Abstract
Summary
Analysis of a synthetic ABA agonist uncovers a new family of ABA binding proteins that control signal transduction by directly regulating the activity of type 2C protein phosphatases.
PP2Cs are vital phosphatases that play important roles in abscisic acid (ABA) signaling. Using chemical genetics, we previously identified a synthetic growth inhibitor called pyrabactin. Here we show that pyrabactin is a selective ABA agonist that acts through PYR1, the founding member of a family of START proteins called PYR/PYLs, which are necessary for both pyrabactin and ABA signaling in vivo. We show that ABA binds to PYR1, which in turn binds to and inhibits PP2Cs. We therefore suggest that PYR/PYLs are ABA-receptors that function at the apex of a negative regulatory pathway that controls ABA signaling by inhibiting PP2Cs. Our results illustrate the power of small-molecule approaches for sidestepping the functional redundancy that hampers genetic analysis.
Abscisic acid (ABA) has been the focus of intense investigation since it was identified in the 1960s as an endogenous small molecule growth inhibitor and regulator of plant stress physiology. Genetic analyses have identified many factors involved in ABA signaling (1), including the group A type 2 C protein phosphatases (PP2Cs) that negatively regulate ABA signaling at an early step in the pathway (2) and the SnRK2 kinases that are positive regulators (3–5). Several ABA binding proteins have been reported (6–8), however their roles are not fully understood, as they do not appear to work via the genetically defined signaling pathway (9). Additionally, they show negligible binding to the non-natural stereoisomer (−)-ABA 1 (6–8), which is bioactive (10, 11) and acts through the (+)-ABA 2 signaling pathway (12). Thus, receptors that recognize both ABA stereoisomers and act through the central pathway remain to be discovered.
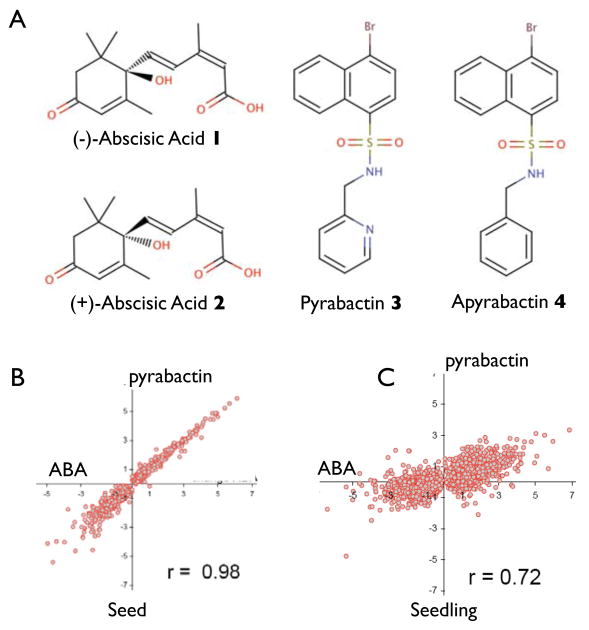
The genetic dissection of ABA perception has not identified proteins resembling receptors, suggesting that the ABA receptor(s) may be functionally redundant or required for viability (13). We therefore pursued a chemical genetic strategy (14) because chemicals can bypass redundancy by inducing phenotypes not revealed by single locus mutations (15). For example, an antagonist with low selectivity can perturb the function of an entire protein family, while a selective agonist can illuminate the function of one member of normally redundant receptors, as we describe here with pyrabactin 3 (Figure 1A). As part of an earlier effort, we identified pyrabactin as a synthetic seed germination inhibitor (14) and noted that an ABA-hypersensitive Arabidopsis accession is hypersensitive to pyrabactin, but not an inactive analog, apyrabactin 4 (Figure 1A). This hinted that pyrabactin might activate the ABA pathway, which plays a role in inhibiting germination in response to environmental stress (16). Aside from ABA analogs, no synthetic agonists of this stress pathway are known. We therefore investigated pyrabactin’s action further. ABA insensitive mutants, but not ABA biosynthesis or GA perception mutants, dramatically reduce pyrabactin sensitivity (SOM Figure 1), consistent with the agonist hypothesis. To investigate further, we compared the ABA and pyrabactin responses of seeds and seedlings using microarrays. In seeds, both compounds induce highly correlated transcriptional responses (r = 0.98; Figure 1B; SOM File 1). Three unrelated germination inhibitors (17) failed to induce ABA-like effects (SOM Figure 1), which demonstrates that an indirect germination effect is not sufficient to account for pyrabactin’s agonist activity in seeds. In contrast to seed responses, seedling ABA and pyrabactin responses show poorer correlation (r = 0.72) (Figure 1C) and few ABA responsive genes significantly respond to pyrabactin (SOM File 1). Collectively, these data suggest pyrabactin’s selectivity is greater than the natural compound it mimics, ABA.
Figure 1. Pyrabactin Is A Seed Selective ABA Agonist.
(A) Structures Of Molecules Described In This Study. (B) ATH1 Microarray Comparison of Pyrabactin and ABA Effects on Seeds and Seedlings. The axes plot log2-transformed values for probe responses to pyrabactin (Y-axis) or ABA (X-axis), relative to control samples. Inset in each scatter plot is the Pearson correlation coefficient for each comparison. Probes selected for analyses were those significantly responsive to either ABA or pyrabactin. Germination responsive transcripts were removed for seed analyses. Detailed methods are provided in SOM.
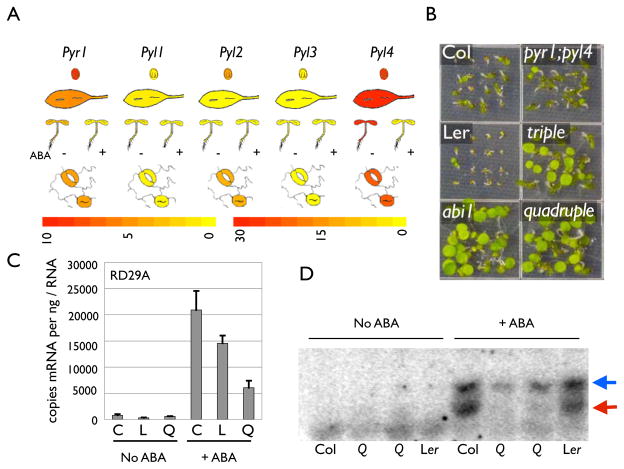
Selective agonists have a long history as reagents for receptor identification, so we used pyrabactin for genetic dissection and isolated 12 PYRABACTIN RESISTANCE 1 (Pyr1) mutant alleles. Pyr1 was isolated by map based cloning, which revealed that it encodes a member of the cyclase subfamily of the START/Bet v I superfamily. START proteins share a conserved hydrophobic ligand-binding pocket (18,19), making PYR1 a candidate cellular target of pyrabactin. There are 13 genes in the Arabidopsis genome possessing significant similarity to Pyr1, which we have named Pyl1 -Pyl13 (for PYR1-Like). Pyl9 has been independently identified as an ABI1 interacting protein (called RCAR1), and its role in ABA signaling is described in an accompanying manuscript (Grill manuscript). The pyr1 mutant alleles isolated are predicted to produce a variety of defects in PYR1 (SOM Figure 2). Gene expression databases (20–24) show that Pyr/Pyl mRNAs are expressed at high levels in seeds and guard cells and respond to ABA (Figure 2A), consistent with a role in ABA signaling. However, all of the pyr1 mutants isolated, including putative null alleles, reduce pyrabactin but not ABA sensitivity. To examine if functional redundancy might obscure a role for Pyr1 in ABA signaling, we isolated Pyl1, Py12 and Py14 insertion alleles and constructed multi-locus mutants. Triple (pyr1;pyl1;pyl4) and quadruple (pyr1;pyl1;pyl2;pyl4) mutant lines display strong ABA insensitivity (Figure 2B), which introduction of PYR1 or PYL4 expressing transgenes (SOM Figure 2) reverses. The quadruple mutant is less sensitive to (+)-ABA as measured using seed germination, root growth, qRT-PCR (Figure 2C) and SnRK2 kinase assays (Figure 2D, SOM Figure 3). Collectively, our genetic data show that Pyr/Pyl genes are necessary for multiple ABA responses in vivo and illustrate the differential behavior that natural and synthetic agonists can display in genetic screens.
Figure 2. Pyr/Pyls Are Necessary For ABA Signaling.
(A) Pyr1 And Pyl1 - Pyl4 Expression Levels. Plots were made using the EFP browser (23); these heatmaps show normalized ATH1 microarray expression values (multiplied by 100) according to the color scales shown, the color bar is for the guard cell data only. (B) PYR/PYL Genes Act Redundantly In ABA Signaling. Genotypes shown were germinated on media containing 0.9 μ,M (+)-ABA and documented 7 days post-imbibition. (C) PYR/PYL Genes Are Required For Normal ABA-lnduced Gene Expression In Seedlings. Shown are qRT-PCR results for the ABA-responsive gene RD29. L= Ler, C = Col, Q = quadruple mutant. (D) The PYR/PYL Genes Are Required For Normal SnRK2 Kinase Activity. In-gel kinase assays were conducted on extracts made from control or ABA treated plants of the genotypes shown; extracts from two separate quadruple (Q) mutant lines are shown. Red arrow corresponds to SnRK2.2 and 2.3; Blue to SnRK2.6/OST1.
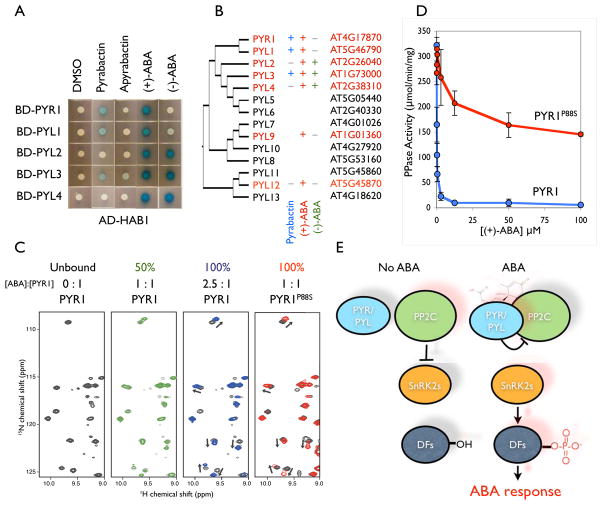
Since PYR1 is a predicted ligand-binding START protein necessary for pyrabactin activity, we hypothesized that pyrabactin might promote a protein-protein interaction between PYR1 and a downstream effector. To test this, ~2 million prey cDNA clones were screened against PYR1 Y2H bait on media containing pyrabactin. This revealed that HAB1 (25, 26) interacts with PYR1 in response to both pyrabactin and (+)-ABA (Figure 3A). HAB1 resides in the group A subfamily of plant PP2Cs, which contains 9 partially redundant members that negatively regulate ABA signaling (27, 28). To examine the relevance of the Y2H data, we next performed a series of control experiments. First, we investigated the effects of defective PYR1 and PP2C amino acid substitution mutants on ABA-responsive interactions in the Y2H. PYR1S152L and PYR1P88S, isolated because they cause strong pyrabactin insensitivity, reduce ABA induced PYR1-PP2C interactions (Figure 3A); similarly the dominant ABA insensitive ABI2G168D mutant (Figure 3B) disrupts the PYR1-ABI2 interaction. Second, we investigated in planta interactions by co-expressing a panel of YFP-tagged PP2Cs and HA-tagged PYR1 constructs in N. benthamiana. Co-immunoprecipitation experiments performed on ABA or mock-treated plants recapitulate the PYR1-PP2C interactions observed in yeast and also show that PYR1 does not interact with ABI1G180D, encoded by abi1-1 (Figure 3C). Third, we queried PYR1 and ABI1 against an ABA-orfeome collection using the Y2H assay and observed that PYR1 only interacts with group A PP2C proteins in response to ABA (Figure 3D; SOM File 2), showing it is not a “sticky” interactor. Fourth, we successfully reconstituted the ABA induced PYR1-PP2C interactions in vitro using recombinant proteins (Figure 3E). These results show that an ABA response system can be reconstituted using only PYR1 and a PP2C. Collectively, our yeast, in planta and in vitro experiments support the hypothesis that ABA promotes a biologically meaningful interaction between PYR1 and group A PP2Cs.
Figure 3. PYR/PYLs Bind To Group A PP2Cs In Response To ABA.
(A) Wild Type and mutant PYR1-PP2C interactions. PYR1 and 3 different substitution mutants were constructed as activation domain (AD) fusion proteins and tested for their interactions with BD-HAB1 (tope panel) using the Y2H assay using the compounds shown at top. In the 2 lower panels, BD fusions of HAB1, ABI1, ABI2 or ABI2G168D were tested for ABA/pyrabactin induced interactions with AD-PYR1. (B) ABA Promotes PYR1 - PP2C Interactions In Planta. Total protein extracts (input) were made from N. benthamiana leaves transformed with the indicated constructs/treatments, immunoprecipitated with anti-HA-agarose and immuno-detected using anti-GFP or ant-HA antibodies. (C) ABA-Orfeome Analysis Of ABI1 Interactions. Shown are subsets of an ABA-orfeome queried with ABI1; auto-activators circled. (D) Reconstitution Of ABA-Responses In Vitro. Pull-down assays using GST-HAB1 and 6×His-PYR1 were conducted using purified recombinant proteins. GST-ABI1 and ABI2 tests were done with crude lysates. 10 μM (+)-ABA was used in (A,D); (B,C) 100 μM ABA (mixed stereoisomers).
Given the redundancy observed in our genetic analyses, it is likely that other PYR/PYLs interact with PP2Cs in response to ABA. We therefore used the Y2H assay to explore interactions of HAB1 and PP2CA/AHG3 with a panel of 12 PYR/PYLs, which shows that (+)-ABA promotes interactions between PYR1, PYL1 - PYL4 and HAB1 (Figure 4A). We next used this PYR/PYL panel to examine ligand response selectivities, which shows that these 5 (+)-ABA responsive PYR/PYLs do not all bind HAB1 in response to non-natural agonists. For example, PYL2, PYL3 and PYL4 respond to both (+)-ABA and (−)-ABA (Figure 4A), making these proteins candidates for the dual stereoisomer receptors predicted by earlier studies (12). Consistent with this hypothesis, our quadruple mutant possesses greatly reduced (−)-ABA sensitivity (SOM Figure 3). Ligand-selective interactions are also observed for pyrabactin, which promotes interactions between HAB1 and PYR1, PYL1 or PYL3 (Figure 4A). Of these, only Pyr1 shows high-level mRNA expression in seeds (Figure 2B), which probably explains why Pyr1 mutates to yield seed pyrabactin insensitivity. Lastly, we observe that PYL12 interacts with PP2CA/AHG3 in response to ABA (SOM Fig 3). Thus, at least 6 of the 14 PYR/PYLs confer ABA responsiveness to yeast. We hypothesize that the entire family participates in ABA-promoted interactions, as these 6 genes are distributed across the PYR/PYL phylogenetic tree (Figure 4B). Additionally, since PYR/PYLs control which ligands trigger ABA signaling, they may be desirable targets for drugging the ABA pathway, which is of considerable interest, given the high cost and poor stability of ABA.
Figure 4. PYR1 Is An ABA Binding Protein That Regulates PP2C Activity.
(A) PYR/PYL Proteins Determine Selectivities For Responses To Different Ligands. A panel of PYR/PYL genes were constructed as AD-fusions and tested in yeast for interactions with HAB1 and AHG3 in response to (+)-ABA, (−)-ABA, pyrabactin or apyrabactin. Shown are results for 5 PYR/PYLs that interact with HAB1 in response to ABA. (B) ABA-Response Activity Is Distributed Throughout The PYR/PYL Family. Shown is a neighbor-joining tree of the PYR/PYL family. The middle panel summarizes ligand selectivity data derived from Y2H experiments. (+)-ABA responsive PYR/PYLs are colored red, AGIs shown at right. (C) ABA Binds To PYR1 And PYR1P88S. Shown are sub-regions of HSQC spectra for 15N-labelled PYR1 and PYR1P88S in response to increasing amounts of ABA. Arrows indicate amide protons whose chemical environments shift in response to ABA. (D) PYR1 Inhibits PP2C Activity In The Presence Of ABA. Initial reaction velocities of recombinant GST-HAB1 were tested in the presence of PYR1 or PYR1P88S and differing ABA concentrations using the substrate pNPP. The measured IC50s are 125 nM for PYR1 and 50 μM for PYR1P88S. (E) Hypothesized Model For PYR/PYL Control Of ABA Signaling. We propose the following model: in the absence of ABA (left), PYR/PYL proteins are not bound to PP2Cs and therefore PP2C activity is high, which prevents phosphorylation and activation of SnRK2s and downstream factors (DFs). In the presence of ABA, PYR/PYLs bind and inhibit PP2Cs. This then allows accumulation of phosphorylated downstream factors and ABA transcriptional responses.
Given the role of PYR/PYLs in controlling ligand responses, we explored if (+)-ABA binds PYR1 using 15N-labeled PYR1 and PYR1P88S in HSQC NMR experiments, which probe chemical shifts of protein amide-NH bonds in response to ligands (29). Since START proteins contain a conserved ligand-binding cavity (30), binding should selectively perturb residues lining this cavity. Addition of (+)-ABA alters the HSQC signals for many PYR1 and PYR1P88S residues (Figure 4C, SOM Figures 4 and 5), which shows that ABA binds PYR1 and likely induces a conformational change. We also investigated the PYR1-HAB1 interaction in the presence of ABA, since our NMR experiments show that PYR1P88S is not defective in ABA binding. Addition of unlabelled HAB1 causes strong peak broadening for PYR1 but not PYR1P88S HSQC signals (SOM Figure 6), which localizes the PYR1P88S defect to binding HAB1 after ABA perception.
Since PP2Cs are negative regulators of ABA signaling, we hypothesized that ABA-promoted PYR/PYL-PP2C interactions would inhibit phosphatase activity. To test this, we examined the effects of (+)-ABA on PP2C enzyme kinetics using recombinant HAB1, PYR1 or PYR1P88S. These experiments show that (+)-ABA acts as a potent saturable inhibitor of phosphatase activity in the presence of PYR1 (IC50 = 125 nM), but not PYR1P88S (IC50 = 50 μM; Figure 4D, SOM Figure 7). Similarly, ABA displays saturable inhibition of HAB1 PP2C activity in the presence of recombinant PYL4 (SOM Figure 7). Thus, PYR/PYL regulation of PP2Cs is ABA-responsive, which defines an unprecedented mechanism for PP2C regulation.
Collectively, we have shown that PYR1 binds (+)-ABA, PYR/PYLs bind to and inhibit PP2Cs in response to (+)-ABA, and that PYR/PYLs control which ligands trigger PP2C interactions. The simplest conclusion from our data is that the PYR/PYLs are a new family of ABA receptors. However, the precise site of ABA binding is currently unknown and could be shared with the PP2C. Discriminating between the receptor/co-receptor models will require atomic level structural studies of co-crystallized PYR/PYLs, PP2Cs and ligands. Importantly, the PYR/PYLs interact directly with PP2Cs, which are core components of the ABA signaling pathway. Since SnRK2 activity is decreased in the PYR/PYL quadruple mutant, we propose a hypothetical model (Figure 4D) for ABA action in which ABA and PYR/PYLs inhibit PP2Cs, which in turn relieves repression of positive factors such as the SnRK2s. Consistent with this model, we observe interaction of SnRK2.2 with PP2CA/AHG3 and AHG1 using the Y2H assay (SOM Figure 3). This suggests that the low SnRK2 activity observed in the PYR/PYL quadruple mutant may be a direct consequence of PP2C-SnRK2 interactions, however this needs further investigation. The role of PP2Cs in ABA signaling has been complicated by abi1-1 and abi2-1. Their dominant phenotypes suggest they encode hypermorphic proteins (31), but they paradoxically reduce, but do not abolish, PP2C activity (32). Our data show that these mutants do not bind PYR1 in response to ABA. We therefore hypothesize that ABA normally lowers wild type PP2C activity via PYR/PYL proteins, but abi PP2Cs escape this and disrupt signaling due to their residual activity. Consistent with this model, a second site mutation that abolishes abi1-1’s catalytic activity suppresses its dominant ABA insensitive phenotype (32).
The ABA signaling pathway has been the subject of genetic analysis for almost 30 years, but the Pyr/Pyls never emerged as factors necessary for ABA response. This appears to be a consequence of the necessity of a multi-gene mutant to observe an ABA-insensitive phenotype, and is thus a consequence of the redundancy typical of most plant genes. When using pyrabactin as a synthetic agonist to probe the pathway genetically, Pyr1 emerged with ease. The reason for this appears to be due to pyrabactin’s selectivity for a subset of the entire PYR/PYL family, which enabled us to bypass the genetic redundancy that masks ABA phenotypes in single mutants. Thus, our results demonstrate the power of the chemical genetic approach to reveal phenotypes for normally redundant genes. Because many eukaryotic genomes are highly redundant, we expect that small molecule approaches will provide a powerful addendum to classical genetic analysis as a tool for pathway dissection.
Supplementary Material
References
- 1.Finkelstein RR, Gampala SSL, Rock CD. The Plant Cell. 2002;14:S15. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen GJ, Kuchitsu K, Chu SP, Murata Y, Schroeder JI. Plant Cell. 1999;11:1785. doi: 10.1105/tpc.11.9.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujii H, Verslues PE, Zhu JK. Plant Cell. 2007 Feb;19:485. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Plant Cell. 2002;14:3089. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida R, et al. Plant Cell Physiol. 2002;43:1473. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, et al. Science. 2007;315:1712. doi: 10.1126/science.1135882. [DOI] [PubMed] [Google Scholar]
- 7.Shen YY, et al. Nature. 2006;443:823. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- 8.Pandey S, Nelson DC, Assmann SM. Cell. 2009;136:136. doi: 10.1016/j.cell.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 9.McCourt P, Creelman R. Current Opinion in Plant Biology. 2008;11:474. doi: 10.1016/j.pbi.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 10.Huang D, et al. The Plant Journal. 2007;50:414. doi: 10.1111/j.1365-313X.2007.03056.x. [DOI] [PubMed] [Google Scholar]
- 11.Sondheimer E, Galson EC, Chang YP, Walton DC. Science. 1971;174:829. doi: 10.1126/science.174.4011.829. [DOI] [PubMed] [Google Scholar]
- 12.Nambara E, et al. Genetics. 2002;161:1247. doi: 10.1093/genetics/161.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCourt P. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:219. doi: 10.1146/annurev.arplant.50.1.219. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, et al. Nat Chem Biol. 2007;3:716. doi: 10.1038/nchembio.2007.32. [DOI] [PubMed] [Google Scholar]
- 15.Cutler S, McCourt P. Plant Physiol. 2005;138:558. doi: 10.1104/pp.104.900152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toh S, et al. Plant Physiol. 2008;146:1368. doi: 10.1104/pp.107.113738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bassel GW, et al. Plant Physiol. 2008;147:143. doi: 10.1104/pp.107.110841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyer LM, Koonin EV, Aravind L. Proteins: Structure, Function, and Genetics. 2001;43:134. doi: 10.1002/1097-0134(20010501)43:2<134::aid-prot1025>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 19.Radauer C, Lackner P, Breiteneder H. BMC Evol Biol. 2008;8:286. doi: 10.1186/1471-2148-8-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmid M, et al. Nat Genet. 2005;37:501. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 21.Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. Plant J. 2005;41:697. doi: 10.1111/j.1365-313X.2005.02337.x. [DOI] [PubMed] [Google Scholar]
- 22.Goda H, et al. Plant J. 2008;55:526. doi: 10.1111/j.0960-7412.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- 23.Winter D, et al. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI. Plant Methods. 2008;4:6. doi: 10.1186/1746-4811-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saez A, et al. The Plant Journal. 2004;37:354. doi: 10.1046/j.1365-313x.2003.01966.x. [DOI] [PubMed] [Google Scholar]
- 26.Leonhardt N, et al. THE PLANT CELL. 2004;16:596. doi: 10.1105/tpc.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishimura N, et al. The Plant Journal. 2007;50:935. doi: 10.1111/j.1365-313X.2007.03107.x. [DOI] [PubMed] [Google Scholar]
- 28.Saez A, et al. Plant Physiol. 2006 Aug;141:1389. doi: 10.1104/pp.106.081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer B, Peters T. Angewandte Chemie International Edition. 2003;42:864. doi: 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- 30.Liu JJ, Ekramoddoullah AKM. Physiological and Molecular Plant Pathology. 2006;68:3. [Google Scholar]
- 31.Robert N, Merlot S, N’Guyen V, Boisson-Dernier A, Schroeder JI. FEBS Letters. 2006;580:4691. doi: 10.1016/j.febslet.2006.07.047. [DOI] [PubMed] [Google Scholar]
- 32.Gosti F, et al. THE PLANT CELL. 1999;11:1897. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.We thank N. Raikhel, J. Bailey-Serres and C. Larive for comments; K. Nito and T. Demura for materials. Supported by CRC (SRC, PM, DD), NSERC (SRC, DD, NJP), NSF awards IOS0820508 (SRC), MCB0417118 (JIS), NIH awards R01GM060396 (JIS), R01GM59138 (JKZ) and U54GM074901 (BV). SRC thanks J.R. Coleman for early support of Pyr1 research.
- 34.Author contributions are as follow. Microarrays: PF, NJP. pyr1 mutants: PF. Mutant analyses: PF, TFC. Y2H, biochemistry, Pyr/Pyl genetics: SYP. In planta pull-downs: NN, JIS. PYL12-AHG3 interaction: RF. HSQC experiments: DJ, BV. SnRK2 assays: HF, JKZ. GFP-PYR1 Localization: SEA. PP2C assays: SYP, JS, AR, PR. EMS seed: DB. ABA Orfeome: SL, DD, PM. Project conception, positional cloning, phone calls, freak-outs and writing: SRC.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.