Abstract
Cell motility of amoeboid cells is mediated by localized F-actin polymerization that drives the extension of membrane protrusions to promote forward movements. We show that deletion of either of two members of the Dictyostelium Dock180 family of RacGEFs, DockA and DockD, causes decreased speed of chemotaxing cells. The phenotype is enhanced in the double mutant and expression of DockA or DockD complements the reduced speed of randomly moving DockD null cells' phenotype, suggesting that DockA and DockD are likely to act redundantly and to have similar functions in regulating cell movement. In this regard, we find that overexpressing DockD causes increased cell speed by enhancing F-actin polymerization at the sites of pseudopod extension. DockD localizes to the cell cortex upon chemoattractant stimulation and at the leading edge of migrating cells and this localization is dependent on PI3K activity, suggesting that DockD might be part of the pathway that links PtdIns(3,4,5)P3 production to F-actin polymerization. Using a proteomic approach, we found that DdELMO1 is associated with DockD and that Rac1A and RacC are possible in vivo DockD substrates. In conclusion, our work provides a further understanding of how cell motility is controlled and provides evidence that the molecular mechanism underlying Dock180-related protein function is evolutionarily conserved.
INTRODUCTION
In eukaryotes, cell migration is essential for many biological processes such as embryonic development and tissue renewal and in humans it is also linked to numerous pathologies including cancer. Cells are set in motion through reorganization of the actin cytoskeleton that creates cytoplasmic protrusions and promotes forward movement. Members of the Rac subfamily of Rho small GTPases are key regulators of cytoskeletal dynamics in virtually all eukaryotes. Activated Rac proteins relay directional signals from the leading edge of migrating cells to downstream effectors such as SCAR/WAVE and WASP proteins that mediate F-actin polymerization and power cell motility (Jaffe and Hall, 2005; Ridley, 2006; Charest and Firtel, 2007; Kolsch et al., 2008; Ladwein and Rottner, 2008). Despite their importance, the upstream signaling mechanisms that mediate Rac activation during cell migration are still unclear.
The activity of the small GTPases is regulated through a GDP/GTP exchange mechanism that is catalyzed by the guanine nucleotide exchange factors (GEF), GTPase-activating proteins (GAPs), and guanine nucleotide-dissociation inhibitors (GDIs). GEFs activate GTPases by catalyzing the dissociation of GDP from the protein, thereby allowing the binding of GTP and their switch to an active state. Members of the classical Dbl homology-pleckstrin homology (DH-PH) domain–containing family were thought to be the universal activators of Rho GTPases until the CZH (CDM-zizimin homology) family of unconventional Rho-GEFs was discovered (Côté and Vuori, 2002; Meller et al., 2005; Côté and Vuori, 2007). In the CZH proteins, a CZH2 domain, rather than a DH domain, interacts with Rac proteins and mediates nucleotide exchange (Côté et al., 2006; Côté and Vuori, 2006). Most CZH proteins also feature a CZH1 domain that can bind phospholipids (Kobayashi et al., 2001; Côté et al., 2005). Dock180 and Dock180-related proteins belong to this novel class of RhoGEFs (Côté and Vuori, 2002). In addition to the CZH1 and CZH2 domains, most Dock180-related proteins also contain an N-terminal Src homology 3 (SH3) domain that mediates intra- or intermolecular protein interaction via binding to proline-rich motifs (Mayer, 2001). Recent findings suggest that Dock180 and Dock180-related proteins play a pivotal role in a wide variety of fundamentally important biological functions including cell migration, phagocytosis of apoptotic cells, myoblast fusion, and neuronal polarization (Hiramoto et al., 2006; Kunisaki et al., 2006; Watabe-Uchida et al., 2006; Bianco et al., 2007; Moore et al., 2007; Geisbrecht et al., 2008). In addition, Dock180 was implicated in the invasive phenotype of glioma cells, suggesting that cancer cell migration might be promoted through this unconventional RhoGEF (Jarzynka et al., 2007). In spite of the important roles for these proteins in normal and pathological conditions, the Dock180-related proteins remain poorly characterized.
Four Dock180-related proteins, DockA to DockD, have been identified in Dictyostelium based on the similarity to their metazoan counterparts (Meller et al., 2005). Two of them, DockA and DockD, are the Dock proteins most homologous to human Dock180 by sequence as well as domain composition and organization. In this article, we demonstrate that Dictyostelium DockA and DockD are important regulators of cell movement, as cells lacking these proteins have severely reduced cell motility. Moreover, overexpression of DockD results in a significant enhancement of F-actin polymerization at the leading edge that causes an increase in membrane protrusion. DockD localizes to the cell cortex in response to global chemoattractant stimulation and to the leading edge of chemotaxing cells via a mechanism that is dependent on PI3K. We also identified DdELMO1 as one of the interactors of DockD by purification of the DockD associated proteins in vegetative and stimulated cells and Rac1 as a putative substrate of DockD. These data indicate that Dock180 family members may constitute a direct link between PI3K activity and F-actin polymerization at the leading edge through a molecular mechanism that seems to be conserved between Dictyostelium and other organisms.
MATERIALS AND METHODS
Cell Culture, Strains, and Plasmids
All Dictyostelium discoideum cell lines were cultured axenically in HL5 medium at 21°C.
Wild-type axenic strain Ax2 was used to generate single and double knockouts of DockA and DockD. To knock out DockD, we made a knockout construct by amplifying positions 1007-1997 of the DockD genomic DNA by PCR and inserting a BamHI site at position 1450. The blasticidin resistance cassette was inserted into the created BamHI site, and the construct was used for gene replacement in the Ax2 parental Dictyostelium strain. To create a DockA construct for gene replacement, the DockA sequence from position 2026-3046 was amplified by PCR and the internal BamHI site at position 2476 was used to insert the blasticidin resistance cassette.
To create dckA−/dckD− cells, the same constructs were used as for creation of the single knockouts. The blasticidin-resistance gene in dckD− cells was removed using the Cre-Lox system and the DockA gene disruption was performed afterward (Kimmel and Faix, 2006). Randomly selected clones were screened for gene disruption by PCR, which was then confirmed by Southern and Northern blot analyses.
The pten null strain was obtained from P. Devreotes (Johns Hopkins University School of Medicine, Baltimore, MD). pi3k1−/2−/3− and pi3k1−/2− cells have been described previously (Funamoto et al., 2002; Takeda et al., 2007).
Full-length DockA and DockD DNA was obtained by PCR amplification of three different pieces from Dictyostelium strain AX2 genomic DNA with primers based on the predicted gene sequences DockA (DDB0201649) and DockD (DDB0233625) in the Dictyostelium Genome Project database (http://dicty.sdsc.edu). To combine the three fragments, the following internal restriction sites were used: for DockA, BamHI at position 2476 and NsiI at position 4584; and for DockD, AlwNI at position 2540 and PvuI at position 4380. The full-length sequences were ligated in-frame into the BglII-XhoI site of GFP-EXP4(+) or V5-EXP4(+). All constructs were sequenced. Overexpression constructs were transformed into wild-type Ax2 cells by electroporation, and cells were selected in the presence of G418 (20 or 40 μg/ml).
Cell Movement and Image Acquisition
The analyses of chemotaxis toward cAMP were performed as described previously (Chung et al., 2000; Park et al., 2004; Mendoza and Firtel, 2006). Briefly, cells were pulsed with 30 nM cAMP at 6-min intervals for 6 h and plated on glass-bottomed microwell plates. A micropipette containing 150 mM cAMP was positioned to stimulate the cells, using a micromanipulator (Eppendorf-Netheler-Hinz GmbH, Hamburg, Germany), and the response of the cells was recorded using a time-lapse video recorder and NIH Image software (one image every 6 s; http://rsb.info.nih.gov/ij/). For a random movement assay, vegetative cells growing on plates were harvested and seeded onto a chambered cover glass in HL5 medium. Cells were rinsed three times with an excess amount of Na-K phosphate buffer at 10 min after seeding and then sat for 1 h before recording. Images were collected on a microscope (model TE300; Nikon, Melville, NY) with DIC and 20×/0.60 NA objectives. Computer-assisted analysis of cell movement and shape change was performed using the DIAS program (Wessels et al., 1988). The pi3k1/2− and pi3k1/2/3− cells expressing GFP-DockD were pulsed with cAMP for 7–8 h before performing global stimulation and chemotaxis experiments conducted on the same pulsed cells confirmed that they were able to polarize and move in a cAMP gradient (data not shown).
Global responses to cAMP were observed as described previously (Sasaki et al., 2004). Quantitation of membrane or cortical localization of GFP fusion proteins and the analysis of chemotaxis of each strain represent the averages of at least five cells from at least three separate experiments. To quantify the relative changes in GFP-DockD at the cell cortex, we used the following protocol. For each cell, we calculated the relative level of DockD at the cell cortex at the 0 time point by quantifying the relative value of DockD fluorescence between the cortex and cytosol at 5–10 different positions along the plasma membrane using NIH Image software and averaged those values. These 0-time point values for multiple cells were then averaged to obtain the 0 value for each strain as plotted on the graph. For each cell at each time point after stimulation, we measured the average intensity across the cell cortex (average of 5–10 measurements for each cell at each time point) and divided that value by the average intensity for the cortex at the 0-time point for that same cell, thus providing a relative change in cortical intensity for each cell at that time point. The values for the multiple cells were then averaged to obtain the value at a specific time point plotted on the graph. This procedure was repeated for each time point producing a relative change in DockD at the cell cortex for each strain. This approach allows calculation of average values for a strain although the level of GFP-DockD expression varies from cell to cell. For the latrunculin B (LatB) experiments, cells were pretreated with 3 μM of LatB (Sigma-Aldrich, St. Louis, MO) for 30 min before assay. For the LY294002 experiments, cells were pretreated with 60 μM of LY294002 (Sigma-Aldrich) for 1 h before assay.
Quantification of F-Actin Polymerization
F-actin polymerization was assayed as described previously (Park et al., 2004). Briefly, cells were starved for 2 h, pulsed with 30 nM cAMP for 5 h and treated with 1 mM caffeine for 30 min before stimulation with 1 mM cAMP. At the indicated time points, the reaction was stopped by lysing the cells in lysis buffer (20 mM TES, 1% Triton-X-100, 2 mM MgCl2, 5 mM EGTA, 5 mg/μl leupeptin, 5 mg/μl aprotinin). Cytoskeletal proteins were isolated as proteins insoluble in the detergent Triton X-100. The protein pellets were dissolved by being boiled in SDS-PAGE sample buffer (120 mM Tris, pH 6.8, 4% SDS, 20% glycerol, and 200 mM DTT), run on 8% acrylamide gels, and stained with Coomassie blue. Protein bands were scanned, and changes in actin content were quantitated using Image Gauge software (Fuji Film, Tokyo, Japan).
For F-actin staining, vegetative cells were taken from plates and allowed to adhere to cover slips. Cells were fixed for 10 min with 3.7% formaldehyde in NaK phosphate buffer, washed three times with buffer, and then permeabilized for 10 min with 0.2% Triton X-100 in NaK phosphate buffer. F-actin was stained with 250 nM TRITC-phalloidin (Sigma). Images were acquired with a Leica inverted DMIRE2 microscope (Deerfield, IL) with a 63×/1.4 NA objective using an ORCA-ER camera (Hamamatsu, Bridgewater, NJ).
In Vivo Pulldown and Purification of DockD-interacting Proteins and MS/MS Analyses
Vegetative and aggregation-competent cells were washed with Na/K phosphate buffer and resuspended at a density of 1 × 108 cells/ml in Na/K phosphate buffer. Cells were lysed (1% NP-40, 300 mM NaCl, 40 mM MOPS, pH 7.0, 20% glycerol, 2 mM Na3VO4, 2 μg/ml leupeptin, and 5 μg/ml aprotinin) and incubated in 2.5 mg/ml DSP (ditho-bis-succinimidylpropionate, EMD Chemicals, La Jolla, CA) for 10 min. The cross-linking reactions were quenched with 200 mM Tris-HCl (pH 7.4). Proteins in total cell extracts were left to solubilize overnight before being subjected to immunoprecipitation with 25 μl resin of anti-V5 (V5-10) antibody agarose conjugate (Sigma). The coimmunoprecipitated proteins were eluted with 0.1 M glycine (pH 2.5), analyzed by silver staining, and subsequently precipitated with methanol/chloroform. The protocol for MS/MS analyses is provided in the Supplementary Methods.
RESULTS
DockA and DockD Play a Role in Cell Motility and Are Functionally Redundant
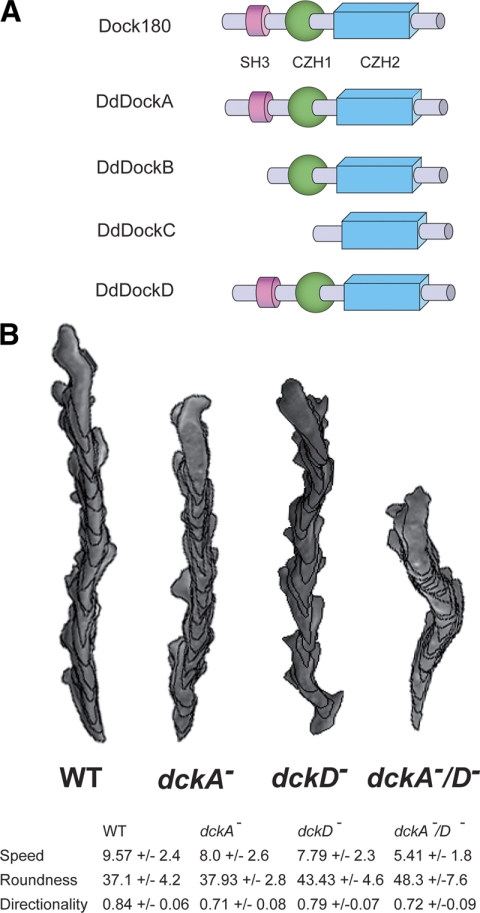
The Dictyostelium genome encodes eight CZH genes: four belong to the Dock180-related subfamily and four to the Zizimin-related group. Dictyostelium Dock180-related proteins DockA and DockD are the Dictyostelium Dock proteins most homologous to human Dock180 by sequence and domain structure, as they contain an N-terminal SH3 domain, a putative lipid-binding CZH1 domain, and a Rac-binding CZH2 domain (Figure 1A). These domains are conserved in metazoan CDM proteins and are essential for the function of these proteins (Meller et al., 2005).
Figure 1.
(A) Schematic representation of the Dictyostelium Dock180 protein family. The domain organization of the Homo sapiens Dock180 protein and the Dictyostelium Dock180 protein family is shown. (B) Computer-assisted analysis of chemotaxis (DIAS) of aggregation-competent wild-type (strain Ax2), dckA−, dckD−, and dckA−/D− cells in a cAMP gradient. Speed refers to the speed of the cell's centroid movement along the total path. Roundness is an indication of the polarity of the cells. Directionality is the distance from the start to finish divided by the total distance moved.
To investigate the role of Dictyostelium Dock180-related proteins in cell migration, we created single and double knockout mutants of the DockA and DockD (dckA− and dckD−) and examined the effects of these mutations on cell growth, morphogenesis, and cell migration. The single knockouts exhibit no growth or developmental defects, whereas the development of dckA−/D− is delayed and the fruiting bodies are abnormal, with shorter and sometimes thicker stalks (Supplemental Figure S1).
Next, we examined the ability of the single and double null strains to migrate toward the chemoattractant cAMP. Developmentally competent dckA− cells chemotax with speed and directionality slightly lower than those of wild-type cells, whereas dckD− cells exhibit moderately reduced speed and polarity but almost wild-type directionality (Figure 1B). Interestingly, the double mutant dckA−/D− exhibit a further reduction in speed and polarity. This finding indicates that both DockA and DockD play a role in mediating chemotaxis toward cAMP in aggregation-competent cells and suggests that these two RacGEFs may act redundantly or control parallel pathways required for proper chemotaxis.
To determine whether the observed chemotaxis phenotypes were the results of impaired cell motility, we studied the random motility of vegetative cells, which do not produce or respond to the chemoattractant cAMP. As shown in Figure 2A, dckD− cells move significantly slower than wild-type cells, whereas vegetative dckA− cells show no significant decrease in random motility and dckA−/D−cells exhibit only a slight decrease in speed compared with dckD− cells. These observations suggest that DockD (but not DockA) participates in random cell movement during the growth phase. However, expression of either DockA or DockD in the double knockout background is able to rescue the dckA−/D− cell motility phenotype, corroborating the hypothesis that these proteins might have redundant functions (Figure 2A). As dckD− cells display a stronger phenotype than dckA−, we decided to focus our analyses on the function of DockD, because it is more likely to play an extensive role in cell motility.
Figure 2.
DockA and DockD roles in random cell movement. (A) Vegetative grown cells were plated in Na/K phosphate buffer and starved on plates for 1 h. Cell movement was monitored by DIC imaging for 30 min at 24-s intervals. Speed and roundness were analyzed with the DIAS program (Soll and Voss, 1998). (a) wild-type, (b) dckA−,(c) dckD−, and dckAA−/D−. (B) Overexpression of either DockA or DockD rescues the motility phenotype of dckA−/D−. (a) DockA in dckA−/D− and DockD in dckA−/D−. (C) Overexpression of GFP-DockD in wild-type cells increases cell motility.
DockD Triggers F-Actin Polymerization in Randomly Moving and Chemotaxing Cells
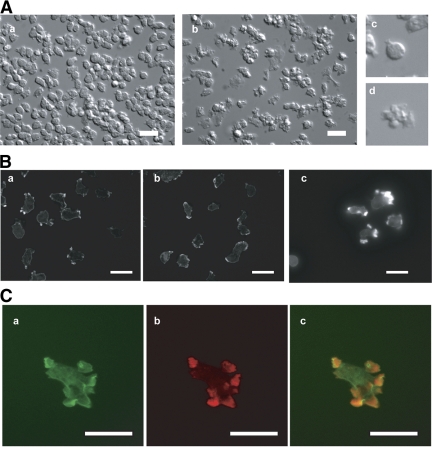
To further elucidate the function of the Dock180-related family, we evaluated the effect of increased levels of DockD in wild-type cells. In random movement assays, cells overexpressing DockD (GFP-DockD) move faster than wild-type cells (Figure 2B). In addition, they exhibit multiple pseudopodial protrusions compared with Dictyostelium vegetative wild-type cells (Figure 3A).
Figure 3.
Increased levels of DockD produce multiple cell protrusions and trigger F-actin polymerization in vegetative cells and chemotaxing cells. (A) Cell shape of vegetative cells: (a) wild-type; (b) GFP-DockD, (c) detail of a; and (d) detail of b. (B) Phalloidin staining of vegetative (a) wild-type, (b) dckA−/D−, and (c) wild-type cells overexpressing GFP-DockD. (C) (a) GFP localization, (b) phalloidin staining, and (c) overlay of the same cells as in B, panel c. Scale bar, 10 μm.
To examine the structure of the cell protrusions in DockD overexpressing cells, we stained GFP-DockD-expressing and wild-type cells with phalloidin and used the same camera settings for collecting the images for both strains. GFP-DockD cells show much stronger phalloidin staining than wild-type cells, and the signal is associated with membrane protrusions that are often broad, resembling the lamellipodia observed in neutrophils rather than the narrower pseudopodia typical of Dictyostelium cells (Figure 3B, a and c). These observations suggest that increased levels of DockD enhance F-actin polymerization and the effect is likely to be direct, as GFP-DockD and TRITC-phalloidin staining colocalize to sites of F-actin accumulation (Figure 3C). In dckA−/D− mutant cells, the region of phalloidin staining is significantly broader than in wild-type cells and is not localized to distinct foci as in the wild type (Figure 3Bb). This observation suggests that loss of both DockA and DockD affect F-actin accumulation by influencing the distribution rather than the amount of F-actin polymerization. This conclusion is supported by the quantification of F-actin polymerization that revealed a comparable amount of F-actin in the wild-type and the single and double mutant strains (Supplemental Figure S2).
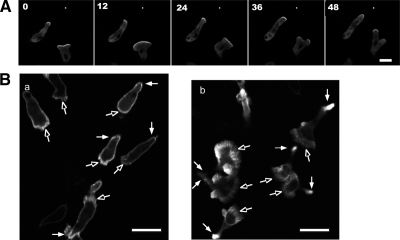
Because DockD overexpression triggers F-actin formation in vegetative cells, we investigated F-actin levels during chemotaxis of aggregation-competent cells using the GFP fusion of F-actin–binding protein ABP as a reporter for the sites of newly synthesized F-actin (Pang et al., 1998). In chemotaxing cells, GFP-DockD preferentially localizes to the leading edge (Figure 4A). In wild-type cells that coexpress V5-tagged DockD and GFP-ABP, we observe an enlarged leading edge with a higher level of GFP-ABP than seen in the wild-type cells, as depicted in Figure 4B and Supplementary Movies M1 and M2. The dynamic nature of the GFP-DockD cell leading edge can be seen in Supplementary Movie M1.
Figure 4.
DockD localizes at the leading edge of migrating cells and colocalizes with sites of F-actin polymerization. (A) Time-lapse images of GFP-DockD in aggregation-competent wild-type cells moving toward a micropipette filled with cAMP. An asterisk indicates the position of the micropipette. Scale bar, 10 μm. (B) ABP-GFP in wild-type cells (a) and wild-type/V5-DockD cells (b). Open arrowhead points to the lamellipod.
Taken together, these results indicate that DockD mediates F-actin polymerization and therefore participates in the formation of the cell protrusions that sustain cell movement.
DockD Translocates to the Cell Cortex in Response to Chemoattractant Stimulation
The key cellular events that mediate the initial steps of cell migration take place at the plasma membrane. In particular, cell motility is thought to be powered by the polymerization of actin near the cell cortex (Rafelski and Theriot, 2004; Upadhyaya and van Oudenaarden, 2004; Disanza et al., 2005; Ridley, 2006; Papakonstanti and Stournaras, 2008). To further elucidate the role of Dictyostelium Dock180-related family members in cell motility, we investigated the relationship between DockD distribution and F-actin polymerization in aggregation-competent cells.
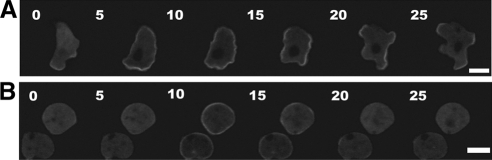
In response to global chemoattractant stimulation, GFP-DockD rapidly translocates to the cortex with a peak at 6–8 s after addition of cAMP and then delocalizes from much of the cell cortex by 15–20 s (Figure 5A and B). Interestingly, DockD remains localized at sites where new pseudopodia start to extend. These long, prominent pseudopodia are not observed in similarly stimulated wild-type cells within the same time frame, appearing only after several minutes (wild-type cells expressing GFP-LimE, a reporter for F-actin, Supplementary Movie M4; Diez et al., 2005; Figure 5, A and B, and Supplementary Movie M3). Inhibition of F-actin by pretreatment with LatB does not affect the cortical localization of GFP-DockD in unstimulated cells (Figure 5B, time 0, 10, and 15 s, respectively). However, most of DockD delocalizes from the cortex in LatB-treated cells within 15–20 s after stimulation (Figure 5, A and B). In summary, after the initial localization of DockD to the cell cortex upon chemoattractant stimulation, DockD is retained at the sites of the cell cortex where pseudopodia are emerging whereas it detaches from the cell cortex in cells pretreated with LatB, which are unable to produce pseudopodia due to inhibition of actin polymerization.
Figure 5.
DockD translocation to the membrane is not impaired by inhibition of F-actin polymerization. Wild-type cells overexpressing GFP-DockD without (A) and with LatB (B). Aggregation-competent cells were treated with LatB before being subjected to uniform stimulation with cAMP. Time points are shown in seconds. Scale bar, 10 μm.
DockD Translocation Is Regulated by PtdIns(3,4,5)P3
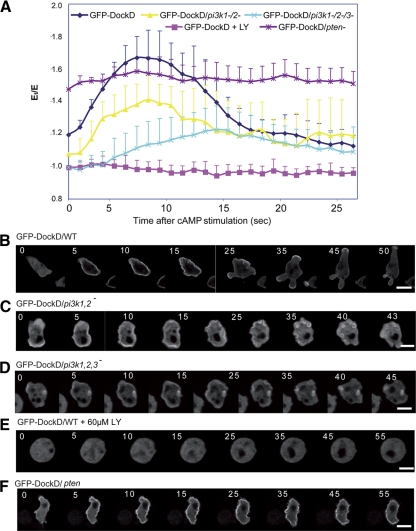
In Dictyostelium, as in other systems, localized production of PtdIns(3,4,5)P3 by class I PI3Ks has been linked to localized F-actin polymerization and cell movement (Van Haastert and Veltman, 2007; Janetopoulos and Firtel, 2008). To investigate whether DockD may provide a link between PtdIns(3,4,5)P3 and leading edge formation in Dictyostelium, we investigated whether DockD localization is dependent on PtdIns(3,4,5)P3 production. Indeed, the translocation of GFP-DockD is impaired in pi3k1−/2− in which ∼85% of PI3K activity is lost (Figure 6, B and C; Takeda et al., 2007). Moreover, DockD localization is not only reduced but also delayed a pi3k1−/2−/3− background in which 95% of PI3K activity is lost (Figure 6, B and D; Takeda et al., 2007). We observed a GFP-DockD signal in vesicles upon global stimulation as well as in chemotaxing cells (data not shown); therefore we concluded that these structures are not hallmarks of underdeveloped cells but rather a feature of the localization of GFP-DockD in pi3k1/2− and pi3k1/2/3− cells.
Figure 6.
DockD translocation to the membrane is regulated by PtdIns(3,4,5)P3. (A) Translocation kinetics. Fluorescent images at the times after stimulation are shown. GFP-DockD in wild-type cells (B), pi3k1−/2− cells (C), pi3k1−/2−/3− cells (D), wild-type cells treated for 1 h with 30 μM LY294002 (E), and pten− cells (F) after uniform stimulation with cAMP. Scale bar, 10 μm. See Materials and Methods for description of data analysis.
Accordingly, pretreatment with the PI3K inhibitor LY294002 completely blocks DockD translocation to the cortex (Figure 6E). Consistent with these findings, cells lacking the PtdIns(3,4,5)P3 phosphatase PTEN exhibit a high basal level of GFP-DockD at the cell cortex, mostly likely as a consequence of the elevated PtdIns(3,4,5)P3 (Funamoto et al., 2002; Iijima and Devreotes, 2002; Figure 6F). Moreover, the amount of GFP-DockD at the plasma membrane in pten− cells is highly elevated even in unstimulated cells, presumably because the increased level of PtdIns(3,4,5)P3 promotes maximal DockD cortical localization. Note that although the pten− cell shown in Figure 6F exhibits a small response, the average of a large number of cells, as graphed in Figure 6A, demonstrates that there is a high level of GFP-DockD at the cell cortex in unstimulated pten− cells and that, on the average, the level does not change significantly upon stimulation (see Materials and Methods). These findings suggest that the localization of DockD to the plasma membrane is dependent on localized production of PtdIns(3,4,5)P3.
DockD Forms a Complex with Dictyostelium ELMO1 and Rac GTPases
To shed light on the molecular mechanism underlying DockD function in cell motility, we attempted to identify its interacting partners through an in vivo pulldown assay and purification of DockD-associated proteins. A V5-tagged version of DockD was expressed in Dictyostelium wild-type cells and immunoprecipitated with an anti-V5 antibody in both vegetative and aggregation-competent cells at different time points after stimulation. Visualization of the coimmunoprecipitated proteins by silver staining revealed the presence of a few bands that were unique to the DockD sample, indicating that they were likely to represent specific DockD-interacting proteins (Figure 7). Mass spectrometry analysis of the sample obtained from V5-DockD aggregation-competent cells before induction uncovered additional proteins that coimmunopurified with DockD and that were not found in the samples from wild-type cells (Table 1; data not shown). Based on spectral count and silver staining, the most abundant component of the DockD immunoprecipitates was a 110-kDa protein that corresponds to Dictybase accession no. DDB0233912. Database homology searches and domain architecture prediction indicate that this accession features an ELM domain and has the highest homology to the human ELMO1 protein (GenBank accession no. AAL14466); therefore, we named it DdELMO1. In addition to DdELMO1, four other ELMO domain–containing proteins are encoded by the Dictyostelium genome (DdELMO2–5), but DdELMO1 is the only one that exhibits a PxxP motif at the C-terminus like the human ELMO1 (Supplemental Figure S3).
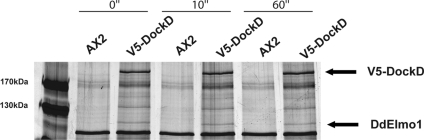
Figure 7.
DdELMO1 is associated with the DockD complex. DockD-associated proteins were coimmunoprecipitated from V5-DockD aggregation-competent cells at 0, 10, and 60 s after stimulation with cAMP, and wild-type Ax2 cells were used as a control. The purified proteins were separated on an acrylamide gel and detected by silver nitrate staining. The upper portion of the gel contains the DockD band (upper arrow) and a band at 109 kDa (lower arrow) that corresponds to DdELMO1.
Table 1.
Partial list of proteins identified by mass spectrometry analysis of DockD associated proteins
| Ax2No. of spectra | V5-DockDNo. of spectra | Distinct peptides (n) | Protein name |
|---|---|---|---|
| 0 | 230 | 68 | DockD |
| 0 | 30 | 20 | DdELMO1 |
| 0 | 3 | 2 | Rac1a |
| 0 | 1 | 1 | RacC |
The DockD-interacting proteins were immunoprecipitated and analyzed by mass spectrometry. Wild-type Ax2 cells were used as a control for the V5-DockD cells. Some of the proteins that are uniquely identified in the V5-DockD cells are listed in the table. Both spectra count and total number of identified distinct peptides are listed for each protein in each sample. The most abundant peptides are from the Dictybase accession no. DDB0233912 that we named DdELMO1.
The band that corresponds to the DdELMO1 protein was detected in a sample from vegetative cells expressing V5-DockD (data not shown) as well as developed cells before and after global stimulation with cAMP, suggesting that DockD forms a stable complex with DdELMO1 (Figure 7).
Two Rac GTPases, Rac1A and RacC, were also found among the proteins that coimmunoprecipitate with DockD in aggregation-competent cells (Table 1). Rac1A was identified by two distinct peptides, whereas RacC was distinguished by one peptide. The low spectral count is in agreement with the notion that interaction between GTPases and their activating protein is known to be transient. Given the relatively low abundance of Rac1A and RacC in the complex, the corresponding band is not detectable by silver staining (data not shown).
In summary, we used a proteomic approach that combines complex purification and mass spectrometry to identify DockD-interacting proteins. This approach has led us to the identification of the Dictyostelium homologue of a known regulator of Dock180 proteins, ELMO1, and two possible targets, Rac1A and RacC, as members of the DockD complex in Dictyostelium cells, strongly suggesting that Dictyostelium Dock180-related proteins are likely to function in a molecular context similar to that of their metazoan homologues.
DISCUSSION
DockA and DockD Participate in Controlling Cell Movement
In this work, we show that the Dictyostelium Dock180 family members DockA and DockD play a role in mediating cell motility. dckA− and dckD− strains exhibit only a modest decrease in speed, but the defect becomes more severe in cells lacking both proteins, suggesting that DockA and DockD may act redundantly during chemotaxis in aggregation-competent cells or control parallel pathways. As both DockA and DockD can complement the dckA/D− cell random movement phenotype, we concluded that these proteins are likely to have similar functions in regulating cell motility. However, DockD appears to play a more general role in this process as indicated by the defect in random movements of dckD− cells that neither secrete nor respond to cAMP. This observation is also in agreement with the DockD expression profile, as the gene is expressed at all stages of development through early culmination (our unpublished observation).
The phenotype that resulted from increased levels of DockD hints at its cellular function, as overexpression of DockD triggers the formation of multiple pseudopodia. We observed that DockD localized at the cortical sites of F-actin polymerization, suggesting that this protein is closely involved in the cytoskeleton remodeling on or near to membranes that produce plasma membrane protrusion. As a consequence, these cells move with a higher speed, suggesting that DockD is capable of regulating forward movement by generating the protrusive force required for cell motility. This is in agreement with the observation that the pattern of F-actin is altered as a consequence of the loss of both DockA and DockD, because the vegetative double null cells appeared incapable of localizing F-actin accumulation at restricted regions of the cortex to create effective membrane protrusions. Thus, the reduction in speed of motility observed in dckA/D− cells can be interpreted as a deficiency in the ability to extend pseudopodia. As overexpression of DockD leads to extended F-actin accumulation at the leading edge of chemotaxing cells, we believe that DockD does directly regulate F-actin polymerization even though one does not observe a distinguishable difference in the second peak of F-actin polymerization between wild-type and dckA−/D−cells.
DockD Is Part of a Positive Feedback Loop That Regulates Chemotaxis
Upon uniform stimulation, DockD rapidly translocates to the cortex and then persists at the sites where cell protrusions are formed. Blocking F-actin polymerization does not prevent the initial chemoattractant-mediated translocation of DockD but results in uniform delocalization of the protein from the plasma membrane. Thus, the persistence of DockD localization relies on F-actin polymerization. Interestingly, we observed that translocation of DockD is significantly reduced in mutants that disrupt the genes encoding class I PI3Ks and is blocked by LY294002, suggesting that DockD localization is dependent on PI3K activity. This observation is in agreement with previous findings that some mammalian Dock180 family members localize to the leading edge through the binding of the CZH1 domain to PtdIns(3,4,5)P3 (Kobayashi et al., 2001; Côté et al., 2005; Kunisaki et al., 2006). Hence, the regulation of Dock180 family members by the PI3K pathway is evolutionarily conserved and is likely to represent one of the mechanisms by which PtdIns(3,4,5)P3 levels are linked to localized F-actin polymerization.
We previously showed that F-actin and PI3K function in a positive feedback loop to amplify leading edge signaling and to stabilize the newly formed pseudopod in response to chemoattractant stimulation (Sasaki et al., 2004; Sasaki and Firtel, 2006). PI3K is present at low levels at the cell cortex in unstimulated cells and additional PI3K is rapidly recruited to the cortex via an F-actin–dependent pathway (Sasaki et al., 2004). As chemoattractant-mediated PI3K translocation to the plasma membrane is dependent on F-actin polymerization, our data suggest that DockD might be part of the arm of the loop that leads to the activation of Rac through PtdIns(3,4,5)P3. In turn, Rac activation is thought to stimulate F-actin polymerization that increases the translocation of PI3K to the plasma membrane. Therefore, the persistence of DockD at the plasma membrane upon chemoattractant stimulation is likely to result from PI3K activity at the site of the membrane where the enzyme localizes through F-actin.
DockD Forms a Complex with Dictyostelium ELMO1 and Rac GTPases
Dock180 was shown previously to interact with several proteins (Hasegawa et al., 1996; Grimsley et al., 2004; Hiramoto et al., 2006; Geisbrecht et al., 2008). In particular, the binding of Dock180 to the scaffold protein ELMO appears to be critical for the function of Dock180, as it stimulates its catalytic activity by increasing the affinity of the complex toward Rac (Brugnera et al., 2002; Grimsley et al., 2004; Lu et al., 2006). To provide insight into the molecular context in which Dictyostelium Dock180-related proteins function, we isolated the DockD complex and uncovered DdELMO1, a Dictyostelium ELMO protein. Among the five ELMO domain-containing proteins that are encoded by the Dictyostelium genome, DdELMO1 is the only one that contains the proline-rich region at the C-terminus that mediates Dock180-ELMO interaction (Supplemental Figure S2; Lu et al., 2004). According to the steric-inhibition model proposed by Lu et al., at the basal state, the N-terminal SH3 domain of Dock180 binds to the distant CZH2 domain and negatively regulates the function of the protein as it prevents the interaction with Rac. ELMO is thought to contribute to the GEF activity of the Dock180/ELMO complex by facilitating Rac access to the CZH2 domain through binding to the SH3 domain of Dock180, therefore disrupting the SH3-CZH2 domain interaction. Interestingly, DdELMO1 was found to be associated with the DockD complex in both vegetative and aggregation-competent Dictyostelium cells either before or after chemoattractant stimulation, suggesting that the DockD/DdELMO1 interaction is stable and that additional regulatory mechanisms might be required to modulate Dock180 activity, possibly through its recruitment to the plasma membrane.
Dock180 interacts with mammalian Rac1 through the CZH2 domain and mediates its activation (Kiyokawa et al., 1998; Nolan et al., 1998; Fukui et al., 2001; Meller et al., 2005; Côté et al., 2006; Watabe-Uchida et al., 2006). In Dictyostelium, a few members of the Rho family can be loosely grouped into the Rac subfamily, and Rac1A/B/C are the closest homologues to the mammalian Rac (they show >85% identity to human Rac1; Rivero et al., 2001; Rivero and Somesh, 2002; Vlahou and Rivero, 2006). As previously reported in other systems, Dictyostelium Racs regulate the basal levels of F-actin assembly, therefore playing a crucial role in an array of cellular activities that depend on the remodeling of the actin cytoskeleton (Ridley, 2006; Ladwein and Rottner, 2008). In particular, Rac1 homologues elicit the dynamic cytoskeletal reorganization that initiates pseudopod extension in the direction of the chemoattractant source (Chung et al., 2000; Dumontier et al., 2000; Palmieri et al., 2000; Han et al., 2006).
We have reported that Rac1A coimmunoprecipitates with DockD. Given the high similarity among the members of this group and the fact that they exert the same effect on actin polymerization, we speculate that Rac1B and Rac1C might be a substrate of DockD GEF activity. In addition, we found RacC in the same complex, suggesting that DockD might control the actin cytoskeleton through the activation of multiple Racs. Interestingly, Han et al. (2006) showed that RacC plays a role in PI3K activation and cell motility, making it tempting to hypothesize a possible feedback loop between DockD and PI3K activity as part of the mechanism controlling cell motility. In conclusion, these findings corroborate the importance of this known interaction and provide a strong indication that the molecular mechanism underlying Dock180-related protein function is conserved between Dictyostelium and other organisms.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Firtel laboratory for their assistance. This work was supported by US Public Health Service Grant GM037830 to R.A.F. and by National Science Foundation Grant IBN 0619411 to S.B.
Abbreviations used:
- GEF
guanine nucleotide exchange factor
- DHR
Dock-homology region
- CZH
CDM(Ced-5 Dock180 Myoblast city)-zizimin homology
- LatB
latrunculin B.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-09-0899) on November 26, 2008.
REFERENCES
- Bianco A., Poukkula M., Cliffe A., Mathieu J., Luque C. M., Fulga T. A., Rorth P. Two distinct modes of guidance signalling during collective migration of border cells. Nature. 2007;448:362–365. doi: 10.1038/nature05965. [DOI] [PubMed] [Google Scholar]
- Brugnera E., Haney L., Grimsley C., Lu M., Walk S. F., Tosello-Trampont A. C., Macara I. G., Madhani H., Fink G. R., Ravichandran K. S. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 2002;4:574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- Charest P. G., Firtel R. A. Big roles for small GTPases in the control of directed cell movement. Biochem. J. 2007;401:377–390. doi: 10.1042/BJ20061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. Y., Lee S., Briscoe C., Ellsworth C., Firtel R. A. Role of Rac in controlling the actin cytoskeleton and chemotaxis in motile cells. Proc. Natl. Acad. Sci. USA. 2000;97:5225–5230. doi: 10.1073/pnas.97.10.5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J., Vuori K., William E., Balch C.J.D., Alan H. Methods in Enzymology. Vol. 406. New York: Academic Press; 2006. In vitro guanine nucleotide exchange activity of DHR:]2/DOCKER/CZH2 domains; pp. 41–57. [DOI] [PubMed] [Google Scholar]
- Côté J.-F., Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 2007;17:383–393. doi: 10.1016/j.tcb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J. F., Motoyama A. B., Bush J. A., Vuori K. A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat. Cell Biol. 2005;7:797–807. doi: 10.1038/ncb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J. F., Vuori K. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 2002;115:4901–4913. doi: 10.1242/jcs.00219. [DOI] [PubMed] [Google Scholar]
- Côté J. F., Vuori K. In vitro guanine nucleotide exchange activity of DHR-2/DOCKER/CZH2 domains. Methods Enzymol. 2006;406:41–57. doi: 10.1016/S0076-6879(06)06004-6. [DOI] [PubMed] [Google Scholar]
- Diez S., Gerisch G., Anderson K., Muller-Taubenberger A., Bretschneider T. Subsecond reorganization of the actin network in cell motility and chemotaxis. Proc. Natl. Acad. Sci. USA. 2005;102:7601–7606. doi: 10.1073/pnas.0408546102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disanza A., Steffen A., Hertzog M., Frittoli E., Rottner K., Scita G. Actin polymerization machinery: the finish line of signaling networks, the starting point of cellular movement. Cell Mol. Life Sci. 2005;62:955–970. doi: 10.1007/s00018-004-4472-6. [DOI] [PubMed] [Google Scholar]
- Dumontier M., Hocht P., Mintert U., Faix J. Rac1 GTPases control filopodia formation, cell motility, endocytosis, cytokinesis and development in Dictyostelium. J. Cell Sci. 2000;113:2253–2265. doi: 10.1242/jcs.113.12.2253. [DOI] [PubMed] [Google Scholar]
- Fukui Y., Hashimoto O., Sanui T., Oono T., Koga H., Abe M., Inayoshi A., Noda M., Oike M., Shirai T., Sasazuki T. Haematopoietic cell-specific CDM family protein DOCK2 is essential for lymphocyte migration. Nature. 2001;412:826–831. doi: 10.1038/35090591. [DOI] [PubMed] [Google Scholar]
- Funamoto S., Meili R., Lee S., Parry L., Firtel R. A. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- Geisbrecht E. R., Haralalka S., Swanson S. K., Florens L., Washburn M. P., Abmayr S. M. Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Dev. Biol. 2008;314:137–149. doi: 10.1016/j.ydbio.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsley C. M., Kinchen J. M., Tosello-Trampont A. C., Brugnera E., Haney L. B., Lu M., Chen Q., Klingele D., Hengartner M. O., Ravichandran K. S. Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. J. Biol. Chem. 2004;279:6087–6097. doi: 10.1074/jbc.M307087200. [DOI] [PubMed] [Google Scholar]
- Han J. W., Leeper L., Rivero F., Chung C. Y. Role of RacC for the regulation of WASP and PI3 kinase during chemotaxis of Dictyostelium. J. Biol. Chem. 2006;281:35224–35234. doi: 10.1074/jbc.M605997200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H., Kiyokawa E., Tanaka S., Nagashima K., Gotoh N., Shibuya M., Kurata T., Matsuda M. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Mol. Cell Biol. 1996;16:1770–1776. doi: 10.1128/mcb.16.4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramoto K., Negishi M., Katoh H. Dock4 is regulated by RhoG and promotes Rac-dependent cell migration. Exp. Cell Res. 2006;312:4205–4216. doi: 10.1016/j.yexcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Iijima M., Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- Jaffe A. B., Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Janetopoulos C., Firtel R. A. Directional sensing during chemotaxis. FEBS Lett. 2008;582:2075–2085. doi: 10.1016/j.febslet.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarzynka M. J., Hu B., Hui K. M., Bar-Joseph I., Gu W., Hirose T., Haney L. B., Ravichandran K. S., Nishikawa R., Cheng S. Y. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007;67:7203–7211. doi: 10.1158/0008-5472.CAN-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel A. R., Faix J. Generation of multiple knockout mutants using the Cre-loxP system. Methods Mol. Biol. 2006;346:187–199. doi: 10.1385/1-59745-144-4:187. [DOI] [PubMed] [Google Scholar]
- Kiyokawa E., Hashimoto Y., Kobayashi S., Sugimura H., Kurata T., Matsuda M. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev. 1998;12:3331–3336. doi: 10.1101/gad.12.21.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Shirai T., Kiyokawa E., Mochizuki N., Matsuda M., Fukui Y. Membrane recruitment of DOCK180 by binding to PtdIns(3,4,5)P3. Biochem. J. 2001;354:73–78. doi: 10.1042/0264-6021:3540073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolsch V., Charest P. G., Firtel R. A. The regulation of cell motility and chemotaxis by phospholipid signaling. J. Cell Sci. 2008;121:551–559. doi: 10.1242/jcs.023333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisaki Y., Nishikimi A., Tanaka Y., Takii R., Noda M., Inayoshi A., Watanabe K., Sanematsu F., Sasazuki T., Sasaki T., Fukui Y. DOCK2 is a Rac activator that regulates motility and polarity during neutrophil chemotaxis. J. Cell Biol. 2006;174:647–652. doi: 10.1083/jcb.200602142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladwein M., Rottner K. On the Rho'd: the regulation of membrane protrusions by Rho-GTPases. FEBS Lett. 2008;582:2066–2074. doi: 10.1016/j.febslet.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Lu M., et al. PH domain of ELMO functions in trans to regulate Rac activation via Dock180. Nat. Struct. Mol. Biol. 2004;11:756–762. doi: 10.1038/nsmb800. [DOI] [PubMed] [Google Scholar]
- Lu M., Ravichandran K. S., William Balch E.C.J.D., Alan H. Methods in Enzymology. Vol. 406. New York: Academic Press; 2006. Dock180-ELMO cooperation in Rac activation; pp. 388–402. [DOI] [PubMed] [Google Scholar]
- Mayer B. J. SH3 domains: complexity in moderation. J. Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- Meller N., Merlot S., Guda C. CZH proteins: a new family of Rho-GEFs. J. Cell Sci. 2005;118:4937–4946. doi: 10.1242/jcs.02671. [DOI] [PubMed] [Google Scholar]
- Mendoza M. C., Firtel R. A. Assaying chemotaxis of Dictyostelium cells. Methods Mol. Biol. 2006;346:393–405. doi: 10.1385/1-59745-144-4:393. [DOI] [PubMed] [Google Scholar]
- Moore C. A., Parkin C. A., Bidet Y., Ingham P. W. A role for the myoblast city homologues Dock1 and Dock5 and the adaptor proteins Crk and Crk-like in zebrafish myoblast fusion. Development. 2007;134:3145–3153. doi: 10.1242/dev.001214. [DOI] [PubMed] [Google Scholar]
- Nolan K. M., Barrett K., Lu Y., Hu K. Q., Vincent S., Settleman J. Myoblast city, the Drosophila homolog of DOCK180/CED-5, is required in a Rac signaling pathway utilized for multiple developmental processes. Genes Dev. 1998;12:3337–3342. doi: 10.1101/gad.12.21.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri S. J., Nebl T., Pope R. K., Seastone D. J., Lee E., Hinchcliffe E. H., Sluder G., Knecht D., Cardelli J., Luna E. J. Mutant Rac1B expression in Dictyostelium: effects on morphology, growth, endocytosis, development, and the actin cytoskeleton. Cell Motil. Cytoskelet. 2000;46:285–304. doi: 10.1002/1097-0169(200008)46:4<285::AID-CM6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Pang K. M., Lee E., Knecht D. A. Use of a fusion protein between GFP and an actin-binding domain to visualize transient filamentous-actin structures. Curr. Biol. 1998;8:405–408. doi: 10.1016/s0960-9822(98)70159-9. [DOI] [PubMed] [Google Scholar]
- Papakonstanti E. A., Stournaras C. Cell responses regulated by early reorganization of actin cytoskeleton. FEBS Lett. 2008;582:2120–2127. doi: 10.1016/j.febslet.2008.02.064. [DOI] [PubMed] [Google Scholar]
- Park K. C., Rivero F., Meili R., Lee S., Apone F., Firtel R. A. Rac regulation of chemotaxis and morphogenesis in Dictyostelium. EMBO J. 2004;23:4177–4189. doi: 10.1038/sj.emboj.7600368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafelski S. M., Theriot J. A. Crawling toward a unified model of cell mobility: spatial and temporal regulation of actin dynamics. Annu. Rev. Biochem. 2004;73:209–239. doi: 10.1146/annurev.biochem.73.011303.073844. [DOI] [PubMed] [Google Scholar]
- Ridley A. J. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Rivero F., Dislich H., Glockner G., Noegel A. A. The Dictyostelium discoideum family of Rho-related proteins. Nucleic Acids Res. 2001;29:1068–1079. doi: 10.1093/nar/29.5.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero F., Somesh B. P. Signal transduction pathways regulated by Rho GTPases in Dictyostelium. J. Muscle Res. Cell Motil. 2002;23:737–749. doi: 10.1023/a:1024423611223. [DOI] [PubMed] [Google Scholar]
- Sasaki A. T., Chun C., Takeda K., Firtel R. A. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J. Cell Biol. 2004;167:505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A. T., Firtel R. A. Regulation of chemotaxis by the orchestrated activation of Ras, PI3K, and TOR. Eur. J. Cell Biol. 2006;85:873–895. doi: 10.1016/j.ejcb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Soll D. R., Voss E. Two- and three-dimensional computer systems for analyzing how animal cells crawl. In: Soll D.R., Wessels D., editors. Motion Analysis of Living Cells. New York: Wiley-Liss; 1998. pp. 25–52. [Google Scholar]
- Takeda K., Sasaki A. T., Ha H., Hyun-A Seung H.-A., Firtel R. A. Role of PI3 kinases in chemotaxis in Dictyostelium. J. Biol. Chem. 2007;282:11874–11884. doi: 10.1074/jbc.M610984200. [DOI] [PubMed] [Google Scholar]
- Upadhyaya A., van Oudenaarden A. Actin polymerization: forcing flat faces forward. Curr. Biol. 2004;14:R467–R469. doi: 10.1016/j.cub.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Van Haastert P. J., Veltman D. M. Chemotaxis: navigating by multiple signaling pathways. Sci. STKE 2007. 2007:pe40. doi: 10.1126/stke.3962007pe40. [DOI] [PubMed] [Google Scholar]
- Vlahou G., Rivero F. Rho GTPase signaling in Dictyostelium discoideum: Insights from the genome. Eur. J. Cell Biol. 2006;85:947–959. doi: 10.1016/j.ejcb.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Watabe-Uchida M., John K. A., Janas J. A., Newey S. E., Van Aelst L. The Rac activator DOCK7 regulates neuronal polarity through local phosphorylation of Stathmin/Op18. Neuron. 2006;51:727–739. doi: 10.1016/j.neuron.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Wessels D., Soll D. R., Knecht D., Loomis W. F., De Lozanne A., Spudich J. Cell motility and chemotaxis in Dictyostelium amebae lacking myosin heavy chain. Dev. Biol. 1988;128:164–177. doi: 10.1016/0012-1606(88)90279-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.