Abstract
Cytoplasmic lipid droplets (CLDs) are cellular structures composed of a neutral lipid core surrounded by a phospholipid monolayer of amphipathic lipids and a variety of proteins. CLDs have classically been regarded as cellular energy storage structures. However, recent proteomic studies reveal that, although many of the proteins found to associate with CLDs are connected to lipid metabolism, storage, and homeostasis, there are also proteins with no obvious connection to the classical function and typically associated with other cellular compartments. Such proteins are termed refugee proteins, and their presence suggests that CLDs may serve an expanded role as a dynamic protein storage site, providing a novel mechanism for the regulation of protein function and transport.
Keywords: adipose differentiation related protein, adipophilin, PAT-family proteins, protein sequestration, mass spectrometry
Previously, cytoplasmic lipid droplets (CLDs) were considered to be relatively simple passive energy storage structures. However, recent studies reveal the intriguing possibility that CLDs are dynamic cellular structures and may play a role in protein sequestration as a mechanism to regulate the protein abundance and function within cells as well as lipid homeostasis. Furthermore, there is evidence that CLDs may also play a role in the regulation of membrane trafficking. This review seeks to analyze the contribution of proteomic studies to the growing body of knowledge over the last decade. [Please refer to recent comprehensive reviews for the cell biology and formation of CLDs (1–11).]
CLDs are composed of a neutral lipid core surrounded by a phospholipid monolayer. Lipids in the neutral core are primarily triacylglycerols and sterol esters, although significant quantities of ether-linked glycerolipids have also been found (12). The phospholipid species in the monolayer reflect the composition of the endoplasmic reticulum (ER) (12, 13), despite some differences observed in fatty acid composition (13).
Although CLDs can be found in virtually every cell type, some intrinsically produce more (e.g., cells involved in lipid metabolism such as adipocytes and hepatocytes, mammary epithelial cells during pregnancy and lactation (14), and fat body tissue from Drosophila embryos) (15, 16). Disease phenotypes can also dramatically alter the total numbers and size of CLDs (e.g., type II diabetes, obesity, neutral lipid storage diseases like Chanarin-Dorfman Syndrome, liver diseases such as steatohepatitis or liver cirrhosis, and cardiovascular diseases such as atherosclerosis). For cells in culture, CLD formation can be stimulated by adding fatty acids bound to albumin and/or cholesterol to promote the production of CLDs (17).
To date, the CLD proteomes of a variety of different cell types and tissues have been characterized, including yeast (Saccharomyces cerevisiae) (18, 19), Drosophila (15, 16), mouse mammary epithelial cells (14), Chinese hamster ovary K2 cell lines (20, 21), 3T3-L1 adipocytes (22, 23), cultured human A431 epithelial cells (24), HuH7 human hepatoma cell line (25), cultured human hepatocyte HepG2 cell lines (26), liver tissue from Sprague-Dawley rats (27), and human lymphoblast U937 cells from lung tissue (28). Although the overall protein compositions are similar, variations between the CLD proteomes seem to depend on cell type and metabolic state.
Two general categories of proteomic platforms have been used to analyze fractionated CLDs. The first approach separates proteins according to molecular weight using gel electrophoresis (1D or 2D) followed by in-gel protease digestion. The peptides are extracted from the gel and separated by chromatography followed by MS analysis (29). The other approach is sometimes termed “shotgun” proteomic analysis, whereby complex protein samples are digested to peptides in solution, and the peptides are then separated by chromatography followed by MS analysis. To date, most of the proteomic analyses of CLDs have used the first approach (14, 16, 18, 20–25, 28) whereas only a few have used either a combined approach (15, 27) or a stand-alone shotgun proteomic approach (19, 26).
All of the proteomic analyses have utilized chromatographic separations of peptides at ambient temperatures. Recently, it was demonstrated that elevated temperatures dramatically improve the recovery of hydrophobic proteins from reverse-phase chromatographic columns (30, 31). The hydrophobic nature of the neutral lipid core of CLDs implies that proteins embedded and/or attached to CLDs are generally hydrophobic in nature, or at a minimum have one or more hydrophobic domains. Proteins are thought to attach to lipid droplets through surface interactions or through helical hairpins of hydrophobic peptides that extend into the lipid core of the LD (32–34). [Note: Hydrophilic proteins have also been observed to be embedded within the lipid core (35).] Therefore, the incorporation of this analytical modification during proteomic analyses may have the potential to improve the comprehensiveness of the current CLD proteomes.
The issue of minimal overlap of proteins identified from different samples derived from CLDs poses a significant challenge in developing a comprehensive CLD proteome. Actual differences between the proteins associated with CLDs in different samples is one reason that proteins may be detected in one sample but not another, but this is not the only reason for disparities in proteins detected. Other reasons may include differences in metabolic state of cells used to isolate CLDs in addition to differences in sample preparation, differences in proteomic methods used (i.e., gel based methods versus shotgun methods), limitations of instrumentation, and differences in searching algorithms or databases used for protein identification.
Sample preparation and the choice of proteomic method can affect which proteins are observed from different samples. Like other biological samples, CLDs contain a wide range of protein abundances, and proteins most likely to be identified using proteomic methods are those present at high abundance, whereas low abundance proteins are more difficult to detect. Within proteomic approaches, there are sample preparation factors that may lead to differences in the proteins detected. For example, in gel-based proteomic methods covered in this review, some investigators analyzed entire 1D-SDS PAGE gel lanes cut into small pieces (16, 21) versus analyzing only gel bands stained in 1D-SDS PAGE gels (18, 20, 22, 24, 25, 28), with the goal of identifying more proteins from a CLD sample. For shotgun approaches, choosing a multidimensional protein analysis involving several steps of chromatography may lead to a greater number of identifications versus single phase chromatographic runs at the cost of requiring increased analysis time and a more complicated experimental approach (36). Altering chromatographic conditions (solid phases, mobile phases, gradients, heated columns, and column lengths) can affect the peptides ionized for detection by tandem MS. Mass spectrometers have different capabilities in terms of resolution, ionization method, and analysis speed (duty cycle) that affect the depth of profiling that can be achieved.
One very important factor to consider when comparing CLD proteomes from different studies is the choice of database search algorithms and databases. This topic has been addressed in recent studies aimed at improving standardization between methods of proteomic analysis and database searches (37–39). False discovery rate is a measure of the statistical significance analysis as it applies to mass spectrometric peptide identification and this subject is given a more in-depth treatment in some recent papers (40–42).
PROTEOMIC STUDIES
The application of proteomic analyses to CLDs within the last decade have revealed a great deal about the proteins associated with CLDs in a variety of mammals, insects, and fungi. The following sections summarize the major findings in each study and are organized according to cell/tissue origin.
SACCHAROMYCES CEREVISIAE
Saccharomyces cerevisiae was used for two proteomic studies and the results are summarized in supplementary Table I. CLDs from wild-type and deletion yeast strains were analyzed by 1D gel/MS analysis by Athenstaedt et al. (18) and proteins identified were those involved in sterol formation, lipid synthesis and lipid metabolism, enzymes, proteins associated with the ER, and several proteins with unknown functions.
A second study showed a relationship between yeast CLDs and peroxisomes (19). Proteins identified using proteomics were localized using electron microscopy (EM), immunoEM, and fluorescence microscopy in intact S. cerevisiae cultured in either control glucose (2%) or in oleic acid. In cells cultured with oleic acid, there were increased numbers of CLDs and peroxisomes. Although only observed in a small number of cases (3.2% of the cell sections where both CLDs and peroxisomes were visible), the peroxisomes were found to extend “pexopodia” into adjacent lipid droplets. Formation of pexopodia was correlated to the nutritional state of the cells, with pexopodia formation being promoted when cells were forced to scavenge fatty acids from CLDs. CLDs from these oleic acid cultures were fractionated and a shotgun approach was employed for proteomic analysis. Proteins identified were classified into three categories: 1) proteins that had previously been identified as LD proteins by Athenstaedt et al. (18), 2) mitochondrial, peroxisomal and/or ER-associated proteins, and 3) cytosolic proteins. The peroxisomal proteins identified were all matrix components that are involved in the β-oxidation of oleic acid. Many mitochondrial proteins were identified and these were further classified as either matrix proteins, matrix proteins localized to nucleoids, or inner/outer membrane or nucleoid proteins. Numerous resident proteins from the ER as well as membrane trafficking proteins, nuclear proteins, chaperones, enzymes, plasma membrane-associated proteins, and two histones, and numerous cytosolic proteins were observed.
DROSOPHILA
Two recent studies were carried out in Drosophila and their results are summarized in supplementary Table II. Ceremelli et al. (15) characterized CLDs isolated from the embryos of Drosophila followed by shotgun proteomic analysis to identify 127 proteins that appeared in more than three separate studies. [Note: 453 additional proteins were identified in only one study (397 proteins) or two studies (56 proteins)]. Proteins were classified into several groups. Lipid metabolism proteins, membrane trafficking proteins (Rab 8, and Rab 11), heat shock proteins, and other organellar proteins were observed. Surprisingly, several histone proteins (H2A, H2B, and H2Av) were identified repeatedly and at high abundances within these CLD preparations. Further orthogonal studies demonstrated that during the first few hours of embryonic development, the protein levels of H2A and H2B increased but not in direct proportion to the number of nuclei, indicating that the embryos have enough H2A and H2B to package chromatin for thousands of nuclei at the time of fertilization. Importantly, Ceremelli et al. (15) demonstrated that the large deposits of maternal histones can be stored on CLDs and rapidly and efficiently transferred to chromatin as they are needed, thereby providing a new embryo with a source of preexpressed histones for rapid embryonic development. Furthermore, by storing the proteins in an inactive form on CLDs, retardation of embryonic growth that might result from the unregulated presence of large quantities of free histones is also avoided.
The second proteomic study of Drosophila using 1D gel/MS analysis, by Beller et al. (16), focuses on lipid droplets from Drosophila fat body tissue to examine the lipid droplet subproteome from Drosophila predisposed to obesity (adp60 and induced Lsd-2:EGFP) or leanness (Lsd-251). This group identified 248 proteins, some that were labeled using GFP to localize the proteins within intact cells. EM studies of the cells and Western blot analyses were performed. Of the total 248 proteins identified, 60 proteins were identified in all four genotypes studied, 127 were found in wild-type larvae, 137 proteins were found in 60 larvae, 153 were found in induced Lsd-2:EGFP, and 159 were found in Lsd-251 mutant larvae. Proteins identified were separated into two classes: class A, composed of proteins that were identified reproducibly in separate lipid droplet preparations, and class B, which was composed of proteins identified only once. In class A, 60 proteins were identified as being common to all four genotypes with smaller numbers of proteins identified that overlapped just one or two genotypes, with a very small number of proteins identified that were genotype specific. The lack of large qualitative differences between these genotypes indicates that genotype characteristics producing leanness or obesity of adult Drosophila may not be reflected in the lipid droplet subproteome of the Drosophila embryos at this early stage of their development. One exception to this finding was the observation of the Regucalcin protein in adp60 (obesity predisposed genotype), a homolog of the vertebrate protein senescence marker protein-30 (SMP30) that has been shown in mice to affect cellular lipid droplets, body weight, and lifespan. Other proteins identified repeatedly in different CLD preparations are proteins involved in fatty acid/lipid metabolism, enzymes modifying short- and long-chain fatty acids, membrane trafficking proteins (Rabs 2, 5, and 6), chaperones, and ER-resident proteins.
MAMMALIAN CELLS AND TISSUES
Mammalian cells in culture
CHO K2 cells.
Analysis of Chinese hamster ovary (CHO) K2 cells using 1D gel/MS analysis identified many novel CLD proteins (20). Cultured CHO K2 cells grown in normal media were homogenized, then fractionated and washed repeatedly to remove contaminants from cytosol or membranes. Samples were imaged using EM and light microscopy. Thirty-seven proteins were identified and can be categorized into four groups: lipid metabolism, membrane trafficking, signaling, and miscellaneous. The full protein list can be found incorporated into supplementary Table III. Lipid metabolism is the largest grouping, with 13 proteins (35% of proteins identified). Of these, only four [S3-12, adipose differentiation related protein or adipophilin (ADRP), PDI, and BIP] were previously identified as lipid droplet associated proteins. Several of these proteins were related either by sequence or function to yeast proteins identified by Athenstaedt et al. (18) Proteins in the lipid metabolism category were typical of those found in other mammalian CLD studies. The membrane trafficking protein group was composed of 11 proteins (30% of identified proteins). The membrane trafficking proteins identified include numerous members of the Rab family of small GTPases (Rabs 1, 2, 5c, 7, 10, 11a, 11b, 14, and 18). The signaling protein group was small, containing only three of the total proteins identified (8%). Western blot analyses were carried out to verify the results obtained from the MS experiments. A second set of CHO K2 cells were incubated with oleic acid to promote CLD formation. The biggest change observed in the cells exposed to oleic acid came from a marked increase of ADRP, along with the identification of an additional gel band (∼40 kDa) containing CGI-58 protein.
The second study to use CHO K2 cells was published in 2007 by Bartz et al. (12) and emphasized several goals. The first goal of this study was to identify more of the proteins from CLDs by analyzing both the unstained and stained regions of the gels. The second goal was to identify phosphorylated proteins isolated from HeLa cell CLDs. The third goal of this study was to examine purified lipid droplets incubated in cytosol either in the presence or absence of GTPγs to determine whether lipid droplets are capable of recruiting effector proteins from the cytosol.
The complete proteome of CLDs from the cultured CHO K2 cells showed 125 proteins with 70 proteins newly recognized as being associated with CLDs (see supplementary Table III). The global analysis by 1-D gel/MS analysis of the CHO K2 proteome revealed numerous proteins previously identified as CLD-associated proteins either in CHO K2 CLDs (20) or in CLDs from other cell types (22, 24–28, 43, 44).
Numerous novel CLD proteins identified by Bartz et al. included lipid metabolism proteins, novel membrane trafficking proteins [including many Rab family proteins (Rabs 19b, 21, 24, 33b, 34, 35, and 41)] as well as some other small GTPase proteins (ArfGAP1 and ARL10C), vesicle transport-related proteins, signaling proteins, cytoskeletal proteins, endoplasmic reticulum proteins, and lysozyme proteins, chaperones, and one protein each from the Golgi and mitochondria. Three unknown proteins were identified, and these were classed as hypothetical protein DKFZp434P0316, KIAA0242 protein, and p25.
Phosphorylation is the control mechanism used by some proteins involved in lipolysis such as perilipin (45, 46) or hormone sensitive lipase (47). Bartz and coworkers used the 1D gel/MS proteomic platform to analyze CLDs isolated from HeLa cells to identify phosphorylation sites on acetyl CoA carboxylase 1, ADRP, NADH dehydrogenase precursor, phospholiase A2 (group IVA), Rab 5, Rab 8, and adipose triglyceride lipase (TTS-2.1 or iPLA2-zeta). Furthermore, this group also identified some additional small GTPases and cofactors that may be selectively recruited to CLDs by incubating purified CLDs with cytosol in the presence or absence of GTPγs in CHO K2 cells.
Mouse 3T3-L1 adipocyte cells.
Lipid droplets from cultured adipocytes of the mouse strain 3T3-L1 were analyzed in two studies and both are summarized in supplementary Table III (22, 23). The initial study by Brasaemle et al. (22) compared the proteomes of CLDs isolated from basal state adipocytes versus those that were lipolytically stimulated using isoproterenol, an agonist of β-adrenergic receptors, and isobutylmethylxanthine (IBMX), a phosphodiesterase inhibitor, using 1D gel/MS analysis. In addition to proteomic analyses, this study also used fluorescence microscopy to confirm the localizations of identified proteins within the cells. PAT family proteins perilipin, S3-12, and TIP47 were found on both control and lipolytically stimulated cells but adipophilin (ADRP) was found selectively on droplets that were lipolytically stimulated. Lipid metabolism proteins, ER-resident proteins, cytoskeletal proteins and several chaperone proteins were found in both preparations of CLDs from control and lipolytically stimulated cells. Proteins identified only in CLDs from lipolytically stimulated adipocytes included PAT-family protein adipophilin, lipid metabolism proteins, cytoskeletal proteins, membrane trafficking proteins (several Rab GTPases) and several proteins of unknown function.
The second study of 3T3-L1 adipocyte CLDs was undertaken with the goal of exploring proteins involved in lipid droplet biogenesis using 2D gel/MS analysis (23). The 3T3-L1 cells were cultured for 10 days after differentiation before cells were homogenized and CLDs were enriched by centrifugation. Although many of the proteins observed by this group were identified in previous proteomic studies, several new proteins were identified and included a lipid metabolism protein, four membrane trafficking proteins, and several proteins of unknown function/localization [a 54.10 kDa protein PRP19 (homolog of S. cerevisiae PSO4) that was selected for further study].
The 504 amino acid protein Prp19p is encoded by the murine Prp19 gene (mPrp19). In S. cerevisiae and humans, Prp19p has functions in DNA combination and error prone repair and previously had not been observed to have functions outside the nucleus (23). This protein acts as a scaffold for protein-protein interactions in the nucleus, and the authors suggest that Prp19p may play a role in the formation or stabilization of CLDs through the ability to make protein-protein interactions. Stable 3T3-L1 cell lines were developed to either overexpress mouse Prp19 (pCDNA3.1-mPrp19) or downregulate mouse Prp19 (p-mPrp19RNAi). RNAi downregulation of Prp19p decreased intracellular fat accretion, thereby repressing lipid droplet biogenesis and expression of SCD1, DGAT-1, GPAT, perilipin, and S3-12, but it did not affect other genes important to adipocyte differentiation. Unlike perilipin, Prp19p does not appear essential for cAMP- and hormone sensitive lipase-dependent lipolysis pathways.
Mammalian cells from tissue
Mouse mammary epithelial cells.
An early study of mammalian tissues focused on CLDs obtained from mammary epithelial cells from the inguinal mammary glands, livers, and milk taken from the same lactating animal. Milk fat globules (MFGs) and milk fat globule membranes (MFGMs) were isolated from the mouse milk (14). Proteomic analysis was performed on these four sample types using 2D gel/MS analysis. Discontinuous sucrose gradients were used for subcellular fractionation of CLDs, MFGs, and MFGMs, followed by washing to de-enrich for proteins not strongly associated with the lipid-rich CLDs, MFGs, or MFGMs. This study employed orthogonal methods (EM and Western blotting) to verify the enrichment of the CLDs and deenrichment of other proteins.
The mammary CLD proteins identified through tandem MS were largely similar to those found in the MFGM sample (see supplementary Table III for full protein list). Only four proteins were common to the liver and mammary CLDs (ADRP, FABP, and probable contaminant serum proteins transferrin and albumin). Proteins observed in the mammary sample included chaperones, ER-resident proteins, and cytoskeletal proteins in addition to proteins known to be associated with CLDs (ADPH, FABP). The MFGM samples contained all the proteins identified in the mammary CLD fraction, but contained additional proteins involved in lipid metabolism and milk proteins, along with some apolipoproteins.
Rat liver cells after traumatic liver injury.
In 2006, Turró et al. (27) studied the CLDs produced in liver tissue after traumatic liver injury (a partial hepatectomy) in Sprague Dawley rats. A dramatic increase of fat was observed in the cytoplasm 24 h after a partial hepatectomy. Enriched CLDs were analyzed using a combined approach of 2D gel/MS analysis and shotgun proteomics resulting in 24 and 27 proteins identified, respectively. Approximately 30% of the total proteins observed were identified by both methods (see supplementary Table III for a full protein list). Four ER-resident proteins, two transport-related proteins, four cellular enzymes, and one protein of unknown function [0610006F02Rik protein- later renamed methyltransferase like protein 7B (Uniprot: Q6UX53)] were uniquely identified using the 2D gel/MS analysis approach. Five proteins not previously associated with CLDs were identified using shotgun analysis, including an ER-resident protein, three cellular enzymes and one protein of unknown function [610006F02Rik (methyltransferase like protein 7B)]. The 0610006F02Rik protein (methyltransferase like protein 7B) was subsequently expressed as a recombinant GFP-tagged protein and expressed in COS cells. Fluorescence microscopy localized the protein to CLDs, and to a smaller extent, the ER.
Human cells in culture
Human cultured liver cell lines.
Two studies have analyzed CLDs from two cultured human liver cell lines, hepatocyte cell line HuH7 (25) and hepatoma cell line HepG2 (26). Results are included in supplementary Table III. Fujimoto et al. (25) examined CLDs isolated from HuH7 hepatocyte cells by subcellular fractionation using sucrose gradients followed by 1D gel/MS analysis. Seventeen proteins were identified with two of those being proteins that had been previously observed to associate with CLDs (ADRP and TIP47). The most abundant proteins identified were ADRP and two other lipid metabolism proteins. A phospholipid binding protein and five lipid metabolizing enzymes were identified. Four ER-resident proteins and one cytosolic protein were identified in the CLD samples but no other organellar proteins were identified within the CLD samples. GAPDH, two acyl-CoA synthetases, lanosterol synthestase, and an SDR family protein had been observed to be associated with CLDs of S. cerevisiae previously, indicating that there may be functional conservation of these proteins within eukaryotic species (18).
In 2006, Sato et al. (26) studied CLDs originating from hepatitis C virus (HCV)-expressing human hepatoma HepG2 cell lines and a control cell line using both a modified version of 1D gel/MS analysis and a shotgun approach. A total of 48 CLD proteins were identified, with 38 CLD proteins identified from HCV-expressing cells, 30 CLD proteins from the control cells, and 20 proteins identified in CLDs from both cell lines. There were 25 CLD proteins identified using 1D gel/MS analysis, with 23 CLD proteins identified from the HCV-expressing cells and 15 CLD proteins identified from the control cells. Thirteen of the proteins identified in the control cells overlapped with proteins identified in the HCV-expressing cells. Among the proteins observed using 1D gel/MS analysis were two PAT family proteins, seven proteins involved in lipid metabolism, membrane trafficking proteins (Rabs 1A, 1B, 5C, and 7), three proteins involved in RNA metabolism and binding, one mitochondrial protein, and several proteins of unknown functions or that do not fit into other categories.
Using the shotgun approach, Sato and coworkers identified 36 total CLD proteins, with 27 CLD proteins found in the HCV-expressing cells and 24 CLD proteins found in the control cells with 15 CLD proteins observed in both cell types. Twenty-two CLD-associated proteins overlapped with proteins identified using 1D gel/MS analysis. Proteins identified via the shotgun approach but not identified using 1D gel/MS analysis were five lipid metabolism proteins, three additional membrane trafficking proteins (Rabs 8, 10, and 11), two RNA metabolism/binding proteins and proteins that did not fit within these categories. Interestingly, both the 1D gel/MS analysis and shotgun analysis showed that RNA metabolism/binding proteins were only found in the HCV-expressing cells and these were not observed in the control cells. Several proteins were observed to be associated with CLDs for the first time in this study (sterol carrier protein 2-related form, 58.85K, FABP5, and apoptosis-inducing factor homologous mitochondrion-associated inducer of death).
Human epithelial and MDCK cultured cell lines.
Cultured cell lines of human epithelial cells (UAC and A431) and Madin Darby Canine Kidney (MDCK) cells were studied in 2004 by Umlauf et al. (24) to identify CLD-associated proteins, and specifically to examine stomatin, due to its biochemical and topological similarity with caveolins (results included in supplementary Table III). Caveolins are known to associate with CLDs (22, 48–50). Transfected cells were cultured to express GFP-tagged stomatin, and immunofluorescence was used to localize stomatin to CLDs within the cultured cells. Western blot analyses were used to identify proteins and enrichment of CLD-associated proteins in the CLD samples. Proteomic 1D gel/MS analysis was employed to study the A431 squaemous epithelial carcinoma cells.
Through immunofluorescence microscopy, stomatin was found to associate with CLDs when expressed exogenously at high levels or expressed endogenously in the presence of oleic acid. If vesicular transport was blocked, the levels of endogenous stomatin were increased causing an increase in association with CLDs. Cycloheximide treatment caused stomatin dissociate from CLDs and stomatin subsequently associated with acidic vesicles. Time lapse video microscopy of live cells was used to show that CLDs interact with small vesicles and occasionally CLDs show high motility.
Proteomic analysis of CLDs from the A431 cells revealed 33 CLD-associated proteins. Studies were carried out on either A431 cells with low endogeneous stomatin levels (called stomatin-negative) or on A431 cells stably overexpressing wild-type stomatin (termed stomatin-positive cells). Comparing the 1D SDS-PAGE bands from stomatin-positive and stomatin-negative cells did not show differences in the protein bands observed through staining of the gels with the exception of the stomatin band. Proteins identified included those previously associated with CLDs, including PAT family proteins, several Rab GTPases (Rab 1B, 6A, 7, 10, and 18), four lipid metabolism proteins, a fatty acid synthesis protein, four proteins involved in steroid synthesis or vitamin A homeostasis, four vesicle transport proteins, seven chaperones or chaperonins, three cytoskeletal proteins, and two uncategorized proteins.
SUMMARY OF PROTEOMIC STUDIES
Over two hundred mammalian proteins have been identified across a number of proteomic analyses of enriched CLDs. [A comprehensive list of all identified CLD proteins in yeast (110 proteins), Drosophila (296 proteins), and mammals (229 proteins) organized according to the function/cellular localization can be found in supplementary Tables I, II, and III, respectively.] Table 1 summarizes the CLD proteins identified in at least three proteomic studies from mammalian cells/tissues. These proteins seem to fall into several distinct functional categories.
TABLE 1.
Mammalian CLD proteins identified in at least three proteomic studies
| Protein Grouping | Uniprot Accession Number | Gene Name | Short Name | Protein Name | Reference |
|---|---|---|---|---|---|
| PAT Family | Q99541 | ADRP or ADFP | Adipose differentiation -related protein (ADRP/ADFP/ADPH) | 14, 20, 21, 26, 27, 28 | |
| O60664 | M6PRBP1 or TIP47 | TIP47 | Cargo selection protein(TIP47)/Mannose-6-phosphate receptor-binding protein 1 | 23, 24, 25, 26, 27, 28 | |
| Lipid Metabolism | Q8NBQ5 | HSD17B11 | 17-β-HSD 11/ retSDR2 | 17-β hydroxysteroid dehydrogenase 11/Retinal short- chain dehydrogenase/reductase retSDR2 | 20, 21, 25, 26, 27, 28 |
| P56937 | HSD17B7 | 17-β-HSD 7 | 17-β hydroxysteroid dehydrogenase 7/3-keto-steroid reductase | 20, 21, 24, 25, 28 | |
| Q13085 | ACACA | ACC-α | acetyl-CoA carboxylase α | 20, 21, 24, 28 | |
| Q8NBX0 | SCCPDH | CGI-49 | CGI-49 protein/Probable saccharopine dehydrogenase | 21, 22, 25, 26, 28 | |
| Q8WTS1 | ABHD5 | CGI-58 | CGI-58 protein/1-acylglycerol-3-phosphate O-acyltransferase ABHD5 | 20, 21, 22, 23, 24, 28 | |
| Q96LJ7 | DHRS1 | dehydrogenase/reductase (SDR family) member 1 | 20, 21, 22, 23 | ||
| P48449 | LSS | Lanosterol synthase | 23, 24, 25, 26, 27, 28 | ||
| O95573 | ACSL3 | LACS 3 | Long-chain-fatty-acid–CoA ligase 3/acyl-CoA synthetase 3 | 20, 21, 22, 24, 25, 26, 28 | |
| O60488 | ACSL4 | LACS 4 | Long-chain-fatty-acid–CoA ligase 4/acyl-CoA synthetase 4 | 20, 21, 22, 23, 25, 26 | |
| P00387 | CYB5R3 | B5R | NADH-cytochrome b5 reductase 3/cytochrome b5 reductase/diaphorase-1 | 20, 21, 23, 24, 25, 26, 28 | |
| Q96AD5 | PNPLA2 | TTS-2 OR IPLA2-zeta | Patatin-like phospholipase domain-containing protein 2/Adipocyte Triglyceride Lipase (TTS-2.1) (iPLA2ξ) | 20, 21, 24, 26, 28 | |
| P30101 | PDIA3 | Protein disulfide-isomerase A3/PLCα/ERp60 | 21, 26, 27 | ||
| Q14534 | Sqle | SE | Squalene monooxygenase/ Squalene epoxidase | 20, 24, 28 | |
| Q15738 | NSDHL | H105e3 | Sterol-4-α-carboxylate 3-dehydrogenase, decarboxylating/NAD(P)- dependent steroid dehydrogenase- like; H105e3 | 25, 26, 27, 28 | |
| Chaperones | P14625 | HSP90B1 | Endoplasmin/Heat shock protein 90 kDa β member 1/94 kDa glucose-regulated protein/gp96 homolog | 14, 24, 26, 27 | |
| P11021 | HSPA5 | BiP | GRP 78 (BIP)/78 kDa glucose- regulated protein/Heat shock 70 kDa protein 5 | 20, 21, 22, 23, 24, 26, 27, 28 | |
| Q61696 | Hspa1a | Heat shock 70 kDa protein 1A/HSP 70 / Heat shock 70 kDa protein 3 / HSP70.3 / Hsp68 | 14, 21, 22 | ||
| Q96CS3 | FAF2 | KIAA0887 | FAS-associated factor 2/UBXD8 / KIAA0887 protein | 20, 21, 25, 26 | |
| Cytoskeleton | P60709 | ACTB | Actin, cytoplasmic 1/β-Actin | 21, 24, 27, 28 | |
| P08670 | VIM | Vimentin | 21, 22, 23 | ||
| Endoplasmic Reticulum | P27824 | CANX | p90 or IP90 | Calnexin | 21, 22, 23, 27, 28 |
| Q9Y673/ Q9HC03 | ALG5 | DoIP- glucosyltransferase | Dolichyl-phosphate β- glucosyltransfrase/Asparagine- linked glycosylation protein 5 | 20, 21, 28 | |
| Q15084 | PDIA6 | protein disulfide isomerase/ Protein disulfide-isomerase A6 | 20, 21, 24, 27, 28 | ||
| P07237 | P4HB | PDI or p55 | Protein disulfide-isomerase/ Prolyl 4-hydroxylase subunit β/ p55 | 24, 26, 27 | |
| P04843 | RPN1 | RPN-1 | ribophorin I/Dolichyl-diphosphooligosaccharide– protein glycosyltransferase subunit 1 | 21, 22, 23 | |
| O43399 | TPD52L2 | hD54 or D52- like 2 | Tumor protein D54/Tumor protein D52-like 2 | 23, 24, 26, 28 | |
| Membrane Trafficking | P04406 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase/GAPDH | 21, 25, 28 | |
| P61026 | RAB10 | Rab10 | Ras-related protein Rab-10 | 20, 21, 24, 26, 28 | |
| P62491 | RAB11A | Rab 11A | Ras-related protein Rab-11A | 20, 21, 28 | |
| P61106 | RAB14 | Rab 14 | Ras-related protein Rab-14 | 20, 21, 22, 23, 28 | |
| Q9NP72 | RAB18 | Rab 18 | Ras-related protein Rab-18 | 20, 21, 22, 24, 26, 28 | |
| P62820 | RAB1A | Rab 1a | Ras-related protein Rab-1a | 20, 21, 24, 26, 28 | |
| Q9H0U4 | RAB1B | Rab 1b | Ras-related protein Rab-1B | 24, 26, 28 | |
| P61020 | RAB5B | Rab 5b | Ras-related protein Rab-5b (a and c) | 20, 21, 22, 25 | |
| P51149 | RAB7A | Rab 7a | Ras-related protein Rab-7a | 20, 21, 22, 23, 24, 26, 28 | |
| Q07960 | ARHGAP1 | Rho GTPase activating protein 1 | 20, 21, 23 | ||
| Q99536 | VAT1 | Synaptic vesicle membrane protein VAT-1 homolog | 23, 26, 27, 28 | ||
| Mitochondrial Protein | Q9BRQ8 | AIFM2 | Apoptosis-inducing factor 2/ Apoptosis-inducing factor–homologous mitochondrion- associated inducer of death | 24, 25, 26, 28 | |
| Q99623 | PHB2 | Prohibitin-2 | 20, 21, 22 | ||
| Signaling Proteins | Q99653 | CHP | calcium binding protein P22 | 21, 24, 28 | |
| Q03135 | CAV1 | caveolin-1 | 20, 21, 22 | ||
| Q9H8H3 | METTL7A | AAM-B | Methyltransferase-like protein 7A (AAM-B) | 20, 21, 26 | |
| Miscellaneous/Serum | P07724 | Alb | Serum albumin | 14, 27, 28 |
The noted “gold standard” marker for CLDs are members of the PAT family [named for the first three members perilipin, adipophilin (also known as adipocyte differentiation related protein, ADRP, or ADFP), and tail interacting protein 47 (TIP47), but the PAT-family also includes lipid storage droplet protein 5 (LSDP5, also known as OXPAT), myocardial lipid droplet protein (MLDP, also called PAT-1), and S3-12] (51). Though PAT proteins are all known to associate with CLDs, they are not all found in every cell. ADRP has been localized to lipid droplets in cultured murine 3T3-Ll adipocytes, murine MA-10 Leydig cells, CHO fibroblasts, and human HepG2 hepatoma cells (43) and TIP47 is similarly found in many cell types (52). However, perilipin has only been found in adipocytes and steroidogenic cells (53). The LSDP5 (OXPAT) protein is found in tissues that rely on β-oxidation of fatty acids for energy production such as heart muscle, brown adipose tissue, and liver (54–56), and S3-12 is found in white adipose tissue (57). Two related proteins, LSD1 and LSD2, are expressed in insects and found in the fat body, a tissue that has similarities to both liver and adipose tissue (58–61).
PAT proteins are thought to be responsible for lipid homeostasis within cells, but the distinctions and specificities of the family members are not yet fully understood. PAT proteins share a conserved 100 amino acid homologous N-terminal PAT domain (the exception is S3-12) (51) and repeating sequences of 11 amino acids thought to form amphipathic α-helices that may assist in embedding the proteins within lipid droplets (33, 34, 51). Phosphorylation controls the functions of some PAT family proteins like perilipin, and the Drosophila PAT family homologs LSD1 and LSD2 in facilitating lipolysis and CLD motility (60, 62). Bickel et al. (63) very recently published a helpful review of PAT proteins.
The role of CLDs in lipid metabolism is critical to maintaining lipid homeostasis and, not surprisingly, proteins involved in lipid metabolism are observed ubiquitously in the CLDs. These include two 17-β hydroxysteroid dehydrogenases, acetyl-CoA carboxylase α, CGI-49 protein, CGI-58 protein (lipase modulator), dehydrogenase/reductase (SDR family) member 1, lanosterol synthase, long-chain-fatty-acid-CoA ligases, NADH-cytochrome b5 reductase 3, patatin-like phospholipase domain-containing protein 2 [also called adipocyte triglyceride lipase (TTS-2.1) (iPLA2ξ)], protein disulfide-isomerase A3, squalene epoxidase, and NAD(P)-dependent steroid dehydrogenase-like protein. These proteins play a variety of roles in lipid synthesis (long-chain-fatty-acid-CoA ligases, lanosterol synthase, squalene epoxidase), lipid homeostasis (long-chain-fatty-acid-CoA ligases), and lipid degradation (patatin-like phospholipase domain-containing protein 2, CGI-58 (64).)
Aside from the PAT family proteins and those involved in lipid metabolism, several proteomic studies have also identified so-called “refugee proteins” that seem to be unrelated to known lipid droplet functions (4) that fall into several categories: membrane trafficking proteins [Arf1, Rab, and Rho proteins (21)], signaling proteins (PKC, Ras, AKAP), cytoskeletal proteins [actinin, filamin A, myosin heavy chain (21)], enzymes (alcohol dehydrogenase), chaperones [pyruvate kinase, peroxiredoxin 1 (21)], and proteins associated with cellular organelles.
Membrane trafficking proteins commonly observed in CLD preparations include proteins from several small GTPase families: Ras-related Rab proteins, ADP Ribosylation factor (ARF) proteins, as well as related proteins phospholipases and caveolins. Though the relationship between CLDs and the small GTPases is unclear, in vivo colocalization studies using immunofluorescence seem to suggest that these interactions are not due to contamination during fractionation (44, 65).
In particular, the Rab protein family of small Ras-related GTPase proteins is well represented in the proteomic studies of CLDs, whereas other members of the small GTPase families such as Rho and Arf proteins are less frequently observed. Rab proteins are known to be important for vesicular trafficking, vesicle budding, and fusion. Although twenty Rab proteins were identified in mammalian studies, Rab effector proteins were not observed in large numbers. The presence of both the small GTPase proteins and their effector proteins is needed for synthesis of a complete pathway. Effector proteins may simply not be present in CLDs or they could be present at such low abundances in CLDs prior to enrichment and purification that they are either lost during enrichment or are below the limit of detection of the analysis. Another possibility is that the effector proteins are only recruited to CLDs under specific conditions (21).
Alternatively, the presence of incomplete and fragmented protein pathways on CLDs in vivo could be the result of CLDs acting as a reservoir of hydrophobic proteins within the cell that functions to transport and assist with synthesis of protein pathways (21). Transfer of substrates and products between CLDs and organelles or other cellular structures could be facilitated by the Rab proteins and other membrane trafficking proteins.
The phospholipid monolayer surrounding CLDs contain abundant phosphatidylcholine and lysophosphatidylcholine (13) that are the substrates for phospholipase D (PLD) (reviewed in ref. 66). PLD1 is involved in many cellular processes like membrane fusion and vesicular trafficking (reviewed in ref. 66). The small protein Arf1 directly activates PLD1, and has received particular attention with respect to its interactions with lipid droplets. Using immunofluorescence, Arf1 and PLD1 have been shown to localize to CLDs (44) and Arf1 was shown to directly bind ADRP (65). A GDP-restricted form of Arf1 caused ADRP dissociation from CLDs and did not inhibit triacylglycerol formation but did prevent formation of new CLDs, which indicates a prevention of CLD budding (65). Arf1 was found in one proteomic study by Bartz et al. (21) along with the ADP-ribosylation factor GTPase-activating protein 1 (ArfGAP1, Uniprot: Q8N6T3).
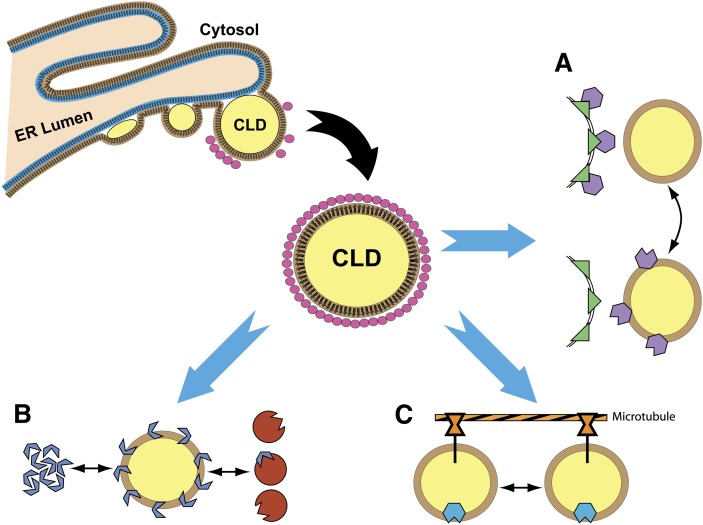
Proteins associated with the ER are also commonly observed in CLD preparations. This is not surprising in light of the fact that lipid droplet biogenesis is thought to occur at the surface of the ER (reviewed in refs. 1, 2, 5, 7, 8, and 67). Numerous mechanisms for LD biosynthesis have been proposed (68–72). Currently, the most widely accepted model is that CLDs are formed by generation of lipid esters by the ER that are deposited into the membrane. As the quantity of lipid esters exceeds the molar proportion that can be assimilated within the phospholipid bilayer, the lipid ester begins to segregate between the two leaflets. This process is represented in Fig. 1. A budding process occurs whereby the lipid droplet begins to separate away from the membrane to form an independent lipid droplet in the cytoplasm (7). Micro-droplets of lipids are formed at the cisternae of the ER and aggregation of those micro-droplets forms larger CLDs (10). Organelles other than the ER are also commonly found in close proximity to CLDs and CLDs have been found to contain proteins associated with the Golgi apparatus (73), peroxisomes (11, 19), mitochondria (22), and the plasma membrane (24), and CLDs are frequently found in close physical contact with these organelles.
Fig. 1.
Proposed cellular functions of CLDs. CLD formation begins in the smooth endoplasmic reticulum. Lipid esters are deposited within the phospholipid bilayer and microlipid droplets form, bud, and are released into the cytoplasm (7) associated with PAT proteins (pink circles). Three potential CLD functional roles are: A, Sequestration: proteins may be reversibly inactivated by CLD sequestration of one binding partner. Active state and sequestered states are shown. B, Chaperone: aggregation prone proteins without binding partners are sequestered until appropriate binding partners are available. C, Protein transport: CLDs move along cytoskeletal components carrying sequestered proteins from one cellular location to another.
CONCLUSIONS AND PERSPECTIVES
The most interesting categories of proteins identified through proteomic analysis are those that do not have known functions within the classical view of the CLD. These “refugee proteins” (4) have prompted an ongoing debate questioning whether these proteins are bona fide CLD proteins. It has been suggested that CLDs may be more susceptible than other cellular structures or organelles to poor isolation due to the intrinsic hydrophobicity of the CLD structure (4, 74). This potential for in vitro contamination is intrinsic to every fractionation protocol. Therefore, in vivo localization studies (immuno-electron microscopy and/or fluorescence microscopy) are required for confirmation of bona fide CLD proteins.
Ideally, genetic studies would follow to validate functional significance (58, 61). Some of the CLD proteomic studies discussed above incorporated genetic knockout or knockdown strategies for functional validation in S. cerevisiae and Drosophila (16, 18, 19). A mouse variant knockout of perilipin has proven to be an extremely valuable tool for demonstrating that perilipin acts as a phosphorylation-controlled gatekeeper that can regulate access to the neutral lipid core of the CLD for lipases, thereby acting as a control on the lipolysis of CLDs (45, 46). Knockouts have been reported for adipose differentiation-related protein (also known as adipophilin, ADRP, or ADFP) (75) as have knockdown studies of ADRP using antisense oligonucleotides (76). Unfortunately, knockout or knockdown studies may sometimes produce ambiguous results that may not clearly define a phenotype related to the reduction in a protein, and this case has been reported for the ADRP phenotype. [(75, 76) and reviewed in (63)]
Knockouts of other CLD-related proteins have been reported for the senescence marker protein-30 (SMP30) (77), adipocyte triglyceride lipase (ATGL) (78), apoA-II (79), hepatic lipases (79), α-synuclein (80, 81), hormone sensitive lipase (82, 83), caveolin-1 (84), and vimentin (85). Knockouts of CLD-related proteins for other species have also been reported, such as the Drosophila adipose gene (adp) (86), and the Drosophila PAT protein analog LSD2 (58, 61).
Sequestration of proteins on CLDs
One emerging idea is that CLDs may serve in a broad cellular role as a protein storage depot for sequestering proteins (15). In this capacity, CLDs would protect proteins from degradation while keeping aggregation-prone proteins physically separated from the cytoplasm. There is evidence in Drosophila that maternal histones, found in high abundance in embryos, are sequestered on the surface of CLDs and transferred rapidly on and off as they are needed (15). This mechanism has also been observed when the levels of protein expression are higher than normal due to unusual cellular metabolic conditions, as in the case of stomatin (24). Protein sequestration is a tantalizing idea because it could explain the many refugee proteins that have been found to associate with CLDs and provide a dynamic regulatory mechanism for their storage and transport (4). This idea fits appealingly with the known regulation of lipids that have been attributed to CLDs for many years. The CLD system could potentially act as a regulatory mechanism for both proteins and lipids within cells. Figure 1A illustrates the sequestration of binding partners, showing the sequestered and un-sequestered state.
Inactivation of proteins through sequestration to CLDs could happen either indirectly by separating proteins from binding partners, cofactors, or substrates, or directly through changing the binding of an enzyme by actively blocking a catalytic site or inducing a conformational change in the enzyme when it is bound to the CLD. Sequestration of proteins within CLDs may provide a buffering mechanism for proteins within cells, making proteins available as needed but keeping proteins separated from binding partners and preventing aggregation when proteins are not needed. Figure 1B illustrates the sequestration of binding partners, showing the sequestration on a CLD of an aggregation-prone protein, followed by release of the sequestered protein once an appropriate binding partner is available.
There are several features of CLDs that offer potential utility as a protein storage mechanism. First, the dynamic nature of CLDs’ size and the number of CLDs can be adjusted based on external stimuli and metabolic state, allowing for a variable surface area and internal hydrophobic core. This ability would provide the capacity for CLDs to dynamically adjust when greater quantities of CLDs might be needed for storage of proteins. Second, the structure of CLDs with a hydrophobic core of neutral lipids surrounded by a phospholipid monolayer may provide an excellent storage site for proteins that have exposed hydrophobic surfaces. Third, the complement of proteins associated with CLDs can respond dynamically to physiological changes around the cells (19, 22). There is some evidence that proteins may localize to specific CLDs within a single cell (16, 87–90). Selective sequestration of proteins could potentially act to distribute proteins throughout cells using CLDs as transport vehicles, but evidence for this is limited at present. Figure 1C shows the hypothesized transport function of CLDs, with the CLD temporarily sequestering a protein for transport, and motion of CLDs along a cytoskeletal component (here, a microtubule).
The current CLD literature is growing, and proteomics has contributed several enticing functional leads. However, much remains to be done and the systematic validation of identified CLD proteins promises to elucidate the full range of functions of CLDs within cells.
Supplementary Material
Footnotes
Abbreviations:
- ADRP
- adipose differentiation related protein or adipophilin
- ARF
- ADP ribosylation factor
- CLD
- cytoplasmic lipid droplet
- CHO
- Chinese hamster ovary
- 1D
- one dimensional
- 2D
- two dimensional
- EM
- electron microscopy
- ER
- endoplasmic reticulum
- HCV
- hepatitis C virus
- MFG
- milk fat globule
- MFGM
- milk fat globule membrane
- PLD
- phospholipase D
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three tables.
REFERENCES
- 1.Murphy D. J. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40: 325–438. [DOI] [PubMed] [Google Scholar]
- 2.Martin S., Parton R. G. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7: 373–378. [DOI] [PubMed] [Google Scholar]
- 3.Dugail I., Hajduch E. 2007. A new look at adipocyte lipid droplets: towards a role in the sensing of triacylglycerol stores? Cell. Mol. Life Sci. 63: 2452–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welte M. A. 2007. Proteins under new management: lipid droplets deliver. Trends Cell Biol. 17: 363–369. [DOI] [PubMed] [Google Scholar]
- 5.Olofsson S-O., Boström P., Andersson L., Rutberg M., Levin M., Perman J., Borén J. 2008. Triglyceride containing lipid droplets and lipid droplet-associated proteins. Curr. Opin. Lipidol. 19: 441–447. [DOI] [PubMed] [Google Scholar]
- 6.Paul A., Chan L., Bickel P. E. 2008. The PAT family of lipid droplet proteins in heart and vascular cells. Curr. Hypertens. Rep. 10: 461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujimoto T., Ohsaki Y., Cheng J., Suzuki M., Shinohara Y. 2008. Lipid droplets: a classic organelle with new outfits. Histochem. Cell Biol. 130: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodman J. M. 2008. The gregarious lipid droplet. J. Biol. Chem. 283: 28005–28009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granneman J. G., Moore H-P. H. 2008. Location, location: protein trafficking and lipolysis in adipocytes. Trends Endocrinol. Metab. 19: 3–9. [DOI] [PubMed] [Google Scholar]
- 10.Olofsson S-O., Boström P., Andersson L., Rutberg M., Perman J., Borén J. 2009. Lipid droplets as dynamic organelles connecting storage and efflux of lipids. Biochim. Biophys. Acta. Mol. Cell Biol. Lipids. 1791: 448–458. [DOI] [PubMed] [Google Scholar]
- 11.Zehmer J. K., Huang Y., Peng G., Pu J., Anderson R. G. W., Liu P. 2009. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 9: 914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartz R., Li W-H., Venables B., Zehmer J. K., Roth M. R., Welti R., Anderson R. G. W., Liu P., Chapman K. D. 2007. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J. Lipid Res. 48: 837–847. [DOI] [PubMed] [Google Scholar]
- 13.Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. 2002. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 277: 44507–44512. [DOI] [PubMed] [Google Scholar]
- 14.Wu C. C., Howell K. E., Neville M. C., Yates J. R. I., McManaman J. L. 2000. Proteomics reveal a link between the endoplasmic reticulum and lipid secretory mechanisms in mammary epithelial cells. Electrophoresis. 21: 3470–3482. [DOI] [PubMed] [Google Scholar]
- 15.Cermelli S., Guo Y., Gross S. P., Welte M. A. 2006. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16: 1783–1795. [DOI] [PubMed] [Google Scholar]
- 16.Beller M., Riedel D., Jansch L., Dieterich G., Wehland J., Jackle H., Kuhnlein R. P. 2006. Characterization of the drosophila lipid droplet subproteome. Mol. Cell. Proteomics. 5: 1082–1094. [DOI] [PubMed] [Google Scholar]
- 17.Brasaemle D. L., Wolins N. E. 2006. UNIT 3.15 Isolation of lipid droplets from cells by density gradient centrifugation. Current Protocols in Cell Biology. Bonifacino J. S., Dasso M., Harford J. B., Lippincott-Schwartz J, Yamada K. M. John Wiley & Sons; Hoboken, NJ: p 3.15.1–3.15.12. [DOI] [PubMed] [Google Scholar]
- 18.Athenstaedt K., Zweytick D., Jandrositz A., Kohlwein S. D., Daum G. 1999. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol. 181: 6441–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Binns D., Januszewski T., Chen Y., Hill J., Markin V. S., Zhao Y., Gilpin C., Chapman K. D., Anderson R. G. W., Goodman J. M. 2006. An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173: 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu P., Ying Y., Zhao Y., Mundy D. I., Zhu M., Anderson R. G. W. 2004. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 279: 3787–3792. [DOI] [PubMed] [Google Scholar]
- 21.Bartz R., Zehmer J. K., Zhu M., Chen Y., Serrero G., Zhao Y., Liu P. 2007. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J. Proteome Res. 6: 3256–3265. [DOI] [PubMed] [Google Scholar]
- 22.Brasaemle D. L., Dolios G., Shapiro L., Wang R. 2004. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3–L1 adipocytes. J. Biol. Chem. 279: 46835–46842. [DOI] [PubMed] [Google Scholar]
- 23.Cho S. Y., Shin E. S., Park P. J., Shin D. W., Chang H. K., Kim D., Lee H. H., Lee J. H., Kim S. H., Song M. J., et al. 2007. Identification of mouse Prp19p as a lipid droplet-associated protein and its possible involvement in the biogenesis of lipid droplets. J. Biol. Chem. 282: 2456–2465. [DOI] [PubMed] [Google Scholar]
- 24.Umlauf E., Csaszar E., Moertelmaier M., Schuetz G. J., Parton R. G., Prohaska R. 2004. Association of stomatin with lipid bodies. J. Biol. Chem. 279: 23699–23709. [DOI] [PubMed] [Google Scholar]
- 25.Fujimoto Y., Itabe H., Sakai J., Makita M., Noda J., Mori M., Higashi Y., Kojima S., Takano T. 2004. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim. Biophys. Acta Mol. Cell Res. 1644: 47–59. [DOI] [PubMed] [Google Scholar]
- 26.Sato S., Fukasawa M., Yamakawa Y., Natsume T., Suzuki T., Shoji I., Aizaki H., Miyamura T., Nishijima M. 2006. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J Biochem. 139: 921–930. [DOI] [PubMed] [Google Scholar]
- 27.Turró S., Ingelmo-Torres M., Estanyol J. M., Tebar F., Fernández M. A., Albor C. V., Gaus K., Grewal T., Enrich C., Pol A. 2006. Identification and characterization of associated with lipid droplet protein 1: a novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 7: 1254–1269. [DOI] [PubMed] [Google Scholar]
- 28.Wan H-C., Melo R. C. N., Jin Z., Dvorak A. M., Weller P. F. 2007. Roles and origins of leukocyte lipid bodies: proteomic and ultrastructural studies. FASEB J. 21: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shevchenko A., Wilm M., Vorm O., Mann M. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858. [DOI] [PubMed] [Google Scholar]
- 30.Speers A. E., Blackler A. R., Wu C. C. 2007. Shotgun analysis of integral membrane proteins facilitated by elevated temperature. Anal. Chem. 79: 4613–4620. [DOI] [PubMed] [Google Scholar]
- 31.Speers A. E., Wu C. C. 2007. Proteomics of integral membrane proteins–theory and application. Chem. Rev. 107: 3687–3714. [DOI] [PubMed] [Google Scholar]
- 32.Capuano F., Beaudoin F., Napier J. A., Shewry P. R. 2007. Properties and exploitation of oleosins. Biotechnol. Adv. 25: 203–206. [DOI] [PubMed] [Google Scholar]
- 33.Bussell R., Jr., Eliezer D. 2003. A structural and functional role for 11-mer repeats in [alpha]-synuclein and other exchangeable lipid binding proteins. J. Mol. Biol. 329: 763–778. [DOI] [PubMed] [Google Scholar]
- 34.Chandra S., Chen X., Rizo J., Jahn R., Sudhof T. C. 2003. A broken alpha -helix in folded alpha -synuclein. J. Biol. Chem. 278: 15313–15318. [DOI] [PubMed] [Google Scholar]
- 35.Robenek H., Robenek M. J., Troyer D. 2005. PAT family proteins pervade lipid droplet cores. J. Lipid Res. 46: 1331–1338. [DOI] [PubMed] [Google Scholar]
- 36.Washburn M. P., Wolters D., Yates J. R. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19: 242–247. [DOI] [PubMed] [Google Scholar]
- 37.Balgley B. M., Laudeman T., Yang L., Song T., Lee C. S. 2007. Comparative evaluation of tandem ms search algorithms using a target-decoy search strategy. Mol. Cell. Proteomics. 6: 1599–1608. [DOI] [PubMed] [Google Scholar]
- 38.Aebersold R. 2009. A stress test for mass spectrometry-based proteomics. Nat. Methods. 6: 411–412. [DOI] [PubMed] [Google Scholar]
- 39.Bell A. W., Deutsch E. W., Au C. E., Kearney R. E., Beavis R., Sechi S., Nilsson T., Bergeron J. J. M. 2009. A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nat. Methods. 6: 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi H., Nesvizhskii A. I. 2007. False discovery rates and related statistical concepts in mass spectrometry-based proteomics. J. Proteome Res. 7: 47–50. [DOI] [PubMed] [Google Scholar]
- 41.Käll L., Storey J. D., MacCoss M. J., Noble W. S. 2008. Posterior error probabilities and false discovery rates: two sides of the same coin. J. Proteome Res. 7: 40–44. [DOI] [PubMed] [Google Scholar]
- 42.Käll L., Storey J. D., MacCoss M. J., Noble W. S. 2008. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 7: 29–34. [DOI] [PubMed] [Google Scholar]
- 43.Brasaemle D. L., Barber T., Kimmel A. R., Londos C. 1997. Post-translational regulation of perilipin expression. Stabilization by stored intracellular neutral lipids. J. Biol. Chem. 272: 9378–9387. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura N., Banno Y., Tamiya-Koizumi K. 2005. Arf1-dependent PLD1 is localized to oleic acid-induced lipid droplets in NIH3T3 cells. Biochem. Biophys. Res. Commun. 335: 117–123. [DOI] [PubMed] [Google Scholar]
- 45.Martinez-Botas J., Anderson J. B., Tessier D., Lapillonne A., Chang B. H-J., Quast M. J., Gorenstein D., Chen K-H., Chan L. 2000. Absence of perilipin results in leanness and reverses obesity in Leprdb/db mice. Nat. Genet. 26: 474–479. [DOI] [PubMed] [Google Scholar]
- 46.Tansey J. T., Sztalryd C., Gruia-Gray J., Roush D. L., Zee J. V., Gavrilova O., Reitman M. L., Deng C. X., Li C., Kimmel A. R., et al. 2001. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc. Natl. Acad. Sci. USA. 98: 6494–6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su C-L., Sztalryd C., Contreras J. A., Holm C., Kimmel A. R., Londos C. 2003. Mutational analysis of the hormone-sensitive lipase translocation reaction in adipocytes. J. Biol. Chem. 278: 43615–43619. [DOI] [PubMed] [Google Scholar]
- 48.Ostermeyer A. G., Paci J. M., Zeng Y., Lublin D. M., Munro S., Brown D. A. 2001. Accumulation of caveolin in the endoplasmic reticulum redirects the protein to lipid storage droplets. J. Cell Biol. 152: 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujimoto T., Kogo H., Ishiguro K., Tauchi K., Nomura R. 2001. Caveolin-2 is targeted to lipid droplets, a new “membrane domain” in the cell. J. Cell Biol. 152: 1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pol A., Martin S., Fernandez M. A., Ingelmo-Torres M., Ferguson C., Enrich C., Parton R. G. 2005. Cholesterol and fatty acids regulate dynamic caveolin trafficking through the golgi complex and between the cell surface and lipid bodies. Mol. Biol. Cell. 16: 2091–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brasaemle D. L., Subramanian V., Garcia A., Marcinkiewicz A., Rothenberg A. 2009. Perilipin A and the control of triacylglycerol metabolism. Mol. Cell. Biochem. 326: 15–21. [DOI] [PubMed] [Google Scholar]
- 52.Wolins N. E., Quaynor B. K., Skinner J. R., Schoenfih M. J., Tzekov A., Bickel P. E. 2005. S3–12, adipophilin, and TIP47 package lipid in adipocytes. J. Biol. Chem. 280: 19146–19155. [DOI] [PubMed] [Google Scholar]
- 53.Londos C., Brasaemle D., Gruia-Gray J., Servetnick D., Schultz C., Levin D., Kimmel A. 1995. Perilipin: unique proteins associated with intracellular neutral lipid droplets in adipocytes and steroidogenic cells. Biochem. Soc. Trans. 3: 611–615. [DOI] [PubMed] [Google Scholar]
- 54.Dalen K. T., Dahl T., Holter E., Arntsen B., Londos C., Sztalryd C., Nebb H. I. 2007. LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1771: 210–227. [DOI] [PubMed] [Google Scholar]
- 55.Wolins N. E., Quaynor B. K., Skinner J. R., Tzekov A., Croce M. A., Gropler M. C., Varma V., Yao-Borengasser A., Rasouli N., Kern P. A., et al. 2006. OXPAT/PAT-1 is a PPAR-induced lipid droplet protein that promotes fatty acid utilization. Diabetes. 55: 3418–3428. [DOI] [PubMed] [Google Scholar]
- 56.Yamaguchi T., Matsushita S., Motojima K., Hirose F., Osumi T. 2006. MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptor {alpha}. J. Biol. Chem. 281: 14232–14240. [DOI] [PubMed] [Google Scholar]
- 57.Wolins N. E., Skinner J. R., Schoenfish M. J., Tzekov A., Bensch K. G., Bickel P. E. 2003. Adipocyte protein S3–12 coats nascent lipid droplets. J. Biol. Chem. 278: 37713–37721. [DOI] [PubMed] [Google Scholar]
- 58.Grönke S., Beller M., Fellert S., Ramakrishnan H., Jäckle H., Kühnlein R. P. 2003. Control of fat storage by a Drosophila PAT domain protein. Curr. Biol. 13: 603–606. [DOI] [PubMed] [Google Scholar]
- 59.Grönke S., Müller G., Hirsch J., Fellert S., Andreou A., Haase T., Jäckle H., Kühnlein R. P. 2007. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 5: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patel R. T., Soulages J. L., Hariharasundaram B., Arrese E. L. 2005. Activation of the lipid droplet controls the rate of lipolysis of triglycerides in the insect fat body. J. Biol. Chem. 280: 22624–22631. [DOI] [PubMed] [Google Scholar]
- 61.Teixeira L., Rabouille C., Rørth P., Ephrussi A., Vanzo N. F. 2003. Drosophila perilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mech. Dev. 120: 1071–1081. [DOI] [PubMed] [Google Scholar]
- 62.Welte M. A., Cermelli S., Griner J., Viera A., Guo Y., Kim D-H., Gindhart J. G., Gross S. P. 2005. Regulation of lipid-droplet transport by the perilipin homolog LSD2. Curr. Biol. 15: 1266–1275. [DOI] [PubMed] [Google Scholar]
- 63.Bickel P. E., Tansey J. T., Welte M. A. 2009. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta. Mol. Cell Biol. Lipids. 1791: 419–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., Zechner R. 2006. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 3: 309–319. [DOI] [PubMed] [Google Scholar]
- 65.Nakamura N., Akashi T., Taneda T., Kogo H., Kikuchi A., Fujimoto T. 2004. ADRP is dissociated from lipid droplets by ARF1-dependent mechanism. Biochem. Biophys. Res. Commun. 322: 957–965. [DOI] [PubMed] [Google Scholar]
- 66.Jenkins G., Frohman M. 2005. Phospholipase D: a lipid centric review. Cell. Mol. Life Sci. 62: 2305–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brasaemle D. L. 2007. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 48: 2547–2559. [DOI] [PubMed] [Google Scholar]
- 68.Wanner G., Formanek H., Theimer R. R. 1981. The ontogeny fo lipid bodies (spherosomes) in plant cells: ultrasturctural evidence. Planta. 151: 109–123. [DOI] [PubMed] [Google Scholar]
- 69.Murphy D. J., Vance J. 1999. Mechanisms of lipid-body formation. Trends Biochem. Sci. 24: 109–115. [DOI] [PubMed] [Google Scholar]
- 70.Ploegh H. L. 2007. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 448: 435–438. [DOI] [PubMed] [Google Scholar]
- 71.Waltermann M., Steinbuchel A. 2005. Neutral lipid bodies in prokaryotes: recent insights into structure, formation, and relationship to eukaryotic lipid depots. J. Bacteriol. 187: 3607–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zweytick D., Athenstaedt K., Daum G. 2000. Intracellular lipid particles of eukaryotic cells. Biochim. Biophys. Acta Rev. Biomembranes. 1469: 101–120. [DOI] [PubMed] [Google Scholar]
- 73.Marchesan D., Rutberg M., Andersson L., Asp L., Larsson T., Boren J., Johansson B. R., Olofsson S-O. 2003. A phospholipase D-dependent process forms lipid droplets containing caveolin, adipocyte differentiation-related protein, and vimentin in a cell-free system. J. Biol. Chem. 278: 27293–27300. [DOI] [PubMed] [Google Scholar]
- 74.Fujimoto T., Ohsaki Y. 2006. Cytoplasmic lipid droplets: rediscovery of an old structure as a unique platform. Ann. N. Y. Acad. Sci. 1086: 104–115. [DOI] [PubMed] [Google Scholar]
- 75.Chang B. H-J., Li L., Paul A., Taniguchi S., Nannegari V., Heird W. C., Chan L. 2006. Protection against fatty liver but normal adipogenesis in mice lacking adipose differentiation-related protein. Mol. Cell. Biol. 26: 1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Imai Y., Varela G. M., Jackson M. B., Graham M. J., Crooke R. M., Ahima R. S. 2007. Reduction of hepatosteatosis and lipid levels by an adipose differentiation-related protein antisense oligonucleotide. Gastroenterology. 132: 1947–1954. [DOI] [PubMed] [Google Scholar]
- 77.Ishigami A., Kondo Y., Nanba R., Ohsawa T., Handa S., Kubo S., Akita M., Maruyama N. 2004. SMP30 deficiency in mice causes an accumulation of neutral lipids and phospholipids in the liver and shortens the life span. Biochem. Biophys. Res. Commun. 315: 575–580. [DOI] [PubMed] [Google Scholar]
- 78.Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., et al. 2006. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 312: 734–737. [DOI] [PubMed] [Google Scholar]
- 79.Weng W., Brandenburg N. A., Zhong S., Halkias J., Wu L., Jiang X-c., Tall A., Breslow J. L. 1999. ApoA-II maintains HDL levels in part by inhibition of hepatic lipase: studies in apoA-II and hepatic lipase double knockout mice. J. Lipid Res. 40: 1064–1070. [PubMed] [Google Scholar]
- 80.Abeliovich A., Schmitz Y., Fariñas I., Choi-Lundberg D., Ho W.-H., Castillo P. E., Shinsky N., Verdugo J. M. G., Armanini M., Ryan A., Hynes M., Phillips H., Sulzer D., Rosenthal A. 2000. Mice lacking [alpha]-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron. 25: 239–252. [DOI] [PubMed] [Google Scholar]
- 81.Cabin D. E., Shimazu K., Murphy D., Cole N. B., Gottschalk W., McIlwain K. L., Orrison B., Chen A., Ellis C. E., Paylor R., et al. 2002. Synaptic vesicle depletion correlates with attenuated synaptic responses to prolonged repetitive stimulation in mice lacking alpha -synuclein. J. Neurosci. 22: 8797–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kraemer F., Shen W-J. 2006. Hormone-sensitive lipase knockouts. Nutr Metab. (Lond). 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Osuga J., Ishibashi S., Oka T., Yagyu H., Tozawa R., Fujimoto A., Shionoiri F., Yahagi N., Kraemer F. B., Tsutsumi O., et al. 2000. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc. Natl. Acad. Sci. USA. 97: 787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cohen A. W., Razani B., Schubert W., Williams T. M., Wang X. B., Iyengar P., Brasaemle D. L., Scherer P. E., Lisanti M. P. 2004. Role of caveolin-1 in the modulation of lipolysis and lipid droplet formation. Diabetes. 53: 1261–1270. [DOI] [PubMed] [Google Scholar]
- 85.Lieber J. G., Evans R. M. 1996. Disruption of the vimentin intermediate filament system during adipose conversion of 3T3–L1 cells inhibits lipid droplet accumulation. J. Cell Sci. 109: 3047–3058. [DOI] [PubMed] [Google Scholar]
- 86.Häder T., Müller S., Aguilera M., Eulenberg K. G., Steuernagel A., Ciossek T., Kühnlein R. P., Lemaire L., Fritsch R., Dohrmann C., et al. 2003. Control of triglyceride storage by a WD40/TPR-domain protein. EMBO Rep. 4: 511–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Whitehead J. P., Simpson F., Hill M. M., Thomas E. C., Connolly L. M., Collart F., Simpson R. J., James D. E. 2004. Insulin and oleate promote translocation of inosine-5′ monophosphate dehydrogenase to lipid bodies. Traffic. 5: 739–749. [DOI] [PubMed] [Google Scholar]
- 88.Ozeki S., Cheng J., Tauchi-Sato K., Hatano N., Taniguchi H., Fujimoto T. 2005. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 118: 2601–2611. [DOI] [PubMed] [Google Scholar]
- 89.Wolins N. E., Brasaemle D. L., Bickel P. E. 2006. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 580: 5484–5491. [DOI] [PubMed] [Google Scholar]
- 90.Ohsaki Y., Cheng J., Fujita A., Tokumoto T., Fujimoto T. 2006. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol. Biol. Cell. 17: 2674–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.