Abstract
Sterol-induced binding of endoplasmic reticulum (ER) membrane proteins Insig-1 and Insig-2 to SREBP cleavage-activating protein (Scap) and HMG-CoA reductase triggers regulatory events that limit cholesterol synthesis in animal cells. Binding of Insigs to Scap prevents proteolytic activation of sterol-regulatory element binding proteins (SREBPs), membrane-bound transcription factors that enhance cholesterol synthesis, by trapping Scap-SREBP complexes in the ER. Insig binding to reductase causes ubiquitination and subsequent proteasome-mediated degradation of the enzyme from ER membranes, slowing a rate-limiting step in cholesterol synthesis. Here, we report the characterization of mutant Chinese hamster ovary cells, designated SRD-20, that are resistant to 25-hydroxycholesterol, which potently inhibits SREBP activation and stimulates degradation of reductase. SRD-20 cells were produced by mutagenesis of Insig-1-deficient SRD-14 cells, followed by selection in 25-hydroxycholesterol. DNA sequencing reveals that SRD-20 cells harbor a point mutation in one Insig-2 allele that results in production of a truncated, nonfunctional protein, whereas the other allele contains a point mutation that results in substitution of glutamic acid for glycine-39. This glycine residue localizes to the first membrane-spanning segment of Insig-2 and is also present in the corresponding region of Insig-1. Mutant forms of Insig-1 and Insig-2 containing the Glu-to-Gly substitution fail to confer sterol regulation upon overexpressed Scap and reductase. These studies identify the intramembrane glycine as a key residue for normal sterol regulation in animal cells.
Keywords: cholesterol, sterol-regulatory element binding protein, SREBP cleavage-activating protein, HMG CoA reductase
Insig-1 and Insig-2, closely related membrane proteins of the endoplasmic reticulum (ER), control cholesterol synthesis in mammalian cells through their regulated binding to the membrane proteins SREBP cleavage-activating protein (Scap) and HMG-CoA reductase (1). Scap mediates the activation of sterol-regulatory element binding proteins (SREBPs), membrane-bound transcription factors that enhance transcription of genes encoding cholesterol biosynthetic enzymes (2). In sterol-deprived cells, Scap escorts SREBPs from the ER to Golgi, where active fragments of SREBPs become released from membranes by proteolysis (3, 4). Sterol accumulation causes Insigs to bind Scap, which traps the protein in the ER and prevents delivery of its associated SREBP to the Golgi for proteolytic release. In the absence of this release, expression of SREBP target genes falls and cholesterol synthesis declines. HMG-CoA reductase catalyzes conversion of HMG-CoA to mevalonate, a rate-determining step in synthesis of cholesterol and nonsterol isoprenoids (5). Excess sterols trigger binding of Insigs to reductase, resulting in recruitment of a membrane-bound ubiquitin ligase called gp78 that initiates ubiquitination and subsequent proteasomal degradation of reductase from ER membranes (6, 7). This sterol-accelerated degradation markedly slows the rate at which mevalonate is produced. Insig-mediated regulation of Scap and reductase ensures that essential nonsterol isoprenoids derived from mevalonate are constantly supplied without risking overproduction of cholesterol.
Scap and reductase are comprised of a hydrophobic NH2-terminal domain that spans the ER membrane eight times and a hydrophilic COOH-terminal domain that projects into the cytosol (8–10). The COOH-terminal domain of Scap mediates association with SREBPs (11), whereas the COOH-terminal domain of reductase exerts catalytic activity (12). The membrane domains of Scap and reductase exhibit significant amino acid identity, especially within a region known as the sterol-sensing domain that comprises transmembrane helices 2–6 of both proteins (13, 14). The sterol-sensing domain mediates sterol-induced binding of Scap and reductase to Insigs; mutations within the region disrupt Insig binding and prevent sterol-mediated ER retention of Scap-SREBP and sterol-induced ubiquitination of reductase (15–18).
The two human Insig proteins are very hydrophobic; topology studies indicate they consist of six membrane-spanning segments separated by extremely short loops (19). The membrane domains of Insig-1 and Insig-2 are 85% identical in amino acid sequence. Despite the redundant roles of Insigs in mediating sterol regulation of Scap and reductase, their regulation is strikingly different (20, 21). The Insig-1 gene is a target of nuclear SREBPs. Thus, Insig-1 mRNA is only produced when cells are deprived of sterols and declines under conditions of sterol overload. On the other hand, Insig-2 mRNA is produced constitutively in cultured cells and is not regulated by SREBPs (21). The level of Insig-1 protein contrasts that of its mRNA, owing to gp78-mediated ubiquitination and subsequent proteasomal degradation in sterol-depleted cells (22, 23). Sterols stabilize Insig-1 by causing the protein to bind to Scap, which displaces gp78 and prevents Insig-1 ubiquitination. In contrast, the Insig-2 protein is stable due in part to its reduced affinity for gp78 as compared with that of Insig-1 (22).
The necessity of both Insigs for normal sterol regulation of Scap and reductase has been revealed through the study of mutant Chinese hamster ovary (CHO) cells. We recently characterized a mutant CHO cell line, designated sterol regulatory deficient (SRD)-14, that harbors a partial deletion in the Insig-1 gene causing deficiencies in Insig-1 mRNA and protein levels (24). As a result of their Insig-1 deficiency, SRD-14 cells fail to ubiquitinate and degrade reductase in the presence of the regulatory oxysterol 25-hydroxycholesterol (25-HC). However, processing of SREBPs in SRD-14 cells is sensitive to 25-HC, although the effect requires extended exposure to the oxysterol compared with wild-type cells. We proposed that the remaining Insig-2 in SRD-14 cells (which accounts for 10% of total Insig in wild-type cells) was sufficient to maintain this residual sterol regulation. This hypothesis was examined by subjecting SRD-14 cells to an additional round of mutagenesis, followed by selection for growth in cholesterol-free medium containing a high level of 25-HC that blocks growth of SRD-14 cells through accelerating degradation of reductase and blocking processing of SREBPs. Through this procedure, we isolated SRD-15 cells, which are deficient in Insig-2 as well as Insig-1 (25). As a result of their combined Insig deficiency, SRD-15 cells resist 25-HC-mediated suppression of SREBP processing and acceleration of reductase degradation, which leads to the massive overproduction of cholesterol and sterol synthesis intermediates (26).
In this study, we describe the characterization of SRD-20 cells, which were produced by mutagenizing Insig-1-deficient SRD-14 cells with ethylmethane sulfonate (EMS), followed by selection in 25-HC. Consistent with the propensity of EMS to generate point mutations (27), the 25-HC resistance of SRD-20 cells was traced to a point mutation in one Insig-2 allele that results in production of a truncated, nonfunctional protein. Another point mutation was found in the other Insig-2 allele that resulted in the substitution of glutamic acid for glycine-39, which localizes to the first membrane segment of Insig-2 and is present in the corresponding region of Insig-1 (G95). Overexpression of Insig-2 (G39E) or Insig-1 (G95E) failed to confer regulation upon overexpressed Scap and reductase, indentifying the intramembrane glycine as a key residue for sterol-mediated regulation in animal cells.
MATERIALS AND METHODS
Materials
We obtained sterols (25-HC and cholesterol) from Steraloids (Newport, RI); MG-132, digitonin, and Nonidet P-40 from Calbiochem; EMS from Sigma; protein A/G-coupled agarose beads from Santa Cruz Biotechnology; 2-hydroxypropyl-β-cyclodextrin from Cyclodextrin Technologies Development; polyclonal anti-Myc from Bethyl Laboratories; and donkey anti-mouse and anti-rabbit IgGs from Jackson ImmunoResearch. Lipoprotein-deficient serum (LPDS) (d > 1.215 g/ml) was prepared from newborn calf serum by ultracentrifugation. Other reagents were obtained from sources described previously (4).
Cell culture
All cell lines used in this study were grown in monolayer culture at 37°C in 8–9% CO2. CHO-7 cells are a subline of CHO-K1 cells selected for growth in LPDS (28). SRD-13A (Scap deficient), SRD-14 (Insig-1 deficient), and SRD-15 (Insig-1 and Insig-2 deficient) are mutant cells derived from CHO-7 cells (24, 25, 29). Stock cultures of CHO-7 cells were maintained in medium A (a 1:1 mixture of Ham's F-12 medium and DMEM containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate) supplemented with 5% (v/v) LPDS. SRD-13A cells were maintained in medium A containing 5% fetal calf serum, 5 µg/ml cholesterol, 1 mM sodium mevalonate, 20 µM sodium oleate, and 0.5 mg/ml G418 (29). SRD-14 and SRD-15 cells were grown in medium A supplemented with 5% LPDS and 10 µM SR-12813 or 2.5 µM 25-HC, respectively (24, 25).
Mutagenesis and isolation of 25-HC-resistant SRD-20 cells
SRD-14 cells (2.5 × 107) were plated on day 0 at 1 × 106 cells/100 mm dish in medium A supplemented with 5% LPDS. On day 1, the medium was replaced with fresh medium A supplemented with 0.3 mg/ml EMS. After 16 h, the cells were washed twice with PBS, trypsinized, and split 1:10 in medium A supplemented with 5% LPDS. The cells were cultured for 3 days to allow expression of altered phenotypes. On day 5, cells were washed and refed medium A containing 5% LPDS and 2.5 µM 25-HC. Fresh medium was added to the cells every 2 days until colonies formed. On day 30, the surviving colonies were isolated with cloning cylinders and allowed to proliferate. Seven 25-HC-resistant colonies were isolated; one colony was cloned by limiting dilution and designated SRD-20 cells.
PCR amplification and cloning of Insig-2 from SRD-20 RNA and genomic DNA
Total RNA was isolated from SRD-14 and SRD-20 cells using the RNeasy kit (Qiagen) according to the manufacturer's instructions and subjected to reverse transcription reactions. First-strand cDNA was used to obtain PCR-amplified Insig-2 cDNA with the following primers: 5′-ATGGCAGAAGGAAAGACCGAGTCAC-3′ and 5′-TTCTTGATGAGATTTTTCAGGAATAAC-3′. The resulting PCR products (full-length and truncated Insig-2) were subcloned into the pCRII vector (Invitrogen), and nine individual clones were subjected to DNA sequencing to identify potential mutations in Insig-2. Two forms of Insig-2 were also confirmed from genomic DNA isolated from the same cell lines.
Plasmids
The following recombinant expression plasmids have been described in the indicated reference: pCMV-Scap, which encodes hamster Scap under transcriptional control of the cytomegalovirus (CMV) promoter (16); pCMV-HMG-Red(TM 1-8)-T7, which encodes transmembrane 1-8 hamster HMG-CoA reductase followed by three tandem copies of the T7 epitope tag under control of the CMV promoter (6); pCMV-Insig-1-Myc and pCMV-Insig-1-T7, which encode human Insig-1, followed by six tandem copies of a c-Myc epitope tag or three copies of a T7 epitope tag under control of the CMV promoter (15, 23); pCMV-Insig-2-Myc or pCMV-Insig-2-T7, encoding human Insig-2, followed by six tandem copies of a c-Myc epitope tag or three copies of a T7 epitope tag under control of the CMV promoter (21, 30). Point mutations in Insig-1 and Insig-2 proteins were generated by site-directed mutagenesis using the QuikChange kit (Stratagene). The coding regions of all plasmids were verified by DNA sequencing prior to use.
Transient transfection
Cells were transiently transfected with FuGENE 6 reagent (Roche Applied Science) according to the manufacturer's protocol. The total amount of DNA in each transfection was adjusted to 3 µg per dish with pcDNA3 (Invitrogen). Conditions of the incubations are described in the figure legends. At the end of the incubations, triplicate dishes of cells for each variable were harvested and pooled for analysis.
Cell fractionation and immunoblot analysis
Triplicate dishes of cells were used to isolate membrane fractions and nuclear extract fractions as described previously (6). Aliquots of nuclear extract and membrane fractions were subjected to 8% SDS-PAGE; the proteins were transferred to Hybond-C extra nitrocellulose filters (Millipore), and immunoblot analysis was carried out as described (6). Primary antibodies used for immunoblotting are as follows: IgG-7D4, a mouse monoclonal antibody against hamster SREBP-2 (amino acids 32–250) (31); IgG-9D5, a mouse monoclonal antibody against amino acids 54–207 of hamster Scap and IgG-R139, a rabbit polyclonal antibody against amino acids 54–207 and 540–707 of hamster Scap (11); IgG-9E10, a mouse monoclonal antibody against c-Myc purified from the culture medium of hybridoma clone 9E10 (American Type Culture Collection); IgG-2b, mouse monoclonal anti-T7-Tag (IgG2b) (Novagen); and IgG-HSV, mouse monoclonal antiherpes simplex virus (Novagen).
Immunoprecipitation
SRD-13A (HMG-CoA reductase and Scap) or CHO-7 and SRD-15 (Insigs) cells were transfected in medium A supplemented with 5% LPDS as described above. Conditions of incubation are described in the figure legends. At the end of the incubations, cells were harvested and lysed in 1 ml of PBS containing either 1% digitonin (for reductase immunoprecipitations) or 0.1% Nonidet P40 (for Scap and Insig immunoprecipitations) supplemented with 5 mM EDTA, 5 mM EGTA, 20 µM leupeptin, 25 µg/ml N-acetyl-leucinal-leucinal-norleucinal, 5 µg/ml pepstatin A, and 2 µg/ml aprotinin and clarified by centrifugation at 100,000 g for 10 min at 4°C. Immunoprecipitation with anti-T7-coupled agarose beads or 20 µg anti-Myc polyclonal antibody, together with 100 µl of protein A/G agarose beads (Scap), was carried out as described previously (6).
RESULTS
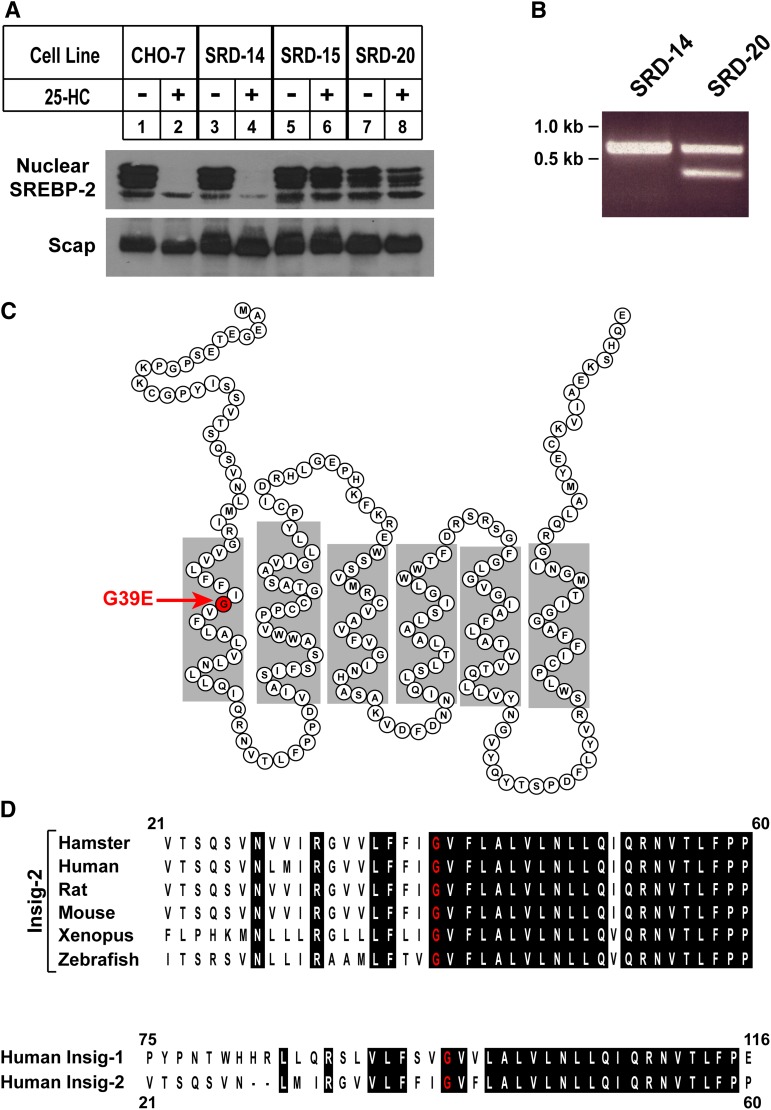
To identify amino acids in Insig-2 that are important for sterol-mediated effects on Scap and reductase, Insig-1-deficient SRD-14 cells were mutagenized with EMS, followed by selection for growth in medium containing 5% lipoprotein-deficient serum and 1 μg/ml 25-HC. This procedure yielded seven 25-HC-resistant colonies, one of which was isolated, expanded, and designated SRD-20. Figure 1A compares levels of nuclear SREBP-2 [the SREBP isoform that preferentially activates genes encoding cholesterol biosynthetic enzymes (32)] in wild-type CHO-7, Insig-1-deficient SRD-14, Insig-1- and Insig-2-deficient SRD-15, and SRD-20 cells. The cells were incubated in sterol-depleting medium containing lipoprotein-deficient serum, the reductase inhibitor compactin, and a low level of mevalonate that permits synthesis of essential nonsterol isoprenoids, but not cholesterol (33). Some of the dishes also received 1 μg/ml 25-HC. Following incubation for 16 h, the cells were harvested, lysed, and subjected to cell fractionation to generate membrane and nuclear extract fractions. Aliquots of each fraction were subjected to SDS-PAGE and immunoblotted with antibodies against Scap and SREBP-2. Processed forms of SREBP-2 accumulated in the nuclear extracts of sterol-depleted CHO-7, SRD-14, SRD-15, and SRD-20 cells (top panel, lanes 1, 3, 5, and 7, respectively). The addition of 25-HC blocked the appearance of nuclear SREBP-2 in CHO-7 and SRD-14 cells (lanes 2 and 4) but not in SRD-15 and SRD-20 cells (lanes 6 and 8, respectively). Suppression of SREBP processing in SRD-14 cells following extended sterol treatment (>16 h) is consistent with our previous findings (24). All of the cells produced similar amounts of Scap protein in membranes, regardless of the absence or presence of 25-HC (bottom panel).
Fig. 1.
Characterization of 25-HC-resistant SRD-20 cells. A: CHO-7, SRD-14, SRD-15, and SRD-20 cells were set up on day 0 at 5 × 105 cells per 100 mm dish in medium A containing 5% LPDS. On day 2, cells were switched to medium A supplemented with 5% LPDS, 10 µM sodium compactin, and 10 μM sodium mevalonate in the absence or presence of 2.5 µM 25-HC. After incubation at 37°C for 16 h, cells were harvested and subjected to cell fractionation as described in Experimental Procedures. Aliquots of the membrane (25 µg of protein/lane) and nuclear extract fractions (12 µg of protein/lane) were subjected to SDS-PAGE, the proteins were transferred to nylon membranes, and immunoblot analysis was carried out with 5 µg/ml IgG-7D4 (against SREBP-2) and 5 µg/ml IgG-9D5 (against Scap). Filters were exposed to film for 5 s to 1 min at room temperature. B: Aliquots of total RNA isolated from SRD-14 and SRD-20 cells were subjected to reverse transcription reactions; the resulting first-strand cDNA was used to amplify the Insig-2 cDNA. PCR products were subjected to electrophoresis on an agarose gel, which was subsequently stained with ethidium bromide and photographed. C: Amino acid sequence and membrane topology of human Insig-2. Glycine 39, which is found mutated to glutamic acid in SRD-20 cells, is highlighted in red. D: Comparison of amino acid residues in hamster, human, rat, mouse, xenopus, and zebrafish Insig-2. Identical residues are highlighted in black, and the conserved glycine-39 residue is highlighted in red. The lower shows the amino acid alignment between human Insig-1 and Insig-2. The residue in human Insig-1 corresponding to Gly-39 in human Insig-1 is Gly-95. All sequence alignments were performed using the CLUSTALW method (DNASTAR). GenBank accession numbers are as follows: AY112745 (human Insig-1); AF527632 (human Insig-2); AF527631 (mouse Insig-2); AF527629 (hamster Insig-2); AF527627 (zebrafish Insig-2); AY152392 (rat Insig-2); and NM_001011169 (xenopus Insig-2).
The failure of SRD-20 cells to respond to the suppressive effects of 25-HC treatment suggested they may be deficient in Insig-2 as well as Insig-1. Thus, total RNA was isolated from SRD-14 and SRD-20 cells and subjected to reverse transcription reactions. PCR amplification of first-strand cDNA from SRD-14 cells produced a single 678 bp band for Insig-2 (Fig. 1B). A band of similar size was generated from SRD-20 first-strand cDNA, but an additional 368 bp band was also observed. Both forms of the Insig-2 cDNA from SRD-20 cells were subcloned, and 10–30 individual clones were subjected to DNA sequencing, which revealed the 678 bp fragment corresponds to full-length Insig-2 containing a G-to-A substitution that changes Gly-39 to Glu. It should be noted that the G-to-A substitution was not present in the Insig-2 cDNA from SRD-14 cells. As shown in Fig. 1C, Gly-39 is predicted to lie in the middle of the first of six transmembrane helices of Insig-2 (19). Sequence alignments revealed that Gly-39 is completely conserved in hamster, human, mouse, rat, xenopus, and zebrafish Insig-2 (Fig. 1D); it is also present in human Insig-1. DNA sequencing revealed that the 386 bp fragment encodes a splice variant of Insig-2 in which Exon 2 is fused to Exon 5, creating a frame shift. The resulting cDNA encodes amino acids 1–81 of Insig-2, followed by 22 novel amino acids and a stop codon; this fusion protein is likely nonfunctional.
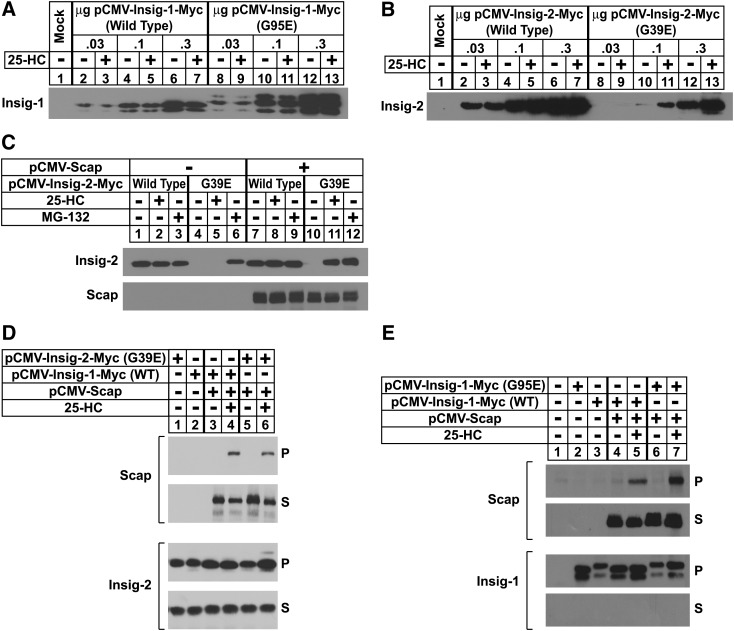
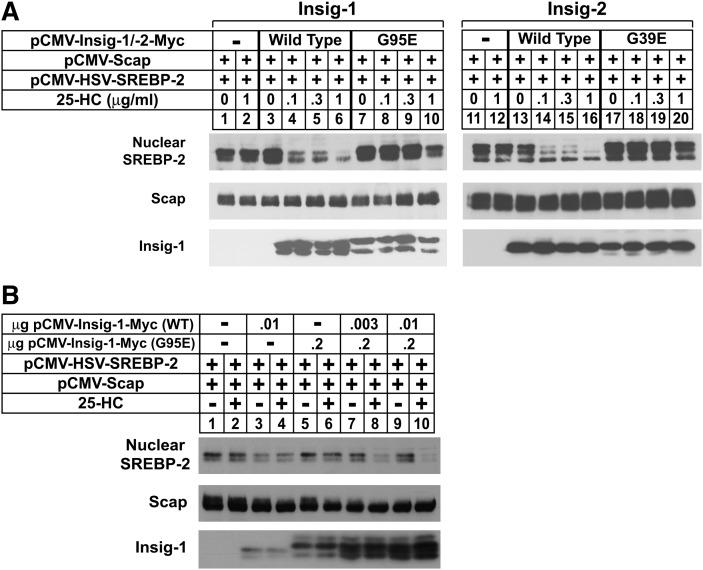
In the experiment of Fig. 2A, B, CHO-7 cells were transfected with wild-type or mutant versions (G95E for Insig-1 and G39E for Insig-2) of pCMV-Insig-1/-2-Myc, expression plasmids that encode full-length human Insig-1 or Insig-2 followed by six copies of a c-Myc epitope. Following sterol depletion, the cells were incubated for 5 h in the absence or presence of 25-HC, after which they were harvested and subjected to cell fractionation. Immunoblots of the resulting membrane fractions with anti-Myc revealed dose-dependent expression of wild-type and mutant Insig-1; similar amounts of expression were observed in the absence and presence of 25-HC (Fig. 2A, compare lanes 2–7 with 8–13). We observed similar dose-dependent expression of wild-type Insig-2-Myc (Fig. 2B, lanes 2–7), but a different result was obtained for the G39E mutant. At low plasmid levels (0.03 μg), G39E Insig-2 was not detected in membranes (lanes 8 and 9). However, the mutant protein was visualized when 0.1 μg plasmid was transfected, but only when the cells were treated with 25-HC (compare lanes 10 and 11). At the highest level of plasmid (0.3 μg), G39E Insig-2 was detected in sterol-depleted cells and expression was marginally increased in the presence of 25-HC (lanes 12 and 13).
Fig. 2.
Expression of Insig-2 (G39E) and Insig-1 (G95E) in transfected CHO cells. A, B: CHO-7 cells were set up on day 0 at 5 × 105 cells per 60 mm dish. On day 1, the cells were transfected with the indicated amount of wild-type or mutant Insig-1 and Insig-2 in 2 ml of medium A containing 5% LPDS. The total DNA in each dish was adjusted to 3 µg by the addition of pcDNA3 empty vector. Six hours after transfection, the cells were depleted of sterols by the direct addition of medium A containing 5% LPDS, 10 µM compactin, and 50 µM mevalonate (final concentrations). After 16 h at 37°C, cells were switched to medium A containing 5% LPDS and 10 µM compactin in the absence or presence of 2.5 µM 25-HC. Following incubation for 5 h, the cells were harvested and aliquots of the resulting membrane fractions (18 µg) were subjected to SDS-PAGE, and immunoblot analysis was carried out with 1 µg/ml monoclonal IgG-Myc (against Insigs). C: On day 0, SRD-13A cells were set up at 7 × 105 cells per 60 mm dish. On day 1, the cells were transfected in 3 ml medium A containing 5% LPDS with 2 µg of pCMV-Scap and 0.1 µg of wild-type or 0.3 µg of mutant pCMV-Insig-2-Myc. After sterol depletion for 16 h at 37°C, the cells were switched to medium A containing 5% LPDS in the absence or presence of 2.5 µM 25-HC; some of the dishes also received 10 µM MG-132. Following incubation for 5 h, the cells were harvested, lysed, and subjected to SDS-PAGE. Immunoblot analysis was carried out with 1 μg/ml monoclonal IgG-9E10 and 1 μg/ml polyclonal IgG-R139 to detect Insig-2 and Scap, respectively. D, E: SRD-13A cells were set up on day 0 as in C and transfected on day 1 with 0.8 µg of pCMV-Scap and 0.05 µg of wild-type or 0.1 µg of mutant pCMV-Insig-1-Myc (E) 1.0 µg of wild-type or 3.0 µg of mutant pCMV-Insig-2-Myc (D). After incubation for 6 h 37°C, the cells were depleted of sterols. After 16 h, the cells were switched on day 2 to medium A containing 5% LPDS in the absence or presence of 1.25 µM 25-HC. After incubation for 5 h, cells were harvested, lysed, and immunoprecipitated with polyclonal anti-Myc as described in Experimental Procedures to precipitate Insig-1. Aliquots of the pellet (P; representing 0.25 dish of cells) and supernatant (S; representing 0.05 dish of cells) fractions of the immunoprecipitate were subjected to SDS-PAGE, and immunoblot analysis was carried out with 1 μg/ml polyclonal IgG-R139 (against Scap) and 1 μg/ml monoclonal IgG-9E10 (against Insig-1). All filters were exposed to film at room temperature for 3 s to 3 min.
We next conducted an experiment to determine whether Scap mediates sterol-dependent expression of G39E Insig-2 (Fig. 2C). Scap-deficient SRD-13A cells (29) were transfected with low levels of G39E Insig-2 in the absence or presence of an expression plasmid encoding hamster Scap (pCMV-Scap). Following sterol depletion, some of the cells were treated with 25-HC or the proteasome inhibitor MG-132 and harvested for cell fractionation after 5 h. Immunoblotting revealed constant expression of wild-type Insig-2 that was not influenced by the presence of 25-HC or MG-132 (Fig. 2C, top panel, lanes 1–3); coexpression of Scap had little effect on this expression (top and bottom panels, lanes 7–9). In the absence of Scap, G39E Insig-2 was not detected in untreated cells or cells treated with 25-HC (lanes 4 and 5); however, the protein was readily visualized upon the addition of MG-132 (lane 6). With Scap coexpression, the mutant Insig protein was stabilized to a similar extent by the addition of 25-HC or MG-132 (lanes 10–12). These findings indicate that G39E Insig-2 undergoes proteasome-mediated degradation in sterol-depleted cells and becomes stabilized upon sterol repletion, which likely results from sterol-induced binding of the protein to Scap. This conclusion is supported by the coimmunoprecipitation experiment of Fig. 2D in which SRD-13A cells were transfected with Scap and either wild-type or G39E Insig-2. After sterol depletion for 16 h, the cells were then incubated in the absence or presence of 25-HC, after which detergent lysates were prepared and transfected Insig was immunoprecipitated with anti-Myc. Immunoblotting of the resulting pellet and supernatant fractions of the precipitation revealed that both wild-type and mutant Insig-2 form a sterol-induced complex with Scap (lanes 5 and 7, respectively). Similar results were obtained for wild-type and mutant versions of Insig-1 (Fig. 2E).
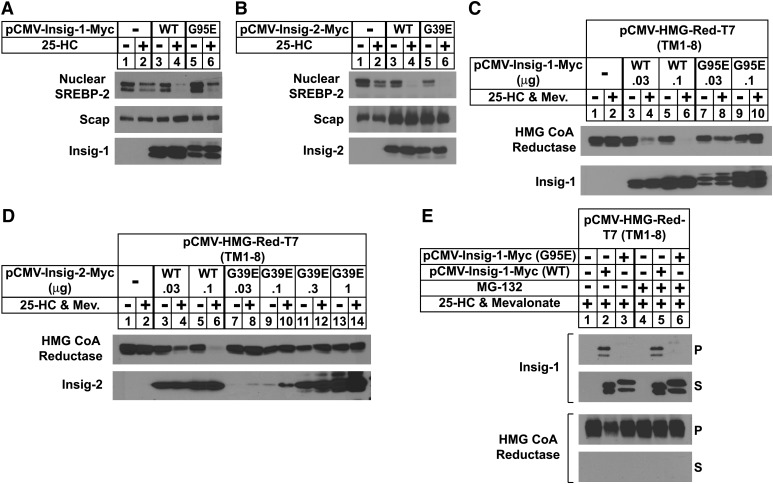
Considering that the Gly to Glu mutants of Insigs continue to bind Scap in the presence of 25-HC, we next designed an experiment to determine whether these mutants are capable of mediating sterol regulation of SREBP processing and reductase degradation. SRD-13A cells were transfected with wild-type or mutant Insig-1 and Insig-2, Scap, and pTK-HSV-SREBP-2, which encodes human SREBP-2 preceded by three copies of an HSV epitope. The cells were acutely depleted of sterols in medium containing LPDS, compactin, and hydroxypropyl cyclodextrin, a reagent that removes cholesterol from the plasma membrane of intact cells. The cells were then further incubated in the absence or presence of 25-HC for 5 h prior to harvesting and cell fractionation. In the absence of wild-type Insig-1 or Insig-2, we observed a band for processed, nuclear SREBP-2 that persisted in the presence of 25-HC (Fig. 3A, B, top panel, lanes 1 and 2). The coexpression of wild-type Insig-1 or Insig-2 led to the disappearance of this band upon 25-HC treatment (lanes 3 and 4); similar results were obtained with the Gly to Glu mutants of both Insigs (lanes 5 and 6). In contrast, G95E Insig-1 and G39E Insig-2 failed to mediate sterol-accelerated degradation of reductase as shown in Fig. 3C, D. CHO-7 cells were transfected with pCMV-HMG-Red-T7 (TM1-8), which encodes transmembrane helices 1–8 of hamster reductase (amino acids 1–348) followed by three copies of the T7 epitope; this region of reductase has been shown to be both necessary and sufficient for Insig-mediated degradation (6, 17). The membrane domain of reductase was completely refractory to sterol-induced degradation when transfected alone, owing to the saturation of endogenous Insigs (Fig. 3C, D, top panel, lanes 1 and 2). Coexpression of increasing amounts of wild-type, but not the Gly to Glu mutants of Insigs, restored sterol-accelerated degradation upon overexpressed reductase (compare lanes 3–6 with lanes 7–10 for Insig-1 and lanes 7–14 for Insig-2). The failure of the mutant Insigs to mediate reductase degradation is consistent with the coimmunoprecipitation experiment of Fig. 3E. CHO-7 cells transfected with the membrane domain of reductase together with either wild-type or G95E Insig-1 were subjected to a brief 25-HC treatment in the absence or presence of MG-132. Subsequently, the reductase membrane domain was immunoprecipitated from detergent lysates. Immunoblots of precipitated material revealed the presence of wild-type, but not G95E, Insig-1 in untreated and MG-132-treated cells (compare lanes 2 and 5 with 3 and 6).
Fig. 3.
Insig-1 (G95E) and Insig-2 (G39E) mediate sterol-regulated inhibition of SREBP-2 processing but not sterol-accelerated degradation of HMG-CoA reductase. A, B: SRD-13A cells were set up on day 0 as in Fig. 2 and transfected on day 1 with 0.5 µg of pCMV-Scap and 2 µg of pTK-HSV-SREBP-2. Cells in A were also transfected with either wild-type (0.1 µg) or mutant (0.2 µg) pCMV-Insig-1-Myc; cells in B were transfected with either wild-type (1 µg) or mutant (3 µg) pCMV-Insig-2-Myc in medium A containing 5% LPDS. The total DNA in each lane was adjusted to 5.5 µg per dish by the addition of pcDNA3 empty vector. On day 2, cells were switched to medium A containing 5% LPDS, 10 µM compactin, and 1% hydroxypropyl-β-cyclodextrin. After 1 h at 37°C, cells were washed twice with PBS and switched to medium A supplemented with 5% LPDS and 10 µM compactin in the absence or presence of 2.5 µM 25-HC. After 5 h, the cells were harvested, and membrane and nuclear extract fractions were prepared and subjected to SDS-PAGE followed by immunoblot analysis with 3 µg/ml monoclonal IgG-Myc (against Insig-1 or Insig-2) and 5 µg/ml IgG-9D5 (against Scap). The nuclear extract fractions were immunoblotted with a 1:10,000 dilution of monoclonal anti-HSV. Filters were exposed to film at room temperature for 1 to 15 s. C, D: CHO-7 cells were set up on day 0 as in Fig. 2 and transfected on day 1 with 1 µg of pCMV-HMG-Red-T7 (TM1-8) together with the indicated amount of wild-type and mutant pCMV-Insig-1-Myc (C) or pCMV-Insig-2-Myc (D). Six hours after transfection, cells were depleted of sterols for 16 h and subsequently switched to medium A containing 5% LPDS and 10 µM compactin in the absence or presence of 2.5 µM 25-HC plus 10 mM mevalonate. After 5 h, the cells were harvested; membrane fractions were isolated and subsequently immunoblotted with 1 µg/ml monoclonal IgG-T7 (against reductase) and 3 µg/ml monoclonal IgG-Myc (against Insig-1 or Insig-2). E: SRD-13A cells were set up on day 0 as in C and transfected on day 1 with 1 µg of pCMV-HMG-Red-T7 (TM1-8) and 0.01 µg of wild-type or 0.02 µg of mutant pCMV-Insig-1-Myc. After 6 h at 37°C, cells were depleted of sterols for 16 h. Following this incubation, the cells were switched to medium A containing 5% LPDS and 10 µM compactin in the absence or presence of 2.5 µM 25-HC plus 10 mM mevalonate; some of the cells also received 10 µM MG-132. After incubation for 20 min at 37°C, cells were harvested and lysed, and HMG-CoA reductase was immunoprecipitated with polyclonal anti-T7 coupled agarose beads as described in Experimental Procedures. Aliquots of the pellet (P; representing 0.25 dish of cells) and supernatant (S; representing 0.05 dish of cells) fractions of the immunoprecipitate were subjected to SDS-PAGE, and immunoblot analysis was carried out with 1 μg/ml anti-T7 (against HMG-CoA reductase) and 1 μg/ml monoclonal IgG-9E10 (against Insig-1). Filters were exposed to film at room temperature for 5 to 30 s.
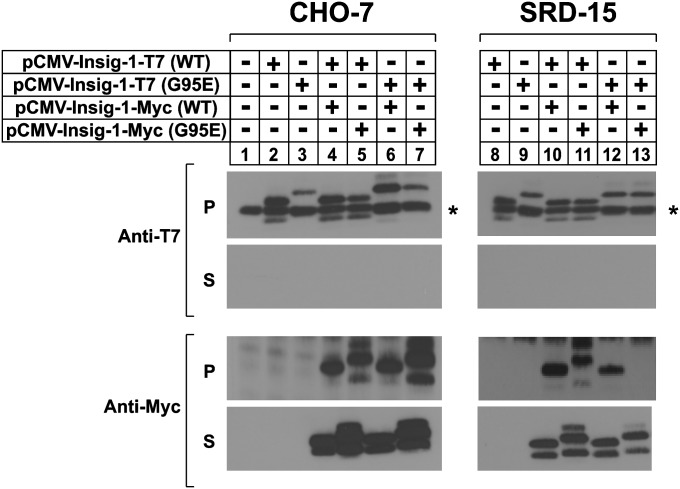
Results presented thus far present an interesting conundrum in that G95E Insig-1 and G39E Insig-2 retain the capacity to mediate sterol regulation of Scap-SREBP, but not reductase. A recent study reported that purified Insigs bind to 25-HC as a dimer (34), which raises the question as to whether the proteins act as dimers (or multimers) in intact cells to regulate SREBP processing. Thus, we next conducted a coimmunoprecipitation experiment to probe for Insig multimerization in CHO-7 and SRD-15 cells, which provide the Insig-null background that is required to test our hypothesis (Fig. 4). For this purpose, the cells were transfected with various combinations of wild-type and mutant Insig-1 plasmids that encoded proteins fused to either T7 or Myc epitope tags. We used Insig-1 rather than Insig-2 to avoid effects that may be caused by Scap-dependent stabilization of mutant Insig-2 (see Fig. 2). Following transfection, detergent lysates were prepared and subjected to immunoprecipitation with anti-T7 antibody. Immunoblot analysis of the resulting pellet fractions with anti-T7 and anti-Myc antibodies revealed that the wild-type and mutant Insig-1-Myc coprecipitated with both version of Insig-1-T7 in CHO-7 cells (Fig. 5B, lanes 4–7). In SRD-15 cells, wild-type Insig-1-T7 precipitated both versions of the Myc-tagged protein (lanes 10 and 11). Similarly, the G95E Insig-1-T7 protein formed a complex with wild-type Insig-1-Myc (lane 12); however, it failed to associate with the G95E mutant (lane 13).
Fig. 4.
Glycine-39 is required for the Insig-Insig coimmunoprecipitation. CHO-7 (left panel) or SRD-15 (right panel) cells were set up on day 0 at 5 × 105 cells per 60 mm dish in medium A containing 5% LPDS. On day 1, the cells were transfected with various combinations of wild-type (0.1 µg) or mutant (0.4 µg) pCMV-Insig-1-Myc and pCMV-Insig-1-T7 in medium A supplemented with 5% LPDS. After incubation for 16 h at 37°C, the cells were harvested, lysed, and immunoprecipitated with polyclonal anti-T7 beads. The pellet (P; representing 0.25 dish of cells) and supernatant (S; representing 0.05 dish of cells) fractions of the immunoprecipitate were subjected to SDS-PAGE, and immunoblot analysis was carried out with 1 µg/ml monoclonal IgG-T7 and 3 µg/ml monoclonal IgG-Myc. Filters were exposed to film for 1 to 3 s.
Fig. 5.
Insig-1 (G95E) and Insig-2 (G39E) are defective in mediating sterol regulation of Scap and HMG-CoA reductase in SRD-15 cells. A: SRD-15 cells were set up on day 0 at 5 × 105 cells per 60 mm dish in medium A supplemented with 5% LPDS. On day 1, cells were transfected with 0.5 µg of pCMV-Scap, 2 µg of pTK-HSV-SREBP-2, and either 0.1 µg wild-type pCMV-Insig-1-Myc, 0.2µg mutant pCMV-Insig-1-Myc, 1 µg wild-type pCMV-Insig-2-Myc, or 3 µg mutant pCMV-Insig-2-Myc in 5% LPDS as indicated. The total DNA in each lane was adjusted to 5.5 µg per dish by the addition of pcDNA3 empty vector. After 6 h at 37°C, cells were changed to sterol-depleting medium. On day 2, cells were switched to medium A containing 5% LPDS, 50 µM compactin, and 1% hydroxypropyl-β-cyclodextrin. After 1 h at 37°C, cells were washed twice with PBS and switched to medium A with 5% LPDS, 10 µM compactin, and the indicated concentration of 25-HC. After incubation for 5 h, cells were harvested and fractionated into membrane and nuclear extract fractions, which were subsequently subjected to SDS-PAGE. Immunoblot analysis was carried out with 3 µg/ml monoclonal IgG-Myc (against Insig-1 and Insig-2) and 5 µg/ml IgG-9D5 (against Scap). The nuclear extracts were immunoblotted with a 1:1,000 dilution of monoclonal anti-HSV IgG. Filters were exposed to film at room temperature for 3 to 15 s. B: SRD-15 cells were set up on day 0 and transfected on day 1 as described in A with 0.5 µg pCMV-Scap, 2 µg of pTK-HSV-SREBP-2, and the indicated amount of wild-type and/or mutant pCMV-Insig-1-Myc. Following sterol depletion for 16 h at 37°C, the cells were switched to medium A containing 5% LPDS, 50 µM compactin, and 1% hydroxypropyl-β-cyclodextrin. Following incubation for 1 h at 37°C, the cells were washed twice with PBS and switched to medium A with 5% LPDS and 50 µM compactin in the absence or presence of 25-HC. After 5 h, the cells were harvested; membrane and nuclear extract fraction were prepared and subsequently subjected to SDS-PAGE. Immunoblot analysis was carried out with 3 µg/ml monoclonal IgG-Myc (against Insig-1 and Insig-2) and 5 µg/ml IgG-9D5 (against Scap). The nuclear extract fractions were immunoblotted with a 1:10,000 dilution of monoclonal anti-HSV IgG. Filters were exposed to film at room temperature for 3 to 15 s.
To correlate Insig-Insig coimmunoprecipitation shown in Fig. 4 with sterol-regulated processing of SREBP-2, we conducted the experiments shown in Fig. 5. The results of Fig. 5A show that as expected, wild-type forms of Insig-1 and Insig-2 conferred sterol regulation upon processing of overexpressed SREBP-2 in SRD-15 cells (top panel, compare lanes 1 and 2 with 3–6 for Insig-1 and lanes 11 and 12 with 13–16 for Insig-2). However, sterols failed to block processing of SREBP-2 in transfected SRD-15 cells when the Gly to Glu mutants of Insigs were coexpressed (lanes 7–10 for Insig-1 and 17–20 for Insig-2). Similar to the results in CHO-7 cells, the mutant forms of both Insigs did not mediate sterol-accelerated degradation of the reductase membrane domain in SRD-15 cells (data not shown). In the experiment of Fig. 5B, SRD-15 cells were transfected with pTK-HSV-SREBP-2 and either a low level (10 ng) of wild-type Insig-1 plasmid or a high level (0.2 μg) of the inactive mutant. Both forms of Insig-1 failed to mediate sterol suppression of nuclear SREBP-2 when transfected alone (top panel, lanes 3–6); however, coexpression of as little as 3 ng of wild-type Insig-1 together with the inactive mutant led to 25-HC-mediated inhibition of SREBP-2 processing (lanes 7 and 8). Similar effects were observed with 10 ng of wild-type Insig-1 (lanes 9 and 10). Considered together, the results of Figs. 4 and 5 support the hypothesis that Insig-1 functions as a multimer, the formation of which requires an intramembrane glycine residue at position 95.
DISCUSSION
Our previous studies show that Insig-1-deficient SRD-14 cells fail to grow indefinitely when cultured in the presence of 25-HC (24). We attributed this failure to residual sterol regulation, owing to the remaining Insig-2 in SRD-14 cells. This hypothesis was substantiated in subsequent experiments in which mutagenized SRD-14 cells were subjected to 25-HC selection, yielding SRD-15 cells that are deficient in Insig-2 as well as Insig-1 (25). In this study, we exploited the 25-HC sensitivity of SRD-14 cells to isolate a new line of mutant CHO cells designated SRD-20. Consistent with their growth phenotype, SRD-20 cells were fully resistant to 25-HC-mediated suppression of SREBP processing (Fig. 1A). DNA sequencing revealed that SRD-20 cells harbor a point mutation in one Insig-2 allele that produces a truncated, nonfunctional protein and another point mutation in the other Insig-2 allele that yields a glutamic acid for glycine-39 substitution. We failed to find mutations in any of the other genes known to participate in the SREBP pathway, including SREBP-1, SREBP-2, and Scap (data not shown). Gly-39 in Insig-2 corresponds to Gly-95 in Insig-1 and is predicted to lie in the middle of the first membrane-spanning segment of Insigs (Fig. 1C). Transfection experiments reveal that in sterol-depleted cells, G39E Insig-2 is rapidly degraded in a saturable fashion (Fig. 2B). This degradation is blocked by the addition of the proteasome inhibitor MG-132 or sterols (Fig. 2C). Sterol-dependent stabilization of G39E Insig-2 requires the presence of Scap, resembling that of wild-type Insig-1 when expressed at near endogenous levels (23). In sterol-deprived cells, Insig-1 binds the ubiquitin ligase gp78, which initiates Insig-1 ubiquitination and subsequent degradation (22). Sterols prevent degradation of Insig-1 by displacing gp78, which results from sterol-induced binding of Scap to Insig-1. In contrast, Insig-2 is a stable protein in both the absence and presence of sterols. The current data indicate that substitution of Glu for Gly-39 in Insig-2 destabilizes the protein, causing its recognition and subsequent ubiquitination by gp78. However, the mechanism by which G39E Insig-2 is recognized for ubiquitination by gp78 and whether this selection is similar to that of Insig-1 is presently unknown. It should be noted that destabilization by the Gly to Glu substitution does not destabilize Insig-1 (Fig. 2A), indicating that the effect is Insig-2 specific.
In wild-type CHO-7 cells, Scap-dependent stabilization of G39E Insig-2 correlated with the ability of the mutant protein to bind Scap and mediate sterol regulation of SREBP processing (Fig. 3A, B). These results contrast those obtained for reductase degradation in that G95E Insig-1 and G39E Insig-2 failed to mediate sterol-accelerated degradation of reductase and did not bind the enzyme in the presence of sterols (Fig. 3C–E). A plausible explanation for this discrepancy can be provided by the recent finding that purified Insig-2 binds 25-HC as a dimer (34). Thus, we postulated that when overexpressed, the mutant Insigs can associate with their endogenous equivalent to form active complexes capable of sterol-induced binding to Scap. When experiments were conduct in SRD-15 cells, both mutant Insigs failed to mediate sterol regulation of Scap and reductase (Fig. 4). However, sterol regulation of SREBP processing was restored in SRD-15 cells when substoichiometric amounts of wild-type Insig-1 were coexpressed with the inactive mutant (Fig. 5A). Coimmunoprecipitation experiments indicate that G95E Insig-1 can only form a multimeric complex in the presence of wild-type Insig-1 (Fig. 5B). These findings indicate that Insigs function as multimers to mediate sterol regulated processing of SREBPs and that one member of the complex must contain a glycine residue at position 39.
Although the combination of wild-type and mutant Insigs can bind Scap in the presence of sterols and prevent SREBP processing, the complex cannot bind reductase and accelerate its degradation. Although Insigs bind to both Scap and reductase, the current data suggest that formation of these sterol-dependent complexes occur through distinct sites on Insigs. It is notable that the Scap and reductase binding sites on Insigs exhibit some degree of overlap as indicated by the inhibition of reductase degradation by overexpression of the Scap sterol-sensing domain (6). However, complete resolution of this requires comparisons of detailed structural analyses of the sterol-dependent reductase-Insig and Scap-Insig complexes.
Acknowledgments
The authors thank Drs. Michael S. Brown and Joseph L. Goldstein for their continued encouragement and advice. We also thank Tammy Dinh and Kristi Garland for excellent technical assistance, Lisa Beatty for help with tissue culture, and Dr. Andy Nguyen for critical reading of the manuscript.
Footnotes
Abbreviations:
- 25-HC
- 25-hydroxycholesterol
- CHO
- Chinese hamster ovary
- CMV
- cytomegalovirus
- EMS
- ethylmethane sulfonate
- ER
- endoplasmic reticulum
- LPDS
- lipoprotein-deficient serum
- Scap
- SREBP cleavage-activating protein
- SRD
- sterol regulatory deficient
- SREBP
- sterol-regulatory element binding protein
This work was supported by grants from the National Institutes of Health (HL-20948) and the Perot Family Foundation. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. R.A.D-B. is the recipient of an Early Career Scientist Award from the Howard Hughes Medical Institute, an Established Investigator Award from the American Heart Association (0540128N), and a Distinguished Young Investigator in Medical Research Award from the W.M. Keck Foundation.
REFERENCES
- 1.Goldstein J. L., DeBose-Boyd R. A., Brown M. S. 2006. Protein sensors for membrane sterols. Cell. 124: 35–46. [DOI] [PubMed] [Google Scholar]
- 2.Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. 2003. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA 100: 12027–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nohturfft A., Yabe D., Goldstein J. L., Brown M. S., Espenshade P. J. 2000. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 102: 315–323. [DOI] [PubMed] [Google Scholar]
- 4.DeBose-Boyd R. A., Brown M. S., Li W. P., Nohturfft A., Goldstein J. L., Espenshade P. J. 1999. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 99: 703–712. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein J. L., Brown M. S. 1990. Regulation of the mevalonate pathway. Nature. 343: 425–430. [DOI] [PubMed] [Google Scholar]
- 6.Sever N., Yang T., Brown M. S., Goldstein J. L., DeBose-Boyd R. A. 2003. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol. Cell. 11: 25–33. [DOI] [PubMed] [Google Scholar]
- 7.Song B. L., Sever N., DeBose-Boyd R. A. 2005. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol. Cell. 19: 829–840. [DOI] [PubMed] [Google Scholar]
- 8.Nohturfft A., Brown M. S., Goldstein J. L. 1998. Topology of SREBP cleavage-activating protein, a polytopic membrane protein with a sterol-sensing domain. J. Biol. Chem. 273: 17243–17250. [DOI] [PubMed] [Google Scholar]
- 9.Liscum L., Finer-Moore J., Stroud R. M., Luskey K. L., Brown M. S., Goldstein J. L. 1985. Domain structure of 3-hydroxy-3-methylglutaryl coenzyme A reductase, a glycoprotein of the endoplasmic reticulum. J. Biol. Chem. 260: 522–530. [PubMed] [Google Scholar]
- 10.Roitelman J., Olender E. H., Bar-Nun S., Dunn W. A., Jr., Simoni R. D. 1992. Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for enzyme degradation in the endoplasmic reticulum. J. Cell Biol. 117: 959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakai J., Nohturfft A., Cheng D., Ho Y. K., Brown M. S., Goldstein J. L. 1997. Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. J. Biol. Chem. 272: 20213–20221. [DOI] [PubMed] [Google Scholar]
- 12.Gil G., Faust J. R., Chin D. J., Goldstein J. L., Brown M. S. 1985. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell. 41: 249–258. [DOI] [PubMed] [Google Scholar]
- 13.Brown M. S., Goldstein J. L. 1999. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA. 96: 11041–11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuwabara P. E., Labouesse M. 2002. The sterol-sensing domain: multiple families, a unique role? Trends Genet. 18: 193–201. [DOI] [PubMed] [Google Scholar]
- 15.Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., Brown M. S. 2002. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 110: 489–500. [DOI] [PubMed] [Google Scholar]
- 16.Hua X., Nohturfft A., Goldstein J. L., Brown M. S. 1996. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell. 87: 415–426. [DOI] [PubMed] [Google Scholar]
- 17.Lee P. C., Nguyen A. D., DeBose-Boyd R. A. 2007. Mutations within the membrane domain of HMG-CoA reductase confer resistance to sterol-accelerated degradation. J. Lipid Res. 48: 318–327. [DOI] [PubMed] [Google Scholar]
- 18.Sever N., Song B. L., Yabe D., Goldstein J. L., Brown M. S., DeBose-Boyd R. A. 2003. Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. J. Biol. Chem. 278: 52479–52490. [DOI] [PubMed] [Google Scholar]
- 19.Feramisco J. D., Goldstein J. L., Brown M. S. 2004. Membrane topology of human insig-1, a protein regulator of lipid synthesis. J. Biol. Chem. 279: 8487–8496. [DOI] [PubMed] [Google Scholar]
- 20.Yabe D., Komuro R., Liang G., Goldstein J. L., Brown M. S. 2003. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc. Natl. Acad. Sci. USA. 100: 3155–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yabe D., Brown M. S., Goldstein J. L. 2002. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl. Acad. Sci. USA. 99: 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J. N., Song B., DeBose-Boyd R. A., Ye J. 2006. Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase gp78. J. Biol. Chem. 281: 39308–39315. [DOI] [PubMed] [Google Scholar]
- 23.Gong Y., Lee J. N., Lee P. C., Goldstein J. L., Brown M. S., Ye J. 2006. Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell Metab. 3: 15–24. [DOI] [PubMed] [Google Scholar]
- 24.Sever N., Lee P. C. W., Song B. L., Rawson R. B., DeBose-Boyd R. A. 2004. Isolation of mutant cells lacking Insig-1 through selection with SR-12813, an agent that stimulates degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem. 279: 43136–43147. [DOI] [PubMed] [Google Scholar]
- 25.Lee P. C., Sever N., DeBose-Boyd R. A. 2005. Isolation of sterol-resistant Chinese hamster ovary cells with genetic deficiencies in both Insig-1 and Insig-2. J. Biol. Chem. 280: 25242–25249. [DOI] [PubMed] [Google Scholar]
- 26.Lee P. C. W., Liu P., Li W. P., DeBose-Boyd R. A. 2007. Amplification of the gene for SCAP, coupled with Insig-1 deficiency, confers sterol resistance in mutant Chinese hamster ovary cells. J. Lipid Res. 48: 1944–1954. [DOI] [PubMed] [Google Scholar]
- 27.Sega G. A. 1984. A review of the genetic effects of ethyl methanesulfonate. Mutat. Res. 134: 113–142. [DOI] [PubMed] [Google Scholar]
- 28.Metherall J. E., Goldstein J. L., Luskey K. L., Brown M. S. 1989. Loss of transcriptional repression of three sterol-regulated genes in mutant hamster cells. J. Biol. Chem. 264: 15634–15641. [PubMed] [Google Scholar]
- 29.Rawson R. B., DeBose-Boyd R., Goldstein J. L., Brown M. S. 1999. Failure to cleave sterol regulatory element-binding proteins (SREBPs) causes cholesterol auxotrophy in Chinese hamster ovary cells with genetic absence of SREBP cleavage-activating protein. J. Biol. Chem. 274: 28549–28556. [DOI] [PubMed] [Google Scholar]
- 30.Lee J. N., Gong Y., Zhang X., Ye J. 2006. Proteasomal degradation of ubiquitinated Insig proteins is determined by serine residues flanking ubiquitinated lysines. Proc. Natl. Acad. Sci. USA. 103: 4958–4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J., Brown M. S., Ho Y. K., Goldstein J. L. 1995. Three different rearrangements in a single intron truncate sterol regulatory element binding protein-2 and produce sterol-resistant phenotype in three cell lines. Role of introns in protein evolution. J. Biol. Chem. 270: 12152–12161. [DOI] [PubMed] [Google Scholar]
- 32.Horton J. D., Goldstein J. L., Brown M. S. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109: 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown M. S., Goldstein J. L. 1980. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J. Lipid Res. 21: 505–517. [PubMed] [Google Scholar]
- 34.Radhakrishnan A., Ikeda Y., Kwon H. J., Brown M. S., Goldstein J. L. 2007. From the Cover: Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc. Natl. Acad. Sci. USA. 104: 6511–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]