Abstract
Telomeric repeat binding factor 1 (TRF1) is a component of the multiprotein complex “shelterin,” which organizes the telomere into a high-order structure. TRF1 knockout embryos suffer from severe growth defects without apparent telomere dysfunction, suggesting an obligatory role for TRF1 in cell cycle control. To date, the mechanism regulating the mitotic increase in TRF1 protein expression and its function in mitosis remains unclear. Here, we identify guanine nucleotide-binding protein-like 3 (GNL3L), a GTP-binding protein most similar to nucleostemin, as a novel TRF1-interacting protein in vivo. GNL3L binds TRF1 in the nucleoplasm and is capable of promoting the homodimerization and telomeric association of TRF1, preventing promyelocytic leukemia body recruitment of telomere-bound TRF1, and stabilizing TRF1 protein by inhibiting its ubiquitylation and binding to FBX4, an E3 ubiquitin ligase for TRF1. Most importantly, the TRF1 protein-stabilizing activity of GNL3L mediates the mitotic increase of TRF1 protein and promotes the metaphase-to-anaphase transition. This work reveals novel aspects of TRF1 modulation by GNL3L.
Introduction
Chromosomal ends are protected from degradation and DNA repair activities by telomeres, formed by long double-stranded DNA (dsDNA) repeats and short single-stranded DNA (ssDNA) overhangs (Henderson et al., 1987; Henderson and Blackburn, 1989). The length and structure of telomeres is maintained by two elongation mechanisms and six telomere-capping proteins. Telomere lengthening is mediated either by the telomerase, a ribonucleoprotein complex composed of the TERC RNA and the TERT reverse transcription (Greider and Blackburn, 1985), or by a telomerase-independent alternative lengthening of telomeres (ALT) mechanism based on homologous recombination between sister telomeres (Bryan et al., 1995; Henson et al., 2002). The telomere-capping proteins, i.e., TRF1, TRF2, RAP1, TIN2, POT1, and TPP1 (de Lange, 2005; Songyang and Liu, 2006), organize the telomere into a high-order structure. TRF1 and TRF2 form homodimers that bind the dsDNA repeats via their myb domains (Bilaud et al., 1997; Broccoli et al., 1997). The ssDNA overhangs are protected by POT1 and TPP1 (Horvath et al., 1998; Wang et al., 2007; Xin et al., 2007). Within the complex, TRF1 and TRF2 are connected to POT1 via TIN2 and TPP1. TRF1 demonstrates the abilities to negatively control the telomere length (van Steensel and de Lange, 1997) and to regulate the mitotic entry of dividing cells (Shen et al., 1997). Its biological importance is highlighted by the early embryonic lethal phenotype of TRF1-deficient mice, which curiously shows no defect in telomere length (Karlseder et al., 2003).
The other side of this story is represented by GNL3L (guanine nucleotide-binding protein-like 3), which belongs to a protein family of three, i.e., nucleostemin (NS), GNL3L, and Ngp-1. The common features of this family include an MMR1_HSR1 domain and nucleolar distribution (Daigle et al., 2002; Leipe et al., 2002). NS was initially identified as an cancer and stem cell–enriched protein (Tsai and McKay, 2002). GNL3L and NS share the same invertebrate orthologue, whereas Ngp-1 exists as a single-gene subfamily from yeast to human (Meng et al., 2007). All three proteins in the NS subfamily shuttle between the nucleolus and the nucleoplasm by a GTP-driven mechanism (Meng et al., 2007), which allows them to interact with proteins found in different subnuclear compartments. Despite their resemblance in primary protein sequences, NS, GNL3L, and Ngp-1 display distinct dynamic properties (Meng et al., 2007) and interactive proteins (Yasumoto et al., 2007), indicative of their different biological activities.
We have previously shown that NS binds and facilitates the degradation of TRF1 protein (Zhu et al., 2006), and therefore are interested in knowing whether GNL3L or Ngp-1 also regulates TRF1. More recently the GNL3L’s role in telomere maintenance was reported by a study showing its isolation from the human telomerase complex and its overexpression effect in reducing the telomere length without affecting the telomerase activity (Fu and Collins, 2007). Here, we demonstrate that GNL3L is capable of binding TRF1 independently of TERT. Evidence is presented for the activity of GNL3L to promote the homodimerization and telomeric retention of TRF1, reduce APB (ALT-associated PML body) formation, and increase TRF1 protein stability. The first two findings correlate with GNL3L’s ability to reduce the telomere length (Fu and Collins, 2007). The last result reveals a novel mechanism that stabilizes TRF1 protein during mitosis and safeguards mitotic transition.
Results
TRF1 interacts with NS and GNL3L, but not with Ngp-1
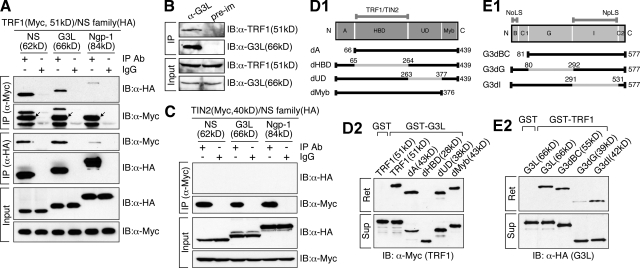
To test whether GNL3L or Ngp-1 interacts with TRF1, coimmunoprecipitation (coIP) experiments of Myc-tagged TRF1 and HA-tagged NS family proteins were performed in HEK293 cells (Fig. 1 A). The results showed that both NS and GNL3L, but not Ngp-1, can be coimmunoprecipitated with TRF1 by anti-Myc (row 1 and 2) or anti-HA antibody (row 3 and 4), and that none of the NS family proteins binds TIN2 (Fig. 1 C). Endogenous coIP results showed that GNL3L and TRF1 coexist in the same complex in vivo, captured from HeLa cell lysates by anti-GNL3L antibody (Fig. 1 B), and that their interaction does not depend on DNA or RNA (Fig. S1 A). To map the interactive domains of these two proteins, deletions of TRF1 were made on its N-terminal acidic domain (A), the homodimerization domain (HBD), an undefined region (UD), and the C-terminal myb domain (Fig. 1 D1). Affinity pull-down assays showed that HBD domain deletion of TRF1 abolishes its binding to the GST-fused GNL3L (Fig. 1 D2). To define the TRF1-interactive domain of GNL3L, mutants lacking the N-terminal basic coiled-coil domain (B-C1), the GTP-binding domain (G), or the intermediate domain (I) of GNL3L were generated (Fig. 1 E1). GST-TRF1 binding assays revealed that interaction between TRF1 and GNL3L relies mostly on the G domain of GNL3L (Fig. 1 E2). These results demonstrate the importance of the HBD domain of TRF1 and the G domain of GNL3L in conferring their interaction.
Figure 1.
Interaction between TRF1 and GNL3L is mediated by the homodimerization domain of TRF1 and the GTP-binding domain of GNL3L. (A) Cell lysates were extracted from HEK293 cells cotransfected with Myc-tagged TRF1 and HA-tagged nucleostemin (NS), GNL3L (G3L), or Ngp-1. Protein complexes were immunoprecipitated (IP) by anti-tag antibodies and immunoblotted (IB) by the indicated antibodies. TRF1 interacts with NS and GNL3L, but not with Ngp-1. Arrows mark the precipitated TRF1. (B) Endogenous coIP confirmed that TRF1 and GNL3L co-reside in the same protein complex immunoprecipitated by anti-GNL3L antibody (α-G3L) from HeLa cell lysates. Pre-im: preimmune serum. (C) None of the NS family proteins interacts with TIN2. (D1, E1) Diagrams of TRF1 and GNL3L deletion mutants. Gray lines mark the deleted regions; numbers indicate amino acid positions. Domain abbreviations: A, acidic; HBD, homodimerization; UD, undefined; B, basic; C1 and C2, coiled-coil-1 and -2; G, GTP-binding; I, intermediate. GST pull-down assays demonstrated that the HBD domain of TRF1 (D2) and the G domain of GNL3L (E2) are required for the interaction of these two proteins. Agarose-retained (Ret) and supernatant fractions (Sup) are indicated.
GNL3L binding to TRF1 promotes TRF1 homodimerization and is competed by TIN2
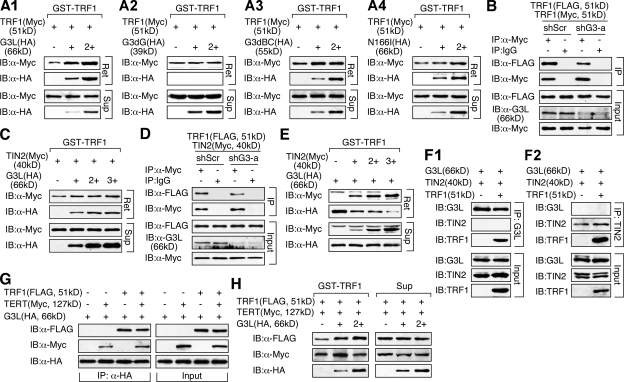
The GNL3L-interactive domain of TRF1 overlaps with its homodimerization and TIN2-binding sites, suggesting a functional connection between GNL3L binding and the formation of TRF1 protein complex. To test this idea, GST fusion proteins of TRF1 were used to pull down lysates containing a fixed amount of Myc-tagged TRF1 admixed with increasing amounts of wild-type or mutant GNL3L separately expressed in HEK293 cells (Fig. 2 A). In each sample, whole cell proteins were adjusted to the same level. The pull-down results showed that GNL3L increases the amounts of Myc-tagged TRF1 proteins bound by GST-TRF1 in a dose-dependent manner (Fig. 2 A1, Ret). This effect of GNL3L was abolished by deletion of its TRF1-interactive G domain (G3dG; Fig. 2 A2) or the BC domain (G3dBC; Fig. 2 A3), but not by a point mutation (N166I) that abrogates its GTP-binding ability (Fig. 2 A4). G3dBC and N166I remain capable of binding TRF1 in the last two experiments. To verify this result in vivo, four microRNA-adapted short hairpin RNA (shRNAmir) constructs (shG3-a to -d) were tested for their efficiencies to deplete the endogenous GNL3L protein (Fig. S2, A and B), and the shG3-a construct was chosen for its highest knockdown efficiency. Confirming the pull-down results, coIP assays showed that knocking down the endogenous GNL3L of HEK293 cells reduces the binding of FLAG- and Myc-tagged TRF1 compared with the shScr (which targets a scrambled sequence) knockdown samples (Fig. 2 B). GST pull-down (Fig. 2 C) and knockdown coIP experiments (Fig. 2 D) showed that GNL3L has no obvious effect on the binding between TIN2 and TRF1. Conversely, GNL3L binding to TRF1 was significantly reduced by TIN2 (Fig. 2 E). To determine whether TRF1 can bind GNL3L and TIN2 simultaneously, coIP experiments of GNL3L (HA), TRF1 (FLAG), and TIN2 (Myc) were conducted in HEK293 cells by anti-HA or anti-Myc antibody. Our data showed that immunoprecipitation of GNL3L copurifies TRF1 but not TIN2 (Fig. 2 F1), and that only TRF1, but not GNL3L, is coimmunoprecipitated with TIN2 (Fig. 2 F2). Therefore, TRF1 binds GNL3L and TIN2 separately. Finally, GNL3L binding to TRF1 or TERT is not affected by overexpression of TERT or TRF1, respectively (Fig. 2 G), and the GNL3L-mediated TRF1 homodimerization is neither enhanced nor reduced by TERT overexpression (compare Fig. 2 H with Fig. 2 A1).
Figure 2.
GNL3L promotes TRF1 homodimerization, and its binding to TRF1 is competed by TIN2. (A) Agarose-bound GST-TRF1 was incubated with cell lysates containing a fixed amount of Myc-tagged TRF1, mixed with increasing amounts of wild-type GNL3L (A1, G3L), the G3dG (A2), G3dBC (A3), or N166I mutant (A4). Wild-type GNL3L enhances the pull-down efficiency of Myc-tagged TRF1 by GST-TRF1 in a dose-dependent manner. Deleting the TRF1-interacting G domain (G3dG) completely abolishes this activity. (B) The coIP efficiency between FLAG- and Myc-tagged TRF1 was examined in control knockdown (shScr) and GNL3L knockdown (shG3-a) HEK293 cells by immunoprecipitation with anti-Myc antibody and immunoblotting with anti-FLAG antibody. The results confirmed that endogenous GNL3L promotes TRF1 homodimerization. (C) A small effect of GNL3L was observed on promoting the binding between Myc-tagged TIN2 and GST-TRF1. (D) This finding was supported by coIP of FLAG-tagged TRF1 and Myc-tagged TIN2 in the control (shScr) and GNL3L (shG3-a) knockdown HEK293 cells. (E) TIN2 binding significantly reduced the pull-down of GNL3L by GST-TRF1. (F) Protein complexes were immunoprecipitated from HEK293 cells cotransfected with GNL3L (HA), TIN2 (Myc), and TRF1 (FLAG) by anti-HA (F1) or anti-Myc antibody (F2). GNL3L, TIN2, and TRF1 proteins were detected by anti-tag antibodies in the IP or input fraction. The results showed that TRF1 does not bind GNL3L and TIN2 simultaneously. (G) Double and triple coIP of TRF1 (FLAG), TERT (Myc), and GNL3L (HA) showed that the coIP efficiency between TERT and GNL3L or between TRF1 and GNL3L is not affected by the coexpression of TRF1 or TERT1, respectively. (H) The ability of GNL3L to promote TRF1 homodimerization, measured by the GST-TRF1 pull-down of Myc-tagged TRF1 and compared with that in Fig. 2 A1, is not changed by TERT overexpression.
GNL3L increases the telomeric retention time of TRF1
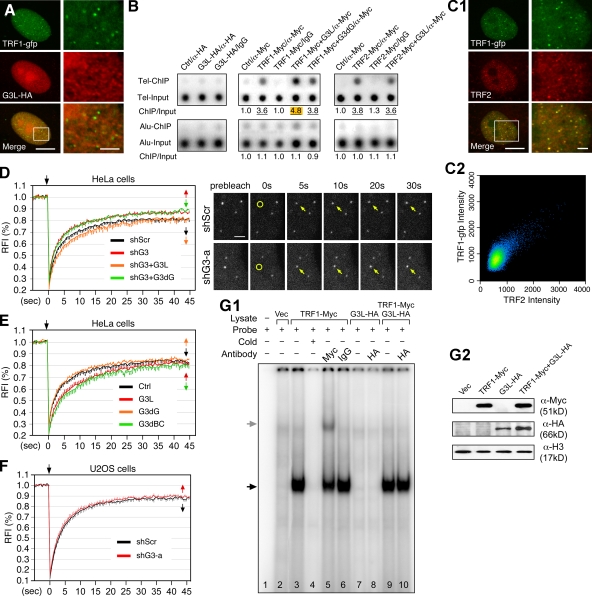
To decide whether GNL3L binds TRF1 at or outside the telomere, the distribution of TRF1 (GFP-tagged) and GNL3L (HA-tagged) was examined in HeLa cells. Confocal studies showed that the TRF1 signal does not colocalize with the GNL3L signal in specific structures (Fig. 3 A), indicating that these two proteins do not form stable complexes at the telomeric foci. Chromatin immunoprecipitation (ChIP) assays directly addressed whether GNL3L binds telomeric DNAs and how it affects the telomeric association of TRF1 at steady state. ChIP results showed that telomeric DNAs are not coimmunoprecipitated with GNL3L compared with the control samples (Fig. 3 B, left). Compared with the TRF1-alone sample (3.58 ± 0.10, n = 3), coexpression of GNL3L (4.82 ± 0.09, P = 0.0009) but not G3dG (3.84 ± 0.04, P = 0.09) shows increased telomeric bindings of TRF1 (Fig. 3 B, middle). This effect of GNL3L is specific for TRF1 but not for TRF2 (Fig. 3 B, right). Next, we used the FRAP approach to investigate how GNL3L regulates the dynamic association between TRF1 and telomere in living cells. Because the telomere-bound TRF1 is constantly exchanged with the unbound TRF1 in the nucleoplasm, the more stable the TRF1–telomere association is, the longer the telomeric retention time of TRF1 will be and hence the slower the recovery rate after photobleaching. The FRAP paradigm was set up such that a single telomere was bleached and its fluorescence recovery was recorded. The validity of using the C-terminally GFP-fused TRF1 to track the movement of endogenous TRF1 protein in living cells was supported by the results showing that its distribution coincides with that of endogenous TRF2 (Fig. 3 C) and that its dynamic property is the same as that of the N-terminally GFP-fused TRF1 (Fig. S3). FRAP analyses showed that GNL3L depletion by shG3-a significantly increases the FRAP recovery rate and the plateau level of TRF1 (P < 0.0001 by repeated measures ANOVA, Fig. 3 D). This GNL3L knockdown FRAP phenotype can be specifically rescued by the shG3-a–resistant full-length GNL3L (shG3+G3L) but not by the shG3-a–resistant G3dG mutant (shG3+G3dG) (Fig. 3 D and Fig. S2 E2). Consistently, the FRAP recovery rate of TRF1 was reduced by coexpression of the wild-type and G3dBC mutant of GNL3L (P < 0.0001) but not by coexpression of the non-TRF1-binding G3dG mutant (P = 0.933) (Fig. 3 E). The ability of GNL3L to enhance the telomeric retention of TRF1 was not observed in telomerase− U2OS cells (Fig. 3 F), indicating that this effect may be cell type– or telomerase-dependent. Finally, we used the electrophoretic mobility shift assay (EMSA) to check how GNL3L affects the DNA-bound TRF1 in vitro. EMSA was conducted with a radiolabeled probe containing six tandem repeats of TTAGGG and nuclear extracts expressing the indicated proteins (Fig. 3 G2). Compared with the probe alone (lane 1) and vector-transfected cell lysate samples (lane 2), the Myc-tagged TRF1-transfected sample (lane 3) yields a specific TRF1–probe complex (black arrow) that can be competed by excess unlabeled probes (lane 4) and supershifted by anti-Myc antibody (lane 5, gray arrow) (Fig. 3 G1). GNL3L by itself does not produce specific shifted complexes (lane 7 and 8). Neither does coexpression of GNL3L supershift or affect the TRF1–probe complex (lane 9 and 10). These results demonstrate that although GNL3L is not a component of the final TRF1 complex at telomeres, it helps stabilize the telomeric association of TRF1 in HeLa cells.
Figure 3.
GNL3L increases telomeric retention of TRF1 in living cells. (A) Lack of nucleolar or telomeric colocalization of GFP-fused TRF1 (green) and HA-tagged GNL3L (red) in HeLa cells was shown by confocal analyses. The rectangular area is enlarged and shown on the right. Bars: 10 µm (left) and 2.5 µm (right). (B; left) ChIP assays showed that immunoprecipitation of HA-tagged GNL3L (G3L-HA/α-HA) does not copurify telomeric DNAs (Tel-ChIP) more than the vector-transfected, α-HA–precipitated sample (Ctrl/α-HA), the GNL3L-transfected, IgG-precipitated sample (G3L-HA/IgG), or the copurified Alu sequence (Alu-ChIP). (Middle) Coexpression of GNL3L increases the association between TRF1 and telomeric DNAs (TRF1-Myc+G3L/α-Myc) compared with the TRF1 alone or TRF1+G3dG transfections. (Right) GNL3L does not affect TRF2 binding to telomeric DNAs. All experiments were repeated three times. (C1) The C-terminally GFP-fused TRF1 (TRF1-gfp) colocalizes with the endogenous TRF2 in HeLa cells. High magnification of the indicated area (rectangle) is shown on the right. Bars: 10 µm (left) and 2 µm (right). (C2) Quantification of TRF1-gfp and TRF2 colocalization. All pixels are plotted based on their red (X-axis) and green (Y-axis) fluorescence intensities, and pseudocolored based on the event frequency, with red representing the highest and blue representing the lowest frequency. (D; left) FRAP analyses showed that GNL3L knockdown in HeLa cells increases the recovery rate and plateau level of TRF1-gfp at the telomere (red; P < 0.0001). Such phenotype can be rescued by the shG3-a–resistant full-length GNL3L (orange) but not by the shG3-a–resistant G3dG (green). RFI, relative fluorescence index. Error bars represent SEM shown on one side indicated by arrows. Top arrows mark the bleaching event. (Right) Time-sequenced images are shown with labels indicating the bleached telomere (yellow circles and arrows) and intervals between image acquisition and the bleaching pulse (in seconds). Bar: 2 µm. (E) The telomeric retention time of TRF1 was increased by overexpression of GNL3L (G3L) or G3dBC (P < 0.0001) but not by overexpression of G3dG. (F) GNL3L knockdown has no effect on the telomeric FRAP of TRF1-gfp in U2OS cells. (G) EMSA was performed using a (TTAGGG)6 probe and nuclear extracts from HEK293 cells expressing the indicated recombinant proteins (G2). A specific TRF1-probe complex was identified in lane 3 (black arrow), which could be competed by excess nonlabeled probes (lane 4) and supershifted by anti-Myc antibody (lane 5, gray arrow). Coexpression of GNL3L had no effect on the TRF1-probe complex (lane 9 and 10).
GNL3L negatively regulates APB formation in ALT (e.g., U2OS) cells
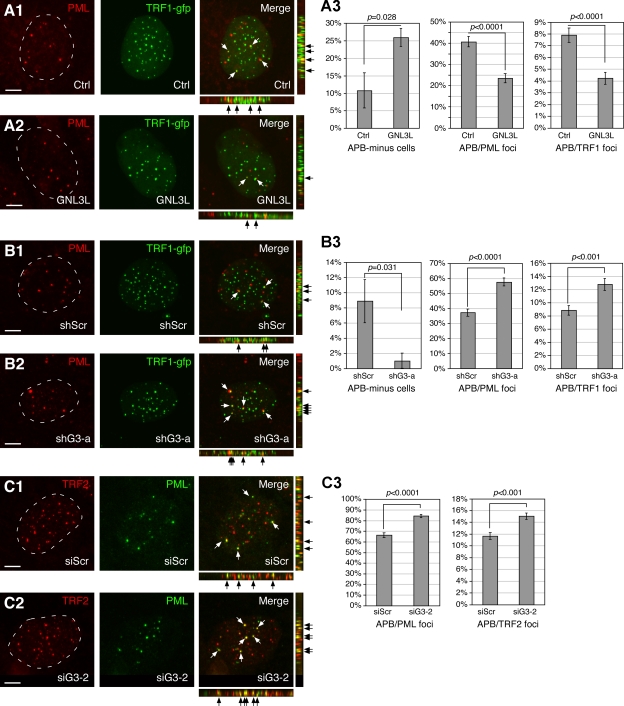
In U2OS cells, telomere elongation does not require telomerase, but instead is associated with the formation of APB. APB contains telomeric DNAs, TRF1, and various other telomere proteins (Yeager et al., 1999; Potts and Yu, 2007). We therefore reason that GNL3L may regulate the APB formation in ALT cells. To test this idea, the telomeric foci and promyelocytic leukemia (PML) bodies of GNL3L-perturbed U2OS cells were labeled by TRF1-GFP and anti-PML (PG-M3) antibody, respectively. Colocalization of TRF1 and PML signals was determined based on 3D reconstruction of confocal images of 1-µm optical thickness serially sampled at 0.5-µm intervals along the Z-axis (Fig. 4, A1–2 and B1–2). Analyses of 95–100 randomly sampled cells from five independent experiments demonstrated that GNL3L coexpression increases the APB-negative cells from 10.8% to 25.9% (P = 0.028, Fig. 4 A3, left), and reduces the percentage of APB/PML body from 40.6% to 23.4% (P < 0.0001, middle) and the percentage of APB/TRF1+ foci from 7.9% to 4.2% (P < 0.0001, right). The total numbers of PML bodies and TRF1+ foci per nucleus are 6.6 (± 0.3, SEM) and 36.2 (± 1.1) in the control cells, and 6.4 (± 0.3, P = 0.63) and 37.1 (± 1.0, P = 0.55) in the GNL3L-overexpressing cells, respectively, which excludes the possibility that the GNL3L-dependent decrease of APB is caused by changes in the number of PML bodies or TRF1+ foci. To confirm that the endogenous GNL3L also shows the same activity, we measured the GNL3L knockdown effect on TRF1-PML colocalization, and showed that GNL3L knockdown by shG3-a decreases the APB-negative cells from 8.9% to 1% (P = 0.031), and increases the percentage of APB/PML body from 37.2 to 57.6% (P < 0.0001) and the percentage of APB/TRF1+ foci from 8.8 to 12.8% (P < 0.001) (Fig. 4 B). The total numbers of PML bodies (7.1 ± 0.2 vs. 6.8 ± 0.2, P = 0.31) and TRF1+ foci (33.8 ± 0.9 vs. 32.9 ± 0.8, P = 0.49) per nucleus show no difference in the shScr and shG3-a–treated cells. We also used TRF2 (4A794) and PML (H-238) colocalization to assess the GN3L effect on APB formation, and showed that GNL3L knockdown by siG3-2 increases the percentage of APB/PML body from 66.2% to 84.2% (P < 0.0001) and the percentage of APB/TRF2+ foci from 11.7% to 15.0% (P < 0.001). Again, the total numbers of PML bodies (6.8 ± 0.2 vs. 6.8 ± 0.2, P = 0.96) and TRF2+ foci (39.7 ± 1.0 vs. 40.1 ± 1.1, P = 0.85) per nucleus remain the same in these two conditions. It should be noted that the baseline APB level is higher in the TRF2-PML than in the TRF1-PML measurement, which may be due to the differences of the antibodies used. Finally, as a previous study showed that the recruitment of TRF1 to PML bodies is related to its SUMOylation (Potts and Yu, 2007), the effect of GNL3L perturbation on TRF1 SUMOylation was examined. In vivo SUMOylation assays showed that GNL3L overexpression does not affect the amount of SUMOylated TRF1 (Fig. S4), suggesting that this APB-reducing activity of GNL3L occurs after the TRF1 SUMOylation step.
Figure 4.
GNL3L negatively regulates APB formation in U2OS cells. (A) APB formation in U2OS cells was scored by colocalization of TRF1-gfp and PML bodies in the control (A1) or GNL3L-overexpression U2OS cells (A2). Serial confocal images of 1-µm thickness were collected at 0.5-µm intervals and reconstructed along the X-Y, X-Z, and Y-Z planes for individual cell. Arrows indicate overlapped spots. (A3) Coexpression of GNL3L increases the percentage of APB-minus cells, and decreases the ratios of APB/PML bodies and APB/TRF1+ foci. (B) Colocalization of TRF1+ foci and PML bodies was measured in control (shScr, B1) or GNL3L knockdown (shG3-a, B2) U2OS cells. (B3) GNL3L knockdown increases colocalization of TRF1 foci and PML bodies. (C) GNL3L knockdown by siG3-2 promotes APB formation in U2OS cells, as scored by colocalization of TRF2+ foci and PML bodies.
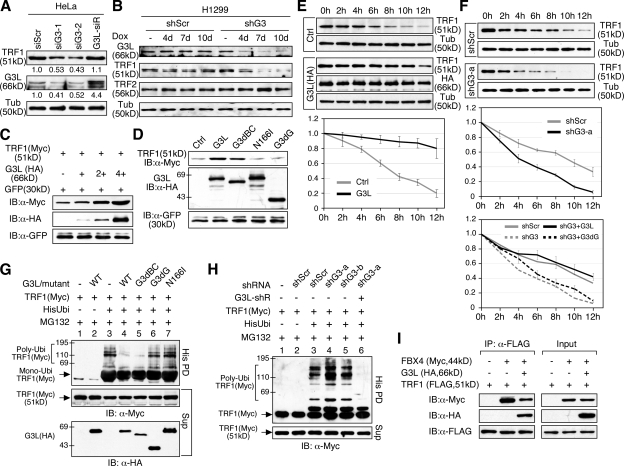
GNL3L stabilizes TRF1 protein by reducing its ubiquitylation and FBX4 binding
To address how GNL3L affects the endogenous protein level of TRF1, HeLa cells were transfected with two previously described GNL3L-targeting siRNA duplexes (siG3-1 and siG3-2) (Yasumoto et al., 2007). Western blots showed that depleting the endogenous GNL3L by either siRNA duplex leads to a decrease in the endogenous TRF1 protein level, and that restoring the expression of GNL3L by a siG3-2–resistant construct (G3L-siR, Fig. S2 E1) can effectively reverse the siG3-2–induced TRF1 protein decrease (Fig. 5 A). To confirm this finding in a different cell type, H1299 stable cell lines with Dox-inducible GNL3L knockdown capabilities were established by cotransfection of a doxycycline (Dox)-inducible shG3-a and a reverse Tet transactivator (rtTA-M2) construct (Fig. S2, C and D). Cell lysates were collected from the control (shScr) and GNL3L knockdown (shG3) cells, receiving either no treatment or Dox treatment for 4, 7, and 10 d. Western analyses confirmed a Dox-dependent reduction of GNL3L protein in the shG3 cells but not in the shScr cells, and most importantly a decrease of the endogenous TRF1 protein in the GNL3L-depleted samples (Fig. 5 B). By contrast, the TRF2 protein level was unchanged by GNL3L knockdown. To determine whether GNL3L increases TRF1 at the transcriptional or posttranscriptional level, HEK293 cells were cotransfected with fixed amounts of Myc-tagged TRF1 and GFP, both driven by the same EF1α promoter, and increasing amounts of HA-tagged GNL3L. Compared with the GFP protein, the levels of exogenously expressed TRF1 protein are also increased by coexpression of GNL3L in a dose-dependent manner (Fig. 5 C), indicating that the GNL3L effect on TRF1 protein occurs posttranscriptionally. This gain-of-function phenotype may depend on the overexpression level of GNL3L because a threefold increase of GNL3L by G3L-siR is not enough to elicit this effect (Fig. 5 A). It should be noted that this activity of GNL3L requires its N166 residue and G domain but not the BC-domain (Fig. 5 D).
Figure 5.
GNL3L stabilizes TRF1 protein by preventing its ubiquitylation and binding to FBX4. (A) Knocking down the endogenous GNL3L expression by siG3-1 or siG3-2 in HeLa cells reduces the endogenous TRF1 protein level, which can be rescued by an siG3-2–resistant GNL3L (G3L-siR). (B) The same result was confirmed in H1299 cells by using the Doxycycline (Dox)-inducible shRNAmir knockdown approach. Western analyses show a time-dependent reduction of GNL3L protein in the Dox-treated GNL3L knockdown (shG3) cells, but not in the Dox-treated shScr cells. Specifically, GNL3L depletion decreases the protein level of TRF1 but not that of TRF2. (C) Overexpression of GNL3L (HA) increases the protein level of TRF1 (Myc) in a dose-dependent manner after normalization by the protein level of GFP cotransfected in the same sample. (D) The GNL3L’s ability to increase TRF1 protein is abolished by deleting its TRF1-interactive G domain and by the N166I mutation, but not by the BC domain deletion. (E) TRF1 protein stability was measured by a protein degradation assay in control (Ctrl) and GNL3L-overexpressing (G3L, HA-tagged) HEK293 cells. The TRF1 protein amounts were measured from three experiments, adjusted based on their α-tubulin (Tub) amounts, and expressed as percentages of the TRF1 protein amount at the 0-h time point. (F) GNL3L depletion by shG3-a increases the protein degradation of TRF1 (top two panels), which can be rescued by coexpression of an shG3-a–resistant full-length GNL3L but not an shG3-a–resistant G3dG mutant (bottom panel). (G) Overexpression of the wild-type (lane 4) or G3dBC mutant (lane 5) of GNL3L decreases TRF1 ubiquitylation compared with the control sample (lane 3), whereas the G3dG (lane 6) and N166I mutants (lane 7) have no such effect. (H) GNL3L depletion by shG3-a (lane 4) increases the ubiquitylation of TRF1, which can be reversed by overexpression of an shG3-a–resistant GNL3L (G3L-shR, lane 6). (I) CoIP experiments showed that GNL3L (HA) binding to TRF1 (FLAG) decreases the coIP efficiency between TRF1 and FBX4 (Myc).
To investigate if GNL3L regulates TRF1 protein stability, HEK293 cells were transfected with TRF1. 36 h later, cells were treated with cycloheximide (CHX, 100 μg/ml), and lysates were collected at 2-h intervals from 0 to 12 h. Western results showed that TRF1 proteins in the GNL3L overexpression cells are degraded much more slowly than that in the control cells (Fig. 5 E, P < 0.0001 by repeated measures ANOVA). In consistence, the protein stability of TRF1 is significantly reduced in the shG3-a–transfected HEK293 cells compared with the shScr-treated samples (Fig. 5 F, top two panels, P < 0.0001). This GNL3L knockdown phenotype of TRF1 destabilization can be specifically rescued by the shG3-a–resistant full-length GNL3L but not by the shG3-a–resistant G3dG (Fig. 5 F, bottom). Because the TRF1 protein is degraded by the ubiquitin-proteasome pathway (Chang et al., 2003), an in vivo ubiquitylation assay was used to decide how GNL3L influences the ubiquitylation of TRF1 (Fig. 5 G). Compared with the control-transfected sample (lane 3), overexpression of wild-type GNL3L (lane 4) or the G3dB mutant (lane 5) significantly reduces TRF1 polyubiquitylation, whereas the G3dG (lane 6) and N166I (lane 7) mutants do not, concurring with the ability of wild-type and mutant GNL3L to increase TRF1 protein (Fig. 5 D). In support of this finding, knocking down the endogenous GNL3L by the shG3-a or shG3-b construct increases the ubiquitylation of TRF1 to a degree compatible with their protein knockdown efficiencies, and the increased TRF1 ubiquitylation by shG3-a can be reversed by coexpression of an shG3-a–resistant GNL3L construct (G3L-shR) (Fig. 5 H). Ubiquitylation of TRF1 requires an E3 ubiquitin ligase component FBX4 (Lee et al., 2006). CoIP assays showed that FBX4 binding to TRF1 is impeded by GNL3L (Fig. 5 I), indicating that GNL3L inhibits TRF1 ubiquitylation by preventing its association with the E3 ubiquitin ligase complex.
GNL3L stabilizes TRF1 protein during mitosis and promotes metaphase-to-anaphase transition
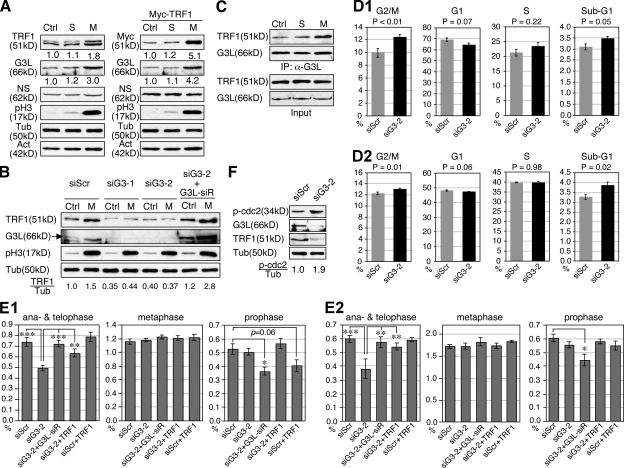
The protein level of TRF1 is up-regulated during mitosis and down-regulated in the G1 phase (Shen et al., 1997). So far, the mechanism responsible for the mitotic increase of TRF1 protein is unknown. To determine whether GNL3L is involved in this process, we first measured the protein level of endogenous TRF1 and GNL3L in the nontreated (Ctrl), S phase–synchronized, and M phase–synchronized HEK293 cells (Fig. 6 A, left). Compared with the control and S phase cells, the endogenous protein levels of TRF1 and GNL3L were both increased in the M phase cells, whereas NS remained unchanged. The same result was observed in HeLa cells (not depicted), as well as with exogenously expressed Myc-tagged TRF1 proteins (Fig. 6 A, right), suggesting that a posttranscriptional mechanism is involved. Importantly, this mitotic increase of TRF1 protein was abolished when GNL3L was depleted by siG3-1 or siG3-2, and restored by coexpression of the siG3-2–resistant GNL3L construct (G3L-siR) (Fig. 6 B). In addition, we found that not only did the protein levels of GNL3L and TRF1 increase, but their binding was also enhanced during mitosis (Fig. 6 C), which is consistent with the idea that the nucleolus is dissembled during mitosis, thereby releasing more GNL3L into the nucleoplasm for TRF1 binding.
Figure 6.
GNL3L stabilizes TRF1 protein during mitosis and promotes the metaphase-to-anaphase transition. (A) The protein amounts of endogenous TRF1 (left) and exogenously expressed TRF1 (Myc-tagged, right) were measured in the nontreated (Ctrl), S phase-synchronized (S), and M phase-synchronized HEK293 cells. (B) GNL3L depletion by siG3-1 and siG3-2 abolishes the mitotic increase of TRF1 protein. This effect of siG3-2 can be reversed by coexpression of a siG3-2–resistant GNL3L (G3L-siR). (C) In addition to the increase in protein level, the binding efficiency between GNL3L and TRF1 is also increased during mitosis. To control for the mitotic increase of TRF1 and GNL3L proteins, their input amounts in each sample were adjusted to the same before coIP. (D) PI-labeled cell cycle analyses showed that GNL3L knockdown in HeLa (D1) or HEK293 cells (D2) increases the percentage of G2/M phase cells. A mild decrease in the G1 cell percentage and an increase in the sub-G1 cell percentage were also noticed. (E) The percentages of cells at different mitotic stages were scored by anti–phospho-Histone H3 (pH3) staining in control and GNL3L knockdown HeLa (E1) and HEK293 cells (E2). Over 1.6 × 104 cells from eight independent experiments were collected for each sample. The results showed that GNL3L depletion triggers cell cycle arrest at the metaphase-to-anaphase transition, and this effect can be rescued by coexpression of the siG3-2–resistant GNL3L (G3L-siR) or TRF1. Cells cotransfected with siG3-2 and G3L-siR (in both HeLa and HEK293 cells) or with siScr and TRF1 (in HeLa cells only) showed decreased prophase cell percentages. *, P < 0.01; **, P < 0.001; ***, P < 0.0001. (F) Silencing GNL3L expression increases the amount of phospho-cdc2 (Tyr15), supporting a role of GNL3L in the prophase entry of dividing cells as well.
To uncover the biological role of GNL3L in cell cycle regulation, its loss-of-function effect was analyzed by flow cytometry in HeLa cells (Fig. 6 D1). The FACS results showed that the main effect of GNL3L knockdown is to increase the G2/M phase cell percentage (P < 0.01, n = 8). Together with a mildly decreased G1 cell percentage (P = 0.07), this finding indicates that GNL3L depletion causes cell cycle arrest at or before the mitotic exit. The same findings of G2/M increase (P = 0.01) and G1 decrease (P = 0.06) were observed in HEK293 cells, although the difference in the G2/M percentage caused by GNL3L knockdown was less than that in HeLa cells (Fig. 6 D2). To further define the time point within the G2/M phase affected by GNL3L knockdown, different phases of mitotic cells were scored by anti-phospho-Histone H3 (pH3) staining in control and GNL3L knockdown HeLa cells (Fig. 6 E1) and HEK293 cells (Fig. 6 E2). Our results consistently showed that GNL3L knockdown causes a significant decrease in the percentages of anaphase and telophase cells compared with the siScr-treated samples (P < 0.0001), and that this GNL3L knockdown phenotype can be completely rescued by coexpression of G3L-siR (P < 0.001). Despite the decrease of anaphase and telophase cells by GNL3L knockdown, a concurrent increase of metaphase or prophase cells is not seen, suggesting a potential blockage before the prophase. This idea is supported by an increased phospho-cdc2 (p-cdc2, Tyr15) level in the GNL3L knockdown samples (Fig. 6 F). To determine whether this function of GNL3L in promoting the metaphase-to-anaphase transition is related to TRF1 stabilization, we also tested the ability of TRF1 to rescue the mitotic phenotype of GNL3L knockdown. First, to minimize the possibility of introducing adverse or additive effects by TRF1 overexpression, we controlled the expression of Myc-tagged TRF1/Pin2 at a level two times that of the endogenous TRF1 (Fig. S2 F), and showed that expressing this amount of TRF1 protein in siScr-treated cells does not alter their anaphase and telophase cell percentage (P = 0.34 for HeLa cells and 0.86 for HEK293 cells). Notably, restoring this amount of TRF1 significantly rescues the stalled metaphase-to-anaphase transition caused by GNL3L knockdown in both HeLa and HEK293 cells (P < 0.01). It is worth noting that although coexpression of TRF1 does not affect the percentages of metaphase, anaphase, and telophase cells, it reduces the prophase cell percentage to some extent in the control knockdown HeLa cells (P = 0.06) but not in the siG3-2 knockdown cells. The same effect is observed with G3L-siR coexpression in GNL3L knockdown HeLa and HEK293 cells (P < 0.01), suggesting that perturbation of GNL3L or TRF1 may dysregulate the prophase entry of dividing cells as well.
Knockdown of GNL3L or TRF1 increases cells with multipolar mitotic spindle
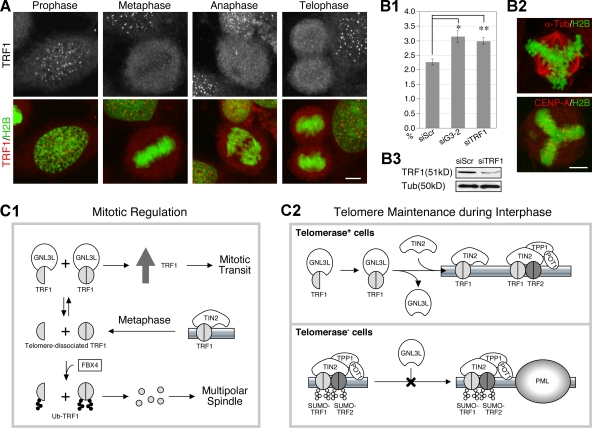
To determine their site of action during the metaphase-to-anaphase transition, protein localization of TRF1 was revealed by high-resolution confocal analysis in HeLa cells stably expressing a GFP-tagged H2B (Kanda et al., 1998). Our results showed that although the total protein level of TRF1 increases during mitosis, its telomeric signal becomes relatively less intense in the metaphase, anaphase, and telophase cells compared with their neighboring interphase and prophase cells imaged in the same field (Fig. 7 A). This phenomenon is also true for the GFP-tagged TRF1 when expressed at a low level (Fig. S5 A), arguing against the possibility that during this mitotic window, the telomeric TRF1 may undergo modification or be masked by its interacting protein, which prevents the epitope recognition of the anti-TRF1 antibody (TRF-78). This conclusion is further bolstered by the staining patterns of another anti-TRF1 antibody (C-19, Fig. S5 B) and anti-TRF2 antibody (Fig. S5 C), which show similar distributions during mitosis, although the sensitivity and specificity of the C-19 anti-TRF1 antibody is less than optimal. It should be noted that the GFP-tagged TRF1 also concentrates at the telomere when its expression level is high, indicating that TRF1 is not completely absent from the telomere during mitosis. On the other hand, the GNL3L signal becomes diffuse and colocalized with TRF1 during this mitotic window (Fig. S5 D). To define the mitotic defect associated with GNL3L knockdown, mitotic spindles and chromosomes were visualized by anti–α-tubulin immunofluorescence and H2B-GFP in GNL3L-depleted cultures. Notably, we have found that both the GNL3L and TRF1 knockdown cultures contain more cells with multipolar spindles than do the control knockdown cultures, and that their centromeres, shown by anti–CENP-A immunostaining, are well aligned along the multipolar metaphase plate (Fig. 7 B).
Figure 7.
Knockdown of GNL3L or TRF1 increases the number of cells with multipolar spindle. (A) Distribution of endogenous TRF1 protein at different stages of mitosis is shown by high-resolution confocal analyses of anti-TRF1 (TRF-78) staining in H2B-GFP HeLa cells. Bar: 5 µm. (B1 and B2) GNL3L or TRF1 knockdown significantly increases the number of cells with multipolar spindle, shown by α-tubulin immunostaining (Tub). Y-axis shows the percentage of metaphase cells with multipolar spindle. *, P < 0.01; **, P < 0.001. The centromeres in these cells, labeled by anti–CENP-A antibody, are aligned along the multipolar metaphase plate. TRF1 knockdown efficiency was shown in B3. (C) Models of GNL3L-mediated regulation of TRF1 protein modification and function in mitotic transition (C1) and telomere maintenance (C2).
Discussion
This study focuses on a novel GNL3L-regulated control of TRF1 complex formation and protein modification (Fig. 7 C). We showed that TRF1 interacts with GNL3L via its HBD domain and the G domain of GNL3L. On one hand, GNL3L decreases the degradation of TRF1 by preventing its FBX4 binding and ubiquitylation, thereby allowing the TRF1 protein to accumulate during mitosis (Fig. 7 C1). On the other hand, GNL3L promotes the homodimerization and telomeric retention of TRF1 in telomerase+ cells (Fig. 7 C2, top), and negatively regulates APB formation in telomerase− cells (bottom). Mutant analyses reveal that loss of GTP binding (i.e., by the N166I mutation) affects the protein stabilization but not the homodimerization activity of GNL3L on TRF1. The differential effects of the N166I mutant on TRF1 stability and homodimerization indicate that TRF1 homodimerization, by itself, is not sufficient to prevent its degradation.
Dynamic interaction between GNL3L and TRF1
In interphase cells, GNL3L resides mainly in the nucleolus and less in the nucleoplasm, whereas TRF1 is concentrated at the telomere. At steady state, there is a constant exchange between the nucleolar and the nucleoplasmic pools of GNL3L (Meng et al., 2007), as well as between the telomeric and the nucleoplasmic pools of TRF1 (Mattern et al., 2004). Several scenarios can be envisioned for the binding of these two proteins and their subsequent journey within the nucleus. After their initial association in the nucleoplasm, GNL3L can either carry TRF1 protein to the nucleolus or simply modify and release it on-site. Alternatively, GNL3L itself may participate in the assembly of the telomere-capping complex. The fact that overexpression of TRF1 and GNL3L does not increase the nucleolar signal of TRF1 or the telomeric signal of GNL3L and that GNL3L is not associated with telomeric DNAs argue against the first and third possibilities. Supporting the notion that GNL3L is not part of the stable telomeric complex, binding and coIP experiments show that TRF1 does not bind simultaneously with GNL3L and TIN2, an essential component of the shelterin complex. We propose that GNL3L may function as a chaperone protein that regulates the complex formation and posttranslational modification of TRF1 protein. Although this regulatory event occurs in the nucleoplasm, it will influence the availability and readiness of TRF1 for its telomere-capping action. This idea somewhat resembles the explanation proposed by Fu and Collins (2007) for why GNL3L overexpression shortens telomere length without apparently affecting the activity of telomerase, that is, binding of GNL3L may sequester the telomerase RNP in the nucleoplasm, rendering it biologically inactive in vivo. One notable difference between this model and ours is that GNL3L increases rather than decreases TRF1 binding to the telomere. As GNL3L is also copurified with the telomerase complex, we provide evidence to show that its abilities to bind TRF1 and to regulate the homodimerization and APB formation of TRF1 are independent of its interaction with TERT. Because the telomerase is a multicomponent complex, the involvement of other telomerase components in the GNL3L–TRF1 interaction cannot be completely ruled out as yet. In addition, the GNL3L’s ability to increase the telomeric retention of TRF1 is seen in HeLa cells but not in U2OS cells, which could mean that either this phenotype requires the presence of telomerase or occurs in a cell type–specific manner.
Roles of GNL3L in the mitotic progression
A much-debated function of TRF1 is in the regulation of mitotic transit, which coincides with a surge of its protein level during mitosis (Shen et al., 1997). TRF1 overexpression was shown to increase the G2/M percentage of cells (Shen et al., 1997) and the apoptotic percentage in cells with short telomeres (Kishi et al., 2001). Our work demonstrates that the protein level and TRF1 binding of GNL3L both increase during mitosis, and that GNL3L is responsible for the mitotic increase of TRF1 protein. In addition, GNL3L is needed for the metaphase-to-anaphase transition, and this activity of GNL3L is partially mediated by TRF1. Initially, our findings may seem contradictory to the idea that increasing TRF1 protein during mitosis creates tethered sister chromatids at the telomere and blocks the transition from metaphase to anaphase (Waizenegger et al., 2000; Dynek and Smith, 2004; Canudas et al., 2007). Detailed confocal analyses help resolve this dilemma by revealing that TRF1 signals in the metaphase, anaphase, and early telophase cells are less concentrated at the telomere despite its protein level increase, compared with that in the interphase cells. This latter finding creates another paradox, as a previous study, based on an in vitro reconstituted system mimicking Xenopus early development, has shown that the telomeric DNA binding of TRF1 is strengthened during mitosis (Nishiyama et al., 2006). Therefore, the possibility of an epitope-masking protein modification or binding that causes failure to detect the telomeric TRF1 during mitosis by the anti-TRF1 antibody needs to be addressed. We do so by showing that the GFP-tagged TRF1 and the endogenous TRF2 display the same pattern of distribution as does the endogenous TRF1 and that the overall TRF1 signal outside the telomere is relatively higher in the metaphase cell than in the interphase cell. It is worth noting that overexpression of TRF1-gfp at high levels still forces its accumulation at the telomere, which may cause tethered chromosomes as reported previously, thereby leading to the same mitotic arrest phenotype as that of TRF1 depletion.
The metaphase-to-anaphase transition may not be the only event affected by GNL3L or TRF1 perturbation. The decrease of anaphase and telophase cell percentages is not accompanied by a concomitant increase of prophase and metaphase cells, suggesting that GNL3L depletion may also block the prophase entry. Indeed, Western blots demonstrate an increased phospho-cdc2 (p-cdc2, Tyr15) level associated with GNL3L knockdown. Furthermore, coexpression of TRF1 in the control knockdown HeLa cells and coexpression of G3L-siR in the GNL3L knockdown HeLa and HEK293 cells both reduce the prophase cell percentage, supporting that the prophase entry of dividing cells may be sensitive to the levels of TRF1 and GNL3L as well. Finally, it should be noted that the cell cycle arrest effect and multipolar spindle phenotype of GNL3L depletion, although significant and consistent, is not absolute. So is the ability of TRF1 to rescue the GNL3L-depleted cell cycle phenotype in HeLa cells. The former finding may be due to incomplete knockdown of the GNL3L protein, and the latter result suggests that the cell cycle activity of GNL3L may involve additional TRF1-independent pathways. Based on our work, we propose that when dividing cells proceed into metaphase, a portion of their TRF1 proteins is dissociated from the telomere to allow proper segregation of sister chromatids. The telomere-dissociated TRF1 also performs a cell cycle regulatory function, but will be degraded unless bound by GNL3L (Fig. 7 C1). Therefore, knocking down the expression of GNL3L favors the ubiquitylation and degradation of TRF1 protein, which then leads to cell cycle arrest at the metaphase-to-anaphase transition and an increased number of cells with multipolar spindles. The increase of metaphase cells with multipolar spindles may suggest a role of TRF1 and GNL3L in regulating the homeostasis of centrosomes or microtubules. A connection between TRF1 and microtubule polymerization has been reported by a previous study (Nakamura et al., 2001).
GNL3L regulates telomeric association and PML body recruitment of TRF1 in interphase cells
A well-known function of TRF1 is to maintain the telomeric structure and negatively control the telomeric length by denying the telomeric access of telomerase (van Steensel and de Lange, 1997). Our study showed that GNL3L increases the homodimerization, the protein stability, and the telomeric association of TRF1. An interesting note of our FRAP findings is that GNL3L knockdown influences both the recovery rate and the plateau level of telomere-bound TRF1, which is consistent with the idea of two populations of TRF1 residing at the telomere (Kim et al., 2008). Because GNL3L overexpression affects the loosely bound (or dynamic) fraction more than the stable fraction, we suggest that the amount of stably bound TRF1 at the telomere may be controlled by other protein(s), such as TIN2 (Fig. 7 C2, top). This effect of GNL3L in retaining telomeric TRF1 is observed mainly in telomerase+ cells and predicts that GNL3L may have a negative impact on telomere elongation in those cells, which indeed is the case in HTC75 cells shown by Fu and Collins (2007).
In telomerase− ALT cells, telomere elongation is achieved by homologous recombination. In those cells, GNL3L is capable of reducing the PML body recruitment of TRF1+ and TRF2+ foci, a process related to the recombination-based mechanism of telomere elongation. GNL3L does so without affecting the level of SUMOylated TRF1 and therefore supports the recruitment model, which depicts that the SUMOylation of TRF1 occurs before its recruitment to the PML body (Potts and Yu, 2007) (Fig. 7 C2, bottom). Thus, our data support the notion that GNL3L stabilizes the telomeric TRF1 complex in telomerase+ cells and prevents APB formation in telomerase− cells, both of which are expected to inhibit telomere elongation. As the telomere plays a key role in limiting the number of cell division, human tumors usually develop an increased telomerase activity or the telomerase-independent ALT mechanism to extend their proliferation without entering into crisis of chromosome fusions and death. Given the ability of GNL3L in modulating TRF1 and telomere length, one would expect that its expression level might be reduced in cancer cells or at least balanced by its ability to promote cell cycle progression. In conclusion, this work exemplifies the great complexity of telomere maintenance and cell cycle regulation.
Materials and methods
Cell culture, transfection, Western blot, and immunofluorescence
All these procedures were described in our previous studies (Tsai and McKay, 2005; Meng et al., 2007). Primary antibodies used in this study are anti-HA (HA.11), Myc (9E10), FLAG (M2), TRF1 (TRF-78, Santa Cruz Biotechnology, Inc.), TRF2 (4A794), PML (PG-M3 and H-238), α-tubulin (Sigma-Aldrich), CENP-A (Millipore), and GNL3L (clone #134).
Coimmunoprecipitation
Cells were harvested in NTEN buffer. Lysates were incubated with primary antibody for 1 h, followed by incubation with protein G–Sepharose beads (GE Healthcare) for an additional 4 h at 4°C. Immunoprecipitates were washed three times with RIPA buffer before SDS-PAGE and Western detection.
GST pull-down assay
GST-fused proteins were expressed in BL21/DE3 by using the pGEX4T-2 vector (Tsai and McKay, 2002). Epitope-tagged proteins were expressed in HEK293 cells and extracted in phosphate-buffered saline (PBS)/Triton X-100 (1%) buffer. Sepharose-bound GST fusion proteins (2 µg) were incubated with cell lysates for 2 h and washed five times with PBS, including two times with high-salt solutions.
siRNA duplex, short hairpin siRNA, and inducible GNL3L knockdown cells
Transient knockdown experiments were performed by transfection of siRNA duplexes or shRNAmir constructs. Control and GNL3L-specific siRNA duplexes (siG3-1 and siG3-2) were described previously (Yasumoto et al., 2007). TRF1-specific siRNA duplex targets 5′-TGCCAGGAACTGCTCGAGT-3′. shRNAmir constructs were generated in the pShag Magic vector (pSM2c) based on a mir-30 hairpin design that targets 21-bp sequences, capped by mir-5′ and mir-3′ sequences, and driven by a U6 promoter. The targeted sequences for GNL3L are: 5′-CCAATCGAGAGGCTGAATTAA-3′ (shG3-a), 5′-GGAGGAGATTTCCAACTATTA-3′ (shG3-b), 5′-GCTGTTCCTGGAATTACCAAA-3′ (shG3-c), and 5′-CCGCCCTAAGAGCAACAGTAT-3′ (shG3-d). The shScr construct targets a scrambled sequence: 5′-TCTCGCTTGGGCGAGAGTAAG-3′. Creation of stable lines with inducible GNL3L knockdown capabilities is described in Fig. S2.
Chromatin immunoprecipitation
Cells were cross-linked by 1% formaldehyde, lysed in 50 mM Tris buffer-1% SDS, and sonicated to obtain chromatin fragments from 300 bp to 1 kb. The resulting lysates were incubated with primary antibodies and protein G–Sepharose for 8 h at 4°C. After extensive washes, chromosomal DNAs were extracted by treatments with Rnase A and proteinase K, and reverse cross-linking at 65°C. DNA samples were dot-blotted onto Hybond membranes, hybridized with a 32P-labeled (TTAGGG)4 or Alu (5′-GGCCGGGCGCGGTGGCTCACGCCTGTAATCCCAGCA-3′) oligonucleotide probe, and exposed to PhosphorImager screen. All lysates were normalized based on protein concentrations. A part of the lysate (1/20th) was used to measure the input DNA amount.
EMSA
EMSA experiments were performed with nuclear extracts prepared from HEK293 cells transfected with the indicated constructs. To generate EMSA probes, the (TTAGGG)6 primer was radiolabeled with γ-P32 ATP in a T4 kinase reaction, annealed with excess amounts of the (CCCTAA)6 primer, and purified through QIAquick nucleotide removal columns (QIAGEN). Reaction and gel electrophoresis conditions follow the same procedure described previously (Yasumoto et al., 2007).
Protein degradation and in vivo ubiquitination assays
Protein degradation assays were performed in cycloheximide-treated HEK293 cells as described previously (Zhu et al., 2006). For in vivo ubiquitination assays, His-tagged ubiquitin and TRF1 expression plasmids were co-expressed in HEK293 cells. 2 d later, cells were treated with MG132 (10 µM) for 6 h before protein extraction in 6M guanidinium buffer. Ubiquitylated proteins were pulled down by Ni2+-chelating Sepharose and washed extensively.
High-resolution confocal imaging of APB, TRF1, and mitotic cells
APBs were scored on the basis of colocalization of TRF1-gfp/PML (PG-M3) or TRF2 (4A794)/PML (H-238). Endogenous TRF1 was detected by a monoclonal (TRF-78) or polyclonal (C-19) antibody. Images were acquired by a confocal microscope (LSM510; Carl Zeiss, Inc.) with a 63X Plan-Apochromat oil objective (1.4 NA), and scanned with ≤1.0-µm optical thickness and 0.5-µm interval along the Z-axis. For APB measurement, stacked images were collected from 95–100 cells of five independent experiments for every experimental condition, and analyzed in a double-blind way using ImageJ 1.36b software. Prophase, metaphase, anaphase, and telophase cells are labeled by anti-pH3 immunofluorescence and H2B-GFP or DAPI, and determined by their condensed chromosomal patterns.
Fluorescence recovery after photobleaching (FRAP)
Bleaching experiments were performed on a confocal microscope (LSM510; Carl Zeiss, Inc.) equipped with a 63X Plan-Apochromat oil objective. The GFP signal was excited with the 488-nm argon laser (21-mW nominal output). Emission above 505 nm was monitored. Cells were maintained at 35°C with a heat blower during the entire course of recording. The FRAP paradigm was designed such that a single telomere was bleached by a short laser pulse administered at 70% of the power of the 488-nm argon laser (21-mW nominal output) for three iterations which lasted for 256 ms. For image acquisition, the laser power was attenuated to 0.6% of the bleach intensity, and cells were scanned with 5X zoom at 0.5-s intervals for 45 s after photobleaching. For quantification, fluorescence intensities of the bleached area, the entire nucleus, and the area outside of the nucleus were measured. The relative fluorescence index (RFI) in the bleached area was normalized to the total intensity in the nucleus after background subtraction by using the following calculation: RFI = (It/I0) × (TN0/TNt), where It and I0 are the background-subtracted intensities of the bleached spot at time point t and before photobleaching, respectively, and TNt and TN0 are the background-subtracted intensities of the entire nucleus at time point t and before photobleaching, respectively. Cells with signal loss of more than 10% during the imaging phase were discarded. 40 cells of four independent experiments were analyzed.
Cell cycle profile analysis and synchronization
Cell cycle profiles were analyzed by counting the PI-labeled cells with a COULTER EPICS XL flow cytometer and the XL System II software (Beckman Coulter). Each cell cycle profile was compiled from 2 × 104 gated events, and analyzed using the Multi Cycle AV software (Phoenix Flow Systems). Early S phase synchronization was achieved by incubating the log-phase cells with 2 mM thymidine for 20 h. G2/M arrest was achieved by incubation with 0.5 µM nocodazole for 20 h.
Online supplemental material
Figure S1 shows control experiments confirming the specific interaction between TRF1 and GNL3L. Figure S2 describes the creation of shRNAmir-based GNL3L knockdown and rescue constructs. Figure S3 validates the use of GFP-fused TRF1 for FRAP studies. Figure S4 summarizes the GNL3L effect on TRF1 SUMOylation. Figure S5 shows the subcellular distributions of TRF1, TRF2, and GNL3L during mitosis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200812121/DC1.
Acknowledgments
We thank Geoff M. Wahl for his generous gift of H2B-GFP cells.
This project is supported by NCI-PHS grant R01 CA113750 to R.Y. Tsai.
Footnotes
Abbreviations used in this paper: ALT, alternative lengthening of telomeres; APB, ALT-associated PML body; coIP, coimmunoprecipitation; EMSA, electrophoretic mobility shift assay; GNL3L, guanine nucleotide-binding protein-like 3; NS, nucleostemin; PML, promyelocytic leukemia; TRF1, telomeric repeat binding factor 1.
References
- Bilaud T., Brun C., Ancelin K., Koering C.E., Laroche T., Gilson E. 1997. Telomeric localization of TRF2, a novel human telobox protein.Nat. Genet. 17:236–239 [DOI] [PubMed] [Google Scholar]
- Broccoli D., Smogorzewska A., Chong L., de Lange T. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2.Nat. Genet. 17:231–235 [DOI] [PubMed] [Google Scholar]
- Bryan T.M., Englezou A., Gupta J., Bacchetti S., Reddel R.R. 1995. Telomere elongation in immortal human cells without detectable telomerase activity.EMBO J. 14:4240–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S., Houghtaling B.R., Kim J.Y., Dynek J.N., Chang W.G., Smith S. 2007. Protein requirements for sister telomere association in human cells.EMBO J. 26:4867–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W., Dynek J.N., Smith S. 2003. TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres.Genes Dev. 17:1328–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle D.M., Rossi L., Berghuis A.M., Aravind L., Koonin E.V., Brown E.D. 2002. YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with circularly permuted G-motifs and marked burst kinetics.Biochemistry. 41:11109–11117 [DOI] [PubMed] [Google Scholar]
- de Lange T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres.Genes Dev. 19:2100–2110 [DOI] [PubMed] [Google Scholar]
- Dynek J.N., Smith S. 2004. Resolution of sister telomere association is required for progression through mitosis.Science. 304:97–100 [DOI] [PubMed] [Google Scholar]
- Fu D., Collins K. 2007. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation.Mol. Cell. 28:773–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C.W., Blackburn E.H. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts.Cell. 43:405–413 [DOI] [PubMed] [Google Scholar]
- Henderson E.R., Blackburn E.H. 1989. An overhanging 3′ terminus is a conserved feature of telomeres.Mol. Cell. Biol. 9:345–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson E., Hardin C.C., Walk S.K., Tinoco I., Jr., Blackburn E.H. 1987. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs.Cell. 51:899–908 [DOI] [PubMed] [Google Scholar]
- Henson J.D., Neumann A.A., Yeager T.R., Reddel R.R. 2002. Alternative lengthening of telomeres in mammalian cells.Oncogene. 21:598–610 [DOI] [PubMed] [Google Scholar]
- Horvath M.P., Schweiker V.L., Bevilacqua J.M., Ruggles J.A., Schultz S.C. 1998. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA.Cell. 95:963–974 [DOI] [PubMed] [Google Scholar]
- Kanda T., Sullivan K.F., Wahl G.M. 1998. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells.Curr. Biol. 8:377–385 [DOI] [PubMed] [Google Scholar]
- Karlseder J., Kachatrian L., Takai H., Mercer K., Hingorani S., Jacks T., de Lange T. 2003. Targeted deletion reveals an essential function for the telomere length regulator Trf1.Mol. Cell. Biol. 23:6533–6541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Davalos A.R., Heo S.J., Rodier F., Zou Y., Beausejour C., Kaminker P., Yannone S.M., Campisi J. 2008. Telomere dysfunction and cell survival: roles for distinct TIN2-containing complexes.J. Cell Biol. 181:447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi S., Wulf G., Nakamura M., Lu K.P. 2001. Telomeric protein Pin2/TRF1 induces mitotic entry and apoptosis in cells with short telomeres and is down-regulated in human breast tumors.Oncogene. 20:1497–1508 [DOI] [PubMed] [Google Scholar]
- Lee T.H., Perrem K., Harper J.W., Lu K.P., Zhou X.Z. 2006. The F-box protein FBX4 targets PIN2/TRF1 for ubiquitin-mediated degradation and regulates telomere maintenance.J. Biol. Chem. 281:759–768 [DOI] [PubMed] [Google Scholar]
- Leipe D.D., Wolf Y.I., Koonin E.V., Aravind L. 2002. Classification and evolution of P-loop GTPases and related ATPases.J. Mol. Biol. 317:41–72 [DOI] [PubMed] [Google Scholar]
- Mattern K.A., Swiggers S.J., Nigg A.L., Lowenberg B., Houtsmuller A.B., Zijlmans J.M. 2004. Dynamics of protein binding to telomeres in living cells: implications for telomere structure and function.Mol. Cell. Biol. 24:5587–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Zhu Q., Tsai R.Y. 2007. Nucleolar trafficking of nucleostemin family proteins: common versus protein-specific mechanisms.Mol. Cell. Biol. 27:8670–8682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Zhou X.Z., Kishi S., Kosugi I., Tsutsui Y., Lu K.P. 2001. A specific interaction between the telomeric protein Pin2/TRF1 and the mitotic spindle.Curr. Biol. 11:1512–1516 [DOI] [PubMed] [Google Scholar]
- Nishiyama A., Muraki K., Saito M., Ohsumi K., Kishimoto T., Ishikawa F. 2006. Cell-cycle-dependent Xenopus TRF1 recruitment to telomere chromatin regulated by Polo-like kinase.EMBO J. 25:575–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts P.R., Yu H. 2007. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins.Nat. Struct. Mol. Biol. 14:581–590 [DOI] [PubMed] [Google Scholar]
- Shen M., Haggblom C., Vogt M., Hunter T., Lu K.P. 1997. Characterization and cell cycle regulation of the related human telomeric proteins Pin2 and TRF1 suggest a role in mitosis.Proc. Natl. Acad. Sci. USA. 94:13618–13623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z., Liu D. 2006. Inside the mammalian telomere interactome: regulation and regulatory activities of telomeres.Crit. Rev. Eukaryot. Gene Expr. 16:103–118 [DOI] [PubMed] [Google Scholar]
- Tsai R.Y., McKay R.D. 2002. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells.Genes Dev. 16:2991–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R.Y., McKay R.D. 2005. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin.J. Cell Biol. 168:179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., de Lange T. 1997. Control of telomere length by the human telomeric protein TRF1.Nature. 385:740–743 [DOI] [PubMed] [Google Scholar]
- Waizenegger I.C., Hauf S., Meinke A., Peters J.M. 2000. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase.Cell. 103:399–410 [DOI] [PubMed] [Google Scholar]
- Wang F., Podell E.R., Zaug A.J., Yang Y., Baciu P., Cech T.R., Lei M. 2007. The POT1-TPP1 telomere complex is a telomerase processivity factor.Nature. 445:506–510 [DOI] [PubMed] [Google Scholar]
- Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., O’Connor M.S., Songyang Z. 2007. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase.Nature. 445:559–562 [DOI] [PubMed] [Google Scholar]
- Yasumoto H., Meng L., Lin T., Zhu Q., Tsai R.Y. 2007. GNL3L inhibits activity of estrogen-related receptor-γ by competing for coactivator binding.J. Cell Sci. 120:2532–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager T.R., Neumann A.A., Englezou A., Huschtscha L.I., Noble J.R., Reddel R.R. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body.Cancer Res. 59:4175–4179 [PubMed] [Google Scholar]
- Zhu Q., Yasumoto H., Tsai R.Y. 2006. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability.Mol. Cell. Biol. 26:9279–9290 [DOI] [PMC free article] [PubMed] [Google Scholar]