Abstract
LGI1 in humans is responsible for a predisposition to autosomal dominant partial epilepsy with auditory features (ADPEAF). However, mechanisms of how LGI1 mutations cause epilepsy remain unclear. We have used a mouse chromosome engineering strategy to create a null mutation for the gene ortholog encoding LGI1. The Lgi1 null mutant mice show no gross overall developmental abnormalities from routine histopathological analysis. After 12–18 days of age, the homozygous mutant mice all exhibit myoclonic seizures accompanied by rapid jumping and running and die shortly thereafter. The heterozygous mutant mice do not develop seizures. Electrophysiological analysis demonstrates an enhanced excitatory synaptic transmission by increasing the release of the excitatory neurotransmitter glutamate, suggesting a basis for the seizure phenotype. This mouse model, therefore, provides novel insights into the mechanism behind ADPEAF and offers a new opportunity to study the mechanism behind the role of LGI1 in susceptibility to myoclonic seizures.
INTRODUCTION
Autosomal dominant partial epilepsy with auditory features (ADPEAF) is a hereditary form of epilepsy (1). This disorder is also referred to (2,3) as autosomal dominant lateral temporal lobe epilepsy (ADLTE). Seizures in patients with this disorder are characterized by auditory auras followed by complex partial seizures and subsequent generalized seizures in many cases. The age of onset of seizures ranges widely between 8 and 50 years of age. Linkage analysis in ADPEAF families identified a locus in 10q24 (1) and subsequently a candidate gene approach identified mutations in the leucine-rich, glioma inactivated-1 (LGI1) gene in affected individuals (4–7). We had originally identified LGI1 from studies of chromosome translocation breakpoints in 10q24 in glioma cell lines (8), which was shown to be inactivated in high-grade brain tumors. The presence of a signal peptide at the N-terminal end indicated it was a secreted protein (9–11). A leucine-rich repeat (LRR) motif was also located in the N-terminal end flanked by cysteine clusters. The C-terminal part of the protein carries a repeat domain that is predicted to form a β-propeller structure indicative of protein–protein binding (12).
The majority of hereditary epilepsy genes encode structural components of ion channels (13–15). LGI1, however, was the first epilepsy predisposition gene which did not possess this function directly. Early studies in glioma cells demonstrated that re-expression of LGI1 suppressed cellular invasion (16), which was later associated with a downregulation of matrix metalloproteinase genes through suppression of signaling through the MEK/ERK pathway (17). In neuroblastoma cells, forced expression of LGI1 resulted in apoptosis (18). We have shown recently that re-expression of LGI1 in glioma cells results in the disregulation of the canonical axon guidance pathway (19). A function in neuronal cells was only recently suggested. Using a heterologous system in Xenopus oocytes, LGI1 was suggested to interact with the Kv1.1 channel (20) which is present on the presynaptic membrane. In other studies (21), LGI1 was shown to co-immunoprecipitate with the ADAM22 receptor protein normally found on the postsynaptic membrane. We have recently confirmed the interaction between LGI1 and ADAM23 (22) and studies by Sagane et al. (23) extended interactions within this family to ADAM11. These observations generally support the role of LGI1 in the seizure phenotype and possibly suggest a role in synapse plasticity/function.
To investigate the mechanism of action of LGI1 further, we have now created mice with a null mutant genotype using a chromosome engineering strategy. These mice develop seizures between 12 and 20 days and die shortly thereafter. Heterozygous mice are seizure-free. Electrophysiological analysis demonstrates an enhanced excitatory synaptic transmission by increasing the release of the excitatory neurotransmitter glutamate, suggesting a basis for the seizure phenotype.
RESULTS
Generation of Lgi1-targeted embryonic stem cells
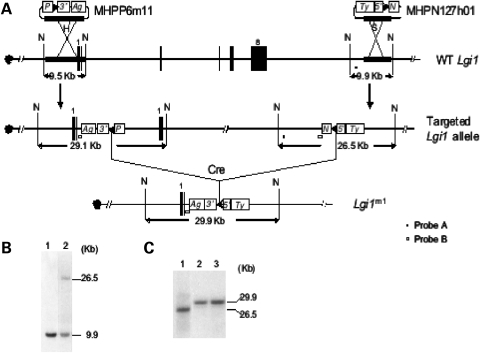
The structural gene of the mouse ortholog of human LGI1 contains eight exons, which spans 44.16 kb (Fig. 1). To reduce the probability of generating a truncated mutant Lgi1 protein after gene targeting, we decided to delete the genomic region extending from exon 3 to exon 8 using Cre/loxP-mediated chromosome engineering (24). The loxP sites were inserted into the desired endpoints using MICER-based targeting vectors (25), available through the Wellcome Trust Sanger Institute (Cambridge, UK). These vectors carry loxP sites and either the 3′- or the 5′-HPRT gene fragments as well as agouti and tyrosinase coat color markers, respectively (Fig. 1). The specific MICER clones selected, MHPP-6m1 and MHPN-127h1, carried genomic inserts which mapped between exon 1 and 2 and exon 8 downstream of the non-coding region of Lgi1, respectively (Fig. 1). To generate the deletion at the Lgi1 locus by cis recombination, AB2.2 embryonic stem (ES) cells were first targeted with MHPN-127h1 (Fig. 1). An ES cell clone with successful targeting of the loxP site using MHPN-127h1 was identified through neomycin selection. This clone was then electroporated with MHPP-6m1, a 3′HPRT targeting vector (Fig. 1), and the ES cells were then selected in puromycin. After 9 days of selection, the cells from the puromycin-resistant clones were pooled and electroporated with pOG231, which carries the Cre recombinase. ES cell clones which had undergone loxP recombination reconstitute the HPRT gene and these cells were then selected using hypoxanthine, aminopterin and thymidine (HAT) medium. Sib-selection of the HAT-resistant clones was performed with G418 and puromycin, where 55% were G418- and puromycin-sensitive, suggesting that they carried the desired deletion. Southern blot analysis of DNA derived from these clones identified the specific restriction enzyme fragment that was predicted to be generated as a result of the deletion of the Lgi1 gene (Fig. 1).
Figure 1.
Generation of the Lgi1 mutation. (A) Strategy to generate the deletion at the Lgi1 locus based on Cre/loxP-mediated recombination. H, HpaI; N, NdeI; S, SmaI; 3′ 3′-HPRT; 5′, 5′-HPRT; Ag, K14-Aguoti gene; Ty, tyrosinase minigene; P, puromycin-resistance gene; N, neomycin-resistance gene; arrowhead, loxP site. Integration sites for MICER clones are shown adjacent to intron 2 and downstream of exon 8. Following Cre-loxP recombination, the HPRT gene is reconstituted from the two fragments located in the vector arms and the two coat color markers are inserted. (B and C) Southern blot analysis of samples from ES cell DNA that were digested with NdeI. DNA isolated from the parental AB2.2 ES cells and an ES cell clone targeted with MHPN-127h1 (B, lanes 1 and 2) was hybridized with Probe A. Cellular DNA isolated from an ES cell clone targeted with MHPN-127h1 (C, lane 1) and two ES cell clones carrying the Lgi1 deletion (C, lanes 2 and 3) hybridized with Probe B.
Fluorescence in situ hybridization analysis of ES clones with Lgi1 deletion
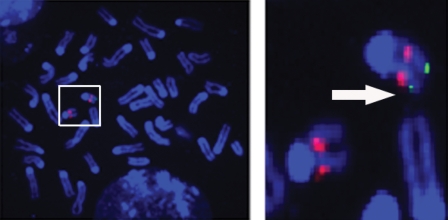
ES cell clones which were shown by Southern blot analysis to carry hemizygous deletions were further analyzed using fluorescence in situ hybridization (FISH). The deleted region spans a <50 Kb genomic interval, which precluded using BACs in the FISH analysis of the deleted region. Instead, individual genomic DNA probes based on the plasmid clones that mapped within the deleted region were obtained from the Mouse 10 Kb plasmid library prepared by Dr R.B. Weiss at the University of Utah. Plasmid DNA from four individual clones was pooled and used to generate the biotinylated probe. This probe was then applied to metaphase chromosome spreads derived from six independently derived ES cell clones. The mouse Lgi1 gene is located on the distal part of chromosome 19. To unequivocally identify chromosome 19 in the metaphase spread, we used BAC RP23-359N19, which is located on the proximal part of chromosome 19. As shown in Figure 2, the control BAC identifies both copies of chromosome 19 within the metaphase spread, but the Lgi1 locus was only present on one of these chromosomes, demonstrating the successful deletion of the homologous region in all of these cell clones.
Figure 2.
FISH analysis of ES cells carrying the heterozygous deletion of the Lgi1 gene. An ES cell metaphase is shown (left). The two copies of chromosome 19 are identified in the box. Individual copies of chromosome 19 (right) were identified by BAC clone RP23-359A49 located close to the centromeric region (red). When co-hybridized with DNA probes from within the deleted region (see text), only one of these chromosomes (arrow) shows the presence of the Lgi1 locus (green).
Generation of the heterozygous null mice
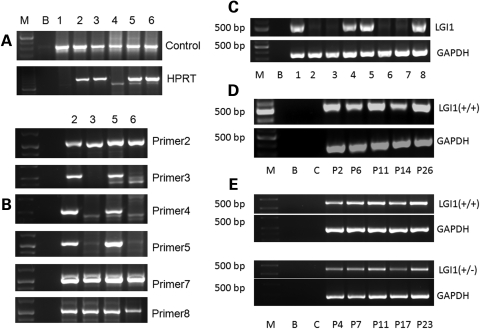
The mutant ES cell clones were used to generate chimeras using standard procedures. Male mice with high chimerism as judged by coat color were then crossed onto albino C57BL/6J mice to generated mice with the constitutional heterozygous deletion. Germline transmission of the Lgi1 mutation was confirmed using a PCR strategy which first demonstrated the constitutional presence of the reconstituted HPRT gene that resulted from the Cre-mediated deletion event (Fig. 3). These heterozygous null mice were then bred to generate homozygous mutant mice, which initially could be identified using the PCR strategy described in Figure 3. In this approach, eight PCR reactions identified the genomic loci, of which two loci lay outside the predicted deletion on either side and four loci were located within the targeted deletion. Analysis of DNA derived from tail snips of 10-day-old mice demonstrated that, as expected, ∼25% of littermates carried the homozygous deletion. From an analysis of >200 mice, there was no apparent sex ratio distortion and the frequency of the Lgi1-targeted alleles showed normal Mendelian inheritance.
Figure 3.
PCR strategy for identification of the homozygous deletion of the Lgi1 gene. (A) Six pups were analyzed, of which four (2, 3, 5 and 6) were positive for the HPRT gene, indicating the presence of the Cre/loxP-mediated recombination product, although this PCR analysis could not distinguish between heterozygous and homozygous deletions. (B) All four of these pups were positive for proximal genomic region 2 (P2) and distal genomic regions 7 and 8 (P7 and P8) which lie outside the deletion (see text). When analyzed for genomic regions within the deletion, pups 3 and 6 were negative, demonstrating that they carry the homozygous deletion and the other two (2 and 5) carry a hemizygous deletion. RT–PCR analysis of wild-type (lanes 1, 4, 5 and 8) and mutant null (lanes 2, 3, 6 and 7) mice demonstrates the absence of PCR products from the hippocampus of the null mice (C). RT–PCR analysis of RNA from hippocampus of the wild-type mice (D) at postnatal days 2–26 demonstrate expression of Lgi1 during early weeks of postnatal life. When wild-type and heterozygous littermates were analyzed using semi-quantitative RT–PCR, evidence for reduced LGI1 expression was seen in the hippocampus, although expression is clearly present at all time points in the heterozygotes (E).
RT–PCR analysis of RNA derived from the hippocampus and cortex of null mutant mice, as expected, showed the absence of Lgi1 gene expression compared with wild-type and heterozygous littermates (Fig. 3). To examine temporal expression of Lgi1, we performed RT–PCR analysis on hippocampal mRNA from wild-type and heterozygous mutant littermates. Lgi1 expression was identified in all animals from 4 days old (P4) through 23 days old (P23) (Fig. 3). In this semi-quantitative analysis, there was evidence for a lower expression level in the heterozygotes as might be expected, although it is still clear that LGI1 expression is present at all time points during the first month of life in these animals.
Cosegregation of coat color markers at the genotypic and phenotypic levels in mutant mice
The MICER clones used to generate the targeted knockout carry either the agouti or tyrosinase coat color markers, which can potentially correlate with specific genotypes within littermates (Fig. 1A). We crossed the original heterozygous targeted mice onto an albino C57BL/6J background where the influence of these genes is often more readily observed in terms of eye and coat color in particular. After six backcrosses, the Lgi1 wild-type mice showed pink eyes and a white coat color, the heterozygous mutants had dark ruby eyes with white coats and the homozygous null mice showed dark ruby eyes with cream-colored coats (Supplementary Material, Fig. S1). These coat-color phenotypes are caused by the presence of one and two copies of the tyrosinase coat color marker in the heterozygous and homozygous mutant mice (Fig. 1A). These phenotypic markers negated the need to genotype mice in this colony in subsequent analyses.
Lgi1 null mice show early-onset seizures
Since LGI1 mutations cause predisposition to ADPEAF in humans, we monitored the mice for seizure activity from birth. Homozygous mutant null mice were viable and developed apparently normally with only a slight reduction in overall body size in some cases. After 12–20 days, however, these mice demonstrated generalized myoclonic seizure phenotypes, which involved uncontrolled jumping and running accompanied by erratic movements which were preceded and followed by periods of relative inactivity involving preening or slow walking. The series of intermittent seizure events were noted to last for ∼20 min (Supplementary Material, videos M1 and M2). Preceding the onset of seizures, these mice were noted to exhibit ‘flagpole’ tail dorsiflexion. During the later stages of the seizures, mice would fall on their sides and exhibit running/galloping motions with the fore and hind limbs. Mice that were followed further would die shortly after an isolated seizure event. These seizure phenotypes were highly consistent between individual mice. Heterozygous null mice have never been observed to exhibit this phenotype and, over a 12–18 month observation period, have not been noted to suffer the premature death experienced by the homozygous null mice.
Hyperexcitability in CA1 hippocampal neurons in Lgi1 null mice
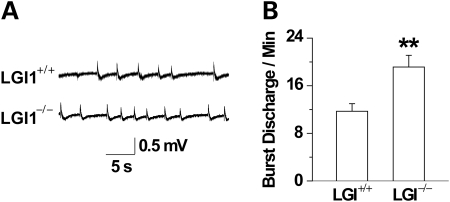
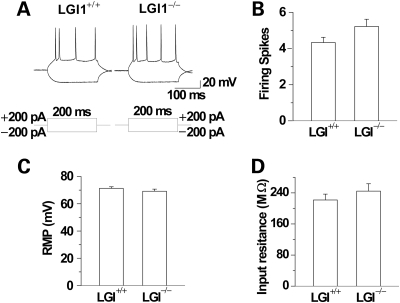
To investigate whether the loss of Lgi1 resulted in altered synaptic function, we investigated extracellular field responses in the CA1 region of the hippocampi from Lgi1-mutant and wild-type mice. When perfused in the modified artificial cerebrospinal fluid (ACSF) (0-Mg2+, low-Ca2+ and high-K+-ACSF), wild-type slices generated characteristic spontaneous epileptiform discharges with burst charges 11.7 ± 1.3 per min (mean ± S.E.M., n = 7). In contrast, the spontaneous burst discharge frequency was significantly increased in the Lgi1-mutant mice (n = 7, P < 0.01; Fig. 4A and B). These results indicate that CA1 neurons in Lgi-mutant mice exhibit higher spontaneous epileptiform-discharge activity in comparison with those in control littermates. To investigate possible mechanisms of the increased discharge activity in mutant mice, we analyzed the intrinsic properties of CA1 pyramidal neurons in current clamp mode. Neurons fired in response to the depolarizing current (+200 pA), but become hyperpolarized when clamped at −200 pA (Fig. 5A). The firing frequency and spikes showed no difference between mutant and wild-type mice (n = 9, P > 0.05; Fig. 5A and B). Moreover, there was no difference in the resting membrane potential (n = 9, P > 0.05; Fig. 5C) and input resistance between the two mice (n = 9, P > 0.05; Fig. 5D). These results indicate that Lgi1 mutation did not change the intrinsic properties of pyramidal neurons in the hippocampal CA1 region. This finding also suggests that the cellular architecture of pyramidal neurons was not altered in the mutant mice, consistent with the gross morphological characterization described below.
Figure 4.
Spontaneous epileptiform-discharge activity in the CA1 regions of hippocampal slices from wild-type (+/ + ) and Lgi1 null (−/ − ) mice. Field responses were recorded extracellularly in a modified ACSF (0 mm MgSO4, 5 mm KCl, 1.6 mm CaCl2). (A) Representative traces of the spontaneous epileptiform burst discharges from mutant and wild-type mice were shown. Quantitative analysis of the burst discharge incidence (B) showed increased frequency in the mutant compared with the wild-type mice (both n = 7, **P < 0.01).
Figure 5.
The intrinsic properties of CA1 pyramidal neurons. (A) Representative response of a CA1 pyramidal neuron to 200 ms depolarizing and hyperpolarizing somatic current pulses. Quantitative analysis showed that the evoked spike frequencies of CA1 pyramidal neurons produced by a 200 pA suprathreshold somatic current injection (B) were similar in wild-type and Lgi1-mutant neurons. The resting membrane potential (RMP) (C) and input resistance (D) of CA1 pyramidal neurons did not vary significantly between wild-type and Lgi1-mutant mice (both n = 9, P > 0.05).
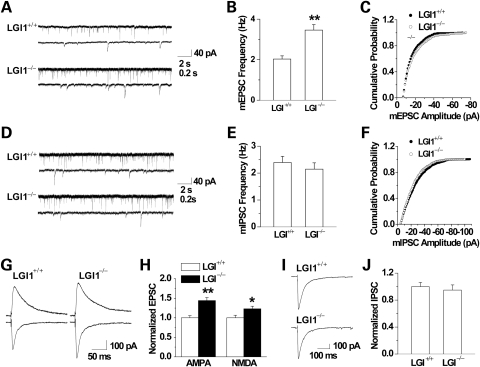
Because there is no difference in the intrinsic properties of pyramidal neurons, we next investigated whether inputs onto pyramidal neurons were altered. First, we recorded miniature excitatory postsynaptic synaptic currents (mEPSCs), a response generated by spontaneous glutamate release. The amplitudes of mEPSCs were similar between mutant and wild-type mice, suggesting the LGI1 mutation did not alter the density and response of postsynaptic receptors (n = 12, P > 0.05; Fig. 6A and C). Remarkably, however, the frequency of mEPSCs was dramatically increased in the mutant mice (n = 12, P < 0.01; Fig. 6A and B), indicating increased release of glutamate. Consistently, AMPA receptor-mediated EPSCs (P < 0.01) and NMDA receptor-mediated EPSCs (P < 0.05) were larger in mutant CA1 pyramidal cells than wild-type controls (n = 7 for wild-type mice and n = 8 for Lgi1-mutant mice, respectively; Fig. 6G and H). We have also investigated the effect of LGI1 mutation on GABAergic transmission by measuring miniature and evoked inhibitory postsynaptic currents (mIPSCs and eIPSCs, respectively). The mIPSC frequency (n = 8, P > 0.05; Fig. 6D and E) and amplitude (P > 0.05; Fig. 6D and F) and the eIPSCs amplitude (n = 7, P > 0.05; Fig. 6I and J) showed no difference between mutant and wild-type mice. These results suggest that Lgi1 mutation had no effect on inhibitory synaptic transmission. Instead, it enhances excitatory synaptic transmission by increasing the release of the excitatory neurotransmitter glutamate. These results provide a potential mechanism of epileptiform activity in mutant mice.
Figure 6.
Analysis of miniature and evoked EPSCs and IPSCs in hippocampal CA1 pyramidal neurons from wild-type (+/ + ) and Lgi1 null (−/ − ) mice. (A) Representative traces of the mEPSCs are shown. Quantitative analysis of mEPSC frequency (B) showed significantly higher frequencies in the mutant compared with the wild-type mouse (both n = 12, **P < 0.01), and cumulative distribution of mEPSC amplitudes (C) showed no significant difference between mutant and wild-type control mice. (D) Representative traces of the mIPSC in CA1 neurons for mutant and wild-type mice are shown. Quantitative analysis of the mIPSC frequency (E) and cumulative distribution of mIPSC amplitudes (F) showed no significant difference between Lgi1-mutant mice and wild-type controls (both n = 8). (G) Representative traces showed that the evoked AMPA (lower) and NMDA (upper) receptor-mediated EPSCs were increased in Lgi1-mutant CA1 pyramidal neurons. (H) The quantification of eEPSC amplitude is shown (n = 7 for wild-type mice and n = 8 for Lgi1-mutant mice, respectively; **P < 0.01, *P < 0.05, compared with wild-type controls). Representative traces (I) and quantification (J) showed that the eIPSC amplitudes were similar in CA1 pyramidal neurons from the wild-type and Lgi1-mutant mice (both n = 7, P > 0.05). The amplitudes for AMPA-eEPSCs, NMDA-eEPSCs and eIPSCs in wild-type mice were 270.6±15.3 pA, 278.0±18.7 pA and 712.9±44.6 pA, respectively.
Gross anatomical analysis of the null mutant mice
To determine whether the Lgi1 null mice showed any gross developmental abnormalities that might explain the seizures, six individual mice from each of the three genotypes underwent extensive routine histopathological and gross anatomy analysis (conducted by the Molecular and Comparative Pathobiology Service at Johns Hopkins University). Overall, no specific abnormalities were observed in the null mutant mice at this level of resolution, with specific attention paid to the hippocampus and cortex as well as other organs demonstrated to express Lgi1 in our previous studies (11).
DISCUSSION
The demonstration here that the null mutant mice for Lgi1 develop myoclonic seizures makes them a representative model for the study of the role of the gene in ADPEAF/ADLTE, with the caveat that human epilepsy patients are only heterozygous for the mutant gene. Loss of function of LGI1 results in early-onset seizures consistent with the phenotype in humans. The seizure phenotype in the null mouse, however, is clearly more severe than that seen in ADPEAF patients, due possibly to the loss of LGI1 function compared with the predicted haplo-insufficiency in humans. The effects of the LGI1 mutations on the function of the protein, however, are still not altogether clear, making it difficult to define the consequences of haploinsufficiency. In a recent review by Nobile et al. (7), 25 mutations were described, of which ∼30% were predicted to create nonsense mutations and ∼70% were missense mutations, presumed to affect protein function. When tested, these missense mutations abrogated or greatly reduced the secretion of the protein. The alleles carrying these particular mutations would be predicted to be non-functional if LGI1 acts on the outer cell membrane as suggested by Fukata et al. (21). If, as suggested by Schulte et al. (20), LGI1 binds to proteins on the inner cell membrane, then it is possible that these missense, or even the truncating, mutations may act in a dominant-negative manner by competing for binding partners in protein complexes, although there is, as yet, no evidence that LGI1 oligomerizes. In our studies there was no evidence that the hemizygous null mice experienced seizures, although we cannot rule out the possibility that they suffer a very mild seizure like activity that cannot be readily detected. There is also no evidence from ADPEAF patients that the nonsense mutations lead to the production of a truncated protein. It might be expected, for example, that mRNA molecules carrying premature chain terminating codons would activate the nonsense mediated decay pathway and, unless they were located in the last exon, degrade the abnormal message (26). In the experiments described by Zhou et al. (27), using knock-in of a nonsense mutation in exon 6 in a transgenic BAC system, a truncated protein was detected supporting the possibility of a dominant-negative effect. A naturally occurring truncated message has also been reported (10) which apparently remains intracellular, which may also affect the function of the full length protein. Thus, although the idea that ADPEAF results from haploinsufficiency due to inactivation of one allele is compelling at this point, until the molecular mechanism of LGI1 function is more clearly understood, it is difficult to interpret the consequences of particular mutations in the gene in the context of the homozygous null mutation.
The identification of proteins that interact with LGI1 has suggested that it is involved in synapse transmission. Thus, Schulte et al. (20) identified an interaction between LGI1 and Kv1.1 (KCNA1) in a heterologous Xeonopus oocyte system, and Fukata et al. (21) identified LGI1 within the PSD95 protein complex which contains ADAM22. Immunoprecipitation (IP) using ADAM22 antibodies demonstrated the presence of LGI1 in this complex. In IP experiments using LGI1 as the bait, we have recently shown that LGI1 interacts with the closely related ADAM23 protein (22). Our mass spectroscopy analysis also identifed syntaxin 1A (SYT1A) and syntaxin binding protein (STXBP1) as interacting protein partners for LGI1 (22). SYT1A also binds to KCNA1 (28). STXBP1 is essential for synaptic vesicle release, and mutations in this gene predispose to infantile epileptic encephalopathy (29). Null mutant mice for KCNA1 (30), ADAM23 (31) and ADAM 22 (32) have been shown to experience seizure/tremor activity, which appears almost identical to that seen in the Lgi1 null mutant mice. The Kv1.2 (KCNA2) protein is commonly found in the same tetramer as KCNA1 and the KCNA2 null mice also develop a phrenetic jumping and running phenotype, with the flagpole tail phenotype preceding the onset of the seizures (33). As in the Lgi1 null mutant mice, these animals die shortly after the seizures begin (32). The emerging evidence, therefore, suggests that the LGI1 protein complex is important for specific neurological function and that loss of any member of this complex results in seizures.
During the preparation of this manuscript, another model for LGI1-related disruption of neuronal function was described by Zhou et al. (27). In this model, transgenic mice were created carrying BACs that resulted in the overexpression of either the wild-type Lgi1 gene or an Lgi1 gene with a truncating mutation in exon 6. In mice overexpressing the exogenous mutant Lgi1 allele, the processes of presynaptic and postsynaptic maturation were arrested. This phenotype was associated with inhibition of dendritic pruning and increased spine density which markedly increased excitatory synaptic transmission. These mice, however, did not demonstrate spontaneous seizures as seen in either human ADPEAF patients or the Lgi1 null mice described here. When mice overexpressing the mutant LGI1 protein were challenged with seizure-inducing pharmacological agents, however, epileptiform discharges were noted and, at high concentrations, mice apparently experienced seizures. Since the human and mouse seizures result from reduced or loss of function, it is not clear at this time to what extent the phenotypes seen in the BAC transgenic model are due to the overexpression of the exogenous genes. It is interesting to note, however, that even though there are differences between the overexpression and null models, both showed increased excitatory synaptic transmission.
In a study by Fukata et al. (21), the frequency of mEPSCs was increased by the presence of LGI1, whereas in this study it was increased by the absence of LGI1. This group (21), however, studied the effect of exogeneous, potentially super physiological concentrations of recombinant LGI1 on neurotransmission in hippocampal slices of P24-29 rats. Because LGI1 had no effect on paired-pulse facilitation, it was concluded that the increase in mEPSC frequency was due to enhanced mEPSC amplitudes. In the present study, neurotransmission was characterized in hippocampal slices of Lgi1 null mutant mice. Owing to neonatal lethality, it was necessary to perform these experiments prior to the age of P14–18. We showed that the mutation had no effect on miniature EPSC (mEPSC) amplitude, but increased its frequency. These results indicate a critical role of LGI1 in the development of synapses, in particular, presynaptic differentiation and/or function. This notion is in general agreement with observations that presynaptic and postsynaptic maturation were arrested in mice overexpressing a mutant Lgi1 allele (27). It would be interesting to investigate the effect of exogenous LGI1 on hippocampal neurotransmission of Lgi1-mutant mice in the future.
From an RT–PCR analysis of mRNA derived essentially from whole brain, previous studies (34) suggested that appreciable levels of LGI1 were only seen after P14, and rising to P28. These observations provided a potential explanation for the age of onset of seizures in the Lgi1 null mice, where the phenotypic effect becomes obvious when the demand for LGI1 function in the CNS becomes highest. A similar observation was reported by Zhou et al. (27) using western blotting with the Santa Cruz antibody SC9583, which we have previously shown to cross-react with other members of the LGI1 family (11). Surprisingly, however, our RT–PCR analysis of the isolated hippocampus from wild-type and heterozygous Lgi1-mutant mice demonstrated an equal expression level from P2 to P28, specifically arguing against a differential temporal expression pattern. Using the BAC transgenic reporter mice for Lgi1 (11), we have also shown that Lgi1 is expressed in the developing hippocampus and cortex between embryonic days E9.5 and 18.5 (manuscript under submission), further supporting a sustained expression in critical cell types during development and into early postnatal life.
The demonstration here that the constitutional inactivation of the Lgi1 gene in mice results in myoclonic seizures supports this as a model for ADPEAF/ADTLE which is due, at least in part, to neuronal hyperexcitability that was specific for excitatory transmission. The availability of the mouse model will facilitate a more detailed understanding of this gene in neuronal dysfunction related to epilepsy, as well as potentially its function in other cell types showing high levels of expression. We have recently shown, for example, that Lgi1, which is also expressed in normal prostate epithelium (11), is inactivated during early-stage hyperplasia of the prostate in a mouse model of prostate cancer, potentially facilitating cellular invasion (35).
MATERIALS AND METHODS
The targeting vectors for generating the targeted Lgi1 mutation
The MICER clone-based targeting vectors were obtained from the Wellcome Trust Sanger Institute (25). The end sequences of the genomic insert of MHPN-127h1 were verified by sequencing using primers TTGGCCGATTCATTAATGCAG and TGAAGAAAGTTGAGGAGAGTTTC, whereas the end sequences of the genomic insert of the MICER clone MHPP-6m1 were verified by sequencing using the primers CGTCCCATTCGCCATTCAGGC and AGACAATAGCAGGCATGCTG.
Gene targeting in ES cells
Culture of the AB2.2 line of ES cell line and the method for gene targeting have been described previously (36,37). For generating the deletion at the Lgi1 locus, MHPN-127h1 and MHPP-6m1 were linearized by digestion with HpaI and SmaI, respectively, prior to transfection. The linearized targeting vectors were transfected into ES cells by electroporation, which were selected in G418 or puromycin. Positive clones were identified by Southern blot analysis with one of the following probes: a PCR product external to the genomic insert of MHPN-127h1 and a 2.3 kb PvuII fragment from the BlueScript plasmid vector backbone.
Generation of the targeted Lgi1 mutation in ES cells and mice
The pOG231 cre-expression vector (38) was electroporated into double-targeted clones. ES cell clones with recombined products were then selected in the HAT medium as described previously (24,38). Clones of ES cells that carried the desired deletion at the Lgi1 locus generated by cis recombination were identified by hybridizing Southern blots of NdeI-digested genomic DNA from HAT-resistant ES cell clones that carried the desired deletion between exon 2 and 3′ non-coding region of Lgi1 to the radiolabeled probe B, a 2.3 kb PvuII fragment that was isolated from the BlueScript plasmid vector backbone (Fig. 1). Germline-transmitting chimeras were generated from ES cell lines carrying the targeted Lgi1 allele by microinjection of blastocysts isolated from albino C57B6/J-Tyrc-Brd females as described previously (39).
Electrophysiological recordings in slices
Brain slices were prepared using standard procedures (40). Briefly, transverse hippocampal slices (0.40 mm) were prepared from P14–P18 Lgi1-mutant mice and wild-type littermates using a Vibroslice (Leica VT 1000S, Leica Instruments, Nussloch, Germany) in an ice-cold solution containing (in mm): 64 NaCl, 2.5 KCl, 1.25 NaH2PO4, 10 MgSO4, 0.5 CaCl2, 26 NaHCO3, 10 glucose and 120 sucrose. Slices were allowed to recover for at least 2 h (1 h at 34°C followed by 1 h at 22°C) in ACSF containing (in mm): 126 NaCl, 2.5 KCl, 1.25 NaH2PO4, 2 MgSO4, 2 CaCl2, 26 NaHCO3 and 10 glucose. Slices were transferred to the recording chamber and superfused with ACSF (2 ml/min) at 34°C that was saturated with 95%O2/5%CO2. CA1 pyramidal neurons were visualized using an infrared-differential interference contrast microscope with a ×40 water-immersion lens (Axioskop2 Fsplus, Carl Zeiss, Germany) and an infrared-sensitive CCD camera. Whole-cell patch-clamp recordings were performed using an Axon Multiclamp 700B amplifier (Molecular Devices, Sunnyvale, CA, USA) and digitized by the pClamp 9.2 software (Molecular Devices). Both mEPSCs and mIPSCs were recorded at a holding potential of −65 mV. For mEPSCs recording, glass pipettes were filled with the solution containing (mm): 120 K-gluconate, 20 KCl, 10 HEPES, 0.1 EGTA, 2 MgCl2, 10 sodium phosphocreatine, 0.2 leupeptin, 4 Mg-ATP and 0.3 Na-GTP (pH 7.3, 280 mOsm with sucrose). K-gluconate in the pipette solution was substituted with 140 mm KCl (final concentration) for mIPSC recording. For eEPSC and eIPSC recording, K-gluconate was substituted with 120 mm CsCH3SO3 and 5 mm lidocaine N-ethylchloride (QX-314) was added in the pipette. The resistance of pipettes was 3–5 MΩ. AMPA and NMDA receptor-mediated EPSCs were recorded at a holding potential of −65 and +40 mV, respectively; eIPSCs were recorded at a holding potential of −65 mV. Tetrodotoxin (1 µm) was included in the perfusion solution for mEPSC and mIPSC recording and 20 µm bicuculline was used to block GABAA receptor-mediated currents to record mEPSCs and eEPSCs. For mIPSC and eIPSC recording, dl-2-amino-5-phosphonovaleric acid (50 µm) and 6-cyano-7-nitroquinoxaline-2,3-dione (20 µm) were supplemented to block NMDA and AMPA receptors, respectively. eEPSCs and eIPSCs were generated with a two-concentric bipolar stimulating electrode (25 mm pole separation) positioned ∼100 µm from the neuron under recording. Single pulses of 0.2 ms were delivered at 0.1 Hz and synchronized using a Mater-8 stimulator (A.M.P.I., Israel). Basic biophysical and firing properties were recorded in current-clamp whole-cell configuration using the K-gluconate-based pipette solution coupled with regular perfusion ACSF. De- and hyperpolarizing current steps (+200 and −200 pA, respectively, 200 ms) were applied for characterization of intrinsic properties. Data were filtered at 2 kHz, and sampled at 10 kHz. Neurons with a resting potential of at least –60 mV and resistance that fluctuated within 15% of initial values (<20 MΩ) were analyzed. Mini events were analyzed with MiniAnalysis software (Synaptosoft). The amplitude histograms were binned at 1 pA. Differences between cumulative amplitude histograms were evaluated using the Kolmogorov–Smirnov test. Spontaneous epileptiform burst discharge was recorded extracellularly in the stratum pyramidal of CA1 in a modified ACSF (0 mm MgSO4, 5 mm KCl, 1.6 mm CaCl2) by using pipettes (2−3 MΩ) filled with modified ACSF.
FISH analysis
FISH was carried out basically as described by Chernova and Cowell (41). DNA probes were derived from BAC clone RP23-359A49 as well as clones 2M0204H23, 1M0418N07, 1M0559N07 and 1M0343M12 where the plasmids were isolated using standard alkaline lysis procedures and were labeled with digoxigenin-11-dUTP using the DIG-Nick Translation Mix (Roche) or with Biotin-16-dUTP using the Biotin-Nick Translation Mix (Roche). The hybridization mixture contained (in a final volume of 20 µl): 2 µg of the labeled probe, a 4-fold excess of human cot-1 DNA (Roche), a 17-fold excess of salmon sperm DNA (Invitrogen) in 50% formamide, 2X SSC and 10% dextran sulfate. The slide was denatured at 70°C in 70% formamide/2X SSC for 1 min, 30 s followed by incubation in ice-cold 70, 80 and 100% ethanol for 2 min each prior to annealing with probe. The probe mixture was denatured at 80°C for 5 min, incubated at 37°C for 45 min, applied to the surface of the slide, overlaid with a coverslip, sealed and then incubated for 20 h at 37°C. The slide was washed three times at 45°C in 50% formamide/2X SSC, two times in 1X SSC and once in 4X SSC/0.1% Tween. The probes hybridized to nuclei were detected using anti-digoxigenin-fluorescein Fab fragments (Roche) or Avidin-Rhodamine (Roche). Slide preparations were viewed using a fluorescent microscope (Nikon) and the images were captured and adjusted for signal intensity with Easyfish software (ASI).
PCR analysis
Total RNA was isolated from cortex and hippocampus of mice from postnatal days 2 (P2), 6 (P6), 11 (P11), 14 (P14) and 26 (P26) using Trizol Reagent RNA isolation system (Invitrogen Corporation, Carlsbad, CA, USA) following standard protocols. First-strand cDNA was carried out using 1–3 mg total RNA and the standard protocol for the Invitrogen SuperScript II Reverse Transcriptase Preamplification System (Invitrogen Corporation). RT–PCR reactions were carried out using specific primers, forward—GAT CCA TTC CAC GCA CCG TTC CTC, reverse—TCT TCT CTA CGT GGT CCC ATT CCA, spanning exons 1–7 to amplify unique fragments homologous to regions of mouse Lgi1 (NCBI: NM_020278) and Gapdh (NCBI: 008084), forward—CAT GTT TGT GAT GGG TGT GAA CCA CGA G, reverse—GAC AAC CTG GTC CTC AGT GTA GCC CAA G. Reverse transcription was performed using cDNA mixture supplemented with 0.3 µl of Taq DNA polymerase, 10 nm of each primer, 2.0 ml of 10X Taq Reaction Buffer, 1.25 mm MgCl2, 125 µm of each DNTP, 100 ng/ml DNA template, 5% DMSO and PCR water to bring the final volume to 20 ml. The reaction mixtures were incubated using a PTC-100 SingleBlock System temperature cycler. PCR reactions were programmed to repeat the following cycles: one cycle with a 5 min denaturation at 95°C, 30 cycles annealing of 30 s at 94°C, 30 s at 61°C and 30 s at 72°C, followed by 5 min extension at 72°C, then bring to and hold at 4°C. The PCR products were electrophoresed on 0.8% agarose DNA grade (Thermo Fisher Scientific, Waltham, MA, USA) gels in 1X TAE. Genotyping was carried out using primers that amplify unique fragments within the mouse Lgi1 locus (Table 1). PCR reactions were performed using genomic DNA mixture supplemented with 0.3 µl of Taq DNA polymerase, 10 nm of each primer, 2.0 ml of 10X Taq Reaction Buffer, 1.25 mm MgCl2, 125 µm of each DNTP, 100 ng/ml genomic DNA template and PCR water to bring the final volume to 20 ml. The reaction mixtures were incubated using a PTC-100 SingleBlock System temperature cycler. PCR reactions were programmed to repeat the following cycles: one cycle with a 5 min denaturation at 95°C, 35 cycles annealing of 30 s at 94°C, 30 s at 55°C and 30 s at 72°C, followed by 5 min extension at 72°C. The PCR products were electrophoresed on 0.8% agarose DNA grade (Thermo Fisher Scientific) gels in 1X TAE.
Table 1.
LGI1 primers used for genotyping
| Primers | Primers sequences |
|---|---|
| HPRT ctrl-F | AGGCTTAGCTGAGGTCACACA |
| HPRT ctrl-R | CCAGTTTCACTAATGACACA |
| HPRT-F | AGGATGTGATACGTGGAAGA |
| HPRT-R | CCAGTTTCACTAATGACACA |
| LGI1 deletion no. 2-F | GGCCCGTAAGCATTTTGATA |
| LGI1 deletion no. 2-R | GCAGCGATGGTGTGAATAAG |
| LGI1 deletion no. 3-F | TGAGAAAAGCTGCCAAGGTT |
| LGI1 deletion no. 3-R | CGTTCACCTAGCAAGCACAA |
| LGI1 deletion no. 4-F | TTTGCAAAGTCCCAAGACCT |
| LGI1 deletion no. 4-R | TACACCTGCATGCCAGAAGA |
| LGI1 deletion no. 5-F | GCCTTGCAAATTGGAACACT |
| LGI1 deletion no. 5-R | ACCGAAATGGCGTTGAGTAG |
| LGI1 deletion no. 7-F | GCCAGTGCAGTCCACAATAA |
| LGI1 deletion no. 7-R | ACTTTCCCAGCCTCACTCAA |
| 127H1 deletion no. 8-F | GATCCCCCACCATGATACAC |
| 127H1 deletion no. 8-R | TGCCCTAGCATTCCCAATAC |
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported in part by grants from the National Institutes of Health NS046706 (J.C.), MH083317 and MH083563 (L.M.), HL091519 (Y.E.Y.).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Mr Jeff LaDuca for preparing the FISH images and to Cory Brayson (Molecular and Comparative Pathobiology Service at Johns Hopkins University) for the histopathological analysis of the Lgi1-mutant and wild-type mice. Dr Cowell is a Georgia Cancer Coalition Distinguished Cancer Scholar.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Ottman R., Risch N., Hauser W.A., Pedley T.A., Lee J.H., Barker-Cummings C., Lustenberger A., Nagle K.J., Lee K.S., Scheuer M.L., et al. Localization of a gene for partial epilepsy to chromosome 10q. Nat. Genet. 1995;10:56–60. doi: 10.1038/ng0595-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michelucci R., Poza J.J., Sofia V., de Feo M.R., Binelli S., Bisulli F., Scudellaro E., Simionati B., Zimbello R., D'Orsi G., et al. Autosomal dominant lateral temporal epilepsy: clinical spectrum, new epitempin mutations, and genetic heterogeneity in seven European families. Epilepsia. 2003;44:1289–1297. doi: 10.1046/j.1528-1157.2003.20003.x. [DOI] [PubMed] [Google Scholar]
- 3.Poza J.J., Sáenz A., Martínez-Gil A., Cheron N., Cobo A.M., Urtasun M., Martí-Massó J.F., Grid D., Beckmann J.S., Prud'homme J.F., et al. Autosomal dominant lateral temporal epilepsy: clinical and genetic study of a large Basque pedigree linked to chromosome 10q. Ann. Neurol. 1999;45:182–188. doi: 10.1002/1531-8249(199902)45:2<182::aid-ana8>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 4.Kalachikov S., Evgrafov O., Ross B., Winawer M., Barker-Cummings C., Martinelli Boneschi F., Choi C., Morozov P., Das K., Teplitskaya E., et al. Mutations in LGI1 cause autosomal-dominant partial epilepsy with auditory features. Nat. Genet. 2002;30:335–341. doi: 10.1038/ng832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morante-Redolate J.M., Gorostidi-Pagola A., Piquer-Sirerol S., Saenz A., Poza J.R., Galan J., Gesk S., Sarafidou T., Mautner V.F., Binelli S., et al. Mutations in the LGI1/Epitempin gene on 10q24 cause autosomal dominant lateral temportal epilepsy. Hum. Mol. Genet. 2002;11:1119–1128. doi: 10.1093/hmg/11.9.1119. [DOI] [PubMed] [Google Scholar]
- 6.Ottman R., Winawer M.R., Kalachikov S., Barker-Cummings C., Gilliam T.C., Pedley T.A., Hauser W.A. LGI1 mutations in autosomal dominant partial epilepsy with auditory features. Neurology. 2004;62:1120–1126. doi: 10.1212/01.wnl.0000120098.39231.6e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nobile C., Michelucci R., Andreazza S., Pasini E., Tosatto S.C., Striano P. LGI1 mutations in autosomal dominant and sporadic lateral temporal epilepsy. Hum. Mutat. 2009;30:530–536. doi: 10.1002/humu.20925. [DOI] [PubMed] [Google Scholar]
- 8.Chernova O., Somerville R.P.T., Cowell J.K. A novel gene, LGI1, from region 10q24, is rearranged and downregulated in malignant brain tumors. Oncogene. 1998;17:2873–2881. doi: 10.1038/sj.onc.1202481. [DOI] [PubMed] [Google Scholar]
- 9.Senechal K.R., Thaller C., Noebels J.L. ADPEAF mutations reduce levels of secreted LGI1, a putative tumor suppressor protein linked to epilepsy. Hum. Mol. Genet. 2005;14:1613–1620. doi: 10.1093/hmg/ddi169. [DOI] [PubMed] [Google Scholar]
- 10.Sirerol-Piquer M.S., Ayerdi-Izquierdo A., Morante-Redolat J.M., Herranz-Pérez V., Favell K., Barker P.A., Pérez-Tur J. The epilepsy gene LGI1 encodes a secreted glycoprotein that binds to the cell surface. Hum. Mol. Genet. 2006;15:3436–3445. doi: 10.1093/hmg/ddl421. [DOI] [PubMed] [Google Scholar]
- 11.Head K., Gong S., Joseph S., Wang C., Burkardt T., Rossi M.R., LaDuca J., Matsui S-I., Vaughan M., Hicks D.G., Heinz N., Cowell J.K. The expression pattern of the LGI1 gene in tissues and organs from BAC transgenic mice demonstrate a neuronal and glial expression pattern as well as other distinct cell types in the adult animal. Mamm. Genome. 2007;18:328–337. doi: 10.1007/s00335-007-9024-6. [DOI] [PubMed] [Google Scholar]
- 12.Staub E., Perez-Tur J., Siebert R., Nobile C., Moschonas N.K., Deloukas P., Hinzmann B. The novel EPTP repeat defines a superfamily of proteins with implications in epileptic disorders. Trends Biochem. Sci. 2002;27:441–444. doi: 10.1016/s0968-0004(02)02163-1. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez-Delicado E., Serratosa J.M. Genetics of the epilepsies. Curr. Opin. Neurol. 2004;17:147–153. doi: 10.1097/00019052-200404000-00011. [DOI] [PubMed] [Google Scholar]
- 14.Lerche H., Jurkat-Rott K., Lehmann-Horn F. Ion channels and epilepsy. Am. J. Med. Genet. 2001;106:146–159. doi: 10.1002/ajmg.1582. [DOI] [PubMed] [Google Scholar]
- 15.Reid C.A., Berkovic S.F., Petrou S. Mechanisms of human inherited epilepsies. Prog. Neurobiol. 2009;87:41–57. doi: 10.1016/j.pneurobio.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 16.Kunapuli P., Chitta K., Cowell J.K. Suppression of the cell proliferation and invasion phenotypes in glioma cells by the LGI1 gene. Oncogene. 2003;22:3985–3991. doi: 10.1038/sj.onc.1206584. [DOI] [PubMed] [Google Scholar]
- 17.Kunapuli P., Kasyapa C., Hawthorn L., Cowell J.K. LGI1, a putative tumor metastasis suppressor gene, controls in vitro invasiveness and expression of matrix metalloproteinases in glioma cells through the Erk1/2 pathway. J. Biol. Chem. 2004;279:23151–23157. doi: 10.1074/jbc.M314192200. [DOI] [PubMed] [Google Scholar]
- 18.Gabellini N., Masola V., Quartesan S., Oselladore B., Nobile C., Michelucci R., Curtarello M., Parolin C., Palù G. Increased expression of LGI1 gene triggers growth inhibition and apoptosis of neuroblastoma cells. J. Cell Physiol. 2006;207:711–721. doi: 10.1002/jcp.20627. [DOI] [PubMed] [Google Scholar]
- 19.Kunapuli P., Lo K., Hawthorn L., Cowell J.K. Reexpression of LGI1 in glioma cells results in dysregulation of genes implicated in the canonical axon guidance pathway. Genomics. 2010;95:93–100. doi: 10.1016/j.ygeno.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulte U., Thumfart J.O., Klöcker N., Sailer C.A., Bildl W., Biniossek B., Dehn D., Deller D., Eble S., Abbass K., et al. The epilepsy-linked Lgi1 protein assembles into presynaptic Kv1 channels and inhibits inactivation by Kvbeta1. Neuron. 2006;49:697–706. doi: 10.1016/j.neuron.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 21.Fukata Y., Adesnik H., Iwanaga T., Bredt D.S., Nicoll R.A., Fukata M. Epilepsy-related ligand/receptor complex LGI1 and ADAM22 regulate synaptic transmission. Science. 2006;313:1792–1795. doi: 10.1126/science.1129947. [DOI] [PubMed] [Google Scholar]
- 22.Kunapuli P., Jang G., Kazim L., Cowell J.K. Mass spectrometry identifies LGI1-interacting proteins that are involved in synaptic vesicle function in the human brain. J. Mol. Neurosci. 2009;39:805–816. doi: 10.1007/s12031-009-9202-y. [DOI] [PubMed] [Google Scholar]
- 23.Sagane K., Ishihama Y., Sugimoto H. LGI1 and LGI4 bind to ADAM22, ADAM23 and ADAM11. Int. J. Biol. Sci. 2008;4:387–396. doi: 10.7150/ijbs.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramirez-Solis R., Liu P., Bradley A. Chromosome engineering in mice. Nature. 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 25.Adams D.J., Biggs P.J., Box T., Davies R., van der Weyden L., Jonkers J., Smith J., Plumb B., Taylor R., Nishijima I., et al. MICER—mutagenic insertion and chromosome engineering resource. Nat. Genet. 2004;36:867–871. doi: 10.1038/ng1388. [DOI] [PubMed] [Google Scholar]
- 26.Neu-Yilik G., Kulozik A.E. NMD: multitasking between mRNA surveillance and modulation of gene expression. Adv. Genet. 2008;62:185–243. doi: 10.1016/S0065-2660(08)00604-4. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y.D., Lee S., Jin Z., Wright M., Smith S.E., Anderson M.P. Arrested maturation of excitatory synapses in autosomal dominant lateral temporal lobe epilepsy. Nat. Med. 2009;15:1208–1214. doi: 10.1038/nm.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fili O., Michaelevski I., Bledi Y., Chikvashvili D., Singer-Lahat D., Boshwitz H., Linial M., Lotan I. Direct interaction of a brain voltage-gated K+ channel with syntaxin 1A: functional impact on channel gating. J. Neurosci. 2001;21:1964–1974. doi: 10.1523/JNEUROSCI.21-06-01964.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitsu H., Kato M., Mizuguchi T., Hamada K., Osaka H., Tohyama J., Uruno K., Kumada S., Nishiyama K., Nishimura A., et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat. Genet. 2008;40:782–788. doi: 10.1038/ng.150. [DOI] [PubMed] [Google Scholar]
- 30.Smart S.L., Lopantsev V., Zhang C.L., Robbins C.A., Wang H., Chiu S.Y., Schwartzkroin P.A., Messing A., Tempel B.L. Deletion of the K(V)1.1 potassium channel causes epilepsy in mice. Neuron. 1998;20:809–819. doi: 10.1016/s0896-6273(00)81018-1. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell K.J., Pinsonm K.I., Kelly O.G., Brennan J., Zupicich J., Scherz P., Leighton P.A., Goodrich L.V., Lu X., Avery B.J., et al. Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat. Genet. 2001;28:241–249. doi: 10.1038/90074. [DOI] [PubMed] [Google Scholar]
- 32.Sagane K., Hayakawa K., Kai J., Hirohashi T., Takahashi E., Miyamoto N., Ino M., Oki T., Yamazaki K., Nagasu T. Ataxia and peripheral nerve hypomyelination in ADAM22-deficient mice. BMC Neurosci. 2005;6:33. doi: 10.1186/1471-2202-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brew H.M., Gittelman J.X., Silverstein R.S., Hanks T.D., Demas V.P., Robinson L.C., Robbins C.A., McKee-Johnson J., Chiu S.Y., Messing A., Tempel B.L. Seizures and reduced life span in mice lacking the potassium channel subunit Kv1.2, but hypoexcitability and enlarged Kv1 currents in auditory neurons. J. Neurophysiol. 2007;98:1501–1525. doi: 10.1152/jn.00640.2006. [DOI] [PubMed] [Google Scholar]
- 34.Ribeiro P.A., Sbragia L., Gilioli R., Langone F., Conte F.F., Lopes-Cendes I. Expression profile of Lgi1 gene in mouse brain during development. J. Mol. Neurosci. 2008;35:323–329. doi: 10.1007/s12031-008-9096-0. [DOI] [PubMed] [Google Scholar]
- 35.Cowell J.K., Head K., Kunapuli P., Vaughan M., Karasik E., Foster B. Inactivation of LGI1 expression accompanies early stage hyperplasia of prostate epithelium in the TRAMP murine model of prostate cancer. Exp. Mol. Pathol. 2009;88:77–81. doi: 10.1016/j.yexmp.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradley A. Production and analysis of chimaeric mice. In: Robertson E., editor. Teratocarcinomas and Embryonic Stem Cells—a Practical Approach. IRL Press; 1987. pp. 113–151. [Google Scholar]
- 37.Ramirez-Solis R., Davis A.C., Bradley A. Gene targeting in embryonic stem cells. Methods Enzymol. 1993;225:855–878. doi: 10.1016/0076-6879(93)25054-6. [DOI] [PubMed] [Google Scholar]
- 38.O'Gorman S., Dagenais N.A., Qian M., Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl Acad. Sci. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu P., Zhang H., McLellan A., Vogel H., Bradley A. Embryonic lethality and tumorigenesis caused by segmental aneuploidy on mouse chromosome 11. Genetics. 1998;150:1155–1168. doi: 10.1093/genetics/150.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woo R.S., Li X.M., Tao Y., Carpenter-Hyland E., Huang Y.Z., Weber J., Neiswender H., Dong X.P., Wu J., Gassmann M., et al. Neuregulin-1 enhances depolarization-induced GABA release. Neuron. 2007;54:599–610. doi: 10.1016/j.neuron.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 41.Chernova O., Cowell J.K. Molecular definition of chromosome translocations involving 10q24 and 19q13 in human malignant glioma cells. Cancer Genet. Cytogenet. 1998;105:60–68. doi: 10.1016/s0165-4608(97)00479-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.