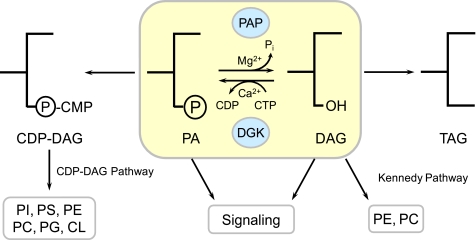
PA2 phosphatase (PAP3; 3-sn-phosphatidate phosphohydrolase, EC 3.1.3.4) catalyzes the Mg2+-dependent dephosphorylation of PA, yielding DAG and Pi (Fig. 1) (1–3). In de novo lipid synthesis, the DAG generated from the PAP reaction is utilized for the synthesis of the phospholipids PE and PC via the Kennedy pathway and for the synthesis of TAG (Fig. 1) (3). The enzyme substrate PA is also used for phospholipid synthesis via CDP-DAG (Fig. 1) (1–3). In lipid signaling, PAP generates a pool of DAG used for protein kinase C activation, and by the nature of its reaction, it attenuates the signaling functions of PA (Fig. 1) (4–8). Thus, the regulation of PAP activity may govern the pathways by which phospholipids are synthesized and control the cellular contents of important signaling lipids. Recent genetic and biochemical studies in yeast and mammalian cells have revealed key roles that PAP plays in lipid metabolism and cell physiology.
FIGURE 1.
Roles of PAP in lipid synthesis and signaling in yeast. The reaction catalyzed by PAH1-encoded PAP (highlighted in yellow) is shown. Regulation of PAP activity plays a role in governing whether cells synthesize membrane phospholipids via CDP-DAG or DAG. The reaction product DAG is also used for the synthesis of the storage lipid TAG. The substrate PA signals growth of the nuclear/ER membrane and the transcriptional regulation of UASINO-containing phospholipid synthesis genes. The signaling roles of PAP are counteracted by the activity of DGK1-encoded DAG kinase (DGK). In mammalian cells, phosphatidylserine (PS), PE, and PC are derived from DAG, and DAG kinase uses ATP instead of CTP as the phosphate donor in its reaction (54, 55). PG, phosphatidylglycerol.
PAP in Yeast and Mammalian Cells
The PAP reaction was first characterized in animal tissues in 1957 by Kennedy and co-workers (9). Subsequent studies during the last half of the 20th century implicated PAP as an important enzyme in lipid metabolism (1, 2, 10). However, studies to establish the roles of PAP in lipid metabolism and cell physiology had been hampered by the lack of genetic and molecular information on the enzyme. In fact, a gene encoding PAP was discovered only recently in 2006 from the yeast Saccharomyces cerevisiae (11). The yeast gene (PAH1; known previously as SMP2) was identified through a reverse genetic approach from amino acid sequence information derived from a purified preparation of the enzyme (11). The heterologous expression of PAH1 in Escherichia coli has confirmed that its protein product is a PAP enzyme with enzymological properties essentially identical to those of enzymes purified from yeast (11–14). The yeast PAH1-encoded PAP is a 95-kDa monomeric protein that migrates upon SDS-PAGE as a 124-kDa protein (11). Smaller molecular mass forms (e.g. 91 kDa) of PAP purified from yeast (12) appear to be proteolytic products of the enzyme (11). PAH1-encoded PAP is associated with the cytosolic and membrane fractions of the cell, and its association with membranes is peripheral in nature (11, 12). The enzyme is specific for PA and catalyzes its dephosphorylation reaction based on a DXDX(T/V) motif within a HAD-like domain (Fig. 2) (11, 12, 15).
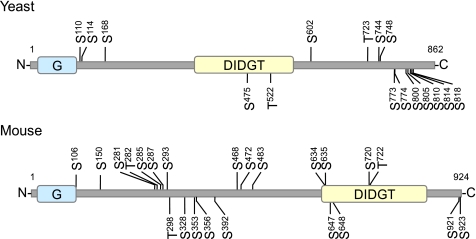
FIGURE 2.
Domain structures and phosphorylation sites of the yeast and mouse PAP enzymes. The diagram shows the positions of the NLIP (blue) and HAD-like (yellow) domains in yeast PAP and mouse lipin 1β. These domains are conserved in lipins 1α, 2, and 3 (not shown) (16). The conserved glycine residue within the NLIP domain and the conserved aspartate residues in the catalytic sequence DIDGT within the HAD-like domain are essential for PAP activity (15, 30, 53). The positions of the phosphorylation sites identified in the yeast (21) and mouse (53) enzymes are indicated. The sites shown on the upper portion of the yeast PAP diagram are targets for Cdk1 phosphorylation (21).
The identification of the yeast PAP-encoding gene has led to the revelation that mammalian lipins (encoded by Lpin1, Lpin2, and Lpin3), which have homology to yeast PAH1-encoded PAP at the N terminus (referred to as the NLIP domain) and within its HAD-like domain (Fig. 2) (16), are also PAP enzymes (11, 17). Lpin1 was first identified by Reue and co-workers (16) through positional cloning as the mutated gene in the fatty liver dystrophy (fld) mouse. In addition, Reue and co-workers (16, 18) had previously established that lipin 1 is a major fat-regulating protein in mice. Loss of lipin 1 prevents normal adipose tissue development, leading to lipodystrophy and insulin resistance, whereas its overexpression leads to obesity (16, 18). Thus, the molecular function of lipin 1 as a PAP enzyme provides a mechanistic basis for why the absence or overexpression of lipin 1 in mice has a major effect on fat metabolism (11, 16, 18). The three lipins exhibit distinct but overlapping expression patterns in many tissues (17). That mammalian lipins 1 and 2 complement the phenotypes (see below) exhibited by yeast pah1Δ mutant cells (19) indicates the evolutionarily conserved functions of PAP.
Roles of PAP in Lipid Metabolism
Insights into the roles that PAP plays in lipid metabolism and cell physiology have been gained through studies using mutants defective in the enzyme (11, 15, 20, 21). The yeast pah1Δ mutant contains elevated levels of PA and decreased levels of DAG and TAG (11, 15). In addition, the contents of the major phospholipids PC, PE, and PI, sterol esters, and fatty acids are affected by the pah1Δ mutation (11, 15), demonstrating that PAP plays a role in the regulation of overall lipid synthesis. The effects of the pah1Δ mutation on phospholipid composition are most pronounced in exponential phase, whereas the effects on the contents of TAG and other neutral lipids are most pronounced in stationary phase (11, 15). In fact, the loss of PAP activity in stationary phase cells results in a >90% decrease in TAG content (11). This phenotype is reminiscent of the lipodystrophy phenotype observed in the fld mouse that lacks Lpin1-encoded PAP activity (16, 17, 22).
The pah1Δ mutation causes the aberrant expansion of the nuclear/ER membrane and the derepression of phospholipid synthesis genes that contain a UASINO in the promoters (11, 15, 20). The derepression of phospholipid synthesis genes is a consequence of elevated PA content at the nuclear/ER membrane (23). Along with the Scs2 protein, PA binds Opi1 at the nuclear/ER membrane and prevents its translocation into the nucleus, where it functions to repress transcription of phospholipid synthesis genes (23).
The membrane expansion phenotype correlates with the increased expression of phospholipid synthesis genes (20). However, data indicate that an increase in phospholipid synthesis alone is not sufficient for nuclear/ER membrane expansion (21), suggesting a specific role of PAP activity in the maintenance of nuclear/ER membrane structure. The effect of the pah1Δ mutation on membrane structure appears to be related to an increased level of PA as opposed to a decreased level of DAG. This hypothesis is supported by the recent identification of the DGK1 gene encoding a novel CTP-dependent DAG kinase whose function counteracts the PAH1-encoded PAP activity (Fig. 1) (24, 25). Overexpression of DGK1 causes the accumulation of PA and the expansion of the nuclear/ER membrane as shown in the pah1Δ mutant (24). The introduction of the dgk1Δ mutation into the pah1Δ mutant causes a decrease in PA content without affecting DAG content and restores normal nuclear/ER membrane structure and expression of UASINO-containing phospholipid synthesis genes (24).
The phenotypes associated with the pah1Δ mutation, which also include slow growth, temperature sensitivity, and respiratory deficiency (11, 20, 26), are specifically due to the loss of PAP activity and not to the loss of some other function associated with the PAP protein (15). Indeed, catalytically inactive PAP enzymes with a mutation in a conserved NLIP domain residue (G80R) or conserved DIDGT catalytic motif residues (D398E or D400E) fail to complement phenotypes caused by the pah1Δ mutation (15).
As indicated above, loss of PAP (i.e. lipin 1) activity in the fld mouse causes loss of body fat (16). In addition, mice lacking lipin 1 exhibit peripheral neuropathy that is characterized by myelin degradation, Schwann cell dedifferentiation and proliferation, and a reduction in nerve conduction velocity (22, 27, 28). These effects are mediated through the MEK (mitogen-activated protein kinase/extracellular signal-regulated kinase kinase)-ERK (extracellular signal-regulated kinase) pathway that is activated by elevated levels of PA due to the loss of PAP activity (22). Moreover, the level and compartmentalization of lipin 1 exert a major impact on the assembly and secretion of hepatic very low density lipoprotein (29). Lipin 1 appears to have dual cellular roles, serving as a PAP enzyme required for the synthesis of lipids and as a transcriptional co-activator in the regulation of lipid metabolism gene expression (30–32).
Regulation of PAP Expression
Expression of the mammalian Lpin-encoded PAP enzymes is regulated at the level of transcription (19, 31, 33–37). For example, Lpin1 expression is induced during adipocyte differentiation (19, 31) and in cells that undergo ER proliferation (38, 39). The induction of Lpin1 during adipogenesis is stimulated by glucocorticoids, and this regulation is mediated by a glucocorticoid-response element within the promoter (36, 37). Interestingly, Lpin2 expression is repressed during adipogenesis, indicating that lipins 1 and 2 have distinct and non-redundant functions in adipocytes (19). In addition, alternative splicing of Lpin1 mRNA gives rise to two lipin 1 isoforms (α and β) that are postulated to play distinct functions in adipogenesis (31). Compared with mammalian PAP, relatively little is known about the transcriptional regulation of the yeast enzyme. Reporter gene analysis indicates that PAH1 expression is elevated in the stationary phase of growth,4 which shows a correlation with increased levels of TAG in stationary phase and with the increased Opi1-mediated repression of phospholipid synthesis genes (40).
Biochemical Regulation of PAP
An understanding of the enzymological properties and biochemical regulation of PAP activity has been facilitated by the purification of the PAH1-encoded enzyme from S. cerevisiae (11, 12). Maximum activity is dependent on Mg2+ and the nonionic detergent Triton X-100 at pH 7.0–7.5 (11, 12). The function of Triton X-100 is to form a mixed micelle with the lipid substrate PA, providing a membrane mimic for catalysis (12, 13, 41). The Triton X-100 micelle serves as a catalytically inert matrix in which PA is dispersed, preventing a high local concentration of substrate at the active site (13). PAP activity is dependent on the molar (e.g. number of micelles containing substrate) and surface (e.g. number of substrate molecules on the micelle surface) concentrations of PA (13). This kinetic property is indicative of surface dilution kinetics where PAP binds to the mixed micelle surface before binding to its substrate and catalysis occurs (41). PAP physically associates with Triton X-100 micelles in the absence of PA; however, the enzyme more tightly associates with micelles when its substrate is present (13). This property is relevant in vivo because the enzyme is associated with both the cytosolic and membrane fractions of the cell (11). PAP presumably associates with the PA at the nuclear/ER membrane used for the synthesis of phospholipids and TAG. The dependence of PAP activity on PA surface concentration is cooperative (11, 14), indicating a regulatory role for the enzyme.
The expression of the mammalian Lpin genes in HEK293 cells has revealed that lipins 1 (α- and β-forms), 2, and 3 are PAP enzymes that are specific for PA and require Mg2+ for activity (17). Like the yeast enzyme, the mammalian PAP activities exhibit cooperative kinetics with respect to the surface concentration of PA (17). Defined studies on the kinetic and biochemical properties of the mammalian PAP enzymes await their purification.
Regulation by Lipids—The yeast PAH1-encoded PAP activity is enhanced by the phospholipids CDP-DAG, PI, and CL (42), whereas the activity is inhibited by the sphingoid bases sphingosine, phytosphingosine, and sphinganine (14). These sphingoid bases are parabolic competitive inhibitors of PAP activity, indicating that more than one inhibitor molecule contributes to the exclusion of PA from the enzyme. CDP-DG, PI, and CL are mixed competitive activators of PAP activity (42). The major effect of the activators is to decrease the Km for PA. Sphinganine antagonizes the activation of PAP activity by CL and PI, whereas it causes an increase in the cooperativity of CL activation (42). Conversely, sphinganine has little effect on the cooperativity of PI activation, but causes an increase in the activation constant for PI (42). Based on the activation/inhibitor constants and cellular concentrations for these lipid effector molecules, their regulatory roles in PAP activity should be physiologically relevant (14, 42). As indicated in Fig. 1, PA is partitioned between CDP-DAG and DAG, and this CDP-DAG is used for the synthesis of PI and CL. Activation of PAP activity by these phospholipids would divert PA toward DAG for PE and PC synthesis via the Kennedy pathway or for the synthesis of TAG. At the same time, activation of PAP lowers PA content and causes the repression of UASINO-containing genes that principally encode enzymes for phospholipid synthesis via CDP-DAG (23). Sphingoid bases are both precursors and turnover products of sphingolipid metabolism in yeast (43). A relationship between the DAG-derived synthesis of PE and sphingolipid synthesis exists through sphingoid base metabolism (40). In addition, DAG is a product of the inositol phosphoceramide synthase enzyme that is responsible for the synthesis of a major yeast sphingolipid inositol phosphoceramide (43). The inhibition of PAP activity by sphingoid bases would cause an increase in PA content, which should stimulate phospholipid synthesis by mechanisms discussed above and thus provide a mechanism for the coordinate regulation of the synthesis of phospholipids and sphingolipids.
Regulation by Nucleotides—The nucleotides ATP and CTP, which are precursors for the synthesis of phospholipids (40), are inhibitors of PAH1-encoded PAP activity (44). Kinetic analyses have revealed that the mechanism of inhibition by ATP and CTP is complex, affecting both the Vmax and Km for PA. In addition, kinetic analysis has shown that ATP and CTP are competitive inhibitors with respect to Mg2+, suggesting that they inhibit PAP activity by a mechanism involving the chelation of the cofactor (44). It is known that cellular ATP levels correlate with the proportional synthesis of phospholipids and TAG (44). High levels of ATP favor elevated PA content and phospholipid synthesis, whereas low levels of ATP favor reduced PA content and an increase in the synthesis of TAG (44). Elevated CTP content favors an increase in PA content and derepression of UASINO-containing phospholipid synthesis genes (45). The regulations of PAP activity by ATP and by CTP are consistent with the regulation of lipid synthesis observed in cells that have fluctuations in ATP and CTP (44, 45).
Regulation by Phosphorylation—PAH1-encoded PAP has been identified as a phosphoprotein by a large-scale analysis of the yeast proteome (46, 47). Proteome-wide in vitro phosphorylation analyses have shown that PAP is a target for multiple protein kinases, including cyclin-dependent kinase Cdk1 (48), Pho85 (49, 50), and Dbf2-Mob1 (51). In synchronized cells, PAP is phosphorylated by Cdk1 in a cell cycle-dependent manner (20). Mass spectrometry analysis of PAP has identified multiple (i.e. 16) sites of phosphorylation (Fig. 2) (21). Of these, seven sites located within the minimal Ser/Thr-Pro motif are targets for Cdk1 phosphorylation (21). Several lines of evidence indicate that the Cdk1 phosphorylation has an inhibitory effect on enzyme activity. First, a purified phosphorylation-deficient (Ser/Thr → Ala) septuple mutant enzyme exhibits elevated PAP activity (21). Second, the overexpression of the mutant enzyme causes inositol auxotrophy by exacerbating the Opi1-mediated repression of the UASINO-containing INO1 gene (21). Third, mutant cells lacking the Nem1-Spo7 phosphatase complex that is responsible for dephosphorylation of PAP exhibit the phenotypes characteristic of the pah1Δ mutant (e.g. derepression of phospholipid synthesis genes and aberrant nuclear/ER membrane expansion) (20). In addition to enzyme activity, the localization of PAP is affected by the Cdk1 phosphorylation. Subcellular fractionation analysis shows that the phosphorylation-deficient mutant enzyme is enriched in the membrane fraction.5
Mammalian lipins 1 and 2 are also phosphoproteins (19, 52, 53), indicating that the phosphorylation of PAP is evolutionarily conserved. Before lipin 1 was known to have PAP activity (11), it had been identified in rat adipocytes as a major protein phosphorylated in response to insulin (52). Phosphorylation of lipin 1, which is dependent on the mTOR signaling pathway (52), occurs at multiple (i.e. 19) sites (Fig. 2) (53). The phosphorylation of lipin 1 in 3T3-L1 adipocytes has no effect on its PAP activity, but affects its subcellular localization between the cytosolic and membrane fractions of the cell (53). Phosphorylation favors a cytosolic association (53), a location that is spatially distinct from its membrane-associated substrate PA. In HeLa cells, phosphorylation regulates the PAP activity of lipins 1 and 2. Like yeast PAP, lipin 1 and 2 proteins are phosphorylated during the mitotic phase of cell growth at sites containing a Cdk1 phosphorylation motif, and their PAP activities are inhibited by the mitotic phosphorylation (19). As shown for lipin 1 in adipocytes, the phosphorylated forms of lipins 1 and 2 in HeLa cells are enriched in the cytosolic fraction, whereas the dephosphorylated forms are enriched in the membrane fraction (19). Overall, these findings indicate that phosphorylation has negative regulatory effects on PAP by inhibiting activity and preventing substrate accessibility.
Perspectives
The identification and characterization of the yeast PAH1 gene and its protein product have established the role of PAP in the synthesis of phospholipids and TAG (11, 12). In addition, PAP activity in yeast plays a major role in controlling cellular PA content, which in turn signals the transcriptional regulation of phospholipid synthesis genes and growth of the nuclear/ER membrane (15, 20, 21, 23). The importance of understanding PAP regulation is underscored by the role this enzyme plays in obesity, lipodystrophy, and peripheral neuropathy in animals. As indicated in this review, studies with yeast PAP have led to an understanding of the basic biochemical properties of the enzyme and how its activity is modulated by membrane- and cytosol-associated effector molecules. Clearly, a key point of PAP regulation is through phosphorylation, which is likely to be mediated by multiple protein kinases known to control various aspects of cell physiology.
The functions of the NLIP and HAD-like domains in yeast and mammalian PAP enzymes are not fully understood. A glycine-to-arginine mutation of the conserved glycine within the NLIP domain has shown that this residue is required for PAP activity (15, 53). In the primary structure of PAP, the NLIP domain is distant from the HAD-like domain that contains the essential catalytic sequence DIDGT (Fig. 2) (15, 30). The conserved glycine residue in the NLIP domain may participate in catalysis if it were in close proximity to the catalytic sequence of the folded protein. Alternatively, the mutation to arginine may simply change protein structure, thereby affecting enzyme activity. These questions can be addressed when the structure of PAP becomes available. In this regard, structural information of the PAP enzymes from yeast and mammalian cells will also permit defined studies into the mechanism of enzyme action and membrane interaction and perhaps lead to the identification of specific enzyme inhibitors and activators that may have pharmaceutical uses in controlling lipid metabolism.
Supplementary Material
Acknowledgments
We thank Hyeon-Son Choi, Jeanelle Morgan, Florencia Pascual, Aníbal Soto, and Wen-Min Su for helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM-28140 and GM-50679 from the United States Public Health Service. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Footnotes
The abbreviations used are: PA, phosphatidic acid; PAP, PA phosphatase; DAG, diacylglycerol; PE, phosphatidylethanolamine; PC, phosphatidylcholine; TAG, triacylglycerol; HAD, haloacid dehalogenase; PI, phosphatidylinositol; ER, endoplasmic reticulum; UASINO, inositol-responsive upstream activating sequence; CL, cardiolipin.
The acronym PAP (also known as PAP1) is used to designate PA phosphatase, the enzyme that is specific for PA and that requires Mg2+ for activity (3). PAP is distinct from lipid phosphate phosphatase (previously known as PAP2), the enzyme that dephosphorylates PA and other lipid phosphates (e.g., lyso-PA, DAG pyrophosphate, sphingoid base phosphates, and isoprenoid phosphates) by a catalytic mechanism that does not require Mg2+ (3). The molecular and biochemical differences between PAP and lipid phosphate phosphatase enzymes are discussed elsewhere (3).
F. Pascual and G. M. Carman, unpublished data.
J. M. Morgan and G. M. Carman, unpublished data.
References
- 1.Brindley, D. N. (1984) Prog. Lipid Res. 23 115–133 [DOI] [PubMed] [Google Scholar]
- 2.Nanjundan, M., and Possmayer, F. (2003) Am. J. Physiol. 284 L1–L23 [DOI] [PubMed] [Google Scholar]
- 3.Carman, G. M., and Han, G.-S. (2006) Trends Biochem. Sci. 31 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Exton, J. H. (1990) J. Biol. Chem. 265 1–4 [PubMed] [Google Scholar]
- 5.Exton, J. H. (1994) Biochim. Biophys. Acta 1212 26–42 [DOI] [PubMed] [Google Scholar]
- 6.Testerink, C., and Munnik, T. (2005) Trends Plant Sci. 10 368–375 [DOI] [PubMed] [Google Scholar]
- 7.Waggoner, D. W., Xu, J., Singh, I., Jasinska, R., Zhang, Q. X., and Brindley, D. N. (1999) Biochim. Biophys. Acta 1439 299–316 [DOI] [PubMed] [Google Scholar]
- 8.Sciorra, V. A., and Morris, A. J. (2002) Biochim. Biophys. Acta 1582 45–51 [DOI] [PubMed] [Google Scholar]
- 9.Smith, S. W., Weiss, S. B., and Kennedy, E. P. (1957) J. Biol. Chem. 228 915–922 [PubMed] [Google Scholar]
- 10.Carman, G. M. (1997) Biochim. Biophys. Acta 1348 45–55 [DOI] [PubMed] [Google Scholar]
- 11.Han, G.-S., Wu, W.-I., and Carman, G. M. (2006) J. Biol. Chem. 281 9210–9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin, Y.-P., and Carman, G. M. (1989) J. Biol. Chem. 264 8641–8645 [PubMed] [Google Scholar]
- 13.Lin, Y.-P., and Carman, G. M. (1990) J. Biol. Chem. 265 166–170 [PubMed] [Google Scholar]
- 14.Wu, W.-I., Lin, Y.-P., Wang, E., Merrill, A. H., Jr., and Carman, G. M. (1993) J. Biol. Chem. 268 13830–13837 [PubMed] [Google Scholar]
- 15.Han, G.-S., Siniossoglou, S., and Carman, G. M. (2007) J. Biol. Chem. 282 37026–37035 [DOI] [PubMed] [Google Scholar]
- 16.Peterfy, M., Phan, J., Xu, P., and Reue, K. (2001) Nat. Genet. 27 121–124 [DOI] [PubMed] [Google Scholar]
- 17.Donkor, J., Sariahmetoglu, M., Dewald, J., Brindley, D. N., and Reue, K. (2007) J. Biol. Chem. 282 3450–3457 [DOI] [PubMed] [Google Scholar]
- 18.Phan, J., and Reue, K. (2005) Cell Metab. 1 73–83 [DOI] [PubMed] [Google Scholar]
- 19.Grimsey, N., Han, G.-S., O'Hara, L., Rochford, J. J., Carman, G. M., and Siniossoglou, S. (2008) J. Biol. Chem. 283 29166–29174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santos-Rosa, H., Leung, J., Grimsey, N., Peak-Chew, S., and Siniossoglou, S. (2005) EMBO J. 24 1931–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Hara, L., Han, G.-S., Peak-Chew, S., Grimsey, N., Carman, G. M., and Siniossoglou, S. (2006) J. Biol. Chem. 281 34537–34548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadra, K., De Preux Charles, A.-S., Medard, J.-J., Hendriks, W. T., Han, G.-S., Gres, S., Carman, G. M., Saulnier-Blache, J.-S., Verheijen, M. H. G., and Chrast, R. (2008) Genes Dev. 22 1647–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carman, G. M., and Henry, S. A. (2007) J. Biol. Chem. 282 37293–37297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han, G.-S., O'Hara, L., Carman, G. M., and Siniossoglou, S. (2008) J. Biol. Chem. 283 20433–20442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han, G.-S., O'Hara, L., Siniossoglou, S., and Carman, G. M. (2008) J. Biol. Chem. 283 20443–20453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irie, K., Takase, M., Araki, H., and Oshima, Y. (1993) Mol. Gen. Genet. 236 283–288 [DOI] [PubMed] [Google Scholar]
- 27.Langner, C. A., Birkenmeier, E. H., Roth, K. A., Bronson, R. T., and Gordon, J. I. (1991) J. Biol. Chem. 266 11955–11964 [PubMed] [Google Scholar]
- 28.Langner, C. A., Birkenmeier, E. H., Ben-Zeev, O., Schotz, M. C., Sweet, H. O., Davisson, M. T., and Gordon, J. I. (1989) J. Biol. Chem. 264 7994–8003 [PubMed] [Google Scholar]
- 29.Bou, K. M., Sundaram, M., Zhang, H. Y., Links, P. H., Raven, J. F., Manmontri, B., Sariahmetoglu, M., Tran, K., Reue, K., Brindley, D. N., and Yao, Z. (2008) J. Lipid Res., in press [DOI] [PubMed]
- 30.Finck, B. N., Gropler, M. C., Chen, Z., Leone, T. C., Croce, M. A., Harris, T. E., Lawrence, J. C., Jr., and Kelly, D. P. (2006) Cell Metab. 4 199–210 [DOI] [PubMed] [Google Scholar]
- 31.Peterfy, M., Phan, J., and Reue, K. (2005) J. Biol. Chem. 280 32883–32889 [DOI] [PubMed] [Google Scholar]
- 32.Reue, K., and Zhang, P. (2008) FEBS Lett. 582 90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao-Borengasser, A., Rasouli, N., Varma, V., Miles, L. M., Phanavanh, B., Starks, T. N., Phan, J., Spencer, H. J., III, McGehee, R. E., Jr., Reue, K., and Kern, P. A. (2006) Diabetes 55 2811–2818 [DOI] [PubMed] [Google Scholar]
- 34.Gowri, P. M., Sengupta, S., Bertera, S., and Katzenellenbogen, B. S. (2007) Endocrinology 148 3685–3693 [DOI] [PubMed] [Google Scholar]
- 35.Donkor, J., Sparks, L. M., Xie, H., Smith, S. R., and Reue, K. (2007) J. Clin. Endocrinol. Metab. 93 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manmontri, B., Sariahmetoglu, M., Donkor, J., Bou, K. M., Sundaram, M., Yao, Z., Reue, K., Lehner, R., and Brindley, D. N. (2008) J. Lipid Res. 49 1056–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, P., O'Loughlin, L., Brindley, D. N., and Reue, K. (2008) J. Lipid Res. 49 1519–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fagone, P., Sriburi, R., Ward-Chapman, C., Frank, M., Wang, J., Gunter, C., Brewer, J. W., and Jackowski, S. (2007) J. Biol. Chem. 282 7591–7605 [DOI] [PubMed] [Google Scholar]
- 39.Sriburi, R., Bommiasamy, H., Buldak, G. L., Robbins, G. R., Frank, M., Jackowski, S., and Brewer, J. W. (2007) J. Biol. Chem. 282 7024–7034 [DOI] [PubMed] [Google Scholar]
- 40.Carman, G. M., and Henry, S. A. (1999) Prog. Lipid Res. 38 361–399 [DOI] [PubMed] [Google Scholar]
- 41.Carman, G. M., Deems, R. A., and Dennis, E. A. (1995) J. Biol. Chem. 270 18711–18714 [DOI] [PubMed] [Google Scholar]
- 42.Wu, W.-I., and Carman, G. M. (1996) Biochemistry 35 3790–3796 [DOI] [PubMed] [Google Scholar]
- 43.Dickson, R. C., and Lester, R. L. (2002) Biochim. Biophys. Acta 1583 13–25 [DOI] [PubMed] [Google Scholar]
- 44.Wu, W.-I., and Carman, G. M. (1994) J. Biol. Chem. 269 29495–29501 [PubMed] [Google Scholar]
- 45.Ostrander, D. B., O'Brien, D. J., Gorman, J. A., and Carman, G. M. (1998) J. Biol. Chem. 273 18992–19001 [DOI] [PubMed] [Google Scholar]
- 46.Chi, A., Huttenhower, C., Geer, L. Y., Coon, J. J., Syka, J. E., Bai, D. L., Shabanowitz, J., Burke, D. J., Troyanskaya, O. G., and Hunt, D. F. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, X., Gerber, S. A., Rudner, A. D., Beausoleil, S. A., Haas, W., Villen, J., Elias, J. E., and Gygi, S. P. (2007) J. Proteome Res. 6 1190–1197 [DOI] [PubMed] [Google Scholar]
- 48.Ubersax, J. A., Woodbury, E. L., Quang, P. N., Paraz, M., Blethrow, J. D., Shah, K., Shokat, K. M., and Morgan, D. O. (2003) Nature 425 859–864 [DOI] [PubMed] [Google Scholar]
- 49.Ptacek, J., Devgan, G., Michaud, G., Zhu, H., Zhu, X., Fasolo, J., Guo, H., Jona, G., Breitkreutz, A., Sopko, R., McCartney, R. R., Schmidt, M. C., Rachidi, N., Lee, S. J., Mah, A. S., Meng, L., Stark, M. J., Stern, D. F., De Virgilio, C., Tyers, M., Andrews, B., Gerstein, M., Schweitzer, B., Predki, P. F., and Snyder, M. (2005) Nature 438 679–684 [DOI] [PubMed] [Google Scholar]
- 50.Dephoure, N., Howson, R. W., Blethrow, J. D., Shokat, K. M., and O'Shea, E. K. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 17940–17945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mah, A. S., Elia, A. E., Devgan, G., Ptacek, J., Schutkowski, M., Snyder, M., Yaffe, M. B., and Deshaies, R. J. (2005) BMC Biochem. 6 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huffman, T. A., Mothe-Satney, I., and Lawrence, J. C., Jr. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harris, T. E., Huffman, T. A., Chi, A., Shabanowitz, J., Hunt, D. F., Kumar, A., and Lawrence, J. C., Jr. (2007) J. Biol. Chem. 282 277–286 [DOI] [PubMed] [Google Scholar]
- 54.Vance, D. E. (1996) in Biochemistry of Lipids, Lipoproteins and Membranes (Vance, D. E., and Vance, J., eds) pp. 153–181, Elsevier Science Publishers B. V., Amsterdam
- 55.Sakane, F., Imai, S., Kai, M., Yasuda, S., and Kanoh, H. (2007) Biochim. Biophys. Acta 1771 793–806 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.