Abstract
Immunosuppression is required for BK viremia and polyomavirus BK–associated nephropathy (PVAN) in kidney transplants (KTs), but the role of viral determinants is unclear. We examined BKV noncoding control regions (NCCR), which coordinate viral gene expression and replication. In 286 day–matched plasma and urine samples from 129 KT patients with BKV viremia, including 70 with PVAN, the majority of viruses contained archetypal (ww-) NCCRs. However, rearranged (rr-) NCCRs were more frequent in plasma than in urine samples (22 vs. 4%; P < 0.001), and were associated with 20-fold higher plasma BKV loads (2.0 × 104/ml vs. 4.4 × 105/ml; P < 0.001). Emergence of rr-NCCR in plasma correlated with duration and peak BKV load (R2 = 0.64; P < 0.001). This was confirmed in a prospective cohort of 733 plasma samples from 227 patients. For 39 PVAN patients with available biopsies, rr-NCCRs were associated with more extensive viral replication and inflammation. Cloning of 10 rr-NCCRs revealed diverse duplications or deletions in different NCCR subregions, but all were sufficient to increase early gene expression, replication capacity, and cytopathology of recombinant BKV in vitro. Thus, rr-NCCR BKV emergence in plasma is linked to increased replication capacity and disease in KTs.
Polyomavirus BK–associated nephropathy (PVAN) has emerged as the most challenging infectious cause of irreversible kidney transplant (KT) failure (1, 2). PVAN is diagnosed in up to 10% of KT patients around the world, causing premature graft loss in the 6–60 mo after transplant (3, 4). Histologically, progression from a mainly cytopathic pattern (PVAN A) to extensive cytopathic/inflammatory changes of interstitial nephritis (PVAN B) is associated with increasing graft loss from <10 to 50%, exceeding 80% for PVAN C when tubular atrophy and fibrosis predominate (5). Histological studies have demonstrated extensive BKV replication in the urothelial cell layer (6); however, unlike in bone marrow transplant patients, BKV-associated hemorrhagic cystitis is rarely encountered in KT patients, despite high urine viral loads (7). The emergence of PVAN is remarkable in view of the 50 yr of experience in kidney transplantation and the basically unchanged high prevalence of BKV infection in the general population (8–10). The “net state of immunosuppression” seems crucial for PVAN pathogenesis and reflects the use of more potent immunosuppressive drugs synergizing with other factors, such as older age, negative BKV recipient and positive BKV donor status, higher number of HLA mismatches, and prior rejection episodes (7, 11–14). The role of viral determinants is presently unclear. As no antiviral drug of proven efficacy is available (15), current treatment is based on reducing immunosuppression to regain immune control over BKV replication and disease (16, 17). This maneuver bears the risk of rejection and entry into a vicious cycle with eventual graft loss (18).
Outside the KT setting, BKV seems well adapted to the human host. BKV asymptomatically infects 60–90% of the world's population during childhood and subsequently persists in the renourinary tract (9, 19). After kidney transplantation, BKV viruria rates increase from 5 to 40%, with the percentage of urine viral loads increasing from <105 to >107 genome equivalents (geq)/ml and decoy cell shedding (1, 5, 20). Renal allograft function is not affected at first, but one third of patients are at high risk of progressing to BKV viremia and overt PVAN (1, 21). In patients with PVAN undergoing allograft nephrectomy, plasma BKV loads drop to undetectable levels with a half-life of <2 h, indicating that BKV viremia is essentially derived from replication in renal allografts (22). Accordingly, >99% of plasma BKV loads in steady state are replaced every day, reflecting loss of at least 106 renal tubular epithelial cells to release progeny virus (22). After reduced immunosuppression, plasma BKV loads decline over 7–13 wk (22), and antiviral cellular immune responses directed against early and late viral proteins become detectable in the peripheral blood (23, 24). Thus, BKV load in plasma is widely accepted as a marker of onset and resolution of PVAN (1, 17).
Polyomavirus genomes are circular dsDNA of 5 kb consisting of the noncoding control region (NCCR) with the origin of replication and promoter/enhancer functions controlling expression of the early proteins, large and small T-antigen, and the late agnoprotein and the capsid proteins VP1-3 (25). BKV strains excreted in urine of immunocompetent individuals have been reported to be of archetype (ww-) NCCR architecture. Upon propagation of BKV in tissue culture, however, ww-NCCR BKV are readily replaced by BKV variants with genetically rearranged (rr-) NCCR, whereas other areas of the BKV genome remain unchanged (26–28). Because of NCCR hypervariability in tissue culture, identification of archetype ww-NCCR requires direct analysis of BKV genomes from biological samples e.g., by PCR amplification and cloning. PCR studies of PVAN biopsies identified rr-NCCR in approximately one fourth of BKV strains, indicating that rr-NCCR BKV were not necessary for histological disease (29–31). Given the rapid dynamics of BKV replication and the close correlation of plasma BKV load with PVAN, we compared the occurrence of rr-NCCR variants in BKV-positive plasma and urine samples, and investigated the functional consequences.
RESULTS
rr-NCCRs are associated with higher plasma BKV load
The NCCR architecture was analyzed by PCR in 129 KT patients, including 70 with PVAN-positive biopsy results, providing a total of 286 same day–matched plasma and urine samples (Table I). The majority of patients had ww-NCCR detectable in plasma (n = 85; 66%) and in urine (n = 119; 92%; Table I, Patients column). rr-NCCRs consisted of larger and shorter amplicons representing insertions (ins-)NCCR in 14%, deletions (del-)NCCR in 42%, and mixtures of ins/ww- and del/ww-NCCR in 6 and 38%, respectively (Table I, rr-NCCR architecture in samples column). The number of patients with rr-NCCR BKV in plasma represented a significant fraction (P < 0.0005, Kolmogorov-Smirnov; see footnote a in Table I, Patients column) compared with the fraction of patients with rr-NCCR in urine (P = 0.45, Kolmogorov-Smirnov). The higher frequency of patients with rr-NCCR in plasma compared with urine was statistically significant (P < 0.0001, χ2 test; see footnote b in Table I, Patients column). The same was true when the analysis was restricted to patients with the histological diagnosis of PVAN in biopsies (n = 70; rr-NCCR in plasma 35, 50%; ww-NCCR in urine 67, 96%; P < 0.0001, χ2 test). When we compared the NCCR architecture in 129 patients with (n = 70) and without histologically defined PVAN (n = 59; Table I, Patients column), the detection of rr-NCCR in plasma was significantly associated with a positive biopsy (35 of 70 vs. 9 of 59; P < 0.0001, χ2 test).
Table I.
Architecture of BKV NCCR in plasma and urine of KT patients
| Patients | Samples | rr-NCCR architecture in samples |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Matched plasma + urine |
NCCR
|
P valuea
|
NCCR
|
P valued
|
||||||||
| ww | rr | ww | rr | ins | ins/ww | del | del/ww | |||||
| Plasma | n = 129 | 85b | 44 b | <0.0005 | n = 286 | 222c | 64c | 9 | 4 | 27 | 24 | |
| Percentage (%) | 66 | 34 | 78 | 22 | 14 | 6 | 42 | 38 | ||||
| PVAN | n = 70 | 35 | 35 | |||||||||
| Percentage (%) | 50 | 50 | <0.0005 | |||||||||
| Viral load (geq/ml) | 1.97 × 104 e | 4.36 × 105 f | <2 × 10−6 | |||||||||
| 25th percentile | 3.93 × 103 | 6.59 × 104 | ||||||||||
| 75th percentile | 1.72 × 105 | 5.47 × 106 | ||||||||||
| Urine | n = 129 | 119b | 10b | =0.45 | 274c | 12c | - | 2 | 5 | 5 | ||
| Percentage (%) | 92 | 8 | 96 | 4 | 0 | 16 | 42 | 42 | ||||
| PVAN | n = 70 | 67 | 3 | =0.5 | ||||||||
| Percentage (%) | 96 | 4 | ||||||||||
| Viral load (geq/ml) | 2.89 × 108 e | 2.56 × 107 f | =0.098 | |||||||||
| 25th percentile | 1.66 × 107 | 2.84 × 106 | ||||||||||
| 75th percentile | 3.38 × 109 | 6.93 × 108 | ||||||||||
| Consecutive plasma (2005–2006) |
n = 227 | 172 | 55 | =0.004 | n = 733 | 620 | 113 | 12 | 15 | 39 | 47 | |
| Percentage (%) | 76 | 24 | 85 | 15 | 11 | 13 | 35 | 41 | ||||
| PVAN | n = 57 | 31 | 26 | <0.002 | ||||||||
| Percentage (%) | 54 | 46 | ||||||||||
| Viral load (geq/ml) | 1.94 × 104 | 4.59 × 105 | <2 × 10−6 | |||||||||
| 25th percentile | 4.55 × 103 | 3.51 × 105 | ||||||||||
| 75th percentile | 1.58 × 105 | 3.04 × 106 | ||||||||||
Kolmogorov-Smirnov Goodness-of-Fit test (one-sided).
χ2 test (two-sided), plasma rr-NCCR versus urine rr-NCCR. P < 10−4.
Mann-Whitney U test (one-sided).
Plasma ww-NCCR versus urine ww-NCCR. P < 10−4.
Plasma rr-NCCR versus urine rr-NCCR. P < 0.002. Viral load is given as median.
When expressed per sample, ww-NCCR BKVs were detected in the majority of plasma and urine, but rr-NCCRs were significantly more frequent in plasma compared with urine samples (64 and 12 of 286, respectively; P < 0.0001, χ2 test; Table I, Samples column). For the difference of 52 samples (18%), we found discordance between rr-NCCR BKV in plasma and ww-NCCR in urine (illustrated for 5 patients in Fig. 1 A). BKV rr-NCCR variants were associated with higher plasma BKV loads compared with ww-NCCR BKV (median 4.36 × 105 vs. 1.97 × 104 geq/ml; P < 0.000002, Mann-Whitney U test; Table I, Samples column). In contrast, no association with ww- or rr-NCCR architecture was apparent for urine BKV loads (median 2.89 × 108 vs. 2.56 × 107 geq/ml; P = 0.098, Mann-Whitney U test). However, urine BKV loads were on average 3,000-fold (range 100–10,000-fold) higher than plasma BKV loads.
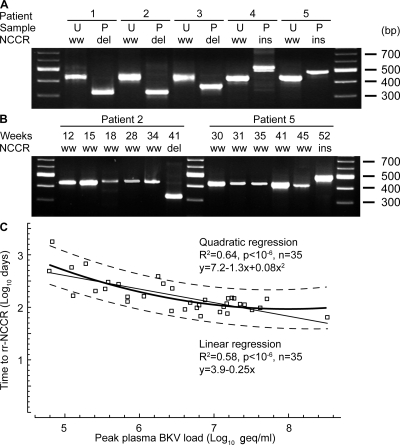
Figure 1.
BKV NCCR in plasma and urine of KT patients. (A) Discordance of NCCR majority species in matched urine (U) and plasma (P) samples shown for 5 patients. Nested PCR targeting the BKV NCCR was resolved on 2% agarose-tris acetate, pH 8.0, electrophoresis gels and stained with ethidium bromide. NCCR ww, archetype; del, deletion, ins, insertion, bp, base pairs of marker. (B) Emergence of rr-NCCR in consecutive plasma shown for patient 2 (del-NCCR) and 5 (ins-NCCR) resolved as in A. (C) Time to rr-NCCR detection and peak plasma BKV load in consecutive plasma samples from 35 KT patients analyzed by linear and quadratic regression. Fine line, linear regression; bold line, quadratic regression; dashed lines, 90% confidence interval.
To corroborate the association of rr-NCCR and plasma BKV loads, we prospectively analyzed 733 serial BKV DNA–positive plasma samples from 227 KT patients, including 57 cases with documented PVAN (Table I). The results identified the overall rr-NCCR prevalence rate, ranging from 15% per sample to 24% per patient, and confirmed the association of rr-NCCR with higher median BKV loads (rr-NCCR 4.59 × 105 geq/ml and ww-NCCR 1.94 × 104 geq/ml; P < 0.00002; Table I). BKV rr-NCCRs were detected in 113/733 plasma samples (15%) consisting of ins-NCCR in 11% and del-NCCR in 35%, as well as mixtures of ins/ww and del/ww NCCRs in 13 and 41%, respectively (Table I, rr-NCCR architecture in samples column). Again, rr-NCCR in plasma was significantly associated with a PVAN-positive biopsy (26 of 57 vs. 19 of 141; P < 0.0001, χ2 test).
We also analyzed 56 BKV-positive urine samples from KT patients without concurrent BKV viremia (median, 14,700 geq/ml; range, 1,000–8.9 × 108) and found ww-NCCR BKV in all cases. When examining 264 healthy blood donors, BKV was only detected in urine, but not in plasma in 26 (9.8%) cases, all of which were of archetype ww-NCCR configuration (median viral load, 2,600 geq/ml; range, 1,000–64,900). We concluded that rr-NCCR BKV variants appeared more frequently in plasma than in urine in viremic KT patients, and were then associated with significantly higher plasma BKV loads. In contrast, the day-matched urine BKV loads were generally 3,000-fold higher than the corresponding plasma BKV loads, regardless of the NCCR architecture. This observation, together with the discordance of rr- and ww-NCCR architecture in day-matched plasma and urine samples provided quantitative and molecular evidence that renal tubular epithelial cells and urothelial cells in ureters and bladder, indeed, reflected different viral replication compartments in immunosuppressed KT patients, thereby extending previous histopathology data (6).
rr-NCCR BKV replace ww-NCCR BKV with persisting viremia
In preceding plasma samples from patients with rr-NCCR variants, we detected ww-NCCR BKV in all but one case. A detailed analysis of 35 PVAN patients with rr-NCCR (Table I) revealed that plasma rr-NCCR BKV variants, indeed, emerged from initial ww-NCCR BKV strains after prolonged viremia (Fig. 1 B). Regression analysis revealed that the time to first rr-NCCR emergence correlated with peak plasma viral load (P < 0.000001; Fig. 1 C). The linear and the quadratic regression models largely overlapped between BKV loads of 5.5–7.5 log10 geq/ml. However, the convex shape of the quadratic regression model provided a better fit of the overall data, suggesting that higher peak plasma BKV loads overproportionally increased the risk of rr-NCCR emergence. Accordingly, rr-NCCRs were detectable in patients with high peak plasma BKV load of 6.5 log10 geq/ml after <4 mo, whereas 100-fold lower viremia (4.5 log10) required >7 times as long. The data indicated that duration and peak replication favored the change from ww- to rr-NCCR BKV variants as the majority species. The only case where rr-NCCR BKV was already present in the first BKV-positive plasma sample was a patient retransplanted after allograft loss and nephrectomy after PVAN. This observation was intriguing, and suggested that impaired BKV-specific cellular immunity is not only critical for curtailing BKV replication (23), but possibly also for recurrence of BKV replication and direct emergence of rr-NCCR variants (24, 32).
rr-NCCR BKV are associated with histological disease progression
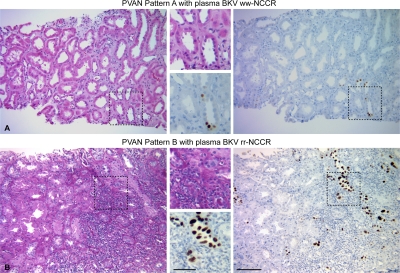
Comparing PVAN histology in needle biopsies from 39 patients with either rr-NCCR (n = 21) or ww-NCCR (n = 18) in plasma, we found that the percentage of surface with tubules showing cytopathic changes staining positive for large T-antigen (LTag) was significantly higher in cases with rr-NCCR BKV variants in plasma compared with ww-NCCR BKV (median 15%, mean 26.6 ± 29.4 vs. median 3.5%, mean 11.3 ± 17.1; P = 0.03; Mann-Whitney U test). In addition, cytopathically altered areas from patients with rr-NCCR BKV were more likely to have lymphocytic inflammatory infiltrates compared with ww-NCCR BKV (60 vs. 23.07%, respectively; P = 0.04, Mann-Whitney U test). Finally, detection of rr-NCCR in plasma was associated with PVAN pattern B (Fig. 2 B), whereas ww-NCCR more readily presented as PVAN pattern A (Fig. 2 A; 15/21 vs. 6/18, respectively; P = 0.03, Fischer's exact test).
Figure 2.
Histological patterns of PVAN in kidney biopsies. (A) Haematoxylin/eosin stain of a representative histological field showing PVAN pattern A consisting of focal viral cytopathic changes with little inflammation or tubular atrophy. Immunohistochemistry of LTag expression using the cross-reacting monoclonal anti–SV40 T-antigen Ab-2 and peroxidase-conjugated anti–mouse. Enlargement of the indicated area is shown. (B) Haematoxylin/eosin stain of a representative histological field showing PVAN pattern B with extensive viral cytopathic changes and inflammatory infiltrates. Immunohistochemistry of LTag expression using the cross-reacting monoclonal anti-SV40 T-antigen Ab-2 and peroxidase conjugated anti–mouse. Bars: (A and B) 200 μm; (A and B, insets) 100 μm.
rr-NCCR are diverse and without corresponding changes in VP1
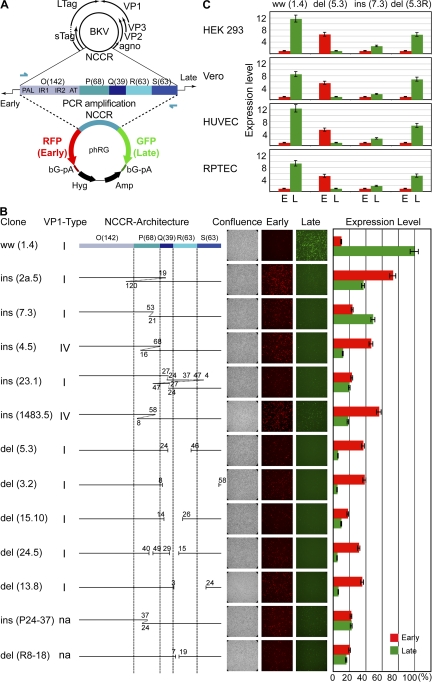
To characterize the architecture of rr-NCCR in more detail, we sequenced 5 ins-NCCR and 5 del-NCCR from 10 different patients with PVAN. Insertions consisted of partial duplication of the P-Q region, whereas deletions preferentially affected the Q-R-S region (Fig. 3). Thus, NCCR rearrangements changed transcription factor-binding sites and enhancer elements, but no common feature was apparent (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20072097/DC1). Sequencing of the antigenic determinants of VP1 revealed BKV serotype I and IV in 8 and 2 rr-NCCR variants, respectively. In all cases, the VP1 sequences were identical with the one detected when the majority species were of archetype ww-NCCR (Fig. 3 B and Fig. S2). Thus, NCCR rearrangements were not linked to changes in VP1 serotype.
Figure 3.
BKV NCCR architecture and expression pattern in cell culture. (A) Architecture of archetype ww-NCCR in BKV genome, with arbitrarily denoted linear blocks (number of base pairs) O(142)-P(68)-Q(39)-R(63)-S(63) and position of primers (arrows). RFP, RFP for early genes; GFP, GFP for late genes; βG-pA, β-globin polyA; Hyg, hygromycin resistance; Amp, ampicillin resistance. (B) Architecture of rr-NCCR clones and gene expression pattern in HEK293 at 2 dat. ins, insertions corresponding to duplications of the numbered base pairs; del, deletions, denoted by gaps and nucleotides. VP1-Type, serotype; n.a., not applicable because generated by in vitro mutagenesis; Confluence, showing phase contrast; Early, red fluorescence; Late, green fluorescence; Expression level, percentage of green and red fluorescence–positive cells normalized to GFP-positive cells of phRG-ww(1.4) bearing the archetype ww-NCCR. (C) NCCR-driven early (E) and late (L) gene expression in different cell types. HEK293, monkey kidney cells (Vero), human umbilical vein endothelial cells (HUVECs), and primary RPTECs were transfected with phRG-bearing archetype ww(1.4)NCCR, del(5.3)NCCR, ins(7.3)NCCR, and del(5.3R)NCCR, which contained the del(5.3)NCCR in reverse orientation relative to RFP and GFP (results as above). Numbers indicate fold expression level for del(5.3)NCCR (green, late) and ww(1.4)NCCR (red, early), arbitrarily set to 1. (B and C) Mean of three independent experiments. Error bars represent the mean ± the SD.
rr-NCCRs are sufficient to increase early reporter gene expression
To investigate functional effects of rr-NCCR in regard to early and late gene expression, we constructed a bidirectional reporter vector phRG, having the red fluorescent protein (RFP) and GFP in the same positions as the early and late genes in the circular BKV DNA genome (Fig. 3 A). Human embryonic kidney (HEK) 293 cells were transfected and assayed for red and green fluorescence. The expression patterns became detectable as early as 1 d after transfection (dat) and remained stable for at least 10 d of observation (all scored at 2 dat; Fig. 3 B). Archetype ww-NCCR conferred a strong late gene expression (green signal) and a relatively weak early gene expression (red signal, at ∼1/10 of the green signal). In contrast, all rr-NCCRs increased early gene expression by two- to sevenfold, and decreased late gene expression, irrespective of insertion or deletion architectures. Simple deletions or duplications had effects similar to more complex ones, suggesting that the architectural context and not the mere number of transcription factor binding sites might be relevant. We therefore generated two NCCR variants by in vitro mutagenesis that reflected small insertions or deletions of the natural NCCR sequences. Both ins(P24-37)NCCR and del(R8-18)NCCR affected early and late gene expression in a manner similar to in vivo–occurring variants ins(23.1)NCCR or del(15.10)NCCR (Fig. 3 B). As NCCR expression might be affected by host cell type and differentiation, we tested two prototype rr-NCCRs (del 5.3, ins 7.3) in addition to the ww-NCCR (ww 1.4) in cells known to support BKV replication in vitro (Vero monkey kidney cells and human umbilical cord vein endothelial cells) and in vivo (primary human renal proximal tubular epithelial cells [RPTECs]). We found that the expression patterns for all three NCCR prototypes were similar in all tested cell types (Fig. 3 C). Northern blot analysis and RNA protection experiments at 2 dat indicated that NCCR-driven RNA expression correlated with RFP and GFP signals (unpublished data). To exclude vector-mediated positional effects on gene expression, the del(5.3)NCCR was inserted in reverse orientation relative to RFP and GFP (phRG-del[5.3R]). Transfection phRG-del(5.3R) into the different cell types revealed strong early but weak late gene expression, as observed for del(5.3)NCCR, but in opposite colors (Fig. 3 C, compare column 2 and 4). We concluded that rr-NCCR BKV variants were of diverse architecture, but all increased early gene expression in vitro as a common hallmark.
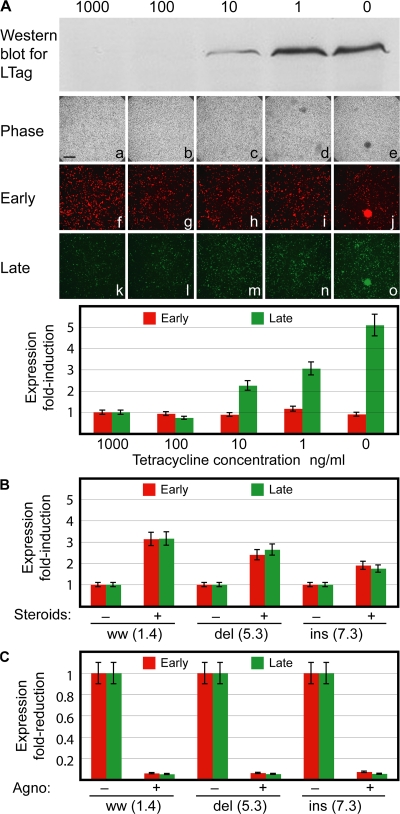
To investigate whether the genomic NCCR rearrangement still allowed for modulatory effects in trans, we examined the responses of the bidirectional reporter vector phRG to BKV LTag, steroids, and agnoprotein expression, which have been reported to positively and negatively regulate NCCR expression (33). As shown for the prototype ins(7.3)NCCR (Fig. 4), increasing LTag expression as detected by Western blot raised late gene expression by 5-fold within 24 h (green, Fig. 4 A). Because glucocorticoid response elements are present in BKV NCCR, we examined whether steroid treatment modulated gene expression in Vero monkey kidney cells that, unlike HEK293, expressed functional glucocorticoid receptors. Treatment with dexamethasone at 10−6 M increased early and late gene expression from the ww-NCCR (3-fold), and to lesser degrees, also from the del(5.3)- (2.5-fold) and ins(7.3)-NCCR (2-fold; Fig. 4 B). When BKV agnoprotein was cotransfected, both early and late reporter gene expression signals were ∼10-fold reduced for all 3 NCCRs (Fig. 4 C). Collectively, these results indicated that rr-NCCR consistently increased early gene expression relative to archetype ww-NCCR, but still remained susceptible to modulatory effects in trans conveyed by LTag, steroids, or agnoprotein.
Figure 4.
Modulation of early and late gene expression. (A) Western blot detecting LTag in HEK293 cells stably expressing BKV LTag with monoclonal anti-SV40 and anti–mouse coupled to horseradish peroxidase at day 5 after transfection. Cells were transfected with phRG7.3 in the presence of Tetracycline (Tet). After 1 d, Tet was replaced by the indicated concentrations. Phase contrast (a–e), RFP (f–j), and GFP (k–o) expression in the presence of decreasing amounts of Tet (repression of LTag at 1,000 ng/ml Tet, maximum expression of LTag at 0 ng/ml Tet). Bar, 400 μm. (B) Effect of steroid treatment on early (RFP) and late (GFP) gene expression in Vero cells. -, absence; +, presence of 10−6 M dexamethasone. Cells were analyzed at 2 dat. (C) Effect of agnoprotein expression on early (RFP) and late (GFP) gene expression in Vero cells. Cells were analyzed at 2 dat. (A–C) Mean of three independent experiments. Error bars represent the mean ± the SD.
rr-NCCR increase BKV replication and cytopathology in vitro
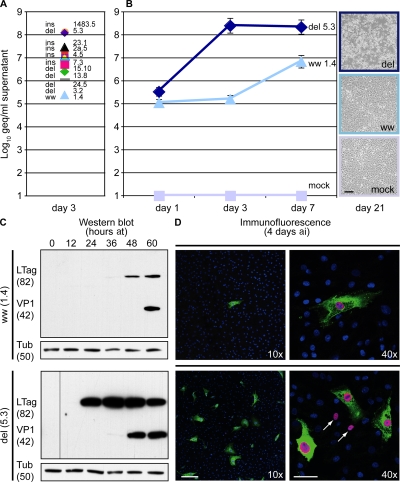
To directly examine the impact of rr-NCCR on BKV gene expression and replication, we generated recombinant BKV strains by cloning each of the indicated 10 rr-NCCR, as well as the archetype ww-NCCR, into the genomic backbone of the BKV Dunlop strain. At 3 dat of recombinant BKV DNA into Vero cells, DNase-protected BKV DNA loads in supernatants were 1–3 log10 higher viral loads for all 10 recombinant rr-NCCR BKV strains compared with ww(1.4)NCCR (Fig. 5 A). More detailed analysis revealed that supernatants of del(5.3)NCCR BKV–transfected cells had 3 and 1.5 log10 higher viral loads at day 3 and 7, respectively, and a pronounced cytopathic effect at day 21 compared with ww(1.4)NCCR BKV (Fig. 5 B). Western blot analysis of the transfected Vero cells demonstrated that LTag expression occurred ∼12–24 h earlier in del(5.3)NCCR BKV, with a corresponding acceleration of VP1 expression compared with ww(1.4)NCCR BKV strains (Fig. 5 C). When Vero cell supernatants were harvested 7 dat and seeded for infection of primary RPTECs, we observed that more cells were infected by the recombinant del(5.3)NCCR BKV than by the ww(1.4)NCCR BKV, with corresponding supernatant viral loads being 100-fold higher (unpublished data). Immunofluorescence staining of RPTECs at day 4 after infection identified a correspondingly higher number of infected cells in del(5.3)NCCR BKV– than in the ww(1.4)NCCR BKV–infected cells (Fig. 5 D, green stain; 10× magnification). Moreover, LTag preceding expression of the late agno gene could be observed at the single-cell level (Fig. 5 D, LTag red nuclear stain, arrows in 40× magnification; agno green cytoplasmic stain) and VP1 expression (not depicted). Thus, we concluded that rr-NCCR were sufficient to confer increased early gene expression, replication, and cytopathology to recombinant BKV in vitro.
Figure 5.
Replication of recombinant rr-NCCR BKV in cell culture. (A) Quantification of DNase-protected BKV load of rr-NCCR variants in Vero cell supernatants collected at indicated times after transfection. Viral load measurements for all 10 recombinant rr-NCCR BKV variants in Vero cells at 3 dat. Time course for archetype ww, ww(1.4)NCCR BKV; del, del(5.3)NCCR BKV). The data display a mean of three independent transfections determined in triplicate. Error bars represent the mean ± SD. (B) Cytopathology of the corresponding cell cultures at 21 dat by phase-contrast microscopy. (C) Western blot was performed with polyclonal anti-LTag, anti-VP1, and anti–rabbit conjugated to peroxidase, respectively. Tubulin was detected with monoclonal mouse anti-tubulin and peroxidase-conjugated anti–mouse. VP1, VP1 capsid protein; Tub, tubulin; at, after transfection. Samples were harvested at indicated times (hours) after transfection. Molecular weight of proteins is shown in kilodaltons. (D) Infection of primary RPTECs. Vero cell supernatants were harvested 7 dat and seeded onto RPTECs. Immunofluorescence for LTag (early gene, red) and agnoprotein (late gene, green) was performed on day 4 after infection using the monoclonal anti-SV40 LTag visualized with anti–mouse Alexa Fluor 568 and polyclonal rabbit anti-agno detected by anti–rabbit Alexa Fluor 488, respectively. Arrows indicate early LTag-positive staining cells without late agnoprotein staining. Cell nuclei are stained with DRAQ5 (blue). Bars: (B) 200 μm; (D, 10×) 200 μm; (D, 40×) 50 μm.
DISCUSSION
The failing balance between BKV replication and antiviral immune control is the hallmark of PVAN in KT recipients (17, 24). Viral factors have been postulated to contribute to PVAN pathogenesis, but determinants, rates, and mechanisms remained speculative (7, 31). In this study, we took advantage of BKV viremia as a noninvasive marker of renal allograft involvement and provide evidence that the NCCR is a relevant viral determinant of BKV replication rate and PVAN pathogenesis. Although BKV variants with rr-NCCR had previously been detected in immunocompromised patients (29–31, 34, 35), we show for the first time that rr-NCCR BKV variants emerge and replace archetype ww-NCCR BKV as majority species in vivo in 24% of KT patients with persisting BKV viremia and in 50% of patients with PVAN. The time to emergence of rr-NCCR BKV variants in plasma was inversely correlated to peak plasma BKV loads. Thus, duration and level of BKV replication increased the risk of rr-NCCR BKV emergence. Compared with ww-NCCR strains, rr-NCCR variants were associated with 20-fold higher median plasma BKV loads, a higher probability of a histologically confirmed disease, and increased viral cytopathology and inflammatory presentation in these patients.
Our in vitro studies revealed complex rr-NCCR emerging in vivo without identifiable common alterations. However, using a novel bidirectional reporter gene replicon that exactly recapitulated the polyomavirus NCCR genome architecture relative to early and late gene organization, we could demonstrate that all rr-NCCR augmented early gene expression compared with ww-NCCR, while decreasing late gene expression. Nevertheless, rr-NCCR remained responsive to up- and down-regulation by LTag-, agnoprotein, and most notably steroids, a clinically defined risk factor of BKV replication and PVAN (1, 36, 37). It has been proposed that rr-NCCR change critical transcription factor binding sites in the ww-NCCR (38), but the differences between various deletions and insertions did not reveal a common alteration. Clearly, further work is needed to unravel the complex molecular organization of BKV NCCR-mediated gene expression.
We demonstrate that rr-NCCR increased viral early gene expression and accelerated viral replication in vitro. The recombinant rr-NCCR BKV strains yielded 1–3 log10 geq/ml higher viral loads in tissue culture compared with archetype ww-NCCR BKV. Detailed analysis indicated an ∼24 h earlier LTag expression with corresponding effects on VP1 expression, a shorter lag phase reducing the BKV generation time from ∼3 to ∼2 d, with prominent cytopathology by day 21. Thus, rr-NCCRs associated with higher plasma BKV loads and accelerated pathology in KTs were sufficient to increase replication capacity and cytopathology of recombinant rr-NCCR BKV in tissue culture.
The molecular mechanisms leading to NCCR rearrangements in a DNA virus at such high frequency are intriguing and as yet unclear. Unlike RNA virus replication generating mutant progeny by lack of proof-reading, DNA virus replication is widely considered a process of high fidelity, particularly for viruses that rely on recruiting host cell DNA polymerases, such as polyomaviruses. The dependence on time and peak plasma BKV load shown here suggests that persistent high-level replication of the circular BKV genome and its associated errors may be crucial for rr-NCCR emergence (27), similar to other viruses (39). Because BKV replication requires LTag with its DNA binding and helicase functions, some genomic rearrangements, and especially deletions of ori or LTag binding sequences, are likely to generate defective NCCR penalized by negative selection. Conversely, NCCR rearrangements conferring increased early gene expression outcompete archetype ww-NCCR BKV strains by increased replication capacity, a phenomenon termed positive selection (39). We noted that NCCR rearrangements were unevenly distributed, being less frequent in the vicinity of the early gene promoter (typically sparing the origin of replication and LTag-binding sites), consisting of partial duplication rather than deletions. In contrast, deletions were more frequent toward the late gene expression, removing Q-R-S sequences. Interestingly, we did not encounter any poorly replicating rr-NCCRs, as had been reported in previous studies not related to kidney transplantation (38, 40, 41). Although these studies support the principle notion that rearrangements as a consequence of replication can also generate defective NCCRs, our results are in line with the fact that we targeted by PCR emerging rr-NCCR majority species that had been positively selected in these immunosuppressed KT patients.
The rapid emergence of rr-NCCR BKV variants with higher replicative capacity in immunosuppressed KT patients is reminiscent of rr-NCCR generation in tissue culture (26, 27, 42), and is in striking contrast to the stable colonization of the world's population with archetype ww-NCCR BKV strains that do not replicate as well (43, 44). Indeed, our ongoing analysis of BKV seropositive healthy blood donors detected asymptomatic BKV viruria in 9.8%, with ww-NCCR in all cases. This is in line with earlier reports implicating urine as a relevant source of BKV transmission and persistence around the world (7, 45). The discordance of rr-NCCR BKV in plasma and ww-NCCR BKV in urine in immunosuppressed KT patients argues that emergence of rr-NCCR in vivo is at least partially defined by host cell properties, i.e., the difference between renal tubular epithelial cells and urothelial cells. A precedent for the role of host cells is documented in tissue culture where rr-NCCR emergence required a longer time in human umbilical vein endothelial cells compared with Vero or in HEK293 cells (26, 27, 42). Histopathology studies demonstrated in KTs that renal tubular epithelial cells, as well as in the urothelial cells support BKV replication (6, 46), but both host cell types differ in anatomical location, function, differentiation and organization as single cell layer versus multicellular transitional cell layer, respectively. However, a higher rr-NCCR replication capacity in renal tubular epithelial cells may not play out to the same extent in the urothelial cells of ureter and bladder because urine BKV loads were, on average, 3,000-fold higher than plasma BKV loads independent of ww- or rr-NCCR architecture. We therefore conclude that the selective advantage of a higher replication capacity may not be operative to the same extent in the urothelial cell compartment. Different, not mutually exclusive, hypotheses may account for this cell type phenomenon: (a) activation of early gene expression is similar between ww- and rr-NCCR BKV in urothelial cells; (b) less LTag is needed to recruit replication factors in continuously dividing urothelial cells; and (c) ww-NCCR strains are reactivated first and saturate susceptible target cells.
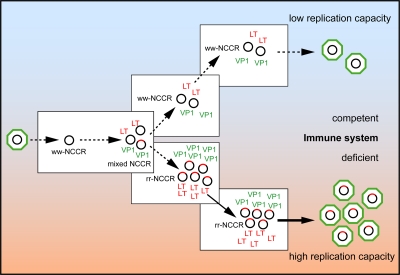
Thus, we suggest that antiviral immunity is another significant barrier that is lacking in tissue culture, as well as in profoundly immunosuppressed KT patients. Because increased early gene expression appears as a common feature of rr-NCCR, cellular immunity directed against early gene encoded epitopes may be the important factor limiting emergence of rr-NCCR BKV in the general immunocompetent population. The rr-NCCR–mediated increase in LTag expression might provide an extended window for cytotoxic immune recognition in immunocompetent hosts, which is lacking in immunosuppressed KT patients, as well as in tissue culture. LTag as a target of cellular immunity has been first identified in experimental SV40 models (47), in healthy individuals (48), and, more recently, in peripheral blood of KT patients with declining BKV viremia (23). The late capsid protein VP1 is also a known target of antiviral immunity, particularly for neutralizing antibodies. As VP1 expression is closely linked to virion assembly and host cell lysis, anti-VP1–directed CD8-T cells may develop their impact at a later stage of the viral life cycle than anti-LTag CD8-T cells (23). We speculate that immune surveillance is likely to be more stringent for renal tubular epithelial cells than for the urothelial cells, as with other mucosal surfaces. This notion is also supported by the strong inflammatory response evolving in the infected renal tubulus and its virtual absence in the heavily BKV replicating transitional cell layer in KT patients. We therefore hypothesize that cellular immunity represents a key barrier to rr-NCCR BKV emergence, rendering less well replicating archetype ww-NCCR BKV evolutionarily more successful in immunocompetent host populations (Fig. 6) (39).
Figure 6.
Emergence of rr-NCCR in the immunocompromised host. Naturally transmitted BKV with archetype ww-NCCR genome (black circles) undergo insertion and deletion errors during replication generating rr-NCCR genomes (red tagged circles). BKV rr-NCCR with increased early gene expression and corresponding higher replication capacity are eliminated in immunocompetent host, but outcompete ww-NCCR BKV in immunodeficient hosts. LT, LTag; VP1, capsid protein 1.
In summary, our study underlines that genetic plasticity of evolutionarily conserved DNA viruses can be observed in vivo in situations permitting unchecked high-level replication, as in immunocompromised hosts, thereby causing increased cytopathic damage. In KT patients, rr-NCCR BKV emergence is a marker of prolonged viral replication and is specifically associated with more advanced PVAN histology. The role of cellular immunity and the prognostic information of rr-NCCR BKV regarding KT function or graft loss has to await properly designed prospective studies.
MATERIALS AND METHODS
Patients.
We characterized BKV NCCR in 1,019 plasma and 342 urine samples from 412 KT patients. Day-matched plasma and urine samples were obtained from KT patients as part of the prospective BKV study protocol approved by the University of Maryland Transplant Center. BKV loads had been determined by real-time PCR in same day–matched plasma and urine samples (n = 286 from 129 patients, including 70 patients with PVAN). The NCCR architecture was determined retrospectively on specimens stored at −20°C. PVAN was graded based on the Banff classification, as recommended by Hirsch et al. (17), as follows: PVAN A, minimal to mild viral cytopathic changes (cy1+–cy2+), insignificant inflammatory infiltrates (<i1+), tubular atrophy (<ct1+), or fibrosis (<ci1+); PVAN B, viral cytopathic changes scoring from mild to severe (cy2+–cy4+) and significant inflammatory infiltrates scoring from moderate to severe (i1+–i3+), but only mild tubular atrophy (≤ci2+) and mild fibrosis (≤ci2+); PVAN C, moderate to severe tubular atrophy (ct3+) and interstitial fibrosis (ci2+–ci3+), variable scores for cytopathic changes (cy1+–cy4+), and for inflammatory infiltrates (i1+–i3+) (17). During the years 2005 and 2006, an additional 733 consecutive BKV DNA–positive plasma samples from 227 patients, including 57 with PVAN, had been submitted for BKV testing to the Molecular Diagnostics Laboratory of the Division of Diagnostics, which is an accredited quality controlled laboratory (EN ISO) of the University of Basel (www.zid.ch). If the samples had detectable BKV loads, BKV NCCR sequences were amplified as an independent confirmatory assay to confirm BKV specificity. Patients with sufficient sampling density defined as >6 consecutive plasma samples collected over >12 wk were identified (n = 35, PVAN in 28) to analyze the dependence of rr-NCCR emergence on time and plasma viral load. NCCR architecture was examined in 56 KT patients (median age 44 yr; range 17–66 yr; 52% female) with urinary BKV shedding, but without concurrent BKV viremia. As controls, we examined BKV in urine and plasma of 264 healthy blood donors (median age 46 yr; range 18–67 yr; 49% female; IRB approval 267/06).
BKV load and NCCR analysis.
BKV DNA was quantified after DNA extraction from 200 μl plasma, urine, or cell culture supernatant using a commercial kit (QIAmp; QIAGEN) and a previously described real-time PCR protocol (1). NCCR amplification used a standard nested PCR approach with the outer primer pairs BKTT5 5′-GAGCTCCATGGATTCTTC-3′ and BKTT6 5′-CCAGTCCAGGTTTTACCA-3′ and the inner primer pair BKTT7 5′-CCCTGTTAAGAACTTTATCCATTT-3′ and BKTT8 5′-AACTTTCACTGAAGCTTGTCGT-3′. The PCR products were compared after 2%TAE-agarose gel electrophoresis with ww-NCCR controls, and excised for sequence analysis, if indicated. Dilution series of increasing ratios of rr-NCCR versus ww-NCCR and cloning into plasmids indicated that the sensitivity of detecting minority species in NCCR mixtures by the PCR protocol was as low as 5%. Patients were scored as bearing rr-NCCR when one or multiple samples showed rr-NCCR. Samples containing rr-NCCR or mixtures of rr- and ww-NCCR (Table I, rr-NCCR architecturein samples column) were scored as rr-NCCR.
Plasmids and primers.
In pTRELTag the cDNA coding for the LTag of BKV is under control of a tetracycline-controlled transactivator-dependent promoter. The intron of the LTag was removed by gene splicing by overlap extension (SOE) (49). Two DNA fragments were generated using primer pairs 1–2 (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.200712097/DC1) and 3–4 and DNA extracted from a BKV-positive urine as template. The two DNA fragments were subsequently joined in a PCR reaction containing primer pair 1–4. The amplification product was digested with ClaI and NheI, followed by ligation into ClaI–NheI-digested pTRE2pur (BD Biosciences). The dual-reporter vector phRG contains hygromycin selectable marker, the enhanced GFP and RFP dsRed2 occurs in opposite directions with both mRNAs, followed by the same β-globin polyA sequence. Plasmid phRG was generated in a three-step procedure. First, the tetracycline response element/minimal CMV promoter cassette was deleted from pTRE2hyg by SOE. Two DNA fragments generated by PCR using primer pairs 5–6 and 7–8 and pTRE2hyg as template were joined in a PCR reaction containing primer pair 5–8. The amplification product was digested with AvrII and BamHI, followed by ligation into AvrII–BamHI-digested pTRE2hyg to yield phyg. Next, BamHI–MluI-cleaved dsRed2 (pdsRed2-C1; BD Biosciences) and ClaI- and SalI-digested EGFP (pEGFP-N1; BD Biosciences) were ligated into BamHI–SalI–restricted pTRE2hyg to obtain pTREdsRed-EGFP. Next, the BamHI–SalI dRed2-EGFP fragment was ligated into phyg cleaved with the same enzymes to yield phdsRed-EGFP. In the final step, the AseI–EcoRV β-globin fragment, blunt ended with T4 polymerase, was subcloned into pGEM-3Zf+ (Promega) to yield pGEMglobin. This plasmid was cleaved with SacI–XbaI, and the resulting fragment was ligated into phdsRed-EGFP digested with SacI–SpeI to obtain phRG. In phRG-NCCR plasmids, the expression of RFP (early) and GFP (late) is under control of the inserted BKV NCCR. NCCR sequences were amplified from patient material by nested PCR using primer pairs 9–10 and 11–12. The MluI–BssHII-cleaved amplicon was ligated into phRG digested with the same enzymes. The plasmids phRGinsP24-37 and phRGdelR8-18 were generated from the archetype ww-NCCR by a duplication of P24-37 or a deletion of R8-18, respectively, by SOE. The two DNA fragments were generated from phRGww by PCR using primer pairs 11–13 and 14–12 (P24-37) or 11–15 and 16–12 (R8-18), respectively, and joined in a PCR reaction containing primer pair 11–12. The amplification products were ligated into MluI–BssHII-digested phRG, as described. To construct recombinant BKV strains, the SacI fragment containing the NCCR was replaced in pBKV Dunlop plasmid (American Type Culture Collection [ATCC] 45025) bearing the full-length genome sequence (50). A synthetic linker (primer 17–18) was ligated into SacI-cleaved pGEM3-Zf+ to yield pGEMSacIlinker. The NCCR released from the indicated phRGww, ins, and del constructs by MluI–BssHII digest was ligated into pGEMSacIlinker cleaved with the same enzymes. Finally, the SacI NCCR fragment was ligated into SacI-restricted pBKV to generate the respective recombinant pBKVww-, ins-, and del-NCCR. pCMVAgno encodes the agnoprotein sequence in the pRC/CMV (Invitrogen). The entire agno sequence was amplified from patient material by PCR using primer pair 19–20. The amplification product was cleaved with BamHI and NotI, followed by ligation into pRC/CMV digested with the same enzymes. The integrity of all plasmid constructs was confirmed by sequencing.
Cell lines.
HUV-EC-C (ATCC; CRL 1730) were grown in Kaighn's FK12 medium (51). HEK293 (ATCC CRL1573) and Vero76 cells (ATCC CRL1587) were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS. Primary RPTECs (Cambrex) were propagated in Renal Epithelial Cell Medium (Cambrex). Infection of RPTECs was performed as previously described (52). HEK293 cells stably expressing BKV LTag were generated by cotransfection of pTET-Off (BD Biosciences) and pTRELTag. Cells were selected and maintained in DME containing 10% FBS in the presence of 500 μg/ml Geneticin (G418; Invitrogen), 1.5 μg/ml Puromycin (Sigma-Aldrich), and 1 μg/ml Tetracycline (Sigma-Aldrich). Transfection of BKV genomes into Vero76 cells was initiated by cleavage of pBKV plasmid DNAs with BamHI, followed by relegation of the diluted DNA. Transfection of cells was performed at 60–80% confluency in 12-well plates and was achieved as specified by the manufacturer using Lipofectamine 2000 or LTX (RPTEC; Invitrogen) at a DNA/Lipofectamine ratio of 0.8:1. Transfection of HUV-EC-C was performed with Transpass D1 reagent (NEB) according to the manufacturer's instructions at a DNA/Transpass D1 ratio of 1.2:1. Medium was replaced after 3–6 h.
Western blot.
Cells were lysed in RIPA-buffer (150 mmol/liter NaCl, 50 mmol/liter Tris-HCl, pH 8.0, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, and protease inhibitors [Roche]). Cell lysates were separated by SDS-PAGE and electrotransferred onto 0.2-μm nitrocellulose membranes (Whatman). Membranes were blocked with phosphate-buffered saline containing 5% nonfat dry milk, incubated with monoclonal mouse anti-SV40 T-antigen, Ab-2 (1:800; Calbiochem), polyclonal rabbit anti-LTag or anti-VP1 (both 1:2,500), or monoclonal anti-tubulin (1:1,000) and washed with phosphate-buffered saline containing 0.1% Tween 20. Anti–mouse or –rabbit horseradish peroxidase–labeled secondary antibody was used at 1:10,000 (Pierce Chemical Co.) or 1:20,000 (GE Healthcare) for detection of bound primary antibody by enhanced chemiluminescence.
Immunofluorescence.
4 d after infection, RPTEC cells were fixed in 4% PFA, and then fixed in methanol. Subsequently, cells were incubated at 37°C with primary and secondary antibodies for 30 min. We used the monoclonal mouse anti-SV40 T-antigen (Ab-2; 1:100) and an IgG-fraction of rabbit antiserum directed against BKV-agnoprotein (1:800) (51). The secondary antibodies were conjugated with Alexa Fluor 568 or 488 (1:500; Invitrogen). Nuclei were labeled in blue with DRAQ5 (Biostatus).
Microscopy and digital image processing.
Microscopy was performed using an epifluorescence microscope (model TE200; Nikon) equipped with suitable filters and a digital camera (Hamamatsu). Red and green cells were counted in a minimum of 3 10× microscopic fields. Images were taken at the times indicated in the figure legends. Digital images were recorded with Openlab 2.2 software and raw images were processed in Photoshop 6.0 (Adobe). Results are shown as the mean of 3 independent experiments yielding a SD of <10%. Confocal microscopy analyses and images were obtained using a microscope (Axiovert 200; Carl Zeiss, Inc.) equipped with a LSM510 confocal module and processed using LSM5 software version 3.2 (Carl Zeiss, Inc.).
Statistics.
The statistics were performed using SPSS-Version 14.0 (SPSS). For categorical values, Fisher's exact test or χ2 test were used. Nonparametric tests were used to compare values lacking normal distribution (Mann-Whitney U test, Wilcoxon) where appropriate. All tests were two-sided, unless otherwise indicated.
Online supplemental material.
Fig. S1 shows NCCR sequences. Fig. S2 shows VP1 serotypes. Fig. S3 shows a list of primers used to amplify BKV sequences and generate plasmid DNAs for cloning. The online version of this article is available at http://www.jem.org/cgi/content/full/jem.20072097/DC1.
Supplemental Material
Acknowledgments
We wish to thank Claudia Mistl, Jaqueline Samaridis, and Kenneth Bowitz Larsen for excellent technical assistance. C.H. Rinaldo performed confocal microscopy at the University of North-Norway/Medical Faculty Bioimaging Core Facility.
This work was founded by the appointment grant of the University of Basel BMM1001 to H.H. Hirsch.
The authors have no conflicting financial interests.
Abbreviations used: dat, days after transfection; geq, genome equivalent; HEK, human embryonic kidney; KT, kidney transplant; LTag, large T-antigen; NCCR, noncoding control region; PVAN, polyomavirus BK–associated nephropathy; RFP, red fluorescent protein; RPTEC, renal proximal tubular epithelial cell.
References
- 1.Hirsch, H.H., W. Knowles, M. Dickenmann, J. Passweg, T. Klimkait, M.J. Mihatsch, and J. Steiger. 2002. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N. Engl. J. Med. 347:488–496. [DOI] [PubMed] [Google Scholar]
- 2.Randhawa, P.S., S. Finkelstein, V. Scantlebury, R. Shapiro, C. Vivas, M. Jordan, M.M. Picken, and A.J. Demetris. 1999. Human polyoma virus-associated interstitial nephritis in the allograft kidney. Transplantation. 67:103–109. [DOI] [PubMed] [Google Scholar]
- 3.Ramos, E., C.B. Drachenberg, J.C. Papadimitriou, O. Hamze, J.C. Fink, D.K. Klassen, R.C. Drachenberg, A. Wiland, R. Wali, C.B. Cangro, et al. 2002. Clinical course of polyoma virus nephropathy in 67 renal transplant patients. J. Am. Soc. Nephrol. 13:2145–2151. [DOI] [PubMed] [Google Scholar]
- 4.Vasudev, B., S. Hariharan, S.A. Hussain, Y.R. Zhu, B.A. Bresnahan, and E.P. Cohen. 2005. BK virus nephritis: risk factors, timing, and outcome in renal transplant recipients. Kidney Int. 68:1834–1839. [DOI] [PubMed] [Google Scholar]
- 5.Drachenberg, C.B., J.C. Papadimitriou, H.H. Hirsch, R. Wali, C. Crowder, J. Nogueira, C.B. Cangro, S. Mendley, A. Mian, and E. Ramos. 2004. Histological patterns of polyomavirus nephropathy: correlation with graft outcome and viral load. Am. J. Transplant. 4:2082–2092. [DOI] [PubMed] [Google Scholar]
- 6.Nickeleit, V., H.H. Hirsch, I.F. Binet, F. Gudat, O. Prince, P. Dalquen, G. Thiel, and M.J. Mihatsch. 1999. Polyomavirus infection of renal allograft recipients: from latent infection to manifest disease. J. Am. Soc. Nephrol. 10:1080–1089. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch, H.H., and J. Steiger. 2003. Polyomavirus BK. Lancet Infect. Dis. 3:611–623. [DOI] [PubMed] [Google Scholar]
- 8.Knowles, W.A., P. Pipkin, N. Andrews, A. Vyse, P. Minor, D.W. Brown, and E. Miller. 2003. Population-based study of antibody to the human polyomaviruses BKV and JCV and the simian polyomavirus SV40. J. Med. Virol. 71:115–123. [DOI] [PubMed] [Google Scholar]
- 9.Shah, K.V., R.W. Daniel, and R.M. Warszawski. 1973. High prevalence of antibodies to BK virus, an SV40-related papovavirus, in residents of Maryland. J. Infect. Dis. 128:784–787. [DOI] [PubMed] [Google Scholar]
- 10.Stolt, A., K. Sasnauskas, P. Koskela, M. Lehtinen, and J. Dillner. 2003. Seroepidemiology of the human polyomaviruses. J. Gen. Virol. 84:1499–1504. [DOI] [PubMed] [Google Scholar]
- 11.Binet, I., V. Nickeleit, H.H. Hirsch, O. Prince, P. Dalquen, F. Gudat, M.J. Mihatsch, and G. Thiel. 1999. Polyomavirus disease under new immunosuppressive drugs: a cause of renal graft dysfunction and graft loss. Transplantation. 67:918–922. [DOI] [PubMed] [Google Scholar]
- 12.Meier-Kriesche, H.U., S. Li, R.W. Gruessner, J.J. Fung, R.T. Bustami, M.L. Barr, and A.B. Leichtman. 2006. Immunosuppression: evolution in practice and trends, 1994-2004. Am. J. Transplant. 6:1111–1131. [DOI] [PubMed] [Google Scholar]
- 13.Rubin, R.H., and N.E. Tolkoff-Rubin. 1983. Viral infection in the renal transplant patient. Proc. Eur. Dial. Transplant Assoc. 19:513–526. [PubMed] [Google Scholar]
- 14.Tantravahi, J., K.L. Womer, and B. Kaplan. 2007. Why hasn't eliminating acute rejection improved graft survival? Annu. Rev. Med. 58:369–385. [DOI] [PubMed] [Google Scholar]
- 15.Rinaldo, C.H., and H.H. Hirsch. 2007. Antivirals for the treatment of polyomavirus BK replication. Expert Rev. Anti Infect. Ther. 5:105–115. [DOI] [PubMed] [Google Scholar]
- 16.Drachenberg, C.B., J.C. Papadimitriou, R. Wali, J. Nogueira, S. Mendley, H.H. Hirsch, C.B. Cangro, D.K. Klassen, M.R. Weir, S.T. Bartlett, and E. Ramos. 2004. Improved outcome of polyoma virus allograft nephropathy with early biopsy. Transplant. Proc. 36:758–759. [DOI] [PubMed] [Google Scholar]
- 17.Hirsch, H.H., D.C. Brennan, C.B. Drachenberg, F. Ginevri, J. Gordon, A.P. Limaye, M.J. Mihatsch, V. Nickeleit, E. Ramos, P. Randhawa, et al. 2005. Polyomavirus-associated nephropathy in renal transplantation: interdisciplinary analyses and recommendations. Transplantation. 79:1277–1286. [DOI] [PubMed] [Google Scholar]
- 18.Fishman, J.A. 2002. BK virus nephropathy-polyomavirus adding insult to injury. N. Engl. J. Med. 347:527–530. [DOI] [PubMed] [Google Scholar]
- 19.Chesters, P.M., J. Heritage, and D.J. McCance. 1983. Persistence of DNA sequences of BK virus and JC virus in normal human tissues and in diseased tissues. J. Infect. Dis. 147:676–684. [DOI] [PubMed] [Google Scholar]
- 20.Brennan, D.C., I. Agha, D.L. Bohl, M.A. Schnitzler, K.L. Hardinger, M. Lockwood, S. Torrence, R. Schuessler, T. Roby, M. Gaudreault-Keener, and G.A. Storch. 2005. Incidence of BK with tacrolimus versus cyclosporine and impact of preemptive immunosuppression reduction. Am. J. Transplant. 5:582–594. [DOI] [PubMed] [Google Scholar]
- 21.Nickeleit, V., T. Klimkait, I.F. Binet, P. Dalquen, V. Del Zenero, G. Thiel, M.J. Mihatsch, and H.H. Hirsch. 2000. Testing for polyomavirus type BK DNA in plasma to identify renal-allograft recipients with viral nephropathy. N. Engl. J. Med. 342:1309–1315. [DOI] [PubMed] [Google Scholar]
- 22.Funk, G.A., J. Steiger, and H.H. Hirsch. 2006. Rapid dynamics of polyomavirus type BK in renal transplant recipients. J. Infect. Dis. 193:80–87. [DOI] [PubMed] [Google Scholar]
- 23.Binggeli, S., A. Egli, S. Schaub, I. Binet, M. Mayr, J. Steiger, and H.H. Hirsch. 2007. Polyomavirus BK-specific cellular immune response to vp1 and large T-antigen in kidney transplant recipients. Am. J. Transplant. 7:1131–1139. [DOI] [PubMed] [Google Scholar]
- 24.Comoli, P., S. Binggeli, F. Ginevri, and H.H. Hirsch. 2006. Polyomavirus-associated nephropathy: update on BK virus-specific immunity. Transpl. Infect. Dis. 8:86–94. [DOI] [PubMed] [Google Scholar]
- 25.Seif, I., G. Khoury, and R. Dhar. 1979. The genome of human papovavirus BKV. Cell. 18:963–977. [DOI] [PubMed] [Google Scholar]
- 26.Rubinstein, R., B.C. Schoonakker, and E.H. Harley. 1991. Recurring theme of changes in the transcriptional control region of BK virus during adaptation to cell culture. J. Virol. 65:1600–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanssen Rinaldo, C., H. Hansen, and T. Traavik. 2005. Human endothelial cells allow passage of an archetypal BK virus (BKV) strain–a tool for cultivation and functional studies of natural BKV strains. Arch. Virol. 150:1449–1458. [DOI] [PubMed] [Google Scholar]
- 28.Sundsfjord, A., T. Flaegstad, R. Flo, A.R. Spein, M. Pedersen, H. Permin, J. Julsrud, and T. Traavik. 1994. BK and JC viruses in human immunodeficiency virus type 1-infected persons: prevalence, excretion, viremia, and viral regulatory regions. J. Infect. Dis. 169:485–490. [DOI] [PubMed] [Google Scholar]
- 29.Randhawa, P., D. Zygmunt, R. Shapiro, A. Vats, K. Weck, P. Swalsky, and S. Finkelstein. 2003. Viral regulatory region sequence variations in kidney tissue obtained from patients with BK virus nephropathy. Kidney Int. 64:743–747. [DOI] [PubMed] [Google Scholar]
- 30.Olsen, G.H., P.A. Andresen, H.T. Hilmarsen, O. Bjorang, H. Scott, K. Midtvedt, and C.H. Rinaldo. 2006. Genetic variability in BK Virus regulatory regions in urine and kidney biopsies from renal-transplant patients. J. Med. Virol. 78:384–393. [DOI] [PubMed] [Google Scholar]
- 31.Chen, C.H., M.C. Wen, M. Wang, J.D. Lian, M.J. Wu, C.H. Cheng, K.H. Shu, and D. Chang. 2001. A regulatory region rearranged BK virus is associated with tubulointerstitial nephritis in a rejected renal allograft. J. Med. Virol. 64:82–88. [DOI] [PubMed] [Google Scholar]
- 32.Comoli, P., A. Azzi, R. Maccario, S. Basso, G. Botti, G. Basile, I. Fontana, M. Labirio, A. Cometa, F. Poli, et al. 2004. Polyomavirus BK-specific immunity after kidney transplantation. Transplantation. 78:1229–1232. [DOI] [PubMed] [Google Scholar]
- 33.Akan, I., I.K. Sariyer, R. Biffi, V. Palermo, S. Woolridge, M.K. White, S. Amini, K. Khalili, and M. Safak. 2006. Human polyomavirus JCV late leader peptide region contains important regulatory elements. Virology. 349:66–78. [DOI] [PubMed] [Google Scholar]
- 34.Bratt, G., A.L. Hammarin, M. Grandien, B.G. Hedquist, I. Nennesmo, B. Sundelin, and S. Seregard. 1999. BK virus as the cause of meningoencephalitis, retinitis and nephritis in a patient with AIDS. AIDS. 13:1071–1075. [DOI] [PubMed] [Google Scholar]
- 35.Smith, R.D., J.H. Galla, K. Skahan, P. Anderson, C.C. Linnemann Jr., G.S. Ault, C.F. Ryschkewitsch, and G.L. Stoner. 1998. Tubulointerstitial nephritis due to a mutant polyomavirus BK virus strain, BKV(Cin), causing end-stage renal disease. J. Clin. Microbiol. 36:1660–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirsch, H.H., S. Friman, A. Wiecek, L. Rostaing, and M. Pescovitz. 2007. Prospective study of polyomavirus BK viruria and viremia in de novo renal transplantation (Abstract #14). Am. J. Transplant. 7:150. [Google Scholar]
- 37.Moens, U., N. Subramaniam, B. Johansen, T. Johansen, and T. Traavik. 1994. A steroid hormone response unit in the late leader of the noncoding control region of the human polyomavirus BK confers enhanced host cell permissivity. J. Virol. 68:2398–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moens, U., and M. Van Ghelue. 2005. Polymorphism in the genome of non-passaged human polyomavirus BK: implications for cell tropism and the pathological role of the virus. Virology. 331:209–231. [DOI] [PubMed] [Google Scholar]
- 39.Domingo, E. 2007. Virus evolution. In Fields Virology. D.M. Knipe and P.M. Howley, editors. Lippincott Williams & Wilkins, Philadelphia, PA. pp. 389–421.
- 40.Sugimoto, C., K. Hara, F. Taguchi, and Y. Yogo. 1989. Growth efficiency of naturally occurring BK virus variants in vivo and in vitro. J. Virol. 63:3195–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin, L., and P.E. Gibson. 1996. Genomic Function and Variation of Human Polyomavirus BK (BKV). Rev. Med. Virol. 6:201–214. [DOI] [PubMed] [Google Scholar]
- 42.Sundsfjord, A., T. Johansen, T. Flaegstad, U. Moens, P. Villand, S. Subramani, and T. Traavik. 1990. At least two types of control regions can be found among naturally occurring BK virus strains. J. Virol. 64:3864–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takasaka, T., N. Goya, H. Ishida, K. Tanabe, H. Toma, T. Fujioka, S. Omori, H.Y. Zheng, Q. Chen, S. Nukuzuma, et al. 2006. Stability of the BK polyomavirus genome in renal-transplant patients without nephropathy. J. Gen. Virol. 87:303–306. [DOI] [PubMed] [Google Scholar]
- 44.Zheng, H.Y., Y. Nishimoto, Q. Chen, M. Hasegawa, S. Zhong, H. Ikegaya, N. Ohno, C. Sugimoto, T. Takasaka, T. Kitamura, and Y. Yogo. 2007. Relationships between BK virus lineages and human populations. Microbes Infect. 9:204–213. [DOI] [PubMed] [Google Scholar]
- 45.Bofill-Mas, S., M. Formiga-Cruz, P. Clemente-Casares, F. Calafell, and R. Girones. 2001. Potential transmission of human polyomaviruses through the gastrointestinal tract after exposure to virions or viral DNA. J. Virol. 75:10290–10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mackenzie, E.F., J.M. Poulding, P.R. Harrison, and B. Amer. 1978. Human polyoma virus (HPV)-a significant pathogen in renal transplantation. Proc. Eur. Dial. Transplant Assoc. 15:352–360. [PubMed] [Google Scholar]
- 47.Tevethia, S.S., L. Mylin, R. Newmaster, M. Epler, J.A. Lednicky, J.S. Butel, and M.J. Tevethia. 1998. Cytotoxic T lymphocyte recognition sequences as markers for distinguishing among tumour antigens encoded by SV40, BKV and JCV. Dev. Biol. Stand. 94:329–339. [PubMed] [Google Scholar]
- 48.Provenzano, M., L. Bracci, S. Wyler, T. Hudolin, G. Sais, R. Gosert, P. Zajac, G. Palu, M. Heberer, H.H. Hirsch, and G.C. Spagnoli. 2006. Characterization of highly frequent epitope-specific CD45RA+/CCR7+/− T lymphocyte responses against p53-binding domains of the human polyomavirus BK large tumor antigen in HLA-A*0201+ BKV-seropositive donors. J. Transl. Med. 4:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horton, R.M., H.D. Hunt, S.N. Ho, J.K. Pullen, and L.R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 77:61–68. [DOI] [PubMed] [Google Scholar]
- 50.Yang, R.C., and R. Wu. 1979. BK virus DNA: complete nucleotide sequence of a human tumor virus. Science. 206:456–462. [DOI] [PubMed] [Google Scholar]
- 51.Rinaldo, C.H., T. Traavik, and A. Hey. 1998. The agnogene of the human polyomavirus BK is expressed. J. Virol. 72:6233–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leuenberger, D., P.A. Andresen, R. Gosert, S. Binggeli, E.H. Strom, S. Bodaghi, C.H. Rinaldo, and H.H. Hirsch. 2007. Human polyomavirus type 1 (BK virus) agnoprotein is abundantly expressed, but immunologically ignored. Clin. Vaccine Immunol. 14:959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.