Abstract
Transcription of retroviruses is initiated at the U3-R region boundary in the integrated provirus and continues unidirectionally to produce genomic and mRNA products of positive polarity. Several studies have recently demonstrated the existence of naturally occurring protein-encoding transcripts of negative polarity in complex retroviruses. We report here on the identification of transcripts of negative polarity in simple murine leukemia virus (MLV). In T-cell and B-cell lymphomas induced by SL3-3 and Akv MLV, antisense transcripts initiated in the U3 region of the proviral 5′ long terminal repeat (LTR) and continued into the cellular proto-oncogenes Jdp2 and Bach2 to create chimeric transcripts consisting of viral and host sequence. The phenomenon was validated in vivo using a knock-in mouse model homozygous for a single LTR at a position known to activate Nras in B-cell lymphomas. A 5′ rapid amplification of cDNA ends (RACE) analysis indicated a broad spectrum of initiation sites within the U3 region of the 5′ LTR. Our data show for the first time transcriptional activity of negative polarity initiating in the U3 region of simple retroviruses and suggest a novel mechanism of insertional activation of host genes. Elucidation of the nature and potential regulatory role of 5′ LTR antisense transcription will be relevant to the design of therapeutic vectors and may contribute to the increasing recognition of pervasive eukaryotic transcription.
The first base of the R region in the 5′ long terminal repeat (LTR) defines the main transcription initiation site of retroviruses. In simple retroviruses, two major RNA species are produced by the actions of the cellular RNA polymerase II: an unspliced transcript, which serves as a template for translation of the viral genes gag and pol as well as for genomic RNA in progeny viruses, and a singly spliced transcript, which allows for production of appropriate levels of env mRNA. In addition to these common species, viral mRNA generated from the usage of an alternative splice donor site and the canonical env acceptor site has been described for Friend and Moloney murine leukemia viruses (MLVs) (11). Also, we have recently identified in Akv MLV an alternative exon generated from usage of cryptic splice acceptor and donor sites (45). Impairment of these alternative exons resulted in an altered disease induction when injected into mice (4, 45), suggesting a role in virus replication. Complex retroviruses such as human T-cell lymphotropic virus type 1 (HTLV-1) and human immunodeficiency virus type 1 (HIV-1) harbor multiple alternative splice sites to generate a variety of different RNA species whose translational products are necessary for optimal replication (18, 20). Early work suggested the existence of viral transcripts of negative polarity in cells infected with HIV-1 (28, 29, 50) and HTLV-1 (23), and more recently, the existence of these antisense transcripts has been conclusively demonstrated (8, 20, 26, 31, 40). In addition, antisense transcripts have been found in cells infected with feline immunodeficiency virus (FIV) (6), suggesting that antisense transcription may be a common feature in complex retroviruses. Initiation sites for antisense transcripts have been mapped to several positions in the R and U5 regions in HTLV-1 (8) and, further upstream in HIV-1, either in the region antisense to the end of the env gene or at the beginning of the U3 region (22, 34). Recently, an antisense HIV-1 transcript complementary to part of the R-U3 region was described (26). In the case of HTLV-1, two isoforms of a basic zipper transcription factor, HBZ, are generated from unspliced and spliced transcripts complementary to the env and tax genes (8, 15, 31). A growing number of studies have shown the importance of the HBZ proteins in HTLV-1 regulation and pathogenicity (2, 3, 15, 24, 39, 40, 53).
MLVs induce tumors of hematopoietic origin when injected into susceptible newborn mice by insertional activation of proto-oncogenes due to promoter/enhancer elements present in the integrated virus (the provirus) (49). Several hundred potential proto-oncogenes from various retroviral tagging screens have currently been identified, many of which are available in the Retrovirus Tagged Cancer Gene Database (http://rtcgd.abcc.ncifcrf.gov/) (1). A provirus inserted in the same transcriptional orientation as a cellular gene may result in the creation of chimeric transcripts consisting of viral and host gene sequence. Typical examples include the so-called promoter and 3′ untranslated region (UTR) stabilization mechanisms of activation. The former term describes a situation in which transcription initiates at the proviral promoter and continues into the host gene, while the latter refers to cases where the provirus confers polyadenylation signals that result in the removal of destabilization elements presumed to reside in the 3′ UTR of many genes. Alternatively, chimeric 3′ UTRs might be selected for due to decoupling of microRNA (miRNA) binding sites, as we recently have shown for the growth factor independence gene 1 (Gfi1) during SL3-3 MLV-induced T-cell lymphomagenesis (10). The ambiguous term “enhancer activation” refers to proviral integrations that activate host genes without the creation of identifiable or predictable chimeric transcripts, typically when the transcriptional orientations of provirus and host genes are opposite. In this case, transcription factors bound to the proviral enhancer are imagined to augment transcription initiation from the cellular promoter.
Here, we report on virus-host chimeric transcripts generated from proviruses situated in “enhancer orientation,” that is, in the opposite transcriptional orientation relative to the host genes. Transcription was found to be initiated in two regions on the negative strand in the U3 region of the 5′ LTR. Using an in vivo model for a single LTR knock-in in the proto-oncogene Nras, we demonstrate that antisense transcription in the U3 requires only the LTR and suggest that it may be a general phenomenon of simple retroviruses. Our data are the first example of MLV transcripts of negative polarity initiating in the U3 region.
MATERIALS AND METHODS
Isolation of RNA.
Total RNA from frozen tumor samples and from cell line cultures was isolated using TRIzol Reagent (Invitrogen) according to the recommendations of the manufacturer. RNA was quantified spectrophotometrically and visualized by agarose gel electrophoresis to ensure high quality.
Reverse transcription-PCR (RT-PCR), quantitative real-time PCR (qRT-PCR), and 5′ rapid amplification of cDNA ends (RACE).
For RT-PCR analyses, 1 or 2 μg of total RNA was reverse transcribed into cDNA using a RevertAid H Minus First Strand cDNA synthesis kit (Fermentas). All PCR analyses were carried out in ABI 2720 thermocyclers (Applied Biosystems) using recombinant Taq polymerase (Invitrogen) with the following program: an initial melting step at 95°C for 5 min; 30 to 35 cycles of amplification, each consisting of a melting step at 95°C for 30 s, annealing at 55 to 62°C for 30 s, and elongation at 72°C for 30 s; and a final elongation step at 72°C for 7 min. The annealing temperature for amplicon 1f-3-4 was 62°C, that for Bach2 antisense U3 (here referred to as asU3) screening was 55°C, and that for canonical Nras RT-PCR was 60°C, while the remaining RT-PCRs were done at 58°C. After a single round of Bach2 asU3 RT-PCR, faint bands appeared, which were subsequently amplified using 1 μl PCR product in a seminested PCR using identical conditions except for only 30 cycles. PCR amplification products from asU3 PCRs were cloned into pCR4-TOPO vector (Invitrogen), and the obtained bacterial colonies were screened by PCR. PCR products were sequenced by using an ABI 7300 Biosystems apparatus with BigDye 3.1 (Applied Biosystems). The following primers were utilized: 47, 5′-CTAGACGAGGAAGAAGAGCGAAG-3′; 51, 5′-GAGGTGAAACTGGGCAAGAG-3′; 57, 5′-gCGGTTGAGCATCAGGATAA-3′; 78, 5′-GTTGGGGCCTCTTGCCCAGTTTCAC-3′; 140, 5′-GTTAGCTGGCTAAGCCTTATGA-3′; 164, 5′-TGTGTTGAAAGGATCTGTCAAGCT-3′; 2620, 5′-GAATTCGATATCGATCCCCGGTCATCTGGG-3′; actb1, 5′-ACACAGTGCTGTCTGGTGGT-3′; actb2, 5′-CTGGAAGGTGGACAGTGAGG-3′; 955, 5′-TGGACACTCACTGATCTATCACAA-3′; 369, 5′-CCAGGCTTCTCATCCACAGA-3′; 705, 5′-ACTGTAGCAGTGGCCCAAAG-3′; 849, 5′-TACAAACTGGTGGTGGTTGGAGCA-3′; 970, 5′-GCAGGTCTCACCATCAATCACCACTTG-3′; and 332, 5′-ACTGGTCTCTCATGGCACTGTACT-3′.
qRT-PCR was done using SYBR green fluorescence (Brilliant II SYBR green; Stratagene) with an Mx3000 apparatus (Stratagene) following the manufacturer's recommendations. All reactions were run in triplicate and analyzed using MxPro 4.1 software (Stratagene). Relative quantification was performed for each amplicon by relating to a 10-fold dilution series of a plasmid cloned with the particular amplicon, which was run simultaneously with the tumor samples. Actb signal was used to normalize total Jdp2 signal. Primers for amplicon Actb (actb1 and actb2), total Jdp2 exon 3-4 (47 and 57), total exon 1f-3-4 (164 and 57), and exon asU3-3-4 (140 and 57) are described above. qRT-PCR amplification conditions consisted of an initial melting step of 95°C for 10 min, and then amplification was conducted through 50 repetitions of a 95°C step for 15 s and a combined annealing and extension step at 60°C for 30 s. In the case of amplicon E1f-E4, the annealing/extension step was done at 62°C.
The 5′RACE analyses were performed using a GeneRacer kit (Invitrogen) for the Nras and a Smart RACE cDNA amplification kit (Clontech) for the Jdp2 according to the manufacturers' recommendations and as described previously (25, 38). The 5′ RACE PCR was done with a touchdown program consisting of an initial melting step at 95°C for 5 min and then six cycles of touchdown in which the annealing temperature was decreased 1°C/cycle, from 74°C to 68°C. This was followed by 30 cycles of amplification with an annealing temperature of 67°C and a final elongation step for 10 min at 72°C. Each touchdown and amplification cycle consisted of a 30-s melting step, a 30-s annealing step, and a 30-s elongation step. Gene-specific reverse primers were located in Jdp2 exon 3 (5′-GCATCTGGCTGCAGCGACTTTGT-3′) and Nras exon 3 (5′-GCACTGTACTCCTCTTGTCCAGCT-3′).
Generation of Nras LTR knock-in mice.
A targeting cassette consisting of the loxP-flanked PGK/Tn5 neomycin selection cassette and the Akv1-99 MLV proviral LTR was inserted by homologous recombination in CJ7 embryonic stem (ES) cells (48) at a specific position in intron 1 of the Nras gene. This position has previously been identified as an Akv1-99 proviral integration site involved in B-cell tumorigenesis (tumor 9 in reference 27). ES cells containing the LTR at this position (in either the same or the opposite transcriptional orientation compared to Nras) were generated and injected into blastocysts which, upon surgical implantation into pseudopregnant mice, developed into chimeric animals. Chimeras were then mated with a wild-type background until heterozygous knock-in offspring were identified. Homozygotes could be obtained at the expected frequency and presented a normal phenotype at birth. At the time of termination (30 to 40 days), no gross enlargement of lymphoid organs could be observed. The PGK/Tn5 neomycin selection cassette was removed by crossing the mice with EIIa-Cre transgenic mice (21).
Northern blotting.
NIH 3T3 cells were either not infected or infected with replication-competent SL3-3 or Akv1-99 (EGFP) MLVs and cultured for 2 weeks. Total RNA was isolated as described above, and 10 μg was subjected to Northern blot analyses using capillary transfer under the conditions described in reference 44. We used oligonucleotides specific for either antisense (5′-TTCATAAGGCTTAGCCAGCTAACTGCAG-3′) or sense (5′-AAATTTGAAACTGTTGTTGTTTTAGCTATTTCTGGGG-3′) transcription within the LTR that were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England BioLabs). Residual nucleotides were removed with Micro Bio-Spin 6 columns (Bio-Rad) before being used for hybridization with buffers and conditions described in reference 44.
RESULTS
Detection of antisense viral RNA in SL3-3-induced T-cell lymphomas in mice.
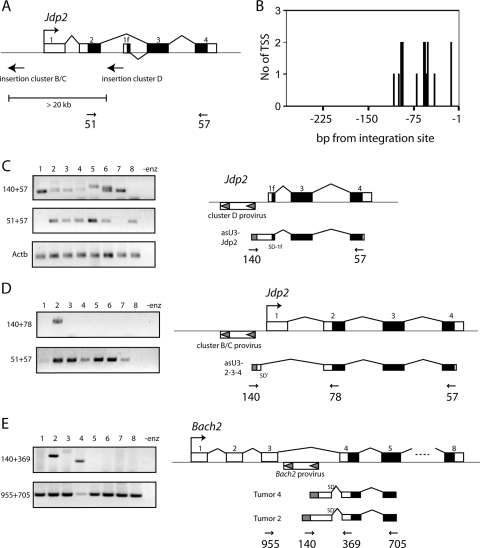
In an effort to identify novel promoters immediately downstream of a common integration site in the gene encoding the AP-1 repressor c-jun dimerization protein (Jdp2), 5′ RACE analyses were performed with total tumor RNA from a murine thymic T-cell lymphoma induced by SL3-3 MLV (38). Surprisingly, we found several chimeric transcripts initiated at the minus strand of the U3 region of a provirus situated in the second intron of Jdp2 in opposite transcriptional orientation (Fig. 1A, integration cluster D). The majority (14 out of 20; 70%) of these transcription start sites (TSS) were within two narrow regions on the negative strand of the viral 5′ LTR (positions −53 to −60 and −95 to −102 with respect to the 5′ LTR U3/host junction) (Fig. 1B). All transcripts continued across the virus-host junction into Jdp2 intron 2 and were spliced from a consensus splice donor site to the splice acceptor site of exon 3. The splice donor site corresponds to the previously reported splice site of the alternative exon 1f (38). Thus, the identified sequences are genuine transcripts and not amplification of contaminating DNA.
FIG. 1.
Detection of antisense transcripts in lymphomas of SL3-3 and Akv MLV-infected mice. (A) Exons and splicing structure of canonical Jdp2 and the alternative 1f-3-4 mRNAs are depicted with coding sequence in black. Positions and the predominant proviral orientations in integration clusters B/C and D are indicated with arrows. Position and orientation of Jdp2 primers 51 and 57 is also shown. (B) The number of individual 5′ RACE transcription start site (TSS) tags on the negative strand of the 5′LTR U3 of a cluster D integrated provirus. (C) RT-PCR on RNA from eight tumor samples using primers that are amplicon specific to asU3-generated Jdp2 chimeras (140+57), canonical Jdp2 using primers in exon 2 and 4 (51+57), and β-Actin (Actb) as the control. In lane 9, no reverse transcriptase enzyme was added in the cDNA preparation (-enz). A schematic representation of the most frequently found asU3-Jdp2 structure was generated using the splice donor site in exon 1f. The transcriptional orientation of the proviral LTR is indicated with gray triangles. (D) RT-PCR of eight cluster B/C tumors using the indicated primers. Legend as for panel B. (E) RT-PCR on RNA from eight B-cell tumors induced by Akv MLV. To the right, a schematic of two of the three identified asU3-Bach2 chimeric transcripts, showing the usage of a cryptic splice donor site (SD′) and the PCR amplicons.
The occurrence of antisense U3 (asU3) transcription could be a special case related to this particular integration event, for instance, as a consequence of aberrant levels of transcription initiation and splicing factors in the tumor sample or mutations within the proviral LTR. We therefore isolated total RNA from a range of tumors with integrations into intron 2 of Jdp2 and performed RT-PCR with viral antisense primer 140 and Jdp2 exon 4 reverse primer 57 (Fig. 1C). PCR products were cloned and amplified in bacteria before being sequenced. We found that chimeric asU3 transcription between viral and Jdp2 exon 3-4 is a common phenomenon in this tumor panel and that in many tumors several distinct integration sites could be found (e.g., tumor 6 in Fig. 1C). Of 25 different tumors, asU3 transcription was observed in 24, with an average of 2.04 independent asU3-Jdp2 mRNA-generating integrations per tumor (M. H. Rasmussen and F. S. Pedersen, unpublished data). Although we isolated transcripts that utilize other splice donor sites in the region, which in all cases splice to the acceptor site of exon 3, splicing from exon 1f was predominant. Also, we never found asU3-Jdp2 transcripts that did not splice in intron 2. In the majority of the tumors, canonical Jdp2 was easily detected (Fig. 1C). We note, however, that in some tumors (tumor 1, 4, 6, 7, and 8), an inverse correlation between canonical and asU3-mediated Jdp2 was seen. The variations in asU3-Jdp2 and canonic Jdp2 were not related to unequal template, as evidenced by similar Actb signal intensities across samples.
In most cases, the integrated viruses were positioned within 1,000 bases from exon 3; hence, it was formally possible that unique cis-acting elements in this local region allowed for asU3 transcription. We therefore isolated total RNA from tumors in which proviral integrations were identified upstream of the RefSeq Jdp2 exon 1 (Fig. 1A, cluster B and cluster C) (37, 38) and screened for asU3-Jdp2 chimeric transcripts by the same RT-PCR strategy as used above but with the Jdp2 exon 2-specific reverse primer 78 situated more than 15 kilobases upstream of exon 3 (Fig. 1D). Among the eight analyzed tumors, a single occurrence of robust asU3 transcription was found. In this case, the integration was situated outside the Jdp2 gene, and asU3 transcription continued into the Jdp2 upstream region and spliced to exon 2 using a cryptic splice donor site. Thus, we conclude that antisense transcription is not unique to a particular tumor sample, nor is it confined to a small genomic region.
Antisense transcription in B-cell lymphomas induced by Akv MLV.
In B-cell lymphomas induced by Akv MLV, the BTB and CNC homology 2 gene Bach2 is a common target of proviral integrations (25). Similar to Jdp2 in T-cell lymphomas induced by SL3-3 MLV, intragenic integrations in Bach2 are mostly inserted in the opposite transcriptional orientation compared to Bach2 (25) (Fig. 1E). To investigate if this locus is also subject to asU3 chimeric transcription, total RNA was isolated from a range of tumors with intragenic Bach2 integrations. By RT-PCR using asU3 primer 140 and the reverse primer 705 in Bach2 exon 5, we could detect in three tumors transcripts generated by the negative strand of Akv MLV, intron 3 sequence, and splicing from a cryptic intron 3 splice donor site (SD′) to the splice acceptor site of Bach2 exon 4 (two cases are depicted in Fig. 1E). Interestingly, in line with what was observed for Jdp2, reduced canonical Bach2 mRNA levels were detected for one tumor with asU3-Bach2 (tumor 4). These results show that antisense transcription is not specific for SL3-3 MLV or T-cell lymphomas.
Antisense U3 transcripts contribute considerably to tumor mRNA levels.
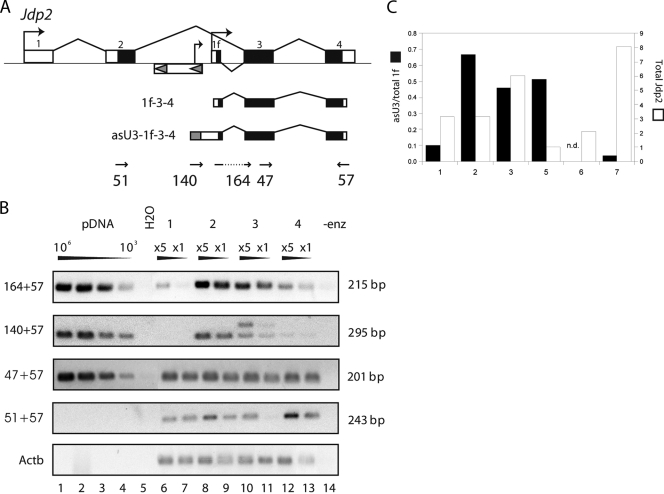
Increased levels of truncated forms of Jdp2, such as those arising from exon 1f-3-4 transcripts, are frequently associated with T-cell lymphomas induced by MLV (38, 47), strongly implying a gain-of-function of truncated Jdp2 mRNA in tumorigenesis. Our data imply that the provirus-induced increment of truncated Jdp2 transcripts in cells harboring a cluster D provirus (in opposite transcriptional orientation) is a sum of two distinct mRNA pools: (i) chimeric transcripts initiating on the negative strand of U3 in the 5′ LTR; and (ii) transcripts initiating from the intronic cellular promoter under more or less influence of proviral cis elements—so-called enhancer activation. In order to estimate the contribution of asU3-1f-3-4 to total Jdp2 1f-3-4 mRNA levels, we employed a semiquantitative RT-PCR strategy (Fig. 2A). Total 1f-3-4 mRNA was measured with forward primer 164 specific for exon 1f-3 splicing together with exon 4 reverse primer 57 (38). Antisense transcription was measured with asU3-specific primer 140 and primer 57. We carefully selected tumors in which we could not detect usage of intron 2 splice events other than exon 1f-3. Furthermore, in order to ensure similar amplification efficiencies, the integration positions were all within 40 bases of the exon 1f splice donor site. The PCR signal from the tumors was compared to the signal from a dilution series of a plasmid containing an asU3-1f-3-4 sequence (Fig. 2B, lanes 1 to 4). Without any identifiable integration in cluster D, no asU3 signal was observed (tumor 1, lanes 6 and 7). In this tumor, most of the signal of total 1f-3-4 mRNA probably stems from the cellular enhancer/promoter. Canonical Jdp2 mRNA was also detected. Conversely, in two tumors expressing large amounts of exon 1f-3-4 (tumors 2 and 3, lanes 8 to 11), the signal from asU3 transcription is not insignificant. For instance, the asU3-1f-3-4 amplification product in tumor 2 (both dilutions) corresponds to the amplification signal generated from between 10,000 and 100,000 plasmid copies, which is comparable to, albeit seemingly a little lower than, the amplification signal for total 1f-3-4. The double band in tumor 3 is due to a second provirus that also utilizes exon 1f, as identified by sequencing of the PCR products. The last tumor (tumor 4, lanes 12 and 13) represents a situation in which the contribution from asU3 initiation appears less predominant. While total Jdp2 (as sampled with amplicon 47+57 across the common exon 3-4) varied little between the tumors, we again observed (for tumors 3 and 4) a tendency toward a negative correlation between asU3-mediated transcription and levels of canonical Jdp2 (compare amplicons 140+57 and 51+57, lanes 10 to 13).
FIG. 2.
Antisense U3-1f chimeric transcript levels are comparable to those initiated in the cellular promoter of exon 1f. (A) Strategy for comparison of the levels of 1f transcripts initiated using asU3 transcription or using the cellular promoter. Structures of the Jdp2 gene and generated mRNAs are shown together with the positions and orientations of the primers used. (B) RT-PCR on RNA from four selected tumor samples at two concentrations (×5 and ×1). PCR amplifications of 10-fold dilutions (from one million to one thousand copies) of a plasmid with an asU3-1f-3-4 insert are loaded to the left (pDNA). The amplicon 164+57 represents total 1f transcript, whereas the amplicon 140+57 measures the contribution from antisense-generated 1f transcript. RT-PCR for total Jdp2 (amplicon 47+57) and canonical Jdp2 (amplicon 51+57) and Actb is also shown. The 140+57 amplicon size (295 bp) is given for the pDNA amplification product. In lane 14, no reverse transcriptase enzyme was added in the cDNA preparation (-enz). Lane numbers are indicated below. (C) Quantitative real-time PCR assay of tumors predominantly activating Jdp2 exon 1f. Tumors 1 to 3 are the same as in panel B. The contribution of asU3-mediated transcription to total 1f mRNA levels is shown with black bars (left axis), while the relative levels of total Jdp2 (amplicon 47+57) is shown with white bars (right axis). In tumor 6, the signal from the asU3 amplicon was below the meaningful detection level (n.d.).
To measure the contribution of asU3-mediated transcription more accurately, we switched to a quantitative real-time PCR strategy using amplicons similar to those in Fig. 2B. In this assay (Fig. 2C), the contribution from asU3-mediated transcription to total 1f-3-4 mRNA was found to be around 50% in some tumors (tumors 2, 3, and 5), while in others (tumors 6 and 7), the contribution was below 5%. We did not observe any correlation across the samples between asU3-mediated transcription and total Jdp2 mRNA levels.
In conclusion, these data are consistent with the notion that asU3-mediated transcription levels in some tumors are comparable to the level mediated by the “enhancer-activated” cellular promoter.
The proviral long terminal repeat alone mediates antisense transcription in vivo.
The results obtained from tumor tissue as described above indicated that asU3 transcription initiated primarily within approximately 100 bp on the negative strand of the proviral 5′ LTR. In order to independently test if the LTR alone is responsible for asU3 transcription, knock-in mice homozygous for a neomycin-LTR cassette in the proto-oncogene Nras were generated (Fig. 3A). The LTR sequence was derived from Akv1-99 and inserted by homologous recombination in either sense or antisense orientation into a position corresponding exactly to that of a previously reported Akv1-99 MLV integration, which was shown to activate Nras by promoter insertion in B-cell lymphomas in mice (27). In this knock-in model, the LTR retains functional activity in adult mice, since Nras expression in different organs correlates with the tissue specificity of the Akv1-99 LTR. For the insertion in sense orientation, Nras expression was found to be high in the spleen relative to the level in wt mice but not in the thymus. Moreover, the presence of chimeric RNA species could be readily detected (B. Ballarín-González, L. B. Lassen, A. Füchtbauer, E.-M. Füchtbauer, and F. S. Pedersen, unpublished data). For studying asU3 transcription, this model has several advantages over transient reporter-based in vitro protocols. First, tumorigenic selection for integrations into Nras seems to favors a promoter activation mechanism since all identified integrations are in the same transcriptional orientation as Nras (data from Retrovirus Tagged Cancer Gene Database, http://rtcgd.abcc.ncifcrf.gov/) (1, 26, 43). Thus, by inverting the LTR orientation at this position, asU3 transcription could be studied at a locus known to be activated by MLV using a mechanism different from that for asU3 transcription. Second, since the LTR will be embedded into chromatin, the model better recapitulates a natural setting than a transient reporter assay. Finally, while we cannot exclude the possibility that asU3 transcription observed in tumors was dependent on secondary mutations in, for instance, genes involved in transcript processing pathways, such mutations are not expected to be selected for in this nontumorigenic model.
FIG. 3.
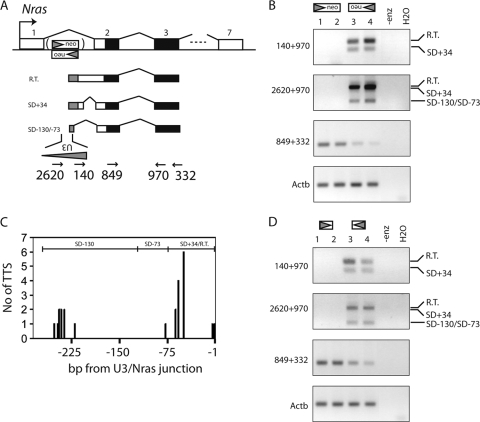
Antisense transcription is dependent only on the LTR. (A) Schematic structure of the Nras gene with coding sequence in black. The transcriptional orientation of the neo-LTR cassettes are indicated with triangles. Also shown are the structures of three asU3-Nras transcripts and the primers used. The relative positions of primers 140 and 2620 on U3 are indicated. (B) RT-PCR on RNA from spleens from two mice with an LTR knock-in transcriptionally identical to Nras (lanes 1 and 2) and two mice with an LTR knock-in transcriptionally opposite to Nras (lanes 3 and 4). The identities of the identified structures are shown to the right, and RT-PCR amplicons are shown to the left. (C) Number of 5′ RACE TSS tags on the negative strand of the 5′LTR U3 isolated from a mouse spleen with a LTR in transcriptionally opposite orientation compared to Nras. The mRNA structures (SD-130, SD-73, and SD+34/R.T.) associated with the TSS regions are indicated on the top. (D) The experiments in panel B were repeated with mice in which the neomycin cassette had been removed. R.T., read-through transcript.
Total RNA was extracted from spleens from two mice homozygous for a LTR knock-in inserted in the same transcriptional orientation as Nras (corresponding to “promoter activation”) and two mice homozygous for a LTR knock-in inserted transcriptionally opposite to that of Nras (“enhancer activation”). All four mice expressed total Nras RNA at detectable levels (Fig. 3B, amplicon 849+332). As expected, RT-PCR using asU3 primer 140 and Nras exon 3 reverse primer 970 did not amplify asU3 transcripts in mice with a LTR knock-in in the same transcriptional orientation as Nras (amplicon 140+970, lanes 1 and 2). However, in mice with the LTR in the opposite transcriptional orientation, a robust level of asU3-Nras chimeric transcripts could be seen (lanes 3 and 4). These RT-PCR products were cloned and sequenced. Analogous to what was observed for the Jdp2 and Bach2 loci, a chimeric mRNA from a donor splice site in intron 1 to the acceptor site of exon 2 was detected (Fig. 3A and B, SD+34). However, the most predominant amplification product corresponded to a read-through transcript in which the intronic splice donor is obviated and the last 73 bp of intron 1 are included (Fig. 3A and B, R.T.). When we used an asU3 primer, oligonucleotide 2620, located further into the U3 region, two additional viral cryptic splice donor sites, SD-130 and SD-73, were identified by bulk cloning and sequencing of PCR products (amplicon 2620+970). Notably, the SD-130 transcript was much more frequent among transformants than was the SD-73 transcript. To further delineate the nature of the asU3 transcripts, 5′ RACE was done with spleen tissue, and the identified TSS tags were mapped onto the virus LTR sequence (Fig. 3C). Interestingly, we found a cluster of tags close to the positions also found in T-cell lymphoma tissue (Fig. 1C). In addition, another cluster of initiation sites, approximately at positions −240 to −250 bp, was identified (Fig. 3C). Furthermore, we note that all TSS tags mapping upstream of the −130 splice donor site utilized this splice site, whereas all read-through transcript and transcripts splicing from the intron (R.T. and SD+34 transcripts, respectively) were found to initiate after the SD-73 splice donor. This result is in agreement with the RT-PCR data in which the most commonly spliced transcript was spliced at position −130. However, the fact that the read-through and SD-73 transcripts also were observed in the RT-PCR analysis indicates that the 5′ RACE did not reach saturation.
To rule out the possibility that the results of the RT-PCR in Fig. 3B were influenced by the presence of the PGK/Tn5 neomycin selection marker, we crossed the mice with Cre-expressing mice in order to excise the loxP-flanked PGK/Tn5 neomycin cassette. We then repeated the RT-PCR on spleen total RNA from these mice (Fig. 3D). Again, we found a robust level of asU3-Nras mRNAs species with structures similar to those in Fig. 3B. The results are in agreement with the hypothesis that asU3 transcription is dependent on the LTR alone. Furthermore, the data suggest that asU3 transcription is a general phenomenon of the LTR of SL3-3 and Akv gammaretroviruses.
Since antisense transcripts initiating in the 3′ LTR and continuing into the env gene are known from complex retroviruses, we wanted to investigate this possibility for MLV. For this purpose, NIH 3T3 cells were either not infected or infected with SL3-3- or Akv1-99-based gammaretroviruses. Total RNA was extracted and subjected to Northern blotting using radioactively labeled oligonucleotides specific for sense LTR transcripts or putative antisense LTR transcripts. However, while we could readily detect spliced and unspliced RNA of positive polarity, we did not find evidence of antisense transcription (Fig. 4). This indicates that the level of putative U3-initiated antisense transcripts in chronically infected cultures is several orders of magnitude lower than that of the plus-strand RNA and is below the sensitivity of the Northern blot.
FIG. 4.
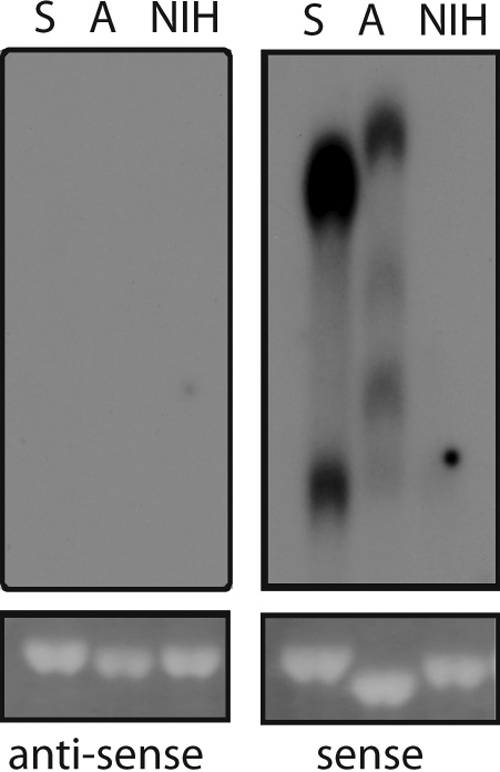
Antisense transcription is not detectable in chronically infected cell cultures. Northern blot analyses of total RNA from uninfected (NIH), or SL3-3 MLV (S)- and Akv1-99 (A)-infected NIH 3T3 cells using an oligonucleotide probe specific for antisense or sense LTR transcription as described in Materials and Methods.
DISCUSSION
Here, we have shown that the simple gammaretroviruses SL3-3 and Akv MLV can initiate transcription on the negative strand of the U3 region to create transcripts of negative polarity (asU3 transcripts). With tumors induced by these viruses, we detected chimeric transcripts consisting of asU3 sequence and the cellular genes Jdp2 (in T-cell lymphomas induced by SL3-3 MLV) or Bach2 (in Akv MLV-induced B-cell lymphomas). These findings were confirmed in an in vivo mouse model for a single LTR knock-in in the Nras gene and hence allow us to conclude that asU3 is not specific to a single genetic locus, nor is it restricted to a tumorigenic condition. One possible implication of our findings is a reassessment of the concept of enhancer activation. Intragenic Bach2 and Jdp2, and, in at least one instance, cluster B/C integrations positioned upstream of Jdp2 appear to activate host gene expression by both enhancer- and asU3-initiated promoter-like mechanisms.
Our data also seem to suggest that antisense transcription lies intrinsic within the LTR, similar to what has been found for the 3′ LTR of complex retroviruses (6, 23, 28), although some reports, including those concerning HTLV-1, have mapped the predominant transcription start site region to R/U5 and not U3 (8, 28). It is possible that the antisense transcriptional activity of the 5′ LTR described here also exists in the 3′ LTR of simple retroviruses. Although we failed to detect antisense transcription by Northern blotting, we cannot exclude the possibility that there is a low level of antisense transcription in MLV-infected cultures. Using more-sensitive PCR based assays, we have not been able to unambiguously confirm a lack of antisense transcription in infected cell cultures. This potential inconsistency may be related to nonspecific priming events by endogenous reverse transcriptase, as noted previously (22), and warrants further investigations. In mice with a LTR knock-in transcriptionally opposite of Nras, RT-PCR using an antisense primer at the U5-R region boundary identified asU3-Nras products that effectively spliced out the U3 region by utilizing a splice donor site at position −370, suggesting antisense transcription across the canonical transcription start site (M. H. Rasmussen, B. Ballarín-González, and F. S. Pedersen, unpublished data). Interestingly, an antisense transcript in HIV-1 overlapping the sense transcription initiation site was reported recently (26) and may show some resemblance to that of MLV. Overlapping transcripts could have a role in controlling initiation of the sense transcript, for instance, in the case of HIV-1 modulating binding of Tat to the TAR element. In this regard, it is interesting that a small antisense RNA targeting a region in U3 205 bp upstream of the cap site was able to suppress Tat-mediated transcription of a HIV-1 LTR-driven reporter in human cells (51). Notably, murine and feline leukemia viruses also transcribe small autonomous noncoding RNAs from the plus strand of the U3 region (9, 14), which have been demonstrated to transactivate cellular genes (9, 16).
Recent findings have shown that eukaryotic genomes are transcribed to a much larger extent than was previously anticipated (5, 7). Both yeast and mammalian promoters have an intrinsic tendency to sustain ongoing bidirectional transcription of noncoding RNAs (13, 32, 35, 52), which is associated with rapid degradation by the nuclear degradation system (32, 35, 52). Although unstable, these different classes of RNA may possess regulatory roles (13, 32, 52). We speculate that asU3-mediated transcription is an inherent feature of the proviral LTR related to transcription of the sense transcript and may have a regulatory impact on the viral promoter similar to cellular genes. While asU3 transcripts under normal circumstances are degraded, we suggest that association with splice sites leads to escape of exosomal degradation (30). Such a hypothesis is in line with our inability to detect robust asU3 transcription in cell cultures and with the tendency of the detected asU3 transcripts to utilize a nearby splice donor site. Specifically, in the Nras model, all 5′ RACE-identified asU3 transcripts starting before the −130 splice donor used this splice site. This result was supported by RT-PCR, in which the SD-130 transcript was the predominant, albeit not the only, transcript. Furthermore, at the Jdp2 locus, retroviral cluster D integrations found more than 200 bp upstream of those analyzed, in general shift from usage of the 1f splice donor site to upstream splice donor sites (M. H. Rasmussen and F. S. Pedersen, unpublished data).
At the current coverage of TSSs, we did not find asU3 transcription beyond position −110 in SL3-3, while in Akv1-99 TSSs were identified in clusters −1 to −80 and −224 to −257. Interestingly, the TSS-less regions coincide with the repeat regions of the proviral core enhancers at positions −129 to −306 and −112 to −210 in SL3-3 and Akv1-99, respectively. These repeats are critical for retroviral replication and tumor induction (12, 19, 33, 46); hence, it would be of interest to investigate asU3-initiated fusion transcripts with intron-containing cellular genes by various enhancer mutants of simple retroviruses.
While our data indicate that antisense transcription is a general phenomenon, we cannot conclude that this is critical for the tumorigenesis process. Akv-mediated transcriptional activation of Bach2 is difficult to assess due to the subclonal nature of these tumors (25), and accordingly, the significance of asU3-Bach2 chimeric transcripts could not be addressed. With regard to asU3-mediated activation of Jdp2, semiquantitative RT-PCR suggested an important contribution to total mRNA levels in some, but not all, tumors. This again may imply that tumorigenic selection for integration sites involves restraints on proviral orientation. In one study using Moloney MLV (M-MLV) to induce T-cell lymphomas in Myc transgenic mice, 10 out of 18 (55%) integrations were transcriptionally opposite of Notch1 as assessed by Southern blotting (17). Interestingly, by using RT-PCR chimeric transcripts created from antisense U3 read-through sequence and cryptic splicing from an intronic donor to Notch1 exons, sequences were observed. Although that study identified a cryptic promoter in Notch1 intron 25 but not in the U3 region, the similarities to Jdp2 and Bach2 strongly suggest that asU3-mediated activation of Notch1 occurs.
In order to integrate our findings of asU3 transcription in tumors with the observation of different proviral orientation biases, we propose that interference effects between cellular and proviral polymerase II complexes be taken into account (41). In essence, we hypothesize that intragenic proviruses inserted transcriptionally opposite, but not transcriptionally similar, to the cellular gene can lead to downregulation of transcription of upstream exons and upregulation of transcription of downstream exons. When the cellular gene and provirus are inserted in the same transcriptional orientation, no transcriptional interference is expected. This would lead to production of a full-length cellular protein or, alternatively, to premature polyadenylation by the viral 5′ LTR, in combination with classical promoter activation of downstream exons. On the other hand, insertions transcriptionally opposite of that of the cellular gene may result in collision of elongating polymerase II complexes from the cellular promoter and the viral LTRs, leading to stalling and abortive elongation (36), with an increasing effect when the distance between the promoters is large (42). In such situation, 5′ LTR-mediated asU3 transcription can initiate transcription of truncated mRNA and protein species. It is conceivable that at loci where intragenic promoters are available, such as in intron 2 of Jdp2, asU3 transcription initially can serve to open up chromatin across the promoter, and subsequently, enhancer activation of the intronic promoter may further augment production of truncated protein. Whether downregulation of full-length transcripts by opposite transcriptional insertions is reflected in the inverse pattern of RT-PCR amplification of canonic and asU3 transcripts found in some Jdp2 and Bach2 tumors is currently unknown, in part owing to the complexity of the tumors.
In summary, we have proven U3-initiated antisense transcription in simple retroviruses of the gammaretroviruses genus, which appears to be different from that of complex retroviruses. Our results may have implications for understanding basic regulatory control mechanisms of pervasive eukaryotic genes, as well as for the design of retrovirally based vector strategies.
Acknowledgments
This project was supported by The Danish Cancer Society, Dansk Kræftforsknings Fond, The Danish Council for Independent Research, The Danish Council for Strategic Research (grant 2101-07-0035), the Danish Genetically Modified Animal Resource (DAGMAR), and the NovoNordisk Foundation. M.H.R. was supported in part by a scholarship from the Danish Research School in Molecular Cancer Research. A.L.N. is supported by a Hallas-Moller fellowship from the NovoNordisk Foundation.
The excellent technical assistance from Lone Højgaard Nielsen and Lisbeth Ahm Hansen is acknowledged.
Footnotes
Published ahead of print on 3 February 2010.
REFERENCES
- 1.Akagi, K., T. Suzuki, R. M. Stephens, N. A. Jenkins, and N. G. Copeland. 2004. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 32:D523-D527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, J., B. Yamamoto, M. Li, A. J. Phipps, I. Younis, M. D. Lairmore, and P. L. Green. 2006. Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1. Blood 107:3976-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold, J., B. Zimmerman, M. Li, M. D. Lairmore, and P. L. Green. 2008. Human T-cell leukemia virus type-1 antisense-encoded gene, Hbz, promotes T-lymphocyte proliferation. Blood 112:3788-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Audit, M., J. Dejardin, B. Hohl, C. Sidobre, T. J. Hope, M. Mougel, and M. Sitbon. 1999. Introduction of a cis-acting mutation in the capsid-coding gene of moloney murine leukemia virus extends its leukemogenic properties. J. Virol. 73:10472-10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birney, E., J. A. Stamatoyannopoulos, A. Dutta, R. Guigo, T. R. Gingeras, E. H. Margulies, Z. Weng, M. Snyder, E. T. Dermitzakis, R. E. Thurman, M. S. Kuehn, C. M. Taylor, S. Neph, C. M. Koch, S. Asthana, A. Malhotra, I. Adzhubei, J. A. Greenbaum, R. M. Andrews, P. Flicek, P. J. Boyle, H. Cao, N. P. Carter, G. K. Clelland, S. Davis, N. Day, P. Dhami, S. C. Dillon, M. O. Dorschner, H. Fiegler, P. G. Giresi, J. Goldy, M. Hawrylycz, A. Haydock, R. Humbert, K. D. James, B. E. Johnson, E. M. Johnson, T. T. Frum, E. R. Rosenzweig, N. Karnani, K. Lee, G. C. Lefebvre, P. A. Navas, F. Neri, S. C. Parker, P. J. Sabo, R. Sandstrom, A. Shafer, D. Vetrie, M. Weaver, S. Wilcox, M. Yu, F. S. Collins, J. Dekker, J. D. Lieb, T. D. Tullius, G. E. Crawford, S. Sunyaev, W. S. Noble, I. Dunham, F. Denoeud, A. Reymond, P. Kapranov, J. Rozowsky, D. Zheng, R. Castelo, A. Frankish, J. Harrow, S. Ghosh, A. Sandelin, I. L. Hofacker, R. Baertsch, D. Keefe, S. Dike, J. Cheng, H. A. Hirsch, E. A. Sekinger, J. Lagarde, J. F. Abril, A. Shahab, C. Flamm, C. Fried, J. Hackermuller, J. Hertel, M. Lindemeyer, K. Missal, A. Tanzer, S. Washietl, J. Korbel, O. Emanuelsson, J. S. Pedersen, N. Holroyd, R. Taylor, D. Swarbreck, N. Matthews, M. C. Dickson, D. J. Thomas, M. T. Weirauch, J. Gilbert, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447:799-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briquet, S., J. Richardson, C. Vanhee-Brossollet, and C. Vaquero. 2001. Natural antisense transcripts are detected in different cell lines and tissues of cats infected with feline immunodeficiency virus. Gene 267:157-164. [DOI] [PubMed] [Google Scholar]
- 7.Carninci, P., A. Sandelin, B. Lenhard, S. Katayama, K. Shimokawa, J. Ponjavic, C. A. Semple, M. S. Taylor, P. G. Engstrom, M. C. Frith, A. R. Forrest, W. B. Alkema, S. L. Tan, C. Plessy, R. Kodzius, T. Ravasi, T. Kasukawa, S. Fukuda, M. Kanamori-Katayama, Y. Kitazume, H. Kawaji, C. Kai, M. Nakamura, H. Konno, K. Nakano, S. Mottagui-Tabar, P. Arner, A. Chesi, S. Gustincich, F. Persichetti, H. Suzuki, S. M. Grimmond, C. A. Wells, V. Orlando, C. Wahlestedt, E. T. Liu, M. Harbers, J. Kawai, V. B. Bajic, D. A. Hume, and Y. Hayashizaki. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38:626-635. [DOI] [PubMed] [Google Scholar]
- 8.Cavanagh, M. H., S. Landry, B. Audet, C. Arpin-Andre, P. Hivin, M. E. Pare, J. Thete, E. Wattel, S. J. Marriott, J. M. Mesnard, and B. Barbeau. 2006. HTLV-I antisense transcripts initiating in the 3′LTR are alternatively spliced and polyadenylated. Retrovirology 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, S. Y., and D. V. Faller. 1995. A transcript from the long terminal repeats of a murine retrovirus associated with trans activation of cellular genes. J. Virol. 69:7054-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dabrowska, M. J., K. Dybkaer, H. E. Johnsen, B. Wang, M. Wabl, and F. S. Pedersen. 2009. Loss of microRNA targets in the 3′-untranslated region as a mechanism of retroviral insertional activation of growth factor independence 1. J. Virol. 83:8051-8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dejardin, J., G. Bompard-Marechal, M. Audit, T. J. Hope, M. Sitbon, and M. Mougel. 2000. A novel subgenomic murine leukemia virus RNA transcript results from alternative splicing. J. Virol. 74:3709-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ejegod, D., K. D. Sorensen, I. Mossbrugger, L. Quintanilla-Martinez, J. Schmidt, and F. S. Pedersen. 2009. Control of pathogenicity and disease specificity of a T-lymphomagenic gammaretrovirus by E-box motifs but not by an overlapping glucocorticoid response element. J. Virol. 83:336-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fejes-Toth, K., V. Sotirova, R. Sachidanandam, G. Assaf, G. J. Hannon, P. Kapranov, S. Foissac, A. T. Willingham, R. Duttagupta, E. Dumais, T. R. Gingeras, et al. 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457:1028-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forman, L. W., R. Pal-Ghosh, R. A. Spanjaard, D. V. Faller, and S. K. Ghosh. 2009. Identification of LTR-specific small non-coding RNA in FeLV infected cells. FEBS Lett. 583:1386-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaudray, G., F. Gachon, J. Basbous, M. Biard-Piechaczyk, C. Devaux, and J. M. Mesnard. 2002. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J. Virol. 76:12813-12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh, S. K., and D. V. Faller. 1999. Feline leukemia virus long terminal repeat activates collagenase IV gene expression through AP-1. J. Virol. 73:4931-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girard, L., Z. Hanna, N. Beaulieu, C. D. Hoemann, C. Simard, C. A. Kozak, and P. Jolicoeur. 1996. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 10:1930-1944. [DOI] [PubMed] [Google Scholar]
- 18.Gramberg, T., N. Sunseri, and N. R. Landau. 2009. Accessories to the crime: recent advances in HIV accessory protein biology. Curr. HIV/AIDS Rep. 6:36-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallberg, B., J. Schmidt, A. Luz, F. S. Pedersen, and T. Grundstrom. 1991. SL3-3 enhancer factor 1 transcriptional activators are required for tumor formation by SL3-3 murine leukemia virus. J. Virol. 65:4177-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kashanchi, F., and J. N. Brady. 2005. Transcriptional and post-transcriptional gene regulation of HTLV-1. Oncogene 24:5938-5951. [DOI] [PubMed] [Google Scholar]
- 21.Lakso, M., J. G. Pichel, J. R. Gorman, B. Sauer, Y. Okamoto, E. Lee, F. W. Alt, and H. Westphal. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. U. S. A. 93:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landry, S., M. Halin, S. Lefort, B. Audet, C. Vaquero, J. M. Mesnard, and B. Barbeau. 2007. Detection, characterization and regulation of antisense transcripts in HIV-1. Retrovirology 4:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larocca, D., L. A. Chao, M. H. Seto, and T. K. Brunck. 1989. Human T-cell leukemia virus minus strand transcription in infected T-cells. Biochem. Biophys. Res. Commun. 163:1006-1013. [DOI] [PubMed] [Google Scholar]
- 24.Lemasson, I., M. R. Lewis, N. Polakowski, P. Hivin, M. H. Cavanagh, S. Thebault, B. Barbeau, J. K. Nyborg, and J. M. Mesnard. 2007. Human T-cell leukemia virus type 1 (HTLV-1) bZIP protein interacts with the cellular transcription factor CREB to inhibit HTLV-1 transcription. J. Virol. 81:1543-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, J., A. B. Sorensen, B. Wang, M. Wabl, A. L. Nielsen, and F. S. Pedersen. 2009. Identification of novel Bach2 transcripts and protein isoforms through tagging analysis of retroviral integrations in B-cell lymphomas. BMC Mol. Biol. 10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig, L. B., J. L. Ambrus, Jr., K. A. Krawczyk, S. Sharma, S. Brooks, C. B. Hsiao, and S. A. Schwartz. 2006. Human Immunodeficiency Virus-Type 1 LTR DNA contains an intrinsic gene producing antisense RNA and protein products. Retrovirology 3:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin-Hernandez, J., A. B. Sorensen, and F. S. Pedersen. 2001. Murine leukemia virus proviral insertions between the N-ras and unr genes in B-cell lymphoma DNA affect the expression of N-ras only. J. Virol. 75:11907-11912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michael, N. L., M. T. Vahey, L. d'Arcy, P. K. Ehrenberg, J. D. Mosca, J. Rappaport, and R. R. Redfield. 1994. Negative-strand RNA transcripts are produced in human immunodeficiency virus type 1-infected cells and patients by a novel promoter downregulated by Tat. J. Virol. 68:979-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, R. H. 1988. Human immunodeficiency virus may encode a novel protein on the genomic DNA plus strand. Science 239:1420-1422. [DOI] [PubMed] [Google Scholar]
- 30.Moore, M. J., and N. J. Proudfoot. 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136:688-700. [DOI] [PubMed] [Google Scholar]
- 31.Murata, K., T. Hayashibara, K. Sugahara, A. Uemura, T. Yamaguchi, H. Harasawa, H. Hasegawa, K. Tsuruda, T. Okazaki, T. Koji, T. Miyanishi, Y. Yamada, and S. Kamihira. 2006. A novel alternative splicing isoform of human T-cell leukemia virus type 1 bZIP factor (HBZ-SI) targets distinct subnuclear localization. J. Virol. 80:2495-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neil, H., C. Malabat, Y. d'Aubenton-Carafa, Z. Xu, L. M. Steinmetz, and A. Jacquier. 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457:1038-1042. [DOI] [PubMed] [Google Scholar]
- 33.Nieves, A., L. S. Levy, and J. Lenz. 1997. Importance of a c-Myb binding site for lymphomagenesis by the retrovirus SL3-3. J. Virol. 71:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peeters, A., P. F. Lambert, and N. J. Deacon. 1996. A fourth Sp1 site in the human immunodeficiency virus type 1 long terminal repeat is essential for negative-sense transcription. J. Virol. 70:6665-6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preker, P., J. Nielsen, S. Kammler, S. Lykke-Andersen, M. S. Christensen, C. K. Mapendano, M. H. Schierup, and T. H. Jensen. 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322:1851-1854. [DOI] [PubMed] [Google Scholar]
- 36.Prescott, E. M., and N. J. Proudfoot. 2002. Transcriptional collision between convergent genes in budding yeast. Proc. Natl. Acad. Sci. U. S. A. 99:8796-8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasmussen, M. H., A. B. Sorensen, D. W. Morris, J. C. Dutra, E. K. Engelhard, C. L. Wang, J. Schmidt, and F. S. Pedersen. 2005. Tumor model-specific proviral insertional mutagenesis of the Fos/Jdp2/Batf locus. Virology 337:353-364. [DOI] [PubMed] [Google Scholar]
- 38.Rasmussen, M. H., B. Wang, M. Wabl, A. L. Nielsen, and F. S. Pedersen. 2009. Activation of alternative Jdp2 promoters and functional protein isoforms in T-cell lymphomas by retroviral insertion mutagenesis. Nucleic Acids Res. 37:4657-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saito, M., T. Matsuzaki, Y. Satou, J. Yasunaga, K. Saito, K. Arimura, M. Matsuoka, and Y. Ohara. 2009. In vivo expression of the HBZ gene of HTLV-1 correlates with proviral load, inflammatory markers and disease severity in HTLV-1 associated myelopathy/tropical spastic paraparesis (HAM/TSP). Retrovirology 6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satou, Y., J. Yasunaga, M. Yoshida, and M. Matsuoka. 2006. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc. Natl. Acad. Sci. U. S. A. 103:720-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shearwin, K. E., B. P. Callen, and J. B. Egan. 2005. Transcriptional interference—a crash course. Trends Genet. 21:339-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sneppen, K., I. B. Dodd, K. E. Shearwin, A. C. Palmer, R. A. Schubert, B. P. Callen, and J. B. Egan. 2005. A mathematical model for transcriptional interference by RNA polymerase traffic in Escherichia coli. J. Mol. Biol. 346:399-409. [DOI] [PubMed] [Google Scholar]
- 43.Sorensen, A. B., M. Duch, H. W. Amtoft, P. Jorgensen, and F. S. Pedersen. 1996. Sequence tags of provirus integration sites in DNAs of tumors induced by the murine retrovirus SL3-3. J. Virol. 70:4063-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sorensen, A. B., A. H. Lund, S. Ethelberg, N. G. Copeland, N. A. Jenkins, and F. S. Pedersen. 2000. Sint1, a common integration site in SL3-3-induced T-cell lymphomas, harbors a putative proto-oncogene with homology to the septin gene family. J. Virol. 74:2161-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorensen, A. B., A. H. Lund, S. Kunder, L. Quintanilla-Martinez, J. Schmidt, B. Wang, M. Wabl, and F. S. Pedersen. 2007. Impairment of alternative splice sites defining a novel gammaretroviral exon within gag modifies the oncogenic properties of Akv murine leukemia virus. Retrovirology 4:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorensen, K. D., S. Kunder, L. Quintanilla-Martinez, J. Sorensen, J. Schmidt, and F. S. Pedersen. 2007. Enhancer mutations of Akv murine leukemia virus inhibit the induction of mature B-cell lymphomas and shift disease specificity towards the more differentiated plasma cell stage. Virology 362:179-191. [DOI] [PubMed] [Google Scholar]
- 47.Stewart, M., N. Mackay, L. Hanlon, K. Blyth, L. Scobie, E. Cameron, and J. C. Neil. 2007. Insertional mutagenesis reveals progression genes and checkpoints in MYC/Runx2 lymphomas. Cancer Res. 67:5126-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swiatek, P. J., and T. Gridley. 1993. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Genes Dev. 7:2071-2084. [DOI] [PubMed] [Google Scholar]
- 49.Uren, A. G., J. Kool, A. Berns, and M. van Lohuizen. 2005. Retroviral insertional mutagenesis: past, present and future. Oncogene 24:7656-7672. [DOI] [PubMed] [Google Scholar]
- 50.Vanhee-Brossollet, C., H. Thoreau, N. Serpente, L. D'Auriol, J. P. Levy, and C. Vaquero. 1995. A natural antisense RNA derived from the HIV-1 env gene encodes a protein which is recognized by circulating antibodies of HIV+ individuals. Virology 206:196-202. [DOI] [PubMed] [Google Scholar]
- 51.Weinberg, M. S., L. M. Villeneuve, A. Ehsani, M. Amarzguioui, L. Aagaard, Z. X. Chen, A. D. Riggs, J. J. Rossi, and K. V. Morris. 2006. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA 12:256-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, Z., W. Wei, J. Gagneur, F. Perocchi, S. Clauder-Munster, J. Camblong, E. Guffanti, F. Stutz, W. Huber, and L. M. Steinmetz. 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao, T., J. Yasunaga, Y. Satou, M. Nakao, M. Takahashi, M. Fujii, and M. Matsuoka. 2009. Human T-cell leukemia virus type 1 bZIP factor selectively suppresses the classical pathway of NF-kappaB. Blood 113:2755-2764. [DOI] [PubMed] [Google Scholar]