breakpoint identification and smoothing of array comparative genomic hybridization data. Bioinformatics. 2004;20(18):3636–3637. doi: 10.1093/bioinformatics/bth355. [DOI] [PubMed] [Google Scholar]
- 35.Coe BP, Lockwood WW, Girard L, et al. Differential disruption of cell cycle pathways in small cell and non-small cell lung cancer. Br J Cancer. 2006;94(12):1927–1935. doi: 10.1038/sj.bjc.6603167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwei KA, Kim YH, Girard L, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27:3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garnis C, Lockwood WW, Vucic E, et al. High resolution analysis of non-small cell lung cancer cell lines by whole genome tiling path array CGH. Int J Cancer. 2006;118(6):1556–1564. doi: 10.1002/ijc.21491. [DOI] [PubMed] [Google Scholar]
- 38.van Staveren WC, Solis DY, Hebrant A, Detours V, Dumont JE, Maenhaut C. Human cancer cell lines: experimental models for cancer cells in situ? For cancer stem cells? Biochim Biophys Acta. 2009;1795(2):92–103. doi: 10.1016/j.bbcan.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Singh A, Greninger P, Rhodes D, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15(6):489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soh J, Okumura N, Lockwood WW, et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One. 2009;4(10) doi: 10.1371/journal.pone.0007464. e7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whang-Peng J, Bunn PA, Jr, Kao-Shan CS, et al. A nonrandom chromosomal abnormality, del 3p(14-23), in human small cell lung cancer (SCLC) Cancer Genet Cytogenet. 1982;6(2):119–134. doi: 10.1016/0165-4608(82)90077-2. [DOI] [PubMed] [Google Scholar]
- 42.Whang-Peng J, Lee EC. Cytogenetics of human small cell lung cancer. Recent results in cancer research. Fortschr Krebsforsch. 1985;97:37–46. doi: 10.1007/978-3-642-82372-5_4. [DOI] [PubMed] [Google Scholar]
- 43.Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983;306(5939):194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- 44.Gazdar AF, Carney DN, Nau MM, Minna JD. Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res. 1985;45(6):2924–2930. [PubMed] [Google Scholar]
- 45.Nau MM, Brooks BJ, Battey J, et al. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature. 1985;318(6041):69–73. doi: 10.1038/318069a0. [DOI] [PubMed] [Google Scholar]
- 46.Johnson BE, Russell EK, Simmons AM, et al. MYC family DNA amplification in 126 tumor cell lines from patients with small cell lung cancer. J Cell Biochem Suppl. 1996;suppl 24:210–217. doi: 10.1002/jcb.240630516. [DOI] [PubMed] [Google Scholar]
- 47.Virmani AK, Fong KM, Kodagoda D, et al. Allelotyping demonstrates common and distinct patterns of chromosomal loss in human lung cancer types. Genes Chromosomes Cancer. 1998;21(4):308–319. doi: 10.1002/(sici)1098-2264(199804)21:4<308::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 48.Lockwood WW, Chari R, Coe BP, et al. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene. 2008;27(33):4615–4624. doi: 10.1038/onc.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450(7171):893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harbour JW, Lai SL, Whang-Peng J, Gazdar AF, Minna JD, Kaye FJ. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science. 1988;241(4863):353–357. doi: 10.1126/science.2838909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otterson GA, Kratzke RA, Coxon A, Kim YW, Kaye FJ. Absence of p16INK4 protein is restricted to the subset of lung cancer lines that retains wildtype RB. Oncogene. 1994;9(11):3375–3378. [PubMed] [Google Scholar]
- 52.Kaye FJ. RB and cyclin dependent kinase pathways: defining a distinction between RB and p16 loss in lung cancer. Oncogene. 2002;21(45):6908–6914. doi: 10.1038/sj.onc.1205834. [DOI] [PubMed] [Google Scholar]
- 53.Sanchez-Cespedes M. A role for LKB1 gene in human cancer beyond the Peutz-Jeghers syndrome. Oncogene. 2007;26(57):7825–7832. doi: 10.1038/sj.onc.1210594. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka H, Yanagisawa K, Shinjo K, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67(13):6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- 55.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 56.Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14(10):2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 57.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28(suppl 1):S24–S31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 59.Ansorge WJ. Next-generation DNA sequencing techniques. N Biotechnol. 2009;25(4):195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 60.Coe BP, Lee EH, Chi B, et al. Gain of a region on 7p22.3, containing MAD1L1, is the most frequent event in small-cell lung cancer cell lines. Genes Chromosomes Cancer. 2006;45(1):11–19. doi: 10.1002/gcc.20260. [DOI] [PubMed] [Google Scholar]
- 61.Shames DS, Girard L, Gao B, et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med. 2006;3(12) doi: 10.1371/journal.pmed.0030486. e486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39(3):347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 63.Hussain M, Rao M, Humphries AE, et al. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res. 2009;69(8):3570–3578. doi: 10.1158/0008-5472.CAN-08-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sos ML, Michel K, Zander T, et al. Predicting drug susceptibility of non-small cell lung cancers based on genetic lesions. J Clin Invest. 2009 doi: 10.1172/JCI37127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whitehurst AW, Bodemann BO, Cardenas J, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446(7137):815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 66.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev. 2006;6(10):813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 67.Weinstein JN. Integromic analysis of the NCI60 cancer cell lines. Breast Dis. 2004;19:11–22. doi: 10.3233/bd-2004-19103. [DOI] [PubMed] [Google Scholar]
- 68.Wang H, Huang S, Shou J, et al. Comparative analysis and integrative classification of NCI60 cell lines and primary tumors using gene expression profiling data. BMC Genomics. 2006;7:166. doi: 10.1186/1471-2164-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alexander J, Bey E, Whitcutt JM, Gear JH. Adaptation of cells derived from human malignant tumours to growth in vitro. S Afr J Med Sci. 1976;41(2):89–98. [PubMed] [Google Scholar]
- 70.Pass HI, Mew DJ. In vitro and in vivo studies of mesothelioma. J Cell Biochem Suppl. 1996;24:142–151. doi: 10.1002/jcb.240630509. [DOI] [PubMed] [Google Scholar]
- 71.Metcalf RA, Welsh JA, Bennett WP, et al. p53 and Kirsten-ras mutations in human mesothelioma cell lines. Cancer Res. 1992;52(9):2610–2615. [PubMed] [Google Scholar]
- 72.Schulten HJ, Perske C, Thelen P, et al. Establishment and characterization of two distinct malignant mesothelioma cell lines with common clonal origin. Cancer Genet Cytogenet. 2007;176(1):35–47. doi: 10.1016/j.cancergencyto.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 73.McClatchey AI, Giovannini M. Membrane organization and tumorigenesis—the NF2 tumor suppressor. Merlin. Genes Dev. 2005;19(19):2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- 74.Xiao GH, Gallagher R, Shetler J, et al. The NF2 tumor suppressor gene product, merlin, inhibits cell proliferation and cell cycle progression by repressing cyclin D1 expression. Mol Cell Biol. 2005;25(6):2384–2394. doi: 10.1128/MCB.25.6.2384-2394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sekido Y, Pass HI, Bader S, et al. Neurofibromatosis type 2 (NF2) gene is somatically mutated in mesothelioma but not in lung cancer. Cancer Res. 1995;55:1227–1231. [PubMed] [Google Scholar]
- 76.Ivanov SV, Ivanova AV, Goparaju CM, Chen Y, Beck A, Pass HI. Tumorigenic properties of alternative osteopontin isoforms in mesothelioma. Biochem Biophys Res Commun. 2009;382(3):514–518. doi: 10.1016/j.bbrc.2009.03.042. [DOI] [PubMed] [Google Scholar]
- 77.Veldwijk MR, Berlinghoff S, Jauch A, et al. Characterization of human mesothelioma cell lines as tumor models for suicide gene therapy. Onkologie. 2008;31(3):91–96. doi: 10.1159/000113504. [DOI] [PubMed] [Google Scholar]
- 78.Gautsch JW, Knowles AF, Jensen FC, Kaplan NO. Highly efficient induction of type C retroviruses by a human tumor in athymic mice. Proc Natl Acad Sci U S A. 1980;77(4):2247–2250. doi: 10.1073/pnas.77.4.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lusso P, di Marzo Veronese F, Ensoli B, et al. Expanded HIV-1 cellular tropism by phenotypic mixing with murine endogenous retroviruses. Science. 1990;247(4944):848–852. doi: 10.1126/science.2305256. [DOI] [PubMed] [Google Scholar]
- 80.Cespedes MV, Casanova I, Parreno M, Mangues R. Mouse models in oncogenesis and cancer therapy. Clin Transl Oncol. 2006;8(5):318–329. doi: 10.1007/s12094-006-0177-7. [DOI] [PubMed] [Google Scholar]
- 81.Man S, Munoz R, Kerbel RS. On the development of models in mice of advanced visceral metastatic disease for anti-cancer drug testing. Cancer Metastasis Rev. 2007;26(3–4):737–747. doi: 10.1007/s10555-007-9087-6. [DOI] [PubMed] [Google Scholar]
- 82.Kondo K, Fujino H, Miyoshi T, Ishikura H, Sakiyama S, Monden Y. Orthotopically implanted SCID mouse model of human lung cancer suitable for investigating metastatic potential and anticancer drug effects. Oncology Rep. 2004;12(5):991–999. [PubMed] [Google Scholar]
- 83.Sodeur S, Ullrich S, Gustke H, Zangemeister-Wittke U, Schumacher U. Increased numbers of spontaneous SCLC metastasis in absence of NK cells after subcutaneous inoculation of different SCLC cell lines into pfp/rag2 double knock out mice. Cancer Lett. 2009;282(2):146–151. doi: 10.1016/j.canlet.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 84.Fine DL, Shoemaker R, Gazdar A, et al. Metastasis models for human tumors in athymic mice: useful models for drug development. Cancer Detect Prev Suppl. 1987;1:291–299. [PubMed] [Google Scholar]
- 85.Gazdar AF, Carney DN, Sims HL, Simmons A. Heterotransplantation of small-cell carcinoma of the lung into nude mice: comparison of intracranial and subcutaneous routes. Int J Cancer. 1981;28(6):777–783. doi: 10.1002/ijc.2910280617. [DOI] [PubMed] [Google Scholar]
- 86.Contero A, Richer E, Gondim A, Mason RP. High-throughput quantitative bioluminescence imaging for assessing tumor burden. Methods Mol Biol. 2009;574:37–45. doi: 10.1007/978-1-60327-321-3_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stell A, Biserni A, Della Torre S, et al. Cancer modeling: modern imaging applications in the generation of novel animal model systems to study cancer progression and therapy. Int J Biochem Cell Biol. 2007;39(7–8):1288–1296. doi: 10.1016/j.biocel.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 88.Gazdar AF. Innovative chemotherapy. Xenografts and in vitro drug sensitivity testing. Chest. 1989;96(suppl 1):56S–59S. doi: 10.1378/chest.96.1.56s. [DOI] [PubMed] [Google Scholar]
- 89.Olive KP, Tuveson DA. The use of targeted mouse models for preclinical testing of novel cancer therapeutics. Clin Cancer Res. 2006;12(18):5277–5287. doi: 10.1158/1078-0432.CCR-06-0436. [DOI] [PubMed] [Google Scholar]
- 90.Talmadge JE, Singh RK, Fidler IJ, Raz A. Murine models to evaluate novel and conventional therapeutic strategies for cancer. Am J Pathol. 2007;170(3):793–804. doi: 10.2353/ajpath.2007.060929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Harlin H, Gajewski TF. Diagnosis and treatment of mycoplasma-contaminated cell cultures. Curr Protoc Cytom. 2008 doi: 10.1002/0471142956.cya03cs43. Appendix 3, Appendix 3C. [DOI] [PubMed] [Google Scholar]
- 92.Schweppe RE, Klopper JP, et al. DNA profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification. J Clin Endocrinol Metab. 2008;93(11):4331–4341. doi: 10.1210/jc.2008-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rojas A, Gonzalez I, Figueroa H, et al. Cell line cross-contamination in biomedical research: a call to prevent unawareness. Acta Pharmacol Sin. 2008;29(7):877–880. doi: 10.1111/j.1745-7254.2008.00809.x. [DOI] [PubMed] [Google Scholar]
- 94.Boonstra JJ, van Marion R, Beer DG, et al. Verification and unmasking of widely used human esophageal adenocarcinoma cell lines. J Natl Cancer Inst. 2010;102(4):271–274. doi: 10.1093/jnci/djp499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lorenzi PL, Reinhold WC, Varma S, et al. DNA fingerprinting of the NCI-60 cell line panel. Mol Cancer Ther. 2009;8(4):713–724. doi: 10.1158/1535-7163.MCT-08-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Masters JR, Thomson JA, Daly-Burns B, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc Natl Acad Sci U S A. 2001;98(14):8012–8017. doi: 10.1073/pnas.121616198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nardone RM. Eradication of cross-contaminated cell lines: a call for action. Cell Biol Toxicol. 2007;23(6):367–372. doi: 10.1007/s10565-007-9019-9. [DOI] [PubMed] [Google Scholar]
- 98.Ringel MD. “Thyroid cancer” cell line misidentification: a time for proactive change. J Clin Endocrinol Metab. 2008;93(11):4226–4227. doi: 10.1210/jc.2008-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Durkin AS, Cedrone E, Sykes G, Boles D, Reid YA. Utility of gender determination in cell line identity. In Vitro Cell Dev Biol Anim. 2000;36(6):344–347. doi: 10.1290/1071-2690(2000)036<0344:UOGDIC>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 100.Capes-Davis A, Theodosopoulos G, Atkin I, et al. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010;127(1):1–8. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]
- 101.Lee W, Jiang Z, Liu J, et al. The mutation spetrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465(7297):473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 102.Whang-Peng J, Kao-Shan CS, Lee EC, et al. Specific chromosome defect associated with human small-cell lung cancer; deletion 3p(14-23) Science. 1982;215(4529):181–182. doi: 10.1126/science.6274023. [DOI] [PubMed] [Google Scholar]
- 103.Li Z, Meng ZH, Sayeed A, Shalaby R, Ljung BM, Dairkee SH. Genome-wide allelotyping of a new in vitro model system reveals early events in breast cancer progression. Cancer Res. 2002;62(20):5980–5987. [PubMed] [Google Scholar]
- 104.Zabarovsky ER, Lerman MI, Minna JD. Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene. 2002;21(45):6915–6935. doi: 10.1038/sj.onc.1205835. [DOI] [PubMed] [Google Scholar]
- 105.Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer Res. 2000;60(21):6116–6133. [PubMed] [Google Scholar]
- 106.Johnson BE, Makuch RW, Simmons AD, Gazdar AF, Burch D, Cashell AW. myc family DNA amplification in small cell lung cancer patients’ tumors and corresponding cell lines. Cancer Res. 1988;48(18):5163–5166. [PubMed] [Google Scholar]
- 107.Takahashi T, Nau MM, Chiba I, et al. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989;246(4929):491–494. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- 108.Burbee DG, Forgacs E, Zochbauer-Muller S, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25(3):315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 110.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 111.Pratilas CA, Hanrahan AJ, Halilovic E, et al. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res. 2008;68(22):9375–9383. doi: 10.1158/0008-5472.CAN-08-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kwak EL, Sordella R, Bell DW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005;102(21):7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3) doi: 10.1371/journal.pmed.0020073. e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 115.Yamamoto H, Shigematsu H, Nomura M, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68(17):6913–6921. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 117.Chiosea S, Jelezcova E, Chandran U, et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67(5):2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 118.Weiss GJ, Bemis LT, Nakajima E, et al. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol. 2008;19(6):1053–1059. doi: 10.1093/annonc/mdn006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.