Abstract
Oxysterol-binding protein (OSBP) and OSBP-related proteins (ORPs) constitute a large gene family that differentially localize to organellar membranes, reflecting a functional role in sterol signaling and/or transport. OSBP partitions between the endoplasmic reticulum (ER) and Golgi apparatus where it imparts sterol-dependent regulation of ceramide transport and sphingomyelin synthesis. ORP9L also is localized to the ER–Golgi, but its role in secretion and lipid transport is unknown. Here we demonstrate that ORP9L partitioning between the trans-Golgi/trans-Golgi network (TGN), and the ER is mediated by a phosphatidylinositol 4-phosphate (PI-4P)-specific PH domain and VAMP-associated protein (VAP), respectively. In vitro, both OSBP and ORP9L mediated PI-4P–dependent cholesterol transport between liposomes, suggesting their primary in vivo function is sterol transfer between the Golgi and ER. Depletion of ORP9L by RNAi caused Golgi fragmentation, inhibition of vesicular somatitus virus glycoprotein transport from the ER and accumulation of cholesterol in endosomes/lysosomes. Complete cessation of protein transport and cell growth inhibition was achieved by inducible overexpression of ORP9S, a dominant negative variant lacking the PH domain. We conclude that ORP9 maintains the integrity of the early secretory pathway by mediating transport of sterols between the ER and trans-Golgi/TGN.
INTRODUCTION
The Golgi apparatus is a highly compartmentalized organelle that serves as a nexus for the processing, sorting, and vesicular transport of protein and lipid cargo between organelles. Regulation of vesicular transport is dependent on coat proteins and factors that control coat assembly/disassembly, as well as membrane dynamics and individual lipid regulators. A continuing challenge is the identification of the proteins responsible for spatial generation of Golgi lipid regulators by localized synthesis and/or transport (Huijbregts et al., 2000; De Matteis et al., 2007). In this study we investigated one such lipid-binding protein, oxysterol-binding protein (OSBP)-related protein 9 (ORP9), and its role in endoplasmic reticulum (ER)-to-Golgi transport.
Lipid synthesis and transport to the Golgi apparatus regulates local and distal cellular functions. For example, synthesis of sphingomyelin (SM) and glycosphingolipids (Huitema et al., 2004) in combination with cholesterol is required for raft assembly in the Golgi and cargo delivery to the apical surface of polarized cells (Hoekstra et al., 2003). SM synthesis in the Golgi apparatus is dependent on delivery of ceramide from the ER by the ceramide transfer protein (CERT; Hanada et al., 2003). Ceramide transport by CERT involves binding of a phosphatidylinositol 4-phosphate (PI-4P)-specific pleckstrin homology (PH) domain to the Golgi apparatus and interaction with vesicle-associated membrane protein (VAMP)-associated-protein (VAP) by the “two phenylalanines in an acidic tract” (FFAT) motif (Hanada et al., 2003). In addition, CERT activates protein kinase D activity and secretory transport by a mechanism that could involve localized diacylglyceride production (Fugmann et al., 2007).
Synthesis of complex glycosphingolipids (D'Angelo et al., 2007; Halter et al., 2007) and generation of Golgi secretory vesicles in the trans-Golgi network (TGN) is dependent on FFAP2, a small PH-domain glucosylceramide (GlcCer)-binding protein (Godi et al., 2004). FFAP2 facilitates the Golgi-to-ER or intra-Golgi delivery of GlcCer to glycolipid synthases in distal Golgi elements (D'Angelo et al., 2007; Halter et al., 2007). The FFAP2 PH domain interacts with Golgi-localized PI-4P and ARF, and a C-terminal glycolipid transfer protein domain binds GlcCer in donor membranes (Godi et al., 2004).
CERT and FAPP2 are involved in interorganelle delivery of lipid substrates to biosynthetic enzymes, but other Golgi-localized lipid- and sterol-binding proteins, such as Nir2 and OSBP, have less clearly defined functions with respect to lipid transfer and/or signaling (Lev, 2004; Perry and Ridgway, 2005). All four of these proteins have unique lipid-binding domains but functionally similar PH and/or FFAT domains, indicating related modes of action involving dual interaction with the ER and Golgi apparatus at membrane contact sites or through long-range targeted interactions (Olkkonen and Levine, 2004). OSBP, the founding member of a 12-gene family in mammals, has a N-terminal PH domain that interacts with PI-4P and ARF in the Golgi apparatus (Levine and Munro, 2002; Lagace et al., 1997), an internal FFAT motif that binds VAP (Wyles et al., 2002; Loewen et al., 2003), and a unique C-terminal sterol-binding domain (Ridgway et al., 1992) termed the OSBP homology (OSH) domain. By virtue of this domain organization, OSBP partitions between the ER and Golgi apparatus in a sterol-dependent manner (Ridgway et al., 1992; Mohammadi et al., 2001). OSBP interaction with sterols and the Golgi apparatus is required to activate CERT-dependent SM synthesis, possibly as a mechanism to coordinate the levels of these two raft components in the Golgi compartment (Perry and Ridgway, 2006). Mechanistically, this could involve OSBP-mediated sterol transport to the Golgi or activation of a signaling pathway by the OSBP–sterol complex. OSBP and other OSBP-related proteins (ORPs) have also been implicated it variety of other cellular functions, such as signal transduction (Wang et al., 2005; Lessmann et al., 2007), cytoskeletal dynamics (Johansson et al., 2007; Wyles et al., 2007), and gene expression (Yan et al., 2007b, 2008), suggesting wide-reaching roles related to sterol sensing or transfer.
Another OSBP family member that specifically localizes to the ER and Golgi apparatus is ORP9/OSBPL9. The ORP9 gene encodes a full-length (ORP9L) and truncated version (ORP9S) that initiates from an alternate promoter and is missing the N-terminal PH domain (Wyles and Ridgway, 2004). Both ORP9S and ORP9L contain a FFAT motif that binds VAP and an OSH domain with undefined sterol-binding specificity (Suchanek et al., 2007). ORP9L partitioned between the ER and Golgi apparatus in a VAP-dependent manner, and enforced overexpression disrupted the organization of the ER and ER–Golgi intermediate compartment (Wyles and Ridgway, 2004). Supporting the concept that ORP9 is involved in the early secretory pathway, heterologous expression of ORP9S in SEC14-null yeast phenocopied the OSBP homologue Osh4p with respect to inhibition of Golgi vesicular transport (Fairn and McMaster, 2005). In the current study we show that ORP9L is a cholesterol-binding/transfer protein that interacts with the trans-Golgi/TGN and ER, and is necessary to maintain Golgi structure, ER–Golgi protein trafficking, and cholesterol distribution in the endosomes and lysosomes. On the other hand, ORP9S is a potent dominant negative inhibitor of ER–Golgi protein transport and cell growth. We conclude that these two products of the ORP9 gene are sterol-binding/transfer proteins that have opposing affects on ER–Golgi transport.
MATERIALS AND METHODS
MATERIALS
G418, hygromycin B, tissue culture media, anti-V5 antibody, and Amplex Red Cholesterol Assay reagents were purchased from Invitrogen (Carlsbad, CA). Doxycycline, FBS, glutathione, and anti-β-actin antibody were obtained from Sigma/Aldrich (Oakville, ON, Canada). Hybond-C Extra, protein A-Sepharose CL-4B, and glutathione-Sepharose 4B were from GE Healthcare (Piscataway, NJ). Complete protease inhibitor cocktail tablets were from Roche Diagnostics (Mannheim, Germany). Phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL), and sterols were from Steraloids (New Port, RI).
ORP9 and VAP-A rabbit polyclonal antibodies were previously described (Wyles et al., 2002; Wyles and Ridgway, 2004). An anti-giantin AlexaFluor-488 conjugate was from Covance (Emeryville, CA). Calnexin, anti-V5 monoclonal, and anti-β-actin rabbit polyclonal antibodies were purchased from Sigma/Aldrich. Myc and PI 4-kianse IIIβ antibodies were from Cell Signaling Technologies (Beverly, MA). Goat anti-mouse– and goat anti-rabbit–conjugated horseradish peroxidase (HRP) secondary antibodies were from Bio-Rad (Mississauga, ON, Canada). ECL substrate was purchased from Millipore (Billerica, MA). AlexaFluor-488 and -594 goat anti-mouse and goat anti-rabbit antibodies were purchased from Molecular Probes (Invitrogen, Eugene, OR).
Plasmids
Tetracycline-inducible expression vectors for ORP9S and ORP9L (pTRE-ORP9L-V5, pTRE-ORP9S-V5), as well as FFAT domain mutants (pTRE-ORP9L-FF/AA-V5 and pTRE-ORP9S-FF/AA-V5) were previously described (Wyles and Ridgway, 2004). The ORP9 PH (pTRE-ORP9L-R22E-V5) and sterol-binding domains (pTRE-ORP9L-Δ375-378-V5) were mutagenized using the QuikChange II XL kit (Stratagene, La Jolla CA). pcDNA3.1-VSVG-tsO45-(myc)3 was kindly given by Dr. Johnny Ngsee (Ottawa Health Research Institute, University of Ottawa, ON, Canada). Bacterial expression and purification of GST-VAP was previously reported (Wyles et al., 2002).
Cell Culture and Transfections
Chinese hamster ovary (CHO)-K1 and CHO Tet-on cells were cultured in DMEM containing 5% FBS and 34 μg proline/ml (medium A). For filipin fluorescence experiments, cells were also cultured in DMEM containing 5% lipoprotein-deficient serum (LPDS) and proline. Expression of ORP9 in Tet-on cells were induced in medium A containing doxycycline (1 μg/ml) for times indicated in figure legends.
ORP9L expression was knocked down by transfection with a pool of three short interfering RNA (siRNA) duplexes (25 nM each, Dharmacon, Lafayette, CO) or a nontargeting siRNA (100 nM) with Trans-IT TKO transfection reagent (Mirus, Madison, WI) for 48 h (Lessmann et al., 2007).
Expression and Purification of Recombinant OSBP and ORP9
A EcoRI-BamHI fragment encoding the ORP9 PH domain (aa 1-104) was generated by PCR amplification, cloned into pGEX-3X, and transformed into BL21 Escherichia coli. Expression of GST-ORP9-PH and GST-OSBP-PH was induced with 100 μM IPTG for 3 h at 25°C and purified by glutathione-Sepharose affinity chromatography (Wyles et al., 2002).
Full-length, C-terminal His-tagged OSBP, ORP9L, ORP9L Δ375-378 (ΔSB), and OSBP PH-RR109,110EE (RR/EE) were expressed and purified from baculovirus-infected Sf21 cells. cDNAs were amplified by PCR and cloned into pENTR/D-Topo (Invitrogen). Inserts were verified by sequencing, inserted into linearized BaculoDirect (C-terminal V5-His tagged) by recombination, and transfected into Sf21 cells. For large-scale expression and purification, Sf21 cells were infected at an MOI of 0.1 for 1 h at 20°C, resuspended in 200–500 ml of Sf-900 II media with 0.5% FCS and incubated with shaking at 27°C. After 72 h, cells were collected by centrifugation at 300 × g for 10 min, resuspended in one-tenth volume of phosphate buffer (50 mM potassium phosphate and 300 mM NaCl, pH 7.4) containing 50 mM imidazole and EDTA-free protease inhibitors, and lysed. OSBP or ORP9L were purified from the supernatant fraction with Talon metal-affinity resin (Clontech, Palo Alto, CA). OSBP required an additional purification step on DEAE-Sepharose. Purified recombinant proteins were stored at −80°C.
Fluorescence Microscopy
After transfections, cells were cultured to 60% confluence on glass coverslips, washed with phosphate-buffered saline (PBS), fixed in 4% (wt/vol) paraformaldehyde for 10 min, and permeabilized in 0.05% (vol/vol) Triton X-100 at −20°C for 10 min before probing with primary and secondary antibodies. Images were captured using a Zeiss LSM 510 Meta laser scanning confocal microscope (0.8-μm sections; Thornwood, NY) using a 100× oil emersion objective (NA 1.4).
CHO cells were fixed as described above, incubated with ORP9L primary and secondary antibodies, and then with filipin (50 μg/ml) for 1 h. Filipin fluorescence images were captured at identical exposure times using a Zeiss Axiovert 200M inverted microscope (Thornwood, NY) equipped with a 63× oil immersion objective (NA 1.4) and axioCam HRm CCD camera. Using NIH Image J software (http://rsb.info.nih.gov/ij/), the background and threshold on image fields of 20–30 cells were normalized and then subjected to fluorescence intensity (integrated density/area) analysis. Intensity values were normalized to cell number and expressed relative to siNT-transfected CHO cells or uninduced CHO Tet-on cells cultured in media A (FCS).
Phospholipid Overlay Assay
Lipid (100 pmol) dissolved in chloroform:methanol:water (1:2:0.8, vol/vol) were spotted on Hybond-C nitrocellulose membrane and dried at room temperature for 1 h. Membranes were incubated with blocking buffer (Tris-buffered saline, 3% fatty acid free BSA, and 0.1% Tween-20 (vol/vol) for 2 h. Membranes were incubated with 100 nM glutathione S-transferase (GST), GST-ORP9-PH, or GST-ORP9-PH-R22E for 1 h in blocking buffer, incubated with an anti-GST monoclonal followed by a goat anti-mouse HRP secondary antibody, and detected by ECL. Full-length OSBP or ORP9L (100 nM) were incubated with HyBond-C–immobilized lipids under the same conditions and detected with monospecific antibodies.
045-Vesicular Stomatitus Virus Glycoprotein EndoH Processing Assay
CHO-K1 cells were transfected with 75 nM siNT or siORP9 for 48 h before transfection with pcDNA3.1-VSV-G-myc3 using Lipofectamine 2000. CHO Tet-on cells were treated with or without doxycycline (1 μg/ml) for 24 h and then transfected with pcDNA3.1-VSV-G-myc3 using Lipofectamine 2000. CHO-K1 or CHO Tet-on cells were then shifted to 40°C for 16 h and incubated with 20 μg/ml cyclohexamide for an addition 20 min before shifting to 32°C for the times indicated in figure legends. Cells were harvested in PBS, collected by centrifugation, sonicated in 10 mM Tris-HCl (pH 7.4), and centrifuged at 100,00 × g for 30 min. Pellets were resuspended in 0.1% SDS, 50 mM sodium citrate (pH 5.5), and incubated for 16 h with 25 mU EndoH at 37°C overnight. Samples were precipitated in 10% trichloroacetic acid and glycosylated forms of vesicular stomatitus virus glycoprotein (VSVG) was analyzed by SDS-PAGE and immunoblotting using an anti-myc monoclonal.
Sterol-binding Assays
25-[3H]Hydroxycholesterol binding by OSBP and ORP9L was assayed by the charcoal-dextran method in the presence or absence of a 40-fold molar excess of unlabeled sterol (Kandutsch and Shown, 1981; Ridgway et al., 1992). [3H]Cholesterol binding to OSBP and ORP9L could not be quantified using the charcoal-dextran method because of high background. Instead, binding assays contained 20 mM HEPES (pH 7.4), 150 mM KCl, and 0.05% Triton X-100 (to disperse [3H]cholesterol). After incubation for 2 h at 20°C, OSBP or ORP9L were bound to Talon metal-affinity resin for 15 min and washed three times with HEPES/KCl buffer, and bound sterols released with 150 mM imidazole. Specific binding was determined in the presence of a 40-fold molar excess of unlabeled cholesterol.
The capacity of OSBP and ORP9L to bind and extract sterols from liposomes was assayed as follows. PC liposomes (0.5 mM) with 1 mol% [3H]cholesterol (specific activity 720 dpm/pmol) were prepared by drying down lipids in chloroform under nitrogen and rehydration in 25 mM HEPES (pH 7.4) and 150 mM NaCl for 1 h at room temperature. Liposomes (400-nm diameter) were prepared by filter extrusion using the Lipofast system (Avestin, Ottawa, ON, Canada) and incubated with recombinant OSBP or ORP9L (1:1; mol protein/mol sterol) for 30 min at 25°C. Liposomes were sedimented by centrifugation at 100,000 × g for 25 min at 4°C and radioactivity in the supernatant was measured by liquid scintillation counting. Specific extraction of [3H]cholesterol from liposomes was determined by subtraction of background radioactivity in the absence of OSBP or ORP9L. The distribution of OSBP and ORP9L in supernatant and pellet fractions were analyzed by SDS-PAGE and Coomassie staining.
[3H]Cholesterol transfer assays were based on previous methods (Kasper and Helmkamp, 1981; Raychaudhuri et al., 2006). Donor PC liposomes (400-nm diameter) containing 10 mol% PE, 1 mol% cholesterol with or without 10 mol% PI-4P, PI-3P, or PI-5P and acceptor PC liposomes (400-nm diameter) containing 10 mol% PE, 10 mol% lactosyl-PE with or without 10 mol% PI4P, PI-3P, or PI-5P were prepared by extrusion. Before the assay, liposomes were cleared by centrifugation at 13,000 rpm for 5 min. The transfer assay consisted of 20 μl of acceptor and donor liposomes (100 pmol cholesterol), recombinant OSBP or ORP9L (100 pmol) in 120 μl of 25 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM EDTA, and 3 μg of fatty acid–free BSA. After incubation at 25°C for 25 min, 30 μg of R. communis agglutinin was added on ice for 15 min and acceptor liposomes were sedimented at 15,000 rpm for 5 min. Radioactivity in the supernatant was measured by scintillation counting and background (absence of OSBP or ORP9L) was subtracted.
RESULTS
ORP9L Is a PI-4P–dependent Cholesterol Transfer Protein
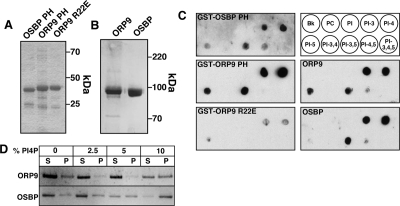
Endogenous ORP9L is primarily on the Golgi apparatus (Wyles and Ridgway, 2004), but whether the PH and sterol-binding domains regulate this interaction is unknown. To establish the PI-phosphate–binding specificity of the OPR9L PH domain, a lipid-overlay assay was used to compare the GST-PH domains of ORP9L and OSBP, as well as purified, full-length His-tagged ORP9L and OSBP expressed and purified from insect cells. Purified GST-fusion proteins (Figure 1A) and ORP9L and OSBP (Figure 1B) had the predicted molecular mass on SDS-PAGE and were incubated with phosphorylated PIs immobilized on nitrocellulose membranes (Figure 1C). GST-ORP9-PH bound to phosphatidylinositol 3-phosphate (PI-3P), PI-4P, and phosphatidylinositol 5-phsophate (PI-5P), but not other 4-phosphorylated derivatives. GST-OSBP-PH displayed a similar pattern but with reduced binding to PI-5P and increased binding of PI-4,5 bisphosphate. The ORP9L PH domain contains several basic residues (R11, R22, and R48) that were implicated in PI-4P and PI-4,5 bisphosphate binding by the related OSBP, CERT, and FAPP PH domain (Perry and Ridgway, 2005). Mutation of one of these arginine residues (ORP9 PH-R22E) dramatically reduced binding to all phosphorylated PIs (Figure 1C). ORP9L and OSBP had similar binding specificities as the corresponding GST-PH domains, suggesting that this activity resides solely in the PH domain. Binding of OSBP and ORP9L to phosphatidylcholine (PC) liposomes containing increasing PI-4P was determined by sedimentation (Figure 1D). By this method, ∼50% of ORP9L was associated with liposomes containing 10 mol% PI-4P, compared with 80–90% for OSBP.
Figure 1.
Phosphatidylinositol phosphate–binding specificity of the ORP9 PH domain. (A) Purified GST fusion proteins of the OSBP, ORP9, and ORP9 R22E PH domains (2 μg) were separated by SDS-12% PAGE and stained with Coomassie Blue. (B) His-tagged ORP9L and OSBP (2 μg) purified from Sf21 cells were resolved on SDS-8% PAGE and stained with Coomassie Blue. (C) GST-PH domain fusion proteins, ORP9L and OSBP (300 pmol each), were incubated with the indicated phospholipids (100 pmol) immobilized on Hybond C filters. GST-fusion protein binding to phospholipids was detected using an anti-GST monoclonal, whereas ORP9 and OSBP were detected with monospecific polyclonal antibodies. (D) OSBP and ORP9 (100 pmol) were incubated with PC liposomes containing the indicated mol% of PI4P for 20 min at 20°C and sedimented at 100,000 × g for 30 min, and the pellet (P) and supernatant (S) fractions were resolved by SDS-8% PAGE and stained with Coomassie Blue.
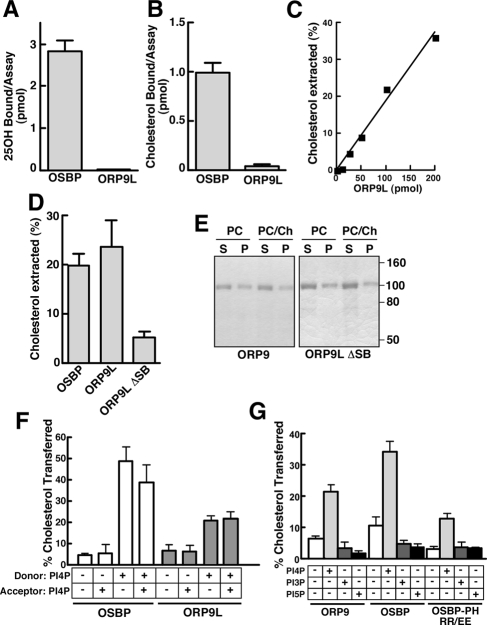
The sterol-binding domain of ORP9 is homologous to OSBP and other ORPs yet was reported not to bind photoderivatives of 25-hydroxycholesterol and cholesterol (Suchanek et al., 2007). As well, it is unclear if ORP9L or OSBP can transfer sterols from donor to acceptor membranes. This was investigated by comparing the in vitro sterol-binding, extraction, and transfer activity of recombinant OSBP and ORP9L (Figure 2). As expected, OSBP bound [3H]25-hydroxycholesterol (Figure 2A) and [3H]cholesterol (Figure 2B) under conditions where the ligand was dispersed in solution or 0.05% Triton X-100, respectively. ORP9L did not bind either sterol under these conditions (Figures 2, A and B) nor over a range of ligand concentrations (results not shown).
Figure 2.
ORP9 preferentially binds and transfers liposomal cholesterol. (A) [3H]25-hydroxycholesterol (100 nM) binding by OSBP and ORP9L (100 pmol) was assayed using a charcoal-dextran method (Ridgway et al., 1992). (B) OSBP and ORP9L (100 pmol) were incubated with 600 nM [3H]cholesterol, and bound sterol was quantified after binding to Talon nickel affinity resin. (C) Extraction of [3H]cholesterol from liposomes was measured in the presence of increasing amounts of ORP9L and quantified as described in Materials in Methods. Results are the mean of two separate experiments. (D) Extraction of cholesterol from PC liposomes by OSBP, ORP9L, and ORP9L ΔSB (100 pmol each). (E) Distribution of ORP9L, OSBP, and ORP9L ΔSB in the supernatant (S) and pellet (P) fraction of a cholesterol extraction assay in the presence and absence of 1 mol% cholesterol liposomes as determined by SDS-PAGE. (F) [3H]Cholesterol transfer from donor to acceptor liposomes by OSBP and ORP9L was assayed as described in the Material and Methods. Liposomes were prepared with or without 10 mol% PI-4P. (G) The sterol transfer activity of ORP9L, OSBP, and OSBP PH-RR/EE (100 pmol) was determined with donor and acceptor vesicles each containing no addition or 10 mol% PI-3P, PI-4P, or PI-5P. Unless indicated, results are the mean and SEM of 3–6 separate experiments.
Next, an assay was used that measured extraction of [3H]cholesterol from PC/cholesterol liposomes by measuring soluble radioactivity after centrifugation. Extraction of cholesterol was linear with increasing ORP9L protein (Figure 2C). Approximately 35% of liposome [3H]cholesterol was extracted into the supernatant by 200 pmol of ORP9L (Figure 2C) compared with extraction of only <5% liposome phospholipid (results not shown). To demonstrate that cholesterol extraction required the sterol-binding domain, an ORP9L mutant with a deletion between the α-helical lid and β1-sheet of the sterol-binding fold (Δ375-378, referred to hereafter as ΔSB) was assayed for [3H]cholesterol extraction activity. A similar mutation in OSBP and ORP4 reduced cholesterol and 25-hydroxycholesterol binding by >90% (Perry and Ridgway, 2006; Wyles et al., 2007). The activity of ORP9L ΔSB was reduced by 80% compared with ORP9L (Figure 2D), but was still present in the soluble fraction similar to the wild-type protein (Figure 2E). OSBP also extracted cholesterol from liposomes to a similar extent as ORP9L (Figure 2D). Attempts to assay extraction of [3H]25-hydroxycholesterol from liposomes by OSBP and ORP9L was not successful because of increased solubility of the oxysterol and resulting high background. Finally, ORP9L- and OSBP-mediated cholesterol transfer between donor and acceptor liposomes was assayed using the lactosyl-phosphatidylethanolamine (PE)/R. communis agglutinin method (Kasper and Helmkamp, 1981; Raychaudhuri et al., 2006; Figure 2F). Both ORP9L and OSBP transferred <5% cholesterol when PI-4P was excluded from the assay or present only in acceptor liposomes. In contrast, ORP9L transferred 20% of donor liposome cholesterol when PI-4P was present in donor liposomes or both donor and acceptor liposomes. OSBP displayed significantly greater cholesterol transfer under the same conditions. Although OSBP and ORP9L bound PI-3P and PI-5P on nitrocellulose (Figure 1C), sterol transfer was not stimulated when either of these lipids was included in donor and acceptor liposomes (Figure 2G). PI-4P–dependent transfer activity required the PH domain as shown by reduced transfer activity of OSBP PH-RR/EE, a mutation that prevents PI-4P binding (Levine and Munro, 2002; Figure 2G). Thus the physical state of sterol ligands presented to OSBP does not affect binding, but ORP9L clearly shows a preference for sterol ligands presented in phospholipid bilayers. More importantly, both proteins display cholesterol transfer activity that was stimulated by PI-4P.
PH and Sterol-binding Domains Control ORP9L Partitioning between the ER and trans-Golgi/TGN
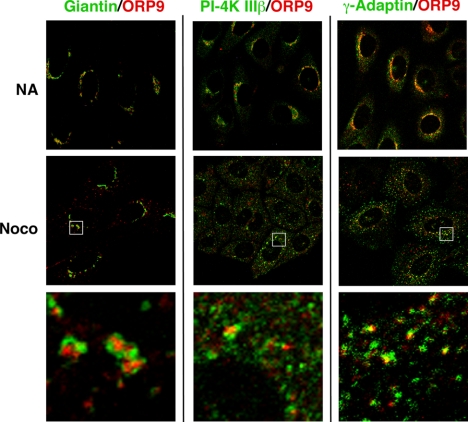
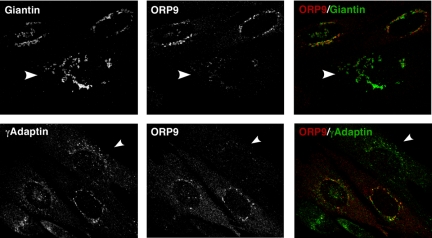
Dual targeting of ORP9L to the ER and Golgi has striking similarity to OSBP and CERT, suggesting a role in sterol trafficking between these compartments. To establish which Golgi compartment contained ORP9L, coimmunofluorescence experiments were conducted with cis (giantin), cis/medial (PI-4 kinase IIIβ), and trans-Golgi/TGN (γ-adaptin) markers (de Graaf et al., 2004; Weixel et al., 2005; Figure 3). Immunofluorescence was performed in control cells and cells treated with nocodazole to fragment the Golgi into mini-stacks to enhance visualization. Giantin was closely associated but did not overlap with endogenous ORP9L in control or nocodazole-treated CHO cells. PI-4 kinase IIIβ generates a pool of PI-4P that targets the isolated PH domains of CERT and OSBP to the trans-Golgi (Balla et al., 2005; Toth et al., 2006). However, PI-4 kinase IIIβ did not colocalize with ORP9L. ORP9L colocalized with γ-adaptin in the TGN, but only partially because there were distinct regions of nonoverlap (see magnified area in Figure 3, bottom panel). This suggests that ORP9L is restricted to distal trans-Golgi/TGN compartments.
Figure 3.
ORP9L localizes to the trans-Golgi/TGN. CHO cells cultured in medium A were treated with nocodazole (2 μg/ml) or solvent control for 1 h and immediately fixed and immunostained with an ORP9 polyclonal antibody and a goat anti-rabbit AlexaFluor594-conjugated secondary antibody. This was followed by a giantin antibody coupled to AlexaFluor488, a PI-4K IIIβ monoclonal and goat anti-mouse AlexaFluor488-conjugated secondary or a γ-adaptin monoclonal antibody, and a goat anti-mouse AlexaFluor488-conjugated secondary antibody. Images are single confocal sections (0.8 μm) taken through the midplane of the cells. Selected high-magnification fields are boxed and are shown in the bottom three panels.
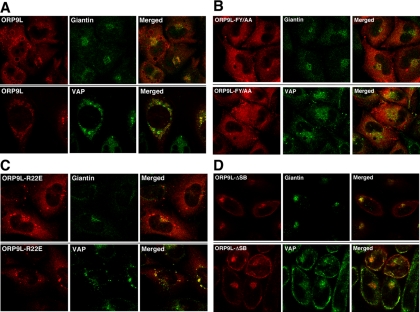
We next identified how the lipid and VAP-binding domains of ORP9L affected partitioning between the ER and Golgi apparatus (Figure 4). To this end, vectors encoding V5-tagged wild-type ORP9L and mutants in the sterol-binding (ORP9L-ΔSB), VAP (ORP9L-FY/AA), or PH domains (ORP9L-R22E), were expressed in CHO cells under the control of the TET-repressor and induced with the doxycycline. To confirm that the PH and sterol-binding domain mutations did not affect the interaction with VAP, cells were induced with doxycycline, and interaction of ORP9L mutants with VAP was determined by GST-VAP pulldown and coimmunoprecipitation. ORP9L, ORP9L-ΔSB, and ORP9L-R22E bound VAP in GST-pulldown and immunoprecipitation assays (Supplementary Figure S1, A and B). As expected, ORP9L-FY/AA did not bind to GST-VAP, but some background binding was detected by coimmunoprecipitation. The effect of inactivating the PH, sterol-binding and FFAT domains of ORP9L on intracellular localization was determined by immunofluorescence (Figure 4). As we previously reported, inducibly overexpressed ORP9L colocalized in CHO cells with VAP in vacuolated ER structures (Figure 4A). There was also limited perinuclear staining that colocalized with giantin but significantly less than observed for endogenous ORP9L, indicating that other factors are limiting for Golgi interaction. Vacuolated ER and Golgi localization was lost when the FFAT motif was mutated (Figure 4B). ORP9L FY/AA was diffusely localized and occasionally in large punctuate structures that contained the ERGIC marker p58 (Wyles and Ridgway, 2004; Figure 3B). The PH domain mutant ORP9L R22E was strongly localized to condensed ER structures containing VAP (Figure 4C) that were reminiscent of OSBP PH domain mutants (Wyles et al., 2002) that caused ER membrane stacking due to weak protein–protein interactions between opposing bilayers (Snapp et al., 2003; Amarilio et al., 2005). Lastly, sterol-binding–defective ORP9L ΔSB was primarily localized to the Golgi apparatus, but was also dispersed to a peripheral ER compartment that did not localize with the perinuclear ER marker calnexin or the PM marker caveolin (results not shown; Figure 3D). ORP9L ΔSB interaction with VAP also resulted in mislocalization of this partner protein to the Golgi apparatus. Collectively this shows that the ORP9L FFAT and sterol-binding domains promote ORP9L retention in the ER, whereas the PH domain promotes partitioning to the trans-Golgi/TGN.
Figure 4.
The ORP9L sterol-binding and PH domains regulate interaction with the Golgi apparatus. CHO cells stably expressing ORP9L (A), OPR9L-FY/AA (B), ORP9L- R22E (C), and ORP9L-ΔSB (D) under the control of the TET promoter were cultured in medium A and induced with 1 μg/ml doxycycline. After 24 h, cells were fixed and immunostained with primary antibodies against V5 (ORP9L), VAP, and/or giantin-conjugated AlexaFluor488, followed by goat anti-rabbit or goat anti-mouse secondary antibodies conjugated to AlexaFluor488 (giantin and VAP) or AlexaFluor594 (V5-tagged ORP9L). Images are single confocal scans (0.8 μm) though the middle plane of the cells.
ORP9L Is Required for Golgi Organization and Protein Transport
Similarities between ORP9L and OSBP domain organization, sterol binding/transfer, and intracellular localization suggested that ORP9L could also be directly or indirectly involved in regulation of CERT and SM synthesis (Perry and Ridgway, 2006). This was tested by depleting CHO cells of ORP9L with siRNA as previously described (Lessmann et al., 2007) and monitoring CERT-dependent SM synthesis by [3H]serine incorporation in the presence and absence of 25-hydroxycholesterol (Figure 5A). As previously reported in CHO and HEK293 cells (Perry and Ridgway, 2006), silencing of OSBP by RNA interference (RNAi) minimally affected basal SM synthesis but profoundly inhibited 25-hydroxycholesterol–activated synthesis. In contrast, depletion of ORP9L caused a slight increase in both basal and 25-hydroxycholesterol–stimulated SM synthesis. SM synthesis in cells depleted of both OSBP and ORP9L was increased slightly compared with OSBP knockdown. ORP9L depletion did not affect synthesis of GlcCer or ceramide, indicating no obvious role in sphingolipid synthesis in the ER or Golgi apparatus. Immunoblots showing the siRNA-mediated knockdown that resulted in 70–90% depletion of OSBP and ORP9L (Figure 5B). Sphingomyelin synthesis was also measured in CHO cells inducibly expressing ORP9L (Figure 5C). In this case, induction of ORP9L expression with doxycycline did not affect basal SM and GlcCer synthesis but blocked stimulation by 25-hydroxycholesterol. Because OSBP and ORP9L share protein and lipid ligands, the inhibitory effect of ORP9L overexpression could be the results of competition for these factors.
Figure 5.
Knockdown of ORP9L expression does not affect oxysterol-regulated SM synthesis. (A) CHO cells cultured in medium A were transiently transfected with siOSBP (75 nM), siORP9L (75 nM), siOSBP and siORP9L (75 nM each), or a nontargeting control siNT (75 nM) for 48 h. Cells then received serine-free medium A containing no addition ( ) or 25-hydroxycholesterol (2.5 μg/ml; ■) for 2 h followed by pulse-labeling with [3H]serine (10 μCi/ml) for 2 h. [3H]Serine-labeled SM, GlcCer and ceramide were extracted from cells, separated by thin-layer chromatography, and quantified as previously described (Perry and Ridgway, 2006). Results are the mean and SE for three separate experiments. (B) Expression of OSBP and ORP9L in CHO cells transiently transfected with control and targeting siRNAs. (C) CHO cells expressing ORP9L under the control of the Tet repressor were cultured in medium A without (−Dox) or with doxycycline (+Dox, 1 μg/ml) for 24 h. Cells then received solvent control (
) or 25-hydroxycholesterol (2.5 μg/ml; ■) for 2 h followed by pulse-labeling with [3H]serine (10 μCi/ml) for 2 h. [3H]Serine-labeled SM, GlcCer and ceramide were extracted from cells, separated by thin-layer chromatography, and quantified as previously described (Perry and Ridgway, 2006). Results are the mean and SE for three separate experiments. (B) Expression of OSBP and ORP9L in CHO cells transiently transfected with control and targeting siRNAs. (C) CHO cells expressing ORP9L under the control of the Tet repressor were cultured in medium A without (−Dox) or with doxycycline (+Dox, 1 μg/ml) for 24 h. Cells then received solvent control ( ) or 25OH (■) and were pulse-labeled with [3H]serine as described in A. Results are the mean and SEM for three separate experiments.
) or 25OH (■) and were pulse-labeled with [3H]serine as described in A. Results are the mean and SEM for three separate experiments.
While analyzing ORP9L depleted cells, we noted that immunofluorescence staining of Golgi resident proteins was frequently dispersed. For example, Figure 6 shows coimmunofluorescence localization of ORP9L with giantin or γ-adaptin in fields of CHO cells transfected with an ORP9L siRNA. Cells with relatively low ORP9L expression (indicated by an arrow) displayed conspicuous dispersion of giantin and γ-adaptin staining compared with adjacent cells that expressed more ORP9L.
Figure 6.
Knockdown of ORP9L expression results in Golgi fragmentation. CHO cells were transfected with siORP9 for 48 h in medium A and processed for immunostaining of ORP9L and giantin or γ-adaptin as described in the legend to Figure 4. Fields were identified in which ORP9L expression displayed variability between individual cells. ORP9L expression was almost undetectable in cells indicated by arrows. Images are single confocal sections (0.8 μm) through the midplane of the cells.
Fragmentation of the giantin-positive cis-Golgi in ORP9L-depleted cells, a compartment in which ORP9L does not reside (Figure 3), suggested that structural changes could be due to defective retrograde or anterograde transport. To test whether anterograde transport was affected, export of myc-tagged, temperature-sensitive 045-VSVG from the ER to the Golgi was monitored by endoH sensitivity. Control and siORP9 transfected cells were maintained at the nonpermissive temperature of 40°C to retain 045-VSVG in the ER, followed by a shift to 33°C to initiate ER export and modification of trimmed N-linked carbohydrates in the Golgi apparatus (Figure 7A). At 40°C, 045-VSVG carbohydrates were completely endoH sensitive in siNT- and siORP9-transfected cells. After a 2-h shift to 32°C, 70% of 045-VSVG in siNT-transfected cells was transported to the Golgi where it acquired endoH resistance, compared with 50% in ORP9L-depleted cells. A comparison of the ratio of resistant to sensitive 045-VSVG (Figure 7B) indicated consistently less 045-VSVG transport to the ER in ORP9L-depleted cells; however, this result did not reach statistical significance.
Figure 7.
Knockdown of ORP9L expression partially suppresses VSVG transport to the Golgi apparatus. (A) CHO cells cultured in medium A were transfected with siORP9 (75 nM) or siNT (75 nM). After 48 h, cells were shifted to 40°C and transiently transfected with pcDNA3.1-VSV-G-tsO45-(myc)3 for a further 16 h. Cells were then maintained at 40°C or shifted to 33°C for 120 min to initiate folding and export of 045-VSVG to the Golgi apparatus. Cells were harvested, total extracts incubated with and without endoH, and sensitive and resistant forms of VSVG were detected by SDS-10%PAGE and immunoblotting as described in Materials and Methods. (B) The distribution of endoH-resistant and -sensitive forms of VSVG was quantified by densitometry. Results are the mean and SEM of three independent experiments.
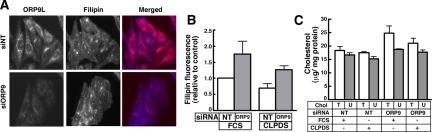
Cholesterol is essential for transport through the secretory pathway (Wang et al., 2000; Ying et al., 2003), where it is progressively enriched from the ER to the plasma membrane (Ridgway, 2000). Based on in vitro cholesterol transfer activity, a primary function of ORP9L could be maintenance of cholesterol distribution within the secretory pathway. RNAi depletion of ORP9L and resulting effects on cholesterol transport could be reflected in altered regulatory responses in the ER. However, ORP9L depletion had no effect on cholesterol or cholesterol ester synthesis, indicating its activity is segregated from sterol-regulatory events in the ER (Supplemental Figure 4A and results not shown). To determine whether ORP9L regulated cholesterol content of other compartments, the cholesterol-binding polyene macrolide filipin was used to visualize cholesterol in control and ORP9L-depleted CHO cells (Figure 8). On visual inspection, it was apparent that, compared with siNT-treated cells, cells transfected with siORP9L and cultured in FCS had increased filipin fluorescence primarily in punctate structures corresponding to endosomes and lysosomes (Figure 8A). There was no apparent change in filipin fluorescence in the PM or other diffusely stained structures. Quantitation of these images showed that knockdown cells cultured in FCS or LPDS had a 50–75% increase in filipin fluorescence relative to siNT controls (Figure 8B). Measurement of cellular cholesterol mass under the same conditions revealed that although ORP9L-depleted cells cultured in FCS or LPDS had a consistent 10–15% increase in total and unesterified cholesterol, this result was not significant (Figure 8C). This indicates that ORP9L maintains cholesterol distribution in the post-Golgi, endosomal compartment rather than influencing regulatory events in the ER.
Figure 8.
Silencing of ORP9L expression promotes cholesterol retention in the endosomal/lysosomal compartment. CHO cells cultured in medium A were transfected with siORP9 (75 nM) or siNT (75 nM) for 32 h. Cells then received medium containing 5% FCS or lipoprotein deficient serum (LPDS) for an additional 16 h. Cells were fixed and incubated with an ORP9L polyclonal antibody, and goat anti-rabbit AlexaFluor594-conjugated secondary antibody and cholesterol was detected with filipin. (A) A representative field of siNT- and siORP9L-transfected cells cultured in FCS and stained with an ORP9L antibody and filipin (cholesterol). (B) Fluorescent intensity of filipin staining was quantified in control and ORP9L knockdown cells cultured in FCS or LPDS for 16 h as described in Material and Methods. Results are the mean and SEM of three independent experiments that quantified 10–12 fields of 20–30 cells. (C) Total (T) and unesterified cholesterol (U) mass was quantified in control and ORP9L-depleted CHO cells cultured in medium with 5% FCS or LPDS. Results are the mean and SEM of three separate experiments.
ORP9S Is a Dominant Inhibitor of Cell Growth and Protein Export from the ER
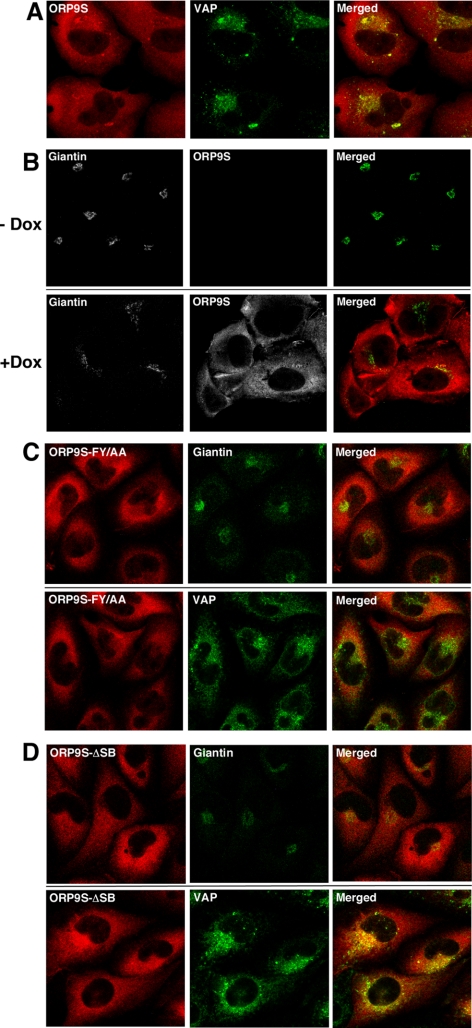
Several ORP genes encode truncated or “short” versions encoding the C-terminal sterol binding and FFAT domains (Lehto and Olkkonen, 2003). Although ORP9S protein and mRNA is expressed in a variety of tissues and cells (Wyles and Ridgway, 2004; Lessmann et al., 2007), its function relative to the full-length proteins is obscure. The absence of a PH domain in ORP9S would restrict it to the ER where it could interfere with the normal function of ORP9L or other VAP-interacting proteins. To test this, CHO cell lines were generated that expressed ORP9S, as well as VAP-binding (ORP9S-FF/AA) and sterol-binding (ORP9S-ΔSB) mutants under the control of a doxycycline-inducible promoter. GST-pulldown and coimmunoprecipitation assays confirmed that wild-type and ORP9S-ΔSB interacted with VAP, but the FF/AA mutant was devoid of VAP-binding activity (Supplementary Figure S1B). Overexpressed ORP9S was dispersed though out the cell but also colocalization with VAP at several foci (Figure 9A). Similar to ORP9L-depleted cells (Figure 6), the Golgi apparatus (visualized with giantin) was fragmented in ORP9S-overexpressing cells (+Dox) compared with uninduced cells (−Dox, Figure 9B). Dispersion of the Golgi apparatus and colocalization with VAP was not evident in cells expressing VAP (ORP9S-FF/AA) and sterol-binding domain (ORP9S-ΔSB) mutants (Figure 9, C and D).
Figure 9.
Inducible expression of ORP9S fragments the Golgi apparatus. CHO cells stably transfected with plasmids encoding the indicated ORP9S proteins were cultured in medium A with or without doxycycline (1 μg/ml) for 24 h. (A) CHO cells expressing ORP9S were induced with doxycycline and localization with VAP was determined. (B) CHO cells expressing ORP9S from the Tet promoter were cultured in the absence (−Dox) or presence of doxycycline (+Dox) and localization of ORP9S and giantin was determined by immunofluorescence as described in the legend to Figure 3. (C) After induction with doxycycline, localization of ORP9S-FF/AA with VAP and giantin was determined as described above. (D) After induction with doxycycline, localization of ORP9S-ΔSB with VAP and giantin was determined as described in above. Images are single confocal sections (0.8 μm) through the midplane of the field of cells.
During initial characterization of overexpressing cells, it became apparent that ORP9S caused growth inhibition. Quantification of cell number over a 72-h induction period revealed that ORP9S expression caused profound inhibition of cell proliferation relative to uninduced controls (Supplemental Figure S2A). This was not due to activation of an irreversible cell death pathway because removal of doxycycline after induction of ORP9S for 36 h led to recovery of cell growth (Supplemental Figure S2B). As well, cells expressing ORP9S did not display any indications of necrosis or apoptosis (result not shown). VAP and sterol binding were required for this phenotype because expression of ORP9S-FY/AA or ORP9S-ΔSB did not affect cell growth (Supplemental Figure S2, C and D). Overexpression experiments with wild-type and mutant ORP9L failed to effect cell growth or viability (Supplementary Figure S3). Interestingly, overexpression of ORP9L-R22E, which had defective PI-4P binding and Golgi localization (Figures 1C and 4D), did not affect cell proliferation, indicating that deletion of the entire interval between amino acids 1-165 is required for the growth arrest phenotype.
Fragmentation of the Golgi apparatus and cessation of cell growth by ORP9S expression suggested that ORP9S was a dominant-negative inhibitor of ORP9L functions related to cholesterol distribution and function in the ER–Golgi pathway. We initially determined whether enforced expression of ORP9S influenced cholesterol homeostasis and cellular distribution. Overexpression of ORP9S, but not ORP9L, caused a two-fold decrease in cholesterol synthesis measured by [3H]acetate incorporation (Supplemental Figure S4A). This was accompanied by a significant reduction in filipin fluorescence in the endosomes/lysosomes of ORP9S cells cultured in FCS or LPDS, but no significant change in cholesterol mass (Supplemental Figure S4, B and C). Enforced expression of ORP9L did not affect either of these parameters (Supplemental Figure S4, D and E).
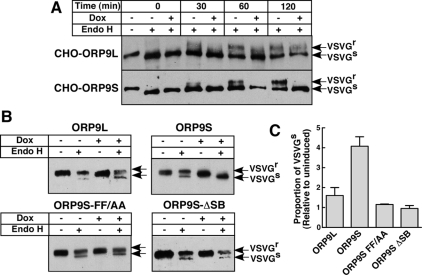
The effect of ORP9S expression on ER–Golgi protein transport was determined by assessing 045-VSVG transport and sensitivity to endoH. For these experiments, CHO cells were cultured in the absence or presence of doxycycline to induce ORP9S, transfected with 045-VSVG at the nonpermissive temperature, and 045-VSVG transport to the Golgi was assessed by endoH sensitivity after shifting to the permissive temperature (Figure 10). At the permissive temperature, 045-VSVG processing to an endoH-resistant form in cells expressing ORP9L was evident after 120 min (Figure 10A), although there appeared to be a slight delay compared with uninduced cells. In contrast, cells expressing ORP9S had a complete block in processing at all time points compared with uninduced cells. In a related experiment, CHO cells expressing ORP9L, ORP9S, and ORP9S mutants were transiently transfected with 045-VSVG and treated without and with doxycycline at 33°C for 16 h to assess steady-state export and carbohydrate processing in the Golgi apparatus (Figure 10B). Under these conditions, 045-VSVG was processed to an endoH-resistant form in ORP9L expressing cells, although there appeared to be slightly less relative to uninduced cells. In contrast, expression of ORP9S prevented transport of 045-VSVG to the Golgi as indicated by a complete lack of the endoH-resistant form. Consistent with the lack of effect of ORP9S FY/AA and ΔSB on cell growth (Supplemental Figure S3), processing of 045-VSVG carbohydrates was similar to matched uninduced cells (Figure 10B). Results from Figure 10B were quantified by densitometry and expressed relative to matched uninduced controls (Figure 10C). It is evident there was a significant block in steady-state processing in cells expressing ORP9S relative to those expressing ORP9L or the ORP9S mutants. Thus ORP9S-mediated inhibition of ER–Golgi transport is correlated with cell growth arrest and requires interaction with VAP and sterol ligand(s).
Figure 10.
Enforced expression of ORP9S prevents ER–Golgi transport of 045-VSVG. (A) CHO cells stably expressing ORP9L and ORP9S under the control the Tet-repressor were transfected with pcDNA3.1-VSV-G-tsO45-(myc)3 for 6 h at 40°C. Cells then received medium A with or without doxycycline (Dox, 1 μg/ml) for 10 h at 40°C. Cells were shifted to 33°C and at the indicated times harvested for analysis of VSVG glycosylation as described in Materials and Methods. (B) CHO cells expressing ORP9L, ORP9S, ORP9S-FF/AA, and ORP9S-ΔSB under the control of the Tet-repressor were transfected with pcDNA3.1-VSV-G-tsO45-(myc)3 for 6 h and then incubated in the presence or absence or doxycycline for 10 h at 33°C before harvesting and analysis of VSVG endoH sensitivity. (C) Quantification of data in B showing the relative proportion of endoH-sensitive O45-VSVG in control and overexpressing cells. Results are the mean and SEM of three experiments.
DISCUSSION
It is well established that cholesterol is required for cargo delivery to and from the Golgi apparatus. The trans-Golgi/TGN is enriched in cholesterol relative to the cis and medial cisternae (Orci et al., 1981), a distribution that coincides with the site of sphingolipid synthesis and raft assembly. This distribution is apparently essential for the organization and function of the Golgi apparatus (Wang et al., 2000; Ying et al., 2003) and is perturbed in the Niemann-Pick C cholesterol storage disorder (Coxey et al., 1993). Delivery of the SREBP-SCAP complex to the Golgi apparatus is stimulated by reduced membrane cholesterol (Nohturfft et al., 2000). In contrast, constitutive COPII-mediated export of cargo from the ER is suppressed by cholesterol depletion (Ridsdale et al., 2006; Runz et al., 2006). Although cholesterol is clearly important for ER–Golgi function, it is unclear whether its distribution is maintained by constitutive membrane transport coupled with protein- and/or lipid-mediated sequestration or by specific cholesterol transport proteins. In this study we have identified ORP9, a cholesterol-binding protein that partitions between the ER and Golgi apparatus, as a potential regulator of sterol homeostasis in the secretory pathway.
Similar to OSBP, ORP9L has PH-, FFAT-, and sterol-binding domains that regulate partitioning between the ER and Golgi apparatus. Analysis of the binding specificity of the ORP9 and OSBP PH domains, both in isolation and in the context of the full-length protein, revealed a preference for monophosphorylated PIs, most notably PI-4P. Interaction of the OSBP PH domain with the Golgi apparatus is dependent on PI-4P synthesized by the PI 4-kinases IIIβ (Balla et al., 2005) and/or IIα (Wang et al., 2003), as well as other factors such as ARF (Levine and Munro, 2002) and Nir2 (Peretti et al., 2008). ORP9L did not localize with PI-4 kinase IIIβ at the Golgi apparatus, and RNAi experiments showed that this kinase was not required for ORP9L localization (results not shown). However, a role for the PH domain in Golgi targeting was supported by the absence of the PI-4P-binding defective ORP9L R22E at the Golgi and its appearance in condensed ER structures with VAP. The lack of influence of ORP9L on SM synthesis indicates it is functionally distinct from OSBP and thus could interact with a different PI-4P pool or with other PH domain-specific lipid or protein ligands.
The influence of sterols on OSBP localization suggests that an intramolecular interaction between the sterol-binding and PH domains is involved in differential targeting to membranes (Ridgway et al., 1992; Mohammadi et al., 2001). Because ORP9L localization was not affected by oxysterols or cholesterol, cholesterol binding was disrupted by a small deletion of conserved residues between the α-helical lid and β-sheet-1. This mutation caused ORP9L to localize to the Golgi apparatus and peripheral ER, suggesting that impairment of sterol binding produces an open conformation that relieves inhibition of the PH domain and enhances interaction with the Golgi apparatus. This is similar to the effect of sterols on OSBP; the apo form is primarily in the cytoplasm and ER but undergoes a shift to the Golgi compartment upon sterol binding, where it stimulates ceramide transfer and SM synthesis (Perry and Ridgway, 2006). Interestingly, ORP9L ΔSB coimmunoprecipitated and colocalized with VAP at the Golgi apparatus and peripheral ER. The presence of VAP at the Golgi apparatus could indicate that ORP9L ΔSB has been trapped at membrane contact sites where sterols are transferred between the ER and Golgi (Olkkonen and Levine, 2004). However, VAP is a predicted C-terminal tail-anchored protein that is inserted into membranes posttranslationally (Egan et al., 1999) and thus could be nonspecifically targeted to membranes enriched in one of its partner proteins.
ORP9L-bound cholesterol by a mechanism that was markedly different from OSBP, ORP4, ORP8, and ORP1, which bound radiolableled sterols in vitro when these ligands were presented as aqueous or detergent dispersions (Lagace et al., 1997; Wyles et al., 2007; Yan et al., 2007a, 2008). Under these assay conditions, recombinant ORP9L (Figure 2) and ORP9L and ORP9S expressed in CHO cells (Wyles and Ridgway, 2004) were completely devoid of cholesterol- and 25-hydroxycholesterol–binding activity. However, both OSBP and ORP9L extracted and transferred cholesterol between liposomes, indicating that ORP9L must interact with membranes before uptake of sterol into the binding fold. Based on the crystal structure and molecular modeling of the yeast OSBP homologue Osh4p (Im et al., 2005; Canagarajah et al., 2008), it is proposed that several conserved basic residues positioned around the entrance to the binding pocket facilitated interaction of the apo form of the receptor with membranes. Once bound, sterol is taken up into the binding pocket, the receptor disengages, and a flexible α-helical lid closes over the pocket. Based on this model, ORP9L could adopt a closed apo structure that does not bind soluble ligands, but interaction with membranes displaces the lid, allowing sterols access to the binding pocket. ORP9 possess the four basic residues implicated in membrane interaction of Osh4p, but the α-helical lid is uniquely enriched in positively and negatively charged amino acids that could also interact with membranes.
These properties of ORP9L indicate it is suited for binding and transfer of hydrophobic ligands between membranes rather than uptake of soluble oxysterols. Indeed, ORP9L and OSBP transferred cholesterol between liposomes in PI-4P–dependent manner. The lack of effect of PI-3P and PI-5P on cholesterol transfer suggests that, in a native membrane environment, PI-4P is the primary activating lipid through interaction with the PH domain. However, OSBP PH-RR/EE, which is devoid of PI-4P–binding activity (Levine and Munro, 2002), had residual cholesterol transfer activity (∼25% of wild type), suggesting that other domains could interact with PI-4P to stimulate transfer. Interestingly, PI-4P did not stimulate transfer when included in acceptor liposomes devoid of cholesterol. Under these conditions, OSBP and ORP9L would be sequestered on PI-4P–enriched liposomes and effectively removed from the transfer reaction (see Figure 1D). Stimulation of transfer by inclusion of PI-4P in donor liposomes indicates that both lipid ligands must be in the same membrane for efficient cholesterol extraction and delivery to the acceptor membrane. During sterol transfer to an acceptor membrane, OSBP or ORP9L could remain tethered to the donor membrane via PI-4P and transfer sterol upon encountering a cholesterol-poor acceptor liposome. Alternatively, sterol loading of OSBP or ORP9L could disengage the PH domain from PI-4P and release the protein for delivery of the sterol ligand to cholesterol-poor acceptor membranes. Similar to several yeast OSH proteins (Raychaudhuri et al., 2006; Prinz, 2007), it appears that the primary function of ORP9L and OSBP is sterol transport.
If ORP9L is a sterol transfer protein, altered expression should influence the cholesterol content of target organelles. ORP9L depletion did not affect ER cholesterol homeostatic responses, potentially indicating no change in ER cholesterol content, but significantly increased cholesterol deposition in the endosomal/lysosomal compartment, as measure by filipin binding (Figure 8). The TGN is an important delivery site for endocytosed cholesterol (Urano et al., 2008) that, in the absence of ORP9L, appeared to backup into the endosomes and lysosomes. This suggests that ORP9L maintains cholesterol flux from the endosomal pathway to the trans-Golgi/TGN by transferring cholesterol to the ER or other organelles. Alterations in Golgi and ER cholesterol content would immediately impact on protein transport and organelle structure. Inhibition of protein export from the TGN and dispersion of the Golgi apparatus were observed when cells were enriched with cholesterol (Ying et al., 2003). Cholesterol depletion with cyclodextrin partially blocked export of apical raft-associated hemagglutinin A but not VSVG (Keller and Simons, 1998); however, only after removal of 50% of cellular cholesterol was Golgi morphology affected (Hansen et al., 2000). On the other hand, depletion of ER cholesterol with statins (Ridsdale et al., 2006) or cyclodextrin (Runz et al., 2006) inhibited export of VSVG either by interfering with lateral mobility or loading into ER exit sites, respectively. These results support the concept that ORP9L could retrieve cholesterol from the trans-Golgi/TGN to the ER. In the absence of ORP9L, cholesterol would be depleted from the ER and accumulate in the trans-Golgi/TGN and endosomes/lysosomes, resulting in disruption of protein transport. On the basis of the relatively minor effect of ORP9L knockdown on VSVG export, we propose that either 1) transfer is localized, affecting the cholesterol content of a restricted membrane environment or 2) the activity is redundant with other ORPs or lipid-binding/transfer proteins. Indeed, enforced expression of ORP9S caused complete cessation of ER export, inhibition of cholesterol synthesis and reduced endosomal/lysosomal cholesterol, suggesting it sequesters VAP and/or sterols from ORP9L as well as other transport proteins that rely on these factors.
Although both ORP9L and OSBP affect distinct Golgi functions, the unifying mechanism appears to be sterol transport between the ER and Golgi apparatus that involves PI-4P– and VAP-dependent interaction with these organelles. We propose that ORP9L specifically modifies the membrane environment by delivery of cholesterol to target membranes and activation of associated proteins. On the other hand, loss of Golgi interaction by N-terminal truncation of ORP9S renders it a dominant inhibitor of this activity.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Mader and Robert Zwicker for excellent technical assistance. Barbara Karten (Dalhousie University) provided assistance in quantifying of filipin fluorescence. This work was supported by an operating grant from the Canadian Institutes for Health Research (MOP-15284) and an Issac Walton Killiam Graduate Studentship (M.N.).
Abbreviations used:
- CERT
ceramide transfer protein
- ER
endoplasmic reticulum
- FFAT
two phenylalanines in an acidic tract
- GlcCer
glucosylceramide
- LPDS
lipoprotein-deficient serum
- OSBP
oxysterol-binding protein
- ORP
OSBP-related protein
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PH
pleckstrin homology
- PI-3P
phosphatidylinositol 3-phosphate
- PI-4P
phosphatidylinositol 4-phosphate
- PI-5P
phosphatidylinositol 5-phosphate
- TGN
trans-Golgi network
- VAP
vesicle-associated membrane protein–associated protein
- VSVG
vesicular stomatitus virus glycoprotein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-09-0905) on January 7, 2009.
REFERENCES
- Amarilio R., Ramachandran S., Sabanay H., Lev S. Differential regulation of endoplasmic reticulum structure through VAP-Nir protein interaction. J. Biol. Chem. 2005;280:5934–5944. doi: 10.1074/jbc.M409566200. [DOI] [PubMed] [Google Scholar]
- Balla A., Tuymetova G., Tsiomenko A., Varnai P., Balla T. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol. Biol. Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canagarajah B. J., Hummer G., Prinz W. A., Hurley J. H. Dynamics of cholesterol exchange in the oxysterol binding protein family. J. Mol. Biol. 2008;378:737–748. doi: 10.1016/j.jmb.2008.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coxey R. A., Pentchev P. G., Campbell G., Blanchette-Mackie E. J. Differential accumulation of cholesterol in Golgi compartments of normal and Niemann-Pick type C fibroblasts incubated with LDL: a cytochemical freeze-fracture study. J. Lipid Res. 1993;34:1165–1176. [PubMed] [Google Scholar]
- D'Angelo G., et al. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature. 2007;449:62–67. doi: 10.1038/nature06097. [DOI] [PubMed] [Google Scholar]
- de Graaf P., et al. Phosphatidylinositol 4-kinasebeta is critical for functional association of rab11 with the Golgi complex. Mol. Biol. Cell. 2004;15:2038–2047. doi: 10.1091/mbc.E03-12-0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis M. A., Di Campli A., D'Angelo G. Lipid-transfer proteins in membrane trafficking at the Golgi complex. Biochim. Biophys. Acta. 2007;1771:761–768. doi: 10.1016/j.bbalip.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Egan B., Beilharz T., George R., Isenmann S., Gratzer S., Wattenberg B., Lithgow T. Targeting of tail-anchored proteins to yeast mitochondria in vivo. FEBS Lett. 1999;451:243–248. doi: 10.1016/s0014-5793(99)00581-5. [DOI] [PubMed] [Google Scholar]
- Fairn G. D., McMaster C. R. The roles of the human lipid-binding proteins ORP9S and ORP10S in vesicular transport. Biochem. Cell Biol. 2005;83:631–636. doi: 10.1139/o05-064. [DOI] [PubMed] [Google Scholar]
- Fugmann T., Hausser A., Schoffler P., Schmid S., Pfizenmaier K., Olayioye M. A. Regulation of secretory transport by protein kinase D-mediated phosphorylation of the ceramide transfer protein. J. Cell Biol. 2007;178:15–22. doi: 10.1083/jcb.200612017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A., Di Campli A., Konstantakopoulos A., Di Tullio G., Alessi D. R., Kular G. S., Daniele T., Marra P., Lucocq J. M., De Matteis M. A. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Halter D., Neumann S., van Dijk S. M., Wolthoorn J., de Maziere A. M., Vieira O. V., Mattjus P., Klumperman J., van Meer G., Sprong H. Pre- and post-Golgi translocation of glucosylceramide in glycosphingolipid synthesis. J. Cell Biol. 2007;179:101–115. doi: 10.1083/jcb.200704091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- Hansen G. H., Niels-Christiansen L. L., Thorsen E., Immerdal L., Danielsen E. M. Cholesterol depletion of enterocytes. Effect on the Golgi complex and apical membrane trafficking. J. Biol. Chem. 2000;275:5136–5142. doi: 10.1074/jbc.275.7.5136. [DOI] [PubMed] [Google Scholar]
- Hoekstra D., Maier O., van der Wouden J. M., Slimane T. A., van I.S.C. Membrane dynamics and cell polarity: the role of sphingolipids. J. Lipid Res. 2003;44:869–877. doi: 10.1194/jlr.R300003-JLR200. [DOI] [PubMed] [Google Scholar]
- Huijbregts R. P., Topalof L., Bankaitis V. A. Lipid metabolism and regulation of membrane trafficking. Traffic. 2000;1:195–202. doi: 10.1034/j.1600-0854.2000.010301.x. [DOI] [PubMed] [Google Scholar]
- Huitema K., van den Dikkenberg J., Brouwers J. F., Holthuis J. C. Identification of a family of animal sphingomyelin synthases. EMBO J. 2004;23:33–44. doi: 10.1038/sj.emboj.7600034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im Y. J., Raychaudhuri S., Prinz W. A., Hurley J. H. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M., Rocha N., Zwart W., Jordens I., Janssen L., Kuijl C., Olkkonen V. M., Neefjes J. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J. Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandutsch A. A., Shown E. P. Assay of oxysterol-binding protein in a mouse fibroblast, cell-free system. Dissociation constant and other properties of the system. J. Biol. Chem. 1981;256:13068–13073. [PubMed] [Google Scholar]
- Kasper A. M., Helmkamp G. M., Jr Intermembrane phospholipid fluxes catalyzed by bovine brain phospholipid exchange protein. Biochim. Biophys. Acta. 1981;664:22–32. doi: 10.1016/0005-2760(81)90025-4. [DOI] [PubMed] [Google Scholar]
- Keller P., Simons K. Cholesterol is required for surface transport of influenza virus hemagglutinin. J. Cell Biol. 1998;140:1357–1367. doi: 10.1083/jcb.140.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace T. A., Byers D. M., Cook H. W., Ridgway N. D. Altered regulation of cholesterol and cholesteryl ester synthesis in Chinese-hamster ovary cells overexpressing the oxysterol-binding protein is dependent on the pleckstrin homology domain. Biochem. J. 1997;326:205–213. doi: 10.1042/bj3260205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehto M., Olkkonen V. M. The OSBP-related proteins: a novel protein family involved in vesicle transport, cellular lipid metabolism, and cell signalling. Biochim. Biophys. Acta. 2003;1631:1–11. doi: 10.1016/s1388-1981(02)00364-5. [DOI] [PubMed] [Google Scholar]
- Lessmann E., Ngo M., Leitges M., Minguet S., Ridgway N. D., Huber M. Oxysterol-binding protein-related protein (ORP) 9 is a PDK-2 substrate and regulates Akt phosphorylation. Cell Signal. 2007;19:384–392. doi: 10.1016/j.cellsig.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Lev S. The role of the Nir/rdgB protein family in membrane trafficking and cytoskeleton remodeling. Exp. Cell Res. 2004;297:1–10. doi: 10.1016/j.yexcr.2004.02.033. [DOI] [PubMed] [Google Scholar]
- Levine T. P., Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr. Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- Loewen C. J., Roy A., Levine T. P. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi A., Perry R. J., Storey M. K., Cook H. W., Byers D. M., Ridgway N. D. Golgi localization and phosphorylation of oxysterol binding protein in Niemann-Pick C and U18666A-treated cells. J. Lipid Res. 2001;42:1062–1071. [PubMed] [Google Scholar]
- Nohturfft A., Yabe D., Goldstein J. L., Brown M. S., Espenshade P. J. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 2000;102:315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- Olkkonen V. M., Levine T. P. Oxysterol binding proteins: in more than one place at one time? Biochem. Cell Biol. 2004;82:87–98. doi: 10.1139/o03-088. [DOI] [PubMed] [Google Scholar]
- Orci L., Montesano R., Meda P., Malaisse-Lagae F., Brown D., Perrelet A., Vassalli P. Heterogeneous distribution of filipin–cholesterol complexes across the cisternae of the Golgi apparatus. Proc. Natl. Acad. Sci. USA. 1981;78:293–297. doi: 10.1073/pnas.78.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretti D., Dahan N., Shimoni E., Hirschberg K., Lev S. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP Proteins and is essential for Golgi-mediated transport. Mol. Biol. Cell. 2008;19:3871–3884. doi: 10.1091/mbc.E08-05-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. J., Ridgway N. D. Molecular mechanisms and regulation of ceramide transport. Biochim. Biophys. Acta. 2005;1734:220–234. doi: 10.1016/j.bbalip.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Perry R. J., Ridgway N. D. Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol. Biol. Cell. 2006;17:2604–2616. doi: 10.1091/mbc.E06-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz W. A. Non-vesicular sterol transport in cells. Prog. Lipid Res. 2007;46:297–314. doi: 10.1016/j.plipres.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S., Im Y. J., Hurley J. H., Prinz W. A. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J. Cell Biol. 2006;173:107–119. doi: 10.1083/jcb.200510084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway N. D. Interactions between metabolism and intracellular distribution of cholesterol and sphingomyelin. Biochim. Biophys. Acta. 2000;1484:129–141. doi: 10.1016/s1388-1981(00)00006-8. [DOI] [PubMed] [Google Scholar]
- Ridgway N. D., Dawson P. A., Ho Y. K., Brown M. S., Goldstein J. L. Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J. Cell Biol. 1992;116:307–319. doi: 10.1083/jcb.116.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridsdale A., Denis M., Gougeon P. Y., Ngsee J. K., Presley J. F., Zha X. Cholesterol is required for efficient endoplasmic reticulum-to-Golgi transport of secretory membrane proteins. Mol. Biol. Cell. 2006;17:1593–1605. doi: 10.1091/mbc.E05-02-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runz H., Miura K., Weiss M., Pepperkok R. Sterols regulate ER-export dynamics of secretory cargo protein ts-O45-G. EMBO J. 2006;25:2953–2965. doi: 10.1038/sj.emboj.7601205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp E. L., Hegde R. S., Francolini M., Lombardo F., Colombo S., Pedrazzini E., Borgese N., Lippincott-Schwartz J. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 2003;163:257–269. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchanek M., Hynynen R., Wohlfahrt G., Lehto M., Johansson M., Saarinen H., Radzikowska A., Thiele C., Olkkonen V. M. The mammalian oxysterol-binding protein-related proteins (ORPs) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. Biochem. J. 2007;405:473–480. doi: 10.1042/BJ20070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth B., Balla A., Ma H., Knight Z. A., Shokat K. M., Balla T. Phosphatidylinositol 4-kinase III beta regulates the transport of ceramide between the endoplasmic reticulum and Golgi. J. Biol. Chem. 2006;281:36369–36377. doi: 10.1074/jbc.M604935200. [DOI] [PubMed] [Google Scholar]
- Urano Y., Watanabe H., Murphy S. R., Shibuya Y., Geng Y., Peden A. A., Chang C. C., Chang T. Y. Transport of LDL-derived cholesterol from the NPC1 compartment to the ER involves the trans-Golgi network and the SNARE protein complex. Proc. Natl. Acad. Sci. USA. 2008;105:16513–16518. doi: 10.1073/pnas.0807450105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. Y., Weng J., Anderson R. G. OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science. 2005;307:1472–1476. doi: 10.1126/science.1107710. [DOI] [PubMed] [Google Scholar]
- Wang Y., Thiele C., Huttner W. B. Cholesterol is required for the formation of regulated and constitutive secretory vesicles from the trans-Golgi network. Traffic. 2000;1:952–962. doi: 10.1034/j.1600-0854.2000.011205.x. [DOI] [PubMed] [Google Scholar]
- Wang Y. J., Wang J., Sun H. Q., Martinez M., Sun Y. X., Macia E., Kirchhausen T., Albanesi J. P., Roth M. G., Yin H. L. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- Weixel K. M., Blumental-Perry A., Watkins S. C., Aridor M., Weisz O. A. Distinct Golgi populations of phosphatidylinositol 4-phosphate regulated by phosphatidylinositol 4-kinases. J. Biol. Chem. 2005;280:10501–10508. doi: 10.1074/jbc.M414304200. [DOI] [PubMed] [Google Scholar]
- Wyles J. P., McMaster C. R., Ridgway N. D. Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J. Biol. Chem. 2002;277:29908–29918. doi: 10.1074/jbc.M201191200. [DOI] [PubMed] [Google Scholar]
- Wyles J. P., Perry R. J., Ridgway N. D. Characterization of the sterol-binding domain of oxysterol-binding protein (OSBP)-related protein 4 reveals a novel role in vimentin organization. Exp. Cell Res. 2007;313:1426–1437. doi: 10.1016/j.yexcr.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Wyles J. P., Ridgway N. D. VAMP-associated protein-A regulates partitioning of oxysterol-binding protein-related protein-9 between the endoplasmic reticulum and Golgi apparatus. Exp. Cell Res. 2004;297:533–547. doi: 10.1016/j.yexcr.2004.03.052. [DOI] [PubMed] [Google Scholar]
- Yan D., Jauhiainen M., Hildebrand R. B., Willems van Dijk K., Van Berkel T. J., Ehnholm C., Van Eck M., Olkkonen V. M. Expression of human OSBP-related protein 1L in macrophages enhances atherosclerotic lesion development in LDL receptor-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2007a;27:1618–1624. doi: 10.1161/ATVBAHA.107.144121. [DOI] [PubMed] [Google Scholar]
- Yan D., Lehto M., Rasilainen L., Metso J., Ehnholm C., Yla-Herttuala S., Jauhiainen M., Olkkonen V. M. Oxysterol binding protein induces upregulation of SREBP-1c and enhances hepatic lipogenesis. Arterioscler. Thromb. Vasc. Biol. 2007b;27:1108–1114. doi: 10.1161/ATVBAHA.106.138545. [DOI] [PubMed] [Google Scholar]
- Yan D., Mayranpaa M. I., Wong J., Perttila J., Lehto M., Jauhiainen M., Kovanen P. T., Ehnholm C., Brown A. J., Olkkonen V. M. OSBP-related protein 8 (ORP8) suppresses ABCA1 expression and cholesterol efflux from macrophages. J. Biol. Chem. 2008;283:332–340. doi: 10.1074/jbc.M705313200. [DOI] [PubMed] [Google Scholar]
- Ying M., Grimmer S., Iversen T. G., Van Deurs B., Sandvig K. Cholesterol loading induces a block in the exit of VSVG from the TGN. Traffic. 2003;4:772–784. doi: 10.1034/j.1600-0854.2003.00134.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.