SUMMARY
Retinoic acid (RA) triggers growth-suppressive effects in tumor cells and therefore RA has and its synthetic analogs have great potential as anti-carcinogenic agent. RA effects are mediated by Retinoic Acid Receptors (RARs), which regulate gene expression in an RA-dependent manner. To define the genetic network regulated by RARs in breast cancer, we identified RAR genomic targets using chromatin immunoprecipitation and expression analysis. We found that RAR binding throughout the genome is highly co-incident with estrogen receptor α (ERα) binding, and identified a widespread crosstalk of RA and estrogen signaling to antagonistically regulate breast cancer-associated genes. ERα and RAR binding sites appear to be co-evolved on a large scale throughout the human genome, allowing for competitive binding between these transcription factors via nearby or overlapping cis-regulatory elements. Together these data indicate the existence of a highly coordinated intersection between these two critical nuclear hormone receptor signaling pathways providing a global mechanism for balancing gene expression output via local regulatory interactions dispersed throughout the genome.
INTRODUCTION
Retinoic acid (RA) plays a major role in physiological processes ranging from embryonic development to homeostasis of adult tissues and organs (Niederreither and Dolle, 2008). Importantly, RA inhibits the growth and survival of cancer cells at pharmacological doses. The potent anti-carcinogenic activity of RA is generally thought to result from direct and indirect effects on gene expression. Therefore a comprehensive analysis of the genomic targets of RA action should provide a better understanding of the mechanism of RA action in the prevention and treatment of cancer, as well as providing a framework that can be extended to other RA functions in organ development and homeostasis.
Previous work has identified two subfamilies of nuclear receptors as major mediators of RA signaling, the retinoic acid receptors (RARs) and the retinoid X receptors (RXRs) (Evans, 1988; Giguere et al., 1987; Mangelsdorf and Evans, 1995; Petkovich et al., 1987). RARs dimerize with RXRs; the resulting heterodimers function as transcription factors, thereby eliciting the transcriptional effects of RA signaling. However, little is known about the genomic targets and effects of the different isoforms of the RARs. There is similarly scant information about the mechanism or extent of crosstalk between RA signaling and other nuclear hormone signaling pathways in a cellular context.
In breast cancer cells, RA and retinoids have been previously shown to be associated with down-regulation of several genes essential for proliferation and survival (Liu et al., 1998; Zhou et al., 1997). However, it has been unclear if such genes are directly or indirectly regulated by RARs, since liganded RAR-RXR heterodimers are thought to function primarily as transcriptional activators while repression is thought to be primarily mediated by unliganded heterodimers interfering with basal transcription (Chambon, 1996; Glass and Rosenfeld, 2000; Hu and Lazar, 2000; Niederreither and Dolle, 2008). Also, RA-induced apoptosis in breast cancer cells has been shown to be associated with up-regulated expression of a handful of proapoptotic genes (Donato and Noy, 2005; Donato et al., 2007). However, although several genes implicated in the negative regulation of breast cancer cell proliferation and survival have been identified as RA-responsive, the knowledge of the mechanism of transcriptional regulation by RARs is fragmentary. Whether and how RA signaling intersects with estrogen signaling, which promotes proliferation, has not been investigated on a genomic scale.
We therefore analyzed the genomic actions of RA through RARα and RARγ, which exert anti-proliferative and apoptotic effects of RA in the breast cancer cell line MCF-7. MCF-7 is the most commonly used experimental system for the study of estrogen receptor α (ERα)-positive breast tumors, affording us the opportunity to compare the genomic effects of RA and estrogen signaling (Levenson and Jordan, 1997). Estrogen signaling, in contrast to RA signaling, drives proliferation and promotes survival and has been extensively studied in this cell line. To uncover the transcriptional networks of RARα and RARγ, we have integrated genome-wide binding site mapping with gene expression profiling. We found that RA signaling regulates the expression of many genes that have been implicated in breast carcinogenesis and/or whose expression is indicative for the clinical outcome of breast cancer. Interestingly, we found that RARα/RARγ exhibit extensive co-localization of their genomic binding regions with ERα in the vicinity of genes that are antagonistically regulated by estrogen and RA.
RESULTS
Identification of genomic RAR target sites
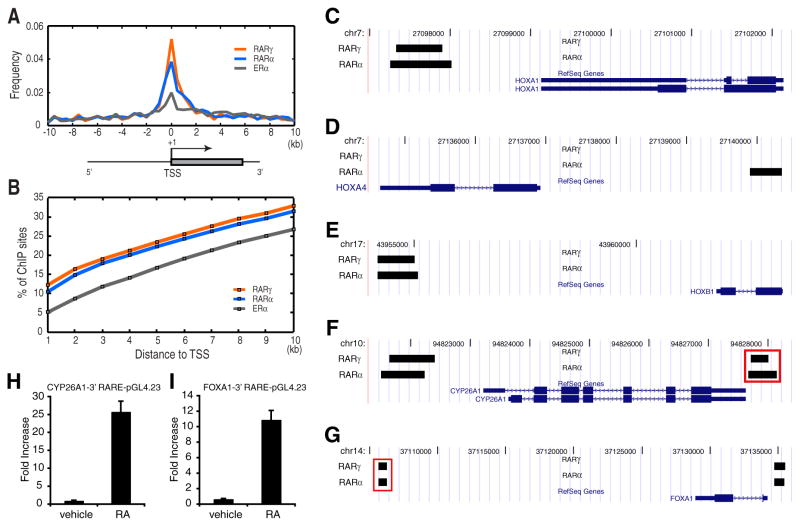
To define the RAR transcriptional network, we first mapped the genomic binding sites of RARα and RARγ. Because none of the commercially available isoform-specific antibodies were adequate to render high quality ChIP (data not shown) we used bacterial artificial chromosome (BAC) transgenesis (Poser et al., 2008) to generate two transgenic MCF-7 lines, which stably express RARα and RARγ tagged with eGFP at their C-termini at physiological levels (Figure S1). A total of 7,346 high-confidence RARα binding sites and 3,916 RARγ sites were identified by ChIP-chip analysis in the transgenic cells treated with synthetic receptor-selective agonists for one hour (Table S1 and Figure S2). We validated 40 randomly selected binding regions by ChIP-qPCR and found all of the tested regions significantly enriched compared to genomic input DNA (Figure S3) indicating a very low number of false-positives in the ChIP-chip experiments. The binding sites of the two isoforms showed a marked overlap as 3,238 (82.7%) of the RARγ sites were found to be within 1 kilobase (kb) to RARα sites. Since RARα is expressed at a higher level than RARγ, the larger number of sites for RARα might reflect an increased binding probability due to the higher abundance of this transcription factor. Overall, the large proportion of RARα/RARγ common target sites indicates a high degree of functional redundancy as suggested by mouse knockout studies (Lohnes et al., 1994).
Identification of RAR binding sites has previously focused on promoter and promoter-proximal regions of RA-regulated genes, and so far only a small number of direct targets are known (Balmer and Blomhoff, 2002; Niederreither and Dolle, 2008). However, we found only a small portion of RAR binding sites mapped to promoter-proximal regions (Figures 1A and 1B). Hence, most RAR binding sites were found in intronic or promoter-distal intergenic regions previously undefined as RAR binding sites. Within the list of RAR binding sites in promoter or in promoter-proximal regions, we confirmed a number of previously characterized functional RAR sites for known RA-inducible genes, including several HOX family genes (Figures 1C–1E), CYP26A1 (Figure 1F), and FOXA1 (Figure 1G) (Balmer and Blomhoff, 2002). Interestingly, we found additional novel RAR binding sites nearby some of these genes, such as 3′ binding sites for FOXA1 and CYP26A1 (Figures 1F and 1G). We tested whether these sites could act as regulatory elements using a luciferase reporter assay, and both were able to drive the reporter gene expression in an RA agonist-dependent manner (Figures 1H and 1I).
Figure 1. Genome-wide identification of RARγ and RARα binding sites in MCF-7 cells.
(A) Distribution of RARγ, RARα and ERα binding sites residing within 10 kb upstream or downstream to annotated transcription start sites (TSSs).
(B) Cumulative frequency of RARγ and RARα binding sites for each 1 kb interval within 10 kb upstream or downstream to known TSSs.
(C–G) Known RAR binding sites identified by ChIP-chip analyses. Black bars depict binding regions for RARγ and RARα. Known promoter-proximal RAR binding sites for HOXA1, HOXA4, HOXB1, CYP26A1, and FOXA1 were identified by genome-wide mapping in MCF-7. In addition, novel RAR binding sites 3′ to FOXA1 (F) and CYP26A1 (G) (denoted by red rectangles) were identified
(H,I) Novel binding regions for CYP26A1 (H) and FOXA1 (I) enhance expression of reporter constructs upon RA treatment. Upon RA agonist treatment these constructs markedly enhanced firefly luciferase expression compared to the original pGL4.23 construct. Error bars represent s.d.
RAR-dependent regulation of gene expression
To correlate the binding site data with the transcriptional effects of the RARs, we performed gene expression profiling after ligand treatment. Because the physiological ligand all-trans retinoic acid (ATRA) can elicit transcriptional effects independent from binding to RARs, e.g. through PPARδ (Schug et al., 2007), we generated expression profiles for ATRA, and RAR-selective agonists AM580 (RARα-specific) and CD437 (RARγ-specific). Comparisons between these expression profiles showed a high degree of correlation (Figure S4). CD437 and AM580 elicited similar transcriptional effects, consistent with the large overlap observed for the binding sites of RARα and RARγ.
To test whether the transcriptional response of the two selective agonists is mediated by RARs, we analyzed gene expression changes upon RAR depletion in the presence and absence of the agonists by RNAi. Knockdown of RARα and RARγ decreased or reverted most transcriptional changes caused by AM580 and CD437 (Figure 2A). This result demonstrates that both activation and repression of most genes in MCF-7 cells by RA agonists require RARs.
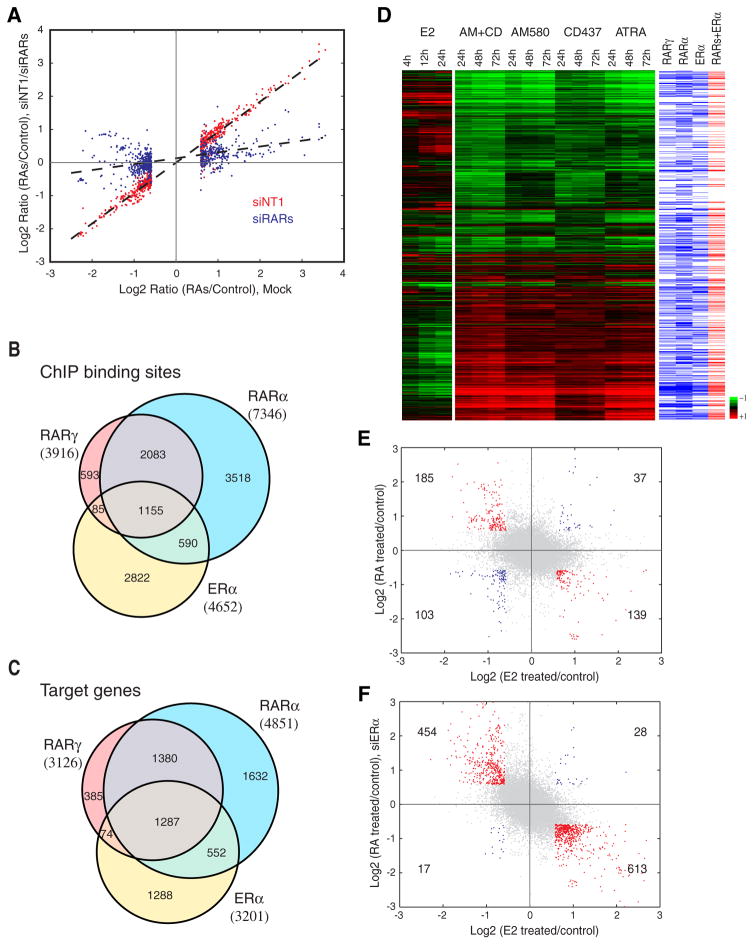
Figure 2. Co-localization of RARα, RARγ and ERα binding regions and antagonistic effects on gene expression between RA and estrogen signaling.
(A) Transcriptional response of RAs in MCF-7 cells is mediated by RARs. X-axis denotes Log2 transformed fold changes in gene expression after RA agonist treatment (100 nM AM580/CD437) relative to vehicle control (DMSO) treatment in mock RNAi experiments. X-axis shows Log2 transformed fold changes in gene expression after RA treatment relative to vehicle control treatment in mock RNAi experiment. Only genes with significant expression changes (1.5 fold change) were shown. Y-axis shows Log2 transformed fold changes in gene expression after RA treatment relative to vehicle control treatment in RARγ and RARα knockdown cells (siRARs, blue spots) and in RNAi control cells (siNT1, red spots).
(B) Venn diagram displaying shared regions bound by RARγ, RARα, and ERα. ERα binding sites are based on the union of two recent genomic studies (Carroll et al., 2006; Hua et al., 2008)
(C) Venn diagram displaying shared putative target genes of RARγ, RARα, and ERα, as defined by the presence of at least one binding region within 50 kb to the TSSs.
(D) Comparison of time-course gene expression profiles induced by estrogen and different RA agonist treatment for 1,413 RA regulated genes. Genes containing binding sites within 50 kb to the TSSs are denoted by blue (RARγ, RARα or ERα) and red bars (RARs and ERα).
(E,F) Comparison of gene expression changes in response to estrogen and RA agonists. X-axis shows Log2 transformed fold changes in gene expression after estrogen (10 nM E2) treatment relative to control (EtOH) treatment for 24 hours. Y-axis shows Log2 transformed fold changes in gene expression after RA agonist treatment (100 nM AM580 and 100 nM CD437) relative to vehicle control (DMSO) treatment for 72 hours in control-treated (siNT1) MCF-7 cells (E) and ERα-depleted (siER) MCF-7 cells (F). Genes with fold changes greater than 1.5 or less than -1.5 for both X- and Y-axes are highlighted in red or blue, respectively.
We analyzed expression changes after treatment with all individual ligands and the combination of AM580 and CD437 in triplicates over a time course (0, 24, 48, 72 hrs). We also compared the gene expression profiles upon ligand treatment in a gene expression time course aimed at identifying early-response direct targets (0, 4, 12, 24 hrs). We observed a relatively small number of significant transcript changes in the 0–24 hr time course compared to the 0–72 hr time course. Overall, we identified a total of 1,413 genes (Benjamini-Hochberg adjusted P <= 0.0005) (Table S3), which were significantly regulated by RA and RA agonists. 306 showed differential expression within the first 24 hours of ligand treatment. For a large proportion of transcripts differentially expressed in the 0–72 hr time course (46.5%) (hypergeometric test, P = 2.30e-140), we observed RAR binding sites within 50 kb to the TSS of the regulated gene, indicating that about half of the RA-regulated genes represent direct effects of liganded RAR rather than secondary effects. Previous work investigating the role of liganded RARs in the regulation of transcription has mainly focused on activation of expression, while the repressive function has been thought to be mediated mainly by unliganded RARs. However, down-regulated transcripts constitute a large fraction (52.8%) of RA-dependent expression changes in MCF-7 cells; and we observed no marked bias of RAR binding toward ligand activated or repressed genes (52.5% and 41.2%, respectively). RAR regions are highly significantly enriched in both up- and down-regulated genes (P = 4.03e-92 and P = 2.20e-50, respectively). Further, we demonstrate for six putative RAR direct target genes, which were significantly down-regulated or up-regulated by RA agonists that both RA-mediated repression and activation do not require de novo protein synthesis (Figure S5). Collectively, these findings support the hypothesis that both activation and repression involves binding of liganded RARs at target genes.
ERα and RAR binding regions co-localize and mediate antagonistic actions on gene expression
We and others have mapped ERα binding genome wide in MCF-7 cells (Carroll et al., 2006; Hua et al., 2008; Lin et al., 2007). When we compared RAR binding regions with ERα regions, we found a marked co-localization. 39.3% of ERα regions were observed within 1 kb of RAR binding regions (Figure 2B). At the gene level there was even a larger overlap; ERα and RARs share 59.8% of their putative target genes as defined by the presence of at least one binding region within 50 kb to the TSS (Figure 2C).
The extensive co-localization of RAR and ERα genomic binding sites suggested potential crosstalk of RA and estrogen signaling in the regulation of gene expression. To systematically identify transcripts that are differentially regulated by RA agonists and estrogen, we analyzed changes in gene expression after treatment with estrogen, and compared these results with our RA agonist data (Figures 2D and 2E). We found 139 genes down-regulated by RA agonists to be up-regulated by estrogen, while 185 estrogen-repressed genes were up-regulated by RA agonists. A considerably smaller number of genes were up-regulated (37) or down-regulated (103) by both estrogen and RA agonists. Thus, the RA and estrogen signaling pathways appear to mainly antagonize each other. We further validated this result by testing the effects of RA agonists before and after the RNAi depletion of ERα. Knockdown of ERα increased the number of both up-regulated and down-regulated transcripts upon RA treatment (Figure 2F). We also analyzed the effects of single treatment with RA agonists and E2, or simultaneously co-treatment on the expression of nine individual target genes that were associated with unique RAR or ERα, and ERα/RAR binding regions (Figure S6). RA agonists and E2 had an antagonistic effect on the expression levels of common target genes but not on unique targets of RARs or ERα, whose expression levels were affected by RA agonists or E2, respectively (Figure S6). Collectively, these findings indicate an extensive crosstalk of ERα and RARs to regulate gene expression. However, despite their opposing effects on the majority of target genes, ERα and RARα appear to activate each other. We observed ERα binding in the proximity of the RARA TSS (Figure S12A) and up-regulated expression of RARA upon estrogen treatment. Likewise RARα bound near the TSS of ESR1 (Figure S12B) and RA agonist treatment led to up-regulated expression of ESR1. This cross-regulation between the two antagonizing transcription factors presents an additional level of control for achieving a balanced regulation of gene expression by the two signaling pathways.
Antagonistic actions of RARs and ERα bound to shared regulatory elements
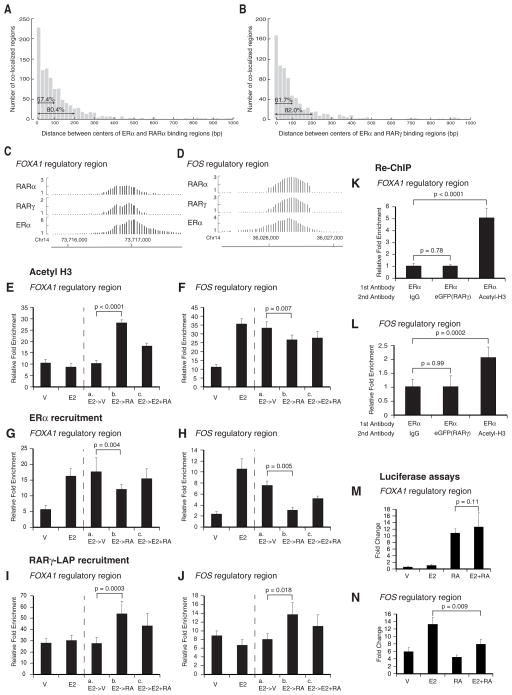
In order to determine the major mechanism of the global ERα/RAR antagonism we performed a series of additional computational analyses and experiments. First, we analyzed the distance between the putative binding sites of ERα and RARs in overlapping binding regions. Using the center of a binding region as the putative binding site, we found that most ERα and RAR binding sites occur within 100 nt (Figure 3A and 3B). Considering the resolution limit of ChIP-chip this finding indicates that most binding sites of ERα and RARs occur very close to each other, overlap or are identical. This finding suggests competitive binding for the same genomic binding sites or steric hindrance between close sites. By manual inspection of ERα and RAR binding events using normalized ChIP-chip intensities we found that many overlapping regions have nearly identical peaks (Figure S7). RARα as well as RARα-RXR dimers bind in vitro to synthetic and natural EREs (IR3) by gel shift assays (Klinge et al., 1997; (Naar et al., 1991), and can compete with ERα for binding to EREs (Joyeux et al., 1996) (Kittler, Hua and White, data not shown). To test the model of competitive binding in vivo we chose two example elements where RARs and ERα overlap: the first was a putative regulatory element of FOXA1, whose gene expression is up-regulated by RA agonists and down-regulated by E2 (Figure 3C). The second was a putative regulatory element of FOS, whose gene expression is up-regulated by E2 and down-regulated by RA agonists (Figure 3D). We tested the effects of RA agonists and E2 treatment on H3 acetylation and ERα and RARγ recruitment at these two ERα/RAR binding regions. E2 decreases and RA agonists increase H3 acetylation for the FOXA1 element (Figure 3E), while opposite effects were observed for the FOS regulatory element (Figure 3F) indicating that the antagonistic effects of RA and E2 on gene expression are mediated through opposite effects on cofactor recruitment to common ERα/RAR sites. These opposite effects on H3 acetylation were also correlated with changes in ERα and RARγ recruitment. Upon initial E2 treatment ERα recruitment was found to be increased for both regions, but subsequent treatment with RA agonists in absence of E2 led to a decrease in ERα binding and an increase in RARγ binding, which could be reverted by simultaneous co-treatment with E2 and RA agonists (Figure 3G–3J). This finding supports a model of competitive binding of ERα and RARγ. To further corroborate this hypothesis, we tested whether ERα and RAR were co-bound to the FOXA1 and FOS elements by Re-ChIP and qPCR. Consistent with a competitive binding mechanism no simultaneous co-binding of RAR and ERα at these regulatory elements was observed in vivo (Figure 3K and 3L).
Figure 3. Antagonistic actions of RARs and ERα bound to shared regulatory elements.
(A,B) Distance between binding region centers of ERα and RARα (A) or RARγ (B).
(C,D) Ratios of normalized ChIP versus input signal intensities for the putative FOXA1 and FOS regulatory regions. Ratios were calculated from three replicates. Coordinates refer to UCSC hg16
(E,F) Histone 3 (H3) acetylation is antagonistically regulated by E2 and RA agonists at FOXA1 and FOS regulatory regions. RARγ-LAP MCF-7 cells grown in medium with charcoal-stripped FBS were either treated with vehicle or E2 (10 nM) for 45 minutes. The medium of E2-treated cells was then changed with medium containing vehicle (a), or CD437 (100 nM) (b), or a mixture of E2 (10 nM) and CD437 (100 nM). RA denotes CD437. Relative fold enrichment was determined by ChIP-qPCR using a pan-specific antibody against Acetyl-H3.
(G–J) ERα and RARγ-LAP recruitment is antagonistically regulated by E2 and RA agonists at FOXA1 and FOS regulatory regions. Relative fold enrichment was determined by ChIP-qPCR using an antibody against ERα or eGFP using the chromatin obtained from the experiment described above (E–F).
(K–L) FOXA1 and FOS regulatory regions do not co-bind ERα and RARγ-LAP. RARγ-LAP MCF-7 cells grown in medium with charcoal-stripped FBS were treated with E2 (10 nM) and CD437 (100 nM) for two hours. The first ChIP was performed with an antibody against ERα. Immunoprecipitated chromatin was eluted and a second ChIP was performed with IgG (negative control), or antibody against eGFP (targeting RARγ-LAP) or Acetyl-H3 (positive control). Relative enrichment was determined for the re-ChIPed chromatin by qPCR.
(M–N) ERα/RAR binding region for FOS exhibits a differential response to estrogen and RA agonists. FOS and FOXA1 regulatory regions (FOS_2, FOXA1_1, Table S9) cloned into Firefly luciferase vector pGL4.23 were co-transfected into MCF-7 cells with the Renilla luciferase vector pGL4.73 used to correct for transfection efficiency. All error bars represent s.d.
Taking these results together, we propose that competition for the same binding element, overlapping or very close elements presents one mechanism for the antagonistic regulation of genes with common ERα/RAR binding regions. Such closely overlapping binding sites are found within the majority of ERα and RAR target genes (71%). In addition, it is notable that 557 out of the 1,913 (29.1%) ERα and RAR common putative target genes do not contain co-localized ERα and RAR binding sites. Thus, while convergent regulation of common target genes by RA and estrogen signaling may occur predominantly through binding shared regulatory regions, it may in a substantial minority of cases also occur through independent regulatory regions via longer range effects.
To determine whether genomic regions with shared ERα and RAR binding have regulatory potential when removed from their genomic context we used a simple luciferase reporter assay. We tested the responsiveness to RA agonists and E2 for seven ERα/RAR binding regions as well as two unique RAR binding regions. RAR only regions were responsive to RA agonists, but not to E2. Two of the ERα/RAR binding regions (a GREB1 element and the FOS element shown in Figure S8 and Figures 3D, 3F, 3H, 3J, 3L and 3N) mediated an antagonistic response to E2 and RA agonists, while the other elements were responsive to either E2 or RA agonists (including the FOXA1 element shown in Figures 3C, 3E, 3G, 3I, 3K and 3M), or did not show any response (Figure S8). These results indicate that co-localized binding elements for ERα and RARs can sometimes be sufficient to cause the antagonizing effect of estrogen and RA on gene expression. However, there might be alternative mechanisms that require the integrated action of multiple cis-regulatory elements to differentially regulate expression by estrogen and RA. In this context, we note that a considerable proportion of putative direct ERα or RAR target genes (25.5% and 35.6% for ERα and RARs, respectively) contain more than one binding site within 50 kb to the TSSs. Also, RAR and/or ERα actions at common regions may require a specific chromatin status that cannot be recapitulated with reporter plasmids.
Enriched HREs and evolutionary conservation of RAR binding sites
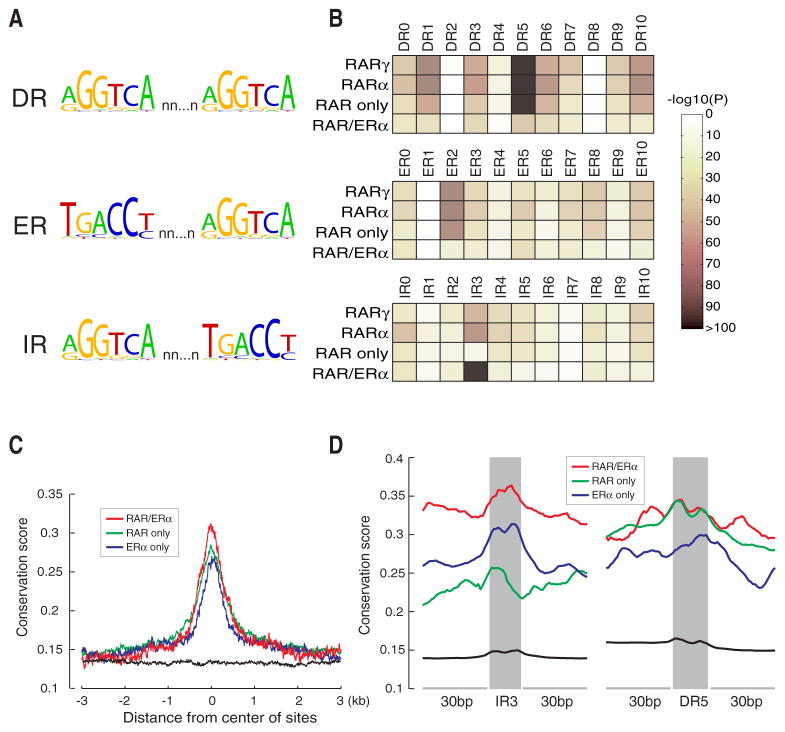
We searched all RAR binding sites for the presence of putative retinoic acid response elements (RAREs). To perform an unbiased analysis, we calculated the enrichment for all canonical hormone response elements (HREs), i.e. two direct (DR), inverted (IR) and everted (ER) hexameric PuGGTCA repeats with half-site spacer lengths from 0 to 10 (Figure 4A). This analysis identified DR5, which is frequently present in known RAR binding sites, as the most significantly enriched HRE in our in vivo RAR binding regions (Figure 4B). We also noticed a significant enrichment for several other types of HREs. For some of them, e.g. DR0, DR1, DR10, IR0 and ER8, there is experimental evidence supporting their role as RAREs (Mangelsdorf, 1994). However, we also found HREs significantly enriched that have not been implicated as RAREs before, e.g. ER2.
Figure 4. Enriched hormone response elements (HREs) and evolutionary conservation of RAR and ERα binding regions.
(A) Canonical HREs are composed of two half-sites (PuGGTCA) separated by a variable-length spacer. HREs can be configured as direct repeats (IR), everted repeats (ER), or inverted repeats (IR).
(B) Motif enrichment analysis for all HREs with spacer lengths from 0 to 10 in RARα or RARγ, binding regions, RAR and ERα common regions (ERα/RAR), and RAR unique regions (RAR only).
(C) Conservation profiles of RAR and ERα common sites (depicted in red), RAR unique sites (depicted in green) and ERα unique sites (depicted in blue). The conservation profile of local genomic background is depicted in black.
(D) Conservation profiles of IR3 and DR5 motifs and 30 bp flanking regions in RAR and ERα common sites (depicted in red), RAR unique sites (depicted in green) and ERα unique sites (depicted in blue). The conservation profile of all predicted IR3 or DR5 motifs and 30 bp flanking regions in the human genome is depicted in black.
Regions with co-localization of ERα and RAR binding showed a significant enrichment for the canonical estrogen response element (ERE) IR3 and several known RAREs such as DR5, indicating that ERα and RARs may bind to canonical response elements in shared binding regions. However, when we compared the enrichment of IR3 and DR5 between unique and co-localizing ERα and RAR binding regions, respectively, we observed a reduced enrichment in the co-localizing regions (Figures S9A and S9B). This latter result indicates that an indirect binding mechanism may play an important role for the recruitment of both RARs and ERα to these elements.
We examined the evolutionary conservation of regions bound by ERα and RAR among vertebrates. Both ERα- and RAR binding regions showed relatively high sequence conservation as compared to genomic background (Figure 4C). Likewise, IR3 and DR5 elements were found to be conserved in these regions (Figure 4D) supporting their putative roles as functional cis-regulatory elements. Interestingly, co-localizing binding regions for ERα- and RARs showed a slight but significantly higher conservation than ERα- or RAR unique sites (P = 1.21e-10 and P = 2.29e-5, respectively), which may indicate a higher functional constraint for the shared cis-regulatory regions. We observed for these regions a markedly higher conservation for IR3 elements than for unique ERα- or RAR binding regions, which may indicate a prevalent role of IR3 for the function of ER/RAR binding regions.
Transcription factor motifs in RAR binding sites
To identify putative transcription factors that specifically facilitate the binding and/or co-regulate transcriptional effects of RARs, we searched all RAR binding regions for enrichment of known transcription factor motifs. This analysis identified a number of putative binding motifs from several transcription factor families (Figure S9C and Table S4). Significantly enriched motifs were found for AP-1, Forkhead and GATA transcription factors, which all have been previously reported to be highly enriched in ERα binding sites (Carroll et al., 2006; Hua et al., 2008).
While motifs for these factors were significantly enriched when we considered all RAR sites, the most significant enrichment of Forkhead and GATA motifs was found in ERα/RAR common binding regions. For the AP-1 (Fos) motif we observed a markedly higher enrichment in both unique RAR binding regions and ERα/RAR common binding regions compared to unique ERα binding regions (Figure S9C).
FoxA1 and GATA3 binding coincides with RAR and ER binding
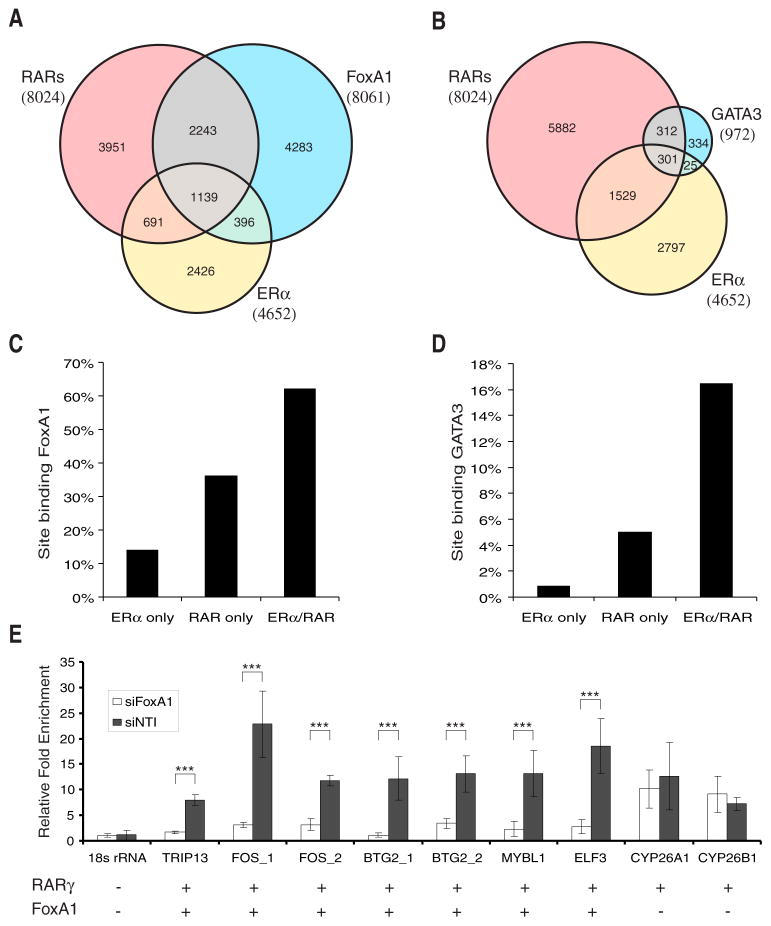
FoxA1 and GATA3 are likely candidates for Forkhead and GATA family members that are binding to the motifs enriched in regions with RAR genomic binding. Transcription factors such as FoxA1 have been proposed to act as pioneering transcription factors that facilitate the binding of ERα to enhancer elements (Carroll et al., 2005; Laganiere et al., 2005; Lupien et al., 2008). GATA transcription factors have been shown to mediate long-range chromatin interactions (Ansel et al., 2006). In particular, GATA3 is an essential regulator of mammary luminal cell fate, is co-expressed with ERα in breast carcinomas, and is a strong predictor of breast cancer differentiation (Kouros-Mehr et al., 2008); (Lacroix and Leclercq, 2004). To validate specific FoxA1 and GATA3 associations with the enriched Forkhead and GATA motifs in RAR binding sites, we performed ChIP-chip analysis using a FoxA1-specific antibody or a GFP antibody with BAC-transgenic GATA3-eGFP cell line, respectively. We identified 8,061 high-confidence FoxA1 and 972 GATA3 binding regions, which we typically found distal to TSSs (Figures S1C and S1D) as observed for ERα and RAR binding regions. These regions significantly co-localized with ERα and RAR binding regions (Figures 5A and 5B). Also, FoxA1 and GATA3 binding regions exhibited a marked co-localization (Figure S10). As predicted by the motif enrichment analysis, FoxA1 and GATA3 binding sites showed the highest overlap with shared ERα/RAR elements (Figures 5C and 5D). Unique RAR binding regions exhibited a lower frequency of FoxA1 and GATA3 binding, which further decreased for unique ERα binding regions. These findings indicate that both FoxA1 and GATA3 may be bona fide co-regulators for RARs and ERα and play in particular an important role for the function of shared ERα/RAR binding elements.
Figure 5. FoxA1 and GATA3 binding coincides with ERα and RAR binding.
(A,B) Venn diagram of FoxA1, ERα, and RARs binding sites (A) or GATA3, ERα, and RARs sites (B).
(C,D) Percentages of ERα unique sites, RAR unique sites, and ERα and RAR common sites co-localized with FoxA1 (C) or GATA3 (D) binding sites.
(E) Effect of FoxA1 knockdown on RAR recruitment. Recruitment of RARγ (defined as fold enrichment relative to input DNA) was quantified by qPCR after ChIP using an eGFP antibody comparing depleted (siFoxA1) and control cells (siNT1). Reduced RARγ recruitment was only observed for seven RARγ sites co-localizing with FoxA1 sites but not for two unique RARγ sites or a negative control site. Error bars represent s.d. ***P < 0.001.
FoxA1 is required for RAR recruitment
FoxA1 was recently shown to facilitate ERα recruitment by inducing chromatin opening at ERα enhancers (Carroll et al., 2005; Lupien et al., 2008). To determine whether there is a similar role of FoxA1 for RAR recruitment, we quantified RARγ binding upon FoxA1 knockdown by quantitative PCR. For this analysis we selected from our ChIP-chip data seven sites that were found to bind both RARγ and FoxA1, two sites that bound RARγ but not FoxA1, and a negative control site that bound neither RARγ nor FoxA1. We found that FoxA1 depletion significantly decreased RARγ binding for all seven RARγ/FoxA1 binding sites tested, but not for the RARγ sites without FoxA1 binding (Figure 5E). Thus, FoxA1 is required for RAR recruitment to specific target sites. We also tested whether GATA3 is required for RAR binding and found that GATA3 depletion had no effect on RAR recruitment (data not shown) suggesting that GATA3 function may be compensated by FoxA1 or other GATA family factors. To further investigate the role of FoxA1 and GATA3 in RA signaling, we profiled the effects of FoxA1 and GATA3 depletion on RA-regulated gene expression. Knockdown of FoxA1 had a significant effect, while GATA3 depletion had only a minor effect on many RA-regulated transcripts (Figure S11). Importantly, FoxA1 depletion affected the expression of genes with adjacent FoxA1/RAR regions but had typically no effect on RA-regulated genes adjacent to unique binding RAR regions.
Together, these findings indicate that the primary interaction of RARs with chromatin might utilize similar mechanisms as have been proposed for ERα. Importantly, the motif enrichment analysis predicts that HRE-independent recruitment requiring FoxA1, GATA3 and AP-1 may play a key role for the binding of ERα and RARs to shared binding elements. In this context it is worthy to note that FOXA1, GATA3 and FOS are also putative direct targets of ERα and RARs with overlapping binding regions (Figures S7C–S7E) that are antagonistically regulated by estrogen and RA. While the expression of FOS and GATA3 (Eeckhoute et al., 2007) is activated by estrogen and according to our data repressed by RA, we found the expression of FOXA1 up-regulated by RA and down-regulated by estrogen. These cross-regulatory loops for putative common co-factors may be a key part of the transcriptional ERα/RAR circuitry mediating the antagonizing effects of estrogen and RA signaling in breast cancer cells.
ERα and RAR-dependent gene regulation in breast cancer
RAR binding site mapping and expression profiling in MCF-7 cells revealed a marked antagonistic crosstalk between RA and estrogen signaling in this breast cancer model. Because estrogen signaling is a key pathway in breast carcinogenesis, we surmised that genes regulated by RA might also play a major role and thus have an important diagnostic and therapeutic value. Indeed, many putative RAR direct targets that are frequently also ERα direct targets have an established role in breast cancer or important cellular pathways (see Table S5 and Figures S12 and S13 for a selection of genes).
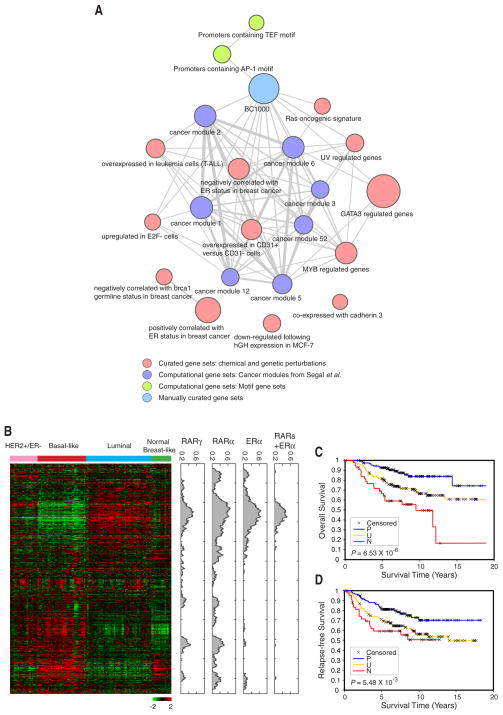
To systematically validate the role of RAR-mediated transcriptional regulation in breast cancer, we analyzed the enrichment of RA-regulated genes in MCF-7 cells in functional modules (Subramanian et al., 2005) or gene signatures that have been previously associated with breast cancer. RA-regulated genes in MCF-7 cells are highly enriched in a number of breast cancer relevant signatures (Figure 6A). Notably, the signature BC1000 comprising 1,347 manually curated genes implicated in breast cancer (Witt et al., 2006), is among the most enriched modules for RA-regulated genes. 229 out of these 1,347 (17.0%, multiple testing adjusted P = 3.03e-19) putative breast cancer genes are significantly regulated by RA in MCF-7 cells and 108 out of those 229 RA-targets have RAR-binding elements within 50 kb of the TSS (Figure 6A and Table S6). We also observed a very significant enrichment (multiple testing adjusted P = 5.32e-22) of the signature composed of genes regulated by GATA3 (Oh et al., 2006). RA-regulated genes in MCF-7 cells were found to be enriched in sets of genes that are both positively and negatively correlated with ER status in breast tumors (P = 1.24e-14 and P = 1.72e-10, respectively). Finally, we observed that RA-regulated genes are enriched in several cancer modules or functionally related genes that are conditionally activated or repressed in a variety of cancer types (Segal et al., 2004), indicating that RAR might act as a direct transcriptional regulator for a subset of genes within these cancer modules.
Figure 6. RAR targets as breast cancer relevant genes.
(A) Network view of functional modules enriched in RA-regulated genes. Each node represents a functional module or set of biologically relevant genes (see also Table S7). The node size is proportional to the minus logarithm of the adjusted P-value for testing the module enrichment of RA-regulated genes. Edge width correlates with the minus logarithm of the adjusted P-value for testing the enrichment between functional modules.
(B) Hierarchical clustering of 146-breast tumor set using the UNC Intrinsic gene set (Hu et al.,2006). The density profiles for RARγ, RARα and ERα putative targets, as well as RAR and ERα common targets, were plotted. The density was calculated as the proportion of transcription factor putative targets in 50 neighbors for each gene in the cluster.
(C,D) RAR targets as prognostic indicators. Kaplan-Meier curves of overall survival (C) and relapse-free survival (D) among the 295 patients (van de Vijver et al., 2002) classified by RA signature values. The patient samples are grouped in three categories based on RA signature scores: P (positive RA score) (n = 73), N (negative RA score) (n = 74), and U (uncorrelated) (n = 148). P-values were obtained from log-rank tests.
The meta-analysis described above suggests that many genes regulated by RA in MCF-7 cells are breast cancer-relevant. We next analyzed the expression of putative direct RAR targets that were derived from our cell line experiments in breast tumor cells from patient samples. For this purpose, we analyzed gene expression profiles of 146 breast cancer patient samples (Hu et al., 2006). These expression profiles were previously used to classify breast tumors into distinct intrinsic subtypes (e.g. Luminal, Basal-like, HER2+/ER−) that differ in their clinical outcome (Hu et al., 2006). When we analyzed RAR and ERα binding in the genes comprising these expression profiles, we observed that a gene cluster with high expression in Luminal type but low expression in more aggressive subtypes (Basal-like and HER2+/ER−) contains a high proportion of RAR targets as well as ERα targets (Figure 6B). This group of genes is characterized by high expression of ERα, its putative co-regulators (e.g. FOXA1, GATA3) and known direct targets (e.g. TFF1, KRT18). Interestingly, we also found gene clusters with high expression in Basal-like and HER2−/ER− subtypes, but low expression in Luminal and Normal-like types to contain a large fraction of genes that are putative direct targets of RARα and RARγ, but not of ERα (Figure 6B).
Our observations that many RAR targets are breast-cancer relevant genes and are specifically expressed in different breast cancer subtypes suggested that these genes might possess a significant prognostic value. We therefore analyzed the clinical outcome for each tumor sample dependent on the expression of putative direct targets of RARs. For this analysis, we defined an RA signature score for each tumor sample, which measures the correlation between RA-dependent gene expression profiles in MCF-7 cells and the gene expression profile in a given tumor sample for putative RAR direct targets (See Experimental Procedures). We first examined the correlation between RA signature scores and clinical outcomes for a cohort of expression profiles from 295 breast tumor patients (van de Vijver et al., 2002). A total of 354 putative RAR direct targets were identified in this dataset. Kaplan-Meier survival analysis demonstrated a highly significant correlation between RA signatures and the patient overall survival (Log-rank test, P = 6.53e-6), and a significant correlation for relapse-free survival (Log-rank test, P = 5.48e-3) (Figures 6C and 6D). Positive RA signature scores indicated good prognosis while negative scores strongly indicated poor clinical outcomes. We also observed significant correlation between RA signature and standard clinical-pathologic indexes, such as tumor grade, tumor size, and ERα status (Table S8). These results were confirmed for two independent breast tumor cohorts (Figure S14).
DISCUSSION
Comparing the overlap of different transcription factors can be a powerful means of inferring functional relationships, particularly when combined with expression data. Our results indicate that RARα and RARγ binding sites frequently overlap in the human genome, and comparisons of gene expression in response to isoform-specific agonists indicate considerable redundant function. Previous models of gene regulation by these crucial physiological and developmental regulators have been restricted by the focus on binding to promoters or promoter-proximal gene regions (Balmer and Blomhoff, 2002; Niederreither and Dolle, 2008). However, we found that the majority of RAR binding sites occur distal to TSSs. These results are reminiscent of recent studies that have revealed a similar tendency for ERα to bind to distal elements (Carroll et al., 2006; Hua et al., 2008; Lin et al., 2007). The co-factor FoxA1 has been implicated in this recruitment of ERα to distal sites (Carroll et al., 2005; Lupien et al., 2008), and our results indicate that FoxA1 is similarly required for RAR recruitment to genomic binding sites. Likewise, GATA3 binding frequently coincides with ERα and RAR binding, although it is not strictly required for RAR recruitment. Strikingly, many of the genomic regions bound by RARs overlapped with those previously identified to bind ERα. Subsequent transcriptional analysis demonstrated that RARs and ERα tend to exhibit antagonistic effects on the transcription of target genes.
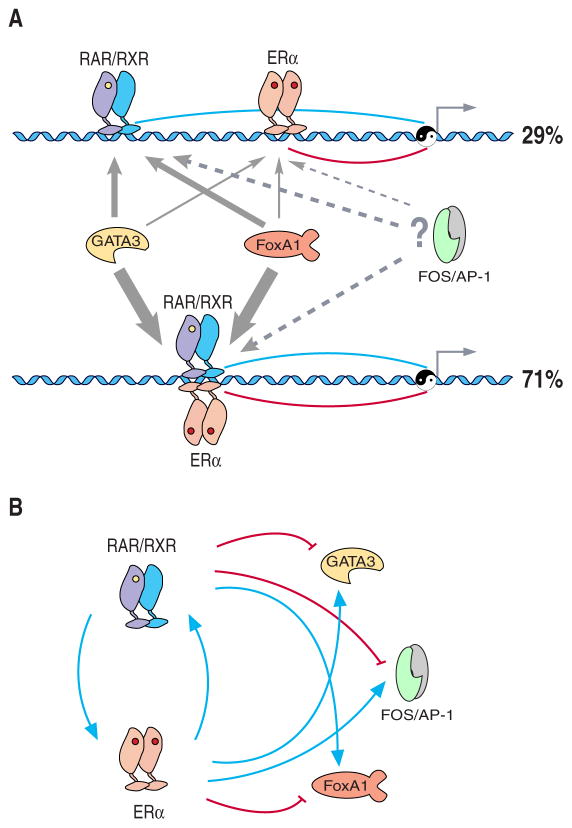
This can occur either through independent cis-regulatory elements, or more frequently, via shared binding regions of ERα and RARs (Figure 7A). In specific instances we tested, ERα and RARs binding was mutually exclusive, indicating competitive binding of the two nuclear receptors to the same element or nearby cis-regulatory elements. Based on the known functions of their target genes in breast cancer, ERα and RARs appear to be “Yin and Yang” for the genetic regulation of proliferation and survival that are promoted by ERα and inhibited by RARs. The finding that binding sites of RARs and ERα are coincident within the same enhancers or located in different enhancers for the same target genes, along with the finding that these two nuclear receptor signalling systems antagonistically regulate their target genes, indicates that these regulatory elements are co-evolving to balance target gene expression. Interestingly, the ERα/RAR antagonism appears to regulate itself through cross-regulatory loops between ERα, RARs and their co-factors (Figure 7B). This balanced control of gene expression regulates fundamental cellular processes that when dysregulated can lead to cancer.
Figure 7. A model for the antagonistic regulation of target genes by RAR and ERα.
(A) The antagonistic regulation of target genes by RAR and ERα can occur either through independent cis-regulatory elements, or as was most frequently found through shared binding regions of ERα and RARs. FoxA1 and GATA3 may be essential for RAR and/or ERα mediated gene regulation. FoxA1 may act as an initial chromatin binding factor and facilitate further recruitment of the RAR/RXR heterodimer, ERα homodimer, and/or other co-factors. The line and arrow width indicates the frequency that FoxA1 or GATA3 participates in different types of RAR or ER regulatory regions. Motif enrichment analysis predicts a potential role for AP-1 in ERα and RAR recruitment to these sites.
(B) Transcriptional regulatory circuits composed of RAR, ERα, and their putative co-factors. The expression of FOXA1, GATA3, and FOS in MCF-7 cells is oppositely regulated by RAR and ERα upon RA or estrogen treatment. A negative feedback is achieved by positive cross-regulation between the two antagonizing transcription factors RAR and ERα
The identification of the genes regulated by RARs in breast cancer cells, and in particular the discovery of their extensive cross-talk with estrogen signalling, may benefit breast cancer diagnostics and therapeutic intervention. Specifically, RAR and ERα binding data can diagnostically differentiate tumor subtypes and patient outcome. Putative direct targets of ERα and RARs in MCF-7 cells are highly expressed in Luminal type breast tumors, indicating that their antagonistic effects may be relevant for primary ER-positive tumors. However, RARs appear to be important regulators of cancer-relevant genes that are not regulated by estrogen. We found such RAR targets expressed at high levels in Basal-like and HER2/ER− tumors that are typically highly aggressive and associated with poor prognosis. Importantly, we demonstrated that in breast tumor samples that the expression of RAR targets identified in MCF-7 cells predicts a positive clinical outcome. Some of these genes may be targets for diagnosis and/or therapeutic intervention. Based on these findings, there is a strong rationale for the use of RA agonists in breast cancer treatment. However, success of RA-based therapies has been limited to treatment of acute promyelocytic leukaemia (Altucci et al., 2007; Soprano et al., 2004), while clinical applications of RA in breast and other solid tumors have shown limited effects due to RA resistance (Freemantle et al., 2003) (Schug et al., 2007). To harness the RA-mediated anti-carcinogenic effects of RARs in breast cancer this resistance must be overcome, perhaps via inhibition of FAB{5 to block metabolism of RA into PPARδ agonists (Schug et al., 2008). Another potential approach would be the use of selective agonists or combination therapy with anti-estrogens in ER+/RAR+ patients. Alternatively, RA resistance could be bypassed by targeting RA-regulated genes and pathways that mediate the anti-neoplastic effects of RA in breast cancer, whose framework we have uncovered in this study.
EXPERIMENTAL PROCEDURES
Generation of BAC transgenic MCF-7 cell lines
The BACs CTD-2343G9 (RARA), CTD-2644H7 (RARG) and RP11-1103A14 (GATA3) were obtained from Invitrogen. A LAP cassette was inserted as a C-terminal fusion using ET cloning, BAC DNA was extracted and transfected into MCF-7 breast cancer cells (ATCC HTB-22) for the generation of stable BAC transgenic cell lines as previously described (Poser et al., 2008).
Luciferase reporter assays
RAR and ERα/RAR binding regions were cloned into pGL4.23 (Promega). MCF-7 cells were transfected with pGL4.23 (containing the binding regions or the empty vector) and pGL4.73 using Lipofectamine LTX (Invitrogen), treated with agonists or vehicle, and assayed using the Dual-Glo Luciferase assay (Promega).
Reverse transfection with siRNAs
Reverse transfection was carried out at a co ncentration of 50 nM of control siRNA or 4 siRNAs (ON-TARGETplus SMARTpool, Dharmacon) directed against the same target gene using Dharmafect 1 transfection reagent (Dharmacon).
Chromatin immunoprecipitation (ChIP) experiments
Cells at 80% confluency (~5x106 cells per ChIP) were subjected to chromatin immunoprecipitation as previously described with the following antibodies: goat anti-GFP (raised against His-tagged full-length eGFP and affinity-purified with GST-tagged full-length eGFP), goat anti-FoxA1 (ab5089) from Abcam, anti-panH3ac (06–599) from Millipore, anti-ERα (MC-20, sc-542x) and normal goat IgG (sc-2028) from Santa Cruz Biotechnologies.
For ChIP-qPCR assays, the fold enrichment of ChIPed DNA relative to input DNA at a given genomic site was determined by comparative CT (ΔΔ CT) method using StepOnePlus Real-Time PCR System and Power SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer’s protocol. An ACTB exonic region or 18S rRNA genomic region was used for normalization. All primer sequences used for qPCR are described in Table S9.
For ChIP-chip, both ChIPed DNA and input DNA were subjected to linker-mediated PCR amplification, fragmentation and end-labeled with biotin using the GeneChip® WT Double-Stranded DNA Terminal Labeling Kit (Affymetrix) as previously described. The resulting labeled samples were hybridized to Affymetrix GeneChip® Human Tiling 2.0R Array Set following the Affymetrix® Chromatin Immunoprecipitation Assay Protocol. Independent biological triplicates were performed for each transcription factor, as well as the control (input DNA).
qRT-PCR and microarray gene expression profiling experiments
qRT-PCR was performed with cDNA generated from total RNA from MCF-7 cells treated with different agonists and/or transfected with siRNAs. Relative expression levels for specific genes was determined using StepOnePlus Real-Time PCR System and Power SYBR Green PCR Master Mix (Applied Biosystems). All primer sequences used for qRT-PCR are described in Table S9. For expression profiling total RNA samples were labeled by direct incorporation of cyanine 3-labeled CTP using the Agilent Low RNA Input Linear Amplification Kit PLUS (One-Color) (Agilent Technologies) and hybridized to Agilent Human Genome Oligo Microarrays (4 X 44K) (Agilent Technologies). Hybridized microarrays were scanned using a GenePix 40040B Scan4ner (Molecular Devices) at 5 μm resolution. All experiments were performed in triplicates.
Data analysis
ChIP-chip tiling array data were normalized and analyzed with Affymetrix Tiling Analysis Software (TAS) as previously described (Bernstein et al., 2005). For the analysis of gene expression data the software package LIMMA (Smyth, 2004) was applied to detect significantly differentially expressed probes. The enrichment of known or predicted transcription factor binding motifs in ChIP-identified RAR or FoxA1 binding regions was were estimated by comparing the number of motifs in binding regions with the number of motifs in randomly selected genomic regions. Associations between RA-responsive genes and clinical and pathologic variables were examined using Chi-Square contingency test with the JMP7 software package (SAS Institute Inc.) Patient sample data grouped into three RA signature categories were used to plot Kaplan-Meier survival curves for patient overall survival or relapse-free survival.
Supplementary Material
Acknowledgments
We thank E. Pendleton and all members of the Functional Genomics Facility of The University of Chicago for technical assistance, T. Hyman and F. Buchholz for providing the LAP-IRES-neo/kan cassette, and D. Drechsel for the production of the goat eGFP antibody. This work was supported by grants 1R01HG004428 and P50GM081892 to K.P.W. from the National Institutes of Health, and by a grant to K.P.W. from the Searle Funds at the Chicago Community Trust from the Chicago Biomedical Consortium. R.K. is supported by a long-term fellowship of the International Human Frontier Science Program Organization.
Footnotes
ACCESSION NUMBERS
Data are available at GEO under GSE15244.
Supplemental data include Supplemental Experimental Procedures, 15 figures, 9 tables, and Supplemental References and can be found with this article online at http://www.cell.com/XXXXX-XXXX(XX)XXXXX-X.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altucci L, Leibowitz MD, Ogilvie KM, de Lera AR, Gronemeyer H. RAR and RXR modulation in cancer and metabolic disease. Nat Rev Drug Discov. 2007;6:793–810. doi: 10.1038/nrd2397. [DOI] [PubMed] [Google Scholar]
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res. 2002;43:1773–1808. doi: 10.1194/jlr.r100015-jlr200. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Chambon P. A decade of molecular biology of retinoic acid receptors. Faseb J. 1996;10:940–954. [PubMed] [Google Scholar]
- Donato LJ, Noy N. Suppression of mammary carcinoma growth by retinoic acid: proapoptotic genes are targets for retinoic acid receptor and cellular retinoic acid-binding protein II signaling. Cancer Res. 2005;65:8193–8199. doi: 10.1158/0008-5472.CAN-05-1177. [DOI] [PubMed] [Google Scholar]
- Donato LJ, Suh JH, Noy N. Suppression of mammary carcinoma cell growth by retinoic acid: the cell cycle control gene Btg2 is a direct target for retinoic acid receptor signaling. Cancer Res. 2007;67:609–615. doi: 10.1158/0008-5472.CAN-06-0989. [DOI] [PubMed] [Google Scholar]
- Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 2007;67:6477–6483. doi: 10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemantle SJ, Spinella MJ, Dmitrovsky E. Retinoids in cancer therapy and chemoprevention: promise meets resistance. Oncogene. 2003;22:7305–7315. doi: 10.1038/sj.onc.1206936. [DOI] [PubMed] [Google Scholar]
- Giguere V, Ong ES, Segui P, Evans RM. Identification of a receptor for the morphogen retinoic acid. Nature. 1987;330:624–629. doi: 10.1038/330624a0. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Hu X, Lazar MA. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol Metab. 2000;11:6–10. doi: 10.1016/s1043-2760(99)00215-5. [DOI] [PubMed] [Google Scholar]
- Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S, Kallen CB, Dhar R, Baquero MT, Mason CE, Russell BA, Shah PK, Liu J, Khramtsov A, Tretiakova MS, et al. Genomic analysis of estrogen cascade reveals histone variant H2A. Z associated with breast cancer progression. Mol Syst Biol. 2008;4:188. doi: 10.1038/msb.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyeux A, Balaguer P, Gagne D, Nicolas JC. In vitro and in vivo interactions between nuclear receptors at estrogen response elements. J Steroid Biochem Mol Biol. 1996;58:507–515. doi: 10.1016/0960-0760(96)00082-9. [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr H, Kim JW, Bechis SK, Werb Z. GATA-3 and the regulation of the mammary luminal cell fate. Curr Opin Cell Biol. 2008;20:164–170. doi: 10.1016/j.ceb.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix M, Leclercq G. About GATA3, HNF3A, and XBP1, three genes co-expressed with the oestrogen receptor-alpha gene (ESR1) in breast cancer. Mol Cell Endocrinol. 2004;219:1–7. doi: 10.1016/j.mce.2004.02.021. [DOI] [PubMed] [Google Scholar]
- Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V. From the Cover: Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci U S A. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson AS, Jordan VC. MCF-7: the first hormone-responsive breast cancer cell line. Cancer Res. 1997;57:3071–3078. [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, et al. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Takayama S, Zheng Y, Froesch B, Chen GQ, Zhang X, Reed JC, Zhang XK. Interaction of BAG-1 with retinoic acid receptor and its inhibition of retinoic acid-induced apoptosis in cancer cells. J Biol Chem. 1998;273:16985–16992. doi: 10.1074/jbc.273.27.16985. [DOI] [PubMed] [Google Scholar]
- Lohnes D, Mark M, Mendelsohn C, Dolle P, Dierich A, Gorry P, Gansmuller A, Chambon P. Function of the retinoic acid receptors (RARs) during development (I). Craniofacial and skeletal abnormalities in RAR double mutants. Development. 1994;120:2723–2748. doi: 10.1242/dev.120.10.2723. [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Umesono K, Evans RM. The retinoid receptors. In: Sporn MB, Roberts AB, Goodman DS, editors. The Retinoids: Biology, Chemistry and Medicine. New York: Raven Press; 1994. pp. 319–349. [Google Scholar]
- Naar AM, Boutin JM, Lipkin SM, Yu VC, Holloway JM, Glass CK, Rosenfeld MG. The orientation and spacing of core DNA-binding motifs dictate selective transcriptional responses to three nuclear receptors. Cell. 1991;65:1267–1279. doi: 10.1016/0092-8674(91)90021-p. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Dolle P. Retinoic acid in development: towards an integrated view. Nat Rev Genet. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- Oh DS, Troester MA, Usary J, Hu Z, He X, Fan C, Wu J, Carey LA, Perou CM. Estrogen-regulated genes predict survival in hormone receptor-positive breast cancers. J Clin Oncol. 2006;24:1656–1664. doi: 10.1200/JCO.2005.03.2755. [DOI] [PubMed] [Google Scholar]
- Petkovich M, Brand NJ, Krust A, Chambon P. A human retinoic acid receptor which belongs to the family of nuclear receptors. Nature. 1987;330:444–450. doi: 10.1038/330444a0. [DOI] [PubMed] [Google Scholar]
- Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129:723–733. doi: 10.1016/j.cell.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug TT, Berry DC, Toshkov IA, Cheng L, Nikitin AY, Noy N. Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARbeta/delta to RAR. Proc Natl Acad Sci U S A. 2008;105:7546–7551. doi: 10.1073/pnas.0709981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Soprano DR, Qin P, Soprano KJ. Retinoic acid receptors and cancers. Annu Rev Nutr. 2004;24:201–221. doi: 10.1146/annurev.nutr.24.012003.132407. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Witt AE, Hines LM, Collins NL, Hu Y, Gunawardane RN, Moreira D, Raphael J, Jepson D, Koundinya M, Rolfs A, et al. Functional proteomics approach to investigate the biological activities of cDNAs implicated in breast cancer. J Proteome Res. 2006;5:599–610. doi: 10.1021/pr050395r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Stetler-Stevenson M, Steeg PS. Inhibition of cyclin D expression in human breast carcinoma cells by retinoids in vitro. Oncogene. 1997;15:107–115. doi: 10.1038/sj.onc.1201142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.