Abstract
During the haploid phase of mammalian spermatogenesis, nucleosomal chromatin is ultimately repackaged by small, highly basic protamines to generate an extremely compact, toroidal chromatin architecture that is critical to normal spermatozoal function. In common with several species, however, the human spermatozoon retains a small proportion of its chromatin packaged in nucleosomes. As nucleosomal chromatin in spermatozoa is structurally more open than protamine-packaged chromatin, we considered it likely to be more accessible to exogenously applied endonucleases. Accordingly, we have used this premise to identify a population of endonuclease-sensitive DNA sequences in human and murine spermatozoa. Our results show unequivocally that, in contrast to the endonuclease-resistant sperm chromatin packaged by protamines, regions of increased endonuclease sensitivity are closely associated with gene regulatory regions, including many promoter sequences and sequences recognized by CCCTC-binding factor (CTCF). Similar differential packaging of promoters is observed in the spermatozoal chromatin of both mouse and man. These observations imply the existence of epigenetic marks that distinguish gene regulatory regions in male germ cells and prevent their repackaging by protamines during spermiogenesis. The ontology of genes under the control of endonuclease-sensitive regulatory regions implies a role for this phenomenon in subsequent embryonic development.
During mammalian spermiogenesis, DNA sequences that are packaged by nucleosomes are sequentially replaced by transition proteins and finally by nucleoprotamines (Tanphaichitr et al. 1978; Ward and Coffey 1991; Braun 2001). This developmental progression facilitates a 10-fold higher order of DNA packaging that cannot be accommodated by nucleosomes alone and leads to a highly condensed chromatin that is transcriptionally inert (Premkumar and Bhargava 1972; Grunewald et al. 2005) and can be accommodated in a volume that is typically <10% that of a somatic cell nucleus (Powell et al. 1990). This repackaging is critically important for normal sperm motility. Following its delivery to the egg, the paternal nucleus is quickly accessed by maternal factors that serve to decondense the chromatin in readiness for pronuclear formation and subsequent syngamy. During this period, protamines are rapidly stripped from paternal chromatin and substituted with maternal histones (Collas and Poccia 1998; McLay and Clarke 2003).
Intriguingly, the spermatozoal DNA of many mammalian species retains a histone-rich compartment (Gusse et al. 1986; Bench et al. 1998). Human spermatozoa, for example, are estimated to retain ∼15% of their total chromatin in a histone-bound, nucleosomal configuration (Gatewood et al. 1987). In the mouse, the nucleosomal content is estimated at <2% (Balhorn et al. 1977). Indeed, the variation in nucleohistone content of mammalian spermatozoa is wide. Certain dasyurid marsupials, for example, repackage <50% of their spermatozoal chromatin into nucleoprotamines, while many vertebrate species do not use nucleoprotamines at all (Soon et al. 1997). The fate of paternal histones introduced into the egg at fertilization is not clear, although recent evidence indicates that at least some are incorporated into the zygotic nucleus (van der Heijden et al. 2008).
Classical DNA footprinting experiments exploit differences in the relative accessibility of euchromatic and heterochromatic regions to exogenously applied endonucleases (typically DNase I and micrococcal nuclease [MNase]) (Davie and Saunders 1981; Sabo et al. 2006), and similar approaches can be taken for the analysis of the nucleosomal domains of mature spermatozoa. One uses the ability of weak salt solutions to disrupt histone–DNA interactions in sperm nuclei permeabilized by detergents, while leaving the much stronger protamine–DNA interactions intact. This treatment depletes accessible histones from the sperm nuclei, and subsequent limited digestion with endonucleases releases the DNA thus exposed. Evidence for the specific and consistent differential packaging of DNA in human sperm nuclei has been presented using this approach (Gatewood et al. 1987; Gardiner-Garden et al. 1998; Wykes and Krawetz 2003). Alternatively, limited digestion of detergent permeabilized sperm nuclei with MNase has been used to facilitate the release of chromatin rich in telomeric and repetitive sequences from mature spermatozoa (Pittoggi et al. 1999; Gineitis et al. 2000). However, these studies did not provide a global insight into the nature and extent of this differential packaging phenomenon.
To address this issue, we have carried out genome-wide analyses of chromatin structure using a microarray-based approach to examine the sequence composition of the DNA released from, or retained by, human sperm using both salt extraction followed by endonuclease digestion, or by MNase digestion methods. Chromatin immunoprecipitation analysis (ChIP-chip) was subsequently used to identify DNA sequences preferentially associated with acetylated histone H4. These procedures yield complementary results and show that the DNA released by salt/endonuclease and MNase treatments is enriched in regulatory sequences. These include regions containing CCCTC-binding factor (CTCF) binding sites and the promoters of genes involved in development. In contrast, genic and intergenic regions as well as many regulatory sequences belonging to olfactory receptor and zinc finger gene clusters are not preferentially accessible to salt or endonuclease extraction. Together, these results suggest that differential nuclease sensitivity is established during the spermiogenic reorganization of the paternal genome and delineates a global epigenetic signature of ontological significance.
Results
Chromatin compositional analysis: DNA analysis
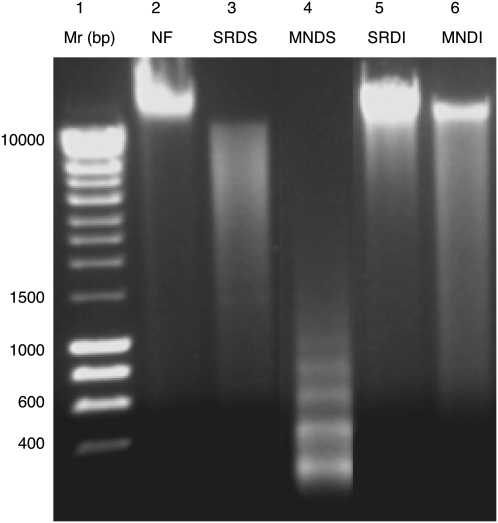
The composition of DNA targets used in the comparative genome hybridization (CGH) analyses, generated following salt extraction and restriction digestion (Gardiner-Garden et al. 1998); salt/restriction endonuclease digestion (SRD) or MNase digestion (Zalenskaya et al. 2000); or micrococcal nuclease digestion (MND) of detergent-permeabilized sperm nuclei, was verified by standard agarose gel electrophoresis (Fig. 1). The DNA released by restriction digestion from 108 salt-washed human sperm nuclei resolved as a smear, consisting of fragments ranging from ∼0.8 to 10 kb in length (Fig. 1, lane 3; salt/restriction endonuclease digested soluble [SRDS]). In contrast, following restriction digestion, the bulk of salt-insoluble DNA remaining in the pellet resolved at >10 kb typical of intact high-molecular-mass DNA (Fig. 1, lane 5; salt/restriction endonuclease digested insoluble [SRDI]). An ∼200-bp ladder was obtained from 108 human sperm nuclei following short exposure to exogenously applied MNase (Fig. 1, lane 4; micrococcal nuclease digested soluble [MNDS]). The repeat length for the ladder in lane 4 at ∼200 bp is in good agreement with the presence of nucleosomes in the soluble fractions. The insoluble pellets remaining after MNase digestion, like the salt-treated, restriction-digested pellets, primarily resolved at >10-kb molecular mass (Fig. 1, lane 6; micrococcal nuclease digested insoluble [MNDI]). Total DNA extracted from sperm nuclei using standard methods without any prior treatment (Fig. 1, lane 2 [NF]) was predominantly >10 kb.
Figure 1.
Analysis of DNA fractions obtained by SRD and MND treatment of sperm nuclei. All DNA samples were resolved on 1.8% agarose gels. (Lane 1) A 0.4–10-kb DNA ladder. (Lane 2) Total (unfractionated) DNA (NF). (Lane 3) The soluble DNA released after extraction of sperm nuclei with 0.65 M NaCl followed by digestion with BamHI and EcoRI (SRDS). (Lane 5) The corresponding insoluble pellet (SRDI). (Lane 4) DNA released from sperm after MNase digestion (MNDS). (Lane 6) The corresponding insoluble pellet (MNDI).
CGH-chip profiling
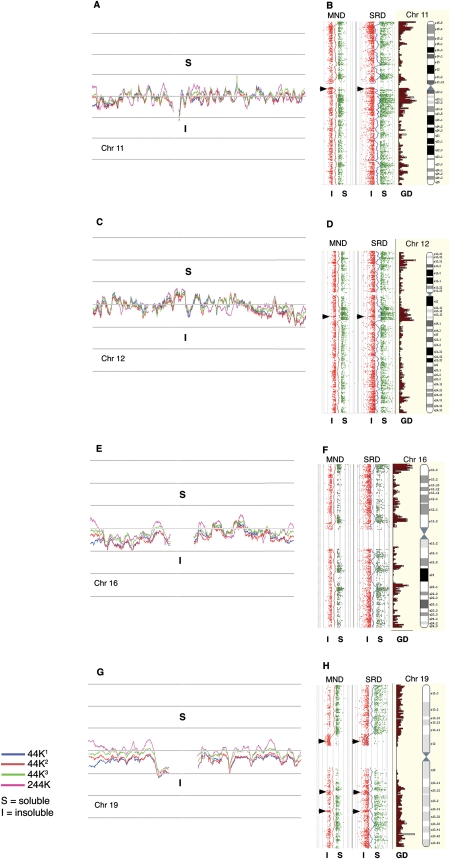
We first used Agilent's whole-genome 44K CGH tiling array with a median probe spacing of 35 kb to examine the DNA composition of human SRD fractions from individual semen samples provided by four unrelated human donors. These were analyzed on 10 arrays, eight of which were duplicates subjected to reciprocal swapping of the Cy3 and Cy5 dyes to control for probe-generated artifacts (none were noted). The remaining arrays were controls using non-fractionated DNA against soluble and insoluble targets (data not shown). Figure 2 shows examples with moving average profiles derived from the Cy3/Cy5 log2 ratios (log2R). Values >0 indicate a comparatively greater abundance of target sequences in the SRDS fraction; conversely, values <0 indicate a comparatively greater abundance of sequences in the SRDI fraction. From the close alignment of the respective traces, obtained from example chromosomes 11, 12, 16, and 19 shown in the left-hand panels (Fig. 2A,C,E,G), it is clear that the SRDS and SRDI fractions from each of the three individual ejaculate samples (44K1, 44K2, and 44K3) displayed have a high level of compositional similarity (rs = 0.95 [441 vs. 442], 0.94 [441 vs. 443], and 0.96 [442 vs. 443]). Fractions from samples provided by two further unrelated donors analyzed on a higher-resolution platform (244K; median 8 kb probe spacing) generated moving average traces aligning closely with the 44K traces (rs = 0.78), showing that the 244K array provides essentially similar output but at higher resolution. Moreover, linear regression analysis showed the 244K MND and SRD data sets were positively correlated (rs = 0.52). These results demonstrate the reproducibility of the techniques used to sample sperm chromatin, the similarity in composition between the samples from different men, and the equivalence in processed data between the 44K and the 244 K CGH platforms.
Figure 2.
Whole chromosome CGH plots (human). (A,C,E,G) The log2R moving average profiles obtained from the 44K CGH analysis of SRDS/SRDI fraction pairs of three individual men (44K1, 44K2, and 44K3) for chromosomes 11, 12, 16, and 19. The moving average profile from an additional fraction pair analyzed on the 244K CGH platform is also shown. (B,D,F,H) The 244K probe plots from SRDS/SRDI and MNDS/MNDI data as individual log2R values >0.5; (green) soluble (S); (red) ≤0.5 insoluble (I) fractions aligned alongside Ensembl gene density profiles and chromosome ideograms (http://www.ensembl.org). Moving average traces that are dynamically equivalent to those drawn in panels A, C, E, and G are drawn between the fraction pairs. (Arrowheads) Olfactory receptor gene clusters on chromosomes 11 and 12 and zinc finger clusters on chromosome 19. With the exception of these cluster types, the tendency for the SRDS/MNDS probes to correspond closely with gene density profiles is clearly evident.
The scatterplots shown in Figure 2, B, D, F, and H, are from the 244K log2Rs generated from target fractions where ratios >0.5 (soluble [S]; green) are plotted to the right of the moving average traces and ratios <−0.5 (insoluble [I]; red) are plotted to the left. Ratios lying between these cutoffs are not plotted. Two profiles are shown representing MND and SRD fractions corresponding to the DNA profiles shown in Figure 1 (i.e., SRDS vs. SRDI and MNDS vs. MNDI). The gene density plots (GDs) for each chromosome are drawn alongside their respective ideograms. The plots suggest that the soluble DNA fractions obtained following the SRD and MND treatments outlined above (Fig. 1, lanes 3,4) are enriched for gene sequences compared with their corresponding insoluble fractions (Fig. 1, lanes 5,6). However, this trend is reversed for olfactory receptor (OR) and zinc finger (ZNF) gene clusters, indicated by arrowheads in Figure 2, B, D, and H, respectively, which are enriched in the insoluble DNA fractions (arrowheads). A similar trend for gene-dense regions to partition preferentially into the MNDS fraction was observed for MND-processed mouse sperm (Fig. 3). Murine olfactory receptor (Olfr) clusters are also found in the MNDI fractions (arrowhead). An analysis of probe signal intensities (Fig. 4) shows that the ranges for SRDS/MNDS and for SRDI/MNDI correspond closely (median soluble/insoluble ratio = 3.57 and 3.43 for SRD and MND, respectively). Moreover, high-intensity signals in each data set are more heavily represented in the soluble fractions, with a far larger number of probes.
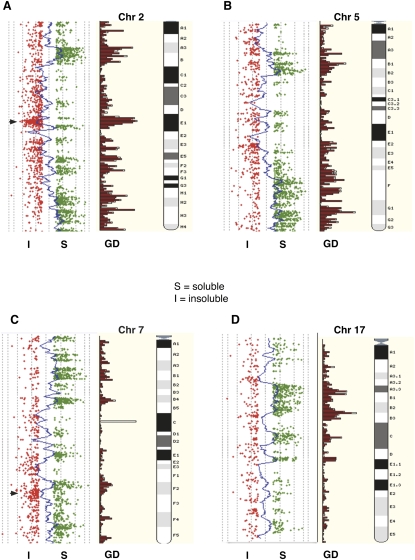
Figure 3.
Whole chromosome CGH scatterplots (mouse). The log2R moving averages from murine MNDS/MNDI data generated on the Agilent mouse 244K CGH platform are shown for chromosome 2, chromosome 5 (B), chromosome 7 (C), and chromosome 17 (D). The probe profiles from MNDS/MNDI fractions as individual log2R values >0.5; (green) soluble (S); (red) ≤0.5 insoluble (I) are aligned alongside Ensembl gene density profiles and chromosome ideograms. As with the human data, the tendency for the MNDS probes to correspond closely with gene density profiles is evident. (Arrowhead) A prominent olfactory receptor gene cluster on chromosome 2 where this trend (as in human sperm chromatin) is reversed.
Figure 4.
Probe signal intensities from MND and SRD fractions. Boxplots showing the range for all probes in all fractions with at least a twofold change in probe signal intensity from the median value (black bars). The plots show clearly that the highest numbers of high signal intensities are in the soluble fractions.
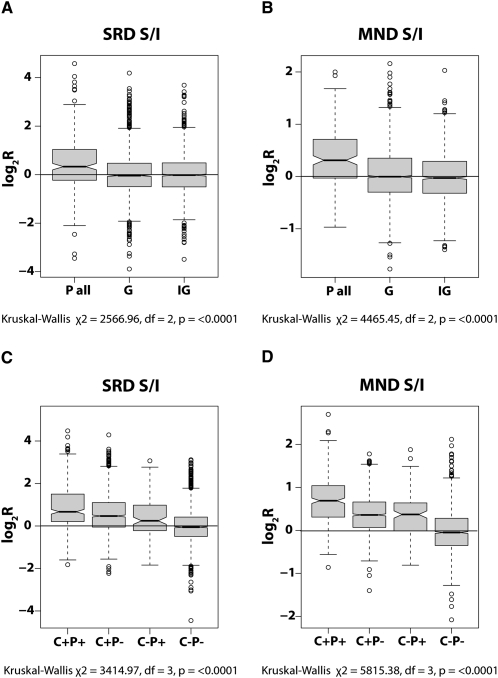
In order to examine the apparent association between packaging preference and gene density in more detail, we used the software tools available at the Galaxy resource (http://galaxy.psu.edu/) to map the CGH probe coordinates to the NimbleGen promoter and gene coordinates for human and mouse. The log2R values of all human probes located within promoter sequences (P-all) were compared with those located within genes (genic; G) or outside both genes and promoters (intergenic; IG) (Fig. 5A,B). Median log2R values of 0.28 (MND) and 0.32 (SRD) for the P-all data contrasted sharply with the median values of −0.01 | −0.03 (MND | SRD) for genic and −0.02 | −0.02 (MND | SRD) for intergenic data. These results indicate that promoter sequences are significantly enriched in the soluble fractions, while surprisingly, genic and intergenic sequences both show no apparent enrichment in either fraction (data summarized in Table 1). A similar, although somewhat less distinct, partitioning is observed in the mouse (Supplemental Table 1), where the median log2R value for probes within promoter sequences is 0.19, as opposed to 0.09 in genic sequences and −0.03 for probes located outside genes and promoters (Kruskal-Wallace, χ2 = 3325.1, degree of freedom [df] = 2, P < 0.0001). Thus, genic and intergenic sequences show little partitioning bias, and the apparent association with gene density shown in Figures 2 and 3 must in part be due to the greater accessibility of promoter sequences to salt/endonuclease and MNase treatments.
Figure 5.
Gene regulatory sequences in MND and SRD fractions. Boxplots showing the partitioning behavior of sequences detected by all probes located within promoters (P all) in the SRD (A) and MND (B) fractions compared with those within genic (G) and intergenic (IG) sequences. The partitioning behavior of sequences detected by probes within CTCF clusters in association with (C+P+) or without (C+P−) promoters is shown for SRD (C) and MND (D) fractions compared with the behavior of promoters alone (C−P+; equivalent to “P all” in A and B) or the absence of both promoters and CTCF clusters (C−P−; equivalent to G/IG in A and B). The center of the boxes indicates the median value with the first and third quartiles drawn on either side. Notches indicate significant differences in median values (Tukey's honest significance difference). In all plots, log2R values >0 indicate solubility and <0 indicate insolubility. Note that the greatest skew toward solubility is for probes detecting sequences located within promoters and CTCF clusters, followed by probes with CTCF clusters alone, and then within promoters alone.
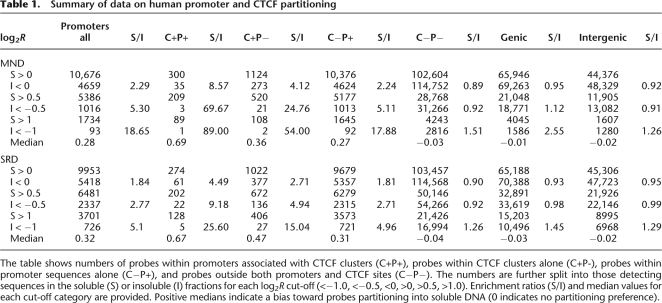
Table 1.
Summary of data on human promoter and CTCF partitioning
The table shows numbers of probes within promoters associated with CTCF clusters (C+P+), probes within CTCF clusters alone (C+P-), probes within promoter sequences alone (C−P+), and probes outside both promoters and CTCF sites (C−P−). The numbers are further split into those detecting sequences in the soluble (S) or insoluble (I) fractions for each log2R cut-off (<−1.0, <−0.5, <0, >0, >0.5, >1.0). Enrichment ratios (S/I) and median values for each cut-off category are provided. Positive medians indicate a bias toward probes partitioning into soluble DNA (0 indicates no partitioning preference).
Notable exceptions to the general accessibility of promoters to our extraction procedures, however, are the promoters for olfactory receptor genes in both humans and mice, and the promoters of ZNF gene clusters in humans. Median log2R values for probes located within human OR promoters were −0.34 (MND) and −0.75 (SRD), and −0.22 in the mouse Olfr promoters. These entire regions, including both promoter and genic sequences in both species, are predominantly located in the most insoluble chromatin (log2R < −1), of which typical clusters are shown for human chromosomes 11 and 12, and mouse chromosomes 2 and 7 (Supplemental Fig. 1).
CTCF binding sites
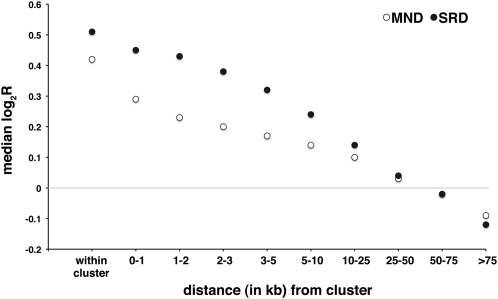
The data suggest that promoter sequences in human and murine sperm nuclei are more accessible to applied endonucleases than other sequences in sperm chromatin. This may explain the observed relationship between the highest soluble fraction signals and gene density profiles. However, regions of open chromatin architecture, as indicated by increased sensitivity to DNase I digestion, exist distal to known and novel genes (The ENCODE Project Consortium 2007). Distal DNase I–hypersensitive sites (DHSs) exhibit epigenetic signatures distinct from those of transcription start sites (TSS) and are characterized by high CTCF occupancy, depletion in H3K4me3, and the presence of histone H3 (K9 acetylated) (H3ac) and histone H4 (K12 acetylated) (H4ac) (The ENCODE Project Consortium 2007). In view of the key role played by CTCF in the three-dimensional organization of chromatin and in the establishment and maintenance of imprinting phenomena (Kuhn and Geyer 2003; Ishihara et al. 2006; Kim et al. 2007), we sought to establish the partitioning behavior of sequences located within CTCF binding sites as defined by previous studies (Kim et al. 2007; Chen et al. 2008) following SRD and MND extraction procedures. Interestingly, the median log2R values of soluble versus insoluble fractions for probes that mapped within regions of CTCF binding in the human genome are 0.42 (MND) and 0.51 (SRD), indicating that in human sperm, CTCF-binding sites are significantly enriched in chromatin domains with an open, accessible architecture. Moreover the chromatin flanking CTCF-binding sites is also significantly enriched in the soluble fractions, and this effect extends over several kilobases (Fig. 6). After mapping probes to discrete intervals away from CTCF-binding sites, the interval median log2R values approach the data set median only after moving more than 50 kb away from a CTCF-binding site. Furthermore, probes located more than 75 kb from any CTCF cluster detect sequences enriched in the insoluble fractions (median log2R = −0.09 [MND] and −0.12 [SRD]). The median log2R value for the equivalent mouse MND sample is 0.40, indicating that CTCF-binding sites are likewise enriched in the soluble fraction from mouse sperm. Furthermore, sites of CTCF at the murine H19/Igf2 (Szabo et al. 2004) and the Dlk1/Meg3 (also known as Gtl2) loci (Hiura et al. 2007) are strongly enriched in the soluble fractions from both human and murine spermatozoal chromatin (Supplemental Fig. 2).
Figure 6.
Regions flanking sites of CTCF binding are also enriched in soluble chromatin. The plot shows that the strong preference for CTCF clusters to partition into soluble SRD and MND fractions is shared by sequences flanking CTCF binding sites and extends for several kilobases. Note that median log2R values only return toward the median for the entire data sets after 50 kb and that sequences detected by probes lying in regions more than 75 kb from any sites of CTCF binding are actually enriched in insoluble chromatin (Kruskal-Wallis; χ2 = 534.5, df = 9, P < 0.0001).
Since CTCF-binding sites are also located close to many TSS, we also sought to assess the accessibility of chromatin in the vicinity of both promoters and CTCF sites. We collated the CGH data for unique probes located both in CTCF-binding sites and within promoters (C+P+), within CTCF-binding sites but outside promoters (C+P−), outside CTCF-binding sites but within promoters (C−P+), and probes located outside both CTCF sites and promoters (C−P−). As shown in Figure 5, C and D, and summarized in Table 1, C+P+ probes detect target sequences even more strongly enriched in the soluble fractions than in either domain alone. Median log2R values of 0.69 and 0.67 represent a 4.5–8.5× over-representation of target sequences in the SRD and MND soluble fractions, respectively, while median values of target sequences detected by the C+P− probes (0.47 and 0.36; SRD and MND, respectively) are markedly higher than those detected by the C−P+ probes (0.31 and 0.26; SRD and MND, respectively). Median values for sequences detected by C−P− probes (−0.03 and −0.04; SRD and MND, respectively) are close to those of the whole data set. These results suggest that regions encompassing CTCF binding sequences likely play an important role in determining chromatin accessibility both within and outside known promoters. A similar relationship is also apparent for mouse probes segregated into the same categories (Supplemental Table 1). Median log2R values for sequences detected by C+P+ (0.39) and C+P− (0.41) probes are clearly more enriched in endonuclease-sensitive chromatin than sequences detected by C−P+ probes (0.18).
ChIP-chip profiling
The partitioning data showed an enrichment of gene regulatory sequences in the endonuclease soluble chromatin fractions, and DNA electrophoresis data indicated the presence of nucleosomal DNA in the MNDS fraction. These regions likely contain H4K12ac, a feature of paternal chromatin (Hazzouri et al. 2000a) that persists in mouse zygotes (van der Heijden et al. 2006). To test this tenet, an antibody directed against H4K12ac was used to precipitate H4ac-associated DNA from total sperm chromatin for hybridization to the NimbleGen human HG18 promoter arrays (http://www.nimblegen.com). Genome-wide colocalization of H4K12ac binding and endonuclease accessibility was significantly correlated (rs = 0.42, P < 0.0001), indicating that despite the very different methods of chromatin separation, the CGH and ChIP data sets are substantially equivalent. In agreement with the observed enrichment of OR gene clusters in the insoluble fractions, human OR promoters are also depleted in H4K12ac (Supplemental Fig. 1A,B, center tracks).
Gene Ontology
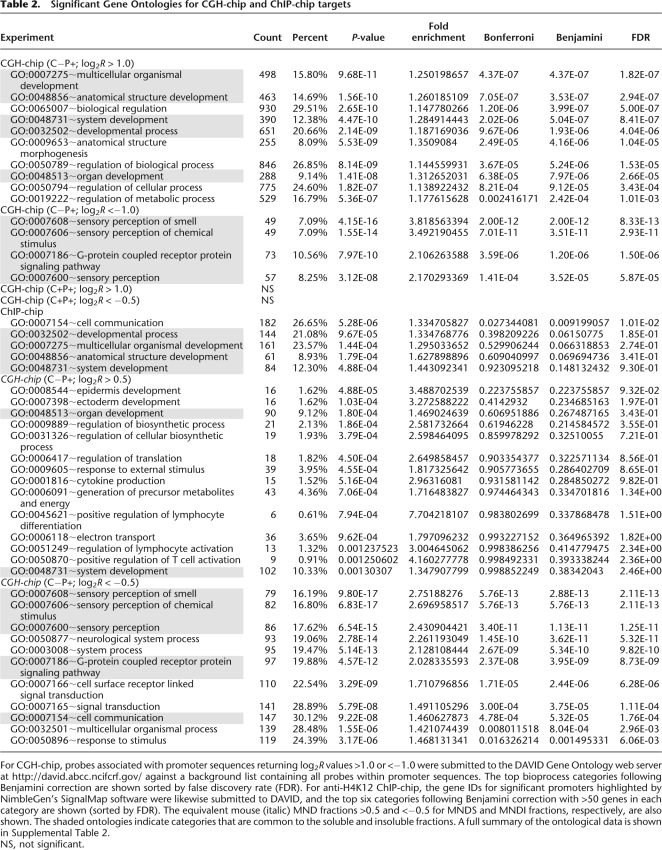
As gene regulatory sequences in sperm chromatin are apparently more sensitive to endonuclease digestion, we considered whether the most soluble (and insoluble) target sequences contain significant ontological information. A list of the highest human log2R > 1 (soluble) and ≤1 (insoluble) signals in the SRD probe subset containing promoter but not CTCF target sequences (C−P+) (see Fig. 6C; Supplemental ontology information file) were submitted to the DAVID Ontology server (http://david.abcc.ncifcrf.gov/) against a background list of all target sequences in this subset. Table 2 shows the main ontological categories after Benjamini correction and false discovery rate (FDR) calculation (the value by which the entries are sorted) to improve the stringency of significance. Genes involved in developmental processes, and not spermatogenesis, dominate the categories in the soluble fractions with an equally strong enrichment for olfactory receptor and ZNF genes in the insoluble fractions. The ontology of outliers in the mouse CGH data set was less pronounced for the soluble fractions, although as with human, genes involved in developmental processes were represented in the dominant categories (Table 2). Also in common with the human ontologies is the observation that genes involved in sensory perception, including the olfactory receptors, are heavily enriched in the mouse insoluble fractions. Although the most soluble targets are associated with both promoter and CTCF binding sequences, these do not contain a significant ontological signature even after removal of the small number of insoluble outliers in this subset. Similarly, outlier targets containing CTCF binding but not promoter sequences (C+P−) have no significant ontology. There is no evidence for either high or low levels of gene expression in any ontological category in data archived on GEO, derived from either embryonic stem cell or trophoblastic cell lines (http://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS2832 and http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2531). Hence, while an intriguing possibility, spermatozoal ontological profiles appear unrelated to differential gene expression in either embryonic or trophectodermal tissues.
Table 2.
Significant Gene Ontologies for CGH-chip and ChIP-chip targets
For CGH-chip, probes associated with promoter sequences returning log2R values >1.0 or <−1.0 were submitted to the DAVID Gene Ontology web server at http://david.abcc.ncifcrf.gov/ against a background list containing all probes within promoter sequences. The top bioprocess categories following Benjamini correction are shown sorted by false discovery rate (FDR). For anti-H4K12 ChIP-chip, the gene IDs for significant promoters highlighted by NimbleGen's SignalMap software were likewise submitted to DAVID, and the top six categories following Benjamini correction with >50 genes in each category are shown (sorted by FDR). The equivalent mouse (italic) MND fractions >0.5 and <−0.5 for MNDS and MNDI fractions, respectively, are also shown. The shaded ontologies indicate categories that are common to the soluble and insoluble fractions. A full summary of the ontological data is shown in Supplemental Table 2.
NS, not significant.
Among the 714 genes whose promoters are most significantly enriched in H4K12ac binding as identified by the ACME algorithm (Scacheri et al. 2006), we found a wider albeit weaker range of significant ontologies than from nuclease-derived targets (see Supplemental ontology data file). However, the most significant categories containing the highest numbers of genes in the ChIP-chip data are dominated by the same developmental ontologies as represented in the CGH data (Table 2), suggesting that developmental processes are dominant features of and common to both data sets.
Discussion
Previous studies have shown that differential DNA packaging in mammalian sperm results from a highly selective and specific partitioning of genomic sequences into nucleosomal and nucleoprotamine compartments, which reportedly coexist at ratios of 15:85 in human and 2:98 in murine spermatozoa (Balhorn et al. 1977; Bench et al. 1996), but vary widely in other mammalian species (Soon et al. 1997). Chromatin packaged by nucleoprotamines is 10 times more condensed than heterochromatin (Balhorn et al. 2000) and achieves a level of compaction necessary to fit the paternal genome into the small spermatozoal nucleus—a structure dynamically optimized to facilitate sperm motility (Ostermeier et al. 2001). There is also good evidence for the existence of chromosomal “territories” in sperm nuclei, suggesting that chromosomes are not arranged randomly (Zalensky et al. 1995; Hazzouri et al. 2000b). Although mature spermatozoa are transcriptionally inert, previous studies (Gatewood et al. 1987; Gardiner-Garden et al. 1998) have shown that the HBE1 and HBG1/2 (hemoglobin gamma 1 and 2) gene members of the beta-globin cluster, which are expressed in the embryo, but not the HBB (hemoglobin beta) and HBD (hemoglobin delta) genes (expressed in fetal and adult life), remain packaged by histones. The promoter sequences of the PRM2 and TNP2 genes have also been reported (Wykes and Krawetz 2003) to be among the histone-packaged regions of the PRM1-PRM2-TNP2 (protamine 1–protamine 2–transition protein 2) locus on human chromosome 16. Indeed, it has been proposed that spermatozoal chromatin may be organized in a fashion analogous to active somatic cell chromatin, although “frozen” in a transcriptionally inactive state (Ward and Zalensky 1996; Kramer et al. 2000).
Nuclease digestion of chromatin is a well-established and widely used tool for the investigation of chromatin architecture and DNA accessibility (Sanders 1978). Recently, a combination of brief nuclease digestion (MNase) followed by washing somatic cell nuclei in increasing concentrations of NaCl has been used successfully to probe the composition of somatic cell chromatin and identify active chromatin domains (Henikoff et al. 2009). Our application of similar tools to analyze the conformational organization of sperm chromatin shows that the composition of soluble and insoluble compartments in samples from six unrelated men is essentially identical (Fig. 2). The location of soluble regions in the sperm nucleus can be inferred from the methods used to generate the fractions. Nucleases are relatively large proteins that only readily gain access to intact nuclear chromatin located close to the nuclear periphery; hence, it is likely that soluble chromatin originated from there. In support of this, Li et al. (2008) used a probe to detect human telomeric DNA, known to be histone-bound (Zalenskaya et al. 2000), and localize it to a narrow zone at the posterior end of the human sperm nucleus. An earlier report provided immunohistochemical evidence that murine spermatozoal histones are located at the nuclear periphery (Pittoggi et al. 1999). However, these observations do not exclude the possibility that sequences packaged by nucleosomes may also be embedded more deeply in the sperm nucleus and protected by protamine-packaged chromatin from nuclease digestion. Our ChIP-chip data, which are independent of positional effects, show a good regional correlation between H4K12ac enrichment and endonuclease accessibility, and H4K12ac is also depleted in many predominantly insoluble regions (Supplemental Fig. 1). The ontologies of genes represented in H4K12ac-associated chromatin and endonuclease-accessible chromatin are also similar, indicating that our biochemical and immunoprecipitation approaches access equivalent chromatin compartments.
Although chromatin accessibility appeared highly correlated with gene density at a chromosomal scale (Figs. 2 and 3), this may have arisen from the known higher nuclease sensitivities of promoters and other regulatory sequences (Davie and Saunders 1981; Sabo et al. 2006). To investigate this in more detail, we used publicly available genome-wide data sets of human and murine CTCF binding (Kim et al. 2007; Chen et al. 2008) and promoter coordinates (http://www.nimblegen.com/) to identify probes in our data set located within and close to these features. We noted a striking enrichment in the soluble fractions of sequences detected by probes located within promoters in both human and mouse chromatin. Equally striking, and perhaps unexpected, is our finding that sequences detected by probes located within genes and intergenic regions show no particular enrichment in either soluble or insoluble chromatin in both species. These results suggest that epigenetic signatures must exist at many promoter sequences that during mammalian spermatogenesis distinguish them for differential packaging.
Recently, it has been shown that in four different mouse cell lines, chromosomal regions actively transcribed by RNA polymerase II bear a characteristic H3K4me3–H3K36me3 signature at promoters and along the transcribed sequences (Mikkelsen et al. 2007; Guttman et al. 2009). It is highly likely that a similar epigenetic signature also marks genes actively transcribed during spermatogenesis. Although it is tempting to assume that the epigenetic signatures of actively transcribed genes may be responsible for the observed differential chromatin packaging in human and mouse spermatozoa, the results of our ontological analysis show that promoters of genes involved in developmental, and not reproductive processes, are enriched in the soluble fractions. Furthermore, many of the promoters enriched in soluble chromatin regulate genes that are not expressed during spermiogenesis. Therefore, epigenetic marks other than H3K4me3–H3K36me3 must operate to direct the differential repackaging of promoter sequences in human and mouse spermatozoa. Since H3ac and H4ac are moderately enriched at both TSS and distal DHSs in humans (The ENCODE Project Consortium 2007) and since our ChIP data show H4ac enrichment at many promoters represented in endonuclease-accessible chromatin and depletion in endonuclease-resistant chromatin, it is possible that histone acetylation may itself be one such epigenetic mark.
Although the observed enrichment of promoter sequences in the soluble fractions is in itself able to explain the correlation between gene density and chromatin accessibility, our results show that chromatin accessible to MND and SRD treatments also exists in the vicinity of CTCF binding sites both within and away from promoters. CTCF plays an essential role as a chromatin insulator in preventing the spread of heterochromatin along chromosomes and ensuring that juxtaposed genes are appropriately expressed (Dunn and Davie 2003). Most importantly, the factor is also closely involved in epigenetic remodeling during male germ cell development (Ishihara et al. 2006) and may therefore influence directly the eventual architecture of the male haploid genome, presumably through direct interactions with its binding sites early in spermatogenesis. Although CTCF has been reported to be present in mature murine spermatozoa (Tang and Chen 2006), CTCF does not bind methylated DNA (Mukhopadhyay et al. 2004). Since the male haploid genome is extensively methylated during spermiogenesis (Houshdaran et al. 2007), including many clusters of CTCF-binding sites enriched in the soluble fractions (Weber et al. 2007), it is unlikely that CTCF occupies its binding sites in mature spermatozoa. However, the testis-specific CTCF isoform CTCFL is expressed at a time when the male haploid genome undergoes extensive methylation (Loukinov et al. 2002) and can bind DNA regardless of its methylation status (Nguyen et al. 2008). It is therefore tempting to speculate that if CTCF is a key mediator of chromatin remodeling, CTCFL may be able to maintain the open chromatin conformations throughout the later, post-methylation stages of spermatogenesis when protamines eventually replace the bulk of nucleosomes packaging the male haploid genome. Once in the ooplasm, paternal CTCF binding sequences would be available to maternal CTCF, depletion of which is known to cause severe developmental abnormalities of the embryo (Wan et al. 2008).
The existence of a significant and complementary developmental ontology of gene-regulatory sequences present in spermatozoal chromatin accessible both to endonuclease digestion and to anti-H4K12ac immunoprecipitation is unlikely to be coincidental. Since spermatozoa are transcriptionally inert, this suggests there may be a function for this phenomenon after fertilization. If such a function exists, it does not appear to relate to a paternally derived preference for gene expression in trophectodermal versus embryonic tissues in the early embryo, as we found no suggestion for enrichment of any ontological category in either lineage (Supplemental Fig. 3). Imprinted genes where parental effects are well understood (Kerjean et al. 2000; Schaefer et al. 2007) tend to reside in endonuclease-accessible DNA; however, olfactory receptor (OR) genes, which tend to reside in endonuclease-inaccessible DNA, are also expressed monoallelically (Chess et al. 1994), but are not influenced by parent of origin. Instead, their expression is randomized for one or other allele by a Polycomb-mediated marking in embryonic stem cells (Alexander et al. 2007). As the likelihood of post zygotic monoallelic expression of OR is randomly determined, their clearly preferential packaging in sperm chromatin is all the more intriguing.
The data presented here suggest that an additional, genome-wide epigenetic signature is delivered by the spermatozoon to the oocyte at fertilization. Indeed, two recent studies showed that spermatozoal nucleosomes persist in the zygote and may carry paternally derived epigenetic information important for the developing embryo (van der Heijden et al. 2006, 2008). Together, these lines of evidence indicate a possible embryological requirement for the retention of nucleosomal chromatin at gene regulatory sites.
Methods
Samples and reagents
Human semen samples (following signed consent and approval by our local ethics review board) were obtained by masturbation into sterile, toxin-free receptacles. Following routine analysis, only normozoospermic samples (World Health Organisation 1999) were washed free of seminal plasma and processed as described below. Murine spermatozoa were obtained from the dissected epididymides of CD1 mice, allowed to swim out into HEPES-buffered synthetic oviduct fluid (HSOF). Cells were collected, washed free of HSOF with phosphate-buffered saline, and processed for chromatin fractionation as described below. Unless indicated otherwise, reagents were purchased from Sigma.
Chromatin fractionation (SRD: salt extraction and endonuclease digestion)
Soluble fraction (SRDS)
The method described originally by Gardiner-Garden et al. (1998) as adapted by Wykes and Krawetz (2003) was used throughout but with some minor modifications. In brief, semen was washed free of seminal plasma by diluting the semen 1:1 with HEPES-buffered HSOF and centrifuging for 10 min at 500g. Pellets were resuspended in HSOF and pelleted as before. This was repeated until the supernatants were clear. Pellets were resuspended in 100 mM NaCl, 10 mM Tris-HCl (pH 8.0), 1 mM EDTA containing 0.2 mM phenylmethylsulphonylfluoride (PMSF) and repelleted to remove HSOF. Pellets were subsequently processed as described previously with sequential extraction in 10% (w/v) mixed CTAB (alklytrimethylammonium bromide), followed by sequential incubation in 1% (w/v) CTAB in 10 mM Tris-HCl (pH 8.0) containing 0.05% (w/v) digitonin; Tris-buffered saline (TBS) containing 0.05% (w/v) digitonin, 10 mM Tris-HCl (pH 8.0) containing 1 mM EDTA, 0.05% (w/v) digitonin, EDTA-free protease inhibitor cocktail (Roche), and finally extraction in 0.65 M NaCl. The pellets containing ∼5 × 108 nuclei were then resuspended in BamHI buffer and digested with 2000 U each of EcoRI and BamHI (Invitrogen) for 90 min at 37°C with gentle agitation, allowing digested DNA to leach out of the nuclei. Following centrifugation, supernatants containing solubilized DNA were removed and treated with proteinase K (200 μg/mL) and SDS (0.5% [w/v]) for 10 h at 55°C. Prior to DNA extraction, RNAse A was added to the digest (10 mg/mL) and incubated for 1 h at 37°C. DNA was then isolated by extraction in phenol chloroform (1:1) followed by ethanol precipitation. Precipitated DNA was dissolved in TE buffer and stored at 4°C.
Insoluble fraction (SRDI)
The residual pellets were resuspended in BamHI buffer containing 0.05% digitonin and digested with ∼150 units each of BamHI and EcoRI for 90 min at 37°C with gentle agitation. Suspensions were centrifuged at 3000g for 5 min and the supernatants removed. Tris-buffered saline containing proteinase K (Bioline: 200 μg/mL), 0.5% (w/v) SDS was added to this suspension and incubated for 10 h at 55°C. DNA was isolated from the pellets as described above.
Chromatin fractionation (MND: micrococcal nuclease digestion)
The method of Zalenskaya et al. (2000) was adapted with minor modifications. Semen was washed free of seminal plasma in HSOF buffer and pellets resuspended in 10 mM Tris-HCl (pH 8.0) containing 0.05% (w/v) PMSF and 1% (w/v) CTAB to detach heads. Following centrifugation, pellets containing about 108 nuclei were resuspended in Mg2+- and Ca2+-free PBS, 0.5% (v/v) Triton X-100, and incubated for 10 min at 4°C. Following centrifugation, the pellets were resuspended in 1 mL of PBS containing 0.5% PMSF and 10 mM DTT and incubated for 30 min at 37°C. CaCl2 was added to a final concentration of 0.6 mM, and 5 units of micrococcal nuclease (Fermentas) was added. Digestion was stopped after 3 min of incubation at 37°C by the addition of EDTA to 5 mM, and samples were gently agitated for 30 min at 37°C to allow digested chromatin to leach out from the nuclei. Following centrifugation at 5000g for 10 min, supernatants (MNDS) and pellets (MNDI) were processed for DNA extraction as described above. A similar procedure was carried out to generate fractions from murine spermatozoa, although targets for CGH analysis were amplified beforehand using the Genomiphi whole-genome amplification (WGA) kit according to the manufacturer's instructions.
Chromatin fractionation (ChIP-chip)
Chromatin immunoprecipitation was performed on the spermatozoa from three fertile normozoospermic individuals using a ChIP assay kit (UpstateMillipore) with modifications due to the relatively low quantity of histones in spermatozoa. In brief, about 107 spermatozoa from each donor were separated from seminal plasma and washed twice in 1× Dulbecco's PBS by 5 min of centrifugation at 300g. Chromatin cross-linking was carried out by addition of formaldehyde to 1% for 10 min at room temperature with constant rotation followed by 10 min of incubation with 125 mM glycine to quench the cross-linking reaction. After fixation, sperm were pelleted (10 min at 300g) and lysed, and the DNA was sonicated to obtain an average fragment length of 200–1000 bp. Chromatin was then diluted 10-fold with the provided dilution buffer. Primary antibodies were purchased from Abcam and included a polyclonal antibody recognizing histone H4 acetylated at lysine 12 (H4K12ac; ab1761); a nonspecific rabbit polyclonal IgG (ab46540) was used as a negative control, and a polyclonal recognizing unmodified histone H3 (Abcam ab1791) was used as a positive control. Cross-links were reversed by addition of 5 M NaCl and incubation for 4 h at 65°C followed by proteinase K digestion for 1 h at 45°C. DNA was extracted by phenol/chloroform using ethanol precipitation with 3 μL of glycogen as inert carrier, resuspended in 30 μL of nuclease-free water, and stored at −20°C.
For the NimbleGen array, purified IP-probe (Qiaquick Purification Kit; QIAGEN) and 10 ng of input material (total chromatin) were prepared by adapting the protocol for whole-genome amplification using the Sigma GenomePlex WGA kit according to the manufacturer's instructions. Amplicons were applied to the NimbleGen HG18 human 5-kb promoter array (http://www.nimblegen.com). Labeling and hybridization of the probes for ChIP-chip analysis was performed by the NimbleGen Service ImaGenes GmbH including data extraction and normalization.
Agarose and protein gel electrophoresis and blotting
Samples of DNA (1–3 μg) were resolved on 0.8% agarose (Bioline) gels in TBE buffer, stained with ethidium bromide, and visualized on a Bio-Rad 500 Gel Documentation platform.
CGH profiling
CGH profiling was carried out according to the manufacturer's (Agilent) instructions. All DNA samples were double redigested with 10 units each of Alu1 and Rsa1 (New England Biolabs). Labeling with Cy3 or Cy5 (GE Healthcare, Amersham UK) was accomplished using a random priming method in the presence of Klenow DNA polymerase (Invitrogen) according to the recommended Agilent protocol. For hybridization, equal quantities of labeled DNA (500 or 1000 ng) were mixed with Cot-1 DNA (1 mg/mL) and incubated for 3 min at 95°C and then reincubated for 30 min at 37°C. The DNAs were applied to either Agilent's 44B (100 μL) or 244A (490 μL) arrays, sealed, and incubated for 40 h (244K arrays) or 24 h (44K arrays) at 65°C with constant rotation (20 rpm). Arrays were washed according to the manufacturer's instructions using Agilent buffers, dried, and scanned on an Agilent BA laser scanner. Data were recovered using Agilent's Feature Extraction software and visualized using CGH Analytics software. All data were transformed using a rank consistency filter method with linear normalization during feature extraction.
Statistical analysis
The linear regression and χ2 analyses of CGH data were based on standard algorithms written into the R suite of open source statistical packages. Graphic output was obtained using both R and Excel software. Probe coordinates from the 44K and 244K CGH data sets that overlapped with promoter sequences obtained from the NimbleGen twin promoter array data set and CTCF coordinates (Kim et al. 2007), were collated using scripts available at Galaxy (http://main.g2.bx.psu.edu/). Galaxy scripts were also used to ensure that each CGH probe was counted only once in any analysis. The gene list used for ontological analysis of ChIP-chip data was generated using the ACME algorithm from each of the three ChIP biological replicates used in this arm of the study (Scacheri et al. 2006).
Acknowledgments
This work was supported by a grant from the BBSRC (BBS/B/04900) to D.M., D.E.I., and M.H.B., and in part by a National Institute of Child Health and Development Grant HD36512 to S.A.K. We gratefully acknowledge the use of the Ensembl (http://www.ensembl.org) and UCSC Genome Browser (http://genome.ucsc.edu/) facilities that were used to contextualize our data. We thank David Westhead (Institute of Molecular and Cellular Biology, University of Leeds), Rupert Quinnel (Institute of Integrative and Comparative Biology, University of Leeds), and Marek Bartkuhn (Justus Liebig-Universität, Institut für Genetik, Giessen, Germany) for guidance on statistical testing.
Footnotes
[Supplemental material is available online at www.genome.org. The microarray data from this study have been submitted to NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession nos. GSM327832–GSM327833, GSM305271–GSM305280, GSM305091–GSM305092, GSM406528–GSM406531, and GSM394743.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.094953.109.
References
- Alexander MK, Mlynarczyk-Evans S, Royce-Tolland M, Plocik A, Kalantry S, Magnuson T, Panning B. Differences between homologous alleles of olfactory receptor genes require the Polycomb Group protein Eed. J Cell Biol. 2007;179:269–276. doi: 10.1083/jcb.200706053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhorn R, Gledhill BL, Wyrobek AJ. Mouse sperm chromatin proteins: Quantitative isolation and partial characterization. Biochemistry. 1977;16:4074–4080. doi: 10.1021/bi00637a021. [DOI] [PubMed] [Google Scholar]
- Balhorn R, Brewer L, Corzett M. DNA condensation by protamine and arginine-rich peptides: Analysis of toroid stability using single DNA molecules. Mol Reprod Dev. 2000;56:230–234. doi: 10.1002/(SICI)1098-2795(200006)56:2+<230::AID-MRD3>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Bench GS, Friz AM, Corzett MH, Morse DH, Balhorn R. DNA and total protamine masses in individual sperm from fertile mammalian subjects. Cytometry. 1996;23:263–271. doi: 10.1002/(SICI)1097-0320(19960401)23:4<263::AID-CYTO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Bench G, Corzett MH, DeYebra L, Oliva R, Balhorn R. Protein and DNA contents in sperm from an infertile human male possessing protamine defects that vary over time. Mol Reprod Dev. 1998;50:345–353. doi: 10.1002/(SICI)1098-2795(199807)50:3<345::AID-MRD11>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Braun RE. Packaging paternal chromosomes with protamine. Nat Genet. 2001;28:10–12. doi: 10.1038/ng0501-10. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Collas P, Poccia D. Remodeling the sperm nucleus into a male pronucleus at fertilization. Theriogenology. 1998;49:67–81. doi: 10.1016/s0093-691x(97)00403-2. [DOI] [PubMed] [Google Scholar]
- Davie JR, Saunders CA. Chemical composition of nucleosomes among domains of calf thymus chromatin differing in micrococcal nuclease accessibility and solubility properties. J Biol Chem. 1981;256:12574–12580. [PubMed] [Google Scholar]
- Dunn KL, Davie JR. The many roles of the transcriptional regulator CTCF. Biochem Cell Biol. 2003;81:161–167. doi: 10.1139/o03-052. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Ballesteros M, Gordon M, Tam PP. Histone- and protamine-DNA association: Conservation of different patterns within the beta-globin domain in human sperm. Mol Cell Biol. 1998;18:3350–3356. doi: 10.1128/mcb.18.6.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236:962–964. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- Gineitis AA, Zalenskaya IA, Yau PM, Bradbury EM, Zalensky AO. Human sperm telomere-binding complex involves histone H2B and secures telomere membrane attachment. J Cell Biol. 2000;151:1591–1598. doi: 10.1083/jcb.151.7.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald S, Paasch U, Glander HJ, Anderegg U. Mature human spermatozoa do not transcribe novel RNA. Andrologia. 2005;37:69–71. doi: 10.1111/j.1439-0272.2005.00656.x. [DOI] [PubMed] [Google Scholar]
- Gusse M, Sautiere P, Belaiche D, Martinage A, Roux C, Dadoune JP, Chevaillier P. Purification and characterization of nuclear basic proteins of human sperm. Biochim Biophys Acta. 1986;884:124–134. doi: 10.1016/0304-4165(86)90235-7. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazzouri M, Pivot-Pajot C, Faure AK, Usson Y, Pelletier R, Sele B, Khochbin S, Rousseaux S. Regulated hyperacetylation of core histones during mouse spermatogenesis: Involvement of histone deacetylases. Eur J Cell Biol. 2000a;79:950–960. doi: 10.1078/0171-9335-00123. [DOI] [PubMed] [Google Scholar]
- Hazzouri M, Rousseaux S, Mongelard F, Usson Y, Pelletier R, Faure AK, Vourc'h C, Sele B. Genome organization in the human sperm nucleus studied by FISH and confocal microscopy. Mol Reprod Dev. 2000b;55:307–315. doi: 10.1002/(SICI)1098-2795(200003)55:3<307::AID-MRD9>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genet Res. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiura H, Komiyama J, Shirai M, Obata Y, Ogawa H, Kono T. DNA methylation imprints on the IG-DMR of the Dlk1-Gtl2 domain in mouse male germline. FEBS Lett. 2007;581:1255–1260. doi: 10.1016/j.febslet.2007.02.034. [DOI] [PubMed] [Google Scholar]
- Houshdaran S, Cortessis VK, Siegmund K, Yang A, Laird PW, Sokol RZ. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS One. 2007;2:e1289. doi: 10.1371/journal.pone.0001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23:733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Kerjean A, Dupont JM, Vasseur C, Le Tessier D, Cuisset L, Paldi A, Jouannet P, Jeanpierre M. Establishment of the paternal methylation imprint of the human H19 and MEST/PEG1 genes during spermatogenesis. Hum Mol Genet. 2000;9:2183–2187. doi: 10.1093/hmg/9.14.2183. [DOI] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JA, McCarrey JR, Djakiew D, Krawetz SA. Human spermatogenesis as a model to examine gene potentiation. Mol Reprod Dev. 2000;56:254–258. doi: 10.1002/(SICI)1098-2795(200006)56:2+<254::AID-MRD9>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Kuhn EJ, Geyer PK. Genomic insulators: Connecting properties to mechanism. Curr Opin Cell Biol. 2003;15:259–265. doi: 10.1016/s0955-0674(03)00039-5. [DOI] [PubMed] [Google Scholar]
- Li Y, Lalancette C, Miller D, Krawetz SA. Characterization of nucleohistone and nucleoprotamine components in the mature human sperm nucleus. Asian J Androl. 2008;10:535–541. doi: 10.1111/j.1745-7262.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukinov DI, Pugacheva E, Vatolin S, Pack SD, Moon H, Chernukhin I, Mannan P, Larsson E, Kanduri C, Vostrov AA, et al. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc Natl Acad Sci. 2002;99:6806–6811. doi: 10.1073/pnas.092123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLay DW, Clarke HJ. Remodelling the paternal chromatin at fertilization in mammals. Reproduction. 2003;125:625–633. doi: 10.1530/rep.0.1250625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Yu W, Whitehead J, Xu J, Lezcano M, Pack S, Kanduri C, Kanduri M, Ginjala V, Vostrov A, et al. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genet Res. 2004;14:1594–1602. doi: 10.1101/gr.2408304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P, Cui H, Bisht KS, Sun L, Patel K, Lee RS, Kugoh H, Oshimura M, Feinberg AP, Gius D. CTCFL/BORIS is a methylation-independent DNA-binding protein that preferentially binds to the paternal H19 differentially methylated region. Cancer Res. 2008;68:5546–5551. doi: 10.1158/0008-5472.CAN-08-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier GC, Sargeant GA, Yandell BS, Evenson DP, Parrish JJ. Relationship of bull fertility to sperm nuclear shape. J Androl. 2001;22:595–603. [PubMed] [Google Scholar]
- Pittoggi C, Renzi L, Zaccagnini G, Cimini D, Degrassi F, Giordano R, Magnano AR, Lorenzini R, Lavia P, Spadafora C. A fraction of mouse sperm chromatin is organized in nucleosomal hypersensitive domains enriched in retroposon DNA. J Cell Sci. 1999;112:3537–3548. doi: 10.1242/jcs.112.20.3537. [DOI] [PubMed] [Google Scholar]
- Powell D, Cran DG, Jennings C, Jones R. Spatial organization of repetitive DNA sequences in the bovine sperm nucleus. J Cell Sci. 1990;97:185–191. doi: 10.1242/jcs.97.1.185. [DOI] [PubMed] [Google Scholar]
- Premkumar EB, Bhargava PM. Transcription and translation in bovine spermatozoa. Nat New Biol. 1972;240:139–143. doi: 10.1038/newbio240139a0. [DOI] [PubMed] [Google Scholar]
- Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- Sanders MM. Fractionation of nucleosomes by salt elution from micrococcal nuclease-digested nuclei. J Cell Biol. 1978;79:97–109. doi: 10.1083/jcb.79.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacheri PC, Crawford GE, Davis S. Statistics for ChIP-chip and DNase hypersensitivity experiments on NimbleGen arrays. Methods Enzymol. 2006;411:270–282. doi: 10.1016/S0076-6879(06)11014-9. [DOI] [PubMed] [Google Scholar]
- Schaefer CB, Ooi SK, Bestor TH, Bourc'his D. Epigenetic decisions in mammalian germ cells. Science. 2007;316:398–399. doi: 10.1126/science.1137544. [DOI] [PubMed] [Google Scholar]
- Soon LL, Ausio J, Breed WG, Power JH, Muller S. Isolation of histones and related chromatin structures from spermatozoa nuclei of a dasyurid marsupial, Sminthopsis crassicaudata. J Exp Zool. 1997;278:322–332. [PubMed] [Google Scholar]
- Szabo PE, Tang SH, Silva FJ, Tsark WM, Mann JR. Role of CTCF binding sites in the Igf2/H19 imprinting control region. Mol Cell Biol. 2004;24:4791–4800. doi: 10.1128/MCB.24.11.4791-4800.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JB, Chen YH. Identification of a tyrosine-phosphorylated CCCTC-binding nuclear factor in capacitated mouse spermatozoa. Proteomics. 2006;6:4800–4807. doi: 10.1002/pmic.200600256. [DOI] [PubMed] [Google Scholar]
- Tanphaichitr N, Sobhon P, Taluppeth N, Chalermisarachai P. Basic nuclear proteins in testicular cells and ejaculated spermatozoa in man. Exp Cell Res. 1978;117:347–356. doi: 10.1016/0014-4827(78)90148-9. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Derijck AA, Ramos L, Giele M, van der Vlag J, de Boer P. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev Biol. 2006;298:458–469. doi: 10.1016/j.ydbio.2006.06.051. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Ramos L, Baart EB, van den Berg IM, Derijck AA, van der Vlag J, Martini E, de Boer P. Sperm-derived histones contribute to zygotic chromatin in humans. BMC Dev Biol. 2008;8:34. doi: 10.1186/1471-213X-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan LB, Pan H, Hannenhalli S, Cheng Y, Ma J, Fedoriw A, Lobanenkov V, Latham KE, Schultz RM, Bartolomei MS. Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Development. 2008;135:2729–2738. doi: 10.1242/dev.024539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward WS, Coffey DS. DNA packaging and organization in mammalian spermatozoa: Comparison with somatic cells. Biol Reprod. 1991;44:569–574. doi: 10.1095/biolreprod44.4.569. [DOI] [PubMed] [Google Scholar]
- Ward WS, Zalensky AO. The unique, complex organization of the transcriptionally silent sperm chromatin. Crit Rev Eukaryot Gene Expr. 1996;6:139–147. doi: 10.1615/critreveukargeneexpr.v6.i2-3.30. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- World Health Organization. WHO laboratory manual for the examination of human semen and sperm–cervical mucus interaction. Cambridge University Press; Cambridge, UK: 1999. [Google Scholar]
- Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J Biol Chem. 2003;278:29471–29477. doi: 10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- Zalenskaya IA, Bradbury EM, Zalensky AO. Chromatin structure of telomere domain in human sperm. Biochem Biophys Res Commun. 2000;279:213–218. doi: 10.1006/bbrc.2000.3917. [DOI] [PubMed] [Google Scholar]
- Zalensky AO, Allen MJ, Kobayashi A, Zalenskaya IA, Balhorn R. Well-defined genome architecture in the human sperm nucleus. Chromosoma. 1995;103:577–590. doi: 10.1007/BF00357684. [DOI] [PubMed] [Google Scholar]