Abstract
Triple‐negative breast cancers (TNBC), characterized by absence of estrogen receptor (ER), progesterone receptor (PR) and lack of overexpression of human epidermal growth factor receptor 2 (HER2), are typically associated with poor prognosis, due to aggressive tumor phenotype(s), only partial response to chemotherapy and present lack of clinically established targeted therapies. Advances in the design of individualized strategies for treatment of TNBC patients require further elucidation, by combined ‘omics’ approaches, of the molecular mechanisms underlying TNBC phenotypic heterogeneity, and the still poorly understood association of TNBC with BRCA1 mutations. An overview is here presented on TNBC profiling in terms of expression signatures, within the functional genomic breast tumor classification, and ongoing efforts toward identification of new therapy targets and bioimaging markers. Due to the complexity of aberrant molecular patterns involved in expression, pathological progression and biological/clinical heterogeneity, the search for novel TNBC biomarkers and therapy targets requires collection of multi‐dimensional data sets, use of robust multivariate data analysis techniques and development of innovative systems biology approaches.
Keywords: Triple-negative breast cancer, Systems biology, BRCA1, BRCA2, Chemotherapy, Targeted therapies
Abbreviations
- aCGH
array comparative genomic hybridization
- CK
cytokeratin
- CI
confidence interval
- EGF
epidermal growth factor
- EGFR (or HER1)
epidermal growth factor receptor
- EMT
epithelial–mesenchymal transition
- ER
estrogen receptor
- HER2 (or Her2/neu, or ErbB2)
human epidermal growth factor receptor 2
- HMMR
hyaluronan-mediated mobility receptor
- HR-MAS
high resolution magic angle spinning
- IHC
immunohistochemistry
- LC
liquid chromatography
- MNI
mode-of-action by network identification
- MRI
magnetic resonance imaging
- MRS
magnetic resonance spectroscopy
- MRSI
magnetic resonance spectroscopic imaging
- ODE
ordinary differential equation
- PARP
poly(ADP-ribose) polymerase
- PCA
principal component analysis
- PET
positron emission tomography
- PR
progesterone receptor
- Rb
retinoblastoma
- ROC
receiver operating characteristic
- TK
tyrosine kinase
- TKI
tyrosine kinase inhibitor
- TNBC
triple-negative breast cancer
1. Introduction
Triple‐negative breast cancers (TNBC), defined as tumors that are negative for estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2), nowadays represent the focus of increasing interest at the clinical, biological and epidemiological level (Irvin and Carey, 2008; Reis‐Filho and Tutt, 2008; Stockmans et al., 2008; Bauer et al., 2007; Dent et al., 2007), due to the aggressive behaviour of the tumor, poor prognosis and present lack of targeted therapies (Mersin et al., 2008; Kaplan et al., 2009; Tan and Swain, 2008). A better understanding of pathological mechanisms of TNBC onset and progression, including the still unclear association with BRCA1 mutations, and the causes of phenotypic heterogeneity may allow improvement in planning prevention and designing novel individualized treatments for this breast cancer subgroup (Goldhirsch et al., 2009).
The new approaches of personalized therapy make use of specific molecular signatures, biology markers and clinico‐pathological features in tumors and patients. For breast cancer, the first clinically used predictive prognostic markers, arising from elucidation of hormonal regulation, were ER/PR which led to the tailoring of endocrine/anti‐hormonal therapy. The first cytogenetic predictor for breast cancer treatment has been the HER2 (HER2/neu, c‐erbB2) gene amplification and protein overexpression. A monoclonal humanized antibody, trastuzumab (Herceptin®), is currently used in the treatment of breast cancer patients presenting with HER2 positivity. Recent scientific and technological advances nowadays provide a large inventory of candidate DNA, RNA, and protein biomarkers, as well as a range of metabolites and networks of cell signaling pathways (Swanton and Caldas, 2009; Bathen et al., 2007; Gast et al., 2009; Chen and Wang, 2009), all potential candidates for disease risk assessment, screening, diagnosis, prognosis, prediction of therapy response and selection of personalized therapy. Nevertheless, such predictors of TNBC prognosis and targeted therapy are presently ill‐defined, making the true challenges of this disease still unmet. A critical step in the expansion of personalized therapy involves development of new molecular imaging tools, adjusted to monitor specific molecules and pathways involved in cancer associated signaling and metabolism. The emerging field of molecular imaging (He et al., 2003; Belkic, 2004; de Vries et al., 2007; Hospers et al., 2008) enables translational medicine from drug discovery via pre‐clinical to clinical research and development and finally, to the clinical practice. Imaging biomarkers have proven utility in the spectrum from intact cells, to experimental tumors in small animals, to patients. Biomarkers are useful in longitudinal quantification of the course of malignancy and therapy response, as well as in early identification of cancer patients and in therapy decision. In conclusion, the success of biomarkers depends on our ability to reveal critical cancer related molecular events and the mechanisms of action of targeted therapy (Tan and Swain, 2008), on how effectively a specific biomarker is related to other biomarkers and to a specific disease condition, as well as on sensitivity and specificity of the available analytical and imaging tools (Dowsett and Dunbier, 2008).
In the search for TNBC biomarkers of diagnosis, prognosis and prediction of therapy response, high dimensional data sets can be generated from different modern ‘omics’ related analyses, such as microarrays in genomics, proteomics and MR‐based metabolomics. The complexity of aberrant molecular patterns involved in expression, pathological progression and biological/clinical heterogeneity of the TNBC phenotype requires collection of multi‐dimensional data sets, use of robust multivariate data analysis techniques (Bishop, 1995; Duda et al., 2001; Vapnik, 2002) and development of innovative systems biology approaches (Chuang et al., 2007; Goh et al., 2007; Pujana et al., 2007; Shen et al., 2009; Fitzgerald et al., 2006; Aebersold et al., 2009). The combination of these progressively more potent technological tools may open new perspectives to the fight against this challenging breast cancer subgroup with worse prognosis and still limited therapy options.
2. Clinical features of TNBC and limitations of current treatment options
2.1. Incidence, recurrence and outcome
According to current estimates, TNBC accounts for 10–17% of all breast carcinomas, depending on thresholds used to define ER and PR positivity and HER2 overexpression (Reis‐Filho and Tutt, 2008). In different series and patient populations TNBC may range 6–28% of breast cancers (Haffty et al., 2006; Rakha et al., 2007; Dent et al., 2007; Kwan et al., 2009), but even higher incidence rates are reported for some ethnical groups such as African Americans and for younger patients (Stead et al., 2009; Trivers et al., 2009; Lund et al., 2009; Morris et al., 2007; Carey et al., 2006).
Despite its relatively small proportion among all breast cancers, TNBC is responsible for a relatively large proportion of breast cancer deaths, due to its generally aggressive clinical course. In a retrospective study on a cohort of 1601 patients with breast cancer (Dent et al., 2007), a subgroup of 180 women with TNBC had a significantly lower mean age at diagnosis (P < 0.0001), increased likelihood of distant recurrence (hazard ratio vs other breast cancer phenotypes equal to 2.6; 95% confidence interval (CI) 2.0–3.5; P < 0.0001) and death (hazard ratio 3.2; 95% CI 2.3–4.5; P < 0.001) within five years of diagnosis, but not thereafter. Also, patients with TNBC were more likely to present with larger mean tumor size (P < 0.0001) and histological tumor grade (P < 0.0001). Although data on differential lymph node spread in TNBC and other breast cancer subgroups are still conflicting (Dent et al., 2007; Reis‐Filho and Tutt, 2008; Rakha et al., 2008), an interesting feature was the substantial lack of correlation between nodal metastasis and tumor size among women with tumors smaller than 50 mm (Dent et al., 2007) (Fig. 1A). A similar trend was also found in BRCA1‐associated tumors (Fig. 1B), as discussed in Section 3.2 (Foulkes et al., 2003a). The patterns of recurrence in TNBC were qualitatively different from the non‐TNBC group. In the former, the risk of distant recurrence peaked at approximately three years and declined rapidly thereafter, whereas in the other breast cancer types the recurrence risk seemed to be constant over time (Dent et al., 2007). TNBC has a preference for visceral metastases (Liedtke et al., 2008).
Figure 1.
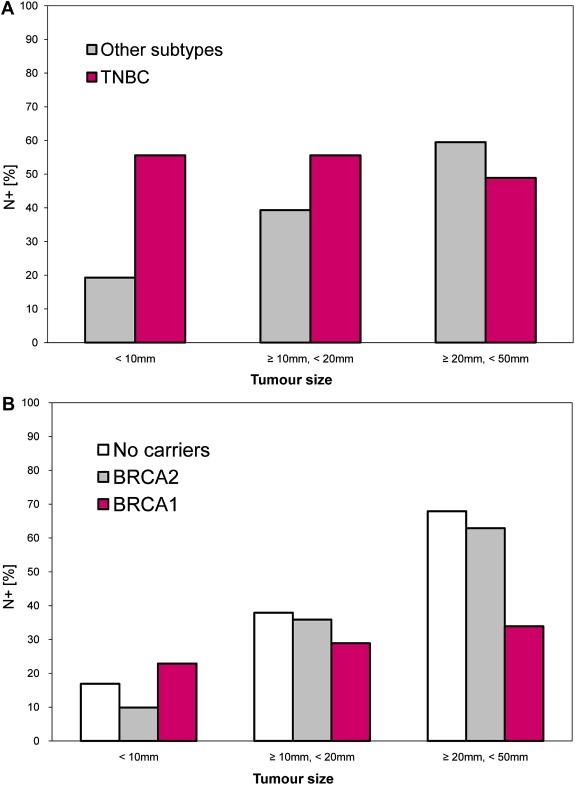
Lack of correlation between tumor size and nodal status among (A) triple‐negative breast cancers (TNBC) with tumor size smaller than 50 mm (χ2 test for trend: P = 0.47 for TNBC; P < 0.0001 for other breast cancer subtypes) and (B) BRCA1‐associated breast cancers (χ2 test for trend: P = 0.183 for BRCA1‐associated tumors; P < 0.0001 for BRCA2‐associated breast tumors and for breast tumors in non‐carriers). N+ positive nodal status (at least one positive lymph node). Graphs adapted from Dent et al. (2007) and (Foulkes et al. 2003a).
It should be emphasized that TNBC currently includes a heterogeneous group of tumors. Already by simple morphology, a group of patients with TNBC can be identified who have a more favourable outcome, for example patients with invasive adenoid cystic, apocrine and typical medullary tumors (Japaze et al., 2005; Azoulay et al., 2005; Orlando et al., 2005). Even within the relatively homogeneous group of patients with triple‐negative invasive ductal carcinoma, patients with higher or lower risk may be identified, based on specific molecular markers (Viale et al., 2009).
2.2. Current cytotoxic treatment options
Several studies support the notion that primary TNBC is a chemosensitive disease. However, although TNBC is associated with enhanced pathological complete response to neoadjuvant chemotherapy (von Minckwitz et al., 2008), these cancers show worse survival due to higher relapse among those with residual disease after chemotherapy (Liedtke et al., 2008; Carey et al., 2007). In fact, despite the high sensitivity to chemotherapy, TNBC patients with truly chemosensitive disease still represent a minority among all TNBC patients. In the metastatic setting, patients progress quickly on first‐, second‐, and third‐line palliative treatment (Kassam et al., 2009).
As triple‐negative disease is often characterized by an impaired DNA repair process, cytotoxic agents inducing DNA damage may be of specific value. Recent small and non‐randomized studies have shown promising results with cisplatinum, both in the neoadjuvant and metastatic setting (Byrski et al., 2009; Sirohi et al., 2008), although the clinical response to paclitaxel and cyclophosphamide may also be high (Rouzier et al., 2005). A new approach under clinical trial includes the use of ixabepilone, a microtubule inhibitor (Thomas et al., 2007; Baselga et al., 2009b).
Larger prospective clinical trials are needed to determine the optimal cytotoxic treatment for TNBC.
3. Molecular profiling of triple‐negative breast cancers
3.1. The relationship between TNBC and basal‐like breast cancer
Molecular profiling of human breast cancers by gene expression assays provided in the last decade new grounds to a clearer understanding of the heterogeneous nature of these tumors with promising trends for outcome prediction and development of individualized therapies (van't Veer et al., 2002; van de Vijver et al., 2002; Perou et al., 2000; Sørlie et al., 2003; Chang et al., 2005). The gene expression based classification proposed by Perou and Sørlie ten years ago originally defined six subtypes (Perou et al., 2000; Sørlie et al., 2003). Three of these were characterized by expression of ER and luminal epithelial cell related genes (Luminal A, Luminal B and Luminal C) while the remaining three groups, basal‐like, ErbB2+ and normal‐like, showed an expression phenotype more similar to myoepithelial/basal epithelial cells. The ErbB2+ tumors were in particular characterized by high expression of ErbB2 and genes located adjacent to the ErbB2 locus and the normal‐like subgroup showed expression patterns similar to normal breast tissue samples. Although this seminal work was based on analyses on neoadjuvantly treated breast carcinomas, the main findings have since been validated in numerous independent cohorts and these intrinsic subtypes show different mutation patterns, prognosis and routes of progression (Sørlie et al., 2003; Calza et al., 2006; Hu et al., 2006; Langerod et al., 2007; Naume et al., 2007; Smid et al., 2008; Parker et al., 2009). Later, using array comparative genomic hybridization (aCGH), several investigators have found that genomic alterations seem to be more frequent in some of the intrinsic subclasses (Chin et al., 2006; Fridlyand et al., 2006; Bergamaschi et al., 2006). Of particular interest to this review, basal‐like tumors frequently have complex rearrangements with higher numbers of gains and losses compared to luminal subtypes (Chin et al., 2006; Fridlyand et al., 2006; Bergamaschi et al., 2006), although there is evidence of a subgroup of basal‐like tumors with low genomic instability (Chin et al., 2007). Such heterogeneity is recognized by gene expression classification as well (Kreike et al., 2007) and multiclonal basal‐like tumors have been described (Navin et al., 2010). Recently a distinct type of rearrangements dominated by genomewide duplications was identified in a selection of basal‐like tumors and cell‐lines (Stephens et al., 2009), suggesting that these types of tumors represent a distinct type of breast carcinomas with a unique path of progression (Navin et al., 2010; Dalgin et al., 2007). In addition, the emerging knowledge of a cellular hierarchy in the breast supports the theory that basal‐like tumors may have a distinct etiology (Villadsen et al., 2007; reviews in: Sims et al., 2007; Polyak, 2007).
Immunohistochemistry (IHC) is frequently used to explore the distribution of the molecular subtypes by using formalin‐fixed, paraffin‐embedded tissues from larger cohorts of breast cancer patients. The ultimate selection of surrogate markers is an ongoing debate and a consensus for an appropriate panel still has to be reached (Rakha et al., 2008). Triple negativity is often used to identify basal‐like tumors (Kreike et al., 2007) although a supplement of additional markers has superior prognostic value (Cheang et al., 2008; Nielsen et al., 2004). Triple negativity as a selection criterion is highly sensitive of basal‐like tumors, but not specific as both luminal, ErbB2+ and normal‐like tumors (as identified by gene expression) can be steroid receptor negative and HER2 non‐amplified (Naume et al., 2007). The heterogeneity of TNBC is acknowledged and includes both basal‐like and non‐basal‐like tumors (Rakha et al., 2009; reviewed in Hurvitz and Finn, 2009). IHC‐based studies use different markers to define their basal‐related tumors and the lack of a systematic classification scheme makes comparison of results difficult. Acknowledging the differences between the two terms is important in clinical studies aiming to identify prognostic markers and targets for therapy for these important subgroups of breast carcinomas.
Morphologically basal‐related tumors are typically of high histological grade with a high mitotic count and they frequently exhibit geographic tumor necrosis, central scar, pushing margings and/or stromal lymphocyte enrichment (Fulford et al., 2006; Livasy et al., 2006; Rakha et al., 2006). Although most TNBCs are classified as ductal carcinomas, tumors of ‘special types’ such as medullary and adenoid cystic carcinoma fall into this category as well (Bertucci et al., 2006; Jacquemier et al., 2005; Vincent‐Salomon et al., 2007; Weigelt et al., 2008). Many of the clinical features of the basal‐like phenotype are similar to those of TNBC (see Section 2.1), including shorter relapse‐free and overall survival times compared with other types of breast cancers, a tendency toward visceral versus bone metastasis (Rodriguez‐Pinilla et al., 2006; Rakha et al., 2008), and over‐representation in BRCA1 mutation carriers (Foulkes et al., 2003b; Haupt et al., 2010).
3.2. The BRCA‐associated triple‐negative breast cancers
Following identification, mapping and cloning of the two major breast cancer predisposing genes, BRCA1 (chromosome 7q21) and BRCA2 (chromosome 13q12) (Hall et al., 1990; Miki et al., 1994; Wooster et al., 1995), increasing attention has been focused on biological and molecular characteristics of breast cancers in BRCA‐ and non‐BRCA1/2 (BRCAX) mutation carriers, in relation to carcinogenesis, disease progression and outcome (Breast Cancer Linkage Consortium, 2007, 1997, 2006, 2007, 1998, 2002, 2008, 2002, 2007, 2004, 2004, 1998).
Major clinical characteristics of breast cancer in BRCA1 mutation carriers, compared with age‐matched patients unselected for family history were envisaged to be younger age at onset (Robson et al., 2004; Cornelis et al., 1995), frequent bilateral occurrence (Hall et al., 1990; Ford et al., 1994; Easton et al., 1995), high frequency of ductal histotype cancer, although with a relative excess of medullary and atypical medullary histotypes, higher overall grade and worse histoprognostic features (Breast Cancer Linkage Consortium, 1997, 1996, 1995, 1998, 2002, 1996, 1998). Histopathological characteristics frequently detected in BRCA1‐associated breast cancers are higher mitotic counts, greater degree of nuclear polymorphism and less tubule formation. However, multifactorial analysis of histopathological differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations (Lakhani et al., 1998) demonstrated that many of these features were linked with each other, the only factors independently associated with BRCA1 being high mitotic count, presence of lymphocytic infiltrate and presence of smooth noninfiltrative pushig border. A similar multifactorial analysis showed that reduction in tubule formation (together with higher overall grade) and presence of continuous pushing margins were also significantly associated with BRCA2‐related breast cancer.
Combined immunohistochemical and molecular analyses of cancer‐associated genes and encoded proteins carried out within a large collaborative study of the Breast Cancer Linkage Consortium (Breast Cancer Linkage Consortium, 1997, 1998, 2002) showed that breast cancers in patients with BRCA1 germline mutation are more often negative for ER, PR and HER2 and are more likely to be positive for p53 protein compared with controls. BRCA2 tumors did not show, instead, any significant difference in the expression of these proteins. In fact, only 10% of BRCA1‐mutated cancers showed positive staining for ER compared with 66% of BRCA2 and control tumors; positivity to PR was 21% for BRCA1‐cancers compared with 55–61% for BRCA2 and control cancers; and a low positivity (3%) was found for HER2 in both BRCA1 and BRCA2 tumors. Multiple logistic regression analysis of the immunohistochemical factors (ER‐negative, PR‐negative and HER2‐negative) and morphological features that were significant predictors of BRCA1 status indicated that the ER status was the most significant risk factor (Lakhani et al., 2002). On the other hand, the expression of ER is known to inversely correlate with tumor grade (Henderson and Patek, 1998) and is reported as one of the most important prognostic and predictive markers for breast cancer (Osborne, 1998).
Unsupervised cluster analysis of non‐BRCA1/2 breast cancer families (50 probands) demonstrated heterogeneity of BRCAX families (Honrado et al., 2007). In fact two main groups were identified, one of high grade and ER‐negative (50%) and one of low‐grade and ER‐positive tumors (50%). These two groups were in turn subdivided into five subgroups: three among the high‐grade and two among the low‐grade groups; one overexpressing HER2 (18%); one with a basal‐like phenotype (14%); one with a normal breast‐like phenotype (18%); a luminal A subgroup (36%) and a luminal B subgroup (14%).
At the molecular level, BRCA1 and BRCA2 proteins are known to be involved in DNA repair (Scully, 2000), cell cycle checkpoint control through regulation of p53 activity (Liu and Kulesz‐Martin, 2001) and maintenance of global chromosome stability (Venkitaraman, 2002).
A more complete molecular portrait has been recently consolidated for the majority of breast tumors in BRCA1‐mutated patients. This general genomic and proteomic profile typically includes lack of (or low) expression of hormone receptors, HER2 and BCL2 and overexpression of p53, EGFR and basal cytokeratines (CK 5/6) (Lacroix and Leclercq, 2005; Honrado et al., 2006), associated at clinical level with high frequency of ductal type, high proliferation rate, high histological grade and manifested lymphocyte infiltration. The reported molecular features are also characteristic to basal‐like carcinomas (Perou et al., 2000; Gorski et al., 2009), although overlap of tumor biological features between BRCA1‐associated and basal‐like cancer subtype is not complete (Dawson et al., 2009). Depletion of BRCA1 affects differentiation and enhances proliferation of mammary epithelial cells, as also confirmed by studies on animal models (Kubista et al., 2002; Furuta et al., 2005; Bradley and Medina, 1998).
Over‐representation of the TNBC phenotype in BRCA1‐associated tumors allows a rational retrospective interpretation of a series of similarities separately reported for the clinico‐pathological features of these two breast cancer subgroups. Of particular interest in this respect are the younger age of the first breast cancer event; high histopathological grade; some common morphological features (e.g. smooth noninfiltrative pushing border); and the above mentioned lack of correlation between tumor size and nodal status (Fig. 1).
Tumors arising from BRCA1 and BRCA2 mutation carriers appear to have specific pathological and gene expression profiles (Honrado et al., 2006), while BRCAX tumors are increasingly believed to originate from multiple distinct genetic events (Lacroix and Leclercq, 2005). The use of microarray technology in the evaluation of the immunophenotypic features of hereditary breast cancer (Palacios et al., 2003) revealed distinct characteristics in BRCA1 and in non‐BRCA1/2 tumors, whereas BRCA2 tumors presented intermediate patterns. No cases of HER2 amplification and/or overexpression were found by the authors, except in sporadic breast cancers.
The BRCA1 tumor suppressor gene and the HER2 oncogene are located in close proximity on the long arm of chromosome 17 (17q11–21), but the aggressive pathological features of BRCA1‐associated tumors appear unrelated to amplification of the adjacent HER2 oncogene (Grushko et al., 2002).
The molecular mechanisms responsible for tissue–specific carcinogenesis in BRCA1‐associated breast cancer are still to be clarified. The existence of common genomic features in the basal‐like phenotype of BRCA1‐associated breast cancers and that of normal breast stem cells recently suggested that BRCA1 may act as a human mammary stem cell fate regulator (Foulkes, 2004; Liu et al., 2008). In particular, the BRCA1 expression was found to be required for the differentiation of ER‐negative stem/progenitor cells to ER‐positive tumoral cells and was proposed to result in the accumulation of genetically unstable breast stem cells, providing prime targets for further carcinogenic events (Liu et al., 2008). On the other hand, the basal‐like molecular subtype can also be found in sporadic, BRCA2‐ and BRCAX‐associated cancers, although with a much lower frequency (10–20% compared with up to 90% in BRCA1‐classified cancers). A critical role of the BRCA1 protein has also been shown in the development of these basal‐like carcinomas (Melchor and Benitez, 2008 and ref. therein).
Attention has been focused on the possible molecular mechanisms underlying the very low or inexistent overexpression (and gene amplification) of HER2 (0–3%) in BRCA1/2‐associated breast cancers (Lakhani et al., 2002; Palacios et al., 2003; Grushko et al., 2002) compared with that (15–20%) in BRCAX‐associated tumors (Honrado et al., 2007). A possible co‐deletion of HER2 and BRCA1 loci (Johannsson et al., 1997) would not explain the lack of HER2 overexpression in BRCA2‐associated cancers. It has been postulated (Melchor and Benitez, 2008) that the defects in the DNA repair system associated with deleterious BRCA1 mutations may not be compatible with the proliferation stress of the aberrant signaling cascade triggered by HER2 tyrosine phosphorylation and therefore, under these conditions, HER2‐overexpressing cancer cells are unlikely to survive.
4. Imaging features
Examinations of women of 50 years and over (Dent et al., 2007) showed that patients with TNBC had a much lower proportion (P = 0.0008) of breast cancers first detected by mammography or ultrasound (19.5%) than patients with other breast cancers (36.5%). This result supported the conclusion that in a mammography screening programme offered to women over 50 years of age, TNBC often presents as an interval cancer, in agreement with a previous study (Collett et al., 2005). This feature, which may relate to differences in breast density, or to a rapid tumor growth in relation to the screening interval, warrants further investigations to optimize multimodality and periodicity of screening events in surveillance programs addressed to women belonging to populations at high risk of TNBC. Mammographic examinations on premenopausal women showed that a circumscribed mass (without spiculated margins) and absence of microcalcifications were most commonly presented features in TNBCs compared with cancers of other subtypes (Yang et al., 2008; Wang et al., 2008). The absence of spiculated masses and pleomorphic microcalcifications suggested that mammography may not be the ideal tool for early detection of TNBCs.
The use of dynamic contrast‐enhanced magnetic resonance imaging (MRI) showed that 97% of TNBC lesions were of mass‐type, with typical malignant signal enhancement kinetics (Chen et al., 2007). Correlations between MRI and pathological findings recently investigated in surgically confirmed TNBC, compared with ER‐positive, PR‐positive and HER2‐negative breast cancers (Uematsu et al., 2009), showed that the former subtype was significantly associated with high histologic grade, unifocal lesions, smooth mass margin, rim enhancement, persistent enhancement pattern, and very high intratumoral signal intensity on T2‐weighted MR images. The last feature was significantly associated with intratumoral necrosis, a characteristics typically associated with poor prognosis.
These radiological imaging methods, and their possible combination with functional imaging approaches, such as diffusion‐weighted MRI (Bogner et al., 2009), may assist in TNBC prognosis and treatment planning.
A study on fluorodeoxy‐glucose‐positron emission tomography (FDG‐PET) characteristics showed a sensitivity of 100% for detection and a higher FDG uptake in TNBC compared with ER‐positive/PR‐positive/HER‐negative tumors, suggesting enhanced glycolysis in the former, more aggressive subgroup (Basu et al., 2008). Further studies should be devoted to better clarify the potential of PET approaches in evaluating prognosis and predicting therapy response in TNBC.
Finally, non invasive in vivo localized magnetic resonance spectroscopy (MRS) examinations and high resolution magic angle spinning (HR‐MAS) MRS analyses on surgical specimens may offer new perspectives to the characterization of metabolic profiles of breast cancers, also in relation to lymphatic spread, grade and hormone receptor status (Bathen et al., 2007; Podo et al., 2007; Sitter et al., 2009; Giskeødegård et al., 2010).
5. Towards pathway‐driven TNBC therapeutics
5.1. Concepts and tools
Classification of tumors by molecular profiles and increasing knowledge on altered regulation of gene expression at the translational, transcriptional and epigenetic levels (Fig. 2) allowed in the last decade the identification of possible novel markers and targets for pathway‐driven therapeutics in tumors (Cleator et al., 2007; Tan and Swain, 2008; Dowsett and Dunbier, 2008; Swanton and Caldas, 2009; Bouchalova et al., 2009; Goldhirsch et al., 2009; Bosch et al., 2010).
Figure 2.
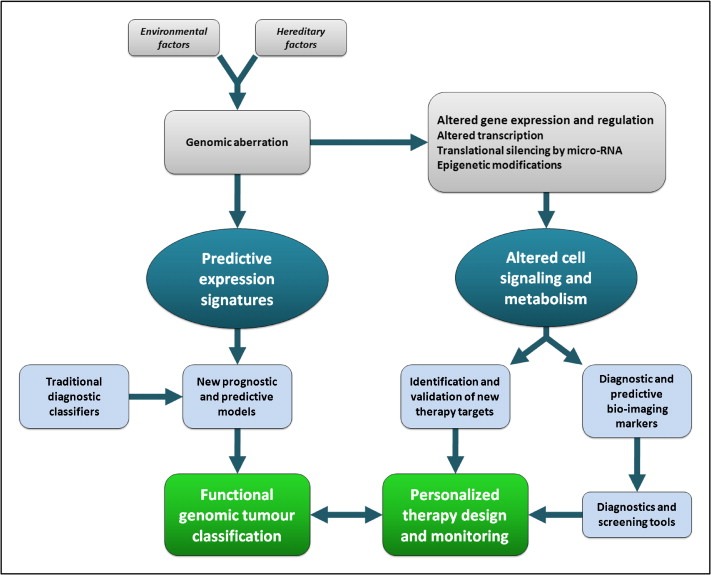
Integration of predictive expression signatures and altered cell signaling patterns in the search for pathway‐driven tumor therapeutics and bioimaging.
Accumulation of defects in cellular regulation mechanisms is the underlying cause for disease heterogeneity and differential response to treatment, also within a subgroup like TNBC. This can be at the level of gene amplification, methylation, expression, mutations, or other as yet unknown regulation mechanisms. Activation of an oncogene can increase activity in downstream pathways without necessarily requiring overexpression of proteins in these pathways. Beyond the wide diversity in the combination of overexpressed pathways, the basal‐like subtype is reported to be associated with increased activity of the HER1, RAS, CTNNB1, TP53 and E2F3 pathways (Bild et al., 2009). Other notable characteristics of TNBC include a high level of proliferative genes, including Ki‐67, basal cytokeratins such as CK5 and CK17, caveolin‐1, alpha B‐crystallin, frequent p53 mutations, Retinoblastoma (Rb) pathway inactivation, high c‐kit expression, reduced DNA repair capability, increased angiogenesis and TGF‐regulated genes (Schneider et al., 2008).
The above mentioned TNBC biomarkers and characteristics are known to have relevant implications on cell signaling pathways, tumor metabolism, evasion of cell‐cycle control, invasion and metastasis, as summarized below (Table 1).
Table 1.
Possible agents for TNBC targeted therapy.
| TNCB biomarkers | Major affected signaling pathways or cellular process | Biological effects | Proposed TNBC therapeutic agents |
|---|---|---|---|
| EGFR (or HER1) tyrosine kinase | MAP kinase (RAS‐RAF‐MEK1/2 – ERK1/2) | Cell proliferation | Cetuximab; Erlotinib; Gefitinib |
| c‐KIT | PI3K‐AKT | Cell survival | Imatinib; Sunitinib; Dasatinib |
| TOP2A (topoisomerase II α) | DNA replication | Chemotherapy response | Antracyclins, Metoxantrone and etoposide |
| c‐Myc | Gene transcription | Cell growth | 10058F4 small molecule compound (potential) |
| Cell‐cycle control | Angiogenesis | ||
| Apoptosis | |||
| PARP (poly(ADP‐ribose) polymerase) BRCA1–associated cancers | DNA repair | Apoptosis | Olaparib |
| VEGF/VEGFR (tumor endothelial cells) | Hypoxia‐induced metabolic pathways | Angiogenesis | Bevacizumab; Sunitinib; Sorafenib |
| p53 | Cell‐cycle control | Proliferation/apoptosis | Not available |
| Src kinase | STAT‐pathway; PI3K; FAK | Cell proliferation; cell survival; angiogenesis; cell invasion, adhesion and migration | Dasatinib |
| TGFβ | T‐reg activation | Repression of antitumor immunity | Anti‐TGF‐β Ab; anti‐sense oligonucleotides; selective TK inhibitors |
| Epithelial‐mesenchimal transition (EMT) | Tumor cell motility, blood borne metastasis | ||
| Alpha B‐crystalline | MAPK/ERK | Anchorage independent cell growth; increased migration and invasion | ERK kinase inhibitors (potential) |
Understanding the relations between these potential markers may help to appreciate the heterogeneity of these tumors and improve the response prediction to single agents and combination treatments.
The multiplicity of ongoing efforts to identify, characterize, and validate new biomarkers and therapy targets for TNBC (Bosch et al., 2010) shows the complexity of such a goal and the need for well designed and rigorous studies, at all levels of investigation, to resolve conflicting results, which halt or slow down the advancement of this field. Systems biology can be an important tool for elucidating crosstalk between pathways and understanding temporal effects of tumor behaviour.
5.2. Targetable pathways and cellular processes
Proliferation and death are uncommon features for non‐tumorigenic, healthy cells and therefore these events are under tight control at many levels. Fluxes through pathways are intricately balanced by stimuli of different nature and checked by multiple feedback control mechanisms at cellular, tissue, organ and organism level. Backup mechanisms to assure proper functioning in case the first level of control goes awry are present in normal cells. Tumor cells have accumulated defects that allow them to bypass the normal control mechanisms that check proliferation. For tumor cells to grow and proliferate, they must evade checks on several cellular controls. The increased energy requirement of tumor cells causes metabolic stress at the level of nutrient and oxygen supply. The accumulation of gene amplifications, translocations and mutations must evade cellular control on DNA damage during cell cycle checkpoints. Whereas most cells can only survive with close contacts to their neighbours, metastatic tumor cells must overcome the apoptosis directed controls associated with anoikis. Within one tumor, large differences exist in local environment that may cause cells at the edge to proliferate, while cells that are deprived of nutrients and oxygen may undergo necrosis, therewith exposing their neighbours to stressful conditions. Histopathology can make this cellular diversity visible, but molecular techniques like gene expression profiling and proteomics determine the average of all these microenvironments in a selected tissue sample.
5.2.1. Metabolism
Rapidly growing tumors need a high supply of nutrients and oxygen. Tumor cells have developed several mechanisms to satisfy their needs in these respects, varying from assuring a good nutrient supply by stimulation of new blood vessels formation, to adjustments in glucose metabolism and methods to survive at low oxygen concentrations. Although debatable whether it is the only cause, hypoxia promotes via the Hypoxia‐inducible factor 1 (HIF‐1) a change in metabolic pathways in tumor cells (Marin‐Hernandez et al., 2009). Tumor cells derive most of their energy from glycolysis rather than from oxidative phosphorylation, even though the yield in ATP per glucose is reduced from 36 for the aerobic oxidative phosphorylation to 2 for the anaerobic process. Furthermore, the formation of lactic acid poses challenges for pH homeostasis in the cell. Glutaminolysis is used to produce sufficient amounts of precursors for synthesis of small molecule building blocks to sustain proliferation, which are usually provided by the TCA cycle. This shift in metabolic pathways makes tumor cells less sensitive to hypoxia and reactive oxygen species.
The flux through metabolic pathways is regulated at different levels, ranging from de novo synthesis of proteins to activation of limited duration of proteins via covalent modification and allosteric activation or inhibition by substrates or products of a pathway. In regulation of the balance between anabolic and catabolic pathways, the protein kinases Akt, with constituents of the Akt pathway like mTor, and AMPK play crucial roles. Akt activity for example is stimulated by a multitude of growth factors via receptor tyrosine kinase induced signaling, transmitted by PI3K, and by small intermediate metabolites like retinoic acid and prostaglandin. AMPK senses a low energy state by the AMP/ATP ratio. Not only do Akt and AMPK directly regulate the balance between the pathways, they also affect levels of tumor suppressor protein p53 by either promoting its degradation or stimulating its synthesis. Discussing the subtilities of the pathways and their regulation is too farfetched for this review. The role of p53 in regulation of metabolism in tumor cells has been recently reviewed (Vousden and Ryan, 2009).
Whereas an abundant energy supply leads to cell proliferation, an insufficient energy supply leads to autophagy and cell death. Autophagy is part of everyday normal cell growth and development eliminating malfunctioning proteins, reshaping membranes, recycling of receptors, etc. During nutrient starvation, autophagy can provide additional energy or building blocks by the breakdown of non‐vital cell components, ensuring that vital processes can continue. mTor plays an important regulatory role in this process. Prolonged starvation will lead to cell death and necrosis, events often found in tumors. The products that result from necrosis, pose additional stress on neighbouring cells and lead to induction of the NFkB pathway.
5.2.2. Evasion of cell‐cycle control
Under conditions of sufficient nutrient supply, proliferation can take place. In most non‐tumorigenic cells, the G0 phase is the default state. Strong stimuli are required to leave this state. When it occurs, the steps of the cell cycle are tightly controlled. In many cancer cells, the control on the cell cycle progression is faulty, permitting cells with errors in DNA to proceed to the next step of the cycle and produce more genetic diversity. TNBC suffers from frequent mutations in the tumor suppressor protein p53 and in Rb pathway inactivation (Schneider et al., 2008), whereas mutations in c‐Myc are relatively rare (Rodriguez‐Pinilla et al., 2007).
The Rb pathway in the G1/S checkpoint is activated by phosphorylation of retinoblastoma by cyclin/CDK complexes, leading to the activation of transcription factor E2F. The activity of cyclin/CDK is stimulated by the proto‐oncogene Myc, which in turn is activated by e.g. the HER1 (or EGFR) pathway. Myc regulates the expression of a multitude of proteins (Bouchalova et al., 2009).
Upregulation of CDNK2A, often observed in TNBC, inhibits the CDK2 activity and therewith promotes inactivation of p53, resulting in blockage of the senescence and apoptosis pathways. The increased expression of CDNK2A and related CDNK2's allows bypassing the checks in cell‐cycle control in the Rb pathways and promotes progression into the G1/S phase (Schneider et al., 2008; Musgrove and Sutherland, 2009). BRCA1 and BRCA2 also have a function in cell‐cycle control via their regulation of p53 activity (Liu and Kulesz‐Martin, 2001). After amplification of DNA, the daughter strands must be separated. Topoisomerases, notably TOP2A, are essential in this process. TOP2A amplification is most frequent in HER2‐overexpressing tumors (Durbecq et al., 2003) and although a triple‐negative phenotype was associated with TOP2A expression, no amplification was found (Tan et al., 2008).
DNA damage repair is not only essential during cell division, but also is a continuous process. Cells are exposed to factors that can cause damage to their DNA. These defects must be repaired (Jackson and Bartek, 2009). When that is not possible, cells go into apoptosis. Exposing cells to chemotherapy or radiation therapy causes irreparable damage in most cells, but some are more resistant than others. BRCA1 and BRCA2 are involved in DNA repair. Cells lacking either of these proteins are more susceptible to poly(ADP‐ribose) polymerase (PARP) inhibitors (see below).
5.2.3. Invasion and metastasis
Cells thrive only when they are in close contact with neighbouring cells and the extracellular matrix. When cells become detached from their natural environment, they die by anoikis. Tumor cells escape this fate when they undergo the epithelial mesenchymal transition (EMT) to acquire a mesenchymal‐like phenotype and can become invasive. The essential features of EMT are the disruption of intercellular contacts and the enhancement of cell motility, thereby leading to the release of cells from the parent epithelial tissue. The EMT enables cells to penetrate vessel endothelium and enter the circulation to form distant metastasis (Guarino et al., 2007). The Basal B subgroup of breast cancer has enhanced invasive properties and a predominantly mesenchymal gene expression signature, distinct from subgroups with predominantly luminal (termed Luminal) or mixed basal/luminal (termed Basal A) features (Guarino et al., 2007). Epithelial mesenchymal transition has long been associated with breast cancer cell invasiveness (Neve et al., 2006). During EMT the abundance of the proteins N‐cadherin, Vimentin, several metalloproteinases and transcription factors like Snail1 (Snail), Snail2 (Slug) increases, while decreased concentrations are observed for E‐cadherin, Desmoplakin, Cytokeratin and Occludin. Several interconnected transduction pathways and a number of signaling molecules potentially involved have been identified. Growth factor driven receptor tyrosine kinases play an important role, but Wnt, NFkB, integrin and TGFβ signaling also contribute (Lee et al., 2006; Giampieri et al., 2009). Most of these pathways converge on Akt (Iliopoulos et al., 2009) and on the downregulation of the epithelial molecule E‐cadherin, an event critical in tumor invasion and a ‘master’ programmer of EMT (Guarino et al., 2007).
EMT is accompanied by changes inside the cells, such as reorganisation of the actin cytoskeleton and regulation of focal adhesion (reviewed by Jiang et al., 2009).
5.2.4. Integration of signals
As illustrated above, different processes take place either simultaneously or sequentially in tumor cells. Signals from the environment can also lead to conflicting stimuli as input. The different routes converge on central signaling hubs. Coordination of the processes requires a tight orchestration of these events, with Akt, mTor, p53, HIF‐1, NFkB and Myc being some of the leading conductors, though at different time scales.
Whereas the Akt/mTor kinases regulate rapid responses of short duration, proteins like p53, Myc and HIF influence the expression of large numbers of genes.
HIF‐1alpha is involved in the activation of numerous cellular processes including resistance against apoptosis, overexpression of drug efflux membrane pumps, vascular remodeling and angiogenesis, as well as EMT and metastasis (Marin‐Hernandez et al., 2009) and in immune reactions and inflammatory response (Hellwig‐Burgel et al., 2005).
5.3. TNBC biomarkers for assessment of prognosis and therapy targeting
Parameters like the proliferating index, ploidity, presence of p53, cytokeratins, HER1, and numerous other molecular alterations, may also be useful for prognostic evaluation, for predicting therapeutic response and for guiding patient management.
Given the poor outcome of TNBC despite cytotoxic treatment, alternative approaches are currently explored for possible application at the clinical level (Table 1).
5.3.1. HER1
HER1 (EGFR) is a receptor tyrosine kinase, that belongs to the HER family of transmembrane receptors. HER1 gene is located on 7q12 and its protein product – 170‐kD glycoprotein – plays an important role in cell proliferation, migration and protection against apoptosis mediated by subsequent activation of intracellular pathways. After binding of epidermal growth factor (EGF), the HER1 receptor can dimerize with other members of the HER family, and it has to create homo– or heterodimers to be functionally active (Hynes and Lane, 2005).
Increased HER1 expression is detected in about 40% of breast carcinomas. Particularly, HER1 expression is higher (up to 80%) in TNBC and metaplastic carcinoma (mostly basal‐like), where it possibly substitutes ineffective, but otherwise major proliferation/survival pathways of breast cancer induced by expression and activation of HER2, ER and PR proteins. Currently, however, HER1 gene status is not used in clinical practice to guide therapy in breast cancer. HER1 protein could be targeted by monoclonal antibodies and/or synthetic tyrosine kinase inhibitors (TKIs). Monoclonal antibodies (cetuximab, panitumumab) are now clinically used in the treatment of colorectal cancer and head and neck carcinoma. TKIs are also important in the therapy of pancreatic and non‐small cell lung cancer.
Given the subset of TNBC which overexpresses EGFR (Viale et al., 2009), targeting EGFR seems to be a rational approach. Although cetuximab monotherapy has little clinical activity, in combination with chemotherapy it may enhance tumor response (Corkery et al., 2009; Carey et al., 2008). HER1 targeted treatment with cetuximab in breast cancer has not produced satisfactory results probably because of the activation of downstream signaling pathways (Shiu et al., 2008) or because of inadequate patient selection. Evaluation of the results of studies testing TKIs (erlotinib, gefitinib) in breast cancer indicated that HER1 protein must be present in targeted tumor tissue to obtain valuable treatment results (Agrawal et al., 2005). Furthermore, it might be better to target more than one of these receptors simultaneously. Thus, HER1 assessment could reveal a particular group of breast cancer patients with probably good response to HER1 targeted therapy.
5.3.2. TOP2A
TOP2A gene is located on 17q21–22, encoding topoisomerase II alpha, which appears to be a molecular target for anthracyclines and hence is predictive of response to anthracycline therapy. Good response to anthracyclines is associated with TOP2A amplification, while deletion may be accompanied by resistance. (Burgess et al. (2008) identified in a nonselected series, TOP2A expression levels as major determinants of response to doxorubicin, which is a topoisomerase II inhibitor, and showed that suppression of TOP2A levels produces resistance to doxorubicin in vitro and in vivo.
Recent publications describe TOP2A amplification in 2.7–8.8% of HER2 non‐amplified breast cancers (Knoop et al., 2005). Adjuvant anthracycline treatment of TNBC patients was shown to be associated with poor response in patients with low expression of TOP2A protein. Microarray expression analysis indicated that in a subgroup of TNBC there is a significant downregulation in PTEN and TOP2A which might partly explain observed differences in response to chemotherapy in TNBC (Weigelt et al., 2009). It should be noted, however, that patients with a pathologic complete response to anthracycline‐based neoadjuvant chemotherapy had a good prognosis regardless of subtype (Carey et al., 2007).
5.3.3. c‐Myc
c‐Myc is a major transcription factor that encodes nuclear DNA binding proteins that regulate cell growth, transformation, angiogenesis, cell‐cycle control and apoptosis; c‐Myc is directly involved in regulating up to 15% of all human genes. c‐Myc amplification is one of the most frequently detected aberrations in breast cancer. Amplification is clearly associated with poor prognosis. In ER‐positive breast cancer cells c‐Myc expression is under the regulation of estrogen, but in ER‐negative/PR‐negative cells c‐Myc constitutive expression level is usually high. C‐MYC protein may affect the response to chemotherapy, probably through DNA damage response regulation (Aulmann et al., 2006; Corzo et al., 2006). BRCA1 is linked to transcriptional regulation through interaction with Myc. Hence, the Myc role in BRCA1‐associated breast cancer makes it an important target in basal‐like/triple‐negative breast cancers.
It was found that only 4% of basal‐like carcinomas showed Myc amplification, compared to 8.75% and 10.7% of luminal and HER2 tumors, respectively (Rodriguez‐Pinilla et al., 2007). Myc amplification displayed a significant association with shorter metastasis‐free and overall survival and proved to be an independent prognostic factor in multivariate survival analysis. Thus, Myc amplification is not associated with “basal‐like” phenotype and proved to be an independent prognostic factor for breast cancer patients treated with anthracycline‐based chemotherapy.
Conflicting results were obtained using microarray technology and performing a detailed kinetic study of genes that respond to MYCN or MYCNDeltaMBII induction in primary human fibroblasts (Chandriani et al., 2009). An overlapping set of 398 genes was designated as “Core MYC Signature” and used for further analysis. Comparison to a panel of breast cancers revealed a strong concordance in gene expression between the Core MYC Signature and the basal‐like breast tumor subtype. This concordance was supported by the higher average level of Myc expression in the same tumor samples. The Core MYC Signature has clinical relevance as this profile can be used to deduce an underlying genetic program that is likely to contribute to a clinical phenotype and may predict clinical responsiveness to drugs that are designed to disrupt Myc‐mediated phenotypes.
5.3.4. VEGF receptor
Targeting angiogenesis by the monocloncal antibody bevacizumab, added to paclitaxel, was shown to be beneficial in terms of prolonged progression free survival (from 5.9 to 11.8 months) in a randomized phase III trial which included patients with both ER‐ and PR‐positive, as well as ER‐ and PR‐negative disease. The majority of patients in this study were HER2‐negative and in the subset analysis ER‐/PR‐positive and negative patients had a similar benefit from bevacizumab. In the neoadjuvant setting preliminary results from a non‐randomized phase II study have been reported which added bevacizumab to cisplatinum. Effects were modest (complete pathological response in 15%) while toxicity was considerable (Ryan et al., 2009). Also, the effects of the small molecule TKI sunitinib, which targets angiogenesis by binding the intracellular domain of cell surface receptors, is modest in metastatic breast cancer and is achieved at the price of significant toxicity (Burstein et al., 2008). In a recently presented randomized phase II trial it was shown that the addition of the another TKI, sorafenib, to capecitabine increased progression free survival from 4.1 to 6.4 months compared to capecitabine monotherapy, again at the cost of considerable toxicity (Baselga et al., 2009a). Of note, these studies with TKIs have not been specifically designed for patients with TNBC.
5.3.5. The role of PARP inhibition
One of the promising new agents for the treatment of TNBC are poly(ADP‐ribose) polymerase (PARP) inhibitors. A subset of PARPs is specifically involved in the detection of single strand breaks and the recruitment of base excision repair elements. In cells with alterations in BRCA function, as is often seen in TNBC, DNA repair processes are largely dependent on PARPs. Inhibition of PARP in these cells ultimately leads to cell death (McCabe et al., 2006; Farmer et al., 2005). Phase I data for the PARP inhibitor olaparib (AZD2281) suggest antitumor effectiveness in cancers associated with the BRCA1 or BRCA2 mutation (Fong et al., 2009). Preliminary results of a recent randomized phase II study with the PARP inhibitor BSI‐201, combined with carboplatin and gemcitabine in metastatic TNBC, showed significantly improved clinical benefit rate, progression free survival and overall survival (O'Shaughnessy et al., 2009).
In conclusion, the development of targeted agents is urgently needed for patients with TNBC. Although promising agents are being developed, no targeted treatment is yet available for routine clinical practice.
6. The promise of multidimension technological approaches
The multiplicity of interacting related factors across the entire genome, the integration of pathways responsible for molecular and pathological factors, and the evolution of imaging approaches, require handling of redundant network interactions (Fig. 2) rather than simple linear systems, to design more robust prognostic and predictive models and treatment algorithms for TNBC and other breast cancer subtypes (Goldhirsch et al., 2009).
Powerful technologies allowing us to take a more comprehensive overview of these multiple events are represented by multivariate data analysis tools and integration of their results into a suitable systems biology approach.
6.1. Multivariate analysis of multi‐modal, multi‐dimensional ‘omics’ data
In the search for biomarkers for breast cancer diagnosis and recognition of subtypes, such as TNBC, for predictive markers of response to treatment, and in the attempt to understand the biological processes/pathways involved in breast cancer (which could also lead to new biomarkers and therapy targets), high dimensional data are frequently used. These data result from the different modern ‘omics’ related analyses (e.g. microarrays in genomics, mass spectrometry combined with some kind of separation, such as liquid chromatography (LC), proteomics or NMR‐based metabolomics). Multivariate data analysis techniques are especially suited for this kind of high and multi‐dimensional data sets, in order to improve medical knowledge discovery and integrate various sources of biomedical information (Fig. 3). However, analysis and integration of (bio)medical data remain a challenging task because of the highly complex nature and the large diversity of the data. Therefore, these approaches, which include data modeling, data visualization and data mining, require a multidisciplinary research environment, unifying expert knowledge from the field of medicine, biochemistry, bioinformatics, biology, chemistry, machine learning and statistics. Data analysis processes and, in particular, the integration of heterogeneous biomedical information provide improved prevention, diagnosis and treatment of disease (e.g. cancer), since the technology enables to better understand the underlying biological relations. Discovery of these relations results in the development of new screening techniques and strategies in the early diagnosis of cancer, including tools for the monitoring and interpretation of disease progression to expand the possibilities and effectiveness of already existing therapies. There is an increasing interest in using biological characteristics to subcategorize cancer within a histological class to provide an improved diagnostic system. For instance, breast tumors could be classified into subtypes, having distinct differences in gene expression patterns, by means of multivariate analysis techniques (Perou et al., 1999, 2000). In addition, evidence has been reported that the incorporation of gene expression signatures into clinical risk stratification can refine prognosis, resulting in optimized therapeutic strategies for breast cancer (Acharya et al., 2008).
Figure 3.
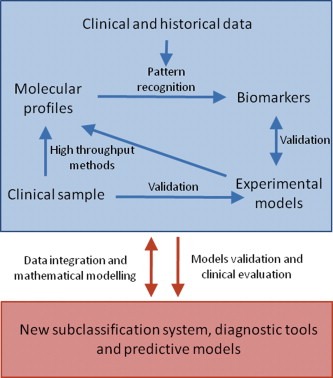
The flow, analysis and integration of multi‐dimensional molecular, biological and clinical data from different biomedical sources leading to a new classification system, diagnostic tools and predictive models in breast cancer.
In the context of breast cancer all kinds of studies are performed using ‘Omic’ data. Most studies involved one type of data, mostly targeted at classification: healthy versus tumor, benign versus tumor, good prognosis versus bad prognosis, etc. Regarding for instance transcriptomic data, Chen et al. (2009) recently combined this data with prior knowledge from already known pathways in order to improve prediction (they applied supervised principal component analysis (PCA) to aggregate genes in so‐called supergenes which are mostly related to outcome). Proteomics data is widely used in breast cancer recognition. An overview has been recently presented by Gast et al. (2009), based on measurements in various matrices like serum, plasma, nipple aspirate fluid, saliva, etc., although the data are multivariate and high dimensional advanced multivariate techniques are rarely used (see e.g. Belluco et al., 2007). An additional problem was the relative low number of samples involved. Furthermore, most of the emphasis was on classification and less on identification of the peaks involved. Metabolites are, in contrast to proteins, much easier to identify. Altered metabolic activities of cancer cells have been observed in many studies. Data in these studies result from mass spectrometry, and in vivo and ex‐vivo MRS (e.g. HR‐MAS MRS of breast cancer tissues). An overview of ex‐vivo MRS studies is presented by Sitter et al. (2009). Again, also in these studies relative simple univariate statistical tests are used to differentiate benign from cancer tissues concentrating on choline related peaks. A broader overview of the applications and perspectives of metabolomics is presented by Claudino et al. (2007). Multivariate techniques (Partial Least Squares, Probabilistic Neural Networks, Support Vector Machines) are applied in e.g. Bathen et al. (2007) on HR‐MAS spectra of breast cancer tissue biopsies, in Woo et al. (2009) using endogenous steroids and nucleosides profiling from GC–MS and LC‐MS measurements of urine, and in Henneges et al. (2009) using a limited set of metabolites from LC‐MS spectra of urine samples. Combining the data and information resulting from the various ‘omics’ techniques is expected to improve prediction and classification. An example of this kind of studies is presented in Chuang et al. (2007). The authors applied a protein‐network‐based approach to analyze the expression profiles of the two cohorts of breast cancer patients previously reported by two other groups. The protein–protein interaction network information was extracted from yeast data from literature. Still mostly statistical techniques were applied to combine the information, after which logistic regression was used to classify the patient data. An improved accuracy was achieved compared to the original articles.
Although having their value in the diagnosis, sub‐classification and prognosis prediction of breast cancer, most of these studies provided just limited or partial additional insight on the complex biological processes involved in breast tumor formation, the biological differences underlying the various types of breast tumor and prognosis, and the processes influencing the response differences of the various therapies. Recent functional genomic and proteomic approaches include protein–protein, protein–DNA or other ‘component–component’ interaction mapping (interactome mapping), systematic phenotypic analyses (phenome mapping) and transcript or protein localization mapping (localizome mapping). ‘Omic’ approaches have already been applied to many biological processes, leading to large lists of genes, proteins and metabolites potentially involved in the corresponding biological processes. As stated earlier, an improvement is expected in combining the data and the results of the various single ‘omics’ approaches leading to an approach within Systems Biology called “top‐down” modeling of the biological processes. Examples of studies more or less aiming at this goal are in Pujana et al. (2007), related to breast cancer and in Ergűn et al. (2007), related to prostate cancer, whereas Nam et al. (2009) is on the border between the application of various data sources to improve classification and the use of these various sources to obtain better insight. In Nam et al. (2009), gene expression data sets of breast cancer and normal subjects, transcriptional regulation information and metabolic pathway information from appropriate databases were combined. Using statistics the authors tried to find altered metabolism (altered pathways) which then lead to a set of potential biomarkers. The latter were then tested in urine samples of various types of breast cancer patients and healthy individuals. Six biomarkers appeared to be statistically different, but the area under the receiver operating characteristic (ROC) curve improved when the biomarkers were combined using multivariate classifiers. Beside this, the candidate biomarkers were tested for potential use as indicators of cancer progression. Pujana et al. (2007) started from four known breast cancer‐associated genes and their products, BRCA1 and BRCA2 and ATM and CHEK2, called ‘reference genes/proteins’, and used data from gene transcript abundance measurements from samples of healthy human tissues or organs and three cell‐lines, to find genes which potentially are functionally associated (using correlation). Next, these potential functional associations were investigated and ranked by integration with functional associations found in largely non‐overlapping ‘Omic’ data sets by using interologous functional relationships from four species. This ultimately has led to genes/proteins which later were proven to have a functional relationship with the reference genes/proteins. Ergűn et al. (2007) used an approach called mode‐of‐action by network identification (MNI). The goal is to find transcript (genes) which are mostly inconsistent with ‘normal’ regulatory gene networks and which could be mediators of a disease. These ‘normal’ regulatory gene networks are first trained using a large set of microarray expression data from 13 projects spanning 7 cancer types by training a relative simple mathematical model. PCA was applied to reduce the massive dimensions of the data. Then using test data from three types of prostate cancer, those genes were identified which could potentially be mediators for each type. Shen et al. (2009) developed a technique which could be used in this context. They developed a latent variable model to allow multiple data types for the purpose of integrative clustering. Additional progress in Systems Biology is expected from bioinformatic and chemometric techniques, which can handle and intelligently explore and apply the complex multi‐modal multi‐dimensional ‘omics’ data.
An essential aspect of the use of multivariate analysis tools in clinical applications is setting up research studies at more than one medical center. A multi‐center approach allows researchers to address a larger population, which is decisive for a better understanding of the disease and the underlying biological relations (Garcia‐Gomez et al., 2009). Depending on the disease, the number of data is often limited. Large scale multi‐center data collection overcomes the problem of sparse data, scattered over local centers' databases and potential bias due to single‐center effects. Therefore, intelligent multivariate analysis should be applied to a centralized collection of multi‐center data or to a distributed database, where each node represents a single center. Furthermore, analysis of multi‐center data, including different types of data, requires the introduction of a common terminology, which is related to the design of ontologies. In addition, the data of the different centers have to be compatible, demanding a fixed protocol for data acquisition and acquisition conditions. In conclusion, results from the analysis of multi‐center studies can potentially unravel the biological dynamics of disease and improve healthcare.
An illustration of the use of multivariate analysis in multi‐center studies is the development of decision support tools within the context of the European projects IOTA, eTUMOUR and HealthAgents (http://www.etumour.net; http://www.healthagents.net; Van Holsbeke et al., 2007; Timmerman et al., 2005). The multi‐center project IOTA focuses on the characterization of ovarian tumors. Ultrasound images and clinical data from 1066 patients were acquired during the period 1999–2002 (Van Holsbeke et al., 2007). Mathematical models to predict the risk of malignancy were developed and were internally and externally validated on nearly 2000 patients with adnexal tumors in 20 centers throughout the world during the periods 2002–2005 and 2005–2007, respectively (Timmerman et al., 2005). Currently, the IOTA project investigates the incorporation of proteomics data in the process of pattern analysis. The multidisciplinary eTUMOUR and HealthAgents projects aimed to develop and validate decision support tools for brain cancer diagnostics by combining multi‐modal multi‐dimensional clinical data, ‘Omic’ data (genomics, metabolomics, transcriptomics) and bioimaging data (in vivo magnetic resonance spectroscopy (MRS), magnetic resonance spectroscopic imaging (MRSI), magnetic resonance imaging (MRI), which were acquired over multiple centers from all over the world and stored in a database system (Lluch‐Ariet et al., 2008). Models for tumor diagnosis were developed from these data and included artificial neural networks, support vector machines, discriminant analysis and nearest neighbour classifiers (Bishop, 1995; Duda et al., 2001; Vapnik, 2002). Furthermore, these models were included in a graphical user interface to make them available for clinicians. The existing breast cancer consortia either focus on ‘Omic’ data only (such as METAcancer (http://www.metacancer‐fp7.eu), MetaBre (http://www.metabre.org), TRANS‐BIG (http://www.breastinternationalgroup.org/Research/TRANSBIG.aspx), or on imaging data only (such as MammoGrid (http://www.cems.uwe.ac.uk/cccs/project.php?name=mammogrid) using mammographic data). This overview enlightens the need for setting up similar European‐wide or worldwide consortia for breast cancer diagnostics by combining all data, from the nano‐level (‘Omic’ data) up to the macro‐level (in vitro, in vivo) so as to further improve breast cancer diagnostics and prognosis.
6.2. The power of systems biology approaches in cancer research
Systems biology is an area that has both provided many promises to support understanding of cellular regulation and that requires new ways of producing and analyzing data. Experimental research and computational modeling of biological processes ranges from small regulatory networks to full‐scale genomic models. Depending on the specific question, they are of different value. Clinically relevant studies need models that provide testable predictions.
Network models comprise interaction information of all compounds of a type such as proteins or genes. Model analysis may reveal insight into hidden relations. A respective study has, for example, linked breast cancer susceptibility with centrosome dysfunction (Pujana et al., 2007): by combining expression profiles with functional genomic and proteomic data, 118 genes were aligned with potential functions. Among those genes is the one coding for hyaluronan‐mediated motility receptor, HMMR, and a functional association with the breast cancer‐associated gene BRCA1 was predicted. Another large‐scale study has systematically linked disorders and genes associated with the diseases (Goh et al., 2007).
Signaling pathways by which cells receive and process external information are a subject of intensive study in experimental and computational systems biology. Amongst the most carefully studied pathways are the EGFR (Samaga et al., 2009; Schoeberl et al., 2002), Wnt (Lee et al., 2003), Jak/STAT (Swameye et al., 2003), TGFβ (Zi and Klipp, 2007; Schmierer et al., 2008), and NFkB pathways. As example for the relevance of signaling pathways for breast cancer, it has been demonstrated recently that TGFβ signaling switches breast cancer cell motility (Giampieri et al., 2009). The models are frequently formulated as sets of ordinary differential equations (ODEs) describing the temporal evolution of the involved components and their activity. They serve various purposes, among them most notably (i) just to understand their architecture and the observed dynamics and (ii) to rationalize the interdependence of independently measured data such as ligand supply, phosphorylation states, protein interactions, and gene expression effects (e.g. Chen et al., 2009). Dynamic features such as the effect of positive or negative feedback (e.g., Bluthgen et al., 2009; Kholodenko, 2000), or crosstalk between different pathways (Borisov et al., 2009) have been studied extensively.
Cell cycle progression is highly regulated and failure to respond to checkpoints or external signals may be a first step to cancer. Mathematical modeling of cell cycle has initially focused on model organisms such as frog eggs or yeast (Chen et al., 2000, 2004), but also on mammalian cells (Alfieri et al., 2009). Again, these models first of all serve to test our understanding of the structure or wiring of cell cycle machinery, but they also analyze detailed questions such as the effect of mutants or of unreplicated DNA on cell cycle progression (Zwolak et al., 2009). While including more and more details of the protein machinery and other components into models, the regulation of cell cycle by nutrition, signaling, or checkpoints becomes more and more predictable based on model simulations.
Development, stem cell differentiation and cell reprogramming have been described with mathematical models on different levels. The Davidson lab has delivered gene regulation networks of Xenopus eggs (Davidson et al., 2003) with ever increasing detailedness over last years, which contain activating and inhibiting influences of developmental genes on each other and also provide a link to regulatory signaling pathways such as Wnt pathway. Translating this static representation into a dynamic model is a challenge on its own, but appears feasible with upcoming data. A computational ODE model describes the dynamics of the core network governing stemcellness and cellular differentiation (Chickarmane and Peterson, 2008).
Target prediction is an important goal of systems biology approaches. Based on sufficiently well described networks and properties of the compounds' interaction, mathematical models can be useful to predict targets for treatment and test the outcome of different target positions, treatment strengths, target combinations or temporal combination scenarios (Fitzgerald et al., 2006; Schulz et al., 2009). This field is only in its infancy, since there is still a lack of sufficiently well understood networks, however there are already very promising examples among signaling pathway models, such as the study of the ErbB network with sensitivity analysis (Schoeberl et al., 2009), which identified ErbB3 as a key node in response to ligands. Boolean modeling (assigning activity values of 0 and 1 to nodes which are updated in time based on their inputs from other nodes) of the ErbB receptor regulated G1/S transition has revealed new potential targets in case of de novo trastuzumab resistance in breast cancer.
Chronotherapy is a field that is based on the fact that living beings exhibit various rhythms, such as cell cycle, circadian or annual rhythms. Circadian rhythms have been investigated, their components identified (Ueda et al., 2005) and their dynamics described with mathematical models (Brown et al., 2008). It has turned out that cancer treatment has different effects if supplied at different times of the day (Levi and Schibler, 2007).
Taken together, different directions of systems biology combining experimental and computational analysis have already made first promising contributions to understand aspects of cellular regulation and dysfunction. Further investigations need precise and reproducible data and sensible mathematical descriptions to produce predictive and helpful models in cancer research (Aebersold et al., 2009).
7. Conclusions
7.1. Challenges and limitations
A better understanding of pathological mechanisms of TNBC onset and progression, including the still unclear association with BRCA1 mutations, and the causes of TNBC phenotypic heterogeneity is expected to improve diagnosis, prognosis, treatment and prevention of these cancers. A major limitation to a robust delineation of differences between basal‐like and triple‐negative breast cancer subtypes is the present lack of consensus on immunohistochemical assays to be used in the clinical setting, and the still unclear relationships between traditional diagnostic classifiers and genomic aberrations.
7.2. Limitations of the present therapeutic options
Larger prospective clinical trials are needed to optimize chemotherapy treatments and predict the response of different TNBC subgroups to cytotoxic agents. Although promising agents are presently being developed and tested, no targeted treatment algorithms are as yet available for TNBC at the clinical level.
7.3. Perspectives
Improved awareness of the significance of gene expression signatures combined with a holistic characterization of cell signaling pathways' aberrations, may lead to more suitable prognostic and predictive models, to more adequate algorithms of therapy targeting, and improved molecular imaging approaches for earlier and non invasive tumor response monitoring. Well designed and rigorous studies are needed to resolve still conflicting or partial results.
A possible breakthrough in the field may derive from application of robust multivariate data analyses on well programmed collections of multi‐modal, multi‐dimensional data sets provided by the simultaneous use of different ‘omics’ at pre‐clinical and clinical level, and processing of the results in the frame of a suitable Systems Biology approach.
Acknowledgements
FP gratefully acknowledges stimulating discussions with Dr Silvana Canevari (Fondazione IRCCS Istituto Nazionale Tumori, Milano, Italy). Partial support was given by the Oncology Screening Programme of the Italian Ministry of Health “Sorveglianza di donne ad alto rischio genetico‐familiare di tumore mammario: National Italian Network” and the Italian Association for Cancer Research, AIRC, Milano, Italy. SWH is funded by GOA Ambiorics, GOA MaNet, CoE EF/05/006, FWO G.0341.07 (Datafusion); IWT: TBM070706‐IOTA3; Belgian Federal Science Policy OfficeIUAP P6/04 (DYSCO).JL is a supported by a PhD grant from IWT.
Podo Franca, Buydens Lutgarde M.C., Degani Hadassa, Hilhorst Riet, Klipp Edda, Gribbestad Ingrid S., Van Huffel Sabine, van Laarhoven Hanneke W.M., Luts Jan, Monleon Daniel, Postma Geert J., Schneiderhan-Marra Nicole, Santoro Filippo, Wouters Hans, Russnes Hege G., Sørlie Therese, Tagliabue Elda, Børresen-Dale Anne-Lise, (2010), Triple‐negative breast cancer: Present challenges and new perspectives, Molecular Oncology, 4, doi: 10.1016/j.molonc.2010.04.006.
References
- Acharya, C.R. , Hsu, D.S. , Anders, C.K. , Anguiano, A. , Salter, K.H. , Walters, K.S. , Redman, R.C. , Tuchman, S.A. , Moylan, C.A. , Mukherjee, S. , Barry, W.T. , Dressman, H.K. , Ginsburg, G.S. , Marcom, K.P. , Garman, K.S. , Lyman, G.H. , Nevins, J.R. , Potti, A. , 2008. Gene expression signatures, clinicopathological features, and individualized therapy in breast cancer. JAMA. 299, 1574–1587. [DOI] [PubMed] [Google Scholar]
- Aebersold, R. , Auffray, C. , Baney, E. , Barillot, E. , Brazma, A. , Brett, C. , Brunak, S. , Butte, A. , Califano, A. , Celis, J. , Cufer, T. , Ferrell, J. , Galas, D. , Gallahan, D. , Gatenby, R. , Goldbeter, A. , Hace, N. , Henney, A. , Hood, L. , Iyengar, R. , Jackson, V. , Kallioniemi, O. , Klingmuller, U. , Kolar, P. , Kolch, W. , Kyriakopoulou, C. , Laplace, F. , Lehrach, H. , Marcus, F. , Matrisian, L. , Nolan, G. , Pelkmans, L. , Potti, A. , Sander, C. , Seljak, M. , Singer, D. , Sorger, P. , Stunnenberg, H. , Superti-Furga, G. , Uhlen, M. , Vidal, M. , Weinstein, J. , Wigle, D. , Williams, M. , Wolkenhauer, O. , Zhivotovsky, B. , Zinovyev, A. , Zupan, B. , 2009. Report on EU-USA workshop: how systems biology can advance cancer research (27 October 2008). Mol. Oncol. 3, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal, A. , Gutteridge, E. , Gee, J.M. , Nicholson, R.I. , Robertson, J.F. , 2005. Overview of tyrosine kinase inhibitors in clinical breast cancer. Endocr. Relat. Cancer. 12, (Suppl. 1) S135–S144. [DOI] [PubMed] [Google Scholar]
- Alfieri, R. , Barberis, M. , Chiaradonna, F. , Gaglio, D. , Milanesi, L. , Vanoni, M. , Klipp, E. , Alberghina, L. , 2009. Towards a systems biology approach to mammalian cell cycle: modeling the entrance into S phase of quiescent fibroblasts after serum stimulation. BMC Bioinformatics. 10, (Suppl. 12) S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulmann, S. , Adler, N. , Rom, J. , Helmchen, B. , Schirmacher, P. , Sinn, H.P. , 2006. c-myc amplifications in primary breast carcinomas and their local recurrences. J. Clin. Pathol. 59, 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azoulay, S. , Lae, M. , Freneaux, P. , Merle, S. , Al Ghuzlan, A. , Chnecker, C. , Rosty, C. , Klijanienko, J. , Sigal-Zafrani, B. , Salmon, R. , Fourquet, A. , Sastre-Garau, X. , Vincent-Salomon, A. , 2005. KIT is highly expressed in adenoid cystic carcinoma of the breast, a basal-like carcinoma associated with a favorable outcome. Mod. Pathol. 18, 1623–1631. [DOI] [PubMed] [Google Scholar]
- Baselga, J. , Segalla, J.G.M. , Roché, H. , del Giglio, A. , Ciruelos, E.M. , Cabral Filho, S. , Gomez, P. , Lluch, A. , Llombart, A. , Costa, F. , 2009. SOLTI-0701: a double-blind, randomized phase 2b study evaluating the efficacy and safety of sorafenib (SOR) compared to placebo (PL) when administered in combination with locally advanced breast cancer. Eur. J. Cancer. Supplements 7, 3 [Google Scholar]
- Baselga, J. , Zambetti, M. , Llombart-Cussac, A. , Manikhas, G. , Kubista, E. , Steger, G.G. , Makhson, A. , Tjulandin, S. , Ludwig, H. , Verrill, M. , Ciruelos, E. , Egyhazi, S. , Xu, L.A. , Zerba, K.E. , Lee, H. , Clark, E. , Galbraith, S. , 2009. Phase II genomics study of ixabepilone as neoadjuvant treatment for breast cancer. J. Clin. Oncol. 27, 526–534. [DOI] [PubMed] [Google Scholar]
- Basu, S. , Chen, W. , Tchou, J. , Mavi, A. , Cermik, T. , Czerniecki, B. , Schnall, M. , Alavi, A. , 2008. Comparison of triple-negative and estrogen receptor-positive/progesterone receptor-positive/HER2-negative breast carcinoma using quantitative fluorine-18 fluorodeoxyglucose/positron emission tomography imaging parameters: a potentially useful method for disease characterization. Cancer. 112, 995–1000. [DOI] [PubMed] [Google Scholar]
- Bathen, T.F. , Jensen, L.R. , Sitter, B. , Fjosne, H.E. , Halgunset, J. , Axelson, D.E. , Gribbestad, I.S. , Lundgren, S. , 2007. MR-determined metabolic phenotype of breast cancer in prediction of lymphatic spread, grade, and hormone status. Breast Cancer Res. Treat. 104, 181–189. [DOI] [PubMed] [Google Scholar]
- Bauer, K.R. , Brown, M. , Cress, R.D. , Parise, C.A. , Caggiano, V. , 2007. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 109, 1721–1728. [DOI] [PubMed] [Google Scholar]
- Belkic, K. , 2004. Current dilemmas and future perspectives for breast cancer screening with a focus on optimization of magnetic resonance spectroscopic imaging by advances in signal processing. Isr. Med. Assoc. J. 6, 610–618. [PubMed] [Google Scholar]
- Belluco, C. , Petricoin, E.F. , Mammano, E. , Facchiano, F. , Ross-Rucker, S. , Nitti, D. , Di Maggio, C. , Liu, C. , Lise, M. , Liotta, L.A. , Whiteley, G. , 2007. Serum proteomic analysis identifies a highly sensitive and specific discriminatory pattern in stage 1 breast cancer. Ann. Surg. Oncol. 14, 2470–2476. [DOI] [PubMed] [Google Scholar]
- Bergamaschi, A. , Kim, Y.H. , Wang, P. , Sørlie, T. , Hernandez-Boussard, T. , Lonning, P.E. , Tibshirani, R. , Børresen-Dale, A.L. , Pollack, J.R. , 2006. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 45, 1033–1040. [DOI] [PubMed] [Google Scholar]
- Bertucci, F. , Finetti, P. , Cervera, N. , Charafe-Jauffret, E. , Mamessier, E. , Adelaide, J. , Debono, S. , Houvenaeghel, G. , Maraninchi, D. , Viens, P. , Charpin, C. , Jacquemier, J. , Birnbaum, D. , 2006. Gene expression profiling shows medullary breast cancer is a subgroup of basal breast cancers. Cancer Res. 66, 4636–4644. [DOI] [PubMed] [Google Scholar]
- Bild, A.H. , Parker, J.S. , Gustafson, A.M. , Acharya, C.R. , Hoadley, K.A. , Anders, C. , Marcom, P.K. , Carey, L.A. , Potti, A. , Nevins, J.R. , Perou, C.M. , 2009. An integration of complementary strategies for gene-expression analysis to reveal novel therapeutic opportunities for breast cancer. Breast Cancer Res. 11, R55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, C.M. , 1995. Neural Networks and Pattern Recognition Oxford University Press; Oxford, UK: [Google Scholar]
- Bluthgen, N. , Legewie, S. , Kielbasa, S.M. , Schramme, A. , Tchernitsa, O. , Keil, J. , Solf, A. , Vingron, M. , Schafer, R. , Herzel, H. , Sers, C. , 2009. A systems biological approach suggests that transcriptional feedback regulation by dual-specificity phosphatase 6 shapes extracellular signal-related kinase activity in RAS-transformed fibroblasts. FEBS J. 276, 1024–1035. [DOI] [PubMed] [Google Scholar]
- Bogner, W. , Gruber, S. , Pinker, K. , Grabner, G. , Stadlbauer, A. , Weber, M. , Moser, E. , Helbich, T.H. , Trattnig, S. , 2009. Diffusion-weighted MR for differentiation of breast lesions at 3.0 T: how does selection of diffusion protocols affect diagnosis?. Radiology. 253, 341–351. [DOI] [PubMed] [Google Scholar]
- Bonadona, V. , Dussart-Moser, S. , Voirin, N. , Sinilnikova, O.M. , Mignotte, H. , Mathevet, P. , Bremond, A. , Treilleux, I. , Martin, A. , Romestaing, P. , Raudrant, D. , Rudigoz, R.C. , Lenoir, G.M. , Lasset, C. , 2007. Prognosis of early-onset breast cancer based on BRCA1/2 mutation status in a French population-based cohort and review. Breast Cancer Res. Treat. 101, 233–245. [DOI] [PubMed] [Google Scholar]
- Borisov, N. , Aksamitiene, E. , Kiyatkin, A. , Legewie, S. , Berkhout, J. , Maiwald, T. , Kaimachnikov, N.P. , Timmer, J. , Hoek, J.B. , Kholodenko, B.N. , 2009. Systems-level interactions between insulin-EGF networks amplify mitogenic signaling. Mol. Syst. Biol. 5, 256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch, A. , Eroles, P. , Zaragoza, R. , Viňa, J.R. , Lluch, A. , 2010 Jan 7. Triple-negative breast cancer: molecular features, pathogenesis, treatment and current lines of research. Cancer Treat. Rev. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Bouchalova, K. , Cizkova, M. , Cwiertka, K. , Trojanec, R. , Hajduch, M. , 2009. Triple negative breast cancer–current status and prospective targeted treatment based on HER1 (EGFR), TOP2A and C-MYC gene assessment. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 153, 13–17. [DOI] [PubMed] [Google Scholar]
- Bradley, A. , Medina, D. , 1998. Introduction: BRCA1 and BRCA2 in mammary gland development and tumorigenesis. J. Mammary. Gland Biol. Neoplasia. 3, 363–364. [DOI] [PubMed] [Google Scholar]
- Breast Cancer Linkage Consortium 1997. Pathology of familial breast cancer: differences between breast cancers in carriers of BRCA1 or BRCA2 mutations and sporadic cases. Lancet. 349, 1505–1510. [PubMed] [Google Scholar]
- Brekelmans, C.T. , Seynaeve, C. , Menke-Pluymers, M. , Bruggenwirth, H.T. , Tilanus-Linthorst, M.M. , Bartels, C.C. , Kriege, M. , van Geel, A.N. , Crepin, C.M. , Blom, J.C. , Meijers-Heijboer, H. , Klijn, J.G. , 2006. Survival and prognostic factors in BRCA1-associated breast cancer. Ann. Oncol. 17, 391–400. [DOI] [PubMed] [Google Scholar]
- Brown, S.A. , Kunz, D. , Dumas, A. , Westermark, P.O. , Vanselow, K. , Tilmann-Wahnschaffe, A. , Herzel, H. , Kramer, A. , 2008. Molecular insights into human daily behavior. Proc. Natl. Acad. Sci. U. S. A. 105, 1602–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, D.J. , Doles, J. , Zender, L. , Xue, W. , Ma, B. , McCombie, W.R. , Hannon, G.J. , Lowe, S.W. , Hemann, M.T. , 2008. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 105, 9053–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein, H.J. , Elias, A.D. , Rugo, H.S. , Cobleigh, M.A. , Wolff, A.C. , Eisenberg, P.D. , Lehman, M. , Adams, B.J. , Bello, C.L. , DePrimo, S.E. , Baum, C.M. , Miller, K.D. , 2008. Phase II study of sunitinib malate, an oral multitargeted tyrosine kinase inhibitor, in patients with metastatic breast cancer previously treated with an anthracycline and a taxane. J. Clin. Oncol. 26, 1810–1816. [DOI] [PubMed] [Google Scholar]
- Byrski, T. , Huzarski, T. , Dent, R. , Gronwald, J. , Zuziak, D. , Cybulski, C. , Kladny, J. , Gorski, B. , Lubinski, J. , Narod, S.A. , 2009. Response to neoadjuvant therapy with cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res. Treat. 115, 359–363. [DOI] [PubMed] [Google Scholar]
- Carey, L.A. , Perou, C.M. , Livasy, C.A. , Dressler, L.G. , Cowan, D. , Conway, K. , Karaca, G. , Troester, M.A. , Tse, C.K. , Edmiston, S. , Deming, S.L. , Geradts, J. , Cheang, M.C. , Nielsen, T.O. , Moorman, P.G. , Earp, H.S. , Millikan, R.C. , 2006. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 295, 2492–2502. [DOI] [PubMed] [Google Scholar]
- Carey, L.A. , Dees, E.C. , Sawyer, L. , Gatti, L. , Moore, D.T. , Collichio, F. , Ollila, D.W. , Sartor, C.I. , Graham, M.L. , Perou, C.M. , 2007. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin. Cancer Res. 13, 2329–2334. [DOI] [PubMed] [Google Scholar]
- Carey, L.A. , Rugo, H.S. , Marcom, P.K. , Irvin, W. , Ferraro, M. , Burrows, E. , He, X. , Perou, C.M. , Winer, E.P. , on behalf of the Translational Breast Cancer Research Consortium2008. TBCRC 001: EGFR inhibition with cetuximab added to carboplatin in metastatic triple-negative (basal-like) breast cancer. J. Clin. Oncol. 26, (Suppl. 15S) abstr 1009 [Google Scholar]
- Calza, S. , Hall, P. , Auer, G. , Bjohle, J. , Klaar, S. , Kronenwett, U. , Liu, E.T. , Miller, L. , Ploner, A. , Smeds, J. , Bergh, J. , Pawitan, Y. , 2006. Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res. 8, R34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandriani, S. , Frengen, E. , Cowling, V.H. , Pendergrass, S.A. , Perou, C.M. , Whitfield, M.L. , Cole, M.D. , 2009. A core MYC gene expression signature is prominent in basal-like breast cancer but only partially overlaps the core serum response. PLoS ONE. 4, e6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H.Y. , Nuyten, D.S. , Sneddon, J.B. , Hastie, T. , Tibshirani, R. , Sørlie, T. , Dai, H. , He, Y.D. , van't Veer, L.J. , Bartelink, H. , van de Rijn, M. , Brown, P.O. , van de Vijver, M.J. , 2005. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc. Natl. Acad. Sci. U. S. A. 102, 3738–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheang, M.C. , Voduc, D. , Bajdik, C. , Leung, S. , McKinney, S. , Chia, S.K. , Perou, C.M. , Nielsen, T.O. , 2008. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin. Cancer Res. 14, 1368–1376. [DOI] [PubMed] [Google Scholar]
- Chen, K.C. , Csikasz-Nagy, A. , Gyorffy, B. , Val, J. , Novak, B. , Tyson, J.J. , 2000. Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol. Biol. Cell. 11, 369–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K.C. , Calzone, L. , Csikasz-Nagy, A. , Cross, F.R. , Novak, B. , Tyson, J.J. , 2004. Integrative analysis of cell cycle control in budding yeast. Mol. Biol. Cell. 15, 3841–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.H. , Agrawal, G. , Feig, B. , Baek, H.M. , Carpenter, P.M. , Mehta, R.S. , Nalcioglu, O. , Su, M.Y. , 2007. Triple-negative breast cancer: MRI features in 29 patients. Ann. Oncol. 18, 2042–2043. [DOI] [PubMed] [Google Scholar]
- Chen, W.W. , Schoeberl, B. , Jasper, P.J. , Niepel, M. , Nielsen, U.B. , Lauffenburger, D.A. , Sorger, P.K. , 2009. Input-output behavior of ErbB signaling pathways as revealed by a mass action model trained against dynamic data. Mol. Syst. Biol. 5, 239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Wang, L. , 2009. Integrating biological knowledge with gene expression profiles for survival prediction of cancer. J. Comput. Biol. 16, 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chickarmane, V. , Peterson, C. , 2008. A computational model for understanding stem cell, trophectoderm and endoderm lineage determination. PLoS ONE. 3, e3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, K. , DeVries, S. , Fridlyand, J. , Spellman, P.T. , Roydasgupta, R. , Kuo, W.L. , Lapuk, A. , Neve, R.M. , Qian, Z. , Ryder, T. , Chen, F. , Feiler, H. , Tokuyasu, T. , Kingsley, C. , Dairkee, S. , Meng, Z. , Chew, K. , Pinkel, D. , Jain, A. , Ljung, B.M. , Esserman, L. , Albertson, D.G. , Waldman, F.M. , Gray, J.W. , 2006. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 10, 529–541. [DOI] [PubMed] [Google Scholar]
- Chin, S.F. , Teschendorff, A.E. , Marioni, J.C. , Wang, Y. , Barbosa-Morais, N.L. , Thorne, N.P. , Costa, J.L. , Pinder, S.E. , van de Wiel, M.A. , Green, A.R. , Ellis, I.O. , Porter, P.L. , Tavare, S. , Brenton, J.D. , Ylstra, B. , Caldas, C. , 2007. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 8, R215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, H.Y. , Lee, E. , Liu, Y.T. , Lee, D. , Ideker, T. , 2007. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 3, 140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudino, W.M. , Quattrone, A. , Biganzoli, L. , Pestrin, M. , Bertini, I. , Di Leo, A. , 2007. Metabolomics: available results, current research projects in breast cancer, and future applications. J. Clin. Oncol. 25, 2840–2846. [DOI] [PubMed] [Google Scholar]
- Cleator, S. , Heller, W. , Coombes, R.C. , 2007. Triple-negative breast cancer: therapeutic options. Lancet Oncol. 8, 235–244. [DOI] [PubMed] [Google Scholar]
- Collett, K. , Stefansson, I.M. , Eide, J. , Braaten, A. , Wang, H. , Eide, G.E. , Thoresen, S.O. , Foulkes, W.D. , Akslen, L.A. , 2005. A basal epithelial phenotype is more frequent in interval breast cancers compared with screen detected tumors. Cancer Epidemiol. Biomarkers Prev. 14, 1108–1112. [DOI] [PubMed] [Google Scholar]
- Corkery, B. , Crown, J. , Clynes, M. , O'Donovan, N. , 2009. Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer. Ann. Oncol. 20, 862–867. [DOI] [PubMed] [Google Scholar]
- Cornelis, R.S. , Vasen, H.F. , Meijers-Heijboer, H. , Ford, D. , van Vliet, M. , van Tilborg, A.A. , Cleton, F.J. , Klijn, J.G. , Menko, F.H. , Meera Khan, P. , 1995. Age at diagnosis as an indicator of eligibility for BRCA1 DNA testing in familial breast cancer. Hum. Genet. 95, 539–544. [DOI] [PubMed] [Google Scholar]
- Corzo, C. , Corominas, J.M. , Tusquets, I. , Salido, M. , Bellet, M. , Fabregat, X. , Serrano, S. , Sole, F. , 2006. The MYC oncogene in breast cancer progression: from benign epithelium to invasive carcinoma. Cancer Genet. Cytogenet. 165, 151–156. [DOI] [PubMed] [Google Scholar]
- Dalgin, G.S. , Alexe, G. , Scanfeld, D. , Tamayo, P. , Mesirov, J.P. , Ganesan, S. , DeLisi, C. , Bhanot, G. , 2007. Portraits of breast cancer progression. BMC Bioinformatics. 8, 291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, E.H. , McClay, D.R. , Hood, L. , 2003. Regulatory gene networks and the properties of the developmental process. Proc. Natl. Acad. Sci. U. S. A. 100, 1475–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, S.J. , Provenzano, E. , Caldas, C. , 2009. Triple negative breast cancers: clinical and prognostic implications. Eur. J. Cancer. 45, (Suppl. 1) 27–40. [DOI] [PubMed] [Google Scholar]
- de Vries, E.F. , Rots, M.G. , Hospers, G.A. , 2007. Nuclear imaging of hormonal receptor status in breast cancer: a tool for guiding endocrine treatment and drug development. Curr. Cancer Drug Targets. 7, 510–519. [DOI] [PubMed] [Google Scholar]
- Dent, R. , Trudeau, M. , Pritchard, K.I. , Hanna, W.M. , Kahn, H.K. , Sawka, C.A. , Lickley, L.A. , Rawlinson, E. , Sun, P. , Narod, S.A. , 2007. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. 13, 4429–4434. [DOI] [PubMed] [Google Scholar]
- Dowsett, M. , Dunbier, A.K. , 2008. Emerging biomarkers and new understanding of traditional markers in personalized therapy for breast cancer. Clin. Cancer Res. 14, 8019–8026. [DOI] [PubMed] [Google Scholar]
- Duda, R. , Hart, P. , Stork, D. , 2001. Pattern Classification Wiley; New York, USA: [Google Scholar]
- Durbecq, V. , Di Leo, A. , Cardoso, F. , Rouas, G. , Leroy, J.Y. , Piccart, M. , Larsimont, D. , 2003. Comparison of topoisomerase-IIalpha gene status between primary breast cancer and corresponding distant metastatic sites. Breast Cancer Res. Treat. 77, 199–204. [DOI] [PubMed] [Google Scholar]
- Easton, D.F. , Ford, D. , Bishop, D.T. , 1995. Breast and ovarian cancer incidence in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Am. J. Hum. Genet. 56, 265–271. [PMC free article] [PubMed] [Google Scholar]
- Eisinger, F. , Stoppa-Lyonnet, D. , Longy, M. , Kerangueven, F. , Noguchi, T. , Bailly, C. , Vincent-Salomon, A. , Jacquemier, J. , Birnbaum, D. , Sobol, H. , 1996. Germ line mutation at BRCA1 affects the histoprognostic grade in hereditary breast cancer. Cancer Res. 56, 471–474. [PubMed] [Google Scholar]
- Ergűn, A. , Lawrence, C.A. , Kohanski, M.A. , Brennan, T.A. , Collins, J.J. , 2007. A network biology approach to prostate cancer. Mol. Syst. Biol. 3, 82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, H. , McCabe, N. , Lord, C.J. , Tutt, A.N. , Johnson, D.A. , Richardson, T.B. , Santarosa, M. , Dillon, K.J. , Hickson, I. , Knights, C. , Martin, N.M. , Jackson, S.P. , Smith, G.C. , Ashworth, A. , 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 434, 917–921. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, J.B. , Schoeberl, B. , Nielsen, U.B. , Sorger, P.K. , 2006. Systems biology and combination therapy in the quest for clinical efficacy. Nat. Chem. Biol. 2, 458–466. [DOI] [PubMed] [Google Scholar]
- Fong, P.C. , Boss, D.S. , Yap, T.A. , Tutt, A. , Wu, P. , Mergui-Roelvink, M. , Mortimer, P. , Swaisland, H. , Lau, A. , O'Connor, M.J. , Ashworth, A. , Carmichael, J. , Kaye, S.B. , Schellens, J.H. , de Bono, J.S. , 2009. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361, 123–134. [DOI] [PubMed] [Google Scholar]
- Ford, D. , Easton, D.F. , Bishop, D.T. , Narod, S.A. , Goldgar, D.E. , 1994. Risks of cancer in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Lancet. 343, 692–695. [DOI] [PubMed] [Google Scholar]
- Foulkes, W.D. , 2004. BRCA1 functions as a breast stem cell regulator. J. Med. Genet. 41, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes, W.D. , Metcalfe, K. , Hanna, W. , Lynch, H.T. , Ghadirian, P. , Tung, N. , Olopade, O. , Weber, B. , McLennan, J. , Olivotto, I.A. , Sun, P. , Chappuis, P.O. , Begin, L.R. , Brunet, J.S. , Narod, S.A. , 2003. Disruption of the expected positive correlation between breast tumor size and lymph node status in BRCA1-related breast carcinoma. Cancer. 98, 1569–1577. [DOI] [PubMed] [Google Scholar]
- Foulkes, W.D. , Stefansson, I.M. , Chappuis, P.O. , Begin, L.R. , Goffin, J.R. , Wong, N. , Trudel, M. , Akslen, L.A. , 2003. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J. Natl. Cancer Inst. 95, 1482–1485. [DOI] [PubMed] [Google Scholar]
- Fridlyand, J. , Snijders, A.M. , Ylstra, B. , Li, H. , Olshen, A. , Segraves, R. , Dairkee, S. , Tokuyasu, T. , Ljung, B.M. , Jain, A.N. , McLennan, J. , Ziegler, J. , Chin, K. , Devries, S. , Feiler, H. , Gray, J.W. , Waldman, F. , Pinkel, D. , Albertson, D.G. , 2006. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 6, 96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulford, L.G. , Easton, D.F. , Reis-Filho, J.S. , Sofronis, A. , Gillett, C.E. , Lakhani, S.R. , Hanby, A. , 2006. Specific morphological features predictive for the basal phenotype in grade 3 invasive ductal carcinoma of breast. Histopathology. 49, 22–34. [DOI] [PubMed] [Google Scholar]
- Furuta, S. , Jiang, X. , Gu, B. , Cheng, E. , Chen, P.L. , Lee, W.H. , 2005. Depletion of BRCA1 impairs differentiation but enhances proliferation of mammary epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 102, 9176–9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gomez, J.M. , Luts, J. , Julia-Sape, M. , Krooshof, P. , Tortajada, S. , Robledo, J.V. , Melssen, W. , Fuster-Garcia, E. , Olier, I. , Postma, G. , Monleon, D. , Moreno-Torres, A. , Pujol, J. , Candiota, A.P. , Martinez-Bisbal, M.C. , Suykens, J. , Buydens, L. , Celda, B. , Van Huffel, S. , Arus, C. , Robles, M. , 2009. Multiproject-multicenter evaluation of automatic brain tumor classification by magnetic resonance spectroscopy. MAGMA. 22, 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gast, M.C. , Schellens, J.H. , Beijnen, J.H. , 2009. Clinical proteomics in breast cancer: a review. Breast Cancer Res. Treat. 116, 17–29. [DOI] [PubMed] [Google Scholar]
- Giampieri, S. , Manning, C. , Hooper, S. , Jones, L. , Hill, C.S. , Sahai, E. , 2009. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat. Cell. Biol. 11, 1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giskeødegård, G.F. , Grinde, M.T. , Sitter, B. , Axelson, D.E. , Lundgren, S. , Fjøsne, H.E. , Dahl, S. , Gribbestad, I.S. , Bathen, T.F. , 2010. Multivariate modeling and prediction of breast cancer prognostic factors using MR metabolomics. J. Proteome Res. 9, 972–979. [DOI] [PubMed] [Google Scholar]
- Goh, K.I. , Cusick, M.E. , Valle, D. , Childs, B. , Vidal, M. , Barabasi, A.L. , 2007. The human disease network. Proc. Natl. Acad. Sci. U. S. A. 104, 8685–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldhirsch, A. , Ingle, J.N. , Gelber, R.D. , Coates, A.S. , Thurlimann, B. , Senn, H.J. , 2009. Thresholds for therapies: highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2009. Ann. Oncol. 20, 1319–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski, J.J. , James, C.R. , Quinn, J.E. , Stewart, G.E. , Staunton, K.C. , Buckley, N.E. , McDyer, F.A. , Kennedy, R.D. , Wilson, R.H. , Mullan, P.B. , Harkin, D.P. , 2009. BRCA1 transcriptionally regulates genes associated with the basal-like phenotype in breast cancer. Breast Cancer Res. Treat. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Grushko, T.A. , Blackwood, M.A. , Schumm, P.L. , Hagos, F.G. , Adeyanju, M.O. , Feldman, M.D. , Sanders, M.O. , Weber, B.L. , Olopade, O.I. , 2002. Molecular-cytogenetic analysis of HER-2/neu gene in BRCA1-associated breast cancers. Cancer Res. 62, 1481–1488. [PubMed] [Google Scholar]
- Guarino, M. , Rubino, B. , Ballabio, G. , 2007. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 39, 305–318. [DOI] [PubMed] [Google Scholar]
- Haffty, B.G. , Yang, Q. , Reiss, M. , Kearney, T. , Higgins, S.A. , Weidhaas, J. , Harris, L. , Hait, W. , Toppmeyer, D. , 2006. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J. Clin. Oncol. 24, 5652–5657. [DOI] [PubMed] [Google Scholar]
- Hall, J.M. , Lee, M.K. , Newman, B. , Morrow, J.E. , Anderson, L.A. , Huey, B. , King, M.C. , 1990. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 250, 1684–1689. [DOI] [PubMed] [Google Scholar]
- Haupt, B. , Ro, J.Y. , Schwartz, M.R. , 2010. Basal-like breast carcinoma: a phenotypically distinct entity. Arch. Pathol. Lab. Med. 134, 130–133. [DOI] [PubMed] [Google Scholar]
- He, Q. , Xu, R.Z. , Shkarin, P. , Pizzorno, G. , Lee-French, C.H. , Rothman, D.L. , Shungu, D.C. , Shim, H. , 2003. Magnetic resonance spectroscopic imaging of tumor metabolic markers for cancer diagnosis, metabolic phenotyping, and characterization of tumor microenvironment. Dis. Markers. 19, 69–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig-Burgel, T. , Stiehl, D.P. , Wagner, A.E. , Metzen, E. , Jelkmann, W. , 2005. Review: hypoxia-inducible factor-1 (HIF-1): a novel transcription factor in immune reactions. J. Interferon Cytokine Res. 25, 297–310. [DOI] [PubMed] [Google Scholar]
- Henderson, I.C. , Patek, A.J. , 1998. The relationship between prognostic and predictive factors in the management of breast cancer. Breast Cancer Res. Treat. 52, 261–288. [DOI] [PubMed] [Google Scholar]
- Henneges, C. , Bullinger, D. , Fux, R. , Friese, N. , Seeger, H. , Neubauer, H. , Laufer, S. , Gleiter, C.H. , Schwab, M. , Zell, A. , Kammerer, B. , 2009. Prediction of breast cancer by profiling of urinary RNA metabolites using Support Vector Machine-based feature selection. BMC Cancer. 9, 104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honrado, E. , Osorio, A. , Palacios, J. , Benitez, J. , 2006. Pathology and gene expression of hereditary breast tumors associated with BRCA1, BRCA2 and CHEK2 gene mutations. Oncogene. 25, 5837–5845. [DOI] [PubMed] [Google Scholar]
- Honrado, E. , Osorio, A. , Milne, R.L. , Paz, M.F. , Melchor, L. , Cascon, A. , Urioste, M. , Cazorla, A. , Diez, O. , Lerma, E. , Esteller, M. , Palacios, J. , Benitez, J. , 2007. Immunohistochemical classification of non-BRCA1/2 tumors identifies different groups that demonstrate the heterogeneity of BRCAX families. Mod. Pathol. 20, 1298–1306. [DOI] [PubMed] [Google Scholar]
- Hospers, G.A. , Helmond, F.A. , de Vries, E.G. , Dierckx, R.A. , de Vries, E.F. , 2008. PET imaging of steroid receptor expression in breast and prostate cancer. Curr. Pharm. Des. 14, 3020–3032. [DOI] [PubMed] [Google Scholar]
- Hu, Z. , Fan, C. , Oh, D.S. , Marron, J.S. , He, X. , Qaqish, B.F. , Livasy, C. , Carey, L.A. , Reynolds, E. , Dressler, L. , Nobel, A. , Parker, J. , Ewend, M.G. , Sawyer, L.R. , Wu, J. , Liu, Y. , Nanda, R. , Tretiakova, M. , Ruiz Orrico, A. , Dreher, D. , Palazzo, J.P. , Perreard, L. , Nelson, E. , Mone, M. , Hansen, H. , Mullins, M. , Quackenbush, J.F. , Ellis, M.J. , Olopade, O.I. , Bernard, P.S. , Perou, C.M. , 2006. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 7, 96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurvitz, S.A. , Finn, R.S. , 2009. What's positive about ‘triple-negative’ breast cancer?. Future Oncol. 5, 1015–1025. [DOI] [PubMed] [Google Scholar]
- Hynes, N.E. , Lane, H.A. , 2005. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer. 5, 341–354. [DOI] [PubMed] [Google Scholar]
- Iliopoulos, D. , Polytarchou, C. , Hatziapostolou, M. , Kottakis, F. , Maroulakou, I.G. , Struhl, K. , Tsichlis, P.N. , 2009. MicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cells. Sci. Signal. 2, ra62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin, W.J. , Carey, L.A. , 2008. What is triple-negative breast cancer?. Eur. J. Cancer. 44, 2799–2805. [DOI] [PubMed] [Google Scholar]
- Jackson, S.P. , Bartek, J. , 2009. The DNA-damage response in human biology and disease. Nature. 461, 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemier, J. , Eisinger, F. , Birnbaum, D. , Sobol, H. , 1995. Histoprognostic grade in BRCA1-associated breast cancer. Lancet. 345, 1503 [DOI] [PubMed] [Google Scholar]
- Jacquemier, J. , Padovani, L. , Rabayrol, L. , Lakhani, S.R. , Penault-Llorca, F. , Denoux, Y. , Fiche, M. , Figueiro, P. , Maisongrosse, V. , Ledoussal, V. , Martinez Penuela, J. , Udvarhely, N. , El Makdissi, G. , Ginestier, C. , Geneix, J. , Charafe-Jauffret, E. , Xerri, L. , Eisinger, F. , Birnbaum, D. , Sobol, H. , 2005. Typical medullary breast carcinomas have a basal/myoepithelial phenotype. J. Pathol. 207, 260–268. [DOI] [PubMed] [Google Scholar]
- Japaze, H. , Emina, J. , Diaz, C. , Schwam, R.J. , Gercovich, N. , Demonty, G. , Morgenfeld, E. , Rivarola, E. , Gil Deza, E. , Gercovich, F.G. , 2005. ‘Pure’ invasive apocrine carcinoma of the breast: a new clinicopathological entity?. Breast. 14, 3–10. [DOI] [PubMed] [Google Scholar]
- Jiang, P. , Enomoto, A. , Takahashi, M. , 2009. Cell biology of the movement of breast cancer cells: intracellular signalling and the actin cytoskeleton. Cancer Lett. 284, 122–130. [DOI] [PubMed] [Google Scholar]
- Johannsson, O.T. , Idvall, I. , Anderson, C. , Borg, A. , Barkardottir, R.B. , Egilsson, V. , Olsson, H. , 1997. Tumour biological features of BRCA1-induced breast and ovarian cancer. Eur. J. Cancer. 33, 362–371. [DOI] [PubMed] [Google Scholar]
- Kaplan, H.G. , Malmgren, J.A. , Atwood, M. , 2009. T1N0 triple negative breast cancer: risk of recurrence and adjuvant chemotherapy. Breast J. 15, 454–460. [DOI] [PubMed] [Google Scholar]
- Kassam, F. , Enright, K. , Dent, R. , Dranitsaris, G. , Myers, J. , Flynn, C. , Fralick, M. , Kumar, R. , Clemons, M. , 2009. Survival outcomes for patients with metastatic triple-negative breast cancer: implications for clinical practice and trial design. Clin. Breast Cancer. 9, 29–33. [DOI] [PubMed] [Google Scholar]
- Kholodenko, B.N. , 2000. Negative feedback and ultrasensitivity can bring about oscillations in the mitogen-activated protein kinase cascades. Eur. J. Biochem. 267, 1583–1588. [DOI] [PubMed] [Google Scholar]
- Knoop, A.S. , Knudsen, H. , Balslev, E. , Rasmussen, B.B. , Overgaard, J. , Nielsen, K.V. , Schonau, A. , Gunnarsdottir, K. , Olsen, K.E. , Mouridsen, H. , Ejlertsen, B. , 2005. retrospective analysis of topoisomerase IIa amplifications and deletions as predictive markers in primary breast cancer patients randomly assigned to cyclophosphamide, methotrexate, and fluorouracil or cyclophosphamide, epirubicin, and fluorouracil: Danish Breast Cancer Cooperative Group. J. Clin. Oncol. 23, 7483–7490. [DOI] [PubMed] [Google Scholar]
- Kreike, B. , van Kouwenhove, M. , Horlings, H. , Weigelt, B. , Peterse, H. , Bartelink, H. , van de Vijver, M.J. , 2007. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 9, R65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubista, M. , Rosner, M. , Kubista, E. , Bernaschek, G. , Hengstschlager, M. , 2002. Brca1 regulates in vitro differentiation of mammary epithelial cells. Oncogene. 21, 4747–4756. [DOI] [PubMed] [Google Scholar]
- Kwan, M.L. , Kushi, L.H. , Weltzien, E. , Maring, B. , Kutner, S.E. , Fulton, R.S. , Lee, M.M. , Ambrosone, C.B. , Caan, B.J. , 2009. Epidemiology of breast cancer subtypes in two prospective cohort studies of breast cancer survivors. Breast Cancer Res. 11, R31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix, M. , Leclercq, G. , 2005. The “portrait” of hereditary breast cancer. Breast Cancer Res. Treat. 89, 297–304. [DOI] [PubMed] [Google Scholar]
- Lakhani, S.R. , Jacquemier, J. , Sloane, J.P. , Gusterson, B.A. , Anderson, T.J. , van de Vijver, M.J. , Farid, L.M. , Venter, D. , Antoniou, A. , Storfer-Isser, A. , Smyth, E. , Steel, C.M. , Haites, N. , Scott, R.J. , Goldgar, D. , Neuhausen, S. , Daly, P.A. , Ormiston, W. , McManus, R. , Scherneck, S. , Ponder, B.A. , Ford, D. , Peto, J. , Stoppa-Lyonnet, D. , Bignon, Y.J. , Struewing, J.P. , Spurr, N.K. , Bishop, D.T. , Klijn, J.G. , Devilee, P. , Cornelisse, C.J. , Lasset, C. , Lenoir, G. , Barkardottir, R.B. , Egilsson, V. , Hamann, U. , Chang-Claude, J. , Sobol, H. , Weber, B. , Stratton, M.R. , Easton, D.F. , 1998. Multifactorial analysis of differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations. J. Natl. Cancer Inst. 90, 1138–1145. [DOI] [PubMed] [Google Scholar]
- Lakhani, S.R. , Van De Vijver, M.J. , Jacquemier, J. , Anderson, T.J. , Osin, P.P. , McGuffog, L. , Easton, D.F. , 2002. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J. Clin. Oncol. 20, 2310–2318. [DOI] [PubMed] [Google Scholar]
- Langerod, A. , Zhao, H. , Borgan, O. , Nesland, J.M. , Bukholm, I.R. , Ikdahl, T. , Karesen, R. , Børresen-Dale, A.L. , Jeffrey, S.S. , 2007. TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res. 9, R30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E. , Salic, A. , Kruger, R. , Heinrich, R. , Kirschner, M.W. , 2003. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol. 1, E10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.M. , Dedhar, S. , Kalluri, R. , Thompson, E.W. , 2006. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J. Cell Biol. 172, 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi, F. , Schibler, U. , 2007. Circadian rhythms: mechanisms and therapeutic implications. Annu. Rev. Pharmacol. Toxicol. 47, 593–628. [DOI] [PubMed] [Google Scholar]
- Liedtke, C. , Mazouni, C. , Hess, K.R. , Andre, F. , Tordai, A. , Mejia, J.A. , Symmans, W.F. , Gonzalez-Angulo, A.M. , Hennessy, B. , Green, M. , Cristofanilli, M. , Hortobagyi, G.N. , Pusztai, L. , 2008. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 26, 1275–1281. [DOI] [PubMed] [Google Scholar]
- Liu, S. , Ginestier, C. , Charafe-Jauffret, E. , Foco, H. , Kleer, C.G. , Merajver, S.D. , Dontu, G. , Wicha, M.S. , 2008. BRCA1 regulates human mammary stem/progenitor cell fate. Proc. Natl. Acad. Sci. U. S. A. 105, 1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Kulesz-Martin, M. , 2001. p53 protein at the hub of cellular DNA damage response pathways through sequence-specific and non-sequence-specific DNA binding. Carcinogenesis. 22, 851–860. [DOI] [PubMed] [Google Scholar]
- Livasy, C.A. , Karaca, G. , Nanda, R. , Tretiakova, M.S. , Olopade, O.I. , Moore, D.T. , Perou, C.M. , 2006. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod. Pathol. 19, 264–271. [DOI] [PubMed] [Google Scholar]
- Lluch-Ariet, M. , Estanyol, F. , Mier, M. , Delgado, C. , Gonzalez-Velez, H. , Dalmas, T. , Robles, M. , Saez, C. , Vicente, J. , Van Huffel, S. , Luts, J. , Arus, C. , Candiota, A.P. , Julia-Sape, M. , Peet, A. , Gibb, A. , Sun, Y. , Celda, B. , Martinez-Bisbal, M.C. , Valsecchi, G. , Dupplaw, D. , Hu, B. , Lewis, P. , 2008. On the implementation of HealthAgents: agent-based brain tumour diagnosis. In Annicchiarico R., Cortes U., Urdiales C.(Eds.), Agent-technology and E-health. Birkhäuser; Basel, Switzerland: 5–24. [Google Scholar]
- Lund, M.J. , Trivers, K.F. , Porter, P.L. , Coates, R.J. , Leyland-Jones, B. , Brawley, O.W. , Flagg, E.W. , O'Regan, R.M. , Gabram, S.G. , Eley, J.W. , 2009. Race and triple negative threats to breast cancer survival: a population-based study in Atlanta, GA. Breast Cancer Res. Treat. 113, 357–370. [DOI] [PubMed] [Google Scholar]
- Marcus, J.N. , Watson, P. , Page, D.L. , Narod, S.A. , Lenoir, G.M. , Tonin, P. , Linder-Stephenson, L. , Salerno, G. , Conway, T.A. , Lynch, H.T. , 1996. Hereditary breast cancer: pathobiology, prognosis, and BRCA1 and BRCA2 gene linkage. Cancer. 77, 697–709. [DOI] [PubMed] [Google Scholar]
- Marin-Hernandez, A. , Gallardo-Perez, J.C. , Ralph, S.J. , Rodriguez-Enriquez, S. , Moreno-Sanchez, R. , 2009. HIF-1alpha modulates energy metabolism in cancer cells by inducing over-expression of specific glycolytic isoforms. Mini Rev. Med. Chem. 9, 1084–1101. [DOI] [PubMed] [Google Scholar]
- McCabe, N. , Turner, N.C. , Lord, C.J. , Kluzek, K. , Bialkowska, A. , Swift, S. , Giavara, S. , O'Connor, M.J. , Tutt, A.N. , Zdzienicka, M.Z. , Smith, G.C. , Ashworth, A. , 2006. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 66, 8109–8115. [DOI] [PubMed] [Google Scholar]
- Melchor, L. , Benitez, J. , 2008. An integrative hypothesis about the origin and development of sporadic and familial breast cancer subtypes. Carcinogenesis. 29, 1475–1482. [DOI] [PubMed] [Google Scholar]
- Mersin, H. , Yildirim, E. , Berberoglu, U. , Gulben, K. , 2008. The prognostic importance of triple negative breast carcinoma. Breast. 17, 341–346. [DOI] [PubMed] [Google Scholar]
- Miki, Y. , Swensen, J. , Shattuck-Eidens, D. , Futreal, P.A. , Harshman, K. , Tavtigian, S. , Liu, Q. , Cochran, C. , Bennett, L.M. , Ding, W. , 1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 266, 66–71. [DOI] [PubMed] [Google Scholar]
- Moller, P. , Borg, A. , Evans, D.G. , Haites, N. , Reis, M.M. , Vasen, H. , Anderson, E. , Steel, C.M. , Apold, J. , Goudie, D. , Howell, A. , Lalloo, F. , Maehle, L. , Gregory, H. , Heimdal, K. , 2002. Survival in prospectively ascertained familial breast cancer: analysis of a series stratified by tumour characteristics, BRCA mutations and oophorectomy. Int. J. Cancer. 101, 555–559. [DOI] [PubMed] [Google Scholar]
- Moller, P. , Evans, D.G. , Reis, M.M. , Gregory, H. , Anderson, E. , Maehle, L. , Lalloo, F. , Howell, A. , Apold, J. , Clark, N. , Lucassen, A. , Steel, C.M. , 2007. Surveillance for familial breast cancer: differences in outcome according to BRCA mutation status. Int. J. Cancer. 121, 1017–1020. [DOI] [PubMed] [Google Scholar]
- Morris, G.J. , Naidu, S. , Topham, A.K. , Guiles, F. , Xu, Y. , McCue, P. , Schwartz, G.F. , Park, P.K. , Rosenberg, A.L. , Brill, K. , Mitchell, E.P. , 2007. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: a single-institution compilation compared with the National Cancer Institute's Surveillance, Epidemiology, and end Results database. Cancer. 110, 876–884. [DOI] [PubMed] [Google Scholar]
- Musgrove, E.A. , Sutherland, R.L. , 2009. Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer. 9, 631–643. [DOI] [PubMed] [Google Scholar]
- Nam, H. , Chung, B.C. , Kim, Y. , Lee, K. , Lee, D. , 2009. Combining tissue transcriptomics and urine metabolomics for breast cancer biomarker identification. Bioinformatics. 25, 3151–3157. [DOI] [PubMed] [Google Scholar]
- Narod, S.A. , Foulkes, W.D. , 2004. BRCA1 and BRCA2: 1994 and beyond. Nat. Rev. Cancer. 4, 665–676. [DOI] [PubMed] [Google Scholar]
- Naume, B. , Zhao, X. , Synnestvedt, M. , Borgen, E. , Russnes, H.G. , Lingjaerde, O.C. , Stromberg, M. , Wiedswang, G. , Kvalheim, G. , Karesen, R. , Nesland, J.M. , Børresen-Dale, A.L. , Sørlie, T. , 2007. Presence of bone marrow micrometastasis is associated with different recurrence risk within molecular subtypes of breast cancer. Mol. Oncol. 1, 160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin, N. , Krasnitz, A. , Rodgers, L. , Cook, K. , Meth, J. , Kendall, J. , Riggs, M. , Eberling, Y. , Troge, J. , Grubor, V. , Levy, D. , Lundin, P. , Maner, S. , Zetterberg, A. , Hicks, J. , Wigler, M. , 2010. Inferring tumor progression from genomic heterogeneity. Genome Res. 20, 68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve, R.M. , Chin, K. , Fridlyand, J. , Yeh, J. , Baehner, F.L. , Fevr, T. , Clark, L. , Bayani, N. , Coppe, J.P. , Tong, F. , Speed, T. , Spellman, P.T. , DeVries, S. , Lapuk, A. , Wang, N.J. , Kuo, W.L. , Stilwell, J.L. , Pinkel, D. , Albertson, D.G. , Waldman, F.M. , McCormick, F. , Dickson, R.B. , Johnson, M.D. , Lippman, M. , Ethier, S. , Gazdar, A. , Gray, J.W. , 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 10, 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, T.O. , Hsu, F.D. , Jensen, K. , Cheang, M. , Karaca, G. , Hu, Z. , Hernandez-Boussard, T. , Livasy, C. , Cowan, D. , Dressler, L. , Akslen, L.A. , Ragaz, J. , Gown, A.M. , Gilks, C.B. , van de Rijn, M. , Perou, C.M. , 2004. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin. Cancer Res. 10, 5367–5374. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy, J. , Osborne, C. , Pippen, J. , Yoffe, M. , Patt, D. , Monaghan, G. , Rocha, C. , Ossovskaya, V. , Sherman, B. , Bradley, C. , 2009. Efficacy of BSI-201, a poly (ADP-ribose) polymerase-1 (PARP1) inhibitor, in combination with gemcitabine/carboplatin (G/C) in patients with metastatic triple-negative breast cancer (TNBC): results of a randomized phase II trial. J. Clin. Oncol. (Suppl. 15S) abstr 3 [Google Scholar]
- Orlando, L. , Renne, G. , Rocca, A. , Curigliano, G. , Colleoni, M. , Severi, G. , Peruzzotti, G. , Cinieri, S. , Viale, G. , Sanna, G. , Goldhirsch, A. , 2005. Are all high-grade breast cancers with no steroid receptor hormone expression alike? The special case of the medullary phenotype. Ann. Oncol. 16, 1094–1099. [DOI] [PubMed] [Google Scholar]
- Osborne, C.K. , 1998. Steroid hormone receptors in breast cancer management. Breast Cancer Res. Treat. 51, 227–238. [DOI] [PubMed] [Google Scholar]
- Palacios, J. , Honrado, E. , Osorio, A. , Cazorla, A. , Sarrio, D. , Barroso, A. , Rodriguez, S. , Cigudosa, J.C. , Diez, O. , Alonso, C. , Lerma, E. , Sanchez, L. , Rivas, C. , Benitez, J. , 2003. Immunohistochemical characteristics defined by tissue microarray of hereditary breast cancer not attributable to BRCA1 or BRCA2 mutations: differences from breast carcinomas arising in BRCA1 and BRCA2 mutation carriers. Clin. Cancer Res. 9, 3606–3614. [PubMed] [Google Scholar]
- Parker, J.S. , Mullins, M. , Cheang, M.C. , Leung, S. , Voduc, D. , Vickery, T. , Davies, S. , Fauron, C. , He, X. , Hu, Z. , Quackenbush, J.F. , Stijleman, I.J. , Palazzo, J. , Marron, J.S. , Nobel, A.B. , Mardis, E. , Nielsen, T.O. , Ellis, M.J. , Perou, C.M. , Bernard, P.S. , 2009. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou, C.M. , Jeffrey, S.S. , van de Rijn, M. , Rees, C.A. , Eisen, M.B. , Ross, D.T. , Pergamenschikov, A. , Williams, C.F. , Zhu, S.X. , Lee, J.C. , Lashkari, D. , Shalon, D. , Brown, P.O. , Botstein, D. , 1999. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc. Natl. Acad. Sci. U. S. A. 96, 9212–9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou, C.M. , Sørlie, T. , Eisen, M.B. , van de Rijn, M. , Jeffrey, S.S. , Rees, C.A. , Pollack, J.R. , Ross, D.T. , Johnsen, H. , Akslen, L.A. , Fluge, O. , Pergamenschikov, A. , Williams, C. , Zhu, S.X. , Lonning, P.E. , Børresen-Dale, A.L. , Brown, P.O. , Botstein, D. , 2000. Molecular portraits of human breast tumours. Nature. 406, 747–752. [DOI] [PubMed] [Google Scholar]
- Podo, F. , Sardanelli, F. , Iorio, E. , Canese, R. , Carpinelli, G. , Fausto, A. , Canevari, S. , 2007. Abnormal choline phospholipid metabolism in breast and ovary cancer: molecularbases for non invasive ima ging approaches. Curr. Med. Imaging Rev. 3, 123–137. [Google Scholar]
- Polyak, K. , 2007. Breast cancer: origins and evolution. J. Clin. Invest. 117, 3155–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujana, M.A. , Han, J.D. , Starita, L.M. , Stevens, K.N. , Tewari, M. , Ahn, J.S. , Rennert, G. , Moreno, V. , Kirchhoff, T. , Gold, B. , Assmann, V. , Elshamy, W.M. , Rual, J.F. , Levine, D. , Rozek, L.S. , Gelman, R.S. , Gunsalus, K.C. , Greenberg, R.A. , Sobhian, B. , Bertin, N. , Venkatesan, K. , Ayivi-Guedehoussou, N. , Sole, X. , Hernandez, P. , Lazaro, C. , Nathanson, K.L. , Weber, B.L. , Cusick, M.E. , Hill, D.E. , Offit, K. , Livingston, D.M. , Gruber, S.B. , Parvin, J.D. , Vidal, M. , 2007. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat. Genet. 39, 1338–1349. [DOI] [PubMed] [Google Scholar]
- Rakha, E.A. , Putti, T.C. , Abd El-Rehim, D.M. , Paish, C. , Green, A.R. , Powe, D.G. , Lee, A.H. , Robertson, J.F. , Ellis, I.O. , 2006. Morphological and immunophenotypic analysis of breast carcinomas with basal and myoepithelial differentiation. J. Pathol. 208, 495–506. [DOI] [PubMed] [Google Scholar]
- Rakha, E.A. , El-Sayed, M.E. , Green, A.R. , Lee, A.H. , Robertson, J.F. , Ellis, I.O. , 2007. Prognostic markers in triple-negative breast cancer. Cancer. 109, 25–32. [DOI] [PubMed] [Google Scholar]
- Rakha, E.A. , Reis-Filho, J.S. , Ellis, I.O. , 2008. Basal-like breast cancer: a critical review. J. Clin. Oncol. 26, 2568–2581. [DOI] [PubMed] [Google Scholar]
- Rakha, E.A. , Elsheikh, S.E. , Aleskandarany, M.A. , Habashi, H.O. , Green, A.R. , Powe, D.G. , El-Sayed, M.E. , Benhasouna, A. , Brunet, J.S. , Akslen, L.A. , Evans, A.J. , Blamey, R. , Reis-Filho, J.S. , Foulkes, W.D. , Ellis, I.O. , 2009. Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin. Cancer Res. 15, 2302–2310. [DOI] [PubMed] [Google Scholar]
- Reis-Filho, J.S. , Tutt, A.N. , 2008. Triple negative tumours: a critical review. Histopathology. 52, 108–118. [DOI] [PubMed] [Google Scholar]
- Robson, M.E. , Chappuis, P.O. , Satagopan, J. , Wong, N. , Boyd, J. , Goffin, J.R. , Hudis, C. , Roberge, D. , Norton, L. , Begin, L.R. , Offit, K. , Foulkes, W.D. , 2004. A combined analysis of outcome following breast cancer: differences in survival based on BRCA1/BRCA2 mutation status and administration of adjuvant treatment. Breast Cancer Res. 6, R8–R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Pinilla, S.M. , Sarrio, D. , Honrado, E. , Hardisson, D. , Calero, F. , Benitez, J. , Palacios, J. , 2006. Prognostic significance of basal-like phenotype and fascin expression in node-negative invasive breast carcinomas. Clin. Cancer Res. 12, 1533–1539. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Pinilla, S.M. , Jones, R.L. , Lambros, M.B. , Arriola, E. , Savage, K. , James, M. , Pinder, S.E. , Reis-Filho, J.S. , 2007. MYC amplification in breast cancer: a chromogenic in situ hybridisation study. J. Clin. Pathol. 60, 1017–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzier, R. , Perou, C.M. , Symmans, W.F. , Ibrahim, N. , Cristofanilli, M. , Anderson, K. , Hess, K.R. , Stec, J. , Ayers, M. , Wagner, P. , Morandi, P. , Fan, C. , Rabiul, I. , Ross, J.S. , Hortobagyi, G.N. , Pusztai, L. , 2005. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin. Cancer Res. 11, 5678–5685. [DOI] [PubMed] [Google Scholar]
- Ryan, P.D. , Tung, N.M. , Isakoff, S.J. , Golshan, M. , Richardson, A. , Corben, A.D. , Smith, B.L. , Gelman, R. , Winer, E.P. , Garber, J.E. , 2009. Neoadjuvant cisplatin and bevacizumab in triple negative breast cancer (TNBC): safety and efficacy. J. Clin. Oncol. 27, (Suppl. 15S) abstr 551 [Google Scholar]
- Samaga, R. , Saez-Rodriguez, J. , Alexopoulos, L.G. , Sorger, P.K. , Klamt, S. , 2009. The logic of EGFR/ErbB signaling: theoretical properties and analysis of high-throughput data. PLoS Comput. Biol. 5, e1000438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmierer, B. , Tournier, A.L. , Bates, P.A. , Hill, C.S. , 2008. Mathematical modeling identifies Smad nucleocytoplasmic shuttling as a dynamic signal-interpreting system. Proc. Natl. Acad. Sci. U. S. A. 105, 6608–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, B.P. , Winer, E.P. , Foulkes, W.D. , Garber, J. , Perou, C.M. , Richardson, A. , Sledge, G.W. , Carey, L.A. , 2008. Triple-negative breast cancer: risk factors to potential targets. Clin. Cancer Res. 14, 8010–8018. [DOI] [PubMed] [Google Scholar]
- Schoeberl, B. , Eichler-Jonsson, C. , Gilles, E.D. , Muller, G. , 2002. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat. Biotechnol. 20, 370–375. [DOI] [PubMed] [Google Scholar]
- Schoeberl, B. , Pace, E.A. , Fitzgerald, J.B. , Harms, B.D. , Xu, L. , Nie, L. , Linggi, B. , Kalra, A. , Paragas, V. , Bukhalid, R. , Grantcharova, V. , Kohli, N. , West, K.A. , Leszczyniecka, M. , Feldhaus, M.J. , Kudla, A.J. , Nielsen, U.B. , 2009. Therapeutically targeting ErbB3: a key node in ligand-induced activation of the ErbB receptor-PI3K axis. Sci. Signal. 2, ra31 [DOI] [PubMed] [Google Scholar]
- Schulz, M. , Bakker, B.M. , Klipp, E. , 2009. TIde: a software for the systematic scanning of drug targets in kinetic network models. BMC Bioinformatics. 10, 344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully, R. , 2000. Role of BRCA gene dysfunction in breast and ovarian cancer predisposition. Breast Cancer Res. 2, 324–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, R. , Olshen, A.B. , Ladanyi, M. , 2009. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics. 25, 2906–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, K.K. , Tan, D.S. , Reis-Filho, J.S. , 2008. Development of therapeutic approaches to ‘triple negative’ phenotype breast cancer. Expert Opin. Ther. Targets. 12, 1123–1137. [DOI] [PubMed] [Google Scholar]
- Sims, A.H. , Howell, A. , Howell, S.J. , Clarke, R.B. , 2007. Origins of breast cancer subtypes and therapeutic implications. Nat. Clin. Pract. Oncol. 4, 516–525. [DOI] [PubMed] [Google Scholar]
- Sirohi, B. , Arnedos, M. , Popat, S. , Ashley, S. , Nerurkar, A. , Walsh, G. , Johnston, S. , Smith, I.E. , 2008. Platinum-based chemotherapy in triple-negative breast cancer. Ann. Oncol. 19, 1847–1852. [DOI] [PubMed] [Google Scholar]
- Sitter, B. , Bathen, T.F. , Tessem, M.B. , Gribbestad, I.S. , 2009. High resolution magic angle spinning (HRMAS) MR spectroscopy in metabolic characterization of human cancer (review). Prog. Nucl. Magn. Reson. Spectrosc. 54, 239–254. [Google Scholar]
- Smid, M. , Wang, Y. , Zhang, Y. , Sieuwerts, A.M. , Yu, J. , Klijn, J.G. , Foekens, J.A. , Martens, J.W. , 2008. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 68, 3108–3114. [DOI] [PubMed] [Google Scholar]
- Sørlie, T. , Tibshirani, R. , Parker, J. , Hastie, T. , Marron, J.S. , Nobel, A. , Deng, S. , Johnsen, H. , Pesich, R. , Geisler, S. , Demeter, J. , Perou, C.M. , Lonning, P.E. , Brown, P.O. , Børresen-Dale, A.L. , Botstein, D. , 2003. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. U. S. A. 100, 8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead, L.A. , Lash, T.L. , Sobieraj, J.E. , Chi, D.D. , Westrup, J.L. , Charlot, M. , Blanchard, R.A. , Lee, J.C. , King, T.C. , Rosenberg, C.L. , 2009. Triple-negative breast cancers are increased in black women regardless of age or body mass index. Breast Cancer Res. 11, R18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, P.J. , McBride, D.J. , Lin, M.L. , Varela, I. , Pleasance, E.D. , Simpson, J.T. , Stebbings, L.A. , Leroy, C. , Edkins, S. , Mudie, L.J. , Greenman, C.D. , Jia, M. , Latimer, C. , Teague, J.W. , Lau, K.W. , Burton, J. , Quail, M.A. , Swerdlow, H. , Churcher, C. , Natrajan, R. , Sieuwerts, A.M. , Martens, J.W. , Silver, D.P. , Langerod, A. , Russnes, H.E. , Foekens, J.A. , Reis-Filho, J.S. , van't Veer, L. , Richardson, A.L. , Børresen-Dale, A.L. , Campbell, P.J. , Futreal, P.A. , Stratton, M.R. , 2009. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 462, 1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockmans, G. , Deraedt, K. , Wildiers, H. , Moerman, P. , Paridaens, R. , 2008. Triple-negative breast cancer. Curr. Opin. Oncol. 20, 614–620. [DOI] [PubMed] [Google Scholar]
- Swameye, I. , Muller, T.G. , Timmer, J. , Sandra, O. , Klingmuller, U. , 2003. Identification of nucleocytoplasmic cycling as a remote sensor in cellular signaling by databased modeling. Proc. Natl. Acad. Sci. U. S. A. 100, 1028–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton, C. , Caldas, C. , 2009. Molecular classification of solid tumours: towards pathway-driven therapeutics. Br. J. Cancer. 100, 1517–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, D.S. , Marchio, C. , Jones, R.L. , Savage, K. , Smith, I.E. , Dowsett, M. , Reis-Filho, J.S. , 2008. Triple negative breast cancer: molecular profiling and prognostic impact in adjuvant anthracycline-treated patients. Breast Cancer Res. Treat. 111, 27–44. [DOI] [PubMed] [Google Scholar]
- Tan, A.R. , Swain, S.M. , 2008. Therapeutic strategies for triple-negative breast cancer. Cancer J. 14, 343–351. [DOI] [PubMed] [Google Scholar]
- Thomas, E.S. , Gomez, H.L. , Li, R.K. , Chung, H.C. , Fein, L.E. , Chan, V.F. , Jassem, J. , Pivot, X.B. , Klimovsky, J.V. , de Mendoza, F.H. , Xu, B. , Campone, M. , Lerzo, G.L. , Peck, R.A. , Mukhopadhyay, P. , Vahdat, L.T. , Roche, H.H. , 2007. Ixabepilone plus capecitabine for metastatic breast cancer progressing after anthracycline and taxane treatment. J. Clin. Oncol. 25, 5210–5217. [DOI] [PubMed] [Google Scholar]
- Timmerman, D. , Testa, A.C. , Bourne, T. , Ferrazzi, E. , Ameye, L. , Konstantinovic, M.L. , Van Calster, B. , Collins, W.P. , Vergote, I. , Van Huffel, S. , Valentin, L. , 2005. Logistic regression model to distinguish between the benign and malignant adnexal mass before surgery: a multicenter study by the International Ovarian Tumor Analysis Group. J. Clin. Oncol. 23, 8794–8801. [DOI] [PubMed] [Google Scholar]
- Trivers, K.F. , Lund, M.J. , Porter, P.L. , Liff, J.M. , Flagg, E.W. , Coates, R.J. , Eley, J.W. , 2009. The epidemiology of triple-negative breast cancer, including race. Cancer Causes Control. 20, 1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, H.R. , Hayashi, S. , Chen, W. , Sano, M. , Machida, M. , Shigeyoshi, Y. , Iino, M. , Hashimoto, S. , 2005. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37, 187–192. [DOI] [PubMed] [Google Scholar]
- Uematsu, T. , Kasami, M. , Yuen, S. , 2009. Triple-negative breast cancer: correlation between MR imaging and pathologic findings. Radiology. 250, 638–647. [DOI] [PubMed] [Google Scholar]
- Van Holsbeke, C. , Van Calster, B. , Valentin, L. , Testa, A.C. , Ferrazzi, E. , Dimou, I. , Lu, C. , Moerman, P. , Van Huffel, S. , Vergote, I. , Timmerman, D. , 2007. External validation of mathematical models to distinguish between benign and malignant adnexal tumors: a multicenter study by the International Ovarian Tumor Analysis Group. Clin. Cancer Res. 13, 4440–4447. [DOI] [PubMed] [Google Scholar]
- Vapnik, V.N. , 2002. The Nature of Statistical Learning Theory Springer; New York, USA: [Google Scholar]
- van't Veer, L.J. , Dai, H. , van de Vijver, M.J. , He, Y.D. , Hart, A.A. , Mao, M. , Peterse, H.L. , van der Kooy, K. , Marton, M.J. , Witteveen, A.T. , Schreiber, G.J. , Kerkhoven, R.M. , Roberts, C. , Linsley, P.S. , Bernards, R. , Friend, S.H. , 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 415, 530–536. [DOI] [PubMed] [Google Scholar]
- Venkitaraman, A.R. , 2002. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 108, 171–182. [DOI] [PubMed] [Google Scholar]
- Verhoog, L.C. , Brekelmans, C.T. , Seynaeve, C. , van den Bosch, L.M. , Dahmen, G. , van Geel, A.N. , Tilanus-Linthorst, M.M. , Bartels, C.C. , Wagner, A. , van den Ouweland, A. , Devilee, P. , Meijers-Heijboer, E.J. , Klijn, J.G. , 1998. Survival and tumour characteristics of breast-cancer patients with germline mutations of BRCA1. Lancet. 351, 316–321. [DOI] [PubMed] [Google Scholar]
- Viale, G. , Rotmensz, N. , Maisonneuve, P. , Bottiglieri, L. , Montagna, E. , Luini, A. , Veronesi, P. , Intra, M. , Torrisi, R. , Cardillo, A. , Campagnoli, E. , Goldhirsch, A. , Colleoni, M. , 2009. Invasive ductal carcinoma of the breast with the “triple-negative” phenotype: prognostic implications of EGFR immunoreactivity. Breast Cancer Res. Treat. 116, 317–328. [DOI] [PubMed] [Google Scholar]
- van de Vijver, M.J. , He, Y.D. , van't Veer, L.J. , Dai, H. , Hart, A.A. , Voskuil, D.W. , Schreiber, G.J. , Peterse, J.L. , Roberts, C. , Marton, M.J. , Parrish, M. , Atsma, D. , Witteveen, A. , Glas, A. , Delahaye, L. , van der Velde, T. , Bartelink, H. , Rodenhuis, S. , Rutgers, E.T. , Friend, S.H. , Bernards, R. , 2002. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009. [DOI] [PubMed] [Google Scholar]
- Villadsen, R. , Fridriksdottir, A.J. , Ronnov-Jessen, L. , Gudjonsson, T. , Rank, F. , LaBarge, M.A. , Bissell, M.J. , Petersen, O.W. , 2007. Evidence for a stem cell hierarchy in the adult human breast. J. Cell Biol. 177, 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent-Salomon, A. , Gruel, N. , Lucchesi, C. , MacGrogan, G. , Dendale, R. , Sigal-Zafrani, B. , Longy, M. , Raynal, V. , Pierron, G. , de Mascarel, I. , Taris, C. , Stoppa-Lyonnet, D. , Pierga, J.Y. , Salmon, R. , Sastre-Garau, X. , Fourquet, A. , Delattre, O. , de Cremoux, P. , Aurias, A. , 2007. Identification of typical medullary breast carcinoma as a genomic sub-group of basal-like carcinomas, a heterogeneous new molecular entity. Breast Cancer Res. 9, R24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Minckwitz, G., Kaufmann, M., Kűmmel, S., 2008. Integrated meta-analysis on 6402 patients with early breast cancer receiving neoadjuvant anthracycline-taxane +/- trastuzumab containing chemotherapy. In: San Antonio Breast Cancer Symposium, San Antonio, TX abstr 79.
- Vousden, K.H. , Ryan, K.M. , 2009. p53 and metabolism. Nat. Rev. Cancer. 9, 691–700. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Ikeda, D.M. , Narasimhan, B. , Longacre, T.A. , Bleicher, R.J. , Pal, S. , Jackman, R.J. , Jeffrey, S.S. , 2008. Estrogen receptor-negative invasive breast cancer: imaging features of tumors with and without human epidermal growth factor receptor type 2 overexpression. Radiology. 246, 367–375. [DOI] [PubMed] [Google Scholar]
- Weigelt, B. , Horlings, H.M. , Kreike, B. , Hayes, M.M. , Hauptmann, M. , Wessels, L.F. , de Jong, D. , Van de Vijver, M.J. , Van't Veer, L.J. , Peterse, J.L. , 2008. Refinement of breast cancer classification by molecular characterization of histological special types. J. Pathol. 216, 141–150. [DOI] [PubMed] [Google Scholar]
- Weigelt, B. , Kreike, B. , Reis-Filho, J.S. , 2009. Metaplastic breast carcinomas are basal-like breast cancers: a genomic profiling analysis. Breast Cancer Res. Treat. 117, 273–280. [DOI] [PubMed] [Google Scholar]
- Woo, H.M. , Kim, K.M. , Choi, M.H. , Jung, B.H. , Lee, J. , Kong, G. , Nam, S.J. , Kim, S. , Bai, S.W. , Chung, B.C. , 2009. Mass spectrometry based metabolomic approaches in urinary biomarker study of women's cancers. Clin. Chim. Acta. 400, 63–69. [DOI] [PubMed] [Google Scholar]
- Wooster, R. , Bignell, G. , Lancaster, J. , Swift, S. , Seal, S. , Mangion, J. , Collins, N. , Gregory, S. , Gumbs, C. , Micklem, G. , 1995. Identification of the breast cancer susceptibility gene BRCA2. Nature. 378, 789–792. [DOI] [PubMed] [Google Scholar]
- Yang, W.T. , Dryden, M. , Broglio, K. , Gilcrease, M. , Dawood, S. , Dempsey, P.J. , Valero, V. , Hortobagyi, G. , Atchley, D. , Arun, B. , 2008. Mammographic features of triple receptor-negative primary breast cancers in young premenopausal women. Breast Cancer Res. Treat. 111, 405–410. [DOI] [PubMed] [Google Scholar]
- Zi, Z. , Klipp, E. , 2007. Constraint-based modeling and kinetic analysis of the Smad dependent TGF-beta signaling pathway. PLoS ONE. 2, e936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwolak, J. , Adjerid, N. , Bagci, E.Z. , Tyson, J.J. , Sible, J.C. , 2009. A quantitative model of the effect of unreplicated DNA on cell cycle progression in frog egg extracts. J. Theor. Biol. 260, 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]


