Abstract
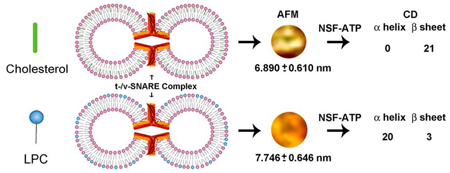
Approximately 11% smaller t-/v-SNARE ring complexes are generated using 50 nm cholesterol-associated vesicles, as opposed to vesicles containing L-α-Lysophosphatidylcholine (LPC), observed using atomic force microscopy (AFM). Circular dichroism (CD) spectroscopy demonstrates that as opposed to cholesterol, in presence of LPC, NSF-ATP induces disassembly of the β-sheet structures, but not the α-helical contents within the t-/v-SNARE complex.
The chemistry of life processes is governed at the molecular level. For example, membrane-directed self-assembly of a supramolecular ring complex is required for the establishment of continuity between opposing membrane compartments in cells1,2. Neurotransmission, and the secretion of hormones or digestive enzymes, all involve fusion of cellular membranes. At the nerve terminal, fusion involves conserved target membrane proteins SNAP-25 and syntaxin 1A, termed t-SNAREs, and synaptic vesicle-associated membrane protein VAMP2 or v-SNARE 3–5. In the presence of Ca2+, when a v-SNARE-reconstituted liposome meets a t-SNARE-reconstituted vesicle, SNAREs in opposing membranes interact and self-assemble in a ring, establishing continuity between the compartments1,2. In the presence of ATP, this highly stable membrane-directed and self-assembled SNARE complex, can undergo disassembly in the presence of the soluble ATPase, N-ethylmaleimide-sensitive factor (NSF) 8,9. Cholesterol and lysophosphatidylcholine (LPC) are known to contribute to the negative and positive curvature of the cell membrane 6,7. Since cholesterol and LPC have been implicated in the promotion and inhibition of membrane fusion respectively10, their influence on membrane-directed assembly and disassembly of the t-/v-SNARE ring complex was hypothesized. To test this hypothesis, structure of the t-/v-SNARE complex at nm resolution using atomic force microscopy (AFM), and at the molecular level, the secondary structure of SNAREs and their complex in membrane containing either cholesterol or LPC, was determined using circular dichroism (CD) spectroscopy9.
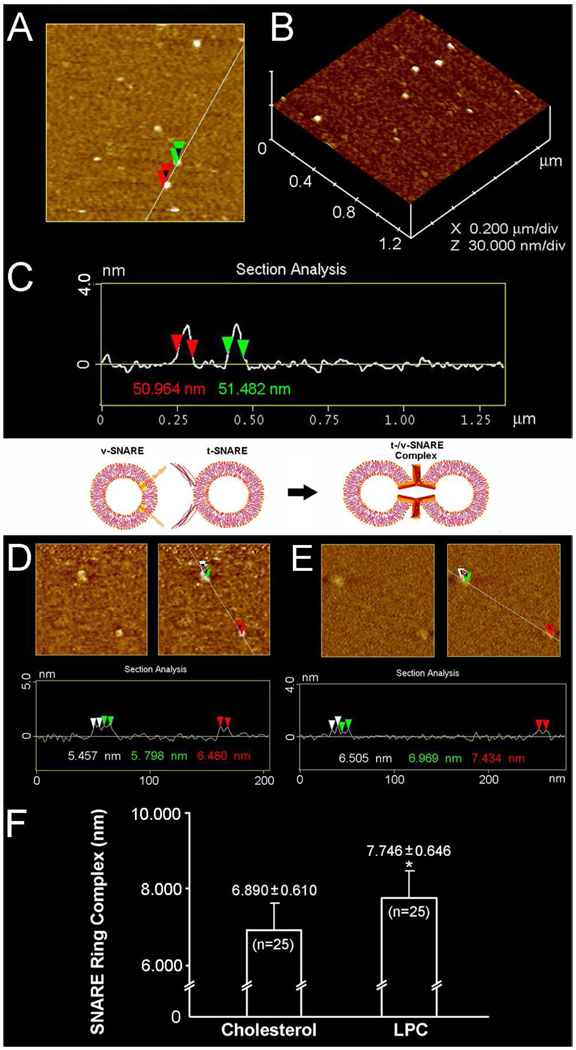
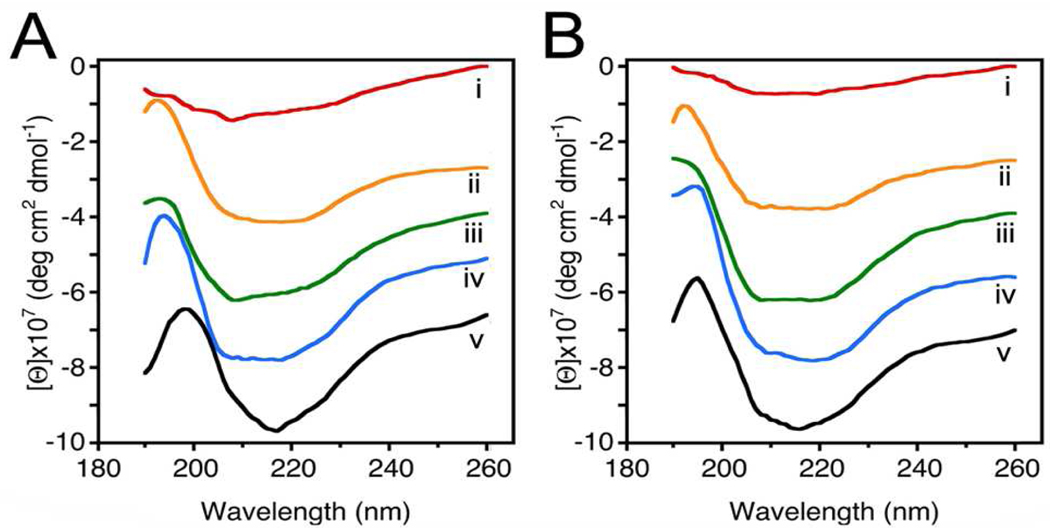
Since vesicle size influences membrane curvature, a uniform vesicle population, prepared using a published2 extrusion method, was used for the entire study. Two sets of 50 nm in diameter liposomes, one set containing cholesterol, and the other LPC, were reconstituted with either t-SNAREs or v-SNARE for use (Figure 1). Surprisingly, examination at the nm level using AFM, demonstrated significant (p< 0.001) differences in SNARE ring size formed in the presence of cholesterol, compared to LPC. SNARE ring complexes formed using cholesterol-associated vesicles were found to be 6.89 nm, approximately 11% smaller than the 7.746 nm formed using LPC containing vesicles. Using CD spectroscopy, SNARE ring complexes formed either in the presence of cholesterol or LPC (Figure 2A,B) further demonstrate profound differences (Table 1). As previously determined9, our results (Table 1) reveal, high α-helical content in t-SNARE and t-/v-SNARE complexes. However, in the presence of NSF and ATP, peaks at 208 and 222 nm, characteristic of α-helical secondary structure, are abolished in the cholesterol groups (Figure 2 and Table 1). CD spectrographs on membrane-associated v-SNARE display little signal, as previously demonstrated9; however, v-SNARE reconstituted in liposomes containing cholesterol display CD signals for α-helical content. In contrast, the LPC groups exhibit no signal for α-helical content. This cholesterol effect on v-SNARE could be due to a weak signal detected by the spectrometer, or due to the limitation of the GlobalWorks software (Olis) in fitting such data. The primary purpose of analyzing and presenting the v-SNARE data is to ensure that the protein was not providing abnormal signals, which could influence CD spectroscopy of the t-/v-SNARE complex.
Figure 1.
Representative AFM micrographs of approximately 50 nm in diameter liposomes and the t-/v-SNARE ring complexes formed when such clolesterol or LPC containing t-SNARE and v-SNARE proteoliposomes meet. Note the 50–53 nm in diameter cholesterol-containing liposomes (A–C). Similar size LPC-containing vesicles were prepared, and observed using the AFM (data not shown). Note the 6.89±0.61 nm t-/v-SNARE ring complexes formed when approximately 50 nm in diameter t-SNARE-cholesterol-liposomes interact with 50 nm v-SNARE-cholesterol-vesicles (D, F). Similarly, 7.746±0.646 nm t-/v-SNARE ring complexes are formed when cholesterol is replaced by LPC (E, F). (*p< 0.001).
Figure 2.
Circular dichroism data reflecting structural changes to SNAREs associated with liposomes containing cholesterol (A) and LPC (B). Structural changes, following the assembly and (NSF-ATP)-induced disassembly of the t-/v-SNARE complex is further shown. (i) v-SNARE; (ii) t-SNAREs; (iii) t-/v-SNARE complex; (iv) t-/v-SNARE + NSF; and (v) t-/v-SNARE + NSF + 2.5 mM ATP, is shown. CD spectra were recorded at 25 °C in 5 mM sodium phosphate buffer (pH 7.5), at a protein concentration of 25 µM. In each experiment, scans were averaged per sample for enhanced signal to noise, and data were acquired on duplicate independent samples to ensure reproducibility. Note the decrease in α-helicity in the cholesterol groups as opposed to the LPC group following exposure of the t-/v-SNARE complex to NSF-ATP (Table 1).
Table 1.
Secondary structural fit parameters of SNARE complex formation and dissociation.
| Cholesterol Group | LPC Group | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Liposome+Cholesterol | Liposome+LPC | |||||||||
| (100 × fa) | (100 × fa) | |||||||||
| Protein | α | β | O | U | Fitc | α | β | O | U | Fitc |
| v-SNARE | 21 | 28 | 0 | 51 | 0.11 | 0 | 23 | 32 | 44 | 0.38 |
| t-SNARE | 20 | 18 | 11 | 51 | 0.17 | 21 | 22 | 0 | 57 | 0.22 |
| t/v-SNARE | 27 | 29 | 0 | 45 | 0.10 | 26 | 20 | 0 | 55 | 0.22 |
| t/v-SNARE +NSF |
18 | 2 | 6 | 75 | 0.26 | 22 | 17 | 3 | 58 | 0.25 |
| t/v-SNARE +NSF+ATP |
0 | 21 | 40 | 39 | 0.20 | 20 | 3 | 0 | 76 | 0.10 |
Abbreviations used: f, fraction of residues is a given conformational class; α, α-helix; β, β-sheet; O, other (sum of turns, distorted helix, distorted sheet); U, unordered.
Protein constructs: v-SNARE (VAMP2); t-SNAREs (SNAP-25 + syntaxin 1A); NSF, N-ethylmaleimide Sensitive Factor. ATP, adenosine triphosphate.
Fit: goodness of fit parameter expressed as Normalized Spectral Fit Standard Deviation (nm).
In both the cholesterol and LPC groups, the overall spectroscopic profile of t-SNAREs and t-/v-SNARE complexes are similar to the previously reported studies9 using t- and t-/v-SNARE control-liposomes (without cholesterol or LPC molecules) 9. However, the NSF-ATP induced disassembly of the SNARE complex is similar in the t-/v-SNARE control-liposome group9 and in the liposome-cholesterol group (Figure 2) compared to the liposome-LPC group. Addition of ATP to the t-/v-SNARE complex in the presence of NSF demonstrated a decrease in α-helicity in the cholesterol groups (Figure 2). Interestingly however, in the presence of LPC, ATP induced SNARE complex disassembly was abrogated. Similarly, in agreement with earlier studies 9, addition of ATP to the t-/v-SNARE complex in the presence of NSF showed little or no change in β-sheet structures in cholesterol groups (Figure 2), however in the LPC groups, NSF-ATP induced near complete disassembly of β-sheet structures within the SNARE complex. Since the presence of LPC blocks NSF-ATP induced disassembly of α-helical contents in the t-/v-SNARE complex, these results are in agreement with previous findings, which support LPC to be a membrane fusion inhibitor 11. Additionally, in presence of LPC, the inability of the SNARE complex to disassemble, and therefore be able to participate in a new round of docking and fusion, would severely limit membrane fusion during cell secretion. Results from this study provide evidence of the critical role of specific lipids on the structure and function of membrane proteins.
In summary, we report for the first time that membrane-curvature-influencing lipids profoundly influence SNARE complex size and its disassembly. This influence is evident in the 11% smaller t-/v-SNARE ring complexes formed using 50 nm cholesterol-associated vesicles, as opposed to LPC. As previously reported 9 in membrane containing no cholesterol or LPC, NSF-ATP induces disassembly of the α-helical contents, not the β-sheet structures in the t-/v-SNARE complex. In contrast, in the presence of LPC, NSF-ATP induces disassembly of the β-sheet structures, and not the α-helical contents in the SNARE complex. Previous studies implicate cholesterol’s role in membrane fusion to be indirect, centered on SNARE formation through cholesterol binding to synaptophysin, a calcium- and cholesterol-dependent vesicle associated protein which forms a complex with synaptobrevin (VAMP), subsequently facilitating v-SNARE interaction with t-SNAREs 12. In the present study however, no synaptophysin was present to influence such interactions of cholesterol with SNAREs, and therefore little or no effect of cholesterol is demonstrated on the α-helical and β-sheet content of membrane-associated SNAREs and the SNARE complex. Our findings further support the existence of direct lipid-protein relationship, to differentially modulate SNARE function within various cellular compartments. Modulating the concentration and distribution of such non-bilayer lipids at various membranes, could regulate the degree and rate of membrane fusion and membrane-directed SNARE complex assembly-disassembly. Cells with higher membrane-cholesterol levels, would promote membrane fusion while cells with increased membrane LPC content would facilitate secretory event longevity by inhibiting SNARE-complex disassembly.
Supplementary Material
Acknowledgment
Supported by grants from NIH and WSU (BPJ).
Footnotes
Supporting Information Available: Experimental procedure. This material is available free of charge via the Internet http://pubs.acs.org
References
- 1.Cho SJ, Kelly M, Rognlien KT, Cho J, Hörber JK, Jena BP. Biophys. J. 2002;83:2522–2527. doi: 10.1016/s0006-3495(02)75263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho WJ, Jeremic A, Jena BP. J. Am. Chem. Soc. 2005;127:10156–10157. doi: 10.1021/ja052442m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oyler GA, Higgins GA, Hart RA, Battenbarg M, Bloom FE, Wilson MC. J. Cell Biol. 1989;109:3039–3052. doi: 10.1083/jcb.109.6.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett K, Calakos N, Scheller RH. Science. 1992;257:255–259. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- 5.Trimble WS, Cowam DM, Scheller RH. PNAS USA. 1988;85:4538–4542. doi: 10.1073/pnas.85.12.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMahon HT, Gallop JL. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 7.Chernomordik L. Chem. Phys. Lipids. 1996;81:203–213. doi: 10.1016/0009-3084(96)02583-2. [DOI] [PubMed] [Google Scholar]
- 8.Jeremic A, Quinn AS, Cho WJ, Taatjes DJ, Jena BP. J. Am. Chem. Soc. 2006;128:26–27. doi: 10.1021/ja056286v. [DOI] [PubMed] [Google Scholar]
- 9.Cook JD, Cho WJ, Stemmler TL, Jena BP. Chem. Phys. Lett. 2008;462:6–9. doi: 10.1016/j.cplett.2008.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W, Yang L, Huang HW. Biophys. J. 2007;92:2819–2830. doi: 10.1529/biophysj.106.097923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stiasny K, Heinz FX. J. Virol. 2004;78:8536–8542. doi: 10.1128/JVI.78.16.8536-8542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitter D, Reisinger C, Hinz B, Hollmann S, Yelamanchili SV, Treiber-Held S, Ohm TG, Herrmann A, Ahnert-Hilger G. J. Neurochem. 2003;84:35–42. doi: 10.1046/j.1471-4159.2003.01258.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.