Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) encodes a cluster of 12 microRNAs (miRNAs) that are processed from a transcript that is embedded within the major latency control region. We have generated a deletion mutation that eliminates 10 of the 12 viral miRNAs from the KSHV bacmid by using recombineering methods. The KSHV miRNA deletion mutant (BAC36 ΔmiR) behaved similarly to wild-type (wt) BAC36 in viral production, latency gene transcription, and viral DNA copy number in 293 and dermal microvascular endothelial cells (DMVECs). However, BAC36 ΔmiR consistently expressed elevated levels of viral lytic genes, including the immediate-early transcriptional activator Rta (ORF50). At least one KSHV microRNA (miRK12-5) was capable of suppressing ORF50 mRNA, but poor seed sequence alignments suggest that these targets may be indirect. Comparison of epigenetic marks in ΔmiR KSHV genomes revealed decreases in histone H3 K9 methylation, increases in histone H3 acetylation, and a striking loss of DNA methylation throughout the viral and cellular genome. One viral miRNA, K12-4-5p, was found to have a sequence targeting retinoblastoma (Rb)-like protein 2 (Rbl2), which is a known repressor of DNA methyl transferase 3a and 3b mRNA transcription. We show that ectopic expression of miR-K12-4-5p reduces Rbl2 protein expression and increases DNMT1, -3a, and -3b mRNA levels relative to the levels for control cells. We conclude that KSHV miRNA targets multiple pathways to maintain the latent state of the KSHV genome, including repression of the viral immediate-early protein Rta and a cellular factor, Rbl2, that regulates global epigenetic reprogramming.
Kaposi's sarcoma-associated herpesvirus (KSHV) is the causative agent of Kaposi's sarcoma (KS), which is the most common malignancy associated with HIV and AIDS (7; reviewed in references 13 and 31). KSHV has also been implicated as a causative agent of pleural effusion lymphoma (PEL) and multicentric Castleman's disease (6, 34). KSHV is a member of the gammaherpesvirus family, with significant sequence similarity to Epstein-Barr virus (EBV) (reviewed in reference 43). Like EBV, KSHV establishes a long-term latent infection in memory B lymphocytes. Like all herpesviruses, spontaneous reactivation from latency is correlated with increased viral load, pathogenesis, and risk of virus-associated disease. The mechanisms that control the switch between latent and lytic replication are highly complex and can be regulated by multiple viral genes and cellular pathways.
During latent infection, KSHV persists as a circular, multicopy minichromosome that is retained in the nuclear compartment (2, 8). The chromatin state of the latent virus is generally similar to the cellular genome, where the majority of viral DNA is nucleosome associated (35; reviewed in reference 26). Virus gene expression during latency is restricted to a subset of viral genes (most of which initiate from a series of multicistronic transcripts) that encode the latency-associated nuclear antigen (LANA) (ORF73), viral cyclin (vCyclin; ORF72), vFLIP (ORF71), kaposin (K12), and a cluster of 12 viral microRNAs (v-miRNAs) (5, 27, 28, 30). The latency-associated proteins are generally required for maintaining the viral genome and promoting host cell survival during latent infection. However, the precise function of the v-miRNAs is not completely understood. At least one of the KSHV v-miRNAs is a mimic of the cellular miR-155, which has been implicated in B-cell lymphomagenesis and control of B-cell differentiation (15, 33). The functional properties of the other KSHV miRNAs have not yet been elucidated.
Regulation of lytic cycle reactivation from latency is thought to be controlled, in part, through epigenetic factors (reviewed in references 25 and 26). Depending on the cell type, latent viral genomes can be reactivated by either inhibitors of histone deacetylases or by inhibitors of DNA methyltransferases (DNMTs). Furthermore, recent studies have suggested that viral genomes are partitioned into active and inactive chromatin domains, which may be organized by chromatin boundary factors, like CTCF (1, 9, 36). It is not known why, during latency, the regions surrounding the latency transcripts are accessible for transcription, while the regions surrounding the lytic proteins, like Rta (ORF50), are generally inaccessible for transcription. A role for noncoding RNAs in chromatin silencing has been demonstrated to occur at the X-chromosome inactivation locus and, more recently, at telomeres (10, 20). In plants and lower eukaryotes, small interfering RNAs have been implicated in heterochromatin formation and gene silencing (4, 22). While viral microRNAs have been shown to suppress translation of viral proteins important for lytic productive infection, no studies have yet demonstrated a direct role for microRNAs in the control of epigenetic states.
In this study, we investigate the role of the KSHV microRNA cluster as a genetic element that helps maintain the latent state of the viral chromosome. We present evidence that the viral miRNAs suppress expression of immediate-early gene transcription through a mechanism that involves the translational suppression of Rta and the maintenance of DNA methylation of the ORF50 promoter as well as other regions of the viral and cellular genome.
MATERIALS AND METHODS
Cells and plasmids.
293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin. Dermal microvascular endothelial cells (DMVECs) were maintained in EGM-MV microvascular endothelial cell growth medium (cc-3125; Lonza). 293-derived cell lines were all cultured identically to 293 cells, except with the addition of 100 μg/ml hygromycin B for the selection of the wild-type (wt) and recombinant viral genomes. The KSHV BAC36 ΔmiR mutant was generated using recombineering methods, essentially as described previously (36). The primers for PCR generation of the Kan-LoxP cassette for site-specific insertion were CCAGTAGAGTGACCCAGCTGGTTTCCATAAATGGATATACTTCCGGAAAAGAATTCCTGCAGCCCAATTCCG (forward [F]) and GGGAGGAGGAAAAAGTACGCGGTTGTTTACGCAGGGTGCGGTGCTGCCCACTAGTGGATCCCCTCGAGGGACCTA (reverse [R]). For ectopic expression of the KSHV miRNA cluster, a region from nucleotide (nt) 118747 to nt 122305 within the KSHV genome was amplified from BCBL-1 genomic DNA and inserted into pcDNA3. The pcDNA3 plasmids expressing individual KSHV miRNAs and the luciferase reporter plasmids as KSHV miRNA sensors were generated previously (29). The 3′ untranslated region (UTR) of ORF50 was amplified by PCR by using primers GCGAGATCTCATTTGGCAGGGTTACACGTTTAC (F) and GATGGCCGGCCCCTCTAGGGCCGTCATATTCTC (R), and the open reading frame (ORF) of ORF50 was amplified by PCR using primers GCGTCTAGAATGAAAGAATGTTCCAAGCTTG (F) and GATGGCCGGCCCATTTGGCAGGGTTACACGTTTA (R). The PCR products were then digested with BglII/FseI (ORF50 3′ UTR) or XbaI/FseI (ORF50) and inserted into the 3′ UTR of pGL3-Promoter. The 3′ UTR of human Rbl2 was amplified by PCR by using primers GCGGCTAGCGGTTAGTCTCTTGTATTAAAC (F) and GATGGCCGGCCTAATTTATAAATGCTTTATTG (R). The PCR product was then digested with NheI/FseI and inserted into the 3′ UTR of pGL3-Promoter. Site-directed mutation of the Rbl2 3′ UTR was generated by using the QuikChange method (Stratagene, Inc.). The 6-mer miR-K12-4-5p seed match site (TTTAGC) was changed to an XhoI restriction enzyme site (CTCGAG). The Renilla internal control vector pGL4.74[hRluc/TK] was purchased from Promega.
Reconstitution of recombinant virus.
Freshly prepared BAC DNA was introduced into 293T cells by using a Qiagen Effectene transfection kit. Briefly, 1 μg of BAC DNA was used to transfect 293T cells of 40 to 60% confluence in a 60-mm dish. Twenty-four hours after transfection, the cells were subcultured in a 100-mm plate with fresh medium containing 100 μg/ml hygromycin B. When the colonies were visible (usually 10 to 14 days after transfection), the cell colonies were dislodged and seeded into new 10-mm plates. When the monolayer reached 80 to 90% confluence, cells from each flask were split into new 150-mm plates. When the monolayer reached 40 to 60% confluence, cells were induced with 20 ng/ml 12-O-tetradecanoylphorbol-13-acetate (TPA) and 1 mM sodium butyrate for 4 days. The induced culture media were collected and filtered through 0.45-μm filters. The virions were then pelleted by ultracentrifugation on a 25% sucrose cushion at 100,000 × g for 1 h with a Beckman SW28 rotor. The pellets were dissolved in 1% of the original volume of DMEM with 10% FBS and stored at −80°C.
Infection.
293T cells or DMVECs plated in 6-well plates were incubated with concentrated virus plus polybrene (4 μg/ml) for 3 to 4 h at 37°C. The inocula were then removed and replaced with fresh media. For reverse transcriptase PCR (RT-PCR), infected cells were harvested at 72 h postinfection. To make stable 293 cell pools, 293T cells were split into new 100-mm plates at 24 h postinfection with fresh medium containing 100 μg/ml hygromycin B. Green fluorescent protein (GFP) expression was used to monitor infection 2 days after infection. When the colonies were visible (usually 14 to 18 days after infection), the cell colonies were dislodged and seeded into new 100-mm plates. Then, the stable cell pools were ready to be used for genome copy number, luciferase, chromatin immunoprecipitation (ChIP), and methyl-DNA immunoprecipitation (MeDIP) assays.
Quantitative RT-PCR.
Briefly, RNA was isolated from 106 cells by using an RNeasy kit (Qiagen) and then further treated with DNase I. RT-PCR was done as previously described. Real-time PCR was performed using a SYBR green probe with an ABI Prism 7000 according to the manufacturer's specified parameters. The primer sequences for RT-PCR were as follows: for ORF50, CCTTCGGCCCGGAGTCT (F) and CGGTTGCAGTTGCGTATACTCT (R); for LANA, TTACCTCCACCGGCACTCTT (F) and GGATGGGATGGAGGGATTG (R); for vFLIP, GGAGGAGGGCAGGTTAACG (F) and TCTGCGACCTGCACGAAA (R); for vCyclin, CATTGCCCGCCTCTATTATCA (F) and TGACGTTGGCAGGAACCA (R); for viral interleukin-6 (vIL-6), ATGCCGGTACGGTAACAGAG (F) and GGTCGGTTCACTGCTGGTAT (R); for PAN, CGGTGTTTTGGCTGGGTTT (F) and AAACCTTGCCGTCTGGTCACT (R); for DNMT1, GCACCTCATTTGCCGAATACA (F) and TCTCCTGCATCAGCCCAAATA (R); for DNMT3a, GCTGAGAGTGGGATTCATCCA (F) and CCGAGGGAGTCTCCTTTTAGAAA (R); for DNMT3b, TGATGACCGGCCGTTCTT (F) and TCTTGTCGCCAACCTTCATG (R); for β-actin, AACCCAGCCACACCACAAAG (F) and CACTGACTTGAGACCAGTTGAATAAAA (R); and for GFP, AGCAAAGACCCCAACGAGAA (F) and GGCGGCGGTCACGAA (R).
Luciferase assay.
293T or 293-derived cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's specifications. 293T cells (2 × 105) were introduced into 800 μg of KSHV miRNA expression vector and 40 ng of reporter DNA with 10 ng Renilla internal control vector pGL4.74 (Promega) and then harvested at 48 h posttransfection. For the luciferase assay of stable 293 cell pools containing BAC, cells were transfected with 200 ng reporter DNA and 10 ng pGL4.74 and then harvested at 48 h posttransfection. Relative luciferase activity was assayed using the Promega dual-luciferase reporter assay system. All data points were averages of results from at least three independent transfections.
Genome copy number assay.
Cells (use approximately 1 × 10 6 cells per sample) were collected and resuspended in 100 μl SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.0.). After brief sonication, immunoprecipitation (IP) dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris, pH 8.0, 167 mM NaCl) was added to 1 ml and then incubated with proteinase K for 2 to 3 h at 50°C. Cell lysate (300 μl) was removed and subjected to phenol-chloroform extraction and ethanol precipitation. Precipitated DNA was then assayed by real-time PCR using primers for the ORF50 region of KSHV and normalized by the cellular DNA signal at the actin gene locus. The primers for the genome copy number assay were as follows: for ORF50, CCTTCGGCCCGGAGTCT (F) and CGGTTGCAGTTGCGTATACTCT (R), and for β-actin, AACCCAGCCACACCACAAAG (F) and CACTGACTTGAGACCAGTTGAATAAAA (R).
Antibodies.
The following rabbit polyclonal antibodies were used for ChIP assays: anti-IgG (Santa Cruz Biotechnology), anti-acetylated histone H3 and H4 (Millipore), and anti-trimethyl histone H3K9 and H3K4 (Millipore). Rabbit polyclonal anti-Rbl2 (Abcam) and anti-DNMT1 (New England Biolabs) and mouse monoclonal anti-actin (Sigma) were used for Western blotting.
ChIP and KSHV genome array.
ChIP assays were performed as described previously. For the KSHV genome array, the entire DNA from one IP (106 cells per IP) was mixed with Power SYBR green PCR master mix and aliquoted completely into one 384-well plate for a total of 12 μl per well (9). Quantification of precipitated DNA was determined using real-time PCR and the Absolute Quantification program (ABI 7900HT fast real-time PCR system; Applied Biosystems). Primers were designed using Primer Express, version 2.0 (Applied Biosystems), in conjunction with a batch algorithm that generated 379 real-time primer sets across the KSHV genome, yielding amplification regions spaced, on average, every 370 bp. PCR data were normalized to input values that were quantified in parallel for each experiment. IPs were performed in triplicate for each antibody, and the PCRs were repeated at least three times. The mean for each identical set of IPs was determined, and values that were greater or less than the mean by more than 10% were excluded.
MeDIP.
Methyl-DNA immunoprecipitation (MeDIP) methods have been described previously (16). Briefly, genomic DNA was purified from 5 × 106 cells by using a Promega Wizard genomic DNA purification kit (A1125). The purified genomic DNA was sonicated to produce DNA fragments between 200 and 800 bp. The sonicated DNA was denatured at 95°C for 10 min. Denatured DNA (1 μg) was saved for the total input control. For each IP, 10 μg denatured DNA was diluted in 500 μl IP buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8.0, 150 mM NaCl) with 12.5 μg bovine serum albumin (BSA) and 5 μl anti-methyl cytidine (ab10805-50; Abcam) and incubated overnight at 4°C. To collect the immune complex, 50 μl of a protein G mixture was added to each IP reaction mixture and incubated at 4°C for 2 to 3 h. Beads were centrifuged and washed for 10 min at 4°C with each of the following: 2× IP buffer, 1× IP buffer plus 500 mM NaCl, 1× LiCl wash (250 mM LiCl, 0.5% NP-40, 0.1% Na deoxycholate, 5 mM EDTA, 10 mM Tris-Cl [pH 8.0]), 2× TE buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA). The immune complexes were eluted by incubation in 200 μl of TE-1% SDS at 65°C for 30 min, and 200 μl of TE and proteinase K (final concentration, 250 μg/ml) was added to the mixture and incubated overnight at 37°C. DNA was purified by phenol-chloroform extraction and ethanol precipitation in the presence of 20 μg of glycogen and then washed in 70% ethanol, dried, and resuspended in 100 μl of TE. The MeDIP DNA was then used for real-time PCR analysis or the KSHV genome array. The primers for MeDIP were as follows: for KSHV 69580, AGGCCGTCAAACTTCGTTGT (F) and GGCGTCGCAGGAAAACC (R); for the ORF50 promoter, CCCGCCCAGAAACCAGTAG (F) and TGCGGAGTAAGGTTGACTTTTTAA (R); for KSHV 124269, TGGCTTTTTTGGGTGGGTAA (F) and CACCCCACAGACCTGGAGTT (R); for α-globin-2, GACAAGTTCCTGGCTTCTGTGA (F) and GCTACCGAGGCTCCAGCTTA (R); and for β-actin, AACCCAGCCACACCACAAAG (F) and CACTGACTTGAGACCAGTTGAATAAAA (R).
RESULTS
Deletion within KSHV miRNA cluster causes derepression of ORF50 transcription.
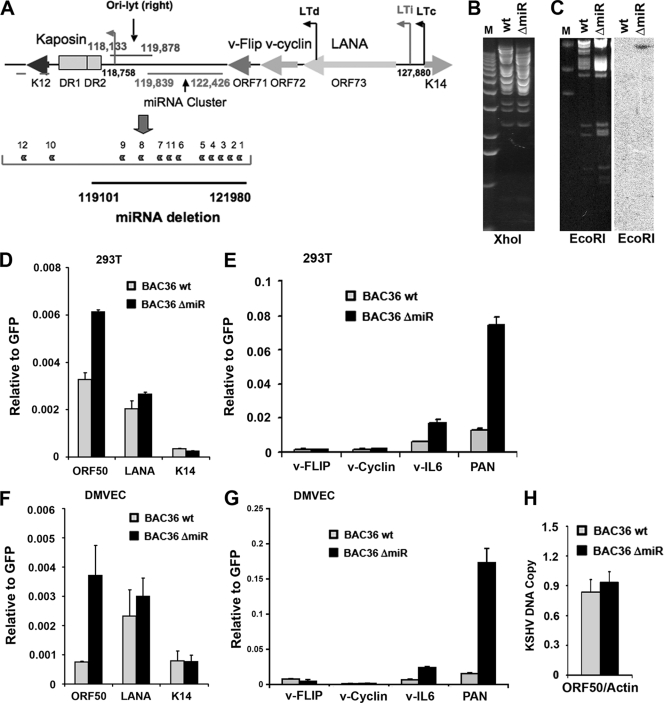
The KSHV miRNA consists of a cluster of 12 miRNAs that are contained within the major latency transcripts and adjacent to Ori-Lyt (R) (Fig. 1A). Two v-miRNAs (miR-K12-10 and -12) are embedded with the K12 ORF, while the others (miR-K12-1 through -9 and miR-K12-11) are downstream of the ORF71 gene. A deletion of miR-K12-1 through -9 and -11 was generated by recombineering of KSHV bacmid (BAC36) (36, 44). The deletion was confirmed by restriction enzyme analysis (Fig. 1B), and Southern blotting was used to demonstrate that a single-site Kan-LoxP cassette was inserted during the recombineering process (Fig. 1C). The final construct was further validated by PCR and sequencing across the deleted region (data not shown). The wild-type (wt) and mutant (ΔmiR) bacmids were transfected into 293 cells and assessed for viral gene expression patterns and production of progeny virus. We found that in transfected 293 cells, wt and ΔmiR bacmids expressed similar levels of LANA, vCyclin, K12, and ORF74 mRNA relative to GFP, which is expressed from the bacmid genome and serves as an internal control for bacmid copy number and expression (data not shown). In general, we observed only minor differences in LANA, vCyclin, K12, and ORF74 viral gene expression but found that ΔmiR-transfected cells expressed consistently higher levels of ORF50 mRNA than wt cells (data not shown). To determine if this difference was physiologically relevant, we generated infectious virus from 293 cells transfected with wt or ΔmiR bacmids. Virus particles were normalized by quantification of GFP-producing units and by real-time PCR analysis of virion DNA (data not shown). Similar numbers of GFP positive cells were formed 72 h after infection of 293 cells (7 to 8% GFP+) (Fig. 1D and E) or DMVECs (3 to 4% GFP+) (Fig. 1F and G). RNA expression was measured by RT-PCR for ORF50, ORF73, and K14 and quantified relative to the level for GFP. As was observed in transfected 293 cells, we found that ORF50 mRNA expression was elevated 2- to 4-fold in ΔmiR bacmid-infected cells relative to the level for cells infected with the wt (Fig. 1D and F). Expression levels of ORF73 (LANA) and K14 were not significantly different. To determine if ORF50 activation corresponded to activation of any other early lytic genes, we assayed mRNA expression of vIL-6 and PAN during infection of 293T cells (Fig. 1E) or DMVECs (Fig. 1G). We found that vIL-6 mRNA levels were increased ∼3- to 4-fold and that PAN mRNA levels were increased ∼6- to 11-fold in BAC36 ΔmiR relative to the levels for the BAC36 wt virus (Fig. 1E and G). In contrast, no change was observed in the mRNA levels of two other latency-associated genes, encoding vFLIP and vCyclin (Fig. 1E and G). To determine if elevated levels of ORF50 correlated with an increase in viral reactivation, we analyzed the average viral DNA copy number in 293 cell pools carrying either the BAC36 wt or BAC36 ΔmiR (Fig. 1H). Real-time PCR analysis of KSHV genome DNA relative to cellular actin DNA did not reveal any significant increase in genome copy number in stable cell lines carrying the ΔmiR mutant relative to the level for cells infected with wt BAC36 virus. This suggests that miRNA deletion (ΔmiR) does not cause robust lytic infection, despite the fact that ORF50 mRNA levels are elevated.
FIG. 1.
Deletion of KSHV miRNA causes derepression of ORF50 mRNA. (A) Schematic of KSHV genome organization surrounding the miRNA cluster and deletion boundaries for BAC36 ΔmiR. (B) Ethidium bromide stain of agarose gel for analysis of the BAC36 wt or BAC36 ΔmiR digested with XhoI. M, molecular mass marker. (C) Ethidium bromide stain (left) and Southern blot (right) for analysis of the Kan-LoxP insertion used to generate BAC36 ΔmiR. The Southern blot indicates that a single Kan-LoxP cassette was inserted into the correct EcoRI fragment. (D) RT-PCR of 293 cells infected with virus generated from the BAC36 wt or BAC36 ΔmiR. RT-PCR was performed to assay for ORF50, LANA, or K14 mRNA relative to the level for virus-encoded GFP. (E) Same as in panel D, except RT-PCR was performed to analyze for vFLIP, vCyclin, vIL-6, or PAN RNA relative to the level for GFP. (F) RT-PCR of DMVECs infected with virus generated from the BAC36 wt or BAC36 ΔmiR. RT-PCR was performed to assay for ORF50, LANA, or K14 mRNA relative to the level for virus-encoded GFP, as in panel D. (G) Same as in panel F, except RT-PCR was performed to analyze for vFLIP, vCyclin, vIL-6, or PAN RNA relative to the level for GFP. (H) DNA copy number was assayed by real-time PCR of KSHV ORF50 DNA relative to the level for cellular actin in stable 293 cell pools containing the BAC36 wt or ΔmiR.
Transcriptional silencing and PTS of ORF50 by v-miRNA.
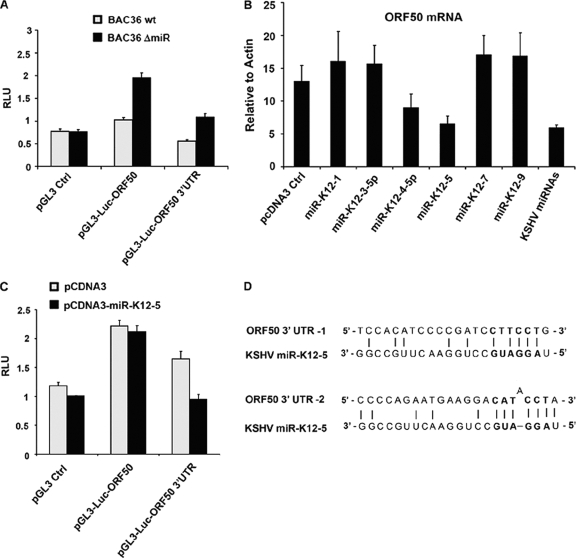
Most miRNAs are thought to function through posttranscriptional silencing (PTS) of target genes mediated by sequence hybridization with 3′ UTRs. To test whether ORF50 was subject to PTS by v-miRNA, we fused the ORF50 3′ UTR and ORF50 coding sequence to a luciferase reporter plasmid. These reporter plasmids were then tested for their relative levels of expression of luciferase in 293 cells stably infected with wt or ΔmiR viral DNA (Fig. 2A). The luciferase control plasmid (pGL3 Ctrl) expressed near-identical levels in 293 cells containing the wt and ΔmiR. In contrast, luciferase fused to the ORF50 3′ UTR or to the ORF50 coding sequence expressed ∼2-fold-higher levels of luciferase in ΔmiR-containing cells (Fig. 2A). This suggests that KSHV v-miRNA is capable of PTS directed against the ORF50 3′ UTR and coding sequence.
FIG. 2.
KSHV miR-K12-5 suppresses translation of the ORF50 3′ UTR. (A) 293 cells containing the BAC36 wt (gray) or BAC36 ΔmiR (black) were transfected with pGL3-Ctrl, pGL3-Luc-ORF50, or pGL3-Luc-ORF50 3′ UTR and assayed for luciferase activity relative to the level for the Renilla internal control (relative luciferase units [RLU]). (B) BAC36 ΔmiR was transfected with control pcDNA3 vector or pcDNA3 expressing individual KSHV miRNA (as indicated). Seventy-two hours after transfection, KSHV ORF50 mRNA was assayed by RT-PCR and quantified relative to the level for β-actin. (C) 293 cells were cotransfected with pGL3-Ctrl, pGL3-Luc-ORF50, or pGL3-Luc-ORF50 3′ UTR and with either pcDNA3 (gray) or pcDNA3-miR-K12-5 (black). Luciferase units were quantified relative to the level for the Renilla internal control. (D) Two potential alignments of the miR-K12-5 seed sequence with the 3′ UTR of ORF50.
ORF50 mRNA expression levels were elevated in the ΔmiR mutant relative to the levels for cells containing the wt bacmid (Fig. 1). To determine if a single miRNA was responsible for ORF50 repression, we examined a panel of KSHV miRNA expression constructs for their ability to repress ORF50. We found that ectopic expression of miR-K12-5 caused an ∼2-fold reduction in ORF50 mRNA relative to the level for vector control-transfected cells (Fig. 2B). This indicates that miR-K12-5 is capable of repressing ORF50 mRNA when introduced in ΔmiR BAC36-containing cells. To determine if miR-K12-5 was capable of inhibiting translation of the ORF50 3′ UTR or the ORF50 coding sequence, we tested its ability to inhibit the luciferase fusion constructs. We found that miR-K12-5 could inhibit luciferase fused to the ORF50 3′ UTR but could not control the vector or ORF50 coding sequence (Fig. 2C). An analysis of the seed sequence of miR-K12-5 revealed several low-probability alignments with the ORF50 3′ UTR, leaving open the possibility that its mechanism of PTS is indirect. Taken together, these results lead us to conclude that KSHV miRNA can suppress ORF50 through posttranslational mechanisms mediated by the ORF and the 3′ UTR and that miR-K12-5 can partially reconstitute this activity.
ΔmiR genomes have altered histone modifications.
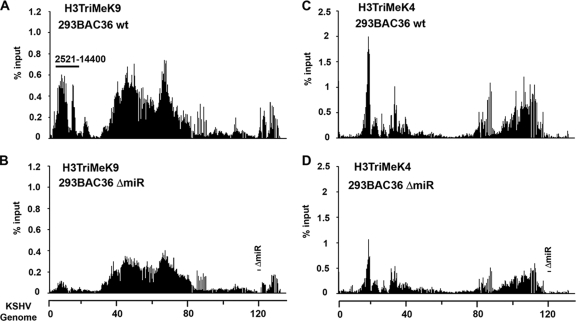
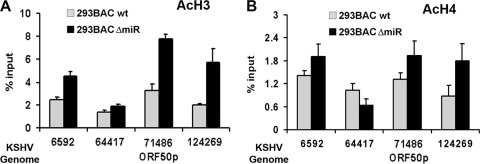
KSHV miRNA may also control ORF50 expression by regulating gene expression and chromatin organization. To investigate whether viral miRNAs have any effect on the viral chromatin structure, we first examined the histone modification patterns of wt and ΔmiR genomes in stable infected 293 cells (Fig. 3). Genome-wide histone modifications were examined by a ChIP assay using a 384-well real-time PCR array covering the genome at a density of ∼400-bp intervals. We assayed histone H3 K4 and K9 methylation patterns, which have provided highly reliable ChIP data in our hands and reflect a general difference in euchromatic (H3 K4 methylation) and heterochromatic (H3 K9 methylation) chromatin domains. Interestingly, we found that the patterns of H3 K4 and K9 were mostly nonoverlapping in cells containing wt BAC36 (compare Fig. 3A and C). A cluster of H3 K4 methylation was found downstream of the major latency transcripts (genome coordinates ∼99,721 to 114,840 in KSHV), and a second cluster was observed at positions ∼15481 to 22320. This second cluster may reflect the expression of vIL-6, which has been shown to be expressed in most latently infected cells. In contrast, H3 K9 methylation was most elevated in a large cluster spanning the regions between kb ∼40 and 80, and additional peaks were detected at positions ∼2521 to 14400 and ∼127876 to 132120. Comparison with ΔmiR genomes revealed a very similar pattern of H3 K4 methylation, although a general reduction in ChIP efficiency was observed across the entire genome. In contrast, the H3 K9 methylation pattern showed a striking loss of signal in the region between kb 2 and 14 in the ΔmiR genomes relative to the level for the wt genomes. This region is in close proximity to the TRs, which have been implicated in heterochromatin formation. We also asked whether histone acetylation was altered significantly at regions where histone methylation was different (Fig. 4). We found that histone H3 and H4 acetylation levels were elevated at genome positions 6592 (near TR), 71486 (ORF50 promoter region), and 124269 (miRNA flanking sequence) but not at position 64417 (Fig. 4A and B). The increase in histone acetylation at these regions mirrors the general loss of histone H3 K9 methylation in the ΔmiR genomes. These findings suggest that miRNA deletion causes an altered pattern of histone H3 K9 methylation and histone H3 acetylation at multiple regions throughout the viral genome.
FIG. 3.
Regional loss of histone H3 K9 trimethylation in miRNA-deleted KSHV genomes. KSHV genome-wide ChIP analysis for 293 cells containing the BAC36 wt (A and C) or BAC36 ΔmiR (B and D) was performed using antibodies specific for histone H3 trimethyl K9 (A and B) or trimethyl K4 (C and D). ChIP assay results were quantified by real-time PCR as percentages of input by using the ΔΔCT method.
FIG. 4.
Alteration in histone H3 acetylation in miRNA-deleted KSHV genomes. ChIP assays using antibodies specific for acetylated histone H3 (AcH3) (A) or AcH4 (B) were performed with 293 cells containing the BAC36 wt (gray) or BAC36 ΔmiR (black). ChIP DNA was quantified using primers specifically centered at KSHV genome position 6592, 64417, 71486 (ORF50p), or 124269, as indicated. ChIP results were quantified as percentages of input DNA by using the ΔΔCT method.
ΔmiR genomes have reduced DNA methylation.
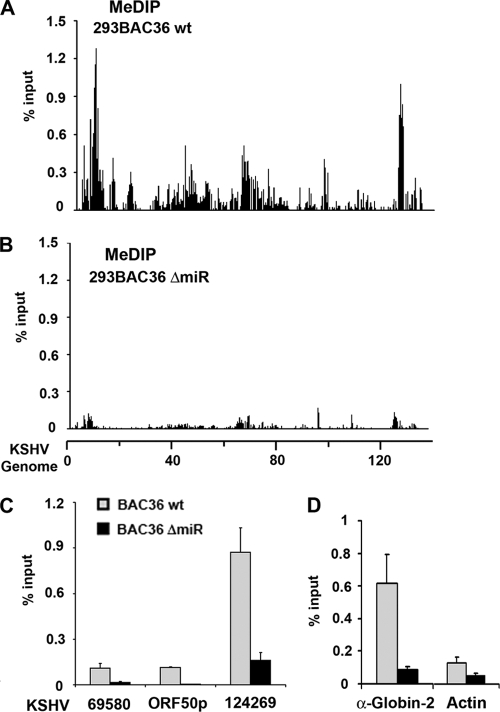
Histone modifications can be coupled with DNA methylation, and some microRNAs have been implicated in regulation of DNA methylation. We therefore investigated whether DNA methylation patterns were altered in ΔmiR genomes relative to the patterns in wt genomes (Fig. 5A). The genome-wide DNA methylation pattern was assessed using a methyl-cytosine DNA immunoprecipitation (MeDIP) assay followed by real-time PCR analysis using the KSHV genome array. We found that cells containing wt BAC36 had specific patterns of DNA methylation, with peak clusters occurring at kb 2 to 6 and at kb 124 to 127. Interestingly, the peak region comprising kb 124 to 127 overlaps the open reading frames of the latency transcripts for ORF73 and ORF72 but does not include the promoter of this gene. Remarkably, almost all of the DNA methylation peaks corresponded to histone H3 K9 methylation peaks. When CpG methylation levels were compared between wt and ΔmiR genomes, we found that miR deletion caused a striking loss of DNA methylation (Fig. 5B). Loss of the MeDIP signal was reexamined by sequence-specific PCR using different primer sets and different 293 cell clones of cells containing the wt and ΔmiR (Fig. 5C). Consistently, the MeDIP signal was reduced ∼4- to 8-fold in ΔmiR genomes relative to the wt level. We also found that a cellular gene locus within the α-globin gene, which is known to be highly methylated in nonerythroid cells, was reduced ∼6-fold for MeDIP signal in cells containing the ΔmiR mutant relative to the level for wt genomes (Fig. 5D). An ∼2-fold loss of DNA methylation was observed at the cellular actin locus (Fig. 5D, right). These data suggest that KSHV miRNAs are required for efficient DNA methylation of both viral and cellular DNA in latently infected cells.
FIG. 5.
Loss of CpG DNA methylation in miRNA-deleted KSHV genomes. (A and B) KSHV genome-wide analysis of MeDIP for the BAC36 wt (A) or BAC36 ΔmiR (B) by use of the ΔΔCT method. (C) MeDIP assay for the BAC36 wt (gray) or BAC36 ΔmiR (black) at KSHV genome position 69580, 71486 (ORF50p), or 124269 by use of the ΔΔCT method for quantification of PCR results. (D) MeDIP assay for quantification of the BAC36 wt (gray) or BAC36 ΔmiR (black) at cellular regions for α-globin-2 and β-actin. PCR was performed using the ΔΔCT method, and the results are presented as percentages of input DNA.
miR-K12-4-5p represses Rbl2 and upregulates DNA methyltransferases.
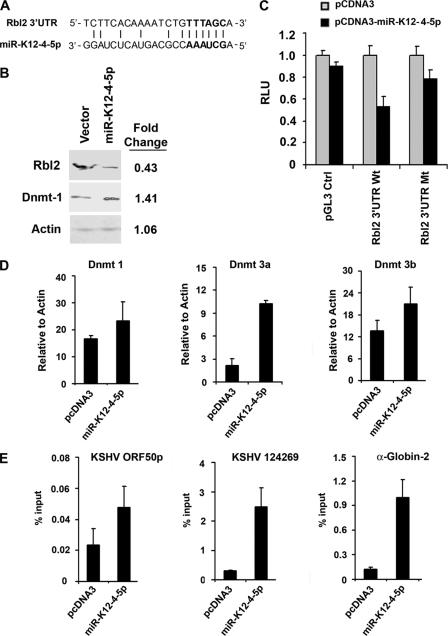
Previous studies with mice have identified a miRNA cluster that affects global DNA methylation patterns (3). In that study, they found that the miRNA targeted a cellular transcriptional repressor, Rbl2, that regulates DNMT3a and -3b mRNA expression. To explore the possibility that KSHV miRNA may function through a similar mechanism, we used bioinformatic methods to search for a KSHV miRNA seed sequence that might target the 3′ UTR of human Rbl2 mRNA (29). We found a high-probability hit for KSHV miR-K12-4-5p and the 3′ UTR of human Rbl2 mRNA (Fig. 6A). To test whether miR-K12-4-5p had any effect on Rbl2 protein levels, we examined the ability of the miR-K12-4-5p expression vector to reduce Rbl2 in ΔmiR-containing 293 cells (Fig. 6B). We found that ectopic expression of miR-K12-4-5p showed a small (∼2-fold) decrease in Rbl2 relative to the level for the control expression vector. Interestingly, we observed a corresponding increase (∼1.4-fold) in DNMT1 protein levels and no significant change in actin protein levels (Fig. 6B, lower panels). To determine if miR-K12-4-5p targeted Rbl2 mRNA directly, we assayed the effect of miR-K12-4-5p expression on luciferase reporter constructs containing either a fusion of the wt Rbl2 3′ UTR or a mutant Rbl2 3′ UTR with a 5-bp substitution mutation in the putative seed target sequence (Fig. 6C). We found that ectopic expression of miR-K12-4-5p reduced the luciferase activity of the wt 3′ UTR fusion but not the mutant fusion or the pGL3 control. This suggests that the 3′ UTR of Rbl2 is a direct target of miR-K12-4-5p. To determine if miR-K12-5p depletion of Rbl2 affected Rbl2-regulated genes involved in DNA methylation, we assayed the mRNA expression of DNMT1, DNMT3a, and DNMT3b in cells transfected with miR-K2-4-5p (Fig. 6D). Examination of mRNA levels by RT-PCR revealed that miR-K12-4-5p-transfected cells resulted in an ∼4-fold increase in DNMT3a and modest increases in DNMT1 and DNMT3b mRNA levels (Fig. 6D). To determine if the changes in DNA methyltransferase levels were functionally significant, we examined the effects of miR-K12-4-5p expression on MeDIP at several viral and cellular loci (Fig. 6E). We observed that ectopic expression of miR-K12-4-5p in ΔmiR-containing cells caused a 2- to 5-fold increase in MeDIP signal relative to the level for the control expression vector at the KSHV loci comprising position 124269 and cellular α-globin-2. Taken together, these results indicate miR12-4-5p can reduce Rbl2 protein levels, increase DNA methyltransferase mRNA levels, and increase cytosine DNA methylation at a several viral and cellular loci.
FIG. 6.
KSHV miR-K12-4-5p targets the Rbl2 3′ UTR and activates DNMT gene transcription. (A) Alignment of the Rbl2 3′ UTR with the seed sequence of KSHV miR-K12-4-5p. (B) Western blot analysis of 293 cells transfected with pcDNA3 or pcDNA3-miR-K12-4-5p was performed using antibodies for Rbl2 (top), DNMT1 (middle), and β-actin (bottom). Quantification of Western blot signals is indicated at the right of each panel. (C) Luciferase reporter assays for the control plasmid pGL3 Ctrl or luciferase fused to the wt Rbl2 3′ UTR or a mutant (Mt) Rbl2 3′ UTR in 293 cells cotransfected with miR-K12-4-5p or a vector control. Luciferase activity was normalized to Renilla activity, and data are presented relative to each luciferase target plasmid. (D) RT-PCR analysis of DNMT1, DNMT3a, and DNMT3b mRNA in BAC36 ΔmiR-containing 293 cells transfected with pcDNA3 or pcDNA3-miR-K12-4-5p for 72 h. RT-PCR was performed for quantification relative to the level for β-actin. (E) MeDIP assay of BAC36 ΔmiR-containing 293 cells transfected with pcDNA3 or pcDNA3-miR-K12-4-5p at KSHV positions 71486 (ORF50p) and 124269 or cellular α-globin-2.
DISCUSSION
In this study, we have shown that KSHV miRNAs contribute to the maintenance of latency by downregulating the lytic activator protein Rta/ORF50 and by increasing the global levels of DNA methyltransferases and CpG methylation. Specifically, we have shown that a KSHV bacmid genome lacking 10 out of 12 miRNAs (BAC36 ΔmiR) expresses ∼2- to 5-fold-higher levels of ORF50 mRNA (Fig. 1). We found that one of the KSHV miRNAs (miR-K12-5) could efficiently repress ORF50 mRNA expression levels and that this was dependent on the ORF50 3′ UTR (Fig. 2). We also found that BAC36 ΔmiR genomes had altered histone modifications relative to the BAC36 wt (Fig. 3 and 4), most notably a loss of histone H3 K9 trimethylation at regions near the terminal repeats, especially at the region spanning positions 2521 to 14400 (Fig. 3). We also found that total DNA CpG methylation was substantially reduced in BAC36 ΔmiR relative to the BAC36 wt level (Fig. 5). The effect on DNA methylation was also observed at viral, as well as some cellular chromosomal, locations (Fig. 5D). One KSHV miRNA, miR-K12-4-5p, was found to target the cellular repressor protein Rbl2, which has been implicated in transcription regulation of DNMTs. We found that miR-K12-4-5p could upregulate DNMT1, -3a, and -3b mRNA expression and increase DNA methylation at several viral and cellular chromosomal sites (Fig. 6). These studies suggest that KSHV mi-RNAs maintain KSHV latency through multiple mechanisms that involve targeting of both viral and cellular factors.
Virus-encoded miRNAs have been shown to modulate viral as well as cellular gene expression. For example, simian virus 40 (SV40)-encoded microRNAs can downregulate T-antigen expression, which limits immune detection during infection (37). Herpes simplex virus (HSV)-encoded miRNAs have been found to downregulate ICP0 and ICP4 immediate protein expression (42). HSV1 miRNAs are expressed during latent infection and function to repress lytic cycle gene activation and replication. HIV has also been reported to encode a miRNA that promotes latency through heterochromatin formation (18). Furthermore, miRNA-processing enzymes have been implicated in HIV gene expression and replication (39), but the effect of HIV infection on cellular miRNA processing remains controversial (21). Our findings are consistent with the general model in which virus-encoded miRNAs function to autoregulate viral gene expression. For viruses capable of latent infection, the viral miRNA may be critical for establishing and maintaining the latent state (42).
Our findings suggest that KSHV-encoded miRNAs can function through multiple mechanisms to help promote and stabilize the latent infection. We have identified KSHV immediate-early mRNA ORF50/Rta as a target of miR-K12-5. However, a relatively low-confidence alignment suggests that miR-K12-5 may inhibit ORF50 mRNA through an indirect mechanism. Although the 3′ UTR of ORF50 was responsive to miR-K12-5 suppression, it is not clear that this is through a direct mechanism mediated by complementary base pairing with the miRNA seed sequence. Future experiments will be required to determine if mutations in the viral seed sequence or the ORF50 3′ UTR account for the suppression of ORF50 mRNA. Despite this lack of a clear mechanism, our data strongly support the conclusion that the ORF50 3′ UTR is downregulated by miR-K12-5. Since we also found that the ORF50 coding sequence could also be targeted by KSHV miRNA, we conclude that additional, as yet unidentified viral miRNAs are involved in targeting the ORF50 coding sequence for posttranscriptional silencing. An ORF50-targeting vmiRNA was recently identified by Bellare and Ganem (2a).
In addition to the targeting of the KSHV immediate-early genes, our data indicate that KSHV miRNAs target cellular factors that regulate epigenetic modifications, particularly CpG DNA methylation. Deletion of 10 miRNAs from KSHV (BAC36 ΔmiR) caused a general loss of CpG DNA methylation across the KSHV genome. KSHV, like the cellular genome, is methylated at specific CpG sites, but the precise mechanism of site-specific DNA methylation is not completely understood. Rb-dependent transcriptional repression, followed by histone deacetylation, H3 K9 methylation, and polycomb-dependent H3 K27 methylation, has been implicated in the recruitment of DNA methyl transferases to specific sites where heterochromatin is combined with DNA methylation (20). We found that KSHV CpG DNA methylation was elevated at the regions near the terminal repeats, especially at positions 2521 to 10,000 and 122000 to 126000 (Fig. 5). These regions were also elevated in H3 K9 trimethylation. Both CpG methylation and H3 K9 trimethylation levels were reduced in BAC36 ΔmiR relative to the levels for the BAC36 wt genomes. The region comprising positions 122000 to 126000 is adjacent to the viral miRNA cluster, suggesting that the miRNA may promote CpG DNA methylation or H3 K9 trimethylation in the region from where they initiate. In plants, DNA methylation can be directly influenced by small interfering RNAs targeting specific genetic loci (22). In mammalian X-chromosome inactivation, noncoding RNAs, like Tsix, can recruit polycomb components and DNA methyltransferases (DNMT3a) to promote CpG methylation to control allele specific X-chromosome silencing (20, 38). It remains to be determined whether viral miRNA may function through a similar mechanism to direct sequence-specific DNA methylation of KSHV.
DNA methyltransferases have been identified as targets of deregulation by several different viruses and through several different mechanisms. Acute HIV infection causes a global increase in de novo DNA methylation and suppression of interferon gamma expression, but the mechanism is not clear (24). Hepatitis B virus-encoded protein X, adenovirus E1a, and BK virus T antigen were found to induce DNMT1 through an Rb-E2F-dependent manner (17, 19, 23). EBV-encoded LMP1 can induce DNMT1 expression and activation through a c-Jun NH2-terminal kinase pathway leading to the methylation-dependent repression of E-cadherin (40, 41). KSHV-encoded LANA can bind directly to DNMT3a to promote de novo DNA methylation of the latent viral genome (32). LANA recruits DNMT3 to both viral and cellular promoters to repress transcription. These findings indicate that viruses can modulate DNMTs and DNA methylation through multiple direct and indirect mechanisms.
Our findings indicate that KSHV miRNAs activate CpG methylation through an indirect mechanism involving modulation of DNMT gene transcription regulation (Fig. 6). This may help to explain how cells expressing viral miRNA can increase cellular as well as viral DNA methylation. Similar mechanisms of regulating DNA methylation by miRNAs have been reported. Rbl2 was identified as a target of a microRNA cluster (miR290) in mouse embryos and was responsible for the increased levels of DNA methyltransferase expression and DNA methylation at subtelomeres during mouse embryogenesis (3). Several human microRNAs, including miR29b and miR148, have been found to target DNMT3b and to cause global hypomethylation and loss of tumor suppressor function (11, 12, 14). We found that KSHV miR-12-4-5p can target the Rbl2 3′ UTR and downregulate Rbl2 protein levels (Fig. 6A and B). This suggests that KSHV miR-12-4-5p is a viral mimic of cellular miRNAs, perhaps analogous to miR-K12-11 as a viral mimic of miR155 (15, 33). The specific targets of miR155 are not completely clear, but miR29 and miR-K12-4-5p are potent silencers of Rbl2. Rbl2 is a member of the Rb family and has global effects on cell cycle and cellular-differentiation control. It may not be that surprising, then, that a viral miRNA targets a global regulator of cell cycle control and epigenetic silencing which may help establish viral latency.
Acknowledgments
We thank S. J. Gao (University of Texas, San Antonio) for generously providing BAC36.
This work was supported by NIH RO1s CA117830 (to P.M.L.) and CA119917 (to R.R.). We also acknowledge support from the Wistar Institute Cancer Center Core Facilities for Genomics and Flow Cytometry.
Footnotes
Published ahead of print on 13 January 2010.
REFERENCES
- 1.Amelio, A. L., P. K. McAnany, and D. C. Bloom. 2006. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J. Virol. 80:2358-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 2a.Bellare, P., and D. Ganem. 2009. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 6:570-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benetti, R., S. Gonzalo, I. Jaco, P. Munoz, S. Gonzalez, S. Schoeftner, E. Murchison, T. Andl, T. Chen, P. Klatt, E. Li, M. Serrano, S. Millar, G. Hannon, and M. A. Blasco. 2008. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat. Struct. Mol. Biol. 15:268-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buhler, M., and D. Moazed. 2007. Transcription and RNAi in heterochromatic gene silencing. Nat. Struct. Mol. Biol. 14:1041-1048. [DOI] [PubMed] [Google Scholar]
- 5.Cai, X., and B. R. Cullen. 2006. Transcriptional origin of Kaposi's sarcoma-associated herpesvirus microRNAs. J. Virol. 80:2234-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 7.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 9.Day, L., C. M. Chau, M. Nebozhyn, A. J. Rennenkamp, M. Showe, and P. M. Lieberman. 2007. Chromatin profiling of Epstein-Barr virus latency control region. J. Virol. 81:6389-6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng, Z., J. Norseen, A. Wiedmer, H. Riethman, and P. M. Lieberman. 2009. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 35:403-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duursma, A. M., M. Kedde, M. Schrier, C. le Sage, and R. Agami. 2008. miR-148 targets human DNMT3b protein coding region. RNA 14:872-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabbri, M., R. Garzon, A. Cimmino, Z. Liu, N. Zanesi, E. Callegari, S. Liu, H. Alder, S. Costinean, C. Fernandez-Cymering, S. Volinia, G. Guler, C. D. Morrison, K. K. Chan, G. Marcucci, G. A. Calin, K. Huebner, and C. M. Croce. 2007. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. U. S. A. 104:15805-15810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganem, D. 2006. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu. Rev. Pathol. 1:273-296. [DOI] [PubMed] [Google Scholar]
- 14.Garzon, R., S. Liu, M. Fabbri, Z. Liu, C. E. Heaphy, E. Callegari, S. Schwind, J. Pang, J. Yu, N. Muthusamy, V. Havelange, S. Volinia, W. Blum, L. J. Rush, D. Perrotti, M. Andreeff, C. D. Bloomfield, J. C. Byrd, K. Chan, L. C. Wu, C. M. Croce, and G. Marcucci. 2009. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 113:6411-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottwein, E., N. Mukherjee, C. Sachse, C. Frenzel, W. H. Majoros, J. T. Chi, R. Braich, M. Manoharan, J. Soutschek, U. Ohler, and B. R. Cullen. 2007. A viral microRNA functions as an orthologue of cellular miR-155. Nature 450:1096-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacinto, F. V., E. Ballestar, and M. Esteller. 2008. Methyl-DNA immunoprecipitation (MeDIP): hunting down the DNA methylome. Biotechniques 44:35-43. [DOI] [PubMed] [Google Scholar]
- 17.Jung, J. K., P. Arora, J. S. Pagano, and K. L. Jang. 2007. Expression of DNA methyltransferase 1 is activated by hepatitis B virus X protein via a regulatory circuit involving the p16INK4a-cyclin D1-CDK 4/6-pRb-E2F1 pathway. Cancer Res. 67:5771-5778. [DOI] [PubMed] [Google Scholar]
- 18.Klase, Z., P. Kale, R. Winograd, M. V. Gupta, M. Heydarian, R. Berro, T. McCaffrey, and F. Kashanchi. 2007. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol. Biol. 8:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, J. O., H. J. Kwun, J. K. Jung, K. H. Choi, D. S. Min, and K. L. Jang. 2005. Hepatitis B virus X protein represses E-cadherin expression via activation of DNA methyltransferase 1. Oncogene 24:6617-6625. [DOI] [PubMed] [Google Scholar]
- 20.Lee, J. T. 2009. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 23:1831-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, J., and B. R. Cullen. 2007. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J. Virol. 81:12218-12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matzke, M., T. Kanno, L. Daxinger, B. Huettel, and A. J. Matzke. 2009. RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 21:367-376. [DOI] [PubMed] [Google Scholar]
- 23.McCabe, M. T., J. A. Low, M. J. Imperiale, and M. L. Day. 2006. Human polyomavirus BKV transcriptionally activates DNA methyltransferase 1 through the pRb/E2F pathway. Oncogene 25:2727-2735. [DOI] [PubMed] [Google Scholar]
- 24.Mikovits, J. A., H. A. Young, P. Vertino, J. P. Issa, P. M. Pitha, S. Turcoski-Corrales, D. D. Taub, C. L. Petrow, S. B. Baylin, and F. W. Ruscetti. 1998. Infection with human immunodeficiency virus type 1 upregulates DNA methyltransferase, resulting in de novo methylation of the gamma interferon (IFN-gamma) promoter and subsequent downregulation of IFN-gamma production. Mol. Cell. Biol. 18:5166-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, G., A. El-Guindy, J. Countryman, J. Ye, and L. Gradoville. 2007. Lytic cycle switches of oncogenic human gammaherpesviruses. Adv. Cancer Res. 97:81-109. [DOI] [PubMed] [Google Scholar]
- 26.Pantry, S. N., and P. G. Medveczky. 2009. Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus replication. Semin. Cancer Biol. 19:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearce, M., S. Matsumura, and A. C. Wilson. 2005. Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi's sarcoma-associated herpesvirus originate from a common promoter. J. Virol. 79:14457-14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samols, M. A., J. Hu, R. L. Skalsky, and R. Renne. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:9301-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samols, M. A., R. L. Skalsky, A. M. Maldonado, A. Riva, M. C. Lopez, H. V. Baker, and R. Renne. 2007. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 3:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schulz, T. F. 2006. The pleiotropic effects of Kaposi's sarcoma herpesvirus. J. Pathol. 208:187-198. [DOI] [PubMed] [Google Scholar]
- 32.Shamay, M., A. Krithivas, J. Zhang, and S. D. Hayward. 2006. Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi's sarcoma-associated herpesvirus LANA. Proc. Natl. Acad. Sci. U. S. A. 103:14554-14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skalsky, R. L., M. A. Samols, K. B. Plaisance, I. W. Boss, A. Riva, M. C. Lopez, H. V. Baker, and R. Renne. 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 81:12836-12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 35.Stedman, W., Z. Deng, F. Lu, and P. M. Lieberman. 2004. ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J. Virol. 78:12566-12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stedman, W., H. Kang, S. Lin, J. L. Kissil, M. S. Bartolomei, and P. M. Lieberman. 2008. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 27:654-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullivan, C. S., A. T. Grundhoff, S. Tevethia, J. M. Pipas, and D. Ganem. 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435:682-686. [DOI] [PubMed] [Google Scholar]
- 38.Sun, B. K., A. M. Deaton, and J. T. Lee. 2006. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol. Cell 21:617-628. [DOI] [PubMed] [Google Scholar]
- 39.Triboulet, R., B. Mari, Y. L. Lin, C. Chable-Bessia, Y. Bennasser, K. Lebrigand, B. Cardinaud, T. Maurin, P. Barbry, V. Baillat, J. Reynes, P. Corbeau, K. T. Jeang, and M. Benkirane. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315:1579-1582. [DOI] [PubMed] [Google Scholar]
- 40.Tsai, C. L., H. P. Li, Y. J. Lu, C. Hsueh, Y. Liang, C. L. Chen, S. W. Tsao, K. P. Tse, J. S. Yu, and Y. S. Chang. 2006. Activation of DNA methyltransferase 1 by EBV LMP1 involves c-Jun NH(2)-terminal kinase signaling. Cancer Res. 66:11668-11676. [DOI] [PubMed] [Google Scholar]
- 41.Tsai, C. N., C. L. Tsai, K. P. Tse, H. Y. Chang, and Y. S. Chang. 2002. The Epstein-Barr virus oncogene product, latent membrane protein 1, induces the downregulation of E-cadherin gene expression via activation of DNA methyltransferases. Proc. Natl. Acad. Sci. U. S. A. 99:10084-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umbach, J. L., M. F. Kramer, I. Jurak, H. W. Karnowski, D. M. Coen, and B. R. Cullen. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wen, K. W., and B. Damania. 2009. Kaposi sarcoma-associated herpesvirus (KSHV): molecular biology and oncogenesis. Cancer Lett. [Epub ahead of print.] doi: 10.1016/j.canlet.2009.07.004. [DOI] [PMC free article] [PubMed]
- 44.Zhou, F. C., Y. J. Zhang, J. H. Deng, X. P. Wang, H. Y. Pan, E. Hettler, and S. J. Gao. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]