Abstract
Consistent with the role of microRNAs (miRNAs) in down-regulating gene expression by reducing translation and/or stability of target mRNAs1, the levels of specific miRNAs are important for correct embryonic development and have been linked to several forms of cancer2-4. However, the regulatory mechanisms by which primary miRNAs (pri-miRNAs) are processed first to precursor miRNAs (pre-miRNAs) and then to mature miRNAs by the multiprotein Drosha and Dicer complexes5-8, respectively, remain largely unknown. The KH-type splicing regulatory protein (KSRP) interacts with single strand AU-rich elements (ARE)-containing mRNAs and is a key mediator of mRNA decay9,10. Here, we show that KSRP also serves as a component of both Drosha and Dicer complexes and regulates the biogenesis of a subset of miRNAs. KSRP binds with high affinity to the terminal loop (TL) of the target miRNA precursors and promotes their maturation. This mechanism is required for specific changes in target mRNA expression that affects specific biological programs, including proliferation, apoptosis and differentiation. These findings reveal an unexpected mechanism that links KSRP to the machinery regulating maturation of a cohort of miRNAs, that, in addition to its role in promoting mRNA decay, independently serves to integrate specific regulatory programs of protein expression.
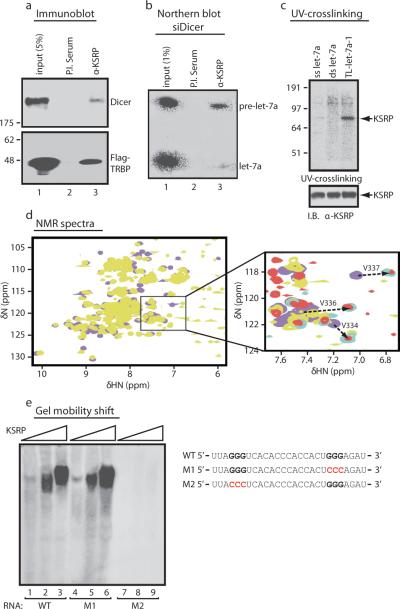
We analysed by mass spectroscopy the immunopurified Dicer-containing complex11 and identified, amongst others, KSRP (Supplementary Fig. 1a), a highly conserved nucleo-cytoplasmic RNA-binding protein regulating distinct steps of mRNA life (12,13 and Supplementary Fig. 1b-d). KH domains 3 and 4 of KSRP (KH3-4) are required to promote ARE-containing labile mRNAs decay9,10. Coimmunoprecipitation revealed that KSRP is an integral component of the Dicer complex in HeLa cells (Fig. 1a). Upon Dicer knock-down—induced (Supplementary Fig. 2a) pre-miRNAs upregulation, an anti-KSRP antibody immunoprecipitated pre-let-7a14 (Fig. 1b). Recombinant KSRP directly interacted with pre-let-7a-1, and KH3-4 accounted for the RNA high affinity binding (Supplementary Fig. 3a). Interestingly, KSRP interacted with the TL of pre-let-7a-1 (TL-let-7a-1) while did not associate with either single- or double-stranded mature let-7a (Fig. 1c) with KH3-4 accounting for KSRP binding to TL-let-7a-1 (Supplementary Fig. 3b).
Figure 1. KSRP, a component of Dicer complex, interacts with the TL of pre-let-7a-1.
a, Coimmunoprecipitation of endogenous KSRP and either Dicer or Flag-TRBP in HeLa cell extracts. b, Coimmunoprecipitation of KSRP and pre-let-7a in HeLa cells transiently transfected with siDicer. c, KSRP (300 nM) binds to TL-let-7a-1 but not to single-strand (ss) or double-strand (ds) let-7a. d, Superimposition of 15N-1H HSQC spectra of KH3-4 free (violet) and bound to TL-let-7a-1 (yellow). In the blow-up, spectra of bound KH3 (cyan) and KH2-3 (red). Arrows highlight the shift of a few selected peaks in the core of the RNA-binding groove. e, Interaction of KSRP (50-300 nM) with either wild-type (WT) TL-let-7a-1 or two distinct mutants.
We titrated the protein with increasing amounts of TL-let-7a-1 and TL-let-7a-1 with increasing amount of protein while monitoring the binding by NMR and CD, respectively. KH3-4 binds to TL-let-7a-1 with a 1:1 stoichiometry and ~50 nM Kd (Supplementary Fig. 3c) while single KH3 and KH4 domains bind to TL-let-7a-1 with ~500 nM and ~ 40 μM Kd, respectively (Supplementary Fig. 3d, e). In contrast to TNFα ARE interaction10, KH3 recognises a specific site in the TL-let-7a-1 and contributes most of the binding affinity in the KH3-4-RNA interaction, while KH4 plays an auxiliary role. Comparison of the chemical shift changes undergone by KH3 amide resonances upon RNA binding in the isolated KH3 and within the two-domain KH2-3 and KH3-4 constructs showed that the bound position of the resonance affected by TL-let-7a-1 binding is the same whether or not KH2 or KH4 are present (Fig. 1d and Supplementary Fig. 3f). NMR spectra showed that the position of nearly all of the resonances of the RNA-bound KH3 is the same for the TL-let-7a-1 and the entire pre-let-7a-1 (Supplementary Fig. 3g).
Our recent work indicated that KH3 recognises short G-rich stretches with high specificity and affinity15. The TL-let-7a-1 presents two GGG triplets, thus supporting the idea that KH3 docks KSRP on a specific site and that KSRP-prelet-7a-1 recognition takes place using a very different mode and a considerably higher affinity compared with ARE mRNA targets10,16. Mutational analysis of TL-let-7a-1 revealed that the 5' GGG triplet accounts for high affinity binding to KSRP (Fig. 1e).
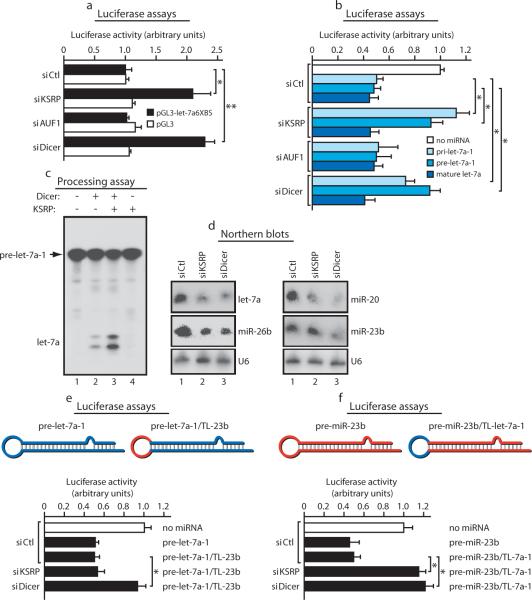
KSRP knock-down in both HeLa and NIH-3T3 cells (Supplementary Fig. 2b, c) abrogated the endogenous let-7a-mediated post-transcriptional silencing of a reporter construct containing six let-7a binding sites (let-7a6XBS) (Fig. 2a). Importantly, KSRP knock-down inhibited the effect of both transfected pri- and pre-let-7a-1 on let-7a6XBS reporter but left transfected mature let-7a function unaffected (Fig. 2b and Supplementary Fig. 4a, b). Control AUF1 knock-down (Supplementary Fig. 2d) had no effect (Fig. 2a, b).
Figure 2. KSRP regulates pre-let-7a-1 processing and controls the expression of select miRNAs.
a, KSRP knock-down reduces the effect of endogenous let-7a on the activity of pGL3-let-7a6XBS. Student's t-test: * p-value < 0.05, ** p-value < 0.01. b, KSRP knock-down reduces the effect of either prilet-7a-1 or pre-let-7a-1, but not of mature let-7a, on pGL3-let-7a6XBS activity. * p-value < 0.05. c, recombinant KSRP increases the processing activity of recombinant Dicer on pre-let-7a. d, Analysis of total RNA from either control (siCtrl), KSRP, or Dicer knock-down HeLa cells. e, f, KSRP knock-down does not reduce the effect of a chimaeric pre-let-7a-1 comprising the TL of miR-23b on the activity of pGL3-let-7a6XBS (e) while impairing the effect of a chimaeric pre-miR-23b comprising the TL of pre-let-7a-1 on the activity of pGL3-miR-23b3XBS (f). * p-value < 0.05. All data are presented as mean ± s.d. (n=4).
Recombinant KSRP increased the processing activity of Dicer (Fig. 2c) while KSRP immunodepletion from 293T extracts (Supplementary Fig. 2e) removed the pre-let-7a-1 processing activity (Supplementary Fig. 4c) leaving unaffected mature let-7a (Supplementary Fig. 4d). Finally, immunopurified KSRP-containing complexes specifically processed pre-let-7a-1 into mature let-7a (Supplementary Fig. 4e-g).
To investigate whether all pre-miRNAs are regulated by KSRP, we performed miRNA microarray analysis. Transient KSRP knock-down in HeLa cells significantly reduced (>1.5 fold) the expression of 14 miRNAs (Supplementary Fig. 5a) and reduced by 1.2-1.5 fold the expression of additional 20 miRNAs (Supplementary Table I). Northern blot analysis in both HeLa and NIH-3T3 cells confirmed that let-7a, miR-26b, miR-20, miR-106a, miR-21, and miR-16 were reduced by 40-70% upon KSRP knock-down while miR-23b and miR-24 were unaffected (Fig. 2d; Supplementary Fig. 5b-e, and data not shown). UV-crosslinking and in vitro processing experiments showed selectivity of KSRP binding and KSRP-induced processing for those miRNAs whose expression was regulated by KSRP (Supplementary Fig. 6a-c, and data not shown). Interestingly, the TL-miR-21 does not contain any GGG triplets but displays two potential nonoptimal binding sites for KSRP KH3 and KH4 (Supplementary Fig. 6d; 15). Mutation of the two G residues in the GUUG 5' element abrogated KSRP interaction while mutation of the 3' GG doublet only reduced the binding affinity (Supplementary Fig. 6d).
To investigate the function of TL-KSRP interaction, we utilized chimaeric pre-miRNAs with swapped loop sequences. KSRP knock-down did not affect the expression of a reporter containing let-7a binding sites when a chimaeric pre-let-7a-1 comprising the TL of miR-23b was expressed in HeLa cells (Fig. 2e). Conversely, expression of a reporter containing miR-23b binding sites was impaired by KSRP knock-down in HeLa cells expressing a chimaeric pre-miR-23b containing the TL of let-7a-1 (Fig. 2f, Supplementary Fig. 6e).
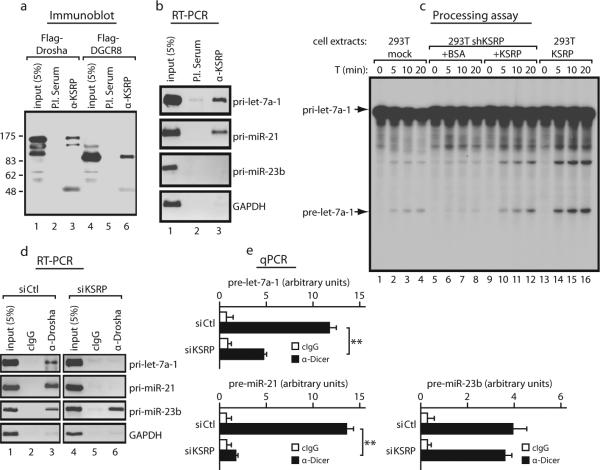
Since pre-miRNA accumulation induced by KSRP knock-down was low in comparison to that induced by Dicer knock-down (Supplementary Fig. 7a) and KSRP knock-down increased the levels of pri-let-7a-1 and pri-miR-21 (Supplementary Fig. 7b), we hypothesized an involvement of KSRP in pri-miRNA processing. KSRP coimmunoprecipitated with Flag-tagged Drosha and DGCR8 (Fig. 3a) and anti-KSRP antibody immunoprecipitated pri-let-7a-1 and pri-miR-21 but not pri-miR23b, pri-miR-24, and pri-miR-17 (Fig. 3b and Supplementary Fig. 7c). Endogenous, transfected and recombinant KSRP specifically interacted with pri-let-7a-1 (Supplementary Fig. 8a-c). Immunopurified KSRP-containing complexes processed pri-let-7a-1 similarly to Drosha-containing immunopurified complexes (Supplementary Fig. 9a). Either stable or transient KSRP knock-down in 293T, HeLa, and NIH-3T3 cells reduced the pri-let-7a-1 processing (Supplementary Fig. 9b-d) leaving unaffected the processing of pri-miR-23b (Supplementary Fig. 9b right panel and data not shown). Addition of recombinant KSRP to 293T shKSRP extracts restored pri-let-7a-1 processing while KSRP overexpression in 293T cells strongly increased pri-let-7a-1 processing (Fig. 3c and Supplementary Fig. 9e).
Figure 3. KSRP is a component of the Microprocessor complex, interacts with pri-let-7a-1 favouring its processing, and is required for both Drosha and Dicer complexes interaction with let-7a precursors.
a, Coimmunoprecipitation of endogenous KSRP with either Flag-Drosha or Flag-DGCR8. b, anti-KSRP immunoprecipitates select pri-miRNAs. c, KSRP (50 nM) restores pri-let-7a-1 processing when added to extracts from shKSRP stably transfected cells while KSRP overexpression enhances pri-let-7a-1 processing. d, e, KSRP knock-down impairs the interaction of Drosha and Dicer with select miRNA precursors. ** p-value < 0.01. All data are presented as mean ± s.d. (n=4).
We explored the possibility that KSRP favours the association of the enzymatic complexes with select miRNA precursors. Indeed, KSRP knock-down abrogated the interaction of Drosha with pri-let-7a-1, and pri-miR-21 (Fig. 3d) and strongly reduced the binding of Dicer to pre-Let-7a-1 and pre-miR-21 (Fig. 3e). In contrast, KSRP knock-down did not affect the interaction of the same pri-miRNAs and pre-miRNAs with either DGCR8 or TRBP (Supplementary Fig. 10a and data not shown)5,11.
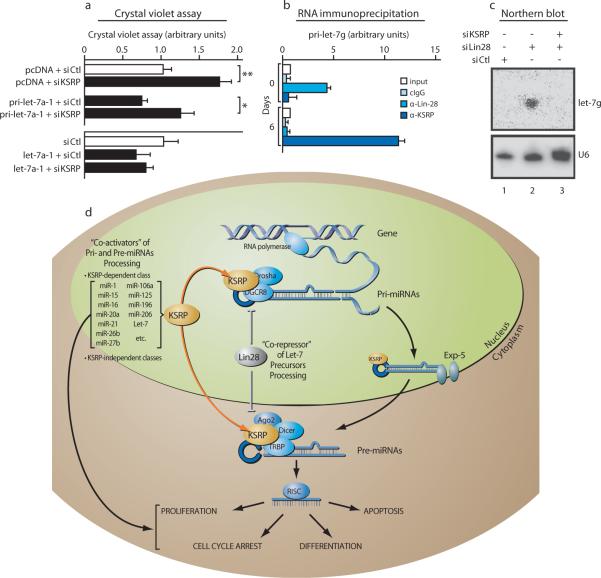
KSRP knock-down increased mRNA levels of two let-7 targets, NRAS and MYC (17,18, Supplementary Fig. 10b), and specificity was established because cotransfection of mature let-7a abolished this effect (Supplementary Fig. 10c, d). Furthermore, KSRP knock-down in U2OS osteosarcoma cells reduced the expression of mature let-7a, significantly upregulated cell proliferation19 (Fig. 4a, and data not shown), and reduced the anti-proliferative effect of transfected pri-let-7a-1 but not of mature let-7a (Fig. 4a, Supplementary Fig. 11a). Similarly, KSRP knock-down prevented the pri-miR-16-1—induced apoptosis20 while did not affect the activity of transfected mature miR-16 (Supplementary Fig. 11b, c). Recently, an essential role of select miRNAs (miR-1, miR-133a, miR-206) in C2C12 myoblasts differentiation has been reported21,22. KSRP knock-down in C2C12 reduced the maturation of “myogenic” miRNAs (Supplementary Fig. 12a-c). KSRP interaction with pri-miR-206, pri-miR-1-1, and pri-miR-1-2 was increased by pro-differentiative stimuli (DM, Supplementary Fig. 12d). Finally, KSRP knock-down inhibited the miR-206-induced down-regulation of direct target mRNAs, including Connexin 43 and DNA pol α22, impairing C2C12 differentiation (Supplementary Fig. 12e, f).
Figure 4. KSRP affects let-7-regulated cell proliferation and is involved in Lin28-regulated maturation of let-7g in P19 cells.
a, KSRP knock-down increases proliferation while inhibits the antiproliferative effect of transfected prilet-7a-1 but not of mature let-7a in U2OS cells. * p-value < 0.05, ** p-value < 0.01. b, KSRP interacts with pri-let-7g only in differentiated P19 cells. c, Lin28 knock-down in undifferentiated P19 cells induces the expression of let-7g. Concomitant Lin28 and KSRP knock-down abolishes let-7g expression. f, A model for KSRP-dependent regulation of processing of select miRNA. All data are presented as mean ± s.d. (n=4).
Recently, four papers8,23-25 demonstrated that the maturation of let-7 is blocked by Lin28 in undifferentiated embryonic stem cells and P19 cells. We observed that KSRP interacts with pri-Let-7g in P19 cells upon retinoic acid-induced differentiation (Fig. 4b). Lin28 knock-down in undifferentiated P19 cells induced let-7g expression while concomitant KSRP knock-down abolished this effect (Fig. 4c) suggesting that, upon Lin28 knock-down, KSRP promotes let-7g precursors maturation. Similarly, upon P19 differentiation, Lin28 expression is abrogated8,23-25 thus allowing KSRP to promote processing of let-7g precursors. This is also compatible with the recently reported mechanism of action of Lin2826. We suggest that TL is a pivotal structure where miRNA processing “activators” (e.g. KSRP) as well as “repressors” (e.g. Lin28) function in a coordinated way to convey proliferating, apoptotic or differentiating cues into changes of miRNA expression (Fig. 4d).
In conclusion, KSRP is a key regulator of the processing of a sizeable subset of miRNA precursors based on its high affinity binding to their TL. The TLs of the majority of KSRP-regulated microRNAs (let-7-a, -b, -c, -d, -f, -i, and miR-196a) contain short G-rich stretches of at least 3 Gs that represent the optimal binding site for KH315. However, the TLs of the other KSRP target miRNAs contain instead two sequential or isolated Gs and our data on KSRPTL-miR-21 interaction show that a significant, albeit different, contribution to the binding is provided by both Gs motifs (Supplementary Fig. 6d). These data underscore the adaptability of the protein to a broad range of single-strand RNA sequences15,16.
Upon binding, KSRP could optimise the positioning/recruitment of both the miRNA precursors processing complexes through protein-protein interactions (Supplementary Fig. 13a-c). The RNase sensitivity of KSRP—exportin-5 (Exp5), interaction suggests that KSRP is associated with the terminal loop of target miRNA precursors during nucleo-cytoplasmic transit (Supplementary Fig. 13a). Sequential immunoprecipitation experiments indicate that at least two pools of miRNA precursors exist, one associated with processing complexes including KSRP and the other associated with processing complexes that do not include KSRP (Supplementary Fig. 14). For example, Caceres and coworkers27 reported that hnRNPA1 binds to the TL of miRNAs whose processing is not affected by KSRP.
Altogether, our findings uncover an additional level of complexity for miRNA-dependent regulation of gene expression that contributes to the modulation of different biological programs.
METHODS SUMMARY
Ribonucleoprotein complexes immunoprecipitation (RIP)
RIP was performed as previously described by Chen et al28 with minor modifications. Briefly, cells lysates were immunoprecipitated with either Protein A- or ProteinA/ProteinG-Sepharose-coupled antibodies at 4° C overnight. Pellets were sequentially washed with the following buffers: Buffer I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl); Buffer II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 500 mM NaCl); and Buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1). Total RNA was prepared using Trizol®, retrotranscribed using random primers and amplified by PCR. The primer sequences are detailed in Supplementary Table II.
METHODS
Identification of Dicer-interacting proteins by liquid chromatography tandem mass spectrometry (LC-MSMS) analysis
Anti-Dicer mAbs 33, 73, and 83 and control mAbs11 were crosslinked to Protein G Sepharose 4 Fast Flow (Amersham Bioscience) and used to purify Dicer complexes from HEK293T (293T) cytoplasmic extracts (S1011). Coimmunoprecipitates were washed five times with lysis buffer (20 mM Tris-HCl [pH 7.5], 300 mM NaCl, 0.5% NP-40, 2.5 mM MgCl2). Proteins were separated by 10% SDS PAGE. Protein containing gel fragments, were digested with trypsin according to Schrimpf et al29 and analysed by LC-MSMS (LCQ Deca XP, Thermo 7 Finnigan). Proteins were identified using Turbo Sequest and MASCOT, searching SwissProt database restricted to human proteins. A protein was considered as identified, if at least two peptides were in the first rank, concerning the correlation of experimental with theoretical data, with an ion score greater than 25. Peptides with ion scores between 20 and 40 were peer reviewed for their quality of alignment. No KSRP-specific peptides were identified in immunoprecipitations with isotype-control mAbs.
NMR and CD spectroscopy
All CD spectra were recorded on Jasco J-715 spectropolarimeter (Jasco) equipped with a PTC-348 Peltier temperature-control system. RNA binding was monitored by adding increasing amounts of protein to 2 μM TL-let-7a-1 RNA in 10 mM Tris-HCl, pH 7.4, 100 mM NaCl, 0.5 mM TCEP. A temperature of 5 °C was chosen to optimise the signal change upon protein binding. The integral of the signal between 255 and 265 nm was fitted against the protein concentration using in house programs previously described30 and the values of the Kds were extracted. All NMR spectra were recorded on Bruker Avance spectrometers operating at 600 and 700 MHz 1H frequencies fitted with a supercooled probe. The spectra were processed with the NMRPipe package31 and analysed with Sparky32. Solutions of 25 μM 15N- labeled samples of KH3, KH4 and KH3-4 in 10 mM Tris-HCl buffer, 50 mM NaCl, 1 mM TCEP, pH 7.4, were titrated with TL-let-7a-1 RNA oligonucleotides. 15N-1H HSQC spectra were recorded at each point of the titration at 27 °C. Amide chemical shift changes as a function of protein/RNA ratio were fitted to obtain the Kd values for the complexes using in-house software as described by Martin et al30. Weighted average values of 15N and 1H chemical shift variations have been calculated as follows: Δδav = [(Δδ1H)2 + (Δδ15N)2/10]1/2 and used to map TL-let-7a-1 binding on a MolMol-generated molecular surface33.
Preparation of RNA substrates and in vitro processing assays
For pri-miRNAs processing assays, total cell extracts were prepared in 50mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% Triton X-100, 1X Complete®, 10% glycerol) from either HeLa or 293T cells and incubated (typically 40 μg per 25 μl reaction at 37° C for the indicated times) with in-vitro synthesised and uniformly labeled pri-miRNA (5 fmol) in processing buffer containing 100 mM KCH3COOH, 2 mM Mg (CH3COOH)2, 10 mM Tris-Cl (pH 7.6), 2 mM DTT, 10 mM creatine phosphate, 1 μg of creatine phosphokinase, 1 mM ATP, 0.4 mM GTP, 0.1 mM spermine, 2 units of RNasin®. Pre-miRNA processing assays were performed as previously described by Haase et al11.
miRNA profiling
HeLa cells were transiently transfected with either siRNA against KSRP or siRNA against Luciferase. Total RNA was prepared using Trizol® (Invitrogen) and enriched with RNA smaller than 40 nt using PureLink® miRNA Isolation Kit. RNA was labeled with either Cy-3 or Cy-5 using the NCode miRNA Labeling System®. A dye-swap design was employed. Labeled miRNAs were hybridized, in triplicate, to the Invitrogen NCode MultiSpecies miRNA Microarray V1®. Data were collected using the GenePix®Pro 5.0 Agilent Software (Axon Instruments). Normalization and data analysis were performed using a bootstrapping method34.
Recombinant proteins and antibodies
Recombinant Dicer was purified as described by Zhang et al35. MBPTRBP2 was expressed in bacteria and purified as described by Dorin et al36. Production of recombinant KSRP and its deletion mutants as well as of p37AUF1 have been described previously9. Affinity-purified rabbit polyclonal anti-KSRP antibody, and rabbit polyclonal anti-Dicer (349) were previously described9,37. Mouse monoclonal anti-GST was purchased from Chemicon. Mouse monoclonal anti-Flag (M2) and anti-alpha and beta-Tubulin were from Sigma. Rabbit polyclonal anti-Drosha (07-717) was purchased from Upstate, anti-DGCR8 (N-19) goat polyclonal IgG was purchased from Santa Cruz, while anti-Lin28 goat polyclonal IgG was purchased from R&D Systems.
Plasmids
Human pre-let-7a-1, and pre-miR-23b were cloned into pSUPER-gfp-neo plasmid (Oligoengine). Chimeric constructs including either the terminal loop sequence of miR-23b in the backbone of pre-let-7a-1 (UGAGGUAGUAGGUUGUAUAGUUGUGACUUAAGAUUAAACUAUACAAUCU ACUGUCUUUC) or the terminal loop sequence of let-7a-1 in the backbone of pre-miR-23b(UGGGUUCCUGGCAUGCUGAUUUUUAGGGUCACACCC ACCACUGGGAGAUAAAUCACAUUGCCAGGGAUUACC) were generated and cloned into pSUPER-gfp-neo. pCY vector38 containing part of the sequence (encompassing the mature miRNA) of either primary-let-7a-1 or primary-miR-23b were generated. Three miR-23b binding sites from a region between 360 and 385 nt of semaphorin 6D 3' UTR were cloned in pMIR-REPORT plasmid (Ambion). This sequence is a potential target of miR-23b according to TargetScan prediction program (http://www.targetscan.org). Flag-Dicer (kindly provided by Dr. M. Doyle, Friedrich Miescher Institute), and Flag-TRBP contain triple Flag tag at the N-terminus of proteins expressed from the pCIneo vector (Haase, unpublished). Flag-KSRP was previously described39. Luciferase reporter gene plasmid containing 6 let-7a binding sites and a plasmid containing genomic sequence encoding primary-let-7a were kind gifs of Dr. J.G. Belasco. Flag-Drosha and Flag-DGCR8 were generously provided by Dr V. Narry Kim. Myc tagged-Exportin-5 was provided by Addgene (Addgene plasmid # 12552). Part of the genomic sequence encompassing mouse pre-miR-1-2 (pri-miR-1-2) was cloned into pcDNA3 vector (Invitrogen). Mouse pre-miR-1-2 was cloned into pSUPER-gfp-neo plasmid (Oligoengine). Luciferase reporter gene plasmid containing 4 miR-1 binding sites was kindly provided by Dr. D. Srivastava.
Cell transfection, coimmunoprecipitation, and immunoblotting
Either HeLa, U2OS, P19 or NIH-3T3 cells were transiently transfected with Lipofectamine 2000 (Invitrogen). After either 48 or 72 hrs, cells were collected, washed with PBS, resuspended in lysis buffer, and the protein concentration of cell extracts determined by the Dc Protein assay (Bio-Rad). When required, cell lysates were incubated at room temperature with either RNase A (10 μg/ml, Ambion) or RNase V1 (1 U/ml, Ambion) for 30 or 15 min, respectively. 300 μg of protein were immunoprecipitated with Protein A-Sepharose-bound anti-KSRP antibody for 16 hr at 4°C with rotation. Immunoprecipitates were washed four times with lysis buffer and resuspended in SDS protein loading buffer. Proteins were subjected to SDS-PAGE, electroblotted onto PVDF membranes, and probed with antibodies as indicated. Anti-connexin 43 antibody was from Sigma, anti-DNA pol α (G-16) was from Santa Cruz, and anti-Myogenin (F5D) was from Iowa Hybridoma Bank. C2C12 myoblasts were cultured and treated as previously described40 and transfected using Lipofectamine 2000.
Northern blot analysis
RNA was extracted using Trizol®, resolved on 15% polyacrylamide-urea gels, and electroblotted onto HyBond N+® membranes. Membranes were hybridized overnight with radiolabeled antisense miRNAs in ExpressHyb® solution (Clontech). After hybridization, membranes were washed three times with 2X SSC and 0.05% SDS, twice with 0.1X SSC and 0.1% SDS, exposed overnight to imaging screens, and analysed using a Storm 860 PhosphorImager®. Signals were quantitated using Imagequant V1.2®. The same blot was hybridized (upon stripping in boiling 0.1% SDS) with three distinct probes, including control U6 RNA.
Luciferase assay
Either HeLa or NIH-3T3 cells (80% confluence in 12-well plates) were transfected with Luc reporters containing either 6 let-7a binding sites (pGL3-let-7a6XBS) or 3 miR-23b binding sites (pGL3-miR-23b3XBS), or empty pGL3 plasmid together with siRNAs using Lipofectamine 2000®. For some experiments cells were co-transfected with either pri-let-7a-1, or pre-let-7a-1 expression plasmids or mature let-7a. A co-transfected beta-galactosidase-containing plasmid was used to normalize firefly luciferase activity.
EdU incorporation for proliferation assay
72 hrs after transfection, cells were incubated for 2 hrs with EdU-containing medium. Nuclear incorporation of EdU was determined using Click-iT™ EdU imaging Kit (Invitrogen).
Crystal violet proliferation assay
72 hrs after transfection U-2OS cells, were fixed and stained with crystal violet solution. After two washes with water crystal violet staining was measured by spectrophotometer at λ 590 nm.
Tunel assay
Tunel-fluorescent staining of apoptotic cells was performed on U-2OS after 72 hrs from the transfection according to the manufacturer's protocol (Roche).
UV-cross linking experiments and gel mobility shift assays
UV-crosslinking experiments and gel mobility shift assays were performed essentially as described9,38.
Expression and purification of the recombinant proteins used for biophysical studies.
KSRP KH3, KH4 and KH3-4 proteins were obtained as previously described10. Briefly, 15N labelled proteins were expressed in Escherichia coli BL21 (DE3), as His-GST-fusion protein and initially purified using nickel affinity chromatography according to the manufacturer instructions. The His-GST fusion tags were then cleaved with TEV protease and removed using a second nickel affinity step. Proteins were further purified and buffer exchanged by gel filtration (Superdex 75 16/60 column, Pharmacia). Protein purity (always >95%) was assessed using SDS-PAGE and Coomassie blue staining. Protein quantification was achieved by a combination of spectrophotometry using predicted extinction coefficients and ninhydrin analysis of protein hydrolysates. All RNA oligonucleotides were chemically synthesized (Dharmacon).
shRNA-mediated KSRP knock-down
In order to stably knock-down KSRP the following oligonucleatides 5'-GATCAACCGGAGAGCAAGA-3' and 5'-GGACAGTTTCACGACAACG-3' for human and mouse proteins, respectively, were cloned into pSUPER-Puro (Oligoengine). Either 293T, NIH-3T3, or C2C12 cells were transfected using Lipofectamine Plus (Invitrogen). Transfectant pools were kept under selection in medium containing either 3 μg/ml (293T) or 1.2 μg/ml puromycin (NIH-3T3).
RT-PCR and Q-PCR
RT-PCR and q-PCR were performed as described in Gherzi et al41.
siRNAs
In order to knock down the following human proteins, these siRNAs were synthesized by Qiagen:
− human KSRP 5'- GAUCAACCGGAGAGCAAGAUU -3'
− mouse KSRP 5'-GGACAGUUUCACGACAACG-3'
− mouse Lin28 5'- GGGUUGUGAUGACAGGCAA-3'
− human Dicer 5'- GAAUCAGCCUCGCAACAAAUU -3'
− Luciferase 5'- CGUACGCGGAAUACUUCGAUU -3'
Supplementary Material
Acknowledgments
We are indebted to Drs. V. Narry Kim, T.C. Hobman, Ian G. Macara, D. Srivastava and J.G. Belasco for reagents, Dr. G. Corte for sharing lab facilities and critical reading, Dr. Y.T. Liu for microarray facility, Dr. M. Ponassi, and C. Nelson for technical assistance, to Drs. I. Diaz-Moreno, M. Doyle, S. Martin, and D. Hollingworth for helpful discussions and reagents, and to Drs. A. Pasquinelli and A. De Flora for helpful comments and discussions. Part of the studies have been conducted in the laboratories and facilities of the Centro Biotecnologie Avanzate (CBA, Genova, Italy). M.G.R. is an investigator with the Howard Hughes Medical Institute. M.T. is supported by a post-doctoral fellowship from the Italian Telethon Foundation. This work has been partly supported by grants from Italian I.S.S. (# 527B/2B/6), AIRC, and CIPE 2007 to R.G., I.S.S. (# 526D/39) to P.B., the EC FP6 Program Sirocco to W.F. and by NIH grants DK018477, DK39949, HL065445 to M.G.R. P.B. is recipient of a Senior Scholar Consultancy grant from A.I.C.F. Structural work on KSRP-RNA interaction is supported by grant WT022088MA from the Wellcome Trust. NMR spectra were recorded at the MRC Biomedical NMR Centre, London. The Friedrich Miescher Institute is supported by the Novartis Research Foundation.
Footnotes
Full methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Pillai RS, Bhattacharyya SN, Filipowicz W. Trends in cell biology. 2007;17(3):118. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Kumar MS, Lu J, Mercer KL, et al. Nature genetics. 2007;39(5):673. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 3.Thomson JM, Newman M, Parker JS, et al. Genes & development. 2006;20(16):2202. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calin GA, Croce CM. Nature reviews cancer. 2006;6(11):857. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 5.Kim VN, Han J, Siomi MC. Nat Rev Mol Cell Biol. 2009;10(2):126. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 6.Obernosterer G, Leuschner PJ, Alenius M, et al. RNA (New York, N.Y. 2006;12(7):1161. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guil S, Caceres JF. Nature structural & molecular biology. 2007;14(7):591. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 8.Viswanathan SR, Daley GQ, Gregory RI. Science (New York, N.Y. 2008;320(5872):97. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gherzi R, Lee KY, Briata P, et al. Molecular cell. 2004;14(5):571. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Mayoral MF, Hollingworth D, Masino L, et al. Structure. 2007;15(4):485. doi: 10.1016/j.str.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Haase AD, Jaskiewicz L, Zhang H, et al. EMBO reports. 2005;6(10):961. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Min M, Turck CW, Nikolic JM, et al. Genes & development. 1997;11(8):1023. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 13.Kroll TT, Zhao WM, Jiang C, et al. Development (Cambridge, England) 2002;129(24):5609. doi: 10.1242/dev.00160. [DOI] [PubMed] [Google Scholar]
- 14.Roush S, Slack FJ. Trends in cell biology. 2008;18(10):505. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Mayoral MF, Diaz-Moreno I, Hollingworth D, et al. Nucleic acids research. 2008 doi: 10.1093/nar/gkn509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruggiero T, Trabucchi M, Ponassi M, et al. BMC molecular biology. 2007;8:28. doi: 10.1186/1471-2199-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson SM, Grosshans H, Shingara J, et al. Cell. 2005;120(5):635. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Sampson VB, Rong NH, Han J, et al. Cancer research. 2007;67(20):9762. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 19.Lee YS, Dutta A. Genes & development. 2007;21(9):1025. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cimmino A, Calin GA, Fabbri M, et al. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(39):13944. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen JF, Mandel EM, Thomson JM, et al. Nature genetics. 2006;38(2):228. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HK, Lee YS, Sivaprasad U, et al. The Journal of cell biology. 2006;174(5):677. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman MA, Thomson JM, Hammond SM. RNA (New York, N.Y) 2008;14(8):1539. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rybak A, Fuchs H, Smirnova L, et al. Nature cell biology. 2008;10(8):987. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 25.Piskounova E, Viswanathan SR, Janas M, et al. The Journal of biological chemistry. 2008;283(31):21310. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 26.Heo I, Joo C, Cho J, et al. Molecular cell. 2008;32(2):276. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 27.Michlewski G, Guil S, Semple CA, et al. Molecular cell. 2008;32(3):383. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CY, Gherzi R, Andersen JS, et al. Genes & development. 2000;14(10):1236. [PMC free article] [PubMed] [Google Scholar]
- 29.Schrimpf SP, Langen H, Gomes AV, et al. Electrophoresis. 2001;22(6):1224. doi: 10.1002/1522-2683()22:6<1224::AID-ELPS1224>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 30.Martin SR, Biekofsky RR, Skinner MA, et al. FEBS letters. 2004;577(1-2):284. doi: 10.1016/j.febslet.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 31.Delaglio F, Grzesiek S, Vuister GW, et al. Journal of biomolecular NMR. 1995;6(3):277. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 32.Goddard TD, Kneller DG. SPARKY. University of California; San Francisco: [Google Scholar]
- 33.Koradi R, Billeter M, Wuthrich K. Journal of molecular graphics. 1996;14(1):51. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 34.Kerr MK, Churchill GA. Biostatistics. 2001;2(2):183. doi: 10.1093/biostatistics/2.2.183. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Kolb FA, Brondani V, et al. The EMBO journal. 2002;21(21):5875. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorin D, Bonnet MC, Bannwarth S, et al. The Journal of biological chemistry. 2003;278(7):4440. doi: 10.1074/jbc.M208954200. [DOI] [PubMed] [Google Scholar]
- 37.Kotaja N, Bhattacharyya SN, Jaskiewicz L, et al. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(8):2647. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen CY, Gherzi R, Ong SE, et al. Cell. 2001;107(4):451. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 39.Briata P, Forcales SV, Ponassi M, et al. Molecular cell. 2005;20(6):891. doi: 10.1016/j.molcel.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 40.Wu Z, Woodring PJ, Bhakta KS, et al. Molecular and cellular biology. 2000;20(11):3951. doi: 10.1128/mcb.20.11.3951-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gherzi R, Trabucchi M, Ponassi M, et al. PLoS biology. 2006;5(1):e5. doi: 10.1371/journal.pbio.0050005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.