Abstract
Translesion DNA synthesis (TLS) is a process whereby specialized DNA polymerases are recruited to bypass DNA lesions that would otherwise stall high-fidelity polymerases. We provide evidence that TLS across cisplatin intrastrand cross-links is performed by multiple translesion DNA polymerases. First, we determined that PCNA monoubiquitination by RAD18 is necessary for efficient bypass of cisplatin adducts by the TLS polymerases eta (Polη), REV1, and zeta (Polζ) based on the observations that depletion of these proteins individually leads to decreased cell survival, cell cycle arrest in S phase, and activation of the DNA damage response. Second, we showed that in addition to PCNA monoubiquitination by RAD18, the Fanconi anemia core complex is also important for recruitment of REV1 to stalled replication forks in cisplatin treated cells. Third, we present evidence that REV1 and Polζ are uniquely associated with protection against cisplatin and mitomycin C-induced chromosomal aberrations, and both are necessary for the timely resolution of DNA double-strand breaks associated with repair of DNA interstrand cross-links. Together, our findings indicate that REV1 and Polζ facilitate repair of interstrand cross-links independently of PCNA monoubiquitination and Polη, whereas RAD18 plus Polη, REV1, and Polζ are all necessary for replicative bypass of cisplatin intrastrand DNA cross-links.
Maintenance of genomic integrity involves the activation of cell cycle checkpoints coupled with DNA repair. Despite these sophisticated mechanisms to remove DNA lesions prior to DNA replication, replication forks may inevitably encounter nonrepaired lesions that block high fidelity polymerases, potentially leading to replication fork instability, gaps in replicated DNA, and the generation of DNA double-strand breaks (DSBs). In order to preserve replication fork stability by allowing replication through polymerase blocking lesions, template DNA containing a damaged base or abasic site can be replicated through the actions of specialized translesion DNA synthesis (TLS) polymerases (61). A key event in the regulation of TLS is the monoubiquitination of PCNA, a homotrimeric protein that functions as an auxiliary factor for DNA polymerases (28, 31, 57, 60). The RAD6 (E2)-RAD18 (E3) complex specifically monoubiquitinates PCNA on Lys-164 in response to replication fork stalling. This event is thought to operate as a molecular switch from normal DNA replication to the TLS pathway based on the observations that association of Y-family TLS polymerases with monoubiquitinated PCNA is strengthened through the cooperative binding of one or more ubiquitin-binding domains (UBM or UBZ) plus a PCNA-interacting domain (6, 25).
Extensive biochemical evidence suggests that replication through a large variety of lesions requires the sequential action of two TLS polymerases (44). The Y-family polymerase eta (Polη) plays a key role in the efficient and error-free bypass of cyclobutane pyrimidine (TT) dimers, one of the major lesions resulting from exposure to UV radiation (45). In contrast, Polη can only insert a nucleotide directly opposite other lesions and requires an additional TLS polymerase, such as Polζ, to extend beyond the insertion (45). Polζ is comprised of the REV3 catalytic subunit that shares homology with B-family polymerases plus the REV7 accessory subunit (34). Polζ is unusual compared to other TLS polymerases due to the fact that it is relatively efficient at extending beyond mispaired primer termini and nucleotides inserted opposite a variety of DNA lesions, although this may occur in a potentially mutagenic manner (45). Genetic evidence in yeast suggest that Polζ activity is regulated by the Y family REV1 polymerase (21). In addition to a UBM domain that directly interacts with monoubiquitinated PCNA, REV1 possesses an N-terminal BRCT motif that directly contacts PCNA and potentially other proteins (24, 25). In addition, REV1 possesses a unique protein interaction domain in its carboxy terminus that interacts with the Polζ accessory subunit, REV7, and other TLS polymerases, including Polη and the Polζ catalytic subunit, REV3 (1, 18, 23, 40, 58). The characterization of these protein-protein interaction domains has led to the proposal that REV1 facilitates polymerase switching from a polymerase that directly inserts a nucleotide opposite a damaged base and Polζ, which subsequently performs the extension step beyond the inserted nucleotide opposite the damaged base (21).
In addition to facilitating direct lesion bypass and filling in postreplicative gaps in DNA, REV1 and Polζ may also play an important role in the repair of interstrand cross-links (46, 63). Deletion of REV1, REV3, or REV7 in chicken DT40 cells leads to remarkable hypersensitivity to a wide variety of genotoxic stresses, most notably cisplatin and other DNA cross-linking agents such as mitomycin C (MMC) (38, 41, 55, 56). The genetic epistasis observed between REV1, REV3, and the Fanconi anemia (FA) complementation group C (FANCC) gene for cisplatin sensitivity further implicates TLS in the interstrand cross-link repair pathway (38). Current models suggest that when two replication forks converge upon an interstrand cross-link, the MUS81-EME1 endonuclease recognizes and cleaves the resulting branched DNA structure by making an incision at one side of the interstrand cross-link creating a replication-associated DSB (26). The XPF-ERCC1 endonuclease uncouples the cross-linked cDNA strands by making an incision on the other side of the interstrand cross-link (37). Recent biochemical evidence suggests that Polζ performs DNA synthesis opposite the DNA strand containing the residual cross-link and this process may be necessary to prepare the daughter strand for subsequent homologous recombination repair of the replication-associated DSB (46).
Agents that introduce intra- and interstrand cross-links are widely used in cancer chemotherapy, and thus understanding the means by which cells repair or cope with these lesions will be instrumental in identifying novel mechanisms leading to drug resistance and designing new agents refractory to DNA damage tolerance mechanisms. Polη, REV1, and Polζ have all been implicated in mediating TLS past cisplatin intrastrand cross-links since lowering their expression increases sensitivity and reduces cisplatin-induced mutagenesis in human cancer cells (2, 5, 12, 42, 62). Furthermore, biochemical and structural analyses of Polη identified this polymerase as being capable of efficiently inserting dCTP opposite the 3′dG of a 1,2-d(GpG) cisplatin intrastrand cross-link (3). Here, we demonstrate that RAD18, Polη, and REV1 all localized to sites of replication stress marked by PCNA and γ-H2AX foci after treatment of cells with cisplatin. However, REV1 focus formation is specifically dependent upon both RAD18 and a functional FA core complex, suggesting FA core proteins are also necessary for directing REV1 to cisplatin-induced stalled replication forks. In addition, depletion of RAD18, Polη, REV1, or Polζ proteins lead to the induction of cellular responses indicative of inefficient lesion bypass of cisplatin adducts. Unexpectedly, we found that REV1- or Polζ-depleted cells displayed a greater loss in cell viability and the accumulation of chromosome aberrations and failed to resolve DSBs after cisplatin treatment. These results lead us to hypothesize that REV1 and Polζ may be necessary for the repair of cisplatin interstrand cross-links in addition to performing lesion bypass of cisplatin intrastrand cross-links. In agreement with this concept, we found that REV1 and Polζ-depleted cells were uniquely hypersensitive to MMC, accumulated greater numbers of chromosome aberrations, and failed to resolve replication-associated DSBs induced by MMC treatment.
Together our findings support a model where replicative bypass of cisplatin intrastrand cross-links requires cooperation of multiple TLS polymerases in mammalian cells and is triggered by PCNA monoubiquitination. Our results also provide evidence that REV1 and Polζ facilitate repair of interstrand cross-links in human cells, and this process is likely independent of PCNA monoubiquitination.
MATERIALS AND METHODS
Cell lines and culture conditions.
HeLa, U2OS, and 293T/17 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. The BL2 human Burkitt's lymphoma cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum. Construction of POLI, POLH, and REV3L−/− BL2 lines have been described (19, 22). Cisplatin [cis-diammineplatinum(II)dichloride] was purchased from Sigma-Aldrich and dissolved in 0.9% NaCl as a 3.3 mM stock solution. MMC was purchased from Roche, dissolved in 95% ethanol at a concentration of 0.2 mg/ml, and stored at −20°C.
RNA interference.
HeLa cells were seeded at a density of 105 per well of a six-well plate. The introduction of small interfering RNA (siRNA) into HeLa or U2OS cells was carried out with X-tremeGENE (Roche). For single transfections, cells were exposed to 50 nM siRNA plus 5 μl of X-tremeGENE overnight in the presence of serum, followed by a change in medium the next morning. For double siRNA transfections, U2OS cells were transfected during the day, the medium was exchanged, and then the transfection was repeated the next day. For most experiments, cells were treated with cisplatin or MMC approximately 40 h after the addition of siRNA. When delivering siRNA into cells cultured in 12-well plates, the conditions for transfection was reduced by one-half. siRNAs directed against RAD18, Polη, REV1, REV3, REV7, FANCA, FANCD2, or RAD51 were obtained from Qiagen with the exception of REV1-B, REV1-C, and REV3-C, which were obtained from Ambion. The gene-specific target sequences were as follows: RAD18-3 (GAGCATGGATTATCTATTCAA), RAD18-4 (TGCGATGCTTTGCATCCTAAA), RAD18-6 (ATGGTTGTTGCCCGAGGTTAA), Polη-1 (CTGGTTGTGAGCATTCGTGTA), Polη-3 (CAGCCAAATGCCCATTCGCAA), Polη-6 (CCCGCTATGATGCTCACAAGA), REV1-1 (AGGAGATATGTCAGTATTGAA), REV1-2 (CAGCGCATCTGTGCCAAAGAA), REV1-3 (CTGCCAGGTCCAAGCAATATA), REV1-4 (ATCGGTGGAATCGGTTTGGAA), REV1-B (AAGCATCAAAGCTGGACGACT), REV1-C (AACCAGTAAATGGCTGTAATA), REV3-1 (CGGGATGTAGTCAAACTGCAA), REV3-2 (CCCACTGGAATTAATGCACAA), REV3-3 (CCCATTATCAACAGAACCAAA), REV3-4 (ATGAGTATGGATCATATACAA), REV3-C (AAGCAATTTTGAACCTTATGG), REV7-1 (GTGGAAGAGCGCGCTCATAAA), REV7-3 (AAGATGCAGCTTTACGTGGAA), REV7-4 (CACCCGGAGCTGAATCAGTAT), FANCA-6 (AGGCCTATGCTAATCATTCTA), FANCA-7 (CAGGGCCATGCTTTCTGATTT), FANCD2-2 (CAGAGTTTGCTTCACTCTCTA), FANCD2-5 (AAGCAGCTCTCTAGCACCGTA), and RAD51 (AAGCTGAAGCTATGTTCGCCA). The negative control Non-si sequence (AATTCTCCGAACGTGTCACGT) was purchased from Qiagen. To validate siRNA sequences as to their ability to downregulate expression of REV1 or REV3 in 293T/17 cells, Flag-tagged REV1 and V5-tagged REV3 expression constructs were generated. cDNA encoding REV1 (Enzymax) was subcloned into the pSG5 vector (Stratagene) such that the Flag epitope was placed at the amino terminus of the REV1 open reading frame. cDNA encoding human REV3L was amplified through reverse transcription-PCR (RT-PCR) from HeLa total RNA by using Pfu DNA polymerase (Stratagene). Primers for cDNA amplification were designed such that the entire coding frame for REV3L was subcloned in frame with the carboxy-terminal V5-His epitope tag of pEF6/V5-HisA (Invitrogen), and the insert was verified by DNA sequence analysis. For the data shown in Fig. 1E, 293T/17 cells were cotransfected with either Flag-REV1 or V5-REV3 expression plasmids plus the indicated siRNAs using X-tremeGENE (Roche) according to the manufacturer's suggestions. REV1-1,2 and REV3-1,2 indicate that a combination of REV1-1 and REV1-2 or REV3-1 and REV3-2 siRNAs were used together at a 1:1 ratio. Total cell lysates were separated by SDS-PAGE, transferred to nitrocellulose membrane and probed with anti-Flag (M2) antibody or anti-V5 monoclonal antibody (Invitrogen). Membranes were immunoblotted for topoisomerase 1 as a loading control. To create stable RAD18 knockdowns, U2OS cells were infected with pLKO.1 lentivirus (Sigma-Aldrich) that encodes no shRNA (control) or shRNA targeting the following sequences: RAD18-2 (TGCTTCGAGTATTTCAACATT) or RAD18-6 (ATGGTTGTTGCCCGAGGTTAA). Cells were selected in puromycin and used within 1 week after drug selection.
FIG. 1.
Model for lesion bypass of cisplatin intrastrand cross-links and validation of siRNAs used to deplete individual components of the TLS pathway. (A) RAD18 responds to stalled replication forks by monoubiquitinating PCNA at K164. This event serves as a molecular switch by increasing the affinity of Y family TLS polymerases for PCNA via their ubiquitin-binding domains. Evidence suggests that Polη performs the initial nucleotide insertion step opposite the cisplatin intrastrand cross-link while Polζ (composed of the catalytic REV3 domain and the REV7 accessory subunit) performs the extension step beyond the initial insertion. Polζ activity requires REV1, a Y family polymerase thought to promote TLS polymerase switching events. See the introduction for details. (B through D) HeLa cells were transfected with the indicated siRNAs as described in Materials and Methods. After 48 h, total cellular RNA was subjected to RT-PCR using primers specific for either REV3 or GAPDH (B) or whole-cell lysates were separated by SDS-PAGE and subjected to immunoblot analysis using the indicated primary antibodies (C and D). (D) Depletion of RAD18 prevents monoubiquitination of PCNA. Control or RAD18-depleted HeLa cells were exposed to 40 J of UV-C/m2 or 150 μM cisplatin for 2 h and then harvested 8 h later. Whole-cell lysates were subjected to immunoblot analysis with anti-PCNA antibody. (E) Multiple siRNAs were individually tested as to their ability to downregulate ectopically expressed flag-tagged REV1- or V5-tagged REV3 in 293T/17 cells. Non-si indicates a nonspecific control siRNA.
Assessment of cell viability.
For clonogenic survival assays, HeLa cells were transfected overnight with siRNA as described above. When assessing survival after cisplatin treatment, cells were seeded at known densities the next day, allowed to attach to culture plates overnight, and then treated with cisplatin for 2 h. Cells were washed and allowed to form colonies for approximately 12 days. When assessing survival after MMC treatment, HeLa cells were transfected with siRNA overnight and, the following afternoon, the cells were treated with MMC for 24 h and then seeded at known cell densities for colony formation. Twelve days later, colonies were simultaneously stained and fixed in a solution containing 3:1 methanol and glacial acetic acid plus 1% trypan blue (Sigma). Colonies of 50 cells or greater were counted, and the surviving fractions for each siRNA treatment group represent the plating efficiency for each treatment divided by the plating efficiency of the corresponding untreated control. The BL2 lymphoma lines were treated with cisplatin for 2 h and then harvested 48 h later or treated with MMC continuously for 48 h. The cells were then washed with phosphate-buffered saline (PBS) and assessed as to their ability to exclude the trypan blue as a measure of viability.
Antibodies.
Rabbit polyclonal anti-Polη (H-300), anti-53BP1 (H-300), anti-GAPDH (FL-335), anti-RAD51 (H92), and anti-REV1 (H-300) antibodies were purchased from Santa Cruz Biotechnology. Rabbit polyclonal anti-phospho-CHK1 (ser345) and anti-phospho-Ser139 Histone H2AX were purchased from Cell Signaling and Active Motif, respectively. Rabbit polyclonal anti-FANCA and anti-FANCD2 were purchased from Bethyl Laboratories and GeneTex, respectively. The following mouse monoclonal antibodies were used in these studies: anti-phospho-ATM (Ser 1981; Rockland), anti-PCNA PC10 (Ab-1; Oncogene Research Products), anti-RAD18 (3H7; Abnova), anti-phospho-histone H2A.X (Ser139, clone JBW301; Millipore), anti-Topoisomerase I (BD Pharmingen), and anti-MAD2B/REV7 (BD Transduction Laboratories). The anti-β-tubulin (TUB 2.1) and anti-Flag M2 monoclonal antibodies were purchased from Sigma-Aldrich.
Immunoblotting and RT-PCR.
Cells were lysed in a sodium dodecyl sulfate (SDS) sample buffer (10 mM Tris [pH 8.0], 2% SDS, 1× protease inhibitor cocktail [Roche], and 1× phosphatase inhibitor cocktails 1 and 2 [Sigma-Aldrich]), sonicated, heated at 95°C, and equal amounts of protein were separated on SDS-PAGE gels. Proteins were transferred onto a nitrocellulose membrane and probed with the appropriate primary antibodies described above, followed by secondary horseradish peroxidase (HRP)-conjugated goat anti-rabbit or mouse antibody (Thermo Scientific). Proteins were visualized by using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). To detect REV3L mRNA, total cellular RNA was extracted with TRIzol reagent (Invitrogen), and 1 μg of RNA was reverse transcribed with a high-capacity RT kit (Applied Biosystems) according to the manufacturer's protocol. REV3L cDNA were PCR amplified by using primers flanking either side of the REV3L siRNA targeting sequence. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA primers were included in each RT-PCR reaction as an internal control. The following PCR primers were used in these studies: REV3L forward (5′-CTT TCT CAG ATG GCA TTC AG-3′) and reverse (5′-TTT CGG AAC TTG ACA GCA GC-3′) and GAPDH forward (5′-AAG GTC GGA GTC AAC GGA TTT GGT-3′) and reverse (5′-AGT GAT GGC ATG GAC TGT GGT CAT-3′). The PCR protocol was as follows: 94°C for 5 min, followed by 25 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 2 min. The RT-PCR products were separated on 1% agarose gels and visualized by ethidium bromide staining.
Immunofluorescence.
For γ-H2AX, S1981P-ATM, and 53BP1 immunofluorescence, cells cultured on glass coverslips were washed in PBS and then fixed in ice-cold 100% methanol for 10 min. Samples were blocked with 5% fetal bovine serum, 0.05% Triton X-100, and 1% goat serum and then incubated with primary antibodies for 45 min. Coverslips were washed three times with PBS and then incubated with the appropriate secondary goat anti-rabbit or goat anti-mouse Alexa Fluor dye-conjugated secondary antibody (Molecular Probes) for 45 min, washed with PBS, counterstained with DAPI to visualize nuclear DNA, and then mounted onto slides with ProLong Gold antifade reagent (Invitrogen). For experiments assessing PCNA colocalization with enhanced green fluorescent protein (EGFP), soluble proteins were first extracted with a Triton X-100 containing buffer (0.5% Triton X-100, 20 mM HEPES [pH 7.4], 3 mM MgCl2, 50 mM NaCl, and 300 mM sucrose) for 5 min at 4°C in order to reveal detergent extraction-resistant proteins in the nucleus. Cells were then fixed with a 3.7% paraformaldehyde solution for 20 min at 4°C, washed once with PBS, and then fixed again in ice-cold 100% methanol to expose the PCNA (PC-10) epitope. Immunofluorescence staining for PCNA was performed as described above. To visualize EGFP-tagged proteins and FANCD2 in lentiviral transduced U2OS cells, cells were fixed in buffered 3.7% paraformaldehyde solution containing 0.5% Triton X-100 for 20 min, permeabilized in PBS containing 0.5% Triton X-100, and then stained for FANCD2 protein as described above. Imaging was performed on either an Olympus BX-51 fluorescence microscope or Olympus FV-500 confocal microscope. Images were further processed and merged by using Adobe Photoshop CS.
Flow cytometry.
For single parameter flow cytometry, cells were fixed with ice-cold 70% ethanol and then resuspended in PBS containing RNase A and propidium iodide to determine DNA content. For two parameter flow cytometry, cells were fixed with 70% ice-cold ethanol, blocked with 5% fetal bovine serum, 1% goat serum plus 0.05% Tween 20, and then stained with anti-γ-H2AX monoclonal antibody. Cells were washed, incubated with goat anti-mouse fluorescein isothiocyanate-conjugated secondary antibody, and then counterstained with propidium iodide in PBS containing RNase A. Cells were acquired on a BD FACSCalibur system by using CellQuest software.
Analysis of chromosomal aberrations.
HeLa or BL2 cells were treated with either cisplatin for 1 h or MMC continuously. After 24 h, mitotic cells were enriched by the addition of 50 ng of Colcemid (Gibco)/ml for 45 min prior to cell harvesting. Cells were treated for 18 min at 37°C with a hypotonic solution consisting of 0.075 M KCl and then fixed in Carnoy's fixative (methanol-glacial acetic acid [3:1]). Cells were dropped onto slides and allowed to dry for a day, and then the chromosomes were stained with Giemsa prior to analysis as described previously (10). A total of 50 mitotic spreads were analyzed for each treatment. For comparisons of the average number of gaps and breaks per metaphase between control and drug-treated treatment groups, a Student t test was used.
Lentiviral vectors and virus production.
The cDNA for human POLH (Polη) and REV1L were purchased from Enzymax. Human RAD18 cDNA was PCR amplified from IMAGE clone 3451960 (American Tissue Culture Collection). POLH, REV1, and RAD18 cDNAs were subcloned into pEGFP-C1 (Clontech) such that EGFP was fused in frame at the amino-terminal end of each protein. The entire cDNA encoding EGFP fused with the open reading frame of Polη, Rev1, or Rad18 was excised and then subcloned into pLenti EV (University of Michigan Vector core facility) to generate pLLEV-EGFP-Polη, pLLEV-EGFP-REV1, and pLLEV-EGFP-RAD18. To generate lentivirus, 293T/17 cells were cotransfected with individual lentiviral vectors plus the following packaging plasmids at a 1:1:1:1 ratio (pLP1, pLP2, and pLP/VSVG, Invitrogen) using the calcium phosphate method to deliver plasmid DNA into cells. U2OS cells were seeded onto coverslips in 12-well plates and then infected with 293T/17 supernatant in the presence of 4 μg of Polybrene/ml overnight. The following morning, the medium was changed, and treatments were initiated 24 h later.
RESULTS
Replicative bypass of cisplatin intrastrand cross-links requires PCNA monoubiquitination, Polη, and the REV1/Polζ functional complex.
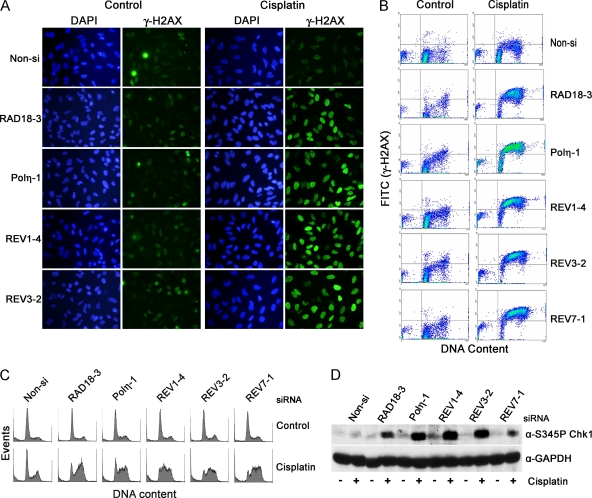
Upon entering a cell, cisplatin forms covalent bonds with the N7 position of adjacent guanines within the same DNA strand (major adduct) or on opposite strands to form interstrand cross-links (minor adduct) (32). Both cisplatin intrastrand and interstrand cross-links have been proposed to contribute to the cytotoxicity of this anticancer drug. To assess the importance of each component of the TLS pathway hypothesized as being necessary for replicative bypass of cisplatin DNA adducts (a simplistic model is shown in Fig. 1A), we transiently depleted HeLa cells of RAD18, Polη, REV1, REV3, or REV7 using siRNA and then analyzed the ability of cisplatin-treated cells to progress through the cell cycle along with their cellular response to DNA damage. Our ability to specifically deplete endogenous proteins with siRNA in HeLa cells was determined by immunoblot analysis (RAD18, Polη, REV1, or REV7) (Fig. 1C and D). Since antibodies are not available to detect endogenous REV3, RT-PCR was used to confirm reduction of REV3 mRNA after transfection with REV3-specific siRNA (Fig. 1B). We further validated the specificity and potency of the siRNAs used in the present study by examining the ability of different siRNA sequences targeting REV1 or REV3 to deplete ectopically expressed proteins in 293T cells (Fig. 1E). In addition, depletion of RAD18 via siRNA prevented efficient monoubiquitination of PCNA in response to UV-C or cisplatin demonstrating effective knockdown of the principle regulator of the TLS pathway (Fig. 1D).
Phosphorylation of the histone variant H2AX at serine 139 occurs in response to DNA damage such as DSBs or replication fork stalling, is primarily mediated by ATM or ATR, respectively (20), and is an extremely sensitive marker for the presence of DNA damage or perturbation of DNA replication. We therefore determined whether RAD18, Polη, REV1, or REV3 knockdown in HeLa cells leads to enhanced γ-H2AX staining by immunofluorescence after cisplatin treatment (Fig. 2A). Rather than seeing individual foci in cisplatin-treated cells, the nuclei of RAD18, Polη, REV1, or REV3 siRNA-transfected cells exhibited indistinguishable, intense, and almost diffuse staining patterns 24 h after drug exposure. In comparison, nonspecific (Non-si) siRNA-transfected cells displayed little staining, suggesting that these cells were capable of replicating DNA containing cisplatin intrastrand cross-links in the presence of an intact TLS pathway. Similar results were found when HeLa cells were transfected with several different siRNAs targeting RAD18, Polη, REV1, REV3, or REV7, thus demonstrating that the enhanced γ-H2AX response induced by cisplatin treatment is not due to siRNA off-target effects. (data not shown). In response to ionizing radiation, γ-H2AX staining patterns present as individual foci directly correlating with the number of DSBs (47). Since it is unlikely that the intense γ-H2AX staining observed in RAD18, Polη, REV1, or REV3 knockdown cells represents mass accumulations of DSBs, we interpret this staining pattern as an indication of extensive DNA replication stalling due to the inability to perform lesion bypass, and this in turn results in the induction of a sustained checkpoint response mediated by ATR (7, 9).
FIG. 2.
Lesion bypass of cisplatin adducts in HeLa cells requires PCNA monoubiquitination by RAD18 and the polymerase activities of Polη and the REV1/Polζ functional complex. HeLa cells transfected with the indicated siRNAs were treated with 10 μM cisplatin for 1 h. After 24 h, the cells were fixed, stained for γ-H2AX (green), and then imaged by fluorescence microscopy (A) or analyzed by flow cytometry (B and C). Dot plots showing the level of γ-H2AX staining versus DNA content are shown in panel B and corresponding histograms from the same experiment showing DNA content per event are shown in panel C. (D) RAD18-, Polη-, REV1-, REV3-, or REV7-depleted HeLa cells were treated with 10 μM cisplatin as described above. Whole-cell lysates were subjected to immunoblot analysis for CHK1 specifically phosphorylated at Ser 345 and GAPDH as a loading control. Depletion of RAD18, Polη, REV1, REV3, or REV7 leads to an enhanced DNA damage response indicative of incomplete DNA replication in cisplatin-treated cells.
To determine whether the increased γ-H2AX staining detected by immunofluorescence was specifically associated with cells residing in S phase, HeLa cells were costained with propidium iodide for DNA content along with γ-H2AX and analyzed individually by flow cytometry. We also examined whether REV7 depletion gives rise to the same phenotype as observed with the knockdown of RAD18, Polη, REV1, or REV3. Transfection of siRNA specific for each component of the TLS pathway, including REV7 (Fig. 1A), was associated with essentially identical γ-H2AX responses in that the majority of cells arrested primarily in S and G2 phases of the cell cycle and stained positive for γ-H2AX after cisplatin treatment (Fig. 2B and C). This result also demonstrates that the gene identified as human REV7 (MAD2L2 or MAD2B) is likely to be the true homologue of yeast REV7. Depletion of TLS proteins using several different siRNA sequences targeting RAD18, Polη, REV1, REV3, or REV7 lead to similar cell cycle arrest profiles after cisplatin treatment demonstrating that the effects observed here are gene specific (data not shown). Time course studies analyzing γ-H2AX formation by immunoblot analysis indicate that the γ-H2AX response developed slowly over time and became prominent by 16 h after cisplatin treatment as cells accumulated in S phase (see Fig. S1 in the supplemental material). Consistent with enhanced ATR-dependent checkpoint activation in response to replication fork stalling, depletion of RAD18, Polη, REV1, REV3, or REV7 led to increased phosphorylation of the CHK1 cell cycle checkpoint kinase, a direct and specific target of the ATR kinase (Fig. 2D) (13).
RAD18, Polη, and REV1 colocalize with PCNA at sites of cisplatin-induced replication stress.
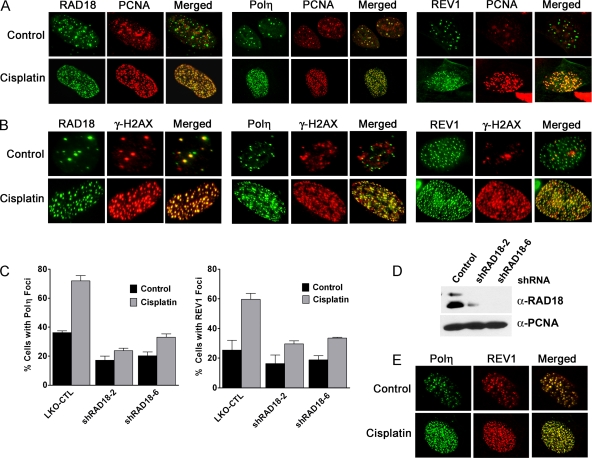
To gain additional evidence that RAD18, Polη, and REV1 are involved in replicative bypass of cisplatin intrastrand cross-links, we assessed whether these proteins colocalize with PCNA foci in a cisplatin-dependent manner. EGFP-tagged RAD18, Polη, or REV1 were delivered into cells via lentivirus such that nearly 100% of cells express EGFP-tagged protein. Since HeLa cells expressed extremely low levels of EGFP-REV1 protein, U2OS osteosarcoma cells were used for these studies. In agreement with previous studies, untreated cells exhibited EGFP-Polη foci that colocalize nearly 1 to 1 with PCNA foci (2, 30). After cisplatin treatment, the number of EGFP-Polη foci within in a given cell increased dramatically (Fig. 3A). In contrast, a significant number of untreated cells exhibited EGFP-REV1 or RAD18 foci that failed to show colocalization with PCNA, suggesting these two proteins do not strictly localize to replication factories. In response to cisplatin treatment, the majority of cisplatin-induced EGFP-REV1 or RAD18 foci colocalized with PCNA, similar to EGFP-Polη (Fig. 3A).
FIG. 3.
RAD18 regulates Polη and REV1 localization to sites of cisplatin-induced replication stress marked by PCNA and γ-H2AX. (A and B) Cisplatin induces RAD18, Polη, and REV1 foci that extensively colocalize with PCNA and γ-H2AX. U2OS cells grown on coverslips were infected with lentivirus designed to express EGFP-tagged RAD18, Polη, or REV1. Cells were treated with 33 μM cisplatin for 2 h, fixed 6 h later, and then stained with for PCNA or γ-H2AX as described in Materials and Methods. Cells stained with anti-PCNA antibody were imaged using confocal microscopy (A), and cells stained with anti-γ-H2AX antibody were imaged using an Olympus BX-51 fluorescence microscope (B). (C) REV1 and Polη focus formation is dependent upon RAD18. U2OS cells were infected with lentivirus encoding one of two different shRNAs specific for RAD18. Cells were then infected with EGFP-Polη or EGFP-REV1 lentivirus as described above. The percentage of EGFP-positive cells exhibiting 10 or more foci was determined. The results are expressed as means ± the standard errors of the mean (SEM) from three independent experiments. (D) Expression of shRNA effectively depletes RAD18 protein in U2OS cells as determined by immunoblotting of whole-cell extracts. (E) Cisplatin induces the formation of Polη and REV1 foci that colocalize. U2OS cells were first infected with lentivirus encoding EGFP-tagged Polη, followed by lentivirus encoding Flag-tagged REV1. Cells were treated with cisplatin, fixed, and then stained with anti-Flag antibody. Cells were imaged by using confocal microscopy.
To distinguish whether cisplatin-induced PCNA foci represent sites of replication stress (e.g., stalled replication forks) as opposed to unperturbed DNA replication, we determined whether cisplatin-induced PCNA foci colocalize with γ-H2AX. As expected, untreated U2OS cells exhibited both PCNA and γ-H2AX foci; however, these foci rarely colocalized (see Fig. S2 in the supplemental material). In response to agents known to cause replicative fork stalling and monoubiquitination of PCNA, such as UV-C or cisplatin, PCNA focus formation greatly increased and exhibited extensive colocalization with γ-H2AX (see Fig. S2 in the supplemental material). Cisplatin-induced EGFP-Polη, REV1, and RAD18 foci also exhibited extensive colocalization with γ-H2AX (Fig. 3B). In addition, cisplatin-induced Polη or REV1 focus formation was abrogated in U2OS cells depleted of RAD18 protein (Fig. 3C and D). Consistent with the model that Polη and REV1 cooperate to perform lesion bypass of cisplatin-DNA adducts, cisplatin induced the formation of foci consisting of EGFP-Polη and Flag-REV1 colocalized together (Fig. 3E).
The Fanconi anemia (FA) core complex recruits REV1, but not Polη, to replication foci in cisplatin-treated cells.
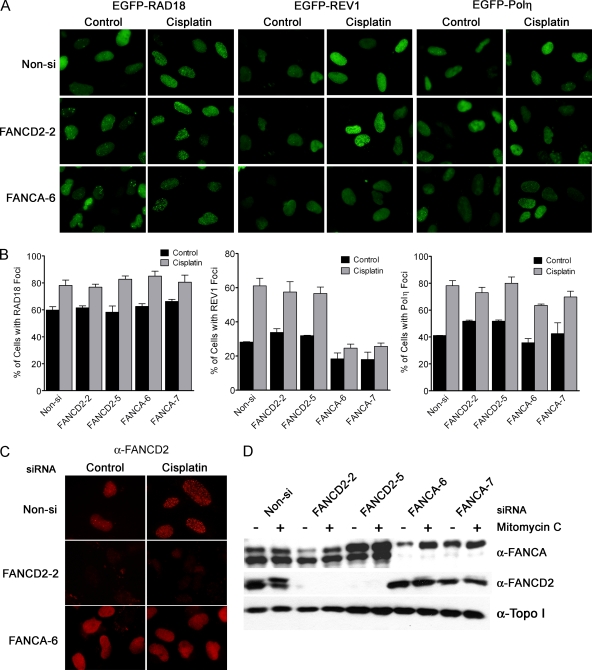
Several lines of evidence implicate the FA pathway in regulating REV1 and Polζ-dependent TLS. First, cells deficient in components of the FA pathway have been characterized as being hypomutagenic at the HPRT locus (27, 33, 43). In addition to the genetic epistasis observed between FANCC and REV1 or REV3 for cisplatin sensitivity in DT40 cells, FANCC-deleted cells are also deficient for somatic hypermutation of abasic sites at the IgM locus, a process dependent upon the TLS pathway (38). Mirchandani et al. have recently shown that the FA core complex is necessary for both spontaneous and UV-C induced mutagenesis, as well as the proper recruitment of ectopically expressed REV1 protein to nuclear foci upon exposure to UV-C irradiation (35). Interestingly, regulation of REV1 localization into UV-C induced foci does not require the FA proteins, FANCI or FANCD2, which are believed to act downstream of the FA core complex to direct checkpoint signaling and DNA repair. Regulation of REV1 focus formation by the FA core complex appears to be a parallel pathway to RAD18-dependent PCNA monoubiquitination since localization of RAD18 and its activity are unperturbed in UV-C irradiated cells deficient in FA core complex function (35). Based on these observations, we tested whether the FA pathway is necessary for the recruitment of REV1 or Polη into foci after cisplatin treatment.
U2OS cells were transfected twice with two different siRNAs specific for either FANCA or FANCD2 mRNA, and then infected with lentivirus encoding EGFP-RAD18, EGFP-Polη, or EGFP-REV1. Cells were exposed to cisplatin, fixed, and then analyzed for focus formation as in Fig. 3. Eliminating FANCA or FANCD2 expression had little impact on cisplatin-induced RAD18 or Polη focus formation (Fig. 4A and B). However, cells depleted of FANCA protein failed to exhibit cisplatin-induced EGFP-REV1 foci demonstrating that the FA core complex specifically regulates REV1 localization at stalled replication forks. The efficiency of FANCA knockdown was confirmed by assessing disruption of cisplatin-induced FANCD2 focus formation (Fig. 4C) and also by demonstrating that FANCD2 monoubiquitination was abrogated in FANCA siRNA-transfected cells (Fig. 4D). Both FANCD2 focus formation and monoubiquitination are known to require a functional FA core complex (36).
FIG. 4.
The FA core complex is necessary for cisplatin-induced REV1 focus formation. U2OS cells were transfected twice with Non-si siRNA or siRNA specific for FANCA or FANCD2 mRNA. After the second transfection, cells were infected with lentivirus encoding EGFP-RAD18, Polη, or REV1 as described in Materials and Methods. At 40 h after infection, cells were treated with 33 μM cisplatin for 2 and 6 h later the samples were fixed in paraformaldehyde plus 0.5% Triton X-100 and then stained for FANCD2 protein. Cells were imaged by using an Olympus BX-51 fluorescence microscope. (A) FANCA-deficient cells are defective in REV1 focus formation in response to cisplatin treatment. (B) The percentage of EGFP-positive cells exhibiting 10 or more foci was determined. The results represent the means ± the SEM of two independent experiments. Effective knockdown of FANCA or FANCD2 protein was assessed by analyzing FANCD2 focus formation by immunofluorescence (C) and FANCD2 monoubiquitination by immunoblotting (D). Depletion of FANCA protein abrogates both cisplatin-induced FANCD2 focus formation and MMC-induced FANCD2 monoubiquitination.
REV1, REV3, and REV7 protect against cisplatin-induced cytotoxicity and formation of chromosomal aberrations.
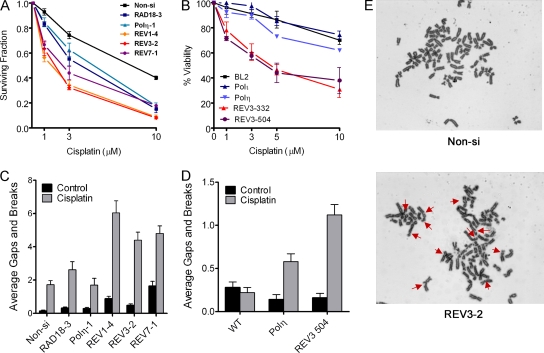
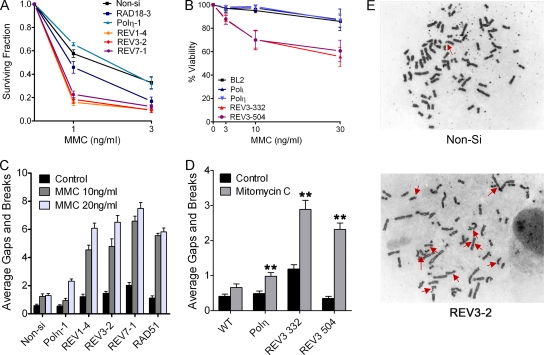
Since depletion of RAD18, Polη, REV1, REV3, or REV7 lead to indistinguishable responses with regard to phosphorylation of H2AX and cell cycle redistribution (Fig. 2), we expected that knockdown of these proteins in HeLa cells would result in equivalent hypersensitivity to cisplatin. Instead, we found that REV1, REV3, or REV7 siRNA-transfected HeLa cells exhibited greater sensitivity to cisplatin-induced loss in clonogenicity at lower drug concentrations (e.g., 1 and 3 μM), suggesting that REV1 and Polζ may perform additional roles in promoting tolerance to cisplatin (Fig. 5A). To confirm this observation, we treated human Burkitt's lymphoma BL2 cells deleted of POLH (eta), POLI (iota), or REV3L with various concentrations of cisplatin for 2 h and measured viability 48 h later by trypan blue dye exclusion (Fig. 5B). Consistent with the results observed in HeLa cells transfected with REV3 siRNA, REV3 knockout BL2 cells were significantly more sensitive to cisplatin cytotoxicity compared to Polη or Polι knockout cells.
FIG. 5.
Depletion of REV1, REV3, or REV7 renders HeLa or BL2 cells hypersensitive to cisplatin-induced cytotoxicity and genomic instability. (A) HeLa cells were transfected with control (Non-si) siRNA or siRNA targeting RAD18, Polη, REV1, REV3, or REV7. Cells were seeded at known densities, treated with cisplatin for 2 h the following day, and then allowed to form colonies for approximately 12 days. The data are expressed as the average surviving fraction ± the SEM for each dose of cisplatin. (B) Wild-type BL2 cells or BL2 cells lacking expression of polymerase Iota (Polι), Polη, or REV3 (clones 332 and 504) were treated with various doses of cisplatin for 2 h. Two days later, the cells were harvested and subjected to the trypan blue exclusion assay to assess viability. The data are expressed as the average percentage of cells stained negative for trypan blue, normalized to the untreated control. At least three independent experiments were performed. Error bars represent the SEM. (C) siRNA-transfected HeLa cells were treated with 10 μM cisplatin for 1 h, and metaphase cells were assessed for chromosomal gaps and breaks 24 h later as described in Materials and Methods. The data are expressed as the average gaps and breaks per metaphase ± the SEM (n = 50). (D) Wild-type BL2 or BL2 knockout cells were treated with 30 μM cisplatin for 1 h and then analyzed for chromosomal aberrations 24 h later. The data are expressed as the average gaps and breaks per metaphase ± SEM (n = 50). P values were calculated by using the Student t test, where “**” represents P values of <0.001. (E) Representative images of chromosomal aberrations observed in Non-si or REV3-2 siRNA-transfected HeLa cells treated with 10 μM cisplatin. Red arrows indicate chromatid gaps and breaks.
Cisplatin is known to create a small percentage of interstrand DNA cross-links (1 to 3%) in addition to intrastrand cross-links (32). Given the proposed role of REV1 and Polζ in interstrand DNA cross-link repair and the genetic epistasis observed between the FANCC gene and REV1 and REV3 in cisplatin resistance in DT40 cells, we investigated whether REV1 or Polζ-deficient cells are more sensitive to cisplatin-induced chromosome aberrations, a hallmark of interstrand DNA cross-link repair deficiency. Depletion of RAD18 or Polη in HeLa cells did not result in a significant increase in cisplatin-induced chromatid gaps and breaks per metaphase compared to controls, (Fig. 5C). In contrast, depletion of REV1, REV3, or REV7 resulted in a three- to fourfold increase in chromatid gaps and breaks per metaphase in cisplatin-treated cells, a finding consistent with the hypothesis that REV1 and Polζ play an important role in interstrand DNA cross-link repair. Similar results were observed in BL2 cells where REV3L knockout cells accumulated twice as many chromosomal aberrations compared to POLH knockout cells treated with cisplatin (Fig. 5D). Representative images of chromosomal aberrations observed in control or Rev3-depleted HeLa cells are displayed in Fig. 5E.
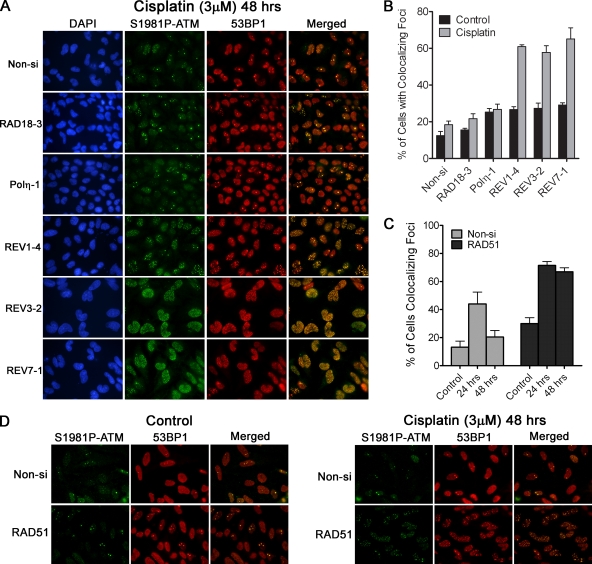
To further test our hypothesis that REV1, REV3, and REV7 contribute to interstrand DNA cross-link repair in human cells, we examined the formation and resolution of DSBs in cisplatin-treated cells as an indirect measurement of interstrand DNA cross-link repair (16, 26, 37, 48). Since γ-H2AX is not a reliable surrogate marker for the presence of DSBs under conditions of pronounced replication stalling (Fig. 2), we examined the formation of foci marked by activated ATM colocalized with 53BP1 in cisplatin-treated cells. ATM is a central cell cycle checkpoint kinase that is specifically activated in response to DSBs. Activation of ATM is associated with rapid transphosphorylation on S1981 and localization to sites of DSBs (4). 53BP1 is involved in DNA damage signaling and repair and is well characterized for its ability to localize to DSBs in cells exposed to DNA-damaging agents (51, 59). By examining colocalization of S1981P-ATM and 53BP1, we reasoned this would be a more specific surrogate marker of DSBs under conditions of DNA replication arrest. HeLa cells were transfected with individual siRNAs, treated with 3 μM cisplatin for 1 h, and then allowed to recover for 24 or 48 h before fixation and imaging of S1981P-ATM and 53BP1 by indirect immunofluorescence. At 24 h, all cisplatin-treated cells appeared to accumulate similar numbers of foci containing S1981P-ATM and 53BP1 regardless of which gene the siRNA was targeting (including the nonspecific Non-si siRNA) (see Fig. S4 in the supplemental material). However, at 48 h, ca. 60% of HeLa cells depleted of REV1, REV3, or REV7 still exhibited more than 10 foci per cell (Fig. 6A and B). In contrast, the majority of Non-si, RAD18 or Polη siRNA-transfected cells contained fewer than 10 foci per cell 48 h after cisplatin treatment, suggesting that interstrand cross-link repair was not impacted to a significant degree. These results were again confirmed by using two additional siRNA sequences targeting Rad18, Polη, REV1, REV3, or REV7 (data not shown). To confirm the validity of the focus retention assay, we also analyzed the effect of depletion of RAD51 on focus resolution after cisplatin. Similar to REV1-, REV3-, or REV7-depleted cells, RAD51-deficient cells (i.e., homologous recombination repair deficient cells) failed to resolve S1981P-ATM and 53BP1 foci in a timely manner (Fig. 6C and D).
FIG. 6.
REV1 and Polζ (REV3 and REV7) are necessary for repair of cisplatin-induced interstrand cross-links. (A) RAD18-, Polη, REV1-, REV3-, or REV7-depleted HeLa cells were treated with 3 μM cisplatin for 1 h and fixed 48 h later. Cells were then stained for S1981P-ATM and 53BP1 as surrogate markers of DNA DSBs. Nuclear DNA was stained with DAPI. (B) The percentage of cells exhibiting 10 or more colocalized foci containing both phospho-ATM and 53BP1 was determined. The data represent the means ± the SEM of three independent experiments where >300 cells were counted in each experiment. REV1-, REV3-, or REV7-depleted cells fail to resolve cisplatin-induced DSBs in a timely manner. (C) Inhibition of homologous recombination repair leads to phospho-ATM and 53BP1 focus retention in cisplatin-treated cells. Control or RAD51-depleted HeLa cells were treated with 3 μM cisplatin for 1 h and then fixed 24 or 48 h later. The percentage of cells exhibiting 10 or more phospho-ATM and 53BP1 colocalized foci are shown. The data represent the means ± the SEM of two independent experiments. Representative images of Non-si or Rad51 siRNA-transfected cells treated with cisplatin are shown in panel D.
The REV1/Polζ functional complex is necessary for efficient repair of MMC interstrand cross-links.
The observation that REV1-, REV3-, or REV7-depleted HeLa cells accumulated or failed to resolve greater numbers of cisplatin-induced DSBs could be explained by increased replication fork instability and collapse due to the inability to complete replicative bypass of cisplatin intrastrand cross-links rather than deficient interstrand DNA cross-link repair. We therefore tested whether REV1 or REV3 depleted cells are uniquely sensitive to MMC, a clinically relevant anticancer agent that produces monoadducts, intrastrand cross-links, and a far greater percentage of interstrand DNA cross-links (20%) compared to cisplatin (17). As shown in Fig. 7A, REV1, REV3, or REV7 siRNA-transfected HeLa cells are significantly more sensitive to loss in clonogenic cell survival compared to RAD18 or Polη siRNA transfected cells. Similarly, REV3L knockout BL2 cells were also more sensitive to loss in viability when exposed to relatively low doses of MMC compared to Polη or Polι knockout cells (Fig. 7B). Consistent with a deficiency in the repair of interstrand DNA cross-links, REV1-, REV3-, or REV7-depleted HeLa cells displayed greater numbers of MMC-induced chromosomal aberrations per metaphase that primarily consisted of chromatid gaps and breaks compared to Polη-depleted cells (Fig. 7C). For comparative purposes, we also depleted HeLa cells of RAD51 via siRNA. As expected, RAD51-depleted cells exhibited similar numbers of chromatid gaps and breaks per metaphase compared to REV1-, REV3-, or REV7-depleted cells, a finding consistent with the model that both translesion DNA synthesis and homologous recombination repair are necessary for efficient repair of interstrand DNA cross-links. We also confirmed the requirement of REV3 for protection against MMC-induced chromosome instability in the BL2 model system, where we consistently observed greater numbers of chromosomal aberrations in REV3L knockout cells compared to wild-type or POLH knockout cells (Fig. 7D). Representative images of chromosomal aberrations observed in mitotic spreads isolated from Non-si or REV3-2 siRNA-transfected HeLa cells treated with MMC are displayed in Fig. 7E.
FIG. 7.
The REV1/Polζ functional complex protects cells from MMC-induced cytotoxicity and genomic instability. (A) HeLa cells depleted of RAD18, Polη, REV1, REV3, or REV7 were treated with MMC for 24 h at the indicated doses and then subjected to the clonogenic survival assay. The average surviving fraction of at least three independent experiments are shown. Error bars represent the SEM. (B) Wild-type BL2 or BL2 cells deficient in polymerase Iota (Polι), Polη, or REV3 (clones 332 and 504) were treated with MMC for 48 h and then subjected to the trypan blue exclusion assay. The average percentage of cells excluding trypan blue is shown (n ≥ 3). Error bars represent the SEM. (C) Control (Non-si) or Polη-, REV1-, REV3-, REV7-, or RAD51-depleted HeLa cells were treated with 10 or 20 ng of MMC/ml for 24 h, fixed, and then analyzed for chromosomal aberrations. (D) BL2 or BL2 knockout cells were treated with 40 ng of MMC/ml for 24 h and then analyzed for chromosomal aberrations. The data are expressed as the average gaps and breaks per metaphase ± the SEM (n = 50). P values were calculated by the Student t test, where “**” represents P values of <0.001. (E) Representative images of chromosomal aberrations observed in Non-si and REV3-2 siRNA transfected HeLa cells treated with 10 ng of MMC/ml. Red arrows point to chromatid gaps and breaks.
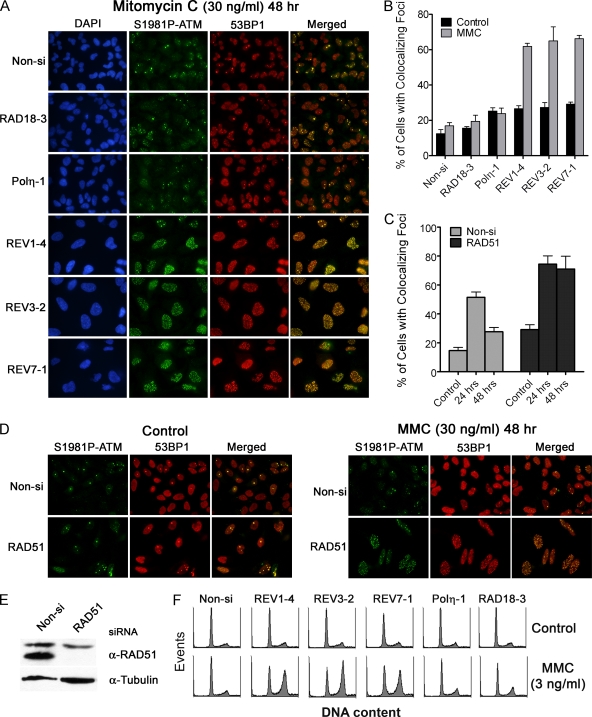
To further test the hypothesis that REV1 and Polζ are necessary for efficient interstrand DNA cross-link repair, we assessed whether siRNA-transfected HeLa cells exhibit residual DSBs after a brief exposure to MMC. At 24 h, all MMC-treated cells accumulated similar numbers of foci containing S1981P-ATM and 53BP1 regardless of which gene the siRNA was targeting (including the nonspecific Non-si siRNA) (see Fig. S5 in the supplemental material). However, REV1-, REV3-, or REV7-depleted HeLa cells, but not RAD18- or Polη-depleted cells, were deficient in resolving DSBs marked by foci containing S1981P-ATM colocalized with 53BP1 48 h after MMC treatment (Fig. 8A and B). Similar results were found using two additional siRNA sequences to deplete Rad18, Polη, REV1, REV3, or REV7 protein, confirming that deficiencies in resolving MMC-induced DSBs are not due to off-target effects (data not shown). Again, we validated this assay by demonstrating that focus retention after MMC treatment is also a major phenotype in RAD51-depleted cells (Fig. 8C through E). Finally, HeLa cells deficient in REV1, REV3, or REV7, but not RAD18 or Polη, displayed prominent cell cycle arrests in late S and G2 phases of the cell cycle indicative of prolonged cell cycle checkpoint induction after treatment with a relatively low dose of MMC (3 ng/ml), a phenotype characteristically displayed by cells deficient in components of the FA pathway (Fig. 8F).
FIG. 8.
The REV1/Polζ functional complex is necessary for efficient repair of interstrand DNA cross-links induced by MMC. (A) RAD18-, Polη-, REV1-, REV3-, or REV7-depleted HeLa cells grown on coverslips were treated with 30 ng of MMC/ml for 1 h and then allowed to recover for 48 h. Cells were fixed in 100% methanol and then stained for S1981P-ATM and 53BP1 as surrogate markers of DSBs. Nuclear DNA was stained with DAPI. (B) The average percentage of cells exhibiting 10 or more foci containing both phospho-ATM and 53BP1 was determined. Error bars represent SEM (n = 3). (C) Inhibition of homologous recombination repair lead to phospho-ATM and 53BP1 focus retention in MMC-treated cells. Control or RAD51-depleted HeLa cells were treated with 30 ng of MMC/ml for 1 h and then fixed 24 or 48 h later. The average percentage of cells exhibiting 10 or more phospho-ATM and 53BP1 colocalized foci was determined. The data represent the means ± the SEM of three independent experiments. Representative images of Non-si or Rad51 siRNA transfected cells treated with MMC are shown in panel D. Effective knockdown of RAD51 protein in HeLa cells is shown in panel E. (F) siRNA-transfected HeLa cells were treated with 3 ng of MMC/ml for 48 h. Cells were fixed, stained for DNA content with propidium iodide, and then analyzed by flow cytometry. REV1-, REV3-, or REV7-depleted cells exhibited a prolonged G2 cell cycle checkpoint suggestive of a defect in interstrand DNA cross-link repair.
DISCUSSION
Here we demonstrate that elimination of TLS activity can significantly alter the ability of cells to progress through the cell cycle when treated with the anti-cancer drug, cisplatin. In order to fully comprehend the complete molecular apparatus that performs TLS across cisplatin DNA adducts in intact cells, we individually depleted RAD18, Polη, REV1, REV3, and REV7 protein via siRNA in HeLa cells and measured their individual responses to cisplatin. Our results demonstrate that PCNA monoubiquitination by RAD18 is necessary to promote TLS across these lesions. Eliminating this essential first step in the TLS pathway lead to the accumulation of cells in S and G2 phases of the cell cycle and this block of cell cycle progression was associated with a markedly enhanced cell cycle checkpoint response, as measured by the phosphorylation of H2AX and the CHK1 kinase. We interpret these observations as an indication that RAD18-depleted cells experienced profound replication stalling due to the inability to bypass cisplatin adducts during DNA replication. The fact that Polη-, REV1-, REV3-, or REV7-depleted cells exhibited essentially identical responses to cisplatin as RAD18-depleted cells, strongly suggests that Polη, REV1, and Polζ cooperate in order to achieve efficient bypass of cisplatin intrastrand cross-links, the major cisplatin adduct formed on DNA.
The Y-family polymerase kappa has also been implicated in performing the insertion step during error-prone replicative bypass of cisplatin adducts (52). Our data are consistent with the model that Polη is the primary TLS polymerase that performs the insertion step opposite these lesions during replication of genomic DNA, at least in HeLa cells. This is based on our observation that eliminating Polη expression is just as effective as eliminating REV3 or REV7 with regard to the ability of cisplatin-treated cells to progress through S phase. Overall, our findings agree with those of Shachar et al. (52) and support the multiple polymerase insertion and extension model for TLS across cisplatin intrastrand cross-links in mammalian cells. Together, these observations are consistent with a model where Polη performs the insertion step across a 1,2-d(GpG) cisplatin adduct and Polζ completes TLS by extending beyond the initial insertion, with the latter Polζ step facilitated through REV1 interactions (Fig. 1A).
Consistent with this model, Polη and REV1 form distinct foci in response to cisplatin treatment which colocalize with PCNA and focus formation is dependent upon monoubiquitination of PCNA. Whether or not TLS polymerase focus formation truly represents an increase in actual TLS activity at stalled replication forks is not known; however, these visible foci likely represent increased residency times of Y-family TLS polymerases associated with monoubiquitinated PCNA (49). The fact that cisplatin-induced PCNA foci also colocalized with γ-H2AX strongly suggests that these sites coincide with a replication stress response that effectively results in the localized phosphorylation of H2AX. It is interesting that TLS polymerase focus formation appears to occur only in response to DNA-damaging agents that are thought to cause uncoupling of replicative helicase and polymerase activities as an indirect consequence of replication fork stalling (9). The generation of RPA-primed single-stranded DNA, a result of extensive DNA unwinding at a stalled replication fork, is essential for triggering RAD18-dependent monoubiquitination of PCNA (11, 14). Consistent with this model, DNA damage-induced Polη and REV1 focus formation occurs in response to agents that strongly induce PCNA monoubiquitination, such as UV-C irradiation, hydroxyurea, aphidicolin, and cisplatin (8, 15, 29, 39). Treatments which predominantly introduce interstrand cross-links (e.g., MMC) or alternatively, DSBs (e.g., ionizing radiation and camptothecin, a topoisomerase I poison), do not effectively induce PCNA monubiquitination or TLS polymerase focus formation (see Fig. S3 in the supplemental material; also, data not shown) (30, 39, 54). Thus, a major question is how the REV1/Polζ functional complex is regulated with respect to its recruitment to an active replication fork blocked by an interstrand DNA cross-link and the subsequent bypass replication across an unhooked interstrand DNA cross-link adduct.
Our observation that cisplatin-induced REV1 focus formation is deficient in FANCA depleted cells, but not FANCD-depleted cells, is consistent with the model that the FA core complex facilitates REV1 localization to stalled replication forks and presumably Polζ-dependent TLS (35). Although RAD18-mediated PCNA monoubiquitination may be necessary for TLS across cisplatin intrastrand cross-links or other distorting DNA adducts, depletion of RAD18 in our experimental system does not appear to have a significant impact on the resolution of replication-associated DSBs presumably associated with interstrand DNA cross-link repair (Fig. 6 and 8). This highlights major differences between how interstrand DNA cross-link repair is regulated in G1 versus the predominant interstrand DNA cross-link repair pathway that is operational during S phase and dependent upon homologous recombination. For example, in yeast and mammals, recombination-independent interstrand DNA cross-link repair involving Polζ-dependent TLS is dependent upon both RAD18-mediated PCNA monoubiquitination and the nucleotide excision repair apparatus (50, 53). In contrast, recent evidence suggests that recombination-dependent interstrand DNA cross-link repair, as measured by a sophisticated reporter system in mammalian cells, requires both the FA core complex and FANCD2, as well as Polζ, the MSH2 mismatch repair protein, and a functional homologous recombination repair complex (63).
Resistance to MMC-induced chromosomal aberrations and cytotoxicity did not appear to require RAD18 in our experimental system, suggesting that PCNA monoubiquitination is not an important regulatory element directing TLS during interstrand cross-link repair during S phase. This conclusion is consistent with our observations that MMC is a relatively weak inducer of PCNA monoubiquitination (see Fig. S3 in the supplemental material). We therefore speculate that the FA core complex performs at least two distinct functions when responding to replication stress: (i) FA core proteins regulate REV1-dependent TLS in response to replication fork stalling via a FANCD2-independent process that is parallel to Rad18-mediated monoubiquitination of PCNA, and (ii) FA core proteins promote REV1 and Polζ-dependent TLS during replication-associated interstrand cross-link repair in a FANCD2-dependent manner (36, 63). Overall, it is becoming increasingly clear that the FA core complex orchestrates multiple responses to replication stress, including regulation of REV1-dependent TLS and directing interstrand DNA cross-link repair, and both of these responses are important for maintaining genomic integrity. The observations reported here and elsewhere (46, 52) provide novel insight into the crucial role REV1 and Polζ play in providing tolerance to a variety of DNA lesions during replication of genomic DNA and in facilitating repair of interstrand DNA cross-links.
Supplementary Material
Acknowledgments
We thank Martin Arlt, David Ferguson, JoAnn Sekiguchi, and Mats Ljungman for helpful discussions and Julia Brennan for technical assistance.
This research was supported by a grant from the National Institutes of Health (CA133046 to C.E.C.), through a University of Michigan's Cancer Center Support Grant (5 P30 CA46592), and by a grant from NIGMS (GM007767 to J.K.H.).
The contents of this study are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of General Medical Sciences.
Footnotes
Published ahead of print on 22 December 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Acharya, N., R. E. Johnson, S. Prakash, and L. Prakash. 2006. Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase ζ for mismatch extension and for extension opposite from DNA lesions. Mol. Cell. Biol. 26:9555-9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albertella, M. R., C. M. Green, A. R. Lehmann, and M. J. O'Connor. 2005. A role for polymerase η in the cellular tolerance to cisplatin-induced damage. Cancer Res. 65:9799-9806. [DOI] [PubMed] [Google Scholar]
- 3.Alt, A., K. Lammens, C. Chiocchini, A. Lammens, J. C. Pieck, D. Kuch, K. P. Hopfner, and T. Carell. 2007. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase η. Science 318:967-970. [DOI] [PubMed] [Google Scholar]
- 4.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499-506. [DOI] [PubMed] [Google Scholar]
- 5.Bassett, E., N. M. King, M. F. Bryant, S. Hector, L. Pendyala, S. G. Chaney, and M. Cordeiro-Stone. 2004. The role of DNA polymerase η in translesion synthesis past Platinum-DNA adducts in human fibroblasts. Cancer Res. 64:6469-6475. [DOI] [PubMed] [Google Scholar]
- 6.Bienko, M., C. M. Green, N. Crosetto, F. Rudolf, G. Zapart, B. Coull, P. Kannouche, G. Wider, M. Peter, A. R. Lehmann, K. Hofmann, and I. Dikic. 2005. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310:1821-1824. [DOI] [PubMed] [Google Scholar]
- 7.Bomgarden, R. D., P. J. Lupardus, D. V. Soni, M.-C. Yee, J. M. Ford, and K. A. Cimprich. 2006. Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase η on ATR checkpoint signaling. EMBO J. 25:2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, S., A. Niimi, and A. R. Lehmann. 2009. Ubiquitination and deubiquitination of PCNA in response to stalling of the replication fork. Cell Cycle. 8:689-692. [DOI] [PubMed] [Google Scholar]
- 9.Byun, T. S., M. Pacek, M. c. Yee, J. C. Walter, and K. A. Cimprich. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19:1040-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casper, A. M., P. Nghiem, M. F. Arlt, and T. W. Glover. 2002. ATR regulates fragile site stability. Cell 111:779-789. [DOI] [PubMed] [Google Scholar]
- 11.Chang, D. J., P. J. Lupardus, and K. A. Cimprich. 2006. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. J. Biol. Chem. 281:32081-32088. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Y. w., J. E. Cleaver, F. Hanaoka, C. f. Chang, and K. m. Chou. 2006. A novel role of DNA polymerase η in modulating cellular sensitivity to chemotherapeutic agents. Mol. Cancer Res. 4:257-265. [DOI] [PubMed] [Google Scholar]
- 13.Cimprich, K. A., and D. Cortez. 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell. Biol. 9:616-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies, A. A., D. Huttner, Y. Daigaku, S. Chen, and H. D. Ulrich. 2008. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol. Cell 29:625-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Feraudy, S., C. L. Limoli, E. Gledzinski, D. Karentz, T. M. Marti, L. Feeney, and J. E. Cleaver. 2007. Pol eta is required for DNA replication during nucleotide deprivation by hydroxyurea. Oncogene 26:5713-5721. [DOI] [PubMed] [Google Scholar]
- 16.De Silva, I. U., P. J. McHugh, P. H. Clingen, and J. A. Hartley. 2000. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell. Biol. 20:7980-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dronkert, M. L. G., and R. Kanaar. 2001. Repair of DNA interstrand cross-links. Mutat. Res./DNA Repair 486:217-247. [DOI] [PubMed] [Google Scholar]
- 18.D'Souza, S., and G. C. Walker. 2006. Novel role for the C terminus of Saccharomyces cerevisiae Rev1 in mediating protein-protein interactions. Mol. Cell. Biol. 26:8173-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faili, A., S. Aoufouchi, E. Flatter, Q. Gueranger, C. A. Reynaud, and J. C. Weill. 2002. Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase iota. Nature 419:944-947. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Capetillo, O., A. Lee, M. Nussenzweig, and A. Nussenzweig. 2004. H2AX: the histone guardian of the genome. DNA Repair 3:959-967. [DOI] [PubMed] [Google Scholar]
- 21.Friedberg, E. C., A. R. Lehmann, and R. P. P. Fuchs. 2005. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell 18:499-505. [DOI] [PubMed] [Google Scholar]
- 22.Gueranger, Q., A. Stary, S. Aoufouchi, A. Faili, A. Sarasin, C. A. Reynaud, and J. C. Weill. 2008. Role of DNA polymerases η, ι, and ζ in UV resistance and UV-induced mutagenesis in a human cell line. DNA Repair 7:1551-1562. [DOI] [PubMed] [Google Scholar]
- 23.Guo, C., P. L. Fischhaber, M. J. Luk-Paszyc, Y. Masuda, J. Zhou, K. Kamiya, C. Kisker, and E. C. Friedberg. 2003. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 22:6621-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo, C., E. Sonoda, T. S. Tang, J. L. Parker, A. B. Bielen, S. Takeda, H. D. Ulrich, and E. C. Friedberg. 2006. REV1 protein interacts with PCNA: significance of the REV1 BRCT domain in vitro and in vivo. Mol. Cell 23:265-271. [DOI] [PubMed] [Google Scholar]
- 25.Guo, C., T. S. Tang, M. Bienko, J. L. Parker, A. B. Bielen, E. Sonoda, S. Takeda, H. D. Ulrich, I. Dikic, and E. C. Friedberg. 2006. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol. Cell. Biol. 26:8892-8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanada, K., M. Budzowska, M. Modesti, A. Maas, C. Wyman, J. Essers, and R. Kanaar. 2006. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strand breaks. EMBO J. 25:4921-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinz, J. M., P. B. Nham, E. P. Salazar, and L. H. Thompson. 2006. The Fanconi anemia pathway limits the severity of mutagenesis. DNA Repair 5:875-884. [DOI] [PubMed] [Google Scholar]
- 28.Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135-141. [DOI] [PubMed] [Google Scholar]
- 29.Howlett, N. G., T. Taniguchi, S. G. Durkin, A. D. D'Andrea, and T. W. Glover. 2005. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum. Mol. Genet. 14:693-701. [DOI] [PubMed] [Google Scholar]
- 30.Kannouche, P., B. C. Broughton, M. Volkner, F. Hanoaka, L. H. F. Mullenders, and A. R. Lehmann. 2001. Domain structure, localization, and function of DNA polymerase η, defective in xeroderma pigmentosum variant cells. Genes Dev. 15:158-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kannouche, P. L., J. Wing, and A. R. Lehmann. 2004. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14:491-500. [DOI] [PubMed] [Google Scholar]
- 32.Kelland, L. 2007. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 7:573-584. [DOI] [PubMed] [Google Scholar]
- 33.Laquerbe, A., M. Sala-Trepat, C. Vives, M. Escarceller, and D. Papadopoulo. 1999. Molecular spectra of HPRT deletion mutations in circulating T-lymphocytes in Fanconi anemia patients. Mutat. Res./Fund. Mol. Mech. Mutagen. 431:341-350. [DOI] [PubMed] [Google Scholar]
- 34.Lawrence, C. W., and D. C. Hinkle. 1996. DNA polymerase zeta and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surv. 28:21-31. [PubMed] [Google Scholar]
- 35.Mirchandani, K. D., R. M. McCaffrey, and A. D. D'Andrea. 2008. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair 7:902-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moldovan, G. L., and A. D. D'Andrea. 2009. How the Fanconi anemia pathway guards the genome. Annu. Rev. Gen. 43:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niedernhofer, L. J., H. Odijk, M. Budzowska, E. van Drunen, A. Maas, A. F. Theil, J. de Wit, N. G. J. Jaspers, H. B. Beverloo, J. H. J. Hoeijmakers, and R. Kanaar. 2004. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 24:5776-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niedzwiedz, W., G. Mosedale, M. Johnson, C. Y. Ong, P. Pace, and K. J. Patel. 2004. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell 15:607-620. [DOI] [PubMed] [Google Scholar]
- 39.Niimi, A., S. Brown, S. Sabbioneda, P. L. Kannouche, A. Scott, A. Yasui, C. M. Green, and A. R. Lehmann. 2008. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 105:16125-16130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohashi, E., Y. Murakumo, N. Kanjo, J. Akagi, C. Masutani, F. Hanaoka, and H. Ohmori. 2004. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells 9:523-531. [DOI] [PubMed] [Google Scholar]
- 41.Okada, T., E. Sonoda, M. Yoshimura, Y. Kawano, H. Saya, M. Kohzaki, and S. Takeda. 2005. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol. Cell. Biol. 25:6103-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okuda, T., X. Lin, J. Trang, and S. B. Howell. 2005. Suppression of hREV1 expression reduces the rate at which human ovarian carcinoma cells acquire resistance to cisplatin. Mol. Pharmacol. 67:1852-1860. [DOI] [PubMed] [Google Scholar]
- 43.Papadopoulo, D., C. Guillouf, H. Mohrenweiser, and E. Moustacchi. 1990. Hypomutability in Fanconi anemia cells is associated with increased deletion frequency at the HPRT locus. Proc. Natl. Acad. Sci. U. S. A. 87:8383-8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prakash, S., and L. Prakash. 2002. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 16:1872-1883. [DOI] [PubMed] [Google Scholar]
- 45.Prakash, S., R. E. Johnson, and L. Prakash. 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74:317-353. [DOI] [PubMed] [Google Scholar]
- 46.Räschle, M., P. Knipscheer, M. Enoiu, T. Angelov, J. Sun, J. D. Griffith, T. E. Ellenberger, O. D. Schärer, and J. C. Walter. 2008. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134:969-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rothfuss, A., and M. Grompe. 2004. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol. Cell. Biol. 24:123-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sabbioneda, S., A. M. Gourdin, C. M. Green, A. Zotter, G. Giglia-Mari, A. Houtsmuller, W. Vermeulen, and A. R. Lehmann. 2008. Effect of proliferating cell nuclear antigen ubiquitination and chromatin structure on the dynamic properties of the Y-family DNA polymerases. Mol. Biol. Cell 19:5193-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarkar, S., A. A. Davies, H. D. Ulrich, and P. J. Mchugh. 2006. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. EMBO J. 25:1285-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schultz, L. B., N. H. Chehab, A. Malikzay, and T. D. Halazonetis. 2000. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151:1381-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shachar, S., O. Ziv, S. Avkin, S. Adar, J. Wittschieben, T. Reiβner, S. Chaney, E. C. Friedberg, Z. Wang, T. Carell, N. Geacintov, and Z. Livneh. 2009. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 28:383-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen, X., S. Jun, L. E. O'Neal, E. Sonoda, M. Bemark, J. E. Sale, and L. Li. 2006. REV3 and REV1 play major roles in recombination-independent repair of DNA interstrand cross-links mediated by monoubiquitinated proliferating cell nuclear antigen (PCNA). J. Biol. Chem. 281:13869-13872. [DOI] [PubMed] [Google Scholar]
- 54.Shiomi, N., M. Mori, H. Tsuji, T. Imai, H. Inoue, S. Tateishi, M. Yamaizumi, and T. Shiomi. 2007. Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 35:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simpson, L. J., and J. E. Sale. 2003. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 22:1654-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sonoda, E., T. Okada, G. Y. Zhao, S. Tateishi, K. Araki, M. Yamaizumi, T. Yagi, N. S. Verkaik, G. van, M. Takata, and S. Takeda. 2003. Multiple roles of Rev3, the catalytic subunit of polζ in maintaining genome stability in vertebrates. EMBO J. 22:3188-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stelter, P., and H. D. Ulrich. 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425:188-191. [DOI] [PubMed] [Google Scholar]
- 58.Tissier, A., P. Kannouche, M. P. Reck, A. R. Lehmann, R. P. Fuchs, and A. Cordonnier. 2004. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REV1 protein. DNA Repair 3:1503-1514. [DOI] [PubMed] [Google Scholar]
- 59.Wang, B., S. Matsuoka, P. B. Carpenter, and S. J. Elledge. 2002. 53BP1, a mediator of the DNA damage checkpoint. Science 298:1435-1438. [DOI] [PubMed] [Google Scholar]
- 60.Watanabe, K., S. Tateishi, M. Kawasuji, T. Tsurimoto, H. Inoue, and M. Yamaizumi. 2004. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 23:3886-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waters, L. S., B. K. Minesinger, M. E. Wiltrout, S. D'Souza, R. V. Woodruff, and G. C. Walker. 2009. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 73:134-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, F., X. Lin, T. Okuda, and S. B. Howell. 2004. DNA polymerase zeta regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance. Cancer Res. 64:8029-8035. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, N., X. Liu, L. Li, and R. Legerski. 2007. Double-strand breaks induce homologous recombinational repair of interstrand cross-links via cooperation of MSH2, ERCC1-XPF, REV3, and the Fanconi anemia pathway. DNA Repair 6:1670-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.